Abstract
Two strains, D5088T and D5095, representing a novel yeast species belonging to the genus Saccharomyces were isolated from oak tree bark and surrounding soil located at an altitude of 1000 m above sea level in Saint Auban, France. Sequence analyses of the internal transcribed spacer (ITS) region and 26S rRNA D1/D2 domains indicated that the two strains were most closely related to Saccharomyces mikatae and Saccharomyces paradoxus. Genetic hybridization analyses showed that both strains are reproductively isolated from all other Saccharomyces species and, therefore, represent a distinct biological species. The species name Saccharomyces jurei sp. nov. is proposed to accommodate these two strains, with D5088T (=CBS 14759T=NCYC 3947T) designated as the type strain.
Keywords: Saccharomyces, yeast, new species, Saccharomyces jurei, isolation protocol
Introduction
The Saccharomyces sensu stricto group is composed of eight biologically distinct yeast species, namely Saccharomyces cerevisiae, S. paradoxus, S. cariocanus, S. uvarum, S. mikatae, S. kudriavzevii, S. arboricola and S. eubayanus [1–6], and two natural hybrids, namely S. pastorianus [7, 8] and S. bayanus [9]. S. cariocanus was initially included in the genus based on karyotyping and reproductive isolation [3]. However, subsequent genome sequence analysis of the only two known strains (of S. cariocanus) showed them to belong to one of three geographically well-defined populations of S. paradoxus (i.e. American population) [10, 11]. The most recent phylogenetic analyses of the genus excluded both S. cariocanus and S. bayanus, the latter due to it being of hybrid origin [12, 13]. The cryotolerant yeast S. eubayanus is the latest addition to the genus. This species was first isolated in Nothofagus (southern beech) forests in Patagonia, Argentina [6], but has since been found in North America, on the Tibetan Plateau and most recently on the North Island of New Zealand [14–16].
The Saccharomyces species are defined by the biological species concept since they are reproductively isolated via postzygotic barriers [3, 10, 17]. All of these species possess typical budding shape morphology, have the same number of 16 chromosomes [18], and they can be differentiated from one another based on the sequences of their internal transcribed spacer (ITS) and 26S rRNA D1/D2 regions [19, 20]. It has been shown that the majority of yeast species can be identified from sequence divergence of the D1/D2 domain [21]. Sequencing of the ITS1 and D1/D2 regions is therefore routinely used for identifying yeast strains [3, 22–25].
Saccharomyces yeasts have been isolated from a wide variety of different substrates including deciduous tree bark, surrounding soil, tree exudates (sap), fruits, insects and vineyard grapes [2, 26–28]. S. paradoxus is the most commonly isolated species in nature and has been found globally from natural resources, and most notably from oak trees (Quercus spp.) and surrounding soil [11, 29]. Moreover, S. cerevisae and S. paradoxus have been isolated from the same locations, indicating that populations of the two species coexist in nature [30, 31]. Saccharomyces hybrids have been often isolated from domesticated environments such as vineyards [32] and breweries, and are known to be associated with fermentation processes for the production of wine and beer. The best example of this is the lager yeast S. pastorianus (syn. S. carlsbergensis), a cold-adapted S. cerevisiae × S. eubayanus alloploid hybrid [6].
To date, most of the Saccharomyces strains held in international yeast collections (e.g. the Westerdijk Fungal Biodiversity Institute, CBS) have been isolated from substrates collected and sampled at low altitudes [26, 30, 33, 34]. Some species found from higher altitude include S. eubayanus isolated from the Tibetan Plateau [14] and S. arboricola from the Qinling Mountains [2]. Consequently, very little is known about the ecology and geographical distribution of Saccharomyces yeasts found at higher altitudes and cooler conditions. Thus, sampling substrates such as soil and trees at higher altitudes may lead to the discovery of new cryotolerant yeast strains and species. In this study, we sampled oak tree bark and surrounding soil at an altitude of 1000 m above sea level in Saint Auban, France. The yeast community was isolated and the species identities were determined by standard ITS and D1/D2 sequencing. Whilst the majority of isolated Saccharomyces were identified as S. paradoxus, two strains (D5088T and D5095) were recovered and found to represent a novel species belonging to the genus Saccharomyces. The novel species is named Saccharomyces jurei sp. nov., in memory of the yeast researcher Professor Jure Piškur. We show here that S. jurei is reproductively isolated from other Saccharomyces species by performing genetic crosses and testing for hybrid sterility. Both strains formed viable hybrids with all other Saccharomyces species and were, as expected from crosses between different biological species, predominantly sterile (with a spore viability ranging from 0 to 3 %).
