Abstract
Rationale: PIWI-interacting RNAs (piRNAs), a class of newly discovered small RNA molecules that function by binding to the Argonaute protein family (i.e., the PIWIL protein subfamily), and long noncoding RNAs (lncRNA) are implicated in several cancers. However, the detailed roles of ncRNAs in glioma remain unclear.
Methods: The expression of PIWIL3, piR-30188, OIP5-AS1, miR-367, CEBPA and TRAF4 were measured in glioma tissues and cells. The role of PIWIL3/OIP5-AS1/miR-367-3p/CEBPA feedback loop was evaluated in cell and animal models. The association of the above molecules was analyzed.
Results: Over-expression of PIWIL3, piR-30188 and miR-367-3p or knockdown of OIP5-AS1 resulted in inhibition of glioma cells progression. Binding sites between piR-30188 and OIP5-AS1 as well as between OIP5-AS1 and miR-367-3p were confirmed by RNA immunoprecipitation and luciferase assays. OIP5-AS1 knockdown or miR-367-3p over-expression contributed to a decrease in CEBPA (CCAAT/enhancer binding protein alpha) protein. Furthermore, CEBPA was detected as a target of miR-367-3p and played an oncogenic role in glioma. Treatment with CEBPA and miR-367-3p resulted in the modulation of downstream TRAF4 (TNF receptor-associated factor 4). PIWIL3 was also a target of CEBPA, forming a positive feedback loop in the growth regulation of glioma cells. Significantly, knockdown of OIP5-AS1 combined with over-expression of PIWIL3 and miR-367-3p resulted in tumor regression and extended survival in vivo.
Conclusion: These results identified a novel molecular pathway in glioma cells that may provide a potential innovative approach for tumor therapy.
Introduction
Glioblastoma is neuroepithelium-derived and the most malignant and invasive form of glioma according to the pathological classification of World Health Organization 1. Despite aggressive surgery followed by adjuvant postoperative radiotherapy and chemotherapy, glioblastoma patients have a 5-year survival rate of less than 5% 2, 3. Resistance of glioblastoma to chemotherapy and radiotherapy is a crucial factor affecting therapeutic efficacy. The pathogenesis of glioblastoma has been a great challenge in recent years requiring novel approaches for diagnosis and targeted therapy.
The transcription products of most genes in the human genome are non-coding RNAs (ncRNAs) 4. Based on the length of ncRNAs fragments, they are divided into short non-coding RNAs and long non-coding RNAs (lncRNAs). The short non-coding RNAs mainly include microRNAs (miRNAs), short-interfering RNAs (siRNAs), and PIWI-interacting RNAs (piRNAs) 5. piRNAs are a class of newly discovered small RNA molecules of 26-31nt in length and generally function by binding to the Argonaute protein family (i.e., the PIWIL protein subfamily) for gene silencing and regulation as well as maintenance of the germline DNA stability. Human PIWI protein members include: PIWIL1, PIWIL2, PIWIL3 and PIWIL4 6. PIWI proteins find their target RNAs by using piRNAs as guides and cleave the targets through the RNase (slicer) activity of the PIWI domain. PIWIL3 expression is deregulated in multiple cancers. Recent studies of single-cell proteomics have demonstrated that PIWIL3 is highly expressed in human oocytes and is associated with development and maturation 7. It has been reported that PIWIL3 is aberrantly expressed in breast and gastric carcinoma 8, 9 and is related to the prognosis of breast cancer and may serve as a new indicator for prognosis 8.PIWIL3 has been shown to be highly expressed in stage III human ovarian epithelial cancer and its metastatic tissues 10. Also, the human protein atlas (http://www.proteinatlas.org/ENSG00000184571-PIWIL3/pathology) shows that PIWIL3 is down-regulated in gliomas. Moreover, piRNA microarray analysis showed that piRNADQ570076 (referred to as piR-30188 in NCBI genebank) is down-regulated in U87 and U251 glioma cells. However, the regulatory role and potential mechanisms of PIWIL3 and piR-30188 affecting the biological behavior of glioblastoma have not been investigated.
piRNAs can bind to lncRNAs in a sequence-specific manner. The major biological functions of lncRNAs include regulation of gene activation and inhibition 11, 12.Current studies have shown that lncRNAs play an important role in tumorigenesis. When piR-30188 was over-expressed in U87 and U251 cells, microarray analysis showed down-regulation of several lncRNAs, and many of these showing a 1-fold change were validated by qRT-PCR. By using piRNA Bank and piRNA predictor, we discovered that piR-30188 has a putative binding site on a highly conserved lncRNA in mammals, OIP5-AS1, which is expressed predominantly in cytoplasm 13. Its homologous gene in zebrafish, Cyrano, is mainly involved in brain and eye development. Knocking out Cyrano leads to a decrease in brain and eye volume and neural tube opening defects in zebrafish 13. It has been reported that OIP5-AS1 may play a biological role as an endogenous competition RNAs (ceRNAs) sponge preventing RNA binding protein HuR (one of the human embryonic lethal abnormal visual RBP family members) from binding mRNA, thereby inhibiting HuR expression and thus participating in phenotypic regulation 14. Whether OIP5-AS1 regulates the biological behavior of tumor cells has yet to be elucidated.
miRNAs have a crucial role in the regulation of gene expression and cell function, and their involvement in tumorigenesis has been widely reported. The regulatory network of non-coding RNAs is related to the development of various tumors. Studies have shown that transcripts such as lncRNAs can recognize miRNAs through miRNA recognition components acting as ceRNAs 15, 16. It has been reported that LncRNA-RSU1P2 binds to let-7a, inhibits its expression, and plays an oncogenic role in cervical cancer cells 17. It also promotes the development of non-small cell lung cancer by modulating miR-377-3p/E2F3 pathway as a ceRNA 18. LncRNA-XIST and HCP5 promote the proliferation, migration, and invasion of glioma cells by inhibiting miR-152 and miR-139, respectively 19, 20. When OIP5-AS1 was knocked down in U87 and U251 cells, microarray results showed that miR-367-3p was one of the up-regulated miRNAs. Also, the bioinformatics database Starbase shows that OIP5-AS1 harbors a putative binding site of miR-367-3p. It has been shown that miR-367-3p acts as a tumor suppressor in gastric cancer. The over-expression of miR-367-3p inhibits invasion and metastasis of gastric cancer cells 21, while decreased expression of miR-367-3p is associated with poor prognosis in ependymoma and high-grade glioma patients 22, 23. Yet, the expression and function of miR-367-3p in glioma cells are still unresolved.
In our preliminary experiments, we detected that CEBPA (CCAAT/enhancer binding protein alpha) was up-regulated in gliomas. Remarkably, by searching bioinformatics databases miRanda and Targetscan, we noticed that CEBPA harbors a putative binding site for miR-367-3p. CEBPA is a member of the CEBP transcription factor family that contains basic leucine zipper (bZIP) 24 and participates in differentiation and energy metabolism in various cells. CEBPA regulates proliferation and apoptosis of zebrafish hepatocytes 25. It is highly expressed in human hepatocellular carcinoma tissues. The knockdown of CEBPA in human hepatocellular carcinoma suppresses the colony formation and cell growth of Hep3B and Huh7 hepatocellular carcinoma cells via lowering the expression of cyclin A and CDK4, suggesting that CEBPA plays an oncogenic role in hepatocellular carcinoma 26, 27. Earlier studies have shown that CEBPA mRNA expression in ovarian cancer is significantly increased. The elevated mRNA was negatively correlated with the survival of patients and the efficacy of platinum/cyclophosphamide chemotherapy 28. Mutations in CEBPA have also been found in acute myeloid leukemia 29. Consistent with the previous findings that miRNAs can inhibit the expression of target genes by binding to transcription factors, miR-381 was shown to regulate CEBPA expression, thus restraining metastasis of breast cancer cells 30.
By searching the bioinformatics database Oncomine (https://www.oncomine.org), we noticed that TNF receptor associated factor 4 (TRAF4) is up-regulated in glioblastomas. We also predicted that TRAF4 promoter might harbor two upstream putative binding sites for CEBPA by using bioinformatics software JASPAR. TRAF4 is highly expressed in colon cancer cells 31. It is also up-regulated in oral squamous cell carcinoma and promotes cell growth, migration, and invasion viaWnt-β-catenin signaling 32. However, it is not known how CEBPA and TRAF4 might affect the biological behavior of gliomas.
In this study, we explored the possible intrinsic relationship between PIWIL3, piR-30188, OIP5-AS1, miR-367-3p and CEBPA and their possible mechanisms of action on the biological behavior of glioma. The ultimate goal of this study was to identify a potential new molecular target for the treatment of gliomas.
Materials and Methods
Human tissue samples
Human glioma specimens and normal brain tissues were obtained from patients diagnosed with glioma who received surgery at the Department of Neurosurgery of Shengjing Hospital, China Medical University, from January 2014 to January 2017. The research methods in our study were approved by the Institutional Review Board at Shengjing Hospital of China Medical University. Informed consent was obtained from all patients, and the study was approved by the Ethics Committee of Shengjing Hospital of China Medical University. All specimens were immediately frozen and preserved in liquid nitrogen after surgical resection for the ensuing experiments. Glioma specimens were divided into four grades (pathological grade and molecular subtype) by three experienced clinical pathologists according to the WHO classification of tumors in the central nervous system (2007): low-grade glioma (WHO I-II, WHO I: n=3; WHO II: n=5) and high-grade glioma (WHO III-IV, WHO III: n=9; WHO IV: n=16). Detailed information about the glioma patients is shown in Table S1. Specimens of normal brain tissues obtained from fresh autopsy material (donation from individuals who died in a traffic accident and confirmed to be free of any prior pathologically detectable conditions) were used as negative controls (n= 15).
Human piRNA, lncRNA and miRNA microarrays
The Arraystar HG19 piRNA array (ArrayStar, Rockville, MD), was used for this study. For lncRNA and miRNA analysis, Sample preparation and microarray hybridization were performed by Kangchen Bio-tech, (Shanghai P.R. China).
Cell culture
Human U87 and U251 glioma cell lines, were purchased from Chinese Academy of Medical Sciences (Beijing, People's Republic of China). Human embryonic kidney (HEK) 293T was purchased from Shanghai Institutes for Biological Sciences Cell Resource Center.U87 and HEK-293T cells were cultured in Dulbecco's modified Eagle medium (DMEM)/high glucose supplemented with10% FBS. U251 cells were cultured in DMEM/F12 medium supplemented with10% FBS. All cells were maintained in a humidified incubator at 37 °C with 5% CO2.Primary NHA were purchased from the ScienCell Research Laboratories (Carlsbad, CA) and cultured under the conditions instructed by the manufacturer.
Reverse transcription and quantitative real-time PCR
Total RNA was extracted from the glioma tissues and NHA, U87, and U251 cells using Trizol reagent (Life Technologies Corporation, Carlsbad, CA). The RNA concentration and quality were detected at the 260/280 nm ratio using a Nanodrop Spectrophotometer (ND-100, Thermo, Waltham, MA). The primers of piR-30188, OIP5-AS1, PIWIL3, GAPDH, hsa-miR-367-3p, and U6 were synthesized by Thermo Fisher. TaqMan miRNA Reverse Transcription kit (Thermo Fisher) was used to synthesize cDNA from miRNA. The primers for glyceraldehyde-3-phosphate dehydrogenase (GAPDH) were: (F)5'-AAATCCCATCACCATCTTCCAG-3', (R) 5'-TGATGACCCTTTTGGCTCCC-3'. qRT-PCR was performed using One-Step SYBR PrimeScript RT-PCR Kit (Perfect Real Time, RR066A) (Takara Bio, Inc, Japan) for OIP5-AS1, PIWIL3, and GAPDH. TaqMan Universal Master Mix II was used for TaqMan microRNA assays of piR-30188, miR-367-3p and U6 (Applied Biosystems) in the ABI 7500 Fast Real-Time PCR System (Applied Biosystems). Expressions were normalized to endogenous controls, and the relative quantification (2-ΔΔCt) method was used for fold-change calculation.
Lentiviral vector construction and infection
PIWIL3 coding sequence (CDS) and sh-OIP5-AS1 sequence were respectively ligated into the LV5-CMV-GFP-Puro lentiviral vector (GenePharma, Shanghai, China) to generate LV3-CMV-GFP-Puro-PIWIL3 and LV5-CMV-GFP-Puro-sh-OIP5-AS1 lentiviral vectors. Lentivirus was harvested 48 h after the LV5-CMVGFP-Puro-PIWIL3 lentiviral vectors or the empty lentiviral vectors (negative control, NC) were co-transfected with the packaging vectors into human glioma cell lines U87 and U251 using Lipofectamine 3000. Cells were then infected with the lentivirus. GFP-positive cells were pooled as PIWIL3 and PIWIL3-NC and used for subsequent assays. These stably expressing cells were subsequently infected with lentivirus carrying LV5-CMV-GFP-Puro-sh-OIP5-AS1 vector (or sh-NC). The cells were then co-transfected with pre-piR-30188 (or pre-NC) and were divided into 6 groups: control group, pre-NC, pre-piR-30188, PIWIL3(+)-NC, PIWIL3(+), and pre-piR-30188+PIWIL3(+)+OIP5-AS1(-) group (OIP5-AS1-NC stably expressing cells co-transfected with pre-piR-30188).
