Abstract
Objective(s):
Nanobodies, the single domain antigen binding fragments of heavy chain-only antibodies occurring naturally in camelid sera, are the smallest intact antigen binding entities. Their minimal size assists in reaching otherwise largely inaccessible regions of antigens. However, their camelid origin raises a possible concern of immunogenicity when used for human therapy. Humanization is a promising approach to overcome the problem.
Materials and Methods:
Here, we designed a humanized version of previously developed nanobody (anti vascular endothelial growth factor nanobody), evaluated and compared its predicted 3D structure, affinity and biological activity with its original wild type nanobody.
Results:
Our in silico results revealed an identical 3D structure of the humanized nanobody as compare to original nanobody. In vitro studies also demonstrated that the humanization had no significant visible effect on the nanobody affinity or on its biological activity.
Conclusion:
The humanized nanobody could be developed and proposed as a promising lead to target pathologic angiogenesis.
Keywords: Angiogenesis, Antibody engineering, Humanization, Nanobody, VEGF
Introduction
There are currently three drugs on the market that target pathologic angiogenesis. Bevacizumab (Avastin®) is the first anti-angiogenic drug that was approved in 2004 by the Food and Drug Administra-tion (FDA) for the treatment of metastatic colorectal cancer (1, 2). Despite the high specificity and affinity of full-length antibodies to their target, some problems such as their large MW (about 150 kDa) and reduced penetration in tumor tissues, may limit their use in the clinical practice (3). Such problems and progress in the field of antibody engineering have led to the emergence of a new generation of therapeutic molecules that combine the advantages of small molecules as well as of antibodies. In fact, one of the major goals of antibody engineering is the development of various antibody derivatives with improved performance (4). Hence, Ranibizumab (Lucentis), a recombinant humanized Fab fragment of a monoclonal antibody, has been introduced (5). Ranibizumab was approved in 2006 by FDA for the treatment of age-related macular degeneration (5).
Nanobodies or Variable domain of heavy chain antibodies are the smallest intact, natural antigen-binding fragments (about 11-15 kDa). They have several advantages over other antibody fragments such as Fab, Fv and single-chain variable fragment (scFv) (6-9). Nanobodies are able to withstand destructive effects of heat and chemicals, much better than other antibodies. Therefore, they can be used in harsh conditions (10, 11). The main advantage of Nanobodies is related to their simple structure that leads to high levels of properly folded material after bacterial or yeast expression. Such production method is more economical than mammalian expression systems (12-14). Nanobodies have also on average a longer complementarity-determining region 3 (CDR3) loop and four hallmark amino acid substitutions in their framework region-2. The large CDR3 loop often enables the nanobody to recognize different epitopes, such as grooves or concave surfaces that are much less antigenic for conventional antibodies. These unique properties make nanobodies suitable for drug development as an alternatives to mouse or human full-length antibodies (15-17). We have previously developed and characterized various nanobodies to different targets (18-22). So far, there are various nanobodies in different phases of clinical trials, and nearly all of them are humanized (www.ablynx.com). However, the humanization activity of Nanobodies and their effect on the folding, expression and biological activity is poorly reported (23). The main aim of such humanization of camelid nanobodies is to reduce their possible immunogenicity. Several nano-body targeting vascular endothelial growth factors (VEGF) have been produced in a previous study and their in vitro (24) and in vivo (19) biological activity have been evaluated. In this study, we decided to develop the humanized format of the nanobody targeting VEGF and evaluate its function in inhibiting the proliferation of human endothelia cells relative to the original wild type nanobody.
Materials and Methods
Materials
Human endothelial primary cells were isolated from umbilical cord (25). For all in vitro assays, the endothelial cells were between passages 2-4. Bevacizumab was from Roche (Basel, Switzerland). The plasmid of pHEN-6c (26) was our standard vector to express Nanobodies under control of lac promotor. The pHEN-6c vector and WK6 Escherichia coli (E. coli) cells were gifted by Serge Muyldermans(Laboratory of Cellular and Molecular Immunology, Vrije University Brussels, Brussels, Belgium). The nanobody was expressed in the periplasm space of bacteria and comprises a His6 tag for purification purposes. The anti-VEGF nanobody used in this study (referred to as Nb42) originated from a previous report (27).