Methods
Yeast isolation, media and maintenance
Samples of bark and soil were obtained in July 2013 from oak trees (Quercus) growing at an altitude of 1000 m above sea level in the Saint Auban region of south-eastern France (43° 5.2′ N 006° 44′ E). The samples were collected aseptically and stored in sterile bags or Petri dishes. Equal amounts of each bark and soil sample were independently placed into one of two 50 ml sterile Falcon tubes containing Sniegowski enrichment medium consisting of 3 g yeast extract, 3 g malt extract, 5 g peptone, 10 g sucrose, 76 ml EtOH, 1 mg chloramphenicol and 1 ml of 1 M HCl per litre [31]. The Falcon tubes were tightly capped and one set was incubated (without agitation) at 30 °C, while the other was incubated at 20 °C for 20–25 days. The tubes were periodically examined for turbidity and fermentation (i.e. evidence of gas formation). These observations were done after 10 days for the samples incubated at 30 °C and after 20 days for the 20 °C samples. Samples showing signs of either turbidity or fermentation were further examined, for signs of yeast growth, by standard light microscopy. All samples positive for yeast growth were plated onto Sniegowski selection medium (SSE) [31] and incubated at 30 °C for several days. Individual yeast colonies were picked and re-streaked onto fresh SSE plates for further characterization.
Morphological and physiological characterization of yeasts
The two strains were characterized biochemically, morphologically and physiologically according to standard methods described previously [35]. Growth temperature was determined by cultivation on YM (yeast extract-malt extract) agar. Sporulation tests were performed on cornmeal agar, Gorodkowa agar, potassium acetate agar and YM agar, and plates were incubated at 25 °C for 3–4 weeks in individual and mixed cultures.
Images of the asci were taken using an Olympus model BH-2 light microscope and a scanning electron microscope. The asci formed on acetate agar after 5 days at 25 °C, and spontaneously broke as result of the general fixing process.
DNA extraction
For ITS1 and 26S rRNA D1/D2 sequencing, genomic DNA was isolated from cultures freshly grown on plates using the Masterpure Yeast DNA extraction kit (catalogue no. MPY80200) and following the manufacturer’s protocol. DNA yields and A260/A280 ratios were measured using a Nanodrop spectrophotometer (ND-1000), while DNA purity and integrity were checked by 0.8 % agarose gel electrophoresis.
DNA sequencing
The variable D1 and D2 domains of the 26S rRNA gene were amplified and sequenced using primers NL1 and NL4 [36]. The ribosomal ITS region was amplified using primers ITS4 and ITS5, and sequenced using these primers as well as internal primers ITS2 and ITS3 [20, 37]. Translation EF-1αA (TEF1) and RPB2 genes were amplified and sequenced as described previously [23]. Other nuclear genes (CAT8, CYR1, GSY1, MET6 and OPY1) were amplified and sequenced using previously published primers [32]. The PCR fragments were analysed by standard 1 % agarose gel electrophoresis, purified and concentrated using QIAquick PCR purification spin columns (Qiagen) following the manufacturer’s instructions. The purified products were sequenced using the BigDye Terminator Ready Reaction kit, version 3.1 (Applied Biosystems) following the manufacturer’s instructions. Sequence traces were edited manually, and consensus sequences were generated using the program seqman, version 11 (DNASTAR). The sequences were compared pairwise using a fasta similarity search [38] and were aligned with the sequences of closely related taxa, retrieved from the EMBL sequence database, using the multiple alignment program clustal w [39] included in the mega version 6 software package [40]. A phylogenetic tree was reconstructed from the combined sequences of the 26S rRNA D1/D2 and ITS regions (including the 5.8S rRNA) using the neighbour-joining (NJ) program [41] included in mega, with the Kimura two-parameter (K2P) distance measure and Naumovozyma castellii selected as the outgroup species. Bootstrap support for the NJ tree was determined from 1000 replicates.
Construction of stable haploid strains possessing auxotrophic marker
Prototrophic diploid strains D5088T and D5095 of S. jurei were made heterothallic by knocking out the HO gene. The heterozygote HO/hoΔ diploid strains were sporulated and tetrads were dissected to obtain stable Mata and Matα hoΔ haploid strains. The mating types were determined by PCR as described previously [42]. A PCR-mediated gene deletion strategy using drug resistance cassettes was applied to generate ura3 auxotrophic strains of S. jurei [43]. A standard PEG/LiAc heat-shock protocol with some modifications was used for transformation. In our modified protocol, 1.0–3.0 µg of PCR product was transformed and cells were incubated at 30 °C for 30 min followed by heat-shock at 37 °C for 20 min. For the selection of transformants, the cells were incubated overnight at room temperature before being plated on selective media. The verification of gene deletions was performed by diagnostic colony PCR using gene-specific and cassette-specific primers.
