Abstract
The endemicity of avian influenza viruses (AIVs) among Egyptian poultry represents a public health risk. Co-circulation of low pathogenic AIV H9N2 subtype with highly pathogenic AIV H5N1 subtype in Egyptian farms provides a possibility to generate novel reassortant viruses. Here, the genetic characteristics of surface glycoproteins of 59 Egyptian H9N2 viruses, isolated between 2013 and 2015, were analysed. To elucidate the potential of genetic reassortment, 10 H9N2 isolates were selected based on different avian hosts (chickens, ducks, pigeons and quails) and phylogenetic analyses of their full genome sequences were conducted. Additionally, we performed antigenic analysis to further investigate the antigenic evolution of H9N2 viruses isolated during 2011–2015. Different viral characteristics including receptor-binding affinity and drug resistance of representative Egyptian H9N2 viruses were further investigated. The surface glycoproteins of current Egyptian H9N2 viruses were closely related to viruses of the G1-like lineage isolated from Egypt. Several genetic markers that enhance virulence in poultry and transmission to humans were detected. Analysis of the full genome of 10 H9N2 isolates indicated that two pigeon isolates inherited five internal genes from Eurasian AIVs circulating in wild birds. Antigenic conservation of different Egyptian H9N2 isolates from chickens, pigeons and ducks was observed, whereas quail isolates showed antigenic drift. The Egyptian H9N2 viruses preferentially bound to the human-like receptor rather than to the avian-like receptor. Our results suggest that the endemic H9N2 viruses in Egypt contain elements that may favour avian-to-human transmission and thus represent a public health risk.
Keywords: avian influenza viruses, H9N2, Egypt, reassortant, antigenic analysis
Introduction
Low pathogenic avian influenza viruses (LPAIVs) of the H9N2 subtype have spread to several regions of the world including Asia, the Middle East, Africa and Europe. These viruses were detected in different types of poultry [1, 2], pigs [3] and humans [4, 5]. H9N2 LPAIVs are divided into two major lineages, North American and Eurasian. Within the Eurasian lineage, several sublineages have evolved, and the most prevalent is the G1-like sublineage, which is represented by A/quail/Hong Kong/G1/97(H9N2). G1-like sublineage viruses have been circulating in Middle Eastern and Central-Asian countries since 1998 and have further diversified into four distinct groups – A, B, C and D [6].
In Egypt, (AI) avian influenza I H9N2 viruses have been isolated from chickens, turkeys and quails [7–9]. All isolates from 2010 to 2013 were closely related to viruses from other Middle Eastern countries [7, 8]. Poultry infected with Egyptian H9N2 viruses showed no clinical illness or mild respiratory signs [10]. In addition to H9N2, the highly pathogenic H5N1 subtype is widely circulating in Egypt, fostering concerns about genetic reassortment of the two co-circulating viruses. Although active surveillance of AI among poultry in Egypt showed that H5N1/H9N2 co-infection in the same avian host is common [11], reassortment has not yet been reported unlike reports from Asia [12–14]. Vaccination by inactivated H9N2 viruses originating from Egypt or elsewhere in Asia has been implemented in Egypt since 2012. Extensive vaccination of poultry for AI can accelerate antigenic drift leading to immunization failure similar to the situation with H5N1 in Egypt [15, 16].
H9N2 viruses are public health pathogens due to their ability to infect humans. A recent study in Egypt documented a 5 to 7 % seroprevalence of H9N2 antibodies in people exposed to poultry [17]. Only three human infection cases of H9N2 viruses were reported according to the Egyptian Ministry of Health [18]. Genotypic markers associated with increased affinity of AI viruses to the human host have been previously described in Egyptian H9N2 viruses [8].
In order to monitor the genetic evolution of Egyptian H9N2 viruses, the characteristics of surface glycoproteins of 59 H9N2 viruses isolated through active surveillance of AI in Egypt between 2013 and 2015 were analysed. To understand the genetic background of H9N2 viruses and to elucidate potential reassortments, we conducted a phylogenetic analysis of whole-genome sequences of 10 H9N2 viruses from different hosts. Additionally, antigenic analysis to further investigate the antigenic evolution of the Egyptian H9N2 viruses from 2011 to 2015 was performed. Receptor-binding affinity and resistance to antiviral drugs were investigated.
Results
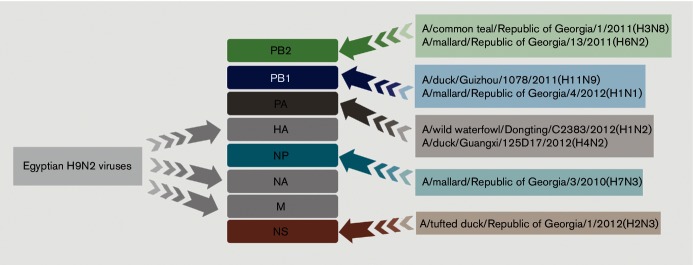
Reassortment in pigeon isolates
Phylogenetic analyses of A/pigeon/Egypt/S10408B/2014(H9N2) and A/pigeon/Egypt/S10409A/2014(H9N2) isolates indicated that PB2, PB1, PA, NP and NS gene segments had more diversified sources than the HA, NA and M genes, suggesting that the two isolated H9N2 viruses had undergone extensive reassortment (Fig. 1). PB2 segments of the two pigeon H9N2 isolates were not related to any PB2 of Egyptian H9N2 isolates and had the highest relationship to the PB2 of A/common teal/Republic of Georgia/1/2011(H3N8) and A/mallard/Republic of Georgia/13/2011(H6N2) viruses, while the PB1 genes were closely related to A/mallard/Republic of Georgia/4/2012(H1N1) viruses. The PA genes of the pigeon viruses were not related to other Egyptian H9N2 isolates but had higher similarity to A/wild water fowl/Dongting/C2383/2012(H1N2), A/duck/Guangxi/125D17/2012(H4N2) and A/mallard/Czech Republic/13438-29K/2010(H11N9). However, blast results showed higher similarity with the Chinese viruses (98 %). NP and NS genes of both H9N2 viruses isolated from pigeons had the highest similarity with A/mallard/Republic of Georgia/3/2010(H7N3) and A/tufted duck/Republic of Georgia/1/2012(H2N3), respectively.
Fig. 1.
Schematic illustration of the reassortment process of H9N2 viruses isolated from pigeons in Egypt.
Genetic analysis of surface glycoproteins
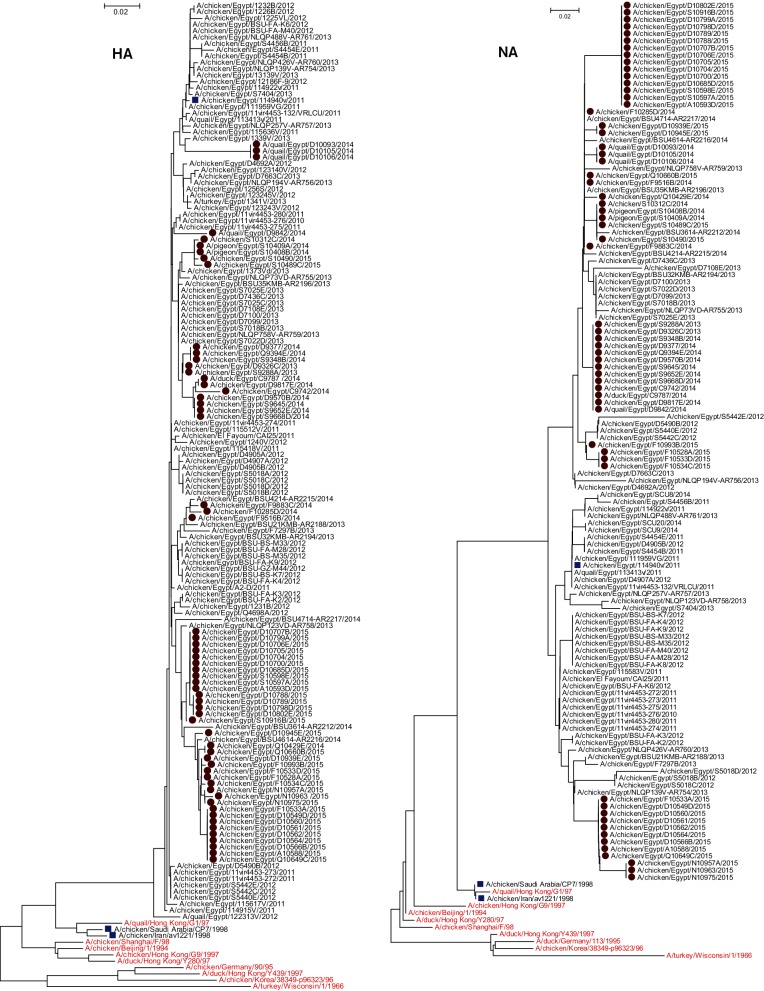
Homologous analyses of HA and NA were performed to compare the nucleotide and deduced amino acid sequences of Egyptian H9N2 viruses with H9N2 representative strains and also with the known commercial vaccine strains used in Egypt. The HA and NA of the current H9N2 isolates showed high sequence homology with other Egyptian H9N2 strains that belonged to G1-like H9N2 viruses (Fig. 2). The nucleotide and deduced amino acid sequence identities among the 59 HAs of Egyptian H9N2 viruses ranged from 94.5 to 100 % and 95 to 100 %, respectively. In comparison with the three H9N2 vaccine strains used in commercial vaccines in Egypt (A/chicken/Saudi Arabia/CP7/1998, A/chicken/Iran/av1221/1998 and A/chicken/Egypt/114940v/2011), ranges of deduced amino acid identities were 92.5 to 93.4 %, 91.6 to 92.9 % and 96.3 to 97.9 %, respectively.
