Abstract
Purpose
The aim of the study was to characterize clinical and environmental Staphylococcus pettenkoferi isolates with regard to genomic diversity and antibiotic susceptibility pattern. Repetitive-sequence-based PCR and core genome phylogenetic analysis of whole-genome sequencing (WGS) data verified the presence of distinct clades comprising closely related S. pettenkoferi isolates from different geographical locations and origins.
Methodology
Phylogenetic relationships between 25 S. pettenkoferi isolates collected from blood cultures and intra-operative air sampling were determined by repetitive-sequence-based PCR typing and analysis of ~157 000 SNPs identified in the core genome after WGS. Antibiotic susceptibility testing and tests for biofilm production (microtitre plate assay) were performed.
Results
Repetitive-sequence-based PCR as well as WGS data demonstrated the close relatedness of clinically significant blood culture isolates to probable contaminants, as well as to environmental isolates. Antibiotic-susceptibility testing demonstrated a low level of antimicrobial resistance. The mecA gene was present in two cefoxitin-resistant isolates. No isolates were found to produce biofilm.
Conclusion
Close genomic relatedness of S. pettenkoferi isolates from different geographical locations and origins were found within clades, but with substantial genomic difference between the two major clades. The ecological niche of S. pettenkoferi remains unconfirmed, but the presence of S. pettenkoferi in the air of the operating field favours the suggestion of a role in skin flora. Identification of S. pettenkoferi in clinical samples should, in a majority of cases, most likely be regarded as a probable contamination, and its role as a possible pathogen in immunocompromised hosts remains to be clarified.
Keywords: Staphylococcus pettenkoferi, genotypic relatedness, repetitive-sequence based PCR typing, whole-genome sequencing
Introduction
Staphylococcus pettenkoferi was first proposed as a novel staphylococcal species by Trülzsch et al. in 2002, after phenotypic and genotypic characterization of two clinical coagulase-negative staphylococci (CoNS) isolates from a German hospital [1]. In the past, S. pettenkoferi was probably often misidentified as other CoNS species, Kocuria sp. or Micrococcus sp. [1–3], because identification by biochemical methods such as analytical profile index (API) has been misleading and the species do not differ from other CoNS in terms of morphology or simple biochemical tests such as for oxidase, novobiocin or coagulase. With the introduction of MALDI-TOF mass spectrometry (MALDI-TOF MS) in clinical practice, species identification of S. pettenkoferi is no longer difficult, and hence correct identification of this novel staphylococcal species in clinical samples is likely to increase [4]. This was illustrated by a recent French study in which 54 of 9847 CoNS isolates were identified as S. pettenkoferi after implementation of MALDI-TOF MS, compared to 0 of 7885 CoNS isolates during a previous time period when phenotypic methods of identification were used [5].
S. pettenkoferi is believed to colonize human skin [4], but little is known about its ecological niche. According to 16S rRNA and rpoB sequencing, S. pettenkoferi is most closely related to S. auricularis [6]. Case reports of occasional clinical findings of S. pettenkoferi isolates include osteomyelitis, bloodstream infections, and bursitis [1–3, 7–10], predominantly in immunocompromised patients. The aim of the present study was to characterize clinical and environmental S. pettenkoferi isolates with regards to diversity and antibiotic susceptibility pattern.
Methods
Bacterial isolates
The clinical isolates (n=16) originated from blood cultures obtained at the Department of Clinical Microbiology, Central Hospital, Växjö, Sweden, in 2011–2013 (n=13; Vaxjo_1–13), at the Department of Clinical Microbiology, Örebro University Hospital, Örebro, Sweden in 2014 (n=1; Orebro_14), and as part of the European arm of the worldwide SENTRY Antimicrobial Surveillance Program during 2006 and 2008 (n=2; JMI_15–16; provided by JMI Laboratories, North Liberty, IA). The clinical isolates from Växjö and Örebro were provided to us together with clinical data noted on the original request form, and the sex and age of the patient, but identification of patients was not possible (Table 1). The microbiological relevance of the isolates was determined according to the European Manual of Clinical Microbiology criteria for interpretation [11]; that is, an isolate was considered a probable contaminant if only one blood culture bottle was positive.
Table 1. Characteristics of S. pettenkoferi isolates (n=25) obtained from blood cultures and air sampling in operating theatres during prosthetic joint surgery.
