Abstract
The diagnostic criteria for CLL rely on morphology and immunophenotype. Current approaches have limitations affecting reproducibility and there is no consensus on the role of new markers. The aim of this project was to identify reproducible criteria and consensus on markers recommended for the diagnosis of CLL. ERIC/ESCCA members classified 14 of 35 potential markers as “required” or “recommended” for CLL diagnosis, consensus being defined as >75% and >50% agreement, respectively. An approach to validate “required” markers using normal peripheral blood was developed. Responses were received from 150 participants with a diagnostic workload >20 CLL cases per week in 23/150 (15%), 5–20 in 82/150 (55%), and <5 cases per week in 45/150 (30%). The consensus for “required” diagnostic markers included: CD19, CD5, CD20, CD23, Kappa, and Lambda. “Recommended” markers potentially useful for differential diagnosis were: CD43, CD79b, CD81, CD200, CD10, and ROR1. Reproducible criteria for component reagents were assessed retrospectively in 14,643 cases from 13 different centers and showed >97% concordance with current approaches. A pilot study to validate staining quality was completed in 11 centers. Markers considered as “required” for the diagnosis of CLL by the participants in this study (CD19, CD5, CD20, CD23, Kappa, and Lambda) are consistent with current diagnostic criteria and practice. Importantly, a reproducible approach to validate and apply these markers in individual laboratories has been identified. Finally, a consensus “recommended” panel of markers to refine diagnosis in borderline cases (CD43, CD79b, CD81, CD200, CD10, and ROR1) has been defined and will be prospectively evaluated. © 2017 International Clinical Cytometry Society
Keywords: chronic lymphocytic leukemia, flow cytometry, diagnosis
BACKGROUND
The WHO, IWCLL, and NCCN diagnostic criteria for CLL is based on the morphology and immunophenotype of the neoplastic B‐cells with co‐expression of CD19, CD5, CD23, with weak CD20 and monoclonal surface immunoglobulin (sIg) expression 1, 2, 3. Although there are several recurrent molecular abnormalities present in CLL, none is specific for CLL 4 and therefore immunophenotyping still plays a central role in the diagnosis of CLL.
The current diagnostic criteria have some limitations affecting reproducibility, in particular relating to flexibility in the requirement for each marker to be present or absent as well as in the required expression level of each marker. The WHO definition states that CLL/SLL cells “usually co‐express CD5 and CD23” and that “using flow cytometry, the tumor cells express dim surface IgM/IgD, CD20, CD22, CD5, CD19, CD79a, CD23, CD43, and CD11c (weak). CD10 is negative and FMC7 and CD79b are usually negative or weakly expressed in typical CLL. It is also considered that “some cases may have an atypical immunophenotype (e.g. CD5‐ or CD23‐, FMC7+ or CD11c+, strong sIg, or CD79b+)” 1. In turn, the current IWCLL guidelines also permit variation in markers expression levels: “CLL cells co‐express the T‐cell antigen CD5 and B‐cell surface antigens CD19, CD20, and CD23. The levels of sIg, CD20, and CD79b are characteristically low compared with those found on normal B cells. Each clone of leukemia cells is restricted to expression of either kappa or lambda immunoglobulin light chains. Variations of the intensity of expression of these markers may exist and do not prevent inclusion of a patient in clinical trials for CLL” 2.
Although some degree of flexibility is required to ensure that CLL diagnostic criteria are widely applicable, this can pose problems to the reproducibility of diagnostic criteria, particular if a scoring system that may permit absence of either CD5 or CD23 is employed 5, 6. In addition, several markers such as CD200 7, 8 and ROR1 (9–12) may contribute to the diagnosis of CLL and related disorders. However, there is no consensus yet on how such markers should be incorporated into diagnostic algorithms. Moreover, although the addition of new markers to established diagnostic panels may improve diagnostic precision, this would require a systematic and well‐designed approach.
The primary aim of this project was to achieve consensus on the minimum set of markers required for the diagnosis of CLL and develop a reproducible approach to validate and apply these markers in different laboratories. A secondary aim was to identify additional markers deserving prospective evaluation.
METHODS
Identification of Consensus on Required and Recommended Marker Panels
ERIC/ESCCA members were invited to participate in a survey to classify 35 potential flow cytometry markers as being “required,” “recommended,” “suggested,” “uninformative,” or “not sure” for the diagnosis of CLL. The full survey is shown in Supporting Information. Results from respondents indicating that they did not work in a diagnostic laboratory or hospital clinic setting, or who indicated that their institution did not perform flow cytometry in the diagnosis or monitoring of CLL were excluded from analysis. The 35 markers were selected based on the inclusion in the diagnostic panels reported in the WHO classification 1, IWCLL guidelines 2, Euroflow B‐cell panel 8, or if a pubmed search for “differential diagnosis chronic lymphocytic leukemia CD” identified a reagent in publications by two or more different groups (Fig. 1). Consensus for a marker to be required for CLL diagnosis needed >75% of participants indicating that the marker was required, while a marker was put forward to review if ≥ 50% of participants considered the marker to be “recommended” or “required.” Positive and negative control populations in normal peripheral blood and the relative signals required for acceptable markers were derived from the previous ERIC project for optimizing CLL MRD 13 or defined through literature search and review by participants.