Fig. 2.
Phylogenetic tree of the nucleotide sequences of HA and NA of H9N2 viruses isolated from different hosts in Egypt. H9N2 isolates sequenced specifically for this study are labelled with red circles. H9N2 vaccine strains are labelled with blue squares. Ancestor H9N2 viruses are labelled with red colour. Phylogenetic analysis was performed by using mega version 6.
The HA cleavage motif sequence of Egyptian isolates was 335RSSR*GLF341 (H9 numbering), which is the signature of low pathogenicity for H9N2 viruses isolated from the Middle East and Asia, and is well adapted to the chicken host [19–21]. Analysis of the N-XT/S motif (X can be any amino acid except proline) revealed that the Egyptian isolates had six potential N-linked glycosylation sites, at positions 29, 105, 141, 298, 305 and 492 within the HA molecule (Table 1). Glycosylation sites 206 and 218 were lost from all Egyptian isolates compared to G1-like viruses (Table 1). Specific glycosylation site was characterized in three quail H9N2 isolates at position 196.
Table 1. Comparison of amino acid sequences of the HA of H9N2 viruses isolated from poultry in Egypt between 2013 and 2015 with ancestor H9N2 viruses (H9 numbering) at receptor binding site (RBS), cleavage site and glycosylation sites.
‘.’ indicates a site at which the amino acid residue is identical to an AI A/quail/Hong Kong/G1/97 (H9N2) virus.
| Virus | RBS | Glycosylation sites | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 166 | 191 | 197 | 198 | 232 | 234 | 235 | 236 | 29 | 105 | 141 | 196 | 206 | 218 | 298 | 305 | 492 | |
| A/quail/Hong Kong/G1/97 | S | H | T | E | N | L | Q | G | NST | NGT | NVT | YTE | NDT | NRT | NST | NIS | NGT |
| A/duck/Hong Kong/Y280/97 | N | N | . | T | . | . | Q | . | . . . | . . L | . . S | D.T | T . . | . . . | . T . | . V . | . . . |
| A/chicken/Egypt/S9288A/2013 | N | . | . | A | . | . | I | . | . . . | . . . | . . . | D.A | T . . | D . . | . . . | . . . | . . . |
| A/chicken/Egypt/D9326C/2013 | N | . | . | A | . | . | I | . | . . . | . . . | . . . | D.A | T . . | D . . | . . . | . . . | . . . |
| A/chicken/Egypt/S9348B/2014 | N | . | . | A | . | . | I | . | . . . | . . . | . . . | D.A | T . . | D . . | . . . | . . . | . . . |
| A/chicken/Egypt/D9377/2014 | N | . | . | A | . | . | I | . | . . . | . . . | . . . | D.A | T . . | D . . | . . . | . . . | . . . |
| A/chicken/Egypt/Q9394E/2014 | N | . | . | A | . | . | I | . | . . . | . . . | . . . | D.A | T . . | D . . | . . . | . . . | . . . |
| A/chicken/Egypt/F9516B/2014 | N | . | . | A | . | . | I | . | . . . | . . . | . . . | D.A | T . . | D . . | . . . | . . . | . . . |
| A/chicken/Egypt/D9570B/2014 | N | . | . | A | . | . | I | . | . . . | . . . | . . . | D.A | T . . | D . . | . . . | . . . | . . . |
| A/chicken/Egypt/S9645/2014 | N | . | . | A | . | . | I | . | . . . | . . . | . . . | D.A | T . . | D . . | . . . | . . . | . . . |
| A/chicken/Egypt/S9652E/2014 | N | . | . | A | . | . | I | . | . . . | . . . | . . . | D.A | T . . | D . . | . . . | . . . | . . . |
| A/chicken/Egypt/S9668D/2014 | N | . | . | A | . | . | I | . | . . . | . . . | . . . | D.A | T . . | D . . | . . . | . . . | . . . |
| A/chicken/Egypt/C9742/2014 | N | . | . | A | . | . | I | . | . . . | . . . | . . . | D.A | T . . | D . . | . . . | . . . | . . . |
| A/duck/Egypt/C9787/2014 | N | . | . | A | . | . | I | . | . . . | . . . | . . . | D.A | T . . | D . . | . . . | . . . | . . . |
| A/chicken/Egypt/D9817E/2014 | N | . | . | A | . | . | I | . | . . . | . . . | . . . | D.A | T . . | D . . | . . . | . . . | . . . |
| A/quail/Egypt/D9842/2014 | N | . | . | T | . | . | I | . | . . . | . . . | . . . | D.T | T . . | D . . | . . . | . . . | . . . |
| A/chicken/Egypt/F9883C/2014 | N | . | . | A | . | . | I | . | . . . | . . . | . . . | D.A | T . . | D . . | . . . | . . . | . . . |
| A/quail/Egypt/D10093/2014 | D | . | . | T | . | . | T | . | . . . | . . . | . . . | N.T | T . . | D . . | . . . | . . . | . . . |
| A/quail/Egypt/D10105/2014 | D | . | . | T | . | . | T | . | . . . | . . . | . . . | N.T | T . . | D . . | . . . | . . . | . . . |
| A/quail/Egypt/D10106/2014 | D | . | . | T | . | . | T | . | . . . | . . . | . . . | N.T | T . . | D . . | . . . | . . . | . . . |
| A/chicken/F10285D/2014 | N | . | . | A | . | . | I | . | . . . | . . . | . . . | D.A | T . . | D . . | . . . | . . . | . . . |
| A/chicken/S10312C/2014 | N | . | . | A | . | . | I | . | . . . | . . . | . . . | D.A | T . . | D . . | . . . | . . . | . . . |
| A/pigeon/Egypt/S10408B/2014 | N | . | . | A | . | . | I | . | . . . | . . . | . . . | D.A | T . . | D . . | . . . | . . . | . . . |
| A/pigeon/Egypt/S10409A/2014 | N | . | . | A | . | . | I | . | . . . | . . . | . . . | D.A | T . . | D . . | . . . | . . . | . . . |
| A/chicken/Egypt/Q10429E/2014 | N | . | . | T | . | . | I | . | . . . | . . . | . . . | D.T | T . . | D . . | . . . | . . . | . . . |
| A/chicken/Egypt/S10489C/2015 | N | . | . | V | . | . | I | . | . . . | . . . | . . . | D.V | T . . | D . . | . . . | . . . | . . . |
| A/chicken/Egypt/S10490/2015 | N | . | . | A | . | . | I | . | . . . | . . . | . . . | D.A | T . . | E . . | . . . | . . . | . . . |
| A/chicken/Egypt/F10528A/2015 | N | . | . | A | . | . | I | . | . . . | . . . | . . . | D.A | T . . | D . . | . . . | . . . | . . . |
| A/chicken/Egypt/F10533A/2015 | N | . | . | T | . | . | I | . | . . . | . . . | . . . | D.T | T . . | D . . | . . . | . . . | . . . |
| A/chicken/Egypt/F10533D/2015 | N | . | . | A | . | . | I | . | . . . | . . . | . . . | D.A | T . . | D . . | . . . | . . . | . . . |
| A/chicken/Egypt/F10534C/2015 | N | . | . | V | . | . | I | . | . . . | . . . | . . . | D.V | T . . | D . . | . . . | . . . | . . . |
| A/chicken/Egypt/D10549D/2015 | N | . | . | T | . | . | I | . | . . . | . . . | . . . | D.T | T . . | D . . | . . . | . . . | . . . |
| A/chicken/Egypt/D10560/2015 | N | . | . | T | . | . | I | . | . . . | . . . | . . . | D.T | T . . | D . . | . . . | . . . | . . . |
| A/chicken/Egypt/D10561/2015 | N | . | . | T | . | . | I | . | . . . | . . . | . . . | D.T | T . . | D . . | . . . | . . . | . . . |
| A/chicken/Egypt/D10562/2015 | N | . | . | T | . | . | I | . | . . . | . . . | . . . | D.T | T . . | D . . | . . . | . . . | . . . |
| A/chicken/Egypt/D10564/2015 | N | . | . | T | . | . | I | . | . . . | . . . | . . . | D.T | T . . | D . . | . . . | . . . | . . . |
| A/chicken/Egypt/D10566B/2015 | N | . | . | T | . | . | I | . | . . . | . . . | . . . | D.T | T . . | D . . | . . . | . . . | . . . |
| A/chicken/Egypt/A10588/2015 | N | . | . | T | . | . | I | . | . . . | . . . | . . . | D.T | T . . | D . . | . . . | . . . | . . . |
| A/chicken/Egypt/A10593D/2015 | N | . | . | A | . | . | I | . | . . . | . . . | . . . | D.A | T . . | D . . | . . . | . . . | . . . |
| A/chicken/Egypt/S10597A/2015 | N | . | . | A | . | . | I | . | . . . | . . . | . . . | D.A | T . . | D . . | . . . | . . . | . . . |
| A/chicken/Egypt/S10598E/2015 | N | . | . | A | . | . | I | . | . . . | . . . | . . . | D.A | T . . | D . . | . . . | . . . | . . . |
