Abstract
Iron sulfur (Fe-S) clusters and the molybdenum cofactor (Moco) are present at enzyme sites, where the active metal facilitates electron transfer. Such enzyme systems are soluble in the mitochondrial matrix, cytosol and nucleus, or embedded in the inner mitochondrial membrane, but virtually absent from the cell secretory pathway. They are of ancient evolutionary origin supporting respiration, DNA replication, transcription, translation, the biosynthesis of steroids, heme, catabolism of purines, hydroxylation of xenobiotics, and cellular sulfur metabolism. Here, Fe-S cluster and Moco biosynthesis in Drosophila melanogaster is reviewed and the multiple biochemical and physiological functions of known Fe-S and Moco enzymes are described. We show that RNA interference of Mocs3 disrupts Moco biosynthesis and the circadian clock. Fe-S-dependent mitochondrial respiration is discussed in the context of germ line and somatic development, stem cell differentiation and aging. The subcellular compartmentalization of the Fe-S and Moco assembly machinery components and their connections to iron sensing mechanisms and intermediary metabolism are emphasized. A biochemically active Fe-S core complex of heterologously expressed fly Nfs1, Isd11, IscU, and human frataxin is presented. Based on the recent demonstration that copper displaces the Fe-S cluster of yeast and human ferredoxin, an explanation for why high dietary copper leads to cytoplasmic iron deficiency in flies is proposed. Another proposal that exosomes contribute to the transport of xanthine dehydrogenase from peripheral tissues to the eye pigment cells is put forward, where the Vps16a subunit of the HOPS complex may have a specialized role in concentrating this enzyme within pigment granules. Finally, we formulate a hypothesis that (i) mitochondrial superoxide mobilizes iron from the Fe-S clusters in aconitase and succinate dehydrogenase; (ii) increased iron transiently displaces manganese on superoxide dismutase, which may function as a mitochondrial iron sensor since it is inactivated by iron; (iii) with the Krebs cycle thus disrupted, citrate is exported to the cytosol for fatty acid synthesis, while succinyl-CoA and the iron are used for heme biosynthesis; (iv) as iron is used for heme biosynthesis its concentration in the matrix drops allowing for manganese to reactivate superoxide dismutase and Fe-S cluster biosynthesis to reestablish the Krebs cycle.
Keywords: aldehyde oxidase, DNA polymerase, electron transport chain, ecdysone, iron regulatory protein, quiescent mitochondria, magnetoreceptor, mitoflashes
Introduction
In the first known biochemical reactions on earth, molybdenum and iron-sulfur (Fe-S) clusters enabled electron transfers turning inorganic molecules into hydrogenated carbon molecules (Mortenson, 1964; Eck and Dayhoff, 1966; Hall et al., 1971; Ochiai, 1978; Wächtershäuser, 1988; Russell and Martin, 2004; Zhang and Gladyshev, 2008; Nitschke and Russell, 2009; Schoepp-Cothenet et al., 2012; Stüeken et al., 2015). Similar biochemistry remains active in living organisms carried out by a variety of metallo-enzymes. In this hypothesis and theory article, we present examples of Fe-S and molybdenum cofactor (Moco) enzymes from the dipteran fly Drosophila melanogaster, a genetically amenable and thoroughly characterized experimental model system (Bellen et al., 2010; St Johnston, 2013; Mohr et al., 2014). By looking at their multiple physiological functions, we propose that Fe-S enzymes are central in the development, life cycle transitions and aging of flies. Given the conservation of these phenomena in the evolution of the animal kingdom, we anticipate that many of our descriptions will be transferable to other organisms.
In the first part of the article, we present how Fe-S clusters are formed, a process that has been studied extensively by biochemists in prokaryotes (Roche et al., 2013; Blanc et al., 2015), yeast (Martinez-Pastor et al., 2017), plants (Balk and Schaedler, 2014), and humans (Paul and Lill, 2015; Rouault and Maio, 2017), but also by the biomedical community intent to find a therapy for patients with Friedreich's ataxia, caused by reduced expression of the frataxin (FXN) gene (Campuzano et al., 1996). Other Fe-S proteins are also implicated in human disease (Rouault, 2012; Beilschmidt and Puccio, 2014; Isaya, 2014). In D. melanogaster, the pioneering work of Maria Moltó and co-workers has almost exclusively focused on the Drosophila frataxin homolog describing what goes wrong when Fe-S biosynthesis is disrupted in flies (reviewed in Mandilaras et al., 2013; Tang and Zhou, 2013b; Zhu et al., 2014; Calap-Quintana et al., 2017). Furthermore, we describe the biosynthesis of Moco (Rajagopalan, 1997; Mendel and Leimkühler, 2015; Leimkühler, 2017), whose basic structure has two sulfur atoms of the tricyclic pyranopterin molecule molybdopterin (MPT) coordinating the Mo atom (Rajagopalan et al., 1982). Work on Moco enzymes in Drosophila started in the fifties and Victoria Finnerty studied the Moco biosynthetic pathway during the last quarter of the twentieth century (Kamdar et al., 1997). Her research program used the molybdoenzyme xanthine dehydrogenase (Xdh) encoded by the rosy gene, whose activity is required for the formation of eye pigments for reasons that are still not fully resolved (Phillips and Forrest, 1980; Wiley and Forrest, 1981; Ferre et al., 1986; Keith et al., 1987; Hilliker et al., 1992), and therefore mutants affecting Moco biosynthesis have evident eye color phenotypes (Kamdar et al., 1997). We complement this review with new discoveries in the role of proteins involved in Fe-S cluster and Moco biosynthesis by showing the original data, which are not published elsewhere (Marelja, 2013), and with a new hypothesis to explain previous observations that dietary copper decreases iron storage in Drosophila (Poulson and Bowen, 1952; Bettedi et al., 2011).
In the second part of the article, we review studies of the Drosophila molybdoenzymes Xdh, aldehyde oxidase (Aox), sulfite oxidase (Suox). Special attention is paid to the problem of how Xdh, a cytosolic enzyme that requires two Fe-S clusters, Moco and flavin for activity (Hughes et al., 1992; Doyle et al., 1996), finds its way into pigment granules of the eye, the only enzyme with such cofactors known to reside in the endomembrane system (Reaume et al., 1989, 1991). Our hypothesis is that exosomes are involved in the process. The fly literature on mitochondrial Fe-S enzymes required for respiration and the biosynthesis of ecdysone, heme, and lipoate is summarized. The role of cytosolic and nuclear Fe-S enzymes in DNA replication, transcription and translation is also reviewed, followed by a brief discussion of the regulation of the Fe-S cluster of iron regulatory protein-1A (IRP-1A) and cellular iron sensing in Drosophila. We then move to the question of how mitochondria sense iron, where we present a new hypothesis suggesting that the mitochondrial superoxide dismutase (Sod2) is a possible mitochondrial iron sensor. Our model of mitochondrial iron sensing also explains the previously observed superoxide bursts in mitochondria (Wang et al., 2008) and the connection between mitochondrial Fe-S cluster biosynthesis and lipogenesis (Tong and Rouault, 2007). Last, we revisit the question of whether Fe-S and/or Moco enzymes are involved in the circadian clock (Mandilaras and Missirlis, 2012).
In the third part of the article, we communicate how profoundly cell physiology depends on Fe-S enzymes. We review the shifts in cell metabolism from glycolysis to aerobic respiration during development (Tennessen et al., 2011, 2014) and during stem cell differentiation (Homem et al., 2014; Sieber et al., 2016), emphasize the requirement of Fe-S clusters for growth through the larval stage and into metamorphosis (Anderson et al., 2005; Uhrigshardt et al., 2013; Llorens et al., 2015; Palandri et al., 2015) and the decline of mitochondrial respiration during aging (Vann and Webster, 1977; Yan et al., 1997; Ferguson et al., 2005). We then discuss the interesting finding that in the female germ line, stem cell differentiation requires the mitochondrial ATP synthase, but not the respiratory chain enzymes (Teixeira et al., 2015). We critically evaluate the possibility that the presence of quiescent mitochondria in the female germ line may serve as a protective hereditary mechanism against the accumulation of mutations in their genome (Allen, 1996). Finally, we describe specific functions of Fe-S or Moco enzymes in the major organs of the fly, concluding that they are a biochemically active component of the complex organization that characterizes living animals.
Fe-S cluster and moco biosynthesis
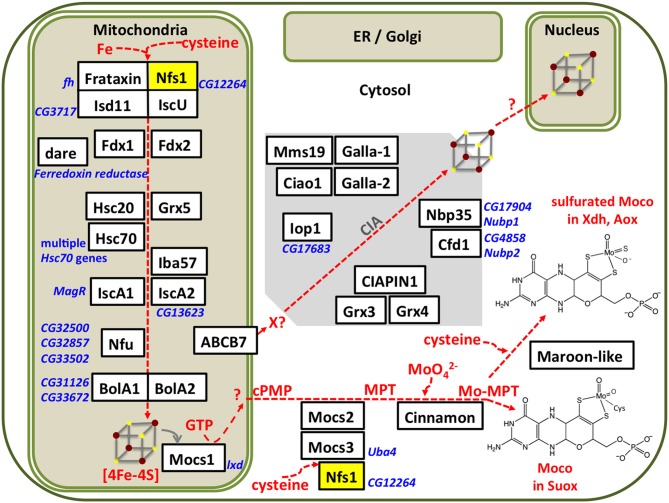
Fe-S cluster assembly initiates in the mitochondrial matrix. We summarize the different steps of the assembly process (Figure 1); for detailed reviews of the pathway the reader is referred elsewhere (Roche et al., 2013; Balk and Schaedler, 2014; Blanc et al., 2015; Paul and Lill, 2015; Martinez-Pastor et al., 2017; Rouault and Maio, 2017). The mitoferrin transporter ensures mitochondrial iron uptake, whereas the L-cysteine desulfurase Nfs1 provides the inorganic sulfide as persulfide. Electrons are required for the cleavage of the persulfide group and assembly of the cluster, which are supplied from ferredoxins Fdx1 and Fdx2. The first assembly protein is IscU, which can accommodate [2Fe-2S] or [4Fe-4S] clusters. Fe-S clusters are transferred from IscU to other scaffold proteins like IscA1, IscA2, Iba57, Nfu, BolA1, or BolA2 that show specificity of cluster delivery to the target enzymes. For Fe-S cluster transfer between the assembly proteins, or between assembly proteins and target enzymes, specialized chaperones (Hsc20 and its cognate partner) and glutaredoxin-5 (Grx5) are required. A cytosolic Fe-S cluster assembly (CIA) pathway has been described, however it is unclear how the first clusters are formed on the CIA complex. The cytosolic electron donors cytokine-induced apoptosis inhibitor-1 (CIAPIN1) and glutaredoxins Grx3 and Grx4 have been identified (Figure 1). Only three genes involved in Fe-S biosynthesis have been individually studied in flies: (i) Drosophila frataxin whose exact biochemical function is still unclear (Cañizares et al., 2000; Anderson et al., 2005, 2008; Llorens et al., 2007; Kondapalli et al., 2008; Runko et al., 2008; Navarro et al., 2010, 2011, 2015; Shidara and Hollenbeck, 2010; Soriano et al., 2013, 2016; Tricoire et al., 2014; Calap-Quintana et al., 2015; Palandri et al., 2015; Chen et al., 2016b; Edenharter et al., 2017), (ii) IscU that encodes a protein assembly platform for Fe-S cluster biosynthesis (Dzul et al., 2017), and iii) Hsc20 encoding one of two chaperones that mobilize the Fe-S cluster from IscU to downstream Fe-S proteins in the mitochondria (Uhrigshardt et al., 2013).
Figure 1.
Subcellular compartmentalization of the Fe-S cluster and Moco assembly processes. Iron and sulfur from cysteine are assembled on IscU in a process that requires electrons provided by ferredoxins (Fdx1, Fdx2, which are themselves Fe-S proteins). Chaperones and Grx5 are required for transfer of the Fe-S cluster to further scaffold proteins. The Drosophila homologs were identified by Blast searches, proposed names for the gene products (in white boxes) were based on the corresponding nomenclature in humans, current Flybase annotations are shown in blue font. Our own results (Marelja, 2013) confirm that the Drosophila genes CG12264 and CG3717 are the homologs of the human genes NFS1 and ISD11, respectively. Multiple Fe-S enzymes are present in mitochondria, cytosol, and nucleus, but so far none have been reported in the secretory pathway. All known components of the CIA pathway are conserved in Drosophila, but the first steps of CIA remain to be elucidated. Moco biosynthesis initiates in the mitochondria with the Fe-S protein Mocs1 and continues in the cytosol with Mocs2, Mocs3, and Cinnamon producing Moco used in Suox. Maroon-like sulfurates Moco for Xdh and Aox. Mitochondrial and cytosolic forms of the cysteine desulfurase Nfs1 (yellow box) provide sulfide to Fe-S cluster and Moco biosynthesis, respectively.
The Drosophila Nfs1/Isd11/IscU/frataxin complex
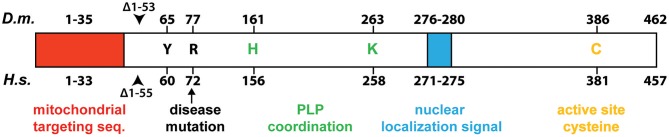
Two Drosophila Fe-S cluster assembly proteins have been purified to date, frataxin (Kondapalli et al., 2008) and IscU (Dzul et al., 2017). We characterized the core mitochondrial protein complex involved in Fe-S cluster assembly in flies (Marelja, 2013). Based on previous work with the human L-cysteine desulfurase NFS1 (Marelja et al., 2008, 2013), the Drosophila Nfs1 homolog (CG12264) was purified. The human and Drosophila enzymes share an amino acid sequence identity of 78% when the mitochondrial targeting sequences are removed from the calculation. All amino acids implicated in enzymatic function of the human protein are conserved in Drosophila Nfs1 (Figure 2).
