Abstract
The broad and variable substrate specificity of cytochrome P450 enzymes makes them a model system for studying the determinants of protein molecular recognition. The archetypal cytochrome P450cam (P450cam) is a relatively specific P450, a feature once attributed to the high rigidity of its active site. However, increasingly studies have provided evidence of the importance of conformational changes to P450cam activity. Here we used infrared (IR) spectroscopy to investigate the molecular recognition of P450cam. Toward this goal, and to assess the influence of a hydrogen bond (H-bond) between active site residue Y96 and substrates, two variants in which Y96 is replaced by a cyanophenyl (Y96CNF) or phenyl (Y96F) group were characterized in complexes with the substrates camphor, isoborneol, and camphane. These combinations allow for a comparison of complexes in which the moieties on both the protein and substrate can serve as a H-bond donor, acceptor, or neither. The IR spectra of heme-bound CO and the site-specifically incorporated CN of Y96CNF were analyzed to characterize the number and nature of environments in each protein, both in the free and bound states. Although the IR spectra do not support the idea that protein–substrate H-bonding is central to P450cam recognition, the data altogether suggest that the differing conformational heterogeneity in the active site of the P450cam variants and changes in heterogeneity upon binding of different substrates likely contribute to their variable affinities via a conformational selection mechanism. This study further extends our understanding of the molecular recognition of archetypal P450cam and demonstrates the application of IR spectroscopy combined with selective protein modification to delineate protein–ligand interactions.
Graphical abstract
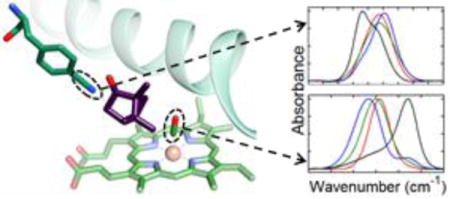
Cytochrome P450 enzymes make up a superfamily of heme enzymes that perform numerous critical metabolic functions, including steroid synthesis1 and drug metabolism,2,3 and they have received considerable attention for biotechnological applications.4 They are also notable for their ability to hydroxylate unactivated hydrocarbons on a wide variety of substrates,5,6 making them attractive models for the study of the determinants of biological molecular recognition.
The archetypal cytochrome P450cam (P450cam) catalyzes the stereo- and regioselective hydroxylation of d-camphor. P450cam shows relatively high substrate specificity in comparison to the specificities of many members of the P450 superfamily, which has been attributed to its relatively small, rigid active site that showed few binding-induced changes in early crystal structures.7–9 However, recently P450cam has been captured in a second conformation by X-ray crystallography in which the active site adopts a more open structure in the absence of a substrate.10 Studies of the enzyme in solution by double electron–electron resonance (DEER) also provide evidence of a conformational change upon binding to camphor,11 and recent nuclear magnetic resonance (NMR) studies suggest that P450cam populates an ensemble of conformations.12,13 Thus, as is recognized for the more flexible P450 homologues,14–18 conformational changes might be important to substrate recognition by P450cam.
Although P450cam is relatively specific for camphor, it also recognizes a number of small, camphor-like substrates, albeit with variable affinity. Early investigations by crystallography8 and site-directed mutagenesis19 implicated a hydrogen bond (H-bond) between residue Y96 and the ketone moiety of camphor in P450cam molecular recognition (Figure 1). Other studies found camphor binding to be largely entropically driven, suggesting the enthalpic contribution from the protein–substrate H-bond to the overall thermodynamics is minor, but not precluding its importance to selectivity for other substrates.20,21 Moreover, removal of this H-bond was reported to lead to some loss of regioselective conversion of d-camphor to 5-exohydroxycamphor, suggesting that the substrate–protein H-bond might restrict the substrate in a conformation conducive to selective catalysis.19 However, our recent studies of the CO/camphor complexes of wild-type (WT) and Y96F P450cam by two-dimensional infrared (2D IR) spectroscopy found the active site dynamics to be independent of the potential for protein–substrate H-bonding, questioning its importance to the conformational landscape of P450cam.22
Figure 1.
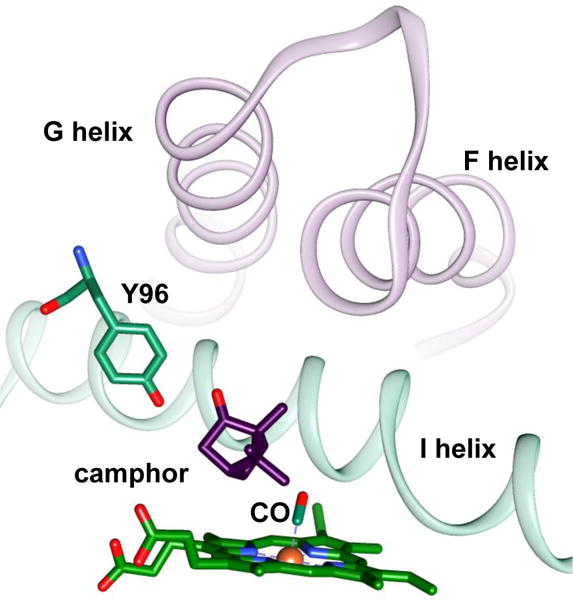
Structural model of the active site of CO-bound P450cam (Protein Data Bank entry 1T87). Shown are the camphor substrate, the side chain of residue Y96, the CO, and the F, G, and I helices.
To improve our understanding of the molecular mechanisms underlying P450cam–substrate recognition, we explored the role of perturbation of the H-bonding between the substrate and Y96 by investigating a series of proteins differing at residue 96 and their complexes with select substrates. We analyzed three P450cam variants: WT, Y96CNF (where CNF is p-cyano-L-phenylalanine), and Y96F (Figure 2), which place a native hydroxyl, a cyano (CN) group, or no functional group, respectively, on the side chain. We then investigated the complexes of the three variants with the substrates camphor, isoborneol, and camphane, which bear a native ketone, a hydroxyl, or no functional group on the substrate. Altogether, these protein and substrate combinations allow for a comparison of complexes in which the moieties that mediate the H-bond are replaced with a donor or an acceptor or are eliminated. We characterized each protein in its free and substrate-bound state by visible spectroscopy and determined the binding affinities. We then characterized each complex by IR spectroscopy with heme-bound CO, and with the CN group in the case of Y96CNF, to elucidate the nature and population of states accessed by the active site.
Figure 2.
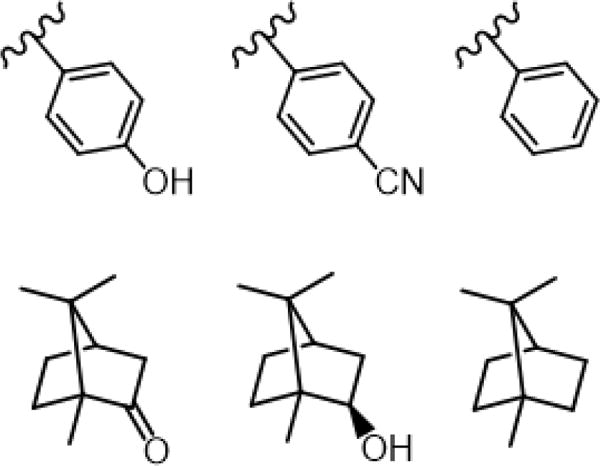
Structures of side chains of residue 96 in WT (left), Y96CNF (center), and Y96F (right) P450cam (top row). Structures of camphor (left), isoborneol (center), and camphane (right) (bottom row).