| A/chicken/Egypt/Q10649C/2015 | N | . | . | T | . | . | I | . | . . . | . . . | . . . | D.T | T . . | D . . | . . . | . . . | . . . |
| A/chicken/Egypt/Q10660B/2015 | N | . | . | T | . | . | I | . | . . . | . . . | . . . | D.T | T . . | D . . | . . . | . . . | . . . |
| A/chicken/Egypt/D10685D/2015 | N | . | . | A | . | . | I | . | . . . | . . . | . . . | D.A | T . . | D . . | . . . | . . . | . . . |
| A/chicken/Egypt/D10700/2015 | N | . | . | A | . | . | I | . | . . . | . . . | . . . | D.A | T . . | D . . | . . . | . . . | . . . |
| A/chicken/Egypt/D10704/2015 | N | . | . | A | . | . | I | . | . . . | . . . | . . . | D.A | T . . | D . . | . . . | . . . | . . . |
| A/chicken/Egypt/D10705/2015 | N | . | . | A | . | . | I | . | . . . | . . . | . . . | D.A | T . . | D . . | . . . | . . . | . . . |
| A/chicken/Egypt/D10706E/2015 | N | . | . | A | . | . | I | . | . . . | . . . | . . . | D.A | T . . | D . . | . . . | . . . | . . . |
| A/chicken/Egypt/D10707B/2015 | N | . | . | A | . | . | I | . | . . . | . . . | . . . | D.A | T . . | D . . | . . . | . . . | . . . |
| A/chicken/Egypt/D10788/2015 | N | . | . | T | . | . | I | . | . . . | . . . | . . . | D.T | T . . | D . . | . . . | . . . | . . . |
| A/chicken/Egypt/D10789/2015 | N | . | . | T | . | . | I | . | . . . | . . . | . . . | D.T | T . . | D . . | . . . | . . . | . . . |
| A/chicken/Egypt/D10798D/2015 | N | . | . | T | . | . | I | . | . . . | . . . | . . . | D.T | T . . | D . . | . . . | . . . | . . . |
| A/chicken/Egypt/D10799A/2015 | N | . | . | A | . | . | I | . | . . . | . . . | . . . | D.A | T . . | D . . | . . . | . . . | . . . |
| A/chicken/Egypt/D10802E/2015 | N | . | . | T | . | . | I | . | . . . | . . . | . . . | D.T | T . . | D . . | . . . | . . . | . . . |
| A/chicken/Egypt/S10916B/2015 | N | . | . | A | . | . | I | . | . . . | . . . | . . . | D.A | T . . | D . . | . . . | . . . | . . . |
| A/chicken/Egypt/D10939E/2015 | N | . | . | A | . | . | I | . | . . . | . . . | . . . | D.A | T . . | D . . | . . . | . . . | . . . |
| A/chicken/Egypt/D10945E/2015 | N | . | . | A | . | . | I | . | . . . | . . . | . . . | D.A | T . . | D . . | . . . | . . . | . . . |
| A/chicken/Egypt/N10957A/2015 | N | . | . | A | . | . | I | . | . . . | . . . | . . . | D.A | T . . | D . . | . . . | . . . | . . . |
| A/chicken/Egypt/N10963/2015 | N | . | . | A | . | . | I | . | . . . | . . . | . . . | D.A | T . . | D . I | . . . | . . . | . . . |
| A/chicken/Egypt/N10975/2015 | N | . | . | A | . | . | I | . | . . . | . . . | . . . | D.A | T . . | D . . | . . . | . . . | . . . |
| A/chicken/Egypt/F10993B/2015 | N | . | . | A | . | . | I | . | . . . | . . . | . . . | D.A | T . . | D . . | . . . | . . . | . . . |
| A/duck/Hong Kong/365/78(H4N6) | D | . | . | . | R | Q | S | . | |||||||||
| A/chicken/Egypt/M7217B/2013(H5N1) | . | . | A | . | . | Q | S | . | |||||||||
| A/Hong Kong/1073/99(H9N2) | . | . | . | . | . | . | Q | . | |||||||||
The receptor-binding site (RBS) is a critical viral factor for host cellular receptor specificity and influences the generation of human viruses from avian sources. Table 1 shows the amino acids at positions 166, 191, 197, 198, 232, 234, 235 and 236, which are components of the RBS of the HA of H9N2 viruses. Within the RBS, all Egyptian H9N2 isolates had H191 and L234 (H9 numbering), which are associated with preferential binding to a cellular receptor present in different respiratory epithelial cells in humans [22]. Avian receptor-specific amino acids were identified at 197T in all Egyptian H9N2 isolates. Four isolates from quails and 14 isolates from chickens carried T at site 198 of RBS similar to A/duck/Hong Kong/Y280/97(H9N2). Only two H9N2 isolates (A/chicken/Egypt/F10534C/2015 and A/chicken/Egypt/S10489C/2015) had V and the remaining isolates had A at this site. Three H9N2 isolates from quails (A/quail/Egypt/D10093/2014, A/quail/Egypt/D10105/2014 and A/quail/Egypt/D10106/2014) were distinguished by D and T at sites 166 and 235 (H9 numbering), respectively, instead of N and I in the other isolates.
Genetic variations were seen in the three H9N2 viruses isolated from quails and were distinguishable from other contemporary H9N2 isolates by D at site 166 instead of N (antigenic site A) and N and A at sites 153 and 201 instead of D and N, respectively (antigenic site B). An AI A/chicken/Egypt/Q10660B/2015(H9N2) virus had G at site 153 in antigenic site B instead of D. Comparing the amino acid residues at the antigenic sites of the HA molecules of the Egyptian H9N2 viruses with vaccine strains used in Egypt showed significant amino acid divergence that may affect vaccine efficacy as shown in Table 2.
Table 2. Comparison of the antigenic sites of the HA of H9N2 viruses isolated from poultry in Egypt between 2013 and 2015 and vaccine strains.
‘.’ indicates a site at which the amino acid residue was not changed.
| Virus | Antigenic site I (A) | Antigenic site II (B) | Overlapping site | ||||||
|---|---|---|---|---|---|---|---|---|---|
| 143T | 166N | 170P | 153D | 201N | 234L | 141N | 197T | 206T | |
| A/quail/Egypt/D10093/2014 | . | D | . | N | A | . | . | . | . |
| A/quail/Egypt/D10105/2014 | . | D | . | N | A | . | . | . | . |
| A/quail/Egypt/D10106/2014 | . | D | . | N | A | . | . | . | . |
| A/chicken/Egypt/Q10660B/2015 | . | . | . | G | . | . | . | . | . |
| The remaining 55 Egyptian H9N2 isolates | . | . | . | . | . | . | . | . | . |
| A/chicken/Saudi Arabia/CP7/98 | . | . | . | G | S | Q | . | . | . |
| A/chicken/Iran/av1221/1998 | . | S | . | G | . | Q | . | . | N |
| A/chicken/Egypt/114940v/2011 | . | . | . | . | . | Q | . | . | . |
Sequence comparisons of the NA of recent Egyptian H9N2 viruses with the three known commercially used H9N2 vaccine strains showed extensive sequence divergence (9.7–11.3 % dissimilarity) with vaccine seeds from Saudi Arabia and Iran, while A/chicken/Egypt/114940v/2011(H9N2) vaccine strain was homologous to Egyptian H9N2 isolates (95.7–97.9 % similarity) based on amino acid analysis. No stalk deletions of any analysed NA segments of H9N2 viruses were detected.
Sequence analysis of binding-pocket residues involved in interactions with antiviral drugs in NA genes revealed no mutations. Analysis of the haemadsorbing site, which is located on the surface of the NA molecule away from the neuraminidase enzyme active site at positions 366–373, 399–404 and 431–433, was performed. A 366IKKDSRAG373 form was present in all isolates. In site 399–404, 399DSDGWS404 form was seen in 46 isolates, 399DSDSWS404 in 11 isolates and 399DSDGRS404 in 2 isolates. At site 431–433, two forms, 431PHE433 and 431PQE433, were found in 12 and 47 isolates, respectively.
All NA genes of the Egyptian H9N2 viruses contained six N-linked glycosylation sites at positions 44, 61, 69, 146, 200 and 234. In addition, the glycosylation site at 86 was missing in the two pigeon and three chicken H9N2 isolates. The glycosylation site at 402, which was described previously as a characteristic of H9N2 viruses [23], was not found in the Egyptian H9N2 isolates.