COPD, chronic obstructive pulmonary disease; CRP, C-reactive protein; TKA, total knee arthroplasty; THA, total hip arthroplasty. –, data not available.
| Isolate ID | Year | Continent | Origin | Positive bottles/bottles cultured | Nosocomial | Clinical data | Age | Sex |
|---|---|---|---|---|---|---|---|---|
| Vaxjo_1 | 2011 | Europe | Blood culture | 1/4 | No | Suspected sepsis | 82 | F |
| Vaxjo_2 | 2011 | Europe | Blood culture | 1/4 | No | Elevated CRP | 90 | F |
| Vaxjo_3 | 2012 | Europe | Blood culture | 1/4 | No | Elevated CRP, fever | 56 | M |
| Vaxjo_4 | 2012 | Europe | Blood culture | 1/4 | Yes | Elevated CRP, shortness of breath | 89 | F |
| Vaxjo_5 | 2012 | Europe | Blood culture | 1/4 | No | Suspected aspiration pneumonia | 72 | M |
| Vaxjo_6 | 2012 | Europe | Blood culture | 1/4 | No | Fever, COPD | 87 | F |
| Vaxjo_7 | 2013 | Europe | Blood culture | 1/4 | No | Suspected sepsis | 60 | M |
| Vaxjo_8 | 2013 | Europe | Blood culture | – | – | – | – | – |
| Vaxjo_9 | 2013 | Europe | Blood culture | 2/4 | No | Elevated CRP | 85 | M |
| Vaxjo_10 | 2013 | Europe | Blood culture | 1/4 | Yes | Immunocompromised, fever | 65 | M |
| Vaxjo_11 | 2013 | Europe | Blood culture | 1/4 | Yes | Fever, elevated CRP | 50 | M |
| Vaxjo_12 | 2013 | Europe | Blood culture | 1/4 | No | Fever | 16 | F |
| Vaxjo_13 | 2013 | Europe | Blood culture | 1/4 | Yes | Elevated CRP | 83 | F |
| Orebro_14 | 2014 | Europe | Blood culture | 2/4 | No | Immunocompromised, fever | 65 | M |
| JMI_15 | 2006 | North America | Blood culture | – | Yes | – | 61 | M |
| JMI_16 | 2008 | Europe | Blood culture | – | No | – | – | – |
| Isolate ID | Year | Continent | Origin | Surgery performed | Median c.f.u./m2 | |||
| Vasteras_17 : 1–8 | 2011 | Europe | Peri-operative air sampling | TKA | 120 | |||
| Vasteras_18 : 1 | 2011 | Europe | Peri-operative air sampling | THA | 15 | |||
The environmental isolates (n=9) originated from active air sampling during one prosthetic knee joint operation (n=8; Vasteras_17 : 1–8) and one prosthetic hip joint operation (n=1; Vasteras_18 : 1) using a Sartorius MD-8 air scanner (Sartorius Mechatronics, Göttingen, Germany) in the operating field at the Department of Orthopaedics, Central Hospital Västerås, Sweden, in November 2011, as described previously [12]. Repetitive-sequence-based PCR (rep-PCR) typing and phylogenetic relationship analysis based on whole-genome sequence (WGS) data, was performed with the inclusion of reference strain CCUG 51270 T (CCUG, Department of Clinical Bacteriology, University of Gothenburg, Sweden).
Species identification
Species identification was performed using direct colony testing with MALDI-TOF MS (Microflex LT and Biotyper 3.1, Bruker Daltonics, Bremen, Germany). The cut-off for species identification was set at a score value ≥2.000. Species identification was verified by nucleotide sequence determination of a segment of the rpoB gene, performed as previously described for S. epidermidis [13].
Rep-PCR typing
DNA from colonies was extracted using an UltraClean Microbial DNA isolation kit (bioMérieux, Marcy-l′Etoile, France) following the manufacturer's instructions. The extracted DNA was amplified using the DiversiLab Staphylococcus DNA fingerprinting kit (bioMérieux). The PCRs were performed on a thermal cycler (GeneAmp PCR System 9700, Applied Biosystem, Foster City, CA) with conditions including an initial pre-incubation at 94 °C for 2 min; 35 cycles of 94 °C for 30 s, 45 °C for 30 s, and 70 °C for 90 s; and a final extension at 70 °C for 3 min.
The amplified rep-PCR products were separated by electrophoresis performed in a microfluidics DNA LabChip (bioMérieux) and detected with an Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA).