Figure 2.
Schematic comparison of Drosophila Nfs1 and human NFS1. Highly conserved residues that play a role for desulfurase catalysis and disease are indicated: the active site cysteine (C) that forms the persulfide intermediate (Zheng et al., 1994; Lauhon et al., 2004); the tyrosine (Y) was shown to be crucial for activity in Saccharomyces cerevisiae Nfs1p (Mühlenhoff et al., 2004); the point mutation p.Arg72Gln (arrow) in human NFS1 was identified to cause infantile mitochondrial complex II/III deficiency (Farhan et al., 2014); whereas histidine (H) and lyine (K) are involved in pyridoxal 5′-phosphate (PLP) coordination (Cupp-Vickery et al., 2003). The proposed N-terminal mitochondrial targeting sequences is shown by a red box, while the nuclear localization signal is shown as a blue box. The beginning of the truncated Nfs1/NFS1 versions used for biochemical purification are indicated by an arrow head (Marelja, 2013). Numbers indicate amino acid position in the Homo sapiens (H.s.) and D. melanogaster (D.m.) proteins.
The first 53 amino acids of the full length Drosophila Nfs1 were removed from an Escherichia coli-driven protein expression construct (for detailed Materials and Methods see Marelja, 2013). Nfs1Δ1–53 was co-expressed with Drosophila Isd11 (CG3717 shows 44% amino acid sequence identity to human ISD11; Adam et al., 2006; Wiedemann et al., 2006) or with Drosophila IscU (Dzul et al., 2017), the scaffold protein where newly formed Fe-S clusters are initially assembled (Zheng et al., 1998; Gerber et al., 2003; Fox et al., 2015; Parent et al., 2015). The respective Nfs1Δ1–53/Isd11 and Nfs1Δ1–53/IscU complexes were affinity purified and recovered in a stable, soluble form. Higher molecular weight complexes were obtained when all three Drosophila proteins were co-expressed and also when human FXN (a gift from Kuanyu Li; Xia et al., 2012; Friemel et al., 2017) was added to the Nfs1Δ1–53/Isd11/IscU complex (Figure 3A). Our own efforts to purify Drosophila frataxin as in Kondapalli et al. (2008) were unsuccessful (Marelja, 2013).
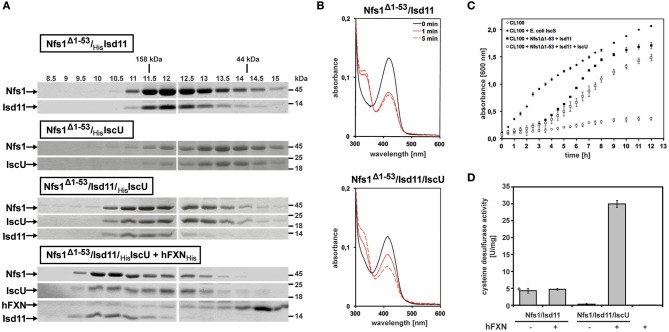
Figure 3.
In vitro characterization of Drosophila Nfs1, Isd11, IscU in complex with human frataxin. (A) Co-expression of Nfs1Δ1–53 with Isd11 and/or IscU in Escherichia coli. After purification of the complexes with affinity chromatography, 30 μM of Nfs1Δ1–53 was applied onto the analytical Superdex200 size exclusion column and compared by SDS-PAGE. The fractions show that the Nfs1Δ1–53/Isd11 complex eluted earlier than the IscU/Nfs1Δ1–53 complex, indicating its smaller size. As expected, the complex between all three proteins was larger. Human frataxin added to the purified Nfs1Δ1–53/Isd11/IscU complex also bound as demonstrated by further increase in size. Gamma globulin (158 kDa) and ovalbumin (44 kDa) were standards (kDa ladder on the top; fractions in ml units). (B) UV-Vis absorption spectra of 10 μM Nfs1Δ1–53 in complex with His6-Isd11 (top panel) and Isd11/His6-IscU (bottom panel) in the absence (solid black line) or 1 min and 5 min after addition of 4 mM L-cysteine (red lines). All spectra were recorded at 23°C in 500 μl 100 mM Tris (pH 8.0), 200 mM NaCl, 1 mM EDTA. (C) Growth curves of 100 ml cultures of the Escherichia coli CL100 strain (ΔiscS) transformed either with vectors pET15b and pACYCDuet-1 or with plasmids containing coding sequences for Drosophila Nfs1Δ1–53, Isd11, IscU, or Escherichia coli IscS were recorded after addition of 100 μM IPTG for 12h at 30°C. (D) Desulfurase activity of Nfs1Δ1–53/Isd11 and Nfs1Δ1–53/Isd11/IscU complexes in absence or presence of human FXN. L-cysteine desulfurase activity was measured by determination of total sulfide produced. One unit is defined as the amount of enzyme producing 1 μmol of sulfide/min. Under anaerobic conditions, Nfs1Δ1–53/Isd11 and Nfs1Δ1–53/Isd11/IscU complexes were mixed in a 1:3 [Nfs1]:[FXN] ratio and incubated for 10 min at 23°C. The last lane (hFXN +) is a negative control to show that the human FXN preparation does not carry contaminant desulfurase activity. Error bars indicate the standard deviation of three measurements.
The characteristic yellow color observed for other L-cysteine desulfurases containing pyridoxal 5′-phosphate (PLP) as a prosthetic group was confirmed in absorption spectra of Drosophila Nfs1 at 420 nm (Figure 3B, solid black lines on both panels). Moreover, addition of the enzyme's substrate L-cysteine induced a decrease of absorbance at 420 nm and an increase of absorbance at 320 nm (Figure 3B, solid and dashed red lines), as reported for Azotobacter vineladii L-cysteine desulfurase NifS, showing that the α-amino group of cysteine binds to the PLP at the enzyme's active site (Zheng et al., 1993). For Nfs1Δ1–53/Isd11, L-cysteine binding reached equilibrium at 1 min (Figure 3B, top panel). In contrast, binding of L-cysteine to the Nfs1Δ1–53/Isd11/IscU complex was slower (Figure 3B, bottom panel), suggesting that the presence of IscU reduced substrate accessibility to Nfs1 in vitro. The Drosophila Nfs1Δ1–53/Isd11 complex was also investigated for its ability to restore the growth deficiency of the E. coli ΔiscS strain CL100 (Figure 3C). As expected, CL100 transformed with the plasmid containing the endogenous iscS gene restored the growth defect. Drosophila Nfs1Δ1–53/Isd11 partially complemented IscS function in the E. coli CL100 strain, whereas expression of IscU along with Nfs1Δ1–53/Isd11 slowed down bacterial growth (Figure 3C). Notably, this in vivo result matches the in vitro observation that L-cysteine binding to Nfs1Δ1–53 is slower in the presence of Isd11 and IscU (Figure 3B). These findings are consistent with the previous demonstration that the L-cysteine desulfurase activity of the human NFS1/ISD11 complex is reduced in the presence of ISCU (Tsai and Barondeau, 2010; Bridwell-Rabb et al., 2014).
To further test this notion, we tested whether IscU altered the desulfurase activity of purified Nfs1 in vitro (Figure 3D). Sulfide production activity was determined with methylene blue in the presence of 1,4 dithiothreitol (Fogo and Popowsky, 1949; Urbina et al., 2001). Only weak activity was detected from the purified Nfs1Δ1–53/Isd11 complex, possibly because the complex was unable to perform reaction turnovers. Addition of IscU, forming the IscU/Nfs1Δ1–53/Isd11 complex, abolished sulfide production, consistent with observations presented above. Addition of human frataxin to the ternary IscU/Nfs1Δ1–53/Isd11 complex led to a six-fold increase in sulfide production activity, showing that frataxin stimulated the desulfurase activity of Nfs1/Isd11 in an IscU-dependent manner (Figure 3D). Steady-state kinetic parameters were determined by varying L-cysteine concentration for Nfs1Δ1–53 activity (after Nfs1Δ1–53/Isd11/IscU/frataxin complex formation) showing a catalytic efficiency of 149 M−1 s−1, kcat 2.6 min−1, and a KM of 0.290 mM. Taken together, these data indicate that Isd11, IscU, and frataxin are required for the activation of Nfs1 in Drosophila, providing experimental evidence that the initial complex of the mitochondrial Fe-S cluster biosynthetic machinery is conserved in Drosophila similar to what is known in other eukaryotes, including species with non-respiring mitochondria (Tovar et al., 2003; Richards and van der Giezen, 2006).
Fe-S cluster biosynthesis occurs in distinct cellular compartments
Like the other proteins of the core complex described above, Drosophila Isd11 is localized in mitochondria (Figure 4A). Two mitochondrial ferredoxins (Palandri et al., 2015) and the Drosophila homolog of ferredoxin reductase (also known as adrenodoxin reductase encoded by the dare gene; Freeman et al., 1999) are required as electron donors for Fe-S cluster formation (Cai et al., 2017). Mitochondrial monothiol glutaredoxin-5 (CG14407) has not been investigated in Drosophila, but is likely required for Fe-S cluster biosynthesis (Rodríguez-Manzaneque et al., 2002; Wingert et al., 2005; Ye et al., 2010; Johansson et al., 2011), participating in the Fe-S cluster transfer to BolA-like proteins (Aldea et al., 1988; Frey et al., 2016; Melber et al., 2016; Uzarska et al., 2016; Nasta et al., 2017). Fe-S clusters are also transferred to Nfu-like (Tong et al., 2003; Melber et al., 2016; Wachnowsky et al., 2016) or Isa-like proteins (Jensen and Culotta, 2000; Kaut et al., 2000; Muhlenhoff et al., 2011; Sheftel et al., 2012) with the chaperone activity of mitochondrial Hsc20 (Uhrigshardt et al., 2010; Sieber et al., 2016) and its cognate Hsc70 [not clear which of several candidate Hsc70 genes present in the fly genome (Adams et al., 2000) functions in Fe-S cluster biosynthesis]. These scaffold proteins deliver the Fe-S clusters to target mitochondrial Fe-S enzymes.
Figure 4.
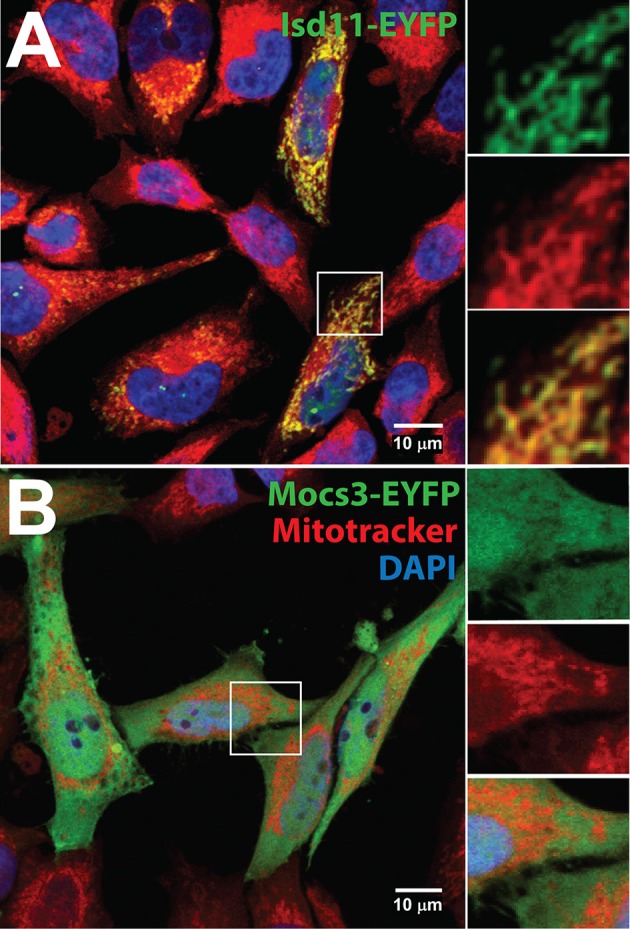
Subcellular localization of Isd11 and Mocs3. Confocal microscopy of (A) Hela cells transfected with a C-terminal fusion of EYFP to Isd11, stained with Mitotracker Red to detect mitochondria and with DAPI to visualize nuclei. Insets show two channels separately to appreciate co-localization of the fluorescent signals from Isd11-EYFP and Mitotracker (merged, bottom inset); (B) Hela cells transfected with Mocs3-EYFP, which localizes in the cytosol and nuclei.
