Abstract
The inhibition of mevalonate kinase (MVK) by downstream metabolites is an important mechanism in the regulation of isoprenoid production in a broad range of organisms. The first feedback-resistant MVK was previously discovered in the methanogenic archaeon Methanosarcinamazei. Here, we report the cloning, expression, purification, kinetic characterization and inhibition analysis of MVKs from two other methanogens, Methanosaetaconcilii and Methanocellapaludicola. Similar to the M. mazei MVK, these enzymes were not inhibited by diphosphomevalonate (DPM), dimethylallyl diphosphate (DMAPP), isopentenyldiphosphate (IPP), geranylpyrophosphate (GPP) or farnesylpyrophosphate (FPP). However, they exhibited significantly higher affinity to mevalonate and higher catalytic efficiency than the previously characterized enzyme.
Keywords: mevalonate kinase, GHMP superfamily, feedback-resistant, mevalonate pathway
Introduction
Isoprenoids, or terpenoids, are a very large class of naturally occurring compounds with diverse structures and functions. All are synthesized from the same five-carbon common precursors, isopentenyl diphosphate (IPP) and dimethylallyl diphosphate (DMAPP). Many isoprenoid compounds exhibit physiological activity. The spectrum of naturally produced isoprenoids, which includes carotenoids, steroid hormones, phytols, redox carriers, secondary metabolites and pheromones, is species-specific.
Two pathways for IPP and DMAPP biosynthesis have been discovered: the mevalonate (MVA) pathway and the 2-C-methyl-d-erythritol 4-phosphate (MEP) pathway ([1–3]; for a review see [4]).13 C NMR studies have shown that both acetate and mevalonate are precursors for isoprenoids in Archaea [5, 6]. These results were consistent with the subsequent analysis of archaeal genomes, which revealed homologues of mevalonate pathway enzymes, including 3-hydroxy-3-methylglutaryl-coenzyme A (HMG-CoA) synthase, HMG-CoA reductase [7–9], mevalonate kinase (MVK) and IPP isomerase [10, 11], but did not find homologous MEP pathway enzymes. Since putative genes encoding phosphomevalonate kinase (PMK) and diphosphomevalonate decarboxylase (DMD) were not identified on the basis of sequence similarity in archaeal genomes, while isopentenyl phosphate kinase (IPK) genes have been found in nearly all archaea, it has been hypothesized that an alternative mevalonate pathway operates in Archaea [12]. In this hypothetical pathway, phosphomevalonate is decarboxylated to yield isopentenyl phosphate (IP), which is then phosphorylated to IPP by the action of IPK. Recently, the missing phosphomevalonate decarboxylase (PMD) enzyme that converts (R)-mevalonate 5-phosphate (MVA-5-P) into IP was discovered in exceptional phyla of bacteria (Chloroflexi) and archaea (Halobacteria) [13–15]. Furthermore, MVA-3-P, but not MVA-5-P, serves as a precursor for IP synthesis in Thermoplasma acidophilum; a novel enzyme, ATP:(R)-MVA 3-phosphotransferase Ta1305, was identified in this species [16].
Organisms use a variety of regulatory mechanisms to ensure the sufficient formation of necessary isoprenoids and prevent the over-production of potentially toxic compounds (e.g. cholesterol). The inhibition of MVK by downstream metabolites such as DMAPP, IPP, GPP and FPP (class I MVKs) [17–20], or by diphosphomevalonate (class II MVKs) [21], plays an important role in the regulation of the mevalonate pathway in different organisms. The strict control of this reaction prevents the accumulation of phosphomevalonate, which can block DNA replication and arrest the cell cycle [22]. The first thermostable archaeal MVK to be biochemically characterized was from Methanocaldococcus jannaschii; inhibition by GPP and FPP, but not IPP, was observed [11].
Exploring the efficient feedback-resistant enzymes of the mevalonate pathway and their introduction into microbial-producing strains is a very important step towards the industrial production of isoprenoids. The discovery of a feedback-resistant MVK in the methanogenic archaeon Methanosarcina mazei [23] not only distinguished a new class of MVKs, but significantly broadened the opportunities for microbial isoprenoid production. Unlike most bacteria and eukaryotes, archaea equip their own membrane phospholipid structure, which is composed of isoprenoid chains condensed with sn-glycerol-1-phosphate by ether linkages (for reviews, see [24–26]). High-performance constitutive biosynthesis of the common precursors of isoprenoids can be expected in Archaea because they are building blocks of abundant components of archaeal membranes, which is why we would expect that the feedback-resistant MVK from M. mazei is not unique. The feedback resistance of the mevalonate pathway enzymes can thus be proposed as a common feature of large taxa belonging to Archaea.
We report here the cloning and overexpression of the mvk genes from Methanosaeta concilii, an archaeon belonging to the same order, Methanosarcinales, as M. mazei, from an evolutionarily distant methanogenic archaeon, Methanocella paludicola, and from Nitrosopumilus maritimus, an unrelated species belonging to Thaumarchaeota, in Escherichia coli; the purification, kinetic characterization and inhibition analysis of the corresponding enzymes are also described. The feedback-regulated Saccharomyces cerevisiae MVK and feedback-resistant M. mazei MVK were recharacterized in this study and used as controls. Our results suggest that M. concilii, M. paludicola and N. maritimus MVKs are feedback-resistant; the catalytic efficiencies of the first and second enzymes are higher than that of the MVK from M. mazei. Hereafter, the MVKs from M. mazei, M. concilii, M. paludicola, N. maritimus and S. cerevisiae will be referred to as MVKmma, MVKmcl, MVKmpd, MVKnmr and MVKsce, respectively.
Methods
Chemicals and reagents
Isopropyl thiogalactoside (IPTG), phosphoenolpyruvate (PEP), MgCl2, NaCl, lactate dehydrogenase, pyruvate kinase, (R)-lithium mevalonate, GPP ammonium salt, FPP ammonium salt, (±)-mevalonic acid 5-diphosphate tetralithium salt (DPM), HEPES and dithiothreitol (DTT) were purchased from Sigma-Aldrich (USA). DMAPP was obtained from Cayman Chemicals. Tris-HCl (pH 7.6) was purchased from WAKO (Japan). NADH and ATP were obtained from Oriental Yeast Co., Ltd. Mevalonate was prepared from mevalonolactone (ADEKA, Japan) as follows: 260 mg of mevalonolactone was mixed well with 5 ml of water and then with 0.6 ml of 10 N KOH. The mixed solution was then incubated at 37 °C for 2 h. After incubation, the pH of the solution was adjusted to 8.0 by neutralization with hydrochloric acid. The solution was then brought up to a final volume of 20 ml to yield a 100 mM potassium mevalonate solution. The obtained mevalonate solution was verified by HPLC. HPLC analysis was performed using 50 mM (R)-lithium mevalonate as the standard. The HPLC used for verification was a Hitachi high-performance liquid chromatograph (L-2000). HPLC was performed using YMC-Pack ODS-A (150×4.6 mm I.D. YMC, Japan) as the stationary phase and phosphoric acid (pH 2.5) as the mobile phase. Detection was performed at 210 nm.