Cell transfections
Two short-hairpin OIP5-AS1 (sh-OIP5-AS1) plasmids miR-367-3p agomir (pre-miR-367-3p) and miR-367-3p antagomir (anti-miR-367-3p) and their respective non-targeting sequences (NC) (pre-NC or anti-NC) were synthesized (GenePharma, Shanghai, People's Republic of China). Short-hairpin CEBPA and TRAF4 plasmids (sh-CEBPA and sh-TRAF4) and their respective non-targeting sequences (NC) were synthesized. Cells were seeded into 24-well plates (Corning), cultured to50-70% confluence and then transfected with Lipofectamine 3000 reagent (Life Technologies Corporation, Carlsbad, CA) according to the manufacturer's instructions. The applicable stably transfected cells were selected by G418 screening. Over-expression and silencing efficiency were analyzed using qRT-PCR. To determine the effect of OIP5-AS1 on glioma, cells were divided into three groups: Control, sh-NC (transfected with sh-NC plasmid), and sh-OIP5-AS1 (transfected with short-hairpin OIP5-AS1 plasmid). Similarly, to determine the effect of miR-367-3p on glioma, cells were divided into five groups: control, pre-NC (transfected with NC), pre-miR-367-3p (transfected with miR-367-3p agomir), anti-NC (transfected with NC), and anti-miR-367-3p (transfected with miR-367-3p antagomir). To explore the underlying mechanism of OIP5-AS1 regulating the biological behavior of glioma cells via decreasing miR-367-3p, cells were divided into five groups: control, sh-NC+pre-NC, sh-NC+anti-NC, sh-OIP5-AS1+anti-miR-367-3p (sh-OIP5-AS1 stably expressing cells co-transfected with anti-miR-367-3p), and sh-OIP5-AS1+pre-miR-367-3p (sh-OIP5-AS1 stably expressing cells co-transfected with pre-miR-367-3p). To determine whether miR-367-3p inhibited the malignant progression of glioma cells via targeting CEBPA 3'UTR, cells were dived into four groups: Control (miR-367-3p-NC+CEBPA-NC), pre-miR-367-3p+CEBPA-NC (pre-miR-367-3p stably expressing cells cotransfected with CEBPA-NC plasmid), pre-miR-367-3p+CEBPA, and pre-miR-367+CEBPA (non-3'UTR).
Cell proliferation assay
Cell Counting Kit-8 assay (CCK-8, Dojin, Japan) was performed to determine the proliferation of U87 and U251 glioma cells. After transfection, U87 and U251 cells were seeded in 96-well plates at a density of 2,000 cells per well. After 72 h, 10 μL of CCK-8 solution was added into each well and incubated for 2 h at 37°C. The absorbance was measured at 450 nm with the SpectraMax M5 microplate reader (Molecular Devices).
Quantization of apoptosis by flow cytometry
Cell apoptosis was quantified by Annexin 7AAD/PE staining (Southern Biotech, Birmingham, AL, USA). After rinsing with PBS and centrifuging twice, cells were resuspended in Annexin-V binding buffer. They were stained with Annexin V-FITC/PI according to the manufacturer's instructions. The cells were then analyzed by flow cytometry (FACScan, BD Biosciences) to acquire the apoptotic fractions.
Cell migration and invasion assay
24-well chambers with 8 μm pore size (Corning) were used for migration and invasion assay of U87 and U251 cells in vitro. Cells were resuspended in 100μL serum-free medium at a density of 105/ml and seeded in the upper chamber (or precoated with 500 ng/ml Matrigel solution (BD, Franklin Lakes, NJ)) for cell migration and invasion assay. 600 μL of 10% FBS medium was placed in the lower chamber. After 48 h of incubation, the cells on the upper membrane surface were physically wiped with a cotton swab. Cells that had migrated or invaded to the lower side of the membrane were fixed with methanol and stained with 10% Giemsa. Five random fields were chosen to count cells for statistics under a microscope and photographs were taken.
Reporter vector constructs and luciferase reporter assay
OIP5-AS1 full-length sequence was amplified by PCR and cloned into a pmirGlo Dual-luciferase miRNA Target Expression Vector (Promega, Madison, WI) to construct luciferase reporter vector (OIP5-AS1-Wt) (GenePharma). The sequences of putative piR-30188and miR-367-3p binding sites were replaced (OIP5-AS1-Mut; for piR-30188 the mutation sequence is: 3'-UUCUUCUCUUUUUUCGGUUUUGUCUUCUGUUU-5'; for miR-367-3p the mutation sequence is: 3'-AUUGCACUA-5') to mutate the putative binding site of OIP5-AS1. HEK-293T cells were seeded in 96-well plates and were co-transfected with OIP5-AS1-Wt (or OIP5-AS1-Mut) and pre-piR-30188 or piR-30188-NC plasmids when they reached 50-70% confluence. Cells co-transfected with OIP5-AS1-Wt (or OIP5-AS1-Mut) and pre-miR-367-3p or pre-NC plasmids were harvested using the same method. The luciferase activity was measured at 48 h after transfection by Dual-Luciferase reporter assay kit (Promega). The cells were divided into five groups: Control, OIP5-AS1-Wt +pre-NC (transfected with OIP5-AS1-Wt and piR-30188-NC), OIP5-AS1-Wt + pre-piR-30188 (transfected with OIP5-AS1-Wt and pre-piR-30188), OIP5-AS1-Mut + pre-NC (transfected with OIP5-AS1-Mut and pre-NC), and OIP5-AS1 Mut+ pre-piR-30188 (transfected with OIP5-AS1-Mut and pre-piR-30188); Control, OIP5-AS1-Wt + pre-NC (transfected with OIP5-AS1-Wt and pre-NC), OIP5-AS1-Wt + pre-miR-367-3p (transfected with OIP5-AS1-Wt and pre-miR-367-3p), OIP5-AS1-Mut + pre-NC (transfected with OIP5-AS1-Mut and pre-NC), and OIP5-AS1-Mut + pre-miR-367-3p (transfected withOIP5-AS1-Mut and pre-miR-367-3p).
RIP assay
RIP was assayed using a Magna RNA-binding protein immunoprecipitation kit according to the instructions provided by the manufacturer. Whole-cell lysate was incubated with RIP buffer containing magnetic beads conjugated with human anti-Ago2 antibody or negative control normal mouse IgG. Samples were incubated with Proteinase K and immunoprecipitated RNA was isolated. The RNA concentration was measured by a spectrophotometer (NanoDrop, Thermo Scientific, Waltham, MA, USA) and the RNA quality was assessed using a bioanalyzer (Agilent, Santa Clara, CA, USA). Furthermore, purified RNAs were extracted and analyzed by quantitative real-time PCR to demonstrate the presence of the binding targets.
RNA pull-down
The interaction between piR-30188 and PIWIL3 protein was examined using Pierce Magnetic RNA-Protein Pull-Down Kit (Thermo fisher) according to the manufacturer's protocols. Biotin-labeled piR-30188 or antisense RNA was co-incubated with protein extract of U87 and U251 cells and magnetic beads. The generated bead-RNA-Protein compound was collected by low-speed centrifugation. After washing with Handee spin columns, bead compound was boiled in SDS buffer, and the retrieved protein was detected by Western blotting with GAPDH as the control.
Western blotting
Total protein was extracted from the frozen cells using RIPA buffer containing 50 mM HEPES and 1 mM EDTA (pH 8.0) on ice. The samples were centrifuged at 16,184rcf4°Cfor 40 min, and the protein concentration of the supernatant extract was determined by BCA protein assay kit (Beyotime, Shanghai, China). Samples were subjected to SDS-PAGE and electrophoretically transferred to PVDF membranes, which were then incubated in tris-buffered saline containing 5% nonfat milk for 2 hat room temperature followed by incubation for 18 h with primary antibodies as follows: PIWIL3 (1:200, Santa Cruz, Dallas, TX), CEBPA (1:2,000, Proteintech, Rosemont, IL), TRAF4 (1:1,000, Abcam, UK), GAPDH(1:5000, Proteintech, Rosemont, IL). The membranes were subsequently subjected to 2 hof incubation with the appropriate corresponding horseradish peroxidase-conjugated secondary antibody (Santa Cruz, Dallas, TX). Immunoblots were visualized by enhanced chemiluminescence (ECL kit, Beyotime, Shanghai, China) and scanned using ChemImager 5,500 V2.03 software (Alpha Innotech, San Leandro, CA). The relative integrated density values (IDVs) were calculated based on GAPDH as an internal control.
Chromatin Immunoprecipitation Assay
Simple ChIP Enzymatic Chromatin IP Kit (Cell Signaling Technology, Danvers, Massachusetts, USA) was used for chromatin immunoprecipitation (ChIP) assays according to the manufacturer's protocol. Briefly, cells were crosslinked with 1% formaldehyde and collected in lysis buffer. Chromatin was then digested with the micrococcal nuclease. Immunoprecipitates were incubated with 3 μg of anti-TRAF4, anti-PIWIL3 antibody, or normal rabbit IgG followed by immunoprecipitation with Protein G agarose beads during an overnight incubation at 4°C with gentle shaking. As an input reference, 2% were removed before antibody supplementation and stored at -20°C. The ChIP DNA was reverse-cross linked with 5 M NaCl and proteinase K and then purified. Immunoprecipitated DNA was amplified by PCR using primers. The primers for each PCR set, the sizes of PCR products, and annealing temperatures are listed in Table S2 and Table S3. For each PCR, the corresponding input was taken in parallel for PCR validation.
Fluorescence in situ hybridization
For identification of piR-30188, miR-367-3p and OIP5-AS1 rearrangement in glioma cells, piR-30188 probe (green-labeled, Boster, Wuhan, China), miR-367-3p probe (green-labeled, Exiqon, Copenhagen, Denmark), and OIP5-AS1 probe (red-labeled, Boster, Wuhan, China) were used. In brief, slides were treated with PCR-grade proteinaseK (Roche Diagnostics, Mannheim, Germany) after blocking with prehybridization buffer (3% BSA in 4× saline-sodium citrate, SSC). The hybridization mix was prepared with the piR-30188 probe, miR-367-3p probe, or OIP5-AS1 probe in the hybridization solution. Then the slides were washed with washing buffer and the sections were stained with anti-digoxin rhodamine conjugate (1:100, Exon Biotech Inc, Guangzhou, China) at 37°C for 1 h in the dark. Subsequently, the sections were stained with 4', 6-diamidino-2-phenylindole (DAPI, Beyotime, China) for nuclear staining. All fluorescence images (100×) were captured using a fluorescence microscope (Leica, Germany).
Tumor xenografts in nude mice
All animal procedures were performed in accordance with the protocols approved by the Animal Care Committee of the Shengjing Hospital. Lentivirus encoding miR-367-3p was generated using pLenti6.3/V5eDEST Gateway Vector Kit (Life Technologies). The miR-367-3p was ligated into the pLenti6.3/V5eDEST vector. Short-hairpin RNAs targeting human OIP5-AS1 and PIWIL3 CDS region were ligated into LV3-CMV-GFP-Puro vector (GenePharma). Additionally, pLenti6.3/V5eDESTmiR-367, LV3-CMV-GFP-Puro-sh-OIP5-AS1, and LV3-CMV-GFP-Puro-PIWIL3 vectors were generated. The ViraPower Packaging Mix was used to generate Lentivirus in 293FT cells. After infection, the U87 and U251 cells stably expressingmiR-367, sh-OIP5-AS1, and PIWIL3 were picked. The lentiviruses of PIWIL3 were transduced in cells stably transfected with sh-OIP5-AS1 to generate PIWIL3+sh-OIP5-AS1 cells. The lentiviruses of miR-367 were transduced in PIWIL3+sh-OIP5-AS1 cells to generate PIWIL3+sh-OIP5-AS1+miR-367 cells. Four-week-old BALB/C athymic nude mice were obtained from the National Laboratory Animal Center (Beijing, People's Republic of China). The animals were fed with autoclaved food and water during the study. The nude mice were divided into five groups: Control (only U87 or U251), PIWIL3 (U87 or U251 cells with stable over-expression of PIWIL3), miR-367 (U87 or U251 cells with stable over-expression of miR-367), sh-OIP5-AS1 (U87 or U251 cells with stable over-expression of sh-OIP5-AS1), and PIWIL3+sh-OIP5-AS1+miR-367 (OIP5-AS1 inhibition and PIWIL3 and miR-367 stable over-expression in U87 or U251 cells). 3x105 cells were subcutaneously injected into the right flanks of the mice. Tumor volume was measured every 4 days when the tumors were obvious, and the volume was calculated by the formula: volume (mm3) = length×width2/2. Forty days after injection, the mice were sacrificed and tumors were isolated. For survival analysis in orthotopic inoculations, 3x105 cells were stereotactically implanted into the right striatum of the mice. The number of surviving nude mice was recorded, and survival analysis was determined using Kaplan-Meier survival curve.
Statistical analysis
Data are presented as mean ± SD. All statistical analyses were evaluated by SPSS 18.0 statistical software (IBM, New York, NY) with the Student's t-test or one-way analysis of variance. Differences were considered to be significant when P < 0.05.