Figure 1.
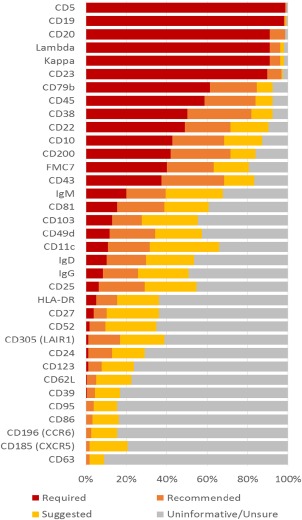
The percentage of participants ranking each marker as required or recommended for evaluation in the diagnosis of CLL. [Color figure can be viewed at wileyonlinelibrary.com]
The definition of “weak” expression requires a threshold that is reproducibly lower than normal expression i.e. lower than acceptable variation due to assay imprecision or due to changes associated with sample stability. The ICCS/ICSH guidelines for validation of cell‐based fluorescence assays 14 recommend assay imprecision ideally below 10% CV (although this may be higher in some settings) while specimen stability is determined by identifying the latest time point at which repeat testing of ≥5 samples shows up to 20% change from baseline. Therefore, difference in fluorescence intensity of <20% may not be reliably determined, so weak expression was defined as a median fluorescence intensity at least 20% lower than the median expression level by normal peripheral blood B‐cells. As laboratories use different reagents, instrumentation and procedures, each laboratory was requested to determine their own reference range to be used for definition of weak expression.
Consensus for the proposed diagnostic panel specification was reached by approval of all participating authors followed by presentation of the proposal at ERIC and ESCCA meetings and open consultation on the final document distributed to all ERIC and ESCCA members three months prior to submission.
Retrospective Evaluation
Survey participants were requested to retrospectively assess the proposed criteria based on the required markers (Table 1) by providing the number of: total B‐LPD cases evaluated; CD5+ B‐LPD cases; cases meeting the proposed criteria and diagnosed as CLL; cases not meeting the proposed criteria and diagnosed with another B‐LPD e.g. mantle cell lymphoma; cases not meeting the proposed criteria and diagnosed with CLL; cases not meeting the proposed criteria with insufficient material to make a final diagnosis or reported to be unclassifiable based on available data.
Table 1.
Required and Recommended Markers for Use in the Diagnosis of CLL with Reagent Specification Based on Expression Patterns in Normal Peripheral Blood.
| Control population in normal peripheral blood | Minimum relative fluorescence intensity of positive and negative control | ||||
|---|---|---|---|---|---|
| Inclusion in diagnostic panel | Antigen | Expression in CLL (% pos vs. control) | Positive | Negative | populations (preferred) |
| Required | CD19 | Positive (>95%) | CD20+ B‐cells | CD3+ T‐cells | ≥10a |
| CD5 | Positive (>20%) | CD3+ T‐cells | CD19+ B‐cells | ≥30 (≥65) | |
| CD23 | Positive (>20%) | CD23+ B‐cells | T‐cells | ≥5a | |
| CD20 | Weak | CD19+ B‐cells | CD3+ T‐cells | ≥10 (≥20) | |
| Igκ Igλ | Weak & restricted | CD20+ B‐cells | CD3+ T‐cells | ≥5a | |
| Recommended | CD43 | Positive (>20%) | CD3+ T‐cells | CD20+ B‐cells | ≥15 (≥40) |
| CD79b | Weak | CD20+ B‐cells | CD3+ T‐cells | ≥15 (≥30) | |
| CD81 | Weak | CD3+ T‐cells | Granulocytes | ≥12 (≥20) | |
| CD200 | Positive (>20%) | CD19+ B‐cells | CD3+ T‐cells | ≥5a | |
| CD10 | Negative (<20%) | Granulocytes | T‐cells | ≥10a | |
| ROR1 | Positive (>20%) | B‐progenitors | T‐cells | ≥5a | |
Required, consensus from >75% of participants. Recommended, consensus from >50% of participants with the following exceptions determined by the steering committee and confirmed by further consensus: exclusion of FMC7 (epitope of CD20) 15, CD38 & CD45 (used for prognostic information and gating orientation but not specifically required for diagnosis), and inclusion of ROR1 which is closely associated with CLL 9, 10 but diagnostic antibodies were not widely available at the time of the survey.
Definition of weak: median fluorescence intensity at least 20% [identified as the minimum measurable difference based on ICSH/ISLH guideline recommendations for acceptable variation due to assay imprecision and specimen stability 14] lower than the median expression level by normal peripheral blood B‐cells. Each laboratory was requested to determine their own reference range.