Fe-S clusters are also assembled on proteins in the cytosol (Figure 1). Early work in this area suggested that core mitochondrial Fe-S cluster assembly proteins were also being directed to the cytosol (Land and Rouault, 1998; Tong and Rouault, 2000, 2006) and protein targeting sequences do not typically result in a unique destination for most proteins (Hegde and Bernstein, 2006). Furthermore, another set of cytosolic Fe-S cluster assembly (CIA) proteins has been described (reviewed in Roy et al., 2003; Hausmann et al., 2005; Paul and Lill, 2015). Briefly, an Fe-S cluster is assembled on CFD1 and NBP35 with electron donors provided by CIAPIN1. The clusters are transferred through IOP1 to a scaffold with CIA1, CIA2, and target proteins. These CIA proteins are conserved in Drosophila (Figure 1), but little work exists in the context of Fe-S cluster assembly. The fly Ciao1 homolog was shown to be required for viability (Radford et al., 2005) prior to assigning its biochemical function as part of the Fe-S scaffold complex in CIA (Balk et al., 2005a). Ciao1 received its name from the word “bridge” in the Chinese language (Johnstone et al., 1998), whereas the yeast homolog was conveniently designated Cia1 when its interaction with the hydrogenase-like Nar1 (IOP1 in the human nomenclature; Huang et al., 2007) was discovered (Balk et al., 2005b). The fly CIA2 scaffold (Zhao et al., 2015; Vo et al., 2017) homologs Galla-1 and Galla-2 (Yeom et al., 2015) associate with target nuclear Fe-S proteins Mms19 (Gari et al., 2012; Papatriantafyllou, 2012; Stehling et al., 2012; Nag et al., 2018) and Xeroderma pigmentosum D (Xpd) (Rudolf et al., 2006). The Drosophila CIAPIN1 homolog is required for oogenesis (Marzuk et al., 2013). CIAPIN1 carries an Fe-S cluster (Zhang et al., 2008) and receives electrons from a cytosolic reductase (Netz et al., 2010). Neither the reductase nor cytosolic monothiol glutaredoxins (Li et al., 2009; Mühlenhoff et al., 2010; Banci et al., 2015) have been characterized in Drosophila. Furthermore, it is important to stress that no experimental data exist to implicate the Drosophila proteins Cfd1, Ciao1, CIAPIN1, Galla-1, Galla-2, Iop1 Mms19, Nbp35 in the assembly of cytosolic Fe-S clusters; there inclusion here and elsewhere (Mandilaras and Missirlis, 2012) as members of the CIA was exclusively based on gene homology searches. Nevertheless, many projects are being undertaken in other systems to better describe the process of Fe-S cluster assembly in cytosol and nucleus and we hope to see contributions from studies in Drosophila in the near future.
Finally, it is noteworthy that no Fe-S enzymes have been reported in the secretory pathway, implying that the presence of thiol-reducing activity in the cytosol, nucleus and mitochondrial matrix is essential for their stability. In insects, such activity rests on the thioredoxin reductase system (Kanzok et al., 2001; Missirlis et al., 2001, 2002).
Excess copper inhibits Fe-S cluster biosynthesis
Copper and iron homeostasis are intimately linked (Fox, 2003). Physiologically, the two metals are required for aerobic respiration, albeit as separate cofactors (Villee, 1948). Poulson and Bowen made an early observation that when Drosophila larvae were fed a diet rich in copper their iron stores were being depleted (Poulson and Bowen, 1952). A key way in which the two metals depend on each other was uncovered when ceruloplasmin and other multicopper oxidases (MCOs) were shown to act as ferroxidases (Osaki et al., 1966) and their activity was linked to iron trafficking across membranes (reviewed in Kosman, 2010). Drosophila MCOs are linked to iron homeostasis in ways that are still not understood (Bettedi et al., 2011; Lang et al., 2012; Peng et al., 2015), but MCO3 mutants fed on copper also show a reduction in ferritin iron content (Bettedi et al., 2011). It was therefore important to identify another hypothesis to explain copper-mediated cellular iron deficiency. The discovery that excess copper displaces the Fe-S cluster of mitochondrial ferredoxin (Vallières et al., 2017), leading to disrupted CIA (Alhebshi et al., 2012), suggests that copper toxicity may directly inhibit Fe-S cluster biosynthesis also in Drosophila. Reducing Fe-S cluster biosynthesis in Drosophila with either RNA interference (RNAi) of frataxin or loss-of-function Hsc20 mutants lead to mitochondrial iron accumulation and reduced ferritin expression (Anderson et al., 2005; Uhrigshardt et al., 2013; Navarro et al., 2015), offering a testable potential explanation of why excess dietary copper affects ferritin iron accumulation. In this respect, we also note that copper chelation ameliorated a fly model of Friedreich's ataxia (Soriano et al., 2016) and that the dithiol Drosophila glutaredoxin-1 was implicated in copper homeostasis (Mercer and Burke, 2016).
A mitochondrial iron sulfur enzyme initiates moco biosynthesis
Turning to Moco biosynthesis, an ancient, ubiquitous and highly conserved pathway underpinning molybdenum biochemistry (Rajagopalan, 1997; Mendel and Leimkühler, 2015; Leimkühler, 2017), it can be divided into three major steps: (i) GTP is converted to cPMP, (ii) cPMP is converted to MPT by generation of the dithiolene group, and (iii) molybdate is then ligated to MPT forming Moco (Figure 1). Three fly genes involved in Moco biosynthesis have attracted individual attention: (i) Mocs1 (formerly known as low xanthine dehydrogenase, lxd) encoding a mitochondrial Fe-S enzyme that converts 5′-guanosine triphosphate (GTP) to cyclic pyranopterin monophosphate (cPMP) (Keller and Glassman, 1964; Courtright, 1975; Duke et al., 1975; Bogaart and Bernini, 1981; Schott et al., 1986; Ho et al., 1992; Tahoe et al., 2002), (ii) cinnamon (cin) encoding a gephyrin homolog that inserts molybdate into MPT (Baker, 1973; Browder and Williamson, 1976; Kamdar et al., 1994; Wittle et al., 1999), and (iii) maroon-like (mal) that encodes a Moco sulfurase (Mitchell and Glassman, 1959; Hubby and Forrest, 1960; Finnerty et al., 1970; Marsh and Wieschaus, 1977; Kamleh et al., 2009).
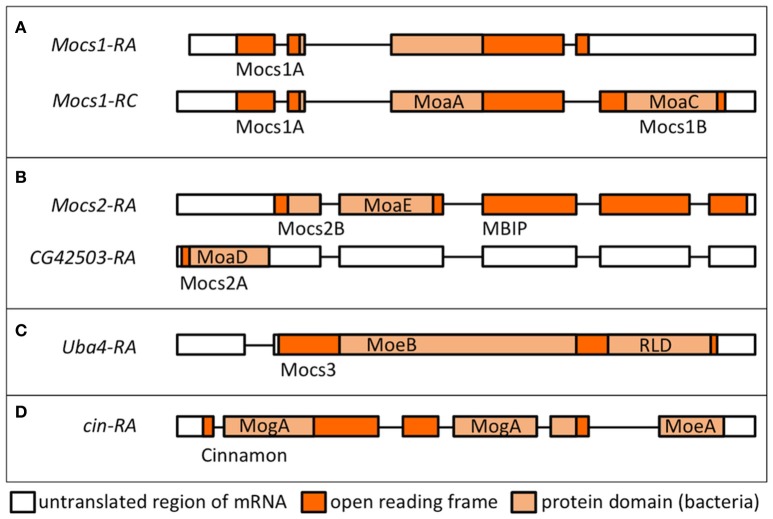
Moco biosynthesis starts within mitochondria with a complex rearrangement reaction in which the C8 atom of the GTP purine is inserted between the 2′ and 3′-ribose carbon atoms (Wuebbens and Rajagopalan, 1993; Hover and Yokoyama, 2015). The human MOCS1 gene is orthologous to the lxd locus (now renamed Mocs1) of D. melanogaster (Gray and Nicholls, 2000). Mutations in lxd affect molybdoenzyme activity in flies (Keller and Glassman, 1964). Alternative splicing at the Mocs1 locus results in short (Mocs1A) and full length (Mocs1A-Mocs1B) proteins (Figure 5A). Mocs1A belongs to the superfamily of SAM-dependent radical enzymes (Hänzelmann and Schindelin, 2004), requiring a [4Fe-4S] cluster for the formation of a substrate radical by reductive cleavage of SAM. Studies on the human and bacterial homologs showed that Mocs1B participates in pyrophosphate cleavage after the formation of the 3′, 8cH2GTP intermediate (Hänzelmann and Schindelin, 2006). Evidence was sought for Mocs1 having a role in lifespan determination of Drosophila, as polymorphisms in the gene were detected between short- and long-lived inbred lines, but the results were inconclusive as the polymorphisms could not be associated with clear effects on enzyme activity (Tahoe et al., 2002). Mutants in Mocs1 showed differential sensitivity to dietary molybdate compared to wild type strains (Duke et al., 1975).
Figure 5.
Transcriptional units and proteins encoded by Drosophila genes involved in Moco biosynthesis. Transcript names are shown as per Flybase. Protein domains in light orange boxes are according to the nomenclature in bacteria (Leimkühler, 2017). Names of the Drosophila proteins as used in text are shown below each transcript. (A) Alternative splicing of Mocs1 results in short (Mocs1A) and long (Mocs1A-Mocs1B) subunits, which complex together to form the active enzyme. (B) A bicistronic transcript results in Mocs2B (which also carries the unrelated to Moco biosynthesis MBIP domain, see text) and Mocs2A. (C) Mocs3 carries the MoeB-like domain and a rhodanese-like domain (RLD), shown to interact with Nfs1 (Marelja et al., 2013). (D) Cinnamon carries the MogA/Gephyrin G domain at the N-terminus prior to the C-terminal MoeA/Gephyrin E domain (in reverse order to the mammalian Gephyrins).
Molybdenum cofactor biosynthesis in the cytosol
In the second step, two sulfur atoms are transferred to cPMP to form MPT. This reaction is catalyzed by the heterotetrameric MPT synthase, which is composed of two small Mocs2A and two large Mocs2B subunits encoded from a single locus (Stallmeyer et al., 1999; Leimkuhler et al., 2003; Figure 5B). The physiological sulfur donor for MPT synthesis is Mocs3 (Matthies et al., 2004, 2005), which resides in the cytosol (Figure 4B). Drosophila Mocs2 and Mocs3 are both required for Aox activity (Figure 6). Based on what is known in human cells (Marelja et al., 2008), Mocs3 is expected to receive sulfur from Nfs1, the same protein that acts as a sulfur donor for Fe-S cluster biosynthesis in mitochondria, to which it binds through a rhodanese-like domain (RLD; Figure 5C). Using Förster resonance energy transfer and a split-EGFP system, NFS1 was shown to interact in the cytosol of human cells with MOCS3 (Marelja et al., 2013). This result was corroborated by showing that human NFS1/ISD11 requires MOCS3 to complement Moco biosynthesis in the E. coli deletion strain used in Figure 3C (Bühning et al., 2017).
Figure 6.
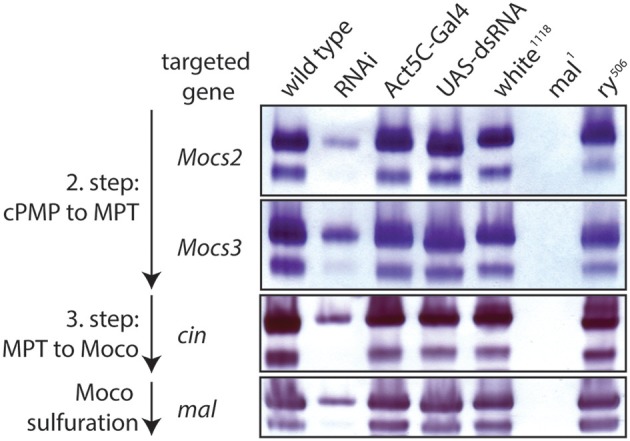
RNAi of Moco biosynthesis genes disrupts Aox activity. In-gel Aox activity is shown as described before (Marelja et al., 2014). Extracts from wild type flies are shown in column 1 of the gels, followed by extracts from RNAi of, respectively, Mocs2, Mocs3, cinnamon, and maroon-like in column 2 (note reduced Aox activity, consistent with a requirement of these genes for Moco biosynthesis). Columns 3 and 4 are extracts from the parental flies of the Gal4/UAS system that show normal Aox activity (controls), as do extracts from white and rosy mutants in columns 5 and 7 (positive controls), whereas extracts from the mal mutant show no detectable Aox activity (negative control, column 6).
Exceptionally, the Drosophila MPT synthase is linked to c-Jun N-terminal kinase (JNK) signaling, since Mocs2B forms a fusion protein with the mitogen activated protein kinase upstream binding inhibitory protein (MBIP; Figure 5B), which is one of 13 subunits of the Ada Two A containing histone acetyltransferase complex (ATAC) transcriptional co-activator (Suganuma et al., 2010). In human cells, Mocs2B/MBIP were shown to regulate ferritin translation through inhibition of PKR, a double-stranded RNA-dependent protein kinase (Suganuma et al., 2012, 2016). Whether this form of regulating iron storage is conserved in Drosophila, whether it serves to regulate the availability of Fe-S clusters for Mocs1, and whether Drosophila Nfs1 has a cytosolic function are questions for future research.
In the last step, molybdate is ligated to the dithiolene group of MPT producing Moco (Figure 1). The first results for the enzyme inserting molybdate to the pterin structure came from the Xdh deficiency of the cin mutation in Drosophila (Baker, 1973). Cin is partially homologous to the two E. coli proteins MogA and MoeA, which are also found as domains G and E of the rat protein Gephyrin, albeit in reverse orientation to the Drosophila protein (Kamdar et al., 1994; Stallmeyer et al., 1995; Feng et al., 1998; Figure 5D). The MogA-like G domain binds MPT and catalyzes the MPT-adenylation from Mg-ATP; MPT-AMP is then transferred to the MoeA-like E domain for hydrolysis and molybdenum insertion (Kuper et al., 2000, 2004; Schwarz et al., 2000; Llamas et al., 2004, 2006). Furthermore, the central domain in Gephyrin binds and anchors inhibitory ligand-gated anion channels in the postsynaptic membrane of neurons (Feng et al., 1998; Stallmeyer et al., 1999). However, it is unknown whether this additional function is conserved for Drosophila Cin.