Results
Functional roles of PIWIL3 and piR-30188 as tumor suppressors and of OIP5-AS1 as an oncogene in glioma tissues and cell lines
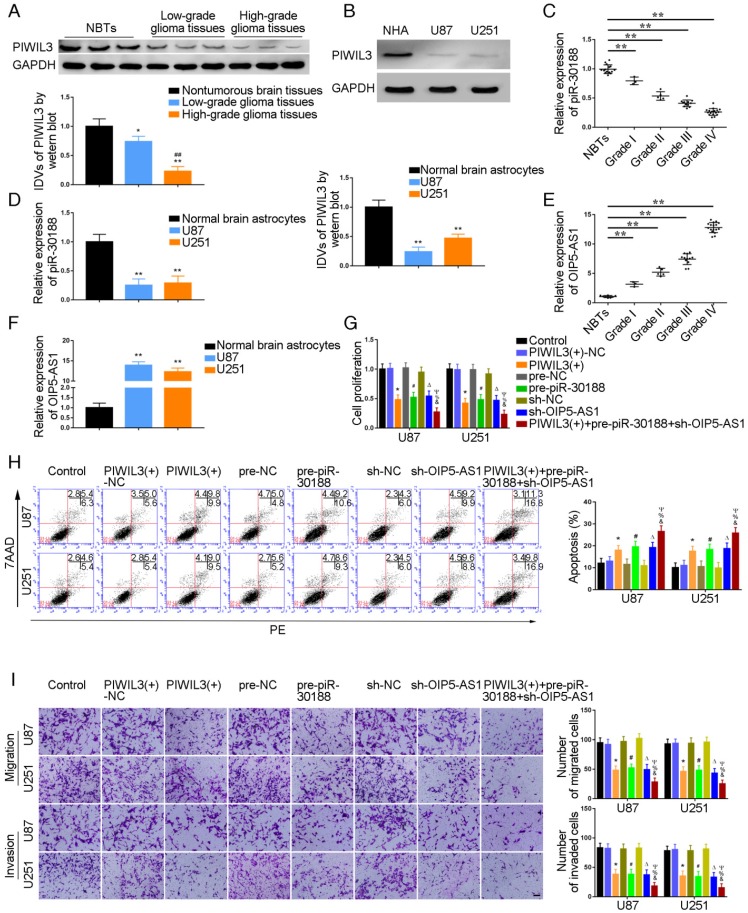
PIWIL3 levels in glioma tissues and U87 and U251 cell lines were analyzed by western blotting (Figure 1A and B). Using microarray analysis and qRT-PCR, piR-30188 expression was found to be down-regulated in U87 and U251 glioma cells (Figure S1A and D). Further, piR-30118 was reintroduced in glioma cells. lncRNA array and qRT-PCR showed that OIP5-AS1 expression was decreased (Figure S1B and E). PiR-30188 and OIP5-AS1 levels in glioma tissues and cell lines were analyzed by quantitative real-time PCR. Fluorescence in situ hybridization (FISH) showed that piR-30188 and OIP5-AS1 were localized in both nucleus and cytoplasm but primarily in the cytoplasm of U87 and U251 cells (Figure S2). As shown in Figure 1A and C, PIWIL3 and piR-30188 expression was decreased and negatively correlated with the pathological grade of glioma; the expression levels were also low in U87 and U251 cells compared with normal human astrocytes (P=0.0016, P=0.0023, respectively) (Figure 1B and D). On the other hand, OIP5-AS1 expression was positively correlated with the pathological grade of glioma (Figure 1E). Compared with normal human astrocytes, OIP5-AS1 was up-regulated in glioma tissues and exhibited high expression levels in U87 and U251 cells (P=0.0014, P=0.0026, respectively) (Figure 1F). To verify the impact of PIWIL3, piR-30188, and OIP5-AS1 on the proliferation of glioma cell lines, CCK-8 assay was performed to assess proliferation. As expected, cell proliferation in PIWIL3(+), pre-piR-30188 and sh-OIP5-AS1 groups was significantly decreased compared with PIWIL3(+)-NC, pre-NC and sh-NC groups (U87 cells: P=0.0017, P=0.0022, P=0.0045, respectively; U251 cells: P=0.0015, P=0.0023, P=0.0030, respectively). Moreover, cell proliferation in the PIWIL3(+)+pre-piR-30188+sh-OIP5-AS1 groups was significantly reduced compared with PIWIL3(+), pre-piR-30188 and sh-OIP5-AS1 groups (U87 cells: P=0.0315, P=0.0193, P=0.0157, respectively; U251 cells: P=0.0440, P=0.0201, P=0.0210, respectively) (Figure 1G). Subsequently, flow cytometry analysis was conducted to measure the impact on cell apoptosis. As displayed in Figure 1H, compared with PIWIL3(+)-NC, pre-NC and sh-NC groups, apoptotic ratios in PIWIL3(+), pre-piR-30188, and sh-OIP5-AS1 groups were significantly increased(U87 cells: P=0.0434, P=0.0184, P=0.0143, respectively; U251 cells: P=0.0276, P=0.0200, P=0.0138, respectively). Moreover, apoptotic ratio was magnified in the PIWIL3(+)+pre-piR-30188+sh-OIP5-AS1 groups compared with PIWIL3(+), pre-piR-30188 and sh-OIP5-AS1 groups(U87 cells: P=0.0121, P=0.0309, P=0.0241, respectively; U251 cells: P=0.0138, P=0.0220, P=0.0279, respectively). Transwell assays revealed that the migrating and invading cell numbers in PIWIL3(+), pre-piR-30188 and sh-OIP5-AS1 groups were reduced compared with PIWIL3(+)-NC, pre-NC and sh-NC groups(Migration analysis, U87: P=0.0026, P=0.0019, P=0.0016, respectively; U251: P=0.0015, P=0.0026, P=0.0014, respectively. Invasion analysis, U87: P=0.0028, P=0.0036, P=0.0027, respectively; U251: P=0.0035, P=0.0039, P=0.0021, respectively). Also, the cotransfection PIWIL3(+)+pre-piR-30188+sh-OIP5-AS1 groups revealed reduced numbers (Migration analysis, U87: P=0.0285, P=0.0134, P=0.0338, respectively; U251: P=0.0254, P=0.0172, P=0.0408, respectively. Invasion analysis, U87: P=0.0294, P=0.0301, P=0.0369, respectively; U251: P=0.0375, P=0.0437, P=0.0444, respectively) (Figure 1I).
Figure 1.
Effect of PIWIL3, piR-30188, and OIP5-AS1 on the biological behavior of glioma cells. (A) PIWIL3 protein levels in normal brain tissue (NBT), low-grade, and high-grade glioma using GAPDH as an endogenous control. Representative protein expressions and their integrated density values (IDVs) of PIWIL3 in NBT (n = 15), low-grade glioma (WHO I-II, WHO I: n=3; WHO II: n=5), and high-grade glioma (WHO III-IV, WHO III: n=9; WHO IV: n=16) are shown (data are presented as the mean ± SD;*P < 0.05 relative to NBTs group; **P < 0.01 relative to NBT group; ##P < 0.01 relative to low-grade glioma tissues group). (B) PIWIL3 protein expression levels in normal human astrocytes (NHA), U87 and U251 cells using GAPDH as an endogenous control. Representative protein expressions and their IDVs in NHA, U87 and U251 are shown (data are presented as the mean ± SD (n = 3, each group);**P < 0.01 relative to NHA group). (C) Expression levels of piR-30188 in glioma tissues of different grades and NBT (data are presented as the mean ± SD (n = 15, each group);**P < 0.01 relative to NBTs group). (D) Expression levels of piR-30188 in NHA and glioma cell lines (data are presented as the mean ± SD (n = 3, each group);**P < 0.01 relative to NHA group). (E) Expression levels of OIP5-AS1 in glioma tissues of different grades and NBT (data are presented as the mean ± SD (n = 15, each group);**P < 0.01 relative to NBT group). (F) Expression levels of OIP5-AS1 in NHA and glioma cell lines (data are presented as the mean ± SD (n = 3, each group); **P < 0.01 relative to NHA group). (G) Cell Counting Kit-8 (CCK-8) assay was used to determine the effect of PIWIL3, piR-30188, and OIP5-AS1 on U87 and U251 cells' proliferation. (H) Flow cytometry analysis of U87 and U251 cells with altered expression of PIWIL3, piR-30188 and OIP5-AS1 (data are presented as the mean ± SD (n =3, each group);* #ΔP < 0.05 relative to NC groups; &%ΨP < 0.05 relative to PIWIL3(+), pre-piR-30188 and sh-OIP5-AS1 groups; NC, negative control). (I) Quantification of number of migrating and invading cells with over-expression of PIWIL3, piR-30188 as well as knockdown of OIP5-AS1. Representative images and corresponding statistical plots are presented (scale bars represent 80μm).
PIWIL3 bound to piR-30188 and OIP5-AS1 was a target of piR-30188
It has been reported that PIWIL3, a member of the Argonaute protein family, binds to piRNA to influence biological functions. To verify this, U87 and U251 cells were cotransfected with PIWIL3 and piR-30188, and their impact on OIP5-AS1 expression was monitored by qRT-PCR. As shown in Figure 2A, OIP5-AS1 expression was significantly attenuated in PIWIL3(+) and pre-piR-30188 groups (U87 cells: P=0.0196, P=0.0245, respectively; U251 cells: P=0.0312, P=0.0103, respectively). The suppressive effect was magnified in the PIWIL3(+)+pre-piR-30188 groups(U87 cells: P=0.0096, P=0.0017,respectively; U251 cells: P=0.0041, P=0.0065, respectively). RNA Binding Protein Immunoprecipitation (RIP) and RNA pulldown experiments confirmed that PIWIL3 binds to piR-30188 (Figure 2B and C). We speculated OIP5-AS1 might harbor one piR-30188 binding site and searched the bioinformatics database Targetscan (Figure 2D). To further verify our speculation, the dual-luciferase reporter assay was performed. As shown in Figure 2E, the luciferase activity was significantly decreased in the OIP5-AS1-Wt+pre-piR-30188 group compared with that in the OIP5-AS1-Wt+pre-NC group (P=0.0013), whereas the luciferase activities of the OIP5-AS1-Mut groups were not affected.
Figure 2.
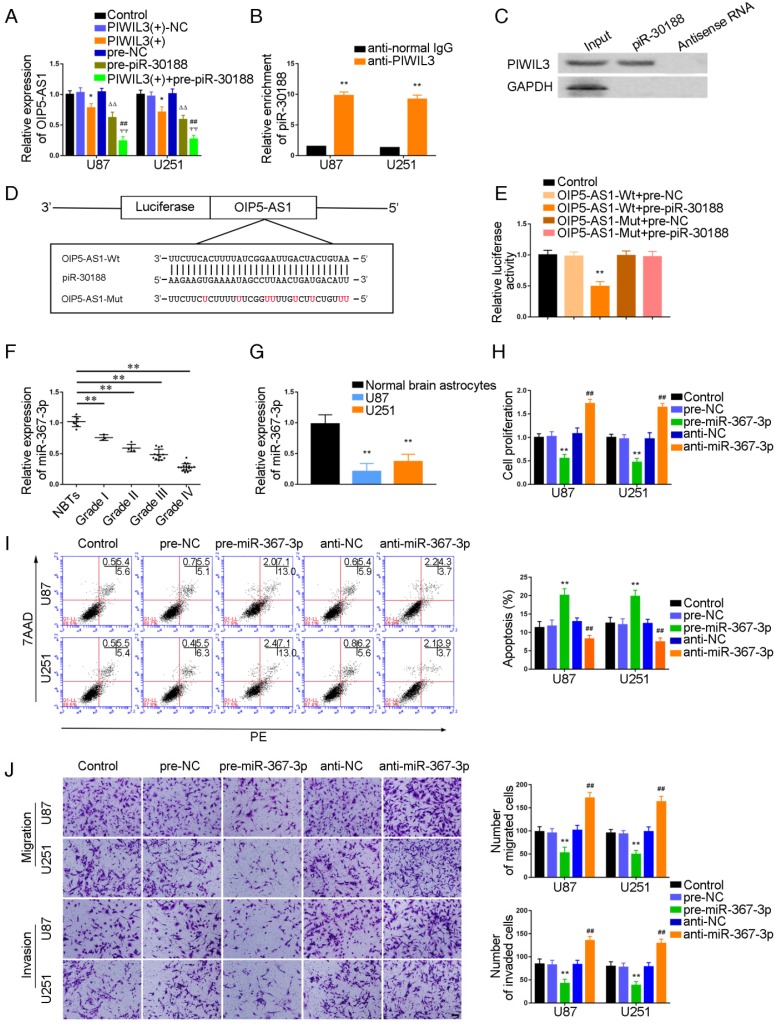
Targeted binding between PIWIL3 and piR-30188, piR-30188, and OIP5-AS1; effect of miR-367-3p on the biological behavior of glioma cells. (A) Quantitative real-time PCR (qRT-PCR) analysis for PIWIL3 and piR-30188 regulating OIP5-AS1 expression in U87 and U251 cells (data are presented as the mean ± SD (n = 3, each group);*P < 0.05 relative to PIWIL3(+)-NC group; ΔΔP< 0.01 relative to pre-NC group; ##ΨΨP< 0.01 relative to PIWIL3(+) and pre-piR-30188 groups). (B, C) RIP and RNA pull-down confirmed that PIWIL3 binds to piR-30188. Relative enrichment of piR-30188 was measured using qRT-PCR (data represent mean ± SD (n= 3, each group);**P< 0.01 relative to anti-normal IgG group). (D) The predicted piR-30188 binding site in OIP5-AS1 (OIP5-AS1-Wt) and the designed mutant sequence (OIP5-AS1-Mut) are illustrated. (E) Luciferase reporter assay of HEK 293T cells co-transfected with OIP5-AS1-Wt or OIP5-AS1-Mut and pre-piR-30188 or pre-NC (data are presented as the mean ± SD (n = 3, each group);**P< 0.01 relative to OIP5-AS1-Wt+pre-NC group). (F) Expression levels of miR-367-3p in glioma tissues of different grades and normal brain tissues (NBT) (data are presented as the mean ± SD (n = 15, each group);**P < 0.01 relative to NBT group). (G) Expression levels of miR-367-3p in normal human astrocytes (NHA) and glioma cell lines (data are presented as the mean ± SD (n = 3, each group);**P <0.01 relative to NHA group). (H) Cell Counting Kit-8 (CCK-8) assay was applied to evaluate the effect of miR-367-3p on U87 and U251 cells proliferation. (I) Flow cytometry analysis of U87 and U251 cells with altered expression of miR-367-3p (data are presented as the mean ± SD (n =3, each group); **P < 0.01 relative to pre-NC group; ##P < 0.01 relative to anti-NC group). (J) Quantification of number of migrating and invading cells with over-expression or knockdown of miR-367-3p. Representative images and corresponding statistical plots are presented (scale bars represent 80μm).
