Abstract
Bedaquiline (BDQ) has been proven to be effective in the treatment of multidrug-resistant tuberculosis. We hypothesized that BDQ could be a potential agent to treat nontuberculous mycobacterial (NTM) infection. The objective of this study was to evaluate the in vitro activity of BDQ against rapidly growing mycobacteria by assessing the minimal inhibitory concentration (MIC) and minimal bactericidal concentration (MBC) against 18 NTM strains. For MIC determination we performed the resazurin microtitre assay broth dilution, and for the MBC the c.f.u. was determined. BDQ exhibited a strong inhibitory effect against most NTM tested; however, for some NTM strains the MBC was significantly higher than the MIC. A new finding is that Mycobacterium flavescens has a mutation in the gene atpE associated with natural resistance to BDQ. These preliminary promising results demonstrate that BDQ could be potentially useful for the treatment of NTM.
Keywords: resistance to bedaquiline, nontuberculous mycobacteria, atpE mutation, Mycobacterium flavescens, minimum inhibitory concentration, minimum bactericidal concentration
The genus Mycobacterium comprises more than 150 different species of mycobacteria with the capacity to cause pathogenicity in humans [1]. Most important among these species, due to their airborne transmission and public health implications, are Mycobacterium tuberculosis and Mycobacterium leprae causing tuberculosis (TB) and leprosy, respectively. Among nontuberculous mycobacteria (NTM), Mycobacterium avium and Mycobacterium abscessus represent prevalent sources of infection not only in immunocompromised individuals but also in other susceptible populations, such as in cystic fibrosis patients [2]. More recently, other emerging NTM, such as Mycobacterium chimaera, have been reported as causes of outbreaks due to heating–cooling devices in surgical rooms [3].
Drug resistance is one of the key issues associated with the current burden of TB around the world, negatively impacting control of the disease [4, 5]. Consequently, efforts have been devoted to the discovery and development of new anti-TB drugs [6]. As a result two new drugs, bedaquiline (BDQ) and delamanid, were recently approved for the treatment of multidrug-resistant TB (MDR-TB) [7, 8]. BDQ has a broad antimycobacterial spectrum and a novel mode of action, inhibiting the ATP synthase [9]. We hypothesized that BDQ could also treat NTM infection.
For infections caused by NTM, combination antimicrobial chemotherapy is the treatment of choice in most cases [10, 11]. Nevertheless, NTM is difficult to eradicate because most of them are naturally resistant to many common antibiotics and in many cases become refractory to the commonly recommended antibiotics [12]. In this context, BDQ has recently been used as off-label for salvage treatment in patients with Mycobacterium intracellulare lung disease, with encouraging results [13]. In order to shed light on the conditions and parameters guiding the potential use of BDQ for NTM infections, we evaluated its in vitro activity against a panel of rapidly growing NTM reference strains and clinical isolates, and explored the possible correlation of single nucleotide polymorphisms in the target gene and natural resistance to the drug.
Eighteen rapidly growing mycobacterial strains were used in this study (Table 1). Seventeen were obtained from the CCUG collection (http://www.ccug.se) and one strain from the UCL collection in Brussels, Belgium. Strains were cultured on Löwenstein–Jensen medium and the inoculum was prepared in distilled water, adjusted to McFarland 0.5 and diluted 1 : 10 in Mueller–Hinton (MH) broth medium. Mycobacterium smegmatis CCUG 28063 was used for quality control since its minimum inhibitory concentration (MIC) for BDQ of 0.015 µg ml−1 is well known [9]. To assess whether BDQ had a bacteriostatic or bactericidal effect, we determined the MIC and minimum bactericidal concentration (MBC) using the resazurin microplate assay (REMA) [14, 15]. Briefly, twofold serial dilutions were made in MH in 96-well polystyrene plates. BDQ concentrations were 2.0–0.0035 µg ml−1 and each experiment was performed in triplicate. An inoculum equal to McFarland 0.5 diluted 1 : 10 was prepared. Growth controls without drug (positive control), a drug control and a sterile control (negative control) were also prepared for each assay. To prevent evaporation during incubation, 200 µl sterile distilled water was added to all perimeter wells. Plates were sealed and incubated at 37 °C for 3 days before adding 30 µl of 0.01 % resazurin to all wells and incubating for a further 24 h. The MIC was determined as the lowest drug concentration that prevented growth and, therefore, a colour change from blue (oxidized state) to pink (reduced state). MIC values were scored for each isolate tested. The same plates were used for MBC determination. At day 4 of incubation and after the MIC reading, four blue wells were chosen to test the viability of the mycobacteria. One hundred microlitres from each well at the MIC, one concentration higher, and the previous two BDQ dilutions, were transferred to a tube and diluted in sterile distilled water to 10−3, 10−4 and 10−5 and plated in duplicate on Luria broth (LB) agar plates to determine the c.f.u. Also, c.f.u. were determined in duplicate for the positive control diluted 10−4, 10−5 and 10−6. The plates were incubated for 4 days. The percentage of killed bacteria was calculated against the control, and the MBC was defined as the lowest drug concentration that killed 99.9 % of bacteria.