MATERIALS AND METHODS
P450cam Expression and Purification
WT, Y96CNF, and Y96F P450cam were expressed using plasmid pDNC334A23 generously provided by T. Pochapsky (Brandeis University, Waltham, MA),24 and each protein was purified as previously described with a few modifications (see the Supporting Information).22 For incorporation of CNF by in vivo amber codon suppression, the plasmid with a TAG codon introduced at position 96 was co-expressed with plasmid pUltraCNF,25,26 generously provided by P. Schultz (Scripps Research Institute, La Jolla, CA), and cultures were supplemented with 1 mM CNF prior to induction. Protein samples with A390/A280 >1.3 for WT, A404/A280 >1.0 for Y96F, and A414/A280 >1.1 for Y96CNF were used in spectroscopic experiments. Purified protein was flash-frozen and stored at −80 °C with 50% (w/v) glycerol. Typical yields were ~10 and ~5 mg of protein/L of cell culture for the WT and for the Y96F and Y96CNF variants, respectively.
Binding Assays
Enantiopure d-camphor was purchased from Sigma-Aldrich, camphane from Apollo Scientific, and racemic isoborneol from Santa Cruz Biotechnology. We note that isoborneol is racemic, but a pure diastereomer, and that the racemic nature is not expected to significantly affect the binding results as previously reported with other camphor analogues.27,28 Dissociation constants were determined by spectrophotometric titrations as described in previous studies,19,29 with modifications as described in the Supporting Information. Briefly, for isoborneol and camphane, visible difference spectra were generated by exchanging a substrate-bound solution of protein into a substrate-free solution (both with the same concentration of protein) to perform a constant-volume spectrophotometric titration. The data were analyzed according to a single-site binding model to determine the KD.
Fourier Transform Infrared (FT IR) Spectroscopy
FT IR spectra were acquired as previously described on an Agilent Cary 670 FT IR spectrometer and a N2(l)-cooled MCT detector.29 For characterization of the CN stretch of Y96CNF, a 4500 ± 500 nm bandpass filter was placed in the beam path after the sample. The instrument was purged with dry N2(g) for 40 min before data were collected. Background transmission spectra were collected with WT P450cam at the same concentration that was used for the samples (0.5–2 mM) in the same buffer (100 mM potassium phosphate, 50 mM KCl, and 20% glycerol). Additional details are provided in the Supporting Information. All spectra shown are representative of at least three independent trials.
Data Processing and Evaluation of Fits to IR Spectra
IR absorption spectra of the CO probe were generated from transmission spectra of the CO-bound complexes and oxidized WT P450cam, while spectra for the CN probe were generated from transmission spectra of Y96CNF and WT protein. The spectra were corrected for a residual slowly varying background absorbance by fitting a region of ~100 or 60 cm−1 around the CO or CN bands, respectively, excluding the band itself, to a polynomial function (nonperiodic residuals that had ±1.5% of the intensity of the band were tolerated). Parameters without fitting (average frequencies at the maximal and half-maximal absorbance and full width at half-maximum) were determined for each of the CN absorptions (Table S3). Each spectrum then was fit to one or a sum of up to three Gaussian functions (Tables 2 and 3 and Tables S4, S5, S7, and S9), as in previous studies.9,30–33 F-tests were performed to determine if inclusion of an additional Gaussian component significantly improved the fit in each case (Tables S6 and S10).
Table 2.
Parameters from Fits of CO Spectra
| substrate | νCO (cm−1) | full width at half-maximum (cm−1) | relative area (%) | statea |
|---|---|---|---|---|
| WT | ||||
| free | 1939.4 ± 0.4 | 17.0 ± 0.1 | 50 ± 4 | |
| 1951.5 ± 0.1 | 13.4 ± 0.4 | 33 ± 4 | ||
| 1962.7 ± 0.1 | 9.8 ± 0.1 | 17 ± 1 | ||
| camphor | 1939.4 ± 0.2 | 12.9 ± 0.5 | ||
| isoborneol | 1946.0 ± 0.06 | 12.4 ± 0.2 | ||
| camphane | 1952.3 ± 0.5 | 11.4 ± 0.2 | ||
| Y96CNF | ||||
| free | 1953.5 ± 0.6 | 22.0 ± 0.3 | 42 ± 1 | S2 |
| 1964.2 ± 0.1 | 9.4 ± 0.1 | 58 ± 1 | S3 | |
| camphor | 1945.6 ± 0.04 | 14.4 ± 0.03 | 82 ± 1 | S1 |
| 1948.9 ± 0.03 | 7.7 ± 0.2 | 9 ± 1 | S2 | |
| 1964.0 ± 0.1 | 9.5 ± 0.1 | 9 ± 1 | S3 | |
| isoborneol | 1949.8 ± 0.07 | 14.4 ± 0.8 | S1/S2 | |
| camphane | 1951.6 ± 0.08 | 11.0 ± 0.2 | S1/S2 | |
| Y96F | ||||
| free | 1956.8 ± 0.4 | 14.4 ± 1.0 | 42 ± 1 | |
| 1962.9 ± 0.07 | 9.0 ± 0.2 | 58 ± 1 | ||
| camphor | 1945.7 ± 0.6 | 13.6 ± 0.1 | ||
| isoborneol | 1949.5 ± 0.2 | 12.9 ± 0.1 | ||
| camphane | 1950.7 ± 0.03 | 11.1 ± 0.1 |
Assigned state based on our model (see the Supporting Information).
Table 3.
Parameters from Fits of CN Spectra
| ligand/substrate | νCN (cm−1) | full width at half-maximum (cm−1) | relative area (%) | statea |
|---|---|---|---|---|
| free | 2232.5 ± 0.2 | 11.2 ± 0.7 | ||
| camphor | 2232.9 ± 0.2 | 12.1 ± 0.5 | ||
| isoborneol | 2228.9 ± 0.2 | 6.6 ± 0.7 | 32 ± 8 | |
| 2234.2 ± 0.2 | 8.8 ± 0.5 | 68 ± 8 | ||
| camphane | 2229.6 ± 0.3 | 5.0 ± 0.4 | 15 ± 2 | |
| 2234.0 ± 0.2 | 6.8 ± 0.4 | 85 ± 2 | ||
| CO | 2227.2 ± 0.1 | 5.7 ± 0.2 | 42 ± 7 | S2 |
| 2232.6 ± 0.4 | 8.4 ± 0.7 | 58 ± 7 | S3 | |
| CO/camphor | 2227.8 ± 0.1 | 6.2 ± 0.1 | 12 ± 1 | S2 |
| 2235.3 ± 0.05 | 8.9 ± 0.1 | 88 ± 1 | S1/S3 | |
| CO/isoborneol | 2228.0 ± 0.4 | 8.3 ± 1.0 | 9 ± 4 | S2 |
| 2234.0 ± 0.3 | 11.3 ± 0.3 | 91 ± 4 | S1 | |
| CO/camphane | 2228.0 ± 0.1 | 6.1 ± 0.2 | 10 ± 0.5 | S2 |
| 2233.9 ± 0.2 | 9.5 ± 0.3 | 90 ± 0.5 | S1 |
Assigned state based on our model. See the Supporting Information.