MiR-367-3p was down-regulated in glioma tissues and cell lines, acted as a tumor-suppressor, and was a target of OIP5-AS1
Having confirmed OIP5-AS1 exerted an oncogenic role in glioma cells, miRNA microarray analysis was performed in glioma cells treated with the OIP5-AS1 knockdown. As shown in Figure S1C and F, miR-367-3p was up-regulated in cells treated with sh-OIP5-AS1. Hence, miR-367-3p might be involved in OIP5-AS1-mediated regulation in glioma cells. miR-367-3p levels in glioma tissues and cell lines were analyzed by qRT-PCR. miR-367-3p was localized in both nucleus and cytoplasm but primarily in the cytoplasm of U87 and U251 cells by fluorescence in situ hybridization (FISH) (Figure S2). MiR-367-3p expression was decreased and negatively correlated with the pathological grade of glioma (Figure 2F) and showed decreased expression in U87 and U251 cells compared with normal human astrocytes(P=0.0026 and P=0.0053,respectively) (Figure 2G). To verify the effect of miR-367-3p on the proliferation of glioma cell lines, U87 and U251 cells were transiently transfected with miR-367-3p. As expected, cell proliferation in the pre-miR-367-3p groups was significantly decreased compared to the pre-NC groups, whereas the anti-miR-367-3p groups exhibited higher proliferation (U87 cells: P=0.0038; U251 cells: P=0.0022) (Figure 2H). Flow cytometry analysis was conducted to measure the impact of miR-367-3p on cell apoptosis. As shown in Figure 2I, the apoptotic ratio in pre-miR-367-3p groups was significantly higher compared with pre-NC groups, whereas anti-miR-367-3p groups exhibited decreased apoptotic ratios compared with anti-NC groups(U87 cells: P=0.0049; U251 cells: P=0.0055). Transwell assays revealed that the migrating and invading cell numbers in pre-miR-367-3p groups were significantly diminished in pre-miR-367-3p groups, whereas anti-miR-367-3p groups exhibited higher numbers compared with anti-NC groups (Migration analysis, U87 cells: P=0.0017; U251 cells: P=0.0022.Invasion analysis, U87 cells: P=0.0023; U251 cells: P=0.0025) (Figure 2J).
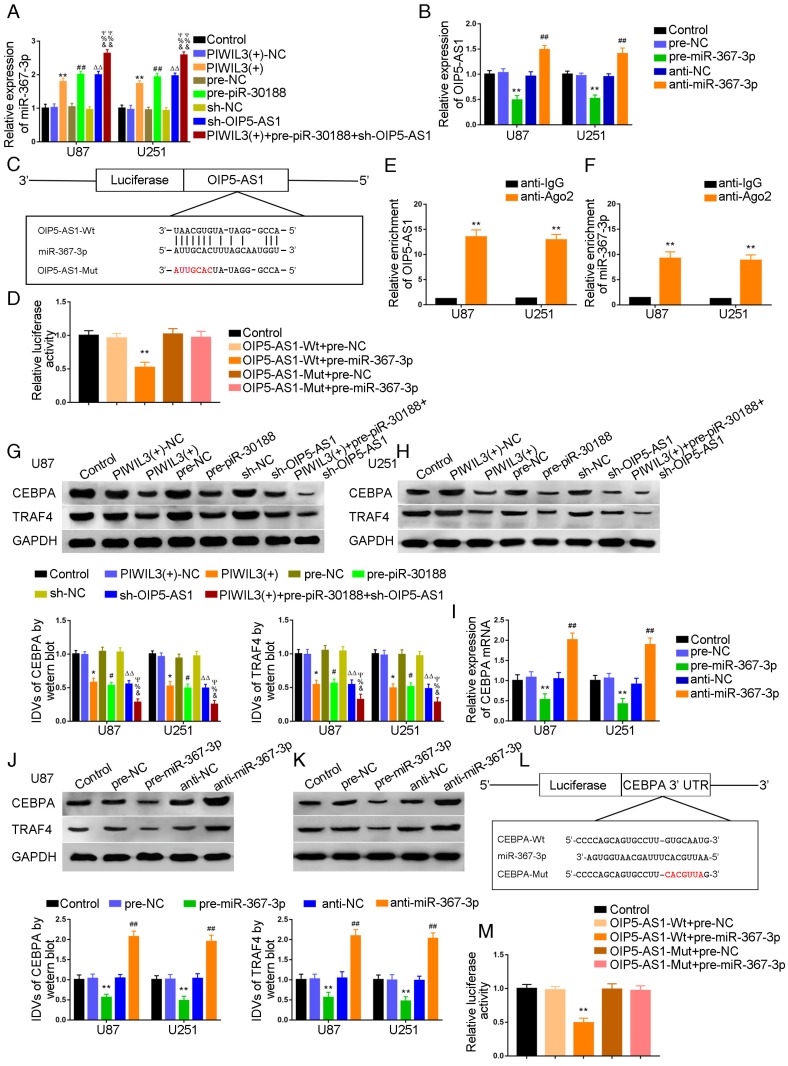
We subsequently studied the effect of PIWIL3, piR-30188, and OIP5-AS1 on the level of miR-367-3p
U87 and U251 cells were transfected with PIWIL3 and piR-30188, respectively, and cotransfected with PIWIL3, piR-30188, and OIP5-AS1. miR-367-3p expression in each group was quantified with qRT-PCR (Figure 3A). The PIWIL3(+), pre-piR-30188, and sh-OIP5-AS1 groups exhibited higher miR-367-3p expression compared with PIWIL3(+)-NC, pre-NC and sh-NC groups, respectively. The effect was magnified in the PIWIL3(+)+pre-piR-30188+sh-OIP5-AS1 groups. In most cases, lncRNA and miRNA reciprocally regulate each other's expression. Therefore, we hypothesized that miR-367-3p exhibits a negative regulatory effect on OIP5-AS1. To confirm our prediction, U87 and U251 cells were transiently transfected with miR-367-3p. OIP5-AS1 expression was analyzed with qRT-PCR. As shown in Figure 3B, OIP5-AS1 level in the pre-miR-367-3p group was significantly lower than that of the pre-NC group, whereas the anti-miR-367-3p groups exhibited higher expression than the anti-NC groups (U87 cells: P=0.0016; U251 cells: P=0.0031). The bioinformatics database Targetscan suggested OIP5-AS1 harbors one miR-367-3p binding site (Figure 3C). A dual-luciferase reporter assay was conducted to determine the binding site of OIP5-AS1 and miR-367-3p. The luciferase activity in the OIP5-AS1-Wt+pre-miR-367-3p group was significantly reduced compared to that in the control group, whereas the luciferase activities in the OIP5-AS1-Mut groups were not affected (P=0.0019) (Figure 3D). Also, RIP results proved that OIP5-AS1 could impair miR-367-3p expression in a sequence-dependent manner (Figure 3E and F).
Figure 3.
Over-expression of PIWIL3, piR-30188and knockdown of OIP5-AS1 inhibited CEBPA expression by down-regulating miR-367-3p. (A) Quantitative real-time PCR (qRT-PCR) analysis for PIWIL3, piR-30188, and OIP5-AS1-regulated miR-367-3p expression in U87 and U251 cells (data are presented as the mean ± SD (n =3, each group); *#ΔP < 0.05 relative to normal control (NC)groups; &#ΨP< 0.05 relative to PIWIL3(+), pre-piR-30188 and sh-OIP5-AS1 groups). (B) OIP5-AS1 expression levels altered by over-expression or knockdown of miR-367-3p (data are presented as the mean ± SD (n =3, each group); **P < 0.01 relative to pre-NC group; ##P < 0.01 relative to anti-NC group). (C) The predicted miR-367-3p binding site in OIP5-AS1 (OIP5-AS1-Wt) and the designed mutant sequence (OIP5-AS1-Mut) are illustrated. (D) Luciferase reporter assay of HEK 293T cells co-transfected with OIP5-AS1-Wt or OIP5-AS1-Mut and pre-miR-367-3p or pre-NC (data are presented as the mean ± SD (n = 3, each group);**P< 0.01 relative to OIP5-AS1-Wt+pre-NC group). (E, F) RIP confirmed that OIP5-AS1 and miR-367-3p were in the RISC complex. (G, H) Effects of over-expression of PIWIL3, piR-30188 as well as knockdown of OIP5-AS1 on CEBPA expression. The integrated density values (IDVs) of CEBPA are shown using GAPDH as an endogenous control (data are presented as the mean ± SD (n =3, each group);*#P < 0.05 relative to NC groups; ΔΔP< 0.01 relative to NC groups; &%ΨP< 0.05 relative to PIWIL3(+), pre-piR-30188, and sh-OIP5-AS1 groups). (I-K) qRT-PCR and Western blot analysis for miR-367-3p-regulated CEBPA expression in U87 and U251 cells. The relative expression of CEBPA is shown using GAPDH as an endogenous control. The IDVs of CEBPA are shown using GAPDH as an endogenous control (data are presented as the mean ± SD (n =3, each group); **P < 0.01 relative to pre-NC group; ##P<0.01 relative to anti-NC group). (L) The predicted miR-367-3p binding site in CEBPA (CEBPA-Wt) and the designed mutant sequence (CEBPA-Mut) are illustrated. (M) Luciferase reporter assay of HEK 293T cells co-transfected with CEBPA-Wt or CEBPA-Mut and pre-miR-367-3p or pre-NC (data are presented as the mean ± SD (n = 3, each group);**P< 0.01 relative to CEBPA-Wt+pre-NC group).
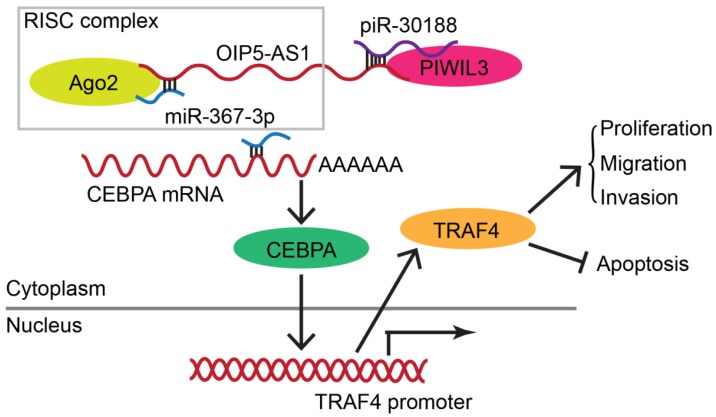
Molecular networks involving PIWIL3, piR-30188, miR-367-3p, and OIP5-AS1 and their downstream targets CEBPA and TRAF4
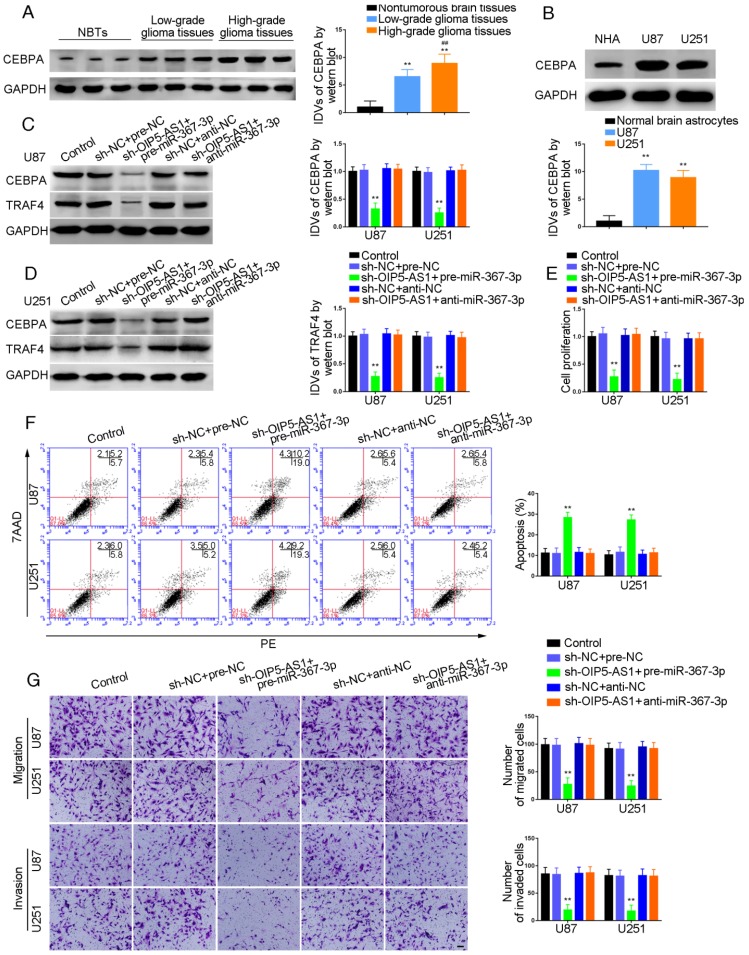
We conducted Western blotting to explore the impact of PIWIL3, piR-30188, OIP5-AS1, and miR-367-3p on the expression of CEBPA and TRAF4. Glioma cell lines U87 and U251 cells were stably transfected with PIWIL3, piR-30188, and OIP5-AS1 or cotransfected with PIWIL3, piR-30188 and OIP5-AS1. Reduced expression of CEBPA and TRAF4 was observed in PIWIL3(+), pre-piR-30188, and sh-OIP5-AS1 groups compared with PIWIL3(+)-NC, pre-NC and sh-NC groups, respectively. The suppressive effect was magnified in the PIWIL3(+)+pre-piR-30188+sh-OIP5-AS1 groups (Figure 3G and H). We then transiently transfected U87 and U251 cells with miR-367-3p. CEBPA mRNA expression was assessed by qRT-PCR and was found to be significantly reduced in the pre-miR-367-3p group compared with the pre-NC group. In contrast, the anti-miR-367-3p groups exhibited significantly higher expression than the anti-NC groups (for U87 cells: P=0.0020; for U251 cells: P=0.0016) (Figure 3I). Western blotting indicated decreased expression of CEBPA and TRAF4 in the pre-miR-367-3p groups than in the pre-NC groups, whereas the anti-miR-367-3p groups revealed higher expression than the anti-NC groups (Figure 3J and K). By searching the bioinformatics database Targetscan, we noticed that the 3'UTR region of CEBPA might harbor one miR-367-3p binding site (Figure 3L). We therefore performed the dual-luciferase reporter assay. As shown in Figure 3M, the luciferase activity was significantly decreased in the CEBPA-Wt+pre-miR-367-3p group compared with that of the CEBPA-Wt+pre-NC group, whereas the luciferase activity of the CEBPA-Mut group was not affected (P=0.0021). Western blot analysis also proved that CEBPA was significantly up-regulated in glioma tissues compared with normal brain tissues and was positively correlated with the pathological grade of glioma. CEBPA also exhibited high expression levels in U87 and U251 cells compared with normal human astrocytes (Figure 4A and B).