Analysis was performed with version 3.6 of the DiversiLab software package (bioMérieux) using the Kullback–Leibler method to calculate similarity. The typing report provided by the software included a dendrogram with a virtual gel image of the samples and a scatter plot (a two-dimensional representation of relative sample similarity). Isolates were categorized as indistinguishable (>97 % similarity and no banding differences), similar (>95 % similarity and 1–2 band differences) or different (<95 % similarity and ≥2 band differences).
Genome sequencing of S. pettenkoferi isolates
DNA from all isolates was extracted using a DNeasy Blood and Tissue kit as described by the manufacturer (Qiagen, Valencia, CA). For sequencing preparation on the MiSeq system, fragment libraries were constructed using the Nextera XT Kit (Illumina, San Diego, CA) followed by 251 bp paired-end sequencing on a MiSeq (Illumina) according to the manufacturer’s instructions. The sequencing reads were assembled using CLC Genomics Workbench 9.5 (Qiagen, Aarhus, Denmark) with default parameters to include only contigs ≥500 nt. The WGS data were aligned against the draft genome of S. pettenkoferi NZ_JVVL00000000.1 (strain 1286_SHAE) from the NCBI Reference Sequence Database (RefSeq) using the short-read alignment component of the Burrows–Wheeler aligner, as were two other available S. pettenkoferi draft genomes, NZ_JVAY00000000.1 (strain 589_SHAE) and NZ_AGUA00000000.1 (strain VCU012), from the RefSeq database using MUMmer. Each alignment was analysed for SNPs using NASP (https://github.com/TGenNorth/NASP), with exclusion of all SNPs that did not meet a minimum coverage of 10 or if the variant was present in ≥90 % of the base calls. SNPs identified in duplicated regions on the reference genome were removed. Phylogenetic relationships were reconstructed using the maximum-likelihood method implemented in PhyML (www.atgc-montpellier.fr/phyml/) using Smart Model Selection with 100 replicates on all included isolates. The Illumina data produced in this study were deposited in the European Nucleotide Archive under study IDs ERS1751294-ERS1751319.
Antibiotic susceptibility testing
Isolates were tested for susceptibility be usinga standardized disk diffusion method as defined by the European Committee on Antimicrobial Susceptibility Testing (EUCAST) [14], using antibiotic disks (Oxoid, Basingstoke, UK), Mueller–Hinton II agar 3.8 % (w/v; BD Diagnostic Systems, Sparks, MD) and the following antibiotics: cefoxitin (30 µg), fusidic acid (10 µg), clindamycin (2 µg), erythromycin (15 µg), gentamicin (10 µg), rifampicin (5 µg), trimethoprim-sulfamethoxazole (25 µg) and norfloxacin (10 µg). Categorization of the isolates into Susceptible Intermediate Resistant (SIR) category was performed according to EUCAST clinical breakpoints (www.eucast.org).
Detection of the mecA gene
All isolates were tested for presence of the mecA gene using a real-time PCR system (Light Cycler 2.0, Roche Diagnostics, GmbH, Mannheim, Germany) as previously described for S. aureus [15].
Determination of biofilm production
A microtitre plate (MTP) assay was used for the detection of biofilm production, as described previously [13]. S. epidermidis RP62A was used as the positive control and S. epidermidis ATCC 12228 as the negative one.
Results
Eleven S. pettenkoferi isolates retrieved from blood cultures were deemed probable contaminants, two (Vaxjo_9 and Orebro_14) were deemed clinically significant and data was not available for three isolates (Table 1).