RESULTS
Visible Spectroscopy
WT, Y96F, and Y96CNF P450cam in complexes with camphor, isoborneol, and camphane, as well as substrate-free, were characterized by visible spectroscopy (Figure S1). The visible spectra for the variants in the substrate-free form are virtually identical, showing the Soret band at 417 nm that is reflective of the low-spin state of the heme. Upon binding d-camphor, the Soret band for WT P450cam shifts to 390 nm, which is reflective of the high-spin state of the heme, in agreement with the literature.20 The visible spectra of the other protein–substrate complexes reflect variable low/high-spin heme content (Table S1). The spin state populations do not show any clear relationship with the affinities of the proteins for the substrates. Although coupling among the spin state, reduction potential, and binding of camphor is found for P450cam,21 a number of studies of wild-type and mutant P450cam and other P450 homologues with a wide variety of substrates show no correlation between spin state and substrate affinity or activity, indicating that their relationship, if any, is complex.9,34–38 When CO is bound, all variants exhibit the expected peak at 446 nm (Figure S2), regardless of the presence or absence of substrate, and none showed >5% of the P420 state.39
Substrate Binding
Binding assays were performed to investigate the differences in affinities of WT, Y96CNF, and Y96F P450cam for camphor, isoborneol, and camphane (Table 1). For WT, the affinity for the different substrates is most variable, with the native substrate camphor binding most tightly and camphane showing much lower affinity, as reported previously,19,28 and the isoborneol showing intermediate affinity. For Y96CNF, the affinity for camphor is also highest, but the affinities for isoborneol and camphane are higher than those of WT. In contrast, for Y96F, the affinity for isoborneol is highest, but like for Y96CNF, the affinities for camphor and camphane are lower and higher, respectively, than those of the WT. Overall, despite the large difference in the nature of the change to Y96, removal of the hydroxyl or replacement with the larger cyano moiety, both modifications to this residue engendered similar changes to the specificity of substrate recognition, decreasing the affinity for camphor and increasing the affinity for both isoborneol and camphane.
Table 1.
Dissociation Constants for P450cam Complexes
| variant | substrate | KD (μM) | ΔG (kcal/mol) |
|---|---|---|---|
| WT | camphor | 1.8 ± 0.8 | −7.8 ± 0.3 |
| WT | isoborneol | 10.9 ± 3.4 | −6.8 ± 0.2 |
| WT | camphane | 18.7 ± 2.5 | −6.4 ± 0.1 |
| Y96CNF | camphor | 3.5 ± 1.0 | −7.4 ± 0.2 |
| Y96CNF | isoborneol | 5.7 ± 1.7 | −7.1 ± 0.2 |
| Y96CNF | camphane | 6.4 ± 1.8 | −7.1 ± 0.2 |
| Y96F | camphor | 3.5 ± 0.6 | −7.4 ± 0.1 |
| Y96F | isoborneol | 1.2 ± 0.3 | −8.1 ± 0.2 |
| Y96F | camphane | 10.7 ± 3.3 | −6.8 ± 0.2 |
Heme-Bound CO Vibrations
CO has long been used as an intense IR probe of heme protein active sites,40 including WT and mutated P450cam-CO when substrate-free and bound to a variety of substrates.9,22,30,31,33,41 Typically, variation in the frequency of heme-bound CO is described well by the influence of the protein electric field via a linear Stark shift model and ascribed to the influence of the distal active site environment on the local electrostatics, with positive or negative electrostatic fields leading to lower or higher frequencies, respectively.42–44 To characterize the changes in the environment at the active site among the three proteins as well as in their complexes with camphor, isoborneol, and camphane, we performed an exhaustive characterization by IR spectroscopy of heme-bound CO in all proteins in the absence and presence of each substrate (Figure 3).
Figure 3.
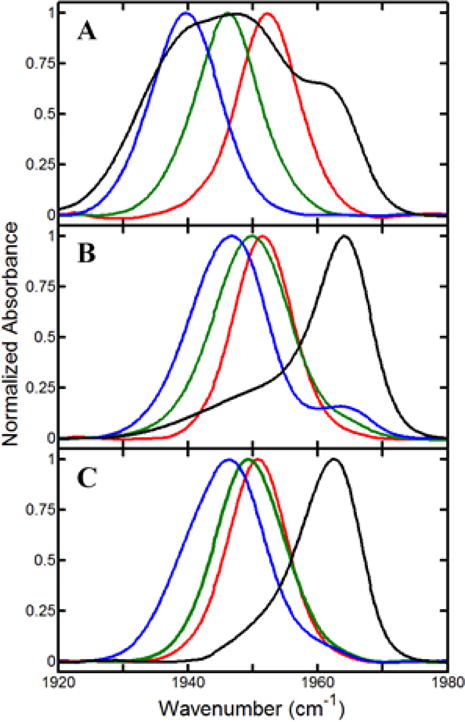
FT IR spectra of CO for (A) WT, (B) Y96CNF, and (C) Y96F, in complexes with camphor (blue), isoborneol (green), and camphane (red), as well as substrate-free (black).
The spectra show large variation among the protein samples in the number and frequencies of the CO absorptions. They were fit to one or more Gaussian functions to deconvolute the contributions of multiple populated states (Table 2). As observed in previous studies,14,16 for the substrate-free WT protein, the CO spectrum is well fit by a superposition of three bands, indicating that the CO experiences three distinct environments, suggestive of three active site conformations. When the WT protein is bound to any of the three substrates, the CO absorption is well fit as a single band, with a center frequency dependent on the particular substrate. In P450cam, the heme-bound CO is next to the I helix and packs within a groove formed by disrupton of the H-bonding within the α-helical structure (Figure 1).12,45 An electrostatic interaction between CO with the I helix has been proposed to account for the lower frequency of the absorption of the camphor complex.9,30 Higher frequencies are found for the CO absorptions of the complexes with isoborneol and camphane, which indicate differences in the electrostatics of the CO environment when bound to the alternate substrates, potentially resulting from differences in the structures that modulate the interaction of CO with the I helix.