Specifically validated otherwise consensus, defined as approval of all contributing authors with no disagreement on open consultation by ERIC/ESCCA members. Values refer to the relative signal on positive versus negative control populations required to achieve optimal separation of CLL cells from normal B‐cells 13.
Assessing Reagent/Instrument Quality
A gating strategy to identify the expression levels of component markers on normal peripheral blood lymphocytes was developed (Fig. 2). Participants providing retrospective evaluation were requested to assess the gating strategy on 10 cases in which the B‐cells were polyclonal. The median fluorescence intensity for the relevant markers on defined positive and negative control populations were recorded by 11 different laboratories and returned for central analysis where the relative fluorescence intensity signal value was calculated. A borderline results was defined as 1–3/10 cases below the minimum acceptable relative signal target value. A suboptimal results was defined as >3/10 cases below the minimum relative signal target value.
Figure 2.
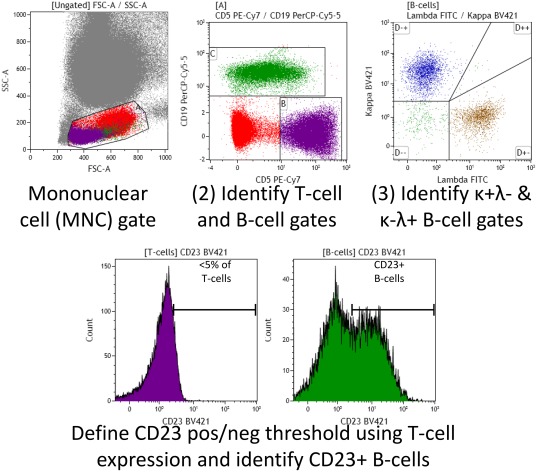
Simple gating strategy for defining positive and negative internal control populations to assess the relative signal on markers required for diagnosis according to the consensus criteria. [Color figure can be viewed at wileyonlinelibrary.com]
RESULTS AND DISCUSSION
Identifying Consensus on the Markers Required or Recommended for the Diagnosis of CLL
ERIC/ESCCA members were invited to identify markers that are “required” or “recommended” for the diagnosis of CLL from a potential panel of 35 markers. Responses were received from 154 members of which 150/154 were involved in CLL diagnosis (100 were diagnostic laboratory staff, 14 were clinicians and 36 were involved in both the laboratory and clinical diagnostic process). Responses from individuals not involved in CLL diagnosis were excluded from further analysis. The diagnostic workload was >20 cases per week in 23/150 (15.3%), 5–20 in 82/150 (54.7%), and <5 cases per week in 45/150 (30%). The survey participant consensus was that the minimum diagnostic panel should include: CD19, CD5, CD20, CD23, Kappa, and Lambda (i.e. “required”). Survey participants recommended that the following markers may also be of value CD22, CD38, CD45, FMC7, CD79b, CD10, CD43, and CD200. The complete list of markers and participant responses are shown in Figure 1.
The minimum required diagnostic panel was put forward for identification of component marker specification that could be used to assess reagent and laboratory quality (see below). The recommended marker panel was reviewed by the steering committee (AR, PH, MH, PG, and EM) and it was proposed that the application of CD22, CD38, CD45, and FMC7 is left to the individual laboratory preference (i.e. “not recommended”) because of a variety of reasons. In detail: FMC7 is an epitope of CD20 (15), the inclusion of both markers being redundant 8; similarly the level of CD22 expression is closely correlated with CD20 (13); CD45 is used for identification of leukocyte subsets and provides a backbone to many gating strategies but is not essential to identify CLL cells 8; CD38 is heterogeneously expressed in CLL 16, 17, 18 difficult to standardize 19 and it is also difficult to identify control populations with stable expression levels; therefore it was proposed that the application of CD38 in diagnosis and prognosis is determined by individual laboratories.
The markers sIgM, CD81, CD103, CD49d, CD11c, IgD, IgG, and CD25 were recommended by 20–40% of participants and therefore their application is best determined by individual laboratories with the exception of CD81 which has been extensively validated in detection of MRD 13, 20, 21, therefore understanding the expression profile prior to treatment may be informative for differential diagnosis. ROR1 was not on the initial survey because at the time of preparation there was limited access to commercial reagents. ROR1 was initially identified by gene expression profiling studies as a CLL‐specific marker and the role of protein expression in diagnosis and prognosis has been analyzed in several different studies 9, 10, 11, 12 and may be particularly informative in the discrimination between CLL and CD5+ post‐germinal center B‐cell disorders 22. Based on all these considerations, the markers recommended for additional analysis were: CD43, CD79b, CD81, CD200, CD10, & ROR1.