Bacterial strains and plasmid vectors
The pET-21a(+) (for the C-terminal His-tag) and pET- 28b(+) (for the N-terminal His-tag) vectors were used to express the recombinant protein. The recombinant plasmids were transformed into E. coli BL21 (DE3) cells for expression. The E. coli BL21 (DE3) strain and expression vectors were purchased from Novagen.
Construction of expression plasmids
DNA fragments containing coding parts of the mvkmma, mvkmpd, mvkmcl and mvknmr genes linked to ribosome-binding sites were chemically synthesized and amplified by PCR using the primers listed in Table 1. The forward and reverse primers contained NdeI and HindIII (EcoRI for mvkmpd) recognition sites (underlined), respectively.
Table 1. Primers used for cloning the mvk genes.
| Plasmid | Gene | Name | Sequence |
|---|---|---|---|
| pET-28b(+) | mvkmcl | P1 | GCAATTCCATATGACGATGGCTTCCGCTCCGGGCAA |
| P2 | CCCAAGCTTACGCGACTTCCAGGCGAACACCTT | ||
| mvkmpd | P3 | GCAATTGCATATGACGATGTGCTCAGCTCCGGGTAA | |
| P4 | GGAATTCACTGAATGAAAATACCTTCCGCCGTCGG | ||
| mvkmma | P5 | GCAATTCCATATGGTATCCTGTTCTGCGCCGG | |
| P6 | CCCAAGCTTAATCTACTTTCAGACCTTGCTCGGTC | ||
| mvksce | P7 | GCAATTGCATATGTCATTACCGTTCTTAACTTCTGC | |
| P8 | CCCAAGCTTATGAAGTCCATGGTAAATTCGTGTT | ||
| mvknmr | P9 | GCAATTCCATATGAAGAGCAAGGCATCTGCGCC | |
| P10 | CCCAAGCTTAAAACGTGTCCAGGCCCTTGAAATC | ||
| pET-21a(+) | mvkmcl | P1 | GCAATTCCATATGACGATGGCTTCCGCTCCGGGCAA |
| P11 | CCCAAGCTTCGCGACTTCCAGGCGAACACCTT | ||
| mvkmpd | P3 | GCAATTCCATATGACGATGTGTTCAGCCCCCGGTAA | |
| P12 | CCCAAGCTTTTGGATGAATATTCCCTCCGCCGTT | ||
| mvkmma | P5 | GCAATTCCATATGGTATCCTGTTCTGCGCCGG | |
| P13 | CCCAAGCTTATCTACTTTCAGACCTTGCTCGGTC | ||
| mvksce | P7 | GCAATTGCATATGTCATTACCGTTCTTAACTTCTGC | |
| P14 | CCCAAGCTTATGAAGTCCATGGTAAATTCGTGTT | ||
| mvknmr | P9 | GCAATTCCATATGAAGAGCAAGGCATCTGCGCC | |
| P15 | CCCAAGCTTAAACGTGTCCAGGCCCTTGAAATC |
The ERG12 mevalonate kinase gene was amplified by PCR with Prime Star polymerase (Takara Bio Inc., Japan) using genomic DNA isolated from S. cerevisiae as a template.
The obtained DNA fragments were digested with NdeI and HindIII (EcoRI for mvkmpd) and inserted into the corresponding sites of pET-21a(+) and pET-28b(+). The expected DNA sequences in the inserted DNA fragments were confirmed by sequencing.
Expression and preliminary mevalonate kinase activity assay
Each transformant was cultured at 20 °C by reciprocal shaking at 140 r.p.m. in Luria–Bertaini (LB) broth [27] containing 0.1 mg ml−1 ampicillin [for pET-21a(+)] or 0.05 mg ml−1 kanamycin [for pET-28b(+)]. When the OD600 reached approximately 0.5, IPTG was added to a final concentration of 1 mM, and cultivation was continued overnight at 20 °C. Cells were harvested by centrifugation and resuspended in buffer solution containing 50 mM sodium phosphate, 0.3 M NaCl and 20 mM imidazole. The cells were then disrupted by an ultrasonic disruptor (TOMY UD-201) at an output level of 3.5. After centrifugation (20 000 g, 20 min), disrupted cells were adsorbed onto a His-SpinTrap column (GE Healthcare). After washing with the same buffer solution, the proteins were eluted with buffer solution containing 50 mM sodium phosphate, 0.3 M NaCl and 0.5 M imidazole (pH 7.5). The eluate was dialyzed overnight at 4 °C, with the external solution containing 20 mM Tris-HCl (pH 7.5) and 50 mM NaCl. The enzyme activity of each eluate in the internal dialysis solution was measured.
Expression and purification of recombinant M. concilii, M. paludicola, M. mazei, N. maritimus and S. cerevisiae MVKs
To analyse the expression of MVKs derived from M. mazei, N. maritimus and S. cerevisiae, each transformant was cultured at 20 °C in a thick test tube containing 20 ml of LB broth by reciprocal shaking at 140 r.p.m. To analyse the expression of MVKs derived from M. concilii and M. paludicola, each transformant was cultured at 20 °C in 2 L LB broth in a 3 L shaking flask with baffles by gyratory culture at 140 r.p.m. When the OD600 reached approximately 0.5, IPTG was added to a final concentration 0.1 mM (for mvkmma, mvknmr and mvksce) or 1 mM IPTG (for mvkmpd and mvkmcl), and the cultures were incubated at 20 °C overnight to induce the target protein. Harvested cells were suspended in buffer solution A (50 mM sodium phosphate, 0.3 M NaCl and 20 mM imidazole), and the cells were disrupted with an ultrasonic disruptor. Ultrasonic disruption of the transformants expressing MVKmma, MVKnmr and MVKsce was performed at an output level of 3.5, while for the transformants expressing MVKmcl and MVKmpd it was performed at an output level of 8. After centrifugation (28 000 g, 30 min), the supernatant was adsorbed onto a His-Trap HP (GE Healthcare) column to elute the target protein by a linear concentration gradient of imidazole (final concentration 0.5 M). The obtained proteins were dialyzed against an external solution containing 20 mM Tris-HCl (pH 8.0), 1 mM DTT and 50 mM NaCl. The dialyzed protein was designated as the purified enzyme. Protein quantity was measured with a Bio-Rad protein assay kit and a BSA standard.
