Abstract
Objective(s):
Cucurbitacins exhibit a range of anti-cancer functions. We investigated the effects of cucurbitacins D, E, and I purified from Ecballium elaterium (L.) A. Rich fruits on some apoptotic and autophagy genes in human gastric cancer cell line AGS.
Materials and Methods:
Using quantitative reverse transcription PCR (qRT-PCR), the expression of LC3, VEGF, BAX, caspase-3, and c-MYC genes were quantified in AGS cells 24 hr after treatment with cucurbitacins D, E, and I at concentrations 0.3, 0.1 and 0.5 μg/ml, respectively. Cell cycle and death were analyzed by flflowcytometry.
Results:
Purified cucurbitacins induced sub-G1 cell-cycle arrest and cell death in AGS cells and upregulated LC3mRNA effectively, but showed a very low effect on BAX, caspase-3, and c-MYC mRNA levels. Also after treatment with cucurbitacin I at concentration 0.5 μg/ml, VEGF mRNA levels were increased about 4.4 times. Pairwise comparison of the effect of cucurbitacins D, E, and I on LC3 mRNA expression showed that the cucurbitacin I effect is 1.3 and 1.1 times that of cucurbitacins E and D, respectively; cucurbitacin D effect is 1.2 times that of cucurbitacin E (P-value <0.05). In silico analysis showed that among autophagy genes, LC3 has an important gastric cancer rank relation.
Conclusion:
Cucurbitacins D, E, and I purified from E. elaterium fruits upregulate LC3 and induce sub-G1 cell-cycle arrest and cell death in human gastric cancer cell line AGS. Cucurbitacin I effect on LC3 mRNA expression is significantly more than that of cucurbitacins E and D.
Keywords: Apoptosis, Autophagy, Cell cycle, Cucurbitacins, Stomach neoplasms
Introduction
Due to a wide variety of biologically effective chemicals in medicinal plants, there is a growing interest in their use as therapeutics (1). The Ecballium elaterium (L.) A. Rich is a wild medicinal herb from the Cucurbi-taceae family, which produce cucurbitacins, a family of highly oxygenated tetracyclic triterpenes (2). The role of cucurbitacins in Janus kinase/signal transducers and activators of transcription (JAK/STAT) pathway inhibition, MAP kinase (MAPK) pathway regulation, and cytoskeleton disruption, suggests their excellent efficacy for cancer treatment and prevention (3, 4).
Recent studies have shown that many anticancer drugs induce both autophagy and apoptosis in various cancer cells. Autophagy is a dynamic multi-step phenomenon in which double-membrane autophago-somes enclose damaged cellular proteins, lipids, and organelles and subsequently deliver them to lysosomes for degradation (5-7). Under normal physiological conditions, autophagic activity is low. However, a range of stimuli can induce autophagy to protect cells from stress (8, 9). The role of autophagy in tumorigenesis is complicated, its role in tumor promotion and suppre-ssion, as well as its contribution to therapeutic resistance, has been reported (10-12).
There are several genes that contribute to auto-phagy and apoptosis. Among them, microtubule-associated protein light chain 3 (LC3) is the key factor in autophagosome formation (13, 14). Also, c-MYC potently induces different types of regulated cell death, including apoptosis (15), and autophagy (16). And BAX and caspase-3 are among main regulators of apoptosis (17). In addition, it is reported that VEGF is one of the participant genes in autophagic cell death (18). Thus, studying LC3, VEGF, BAX, caspase-3, and c-MYC genes’ contribution to autophagy could be a valuable goal for anticancer investigations.
Gastric cancer is considered as the fifth most common cancer in the world and the third leading cause of cancer mortality and morbidity (19). Our previous MTT assay using purified cucurbitacins D, E, and I from E. elaterium showed that these chemicals have cytotoxic effects on human stomach adenocarci- noma cell line AGS (20). The aim of this study was to investigate the effects of cucurbitacins D, E, and I purified from E. elaterium fruits on the expression of BAX, caspase-3, LC3, and VEGF and c-MYC genes in the AGS cell line.