Figure 4.
Effects of OIP5-AS1 and miR-367-3p on the biological behavior of glioma cells: over-expression of OIP5-AS1 elevated CEBPA expression via down-regulating miR-367-3p. (A) CEBPA protein expression levels in normal brain tissues (NBT), low-grade, and high-grade glioma using GAPDH as an endogenous control. Representative protein expression and their integrated density values (IDVs) of CEBPA in NBT, low-grade glioma tissues (WHO I-II), and high-grade glioma (WHO III-IV) are shown (data are presented as the mean ± SD (n = 15, each group);**P < 0.01 relative to NBTs group; ##P < 0.01 relative to low-grade glioma tissues group). (B) CEBPA protein expression levels in normal human astrocytes (NHA), U87 and U251 cells using GAPDH as an endogenous control. Representative protein expressions and their IDVs in NHA, U87 and U251 are shown (data are presented as the mean ± SD (n = 3, each group);**P < 0.01 relative to NHA group). (C, D) Western blot analysis forOIP5-AS1 and miR-367-3p-regulatedCEBPA expression using GAPDH as endogenous control (data are presented as the mean ± SD (n = 3, each group);**P < 0.01 relative to sh-NC+pre-NC group). (E) Cell Counting Kit-8 (CCK-8) assay was used to evaluate the effect of OIP5-AS1 and miR-367-3p on U87 and U251 cells' proliferation. (F) Flow cytometry analysis of U87 and U251 cells with altered expression of OIP5-AS1 and miR-367-3p (data are presented as the mean ± SD (n =3, each group);**P < 0.01 relative to sh-NC+pre-NC group). (G) Quantification of number of migrating and invading cells with altered expression of OIP5-AS1 and miR-367-3p. Representative images and corresponding statistical plots are presented (scale bars represent 80μm).
Knockdown of OIP5-AS1 combined with over-expression of miR-367-3p decreased CEBPA and TRAF4 expression and inhibited glioma cells growth
To investigate whether miR-367-3p intensified the tumor-suppressive effect of OIP5-AS1 knockdown, we transfected OIP5-AS1-silenced U87 and U251 cells with miR-367-3p. As shown in Figure 4C, sh-OIP5-AS1+pre-miR-367-3p groups exhibited lower levels of CEBPA and TRAF4 expression compared with sh-NC+pre-NC groups, whereas there was no significant difference between sh-NC+anti-NC and sh-OIP5-AS1+anti-miR-367-3p groups (Figure 4C and D). Cell proliferation in sh-OIP5-AS1+pre-miR-367-3p groups was attenuated compared with sh-NC+pre-NC groups, while sh-OIP5-AS1+anti-miR-367-3p groups showed no significant difference from sh-NC+anti-NC groups (U87 cells: P=0.0015; U251 cells: P=0.0016) (Figure 4E). Flow cytometry analysis demonstrated that the apoptotic ratios in sh-OIP5-AS1+pre-miR-367-3p groups were higher compared with sh-NC+pre-NC groups, while sh-OIP5-AS1+anti-miR-367-3p groups showed no significant difference from sh-NC+anti-NC groups (U87 cells: P=0.0013; U251 cells: P=0.0016) (Figure 4F). Transwell assays were conducted to assess the migrating and invading cell numbers. The numbers in sh-OIP5-AS1+pre-miR-367-3p groups were attenuated compared with sh-NC+pre-NC groups, while sh-OIP5-AS1+anti-miR-367-3p groups showed no significant difference from sh-NC+anti-NC groups (Migration analysis, U87 cells: P=0.0019; U251 cells: P=0.0018. Invasion analysis, U87 cells: P=0.0020; U251 cells: P=0.0021) (Figure 4G).
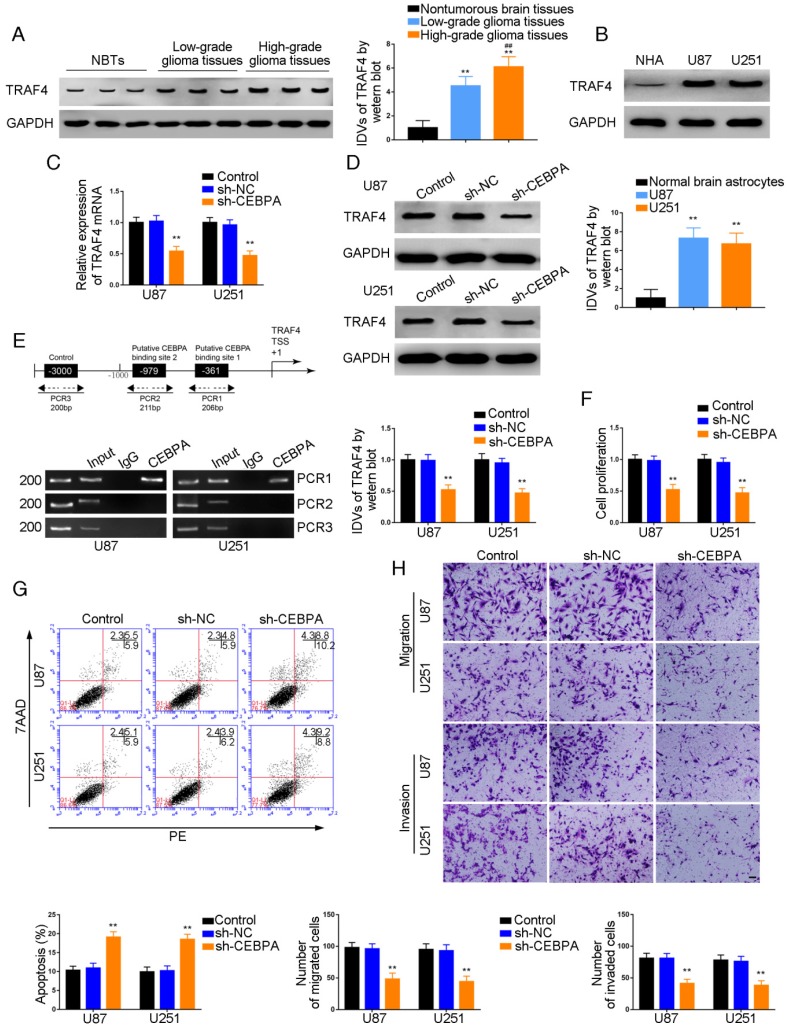
TRAF4 was a target of CEBPA and knockdown of CEBPA restrained malignant growth of glioma
Western blot analysis demonstrated a positive correlation of TRAF4 expression with pathological grade of glioma. TRAF4 was up-regulated in glioma tissues compared with normal brain tissues and also exhibited high expression in U87 and U251 cells compared with normal human astrocytes (P=0.0015, P=0.0025, respectively) (Figure 5A and B). U87 and U251 cells with CEBPA knockdown were established and qRT-PCR was used to assess the expression level of TRAF4 mRNA. As shown in Figure 5C, TRAF4 mRNA expression was attenuated in sh-CEBPA groups compared with sh-NC groups (P=0.0024, P=0.0017, respectively) and TRAF4 expression, as confirmed by Western blotting, was decreased (P=0.0028, P=0.0013, respectively) (Figure 5D). By using the bioinformatics software JASPAR, we predicted that the TRAF4 promoter might harbor two upstream putative binding sites for CEBPA. ChIP assays were performed to verify the putative correlation between TRAF4 promoter and CEBPA. The 3000 bp upstream region of the putative CEBPA binding site was amplified by PCR and used as a negative control. There was a direct association of CEBPA with the putative binding site1 of TRAF4 (Figure 5E), but there were no interactions of CEBPA with the negative control or putative binding site 2. CCK-8 assays revealed cell proliferation in sh-CEBPA groups was significantly suppressed in contrast to sh-NC groups (U87 cells: P=0.0023; U251 cells: P=0.0018) (Figure 5F). Flow cytometry analysis demonstrated that the apoptotic ratio of sh-CEBPA groups was higher compared with that of sh-NC groups (U87 cells: P=0.0017; U251 cells: P=0.0015) (Figure 5G). Transwell assays showed sh-CEBPA groups had lower numbers of migration and invasion in contrast to sh-NC groups (Figure 5H).
Figure 5.
Effect of CEBPA on the biological behavior of glioma cells via up-regulation of TRAF4. (A) TRAF4 protein expression levels in normal brain (NBT), low-grade, and high-grade glioma using GAPDH as an endogenous control. Representative protein expressions and their integrated density values (IDVs) of TRAF4 NBT, low-grade glioma (WHO I-II), and high-grade glioma (WHO III-IV) are shown (data are presented as the mean ± SD (n = 15, each group);**P < 0.01 relative to NBTs group; ##P < 0.01 relative to low-grade glioma group). (B) TRAF4 protein expression levels in normal human astrocytes (NHA), U87 and U251 cells and using GAPDH as an endogenous control. Representative protein expressions and their IDVs in NHA, U87 and U251 are shown (data are presented as the mean ± SD (n = 3, each group);**P < 0.01 relative to NHA group). (C, D) qRT-PCR and Western blot analysis for CEBPA-regulatedTRAF4 expression in U87 and U251 cells. The relative expression of TRAF4 is shown using GAPDH as an endogenous control. The IDVs of TRAF4 are shown using GAPDH as an endogenous control (data are presented as the mean ± SD (n =3, each group; **P < 0.01 relative to sh-NC group). (E) CEBPA bound to the promoters of TRAF4 in U87 and U251 glioma cells. Transcription start site (TSS) was designated as +1. Putative CEBPA binding sites are illustrated. Immunoprecipitated DNA was amplified by PCR. Normal rabbit IgG was used as a negative control. (F) Cell Counting Kit-8 (CCK-8) assay was used to evaluate the effect of CEBPA on U87 and U251 cells' proliferation. (G) Flow cytometry analysis of U87 and U251 cells affected by CEBPA (data are presented as the mean ± SD (n =3, each group);**P < 0.01 relative to sh-NC group). (H) Quantification of number of migrating and invading cells affected by CEBPA. Representative images and corresponding statistical plots are presented (scale bars represent 80μm).
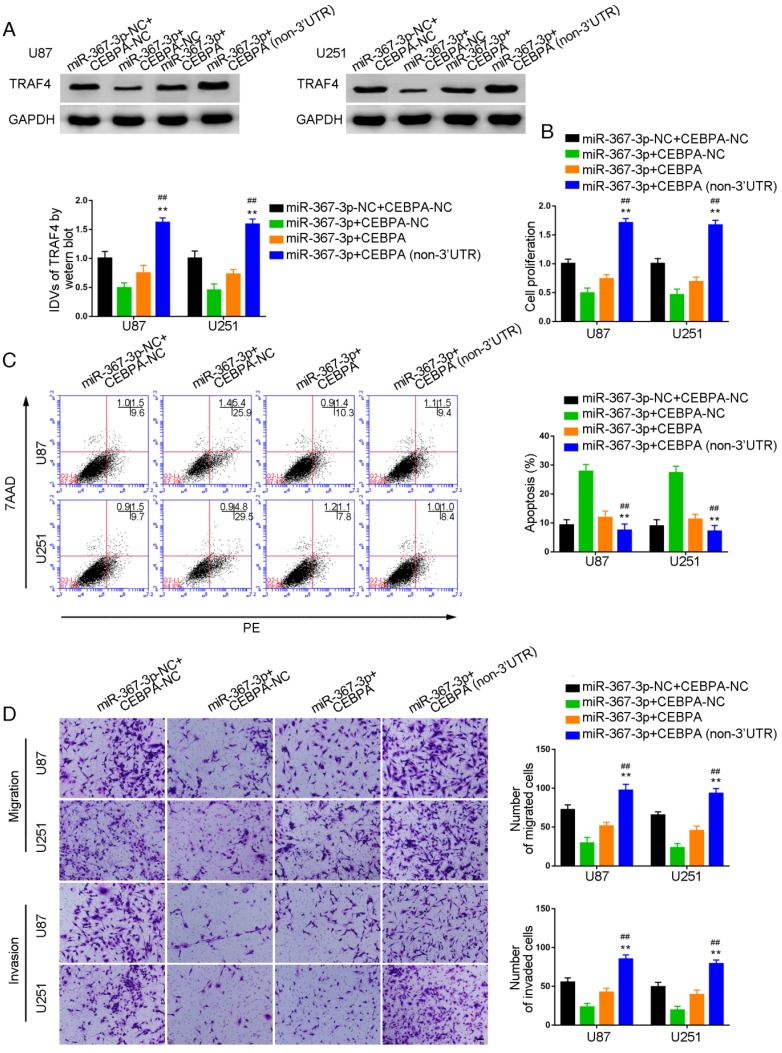
Over-expression of miR-367-3p inhibited CEBPA-induced malignant growth of glioma and promoted apoptosis by targeting CEBPA 3'-UTR
To investigate whether CEBPA could reverse the miR-367-3p-mediated inhibition of malignant biological behavior in glioma, we assessed the effect of expression of TRAF4 on cell proliferation, apoptosis, migration, and invasion in U87 and U251 glioma cells stably expressing miR-367-3p+CEBPA (non-3'UTR). Western blot analysis of TRAF4 expression is illustrated in Figure 6A. TRAF4 was highly expressed and the proliferation of U87 and U251 cells was significantly increased in miR-367-3p+CEBPA (non-3'UTR) groups as compared to miR-367-3p+CEBPA-NC and miR-367-3p+CEBPA groups (Figure 6B). Flow cytometry analysis showed increased apoptotic ratios (Figure 6C) and Transwell assays revealed increased numbers of migrating and invading cells in miR-367-3p+CEBPA (non-3'UTR) groups compared to miR-367-3p+CEBPA-NC and miR-367-3p+CEBPA groups (Figure 6D).