Design, modeling and construction of the Humanized Nanobody
The CDR and framework region of the camel nanobody are numbered according to the international ImMunoGeneTics information system® (IMGT )and the nanobody humanization protocol was according to published methods (28, 29). Briefly, the amino acid sequence of our camel nanobody was aligned with the humanized nanobodies (23, 30) to determine the sequence differences in framework regions. Hallmark amino acids in framework region 2 (amino acids at position 42, 49, 50 and 52) were not substituted because of their critical role. To find protein regions that are possibly antigenic, we used a program that predicts T-cell epitopes. The MHCII peptide binding predictions were made using the Immune Epitope Data Base (IEDB) analysis resource Consensus tool (31, 32). The analysis was made on only 8 common alleles of MHC class II (Because they are present in more general societies as well as in the majority of populations): HLA-DRB1 * 0101, HLA-DRB1 * 0301, HLA-DRB1 * 0401, HLA-DRB1 * 0701, HLA-DRB1 * 0801, HLA-DRB1 * 1101, HLA-DRB1 * 1301, HLA-DRB1 * 1501. The primary structure of camel and humanized nanobody was analyzed with ExPASy ProtParam tool (online version) (http://web.expasy.org/protparam/). The 3D structure of the camel and humanized nanobody were modeled by the online version of I-TASSER - the protein structure and function prediction server (33, 34). The quality of the predicted models was examined by ProSA-the Protein Structure Analysis server (35, 36) and RAMPAGE-Ramachandran plot assessment program (37). The nanobody nucleotide sequence was codon optimized for expression in an E. coli host and then synthesized into pBSK-G standard vector by GENERAY.
Sub-cloning, expression and purification
The nanobody gene was amplified by PCR using forward and reverse primers: A6E (5’-GATGTGCAG CTGCAGGAGTCTGGAGGAGG-3’) and p38 (5’-GGACTAGT GCGGCCGCTGGAGACGGTGACCTGGGT-3’) containing PstI and BstEII restriction enzyme sites (underlined). The standard vector, pBSK-G, containing the syn-thesized nanobody sequence was used as template. The humanized nanobody gene was sub-cloned into pHEN6c expression vector. The condition of ligation was as follows: 7 µl of amplified nanobody gene (50 ng), 6 µl of pHEN6c vector (50 ng), 1 µl of T4-DNA ligase (Fermentas, Canada) and 6 µl of H2O. The recombinant construct transformed by heat shock into WK6 E. coli competent cells. The obtained clones were verified by colony-PCR, double digestion (with PstI and BstEII) as well as by nucleotide sequencing. The colony-PCR was performed by universal reverse primer (5’ TCACACAGGAAACAGCTATGAC 3’) as well as universal forward primer (5’ CGCCAGGGTTTTCC-CAGTCACGAC 3’). Expression of humanized nanobody was induced by Isopropyl β-D-1-thiogalactopyranoside (IPTG) 1 mM in TB medium containing 50 µg/ml ampicillin at 28 °C for 16 hr (shacking at 250 rpm). Due to the presence of PelB leader sequence in pHEN6c vector, the nanobody was expressed in the periplasm of bacterial cells. Therefore, the expressed periplasmic proteins were extracted using an osmotic shock and then further purified by nickel affinity chromate-graphy according to the manufacturer protocol (Qiagen, Germany). The purified nanobody was identified by 15% SDS-PAGE (under reducing condi-tions) and western blot analysis (with anti-His-HRP conjugate (final concentration: 0.5 µg/ml)). The amount of purified protein was evaluated using the Bradford assay (18).
Enzyme-linked immunosorbent assay
The recognition of human VEGF by our humanized nanobody was evaluated using enzyme-linked immunosorbent assay (ELISA). A 96-well was coated with various concentrations (0-1000 ng/ml) of VEGF (R&D, Canada) at 4 °C for 16 hr. The next day, residual protein binding sites in the wells were blocked with 2% bovine serum albumin (BSA) for 2 hr at room temperature. Then 1000 ng/ml of humanized nanobody or Bevacizumab (as control) were added to the wells and incubation was continued for 1 hr at RT. Subsequently, after rinsing the wells, VEGF-associated nanobody and Bevacizumab were detected by anti-His-HRP (final concentration: 0.5 µg/ml) and anti-human IgG1 Fc-HRP (final concentration: 0.5 µg/ml), respectively. Finally, the rate of absorbance and signal intensity was measured at 450 nm (20). The affinity of camel and humanized nanobody was measured according to the method of Beatty (38). Bevacizumab was used as control antibody in affinity analysis (24).
Biological activity
The activity of humanized nanobody was examined by MTT assay on human endothelial cells.