Materials and Methods
Cell culture
In this research, human stomach adenocarcinoma cell line AGS was provided from Iranian Biological Resources Center’s Cell Bank (Tehran, Iran). Cells were cultured in Ham’s F-12 nutrient mix with L-glutamine and sodium bicarbonate (Cat. No. 10-FN1-500, G. Innovative Biotech Co, Iran) medium supple-mented with 10% FBS (Cat. No. FB-st 500, Pasteur Institute of Iran) and were incubated at 37 °C in a water-saturated atmosphere of 5% CO2 and 95% air until confluence.
Cucurbitacins
We obtained cucurbitacins D, E and I from the stock of our previous purification study (20). The methanolic extract of E. elaterium fruits was frac-tionated to petroleum ether, chloroform, and ethyl acetate fractions. The chloroform fraction was chosen for further purification with column chromatography. Finally, cucurbitacins D, E, and I were isolated by column chromatography and identified by NMR spectroscopy (20).
RNA extraction
AGS cells (5×105 cells/well) were seeded into 6-well plates and were grown to 80% confluency. 24 hr after treatment with cucurbitacins D, E, and I at concen-trations 0.3, 0.1 and 0.5 μg/ml, respectively, cells were harvested and total RNA was extracted from the cells using RNeasy Mini Kit (QIAGEN, Germany) according to the manufacturer’s instructions.
Synthesis of cDNA
The cDNA was synthesized using Easy cDNA Synthesis Kit (Cat. No. A101161, pars tous biotechno-logy, Iran) according to the manufacturer’s instructions.
Quantitative reverse transcription polymerase chain reaction (qRT-PCR)
Using quantitative polymerase chain reaction (q-PCR), expression of LC3, VEGF, BAX, caspase-3, and c-MYC genes was quantified in AGS cells 24 hr after treatment with cucurbitacins D, E, and I. All experi-ments were performed at least in duplicate using a 48 well StepOne Real-Time PCR Systems (Applied Biosystems, USA) and specific primers (Table 1) with the following conditions: 95 °C for 15 min, 40 amplification cycles consisting of 95 °C for 15 sec, 60 °C for 30 sec, and 72 °C for 60 sec. Melting curves were then determined with temperature ranging from 60 to 95 °C. GAPDH was chosen as an internal control. SYBR Green reagents were used for all real-time PCR reactions. The expression of the genes was analyzed based on the cycle threshold (Ct) and relative expression levels were determined as 2-[ΔΔC(t)].
Table 1.
Primers used in quantitative reverse transcription polymerase chain reaction (qRT-PCR) and amplicon sizes (bp: base pair)
| Target | Forward primer | Reverse primer | Amplicon size (bp) |
|---|---|---|---|
| BAX | 5’GCCCTTTTGCTTCAGGGTTTC3′ | 5’CATCCTCTGCAGCTCCATGT3′ | 168 |
| caspase-3 | 5’GCGGTTGTAGAAGAGTTTCGTG3′ | 5’CTCACGGCCTGGGATTTCAA3′ | 101 |
| c-MYC | 5’CCCTCCACTCGGAAGGACTA3′ | 5’GCTGGTGCATTTTCGGTTGT3′ | 96 |
| GAPDH | 5′GACCCCTTCATTGACCTCAACTAC3′ | 5′TCGCTCCTGGAAGATGGTGATGG3′ | 138 |
| LC3 | 5’GGACATCTACGAGCAGGAGAAAGACGAG3′ | 5’TCAGAAGCCGAAGGTTTCCTGGGAG3′ | 79 |
| VEGF | 5’TGTCTAATGCCCTGGAGCCT | 5’GCTTGTCACATCTGCAAGTACG | 175 |
Cell cycle and cell death analysis by flflow cytometry
For cell cycle and cell death analysis, AGS cells (5×105cells/well) were seeded into 6-well plates and were grown to 80% confluency. Two wells were chosen as control and received no treatment. Cells were collected 24 hr after treatment with cucurbitacins D, E, and I at concentrations 0.3, 0.1, and 0.5 μg/ml, respectively and were washed with phosphate buffered saline (PBS) and trypsinized with 0.025% trypsin-EDTA to yield single cell suspension. Two groups of cells were used for flowcytometric analysis using a BD FACSCalibur flowcytometer (BD Biosciences, USA). For cell cycle analysis, cells were then fixed in ice-cold 70% ethanol and stained with 50 μg/ml propidium iodide (PI) solution containing 10 μg/ml RNaseA (Takara, Japan). For cell death analysis, cells were stained with AnnexinV/PI. Experiments were repeated at least twice and cell cycle profiles were analyzed using the FlowJo Software (ver. 7.6.1).