Table 1. MIC and MBC range for 18 NTM strains.
|
EMBL accession number
for atpE sequences |
Collection number, strain reference | Species | Bedaquiline | |
|---|---|---|---|---|
| MIC (µg ml−1) | MBC (µg ml−1) | |||
| LT841272 | CCUG 28063, R-50263 | Mycobacterium smegmatis | 0.015 | >2 |
| LT841273 | CCUG 28060, R-50260 | Mycobacterium phlei | 0.07 | >2 |
| LT841274 | CCUG 41352, R-52136 | Mycobacterium duvalii | 2 | >2 |
| LT841275 | CCUG 55442, R-52147 | Mycobacterium cosmeticum | 0.007 | 0.015 |
| LT841276 | CCUG 25238, R-50254 | Mycobacterium mucogenicum | 0.062 | >0.5 |
| LT841277 | CCUG 37665T, R-50255 | Mycobacterium neoaurum | 2 | 2 |
| LT841278 | CCUG 28064, R-50259 | Mycobacterium peregrinum | 0.015 | >2 |
| LT841279 | CCUG 20999T, R-50257 | Mycobacterium parafortuitum | 0.015 | >2 |
| LT841280 | CCUG 29050, R-50269 | Mycobacterium flavescens | >2 | >2 |
| LT841281 | UCL 22248247, R-50337 | Mycobacterum flavescens | >2 | >2 |
| LT841282 | CCUG 31556, R-50246 | Mycobacterium fortuitum | 0.031 | 0.062 |
| LT841283 | CCUG 39181, R-52149 | Mycobacterium mageritense | 0.062 | >2 |
| LT841284 | CCUG 55633, R-52144 | Mycobacterium mageritense | 0.062 | >2 |
| LT841285 | CCGU 52054, R-52130 | Mycobacterium wolinskyi | 0.031 | >2 |
| LT841286 | CCUG 41449, R-50243 | Mycobacterium abscessus | 0.062 | >2 |
| LT841287 | CCUG 27851, R-50244 | Mycobacterium abscessus | 0.25 | >2 |
| LT841288 | ATCC 14472, R-50274 | Mycobacterium chelonae | 0.062 | >2 |
| LT841289 | DSM 45524T, R-50963 | Mycobacterium franklinii | 0.062 | 1 |
For investigation of the mutation in the gene atpE, DNA extraction was carried out according to Perez-Martinez et al. [16]. Briefly, a loopful of mycobacteria from a Löwenstein–Jensen culture was resuspended in 100 µl Milli-Q water, boiled for 5 min, placed on ice for 10 min, centrifuged at room temperature (13 600 g, 5 min) and the supernatants were used for PCR. The atpE gene was amplified using degenerated primers atpE forward (degenerated) 5′-TGTAYTTCAGCCARGCSATGG-3′ and atpE reverse (degenerated) 5′-CCGTTSGGDABGAGGAAGTTG-3′ [17]. However, if these primers did not amplify atpE, a second set of primers was used: atpE forward 5′ -TGTACTTCAGCCAAGCGATGG-3′ and atpE reverse 5′-CCGTTGGGAATGAGGAAGTTG-3′ [18]. For the degenerated primers, the PCR was run with an initial pre-denaturation at 95 °C for 5 min, followed by 30 cycles of denaturation at 95 °C for 1 min, annealing at 57 °C for 1 min and elongation at 72 °C for 1 min. The reaction was finished with 7 min final elongation at 72 °C. Amplicons were detected by agarose (1.5 %) gel electrophoresis and ethidium bromide staining.