After molybdenum insertion into MPT, Moco is either inserted into Suox or further modified by exchanging an oxo ligand by a sulfido group (Hille, 1996, 2002; Hille et al., 2014). The sulfur incorporation was also first discovered in the fly mal mutant, which lacked Xdh and Aox activities (Hadorn and Mitchell, 1951; Glassman and Mitchell, 1959; Hubby and Forrest, 1960; Forrest et al., 1961; Courtright, 1967) but retained or induced Suox activity (Figure 7) and an apparently normal concentration of total Moco (Bogaart and Bernini, 1981; Warner and Finnerty, 1981; Wahl et al., 1982). In vitro reconstitution of Xdh and Aox activities with sulfide/dithionite treatment led to the suggestion that mal affected the sulfur modification after the Mo insertion (Wahl and Rajagopalan, 1982; Wahl et al., 1982). A sequence comparison with the L-cysteine desulfurase gene nifS from Azotobacter vinelandii suggested that the mal gene encodes the enzyme that catalyzes sulfuration of Moco used by Xdh and Aox (Amrani et al., 2000; Ichida et al., 2001). Oxo-containing Moco does not function in Xdh and Aox, but is the cofactor for Suox (Figure 1).
Figure 7.
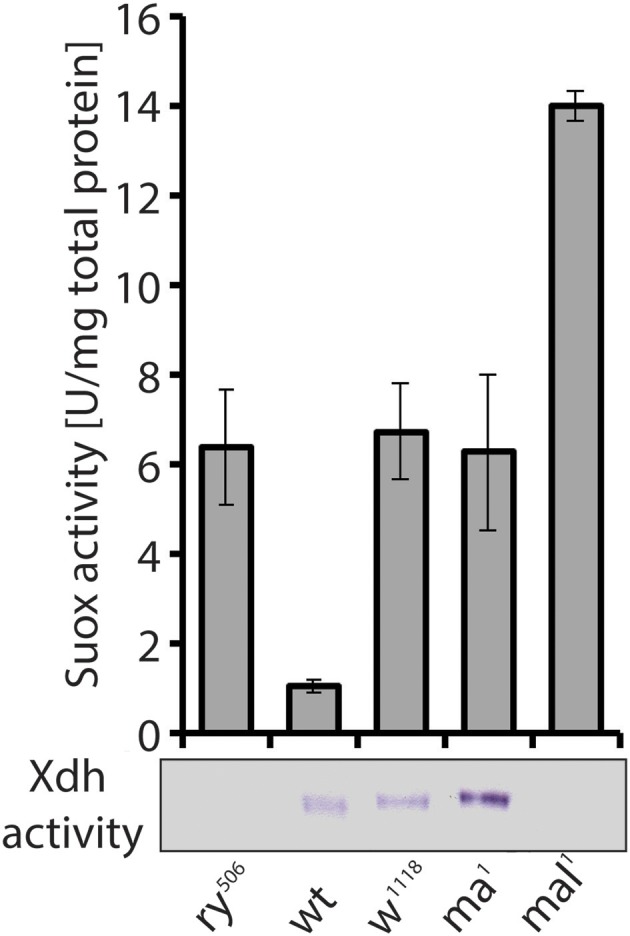
Despite similar eye pigment phenotypes with rosy and mal, the maroon mutant shows increased Xdh activity. Suox activity measured in extracts from mutants (ry, rosy; w, white; ma, maroon; mal, maroon-like) and a wild type (wt) strain is shown in bars. In-gel Xdh activity of the same extracts is also presented. Loss of Xdh activity in rosy and mal explains the phenotypic changes in eye pigmentation, but it was surprising to find increased Xdh activity in maroon mutants, given their eye phenotype.
Fe-S and moco enzymes
The fly offers an ideal system for interdisciplinary research to bridge findings from genetic manipulations and resulting phenotypes with detailed biochemical studies to build functional understanding for animal cell physiology, keeping in mind the diversity of cell types and subcellular microenvironments. In this section, we review the enzymes that use Moco and/or Fe-S clusters. A representative list of these enzymes in D. melanogaster is provided (Table 1). We emphasize the physiological functions and biochemical pathways, as opposed to the detailed biochemistry of the cofactors at the active sites, which only in few cases has been the primary interest of Drosophila researchers.
Table 1.
List of known molybdoenzymes, iron sulfur enzymes and scaffold proteins.
| Protein | Metal Cofactors | Cellular Compartment | Function | Key References |
|---|---|---|---|---|
| MOLYBDO-ENZYMES | ||||
| Aox 1-4 (aldehyde oxidases) | Moco 2 × [2Fe-2S] | Cytosol | Hydroxylation of xenobiotics | Dickinson and Gaughan, 1981; Marelja et al., 2014 |
| mArc (mitochondrial amidoxime reducing component; CG1665) | Moco | Unknown | Unknown | Llamas et al., 2017 |
| Rosy (xanthine dehydrogenase) | Moco 2 × [2Fe-2S] | Cytosol Pigment granule | Purine degradation Eye pigment formation | Keith et al., 1987; Reaume et al., 1991; Hilliker et al., 1992; Hughes et al., 1992; Kamleh et al., 2008 |
| Suox (sulfite oxidase; CG7280) | Moco Heme | Mitochondrial intermembranes | H2S detoxification | Bogaart and Bernini, 1981; Braaten and Bentley, 1993 |
| MITOCHONDRIAL IRON SULFUR ENZYMES AND SCAFFOLD PROTEINS | ||||
| Acon (mitochondrial aconitase) | [4Fe-4S] | Mitoch. matrix | Krebs cycle | Cheng et al., 2013; Esposito et al., 2013 |
| BolA 1-2 | [4Fe-4S] | Mitoch. matrix | 2nd Fe-S assembly scaffold | Uzarska et al., 2016 |
| Fdx 1-2 (ferredoxins) | [2Fe-2S] | Mitoch. matrix | Electron transfer | Palandri et al., 2015 |
| Fech (ferrochelatase) | [2Fe-2S] | Inner membrane | Heme biosynthesis | Sellers et al., 1998 |
| IscA 1-2 (magnetoreceptor?) | [4Fe-4S] | Mitoch. matrix | 2nd Fe-S assembly scaffold | Mandilaras and Missirlis, 2012; Qin et al., 2016 |
| IscU | [2Fe-2S] or [4Fe-4S] | Mitoch. matrix | 1st Fe-S assembly scaffold | Dzul et al., 2017 |
| Las (lipoic acid synthase) | 2 × [4Fe-4S] | Inner membrane | Lipoate cofactor biosynthesis | Harmer et al., 2014 |
| Mocs1 (molybdenum cofactor synthesis 1) | [4Fe-4S] | Mitoch. matrix | Moco biosynthesis | Duke et al., 1975; Gray and Nicholls, 2000 |
| mtDNA-helicase | [2Fe-2S] | Mitoch. matrix | Mitoch. DNA replication | Stiban et al., 2014 |
|
ND-24 (NADH dehydrogenase) ND-51 ND-75 ND-23 ND-20 |
[2Fe-2S] [4Fe-4S] [2Fe-2S] & 2 x [4Fe-4S] 2 x [4Fe-4S] [4Fe-4S] |
Facing matrix Inner membrane | Oxidative phosphorylation Respiratory complex I | Zhang K. et al., 2013; Fiedorczuk et al., 2016; Garcia et al., 2017 |
| NFU 1-3 | [4Fe-4S] | Mitoch. matrix | 2nd Fe-S assembly scaffold | Tong et al., 2003 |
| Nvd (cholesterol 7 desaturase) | [2Fe-2S] | Mitoch. matrix | Steroid biosynthesis | Yoshiyama et al., 2006 |
| RFeSP (Rieske iron-sulfur protein) | [2Fe-2S] | Inner membrane | Respiratory complex III | Gontijo et al., 2011 |
| SdhB (succinate dehydrogenase) | [4Fe-4S] | Inner membrane | Respiratory complex II Krebs cycle | Au and Scheffler, 1994; Kohler et al., 1995; Gray et al., 1996; Walker et al., 2006; Na et al., 2014; Van Vranken et al., 2014 |
| CYTOSOLIC IRON SULFUR ENZYMES | ||||
| Cisd2 (CG1458) mitoNEET | [2Fe-2S] | Outer membrane | Fe-S cluster repair | Jones et al., 2014 |
| Elp3 (lysine acetyltransferase) Elongator complex protein 3 | [4Fe-4S] | Cytosol Nucleus | Acetylates synapse active zone RNA transcript elongation | Miśkiewicz et al., 2011 |
| IRP 1A-1B (cytosolic aconitases/ Iron regulatory proteins) | [4Fe-4S] | Cytosol | Cellular iron homeostasis Intermediary metabolism | Muckenthaler et al., 1998; Lind et al., 2006; Surdej et al., 2008 |
| Pixie (RNAase L inhibitor, ABCE1) | 2 × [2Fe-2S] | Cytosol | Ribosome biogenesis Translation initiation | Andersen and Leevers, 2007; Kashima et al., 2014 |
| Prat 1-2 | [4Fe-4S] | Cytosol | Purine nucleotide biosynthesis | Clark, 1994; Merzetti et al., 2013 |
| Su(r) (CG2194) dihydropyrimidine dehydrogenase | [4Fe-4S] | Cytosol | Pyrimidine degradation | Van Gelder et al., 1995; Rawls, 2006 |
| NUCLEAR IRON SULFUR ENZYMES | ||||
| Chl1 (CG11403) DNA helicase | [4Fe-4S] | Nucleus | Heterochromatin organization | – |
|
DNApol-α180 (DNA polymerase) DNApol-α60 (DNA primase) DNApol-δ (DNA polymerase)yDNApol-ε255 (DNA polymerase) DNA2 (CG2990) DNA helicase |
[4Fe-4S] [4Fe-4S] [4Fe-4S] [4Fe-4S] [4Fe-4S] |
Nucleus | Nuclear DNA replication | Kaguni et al., 1983; Peck et al., 1992; Sahashi et al., 2014 |
| Nthl1 (CG9272) DNA glycosylase | [4Fe-4S] | Nucleus | Base excision repair | – |
| Xpd (xeroderma pigmentosum D) | [4Fe-4S] | Nucleus | Subunit of basal transcription and DNA repair factor TFIIH | Reynaud et al., 1999; Chen et al., 2003; Aguilar-Fuentes et al., 2006; Li et al., 2010 |
The molybdoenzymes: Xdh, Aox, Suox
In D. melanogaster, the rosy gene encodes Xdh (Chovnick et al., 1976; Keith et al., 1987). Rosy mutants accumulate xanthine and hypoxanthine and are devoid of urate (Hadorn and Schwinck, 1956; Morita, 1958; Glassman and Mitchell, 1959) and show a dull reddish-brown eye color because of the lowered concentrations of the red eye pigment drosopterin (bright red), and the increased chromogenic oxidation of the eye pigment dihydroxanthommatin (yellow brown) to xanthommatin (dull dark brown), either due to enzymatic activity (Phillips and Forrest, 1980; Wiley and Forrest, 1981; Ferre et al., 1986) or due to the decreased urate concentration (Hilliker et al., 1992). The use of rosy mutant strains to probe the structure and function of Xdh provides an early, elegant example in the field of biological inorganic chemistry (Hughes, 1992; Hughes et al., 1992; Doyle et al., 1996). Allelic series of mutants affecting residues coordinating the Fe-S clusters, the Moco, the flavin cofactor or the binding sites for NAD+/NADH were used to decipher the complex mechanism of action for this prototypical molybdoenzyme. More recently, metabolic profiling of rosy mutants revealed additional changes in the tryptophan, arginine, pyrimidine, and glycerophospholipid metabolic pathways (Kamleh et al., 2008, 2009).
Two genes, low pyridoxal oxidase (lpo) and aldehyde oxidase (Aldox), were initially recognized to encode Aoxs in D. melanogaster (Courtright, 1967; Collins and Glassman, 1969; Dickinson, 1970; Browder and Williamson, 1976; Dickinson and Gaughan, 1981; Cypher et al., 1982; Nelson and Szauter, 1992). Genome analysis revealed, however, a cluster of four Aox genes (Garattini et al., 2008). We showed previously that (i) the lpo gene encodes Aox1, (ii) Aox2 activity is only present during metamorphosis, (iii) the activities associated with the classic Aldox locus correspond to two splice forms of the Aox3 gene, and (iv) no major activity has been found associated with Aox4, the newest addition to a highly evolving protein family in Drosophilidae (Marelja et al., 2014). Aoxs show specific substrate specificities, but their in vivo substrates and physiological functions remain unclear (Dickinson and Gaughan, 1981; Cypher et al., 1982; Marelja et al., 2014).
Suox contains a cytochrome b5 (heme-containing) domain followed by 12–15 amino acids connecting it to the Moco domain. Suox catalyzes an oxygen atom transfer reaction to sulfite leading to its oxidation (and detoxification) to sulfate (Feng et al., 2007; Hille et al., 2011), an activity previously detected in Drosophila (Bogaart and Bernini, 1981; Braaten and Bentley, 1993). Based on sequence homology, we find that CG7280 encodes for the Drosophila Suox.