Alteration of residue Y96, either by removal of the hydroxyl group or by its replacement by CN, leads to significant, but similar, changes in the CO spectra. The spectra for the substrate-free states of both Y96CNF and Y96F P450cam are well fit by two bands, unlike that of the WT, which clearly shows three. All show a component at ~1963 cm−1 with a relatively narrow line width of <10 cm−1, indicative of a state in which the CO environment is relatively homogeneous. Whereas this is a minor species for the WT, the band is dominant for the modified proteins. In addition, the spectra for neither Y96CNF nor Y96F P450cam show the band at ~1939 cm−1 similar to that found for the WT substrate-free protein and camphor complex. The spectra for the substrate complexes of Y96CNF and Y96F P450cam are similar but show large differences from those of the WT. Specifically, for the camphor and isoborneol complexes, the frequencies of absorptions for the modified proteins are higher than for the WT, whereas for the camphane complex, the frequencies of the bands are lower than for the WT.
CN Absorptions of Y96CNF
The CN group of Y96CNF provides a spectroscopic handle to probe the interactions of the side chain in both the substrate-free and bound states. The CN vibrational frequency is sensitive to the local electric field via a Stark effect, with higher-frequency bands typically found in more polar solvents.46,47 Additionally, the CN vibration is known to be sensitive to local interactions like H-bonding and electron-repulsive interactions due to packing with heteroatoms.29,48–50 These interactions also lead to shifts in the bands to higher frequencies. The multiple contributions to CN frequency shifts mean that knowledge of the exact mechanism underlying specific frequency shifts requires additional investigation. However, if the CN experiences more than one distinct environment, the high sensitivity of the CN to its environment makes it an excellent reporter of conformational heterogeneity. In addition, the relative areas of component bands inform on the relative populations of the corresponding states, and their center frequencies provide information for comparing the nature of the environments within the populated states.
The spectra of the substrate-free and camphor complexes of Y96CNF have been previously reported29 and are reproduced with the spectra obtained for the camphane and isoborneol complexes in Figure 4. All exhibit CN absorption bands with center frequencies within the range of 2227–2236 cm−1, consistent with previous values for CNF incorporated into proteins.26,51–54 Although the spectra for both substrate-free Y96CNF and the camphor complex are adequately fit by single Gaussian functions, the spectra for the isoborneol and camphane complexes clearly require two components (Supporting Information). The two bands of the isoborneol and camphane complexes are relatively narrow and similar in frequency, but their relative areas indicate that the state associated with the higher-frequency band is more highly populated in the camphane complex than in the isoborneol complex. Similarly fitting the spectra for the substrate-free protein and camphor complex with two components yields bands of roughly equal area that are relatively broad (Supporting Information). For the camphor complex, one of these component bands corresponds in frequency (2233.8 cm−1) with the higher-frequency bands found for the isoborneol and camphane complexes, which supports the two-component fit. The other component band occurs relatively high in frequency (2232.4 cm−1) in comparison to those for the isoborneol and camphane complexes. Thus, the camphor complex differs from the isoborneol and camphane complexes in at least one populated state.
Figure 4.
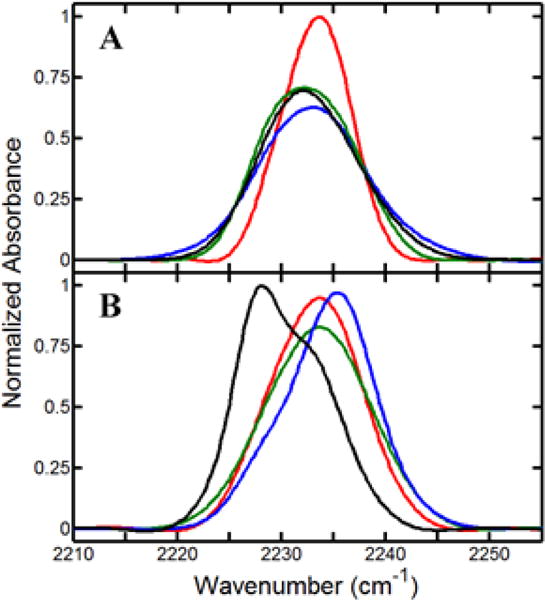
FT IR spectra of CN for substrate-free Y96CNF (black) and complexes with camphor (blue), isoborneol (green), and camphane (red), both (A) without CO and (B) with CO. All spectra are averages of at least three unique trials, and spectra in both panels are normalized by integrated area.
CN Vibrations of CO-Bound Y96CNF
For CO-bound Y96CNF, the CO and CN vibrations may be simultaneously used to characterize the binding pocket. The CNF is on the opposite side of the substrate binding site as CO, so the two probes report on distinct local environments of the active site. As before, the CN spectra were fit to a superposition of Gaussian functions to determine the number and nature of the populated states. The best fits to all spectra include two components, indicating population of two distinct states.
Comparison of the CN spectra in the presence and absence of CO (Figure 5) reveals that CO binding at the heme is sensed by the CNF96 probe, despite it being on the opposite side of the binding pocket. For the camphane and isoborneol complexes, the frequencies of the bands in the presence and absence of CO are approximately the same, but the relative band areas depend on CO, with the presence of CO shifting population of the states toward the state associated with the higher-frequency band. For the substrate-free protein and camphor complex, the spectra clearly show two bands when CO is bound, and analysis of the spectra in the absence and presence of CO yields bands of different frequencies. Therefore, compared to those of the isoborneol and camphane complexes, the spectra for the substrate-free protein and camphor complex are more significantly perturbed by CO binding.
Figure 5.
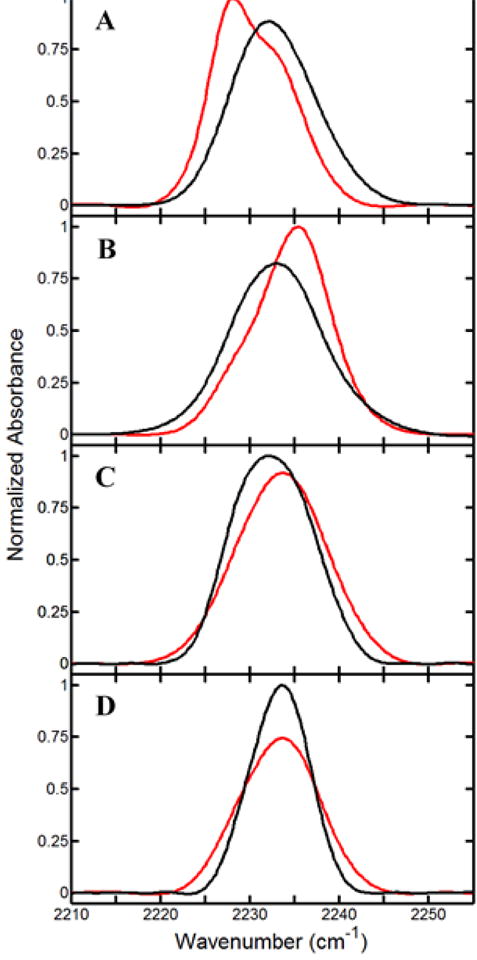
FT IR spectra of CN for (A) substrate-free Y96CNF and complexes with (B) camphor, (C) isoborneol, and (D) camphane in the presence (red) and absence (black) of CO. All spectra are normalized by integrated area.
