Abstract
A newly emerged H5N8 influenza virus was isolated from green-winged teal in Egypt during December 2016. In this study, we provide a detailed characterization of full genomes of Egyptian H5N8 viruses and some virological features. Genetic analysis demonstrated that the Egyptian H5N8 viruses are highly pathogenic avian influenza viruses. Phylogenetic analysis revealed that the genome of the Egyptian H5N8 viruses was related to recently characterized reassortant H5N8 viruses of clade 2.3.4.4 isolated from different Eurasian countries. Multiple peculiar mutations were characterized in the Egyptian H5N8 viruses, which probably permits transmission and virulence of these viruses in mammals. The Egyptian H5N8 viruses preferentially bound to avian-like receptors rather than human-like receptors. Also, the Egyptian H5N8 viruses were fully sensitive to amantadine and neuraminidase inhibitors. Chicken sera raised against commercial inactivated avian influenza-H5 vaccines showed no or very low reactivity with the currently characterized H5N8 viruses in agreement with the genetic dissimilarity. Surveillance of avian influenza in waterfowl provides early warning of specific threats to poultry and human health and hence should be continued.
Keywords: Highly pathogenic avian influenza virus, H5N8 virus, Egypt, Wild birds
Introduction
Since the first detection of highly pathogenic avian influenza virus (HPAI) H5N1 in China in 1996, the virus has evolved into 10 main genetically distinct clades (clades 0–9) and several subclades of the second-, third- and fourth-order groups [1, 2]. HPAI H5N1 viruses spread during the last decade to Asia, Europe, the Middle East and Africa resulting in the culling and deaths of millions of birds as well as hundreds of human infection cases with a case fatality rate of more than 52 % [3–5]. The hemagglutinin (HA) gene of subtype H5 underwent reassortment with different neuraminidase subtypes (1–9) to form AI H5NX viruses.
In Egypt, domestic poultry were infected with the HPAI H5N1 virus in February 2006 after the virus was detected in wild migratory birds in Damietta Governorate [5]. A few months after the introduction of the virus, the national veterinary authorities devised a comprehensive response plan to control the spread of the virus in Egypt. This included increasing public awareness and biosecurity through media, culling infected poultry, commercial life poultry movement restrictions and emergency vaccination [6, 7]. The number of recorded H5N1 outbreaks decreased as a result of administrating H5-inactivated vaccines [6, 8]. However, the AI H5N1 virus continued to circulate, and the virus became endemic in 2008 leading to a drift of the surface immunogenic glycoproteins [9]. Two subclades of the H5N1 viruses, 2.2.1 and 2.2.1.1, co-circulated in poultry from late 2009 to 2011 [10]. Subclade 2.2.1.1 of the H5N1 viruses is thought to have emerged as a vaccine-escape mutant. Subclade 2.2.1 of the H5N1 viruses continued to evolve to form a new phylogenetic cluster named clade 2.2.1.2 [11], which recently evolved into clade 2.2.1.2a [12].
The HPAI H5N8 virus was first detected during 2010 in live bird markets in China [13]. It has a HA segment of H5 clade 2.3.4.4 with the other seven segments from multiple avian influenza viruses (AVIs) circulating in China [13]. Following this, the HA of this clade has been found in combination with different types of neuraminidase segments forming H5NX viruses that co-circulated at the same time in the same sites [14]. Outbreaks of the HPAI subtype H5N8 virus of clade 2.3.4.4 in domestic poultry and wild birds were reported in poultry farms and wetlands in January 2014 in South Korea [15]. By summer 2014, the virus was detected in Japan, Beringia and Siberia [16, 17]. By the end of 2014, waterfowl migration played an essential role in spreading HPAIV H5N8 in Europe, North America and East Asia [18–20].
Egypt is a habitat to an impressive number of bird species. Specifically, the Nile Delta and the North Mediterranean Coast of Egypt serve as a vital stopover for millions of migratory birds during their annual migration between the Palearctic and Afrotropical ecozones [21]. Two migratory birds’ flyways, the Black Sea–Mediterranean and East African–West Asian, overlap in Egypt. Hence, Egypt acts as a bridge between Europe, Asia and Africa where millions of migrating birds pass during their flights annually particularly in winter. We detected an H5N8 virus in a wild migratory bird in Egypt through systematic surveillance for AI in wild birds. This study was carried out to describe Egyptian H5N8 viruses and its relationships with other contemporary H5N8 viruses.
Results
Among 128 collected samples from 64 birds of six species, only two samples from green-winged teals were found positive for HA activity and M gene real-time PCR (RT-PCR). All birds tested in this study were apparently healthy without any clinical signs of AI infection. Full genome sequences of these two H5N8 isolates, A/green-winged teal/Egypt/871/2016 (H5N8) and A/green-winged teal/Egypt/877/2016 (H5N8) were successfully generated (Table S1, available in the online Supplementary Material).
Genetic analysis of the viral genome of Egyptian H5N8 viruses
PB2
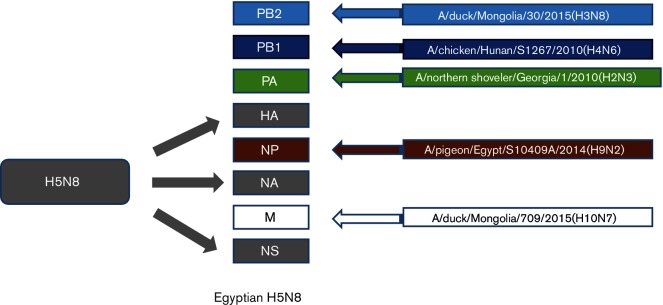
The similarity percentage of amino acid sequences between the currently isolated Egyptian H5N8 viruses was 99.9 %. blastn analysis of the PB2 sequences showed that they had the highest nucleotide identity (98 %) to PB2 of A/duck/Mongolia/30/2015(H3N8), A/duck/Mongolia/118/2015(H4N6) and A/duck/Mongolia/655/2015(H2N3) viruses (Fig. 1), suggesting that Egyptian H5N8 viruses are reassortant viruses. A phylogenetic tree of the PB2 gene of H5N8 viruses showed that the Egyptian H5N8 viruses are closely related to recently characterized reassortant H5N8 viruses from Russia, India and Qinghai Lake in China in 2016 that are of group B and are closely related to various LPAI viruses circulating in aquatic birds in Mongolia in 2015 (Fig. 2). The I504V substitution, which is associated with enhanced activity of the polymerase complex [22], was present in both Egyptian H5N8 isolates. While amino acid residues associated with virulence and virus transmission in mammals, were not found (as shown in Tables S2 and S3).
Fig. 1.
Schematic illustration of the similarity of each gene of the H5N8 viruses isolated from wild birds in Egypt.