About 104 human endothelial cells in DMEM and F12 medium containing 2% fetal bovine serum (FBS) were seeded in 96-well plates and incubated at 37 °C and 5% CO2. Different concentrations of humanized, wild type and irrelevant nanobody (0-10 µg/ml) as well as Bevacizumab in presence or absence of VEGF (50 ng/ml) were added to the wells and incubation was continued for 48 hr under the same conditions. Then, the inhibition of proliferation of VEGF-stimulated human endothelial cells by humanized nanobody was evaluated by the MTT assay. The wells incubated with 50 µl of MTT solution (at 2 mg/ml in PBS) and kept in dark. Then, MTT dye was solubilized with isopropanol and absorbance was measured at 570 nm.
Statistical analysis
Statistical analysis carried out using GraphPad PRISM.v 5.0. Unpaired Student’s t-test was performed for analysis of each group The difference between groups were significant in case of P<0.05 value.
Results
Analysis of humanized format of the nanobody
After designating CDR and framework regions and numbering the amino acids of the nanobody according to IMGT, we aligned the sequence to human variable domain or VH of family III and identified those amino acids that needed to be substituted to obtain the humanized nanobody (Figure 1). The amino acids to be substituted are highlighted in this figure. For MHCII peptide binding prediction, amino acid sequence stretches of framework regions of wild type as well as humanized nanobody were considered and the percentage of possible binding with different HLAs was examined. According to MHCII binding prediction of IEDB, the higher percentile rank is indicative for stronger connections between T-cell epitopes and MHCII and therefore, those regions are predicted to be strongly antigenic. Results of MHCII binding prediction indicated that the humanized nanobody in comparison with the original dromedary nanobody has a reduced antigenicity (data not shown). According to ExPASy ProtParam results, molecular weight and theoretical isoelectric point (pI) of wild type dromedary and humanized nanobody changed from 13853.40 and 8.98 to 13807.36 and 9.01, respectively. The instability index (II) of camel and humanized nanobody was computed as 32.42 and 31.78, respectively which are therefore both classified as stable proteins. The 3D structure of camel and humanized nanobody was modeled using the I-TASSER server and the models with highest C-score were selected and shown in Figure 2. The quality of the predicted models was evaluated using the ProSA server and the Z-score of each model was obtained (Figure 3). The lower Z-score is indicative for predicted struc-tural models that are close to the structure of the native proteins. The Z-score for predicted models of wild type dromedary and humanized nanobody is -5.46 and -6.44, respectively. The Ramachandran plot analysis was performed on wild type dromedary and humanized nanobody using the RAMPAGE program and results showed that about 4% and 7% of residues of camel and humanized nanobody are in the outlier region (Figure 4).
Figure 1.

Numbering and humanization of the amino acid sequences of the nanobody. Camel nanobody (black color), Humanized nanobody (red color). The changed amino acids showed as highlighted
Figure 2.
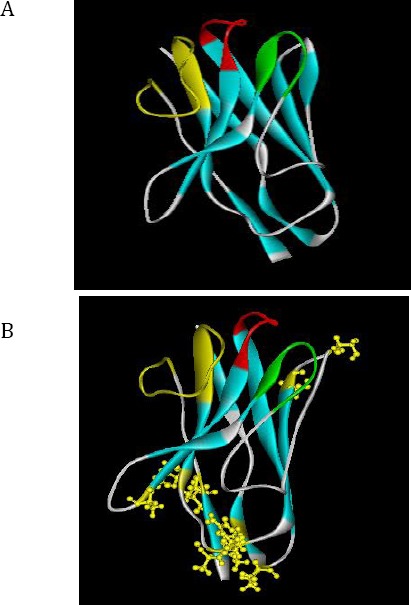
Predicted 3D structures of the nanobody by I-TASSER server. A) Camel nanobody modeled by I-TASSER server. The parameters of prediction: C-score=0.98, Estimated TM-score = 0.85±0.08, Estimated RMSD = 2.5±1.9Å. B) Humanized nanobody modeled by I-TASSER server. The parameters of prediction: C-score=0.95, Estimated TM-score = 0.84±0.08, Estimated RMSD = 2.6±1.9Å. The complementarity-determining region (CDR) regions are shown with different colors: CDR1 (red color), CDR2 (green color) and CDR3 (yellow color)
Figure 3.