Preparation of PMK
The ERG8 gene (NM_001182727.1) encoding the phosphomevalonate kinase in Saccharomyces cerevisiae was amplified by PCR containing PrimeSTAR MAX DNA polymerase premix (Takara Bio, Inc.) using the primers 5′-TCAGAGTTGAGAGCCTTCAGTGCCCCAG-3′ and 5′-GGAATTCTCTTTATCAAGATAAGTTTCCGGATCTTTTT-3′ and genomic DNA from S. cerevisiae as a template. pET21-d(+) was digested with NcoI, treated with the Klenow fragment of DNA polymerase I, and then digested with EcoRI. The PCR-generated DNA fragment was digested with EcoRI and ligated with the described vector fragment. The expected DNA sequence of the inserted fragment was confirmed by sequencing.
E. coli BL21(DE3) was transformed with the obtained plasmid and cultured in 20 ml of LB broth at 30 °C by reciprocal shaking at 140 r.p.m. When the OD600 reached approximately 0.7, 0.1 mM IPTG was added and cultivation was continued overnight under the same conditions. Cells were harvested by centrifugation, resuspended in buffer solution A (50 mM sodium phosphate, 0.3 M NaCl and 20 mM imidazole) and disrupted by ultrasonication. After centrifugation, the resulting supernatant was adsorbed onto a His SpinTrap (GE Healthcare) column and the adsorbed proteins were eluted with eluting solution (buffer solution A containing 0.5 M imidazole). The obtained eluate was dialyzed with 20 mM Tris-HCl (pH 8.0) containing 50 mM NaCl as the external solution.
Enzyme activity and inhibition by DMAPP, GPP, FPP and DPM
The catalytic activities of the MVKs were measured using a modified spectrophotometric assay that couples ADP formation to reverse pyruvate kinase and lactate dehydrogenase reactions [21]. The initial rate of disappearance of NADH served as a measure of the phosphorylation of mevalonate by MVK. The NADH millimolar extinction coefficient of 6.22 mM−1 cm−1 was used in this study.
Each 100 -µl reaction mixture contained 50 mM Tris (pH 7.6), 50 mM NaCl, 0.4 mM phosphoenol pyruvate, 0.05 mM DTT, 0.33 mM NADH, 10 mM MgCl2, 2 units of LDH and 2 units of PK.
The Michaelis constants with respect to mevalonate Km-Mev for MVK from M. concilii, M. paludicola, M. mazei, N. maritimus and S. cerevisiae were determined at a saturating concentration of ATP (5 mM) and variable concentrations of potassium mevalonate. The Michaelis constants to ATP Km-ATP were determined at a saturating concentration of potassium mevalonate (5 mM) and variable concentrations of ATP. Km and kcat values were determined by fitting the Michaelis–Menten equation using SigmaPlot 13.0/Enzyme Kinetics Module 1.3 (Systat Software, Inc.). The error values represent the standard deviation from three replicates. Changes in absorbance associated with the amount of NADH oxidized to NAD+ were monitored continuously at 340 nm and plotted against time to determine the rate of the MVK-coupled reactions.
Protein inhibition studies were performed by adding terpenyl diphosphates (DMAPP, GPP, FPP, or diphosphomevalonate) at various concentrations to the reaction mixture.
Results
Exploring the candidate mvk genes
The first feedback-resistant MVK was found in the methanogenic archaeon Methanosarcina mazei [23]. Methanogenic archaea are divided into seven distant taxonomic orders [28]. The majority of methanogenic archaea use CO2+H2 as substrates but cannot utilize acetate. Only two genera belonging to Methanosarcinales, Methanosarcina and Methanosaeta are able to utilize acetate for growth and methane production [29]; they are responsible for a major part of methane production in the biosphere.
Primak et al. demonstrated a clear evolutionary separation of MVKs from Archaea, Bacteria and Eukarya [23]. They also proposed that alternative regulatory mechanisms are inherent for different MVK phylogenetic branches.
We analysed MVKs from representatives of the class Methanomicrobia. Homologues of the M. mazei MVK were chosen from the database of non-redundant protein sequences of Methanomicrobia (taxid: 224756) using the psi-blast program [30]. clustalw [31] was used for multiple alignment, and a phylogenetic tree of the chosen proteins was constructed using the neighbour-joining (NJ) method [32]. Unexpectedly, the proteins from the genus Methanosaeta formed a distinct evolutionary branch that was proven to be more distant from the Methanosarcina branch than Methanocellales and Methanomicrobiales (Figs S1 and S2, available with the online Supplementary Material). Putative MVKs from M. paludicola SANAE and M. concilii GP6 were chosen for further analysis; both M. paludicola SANAE and M. concilii GP6 are mesophilic micro-organisms of biosafety level 1. Complete annotated genome sequences are available for both strains. M. concilii is a well-studied methanogenic bacterium commonly observed in digested sludge [33–35]. M. paludicola was the first cultivated representative of Methanocellales isolated from rice field soil [36, 37]. M. paludicola is unrelated to M. mazei, but it possesses similar MVK. Conversely, M. concilii belongs to the same family as M. mazei, but the MVK from M. concilii is more evolutionarily distant. We chose MVK from N. maritimus, which is classified in the phylum Thaumarchaeota, as an outgroup of MVK from Methanomicrobia [38].
Cloning of the mvk genes
Nucleotide sequences of mvk genes from M. concilii GP6 (GenBank accession number: NC_015416.1, locus_tag=MCON_2559), M. paludicola SANAE (GenBank accession number: AP011532.1, locus_tag=MCP_1639), M. mazei Go1 (GenBank accession number: NC_003901.1, locus_tag=MM_RS09140) and N. maritimus (GenBank accession number: CP_000866.1, locus_tag=NMAR_0315), and of the ERG12 gene from Saccharomyces cerevisiae (GenBank accession number: X55875.1), were modified for efficient cloning and expression in E. coli. For the mvkmcl, mvkmpd and mvkmma genes, codon optimization (the substitution of codons that are rare in E. coli with synonymous codons that are more frequently used) was performed using OptimumGene provided by GenScript. The resultant nucleotide sequences are shown in Fig. S3 [39]. The DNA fragments were cloned into pET-21a(+) (C-terminal His-tag) and pET-28b(+) (N-terminal His-tag) expression vectors (Novagene), as described in the Methods section.