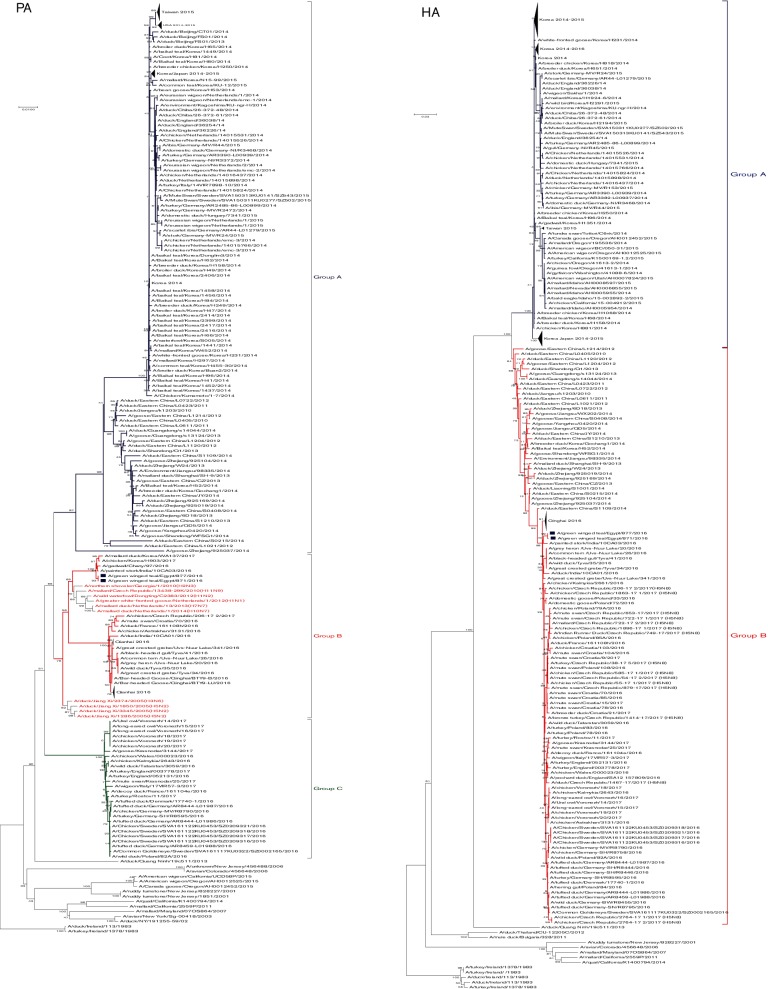
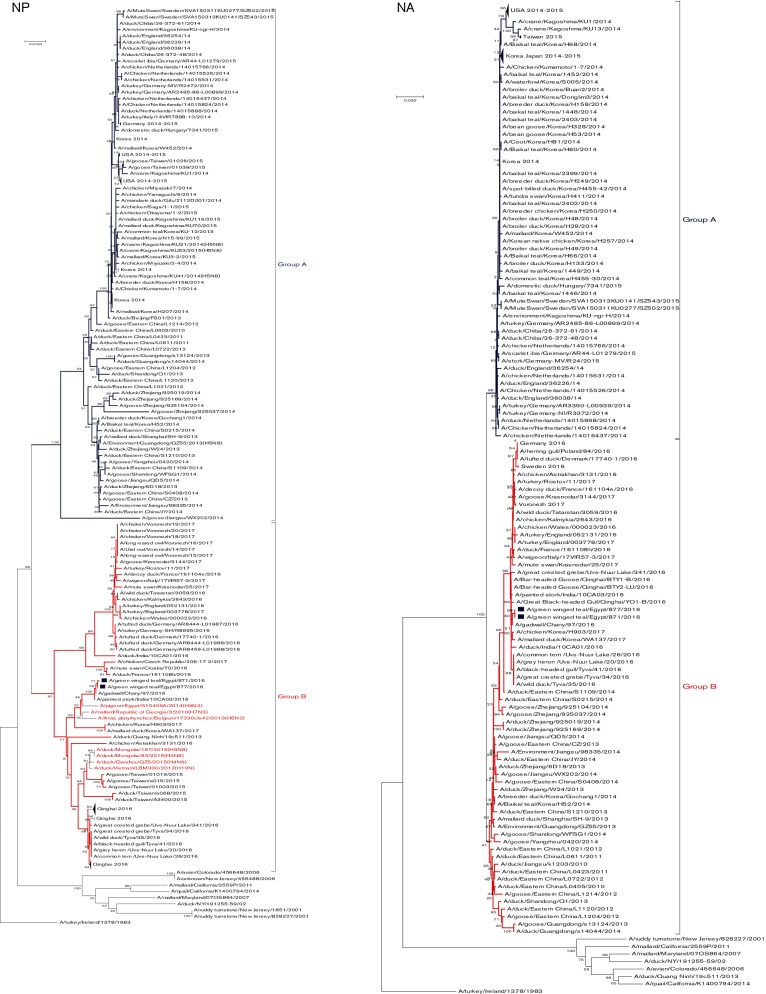
Fig. 2.
Phylogenetic tree of the nucleotide sequences of PB2, PB1, PA, HA, NP, NA, M and NS of the H5N8 viruses isolated in Egypt from wild birds. H5N8 isolates sequenced specifically for this study are labeled with blue squares. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) is shown at the dendrogram nodes. The phylogenetic analysis was performed by using mega version 7. An AI A/turkey/Ireland/1378/1983 (H5N8) virus was used as a root for generated trees.
PB1
The diversity percentage of the nucleotide sequences of the Egyptian H5N8 viruses was 0.6 %. blastn analysis showed that the PB1 genes were closely related to A/chicken/Hunan/S1267/2010(H4N6) and A/duck/Mongolia/179/2015(H3N8) viruses (98 %) (Fig. 1). Like the PB2 gene, the phylogenetic tree of the PB1 gene indicated that the Egyptian H5N8 viruses are closely related to recently characterized reassortant H5N8 viruses from India in 2016 that clustered in H5N8 group B. Among peculiar mutations in the PB1 associated with virulence and virus transmission in mammals, only the mammalian host-specific substitution L13P was identified in both H5N8 isolates (Tables S2 and S3).
PA
Nucleotide sequences of the PA genes of both H5N8 viruses were identical. By blastn analysis, PA genes of the Egyptian H5N8 isolates showed higher similarity to A/northern shoveler/Georgia/1/2010(H2N3) (98 %) as shown in Fig. 1. Phylogenentic analysis showed that there are three distinct groups (A, B and C) of H5N8 PA genes with different ancestors (Fig. 2). PA genes of the Egyptian H5N8 viruses were related to reassortant H5N8 viruses in group B from India, Russia and Korea in 2016 and 2017. Deduced PA amino acid sequences of the two H5N8 isolates indicated that all residues associated with host range are avian-like (Table S2). Amino acid substitutions, R100, L550 and L672 that are associated with virulence, were observed in Egyptian H5N8 isolates (Table S3).
HA
Phylogenetic analysis of the HA gene of the Egyptian H5N8 viruses indicated that they clustered together in group B of H5N8 viruses of clade 2.3.4.4 (Fig. 2). The multibasic cleavage sites of the HA that characterizes the high pathogenicity of AIVs were present in the currently characterized H5N8 viruses in the motif PLREKRRKR↓GLF (↓ denotes cleavage site). Analysis of the N-XT/S motif (X can be any amino acid except proline) using a NetNGlyc 1.0 server revealed that the Egyptian H5N8 isolates have six potential glycosylation sites at positions 10, 23, 165, 286, 483 and 542 within the HA molecule similar to other H5N8 viruses (Table 1). N-linked glycosylation sites of the HA of influenza A viruses affect receptor-binding specificity and/or masking antigenic sites on the globular head.