Despite probing the same protein active sites, the CN and CO spectra do not always require the same number of components for fitting, which means that either the probes experience different levels of conformational heterogeneity or the conformations do not differentiate the environments identically and generate spectrally distinct absorptions for all states. Under the latter assumption, we could rationalize both the CO and CN spectral data for the free and bound states assuming variable population of three conformations (see the Supporting Information for details). The assignments of the CO and CN bands to the individual states are provided in Tables 2 and 3, respectively, and the results of our analysis for their relative populations in the absence or presence of the different substrates are listed in Tables S11–S13.
DISCUSSION
Although P450cam is a highly substrate-specific member of the P450 family with a relatively rigid active site, the IR spectra provide evidence of conformational heterogeneity in both substrate-bound and free proteins. The substrate-free protein appears to be highly heterogeneous, with the CO spectra indicating that its active site populates at least three distinct conformations. Although the CO probe generally shows a single absorption in the substrate complexes, the CN probe of Y96CNF P450cam, which reports on a different location of the active site, shows evidence that multiple distinct environments also are populated in all substrate complexes. One state appears dominant; however, a second shows significant (minimally ~10%) population for all substrates. In the substrate-free protein, two states appear to be populated approximately equally.
The presence of conformational heterogeneity and its dependence on substrate binding bolster the recently developing appreciation for the role of conformational dynamics in substrate recognition by P450cam. Although early crystal structures showed no changes induced by substrate binding,55 a second, more open, state was observed in a crystal structure with large tethered substrates,56 and a similarly open conformation was captured in the substrate-free protein.10 A recent analysis of all P450cam structures finds that they can be clustered into three distinct states: open, closed, and intermediate.57 It is possible that the three CO absorptions observed in the substrate-free protein reflect population of these three states. A major difference among the structures involves a change in the H-bonding within the I helix, which alters the size of the groove into which CO binds. Such a conformational change between an open and closed state in solution was also confirmed by DEER spectroscopy.11 Additionally, a number of NMR studies of the CO-bound protein have indicated that both substrate-free and camphor-bound P450cam are a heterogeneous ensemble of states.12,13,38 In agreement, the CO and CN probes employed here show evidence of conformational heterogeneity at two distinct locations in the active site in the absence and presence of substrates, as well as for the substrate binding-induced population shift among conformations. The conformational change from the open to closed states involves displacement of the F and G helices and intervening loop that covers the active site. Our previous investigation of CNF-labeled P450cam by molecular dynamics simulations found the CN absorptions to be sensitive to changes in the side chain packing that accompany a conformational change in these structural elements.29 Alternately, although most structures of P450cam show the same orientation of the Y96 side chain pointing toward the active site, the multiple bands observed for the CNF probe could reflect co-population of a second side chain orientation wherein the Y96 side chain is flipped out toward the protein surface that is observed in a structure of a closely related P450 homologue.58 A structure of P450cam tethered to its electron transfer partner putidaredoxin showed a similarly large change in the side chain orientation of Y96.59 In addition, a recent structure of P450cam in the open state with camphor bound showed the Y96 side chain was disordered,60 suggestive of high conformational heterogeneity, which agrees with the heterogeneity observed for Y96CNF in solution by IR spectroscopy.
The CN probe of Y96CNF reveals that CO binding induces conformational dynamics, which are also substrate-dependent. Because the CO does not directly contact the CN, the spectral changes are likely modulated via structural changes of the protein and/or substrate. Consistent with this possibility, a recent study by DEER showed that binding of CO to the P450cam–putidaredoxin complex induces a conformational change in P450cam from the open to the closed state.60 For both the isoborneol and camphane complexes, the changes in the relative areas of the CN absorptions suggest a decrease in conformational heterogeneity upon binding of CO. Although the substrate-free protein and camphor complex show single CN bands in the absence of CO, as opposed to clearly showing two in the CO complex, the bands in the absence of CO are much broader. Binding of CO to the substrate-free protein and camphor complex also results in significant changes in the CN frequencies, unlike those of the isoborneol and camphane complexes, and the spectral changes overall reflect more substantial conformational changes upon CO binding.
The changes in the population and nature of the conformations upon binding to different substrates shown by the IR data suggest a conformational selection aspect to substrate molecular recognition, wherein one of the substrate-free states is selectively stabilized by substrate binding. The variation in absorption frequencies found among the substrate complexes implies that the environment at the active site depends on the substrate, which could result from population of distinct conformations in the different substrate complexes. Our previous studies find that multiple distinct conformations are populated for the complexes with the substrates camphor, norcamphor, and thiocamphor.22 Characterization of the camphor and camphane complexes by 2D IR spectroscopy also revealed differences in the dynamics of the active sites,33 which suggests that they also might populate distinct states of the enzyme and thus that the stabilization of different states by substrates within a conformational selection mechanism might underlie the specificity of P450cam molecular recognition.
This possibility, that conformational changes contribute to variable substrate recognition by P450cam, is supported by the combined IR and affinity data. For WT P450cam, the substrate-free protein most highly populates a state that appears to correspond most closely to that of the camphor complex, where the closed state is dominantly populated. In contrast, the higher-frequency bands found for the isoborneol and camphane complexes are more similar to those bands associated with the less populated states of the substrate-free protein. Thus, the spectral data indicate that formation of the camphor complex involves a less extensive shift of population among the conformations than does binding of isoborneol or camphane, which is consistent with the observed variation in binding affinities.
This possibility is further supported by the experiments with Y96CNF and Y96F, which show corresponding differences from WT in both the affinities for each substrate and conformational heterogeneity reflected by their CO spectra. In particular, for the substrate-free states of both variants, the CO spectra show loss of the low-frequency band that is similar to that found in the camphor complex and a greater contribution from higher-frequency bands that are more similar to those found for the isoborneol and camphane complexes. Within a conformational selection model of binding, the substrate-free states of Y96CNF and Y96F are more primed than the WT to bind isoborneol and camphane, which would account for the lower affinity of Y96F and Y96CNF for camphor and higher affinities for isoborneol and camphane.
The set of proteins and substrates selected for study include combinations of H-bond donors and acceptors that modulate the potential for substrate–protein H-bonding, which has been suggested to lock the substrate within the active site.8,30,55 For example, Y96F P450cam has previously been shown to have a lower affinity for camphor and a slight loss of regioselectivity of camphor hydroxylation.19 We, however, found by 2D IR spectroscopy that the dynamics of the active site of the camphor complex were unchanged by Y96F,22 calling into question the importance of the interaction to the conformational landscape. The contribution of the H-bond to the overall thermodynamics of camphor recognition also has been called into question, as the binding was shown by previous temperature-dependent kinetic studies to be entropically driven.20 However, this could be the case either because the H-bonding interaction does not occur or because both the substrate and Y96 are also engaged in H-bonding in their free states, such that the net change in H-bonding is neutral. In either case, the potential of the protein to H-bond could influence the relative interaction of P450cam with different substrates, influencing the specificity of molecular recognition. Thermodynamic “double-mutant” cycles were calculated for the variants to help assess the energetic contribution of the interaction between the polar moiety on residue 96 and that on the substrate (Figures S3–S6). It is noted that such a treatment is valid only if the free energy changes for the individual steps are completely uncorrelated.61 Nevertheless, the thermodynamic cycle analysis yields interaction free energies that are favorable only by −0.7 kcal/mol for the WT and camphor, which is small relative to those typically ascribed to H-bonding interactions,62 and gives interaction free energies that are unfavorable by 1.2 kcal/mol for the side chain–substrate interactions of the Y96CNF and WT with isoborneol.