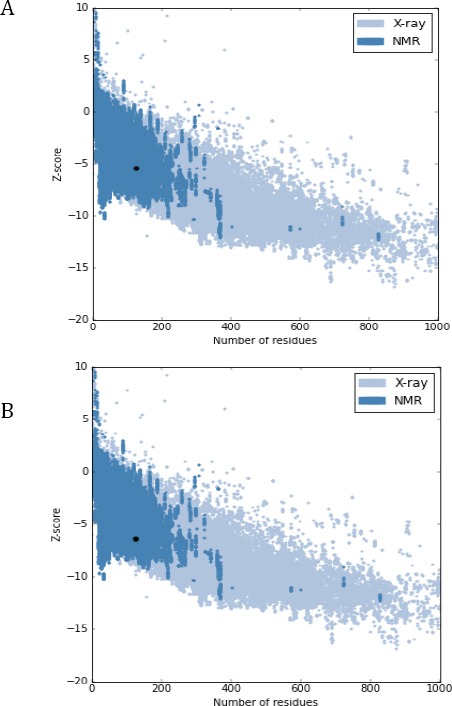
Quality assessment of predicated models. A) Predicted 3D model of camel nanobody was evaluated by ProSA server (Z-score: -5.46). B) Predicted 3D model of humanized nanobody was evaluated by ProSA server (Z-score: -6.44)
Figure 4.
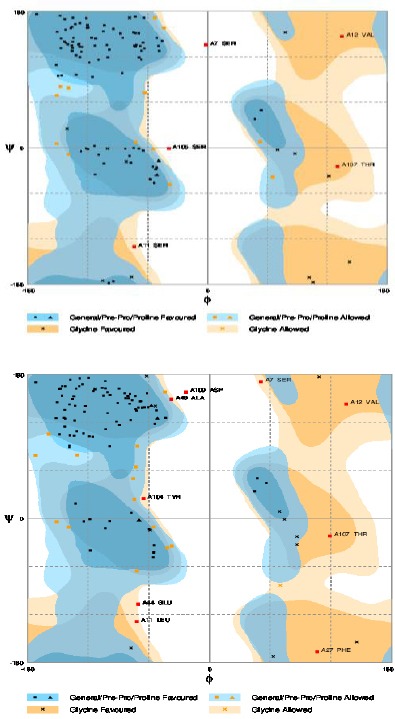
Ramachandran plot assessment. A) Ramachandran plot assessment of predicted model of camel nanobody (about 4% of residues are in outlier region). B) Ramachandran plot assessment of predicted model of humanized nanobody (about 7% of residues are in outlier region)
Construction, sub-cloning, expression and purification results
The humanized sequence of our nanobody was synthesized by GENERAY and cloned in the pBSK-G vector. The nanobody gene was amplified using A6E and p38 primers and PCR amplicon after gel electrophoresis giving a single band of 400 bp, which is shown in Figure 5A. Then, this fragment was ligated into the pHEN6c bacterial expression vector between PstI and BstEII restriction enzyme sites and transformed into WK6 cells. The accuracy of cloning was checked by colony-PCR, restriction enzyme digestion and by nucleotide sequencing. After gel electrophoresis, for 3 out of the 9 clones tested, the amplicon was shown to have a size of 550 bp (Figure 5B), which is the proper size for a nanobody between these primer annealing sites on pHEN6c. The nucleotide sequence also confirmed the faithful cloning. One clone was cultured and the Nanobody gene was expressed. The protein was extracted from the periplasm and purified by nickel affinity chromatography. An aliquot separated by SDS-PAGE demonstrated the homogeneity and purity of humanized nanobody material as eluted from IMAC (Figure 5C). A western blot analysis on the same sample identified the presence of the recombinant His tag on our humanized nanobody protein (Figure 5D). The final yield of purified humanized and wild type Nb was 5 and 3 mg/liter, respectively.
Figure 5.