For the second set of primers, the PCR was run with an initial pre-denaturation at 95 °C for 5 min, followed by 30 cycles of denaturation at 95 °C for 1 min, annealing at 62 °C for 1 min and elongation at 72 °C for 1 min. The reaction was finished with 7 min final elongation at 72 °C.
In both cases, identical primers were used for sequencing PCR (BigDye Terminator Sequencing Kit; Applied Biosystems,). Purified products (BigDye XTerminator kit; Applied Biosystems) were sequenced using the ABI Prism 3130 XL Genetic Analyzer (Applied Biosystems). Sequence assembly was performed with BioNumerics v. 7.0 (Applied Maths). Blast sequence software was used to align the wild-type H37Rv atpE gene (GenBank accession number NC_000962.3 and GeneID:886937) with the sequences obtained. Nucleotide alignment was done using ClustalX version 2.0 software. This alignment was cleaned with GeneDoc Version 2.7.000 and translated into protein sequences and aligned with mega version 7.0.21 software.
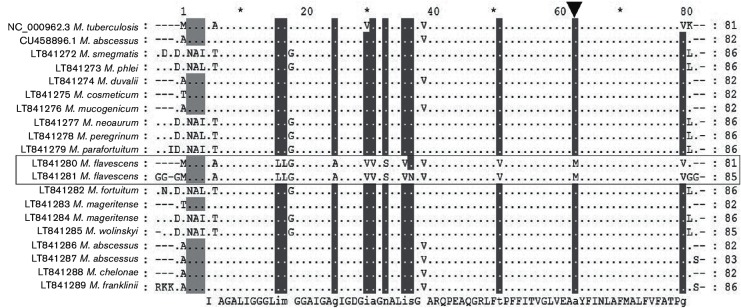
MIC and MBC results are shown in Table 1. For the majority of strains, BDQ had a significantly higher MBC compared to the respective MIC, suggesting a bacteriostatic effect. This occurred for Mycobacterium smegmatis, M. phlei, M. peregrinum, M. parafortuitum, M. mageritense, M. wolinskyi, M. abscessus and M. chelonae. On the other hand, M. duvalii and M. neoaurum had a high MIC (2 µg ml−1) without atpE mutation; however, as the other genes of the ATP synthase operon were not sequenced beside atpE, we cannot exclude a mutation in one of the other seven genes of the ATP synthase operon. Interestingly, the BDQ MBC value was almost equivalent to the MIC for M. cosmeticum and M. fortuitum. BDQ also showed bactericidal activity against M. mucogenicum and M. franklini, with an MBC of 0.5 and 1 µg ml−1, respectively. The MIC of 0.06 µg ml−1 for M. chelonae is the same as previously described by Huitric et al. [17]. In the same study they found a BDQ MIC of 0.13–0.25 µg ml−1 for M. fortuitum while in this study we found a lower MIC of 0.03 µg ml−1 The MIC of BDQ for M. mageritense was similar in both studies, at 0.03 (Huitric et al. [17]) and 0.06 µg ml−1 (this study). On the contrary, M. flavescens was completely resistant to BDQ. Three NTM species (M. xenopi, M. novocastrense and M. shimoidei) with significantly higher MICs for BDQ have been described and are considered naturally resistant to BDQ [17]. Petrella et al. [19] reported that M. xenopi had a high BDQ MIC (4 µg ml−1), most likely attributed to the polymorphism seen at amino acid 63 of AtpE. To investigate whether a similar polymorphism existed and to look into the possible role of gene mutations on the activity of BDQ against NTMs, the gene atpE was sequenced in all strains. Fig. 1 shows the amino acid sequence alignment confirming that the degree of identity at the protein level is very high for all NTM tested compared to M. tuberculosis H37Rv. An interesting finding of this study is that for M. flavescens, as previously reported for M. xenopi, M. shimoidei and M. novocastrense, the alanine at position 63 is replaced by a methionine. In all other known NTM AtpE sequences, this alanine is conserved. We confirmed the presence of this methionine at position 63 by sequencing atpE in one clinical M. flavescens isolate from St Luc Hospital, Brussels. The presence of this specific mutation is clearly associated with resistance to BDQ resulting in a high MIC. Our data show that the mechanism by which the NTM are inhibited in their growth (as reflected by their MIC) may be different from the mechanism by which they are killed (as reflected by their MBC). BDQ did not show bactericidal activity for the majority of strains tested. However, we did find BDQ bactericidal activity for M. cosmeticum, M. mucogenicum, M. fortuitum and M. franklinii. This finding should be confirmed with a larger number of clinical isolates. There is still more research to be done to explain why the NTM strains tested are highly sensitive to BDQ and which other factors besides polymorphisms in AtpE may influence the sensitivity of NTM to BDQ. In addition, more research is needed to understand why in some species the MIC and MBC of BDQ are very close and for other species the MBC is much higher than the MIC, and to elucidate the most promising companion drugs.