Figure 6.
MiR-367-3p regulated the biological behavior of glioma cells via targeting CEBPA 3'UTR. (A) Western blot analysis of miR-367-3p-regulatedTRAF4 expression via targeting CEBPA 3'UTR shown using GAPDH as endogenous control (data are presented as the mean ± SD (n = 3, each group);**P < 0.01 relative to miR-367-3p+CEBPA-NC group; ##P < 0.01 relative to miR-367-3p+CEBPA group). (B) Cell Counting Kit-8 (CCK-8) assay was used to evaluate the effect of miR-367-3p and CEBPA on U87 and U251 cells' proliferation. (C) Flow cytometry analysis of U87 and U251 cells with altered expression of miR-367-3p and CEBPA (data are presented as the mean ± SD (n =3, each group);**P < 0.01 relative to miR-367-3p+CEBPA-NC group; ##P< 0.01 relative to miR-367-3p+CEBPA group). (D) Quantification of number of migrating and invading cells with altered expression of miR-367-3p and CEBPA. Representative images and corresponding statistical plots are presented (scale bars represent 80μm).
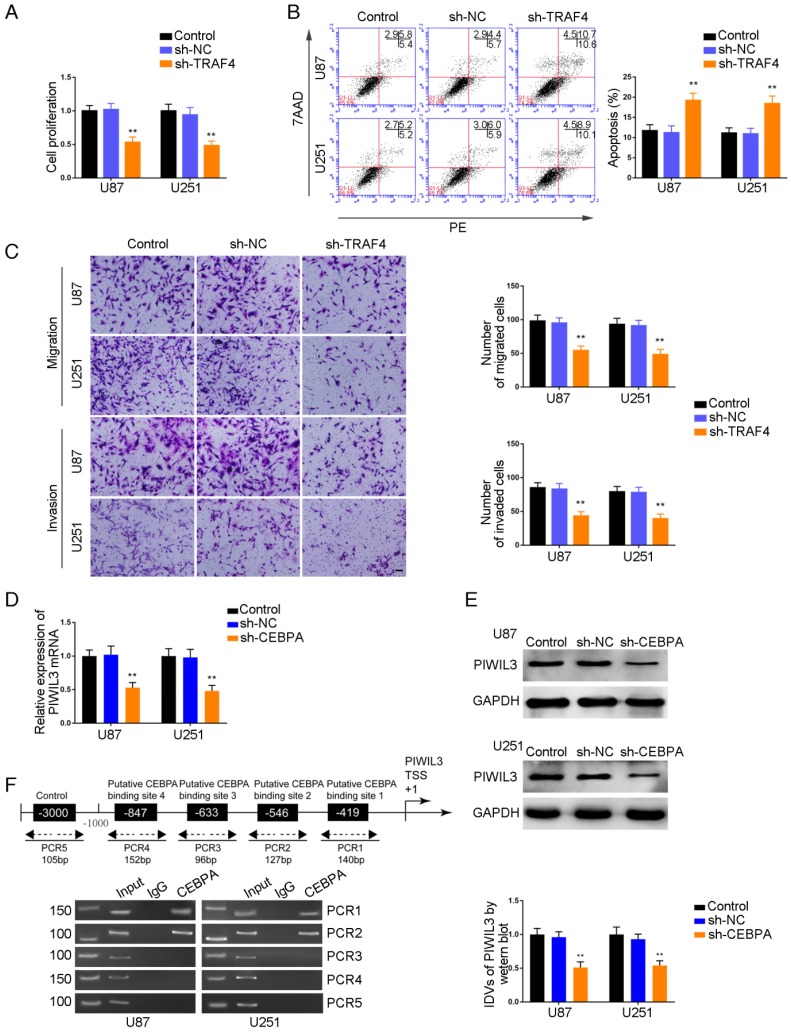
TRAF4 exerted oncogenic function in glioma cells, and PIWIL3 was a target of CEBPA
CCK-8 assays were conducted to assess proliferation in TRAF4-knocked down U87 and U251 cells. As shown in Figure 7A and 7B, respectively, sh-TRAF4 groups showed decreased proliferation (U87 cells: P=0.0021; U251 cells: P=0.0036) and increased apoptotic ratios by flow cytometry (U87 cells: P=0.0050; U251 cells: P=0.0053) compared withsh-NC groups. Transwell assays verified decreased numbers of migrating and invading cells in sh-TRAF4 groups compared with sh-NC groups (Figure 7C).The bioinformatics software JASPAR predicted the putative binding sites in PIWIL3 for CEBPA. We established U87 and U251 cells with knocked-down of CEBPA and showed attenuation of PIWIL3 mRNA expression by qRT-PCR (Figure 7D) and protein expression by Western blotting (Figure 7E) in sh-CEBPA groups compared with sh-NC groups (U87 cells: P=0.0026; U251 cells: P=0.0028). ChIP assays were performed to verify the putative correlation between thePIWIL3 promoter and CEBPA. The 3000 bp upstream region of the putative CEBPA binding, which was not expected to associate with CEBPA, was amplified using PCR and used as a negative control. There was a direct association of CEBPA with putative binding sites1 and 2 of PIWIL3 (Figure 7F), but there were no interactions of CEBPA with the control region or putative binding sites3 and 4. These results suggested CEBPA and PIWIL3 may form a reciprocal feedback loop.
Figure 7.
Effect of TRAF4 on the biological behavior of glioma cells. (A) Cell Counting Kit-8 (CCK-8) assay was used to evaluate the effect of TRAF4 on U87 and U251 cells' proliferation. (B) Flow cytometry analysis of U87 and U251 cells affected by TRAF4 (data are presented as the mean ± SD (n =3, each group);**P < 0.01 relative to sh-NC group). (C) Quantification of number of migrating and invading cells affected by TRAF4. Representative images and corresponding statistical plots are presented (scale bars represent 80μm). (D, E) qRT-PCR and Western blot analysis for miR-367-3p-regulatedCEBPA expression in U87 and U251 cells. The relative expression of CEBPA is shown using GAPDH as an endogenous control. The integrated density values (IDVs) of CEBPA are shown using GAPDH as an endogenous control (data are presented as the mean ± SD (n =3, each group); **P < 0.01 relative to pre-NC group). (F) CEBPA bound to the promoter of PIWIL3 in U87 and U251 glioma cells. Transcription start site (TSS) was designated as +1. Putative CEBPA binding sites are illustrated. Immunoprecipitated DNA was amplified by PCR. Normal rabbit IgG was used as a negative control.
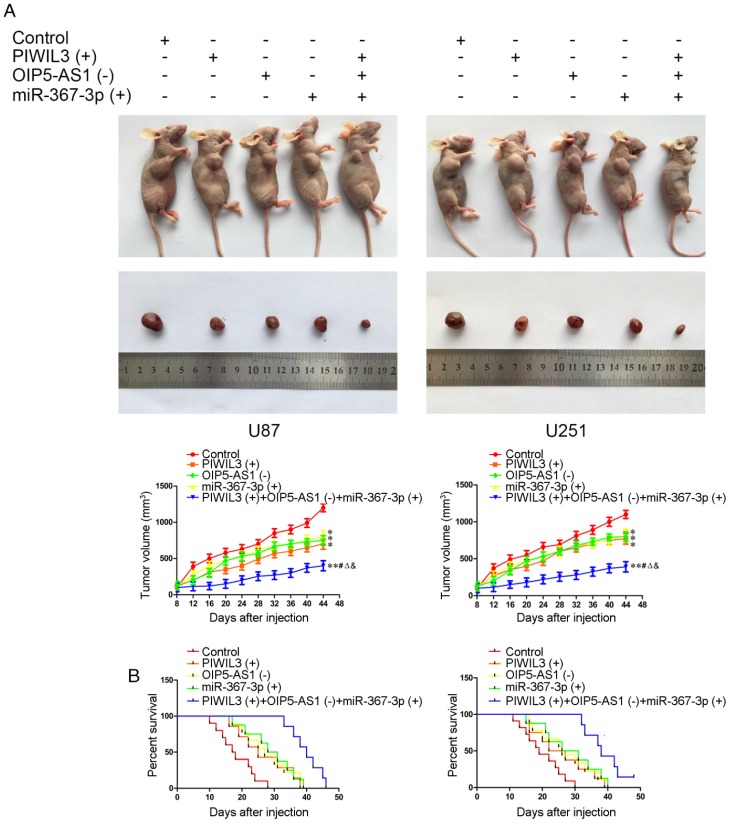
Knockdown of OIP5-AS1 combined with over-expression of PIWIL3 and miR-367-3p restrained tumor growth and prolonged nude mice survival
The in vivo results are illustrated in Figure 8A. In contrast to control groups, PIWIL3(+), OIP5-AS1(-) and miR-367-3p(+) groups produced smaller tumor volumes(U87 cells: P=0.032, P=0.011, P=0.047, respectively; U251 cells: P=0.015, P=0.024, P=0.021, respectively). Also, PIWIL3(+)+OIP5-AS(-)+miR-367-3p(+) groups produced smaller tumors than their respective controls, PIWIL3(+), OIP5-AS1(-) and miR-367-3p(+) groups. The Kaplan-Meier method was further used to estimate the overall survival time for each group. PIWIL3(+), OIP5-AS1(-) and miR-367-3p(+) groups exhibited longer survival compared with control groups. Moreover, PIWIL3(+)+OIP5-AS1(-)+miR-367-3p(+) groups exhibited the longest survival (Figure 8B).
Figure 8.
In vivo study. (A) Cells with stable expression were used for the in vivo study. Nude mice harboring tumors are presented and sample tumors from the respective groups are shown. (B) Tumor volume was calculated every 4 days after injection, and the tumor was excised after 40 days (*P < 0.05 relative to Control group; **P < 0.01 relative to Control group;#P< 0.05 relative to PIWIL3(+) group; ΔP< 0.05 relative to OIP5-AS1(-) group; &P < 0.05 relative to miR-367-3p(+) group).Survival curves of nude mice injected in the right striatum (n = 15;P< 0.05 for PIWIL3(+), OIP5-AS1(-) and miR-367-3p(+) groups relative to Control group; P< 0.01 for PIWIL3(+)+OIP5-AS1(-)+miR-367-3p(+) relative to Control group).
Discussion
This study provided evidence that PIWIL3, piR-30188, and miR-367-3p are expressed at low levels in glioma tissues and are negatively correlated with the pathological grade of glioma. Decreased expression of PIWIL3, piR-30188, and miR-367-3p was also observed in U87 and U251 cell lines, whereas OIP5-AS1 was up-regulated in glioma tissues and cell lines and was positively correlated with the pathological grade of glioma. Over-expression of PIWIL3, piR-30188, and miR-367-3p, and knockdown of OIP5-AS1 resulted in the inhibition of malignant growth of glioma cells. Over-expression of PIWIL3 and piR-30188, together or individually, inhibited OIP5-AS1 expression. PIWIL3 and piR-30188 over-expression as well as OIP5-AS1 knockdown attenuated glioma cells progression, with the effect of the combined application being the most significant. RNA IP and RNA Pull-down experiments demonstrated that piR-30188 bound to PIWIL3. Dual luciferase assays confirmed that piR-30188 targeted and specifically bound to OIP5-AS1 and miR-367-3p. Knockdown of OIP5-AS1 increased miR-367-3p expression, whereas over-expression of miR-367-3p decreased OIP5-AS1 expression in a sequence-dependent manner. PIWIL3 and piR-30188 over-expression as well as the OIP5-AS1 knockdown and their combined application attenuated glioma cells progression by up-regulating miR-367-3p. CEBPA and TRAF4 expression was up-regulated in glioma tissues and cells and was positively correlated with the pathological grade of glioma. Knockdown of CEBPA and TRAF4 significantly attenuated glioma cells malignant growth and CEBPA knockdown reduced TRAF4 mRNA expression. Over-expression of miR-367-3p decreased CEBPA and its mRNA expression, whereas knockdown of miR-367-3p increased CEBPA and its mRNA expression. Dual-luciferase assays showed that miR-367-3p specifically bound to the 3'UTR of CEBPA mRNA. CHIP assays revealed binding of CEBPA to the promoter of TRAF4. PIWIL3 and piR-30188 over-expression as well as the OIP5-AS1 knockdown and their combined application inhibited CEBPA and TRAF4 expression. In vivo studies suggested that over-expression of PIWIL3 and miR-367-3p together with OIP5-AS1 knockdown produced the smallest xenograft volume in contrast to their respective single application, contributing to the longest survival.