Spore viability analysis
S. jurei strains D5088T and D5095 were crossed with other species of Saccharomyces using a micromanipulator. The hybrids were selected on SD plates containing different selective markers [44]. The tetrads were formed by growing the hybrids in pre-sporulation medium at 30 °C for 12 h before plating on minimal sporulation medium. The sporulation plates were incubated at 20 °C for 7–10 days for the formation of tetrads. The tetrads were dissected using a Singer MSM-300 micromanipulator. Spore viability was calculated based on the percentage of viable spores that had grown for each variant of the strain out of a possible 64 dissected tetrads.
Results and Discussion
Isolation of yeast species from Quercus robur
We obtained a total of 284 yeast isolates from oak tree bark and soil samples incubated at 20 and 30 °C (see Table S1, available with the online Supplementary Material). S. paradoxus was by far the most abundant species isolated from the bark and soil samples, with Kazachstania servazzii and Lachancea (Kluyveromyces) thermotolerans being the other species recovered from this site.
DNA sequencing and phylogenetic analysis
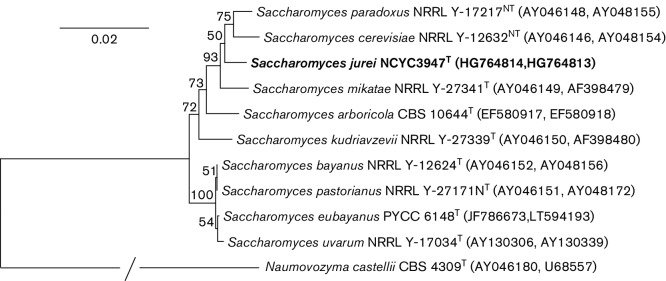
All yeast isolates were initially screened by amplifying the ITS region to distinguish between Saccharomyces and non-Saccharomyces species based on differing fragment size. Of 284 isolates collected, 180 amplified ITS fragments of the correct size for Saccharomyces yeasts (~850 bp), and their species identities were confirmed by sequencing the ITS1 region. Although the majority of isolates were identified as representing S. paradoxus (172 isolates), two isolates, D5088T and D5095, had ITS1 sequences that did not match with any currently described Saccharomyces species. Both isolates had identical ITS1 sequences, and a fasta sequence similarity search of the EMBL fungal sequence database revealed no other yeast taxon, either Saccharomyces or non-Saccharomyces, with an ITS1 sequence identical to these isolates. In terms of pairwise sequence similarity, the closest taxa were S. mikatae (98.1 %; 7 nt substitutions in 360 nt) and S. paradoxus (96.1 %; 12 nt sustitutions and one indel in 362 nt). Indeed, an ITS1 sequence alignment of the novel Saccharomyces taxon, S. cerevisiae, S. mikatae and representatives of the three geographically distinct populations of S. paradoxus (i.e. North American, European and Far Eastern) confirmed that the ITS1 region of strains D5088T and D5095 was unique and possessed four species-specific single nucleotide polymorphisms (Fig. S1). The level of sequence similarity seen between S. jurei and S. paradoxus is comparable to that observed between S. mikatae and S. paradoxus (96.1 % versus 96.4 %). Furthermore, the level of sequence similarity between S. jurei and S. mikatae (98.1 %) is lower than that observed between the three S. paradoxus populations (99.2–99.7 %). In contrast to the ITS1 region, a fasta sequence similarity search with the 26S rRNA D1/D2 sequence revealed that the closest known taxon was S. paradoxus (99.8 %; 1 nt substitution in 579 nt), with S. mikatae displaying only 98.6 % similarity (6 nt substitutions and one indel in 574 nt). Sequence analysis of seven other nuclear genes (CAT8, CYR1, OPY1, GSY1, MET6, TEF1 and RPB2) showed that S. jurei is divergent from S. cerevisiae, S. mikatae and S. paradoxus (Table S2). Moreover, different populations of S. paradoxus and S. cerevisiae possess approximately 98–99 % sequence similarity for the seven nuclear genes analysed in this study. A phylogenetic analysis based on the combined (i.e. concatenated) sequences of the ITS and 26S rRNA D1/D2 regions showed that the novel taxon [as represented by D5088T (=NCYC 3947T)] belonged to the genus Saccharomyces, and is located between S. mikatae and the species pair of S. cerevisiae and S. paradoxus (Fig. 1).
Fig. 1.
NJ dendrogram based on the combined sequences of the LSU D1/D2 and ITS regions (including 5.8S rRNA) of Saccharomyces jurei sp. nov. and its closest relatives. Species names are followed by CBS, NCYC, NRRL or PYCC strain accession numbers and, respectively, the EMBL/GenBank accession numbers for the ITS and LSU D1/D2 regions. Naumovozyma castellii was used as the outgroup species for the analysis. Boostrap values of >50 %, determined from 1000 replicates, are shown at branch nodes. Bar, 2 base substitutions per 100 nt.