A novel molybdoenzyme, mitochondrial amidoxime reducing component (mArc), has been described and has an identifiable Drosophila homolog, CG1665. The enzyme from humans has been proposed to reduce a broad range of N-hydroxylated compounds receiving electrons from cytochrome b5 (Gruenewald et al., 2008) and to reduce nitrite to nitric oxide (Sparacino-Watkins et al., 2014). A number of controversies over the function, subcellular localization and whether the newly discovered enzyme binds to sulfurated or oxo Moco, have been reviewed (Llamas et al., 2017).
Another relatively unexplored area in Drosophila molybdenum biology relates to the trafficking and homeostatic mechanisms for handling dietary molybdate. In wild type flies, Xdh and Aox activities are relatively stable, not responding to concentrations between 1 and 10 mM molybdate (Duke et al., 1975), although more recent studies documented a number of, so far unexplained, sex-specific physiological responses in carbohydrate and thiol metabolism at the same concentrations (Rovenko et al., 2014; Perkhulyn et al., 2017). Interestingly, 50 mM molybdate was tolerated by wild type flies, but was lethal to Mocs1 mutant flies, implicating the Moco biosynthetic pathway as part of the detoxification mechanisms available to the fly (Duke et al., 1975).
The curious case of the Maroon mutant
A second unusual aspect of Xdh, besides its implication in multiple metabolic pathways (see above Kamleh et al., 2008, 2009), relates to the enzyme's trafficking to the eye imaginal disk, where it accumulates in pigment granules (Reaume et al., 1989, 1991). Due to the similar eye color of the classic maroon mutant with that of the rosy and mal mutants (Bridges, 1918), we assayed maroon fly extracts for Xdh and Suox activity, expecting to find decreased Xdh activity consistent with the eye phenotype. To our surprise, increased activities of both enzymes compared to wild type flies were observed in maroon extracts (Figure 7). As Suox utilizes unmodified Moco, the increase in its activity was not entirely unexpected, because this form of Moco might accumulate as in rosy and mal. Accounting for increased Xdh activity was harder, however, given the similar eye color between maroon, rosy, and mal. The demonstration that the maroon gene encodes for Vacuolar protein sorting 16A (Vps16A; Grant et al., 2016), a protein implicated in granule formation (Pulipparacharuvil et al., 2005; Lorincz et al., 2016), suggests a possible defect in the tissue localization of Xdh in maroon mutants, as the enzyme obviously remains functional in whole fly extracts and is even induced (Figure 7). It will be informative to test in maroon mutants whether the Xdh activity is localized in peripheral tissues, such as the Malpighian tubules, and not in the eyes. If this prediction is correct, then Vps16A may represent the first known mutant that blocks the delivery of a vesicular structure to the eye. Another unresolved piece of this puzzle relates to the way in which Xdh, a cytosolic enzyme, is found in pigment granules in the eye (Reaume et al., 1989, 1991). The discovery of exosomes gives a possible answer to this conundrum (Hemler, 2003; Gross et al., 2012; Gradilla et al., 2014; Takeuchi et al., 2015; Beer and Wehman, 2017; Shibata et al., 2017; Tassetto et al., 2017). Clearly more experiments are required to explain how Xdh acts in the formation of eye color in flies, but complex non-cell autonomous processes relating to enzyme maturation, regulation, and transport are involved.
Mitochondrial Fe-S proteins at the heart of mitochondrial bioenergetics
Mitochondria form an important organelle of eukaryotic cells, typically containing their proper genome (Lane and Martin, 2010; Schatz, 2013; Allen, 2015) and performing various functions (Pagliarini and Rutter, 2013; Chandel, 2015; Munro and Treberg, 2017), of which the tricarboxylic acid (TCA) cycle and oxidative phosphorylation are famous (Vakifahmetoglu-Norberg et al., 2017). The idea that iron plays a part in the oxidation reactions of the living cell was firmly established by Warburg (1925). Beinert and Sands interpreted electron paramagnetic resonance spectra of succinate dehydrogenase (Sdh) as “non heme iron” (Beinert and Sands, 1960; Beinert, 2002). Later, Sdh and aconitase (both TCA cycle enzymes) were shown to carry [4Fe-4S] clusters (Ruzicka and Beinert, 1978; Cammack, 1982). The Drosophila enzymes are no exception to the rule (Duke, 1968; Fox et al., 1972; Au and Scheffler, 1994; Vincent et al., 2012; Esposito et al., 2013). Failure to build these Fe-S clusters will inevitably block the TCA cycle, and thereby development and growth (Au and Scheffler, 1994; Yan et al., 1997; Walker et al., 2006; Uhrigshardt et al., 2013; Na et al., 2014). Sdh is also known as Complex II of the respiratory chain that generates the inner mitochondrial membrane potential and proton gradient used by the F-ATPase for the production of ATP during oxidative phosphorylation (Alziari et al., 1985; Sardiello et al., 2005; Liu et al., 2011; Barry and Thummel, 2016). Complex I of the respiratory chain, otherwise known as NADH dehydrogenase, carries eight precisely spaced Fe-S clusters of different reduction-oxidation potentials assembled on five subunits of the complex (Table 1; Zhang K. et al., 2013; Fiedorczuk et al., 2016; Garcia et al., 2017). It is thus plain that without Fe-S clusters ATP cannot be produced in the mitochondrial process of aerobic respiration (Anderson et al., 2005; Llorens et al., 2007; Navarro et al., 2011; Edenharter et al., 2017).
Fe-S enzymes are required for heme, ecdysone, and lipoate biosynthesis
Ferrochelatase is an enzyme that resides in the inner mitochondrial membrane accepting iron from the mitochondrial matrix and protoporphyrin IX from the intermembrane space to generate heme (Wu et al., 2001). Heme is another abundant iron-dependent protein cofactor (Ponka et al., 2017). Many ferrochelatases, including the Drosophila enzyme, carry a [2Fe-2S] cluster (Sellers et al., 1998). Therefore, both major forms of iron cofactors used in biology rest on the mitochondrial Fe-S cluster assembly machinery.
Furthermore, Drosophila ferredoxins carry a [2Fe-2S] cluster required for electron transfer during Fe-S cluster assembly, but also for the production of ecdysone in the larval prothoracic gland and other steroidogenic tissues (Uhrigshardt et al., 2013; Palandri et al., 2015). In this way, iron availability is linked to a key developmental signal that terminates growth and initiates metamorphosis (Yamanaka et al., 2013; Sandoval et al., 2014). Interestingly, glutathione production in the prothoracic gland is also required for steroidogenesis (Enya et al., 2017). The possibility that glutathione supports Fe-S cluster biosynthesis in this tissue should be considered (Song et al., 2006; Auchère et al., 2008; Qi et al., 2013; Ozer et al., 2015).
Lipoic acid or lipoate is a cofactor required in intermediary metabolism enzymes α-oxoglutarate dehydrogenase, pyruvate dehydrogenase, branched-chain oxoacid dehydrogenase, 2-oxoadipate dehydrogenase, and in the glycine cleavage system (Habarou et al., 2017). Biosynthesis of the lipoate cofactor is not well understood beyond prokaryotes (for comprehensive review see Cronan, 2016). Nevertheless, it is clear that lipoic acid synthase is required for the maturation of enzymes dependent on lipoate and uses two [4Fe-4S] clusters for its catalytic activity (Harmer et al., 2014). No study describing the Drosophila lipoic acid synthase has been published despite human disease related to lipoic acid deficiency (Mayr et al., 2014; Cronan, 2016; Habarou et al., 2017).
The central dogma of molecular biology depends on Fe-S enzymes
The central dogma of molecular biology, originally proposed by Crick (1958), radically changed the way biology is understood and taught (Cobb, 2017). DNA replication is the primary mode of information transfer during successive generations, whereas DNA transcription is the primary mechanism for specifying which proteins can be translated on ribosomes assembled in the cytoplasm. In addition to the nucleotide and amino acid building blocks, all three steps require energy and are interdependent, as nucleic acids and proteins are both essential for each process. Likewise, Fe-S clusters are also required at each step as cofactors of the DNA polymerase (see references in Table 1 and Kaguni et al., 1983; Peck et al., 1992; Sahashi et al., 2014; Stiban et al., 2014), of the essential subunit of the basal transcription factor TFIIH Xpd (Reynaud et al., 1999; Chen et al., 2003; Aguilar-Fuentes et al., 2006; Li et al., 2010) and of Pixie, which is required for ribosome biosynthesis and the initiation of translation (Andersen and Leevers, 2007; Kashima et al., 2014). Thus, DNA replication, transcription, and translation rest on the CIA providing Fe-S cluster to DNA polymerase, Xpd, and Pixie, respectively.
Are Fe-S and/or moco enzymes implicated in the circadian clock?
Plants alternate between photosynthesis and respiration during day-night cycles, whereas animal behavior shifts between an active stage that includes foraging, feeding and other motile behaviors, and sleep (Haydon et al., 2013; Mellor, 2016; Dubowy and Sehgal, 2017). Most animals anticipate the periodicity of sunlight and darkness through dedicated neuronal circuits whose rhythmic activity is sometimes referred to as the circadian clock (the genetic basis of which, was first discovered by Konopka and Benzer, 1971). In Drosophila the organization of the circadian circuitry has received considerable attention (Nitabach and Taghert, 2008; Hermann et al., 2013; Simoni et al., 2014). Similar to other animals, the circadian clock is interlinked with physiological functions in flies (Barber et al., 2016; Katewa et al., 2016; Rey et al., 2016; Kijak and Pyza, 2017; Klemz et al., 2017). Given the major role of Fe-S enzymes in intermediary metabolism and aerobic respiration, the question of whether Fe-S clusters are continuously present in key enzymes during the day-night cycle or whether some recycling of iron takes place in a rhythmic function has been posed (Mandilaras, 2012).
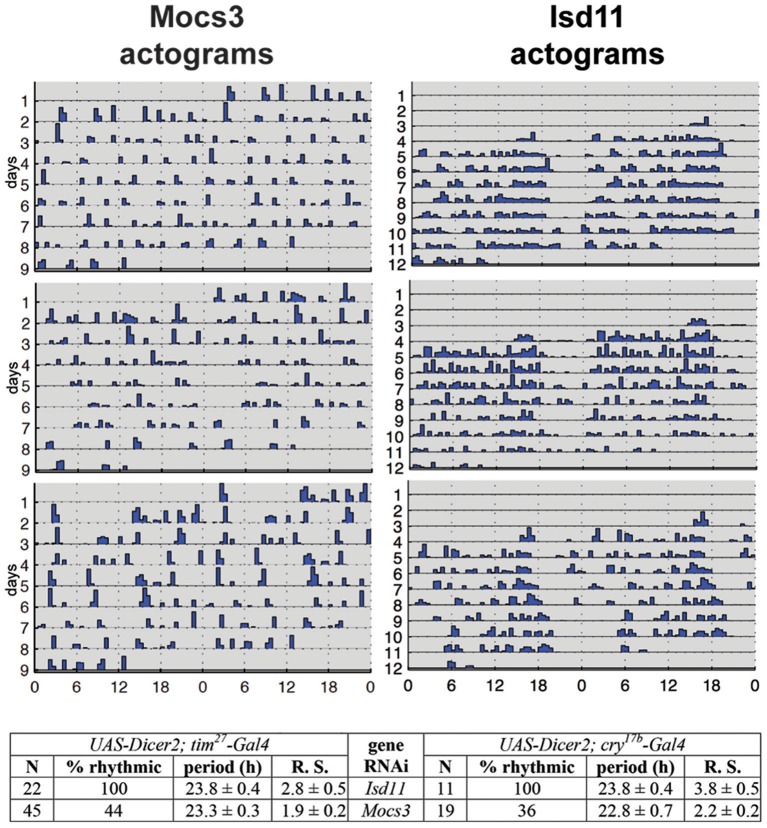
RNAi of Nfs1 (the cysteine desulfurase required for the biosynthesis of Fe-S clusters and Moco; Figure 1) in the circadian clock neurons resulted in loss of rhythmic activity of flies monitored under constant darkness (Mandilaras and Missirlis, 2012). Ubiquitous RNAi of Nfs1 caused lethality and eye-specific RNAi caused photoreceptor cell loss (Marelja, 2013). Two driver lines with overlapping, but not identical, expression patterns in the clock neurons, tim27-Gal4 and cry17b-Gal4 were recombined to the RNAi potentiator UAS-Dicer2 (Dietzl et al., 2007) and used, showing that Nfs1 RNAi driven by tim27-Gal4 resulted in a weaker arrhythmia than when driven by cry17b-Gal4 (Mandilaras and Missirlis, 2012). IscU RNAi driven by cry17b-Gal4 also resulted in arrhythmic flies, IscU RNAi driven by tim27-Gal4, however, resulted in lethality (a more severe phenotype, but one that cannot be tested for rhythmicity). In contrast, frataxin RNAi with both drivers did not show an arrhythmic phenotype (Mandilaras and Missirlis, 2012). To probe these genetic results further, RNAi of Isd11 and Mocs3 using the same drivers and assay was undertaken. Isd11 RNAi in circadian clock neurons showed no discernible phenotype, in contrast to Mocs3 RNAi, which resulted in 56% arrhythmic flies when driven with tim27-Gal4 and 64% arrhythmic flies when driven with cry17b-Gal4 (Figure 8).