In silico analysis: Gene prioritization
After harvesting the results of the experimental part we performed an in silico analysis to show the validity of findings. The hypothesis of LC3 contribution in gastric cancer cell autophagy was tested using a bioinformatics approach: gene prioritization. Using statistical methods, this approach scores and ranks a set of test genes through complex comparison with a set of training genes based on their average similarity in function and interactions. LC3 is one of the main autophagy-associated genes. We obtained the list of these genes from HADb (http://autophagy.lu/clustering/), which is the first human autophagy-dedicated database. The autophagy genes list, containing LC3, was used as test genes set to find LC3 priority among them in gastric cancer. For training gene set, we used the SNPs3D server (http://www.snps3d.org/) to get the list of candidate genes associated with gastric cancer. After that, prioritizing servers: ToppNet (https://toppgene.cchmc.org/network.jsp), which prioritize and rank candidate genes based on topological features in protein-protein interaction network (21), and Endeavour (https://endeavour.esat.kuleuven.be/), which works based on similarity of a candidate gene with a profile derived from genes already known to be involved in the disease (22), were used for finding the LC3 rank among autophagy genes in relation to gastric cancer.
Statistical analysis
One way ANOVA followed by Tukey’s post-hoc test was used to compare the expression level of desired
Results
Effect of cucurbitacins D, E, I on the AGS cell cycle and cell death
The accumulation of the sub-G1 population is considered as a biomarker of DNA damage. As shown in Figure 1, the sub-G1 accumulation of AGS cells treated with cucurbitacins D, E, and I, was increased in comparison to the untreated control cells. Moreover, treatment of AGS cells with cucurbitacins D, E, and I induced cell death (Figure 1).
Figure 1.
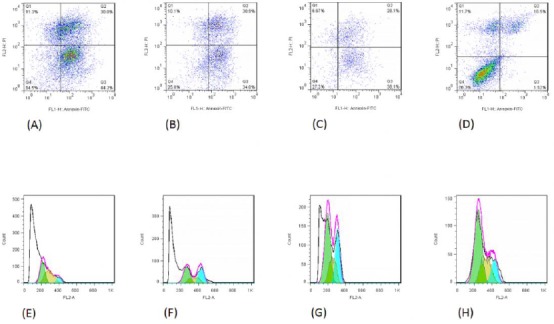
The death of AGS cells treated with cucurbitacins D (panel A), E (panel B), I (panel C), and untreated control cells (panel D). Panels A-D: Top left corner shows the necrotic cells, top right corner shows late apoptotic cells, bottom left corner shows healthy live cells, and bottom right corner shows early apoptotic cells. Panels E-H: Sub-G1 accumulation of AGS cells treated with cucurbitacins D (panel E), E (panel F), and I (panel G), in comparison to the untreated control cells (panel H)
Effect of the cucurbitacins on LC3, VEGF, BAX, caspase-3, and c-MYC mRNA expression
We evaluated the effects of cucurbitacins D, E, and I on the mRNA expression levels of LC3, VEGF, BAX, caspase-3, and c-MYC genes using qRT-PCR. As shown in Figure 2, based on P-value ≤ 0.05 LC3 mRNA levels were increased about 23, 20, and 25 times after treatment with cucurbitacins D, E, and I at concentrations 0.3, 0.1, and 0.5 μg/ml, respectively, for 24 hr. Also, after treat-ment with cucurbitacin I at concentration 0.5 μg/ml, VEGF mRNA levels were increased about 4.4 times. However, BAX, caspase-3, and c-MYC mRNA levels were not considerably changed after treatment with cucurbitacins D, E, and I at aforementioned concen-trations. Effect of cucurbitacin I treatment on LC3 mRNA expression levels was significantly more than that of cucurbitacins E and D. Comparing the effect of cucurbitacin D on LC3 mRNA expression with that of cucurbitacin E showed that cucurbitacin D’s effect is significantly more than that of cucurbitacin E. It was LC3 among studied genes which showed significant induction under the cucurbitacin treatments.