Genetic hybridization analysis
All eight members of the Saccharomyces genus are biological species since they are reproductively isolated from each other [3]. The species of this genus can readily hybridize with each other, although the interspecific hybrids (F1 hybrids) formed are sexually sterile [45, 46]. This sterility is caused by the inability of the two diverged homologous chromosomes to recombine during meiosis [17, 47, 48]. The presence of chromosomal rearrangements also lowers spore viability and contributes to hybrid infertility [10, 49, 50]. In contrast, intraspecifc Saccharomyces hybrids are fertile and yield highly viable ascospores [3]. To establish that S. jurei is a novel biological species of the genus, we performed direct genetic crosses with representative strains from all other Saccharomyces species and tested the fertility of the resulting hybrids.
We first analysed the fertility of strains D5088T and D5095. Both were observed to be homothallic and highly fertile, with ascospore viability ranging from 95 to 100 %. To test the fertility of intra- and interspecific hybrids, we successfully constructed genetically stable haploid strains (a and α mating types) of D5088T and D5095 possessing ura3 auxotrophy and drug resistance markers (Clonat and KanMX, respectively). The hybrids produced from crossing D5088T with D5095 showed high spore viability of ca. 89 %, confirming that both strains belong to the same biological species (Table 1). In contrast, the interspecific hybrids produced from crosses between S. jurei and the other Saccharomyces species although viable displayed extremely low spore viability, ranging from 0 to 0.3 % (Table 1). Collectively, these data confirm the post-zygotic isolation between S. jurei and the other member species of the genus Saccharomyces, and demonstrate that it represents a biologically distinct novel Saccharomyces species.
Table 1. Genetic identification of hybrids between S. jurei (D5088T and D5095), S. cerevisiae (FY3), S. mikatae (IFO 1815T), S. paradoxus (N-44, N-17, YPS138), S. uvarum (CBS 7001), S. kudriavzevii (IFO 1802T), S. arboricola (CBS 10644T) and S. eubayanus (PYCC 6148T).
| Hybrids engineered | No. of tetrads dissected | Percentage of viable ascospores of hybrids |
|---|---|---|
| S. jurei×S. jurei | ||
| D5095×D5088T | 640 | 89 |
| S. jurei×S. cerevisiae | ||
| D5095×FY3 | 624 | 0.30 |
| D5088T×FY3 | 612 | 0.50 |
| S. jurei×S. mikatae | ||
| D5095×IFO 1815T | 648 | 2.0 |
| D5088T×IFO 1815T | 472 | 1.0 |
| S. jurei×S. paradoxus N-44 | ||
| D5095×N-44 | 484 | 0 |
| D5088T×N-44 | 648 | 0.3 |
| S. jurei×S. paradoxus N-17 | ||
| D5095×N17 | 532 | 1.6 |
| D5088T×N-17 | 648 | 0 |
| S. jurei×S. paradoxus YPS138 | ||
| D5095×YPS138 | 640 | 1.2 |
| D5088T×YPS138 | 600 | 0.25 |
| S. jurei×S. uvarum | ||
| D5095×CBS 7001 | 604 | 0.3 |
| D5088T×CBS 7001 | 644 | 0.2 |
| S. jurei×S. kudriavzevii | ||
| D5095×IFO 1802T | 620 | 0 |
| D5088T×IFO 1802T | 614 | 1 |
| S. jurei×S. arboricola | ||
| D5095×CBS 10644T | 632 | 0 |
| D5088T×CBS 10644T | 648 | 0 |
| S. jurei×S. eubayanus | ||
| D5095×PYCC 6148T | 632 | 0.2 |
| D5088T×PYCC 6148T | 648 | 0 |
Phenotypic characterization
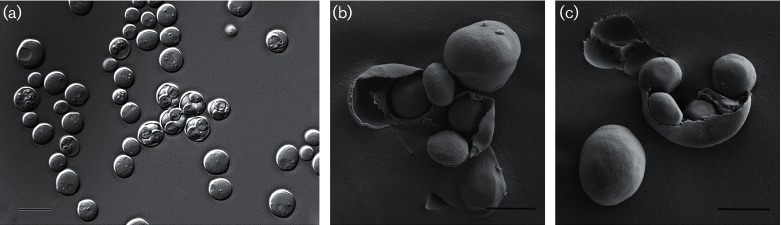
Strains D5088T and D5095 exhibited similar morphological and physiological characters that are typical for species belonging to the genus Saccharomyces [51]. Moderate sporulation was observed for both strains at 25 °C on potassium acetate agar, cornmeal agar and YM agar. The asci had a spherical shape with two to four oval spores per ascus (Fig. 2).
Fig. 2.