Recently, the PIWI protein family has been the subject of intense studies. The expression of PIWIL proteins varies in different tumors and, as reported recently, PIWIL1, PIWIL2, and PIWIL4 are expressed at low levels in human renal cell carcinoma and are related to shortened survival 33. PIWIL2 has weak cytoplasmic but no nuclear expression and is associated with the progression of bladder cancer, suggesting it acts as a tumor suppressor 34. However, PIWIL2 is up-regulated in colorectal tumors and is positively correlated with poor prognosis, indicating it acts as an oncogene 35. PIWIL3 expression is closely related to survival and recurrence of breast cancer 8. It is highly expressed in the primary site and metastatic lymph nodes of human stage III epithelial ovarian cancer 10. Our study demonstrated that expression of PIWIL3 was decreased in glioma tissues and cells, whereas its over-expression attenuated the malignant behavior of glioma cells. These results indicate the complexity and diversity of PIWIL3 functions in various cancers. Our study proved, for the first time, that piR-30188 expression was decreased in U87 and U251 glioma cells and negatively correlated with the pathological grade of glioma. Over-expression of piR-30188 inhibited the proliferation, migration, and invasion of glioma cells, simultaneously promoting apoptosis. However, there are no reports providing mechanistic insights into the role of piR-30188 in tumors.
So far, piRNAs have been found to correlate with various malignancies. PiR-651 is highly expressed in prostate and breast cancer cells 36 whereas PiR-823 has decreased expression in renal cell carcinoma and is correlated with poor prognosis 37. Genome-wide association as well as functional studies showed that piR-598 containing the variant allele at rs147061479 promoted glioma growth and increased its risk 38. piRNAs regulate cell functions via forming piRNA complexes with PIWI proteins from the Argonaute family and silencing their downstream genes 39. Our study confirmed that piR-30188 bound to PIWIL3. Combined over-expression of PIWIL3 and piR-30188 contributed to the attenuation of the proliferation, migration, and invasion of glioma cells while promoting apoptosis, suggesting that piR-30188 was involved in PIWIL3's regulation of the biological behavior of glioma cells.
lncRNAs are involved in the genesis and progression of various tumors. We have previously reported that CRNDE and TUG1 exert regulatory effects on the biological behavior of glioma cells as ceRNAs 40-42. The current study confirmed the up-regulation of OIP5-AS1 in glioma tissues and its positive correlation with the pathological grade of glioma. OIP5-AS1, whose knockdown inhibited the malignant progression of glioma cells, was highly expressed in U87 and U251 cells. These results demonstrated that OIP5-AS1 acts as an oncogenic factor in glioma tissues and cells. Our results have further illustrated that over-expression of PIWIL3 and piR-30188, individual or combined, significantly suppressed OIP5-AS1 expression. This indicated that piR-30188 bound to PIWIL3 exerted an inhibitory effect on OIP5-AS1. Dual luciferase assays further confirmed the binding site between piR-30188 and OIP5-AS1. PIWIL3 and piR-30188 over-expression as well as OIP5-AS1 knockdown and their combined application reduced the malignant progression of glioma cells. Moreover, the combined application showed an additive effect, suggesting the combination of PIWIL3, piR-30188, and OIP5-AS1 played a key role in regulating the malignant behavior of glioma cells.
miR-367-3p, a member of the miR-302/367 cluster, was expressed at a low level in glioma tissues as well as U87 and U251 cells and was negatively correlated with the pathological grade of glioma. Over-expression of miR-367-3p hindered the malignant growth of glioma cells, indicating that it acted as a tumor suppressor. The miR-302/367 cluster is highly expressed in malignant germ cell tumors (GCT). miR-367-3phas been shown to have high sensitivity/specificity for diagnosing pediatric extracranial malignant GCTs 43, 44.It has been reported that miR-367-3p inhibits migration of hepatocellular carcinoma and increases sorafenib chemotherapy efficacy via altering the MDM2/AR/FKBP5/PHLPP/(p-AKT and p-ERK) signals 45. miR-367 is down-regulated in gastric cancer and is related to differentiation, TNM staging, and migration. Over-expression of miR-367 inhibits migration and invasion of gastric cancer cells via regulating the target gene Rab23 21. The expression of miR-367 is also reduced in ependymoma and high-grade glioma, and is related to poor prognosis 23. These results together with our study suggest that miR-367-3p exerts significant effects on a variety of malignant tumors. Our study has shown that miR-367-3p specifically bound to OIP5-AS1, and OIP5-AS1 knockdown increased miR-367-3p expression, attenuating the malignant progression of glioma cells. These results indicated that OIP5-AS1 inhibited the malignant growth of glioma cells via binding to miR-367-3p. Furthermore, we discovered miR-367-3p over-expression lowered OIP5-AS1 expression in U87 and U251 cells. RNA IP experiments confirmed that OIP5-AS1 and miR-367-3p were enriched in Ago2 protein, suggesting OIP5-AS1 decreased miR-367-3p expression in a RISC-dependent manner. There was a reciprocal regulatory effect between OIP5-AS1 and miR-367-3p. A similar pattern was reported in the case of regulatory functions of HOTAIR and miR-141 in renal cancer cells 46. Our results showed that PIWIL3, OIP5-AS1, and miR-367-3p expression was correlated with WHO grading, which is indicative of the prognosis of glioma. Also, patients with PIWIL3-low, miR-367-3p-low or OIP5-AS1-high expression experienced shorter survival. Thus, it is conceivable that PIWIL3, OIP5-AS1 and miR-367-3p are related to glioma progression.
Numerous reports have shown that the CEBPA protein has different expression levels and functions in various tumors. It is down-regulated in clear cell renal cell carcinoma and is an indicator of early diagnosis 47. CEBPA is also decreased in lung adenocarcinoma, showing value for prognostic prediction 48. It serves as both a tumor suppressor and promoter in AML 49 but is highly expressed in hepatocellular carcinoma and exerts oncogenic effects 27. Knockdown of CEBPA down-regulates Cyclin A and CDK4, blocking colony formation and cell growth of Hep3B and Huh7. So far, only one report has discussed CEBPA function in glioma where CEBPA mediates wild-type p53-dependent transcriptional up-regulation of cathepsin L 50. Our study demonstrated that CEBPA was highly expressed in glioma tissues as well as U87 and U251 cells, and was positively correlated with the pathological grade of glioma. Furthermore, silencing of CEBPA inhibited the progression of glioma cells, suggesting its oncogenic effect in this tumor type.
It is well established that miRNAs inhibit transcription of target genes via binding to the 3'UTRof mRNAs. miR-381 has been reported to bind to the 3'UTRof CEBPA mRNA, thus attenuating the downstream molecule Cx43's expression in breast cancer cells and suppressing migration. In our study, the dual luciferase assay showed specific binding of miR-367-3p to 3'UTR of CEBPA. Our results confirmed that miR-367-3p regulated glioma cells malignant grow at the transcriptional and post-transcriptional levels via negatively modulating CEBPA mRNA and protein expression.
TRAF4 is highly expressed in various tumor tissues and cells, and exerts oncogenic functions via activating multiple signaling pathways. It is up-regulated in hepatocellular carcinoma and promotes migration, invasion, and EMT by activating the PI3K/AKT pathway 51. Similarly, TRAF4 is highly expressed in osteosarcoma and enhances migration as well as invasion via activating the Akt pathway 52. TRAF4 upregulation also occurs in lung cancer, where its knockdown significantly inhibits proliferation and migration resulting in decreased xenograft volume. In lung cancer, TRAF4 plays an important role in the activation of Akt through ubiquitination, and its attenuation impairs glucose metabolism mediated by the Akt pathway. These observations suggested that TRAF4 is a candidate molecular target for lung cancer prevention and therapy 53. Upregulation of TRAR4 in breast cancer contributes to the activation of the p70s6k signaling pathway promoting proliferation and is associated with poor prognosis 54. Our study demonstrated that TRAF4 was highly expressed in glioma tissues and cells and attenuation of TRAF4 inhibited the malignant growth of glioma. Thus, TRAF4 functioned as an oncogene in glioma. ChIP analysis further confirmed CEBPA specifically bound to the TRAF4 promoter, whose mRNA and protein were then attenuated, indicating that TRAF4 is a CEBPA target. TRAF4 knockdown inhibited proliferation, migration, and invasion of glioma cells but promoted apoptosis, suggesting its oncogenic effect. CEBPA increased TRAF4 expression through positive regulation, promoting proliferation, migration, and invasion. Interestingly, CEBPA also increased PIWIL3 expression by binding to the PIWIL3 promoter. These results demonstrated that there was a positive feedback loop between PIWIL3 and CEBPA.
As displayed in the Figure 9 schematic, our study revealed that PIWIL3 and piR-30188 over-expression as well as OIP5-AS1 knockdown and their combined application significantly up-regulated miR-367-3p levels, thus inhibiting CEBPA and TRAF4 expression. As a result, the proliferation, migration, and invasion of glioma cells were attenuated while apoptosis was promoted, indicating PIWIL3/piR-30188/OIP5-AS1 regulated the biological behavior of glioma cells via the miR-367/CEBPA/TRAF4 pathway. To explore the potential therapeutic effect of the above targets, we conducted in vivo studies. Our findings demonstrated that up-regulation of PIWIL3 and miR-367-3p and silencing of OIP5-AS1 resulted in significantly smaller xenografts and longer survival of mice. Significantly, simultaneous over-expression of PIWIL3 and miR-367-3p and OIP5-AS1 knockdown resulted in the smallest xenografts and longest survival. Our study strongly suggests that PIWIL3, OIP5-AS1, and miR-367-3p can serve as molecular targets with great therapeutic potential in gliomas and possibly other cancers. A schematic illustration of the pathway is shown in Figure 9.
Figure 9.
Schematic illustration of the mechanism of PIWIL3/piR-30188/OIP5-AS1.
In conclusion, we studied the endogenous expression levels of PIWIL3, piR-30188, OIP5-AS1, and miR-367-3p in glioma cells and elucidated, for the first time, the underlying molecular mechanisms involving CEBPA and TRAF4 (Figure 9). The interactions between PIWIL3 and piR-30188, piR-30188 and OIP5-AS1, between OIP5-AS1 and miR-367-3p, miR-367-3p and CEBPA, as well as between CEBPA and TRAF4 were highlighted. Our study also revealed the novel mechanism by which PIWIL3/piR-30188/OIP5-AS1 regulated the biological behavior of glioma cells via the miR-367-3p/CEBPA/TRAF4 pathway. More significantly, PIWIL3, OIP5-AS1, and miR-367-3p may have a significant therapeutic potential in glioma treatment. Our results provide a new mechanistic insight into the pathogenesis of glioma and identify novel targets for its treatment.
Supplementary Material
Supplementary Figure 1: piRNA, lncRNA and miRNA microarrays data in astrocytes, U87, and U251 cells. Supplementary Figure 2: Location and expression of piR-30188, OIP5-AS1, and miR-367-3p in normal human astrocytes, U87, and U251 cells. Supplementary Table 1: Detailed information about the glioma patients.
Supplementary Table 2.
Supplementary Table 3.
Acknowledgments
This work is supported by grants from the Natural Science Foundation of China (81672511, 81372484 and 81573010), Liaoning Science and Technology Plan Project (No. 2015225007), Shenyang Science and Technology Plan Projects (Nos. F15-199-1-30 and F15-199-1-57), and the outstanding scientific fund of Shengjing hospital (No. 201304).
Abbreviations
- piRNAs
PIWI-interacting RNAs
- lncRNA
long noncoding RNAs
- CEBPA
CCAAT/enhancer binding protein alpha
- TRAF4
TNF receptor-associated factor 4
- miRNAs
microRNAs
- siRNAs
short-interfering RNAs
- ceRNA
endogenous competition RNA
- DMEM
Dulbecco's modified Eagle medium
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- CDS
coding sequence
- NC
negative control
- ChIP
chromatin immunoprecipitation
- DAPI
4', 6-diamidino-2-phenylindole
- qRT-PCR
quantitative Real-time PCR
- FISH
fluorescence in situ hybridization
- IDVs
integrated density values
- NHA
normal human astrocytes
- NBTs
nontumourous brain tissues.