For each marker in the required and recommended panels, a positive and negative control population that could be readily recognized in normal peripheral blood was identified, with the exception of ROR1 for which B‐progenitors in the bone marrow are the only normal positive control. A minimum and recommended relative signal was also determined either by consensus or by using data from the CLL MRD project which identified the relative signal on positive versus negative control populations required to achieve optimal separation of CLL cells from normal B‐cells 13. The required and recommended markers with relevant positive and control populations and expected relative signals are shown in Table 1. This information was distributed to all ERIC and ESCCA members for consultation in order to confirm consensus.
Retrospective Application of the Minimum Required Panel Using the Proposed Specification
Survey participants involved in the diagnosis of CLL were requested to retrospectively assess the proposed criteria shown in Table 1, and 13/150 responded with the results shown in Table 2. The required criteria were assessed retrospectively in 14,643 cases referred for diagnosis of a potential B‐LPD, of which 11,721 were diagnosed with a CD5+ B‐LPD. Central laboratories for clinical trials identified cases which had been submitted for a CLL trial i.e. considered to have a diagnosis of CLL by another center, as “trial” cases (2,427/11,721) while all other cases were classified as “primary referral” (9,294/11,721).
Table 2.
Retrospective Assessment of the Proposed Criteria for Diagnosis of CLL
| Not meeting the proposed criteria | |||||
|---|---|---|---|---|---|
| Requires MDT or trial‐total CD5+ B‐ specific decision | |||||
| Total CD5+ B‐LPD diagnoses | Meeting the proposed criteria and diagnosed with CLL | Other diagnosis, e.g. Mantle cell lymphoma | Not CLL or not specified | Diagnosed with CLL | |
| Primary referral | 9,294 | 7,379 (79.4%) | 1,025 (11%) | 639 (6.9%) | 251 (2.7%) |
| Trial | 2,427 | 2,267 (93.4%) | 54 (2.2%) | 93 (3.8%) | 13 (0.5%) |
The high proportion (7,379/9,294, 79%) of primary referral cases met the proposed criteria and obtained a diagnosis of CLL. A clear alternative diagnosis (e.g. mantle cell lymphoma) was made in 54% (1,025/1,915) of primary referral cases that did not meet the proposed criteria. For primary referrals not meeting the criteria and not having a clear alternative diagnosis, a final diagnosis of CLL was made using the diagnostic unit's current practice in 2.7% of total CD5+ LPD cases (n = 251, 28% of cases not meeting the proposed criteria); there was insufficient material or data on the final diagnosis was not available in 6.9% of total CD5+ LPD cases (n = 639, 72% of the 890 cases not meeting the proposed criteria) of cases. For primary referral cases, there was concordance in 97.2% (9,043/9,294, comprising 7,379 diagnosed with CLL, 1,025 diagnosed with another non‐CLL B‐LPD and 639 non‐diagnostic with both approaches) using the reproducible criteria compared to each laboratory's current practice.
The vast majority (2,267/2,427, 93.4%) of trial referrals i.e. cases previously considered to have a diagnosis of CLL at another center, were confirmed to meet the proposed criteria by the referral center (classed as true positive). There were 160 cases that did not meet the proposed CLL criteria, of which 54/160 had a clear alternative diagnosis and 93/160 were considered ineligible for the trial due to lack of sufficient diagnostic material or non‐specific diagnosis (true negative n = 147/2,427, 6.1%). 13/160 were finally classified as CLL (false negative) and included in the relevant trial (false negative n = 13/2,427, 0.5%). Based on cases referred for entry into a clinical trial and using each center's current practice, the proposed criteria would have a negative predictive value of 92% with a positive predictive value, specificity and sensitivity for the diagnosis of CLL of >99%.
Evaluation of a Pilot Study to Assess Reagent and Instrument Quality
The 13 participants providing retrospective data were invited to assess the proposed specifications for the six markers identified as “required” for diagnosis of which 11/13 responded. Using a simple gating strategy (Fig. 2), participating centers evaluated 10 cases with polyclonal B‐cells. The details and performance characteristics for the individual markers used in the 11 different centers are shown in Table 3. Each center used a different combination of reagents but the performance characteristics were optimal in 525/600 (88%) for individual reagents. Only 1/11 centers obtained optimal results (i.e. the relative signal was above the minimum relative fluorescence intensity of positive and negative control populations) for all six markers in all 10 cases. A further 3/11 centers had some borderline signals (defined as 1–3 cases with results below the relative signal target value), in most cases likely to reflect the samples rather than instrument/reagent quality (e.g. two cases with weak CD23 expression on the polyclonal B‐cells at center 4). Sub‐optimal signals (defined as >3 cases with results below the relative signal target value) were identified with respect to an individual marker in 7/11 centers. In one case, this reflected a limitation of the proposed gating strategy for centers using a multiplex approach (CD20 at center 3). Several centers had sub‐optimal results for the CD5 reagent, which may indicate that this marker requires a more stringent specification. The other sub‐optimal results may be due to one or more of several factors (e.g. clone, fluorochrome, manufacturer, equipment, and operating procedure) and do not necessarily relate to the reagent used. Optimizing and standardizing each component of the process can be labor‐intensive and should be specifically addressed in order to improve the overall quality of CLL diagnosis, even when using the most “basic” markers. The relatively simple global approach developed by ERIC/ESCCA to assess the CLL diagnostic panel is applicable to a variety of reagent and instrument suppliers and can easily identify potential problems or confirm acceptable performance in individual laboratories. In addition, it can be utilized in the future as the basis for a more homogeneous and standardized diagnostic approach in CLL, allowing cross‐center comparison and reproducibility both in clinical trials as well as daily diagnostic procedures.