Phenotypic chatacteristics of Saccharomyces jurei sp. nov. D5088T. Photomicrograph of the asci (a) and scanning electron micrographs of ascospores (b and c) formed on acetate agar after 5 days at 25 °C. Bars, 2 µm
Phenotypically, as shown in Table S3, there appear to be no standard assimilation or fermentation tests which can be used reliably to differentiate between S. jurei and its closest relatives, namely S. cerevisiae, S. mikatae and S. paradoxus. Amongst these four Saccharomyces species, S. cerevisiae and S. paradoxus are the only ones which are able to grow at the elevated temperature of 37 °C, although this trait is somewhat strain-variable [51, 52]. At present, the species descriptions of S. jurei (this study) and S. mikatae [3, 52] are restricted to just two strains each. However, it is quite possible that in time, as additional strains of each of these species are discovered, some of the traits currently listed as positive (e.g. maltose assimilation and fermentation; Table S3) will be found to be variable, as is the case for both S. cerevisiae and S. paradoxus [51–55]. The molecular comparisons showed that strains D5088T and D5095 represent a novel species of the genus Saccharomyces, for which the name Saccharomyces jurii sp. nov. is proposed.
Description of Saccharomyces jurei Naseeb S, James SA, Alsammar H, Michaels C, Gini B, Neuno-Palop C, Bond CJ, McGhie H, Roberts IN, Delneri D., sp. nov.
Saccharomyces jurei (ju’re.i. N.L. gen. n. jurei in memory of Professor Jure Piškur for his considerable contribution to the fields of yeast genetics and molecular biology).
On YM agar, after 3 days incubation at 25 °C, colonies are light cream-coloured, slightly shiny, smooth and with an entire margin. In YM broth, after 2 days of incubation at 25 °C, cells are spherical to ovoid (5.0–8.0×6.0–10.0 µm) and occur singly or in pairs. Budding is multipolar. No pseudohyphae are observed in cultures grown on cornmeal agar or potato agar. Oval asci containing 2–4 smooth round ascospores are formed after incubation for 1–3 weeks at 25 °C on cornmeal agar, potassium acetate agar and YM agar (Fig. 2). Asci are persistent. Glucose, galactose, sucrose, maltose, raffinose, melizitose and methyl α-d-glucoside are fermented, but not lactose, trehalose, melibiose, cellobiose, inulin, soluble starch or d-xylose. Glucose, sucrose, raffinose, galactose, trehalose (latent or weak), maltose, melezitose, methyl α-d-glucoside, ethanol, glycerol (latent), d-mannitol and dl-lactate are assimilated. No growth occurs on inulin, melibiose, lactose, soluble starch, cellobiose, salicin, l-sorbose, l-rhamnose, d-xylose, l-arabinose, d-arabinose, d-ribose, methanol, erythritol, ribitol, xylitol, galactitol, d-glucitol, inositol, succinate or citrate. No growth occurs on cadaverine, lysine, ethylamine hydrochloride or nitrate. Growth occurs at 30 °C, but not at 37 °C. No growth occurs on either YM agar with 10 % (w/v) NaCl or on 100 µg cycloheximide ml−1. Growth occurs on 50 % glucose/yeast extract. Starch-like compounds are not produced.
The type strain, D5088T, was isolated from north-facing oak bark, collected at an altitude of 1000 m above sea level in the Saint Auban region of south-eastern France. This strain has been deposited in the National Collection of Yeast Cultures (NCYC), Norwich, UK, as NCYC 3947T (=CBS 14759T), and is stored in a metabolically inactive form in accordance with the Code. Strain D5095 has also been deposited in the NCYC as NCYC 3962. The MycoBank deposit number is MB 819910.
Funding information
SN is supported through BBSRC funding (BB/L021471/1). HA is supported by a scholarship funded by the Kuwait government through Kuwait University. The NCYC is a BBSRC-supported National Capability.
Acknowledgements
The authors wish to thank Kathryn Cross in the Electron Microscopy facility in the Institute of Food Research, Norwich, UK, for taking the scanning electron micrographs of the asci.
Conflicts of interest
The authors declare no conflicts of interest.
Supplementary Data
Footnotes
Abbreviations: ITS, internal transcribed spacer; NJ, neighbour-joining.
The GenBank/EMBL/DDBJ accession numbers for the 26S rRNA D1/D2 and ITS sequences of D5088T are HG764813 and HG764814, respectively. The MycoBank number for Saccharomyces jurei sp. nov. is MB 819910.
Three supplementary tables and one supplementary figure are available with the online Supplementary Material.