Fig. 1.
Amino acid alignment of AtpE from selected NTM. Conserved amino acids are represented by dots; only differences among amino acids are shown with letters. Sequences of wild-type Mycobacterium tuberculosis H37Rv (GenBank accession number NC_000962.3 and GeneID: 886937) and Mycobacterium abscessus ATCC 19977 (GenBank accession number CU458896.1 and GeneID: CAM61534.1) were used as controls. The last line indicates the consensus sequence. Sequences of M. flavescens strains R-50269 and R-50337 are highlighted.
In conclusion, to the best of our knowledge, this is the first study to have assessed the MIC and MBC values for BDQ against a large number of rapidly growing NTM. We also described for the first time that M. flavescens is naturally resistant to BDQ and its high MIC correlates with the mutation found at amino acid 63 in AtpE (alanine replaced by methionine). However, despite this finding, BDQ exhibited a strong inhibitory effect against all NTM tested, suggesting the potential to treat NTM infections. These preliminary results warrant further research in this area.
Funding information
The authors received no specific grant from any funding agency.
Acknowledgements
Bedaquiline was generously provided by Johnson and Johnson (Beerse). We acknowledge the support of the BOF fellowship (‘Bijzonder Onderzoeksfonds’ register number 01W01514, special foundation for research of Ghent University, Belgium) to D. A. A. The support of the CONACyT (Mexico) scholarship to D. A. A. is gratefully acknowledged.
Conflicts of interest
K. A. has conducted research experiments on bedaquiline activity against M. tuberculosis that has been supported by Janssen Laboratory. All other authors report no potential conflicts.
Footnotes
Abbreviations: BDQ, bedaquiline; MBC, minimum bactericidal concentration; MDR, multidrug-resistant; NTM, nontuberculous mycobacteria; TB, tuberculosis.
The GenBank/EMBL/DDBJ accession numbers for the atpA sequences determined in this study are LT841272-LT841289. Further details, including the respective mycobacterial species and strains, are listed in Table 1.
References
- 1.Tortoli E. Microbiological features and clinical relevance of new species of the genus Mycobacterium. Clin Microbiol Rev. 2014;27:727–752. doi: 10.1128/CMR.00035-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Martiniano SL, Nick JA, Daley CL. Nontuberculous mycobacterial infections in cystic fibrosis. Clin Chest Med. 2016;37:83–96. doi: 10.1016/j.ccm.2015.11.001. [DOI] [PubMed] [Google Scholar]
- 3.Sommerstein R, Schreiber PW, Diekema DJ, Edmond MB, Hasse B, et al. Mycobacterium chimaera outbreak associated with heater-cooler devices: piecing the puzzle together. Infect Control Hosp Epidemiol. 2017;38:103–108. doi: 10.1017/ice.2016.283. [DOI] [PubMed] [Google Scholar]
- 4.Frieden TR, Brudney KF, Harries AD. Global tuberculosis: perspectives, prospects, and priorities. JAMA. 2014;312:1393–1394. doi: 10.1001/jama.2014.11450. [DOI] [PubMed] [Google Scholar]
- 5.Falzon D, Mirzayev F, Wares F, Baena IG, Zignol M, et al. Multidrug-resistant tuberculosis around the world: what progress has been made? Eur Respir J. 2015;45:150–160. doi: 10.1183/09031936.00101814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ma Z, Lienhardt C, Mcilleron H, Nunn AJ, Wang X. Global tuberculosis drug development pipeline: the need and the reality. Lancet. 2010;375:2100–2109. doi: 10.1016/S0140-6736(10)60359-9. [DOI] [PubMed] [Google Scholar]