Purification of MVKs
All constructed plasmids were transformed into the E. coli BL21(DE3) strain. To improve the solubility of MVKs, cultivation of the transformants was carried out at a low temperature (20 °C). The expression levels provided by the pET-21a(+)- and pET-28b(+)-derived plasmids were compared on the basis of the MVK activity assay (see the Methods section). In all cases, the pET-28b(+)-derived plasmids yielded higher or equal activity levels compared to derivatives of pET-21a(+). Thus, the pET-28b(+)-derived plasmids were chosen for further study. These plasmids provided the accumulation of the target recombinant proteins, which contained 20 additional amino acid residues, including a cluster of 6 His residues fused to the N-terminus of MVK, to simplify the purification process.
The recombinant MVKs from M. concilii, M. paludicola, M. mazei, N. maritimus and S. cerevisiae were purified using immobilized metal ion affinity chromatography (see the Methods section) up to >95 % apparent homogeneity, as determined based on SDS-PAGE and Coomassie staining.
MVKs belong to the GHMP kinase family [40]. Monomeric or homodimeric assembly was observed for the previously described members of this protein family. The apparent masses of MVKs from M. concilii and M. paludicola were determined by gel filtration to be 158 and 72 kDa, respectively. The corresponding molecular masses calculated from the primary amino acid sequences were 35.6 and 33.9 kDa, respectively. These results suggest that MVKmpd forms a dimer in solution, similar to other homodimeric GHMP kinases, while MVKmcl has an unusual tetrameric structure (Table S1).
Kinetic characterization of MVKs
For each enzyme, the rates of mevalonate phosphorylation were monitored in coupled reactions with pyruvate kinase and lactate dehydrogenase according to [21] (see the Methods section). The obtained [S]-v plots are indicated in Fig. S4. The apparent Km values with respect to ATP (Km-ATP) and mevalonate (Km-Mev) were calculated for each His-tagged recombinant enzyme using the Michaelis–Menten equation.
Under the experimental conditions, significantly higher affinity to mevalonate and ATP was observed for the MVKs from M. concilii (Km-Mev=17.0±0.5 µM, Km-ATP=74±1 µM) and M. paludicola (Km-Mev=15.0±0.5 µM, Km-ATP=119±2 µM) relative to MVKmma (Km-Mev=83±2 µM, Km-ATP=687±9 µM) or MVKsce (Km-Mev=73±2 µM, Km-ATP=464±8 µM). MVKmpd exhibited the lowest turnover (kcat=7.30±0.04 s−1) among all the studied enzymes, while MVKmcl had a kcat that was comparable to that of MVKmma and MVKsce (14.0±0.1 s−1, 19.0±0.2 s−1 and 16.0±0.1 s−1, respectively). The highest catalytic efficiency (kcat/Km-Mev=0.824 µM s−1) was observed for MVKmcl (Table 2).
Table 2. Kinetic characterization of MVKs.
| MVK | Km-Mev (μM) | Km-ATP (μM) | MW (Da) | Specific activity µmol/min*mg | kcat (s−1) | kcat /Km-Mev (µM/s) |
|---|---|---|---|---|---|---|
| mcl | 17.0±0.5 | 74±1 | 35 626 | 233±2 | 14.0±0.1 | 0.824 |
| mpd | 15.0±0.5 | 119±2 | 33 908 | 129±1 | 7.30±0.04 | 0.487 |
| mma | 83±2 | 687±9 | 33 575 | 348±3 | 19.0±0.2 | 0.229 |
| nmr | 461±7.5 | 1006±20 | 36 594 | 204±2 | 12±0.11 | 0.026 |
| sce | 73±2 | 464±8 | 50 622 | 188±3 | 16.0±0.1 | 0.219 |
Conversion of mevalonate to phospho- and diphosphomevalonate in an in vitro system containing a purified MVK and an S. cerevisiae phosphomevalonate kinase
Mevalonic acid contains two hydroxyl groups (C3 and C5) and a C1 carboxyl group that can be phosphorylated. Mevalonate-5-phosphate is an intermediate of the classical mevalonate pathway. Products of the reactions catalyzed by two other characterized archaeal MVKs from Methanosarcina mazei and Methanocaldococcus jannaschii can be further converted into diphosphomevalonate by S. cerevisiae 5-phosphomevalonate kinase (PMK) [23]. On the other hand, two ATP:(R)-mevalonate 3-phosphotransferases were recently identified in the archaea Thermoplasma acidophilum [16] and Picrophilus torridus [41].
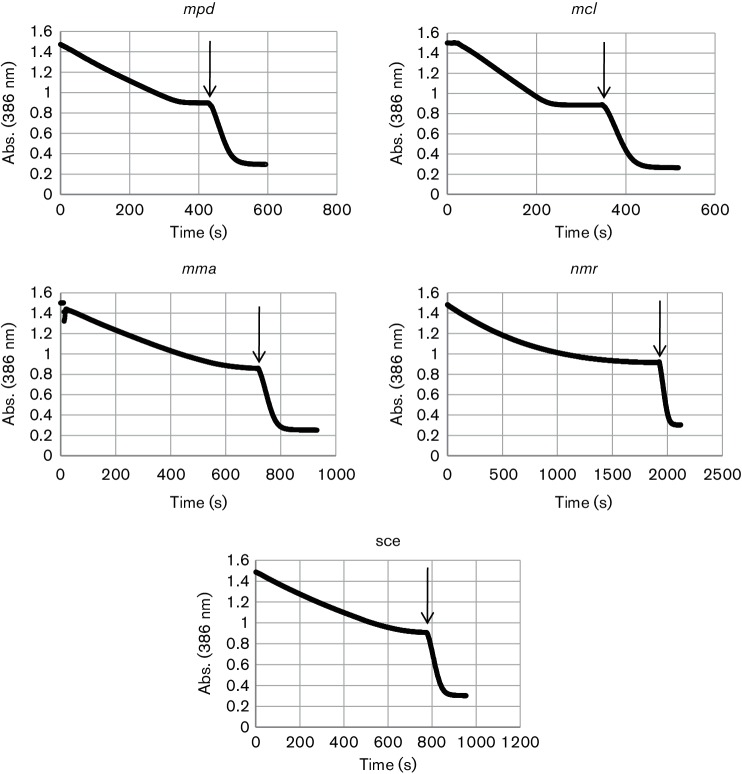
To determine the group that is phosphorylated by MVKmcl and MVKmpd, we applied an in vitro assay previously developed by Andreassi et al. [21] and exploited in [23] to analyse the inhibition of MVKs by diphosphomevalonate. This approach couples the initial phosphorylation of mevalonate by MVK and the subsequent conversion of phosphomevalonate to diphosphomevalonate after PMK addition. In both cases, the rate of ADP formation was monitored as a decrease in absorbance at 386 nm in the spectrophotometric pyruvate kinase and lactate dehydrogenase-coupled assay (0.61 mM−1 cm−1 was designated as the millimolar absorption coefficient of NADH at 386 nm). The obtained curves confirmed the complete conversion of mevalonate to diphosphomevalonate via phosphomevalonate (Fig. 1). The obtained results strongly suggest that MVKmcl and MVKmpd should be considered as mevalonate-5-kinases. Nevertheless, to make sure that these enzymes cannot also produce mevalonate-3-phosphate, the direct identification of products formed in reactions with MVKmcl and MVKmpd is necessary.