Figure 2.
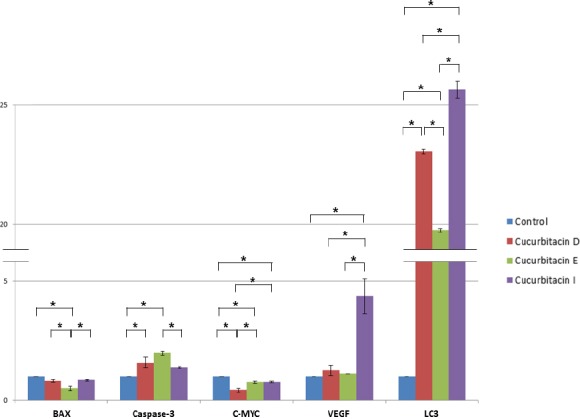
Effects of cucurbitacins D, E, and I on the mRNA expression levels of LC3, VEGF, BAX, caspase-3, and c-MYC genes (P-value ≤ 0.05). GAPDH was chosen as an internal control. The expression of the genes was analyzed based on the cycle threshold (Ct) and relative expression levels were determined as 2-[ΔΔC(t)]
In silico analysis: Gene prioritization
SNPs3D introduced 567 possible gastric cancer-related genes. Also, 232 genes were obtained from HADb as human autophagy-associated genes. Since ToppNet and Endeavour servers apply different approaches to compare training and test gene sets and prioritize candidate genes importance in relation to a disease, the ranks of candidate genes in the result sheets of these servers were different. ToppNet server identified 185 genes from the test genes set and located the LC3 gene in rank 38 among them. It means that according to ToppNet, among 185 autophagy genes LC3 is the 38th gene in ranking, which probably implicates it in gastric cancer. Also, the Endeavour server identified 182 test genes and compared them with 565 identified training genes. Endeavour ranked LC3 as the 92nd gene among 182 analyzed autophagy genes that could be related to gastric cancer. Endea-vour provides a diagram for test genes’ P-values in numerous databases, however, the server stated that this is not a real statistical P-value. But, it could be used as a comparative item to show the place of a special gene among the candidates list. The LC3 Endeavour P-values among autophagy genes are depicted in Figure 3.
Figure 3.
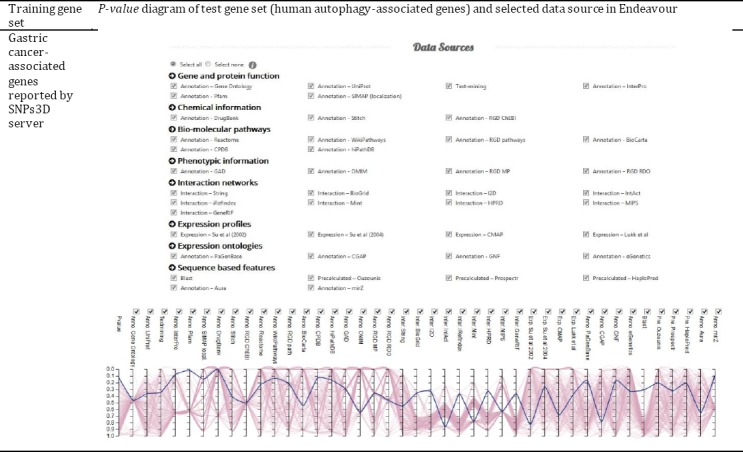
Endeavour prioritization diagram of autophagy genes in relation to gastric cancer. The bold line in the diagram corresponds to LC3 gene’s P-value. The other lines belong to other autophagy-associated genes.
Discussion
There is a 10-fold variation in gastric cancer incidence internationally, with high rates seen in many countries of Eastern Asia, Central and Eastern Europe, and Central and South America, and much lower rates reported from North America, and Africa (19). Indeed, stomach cancer is the third leading cause of cancer death worldwide (23).
Natural therapies based on medicinal plants are becoming increasingly important means of disease treatment. Indeed, more than 70 percent of new pharmaceuticals that have been approved since 1981 have directly or indirectly been derived from natural products (24). The E. elaterium is a wild toxic herb from the Cucurbitaceae family, which produce cucurbitacin molecules and has medicinal importance in traditional treatment prescriptions (25).