Figure 8.
RNAi of Mocs3 disrupts circadian activity in constant darkness. Flies were first entrained on 3 12:12 h light-dark cycles then representative actograms are presented during constant darkness. Statistical analysis is depicted for two independent drivers. N represents number of individuals tested, Rhythmic Statistic (R. S.) values are provided for flies scored as rhythmic: higher values demonstrate more robust rhythmicity (for detailed methodology see Mandilaras and Missirlis, 2012).
The other Fe-S cluster biosynthesis genes that were implicated in the Drosophila circadian clock were Iba57 (CG8043) and Cfd1 (CG4858; also referred to as Nubp2), both encoding components of the CIA machinery, and IscA1, which is an Fe-S scaffold protein predicted to be localized to mitochondria (Mandilaras and Missirlis, 2012). IscA1 has recently found an exciting application in a new, non-invasive, technology to control experimentally the generation of neuronal action potentials, termed magnetogenetics (Long et al., 2015). It was also suggested that IscA1 is a component of fly magnetoreception (Cyranoski, 2015; Qin et al., 2016), but see (Meister, 2016; Pang et al., 2017). The proposed cryptochrome-IscA1 protein complex is predicted to reside in the cytosol or nucleus, given that cryptochrome is a protein localized in the cytosol and nucleus (Stanewsky et al., 1998; Yoshii et al., 2008). Whether, in addition to their predominant presence in mitochondria, IscA1, IscU, and Nfs1 have cytosolic localization in flies, as recently shown for human HSC20 (Kim et al., 2018), requires experimental verification. It is an interesting possibility to keep in mind when considering the result that RNAi against Mocs3, whose product is the proposed cytosolic partner of Nfs1 (Marelja et al., 2008, 2013), resulted in arrhythmic flies (Figure 8). Of note, two Fe-S enzyme encoding genes, dihydropyrimidine dehydrogenase and phosphoribosylamidotransferase, show circadian expression and are localized in the cytosol (Van Gelder et al., 1995; Rey et al., in review).
The new results presented here and the findings in Mandilaras and Missirlis (2012) are only based on RNAi and require independent confirmation. That iron may play a role in the circadian clock and sleep disorders was also proposed based on a Drosophila model of the human Restless Legs Syndrome (Freeman et al., 2012, 2013). This line of research was unfortunately disrupted at Queen Mary University of London (Allen and Missirlis, 2012; Ashworth, 2012; Horton, 2012), but deserves further attention, given the interplay between iron metabolism and the circadian clock in humans (Earley et al., 2014; Furudate et al., 2014; Dye et al., 2016), pigs (Zhang et al., 2017), rodents (Yin et al., 2007; Bianco et al., 2009; Simcox et al., 2015; Janich et al., 2016; Okazaki et al., 2016), plants (Chen et al., 2013; Hong et al., 2013; Salomé et al., 2013), and even diatoms (Botebol et al., 2015).
Cellular iron sensing and regulation is coupled to Fe-S cluster biosynthesis
D. melanogaster larvae or flies grown on diets with different iron content show cell-type specific responses (Poulson and Bowen, 1952; Georgieva et al., 1999; Mehta et al., 2009; Mandilaras et al., 2013). In mammals, IRPs regulate cytosolic iron concentrations (Zhang et al., 2014; Muckenthaler et al., 2017; Papanikolaou and Pantopoulos, 2017; Rouault and Maio, 2017). D. melanogaster has two IRP-like proteins, showing a partial evolutionary conservation in its iron sensing mechanism (Muckenthaler et al., 1998; Lind et al., 2006; Freeman et al., 2013). Another conserved aspect in cellular iron homeostasis is that genetic manipulations that affect mitochondrial Fe-S cluster biosynthesis lead to cytosolic iron depletion (Anderson et al., 2005; Uhrigshardt et al., 2013; Navarro et al., 2015). The mechanism of this response is not fully conserved between yeast (Wofford and Lindahl, 2015) and vertebrates (Wingert et al., 2005; Ye et al., 2010), as the former lack IRPs, and it is not known how Drosophila cytosolic iron is regulated through the mitochondrial Fe-S cluster assembly machinery. Furthermore, transcriptional regulation of ferritin upon iron sensing appears to dominate the Drosophila iron response (Georgieva et al., 1999; Missirlis et al., 2007; Rosas-Arellano et al., 2016), suggesting that an undiscovered transcription factor responds to varying cytosolic iron concentration in insects. Of note, Drosophila ferritin is loaded with iron in the endoplasmic reticulum (Xiao et al., 2014; Xiao and Zhou, 2018) and is an essential gene (González-Morales et al., 2015), as is also the case for IRP-1A (Puri et al., 2008). More work is needed to understand cellular iron homeostasis in Drosophila.
How do mitochondria regulate their iron needs?
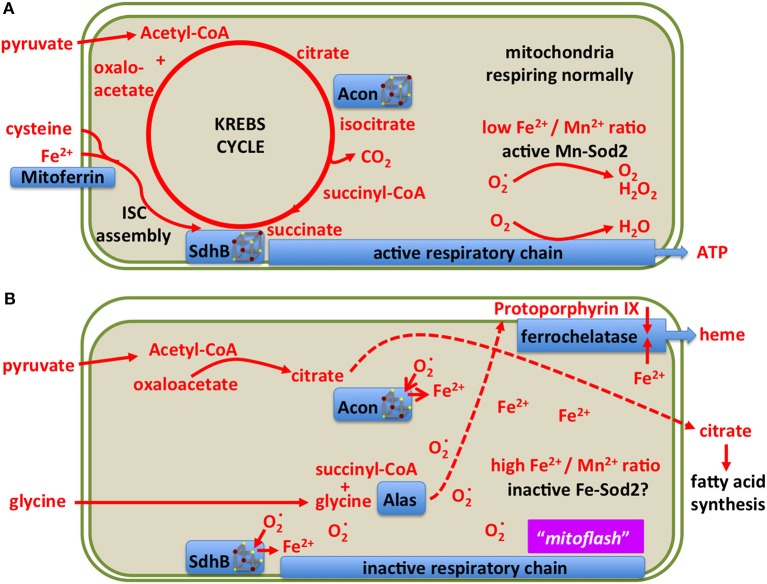
Mitochondria use a specialized ferritin (Missirlis et al., 2006) and at least one specialized transporter for iron import into the mitochondrial matrix, mitoferrin (Metzendorf and Lind, 2010; Navarro et al., 2015; Edenharter et al., 2017). The question how they sense and regulate iron concentration in the matrix is not resolved. One unexplained observation in this respect is that, at least in yeast, the GTP to GDP ratio affects iron concentration within this compartment (Gordon et al., 2006). In what follows, we explore the idea that manganese Sod2 (Kirby et al., 2002; Duttaroy et al., 2003) may serve as a mitochondrial iron sensor (Figure 9). Our view of normally respiring mitochondria is the familiar setting with an active Sod2 protecting Fe-S clusters and maintaining the TCA cycle (Figure 9A; Missirlis et al., 2003a). An increase in superoxide (Wong H. S. et al., 2017) deactivates the Fe-S clusters in aconitase and Sdh (Gardner and Fridovich, 1991; Gardner et al., 1995) leading to an increase in ferrous iron in the mitochondrial matrix (Srinivasan et al., 2000; Jensen et al., 2004; Esposito et al., 2013). The literature on these reactions has been reviewed with a discussion of the accompanying consequences for cell metabolism (see superoxide/aconitase rheostat model; Armstrong et al., 2004). Increased mitochondrial iron can replace manganese on Sod2 and inactivate the enzyme (Yang et al., 2006; Naranuntarat et al., 2009). Sod2 inactivation would result in a positive feedback loop, as more superoxide would accumulate, fully inactivating aconitase and Sdh bringing the TCA cycle to a halt.
Figure 9.
A model showing two different states of mitochondrial metabolism. (A) Conventional state based on the TCA (Krebs) cycle feeding reducing equivalents into the respiratory chain for the production of ATP. Only metabolites and enzymes referred to in the text are shown for simplicity. (B) A superoxide burst is predicted to reduce the [4Fe-4S] clusters of aconitase and Sdh, releasing ferrous iron into the mitochondrial matrix and blocking the TCA cycle. The resulting increase in iron concentration may inactivate manganese Sod2 resulting in a sustained pulse of superoxide, previously referred to as a “mitoflash.” In this state, mitochondria export citrate to the cytosol for fatty acid production, Alas uses up succinyl-CoA for protoporphyrin biosynthesis, ferrochelatase uses protoporphyrin and iron to produce heme. The consequent drop in matrix soluble iron reactivates manganese Sod2, allowing for Fe-S cluster biosynthesis to reactivate the TCA cycle enzymes.
Heping Cheng and co-workers described a superoxide burst in mitochondria, termed mitoflash, lasting for several seconds (Wang et al., 2008; Shen et al., 2014). Some skepticism on whether the phenomenon is real exists, mostly arguing that perhaps the change in the fluorescence of the reporter used relates to pH changes (Schwarzländer et al., 2012, 2014), but the coupling of TCA cycle with oxidative phosphorylation could mean that both claims (a burst in superoxide and alkalinization of the mitochondrial matrix) occur at similar timeframes or in parallel (Wei-LaPierre et al., 2013).
During the inactivation of Sod2, citrate cannot be isomerized to isocitrate and is therefore exported to the cytosol for fatty acid synthesis (see below). Likewise succinate cannot be metabolized, but its precursor, succinyl-CoA is a substrate of aminolevulinate synthase (Alas) in the first step of heme biosynthesis (Ruiz de Mena et al., 1999; Ponka et al., 2017). Thus the superoxide burst couples the redirection of iron and intermediary metabolites to ferrochelatase for heme production (Figure 9B). This way, iron concentration in the mitochondrial matrix drops, manganese binds to and reactivates Sod2, and the mitochondria return to their conventional state. In the above discussion we have not considered the role of hydrogen peroxide, produced by Sod2, which should be treated as a distinct metabolite (Missirlis et al., 2003b; Munro and Treberg, 2017).
Although the metabolites that are affected in the Sod2 mutants remain to be described, a recent paper reported a four-fold induction of aminolevulinic acid in Sod1 mutants, and curiously, a 20-fold induction of citrulline (Doran et al., 2017). Citrulline production depends on (the heme-containing) nitric oxide synthase (Nos), which uses as substrate arginine (Kuzin et al., 1996; Jaszczak et al., 2015). Superoxide-dependent inactivation of another manganese-containing enzyme, arginase, which degrades arginine (Samson, 2000), could lead to increased substrate availability for Nos. Indeed, it has been shown that arginase inactivation induces Nos activity (Caldwell et al., 2015). The above suggest another testable example of a similar hypothesis, where the interaction of superoxide with Fe-S clusters influences cell metabolism by releasing iron, which could transiently displace manganese from the active site of its target enzyme(s).
Fe-S clusters and lipid metabolism
Citrate is a key precursor for fatty acid synthesis (Watson and Lowenstein, 1970; Halperin et al., 1975) and therefore it is not surprising that inactivation of aconitase leads to increased lipogenesis (reviewed in Tong and Rouault, 2007). The same metabolic connection has also been demonstrated in Drosophila, by driving frataxin RNAi in glial cells (Navarro et al., 2010) or in photoreceptor mutant clones (Chen et al., 2016b). Another outcome of increased mitochondrial iron due to loss of frataxin is the activation, in a way that is not yet understood, of sphingolipids. Sphingolipids, in turn, activate signal transduction pathways, like, in the example of photoreceptors, of Phosphoinositide-dependent kinase 1 and Myocyte enhancer factor 2 (Chen et al., 2016a,b). Moreover, murine mutants in Sod2 show increased lipid accumulation in their livers (Li et al., 1995; Chouchani et al., 2017), which we attribute to the inactivation of aconitase (Figure 9). It remains to be shown whether during normal physiology our hypothesis of two alternating mitochondrial states, one driving aerobic respiration, the other driving lipogenesis and heme biosynthesis, is valid or not.
Physiological relevance of Fe-S and moco enzymes in different tissues
In this section we first look at the role of Fe-S enzymes during the life cycle of D. melanogaster, then focus on the main tissues of the adult fly where functions of Fe-S and Moco enzymes are known.
Development, growth, and aging: stem cells and cellular differentiation
The life cycle of Drosophila is separated in distinct stages. Egg-laying follows the insemination of oocytes in the female, embryogenesis gives rise to the larva, which feeds and grows until entry into metamorphosis, at the end of which adult flies emerge (Demerec, 1950). We discussed above that Fe-S enzymes drive aerobic respiration, which changes dramatically with the progress of embryogenesis (Lints et al., 1967; Tennessen et al., 2011, 2014), during larval growth (Heinrich et al., 2011; Merkey et al., 2011; Sen et al., 2013; Da-Ré et al., 2014) and adult aging (Lints and Lints, 1968; Ferguson et al., 2005; Dubessay et al., 2007; Klichko et al., 2014; Wolff et al., 2016). The decline in mitochondrial functions observed in late life, along with experiments showing that genetic manipulations leading to improved mitochondrial functions extended lifespan, suggested that mitochondrial metabolism governs the adult life span (Villee, 1948; Miquel, 1998; Fukagawa, 1999; Ross, 2000; Muller et al., 2007), but the finding that mitochondria isolated from old flies incubated with cytosol from young individuals restore respiration, whereas mitochondria from young individuals incubated with cytosol of old individuals fail to produce ATP questions this view (Vann and Webster, 1977; see also Sanz, 2016). On the other hand, there can be no doubt that defects in Fe-S cluster biosynthesis inhibit growth and dramatically shorten lifespan (Missirlis, 2003). It is also good to remember that the two sexes need to be considered separately when studying mitochondrial metabolism (Camus et al., 2012, 2015; Pomatto et al., 2017).