Fig. 1.
Conversion of phosphomevalonate produced by MVKmma, MVKsce, MVKmcl, MVKmpd and MVKnmr to diphosphomevalonate by S. cerevisiae PMK. The conversion of mevalonate to phospho- and, subsequently, diphosphomevalonate was monitored indirectly by the oxidation of NADH in a pyruvate kinase and lactate dehydrogenase-coupled assay. The kinetics of absorbance at 386 nm is plotted. Purified PMK was added at the points indicated by arrows.
In the reactions with MVKmma, MVKsce, MVKmcl and MVKmpd, an almost linear decrease in absorbance was observed until PMK addition. In contrast, a stepwise decrease in the reaction rate associated with product accumulation was observed for MVKnmr. This result suggests the product inhibition of MVKnmr by phosphomevalonate.
Archaeal MVKs are not inhibited by downstream intermediates of isoprenoid biosynthesis (DPM, DMAPP, GPP and FPP)
The feedback inhibition of the M. concilii, M. paludicola, M. mazei and N. maritimus MVKs by the downstream intermediates of isoprenoid biosynthesis (diphosphomevalonate (DPM), DMAPP, geranyl pyrophosphate (GPP) and farnesyl pyrophosphate FPP) was evaluated. Parallel spectrophotometric activity assays that contained a high concentration (indicated in Table 3) of the potential inhibitor or lacked inhibitors were performed.
Table 3. Influence of the potential inhibitors on mevalonate kinase activity.
nd, not determined. Results of single experiments are represented. Relative catalytic activity (%) of the enzymes is indicated. Activity without potential inhibitors was taken as 100 %.
| MVK | DPM | DMAPP | GPP | FPP | Reference | ||||
|---|---|---|---|---|---|---|---|---|---|
| 1 mM | 5 mM | 5 µM | 10 µM | 100 µM | 5 µM | 10 µM | 100 µM | ||
| sce | 96 | 8 | nd | nd | 9 | nd | nd | 10 | This work |
| mma | 85 | 91 | nd | nd | 96 | nd | nd | 98 | This work |
| mcl | 99 | 122 | nd | nd | 104 | nd | nd | 105 | This work |
| mpd | 105 | 97 | nd | nd | 101 | nd | nd | 101 | This work |
| nmr | 97 | 95 | nd | nd | 100 | nd | nd | 92 | This work |
| mja | nd | nd | 58 | 45 | nd | 46 | 35 | nd | [11] |
Table 3 indicates the relative catalytic activity (%) of each enzyme. The activity in the reaction mixture without inhibitors was taken as 100 %. According to previously published data, S. cerevisiae MVK is strongly inhibited by 5 mM DMAPP, 0.1 mM GPP and 0.1 mM FPP, but is not inhibited by 1 mM DPM. The M. mazei MVK was not inhibited by 0.1 mM GPP or 0.1 mM FPP. A slight decrease in activity was observed for this enzyme when 1 mM DPM or 5 mM DMAPP was added. Neither MVKmcl nor MVKmpd was inhibited by 1 mM DPM, 5 mM DMAPP, 0.1 mM GPP or 0.1 mM FPP. Conversely, the enzyme from M. concilii was even slightly activated by 5 mM DMAPP (its activity increased by 20 % compared to that of the control). Lower concentrations (33.6 or 168 µM) of DMAPP or IPP did not affect the activity of this enzyme.
Discussion
Isoprenoids are produced by all organisms, and their precursors are known as essential compounds [42]. However, the redundant production of terpenoids or their precursors can be toxic for cells and cause various diseases [43, 44], which is why the strict control of isoprenoid biosynthesis is of vital importance. The regulation mode can be species-specific. Clarifying and comparing the biochemical features of archaeal MVKs is important for understanding the control mechanism.
The archaeal MVKs from M. concilii and M. paludicola characterized in this work were not inhibited by DPM, DMAPP, IPP, GPP or FPP, and exhibited very high affinity to mevalonate. The Michaelis constants for (R, S)-mevalonate determined for these enzymes are four- to fivefold lower than those for MVKmma (see [23] and this work) and MVKnmr (this work), or for the MVK from M. jannaschii, a hyperthermophilic, autotrophic and strictly hydrogenotrophic archaeon [11]. A similar Km for (R, S)-mevalonate Km-Mev=19.0 µM was previously observed for pig liver MVK; however, this enzyme is feedback-sensitive [45].
MVKmcl has proven to be an unusual enzyme. It is not inhibited by phosphomevalonate, DPM, GPP or FPP, and its activity is even increased by the addition of DMAPP. Unlike all previously characterized MVKs (Table S1), it has a tetrameric structure. Moreover, the conserved motif 4 of MVKmcl contains two positively charged amino acids – lysine and arginine – that are located between the residues involved in binding of the phosphate moiety of ATP (Fig. S5, triangles). This structural feature may cause the specific biochemical features of MVKmcl.
The biochemical properties of MVKmcl and MVKmpd make them promising candidates for industrial applications to promote isoprenoid compound production through the mevalonate pathway. In particular, our previous experiments confirmed the advantages of MVKmcl for isoprene production [39].