Genetic analysis of internal proteins
PB2
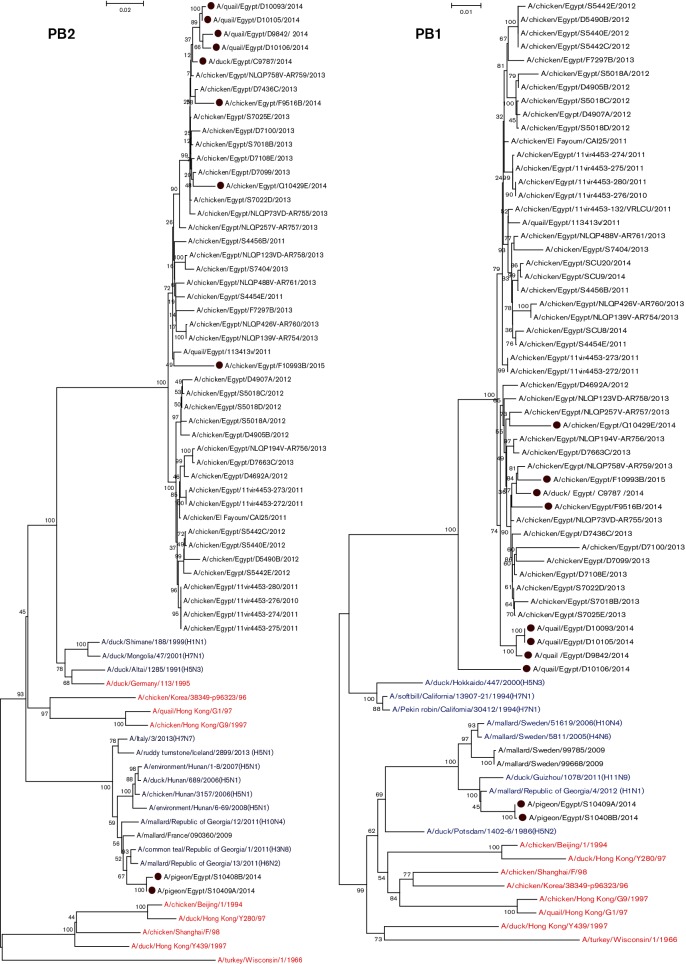
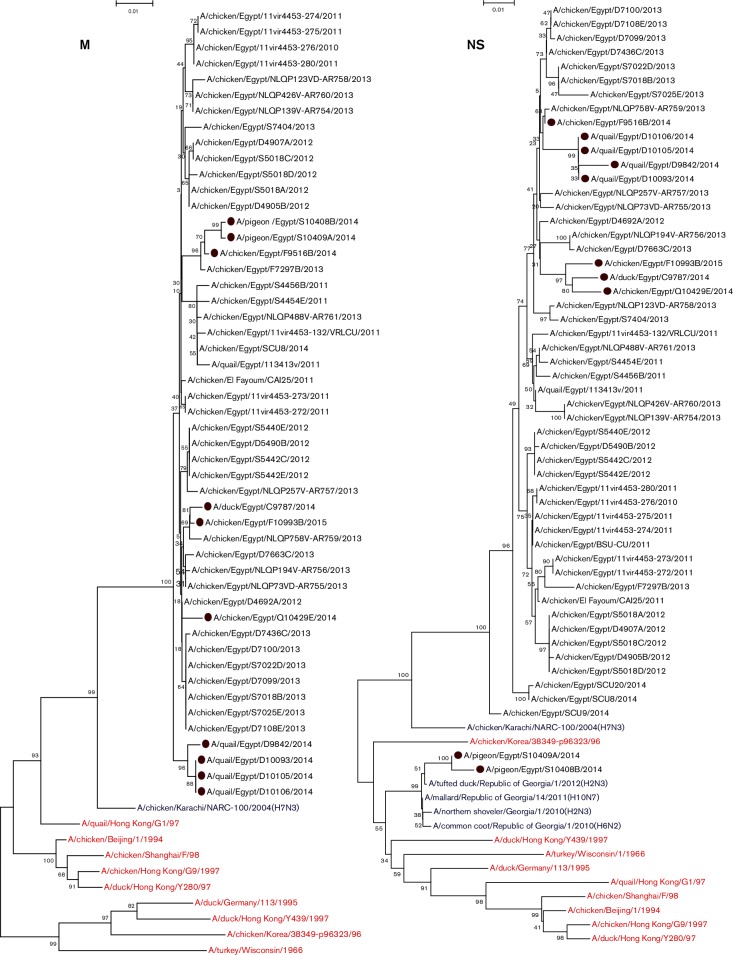
Except for the two pigeon isolates, PB2 genes of Egyptian H9N2 isolates showed higher identity to H9N2 viruses isolated previously from Egypt, which were closely related to A/duck/Shimane/188/1999(H1N1), A/duck/Altai/1285/1991(H5N3), A/duck/Mongolia/47/2001(H7N1) and A/duck/Germany/113/1995(H9N2) viruses rather than to those of other ancestral H9N2 viruses such as G1 (85 %) and Y280 (82 %). Except for the two pigeon isolates, Egyptian viruses were phylogenetically grouped into two clusters: the first included viruses from 2011 to 2015, while the second included viruses from 2010 till 2013 (Fig. 3).
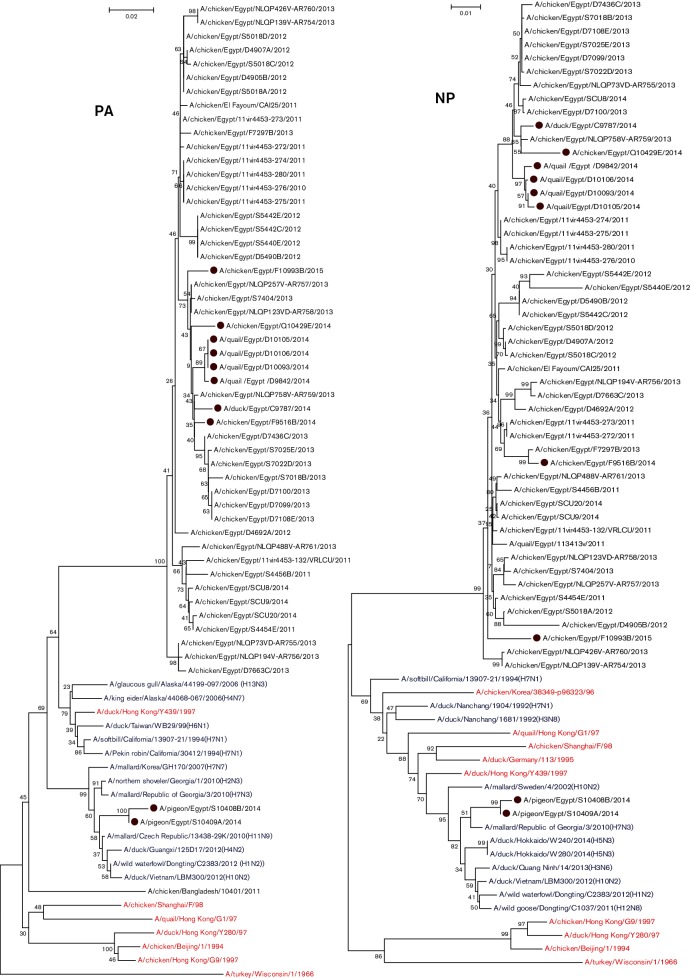
Fig. 3.
Phylogenetic trees of the nucleotide sequences of the PB2, PB1, PA, NP, M and NS genes of H9N2 viruses isolated from different hosts in Egypt. H9N2 isolates sequenced specifically for this study are labelled with red circles. Ancestor H9N2 viruses are labelled with red colour. Non-H9N2 subtypes are labelled with blue colour. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) is shown at the dendrogram nodes. Phylogenetic analysis was performed by using mega version 6.
Genetic analysis of virulence determinants in PB2 segment revealed that only the two pigeon isolates had K591, which is previously known as a virulence marker and significantly enhanced polymerase activity and replication rate in human cells (Table S1, available in the online Supplementary Material).
The I504V substitution is associated with enhanced activity of the polymerase complex [24], and this substitution was observed in all Egyptian isolates. Except for 318R, which was detected in A/pigeon/Egypt/S10408B/2014(H9N2), A/pigeon/Egypt/S10409A/2014(H9N2) and A/chicken/Egypt/F10993B/2015(H9N2) viruses, all other PB2 residues that are associated with host specificity were avian like (Table S2). Substitutions of E to K and D to N at positions 627 and 701, respectively, that were associated with virulence and virus transmission in mammals were not observed [25, 26] (Table S2).
PB1
PB1 genes of Egyptian isolates showed higher identity with H9N2 viruses isolated from Egypt during 2010 to 2013. The PB1 tree topology revealed that, with the exception of pigeon isolates, all isolates were related to A/duck/Hokkaido/447/2000(H5N3), A/Pekin robin/California/30412/1994(H7N1) and A/softbill/California/13907-21/1994(H7N1) rather than to H9N2 ancestors. The PB1 genes of quail isolates were distinct from the other Egyptian H9N2 isolates (Fig. 3).
The N66S mutation in the PB1-F2 protein is important for increasing viral pathogenesis [27]. This substitution was present in PB1-F2 of seven H9N2 isolates (Table S1). The mammalian host-specific substitutions V76A, R79Q and L82S were also identified in three, one and eight H9N2 isolates, respectively. Mammalian host-specific substitution T68I was not identified in any isolate (Table S2). Pigeon isolates had 317M (avirulent form) while the other isolates had I (virulent form) (Table S1). PB1 of the analysed viruses possessed 336V and 375N as avirulence determinants.
PA
The PA genes of the current Egyptian H9N2 isolates were closely related to H9N2 viruses isolated previously from Egypt that are associated with A/softbill/California/13907-21/1994(H7N1), A/Pekin robin/California/30412/1994(H7N1), A/duck/Taiwan/WB29/99(H6N1) and A/duck/Hong Kong/Y439/1997(H9N2) and genetically distant from G1- and Y280-like sublineage viruses (Fig. 3).
The deduced PA amino acid sequences of the two pigeon isolates and the duck isolate had a mammalian host-specific residue, D instead of E (present in all PA of previously known Egyptian H9N2 isolates) at site 382. An H9N2 isolate from duck had A instead of V at site 100, which was previously identified as important for changing host range from avian to human (Table S2). Amino acid substitutions V127, L672 and L550, which are associated with virulence, were observed in all Egyptian H9N2 isolates (Table S1).
NP
The NPs of eight of ten H9N2 isolates were closely related to isolates from Egypt. Sequence analysis showed mammalian host-specific markers at K398Q (ten isolates) and R214K (eight isolates) (Table S2). Phylogenetic analysis showed that all the Egyptian H9N2 NP genes are not related to ancestral H9N2 viruses (Fig. 3).
M
M genes of Egyptian isolates showed higher similarity to H9N2 viruses isolated from Egypt, which were closely related to A/chicken/Karachi/NARC-100/2004(H7N3) (97 %) rather than to those of other ancestral H9N2 viruses (Fig. 3).