References
- 1.Martini AV, Martini A. Three newly delimited species of Saccharomyces sensu stricto. Antonie van Leeuwenhoek. 1987;53:77–84. doi: 10.1007/BF00419503. [DOI] [PubMed] [Google Scholar]
- 2.Wang SA, Bai FY. Saccharomyces arboricolus sp. nov., a yeast species from tree bark. Int J Syst Evol Microbiol. 2008;58:510–514. doi: 10.1099/ijs.0.65331-0. [DOI] [PubMed] [Google Scholar]
- 3.Naumov GI, James SA, Naumova ES, Louis EJ, Roberts IN. Three new species in the Saccharomyces sensu stricto complex: Saccharomyces cariocanus, Saccharomyces kudriavzevii and Saccharomyces mikatae. Int J Syst Evol Microbiol. 2000;50:1931–1942. doi: 10.1099/00207713-50-5-1931. [DOI] [PubMed] [Google Scholar]
- 4.Naumov GI, Naumova ES, Hagler AN, Mendonça-Hagler LC, Louis EJ. A new genetically isolated population of the Saccharomyces sensu stricto complex from Brazil. Antonie van Leeuwenhoek. 1995;67:351–355. doi: 10.1007/BF00872934. [DOI] [PubMed] [Google Scholar]
- 5.Naumov GI, Naumova ES, Louis EJ. Two new genetically isolated populations of the Saccharomyces sensu stricto complex from Japan. J Gen Appl Microbiol. 1995;41:499–505. doi: 10.2323/jgam.41.499. [DOI] [PubMed] [Google Scholar]
- 6.Libkind D, Hittinger CT, Valério E, Gonçalves C, Dover J, et al. Microbe domestication and the identification of the wild genetic stock of lager-brewing yeast. Proc Natl Acad Sci USA. 2011;108:14539–14544. doi: 10.1073/pnas.1105430108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Masneuf I, Hansen J, Groth C, Piskur J, Dubourdieu D. New hybrids between Saccharomyces sensu stricto yeast species found among wine and cider production strains. Appl Environ Microbiol. 1998;64:3887–3892. doi: 10.1128/aem.64.10.3887-3892.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Querol A, Bond U. The complex and dynamic genomes of industrial yeasts. FEMS Microbiol Lett. 2009;293:1–10. doi: 10.1111/j.1574-6968.2008.01480.x. [DOI] [PubMed] [Google Scholar]
- 9.Nguyen HV, Legras JL, Neuvéglise C, Gaillardin C. Deciphering the hybridisation history leading to the lager lineage based on the mosaic genomes of Saccharomyces bayanus strains NBRC1948 and CBS380. PLoS One. 2011;6:e25821. doi: 10.1371/journal.pone.0025821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liti G, Barton DB, Louis EJ. Sequence diversity, reproductive isolation and species concepts in Saccharomyces. Genetics. 2006;174:839–850. doi: 10.1534/genetics.106.062166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liti G, Carter DM, Moses AM, Warringer J, Parts L, et al. Population genomics of domestic and wild yeasts. Nature. 2009;458:337–341. doi: 10.1038/nature07743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hittinger CT. Saccharomyces diversity and evolution: a budding model genus. Trends Genet. 2013;29:309–317. doi: 10.1016/j.tig.2013.01.002. [DOI] [PubMed] [Google Scholar]
- 13.Boynton PJ, Greig D. The ecology and evolution of non-domesticated Saccharomyces species. Yeast. 2014;31:449–462. doi: 10.1002/yea.3040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bing J, Han PJ, Liu WQ, Wang QM, Bai FY. Evidence for a Far East Asian origin of lager beer yeast. Curr Biol. 2014;24:R380–R381. doi: 10.1016/j.cub.2014.04.031. [DOI] [PubMed] [Google Scholar]
- 15.Gayevskiy V, Goddard MR. Saccharomyces eubayanus and Saccharomyces arboricola reside in North Island native New Zealand forests. Environ Microbiol. 2016;18:1137–1147. doi: 10.1111/1462-2920.13107. [DOI] [PubMed] [Google Scholar]
- 16.Peris D, Sylvester K, Libkind D, Gonçalves P, Sampaio JP, et al. Population structure and reticulate evolution of Saccharomyces eubayanus and its lager-brewing hybrids. Mol Ecol. 2014;23:2031–2045. doi: 10.1111/mec.12702. [DOI] [PubMed] [Google Scholar]
- 17.Greig D. Reproductive isolation in Saccharomyces. Heredity. 2009;102:39–44. doi: 10.1038/hdy.2008.73. [DOI] [PubMed] [Google Scholar]