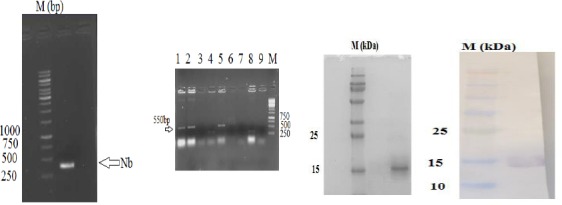
Sub-cloning and purification results of humanized nanobody. A) PCR results of humanized nanobody. As can be observed, the sequence of about 400 bp was amplified. B) The colony-PCR results. The existence of band 550 bp indicated the success in cloning process. C) Purification results. Coomassie brilliant blue stained 15% SDS-PAGE results showed that humanized nanobody was purified using nickel affinity chromatography. D) Western blots results. Western blot was performed with anti-His-HRP conjugated antibody
VEGF binding capacity of the Nanobodies
An ELISA experiment was developed to demonstrate that the humanized nanobody is able to detect VEGF. The VEGF was coated at various concentrations on the wells of the microtiter plate and the nanobody and Bevacizumab concentrations were 1000 ng/ml. ELISA showed that both the original wild type and humanized nanobody like non-humanized nanobody are able to detect VEGF. The affinity of both, the wild type and humanized nanobody, was calculated according to the method of Beatty to be around 60 nM. The calculated affinity for Bevacizumab was around 40 nM by same method.
Biological assay
To examine the biological activity of the humanized nanobody and assessing whether it is capable to inhibit cell proliferation, we performed an MTT assay on cultures of human endothelia cells. Results of MTT assay showed that a concentration of 10 µg/ml of the humanized nanobody, just like the wild type dromedary nanobody, inhibited significantly the proliferation of human endothelial cells through targeting VEGF (Figure 6). However, irrelevant nanobody did not inhibit proliferation of human endothelial cells. The highest inhibition of human endothelial cell proliferation was noticed at a concentration of 10 µg/ml of the nanobody, but this level of inhibition was already attained with 1 µg/ml of Bevacizumab.
Figure 6.
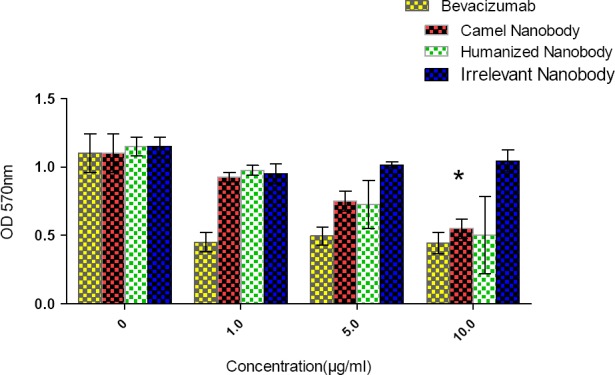
Biological activity results. MTT was performed for biological activity assay of camel and humanized nanobody. As can be observed, the most inhibitory effect of camel and humanized nanobody was observed at concentration of 10 µg/ml (*, P=0.043). The assay was performed in triplicate. The error bar represents for mean ± SD
Discussion
Due to the increasing rate of resistance of cancer cells to common treatment, there is a growing need to research on the discovery of new anticancer agents. Resistance of cancer cells to the chemotherapeutic drugs in addition to the side effects of these drugs is at the origin of the failure of cancer treatment. Thus, finding more effective drugs with fewer side effects is very important (39-42). The small size, monomeric behavior and single domain properties of nanobodies provide beneficial characteristics for development of novel drugs (16). Here, we humanized the framework regions of the nanobody targeting VEGF referred to as Nb42 (24) according to the dromedary antibody humanization protocol (28, 30) to reduce the possible antigenicity of the original dromedary-derived nanobody. Nanobodies are naturally single-domain antibodies with highly identical sequence to human VH domains. For Nb42, we identified nine positions where the amino acids needed to be substituted: in framework region-1 S11L and A14P, in framework region-2 region V40A, in framework region-3 A75S, M78T, K87R, P88A and I93V and in framework region-4 Q121L. The VHH hallmark amino acids in framework region-2 (F42, E49, R50 and A52) were not modified as it would be detrimental for solubility of the protein domain and for antigen-recognition (30). According to the instability index results of ExPASy ProtParam, both camel and humanized nanobody are stable proteins, indicating that the amino acid substitutions in framework regions of the nanobody have no significant effect on protein stability. The obtained Z-score for humanized and wild type dromedary nanobody was in the range of proteins whose structure has been determined by NMR. The structure prediction gave an identical architecture for both proteins, including their antigen binding loops, which is especially important for the antigen recognition.
Conclusion
The bacterial protein expression yield of the humanized (codon optimized) nanobody was very similar to that of the wild type nanobody (24). Finally, the affinity and functional analysis data confirmed that the antigen association and cell proliferation inhibition remains intact upon humanization of wild type dromedary nanobody. The investigation on the effect of amino acid substitutions to humanize camelid nanobodies is very scarce. This current report, on the in silico as well as in vitro studies can be taken as a guideline for such tasks that will lead to nanobody-based therapeutics of potential lower antigenicity.