Table 1. Amino acid characteristics of the HA glycoproteins of the Egyptian H5N8 isolates compared to previously characterized H5N8 viruses.
| H5N8 | Receptor binding sites | N-linked glycosylation site | Antigenic site A | Antigenic site B | Antigenic site E | New antigenic site | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 222 | 224 | 10 | 23 | 165 | 286 | 483 | 542 | 133 | 140 | 141 | 154 | 156 | 184 | 71 | 83 | 86 | 94 | 120 | 162 | 227 | 252 | 263 | 282 | ||
| Egyptian | Q | G | NNST | NVTV | NNTN | NSSM | NGTY | NGSL | A | T | P | N | A | A | I | A | A | S | S | I | D | Y | T | v | |
|
A/turkey/Ireland/ 1378/1983 |
Q | G | NNST | NVTV | SNTN | NSSM | NGTY | NGSL | S | R | S | N | A | A | L | D | V | D | D | R | E | Y | A | I | |
| Group A | Q | G | NNST | NVTV | NNTN | NSSM | NGTY | NGSL | A | A | S | N | A/V | A | I | A | A | T | S | I | D | Y | T | I | |
| Group B | Q | G | NNST | NVTV | NNTN | NSSM | NGTY | NGSL | A | T | P | N | A | A | I | A | A | S | S | I | D | Y | T | V | |
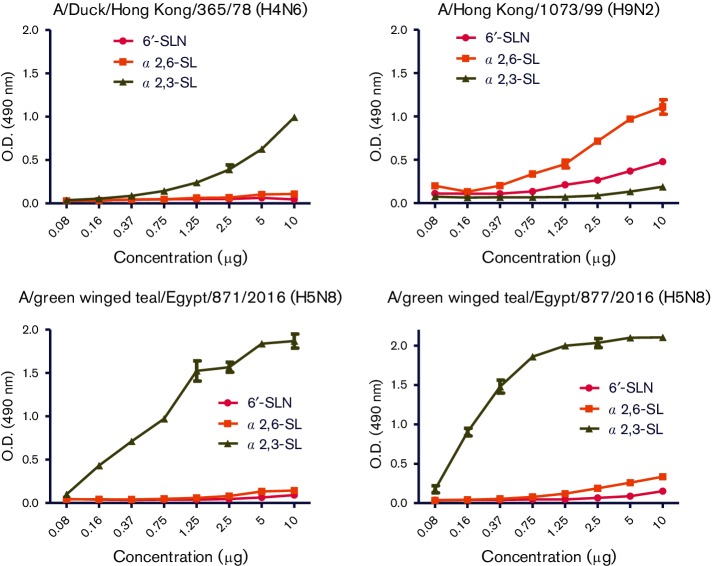
The receptor-binding site (RBS) is a critical viral factor for receptor specificity and influences the generation of human viruses from avian sources. Within the RBS, all Egyptian H5N8 isolates had Q222 and G224 (H5 numbering), indicating preferential binding to sialic acid linked to galactose via 2,3-α-linkages (Sia 2,3-α-Gal) of avian cell-surface receptors. We confirmed the genotypic analysis by the sialic-acid binding preferences of Egyptian H5N8 isolates using a solid phase binding assay. AI (H4N6) and human (H9N2) viruses, which had α2,3-SL and α2,6-SL binding preferences respectively, were used as a point of comparison in this assay. The AI H4N6 virus showed a higher binding preference for α2,3-SL (Fig. 3) while the human A/Hong Kong/1073/99 (H9N2) virus showed a higher binding preference for α2,6-SL and α2,6-SLN (Fig. 3). Egyptian H5N8 isolates had a higher binding preference for α2,3-SL than α2,6-SL and α2,6-SLN, indicating that H5N8 viruses have shown stronger binding specificity toward avian-like receptors.
Fig. 3.
Receptor-binding specificity of Egyptian H5N8 viruses. Binding assay of Egyptian H5N8 viruses to biotinylated sialylglycopolymers containing 3′-sialyllactose (α2,3-SL), 6′-sialyllactose (α2,6-SL) or 6-sialyl-N-acetyllactosamine (6′–SLN) were measured.
No antigenic drift was detected in the antigenic sites in both characterized Egyptian H5N8 viruses that had the same antigenic profile of group B of clade 2.3.4.4 viruses (Table 1). Viruses in group B shared amino acids S94 and V282 (H5 numbering) while those in group A had characteristic T94 and I282 (H5 numbering) amino acids (Table 1).
NP
The diversity percentage of the nucleotide and amino acid sequences of the NP of the currently isolated Egyptian H5N8 viruses was 0.6 and 0.2 %, respectively. blastn analysis showed that the NP genes of the two Egyptian H5N8 were closely related to A/pigeon/Egypt/S10409A/2014(H9N2) (Fig. 1). The phylogenetic tree of the NP gene indicated that the Egyptian H5N8 viruses are closely related to recently characterized reassortant H5N8 viruses from India in 2016 that clustered in H5N8 group B (Fig. 2). Sequence analysis of the NP genes of Egyptian H5N8 viruses showed only one mammalian host–specific marker at K398Q in both isolates (Table S2).
NA
The homology between the amino acid sequences of the NA glycoprotein of both H5N8 isolates was 99.8 %. Analysis of stalk length revealed that no stalk deletions were present in all H5N8 clade 2.3.4.4 viruses. The NA genes of the Egyptian viruses contained five glycosylation sites at positions 54, 67, 84, 144 and 293. The NA of the Egyptian H5N8 viruses had the I314/312V (N2/N8 numbering) substitution, which is a molecular marker for oseltamivir resistance [23]. Other molecular markers of oseltamivir resistance (I117V, E119V, D198N, H274Y, R292K and N294S) and zanamivir resistance (V116A, R118K, E119G/A/D, Q136K, D151E, R152K, R224K, E276D, R292K and R371K) (N2 numbering) were not observed among either Egyptian H5N8 or any H5N8 elsewhere [23]. In agreement with NA gene sequence analysis, both Egyptian H5N8 viruses were sensitive to zanamivir in concentrations ranging from 1 to 20 µM with IC50 (inhibitory concentration 50) <1 µM. Testing the sensitivity of H5N8 viruses toward different concentrations of oseltamivir ranging from 1 to 20 µM showed that H5N8 viruses were sensitive to oseltamivir with IC50 <1 µM contrary to NA sequence findings.
A phylogenetic tree showed that Egyptian viruses clustered together in group B within clade 2.3.4.4 (Fig. 2). Egyptian isolates showed a close relationship to isolates from India, Russia and Korea.
M
M genes of Egyptian isolates showed higher identity to group B H5N8 viruses isolated from India, Cheny lake in Russia and recently isolated H5N8 viruses from Korea in 2017 (Fig. 2). M genes of group B H5N8 viruses were closely related to the A/duck/Mongolia/709/2015(H10N7) virus (98 %) (Fig. 1).
Mammalian-specific residues in M1 and M2 viral proteins were not found in both Egyptian H5N8 isolates (Table S3). Those isolates possessed the virulent form of amino acid residues at positions 64 and 69 in the M2 viral protein (Table S2).
The genetic analysis of the amino acid residues associated with resistance of amantadine in the M2 protein revealed that both isolates had L26, V27, A30, S31 and G34, indicating sensitivity to amantadine. This was confirmed through biological testing as our results showed that both H5N8 viruses were amantadine-susceptible at different concentrations of amantadine (Table 2).
Table 2. Susceptibility of Egyptian H5N8 viruses to amantadine compared to control resistance and sensitive H5N1 viruses.
| H5Nx virus | Amantadine (μg 500 µl−1)* | Phenotype | Susceptibility (S)/resistance (R) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 2 | 20 | 40 | 100 | 26 I | 27A | 31 N | 33A | 34E | ||
| A/green-winged teal/Egypt/871/2016 (H5N8) | 64 | 8 | 8 | 2 | 0 | L | V | S | I | G | S |
| 64 | 8 | 8 | 0 | 0 | |||||||
| 64 | 16 | 8 | 0 | 0 | |||||||
| A/green-winged teal/Egypt/877/2016 (H5N8) | 128 | 16 | 16 | 16 | 0 | L | V | S | I | G | S |
| 128 | 32 | 32 | 8 | 0 | |||||||
| 128 | 32 | 16 | 8 | 0 | |||||||
| A/chicken/Egypt/S10739C/2015(H5N1) | 32 | 32 | 32 | 32 | 16 | L | A | S | I | G | R |
| 32 | 32 | 32 | 16 | 16 | |||||||
| 32 | 32 | 32 | 32 | 16 | |||||||
| A/chicken/Egypt/M2583D/2010 (H5N1) | 16 | 4 | 0 | 0 | 0 | L | V | S | I | G | S |
| 16 | 4 | 0 | 0 | 0 | |||||||
| 16 | 2 | 0 | 0 | 0 | |||||||
| A/duck/Egypt/Q4596D/2012 (H5N1) | 64 | 8 | 8 | 0 | 0 | L | V | S | I | G | S |
| 64 | 8 | 8 | 4 | 0 | |||||||
| 64 | 8 | 8 | 4 | 0 | |||||||
*Haemagglutination titre of the supernatant from infected MDCK cells after 24 h post infection with 0.01 multiplicity of infection (MOI) of indicated virus in the presence or absence of amantadine.