Genetic analysis of the amino acid residues associated with amantadine resistance in the M2 viral protein revealed that all isolates had L26, V27, A30, S31 and G34, suggesting the absence of resistance to amantadine. Mammalian-specific residues (G16 and F55 of M2 and I15 of M1) were found. All 10 Egyptian H9N2 isolates possessed virulent form of residues at positions 64 and 69 in the M2 viral protein (Table S1).
NS
Except for two H9N2 isolates from pigeons, NS genes of eight Egyptian H9N2 isolates showed higher identity to H9N2 viruses isolated previously from Egypt that were closely related to A/chicken/Karachi/NARC-100/2004(H7N3) (93 %) rather than to those of other ancestral H9N2 viruses such as G1 (85 %) and Y280 (82 %) (Fig. 3).
All the current H9N2 isolates had PDZ motif (X-S/T-X-V) at C-terminal of NS1 protein in the form of ESEV in the two pigeon H9N2 isolates and the remaining H9N2 isolates had KSEV motif. Except for the two H9N2 pigeon isolates, the NS1 of the H9N2 isolates harboured the mammalian-specific E227K substitution. The NS1 of all isolates had S42, which is previously known as a virulence determinant [28]. Egyptian H9N2 strains exhibited no substitutions of NS2 virulence determinant residues at positions 31 and 56 in all NS2 amino acid sequences (Table S1).
Antigenic analysis
To characterize the antigenic variation of 37 Egyptian H9N2 viruses isolated from 2011 to 2015, we tested the haemagglutination inhibition (HI) activity of a panel of anti-H9 mAbs against Egyptian H9N2 viruses. All but four isolates from quails reacted well with mAb G1-26. A/quail/Egypt/D10093/2014(H9N2), A/quail/Egypt/D10105/2014(H9N2) and A/quail/Egypt/D10106/2014(H9N2) viruses reacted weakly with mAb G9-25. The remaining H9N2 viruses reacted strongly with mAb G9-25. The results revealed that mAb G9-25 is more reactive than G1-26 with Egyptian H9N2 isolates. The 1073-9 mAb differentiated isolates into three groups according to its reactivity with H9N2 viruses (not reactive: HI titre <100; weakly reactive: 200≥HI titre≤800; reactive: HI titre >800). All Egyptian H9N2 viruses were weakly reactive or not reactive with Y280-18G4 mAb (Table S3). The antigenic cartography generated using the HI titres against the mAbs revealed that Egyptian H9N2 isolates did not cluster chronologically and three isolates from quails, A/quail/Egypt/D10093/2014(H9N2), A/quail/Egypt/D10105/2014(H9N2) and A/quail/Egypt/D10106/2014(H9N2), separated from the Egyptian H9N2 viruses cluster (Fig. 4a). The three quail isolates separated from all other Egyptian H9N2 isolates tested and may be representatives of a new antigenic group. Also, the three quail isolates showed moderate or low reactivity with anti-H9 hyperimmune sera. The remaining Egyptian H9N2 viruses isolated between 2011 and 2015 behaved similarly in reacting with the antisera raised against Egyptian H9N2 viruses. Similarly, the A/turkey/Israel/1567/2004 antiserum reacted well with Egyptian H9N2 viruses except the three isolates from quails. The A/Hong Kong/1073/99 antiserum exhibited moderate or very low HI titres against tested H9N2 isolates (Table S4). Cartography of the HI assay results of hyperimmune sera raised against H9N2 viruses showed extensive overlap indicating cross-reactivity among Egyptian H9N2 viruses. Consistent with mAb results, three isolates from quails [A/quail/Egypt/D10093/2014(H9N2), A/quail/Egypt/D10105/2014(H9N2) and A/quail/Egypt/D10106/2014(H9N2)] drifted away from the other Egyptian H9N2 viruses (Fig. 4b).
Fig. 4.
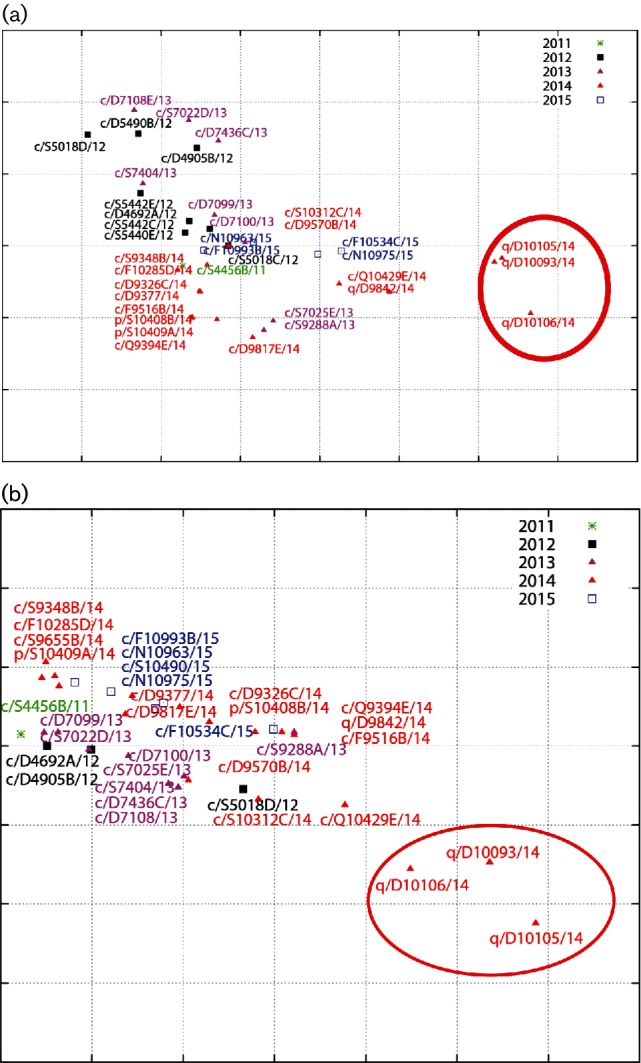
Antigenic cartography representation of the HI data generated by using a panel of mAbs (a) and polyclonal chicken antisera (b). The maps were generated by using AntigenMap (http://sysbio.cvm.msstate.edu/AntigenMap). One grid represents a twofold change in the HI assay results. Three drifted isolates from quails are enclosed by an oval.
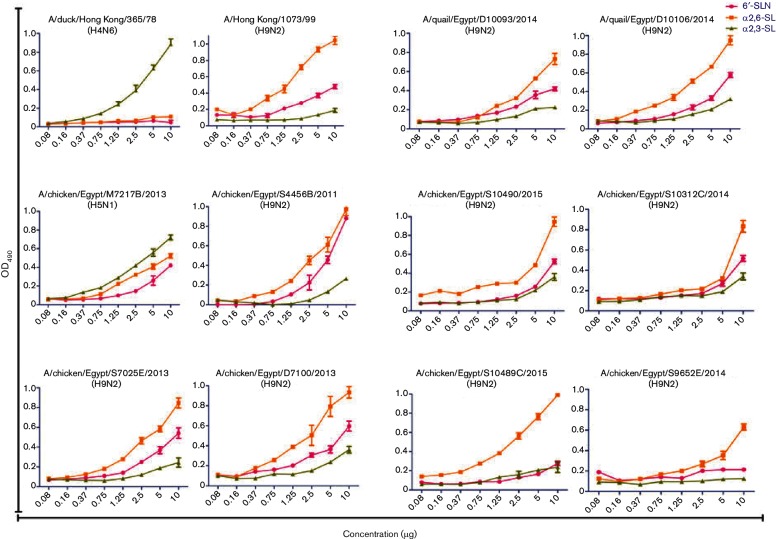
Receptor-binding specificity of H9N2 viruses isolated in Egypt
Since amino acid differences between Egyptian H9N2 viruses were observed around the receptor-binding domain, we tested the sialic acid-binding preferences of Egyptian H9N2 isolates harbouring amino acid diversity at RBS as shown in Table 1. In addition, two AI isolates [A/duck/Hong Kong/365/78(H4N6) and A/chicken/Egypt/M7217B/2013(H5N1)] and one human (A/Hong Kong/1073/1999(H9N2)) isolate, which had α2,3-SL and α2,6-SL binding preference, respectively, were used as a point of comparison in this assay. H4N6 and H5N1 viruses showed higher binding preference for α2,3-SL, while A/Hong Kong/1073/99(H9N2) virus showed a preference for α2,6-SL (Fig. 5). All tested Egyptian H9N2 isolates had higher binding preference for human-like α2,6-SL and α2,6-SLN receptors than α2,3-SL avian-like receptor.
Fig. 5.
Receptor-binding specificity of Egyptian H9N2 viruses. Direct binding of Egyptian H9N2 viruses to different concentrations of biotinylated sialylglycopolymers (X axis) containing 3′-sialyllactose (α2,3-SL), 6′-sialyllactose (α2,6-SL) or 6-sialyl-N-acetyllactosamine (6′-SLN) was measured at 490 nm (Y axis). Influenza A/chicken/Egypt/M7217B/2013(H5N1), A/Hong Kong/1073/99(H9N2) and A/duck/Hong Kong/365/78(H4N6) were used as controls for binding assay.