The tree topology of MVK differs from its representative phylogeny (Fig. S1), suggesting the existence of different biochemical features. Surprisingly, the three MVKs isolated from representatives of different families belonging to Methanomicrobia have proven to be feedback-resistant. Based on these results, we proposed that these organisms might use another mechanism (or other mechanisms) that is (are) distinct from the feedback system to regulate the mevalonate pathway. To elucidate whether this regulation could be realized at the level of transcription, we analysed the surroundings of the mvk orthologues found in the sequenced genomes using Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) v.10 software [46]. All representatives of Methanomicrobia harbour long operons, including three genes of the lower mevalonate pathway coding for MVK, IP kinase and IPP isomerase (Fig. 2). It is likely that the expression of genes involved in the lower mevalonate pathway is co-regulated in these organisms at the transcriptional level. In many other Euryarchaeota, the mvk gene is linked to the IP kinase gene, as shown in Fig. 2. Conversely, an ORF encoding the previously characterized archaeal feedback-sensitive MVK from M. jannaschii is arranged in the genome as an individual gene. Further investigation is necessary to reveal the regulatory mechanisms that control isoprenoid biosynthesis in organisms harbouring feedback-resistant MVKs, and in addition, if a strong correlation between MVK inhibition and the genome arrangement of the corresponding ORF does exist, analysis of the genetic surroundings of mvk genes could help us explore novel feedback-resistant MVKs.
Fig. 2.
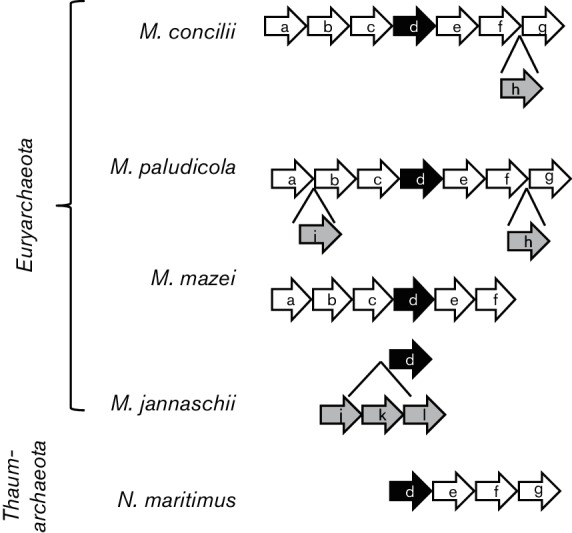
Conserved strings include genes encoding the feedback-resistant mevalonate kinases in Archaea. (a) RpoK, (b) 30S ribosomal protein S2, (c) hypothetical protein, (d) feedback-resistant mevalonate kinase, (D) feedback-sensitive mevalonate kinase, (e) isopentenyl kinase, (f) isopentenyl pyrophosphate isomerase, (g) bifunctional short chain isoprenyl diphosphate synthase, (h) beta-lactamase domain-containing protein, (i) enolase, (j) shikimate 5-dehydrogenase, (k) hypothetical protein and (l) hypothetical protein.
We hope that the obtained recombinant feedback-resistant MVK proteins will facilitate future studies on structure–function relationships. Previously, an X-ray structure of rat MVK in complex with farnesyl thiophosphate or ATP has been resolved; the amino acid residues that interact with the phosphate moieties of ATP or farnesyl thiophosphate have been determined [47]. The structure-based alignment of MVKs from organisms belonging to different domains of life (Fig. S5) revealed the strict conservation of these residues. On the other hand, among the residues of rat MVK involved in the binding of the isoprenoid moiety of farnesyl thiophosphate, only P139 is highly conserved; four residues are located in conserved regions, and five residues are located in a variable spacer between the conserved motifs 1 and 2. In prokaryotic MVKs, this spacer is considerably shorter than in eukaryotic enzymes. Thus, it is likely that the structure of this region determines the spectrum of inhibitors that can be bound by a concrete MVK. Further study of archaeal MVKs and their comparison with bacterial and eukaryotic enzymes can provide new insights into the mechanism of interaction between potential inhibitors and prokaryotic and eukaryotic MVKs. Such an investigation would provide interesting ideas for the relief of MVK inhibition in patients with autoinflammatory disorders caused by a mevalonate kinase (MK) deficiency [48] and for the design of new MVK inhibitors, which are being considered as potential drugs for the treatment of cardiovascular diseases and cancer [49] and as antibacterial agents [50].
Conclusion
In conclusion, cloning, expression, purification, kinetic parameter determination and inhibition analysis of the MVKs from the mesophilic archaea M. concilii and M. paludicola, as well as from the marine thaumarchaeote N. maritimus, were performed. Consistent with the previously reported MVK from M. mazei, the new enzymes were not inhibited by downstream metabolites (DPM, DMAPP, IPP, GPP and FPP). At the same time, the MVKs from M. concilii and M. paludicola exhibited significantly higher affinity to mevalonate and higher catalytic efficiency than the previously characterized enzyme.
Funding information
The authors received no specific grants from any funding agency.
Acknowledgements
We thank Hiroaki Rachi and Kunio Nakata for invaluable discussions. We also thank Yasuhiro Kashima and Takashi Oku at Thermostable Enzyme Laboratory Co., Ltd. for excellent technical support.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Supplementary Data
Footnotes
Abbreviations: DMAPP, dimethylallyl diphosphate; DMD, diphosphomevalonate decarboxylase; DPM, diphosphomevalonate; FPP, farnesyl pyrophosphate; GPP, geranyl pyrophosphate; HMG-CoA, 3-hydroxy-3-methylglutaryl-coenzyme A; IPP, isopentenyl diphosphate; MEP, 2-C-methyl-D-erythritol 4-phosphate; MVA, mevalonate; MVA-3-P, (R)-mevalonate 3-phosphate; MVA-5-P, (R)-mevalonate 5-phosphate; MVK, mevalonate kinase; NADH, reduced nicotinamide adenine dinucleotide; PEP, phosphoenolpyruvate; PMD, phosphomevalonate decarboxylase; PMK, phosphomevalonate kinase.
One supplementary table and five supplementary figures are available with the online Supplementary Material.