Table 3.
Assessment of the Reagents and Instrument Set‐up in Different Centers by Evaluating the Relative Signal of Required Diagnostic Markers on Control Samples
| Antigen | CD19 | CD20 | CD5 | Kappa | Lambda | CD23 |
|---|---|---|---|---|---|---|
| Relative signal target value | ≥10 | ≥10 | ≥30 | ≥5 | ≥5 | ≥5 |
| Center 1 | 225 (123–479) HD37 RPE‐Cy5 | 127 (51.9–183) L27 FITC | 56.3b (16.2–5,892) DK23 APC | 24.4 (12.6–87.6) Polyclonal FITC | 100 (44.8–302) Polyclonal PE | 11 (7.4–17.9) MHM6 FITC |
| Center 2 | 5,462 (4,291‐6,393) LT19 APC | 64.8 (36.6–103) 2H7 APE‐eF780 | 41.1a (17.7–57.2) L17F12 V450 | 17.1a (4.9–37.6) G20–193 APC‐H7 | 2.9b (2.1–4.9) 1–155‐2 APC | 4b (3.1–6.8) Tu1 FITC |
| Center 3 | 12,126 (85.1–14,264) J3–119 PE‐Cy7 | 5.4b (2.5–7.1) L27 V450 | 44.2a (2.8–102) L17F12 PerCP‐Cy5.5 | 20.2 (7.1–55.5) Polyclonal PE | 35.8 (8.4–116) Polyclonal FITC | 43.2a (0.8‐1,670) MHM6 FITC |
| Center 4 | 17.9a (5.6–23.5) SJ25C1 PerCP‐Cy5.5 | 175 (102–306) L27 FITC | 237 (52.8–368) L17F12 PE | 35.6 (12.6–60) TB28‐2 FITC | 430 (148–612) 1–155‐2 PE | 49a (2.5–223) EBVCS‐5 PE |
| Center 5 | 16.5 (11.2–18.8) SJ25C1 PerCP‐Cy5.5 | 24.6 (16.7–30.2) L27 APC‐H7 | 42.9a (15.1–56.7) L17F12 PE‐Cy7 | 22.6 (10.3–65.1) G20–193 BV421 | 17.5 (10.3–24.2) JDC‐12 FITC | 18.7 (8.6–31.7) M‐L233 BV421 |
| Center 6 | 56.8 (32.8–81.9) J3–119 PE‐Cy7 | 2,812 (398‐5,030) 2H7 PacBlue | 37.2a (24.4–105) L17F12 PerCP‐Cy5.5 | 19.7 (11.4–65.6) Polyclonal FITC | 74.4 (13.6–317) Polyclonal PE | 15.4 (9.7–39.3) MHM6 FITC |
| Center 7 | 106 (89.9–175) SJ25C1 APC | 53.6 (41.2–67.4) L27 PerCP | 26.2b (17.9–39) L17F12 FITC | 22.1 (6.9–45.1) TB28‐2 FITC | 149 (72.2–287) 1–155‐2 PE | 16.9 (8.6–35) EBVCS‐5 PE |
| Center 8 | 217 (130–234) J3–119 PE‐Cy7 | 82 (58.8–145) 2H7 Pacific Blue | 88.6 (51–123) BL1a APC | 25.3 (10.7–80.1) Polyclonal PE | 19.6 (7.4–74.8) Polyclonal FITC | 10.3 (5.8–14.1) 9P25 FITC |
| Center 9 | 16.3a (5.5–130) J3–119 PE‐Cy7 | 29.9 (18.3–58.7) B‐Ly1 FITC | 5.4b (2.4–45.6) BL1a PE | 12.3a (4.7–29.7) Polyclonal FITC | 46.6 (6.5–75.5) Polyclonal PE | 19.1 (9.8–48.4) 9P25 FITC |
| Center10 | 31.6 (22.6–41.7) J3–119 ECD (Coulter) | 82.1 (38.4–119) B9E9 Pacific Blue | 16.6b (3–31.5) BL1a APC‐AF750 | 6.1a (1.7–11.2) Polyclonal FITC | 18 (12.3–37) Polyclonal PE | 9.1 (7.1–13.6) 9P25 APC‐AF700 |
| Center 11 | 142 (20.2–10,558) SJ25C1 APC | 99.5 (46.9–240) L27 FITC | 24.1b (9.7–45.8) L17F12 PerCP‐Cy5.5 | 87.1 (33.3‐3,398) TB28‐2 FITC | 160 (55.4‐2,821) 1–155‐2 PE | 70.6 (31.3–290) EBVCS‐5 PE |
The signal for each marker on the internal positive and negative controls was determined using a simple gating strategy applied to ten control cases (see Fig. 2). The table shows the median relative signal (range) for the cases above the clone and fluorochrome (supplier).