Antiviral sensitivity
The analysis of NA and M2 protein sequences revealed that there are no observed resistance mutations of all analysed sequences. The sensitivity was confirmed quantitatively against amantadine, using serially increasing concentrations of the antiviral drug by plaque reduction assay in Madin–Darby canine kidney (MDCK)cells. All tested viruses were sensitive (1.4≥IC50≤110 µM) to amantadine with various IC50 values (drug concentration that resulted in a 50 % reduction of the plaque number). At amantadine concentration of 20 µM, the virus growth of A/chicken/Egypt/S4456B/2011(H9N2), A/quail/Egypt/D10093/2014(H9N2) and A/quail/Egypt/D10106/2014(H9N2) was completely inhibited. In the case of A/pigeon/Egypt/S10408B/2014 and A/pigeon/Egypt/S10409A/2014, the percentage of inhibition at amantadine concentration of 160 µM was 72 % (Fig. S1a). In agreement with NA gene sequence analysis, all tested viruses were sensitive to oseltamivir (Fig. S1b).
Discussion
In Egypt, AI H9N2 infections in domestic poultry are geographically widespread across the country. In addition, H5N1 is enzootic, which has raised the possibility of reassortment between the gene segments of both subtypes.
Phylogenetic analysis revealed that the HA and NA of Egyptian H9N2 isolates are related to the Middle Eastern H9N2 isolates and maintained a direct out-group relationship to the prototype G1-like viruses, forming a distinct cluster. Although Egyptian H9N2 viruses share a close genetic relationship, they diverged into different sublineages. The antigenic relatedness of different sublineages needs further examination to select suitable vaccine strains. No evidence of new introductions of H9N2 strains to Egyptian poultry during 2011–2015 was detected, as shown by analysis of our neighbour-joining phylogenetic tree of the HA and NA genes. These genetic characteristics strongly suggest that these viruses are descendant from the first characterized Egyptian H9N2 isolate in 2010.
Genetic relatedness of PB2, PB1, PA, NP and NS segments of the Egyptian H9N2 viruses with other AI subtypes such as H1N1, H7N1, H7N3 and H5N3 rather than to ancestral H9N2 viruses indicates multiple reassortment events that occurred in Asia prior to the virus emerging in Middle Eastern countries. Since viruses from Georgia were genetically similar, it becomes plausible that the reassortment events occurred in Georgia. Those viruses may have been brought into Egypt by wild birds travelling the Black Sea–Mediterranean route that includes Georgia and Egypt. The continued evolution of these H9N2 viruses with accumulation of mutations in their genomes led to the increasing differentiation of Egyptian H9N2 viruses from other H9N2 viruses. Although Egyptian H9N2 viruses were isolated from a country where H5N1 is enzootic, no evidence of reassortment was identified.
Vaccination by inactivated H9N2 viruses has been implemented in Egypt since 2012. In the present investigation, an AI A/chicken/Egypt/F10993B/2015(H9N2) virus was isolated from dead chickens that were vaccinated with inactivated H9N2 vaccine (Table S5). H9N2 isolates during a certain period or in a certain region were thought to have similar profiles of antigenicity [29–31]. Therefore, several commercial AI vaccines based on strains isolated during the late twentieth century were widely used in domestic poultry [32, 33]. However, recent studies showed that H9N2 viruses were isolated from vaccinated chicken flocks [34, 35]; hence some vaccines did not provide complete protection against viral infection. Accordingly, H9N2 vaccine candidates from local strains that reacted well with the circulating strains should be used and continuously updated based on antigenic analysis of local strains.
In a previous study, we found that H9N2 isolates from 2011 to 2013 underwent no significant antigenic drift [8]. In this study, H9N2 viruses isolated from quails were antigenically distinct from those isolated from chickens as a result of mutations in the antigenic sites A (N166D) and B (D153N and N201A). Substitution of amino acids in the antigenic sites in the HA protein of AI viruses usually results in antigenic drift [36]. Kaverin et al. [37] reported that amino acid changes at any of the nine sites on the HA protein of H9 led to the emergence of a variant. Evidence of rapid antigenic evolution of H9N2 viruses isolated from quails was previously recorded and might be indicative of host selection pressure [38, 39]. Antigenic and genetic changes in H9N2 viruses from quails in Egypt should be monitored carefully as evidence from this work suggests that those viruses are evolving independently of the evolution of H9N2 viruses from other poultry species. Quail viruses were implicated in the transmission of H9N2 to humans in 1999 [4].
The RBS of HA of Egyptian H9N2 viruses had the Q234L substitution, which has been implicated in increasing human-like receptor (α2,6-SL and α2,6-SLN) specificity rather than avian-like receptor specificity (α2,3-SL). In this study, all tested Egyptian H9N2 isolates had higher binding preference for α2,6-SL and α2,6-SLN than α2,3-SL, indicating that H9N2 viruses have shown stronger binding specificity towards human-like receptors. These results are consistent with previous studies that demonstrated that H9N2 viruses carrying HA-L234 (present in all Egyptian H9N2 isolates) preferentially recognize human-like receptors, while H9N2 viruses bearing HA-Q234 (present in H4 and H5) preferentially recognize avian-like receptors [40–42]. The increased affinity of binding with human-like receptors suggests that the risk of humans being infected with H9N2 viruses is present. Amino acid changes at 198 (A/T/V) and 235 (I/T) did not affect the binding specificity of the H9N2 viruses for α2,6-SL. Thus, human-like receptor specificity of the Egyptian H9N2 viruses clearly correlated with the presence of L234 in the RBS. The combination of H191 and L234, which was typical of early human H3N2 isolates, is observed in all Egyptian H9N2 isolates [22]. Moreover, several distinct molecular markers that are associated with virus transmission and adaptation to mammalian host were identified in Egyptian H9N2 isolates, yet whether this enhancement occurred in Egypt is not known.
The wide circulation of H9N2 viruses provides more opportunities for reassortment with other AIV subtypes. Our study indicated that two of the ten H9N2 isolates inherited five internal genes from Eurasian AIVs circulating in wild birds. The two reassortant H9N2 viruses were isolated from pigeons. This reassortment is the first to be reported in Egyptian poultry. Pigeons act as a bridge species in the ecology of AIVs due to their potential to transmit viruses between poultry and migratory water fowl. In Egypt, domestic pigeons are raised in open cages and are free to fly, and they frequently mix with wild pigeons and other wild bird species. The emergence of these reassortant influenza viruses suggests that co-infections with influenza viruses of different subtypes have presumably occurred.
Continuous surveillance of influenza virus in poultry and waterfowl is critical for monitoring the genesis and emergence of potentially pandemic strains in this region. The quail may accelerate antigenic drift rates of influenza viruses. The biological impacts of the newly emerging reassorted H9N2 viruses in pigeons need to be fully examined in poultry.
Methods
Viruses
A total of 59 Egyptian H9N2 viruses were isolated from poultry during active surveillance of AI in seven governorates, in the period from December 2013 to April 2015 (13 H9N2 isolates from Sharqia governorate, 27 from Dakahlia, 4 from Qalyubia, 8 from Faiyum, 3 from Minya, 2 from Cairo and 2 from Asyut). Isolates were collected from healthy, sick and dead poultry. The details of isolation area, health status of the host, date of isolation and site of sampling (commercial farm, market, house and abattoir) of these isolates are provided in Table S5. One isolate from a duck, 4 isolates from quails, 2 isolates from pigeons and 52 isolates from chickens were propagated in the allantoic fluids of 10-day-old specific-pathogen-free embryonated chicken eggs (SPF Eggs Production Farm). Inoculated eggs were incubated for 48 h at 37 °C and then chilled at 4 °C for 4 h before harvesting. Only one isolate was obtained from vaccinated broiler chickens with inactivated AI H9N2 vaccine (CEVA) (Table S5).
Amplification of full genome and sequencing
Viral RNA was extracted from harvested allantoic fluid, using QIAamp Viral RNA Mini kit (Qiagen) according to the manufacturer’s protocol. The first-strand cDNA was synthesized using SuperScript III Reverse Transcriptase (Invitrogen) and Uni-12 primer (5′AGCRAAAGCAGG3′) as per manufacturer’s protocol. Using Phusion Master Mix kit (Thermo Scientific), the desired genes of the H9N2 isolates were amplified using universal primers [43]. Briefly, using gene-specific primers, 2 µl of each reverse transcription reaction was subjected to PCR by an initial denaturation step (98 °C, 30 s), followed by 40 cycles each consisting of 98 °C for 10 s, 57 °C for 30 s, 72 °C for 3 min and final elongation at 72 °C for 10 min. Amplicons of the appropriate sizes were subsequently gel purified using QIAquick Gel Extraction kit (Qiagen). The purified PCR products were directly used for sequencing reactions at the Macrogen sequencing facility. Sequences were assembled using SeqMan DNA Lasergene 7 software (DNASTAR). Sequences were deposited in GenBank under the accession numbers listed in Table S6.
Sequence analysis and phylogenetic tree construction
The assembled sequences were subjected to NCBI blast analysis. BioEdit 7.0 was used for multiple sequence alignment [44]. The nucleotide and amino acid homologies were further assessed by the ClustalW method of MegAlign (DNASTAR). mega 6.0 was used for phylogenetic tree construction by applying the neighbour-joining method with Kimura’s two-parameter distance model and 1000 bootstrap replicates [45]. The trees included all Egyptian H9N2 virus sequences, major ancestral H9N2 strains and other influenza virus subtypes with closely related H9N2 genes, as shown by a blastn analysis. Sequences were obtained from the Global Initiative on Sharing All Influenza Data and GenBank. BioEdit program version 7.0 was used for genomic signature analysis.