Acknowledgment
The current study was supported by Iran National Science Foundation (INSF) [grant number 94012812] and Pasteur Institute of Iran, Tehran, Iran.
Conflict of interest
The authors declare that no conflict of interest exists.
Funding
This study funded by Iran National Science Foundation (INSF) [to F Kazemi-Lomedasht; grant number 94012812].
References
- 1.Ferrara N, Hillan KJ, Gerber H-P, Novotny W. Discovery and development of bevacizumab, an anti-VEGF antibody for treating cancer. Nat Rev Drug Discov. 2004;3:391–400. doi: 10.1038/nrd1381. [DOI] [PubMed] [Google Scholar]
- 2.Hurwitz H, Fehrenbacher L, Novotny W, Cartwright T, Hainsworth J, Heim W, et al. Bevacizumab plus irinotecan, fluorouracil, and leucovorin for metastatic colorectal cancer. N Engl J Med. 2004;350:2335–2342. doi: 10.1056/NEJMoa032691. [DOI] [PubMed] [Google Scholar]
- 3.Chames P, Van Regenmortel M, Weiss E, Baty D. Therapeutic antibodies:successes, limitations and hopes for the future. Br J Pharmacol. 2009;157:220–233. doi: 10.1111/j.1476-5381.2009.00190.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Filpula D. Antibody engineering and modification technologies. Biomolecular engineering. 2007;24:201–215. doi: 10.1016/j.bioeng.2007.03.004. [DOI] [PubMed] [Google Scholar]
- 5.Rosenfeld PJ, Brown DM, Heier JS, Boyer DS, Kaiser PK, Chung CY, et al. Ranibizumab for neovascular age-related macular degeneration. N Engl J Med. 2006;355:1419–1431. doi: 10.1056/NEJMoa054481. [DOI] [PubMed] [Google Scholar]
- 6.Harmsen MM, De Haard HJ. Properties, production, and applications of camelid single-domain antibody fragments. Appl Microbiol Biotechnol. 2007;77:13–22. doi: 10.1007/s00253-007-1142-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dumoulin M, Conrath K, Van Meirhaeghe A, Meersman F, Heremans K, Frenken LG, et al. Single-domain antibody fragments with high conformational stability. Protein Sci. 2002;11:500–515. doi: 10.1110/ps.34602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Desmyter A, Decanniere K, Muyldermans S, Wyns L. Antigen specificity and high affinity binding provided by one single loop of a camel single-domain antibody. J Biol Chem. 2001;276:26285–26290. doi: 10.1074/jbc.M102107200. [DOI] [PubMed] [Google Scholar]
- 9.Nicholls H. The Camel Factor. NewScientist. 2007:50–53. [Google Scholar]
- 10.Dolk E, van der Vaart M, Lutje Hulsik D, Vriend G, de Haard H, Spinelli S, et al. Isolation of llama antibody fragments for prevention of dandruff by phage display in shampoo. Appl Environ Microbiol. 2005;71:442–450. doi: 10.1128/AEM.71.1.442-450.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Olichon A, Schweizer D, Muyldermans S, Marco Ad. Heating as a rapid purification method for recovering correctly-folded thermotolerant VH and VHH domains. BMC Biotechnol. 2007;7:1–8. doi: 10.1186/1472-6750-7-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Arbabi-Ghahroudi M, Tanha J, MacKenzie R. Prokaryotic expression of antibodies. Cancer Metastasis Rev. 2005;24:501–519. doi: 10.1007/s10555-005-6193-1. [DOI] [PubMed] [Google Scholar]
- 13.Frenken LG, van der Linden RH, Hermans PW, Bos JW, Ruuls RC, de Geus B, et al. Isolation of antigen specific llama VHH antibody fragments and their high level secretion by Saccharomyces cerevisiae. J Biotechnol. 2000;78:11–21. doi: 10.1016/s0168-1656(99)00228-x. [DOI] [PubMed] [Google Scholar]
- 14.Rahbarizadeh F, Rasaee MJ, Forouzandeh M, Allameh AA. Over expression of anti-MUC1 single-domain antibody fragments in the yeast Pichia pastoris. Mol Immunol. 2006;43:426–435. doi: 10.1016/j.molimm.2005.03.003. [DOI] [PubMed] [Google Scholar]