Genomic relatedness based on rep-PCR verified by core genome phylogenetic analysis of WGS data
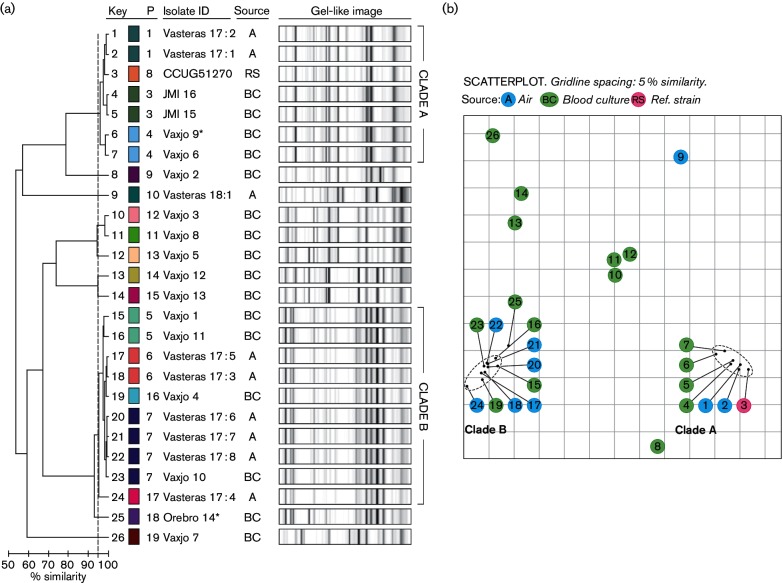
Repetitive-sequence-based typing generated several high intensity peaks at various molecular weights for S. pettenkoferi. The dendrogram (Fig. 1a) and scatterplot (Fig. 1b) depicted two main clades (A and B) comprising 72 % of the isolates. The seven isolates in clade A included two indistinguishable clinical isolates from Europe, two indistinguishable environmental isolates from a prosthetic knee joint operation in Västerås, two indistinguishable clinical isolates from Växjö [including the clinically significant isolate Vaxjo_9, obtained from an 85-year-old man admitted to the emergency ward with worsening of general condition and elevated C-reactive protein (CRP)] and reference strain CCUG51270. Clade B included five environmental isolates and five clinical isolates (including the clinically significant isolate Orebro_14, obtained from a 75-year-old man with chronic lymphocytic leukaemia admitted to the infectious diseases ward due to neutropenic fever). The presence/absence and intensity of peaks differed between clade A and B, as illustrated in Fig. 2. The environmental isolates from operation 1 belonged to both clades, while the single environmental isolate from operation 2 had a unique rep-PCR fingerprint pattern.
Fig. 1.
DiversiLab (DL) rep-PCR profiles of 26 S. pettenkoferi isolates. (a) Profiles and corresponding dendrogram obtained using the Kullback–Leibner method for correlation analysis. The similarity line (95 %) is presented as a light grey dotted vertical line. Each colour represents a single pattern (indistinguishable profiles). Isolates deemed clinically significant are marked with an asterisk. (b) Scatter plot of 26 S. pettenkoferi isolates. Each isolate is represented by its number corresponding to the DL profile, as presented in (a) ('key'). Colour represents origin of isolates [blue sampling (A); green, blood culture (BC); pinkreference strain (RS); pattern (P)].
Fig. 2.
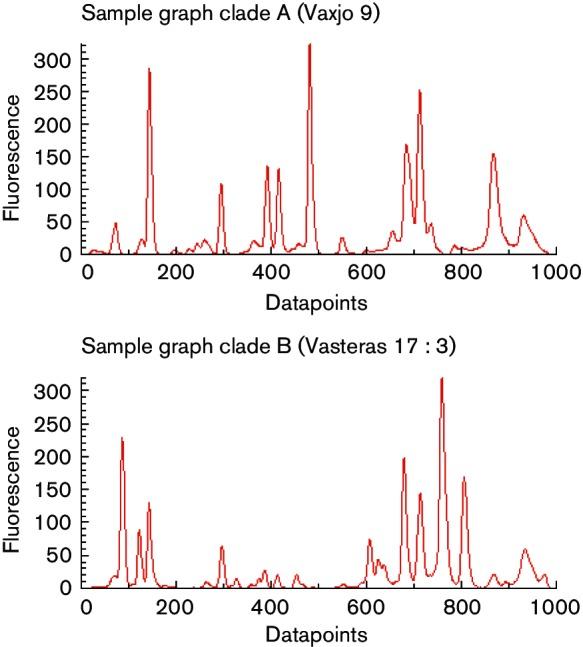
Examples of graphs of isolates belonging to clade A and B. Clade A is represented by the clinically significant isolate Vaxjo_9 and clade B is represented by the environmental isolate Vasteras 17 : 3.