Cucurbitacins are highly oxidized tetracyclic triterpenoids, which have cytotoxic effects on various cancer cell lines. Cucurbitacins are claimed to be inhibitors of the JAK/STAT pathway, however, their cytotoxic effects may involve other mechanisms including the MAPK pathway, PARP cleavage, expression of active caspase-3, decreasing pSTAT3 and JAK3 levels, as well as decreasing various STAT3 downstream targets such as MCL-1, BCL-2, BCL-xL, and CYCLIN D3 (26). However, the anticancer effects and underlying mechanisms of cucurbitacins in human gastric cancer are still elusive.
There are many instances of cell death similar to classical apoptosis that show no evidence of caspase activation. Likewise, increased amounts of phospha-tidylserine in the plasma membrane exoplasmic leaflet as detected by Annexin V binding is not a definite marker of apoptosis. Similarly, extensive DNA fragmentation is frequently assumed a specific marker of apoptosis. However, there are increasing examples of apoptotic or apoptotic-like cell death proceeding without internucleosomal DNA degradation. In these cases, the intensity of cell labeling in TdT-mediated dUTP nick end labeling (TUNEL) assay will not be sufficient to positively identify apoptotic cells (27). It is suggested to restrict the term apoptosis to only the conventional cascade displaying all canonical hallmarks of apoptotic cell death such as (i) activation of caspases as a definite marker of cell death; (ii) high degree of compaction of chromatin; (iii) activation of endonucleases(s) causing internucleosomal DNA cleavage and extensive DNA fragmentation; (iv) appearance of characteristic cellular morphology with maintenance of organelles, (v) cell shrinkage, (vi) plasma membrane blebbing, and (vii) nuclear fragmentation followed by formation of apoptotic bodies (27).
In the present study, we show that cucurbitacins D, E, and I purified from E. elaterium inhibit AGS gastric cancer cell growth by inducing sub-G1 cell-cycle arrest and cell death. We evaluated the effects of cucurbitacins D, E, and I on the mRNA expression levels of LC3, VEGF, BAX, caspase-3, and c-MYC genes using qRT-PCR. As shown in Figure 2, LC3 mRNA levels were significantly increased after treatment with cucurbitacins D, E, and I.
Increased expression of the LC3 gene may be indicative of autophagic activity. Upon induction of autophagy, cytosolic LC3-I is cleaved and conjugated to phosphatidylethanolamine to form LC3-II, which associates with the phagophore (28). The use of autophagy markers such as LC3-II requires assays to estimate overall autophagic flux or flow, to permit an accurate interpretation of the results. Therefore, autophagic substrates should be followed dynamically over time to confirm that they have reached the lysosome, and, when appropriate, are degraded. Also, our in silico analysis revealed that among autophagy genes in humans, LC3 is a promising candidate for studying and analyzing in gastric cancer cellular and molecular studies.
Cucurbitacins D, E, and I induced LC3 mRNA expression, but BAX, caspase-3, and c-MYC mRNA levels were not considerably changed and effect of cucurbitacin I was significantly more than that of cucurbitacins E and D. Researchers (29) reported that cucurbitacin E induced autophagy at least partly via downregulation of mTORC1 signaling and upregulation of AMPK activity. Likewise, another study (30) reported that cucurbitacin B and I induced autophagy by the production of mitochondrial-derived reactive oxygen species (ROS) through a STAT3-independent process. Thus, another potential mechanism(s) may also contribute to cucurbitacin-induced autophagy. Research (31) showed that GBM cells treated with cucurbitacin I for 48 hr significantly upregulated Bax and cleaved caspase-3 (p17) but decreased antiapoptotic proteins such as Bcl-2 and Bcl-xL in a dose-dependent manner. Our findings that cucurbitacins show a very low effect on BAX and caspase-3 mRNA levels in AGS cells, disagree with these results. However, consistent with these results, LC3 mRNA was upregulated. This discrepancy may be explained by the fact that there is a complicated connection between autophagy and apoptosis. These