Indicates that the results were sub‐optimal in 1–3 of the 10 cases, typically reflecting issues with individual samples rather than instrument/reagent quality.
Indicates that the results did not meet the specified criteria in >3/10 cases due to one or more of several factors such as clone, fluorochrome, manufacturer, equipment, or operating procedures as well as factors related to the evaluation procedure such as a limitation in the proposed gating strategy with multiplex approaches (e.g. CD20 at center 3).
SUMMARY
CLL is one of the most common diagnoses made by hematology–oncology laboratories. Flow cytometry plays a central role in diagnosis but differential diagnosis remains an issue in a small proportion of cases. Due to the lack of a pathognomonic molecular abnormality in CLL there is no a gold‐standard for its diagnosis. Also, characteristic immunophenotypic features, such as weak expression of sIg and CD20, are difficult to define in a reproducible fashion, thus making it difficult to ensure consistent diagnosis across laboratories.
This study demonstrates clear consensus on the minimum set of markers required for the diagnosis of CLL: CD19, CD5, CD20, CD23, Kappa, and Lambda. The identification of positive and negative control populations in normal peripheral blood, as well as uniform performance criteria facilitate the evaluation of the diagnostic quality and a reproducible diagnosis. The approach piloted in this study provides a comprehensive evaluation of the components of the diagnostic flow cytometry process including technical equipment and specific combinations and concentrations of reagents allowing a reproducibility and comparability among different laboratories. The results demonstrate that this goal has still to be consistently achieved, particularly with respect to the CD5 reagents.
A further component of this project was to identify a panel of reagents that may improve differential diagnosis. The identification of markers that could contribute to differential diagnosis is confounded not only by the lack of a diagnostic gold‐standard, but also because new markers are often assessed along with others that may also contribute to the differential diagnosis. In this regards, it is recommended that in addition to the minimum panel, reference centers and those involved in research assess CD43, CD79b, CD81 (required also for subsequent disease monitoring and MRD assessment), as well as CD200, CD10, & ROR1 (useful for differential diagnosis of CLL vs. mantle cell lymphoma, and germinal‐center B‐LPD vs. post‐germinal center B‐LPD, respectively) 8, 22, 23. This should provide a stable platform for evaluating the contribution of cellular markers to the diagnosis and prognosis in CLL.
Supporting information
Additional Supporting Information may be found online in the supporting information tab for this article.
Supporting Information
ACKNOWLEDGMENTS
The work was partially supported by MSMT CR project CEITEC 2020 (LQ1601) and project AZV MZCR 15–30015 A.
How to cite this article: Rawstron AC, Kreuzer K‐A, Soosapilla A, Spacek M, Stehlikova O, Gambell P, McIver‐Brown N, Villamor N, Psarra K, Arroz M, Milani R, de la Serna J, Cedena MT, Jaksic O, Nomdedeu J, Moreno C, Rigolin GM, Cuneo A, Johansen P, Johnsen HE, Rosenquist R, Utoft Niemann C, Kern W, Westerman D, Trneny M, Mulligan S, Doubek M, Pospisilova S, Hillmen P, Oscier D, Hallek M, Ghia P, and Montserrat E. Reproducible Diagnosis of Chronic Lymphocytic Leukemia by Flow Cytometry: An European Research Initiative on CLL (ERIC) & European Society for Clinical Cell Analysis (ESCCA) Harmonisation Project. Cytometry Part B 2018; 94B: 121–128.
LITERATURE CITED
- 1. Swerdlow SH, Campo E, Harris NL, Jaffe ES, Pileri SA, Stein H, Thiele J, Vardiman JW. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues. WHO Press, 2008. [Google Scholar]
- 2. Hallek M, Cheson BD, Catovsky D, Caligaris‐Cappio F, Dighiero G, Döhner H, Hillmen P, Keating MJ, Montserrat E, Rai KR, Kipps TJ. Guidelines for the diagnosis and treatment of chronic lymphocytic leukemia: a report from the International Workshop on Chronic Lymphocytic Leukemia updating the National Cancer Institute‐Working Group 1996 guidelines. Blood 2008;111:5446–5456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.NCCN ‐ Evidence‐Based Cancer Guidelines, Oncology Drug Compendium, Oncology Continuing Medical Education. https://www.nccn.org/ (accessed 3 Sep 2016).