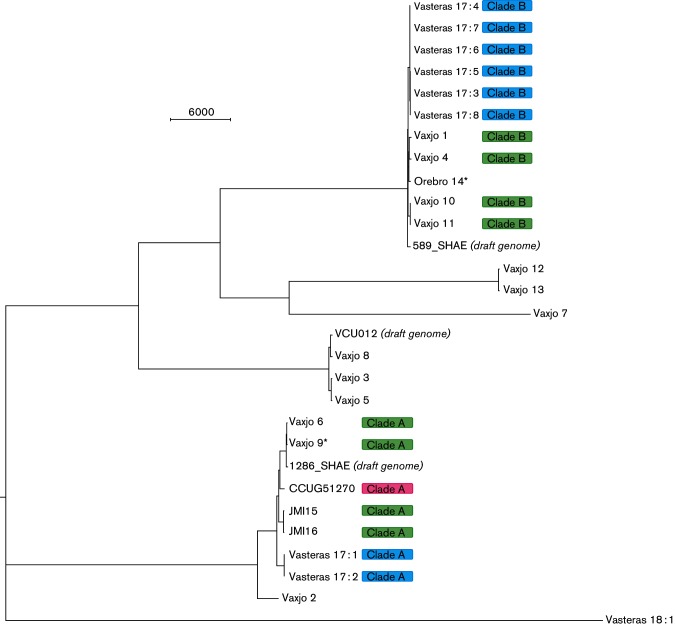
In order to verify the findings of the DiversiLab rep-PCR typing, core genome phylogenetic analysis based on WGS data was performed. About 157 000 SNPs in the core of about 80 % of the selected reference were identified. The inferred relationship using WGS data was in agreement with the dendrogram obtained by the DiversiLab rep-PCR and demonstrated two major clades (A and B), which each comprised isolates that clustered by rep-PCR (Fig. 3). Two smaller clades were also in agreement with the rep-PCR dendrogram, as was the presence of two singletons (Vaxjo_7 and Vasteras_18 : 1). Intra-clade diversity was <2000 and <700 SNPs for A and B, respectively. By contrast, clade A and clade B differed by >58 000 SNPs from each other. Environmental isolate Vasteras_18 : 1, with a unique rep-PCR fingerprint pattern, differed from isolates in clade A and clade B with a minimum of 85 000 SNPs.
Fig. 3.
Midpoint-rooted maximum-likelihood approximation using FastTree 2.1.5 on the 157 438 core SNP within 26 isolates of S. pettenkoferi aligned with three available S. pettenkoferi draft genomes. Isolates deemed clinically significant are marked with an asterisk. Rep-PCR clade is presented to the right of isolate ID. Colour represents origin of isolates (blue, air sampling; green, blood culture; pink, reference strain). Scale bar indicates the number of SNP distances. Range of intra-clade SNP distances: clade A 11–1872, clade B 0–672.
Antibiotic susceptibility testing
Three clinical isolates (including one clinically significant isolate, Vaxjo_9), but no environmental isolates, were resistant to cefoxitin (Table 2). The mecA gene was detected by PCR in two of these cefoxitin-resistant isolates. The mecC gene was not detected in the third isolate by a specific mecC PCR (personal communication M. Bergman Jungeström, Linköping, Sweden) nor according to whole genome sequence data. Three isolates (12.5 %) were multidrug-resistant (i.e. resistant to ≥3 antimicrobial groups); two clinical isolates (both mecA-positive) and one environmental isolate. All isolates were susceptible to rifampicin and gentamicin.
Table 2. Antibiotic susceptibility patterns of S. pettenkoferi isolates (n=25).
Antimicrobial susceptibility testing according to EUCAST guidelines. S, susceptible (white); R, resistant (light grey).