two processes can mutually regulate and interconvert to determine the fate of a cell, depending on the context. Deng et al. (32) showed that cucurbitacin I markedly inhibits gastric cancer cell growth by inducing G2/M phase cell cycle arrest and apoptosis at low nanomolar concentrations via a STAT3-independent mechanism. Our study showed that cucurbitacins D, E, and I inhibit AGS gastric cancer cell growth by inducing sub-G1 cell-cycle arrest at concentrations 0.58, 0.18, and 0.97 μM, respectively. It seems that cucurbitacins inhibit gastric cancer cell cycle progression through different mechanisms at different concentrations. Further-more, comparing our results with the Deng et al. results suggests that cucurbitacins may preferentially induce autophagy at a particular concentration range but induce apoptosis at other concentration ranges. Common upstream signals may trigger both autophagy and apoptosis resulting in the mutual activation of autophagy and apoptosis. Under certain conditions, they may be also mutually exclusive. Recent research has implied that MAPK, particularly p38 MAPK and c-Jun NH2-terminal kinase (JNK), plays a key role in crosstalk between apoptosis and autophagy induced by genotoxic stress (31). Sun et al. reported that JNK activation is essential for up-regulation of LC3 during ceramide-induced autophagy
in human nasopharyngeal carcinoma cells. Furthermore, their findings suggest that c-Jun is essential for LC3 transcription after ceramide treatment (33). Ishdorj et al. (18) demonstrated that in B Leukemic Cells, cucurbitacin I activates the JNK/c-Jun signaling pathway independent of apoptosis and cell cycle arrest, leading to increased VEGF expression. Comparing our results with Sun et al. and Ishdorj et al. results suggests that cucurbitacin I from E. elaterium may activate the JNK/c-Jun signaling pathway in human gastric cancer cell line AGS leading to increased LC3 and VEGF expression.
It is reported that autophagosomal membranes serve as platforms for intracellular death-inducing signaling complex (iDISC) mediated caspase-8 activa-tion and apoptosis. Upon the formation of iDISC, procaspase-8 is recruited to the phagophore by two mechanisms: (1) through Atg12-Atg5 complex inter-action with FADD; and (2) LC3-II: p62 interaction. iDISC mediates apoptosis independent from death receptor signaling and requires LC3-positive autophagic membranes (34). Purified cucurbitacins D, E, and I induced cell death in AGS cells but showed the negligible effect on BAX, caspase-3, and c-MYC mRNA levels. One possibility is that cucurbitacins trigger non-canonical apoptosis. This could be of great importance as the induction of non-canonical apoptosis in cancer cells, could represent a novel and effective strategy for elimination of cancer cells. Another possibility is that cucurbitacins induce autophagy to limit caspase-dependent apoptosis. Yet another possibility is that cucurbitacins limit caspase-dependent apoptosis and at the same time induce non-canonical apoptosis. Since cucurbitacins D, E, and I showed no considerable effect on BAX, caspase-3, and c-MYC genes expression but increased LC3 gene mRNA level in AGS cells, they may preferentially induce autophagy. However, these and other possibilities need further investigation. Also, it must be noted that the concentrations used in the current study were achieved by an in vitro study and the possibility of extrapolating these concentrations to clinical practice needs more investigation.
Conclusion
Treatment with cucurbitacins D, E, and I purified from E. elaterium fruit resulted in AGS cancer cell line death. Cucurbitacins increased expression of the LC3 gene, but BAX, caspase-3, and c-MYC mRNA levels were not considerably changed. Notably, cucurbitacin I effect on expression of the LC3 gene is significantly more than that of cucurbitacins E and D. The present study provides new insights into the molecular mechanisms underlying cucurbitacin-mediated cell death in gastric cancer.
Acknowledgment
This research was supported by Iran National Science Foundation (INSF) grant no. 90004329. The results presented in this paper were part of the Ph.D. thesis of Naser Jafargholizadeh.