- 15.Alvarez-Rueda N, Behar G, Ferre V, Pugniere M, Roquet F, Gastinel L, et al. Generation of llama single-domain antibodies against methotrexate, a prototypical hapten. Mol Immunol. 2007;44:1680–1690. doi: 10.1016/j.molimm.2006.08.007. [DOI] [PubMed] [Google Scholar]
- 16.Kolkman JA, Law DA. Nanobodies–from llamas to therapeutic proteins. Drug Discov Today:Technol. 2010;7:e139–e146. doi: 10.1016/j.ddtec.2010.03.002. [DOI] [PubMed] [Google Scholar]
- 17.Rahbarizadeh F, Rasaee MJ, Forouzandeh Moghadam M, Allameh AA, Sadroddiny E. Production of novel recombinant single-domain antibodies against tandem repeat region of MUC1 mucin. Hybrid Hybridomics. 2004;23:151–159. doi: 10.1089/1536859041224334. [DOI] [PubMed] [Google Scholar]
- 18.Kazemi-Lomedasht F, Behdani M, Rahimpour A, Habibi-Anbouhi M, Poshang-Bagheri K, Shahbazzadeh D. Selection and characterization of specific Nanobody against human immunoglobulin G. Monoclon Antib Immunodiagn Immunother. 2015;34:201–205. doi: 10.1089/mab.2014.0086. [DOI] [PubMed] [Google Scholar]
- 19.Kazemi-Lomedasht F, Pooshang-Bagheri K, Habibi-Anbouhi M, Hajizadeh-Safar E, Shahbazzadeh D, Mirzahosseini H, et al. In vivo immunotherapy of lung cancer using cross-species reactive vascular endothelial growth factor nanobodies. Iran J Basic Med Sci. 2017;20:489–496. doi: 10.22038/IJBMS.2017.8672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kazemi-Lomedasht F, Behdani M, Habibi-Anbouhi M, Shahbazzadeh D. Production and Characterization of Novel Camel Single Domain Antibody Targeting Mouse Vascular Endothelial Growth Factor. Monoclon Antib Immunodiagn Immunother. 2016;35:167–171. doi: 10.1089/mab.2016.0001. [DOI] [PubMed] [Google Scholar]
- 21.Homayouni V, Ganjalikhani-hakemi M, Rezaei A, Khanahmad H, Behdani M, Lomedasht FK. Preparation and characterization of a novel nanobody against T-cell immunoglobulin and mucin-3 (TIM-3) Iran J Basic Med Sci. 2016;19:1201–1208. [PMC free article] [PubMed] [Google Scholar]
- 22.Bagheri M, Babaei E, Shahbazzadeh D, Habibi-Anbouhi M, Alirahimi E, Kazemi-Lomedasht F, et al. Development of a recombinant camelid specific diabody against the heminecrolysin fraction of Hemiscorpius lepturus scorpion. Toxin Rev. 2017;36:7–11. [Google Scholar]
- 23.Vaneycken I, Govaert J, Vincke C, Caveliers V, Lahoutte T, De Baetselier P, et al. In vitro analysis and in vivo tumor targeting of a humanized, grafted nanobody in mice using pinhole SPECT/micro-CT. J Nuc Med. 2010;51:1099–1106. doi: 10.2967/jnumed.109.069823. [DOI] [PubMed] [Google Scholar]
- 24.Kazemi-Lomedasht F, Behdani M, Bagheri KP, Habibi-Anbouhi M, Abolhassani M, Arezumand R, et al. Inhibition of angiogenesis in human endothelial cell using VEGF specific nanobody. Mol Immunol. 2015;65:58–67. doi: 10.1016/j.molimm.2015.01.010. [DOI] [PubMed] [Google Scholar]
- 25.Kazemi-Lomedasht F, Behdani M, Bagheri KP, Anbouhi MH, Abolhassani M, Khanahmad H, et al. Expression and purification of functional human vascular endothelial growth factor-a121;the most important angiogenesis factor. Adv Pharm Bull. 2014;4:323–328. doi: 10.5681/apb.2014.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Conrath KE, Lauwereys M, Wyns L, Muyldermans S. Camel single-domain antibodies as modular building units in bispecific and bivalent antibody constructs. J Biol Chem. 2001;276:7346–7350. doi: 10.1074/jbc.M007734200. [DOI] [PubMed] [Google Scholar]
- 27.Kazemi-Lomedasht Fatemeh BM, Pooshang Bagheri Kamran, Habibi-Anbouhi Mahdi, Abolhassani Mohsen, Arezumand Roghaye, Shahbazzadeh Delavar, Mirzahoseini Hasan. Inhibition of angiogenesis in human endothelial cell using VEGF specific nanobody. Mol Immunol. 2015;65:58–67. doi: 10.1016/j.molimm.2015.01.010. [DOI] [PubMed] [Google Scholar]