- 18.Duina AA, Miller ME, Keeney JB. Budding yeast for budding geneticists: a primer on the Saccharomyces cerevisiae model system. Genetics. 2014;197:33–48. doi: 10.1534/genetics.114.163188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schoch CL, Seifert KA, Huhndorf S, Robert V, Spouge JL, et al. Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. Proc Natl Acad Sci USA. 2012;109:6241–6246. doi: 10.1073/pnas.1117018109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.White TJ, Bruns TD, Lee S, Taylor JW. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis M, Gelfand DH, Sninsky JJ, White TJ, editors. PCR Protocols. San Diego, Calif: Academic Press; 1990. pp. 315–322. (editors) [Google Scholar]
- 21.Kurtzman CP, Robnett CJ. Molecular relationships among hyphal ascomycetous yeasts and yeastlike taxa. Canadian Journal of Botany. 1995;73:824–830. doi: 10.1139/b95-328. [DOI] [Google Scholar]
- 22.Naumov GI, Naumova ES, Masneuf-Pomarède I. Genetic identification of new biological species Saccharomyces arboricolus Wang et Bai. Antonie van Leeuwenhoek. 2010;98:1–7. doi: 10.1007/s10482-010-9441-5. [DOI] [PubMed] [Google Scholar]
- 23.Kurtzman CP, Robnett CJ. Phylogenetic relationships among yeasts of the 'Saccharomyces complex' determined from multigene sequence analyses. FEMS Yeast Res. 2003;3:417–432. doi: 10.1016/S1567-1356(03)00012-6. [DOI] [PubMed] [Google Scholar]
- 24.Ciardo DE, Schär G, Böttger EC, Altwegg M, Bosshard PP. Internal transcribed spacer sequencing versus biochemical profiling for identification of medically important yeasts. J Clin Microbiol. 2006;44:77–84. doi: 10.1128/JCM.44.1.77-84.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vu D, Groenewald M, Szöke S, Cardinali G, Eberhardt U, et al. DNA barcoding analysis of more than 9 000 yeast isolates contributes to quantitative thresholds for yeast species and genera delimitation. Stud Mycol. 2016;85:91–105. doi: 10.1016/j.simyco.2016.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Charron G, Leducq JB, Bertin C, Dubé AK, Landry CR. Exploring the northern limit of the distribution of Saccharomyces cerevisiae and Saccharomyces paradoxus in North America. FEMS Yeast Res. 2014;14:281–288. doi: 10.1111/1567-1364.12100. [DOI] [PubMed] [Google Scholar]
- 27.Naumov GI, Lee CF, Naumova ES. Molecular genetic diversity of the Saccharomyces yeasts in Taiwan: Saccharomyces arboricola, Saccharomyces cerevisiae and Saccharomyces kudriavzevii. Antonie van Leeuwenhoek. 2013;103:217–228. doi: 10.1007/s10482-012-9803-2. [DOI] [PubMed] [Google Scholar]
- 28.Naumov GI, Naumova ES, Hagler AN, Mendonça-Hagler LC, Louis EJ. A new genetically isolated population of the Saccharomyces sensu stricto complex from Brazil. Antonie van Leeuwenhoek. 1995;67:351–355. doi: 10.1007/BF00872934. [DOI] [PubMed] [Google Scholar]
- 29.Johnson LJ, Koufopanou V, Goddard MR, Hetherington R, Schäfer SM, et al. Population genetics of the wild yeast Saccharomyces paradoxus. Genetics. 2004;166:43–52. doi: 10.1534/genetics.166.1.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Naumov GI, Naumova ES, Sniegowski PD. Saccharomyces paradoxus and Saccharomyces cerevisiae are associated with exudates of north American Oaks. Can J Microbiol. 1998;44:1045–1050. [PubMed] [Google Scholar]
- 31.Sniegowski PD, Dombrowski PG, Fingerman E. Saccharomyces cerevisiae and Saccharomyces paradoxus coexist in a natural woodland site in North America and display different levels of reproductive isolation from european conspecifics. FEMS Yeast Res. 2002;1:299–306. doi: 10.1111/j.1567-1364.2002.tb00048.x. [DOI] [PubMed] [Google Scholar]
- 32.González SS, Barrio E, Gafner J, Querol A. Natural hybrids from Saccharomyces cerevisiae, Saccharomyces bayanus and Saccharomyces kudriavzevii in wine fermentations. FEMS Yeast Res. 2006;6:1221–1234. doi: 10.1111/j.1567-1364.2006.00126.x. [DOI] [PubMed] [Google Scholar]
- 33.Hyma KE, Fay JC. Mixing of vineyard and oak-tree ecotypes of Saccharomyces cerevisiae in North American vineyards. Mol Ecol. 2013;22:2917–2930. doi: 10.1111/mec.12155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Naumov GI, Naumova ES, Sancho ED. Genetic reidentification of Saccharomyces strains associated with black knot disease of trees in Ontario and Drosophila species in California. Can J Microbiol. 1996;42:335–339. doi: 10.1139/m96-049. [DOI] [Google Scholar]