Increased glycolysis uncoupled from aerobic respiration is a characteristic of proliferative cells, for example during the early stages of Drosophila embryogenesis (Tennessen et al., 2011, 2014). This major metabolic switch is mediated through cell signaling (Thörig et al., 1981a,b; Markopoulou and Artavanis-Tsakonas, 1989; Homem et al., 2014; Barry and Thummel, 2016; Sieber et al., 2016; Slaninova et al., 2016; Mattila and Hietakangas, 2017). We already discussed that exit from the growth stage requires the concerted activity of Fe-S and heme enzymes in the prothoracic gland for ecdysone synthesis (Llorens et al., 2015; Palandri et al., 2015). Further, ecdysone is one of the signals inducing oxidative phosphorylation through the mitochondrial respiratory chain, sensed by neuroblasts and leading to their terminal differentiation (Homem et al., 2014) (see also Sen et al., 2013). Iron itself can directly influence stem cell differentiation, exemplified by hemocyte production in the lymph gland (Yoon et al., 2017). Thus, primary cell metabolism can define the fate of stem cells in a developing organism.
The germ line: is a lineage of quiescent mitochondria set aside for reproduction?
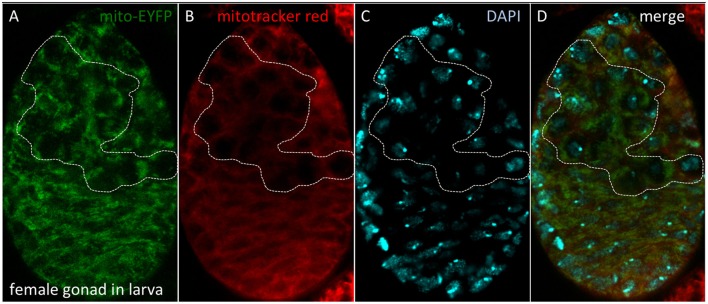
Stem cell differentiation in the ovary was found to depend on the mitochondrial ATP synthase (Teixeira et al., 2015). Surprisingly, in the female germarium, oxidative phosphorylation played no role in the early differentiation steps of the female germ line (Teixeira et al., 2015). Why are differentiating stem cells in the ovary and the larval brain different? One idea, first proposed by Allen (1996), is that the female germ line is defined as a carrier of quiescent (non-respiring) mitochondria (Cox and Spradling, 2003; de Paula et al., 2013a,b; Sieber et al., 2016). A key experiment to test this hypothesis is whether quiescent mitochondria can be detected throughout the life cycle in the female germ line (Allen and de Paula, 2013). We showed that quiescent mitochondria are observed in the female germ line within gonads of third instar larvae (Figure 10). However, similar experiments have proven harder to perform in embryos due to impermeability of the Mitotracker Red and reduced spaghetti squash-EYFP fluorescence (LaJeunesse et al., 2004), although the laboratory of Richa Rikhy recently succeeded to image mitochondria in living embryos using new constructs (Chowdhary et al., 2017). Of note, these quiescent mitochondria would constitute a third mitochondrial state, not described in Figure 9, since they lack the oxidative phosphorylation complexes and hence a source of superoxide to protect the mitochondrial DNA. This line of research was also unfortunately disrupted at Queen Mary University of London (Allen and Missirlis, 2012; Ashworth, 2012; Horton, 2012); for further insights see (Burrows, 2012; Mahul-Mellier et al., 2015; Lawrence, 2016; Edwards and Roy, 2017; Tsimilli-Michael and Haldimann, 2017).
Figure 10.
Quiescent mitochondria in female gonads of D. melanogaster larvae. (A) Bloomington stock #7194 carries a P-element insertion including the spaghetti squash promoter driving expression of EYFP tagged at the N-terminal end with a mitochondrial targeting sequence. (B) Dissected gonads were incubated with Mitotracker Red, which accumulates in mitochondria with active membrane potential. (C) Nuclei were observed with DAPI. (D) Merged image. The dotted line depicts the female germ line, where quiescent mitochondria are observed. Methods as described in Cox and Spradling (2003).
Alan Spradling and co-workers have also described quiescent mitochondria in the oocyte (Cox and Spradling, 2003), but they have shown in addition an active mitochondrial membrane potential during stages 4–8 of oogenesis (Sieber et al., 2016). Insulin signaling acting through glycogen synthase kinase 3 is required to set mitochondria into a quiescent phase from stage 10 of oogenesis (Sieber et al., 2016). Is the finding of an active membrane potential during oogenesis sufficient to refute the theory of quiescent mitochondria? The first issue to resolve is whether mitochondria in the germarium respire or are quiescent. Our interpretation of the limited published data (Sieber et al., 2016) is that mitochondria in female germ cells are quiescent and this would offer an explanation for the unexpected finding that the early steps of the female germ line differentiation is independent of oxidative phosphorylation (Teixeira et al., 2015). However, another well-known phenomenon in the transmission of mitochondria through successive generations is bottleneck selection of functional mitochondria, which occurs during Drosophila oogenesis (Cox and Spradling, 2003, 2006; Hill et al., 2014; Ma et al., 2014; Chen Z. et al., 2015). It is therefore possible that the discovery by Ruth Lehmann and co-workers of ATP synthase induction in the first steps of such a differentiation program proceeds in order for mitochondria to test their ability to sustain a membrane potential during stages 4–8 of oogenesis as a selection of functional mitochondria that can be safely transmitted to the oocyte.
If Allen's hypothesis is proven correct, he may have resolved Darwin's still-standing question of why it is that two separate sexes evolved (Venton, 2013). The two sexes were required to keep a quiescent form of mitochondria in the resting gamete, while dispensing the mitochondria used by the motile gamete (DeLuca and O'Farrell, 2012; Politi et al., 2014). The mitochondrial DNA polymerase (an Fe-S enzyme, Stiban et al., 2014) participates in the elimination of paternal mitochondrial genomes in D. melanogaster by a mechanism that is not yet understood (Yu et al., 2017).
Eye pigment formation
The study of heredity defines the birth of the field of Drosophila research, famously through observations of the color of their eyes (Morgan, 1910, 1911). The study of how eye pigments are formed, inspired by the drive to understand the genetic control of development (Beadle and Ephrussi, 1936; Lewis, 1978), gave rise to biochemical genetics. Progress in biochemistry (i.e., Hadorn and Mitchell, 1951) made clear early on that the Moco is required for pigment biosynthesis (Glassman and Mitchell, 1959; Hubby and Forrest, 1960). Progress in electron microscopy revealed impressive changes in cell biology occurring in eye color mutants (Nolte, 1961; Shoup, 1966; Sullivan and Sullivan, 1975) and, as discussed above, how Xdh reaches the pigment cells of the eye remains unsolved (Reaume et al., 1989, 1991).
Malpighian tubules and urate excretion
Malpighian tubules are the major excretory organ in flies (Beyenbach et al., 2010). The primary role of Xdh is in purine catabolism, mostly taking place in the Malpighian tubules (Dickinson and Gaughan, 1981; Reaume et al., 1989). Zinc ions are implicated in mineral excretion through this organ (Chi et al., 2015; Yin et al., 2017), while the same genes that govern pigment granule formation in the eye are also required for the formation of zinc storage granules in the Malpighian tubules (Tejeda-Guzman et al., 2018). Finally, the Malpighian tubules also show strong Aox activity (Dickinson and Gaughan, 1981), although the physiological function of this activity remains to be shown (Marelja et al., 2014).
Muscles for flight and heartbeat
D. melanogaster is able to fly several kilometers in the open desert (Dickinson, 2014). To do so, it uses specialized muscles (Iwamoto, 2011), which receive oxygen directly through the trachea (Lehmann and Schützner, 2010), respiring over 90% of the oxygen to sustain flight (Suarez, 2000). Mitochondria are key to this action (Levenbook and Williams, 1956) and ambient oxygen concentrations alter flight performance (Skandalis et al., 2011; Bosco et al., 2015; Shiehzadegan et al., 2017). If Fe-S respiratory enzymes are affected either by aging (Ferguson et al., 2005) or by mutation (Walker et al., 2006; Godenschwege et al., 2009; Martin et al., 2009; Vrailas-Mortimer et al., 2011; Oka et al., 2015), muscle pathology ensues. On the other hand, flies are highly resistant to hypoxia and indeed lacking manganese Sod2 results in short-lived adults (the majority dying within the first couple of days from pupal eclosion; Godenschwege et al., 2009), but this mortality is rescued by moving the flies in a hypoxic environment (Wicks et al., 2009). A survey into the genetic factors regulating natural variation in mitochondrial function in the Drosophila muscle revealed nuclear genomic control of naturally occurring variation in mitochondrial respiration (Correa et al., 2012; Jumbo-Lucioni et al., 2012), a process also regulated by calcineurin (Pfluger et al., 2015) and the mitochondrial contact site and cristae junction organizing system (Guarani et al., 2015). Finally, the requirement of Fe-S clusters and the respiratory chain has also been demonstrated in the Drosophila heart muscle (Tricoire et al., 2014; Martínez-Morentin et al., 2015).
The insect intestine functions beyond nutrient absorption
The primary function of the intestine lies in digestion of food and absorption of nutrients (Shanbhag and Tripathi, 2009; Lemaitre and Miguel-Aliaga, 2013). As this epithelium is continuously renewed, Drosophila researchers have paid more attention to the regulation of stem cells (Ohlstein and Spradling, 2006, 2007; Biteau et al., 2008; Lin et al., 2008; Takashima et al., 2008; Jiang et al., 2009; Scopelliti et al., 2014; Reiff et al., 2015; Brand et al., 2016; Hudry et al., 2016; Resnik-Docampo et al., 2017). Furthermore, many microbes reside in the intestine (Leulier and Royet, 2009; Shin et al., 2011; Buchon et al., 2013) and their activity can influence fly behavior and physiology (Sharon et al., 2010; Hang et al., 2014; Fischer et al., 2017; Leitão-Gonçalves et al., 2017; Mistry et al., 2017; Wong A. C. et al., 2017). One of the unique functions of the fly intestine is its role in copper (Filshie et al., 1971; Dubreuil, 2004; Burke et al., 2008) and iron (Tang and Zhou, 2013a; Rosas-Arellano et al., 2016) homeostasis. The specific roles of Fe-S and Moco enzymes in this tissue remain to be shown (Uhrigshardt et al., 2013).
Secretory glands, fat bodies, and nephrocytes
The role of Fe-S proteins in secretory tissues, like the salivary glands and the fat bodies, and in hemolymph filtering tissues, like the Garland and pericardial nephrocytes is also not resolved. One common property of these tissues is that their nuclei undergo polyploidy (Nordman et al., 2011), hence nuclear Fe-S enzymes involved in DNA replication are expected to have an enhanced role.
The nervous system
The ways in which iron and other metals relate to neurodegeneration have been reviewed (Zhu et al., 2014; Calap-Quintana et al., 2017), therefore a discussion on this topic will not be included here, except for the following points. Despite general agreement that frataxin is required for a functional nervous system, disagreement has been expressed on the cause, with different authors favoring oxidative stress (Llorens et al., 2007; Anderson et al., 2008; Kondapalli et al., 2008), iron toxicity (Soriano et al., 2013; Navarro et al., 2015), altered mitochondrial metabolism (Navarro et al., 2010; Tricoire et al., 2014; Calap-Quintana et al., 2015; Soriano et al., 2016), sphingolipid signaling (Chen et al., 2016b), and failure to maintain neuronal membrane potential (Shidara and Hollenbeck, 2010). We do not see any contradiction in the various positive claims made in the above-cited literature, whereas the negative claim that is often repeated—refuting a role for oxidative stress in explaining the phenotypes—normally arises because of failure to rescue the phenotypes with some transgenes as opposed to others. Similar failure could have various explanations: for example overexpression of Sod2 (Mockett et al., 1999) does not guarantee that the enzyme will be active in mitochondria with iron overload, at least our discussion of mitochondrial metabolism above (Figure 9) predicts otherwise. Furthermore, not all reactive oxygen species will act in the same way and their source and subcellular localization is also important to the effects they cause (Missirlis et al., 2003b). Last, cells handle iron in different ways: neurons and glia for example differ dramatically in their ability to store iron (Kosmidis et al., 2011, 2014) for reasons that are not understood. When ferritin mutants progress through early embryogenesis (they do so thanks to maternal contribution of iron-loaded ferritin to the oocyte) severe defects in the development of the nervous system ensue (González-Morales et al., 2015). Which step in brain development is most sensitive to the lack of ferritin has not been resolved. The blood-brain barrier regulates iron entry into the brain (Mehta et al., 2009), but we still do not know how iron traffics in the peripheral and central nervous systems or how Fe-S and/or Moco enzymes affect the circadian clock. Answers to questions of the basic cell biology of metal homeostasis are prerequisite for proposing better therapeutics when neuronal functions are compromised in disease (Zhu et al., 2014; Calap-Quintana et al., 2017; Ruland et al., 2017; Poetini et al., 2018).