- 28.Conrath K, Vincke C, Stijlemans B, Schymkowitz J, Decanniere K, Wyns L, et al. Antigen binding and solubility effects upon the veneering of a camel VHH in framework-2 to mimic a VH. J Mol Biol. 2005;350:112–125. doi: 10.1016/j.jmb.2005.04.050. [DOI] [PubMed] [Google Scholar]
- 29.Abderrazek RB, Vincke C, Hmila I, Saerens D, Abidi N, El Ayeb M, et al. Development of Cys38 knock-out and humanized version of NbAahII10 nanobody with improved neutralization of AahII scorpion toxin. Protein Eng Des Sel. 2011;24:727–735. doi: 10.1093/protein/gzr037. [DOI] [PubMed] [Google Scholar]
- 30.Vincke C, Loris R, Saerens D, Martinez-Rodriguez S, Muyldermans S, Conrath K. General strategy to humanize a camelid single-domain antibody and identification of a universal humanized nanobody scaffold. J Biol Chem. 2009;284:3273–3284. doi: 10.1074/jbc.M806889200. [DOI] [PubMed] [Google Scholar]
- 31.Wang P, Sidney J, Dow C, Mothe B, Sette A, Peters B. A systematic assessment of MHC class II peptide binding predictions and evaluation of a consensus approach. PLoS Comput Biol. 2008;4:e1000048. doi: 10.1371/journal.pcbi.1000048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang P, Sidney J, Kim Y, Sette A, Lund O, Nielsen M, et al. Peptide binding predictions for HLA DR, DP and DQ molecules. BMC Bioinformatics. 2010;11:568. doi: 10.1186/1471-2105-11-568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang J, Yan R, Roy A, Xu D, Poisson J, Zhang Y. The I-TASSER Suite:protein structure and function prediction. Nat Methods. 2015;12:7–8. doi: 10.1038/nmeth.3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roy A, Kucukural A, Zhang Y. I-TASSER:a unified platform for automated protein structure and function prediction. Nat Protoc. 2010;5:725–738. doi: 10.1038/nprot.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wiederstein M, Sippl MJ. ProSA-web:interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res. 2007;35:W407–W410. doi: 10.1093/nar/gkm290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sippl MJ. Recognition of errors in three-dimensional structures of proteins. Proteins. 1993;17:355–362. doi: 10.1002/prot.340170404. [DOI] [PubMed] [Google Scholar]
- 37.Lovell SC, Davis IW, Arendall WB, de Bakker PI, Word JM, Prisant MG, et al. Structure validation by Cαgeometry:ϕ, ψand Cβdeviation. Proteins. 2003;50:437–450. doi: 10.1002/prot.10286. [DOI] [PubMed] [Google Scholar]
- 38.Beatty JD, Beatty BG, Vlahos WG. Measurement of monoclonal antibody affinity by non-competitive enzyme immunoassay. J Immunol Methods. 1987;100:173–179. doi: 10.1016/0022-1759(87)90187-6. [DOI] [PubMed] [Google Scholar]
- 39.Kluetz PG, Figg WD, Dahut WL. Angiogenesis inhibitors in the treatment of prostate cancer. Expert Opin Pharmacother. 2010;11:233–247. doi: 10.1517/14656560903451716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vermeulen PB, van Golen KL, Dirix LY. Angiogenesis, lymphangiogenesis, growth pattern, and tumor emboli in inflammatory breast cancer:a review of the current knowledge. Cancer. 2010;116:2748–2754. doi: 10.1002/cncr.25169. [DOI] [PubMed] [Google Scholar]
- 41.Szala S, Mitrus I, Sochanik A. Can inhibition of angiogenesis and stimulation of immune response be combined into a more effective antitumor therapy? Cancer Immunol Immunother. 2010;59:1449–1455. doi: 10.1007/s00262-010-0873-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Monk BJ, Willmott LJ, Sumner DA. Anti-angiogenesis agents in metastatic or recurrent cervical cancer. Gynecol Oncol. 2010;116:181–186. doi: 10.1016/j.ygyno.2009.09.033. [DOI] [PubMed] [Google Scholar]