Edited by: S-V. Albers and F. Sargent
References
- 1.Katsuki H, Bloch K. Studies on the biosynthesis of ergosterol in yeast. Formation of methylated intermediates. J Biol Chem. 1967;242:222–227. [PubMed] [Google Scholar]
- 2.Lynen F. Biosynthetic pathways from acetate to natural products. Pure Appl Chem. 1967;14:137–167. doi: 10.1351/pac196714010137. [DOI] [PubMed] [Google Scholar]
- 3.Rohmer M, Seemann M, Horbach S, Bringer-Meyer S, Sahm H. Glyceraldehyde 3-phosphate and pyruvate as precursors of isoprenic units in an alternative non-mevalonate pathway for terpenoid biosynthesis. J Am Chem Soc. 1996;118:2564–2566. doi: 10.1021/ja9538344. [DOI] [Google Scholar]
- 4.Kuzuyama T, Seto H. Two distinct pathways for essential metabolic precursors for isoprenoid biosynthesis. Proc Jpn Acad Ser B Phys Biol Sci. 2012;88:41–52. doi: 10.2183/pjab.88.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ekiel I, Smith IC, Sprott GD. Biosynthetic pathways in Methanospirillum hungatei as determined by 13C nuclear magnetic resonance. J Bacteriol. 1983;156:316–326. doi: 10.1128/jb.156.1.316-326.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ekiel I, Sprott GD, Smith IC. Mevalonic acid is partially synthesized from amino acids in Halobacterium cutirubrum: a C13 nuclear magnetic resonance study. J Bacteriol. 1986;166:559–564. doi: 10.1128/jb.166.2.559-564.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bult CJ, White O, Olsen GJ, Zhou L, Fleischmann RD, et al. Complete genome sequence of the methanogenic archaeon, Methanococcus jannaschii. Science. 1996;273:1058–1073. doi: 10.1126/science.273.5278.1058. [DOI] [PubMed] [Google Scholar]
- 8.Kyrpides NC, Olsen GJ, Klenk HP, White O, Woese CR. Methanococcus Jannaschii genome: revisited. Microb Comp Genomics. 1996;1:329–338. [PubMed] [Google Scholar]
- 9.Selkov E, Maltsev N, Olsen GJ, Overbeek R, Whitman WB. A reconstruction of the metabolism of Methanococcus jannaschii from sequence data. Gene. 1997;197:GC11–GC26. doi: 10.1016/S0378-1119(97)00307-7. [DOI] [PubMed] [Google Scholar]
- 10.Barkley SJ, Cornish RM, Poulter CD. Identification of an Archaeal type II isopentenyl diphosphate isomerase in Methanothermobacter thermautotrophicus. J Bacteriol. 2004;186:1811–1817. doi: 10.1128/JB.186.6.1811-1817.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang KX, Scott AI, Bennett GN. Overexpression, purification, and characterization of the thermostable mevalonate kinase from Methanococcus jannaschii. Protein Expr Purif. 1999;17:33–40. doi: 10.1006/prep.1999.1106. [DOI] [PubMed] [Google Scholar]
- 12.Smit A, Mushegian A. Biosynthesis of isoprenoids via mevalonate in Archaea: the lost pathway. Genome Res. 2000;10:1468–1484. doi: 10.1101/gr.145600. [DOI] [PubMed] [Google Scholar]
- 13.Dellas N, Thomas ST, Manning G, Noel JP. Discovery of a metabolic alternative to the classical mevalonate pathway. Elife. 2013;2:e00672. doi: 10.7554/eLife.00672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vannice JC, Skaff DA, Keightley A, Addo JK, Wyckoff GJ, et al. Identification in Haloferax volcanii of phosphomevalonate decarboxylase and isopentenyl phosphate kinase as catalysts of the terminal enzyme reactions in an archaeal alternate mevalonate pathway. J Bacteriol. 2014;196:1055–1063. doi: 10.1128/JB.01230-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Beck ZQ, Mampel J, Meurer G, Miller MC, Sanford KJ, et al. United States Patent Application. Production of isoprene, isoprenoid, and isoprenoid precursors using an alternative lower mevalonate pathway. 20160002672(A1) [Google Scholar]
- 16.Azami Y, Hattori A, Nishimura H, Kawaide H, Yoshimura T, et al. (R)-mevalonate 3-phosphate is an intermediate of the mevalonate pathway in Thermoplasma acidophilum. J Biol Chem. 2014;289:15957–15967. doi: 10.1074/jbc.M114.562686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dorsey JK, Porter JW. The inhibition of mevalonic kinase by geranyl and farnesyl pyrophosphates. J Biol Chem. 1968;243:4667–4670. [PubMed] [Google Scholar]
- 18.Gray JC, Kekwick RG. The inhibition of plant mevalonate kinase preparations by prenyl pyrophosphates. Biochim Biophys Acta. 1972;279:290–296. doi: 10.1016/0304-4165(72)90145-6. [DOI] [PubMed] [Google Scholar]
- 19.Hinson DD, Chambliss KL, Toth MJ, Tanaka RD, Gibson KM. Post-translational regulation of mevalonate kinase by intermediates of the cholesterol and nonsterol isoprene biosynthetic pathways. J Lipid Res. 1997;38:2216–2223. [PubMed] [Google Scholar]
- 20.Voynova NE, Rios SE, Miziorko HM. Staphylococcus aureus mevalonate kinase: isolation and characterization of an enzyme of the isoprenoid biosynthetic pathway. J Bacteriol. 2004;186:61–67. doi: 10.1128/JB.186.1.61-67.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Andreassi JL, Dabovic K, Leyh TS. Streptococcus pneumoniae isoprenoid biosynthesis is downregulated by diphosphomevalonate: an antimicrobial target. Biochemistry. 2004;43:16461–16466. doi: 10.1021/bi048075t. [DOI] [PubMed] [Google Scholar]
- 22.Martín Sánchez C, Pérez Martín JM, Jin JS, Dávalos A, Zhang W, et al. Disruption of the mevalonate pathway induces dNTP depletion and DNA damage. Biochim Biophys Acta. 2015;1851:1240–1253. doi: 10.1016/j.bbalip.2015.06.001. [DOI] [PubMed] [Google Scholar]