The IR data also do not support the possibility that H-bond formation is important to recognition specificity. The CN frequencies of Y96CNF for the different substrate complexes do not show large differences. In particular, the similarity in the frequencies between the isoborneol and both the camphor and camphane complexes is not consistent with engagement of the isoborneol hydroxyl group and CN in a H-bond, as such an interaction is expected to lead to a significantly higher CN frequency.50,63 It is noted that the data cannot rule out the possibility that a H-bond occurs between camphor and Y96 of the WT protein. However, if H-bonds do form between the substrate and proteins, the spectral and binding analysis indicates that their contribution to the specificity of molecular recognition, and in particular to the active site conformational landscape, appears to be minor.
CONCLUSIONS
We performed a thorough spectroscopic analysis of three P450cam variants and three substrates to investigate the mechanism of molecular recognition. The IR spectral data provide evidence of conformational heterogeneity in all states, as well as its restriction upon binding to substrates, supporting a conformational selection aspect to the binding mechanism. In addition, the correspondence of the number and frequencies of IR bands to the affinity data supports the idea that conformational entropy changes upon binding likely play a substantial role in the specificity of P450cam recognition. Although previous structural studies show a H-bond occurring between the ketone of the camphor substrate and Y96, the interaction does not appear to be critical in stabilizing particular conformational states of the substrate complexes. This study further advances our continually evolving understanding of substrate molecular recognition by the archetypal P450cam and demonstrates the application of FT IR spectroscopy with selectively incorporated CN probes for assessment of possible hydrogen bonding interactions.
Supplementary Material
Acknowledgments
The authors thank Thomas Pochapsky at Brandeis University for providing the pDNC334A plasmid to express cytochrome P450cam and Peter Schultz at the Scripps Research Institute for providing the pUltraCNF plasmid for in vivo amber codon suppression and incorporation of CNF.
Funding
We thank Indiana University and the National Institutes of Health (1R01GM114500) for support. E.J.B. was also supported by the Graduate Training Program in Quantitative and Chemical Biology (T32 GM109825).
Footnotes
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.biochem.7b00238.
Additional experimental methods regarding P450cam expression and purification, visible spectroscopy, substrate binding assays, FT IR sample preparation, and evaluation of fits to IR spectra as well as discussion of spin states in P450cam complexes and details of our model for population of multiple states accounting for CN and CO bands (PDF)
ORCID
Megan C. Thielges: 0000-0002-4520-6673
Notes
The authors declare no competing financial interest.
References
- 1.Auchus RJ, Miller WL. P450 Enzymes in Steroid Processing. In: Ortiz de Montellano PR, editor. Cytochrome P450: : Structure, Mechanism, and Biochemistry. 4th. Springer International Publishing; Cham, Switzerland: 2015. pp. 851–879. [Google Scholar]
- 2.Denisov IG, Makris TM, Sligar SG, Schlichting I. Structure and Chemistry of Cytochrome P450. Chem Rev. 2005;105:2253–2277. doi: 10.1021/cr0307143. [DOI] [PubMed] [Google Scholar]
- 3.Zanger UM, Schwab M. Cytochrome P450 enzymes in drug metabolism: Regulation of gene expression, enzyme activities, and impact of genetic variation. Pharmacol Ther. 2013;138:103–141. doi: 10.1016/j.pharmthera.2012.12.007. [DOI] [PubMed] [Google Scholar]
- 4.Girvan HM, Munro AW. Applications of microbial cytochrome P450 enzymes in biotechnology and synthetic biology. Curr Opin Chem Biol. 2016;31:136–145. doi: 10.1016/j.cbpa.2016.02.018. [DOI] [PubMed] [Google Scholar]
- 5.Ortiz de Montellano PR. Hydrocarbon Hydroxylation by Cytochrome P450 Enzymes. Chem Rev. 2010;110:932–948. doi: 10.1021/cr9002193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sigel A, Sigel H, Sigel RKO. The Ubiquitous Roles of Cytochrome P450: Proteins: Metal Ions in Life Sciences. Wiley; New York: 2007. [Google Scholar]
- 7.Poulos TL, Finzel BC, Howard AJ. High-resolution Crystal Structure of Cytochrome P450cam. J Mol Biol. 1987;195:687–700. doi: 10.1016/0022-2836(87)90190-2. [DOI] [PubMed] [Google Scholar]
- 8.Raag R, Poulos TL. Crystal Structures of Cytochrome P-450cam Complexed with Camphane, Thiocamphor, and Adamantane: Factors Controlling P-450 Substrate Hydroxylation. Biochemistry. 1991;30:2674–2684. doi: 10.1021/bi00224a016. [DOI] [PubMed] [Google Scholar]
- 9.Schulze H, Hui Bon Hoa G, Helms V, Wade RC, Jung C. Structural Changes in Cytochrome P-450cam Effected by the Binding of the Enantiomers (1R)-Camphor and (1S)-Camphor. Biochemistry. 1996;35:14127–14138. doi: 10.1021/bi9527303. [DOI] [PubMed] [Google Scholar]
- 10.Lee YT, Wilson RF, Rupniewski I, Goodin DB. P450cam Visits an Open Conformation in the Absence of Substrate. Biochemistry. 2010;49:3412–3419. doi: 10.1021/bi100183g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stoll S, Lee YT, Zhang M, Wilson RF, Britt RD, Goodin DB. Double electron–electron resonance shows cytochrome P450cam undergoes a conformational change in solution upon binding substrate. Proc Natl Acad Sci U S A. 2012;109:12888–12893. doi: 10.1073/pnas.1207123109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Asciutto EK, Dang M, Pochapsky SS, Madura JD, Pochapsky TC. Experimentally Restrained Molecular Dynamics Simulations for Characterizing the Open States of Cytochrome P450cam. Biochemistry. 2011;50:1664–1671. doi: 10.1021/bi101820d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Asciutto EK, Young MJ, Madura J, Pochapsky SS, Pochapsky TC. Solution Structural Ensembles of Substrate-Free Cytochrome P450cam. Biochemistry. 