NS
NS segment of the two Egyptian H5N8 isolates showed higher similarity to H5N8 viruses isolated previously from China that are closely related to group B of clade 2.3.4.4 (Fig. 2). All the current H5N8 isolates had PDZ motif (X-S/T-X-V) at the C-terminal of the NS1 protein in the form of GSEV. The NS1 of Egyptian H5N8 viruses had virulence determinants S42. Mammalian-specific residues in NS1 and NS2 viral proteins were not found in both Egyptian H5N8 isolates. Egyptian H5N8 strains exhibited no substitutions of NS2 virulence determinant residues at position 31 and 56 (Table S2).
Antigenic analysis and efficacy of commercial H5-inactivated vaccines
All Egyptian H5N8 viruses were not reactive with H5 mAbs that were prepared against different antigenic epitopes of the A/Vietnam/1203/2004 (H5N1, clade 1) virus and the A/bar-headed goose/QH/1A/05 (H5N1, clade 2.2.1) virus (Table 3). Also, the two H5N8 isolates showed moderate or low reactivity with anti-H5 hyperimmune sera raised against H5N1 viruses of clades 2.2.1, 2.2.1.1a, 2.2.1.2 and 2.3.2.1c (Table 4). Similarly, rat sera raised against inactivated H5N8 vaccines showed low reactivity with heterologous H5 antigens of clades 2.2.1, 2.2.1.1a, 2.2.1.2 and 2.3.2.1c.
Table 3. Haemagglutination inhibition reactions of the Egyptian H5N8 viruses compared to H5N1 viruses from different clades with a panel of H5-mAbs using 0.5 % chicken erythrocytes.
| Viruses | Clade | VN04-2 | VN04-15 | VN04-8 | VN04-9 | VN04-10 | VN04-13 | VN04-16 | BHG05-1 | BHG05-2 | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| A/green-winged teal/Egypt/871/2016 (H5N8) | 2.3.4.4 | <100 | <100 | <100 | <100 | <100 | <100 | <100 | <100 | <100 | |
| A/green-winged teal/Egypt/877/2016 (H5N8) | 2.3.4.4 | <100 | <100 | <100 | <100 | <100 | <100 | <100 | <100 | <100 | |
| A/turkey/Egypt/7/2007 (H5N1) | 2.2.1 | 400 | 3200 | 6400 | <100 | 800 | 1600 | 200 | 400 | <100 | |
| A/chicken/Egypt/Q1995A/2010 (H5N1) | 2.2.1.1a | <100 | <100 | <100 | <100 | 6400 | 200 | 6400 | <100 | <100 | |
| A/duck/Egypt/M2583D/2010 (H5N1) | 2.2.1.2 | <100 | 1600 | 12 800 | <100 | <100 | 3200 | <100 | 6400 | <100 | |
| A/chicken/Egypt/10552B/2015(H5N1) | 2.2.1.2 | <100 | 6400 | 12 800 | <100 | <100 | 12 800 | <100 | 800 | <100 | |
| A/chicken/Lebanon/157/2016(H5N1) | 2.3.2.1c | <100 | <100 | <100 | 200 | 800 | <100 | 1600 | 1600 | <100 | |
| A/Vietnam/1203/2004 (H5N1) | 1 | 6400 | nd* | 1600 | 12 800 | 3200 | 6400 | 1600 | nd | nd | |
| A/chicken/Indonesia/PA03/03 (H5N1) | 2 | 800 | nd | 3200 | 200 | 3200 | 1600 | 1600 | nd | nd | |
| A/duck/Singapore/3/1997 (H5N1) | 2.3 | 200 | nd | <100 | 200 | 800 | 6400 | 200 | nd | nd | |
*nd, Not done.
Table 4. Cross-reactivity of chicken and rat antisera of commercial and experimental inactivated H5 vaccines against the current Egyptian H5N8 isolates compared to representative H5N1 viruses from different clades. Data represent log2 titres of the hemagglutination inhibition assay using 0.5 % chicken red blood cells (RBCs).
| Viruses | Clade | A/bar-headed goose/Qinghai/1A/2005 (H5N1) | A/whooper swan/Mongolia/244/2005 (H5N1) | A/chicken/Egypt/1C/2006 (H5N1) | A/Egypt/321/07 (H5N1) | A/turkey/Qalubia-Egypt/7/07 (H5N1) | A/Egypt/3300-NAMRU/2008 (H5N1) | A/duck/Egypt/M2583D/2010 (H5N1) | A/Egypt/NO3072/2010 (H5N1) | A/chicken/Egypt/10552B/2015 (H5N1) | A/chicken/Lebanon/157/2016 (H5N1) | A/green winged teal/Egypt/877/2016 (H5N8) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2.2 | 2.2 | 2.2.1 | 2.2.1 | 2.2.1 | 2.2.1 | 2.2.1.2. | 2.2.1 | 2.2.1.2 | 2.3.2.1c | 2.3.4.4 | ||
| A/green-winged teal/Egypt/871/2016 (H5N8) | 2.3.4.4 | 4 | >3 | >3 | >3 | 4 | >3 | 4 | >3 | >3 | 4 | 7 |
| A/green-winged teal/Egypt/877/2016 (H5N8) | 2.3.4.4 | 5 | >3 | >3 | >3 | 4 | >3 | 5 | >3 | 3 | 4 | 7 |
| A/turkey/Egypt/7/2007 (H5N1) | 2.2.1 | 8 | 9 | 10 | 6 | 8 | 7 | 6 | 7 | 4 | 7 | 4 |
| A/chicken/Egypt/Q1995A/2010 (H5N1) | 2.2.1.1a | 7 | 3 | 8 | 6 | 4 | 8 | >3 | >3 | 5 | 7 | >3 |
| A/duck/Egypt/M2583D/2010 (H5N1) | 2.2.1.2 | 8 | 7 | 8 | 7 | 6 | 5 | 8 | 6 | 6 | 6 | 3 |
| A/chicken/Egypt/10552B/2015 (H5N1) | 2.2.1.2 | 9 | 7 | 9 | 7 | 7 | 7 | 8 | 8 | 8 | 6 | 4 |
| A/chicken/Lebanon/157/2016 (H5N1) | 2.3.2.1c | 6 | 3 | 5 | >3 | 5 | 5 | 7 | 3 | 3 | 7 | >3 |
Sera obtained from vaccinated chickens with different commercial inactivated H5-based vaccines showed no or very low hemagglutination inhibition (HI) titres with H5N8 viruses in agreement with the genetic dissimilarity (Table 5).