| Isolate ID | Year | Cefoxitin | Fusidic acid | Clindamycin | Erythromycin | Gentamicin | Rifampicin | TMP/SMX | Norfloxacin | |
|---|---|---|---|---|---|---|---|---|---|---|
| Vaxjo_1 | 2011 | R | S | S | S | S | S | S | S | |
| Vaxjo_2 | 2011 | S | S | S | S | S | S | S | S | |
| Vaxjo_3 | 2012 | S | R | S | S | S | S | S | S | |
| Vaxjo_4 | 2012 | S | S | S | S | S | S | S | S | |
| Vaxjo_5 | 2012 | S | S | S | S | S | S | R | S | |
| Vaxjo_6 | 2012 | R* | R | R | R | S | S | S | R | |
| Vaxjo_7 | 2013 | S | S | S | R | S | S | S | S | |
| Vaxjo_8 | 2013 | S | R | S | S | S | S | S | S | |
| Vaxjo_9 | 2013 | R* | S | R | R | S | S | S | R | |
| Vaxjo_10 | 2013 | S | S | S | S | S | S | S | S | |
| Vaxjo_11 | 2013 | S | S | S | S | S | S | S | S | |
| Vaxjo_12 | 2013 | S | S | S | S | S | S | S | S | |
| Vaxjo_13 | 2013 | S | S | S | S | S | S | S | S | |
| Orebro_14 | 2014 | S | S | S | S | S | S | S | S | |
| JMI_15 | 2006 | S | S | S | S | S | S | S | S | |
| JMI_16 | 2008 | S | S | S | S | S | S | S | S | |
| Vasteras_17 : 1 | 2011 | S | R | S | S | S | S | S | S | |
| Vasteras_17 : 2 | 2011 | S | R | R | R | S | S | S | S | |
| Vasteras_17 : 3 | 2011 | S | S | S | S | S | S | S | S | |
| Vasteras_17 : 4 | 2011 | S | S | S | S | S | S | S | S | |
| Vasteras_17 : 5 | 2011 | S | S | S | S | S | S | S | S | |
| Vasteras_17 : 6 | 2011 | S | S | S | S | S | S | S | S | |
| Vasteras_17 : 7 | 2011 | S | S | S | S | S | S | S | S | |
| Vasteras_17 : 8 | 2011 | S | S | S | S | S | S | S | S | |
| Vasteras_18 : 1 | 2011 | S | S | S | S | S | S | S | S | |
*Isolate mecA-positive.
Biofilm production
None of the clinical or environmental S. pettenkoferi isolates, nor the reference strain, were found to be biofilm-producing by the MTP assay.
Discussion
The aim of the study was to characterize clinical and environmental S. pettenkoferi isolates with regard to diversity and antibiotic susceptibility pattern. Rep-PCR and core genome phylogenetic analysis obtained by WGS data verified the presence of distinct clades, each comprising closely related S. pettenkoferi isolates from different geographical locations and origins. Substantial genomic differences were observed between the two major clades, but the overall resemblance to the reference chromosome of S. pettenkoferi in the NCBI Reference Sequence Database was high (~87 %, data not shown).
In this study, S. pettenkoferi was regarded as a probable pathogen in the minority of patients with positive cultures. This is in accordance with previous studies. Argemi et al. [5] found that 45 of 50 S. pettenkoferi isolates obtained from various cultures could be determined as not microbiologically relevant and only one isolate was regarded as clinically relevant. In another study from Korea [10], where six blood culture isolates of S. pettenkoferi were characterized, S. pettenkoferi was interpreted as a probable pathogen in only one case (septic shock and aspiration pneumonia in a patient with chronic alcohol abuse and diabetes mellitus). The other five findings were categorized either as contaminants or as colonization of indwelling catheters. Consequently, although the pathogenic potential of S. pettenkoferi may not yet be fully understood, it is likely to be low.
The clinically significant isolates in this study were closely related, by rep-PCR and analysis of WGS data, to isolates regarded as contaminants. This may indicate that they represent clones present as part of the normal flora on the skin. In the present study, several of the blood culture isolates deemed as contaminants were nosocomial. Genetically diverse S. pettenkoferi isolates were retrieved from intra-operative air sampling: two isolates (Vasteras_17 : 1–2) were closely related to the reference strain CCUG 51270 T and three isolates (Vasteras_17 : 6–8) were indistinguishable from a blood culture isolate (Vaxjo_10) from a different hospital and year, according to rep-PCR. Nosocomial spread of S. pettenkoferi has previously been reported from a French ICU [7].
Staphylococcus spp. preferably colonize moist areas of the skin [16], but the ecological niche of S. pettenkoferi remains to be clarified. The genetically related S. auricularis, which is believed to have the external auditory canal as its principal habitat [4], was not encountered during air sampling in operating theatres [12], which might have been expected if these species share habitats. In a study in a small animal clinic, aiming to investigate the potential transmission of methicillin-resistant staphylococci, two multidrug-resistant isolates of S. pettenkoferi were isolated from swabs taken from a cat cage, but none were found in swabs taken from pets (n=10) or employees (n=4) [17]. S. pettenkoferi has also been isolated from sampling of public restrooms in London [18]. Since S. pettenkoferi occurs as a rare contaminant of blood cultures obtained via puncture of the skin as well as in the air in operating theatres during surgery, it is probably part of the normal flora of the skin, at least in some individuals. However, an environmental source may also be possible.