References
- 1.Ostrom QT, Gittleman H, Farah P, Ondracek A, Chen Y, Wolinsky Y. et al. CBTRUS statistical report: Primary brain and central nervous system tumors diagnosed in the United States in 2006-2010. Neuro Oncol. 2013;15(Suppl 2):ii1–56. doi: 10.1093/neuonc/not151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dunn GP, Rinne ML, Wykosky J, Genovese G, Quayle SN, Dunn IF. et al. Emerging insights into the molecular and cellular basis of glioblastoma. Gene Dev. 2012;26:756–84. doi: 10.1101/gad.187922.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bartek J, Ng K, Bartek J, Fischer W, Carter B, Chen CC. Key concepts in glioblastoma therapy. J Neurol Neurosurg Psychiatry. 2012;83:753–60. doi: 10.1136/jnnp-2011-300709. [DOI] [PubMed] [Google Scholar]
- 4.Shi XF, Sun M, Liu HB, Yao YW, Song Y. Long non-coding RNAs: A new frontier in the study of human diseases. Cancer Lett. 2013;339:159–66. doi: 10.1016/j.canlet.2013.06.013. [DOI] [PubMed] [Google Scholar]
- 5.Ghildiyal M, Zamore PD. Small silencing RNAs: an expanding universe. Nat Rev Genet. 2009;10:94–108. doi: 10.1038/nrg2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sasaki T, Shiohama A, Minoshima S, Shimizu N. Identification of eight members of the Argonaute family in the human genome. Genomics. 2003;82:323–30. doi: 10.1016/s0888-7543(03)00129-0. [DOI] [PubMed] [Google Scholar]
- 7.Virant-Klun I, Leicht S, Hughes C, Krijgsveld J. Identification of Maturation-Specific Proteins by Single-Cell Proteomics of Human Oocytes. Mol Cell Proteomics. 2016;15:2616–27. doi: 10.1074/mcp.M115.056887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Krishnan P, Ghosh S, Graham K, Mackey JR, Kovalchuk O, Damaraju S. Piwi-interacting RNAs and PIWI genes as novel prognostic markers for breast cancer. Oncotarget. 2016;7:37944–56. doi: 10.18632/oncotarget.9272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Suzuki R, Honda S, Kirino Y. PIWI Expression and Function in Cancer. Front Genet. 2012;3:204. doi: 10.3389/fgene.2012.00204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen C, Liu JH, Xu GX. Overexpression of PIWI proteins in human stage III epithelial ovarian cancer with lymph node metastasis. Cancer Biomark. 2013;13:315–21. doi: 10.3233/CBM-130360. [DOI] [PubMed] [Google Scholar]
- 11.Wahlestedt C. Targeting long non-coding RNA to therapeutically upregulate gene expression. Nat Rev Drug Discov. 2013;12:433–46. doi: 10.1038/nrd4018. [DOI] [PubMed] [Google Scholar]
- 12.Geisler S, Coller J. RNA in unexpected places: long non-coding RNA functions in diverse cellular contexts. Nat Rev Mol Cell Bio. 2013;14:699–712. doi: 10.1038/nrm3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ulitsky I, Shkumatava A, Jan CH, Sive H, Bartel DP. Conserved Function of lincRNAs in Vertebrate Embryonic Development despite Rapid Sequence Evolution. Cell. 2011;147:1537–50. doi: 10.1016/j.cell.2011.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim J, Abdelmohsen K, Yang XL, De S, Grammatikakis I, Noh JH. et al. LncRNA OIP5-AS1/cyrano sponges RNA-binding protein HuR. Nucleic Acids Res. 2016;44:2378–92. doi: 10.1093/nar/gkw017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Karreth FA, Pandolfi PP. ceRNA Cross-Talk in Cancer: When ce-bling Rivalries Go Awry. Cancer Discov. 2013;3:1113–21. doi: 10.1158/2159-8290.CD-13-0202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu XH, Sun M, Nie FQ, Ge YB, Zhang EB, Yin DD. et al. Lnc RNA HOTAIR functions as a competing endogenous RNA to regulate HER2 expression by sponging miR-331-3p in gastric cancer. Mol Cancer. 2014;13:92. doi: 10.1186/1476-4598-13-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu Q, Guo X, Que SS, Yang X, Fan HX, Liu M. et al. LncRNA RSU1P2 contributes to tumorigenesis by acting as a ceRNA against let-7a in cervical cancer cells. Oncotarget. 2017;8:43768–81. doi: 10.18632/oncotarget.10844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sun CC, Li SJ, Zhang F, Xi YY, Wang L, Bi YY. et al. Long non-coding RNA NEAT1 promotes non-small cell lung cancer progression through regulation of miR-377-3p-E2F3 pathway. Oncotarget. 2016;7:51784–814. doi: 10.18632/oncotarget.10108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Teng H, Wang P, Xue Y, Liu X, Ma J, Cai H. et al. Role of HCP5-miR-139-RUNX1 Feedback Loop in Regulating Malignant Behavior of Glioma Cells. Mol Ther. 2016;24:1806–22. doi: 10.1038/mt.2016.103. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 20.Yao Y, Ma J, Xue Y, Wang P, Li Z, Li Z. et al. MiR-449a exerts tumor-suppressive functions in human glioblastoma by targeting Myc-associated zinc-finger protein. Mol Oncol. 2015;9:640–56. doi: 10.1016/j.molonc.2014.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang B, He DD, Fang XJ, Xu HW, Yang QH. The microRNA-367 Inhibits the Invasion and Metastasis of Gastric Cancer by Directly Repressing Rab23. Genet Test Mol Bioma. 2015;19:69–74. doi: 10.1089/gtmb.2014.0210. [DOI] [PubMed] [Google Scholar]
- 22.Guan YL, Chen L, Bao YJ, Qiu B, Pang C, Cui R. et al. High miR-196a and low miR-367 cooperatively correlate with unfavorable prognosis of high-grade glioma. Int J Clin Exp Patho. 2015;8:6576–88. [PMC free article] [PubMed] [Google Scholar]
- 23.Costa FF, Bischof JM, Vanin EF, Lulla RR, Wang M, Sredni ST. et al. Identification of MicroRNAs as Potential Prognostic Markers in Ependymoma. Plos One. 2011;6:e25114. doi: 10.1371/journal.pone.0025114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nerlov C. The C/EBP family of transcription factors: a paradigm for interaction between gene expression and proliferation control. Trends Cell Biol. 2007;17:318–24. doi: 10.1016/j.tcb.2007.07.004. [DOI] [PubMed] [Google Scholar]
- 25.Yuan H, Wen B, Liu XH, Gao C, Yang RM, Wang LX. et al. CCAAT/enhancer-binding protein alpha is required for hepatic outgrowth via the p53 pathway in zebrafish. Sci Rep. 2015;5:15838. doi: 10.1038/srep15838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lu GD, Ang YH, Zhou J, Tamilarasi J, Yan B, Lim YC. et al. CCAAT/Enhancer Binding Protein alpha Predicts Poorer Prognosis and Prevents Energy Starvation-Induced Cell Death in Hepatocellular Carcinoma. Hepatology. 2015;61:965–78. doi: 10.1002/hep.27593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lu GD, Leung CHW, Yan B, Tan CMY, Low SY, Aung MO. et al. C/EBP alpha Is Up-regulated in a Subset of Hepatocellular Carcinomas and Plays a Role in Cell Growth and Proliferation. Gastroenterology. 2010;139:632–43. doi: 10.1053/j.gastro.2010.03.051. [DOI] [PubMed] [Google Scholar]
- 28.Konopka B, Szafron LM, Kwiatkowska E, Podgorska A, Zolocinska A, Pienkowska-Grela B. et al. The significance of c.690G>T polymorphism (rs34529039) and expression of the CEBPA gene in ovarian cancer outcome. Oncotarget. 2016;7:67412–24. doi: 10.18632/oncotarget.11822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Su L, Gao SJ, Liu XL, Tan YH, Wang L, Li W. CEBPA mutations in patients with de novo acute myeloid leukemia: data analysis in a Chinese population. Onco Targets Ther. 2016;9:3399–403. doi: 10.2147/OTT.S94975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ming J, Zhou Y, Du JZ, Fan SG, Pan BB, Wang YH. et al. miR-381 suppresses C/EBP alpha-dependent Cx43 expression in breast cancer cells. Biosci Rep. 2015;35:e00266. doi: 10.1042/BSR20150167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meerson A, Yehuda H. Leptin and insulin up-regulate miR-4443 to suppress NCOA1 and TRAF4, and decrease the invasiveness of human colon cancer cells. BMC Cancer. 2016;16:882. doi: 10.1186/s12885-016-2938-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang J, Wei D, Wang W, Shen B, Xu S, Cao Y. TRAF4 enhances oral squamous cell carcinoma cell growth, invasion and migration by Wnt-beta-catenin signaling pathway. Int J Clin Exp Pathol. 2015;8:11837–46. [PMC free article] [PubMed] [Google Scholar]
- 33.Iliev R, Stanik M, Fedorko M, Poprach A, Vychytilova-Faltejskova P, Slaba K. et al. Decreased expression levels of PIWIL1, PIWIL2, and PIWIL4 are associated with worse survival in renal cell carcinoma patients. Onco Targets Ther. 2016;9:217–22. doi: 10.2147/OTT.S91295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Taubert H, Wach S, Jung R, Pugia M, Keck B, Bertz S. et al. Piwil 2 Expression Is Correlated with Disease-Specific and Progression-Free Survival of Chemotherapy-Treated Bladder Cancer Patients. Mol Med. 2015;21:371–80. doi: 10.2119/molmed.2014.00250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Oh SJ, Kim SM, Kim YO, Chang HK. Clinicopathologic Implications of PIWIL2 Expression in Colorectal Cancer. Korean J Pathol. 2012;46:318–23. doi: 10.4132/KoreanJPathol.2012.46.4.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Oner C, Cosan DT, Colak E. Estrogen and Androgen Hormone Levels Modulate the Expression of PIWI Interacting RNA in Prostate and Breast Cancer. Plos One. 2016;11:e0159044. doi: 10.1371/journal.pone.0159044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Iliev R, Fedorko M, Machackova T, Mlcochova H, Svoboda M, Pacik D. et al. Expression Levels of PIWI-interacting RNA, piR-823, Are Deregulated in Tumor Tissue, Blood Serum and Urine of Patients with Renal Cell Carcinoma. Anticancer Res. 2016;36:6419–23. doi: 10.21873/anticanres.11239. [DOI] [PubMed] [Google Scholar]
- 38.Jacobs DI, Qin Q, Lerro MC, Fu A, Dubrow R, Claus EB. et al. PIWI-Interacting RNAs in Gliomagenesis: Evidence from Post-GWAS and Functional Analyses. Cancer Epidemiol Biomarkers Prev. 2016;25:1073–80. doi: 10.1158/1055-9965.EPI-16-0047. [DOI] [PubMed] [Google Scholar]
- 39.Ng KW, Anderson C, Marshall EA, Minatel BC, Enfield KS, Saprunoff HL. et al. Piwi-interacting RNAs in cancer: emerging functions and clinical utility. Mol Cancer. 2016;15:5. doi: 10.1186/s12943-016-0491-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zheng J, Li XD, Wang P, Liu XB, Xue YX, Hu Y. et al. CRNDE affects the malignant biological characteristics of human glioma stem cells by negatively regulating miR-186. Oncotarget. 2015;6:25339–55. doi: 10.18632/oncotarget.4509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zheng J, Liu X, Wang P, Xue Y, Ma J, Qu C. et al. CRNDE Promotes Malignant Progression of Glioma by Attenuating miR-384/PIWIL4/STAT3 Axis. Mol Ther. 2016;24:1199–215. doi: 10.1038/mt.2016.71. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 42.Cai H, Liu X, Zheng J, Xue Y, Ma J, Li Z. et al. Long non-coding RNA taurine upregulated 1 enhances tumor-induced angiogenesis through inhibiting microRNA-299 in human glioblastoma. Oncogene. 2017;36:318–31. doi: 10.1038/onc.2016.212. [DOI] [PubMed] [Google Scholar]
- 43.Murray MJ, Bell E, Raby KL, Rijlaarsdam MA, Gillis AJ, Looijenga LH. et al. A pipeline to quantify serum and cerebrospinal fluid microRNAs for diagnosis and detection of relapse in paediatric malignant germ-cell tumours. Br J Cancer. 2016;114:151–62. doi: 10.1038/bjc.2015.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Murray MJ, Huddart RA, Coleman N. The present and future of serum diagnostic tests for testicular germ cell tumours. Nat Rev Urol. 2016;13:715–25. doi: 10.1038/nrurol.2016.170. [DOI] [PubMed] [Google Scholar]
- 45.Xu J, Lin H, Li G, Sun Y, Chen J, Shi L. et al. The miR-367-3p Increases Sorafenib Chemotherapy Efficacy to Suppress Hepatocellular Carcinoma Metastasis through Altering the Androgen Receptor Signals. EBioMedicine. 2016;12:55–67. doi: 10.1016/j.ebiom.2016.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chiyomaru T, Fukuhara S, Saini S, Majid S, Deng GR, Shahryari V. et al. Long Non-coding RNA HOTAIR Is Targeted and Regulated by miR-141 in Human Cancer Cells. J Biol Chem. 2014;289:12550–65. doi: 10.1074/jbc.M113.488593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.De Palma G, Sallustio F, Curci C, Galleggiante V, Rutigliano M, Serino G. et al. The Three-Gene Signature in Urinary Extracellular Vesicles from Patients with Clear Cell Renal Cell Carcinoma. J Cancer. 2016;7:1960–7. doi: 10.7150/jca.16123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yong KJ, Basseres DS, Welner RS, Zhang WC, Yang H, Yan B. et al. Targeted BMI1 inhibition impairs tumor growth in lung adenocarcinomas with low CEBP alpha expression. Sci Transl Med. 2016;8:350ra104. doi: 10.1126/scitranslmed.aad6066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ohlsson E, Schuster MB, Hasemann M, Porse BT. The multifaceted functions of C/EBP alpha in normal and malignant haematopoiesis. Leukemia. 2016;30:767–75. doi: 10.1038/leu.2015.324. [DOI] [PubMed] [Google Scholar]
- 50.Katara R, Mir RA, Shukla AA, Tiwari A, Singh N, Chauhan SS. Wild type p53-dependent transcriptional upregulation of cathepsin L expression is mediated by C/EBP alpha in human glioblastoma cells. Biol Chem. 2010;391:1031–40. doi: 10.1515/BC.2010.103. [DOI] [PubMed] [Google Scholar]
- 51.Liu K, Wu X, Zang X, Huang Z, Lin Z, Tan W. et al. TRAF4 regulates migration, invasion and the epithelial-mesenchymal transition via PI3K/AKT signaling in hepatocellular carcinoma. Oncol Res. 2017;25:1329–40. doi: 10.3727/096504017X14876227286564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yao W, Wang X, Cai Q, Gao S, Wang J, Zhang P. TRAF4 enhances osteosarcoma cell proliferation and invasion by Akt signaling pathway. Oncol Res. 2014;22:21–8. doi: 10.3727/096504014X14077751730351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li W, Peng C, Lee MH, Lim D, Zhu F, Fu Y. et al. TRAF4 Is a Critical Molecule for Akt Activation in Lung Cancer. Cancer Res. 2013;73:6938–50. doi: 10.1158/0008-5472.CAN-13-0913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ren HY, Wang J, Yang F, Zhang XL, Wang AL, Sun LL. et al. Cytoplasmic TRAF4 contributes to the activation of p70s6k signaling pathway in breast cancer. Oncotarget. 2015;6:4080–96. doi: 10.18632/oncotarget.2977. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1: piRNA, lncRNA and miRNA microarrays data in astrocytes, U87, and U251 cells. Supplementary Figure 2: Location and expression of piR-30188, OIP5-AS1, and miR-367-3p in normal human astrocytes, U87, and U251 cells. Supplementary Table 1: Detailed information about the glioma patients.
Supplementary Table 2.
Supplementary Table 3.