Table 5. Cross-reactivity of antisera of commercial inactivated H5 AIV vaccines to Egyptian H5N8 viruses compared to H5N1 viruses from different clades. Data represent log2 titres of the hemagglutination inhibition assay using 0.5 % chicken RBCs.
| Commercial name | FLUKEM | LOHMAN | VOLVAC | Yebio | Yebio Re-1 | Egyflu | |
|---|---|---|---|---|---|---|---|
| Viruses | Clade | A/chicken/Mexico/232/1994 (H5N2) | A/turkey/Minnesota/3689–1551/1981(H5N2) | A/chicken/Mexico/232/94(H5N2) | A/turkey/England/N28/73 (H5N2) | A/goose/Guangdong/96 (H5N1) | A/chicken/Egypt/18 h/2009 (H5N1) |
| A/green-winged teal/Egypt/871/2016 (H5N8) | 2.3.4.4 | <3 | <3 | <3 | <3 | <3 | <3 |
| A/green-winged teal/Egypt/877/2016 (H5N8) | 2.3.4.4 | <3 | <3 | <3 | 3 | 3 | 3 |
| A/turkey/Egypt/7/2007 (H5N1) | 2.2.1 | 4 | 4 | 3 | 6 | 6 | 7 |
| A/chicken/Egypt/Q1995A/2010 (H5N1) | 2.2.1.1a | 3 | 4 | 6 | 5 | 6 | 6 |
| A/duck/Egypt/M2583D/2010 (H5N1) | 2.2.1.2 | 5 | 5 | 3 | 5 | 5 | 6 |
| A/chicken/Egypt/10552B/2015 (H5N1) | 2.2.1.2 | 5 | 5 | 5 | 6 | 6 | 6 |
| A/chicken/Lebanon/157/2016 (H5N1) | 2.3.2.1c | <3 | <3 | <3 | 3 | 4 | <3 |
Discussion
Wild birds are known to play a major role in the evolution and spread of AIVs during seasonal migration. Wild aquatic birds migrate for breeding, better ecological conditions and molting. The majority of wild aquatic birds migrate from northern breeding areas to southern wintering grounds using different migration flyways. This unique ecosystem played an essential role in the dissemination of HPAIV H5N1 clade 2.2 during 2005–2006 from Asia to Europe and Africa [24]. Egypt serves as a vital stopover for millions of wild migratory birds during their annual migration between the Palearctic and Afrotropical ecozones [21]. Saad et al. [5] reported the possibility of emergence of HPAIV H5N1 through common teal [5]. Also in this study, we provided a detailed characterization of full genomes of HPAI H5N8 viruses isolated from green-winged teal in Egypt in 2016. Several outbreaks of H5N8 were reported to the World Organization for Animal Health (OIE) in backyard and non-regulated farms in different Egyptian governorates during the first quarter of this year following the detection of H5N8 viruses in wild birds [25].
No clinical signs of HPAI infection were observed during sampling from different wild birds’ species. A previous study showed that there is a significant species-related variation in susceptibility and clinical signs to H5N1 virus infection in wild birds [26].
To understand the genetic relationship between Egyptian H5N8 isolates and related viruses, the eight segments of the current viruses were phylogenetically analysed with counterparts from all available sequences. Phylogenetic analysis revealed that the HA of Egyptian H5N8 isolates are related to clade 2.3.4.4 H5N8 viruses. Except for the PA segment, we found that the genes were genetically divided into two main groups A and B. Egyptian H5N8 viruses fell into a phylogenetic cluster together with Indian and Russian H5N8 isolates (group B, indicated in red in the phylogenetic figures).
Genetic relatedness of PB2, PB1, PA, NP and M segments of group B including Egyptian H5N8 viruses with other AI subtypes such as H3N9, H4N6, H2N3, H10N7 and H9N2, rather than to ancestral H5N8 viruses indicates intra-reassortments among different AI subtypes to form the new group B of H5N8 viruses. Our report suggests that the current Egyptian H5N8 viruses have emerged from multiple reassortment events that occurred before the virus was detected in Egypt. Recently, several studies showed novel reassortment of clade 2.3.4.4 (H5N8) viruses in wild aquatic birds and domestic poultry during 2016 [27–30]. Phylogenetic analysis of full genome sequences revealed that Egyptian H5N8 viruses clustered with clade 2.3.4.4B with recent viruses detected in Russia, India, China and Germany. This reassortment constellation probably emerged in Asia.
The current Egyptian H5N8 viruses have many molecular markers associated with virulence in mammals and adaptation to mammalian hosts in the PB2 (504V), PB1 (13P), PA (100R, 550L and 672L) and NP (398Q). Both Egyptian H5N8 isolates have S42 in NS1, which can increase the virulence of AIV in mammalian models [31]. NS1 viral protein of some influenza A viruses possess PDZ motifs at the C-terminal sequences [32]. Previous studies showed that this motif plays an important role in the virulence of the influenza virus [33, 34]. Egyptian H5N1 showed four forms (ESKV, EPKV, ESEV and ESEI) of PDZ motif. All the current H5N8 isolates had PDZ motif at the C-terminal of NS1 protein in the form of GSEV.
A major determinant of influenza virus virulence and host range is the viral surface HA glycoprotein. Presence of polybasic sequences at the HA1/HA2 cleavage site in both HAs of two H5N8 isolates indicate the virulence of these isolates. HA receptor-binding characteristics affect influenza virus host range [35]. Analysis of the RBS of the Egyptian H5N8 viruses indicated that these viruses harbour AVI-like RBS and do not possess any of the residues known to be specific for pandemic H1, H2 and H3 viruses [36]. A previous study showed that four mutations (Q222L, G224S, T156A and H103Y) in HA glycoprotein of the H5N1 virus can render it transmissible between ferrets via respiratory droplets [37]. Only mutation T156A is found in all HAs of H5N8 viruses. Most of the H5N1 viruses have a glycosylation site at 154–156 so that this mutation leads to the loss of a glycosylation site at position 154–156, hence increasing the ability of the H5N1 virus transmission to mammals [37]. Our results of the receptor specificity assay reveal that Egyptian H5N8 viruses have a strict avian receptor-binding preference.
HI analysis revealed a low reactivity of sera raised against these H5N8 viruses to clades 2.2.1, 2.2.1.1, 2.2.1.2 and 2.3.2.1c and vice versa. Although Egypt has used AI H5 vaccines for more than 10 years, AI H5N1 viruses have been undergoing antigenic drift due to the presence of immune pressure and could therefore escape from vaccine protection [6]. Several commercial inactivated AI vaccines were widely used in domestic poultry [38, 39]. Contrary to our results, a previous study revealed the protective efficacy of a stockpiled vaccine based on A/duck/Hokkaido/Vac-1/2004 (H5N1, classical clade) against the HPAI H5N8 virus [40]. We showed the poor reactivity of all commercial vaccines against recently characterized H5N8 viruses. The genetic dissimilarity and poor reactivity between H5 commercial vaccines used in Egypt and currently circulating H5N8 viruses indicates that the vaccines might not be effective in the field or at least introduce partial protection and thus might lead to vaccine-induced escape mutant strains. Improper antigenic matching between vaccines and circulating AIVs might reduce vaccine efficacy and accelerate antigenic drift of circulating viruses [41].
Given the previous experience with HPAI H5N1 in Egypt, and given our current findings concerning the lack of cross-reactivity between current vaccines used in Egypt and H5N8 viruses, we strongly recommend that Egyptian authorities prepare for the scenario when H5N8 is spreading in domestic poultry. A comprehensive preparedness and response plan must be devised focusing on increased surveillance to detect the virus early and the actions to be taken to prevent its spread. It is important to note that if emergency vaccination will be adopted, H5N8 vaccine candidates from local strains should be used to reduce the risk of emergence of vaccine-escape mutants.