Previous reports of clinical findings of S. pettenkoferi include a limited number of cases of osteomyelitis, bloodstream infections in immunocompromised hosts and bursitis [1–3, 7–10, 19]. S. pettenkoferi has not yet been reported to be associated with implant infections and then S. pettenkoferi isolates included in this study did not form biofilm phenotypically. Variable antibiotic susceptibility profiles for S. pettenkoferi have been reported. In a collection of 44 isolates, 43.2 % were resistant to oxacillin [5], but the reported frequency of resistance to oxacillin varies between 12.5 % (present study; cefoxitin screen) and 83 % [10]. In the present study, antimicrobial resistance of S. pettenkoferi was low, with only a few multidrug-resistant isolates found.
A limitation of this study is that it includes a relatively small number of isolates that were almost all obtained from the same country. However, the isolates originated from different counties, and comprised both clinical and environmental samples. To our knowledge, this is the first study of the genetic diversity of S. pettenkoferi isolates.
To conclude, in this study, S. pettenkoferi isolated from different geographical locations and origins demonstrated close genomic relatedness within clades, but with substantial genomic differences between the major clades. Isolates interpreted as clinically significant were closely related to environmental isolates, irrespective of geographic origin, as well as to clinical isolates regarded as contaminants. Identification of S. pettenkoferi in clinical samples should, in a majority of cases, most likely be regarded as a probable contamination, and its role as a possible pathogen in immunocompromised hosts remains to be clarified.
Funding information
The study was funded by Örebro County Council Research Committee, Örebro, Sweden, Centre for Clinical Research, Västerås and the County Council of Västmanland Research Fund. The funders played no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Acknowledgements
The authors would like to thank Erika Matuschek, Dept of Clinical Microbiology, Växjö, and Paul Rhomberg, JMI Laboratories (North Liberty, Iowa, USA), for their contribution with clinical isolates, and Christer Häggström, Hospital of Västmanland Västerås, for help with air sampling.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Ethical statement
Clinical bacterial isolates from human blood cultures were included in this study. Only subcultured bacterial isolates were stored, no tissue material or other biological material from patients. Swedish law does not require ethical approval for work with bacterial isolates from humans. Sparse clinical data was provided together with the isolates, but identification of patients was not possible.
Footnotes
Abbreviations: CoNS, coagulase-negative staphylococci; MTP, microtitre plate; rep-PCR, repetitive-sequence-based PCR; WGS, whole-genome sequence.
References
- 1.Trülzsch K, Rinder H, Trcek J, Bader L, Wilhelm U, et al. "Staphylococcus pettenkoferi," a novel staphylococcal species isolated from clinical specimens. Diagn Microbiol Infect Dis. 2002;43:175–182. doi: 10.1016/S0732-8893(02)00399-1. [DOI] [PubMed] [Google Scholar]
- 2.Loïez C, Wallet F, Pischedda P, Renaux E, Senneville E, et al. First case of osteomyelitis caused by "Staphylococcus pettenkoferi". J Clin Microbiol. 2007;45:1069–1071. doi: 10.1128/JCM.02328-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Morfin-Otero R, Martínez-Vázquez MA, López D, Rodríguez-Noriega E, Garza-González E. Isolation of rare coagulase-negative isolates in immunocompromised patients: Staphylococcus gallinarum, Staphylococcus pettenkoferi and Staphylococcus pasteuri. Ann Clin Lab Sci. 2012;42:182–185. [PubMed] [Google Scholar]