2012;51:3383–3393. doi: 10.1021/bi300007r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Scott EE, He YA, Wester MR, White MA, Chin CC, Halpert JR, Johnson EF, Stout CD. An open conformation of mammalian cytochrome P450 2B4 at 1.6-Å resolution. Proc Natl Acad Sci U S A. 2003;100:13196–13201. doi: 10.1073/pnas.2133986100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ouellet H, Podust LM, Ortiz de Montellano PR. Mycobacterium tuberculosis CYP130: crystal structure, biophysical characterization, and interactions with antifungal azole drugs. J Biol Chem. 2008;283:5069–5080. doi: 10.1074/jbc.M708734200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sevrioukova IF, Poulos TL. Understanding the mechanism of cytochrome P450 3A4: recent advances and remaining problems. Dalton Trans. 2013;42:3116–3126. doi: 10.1039/c2dt31833d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Savino C, Montemiglio LC, Sciara G, Miele AE, Kendrew SG, Jemth P, Gianni S, Vallone B. Investigating the structural plasticity of a cytochrome P450: three-dimensional structures of P450 EryK and binding to its physiological substrate. J Biol Chem. 2009;284:29170–29179. doi: 10.1074/jbc.M109.003590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dubey KD, Wang B, Shaik S. Molecular Dynamics and QM/MM Calculations Predict the Substrate-Induced Gating of Cytochrome P450 BM3 and the Regio- and Stereoselectivity of Fatty Acid Hydroxylation. J Am Chem Soc. 2016;138:837–845. doi: 10.1021/jacs.5b08737. [DOI] [PubMed] [Google Scholar]
- 19.Atkins WM, Sligar SG. The Roles of Active Site Hydrogen Bonding in Cytochrome P-450cam as Revealed by Site-directed Mutagenesis. J Biol Chem. 1988;263:18842–18849. [PubMed] [Google Scholar]
- 20.Griffin BW, Peterson JA. Camphor Binding by Psuedomonas Putida Cytochrome P450. Kinetics and Thermodynamics of the Reaction. Biochemistry. 1972;11:4740–4746. doi: 10.1021/bi00775a017. [DOI] [PubMed] [Google Scholar]
- 21.Sligar SG. Coupling of Spin, Substrate, and Redox Equilibria in Cytochrome P450. Biochemistry. 1976;15:5399–5406. doi: 10.1021/bi00669a029. [DOI] [PubMed] [Google Scholar]
- 22.Basom EJ, Spearman JW, Thielges MC. Conformational Landscape and the Selectivity of Cytochrome P450cam. J Phys Chem B. 2015;119:6620–6627. doi: 10.1021/acs.jpcb.5b03896. [DOI] [PubMed] [Google Scholar]
- 23.Nickerson DP, Wong LL. The dimerization of Pseudomonas putida cytochrome P450cam: practical consequences and engineering of a monomeric enzyme. Protein Eng, Des Sel. 1997;10:1357–1361. doi: 10.1093/protein/10.12.1357. [DOI] [PubMed] [Google Scholar]
- 24.Ouyang B, Pochapsky SS, Pagani GM, Pochapsky TC. Specific Effects of Potassium Ion Binding on Wild-Type and L358P Cytochrome P450cam. Biochemistry. 2006;45:14379–14388. doi: 10.1021/bi0617355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chatterjee A, Sun SB, Furman JL, Xiao H, Schultz PG. A Versatile Platform for Single- and Multiple-Unnatural Amino Acid Mutagenesis in Escherichia coli. Biochemistry. 2013;52:1828–1837. doi: 10.1021/bi4000244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schultz KC, Supekova L, Ryu Y, Xie J, Perera R, Schultz PG. A Genetically Encoded Infrared Probe. J Am Chem Soc. 2006;128:13984–13985. doi: 10.1021/ja0636690. [DOI] [PubMed] [Google Scholar]
- 27.Atkins WM, Sligar SG. Metabolic Switching in Cytochrome P-450cam: Deuterium Isotope Effects on Regiospecificity and the Monooxygenase/Oxidase Ratio. J Am Chem Soc. 1987;109:3754–3760. [Google Scholar]
- 28.Schulze H, Hui Bon Hoa G, Jung C. Mobility of norbornane-type substrates and water accessibility in cytochrome P450cam. Biochim Biophys Acta, Protein Struct Mol Enzymol. 1997;1338:77–92. doi: 10.1016/s0167-4838(96)00192-6. [DOI] [PubMed] [Google Scholar]
- 29.Basom EJ, Maj M, Cho M, Thielges MC. Site-Specific Characterization of Cytochrome P450cam Conformations by Infrared Spectroscopy. Anal Chem. 2016;88:6598–6606. doi: 10.1021/acs.analchem.6b01520. [DOI] [PubMed] [Google Scholar]
- 30.Jung C, Hui Bon Hoa G, Schröder KL, Simon M, Doucet JP. Substrate Analogue Induced Changes of the CO-Stretching Mode in the Cytochrome P450cam-Carbon Monoxide Complex. Biochemistry. 1992;31:12855–12862. doi: 10.1021/bi00166a021. [DOI] [PubMed] [Google Scholar]
- 31.Jung C, Ristau O, Schulze H, Sligar SG. The CO stretching mode infrared spectrum of substrate-free cytochrome P-450cam-CO The effect of solvent conditions, temperature, and pressure. Eur J Biochem. 1996;235:660–669. doi: 10.1111/j.1432-1033.1996.00660.x. [DOI] [PubMed] [Google Scholar]
- 32.Nagano S, Shimada H, Tarumi A, Hishiki T, Kimata-Ariga Y, Egawa T, Suematsu M, Park SY, Adachi S, Shiro Y, Ishimura Y. Infrared Spectroscopic and Mutational Studies on Putidaredoxin-Induced Conformational Changes in Ferrous COP450cam. Biochemistry. 2003;42:14507–14514. doi: 10.1021/bi035410p. [DOI] [PubMed] [Google Scholar]
- 33.Thielges MC, Chung JK, Fayer MD. Protein Dynamics in Cytochrome P450 Molecular Recognition and Substrate Specificity Using 2D IR Vibrational Echo Spectroscopy. J Am Chem Soc. 2011;133:3995–4004. doi: 10.1021/ja109168h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guengerich FP. Oxidation-Reduction Properties of Rat Liver Cytochromes P-450 and NADPH-Cytochrome P-450 Reductase Related to Catalysis in Reconstituted Systems. Biochemistry. 1983;22:2811–2820. doi: 10.1021/bi00281a007. [DOI] [PubMed] [Google Scholar]
- 35.De Voss JJ, Sibbesen O, Zhang Z, Ortiz de Montellano PR. Substrate Docking Algorithms and Prediction of the Substrate Specificity of Cytochrome P450cam and Its L244A Mutant. J Am Chem Soc. 1997;119:5489–5498. [Google Scholar]
- 36.Zhang Z, Sibbesen O, Johnson RA, Ortiz de Montellano PR. The Substrate Specificity of Cytochrome P450cam. Bioorg Med Chem. 1998;6:1501–1508. doi: 10.1016/s0968-0896(98)00091-1. [DOI] [PubMed] [Google Scholar]
- 37.French KJ, Rock DA, Rock DA, Manchester JI, Goldstein BM, Jones JP. Active Site Mutations of Cytochrome P450cam Alter the Binding, Coupling, and Oxidation of the Foreign Substrates (R)- and (S)-2-Ethylhexanol. Arch Biochem Biophys. 2002;398:188–197. doi: 10.1006/abbi.2001.2732. [DOI] [PubMed] [Google Scholar]