Methods
Samples and virus isolation
In total, 128 oral and cloacal samples were collected from 64 live birds in a live bird market located in Port Said City north of Egypt on 8 December 2016. Samples were collected from 33 green-winged teals (Anas carolinensis), 24 shovelers (Anas clypeata), three common coots (Fulica atra), two pintails (Anas acuta), one mallard (Anas platyrhynchos) and one moorhen (Gallinula chloropus). No clinical signs of AIV infection were observed in birds during sampling. Oral and cloacal swabs were inoculated into 10-day-old SPF-embryonated chicken eggs for virus isolation. The allantoic fluids of the inoculated eggs were tested for agglutination activity via hemagglutination assay (HA). HA was performed using 0.5 % chicken erythrocytes according to WHO protocol [42]. The positive HA samples were subjected to viral RNA extraction using a QIAamp viral RNA mini kit (Qiagen, Germany) according to the manufacturer's protocol, then typed by M gene using RT-PCR [43]. The positive M gene samples were further subtyped for HA and NA as previously described [44, 45].
Full genome amplification and sequencing
The cDNA was synthesized using Superscript III reverse transcriptase (Invitrogen, Carlsbad, CA, USA) and Uni-12 primer (5′-AGCRAAAGCAGG-3′) according to the manufacturer’s protocol. Genes were amplified using the Phusion master mix kit (Thermo, MA, USA) and universal primers [46]. Amplicons of appropriate sizes were subsequently gel-purified using QIAGEN gel extraction kit (Qiagen). The purified PCR products were directly used for sequencing reactions at Macrogen Sequencing Facility (Macrogen, South Korea). Full genome sequences were assembled using SeqMan DNA Lasergene 7 software (DNASTAR, Madison, WI, USA). Sequences were deposited in GenBank under accession numbers listed in Table S1.
Sequence analysis and phylogenetic tree construction
The assembled sequences were subjected to NCBI blastn analysis (https://blast.ncbi.nlm.nih.gov/Blast.cgi). BioEdit 7.0 was used for multiple sequence alignment [47]. The nucleotide and amino acid homologies were further assessed by the ClustalW method of MegAlign (DNASTAR). mega 7.0 was used for phylogenetic tree construction by applying the neighbour-joining method with Kimura’s two-parameter distance model and 1000 bootstrap replicates [48]. The trees included the currently assembled Egyptian virus sequences; all related viruses to our isolates are available on the Global Initiative on Sharing All Influenza Data (GISAID) and GenBank till 14/3/2017. BioEdit program version 7.0 was used for genomic signature analysis.
Antigenic analysis
Antigenic analyses of Egyptian H5N8 viruses were performed by HI test using a panel of antisera generated either by vaccination of SPF chicken groups with different reverse genetics (Rg) H5N1-inactivated vaccines A/chicken/Lebanon/157/2016 (H5N1, clade 2.3.2.1c) and A/chicken/Egypt/10552B/2015 (H5N1, clade 2.2.1.2) or infection of ferrets with Rg viruses [A/bar-headed goose/Qinghai/1A/2005 (H5N1, clade 2.2), A/whooper swan/Mongolia/244/2005 (H5N1, clade 2.2), A/Egypt/321/2007 (H5N1, clade 2.2.1), A/turkey/Qalubia-Egypt/7/07 (H5N1, clade 2.2.1), A/Egypt/3300-NAMRU/2008 (H5N1, clade 2.2.1), A/chicken/Egypt/1C/2006 (H5N1, clade 2.2.1), A/Egypt/NO3072/2010 (H5N1, clade 2.2.1), A/duck/Egypt/M2583D/2010 (H5N1, clade 2.2.1.2)]. Rats were used for production of antisera for the A/green-winged teal/Egypt/871/2016 (H5N8) virus using inactivated vaccine. H5N1- and H5N8-inactivated vaccines were individually prepared using viruses inactivated with 0.1 % formalin [49] and mixed with Montanide ISA 70 VG (Seppic, France) in the ratio 30 viral antigen: 70 adjuvant according to the manufacturer’s instructions. Four weeks post vaccination of chickens and rats, and 3 weeks post infection of ferrets, sera were collected from immunized animals and were tested for antibodies against homologous H5N1 and H5N8 antigens using HI [42].
A panel of anti-H5 monoclonal antibodies (mAbs) (VN04-2, VN04-8, VN04-9, VN04-10, VN04-13, VN0415 and VN04-16) prepared against different antigenic epitopes of the A/Viet Nam/1203/2004 (H5N1) virus and two mAbs (BHG05-1 and BHG05-2) raised against the A/bar-headed goose/QH/1A/05 (H5N1) virus were used to antigenically characterize the different H5N1 isolates using the HI assay.
Archived sera obtained from chickens vaccinated with commercial vaccines used in Egypt that were produced for a previous study [7] and an experimental Rg vaccine based on clade 2.2.1.2 A/chicken/Egypt/10552B/2015(H5N1) were tested against Egyptian H5N8 isolates and different H5N1 viruses from clades 2.2.1, 2.2.1.1a, 2.2.1.2 and 2.3.2.1c.
Receptor specificity assay
Virus receptor specificity was determined as previously described [50]. The 96-well fetuin-coated (10 µg ml−1) plates were washed with ice cold washing buffer (0.01 % Tween 80 in 0.23X PBS), blocked with PBS containing 1 % bovine serum albumin (BSA), and incubated overnight with 32 hemagglutination (HA) units of influenza viruses at 4 °C. Plates were washed with washing buffer four times. Biotinylated sialylglycopolymers, 3′-sialyllactose (α2,3-SL, Neu5Acα2-3Galβ1-4Glc), 6′-sialyllactose (α2,6-SL, Neu5Acα2-6Galβ1-4Glc) and 6-sialyl-N-acetyllactosamine (6′–SLN, Neu5Acα2-6Galβ1-4GlcNAc) (Glycotech,Gaithersburg, MD, USA) were serially diluted in reaction buffer [0.02 % Tween 80, 0.02 % BSA, 1 µM sialdase inhibitor (Zanamivir) and 1X PBS] and were added and incubated at 4 °C for 2 h. The plates were washed (4X) and incubated with 100 µl of horse-radish peroxidase–conjugated streptavidin (1 : 2000) at 4 °C for 1 h. After a final wash, a volume of 50 µl of the o-Phenylenediamine (OPD) substrate was added to each well and incubated for 10 min at room temperature. The reaction was stopped using 1 N sulfuric acid, and absorbance was measured at 490 nm.
Antiviral sensitivity assays
The susceptibility of Egyptian H5N8 viruses to amantadine was performed as previously described [51]. Briefly, stocks of amantadine hydrochloride (Amantadine, Sigma–Aldrich, St. Louis, MO, USA) were prepared in distilled water. Monolayers of MDCK cells in a 12-well tissue culture plate were pre-treated with 400 µl of Dulbecco's modified Eagle's medium supplemented with 0.2 % bovine serum albumin containing 0, 0.2, 2.0, 20, 40 or 100 µg of amantadine, for 45 min at 37 °C in 5 % CO2. Cells were then washed with PBS and infected with 0.01 multiplicity of infection (MOI) for 1 h at 37 °C. Culture medium (1 ml) containing the respective concentration of amantadine was added to each well. Plates were incubated at 37 °C with 5 % CO2 for 24 h. Virus replication was assessed by measuring hemagglutinin titres in the supernatant.
Resistance of the Egyptian H9N2 viruses to Zanamivir (Sigma) and oseltamivir (TRC, Canada) was determined in MDCK cells by plaque reduction assay as described previously [52]. Cultured MDCK cells in six-well plates were inoculated with the current isolated viruses diluted in infection medium to give 30 to 100 plaques per well. The Egyptian viruses were tested for sensitivity against antiviral drug concentrations of 1, 5, 10 or 20 µM. Cells were incubated for 1 h at 37 °C and then overlaid with a DMEM overlay medium containing 1 % agarose, 4 % BSA and 1 % antibiotic-antimycotic mixture (Worthington Diagnostics, Freehold, NJ, USA). After 4 days of incubation at 37 °C, plaques were visualized by staining with 0.1 % crystal violet containing 10 % formaldehyde. The percentages of viral inhibition relative to untreated control viruses were calculated at each concentration.