- 4.Becker K, Heilmann C, Peters G. Coagulase-negative staphylococci. Clin Microbiol Rev. 2014;27:870–926. doi: 10.1128/CMR.00109-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Argemi X, Riegel P, Lavigne T, Lefebvre N, Grandpré N, et al. Implementation of matrix-assisted laser desorption ionization-time of flight mass spectrometry in routine clinical laboratories improves identification of coagulase-negative staphylococci and reveals the pathogenic role of Staphylococcus lugdunensis. J Clin Microbiol. 2015;53:2030–2036. doi: 10.1128/JCM.00177-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Trülzsch K, Grabein B, Schumann P, Mellmann A, Antonenka U, et al. Staphylococcus pettenkoferi sp. nov., a novel coagulase-negative staphylococcal species isolated from human clinical specimens. Int J Syst Evol Microbiol. 2007;57:1543–1548. doi: 10.1099/ijs.0.64381-0. [DOI] [PubMed] [Google Scholar]
- 7.Mihaila L, Defrance G, Levesque E, Ichai P, Garnier F, et al. A dual outbreak of bloodstream infections with linezolid-resistant Staphylococcus epidermidis and Staphylococcus pettenkoferi in a liver intensive care unit. Int J Antimicrob Agents. 2012;40:472–474. doi: 10.1016/j.ijantimicag.2012.06.014. [DOI] [PubMed] [Google Scholar]
- 8.Mammina C, Bonura C, Verde MS, Fasciana T, Palma DM. A fatal bloodstream infection by Staphylococcus pettenkoferi in an intensive care unit patient. Case Rep Crit Care. 2011;2011:1–3. doi: 10.1155/2011/612732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Song SH, Park JS, Kwon HR, Kim SH, Kim HB, et al. Human bloodstream infection caused by Staphylococcus pettenkoferi. J Med Microbiol. 2009;58:270–272. doi: 10.1099/jmm.0.004697-0. [DOI] [PubMed] [Google Scholar]
- 10.Park S, Chung HS, Lee M. Clinical and microbiological characteristics of six Staphylococcus pettenkoferi isolates from blood samples. Ann Lab Med. 2015;35:250–253. doi: 10.3343/alm.2015.35.2.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cornaglia G, Courcol R, Herrman J-L, Kahlmeter G, Peigue-Lafeuille H, et al. European Manual of Clinical Microbiology. 1st ed. Epernay, France: European Society of Clinical Microbiology and Infectious Diseases; 2012. (editors) [Google Scholar]
- 12.Månsson E, Hellmark B, Sundqvist M, Söderquist B. Sequence types of Staphylococcus epidermidis associated with prosthetic joint infections are not present in the laminar airflow during prosthetic joint surgery. APMIS. 2015;123:589–595. doi: 10.1111/apm.12392. [DOI] [PubMed] [Google Scholar]
- 13.Hellmark B, Söderquist B, Unemo M, Nilsdotter-Augustinsson Å. Comparison of Staphylococcus epidermidis isolated from prosthetic joint infections and commensal isolates in regard to antibiotic susceptibility, agr type, biofilm production, and epidemiology. Int J Med Microbiol. 2013;303:32–39. doi: 10.1016/j.ijmm.2012.11.001. [DOI] [PubMed] [Google Scholar]
- 14.Matuschek E, Brown DF, Kahlmeter G. Development of the EUCAST disk diffusion antimicrobial susceptibility testing method and its implementation in routine microbiology laboratories. Clin Microbiol Infect. 2014;20:O255–O266. doi: 10.1111/1469-0691.12373. [DOI] [PubMed] [Google Scholar]
- 15.Berglund C, Mölling P, Sjöberg L, Söderquist B. Predominance of staphylococcal cassette chromosome mec (SCCmec) type IV among methicillin-resistant Staphylococcus aureus (MRSA) in a Swedish county and presence of unknown SCCmec types with Panton-Valentine leukocidin genes. Clin Microbiol Infect. 2005;11:447–456. doi: 10.1111/j.1469-0691.2005.01150.x. [DOI] [PubMed] [Google Scholar]
- 16.Grice EA, Segre JA. The skin microbiome. Nat Rev Microbiol. 2011;9:244–253. doi: 10.1038/nrmicro2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weiss S, Kadlec K, Fessler AT, Schwarz S. Identification and characterization of methicillin-resistant Staphylococcus aureus, Staphylococcus epidermidis, Staphylococcus haemolyticus and Staphylococcus pettenkoferi from a small animal clinic. Vet Microbiol. 2013;167:680–685. doi: 10.1016/j.vetmic.2013.07.036. [DOI] [PubMed] [Google Scholar]
- 18.Mkrtchyan HV, Russell CA, Wang N, Cutler RR. Could public restrooms be an environment for bacterial resistomes? PLoS One. 2013;8:e54223. doi: 10.1371/journal.pone.0054223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.D'Azevedo PA, Comin G, Cantarelli V. Characterization of a new coagulase-negative Staphylococcus species (Staphylococcus pettenkoferi) isolated from blood cultures from a hospitalized patient in Porto Alegre, Brazil. Rev Soc Bras Med Trop. 2010;43:331–332. doi: 10.1590/S0037-86822010000300023. [DOI] [PubMed] [Google Scholar]