- 35.Kurtzman CP, Fell JW, Boekhout T, Robert V. Methods for isolation, phenotypic characterization and maintenance of yeasts. In: Kurtzman CP, Fell JW, Boekhout T, editors. The Yeasts, a Taxonomic Study. 5th ed. Amsterdam: Elsevier; 2011. pp. 87–110. (editors) [Google Scholar]
- 36.O’Donnell K. Fusarium and its near relatives. In: Reynolds DR, Taylor JW, editors. The Fungal Holomorph: Mitotic, Meiotic and Pleomorphic Speciation in Fungal Systematics. Wallingford: CAB International; 1993. pp. 225–233. (editors) [Google Scholar]
- 37.James SA, Collins MD, Roberts IN. Use of an rRNA internal transcribed spacer region to distinguish phylogenetically closely related species of the genera Zygosaccharomyces and Torulaspora. Int J Syst Bacteriol. 1996;46:189–194. doi: 10.1099/00207713-46-1-189. [DOI] [PubMed] [Google Scholar]
- 38.Pearson WR, Lipman DJ. Improved tools for biological sequence comparison. Proc Natl Acad Sci USA. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary Genetics analysis version 6.0. Mol Biol Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 42.Huxley C, Green ED, Dunham I. Rapid assessment of S. cerevisiae mating type by PCR. Trends Genet. 1990;6:236. doi: 10.1016/0168-9525(90)90190-h. [DOI] [PubMed] [Google Scholar]
- 43.Güldener U, Heck S, Fielder T, Beinhauer J, Hegemann JH. A new efficient gene disruption cassette for repeated use in budding yeast. Nucleic Acids Res. 1996;24:2519–2524. doi: 10.1093/nar/24.13.2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Delneri D, Colson I, Grammenoudi S, Roberts IN, Louis EJ, et al. Engineering evolution to study speciation in yeasts. Nature. 2003;422:68–72. doi: 10.1038/nature01418. [DOI] [PubMed] [Google Scholar]
- 45.Naumov GI, Naumova ES, Querol A. Genetic study of natural introgression supports delimitation of biological species in the Saccharomyces sensu stricto complex. Syst Appl Microbiol. 1997;20:595–601. doi: 10.1016/S0723-2020(97)80031-8. [DOI] [Google Scholar]
- 46.Naumov GI. Genetic basis for classification and identification of the ascomycetous yeasts. Stud Mycol. 1987;30:469–475. [Google Scholar]
- 47.Roeder GS. Meiotic chromosomes: it takes two to tango. Genes Dev. 1997;11:2600–2621. doi: 10.1101/gad.11.20.2600. [DOI] [PubMed] [Google Scholar]
- 48.Greig D, Travisano M, Louis EJ, Borts RH. A role for the mismatch repair system during incipient speciation in Saccharomyces. J Evol Biol. 2003;16:429–437. doi: 10.1046/j.1420-9101.2003.00546.x. [DOI] [PubMed] [Google Scholar]
- 49.Coyne JA, Orr HA. Speciation. Sunderland: Sinauer; 2004. pp. 256–267. [Google Scholar]
- 50.Hou J, Friedrich A, de Montigny J, Schacherer J. Chromosomal rearrangements as a major mechanism in the onset of reproductive isolation in Saccharomyces cerevisiae. Curr Biol. 2014;24:1153–1159. doi: 10.1016/j.cub.2014.03.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Martini AV. Martini A. Saccharomyces Meyen ex Reess (1870) In: Kurtzman CP, Fell JW, Boekhout T, editors. The Yeasts, a Taxonomic Study. 5th ed. Amsterdam: Elsevier; 2011. pp. 733–746. (editors) [Google Scholar]
- 52.Vaughan-Martini A, Martini A. Saccharomyces Meyen ex Reess (1870) In: Kurtzman CP, Fell JW, Boekhout T, editors. The Yeasts, a Taxonomic Study. 5th ed. Amsterdam: Elsevier; 2011. pp. 733–746. (editors) [Google Scholar]
- 53.Scheda R, Yarrow D. Variation in the fermentative pattern of some Saccharomyces species. Arch Mikrobiol. 1968;61:310–316. doi: 10.1007/BF00446616. [DOI] [PubMed] [Google Scholar]
- 54.Scheda R, Yarrow D. The instability of physiological properties used as criteria in the taxonomy of yeasts. Archiv für Mikrobiologie. 1966;55:209–225. doi: 10.1007/BF00410244. [DOI] [Google Scholar]
- 55.Naumov GI, Naumova ES, Michels CA. Genetic variation of the repeated MAL loci in natural populations of Saccharomyces cerevisiae and Saccharomyces paradoxus. Genetics. 1994;136:803–812. doi: 10.1093/genetics/136.3.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.