Antigenic analysis
Antigenic analyses of Egyptian H9N2 viruses were performed by haemagglutination inhibition (HI) test using chicken antisera generated by vaccination of several chicken groups with different H9N2 antigens. The selected H9N2 virus isolates that were used for vaccine preparation were selected based on host variation and location of sampling. H9N2 inactivated vaccines were individually prepared using AI (A/chicken/Egypt/S4456B/2011, A/chicken/Egypt/D9570B/2014, A/chicken/Egypt/D9817E/2014, A/pigeon/Egypt/S10408B/2014, A/pigeon/Egypt/S10409A/2014, A/Bangladesh/10994/2011, A/quail/Lebanon/272/2010, A/turkey/Israel/1567/2004 and A/Hong Kong/1073/1999) H9N2 viruses inactivated with 0.1 % formalin and mixed with Montanide ISA 70 VG (Seppic) in the ratio recommended by the manufacturer (30 antigen/70 adjuvant). Four weeks post-vaccination, sera were collected from each chicken and were tested for antibodies against homologous H9N2 antigens using HI as previously described [46]. Based on differences in HI titres of the reactivity of H9N2 viruses with generated chicken polyclonal antibodies, antigenic mapping of Egyptian H9N2 viruses was done. A panel of anti-H9 mAbs prepared against different antigenic epitopes of A/chicken/Hong Kong/G9/97(G9-25), A/quail/Hong Kong/G1/97(G1-26), A/Hong Kong/1073/99(1073-9) and A/duck/Hong Kong/Y280/97(Y280-18G4) was also used for antigenic mapping of Egyptian H9N2 viruses [37]. HI data of Egyptian H9N2 viruses with mAbs and polyclonal antibodies were used to construct antigenic cartographies using the integrative matrix completion multi-dimensional scaling method as described previously [47, 48]. Matrix completion was used to remove the data noise in the HI experiment. Multi-dimensional scaling projected the antigens onto a two-dimensional grid.
Receptor specificity assay
Virus receptor specificity was determined as previously described [49]. Ninety-six-well fetuin-coated (10 µg ml−1) plates were washed with ice-cold washing buffer (0.01 % Tween 80 in 0.23× PBS), blocked with PBS containing 1 % BSA and incubated overnight with 32 haemagglutination units of influenza viruses at 4 °C. Plates were washed four times with washing buffer. Biotinylated sialylglycopolymers 3′-sialyllactose (α2,3-SL, Neu5Acα2-3Galβ1-4Glc), 6′-sialyllactose (α2,6-SL, Neu5Acα2-6Galβ1-4Glc) and 6-sialyl-N-acetyllactosamine (6′-SLN, Neu5Acα2-6Galβ1-4GlcNAc) (Glycotech) were serially diluted in reaction buffer [0.02 % Tween 80, 0.02 % BSA, 1 µM sialidase inhibitor (zanamivir) and 1× PBS] and were added and incubated at 4 °C for 2 h. The plates were washed four times and incubated with 100 µl of HRP-conjugated streptavidin (1 : 2000) at 4 °C for 1 h. After a final wash, 50 µl of the o-phenylenediamine substrate was added and incubated for 10 min at room temperature. The reaction was stopped with 0.5 M sulfuric acid, and absorbance was measured at 490 nm.
Antiviral sensitivity assays
Sensitivity of the Egyptian H9N2 viruses to amantadine and oseltamivir was determined in Madin-Darby Canine Kidney (MDCK) cells by plaque reduction assay as described previously [50]. Briefly, MDCK cell monolayers in six-well plates were inoculated with A/chicken/Egypt/S4456B/2011(H9N2), A/quail/Egypt/D10093/2014(H9N2), A/quail/Egypt/D10106/2014(H9N2), A/pigeon/Egypt/S10408B/2014(H9N2) and A/pigeon/Egypt/S10409A/2014(H9N2) viruses diluted in infection medium to give 30 to 100 plaques per well and desired concentration of antiviral drug. The viruses were tested for sensitivity against concentrations of 0, 5, 10, 20, 40, 80 and 160 µM amantadine (1-aminoadamantane hydrochloride; Sigma-Aldrich) and concentrations of 5 and 10 µM oseltamivir (TRC). Cells were incubated for 1 h at 37 °C and then overlaid with Dulbecco modified Eagle's medium overlay containing 1 % agarose, 4 % BSA, 1 % antibiotic/antimycotic mixture and 1 µg ml−1 trypsin-Tosyl phenylalanyl chloromethyl ketone (TPCK) (Worthington Diagnostics). After 3 days incubation at 37 °C, 1 ml of 10 % formaldehyde was added to each well for 1 h for cell fixation and virus inactivation. The formaldehyde was then discarded and the plates rinsed with water and dried. For visualization of the plaques, 1 ml of the staining solution, consisting of 1 % crystal violet and 20 % methanol in distilled water, was added to each well and incubated at room temperature for 5 min; the dye was then discarded and the wells were rinsed with water and dried. The percentages of viral inhibition relative to untreated control viruses were calculated at each concentration.
Funding information
This work was funded by the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services, under contract number HHSN272201400006C; and by the Science and Technology Development Fund (STDF) in Egypt, under contract number 5175; and supported by the American Lebanese Syrian Associated Charities (ALSAC).
Conflicts of interest
The authors declare that there are no conflicts of interest.
Supplementary Data
Footnotes
Abbreviations: AI, avaian influenza; AIV, avian influenza virus; HI, haemagglutination inhibition; LPAIV, low pathogenic avian influenza virus; RBS, receptor-binding site.
The Genbank/EMBL/DDBJ accession numbers for the novel sequences obtained in this study are listed in Table S6, available in the online Supplementary Material.
One supplementary figure and six supplementary tables are available with the online Supplementary Material.
References
- 1.Naeem K, Ullah A, Manvell RJ, Alexander DJ. Avian influenza A subtype H9N2 in poultry in Pakistan. Vet Rec. 1999;145:560. doi: 10.1136/vr.145.19.560. [DOI] [PubMed] [Google Scholar]
- 2.Wu H, Peng X, Peng X, Cheng L, Lu X, et al. Genetic and molecular characterization of H9N2 and H5 avian influenza viruses from live poultry markets in Zhejiang Province, eastern China. Sci Rep. 2015;5:17508. doi: 10.1038/srep17508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guo YJ, Wen LY, Zhang Y, Wan M, Guo JF, et al. [Do pigs play a role in human infection with avian influenza A H9N2 viruses?] Zhonghua Shi Yan He Lin Chuang Bing Du Xue Za Zhi. 2005;19:106–109. (English abstract) [PubMed] [Google Scholar]
- 4.Peiris M, Yuen KY, Leung CW, Chan KH, Ip PL, et al. Human infection with influenza H9N2. Lancet. 1999;354:916–917. doi: 10.1016/S0140-6736(99)03311-5. [DOI] [PubMed] [Google Scholar]
- 5.Saito T, Lim W, Suzuki T, Suzuki Y, Kida H, et al. Characterization of a human H9N2 influenza virus isolated in Hong Kong. Vaccine. 2001;20:125–133. doi: 10.1016/S0264-410X(01)00279-1. [DOI] [PubMed] [Google Scholar]
- 6.Fusaro A, Monne I, Salviato A, Valastro V, Schivo A, et al. Phylogeography and evolutionary history of reassortant H9N2 viruses with potential human health implications. J Virol. 2011;85:8413–8421. doi: 10.1128/JVI.00219-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.El-Zoghby EF, Arafa AS, Hassan MK, Aly MM, Selim A, et al. Isolation of H9N2 avian influenza virus from bobwhite quail (Colinus virginianus) in Egypt. Arch Virol. 2012;157:1167–1172. doi: 10.1007/s00705-012-1269-z. [DOI] [PubMed] [Google Scholar]
- 8.Kandeil A, El-Shesheny R, Maatouq AM, Moatasim Y, Shehata MM, et al. Genetic and antigenic evolution of H9N2 avian influenza viruses circulating in Egypt between 2011 and 2013. Arch Virol. 2014;159:2861–2876. doi: 10.1007/s00705-014-2118-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Naguib MM, Arafa AS, El-Kady MF, Selim AA, Gunalan V, et al. Evolutionary trajectories and diagnostic challenges of potentially zoonotic avian influenza viruses H5N1 and H9N2 co-circulating in Egypt. Infect Genet Evol. 2015;34:278–291. doi: 10.1016/j.meegid.2015.06.004. [DOI] [PubMed] [Google Scholar]
- 10.Kandeil A, Moatasim Y, Gomaa MR, Shehata MM, El-Shesheny R, et al. Generation of a reassortant avian influenza virus H5N2 vaccine strain capable of protecting chickens against infection with Egyptian H5N1 and H9N2 viruses. Vaccine. 2016;34:218–224. doi: 10.1016/j.vaccine.2015.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kayali G, Kandeil A, El-Shesheny R, Kayed AS, Gomaa MM, et al. Active surveillance for avian influenza virus, Egypt, 2010–2012. Emerg Infect Dis. 2014;20:542–551. doi: 10.3201/eid2004.131295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dong G, Xu C, Wang C, Wu B, Luo J, et al. Reassortant H9N2 influenza viruses containing H5N1-like PB1 genes isolated from black-billed magpies in southern China. PLoS One. 2011;6:e25808. doi: 10.1371/journal.pone.0025808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Iqbal M, Yaqub T, Reddy K, Mccauley JW. Novel genotypes of H9N2 influenza A viruses isolated from poultry in Pakistan containing NS genes similar to highly pathogenic H7N3 and H5N1 viruses. PLoS One. 2009;4:e5788. doi: 10.1371/journal.pone.0005788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Monne I, Yamage M, Dauphin G, Claes F, Ahmed G, et al. Reassortant avian influenza A(H5N1) viruses with H9N2-PB1 gene in poultry, Bangladesh. Emerg Infect Dis. 2013;19:1630–1634. doi: 10.3201/eid1910.130534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Abdelwhab EM, Abdel-Moneim AS. Epidemiology, ecology and gene pool of influenza A virus in Egypt: will Egypt be the epicentre of the next influenza pandemic? Virulence. 2015;6:6–18. doi: 10.4161/21505594.2014.992662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kayali G, Kandeil A, El-Shesheny R, Kayed AS, Maatouq AM, et al. Avianinfluenza A(H5N1) virus in Egypt. Emerg Infect Dis J. 2016;22 doi: 10.3201/eid2203.150593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gomaa MR, Kayed AS, Elabd MA, Zeid DA, Zaki SA, et al. Avian influenza A(H5N1) and A(H9N2) seroprevalence and risk factors for infection among Egyptians: a prospective, controlled seroepidemiological study. J Infect Dis. 2015;211:1399–1407. doi: 10.1093/infdis/jiu529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.FAO EMPRES EMPRES animal influenza update. 2015. http://cdn.aphca.org/dmdocuments/Avian%20Infuenza%20Alert/Influenza_update_260215.pdf