- 4. Puente XS, Beà S, Valdés‐Mas R, Villamor N, Gutiérrez‐Abril J, Martín‐Subero JI, Munar M, Rubio‐Pérez C, Jares P, Aymerich M, Baumann T, Beekman R, Belver L, Carrio A, Castellano G, Clot G, Colado E, Colomer D, Costa D, Delgado J, Enjuanes A, Estivill X, Ferrando AA, Gelpí JL, González B, González S, González M, Gut M, Hernández‐Rivas JM, López‐Guerra M, Martín‐García D, Navarro A, Nicolás P, Orozco M, Payer ÁR, Pinyol M, Pisano DG, Puente DA, Queirós AC, Quesada V, Romeo‐Casabona CM, Royo C, Royo R, Rozman M, Russiñol N, Salaverría I, Stamatopoulos K, Stunnenberg HG, Tamborero D, Terol MJ, Valencia A, López‐Bigas N, Torrents D, Gut I, López‐Guillermo A, López‐Otín C, Campo E. Non‐coding recurrent mutations in chronic lymphocytic leukaemia. Nature 2015;526:519–524. [DOI] [PubMed] [Google Scholar]
- 5. Matutes E, Owusu‐Ankomah K, Morilla R, Garcia Marco J, Houlihan A, Que TH, Catovsky D. The immunological profile of B‐cell disorders and proposal of a scoring system for the diagnosis of CLL. Leukemia 1994;8 :1640–1645. [PubMed] [Google Scholar]
- 6. Moreau EJ, Matutes E, A'Hern RP, Morilla AM, Morilla RM, Owusu‐Ankomah KA, Seon BK, Catovsky D. Improvement of the chronic lymphocytic leukemia scoring system with the monoclonal antibody SN8 (CD79b). Am J Clin Pathol 1997;108:378–382. [DOI] [PubMed] [Google Scholar]
- 7. Challagundla P, Medeiros LJ, Kanagal‐Shamanna R, Miranda RN, Jorgensen JL. Differential expression of CD200 in B‐cell neoplasms by flow cytometry can assist in diagnosis, subclassification, and bone marrow staging. Am J Clin Pathol 2014;142 :837–844. [DOI] [PubMed] [Google Scholar]
- 8. van Dongen JJM, Lhermitte L, Böttcher S, Almeida J, van der Velden VHJ, Flores‐Montero J, Rawstron A, Asnafi V, Lécrevisse Q, Lucio P, Mejstrikova E, Szczepański T, Kalina T, de Tute R, Brüggemann M, Sedek L, Cullen M, Langerak AW, Mendonça A, Macintyre E, Martin‐Ayuso M, Hrusak O, Vidriales MB, Orfao A. EuroFlow Consortium (EU‐FP6, LSHB‐CT‐ 2006–018708) EuroFlow antibody panels for standardized n‐dimensional flow cytometric immunophenotyping of normal, reactive and malignant leukocytes. Leukemia 2012;26 :1908–1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Uhrmacher S, Schmidt C, Erdfelder F, Poll‐Wolbeck SJ, Gehrke I, Hallek M, Kreuzer K‐A. Use of the receptor tyrosine kinase‐like orphan receptor 1 (ROR1) as a diagnostic tool in chronic lymphocytic leukemia (CLL). Leuk Res 2011;35 :1360–1366. [DOI] [PubMed] [Google Scholar]
- 10. Baskar S, Kwong KY, Hofer T, Levy JM, Kennedy MG, Lee E, Staudt LM, Wilson WH, Wiestner A, Rader C. Unique cell surface expression of receptor tyrosine kinase ROR1 in human B‐cell chronic lymphocytic leukemia. Clin Cancer Res off J Am Assoc. Cancer Res 2008;14 :396–404. [DOI] [PubMed] [Google Scholar]
- 11. Broome HE, Rassenti LZ, Wang H‐Y, Meyer LM, Kipps TJ. ROR1 is expressed on hematogones (non‐neoplastic human B‐lymphocyte precursors) and a minority of precursor‐B acute lymphoblastic leukemia. Leuk Res 2011; 35 :1390–1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Daneshmanesh AH, Mikaelsson E, Jeddi‐Tehrani M, Bayat AA, Ghods R, Ostadkarampour M, Akhondi M, Lagercrantz S, Larsson C, Osterborg A, Shokri F, Mellstedt H, Rabbani H. Ror1, a cell surface receptor tyrosine kinase is expressed in chronic lymphocytic leukemia and may serve as a putative target for therapy. Int J Cancer 2008;123 :1190–1195. [DOI] [PubMed] [Google Scholar]