References
- 1.Sporn MB, Suh N. Chemoprevention of cancer. Carcinogenesis. 2000;21:525–530. doi: 10.1093/carcin/21.3.525. [DOI] [PubMed] [Google Scholar]
- 2.Chen JC, Chiu MH, Nie RL, Cordell GA, Qiu SX. Cucurbitacins and cucurbitane glycosides:structures and biological activities. Nat Prod Rep. 2005;22:386–399. doi: 10.1039/b418841c. [DOI] [PubMed] [Google Scholar]
- 3.Ríos JL, Escandell JM, Recio MC. New insights into the bioactivity of cucurbitacins. In: Atta-ur-Rahman, editor. Studies in Natural Products Chemistry Volume 32:Bioactive Natural Products Part L. Netherlands: Elsevier Science; 2005. pp. 429–469. [Google Scholar]
- 4.Lee DH, Iwanski GB, Thoennissen NH. Cucurbitacin:ancient compound shedding new light on cancer treatment. Sci World J. 2010;10:413–418. doi: 10.1100/tsw.2010.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hamasaki M, Furuta N, Matsuda A, Nezu A, Yamamoto A, Fujita N, et al. Autophagosomes form at ER mitochondria contact sites. Natur. e2013;495:389–393. doi: 10.1038/nature11910. [DOI] [PubMed] [Google Scholar]
- 6.Bernard A, Klionsky DJ. Autophagosome formation:tracing the source. Dev Cell. 2013;25:116–117. doi: 10.1016/j.devcel.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mizushima N, Yoshimori T, Ohsumi Y. The role of Atg proteins in autophagosome formation. Annu Rev Cell Dev Biol. 2011;27:107–132. doi: 10.1146/annurev-cellbio-092910-154005. [DOI] [PubMed] [Google Scholar]
- 8.Ryter SW, Choi AM. Regulation of autophagy in oxygen-dependent cellular stress. Curr Pharm Des. 2013;19:2747–2756. doi: 10.2174/1381612811319150010. [DOI] [PubMed] [Google Scholar]
- 9.Ryter SW, Cloonan SM, Choi AM. Autophagy:A critical regulator of cellular metabolism and homeostasis. Mol Cells. 2013;36:7–16. doi: 10.1007/s10059-013-0140-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brech A, Ahlquist T, Lothe RA, Stenmark H. Autophagy in tumour suppression and promotion. Mol Oncol. 2009;3:366–375. doi: 10.1016/j.molonc.2009.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schmukler E, Grinboim E, Schokoroy S, Amir A, Wolfson E, Kloog Y, et al. Ras inhibition enhances autophagy, which partially protects cells from death. Oncotarget. 2013;4:142–152. doi: 10.18632/oncotarget.703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.CufíS Vazquez-Martin A, Oliveras-Ferraros C, Corominas-Faja B, Urruticoechea A, Martin-Castillo B, et al. Autophagy-related gene 12 (ATG12) is a novel determinant of primary resistance to HER2-targeted therapies:utility of transcriptome analysis of the autophagy interactome to guide breast cancer treatment. Oncotarget. 2012;3:1600–1614. doi: 10.18632/oncotarget.742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gewirtz D.A. Cytoprotective and nonprotective autophagy in cancer therapy. Autophagy. 2013;9:1263–1265. doi: 10.4161/auto.25233. [DOI] [PubMed] [Google Scholar]
- 14.Chu SC, Hsieh YS, Yu CC, Lai YY, Chen PN. Thymoquinone induces cell death in human squamous carcinoma cells via caspase activation-dependent apoptosis and LC3-II activation-dependent autophagy. PLoS One. 2014;9:e101579. doi: 10.1371/journal.pone.0101579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hoffman B, Liebermann DA. Apoptotic signaling by c-MYC. Oncogene. 2008;27:6462–6472. doi: 10.1038/onc.2008.312. [DOI] [PubMed] [Google Scholar]
- 16.Tsuneoka M, Umata T, Kimura H, Koda Y, Nakajima M, Kosai K, et al. c-myc induces autophagy in rat 3Y1 fibroblast cells. Cell Struct Funct. 2003;28:195–204. doi: 10.1247/csf.28.195. [DOI] [PubMed] [Google Scholar]