- 19.Aamir UB, Wernery U, Ilyushina N, Webster RG. Characterization of avian H9N2 influenza viruses from United Arab Emirates 2000 to 2003. Virology. 2007;361:45–55. doi: 10.1016/j.virol.2006.10.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Golender N, Panshin A, Banet-Noach C, Nagar S, Pokamunski S, et al. Genetic characterization of avian influenza viruses isolated in Israel during 2000–2006. Virus Genes. 2008;37:289–297. doi: 10.1007/s11262-008-0272-7. [DOI] [PubMed] [Google Scholar]
- 21.Tosh C, Nagarajan S, Behera P, Rajukumar K, Purohit K, et al. Genetic analysis of H9N2 avian influenza viruses isolated from India. Arch Virol. 2008;153:1433–1439. doi: 10.1007/s00705-008-0131-9. [DOI] [PubMed] [Google Scholar]
- 22.Sorrell EM, Wan H, Araya Y, Song H, Perez DR. Minimal molecular constraints for respiratory droplet transmission of an avian-human H9N2 influenza A virus. Proc Natl Acad Sci USA. 2009;106:7565–7570. doi: 10.1073/pnas.0900877106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin YP, Shaw M, Gregory V, Cameron K, Lim W, et al. Avian-to-human transmission of H9N2 subtype influenza A viruses: relationship between H9N2 and H5N1 human isolates. Proc Natl Acad Sci USA. 2000;97:9654–9658. doi: 10.1073/pnas.160270697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rolling T, Koerner I, Zimmermann P, Holz K, Haller O, et al. Adaptive mutations resulting in enhanced polymerase activity contribute to high virulence of influenza A virus in mice. J Virol. 2009;83:6673–6680. doi: 10.1128/JVI.00212-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang J, Sun Y, Xu Q, Tan Y, Pu J, et al. Mouse-adapted H9N2 influenza A virus PB2 protein M147L and E627K mutations are critical for high virulence. PLoS One. 2012;7:e40752. doi: 10.1371/journal.pone.0040752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhou B, Pearce MB, Li Y, Wang J, Mason RJ, et al. Asparagine substitution at PB2 residue 701 enhances the replication, pathogenicity, and transmission of the 2009 pandemic H1N1 influenza A virus. PLoS One. 2013;8:e67616. doi: 10.1371/journal.pone.0067616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Conenello GM, Tisoncik JR, Rosenzweig E, Varga ZT, Palese P, et al. A single N66S mutation in the PB1-F2 protein of influenza A virus increases virulence by inhibiting the early interferon response in vivo. J Virol. 2011;85:652–662. doi: 10.1128/JVI.01987-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jiao P, Tian G, Li Y, Deng G, Jiang Y, et al. A single-amino-acid substitution in the NS1 protein changes the pathogenicity of H5N1 avian influenza viruses in mice. J Virol. 2008;82:1146–1154. doi: 10.1128/JVI.01698-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Webster RG, Bean WJ, Gorman OT, Chambers TM, Kawaoka Y. Evolution and ecology of influenza A viruses. Microbiol Rev. 1992;56:152–179. doi: 10.1128/mr.56.1.152-179.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xu KM, Smith GJ, Bahl J, Duan L, Tai H, et al. The genesis and evolution of H9N2 influenza viruses in poultry from southern China, 2000 to 2005. J Virol. 2007;81:10389–10401. doi: 10.1128/JVI.00979-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang P, Tang Y, Liu X, Peng D, Liu W, et al. Characterization of H9N2 influenza viruses isolated from vaccinated flocks in an integrated broiler chicken operation in eastern China during a 5 year period (1998–2002) J Gen Virol. 2008;89:3102–3112. doi: 10.1099/vir.0.2008/005652-0. [DOI] [PubMed] [Google Scholar]
- 32.Li C, Yu K, Tian G, Yu D, Liu L, et al. Evolution of H9N2 influenza viruses from domestic poultry in mainland China. Virology. 2005;340:70–83. doi: 10.1016/j.virol.2005.06.025. [DOI] [PubMed] [Google Scholar]
- 33.Sun Y, Pu J, Fan L, Sun H, Wang J, et al. Evaluation of the protective efficacy of a commercial vaccine against different antigenic groups of H9N2 influenza viruses in chickens. Vet Microbiol. 2012;156:193–199. doi: 10.1016/j.vetmic.2011.10.003. [DOI] [PubMed] [Google Scholar]
- 34.Bahari P, Pourbakhsh SA, Shoushtari H, Bahmaninejad MA. Molecular characterization of H9N2 avian influenza viruses isolated from vaccinated broiler chickens in northeast Iran. Trop Anim Health Prod. 2015;47:1195–1201. doi: 10.1007/s11250-015-0848-x. [DOI] [PubMed] [Google Scholar]
- 35.Shen HQ, Yan ZQ, Zeng FG, Liao CT, Zhou QF, et al. Isolation and phylogenetic analysis of hemagglutinin gene of H9N2 influenza viruses from chickens in South China from 2012 to 2013. J Vet Sci. 2015;16:317–324. doi: 10.4142/jvs.2015.16.3.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Caton AJ, Brownlee GG, Yewdell JW, Gerhard W. The antigenic structure of the influenza virus A/PR/8/34 hemagglutinin (H1 subtype) Cell. 1982;31:417–427. doi: 10.1016/0092-8674(82)90135-0. [DOI] [PubMed] [Google Scholar]
- 37.Kaverin NV, Rudneva IA, Ilyushina NA, Lipatov AS, Krauss S, et al. Structural differences among hemagglutinins of influenza A virus subtypes are reflected in their antigenic architecture: analysis of H9 escape mutants. J Virol. 2004;78:240–249. doi: 10.1128/JVI.78.1.240-249.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guan Y, Smith GJ. Genetic characterisation of H9N2 influenza viruses in southern China. Hong Kong Med J. 2016;22:4–6. [PubMed] [Google Scholar]
- 39.Perez DR, Lim W, Seiler JP, Yi G, Peiris M, et al. Role of quail in the interspecies transmission of H9 influenza A viruses: molecular changes on HA that correspond to adaptation from ducks to chickens. J Virol. 2003;77:3148–3156. doi: 10.1128/JVI.77.5.3148-3156.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sang X, Wang A, Ding J, Kong H, Gao X, et al. Adaptation of H9N2 AIV in guinea pigs enables efficient transmission by direct contact and inefficient transmission by respiratory droplets. Sci Rep. 2015;5:15928. doi: 10.1038/srep15928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Srinivasan K, Raman R, Jayaraman A, Viswanathan K, Sasisekharan R. Quantitative characterization of glycan-receptor binding of H9N2 influenza A virus hemagglutinin. PLoS One. 2013;8:e59550. doi: 10.1371/journal.pone.0059550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wan H, Perez DR. Amino acid 226 in the hemagglutinin of H9N2 influenza viruses determines cell tropism and replication in human airway epithelial cells. J Virol. 2007;81:5181–5191. doi: 10.1128/JVI.02827-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hoffmann E, Stech J, Guan Y, Webster RG, Perez DR. Universal primer set for the full-length amplification of all influenza A viruses. Arch Virol. 2001;146:2275–2289. doi: 10.1007/s007050170002. [DOI] [PubMed] [Google Scholar]
- 44.Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis for Windows 95/98/NT. Nucleic Acids Symp. 1999;41:95–98. [Google Scholar]
- 45.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, et al. mega5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.WHO . WHO Manual on Animal Influenza Diagnosis and Surveillance. World Health Organization: Geneva, Switzerland; 2002. [Google Scholar]
- 47.Cai Z, Zhang T, Wan XF. A computational framework for influenza antigenic cartography. PLoS Comput Biol. 2010;6:e1000949. doi: 10.1371/journal.pcbi.1000949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cai Z, Zhang T, Wan XF. Concepts and applications for influenza antigenic cartography. Influenza Other Respir Viruses. 2011;5:204–207. [PMC free article] [PubMed] [Google Scholar]
- 49.Matrosovich MN, Gambaryan AS. Solid-phase assays of receptor-binding specificity. Methods Mol Biol. 2012;865:71–94. doi: 10.1007/978-1-61779-621-0_5. [DOI] [PubMed] [Google Scholar]
- 50.Hayden FG, Cote KM, Douglas RG., Jr Plaque inhibition assay for drug susceptibility testing of influenza viruses. Antimicrob Agents Chemother. 1980;17:865–870. doi: 10.1128/AAC.17.5.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.