- 13. Rawstron AC, Fazi C, Agathangelidis A, Villamor N, Letestu R, Nomdedeu J, Palacio C, Stehlikova O, Kreuzer K‐A, Liptrot S, O'Brien D, de Tute RM, Marinov I, Hauwel M, Spacek M, Dobber J, Kater AP, Gambell P, Soosapilla A, Lozanski G, Brachtl G, Lin K, Boysen J, Hanson C, Jorgensen JL, Stetler‐Stevenson M, Yuan C, Broome HE, Rassenti L, Craig F, Delgado J, Moreno C, Bosch F, Egle A, Doubek M, Pospisilova S, Mulligan S, Westerman D, Sanders CM, Emerson R, Robins HS, Kirsch I, Shanafelt T, Pettitt A, Kipps TJ, Wierda WG, Cymbalista F, Hallek M, Hillmen P, Montserrat E, Ghia P. A complementary role of multiparameter flow cytometry and high‐throughput sequencing for minimal residual disease detection in chronic lymphocytic leukemia: an European Research Initiative on CLL study. Leukemia 2015;doi:10.1038/leu.2015.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wood B, Jevremovic D, Béné MC, Yan M, Jacobs P, Litwin V, ICSH/ Working Group ICCS. Validation of cell‐based fluorescence assays: practice guidelines from the ICSH and ICCS ‐ part V ‐ assay performance criteria. Cytometry B Clin Cytom 2013;84 :315–323. [DOI] [PubMed] [Google Scholar]
- 15. Deans JP, Polyak MJ. FMC7 is an epitope of CD20. Blood 2008;111:2492. [DOI] [PubMed] [Google Scholar]
- 16. Pepper C, Majid A, Lin TT, Hewamana S, Pratt G, Walewska R, Gesk S, Siebert R, Wagner S, Kennedy B, Miall F, Davis ZA, Tracy I, Gardiner AC, Brennan P, Hills RK, Dyer MJS, Oscier D, Fegan C. Defining the prognosis of early stage chronic lymphocytic leukaemia patients. Br J Haematol 2012;156:499–507. [DOI] [PubMed] [Google Scholar]
- 17. Ghia P, Guida G, Stella S, Gottardi D, Geuna M, Strola G, Scielzo C, Caligaris‐Cappio F. The pattern of CD38 expression defines a distinct subset of chronic lymphocytic leukemia (CLL) patients at risk of disease progression. Blood 2003;101:1262–1269. [DOI] [PubMed] [Google Scholar]
- 18. Medd PG, Clark N, Leyden K, Turner S, Strefford JA, Butler C, Collins GP, Roberts DJ, Atoyebi W, Hatton CSR. A novel scoring system combining expression of CD23, CD20, and CD38 with platelet count predicts for the presence of the t(11;14) translocation of mantle cell lymphoma. Cytometry B Clin Cytom 2011;80 :230–237. [DOI] [PubMed] [Google Scholar]
- 19. Hamblin TJ. Prognostic markers in chronic lymphocytic leukaemia. Best Pract Res Clin Haematol 2007;20 :455–468. [DOI] [PubMed] [Google Scholar]
- 20. Rawstron AC, Böttcher S, Letestu R, Villamor N, Fazi C, Kartsios H, de Tute RM, Shingles J, Ritgen M, Moreno C, Lin K, Pettitt AR, Kneba M, Montserrat E, Cymbalista F, Hallek M, Hillmen P, Ghia P. Improving efficiency and sensitivity: European Research Initiative in CLL (ERIC) update on the international harmonised approach for flow cytometric residual disease monitoring in CLL. Leukemia 2013;27 :142–149. [DOI] [PubMed] [Google Scholar]
- 21. Rawstron AC, Villamor N, Ritgen M, Böttcher S, Ghia P, Zehnder JL, Lozanski G, Colomer D, Moreno C, Geuna M, Evans PAS, Natkunam Y, Coutre SE, Avery ED, Rassenti LZ, Kipps TJ, Caligaris‐Cappio F, Kneba M, Byrd JC, Hallek MJ, Montserrat E, Hillmen P. International standardized approach for flow cytometric residual disease monitoring in chronic lymphocytic leukaemia. Leukemia 2007;21:956–964. [DOI] [PubMed] [Google Scholar]
- 22. Rawstron AC, de Tute RM, Shingles J, Gorman L, Turner K, Evans PAS, Barrans SL, O'Connor SJM, Burton C, Owen RG, Hillmen P. Improving the Differential Diagnosis of CD5+ B‐Lymphoproliferative Disorders. http://learningcenter.ehaweb.org/eha/2016/21st/133474/andy.rawstron.improving.the.differential.diagnosis.of.cd52B.html?f=m1
- 23. Morice WG, Kurtin PJ, Hodnefield JM, Shanafelt TD, Hoyer JD, Remstein ED, Hanson CA. Predictive value of blood and bone marrow flow cytometry in B‐cell lymphoma classification: Comparative analysis of flow cytometry and tissue biopsy in 252 patients. Mayo Clin Proc 2008;83 :776–785. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional Supporting Information may be found online in the supporting information tab for this article.
Supporting Information