Funding information
This work was funded by the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services, under contract number HHSN272201400006C; by the Science and Technology Development Fund (STDF) in Egypt, under contract number 5175; and supported by the American Lebanese Syrian Associated Charities (ALSAC).
Conflicts of interest
The authors declare that there are no conflicts of interest.
Ethical statement
All applicable international, national and/or institutional guidelines for the care and use of animals were followed. This article does not contain any studies with human participants performed by any of the authors.
Supplementary Data
Footnotes
Abbreviations: AIV, Avian influenza virus; HPAI, Highly pathogenic avian influenza; mAbs, Monoclonal antibodies; HI, Hemagglutination inhibition.
Three supplementary tables are available with the online Supplementary Material.
References
- 1.Chen H, Deng G, Li Z, Tian G, Li Y, et al. The evolution of H5N1 influenza viruses in ducks in southern China. Proc Natl Acad Sci USA. 2004;101:10452–10457. doi: 10.1073/pnas.0403212101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.WHO/OIE/FAO . Continued Evolution of Highly Pathogenic Avian Influenza a (H5N1): Updated Nomenclature. 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang G, Zhan D, Li L, Lei F, Liu B, et al. H5N1 avian influenza re-emergence of Lake Qinghai: phylogenetic and antigenic analyses of the newly isolated viruses and roles of migratory birds in virus circulation. J Gen Virol. 2008;89:697–702. doi: 10.1099/vir.0.83419-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kwon HI, Song MS, Pascua PN, Baek YH, Lee JH, et al. Genetic characterization and pathogenicity assessment of highly pathogenic H5N1 avian influenza viruses isolated from migratory wild birds in 2011, South Korea. Virus Res. 2011;160:305–315. doi: 10.1016/j.virusres.2011.07.003. [DOI] [PubMed] [Google Scholar]
- 5.Saad MD, Ahmed LS, Gamal-Eldein MA, Fouda MK, Khalil F, et al. Possible avian influenza (H5N1) from migratory bird, Egypt. Emerg Infect Dis. 2007;13:1120–1121. doi: 10.3201/eid1307.061222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kayali G, Kandeil A, El-Shesheny R, Kayed AS, Maatouq AM, et al. Avian influenza A(H5N1) Virus in Egypt. Emerg Infect Dis. 2016;22:379–388. doi: 10.3201/eid2203.150593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kayali G, Kandeil A, El-Shesheny R, Kayed AS, Gomaa MR, et al. Do commercial avian influenza H5 vaccines induce cross-reactive antibodies against contemporary H5N1 viruses in Egypt? Poult Sci. 2013;92:114–118. doi: 10.3382/ps.2012-02637. [DOI] [PubMed] [Google Scholar]
- 8.Swayne DE. Impact of vaccines and vaccination on global control of avian influenza. Avian Dis. 2012;56:818–828. doi: 10.1637/10183-041012-Review.1. [DOI] [PubMed] [Google Scholar]
- 9.Abdelwhab EM, Hafez HM. Insight into alternative approaches for control of avian influenza in poultry, with emphasis on highly pathogenic H5N1. Viruses. 2012;4:3179–3208. doi: 10.3390/v4113179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kayali G, Webby RJ, Ducatez MF, El Shesheny RA, Kandeil AM, et al. The epidemiological and molecular aspects of influenza H5N1 viruses at the human-animal interface in Egypt. PLoS One. 2011;6:e17730. doi: 10.1371/journal.pone.0017730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.El-Shesheny R, Kandeil A, Bagato O, Maatouq AM, Moatasim Y, et al. Molecular characterization of avian influenza H5N1 virus in Egypt and the emergence of a novel endemic subclade. J Gen Virol. 2014;95:1444–1463. doi: 10.1099/vir.0.063495-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Salaheldin AH, Veits J, Abd El-Hamid HS, Harder TC, Devrishov D, et al. Isolation and genetic characterization of a novel 2.2.1.2a H5N1 virus from a vaccinated meat-turkeys flock in Egypt. Virol J. 2017;14:48. doi: 10.1186/s12985-017-0697-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee YJ, Kang HM, Lee EK, Song BM, Jeong J, et al. Novel reassortant influenza A(H5N8) viruses, South Korea, 2014. Emerg Infect Dis. 2014;20:1087–1089. doi: 10.3201/eid2006.140233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hall JS, Dusek RJ, Spackman E. Rapidly expanding range of highly pathogenic avian influenza viruses. Emerg Infect Dis. 2015;21:1251–1252. doi: 10.3201/eid2107.150403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jeong J, Kang HM, Lee EK, Song BM, Kwon YK, et al. Highly pathogenic avian influenza virus (H5N8) in domestic poultry and its relationship with migratory birds in South Korea during 2014. Vet Microbiol. 2014;173:249–257. doi: 10.1016/j.vetmic.2014.08.002. [DOI] [PubMed] [Google Scholar]
- 16.Kanehira K, Uchida Y, Takemae N, Hikono H, Tsunekuni R, et al. Characterization of an H5N8 influenza A virus isolated from chickens during an outbreak of severe avian influenza in Japan in April 2014. Arch Virol. 2015;160:1629–1643. doi: 10.1007/s00705-015-2428-9. [DOI] [PubMed] [Google Scholar]
- 17.Marchenko VY, Susloparov IM, Kolosova NP, Goncharova NI, Shipovalov AV, et al. Influenza A(H5N8) virus isolation in Russia, 2014. Arch Virol. 2015;160:2857–2860. doi: 10.1007/s00705-015-2570-4. [DOI] [PubMed] [Google Scholar]
- 18.Bouwstra R, Heutink R, Bossers A, Harders F, Koch G, et al. Full-genome sequence of influenza A(H5N8) virus in poultry linked to sequences of strains from Asia, the Netherlands, 2014. Emerg Infect Dis. 2015;21:872–874. doi: 10.3201/eid2105.141839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hanna A, Banks J, Marston DA, Ellis RJ, Brookes SM, et al. Genetic characterization of highly pathogenic avian influenza (H5N8) Virus from domestic ducks, England, November 2014. Emerg Infect Dis. 2015;21:879–882. doi: 10.3201/eid2105.141954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pasick J, Berhane Y, Joseph T, Bowes V, Hisanaga T, et al. Reassortant highly pathogenic influenza A H5N2 virus containing gene segments related to Eurasian H5N8 in British Columbia, Canada, 2014. Sci Rep. 2015;5:9484. doi: 10.1038/srep09484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Denny P. In: Africa. Finlayson M, Moser M, editors. Wetlands, London: International Waterfowl and Wetlands Research Bureau; 1991. (editors) [Google Scholar]
- 22.Rolling T, Koerner I, Zimmermann P, Holz K, Haller O, et al. Adaptive mutations resulting in enhanced polymerase activity contribute to high virulence of influenza A virus in mice. J Virol. 2009;83:6673–6680. doi: 10.1128/JVI.00212-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Orozovic G, Orozovic K, Lennerstrand J, Olsen B. Detection of resistance mutations to antivirals oseltamivir and zanamivir in avian influenza A viruses isolated from wild birds. PLoS One. 2011;6:e16028. doi: 10.1371/journal.pone.0016028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Olsen B, Munster VJ, Wallensten A, Waldenström J, Osterhaus AD, et al. Global patterns of influenza A virus in wild birds. Science. 2006;312:384–388. doi: 10.1126/science.1122438. [DOI] [PubMed] [Google Scholar]