- 38.Colthart AM, Tietz DR, Ni Y, Friedman JL, Dang M, Pochapsky TC. Detection of substrate-dependent conformational changes in the P450 fold by nuclear magnetic resonance. Sci Rep. 2016;6:22035. doi: 10.1038/srep22035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yu CA, Gunsalus IC. Cytochrome P-450cam II. Interconversion with P-420. J Biol Chem. 1974;249:102–106. [PubMed] [Google Scholar]
- 40.Spiro TG, Wasbotten IH. CO as a vibrational probe of heme protein active sites. J Inorg Biochem. 2005;99:34–44. doi: 10.1016/j.jinorgbio.2004.09.026. [DOI] [PubMed] [Google Scholar]
- 41.O’Keefe DH, Ebel RE, Peterson JA, Maxwell JC, Caughey WS. An Infrared Spectroscopic Study of Carbon Monoxide Bonding to Ferrous Cytochrome P-450. Biochemistry. 1978;17:5845–5852. doi: 10.1021/bi00619a036. [DOI] [PubMed] [Google Scholar]
- 42.Park ES, Andrews SS, Hu RB, Boxer SG. Vibrational Stark Spectroscopy in Proteins: A Probe and Calibration for Electrostatic Fields. J Phys Chem B. 1999;103:9813–9817. [Google Scholar]
- 43.Spiro TG, Soldatova AV, Balakrishnan G. CO, NO and O2 as Vibrational Probes of Heme Protein Interactions. Coord Chem Rev. 2013;257:511–527. doi: 10.1016/j.ccr.2012.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Phillips GN, Jr, Teodoro ML, Li T, Smith B, Olson JS. Bound CO Is A Molecular Probe of Electrostatic Potential in the Distal Pocket of Myoglobin. J Phys Chem B. 1999;103:8817–8829. [Google Scholar]
- 45.Raag R, Poulos TL. The Structural Basis for Substrate-Induced Changes in Redox Potential and Spin Equilibrium in Cytochrome P-450cam. Biochemistry. 1989;28:917–922. doi: 10.1021/bi00428a077. [DOI] [PubMed] [Google Scholar]
- 46.Fried SD, Boxer SG. Measuring Electric Fields and Noncovalent Interactions Using the Vibrational Stark Effect. Acc Chem Res. 2015;48:998–1006. doi: 10.1021/ar500464j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Levinson NM, Fried SD, Boxer SG. Solvent-Induced Infrared Frequency Shifts in Aromatic Nitriles Are Quantitatively Described by the Vibrational Stark Effect. J Phys Chem B. 2012;116:10470–10476. doi: 10.1021/jp301054e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Błasiak B, Cho M. Vibrational solvatochromism. II A first-principle theory of solvation-induced vibrational frequency shift based on effective fragment potential method. J Chem Phys. 2014;140:164107. doi: 10.1063/1.4872040. [DOI] [PubMed] [Google Scholar]
- 49.Błasiak B, Cho M. Vibrational solvatochromism. III Rigorous treatment of the dispersion interaction contribution. J Chem Phys. 2015;143:164111. doi: 10.1063/1.4934667. [DOI] [PubMed] [Google Scholar]
- 50.Fafarman AT, Sigala PA, Herschlag D, Boxer SG. Decomposition of Vibrational Shifts of Nitriles into Electrostatic and Hydrogen-Bonding Effects. J Am Chem Soc. 2010;132:12811–12813. doi: 10.1021/ja104573b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Getahun Z, Huang CY, Wang T, De Leon B, DeGrado WF, Gai F. Using Nitrile-Derivatized Amino Acids as Infrared Probes of Local Environment. J Am Chem Soc. 2003;125:405–411. doi: 10.1021/ja0285262. [DOI] [PubMed] [Google Scholar]
- 52.Adhikary R, Zimmermann J, Dawson PE, Romesberg FE. IR Probes of Protein Microenvironments: Utility and Potential for Perturbation. ChemPhysChem. 2014;15:849–853. doi: 10.1002/cphc.201400017. [DOI] [PubMed] [Google Scholar]
- 53.Horness RH, Basom EJ, Thielges MC. Site-selective characterization of Src homology 3 domain molecular recognition with cyanophenylalanine infrared probes. Anal Methods. 2015;7:7234–7241. doi: 10.1039/C5AY00523J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shrestha R, Cardenas AE, Elber R, Webb LJ. Measurement of the Membrane Dipole Electric Field in DMPC Vesicles Using Vibrational Shifts of p-Cyanophenylalanine and Molecular Dynamics Simulations. J Phys Chem B. 2015;119:2869–2876. doi: 10.1021/jp511677j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Poulos TL, Raag R. Cytochrome P450cam: crystallography, oxygen activation, and electron transfer. FASEB J. 1992;6:674–679. doi: 10.1096/fasebj.6.2.1537455. [DOI] [PubMed] [Google Scholar]
- 56.Dunn AR, Dmochowski IJ, Bilwes AM, Gray HB, Crane BR. Probing the open state of cytochrome P450cam with ruthenium-linker substrates. Proc Natl Acad Sci U S A. 2001;98:12420–12425. doi: 10.1073/pnas.221297998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lee YT, Glazer EC, Wilson RF, Stout CD, Goodin DB. Three Clusters of Conformational States in P450cam Reveal a Multistep Pathway for Closing of the Substrate Access Channel. Biochemistry. 2011;50:693–703. doi: 10.1021/bi101726d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yang W, Bell SG, Wang H, Zhou W, Bartlam M, Wong LL, Rao Z. The structure of CYP101D2 unveils a potential path for substrate entry into the active site. Biochem J. 2011;433:85–93. doi: 10.1042/BJ20101017. [DOI] [PubMed] [Google Scholar]
- 59.Tripathi S, Li H, Poulos TL. Structural Basis for Effector Control and Redox Partner Recognition in Cytochrome P450. Science. 2013;340:1227–1230. doi: 10.1126/science.1235797. [DOI] [PubMed] [Google Scholar]
- 60.Liou SH, Mahomed M, Lee YT, Goodin DB. Effector Roles of Putidaredoxin on Cytochrome P450cam Conformational States. J Am Chem Soc. 2016;138:10163–10172. doi: 10.1021/jacs.6b04110. [DOI] [PubMed] [Google Scholar]
- 61.Mark AE, van Gunsteren WF. Decomposition of the Free Energy of a System in Terms of Specific Interactions. J Mol Biol. 1994;240:167–176. doi: 10.1006/jmbi.1994.1430. [DOI] [PubMed] [Google Scholar]
- 62.Jeffrey G. An Introduction to Hydrogen Bonding. Oxford University Press; Oxford, U.K: 1997. [Google Scholar]
- 63.Aschaffenburg DJ, Moog RS. Probing Hydrogen Bonding Environments: Solvatochromic Effects on the CN Vibration of Benzonitrile. J Phys Chem B. 2009;113:12736–12743. doi: 10.1021/jp905802a. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