- 23.Primak YA, du M, Miller MC, Wells DH, Nielsen AT, et al. Characterization of a feedback-resistant mevalonate kinase from the archaeon Methanosarcina mazei. Appl Environ Microbiol. 2011;77:7772–7778. doi: 10.1128/AEM.05761-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.de Rosa M, Gambacorta A, Gliozzi A. Structure, biosynthesis, and physicochemical properties of archaebacterial lipids. Microbiol Rev. 1986;50:70–80. doi: 10.1128/mr.50.1.70-80.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peretó J, López-García P, Moreira D. Ancestral lipid biosynthesis and early membrane evolution. Trends Biochem Sci. 2004;29:469–477. doi: 10.1016/j.tibs.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 26.Lombard J, López-García P, Moreira D. Phylogenomic investigation of phospholipid synthesis in archaea. Archaea. 2012;2012:1–13. doi: 10.1155/2012/630910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bertani G. Studies on lysogenesis. I. The mode of phage liberation by lysogenic Escherichia coli. J Bacteriol. 1951;62:293–300. doi: 10.1128/jb.62.3.293-300.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Welte C, Deppenmeier U. Bioenergetics and anaerobic respiratory chains of aceticlastic methanogens. Biochim Biophys Acta. 2014;1837:1130–1147. doi: 10.1016/j.bbabio.2013.12.002. [DOI] [PubMed] [Google Scholar]
- 29.Ferry JG. How to make a living by exhaling methane. Annu Rev Microbiol. 2010;64:453–473. doi: 10.1146/annurev.micro.112408.134051. [DOI] [PubMed] [Google Scholar]
- 30.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 33.Zehnder AJ, Huser BA, Brock TD, Wuhrmann K. Characterization of an acetate-decarboxylating, non-hydrogen-oxidizing methane bacterium. Arch Microbiol. 1980;124:1–11. doi: 10.1007/BF00407022. [DOI] [PubMed] [Google Scholar]
- 34.Fathepure BZ. Isolation and characterization of an aceticlastic methanogen from a biogas digester. FEMS Microbiol Lett. 1983;19:151–156. doi: 10.1111/j.1574-6968.1983.tb00531.x. [DOI] [Google Scholar]
- 35.Patel GB, Sprott GD. Methanosaeta concilii gen. nov., sp. nov. ("Methanothrix concilii") and Methanosaeta thermoacetophila nom. rev., comb. nov. Int J Syst Bacteriol. 1990;40:79–82. doi: 10.1099/00207713-40-1-79. [DOI] [Google Scholar]
- 36.Sakai S, Imachi H, Hanada S, Ohashi A, Harada H, et al. Methanocella paludicola gen. nov., sp. nov., a methane-producing archaeon, the first isolate of the lineage 'Rice Cluster I', and proposal of the new archaeal order Methanocellales ord. nov. Int J Syst Evol Microbiol. 2008;58:929–936. doi: 10.1099/ijs.0.65571-0. [DOI] [PubMed] [Google Scholar]
- 37.Sakai S, Takaki Y, Shimamura S, Sekine M, Tajima T, et al. Genome sequence of a mesophilic hydrogenotrophic methanogen Methanocella paludicola, the first cultivated representative of the order Methanocellales. PLoS One. 2011;6:e22898. doi: 10.1371/journal.pone.0022898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Walker CB, de La Torre JR, Klotz MG, Urakawa H, Pinel N, et al. Nitrosopumilus maritimus genome reveals unique mechanisms for nitrification and autotrophy in globally distributed marine crenarchaea. Proc Natl Acad Sci USA. 2010;107:8818–8823. doi: 10.1073/pnas.0913533107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mihara Y, Rachi H, Nishio Y, Katashkina JY, Kazieva ED, et al. Method of producing isoprene monomer. Us2015275233(a1)
- 40.Bork P, Sander C, Valencia A. Convergent evolution of similar enzymatic function on different protein folds: the hexokinase, ribokinase, and galactokinase families of sugar kinases. Protein Sci. 1993;2:31–40. doi: 10.1002/pro.5560020104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rossoni L, Hall SJ, Eastham G, Licence P, Stephens G. The putative mevalonate diphosphate decarboxylase from Picrophilus torridus is in reality a mevalonate-3-kinase with high potential for bioproduction of isobutene. Appl Environ Microbiol. 2015;81:2625–2634. doi: 10.1128/AEM.04033-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sauret-Güeto S, Ramos-Valdivia A, Ibáñez E, Boronat A, Rodríguez-Concepción M. Identification of lethal mutations in Escherichia coli genes encoding enzymes of the methylerythritol phosphate pathway. Biochem Biophys Res Commun. 2003;307:408–415. doi: 10.1016/S0006-291X(03)01211-7. [DOI] [PubMed] [Google Scholar]
- 43.Hashemi M, Hoshyar R, Ande SR, Chen QM, Solomon C, et al. Mevalonate cascade and its regulation in cholesterol metabolism in different tissues in health and disease. Curr Mol Pharmacol. 2017;10:13–26. doi: 10.2174/1874467209666160112123746. [DOI] [PubMed] [Google Scholar]
- 44.Sivy TL, Fall R, Rosenstiel TN. Evidence of isoprenoid precursor toxicity in Bacillus subtilis. Biosci Biotechnol Biochem. 2011;75:2376–2383. doi: 10.1271/bbb.110572. [DOI] [PubMed] [Google Scholar]
- 45.Houten SM, Wanders RJ, Waterham HR. Biochemical and genetic aspects of mevalonate kinase and its deficiency. Biochim Biophys Acta. 2000;1529:19–32. doi: 10.1016/S1388-1981(00)00135-9. [DOI] [PubMed] [Google Scholar]
- 46.Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, et al. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43:D447–D452. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fu Z, Voynova NE, Herdendorf TJ, Miziorko HM, Kim JJ. Biochemical and structural basis for feedback inhibition of mevalonate kinase and isoprenoid metabolism. Biochemistry. 2008;47:3715–3724. doi: 10.1021/bi7024386. [DOI] [PubMed] [Google Scholar]
- 48.Schneiders MS, Houten SM, Turkenburg M, Wanders RJ, Waterham HR. Manipulation of isoprenoid biosynthesis as a possible therapeutic option in mevalonate kinase deficiency. Arthritis Rheum. 2006;54:2306–2313. doi: 10.1002/art.21960. [DOI] [PubMed] [Google Scholar]
- 49.Qiu Y, Li D. Bifunctional inhibitors of mevalonate kinase and mevalonate 5-diphosphate decarboxylase. Org Lett. 2006;8:1013–1016. doi: 10.1021/ol052573s. [DOI] [PubMed] [Google Scholar]
- 50.Gharehbeglou M, Arjmand G, Haeri MR, Khazeni M. Nonselective mevalonate kinase inhibitor as a novel class of antibacterial agents. Cholesterol. 2015;2015:147601. doi: 10.1155/2015/147601. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.