- 25.OIE Update on avian influenza in animals. 2017. www.oie.int/wahis_2/public%5C.%5Ctemp%5Creports/en_fup_0000023847_20170522_184813.pdf
- 26.Brown JD, Stallknecht DE, Beck JR, Suarez DL, Swayne DE. Susceptibility of North American ducks and gulls to H5N1 highly pathogenic avian influenza viruses. Emerg Infect Dis. 2006;12:1663–1670. doi: 10.3201/eid1211.060652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee DH, Sharshov K, Swayne DE, Kurskaya O, Sobolev I, et al. Novel reassortant clade 2.3.4.4 avian influenza A(H5N8) virus in wild aquatic birds, Russia, 2016. Emerg Infect Dis. 2017;23:359–360. doi: 10.3201/eid2302.161252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li M, Liu H, Bi Y, Sun J, Wong G, et al. Highly pathogenic avian influenza A(H5N8) virus in wild migratory birds, Qinghai Lake, China. Emerg Infect Dis. 2017;23:637–641. doi: 10.3201/eid2304.161866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nagarajan S, Kumar M, Murugkar HV, Tripathi S, Shukla S, et al. Novel reassortant highly pathogenic avian influenza (H5N8) virus in Zoos, India. Emerg Infect Dis. 2017;23:717–719. doi: 10.3201/eid2304.161886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pohlmann A, Starick E, Harder T, Grund C, Höper D, et al. Outbreaks among wild birds and domestic poultry caused by reassorted influenza A(H5N8) clade 2.3.4.4 viruses, Germany, 2016. Emerg Infect Dis. 2017;23:633–636. doi: 10.3201/eid2304.161949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jiao P, Tian G, Li Y, Deng G, Jiang Y, et al. A single-amino-acid substitution in the NS1 protein changes the pathogenicity of H5N1 avian influenza viruses in mice. J Virol. 2008;82:1146–1154. doi: 10.1128/JVI.01698-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Obenauer JC, Denson J, Mehta PK, Su X, Mukatira S, et al. Large-scale sequence analysis of avian influenza isolates. Science. 2006;311:1576–1580. doi: 10.1126/science.1121586. [DOI] [PubMed] [Google Scholar]
- 33.Soubies SM, Volmer C, Croville G, Loupias J, Peralta B, et al. Species-specific contribution of the four C-terminal amino acids of influenza A virus NS1 protein to virulence. J Virol. 2010;84:6733–6747. doi: 10.1128/JVI.02427-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zielecki F, Semmler I, Kalthoff D, Voss D, Mauel S, et al. Virulence determinants of avian H5N1 influenza A virus in mammalian and avian hosts: role of the C-terminal ESEV motif in the viral NS1 protein. J Virol. 2010;84:10708–10718. doi: 10.1128/JVI.00610-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Neumann G, Kawaoka Y. Host range restriction and pathogenicity in the context of influenza pandemic. Emerg Infect Dis. 2006;12:881–886. doi: 10.3201/eid1206.051336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Matrosovich M, Tuzikov A, Bovin N, Gambaryan A, Klimov A, et al. Early alterations of the receptor-binding properties of H1, H2, and H3 avian influenza virus hemagglutinins after their introduction into mammals. J Virol. 2000;74:8502–8512. doi: 10.1128/JVI.74.18.8502-8512.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Herfst S, Schrauwen EJ, Linster M, Chutinimitkul S, de Wit E, et al. Airborne transmission of influenza A/H5N1 virus between ferrets. Science. 2012;336:1534–1541. doi: 10.1126/science.1213362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li C, Yu K, Tian G, Yu D, Liu L, et al. Evolution of H9N2 influenza viruses from domestic poultry in Mainland China. Virology. 2005;340:70–83. doi: 10.1016/j.virol.2005.06.025. [DOI] [PubMed] [Google Scholar]
- 39.Sun Y, Pu J, Fan L, Sun H, Wang J, et al. Evaluation of the protective efficacy of a commercial vaccine against different antigenic groups of H9N2 influenza viruses in chickens. Vet Microbiol. 2012;156:193–199. doi: 10.1016/j.vetmic.2011.10.003. [DOI] [PubMed] [Google Scholar]
- 40.Gamoh K, Nakamizo M, Okamatsu M, Sakoda Y, Kida H, et al. Protective efficacy of stockpiled vaccine against H5N8 highly pathogenic avian influenza virus isolated from a chicken in Kumamoto prefecture, Japan, in 2014. J Vet Med Sci. 2016;78:139–142. doi: 10.1292/jvms.15-0324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kapczynski DR, Pantin-Jackwood M, Guzman SG, Ricardez Y, Spackman E, et al. Characterization of the 2012 highly pathogenic avian influenza H7N3 virus isolated from poultry in an outbreak in Mexico: pathobiology and vaccine protection. J Virol. 2013;87:9086–9096. doi: 10.1128/JVI.00666-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.WHO Manual on Animal Influenza diagnosis and surveillance. 2002. www.wpro.who.int/emerging_diseases/documents/docs/manualonanimalaidiagnosisandsurveillance.pdf?ua=1
- 43.CDC CDC realtime RTPCR protocol for detection and characterization of influenza. 2009. www.who.int/csr/resources/publications/swineflu/CDCRealtimeRTPCR_SwineH1Assay-2009_20090430.pdf
- 44.Lee MS, Chang PC, Shien JH, Cheng MC, Shieh HK. Identification and subtyping of avian influenza viruses by reverse transcription-PCR. J Virol Methods. 2001;97:13–22. doi: 10.1016/S0166-0934(01)00301-9. [DOI] [PubMed] [Google Scholar]
- 45.Fereidouni SR, Starick E, Grund C, Globig A, Mettenleiter TC, et al. Rapid molecular subtyping by reverse transcription polymerase chain reaction of the neuraminidase gene of avian influenza A viruses. Vet Microbiol. 2009;135:253–260. doi: 10.1016/j.vetmic.2008.09.077. [DOI] [PubMed] [Google Scholar]
- 46.Hoffmann E, Stech J, Guan Y, Webster RG, Perez DR. Universal primer set for the full-length amplification of all influenza A viruses. Arch Virol. 2001;146:2275–2289. doi: 10.1007/s007050170002. [DOI] [PubMed] [Google Scholar]
- 47.Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis for Windows 95/98/NT. Nucleic Acids Symp. 1999;41:95–98. [Google Scholar]
- 48.Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pawar SD, Murtadak VB, Kale SD, Shinde PV, Parkhi SS. Evaluation of different inactivation methods for high and low pathogenic avian influenza viruses in egg-fluids for antigen preparation. J Virol Methods. 2015;222:28–33. doi: 10.1016/j.jviromet.2015.05.004. [DOI] [PubMed] [Google Scholar]
- 50.Matrosovich MN, Gambaryan AS. Solid-phase assays of receptor-binding specificity. Methods Mol Biol. 2012;865:71–94. doi: 10.1007/978-1-61779-621-0_5. [DOI] [PubMed] [Google Scholar]
- 51.Bright RA, Shay DK, Shu B, Cox NJ, Klimov AI. Adamantane resistance among influenza A viruses isolated early during the 2005–2006 influenza season in the United States. JAMA. 2006;295:891–894. doi: 10.1001/jama.295.8.joc60020. [DOI] [PubMed] [Google Scholar]
- 52.Hayden FG, Cote KM, Douglas RG. Plaque inhibition assay for drug susceptibility testing of influenza viruses. Antimicrob Agents Chemother. 1980;17:865–870. doi: 10.1128/AAC.17.5.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.