- 7.Palomino JC, Martin A. TMC207 becomes bedaquiline, a new anti-TB drug. Future Microbiol. 2013;8:1071–1080. doi: 10.2217/fmb.13.85. [DOI] [PubMed] [Google Scholar]
- 8.Sotgiu G, Pontali E, Centis R, D'Ambrosio L, Migliori GB. Delamanid (OPC-67683) for treatment of multi-drug-resistant tuberculosis. Expert Rev Anti Infect Ther. 2015;13:305–315. doi: 10.1586/14787210.2015.1011127. [DOI] [PubMed] [Google Scholar]
- 9.Andries K, Verhasselt P, Guillemont J, Göhlmann HW, Neefs JM, et al. A diarylquinoline drug active on the ATP synthase of Mycobacterium tuberculosis. Science. 2005;307:223–227. doi: 10.1126/science.1106753. [DOI] [PubMed] [Google Scholar]
- 10.Esteban J, García-Pedrazuela M, Muñoz-Egea MC, Alcaide F. Current treatment of nontuberculous mycobacteriosis: an update. Expert Opin Pharmacother. 2012;13:967–986. doi: 10.1517/14656566.2012.677824. [DOI] [PubMed] [Google Scholar]
- 11.Stout JE, Koh WJ, Yew WW. Update on pulmonary disease due to non-tuberculous mycobacteria. Int J Infect Dis. 2016;45:123–134. doi: 10.1016/j.ijid.2016.03.006. [DOI] [PubMed] [Google Scholar]
- 12.Rubio M, March F, Garrigó M, Moreno C, Español M, et al. Inducible and acquired clarithromycin resistance in the Mycobacterium abscessus complex. PLoS One. 2015;10:e0140166. doi: 10.1371/journal.pone.0140166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alexander DC, Vasireddy R, Vasireddy S, Philley JV, Brown-Elliott BA, et al. Emergence of mmpT5 variants during Bedaquiline treatment of Mycobacterium intracellulare lung disease. J Clin Microbiol. 2017;55:574–584. doi: 10.1128/JCM.02087-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Palomino JC, Martin A, Camacho M, Guerra H, Swings J, et al. Resazurin microtiter assay plate: simple and inexpensive method for detection of drug resistance in Mycobacterium tuberculosis. Antimicrob Agents Chemother. 2002;46:2720–2722. doi: 10.1128/AAC.46.8.2720-2722.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martin A, Camacho M, Portaels F, Palomino JC. Resazurin microtiter assay plate testing of Mycobacterium tuberculosis susceptibilities to second-line drugs: rapid, simple, and inexpensive method. Antimicrob Agents Chemother. 2003;47:3616–3619. doi: 10.1128/AAC.47.11.3616-3619.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pérez-Martínez I, Ponce-de-León A, Bobadilla M, Villegas-Sepúlveda N, Pérez-García M, et al. A novel identification scheme for genus Mycobacterium, M. tuberculosis complex, and seven mycobacteria species of human clinical impact. Eur J Clin Microbiol Infect Dis. 2008;27:451–459. doi: 10.1007/s10096-008-0459-9. [DOI] [PubMed] [Google Scholar]
- 17.Huitric E, Verhasselt P, Andries K, Hoffner SE. In vitro antimycobacterial spectrum of a diarylquinoline ATP synthase inhibitor. Antimicrob Agents Chemother. 2007;51:4202–4204. doi: 10.1128/AAC.00181-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huitric E, Verhasselt P, Koul A, Andries K, Hoffner S, et al. Rates and mechanisms of resistance development in Mycobacterium tuberculosis to a novel diarylquinoline ATP synthase inhibitor. Antimicrob Agents Chemother. 2010;54:1022–1028. doi: 10.1128/AAC.01611-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Petrella S, Cambau E, Chauffour A, Andries K, Jarlier V, et al. Genetic basis for natural and acquired resistance to the diarylquinoline R207910 in mycobacteria. Antimicrob Agents Chemother. 2006;50:2853–2856. doi: 10.1128/AAC.00244-06. [DOI] [PMC free article] [PubMed] [Google Scholar]