- 17.Luna-Vargas MP, Chipuk JE. Physiological and pharmacological control of BAK, BAX, and beyond. Trends Cell Biol. 2016;26:906–917. doi: 10.1016/j.tcb.2016.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ishdorj G, Johnston JB, Gibson SB. Cucurbitacin-I (JSI-124) activates the JNK/c-Jun signaling pathway independent of apoptosis and cell cycle arrest in B leukemic cells. BMC cancer. 2011;11:268–279. doi: 10.1186/1471-2407-11-268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, et al. Cancer incidence and mortality worldwide:sources, methods and major patterns in GLOBOCAN, 2012. Int J Cancer. 2015;136:E359–386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 20.Jafargholizadeh N, Zargar SJ, Yassa N, Tavakoli S. Purifcation of Cucurbitacins D, E, and I from Ecballium Elaterium(L.). A. Rich Fruits and Study of Their Cytotoxic Effects on the AGS Cell Line. Asian Pac J Cancer Prev. 2016;17:4631–4635. doi: 10.22034/APJCP.2016.17.10.4631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 22.Chen J, Bardes EE, Aronow BJ, Jegga AG. ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res. 2009;37(suppl_2):W305–W311. doi: 10.1093/nar/gkp427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aerts S, Lambrechts D, Maity S, Van Loo P, Coessens B, De Smet F, et al. Gene prioritization through genomic data fusion. Nat Biotechnol. 2006;24:537–544. doi: 10.1038/nbt1203. [DOI] [PubMed] [Google Scholar]
- 24.Newman DJ, Cragg GM. Natural products as sources of new drugs over the 30 years from 1981 to 2010. J Nat Prod. 2012;75:311–335. doi: 10.1021/np200906s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Attard E, Scicluna-Spiteri A. The cultivation and cucurbitacin content of Ecballium elaterium(L.) A. Rich. Rep Cucurbit Genet Coop. 2003;26:66–69. [Google Scholar]
- 26.Ríos JL, Andújar I, Escandell JM, Giner RM, Recio MC. Cucurbitacins as inducers of cell death and a rich source of potential anticancer compounds. Curr Pharm Des. 2012;18:1663–1676. doi: 10.2174/138161212799958549. [DOI] [PubMed] [Google Scholar]
- 27.Wlodkowic D, Skommer J, Darzynkiewicz Z. Cytometry in cell necrobiology revisited. Recent advances and new vistas. Cytometry A. 2010;77:591–606. doi: 10.1002/cyto.a.20889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gurusamy N, Das DK. Detection of cell death by autophagy. Methods Mol Biol. 2009;559:95–103. doi: 10.1007/978-1-60327-017-5_7. [DOI] [PubMed] [Google Scholar]
- 29.Zha QB, Zhang XY, Lin QR, Xu LH, Zhao GX, Pan H, et al. Cucurbitacin E induces autophagy via downregulating mTORC1 signaling and upregulating AMPK activity. PloS one. 2015;10:e0124355. doi: 10.1371/journal.pone.0124355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang T, Li Y, Park KA, Byun HS, Won M, Jeon J, et al. Cucurbitacin induces autophagy through mitochondrial ROS production which counteracts to limit caspase-dependent apoptosis. Autophagy. 2012;8:559–576. doi: 10.4161/auto.18867. [DOI] [PubMed] [Google Scholar]
- 31.Yuan G, Yan SF, Xue H, Zhang P, Sun JT, Li G. Cucurbitacin I induces protective autophagy in glioblastoma in vitro and in vivo. J Biol Chem. 2014;289:10607–10619. doi: 10.1074/jbc.M113.528760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Deng C, Zhang B, Zhang S, Duan C, Cao Y, Kang W, et al. Low nanomolar concentrations of Cucurbitacin-I induces G2/M phase arrest and apoptosis by perturbing redox homeostasis in gastric cancer cells in vitro and in vivo. Cell Death Dis. 2016;7:e2106. doi: 10.1038/cddis.2016.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sun T, Li D, Wang L, Xia L, Ma J, Guan Z, et al. c-Jun NH2-terminal kinase activation is essential for up-regulation of LC3 during ceramide-induced autophagy in human nasopharyngeal carcinoma cells. J Transl Med. 2011;9:161–170. doi: 10.1186/1479-5876-9-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Young MM, Takahashi Y, Khan O, Park S, Hori T, Yun J, et al. Autophagosomal membrane serves as platform for intracellular death-inducing signaling complex (iDISC)-mediated caspase-8 activation and apoptosis. J Biol Chem. 2012;287:12455–12468. doi: 10.1074/jbc.M111.309104. [DOI] [PMC free article] [PubMed] [Google Scholar]