Conclusion
We cannot think of any biological function for which the fly will not require the biochemical participation of Fe-S clusters. We have attempted to describe our progress in understanding the role of Fe-S enzymes during the past 93 years since Otto Warburg firmly connected iron to respiration (Warburg, 1925) and also the Fe-S and Moco enzymes Xdh and Aox. We have used the fly as an example, but of course knowledge has been acquired from studying all forms of life as these enzymes are universal in character and may have formed at the origin of life (Hall et al., 1971; Russell and Martin, 2004; Nitschke and Russell, 2009; Schoepp-Cothenet et al., 2012; Varma et al., in review). We suggest that bioinorganic contributions to biology and bioenergetics be taken into account not only as having “house-keeping” roles, but also as an active component of the complex organization that characterizes all living systems (Frausto da Silva and Williams, 2001). Renewed attention on the inorganic chemistry underpinning Drosophila biology, together with the new analytical tools and methodologies available, should help integrate cellular metal homeostasis with metabolism (Dow, 2017; Navarro and Schneuwly, 2017). The humble fly has still much to contribute to our understanding of the workings of biology.
Dedication
Dedicated to Stefan Grimm (1963–2014) who discovered, while working at Imperial College London, a moonlighting function for IkBα binding to the outer mitochondrial membrane: protecting cells from suicide.
Author contributions
ZM: Performed the experiments on the mitochondrial Fe-S assembly core complex (Figure 3), determined the subcellular localization of fly Isd11 and Mocs3 (Figure 4), and performed the biochemical assays for aldehyde oxidase and sulfite oxidase (Figures 6, 7) during his Ph.D. thesis with SL; ZM, SL, and FM: Wrote the sections on Fe-S cluster assembly, Moco biosynthesis and the function of molybdoenzymes; FM: Found the quiescent mitochondria in the female gonad of larvae (Figure 10) and is responsible for the hypothesis that Sod2 acts as a mitochondrial iron sensor and other theories expressed in the latter part of the article; All authors have read, reviewed, and endorsed the full content of this publication.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors thank Kuanyu Li from Nanjing University who provided the purified human frataxin (Figure 3), Konstantinos Mandilaras for the circadian behavior tests performed at Queen Mary University of London (Figure 8) and the two reviewers for their helpful comments. Ralf Stanewsky, who had worked with two of the 2017 Nobel Prize in Physiology and Medicine laureates on light entrainment of the circadian clock (Stanewsky et al., 1996, 1997, 1998; Emery et al., 2000) and now focuses on the question how the clock is temperature compensated (Sehadova et al., 2009; Gentile et al., 2013; Simoni et al., 2014; Chen C. et al., 2015; Buhl et al., 2016; Klemz et al., 2017; Tang et al., 2017) introduced FM to circadian clocks; Tracey Rouault to iron biology (Missirlis et al., 2003a, 2006, 2007; Kosmidis et al., 2011, 2014; Uhrigshardt et al., 2013). The 2017 Nobel Prize in Chemistry, on the other hand, was awarded in recognition of cryo-electron microscopy, a new technique Jon Nield had proposed to FM for analyzing iron loading into the Drosophila ferritins (Ruprecht and Nield, 2001; Gutiérrez et al., 2013). Norbert Krauss determined with co-workers the first X-ray crystallographic structures of photosystems I and II (Jordan et al., 2001; Zouni et al., 2001) and later resolved structures of G-coupled protein receptors and cryptochromes (Scheerer et al., 2008; Choe et al., 2011; Zhang F. et al., 2013; Nagano et al., 2016). The experimental work and theories presented here benefited by close interactions with the above-mentioned colleagues and with John F. Allen (Allen, 1996, 2010, 2015; Allen and Missirlis, 2012; Allen and de Paula, 2013; de Paula et al., 2013a,b; Ibrahim et al., 2017), during the period our appointments at Queen Mary University of London coincided (2007–2012).
Footnotes
Funding. This work was supported by the Marie Curie International Reintegration Grant “DrosoFela” (MIRG-CT-2007-204832) and the Consejo Nacional de Ciencia y Tecnología Basic Science Young Investigator Project #179835 to FM, and the Deutsche Forschungsgemeinschaft grant LE1171/11-1 to SL. ZM was supported by the Ph.D. grant of the German National Academic Foundation (Studienstiftung des deutschen Volkes) and was recipient of the short-term European Molecular Biology Laboratory fellowship ASTF 226-2011.
References
- Adam A. C., Bornhövd C., Prokisch H., Neupert W., Hell K. (2006). The Nfs1 interacting protein Isd11 has an essential role in Fe/S cluster biogenesis in mitochondria. EMBO J. 25, 174–183. 10.1038/sj.emboj.7600905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams M. D., Celniker S. E., Holt R. A., Evans C. A., Gocayne J. D., Amanatides P. G., et al. (2000). The genome sequence of Drosophila melanogaster. Science 287, 2185–2195. 10.1126/science.287.5461.2185 [DOI] [PubMed] [Google Scholar]
- Aguilar-Fuentes J., Valadez-Graham V., Reynaud E., Zurita M. (2006). TFIIH trafficking and its nuclear assembly during early Drosophila embryo development. J. Cell Sci. 119, 3866–3875. 10.1242/jcs.03150 [DOI] [PubMed] [Google Scholar]
- Aldea M., Hernández-Chico C., de la Campa A. G., Kushner S. R., Vicente M. (1988). Identification, cloning, and expression of bolA, an ftsZ-dependent morphogene of Escherichia coli. J. Bacteriol. 170, 5169–5176. 10.1128/jb.170.11.5169-5176.1988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alhebshi A., Sideri T. C., Holland S. L., Avery S. V. (2012). The essential iron-sulfur protein Rli1 is an important target accounting for inhibition of cell growth by reactive oxygen species. Mol. Biol. Cell 23, 3582–3590. 10.1091/mbc.E12-05-0413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen J. F. (1996). Separate sexes and the mitochondrial theory of ageing. J. Theor. Biol. 180, 135–140. 10.1006/jtbi.1996.0089 [DOI] [PubMed] [Google Scholar]
- Allen J. F. (2010). Research and how to promote it in a university. Future Med. Chem. 2, 15–20. 10.4155/fmc.09.150 [DOI] [PubMed] [Google Scholar]
- Allen J. F. (2015). Why chloroplasts and mitochondria retain their own genomes and genetic systems: colocation for redox regulation of gene expression. Proc. Natl. Acad. Sci. U.S.A. 112, 10231–10238. 10.1073/pnas.1500012112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen J. F., de Paula W. B. (2013). Mitochondrial genome function and maternal inheritance. Biochem. Soc. Trans. 41, 1298–1304. 10.1042/BST20130106 [DOI] [PubMed] [Google Scholar]
- Allen J. F., Missirlis F. (2012). Queen Mary: nobody expects the Spanish Inquisition. Lancet 379, 1785. 10.1016/S0140-6736(12)60697-7 [DOI] [PubMed] [Google Scholar]
- Alziari S., Berthier F., Touraille S., Stepien G., Durand R. (1985). Mitochondrial DNA expression in Drosophila melanogaster: neosynthesized polypeptides in isolated mitochondria. Biochimie 67, 1023–1034. 10.1016/S0300-9084(85)80297-2 [DOI] [PubMed] [Google Scholar]
- Amrani L., Primus J., Glatigny A., Arcangeli L., Scazzocchio C., Finnerty V. (2000). Comparison of the sequences of the Aspergillus nidulans hxB and Drosophila melanogaster ma-l genes with nifS from Azotobacter vinelandii suggests a mechanism for the insertion of the terminal sulphur atom in the molybdopterin cofactor. Mol. Microbiol. 38, 114–125. 10.1046/j.1365-2958.2000.02119.x [DOI] [PubMed] [Google Scholar]
- Andersen D. S., Leevers S. J. (2007). The essential Drosophila ATP-binding cassette domain protein, pixie, binds the 40 S ribosome in an ATP-dependent manner and is required for translation initiation. J. Biol. Chem. 282, 14752–14760. 10.1074/jbc.M701361200 [DOI] [PubMed] [Google Scholar]
- Anderson P. R., Kirby K., Hilliker A. J., Phillips J. P. (2005). RNAi-mediated suppression of the mitochondrial iron chaperone, frataxin, in Drosophila. Hum. Mol. Genet. 14, 3397–3405. 10.1093/hmg/ddi367 [DOI] [PubMed] [Google Scholar]
- Anderson P. R., Kirby K., Orr W. C., Hilliker A. J., Phillips J. P. (2008). Hydrogen peroxide scavenging rescues frataxin deficiency in a Drosophila model of Friedreich's ataxia. Proc. Natl. Acad. Sci. U.S.A. 105, 611–616. 10.1073/pnas.0709691105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong J. S., Whiteman M., Yang H., Jones D. P. (2004). The redox regulation of intermediary metabolism by a superoxide-aconitase rheostat. Bioessays 26, 894–900. 10.1002/bies.20071 [DOI] [PubMed] [Google Scholar]
- Ashworth R. (2012). Redundancies at Queen Mary,University of London. Lancet 379, 2338–2339. 10.1016/S0140-6736(12)61013-7 [DOI] [PubMed] [Google Scholar]
- Au H. C., Scheffler I. E. (1994). Characterization of the gene encoding the iron-sulfur protein subunit of succinate dehydrogenase from Drosophila melanogaster. Gene 149, 261–265. 10.1016/0378-1119(94)90158-9 [DOI] [PubMed] [Google Scholar]
- Auchère F., Santos R., Planamente S., Lesuisse E., Camadro J. M. (2008). Glutathione-dependent redox status of frataxin-deficient cells in a yeast model of Friedreich's ataxia. Hum. Mol. Genet. 17, 2790–2802. 10.1093/hmg/ddn178 [DOI] [PubMed] [Google Scholar]
- Baker B. S. (1973). The maternal and zygotic control of development by cinnamon, a new mutant in Drosophila melanogaster. Dev. Biol. 33, 429–440. 10.1016/0012-1606(73)90148-6 [DOI] [PubMed] [Google Scholar]
- Balk J., Aguilar Netz D. J., Tepper K., Pierik A. J., Lill R. (2005a). The essential WD40 protein Cia1 is involved in a late step of cytosolic and nuclear iron-sulfur protein assembly. Mol. Cell. Biol. 25, 10833–10841. 10.1128/MCB.25.24.10833-10841.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balk J., Pierik A. J., Aguilar Netz D. J., Mühlenhoff U., Lill R. (2005b). Nar1p, a conserved eukaryotic protein with similarity to Fe-only hydrogenases, functions in cytosolic iron-sulphur protein biogenesis. Biochem. Soc. Trans. 33, 86–89. 10.1042/BST0330086 [DOI] [PubMed] [Google Scholar]
- Balk J., Schaedler T. A. (2014). Iron cofactor assembly in plants. Annu. Rev. Plant Biol. 65, 125–153. 10.1146/annurev-arplant-050213-035759 [DOI] [PubMed] [Google Scholar]
- Banci L., Ciofi-Baffoni S., Gajda K., Muzzioli R., Peruzzini R., Winkelmann J. (2015). N-terminal domains mediate [2Fe-2S] cluster transfer from glutaredoxin-3 to anamorsin. Nat. Chem. Biol. 11, 772–778. 10.1038/nchembio.1892 [DOI] [PubMed] [Google Scholar]
- Barber A. F., Erion R., Holmes T. C., Sehgal A. (2016). Circadian and feeding cues integrate to drive rhythms of physiology in Drosophila insulin-producing cells. Genes Dev. 30, 2596–2606. 10.1101/gad.288258.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barry W. E., Thummel C. S. (2016). The Drosophila HNF4 nuclear receptor promotes glucose-stimulated insulin secretion and mitochondrial function in adults. eLife 5:e11183. 10.7554/eLife.11183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beadle G. W., Ephrussi B. (1936). The differentiation of eye pigments in Drosophila as studied by transplantation. Genetics 21, 225–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beer K. B., Wehman A. M. (2017). Mechanisms and functions of extracellular vesicle release in vivo-What we can learn from flies and worms. Cell Adh. Migr. 11, 135–150. 10.1080/19336918.2016.1236899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beilschmidt L. K., Puccio H. M. (2014). Mammalian Fe-S cluster biogenesis and its implication in disease. Biochimie 100, 48–60. 10.1016/j.biochi.2014.01.009 [DOI] [PubMed] [Google Scholar]
- Beinert H. (2002). Spectroscopy of succinate dehydrogenases, a historical perspective. Biochim. Biophys. Acta 1553, 7–22. 10.1016/S0005-2728(01)00235-3 [DOI] [PubMed] [Google Scholar]
- Beinert H., Sands R. H. (1960). Studies on succinic and DPNH dehydrogenase preparations by paramagnetic resonance (EPR) spectroscopy. Biochem. Biophys. Res. Commun. 3, 41–46. 10.1016/0006-291X(60)90100-5 [DOI] [Google Scholar]
- Bellen H. J., Tong C., Tsuda H. (2010). 100 years of Drosophila research and its impact on vertebrate neuroscience: a history lesson for the future. Nat. Rev. Neurosci. 11, 514–522. 10.1038/nrn2839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bettedi L., Aslam M. F., Szular J., Mandilaras K., Missirlis F. (2011). Iron depletion in the intestines of Malvolio mutant flies does not occur in the absence of a multicopper oxidase. J. Exp. Biol. 214, 971–978. 10.1242/jeb.051664 [DOI] [PubMed] [Google Scholar]
- Beyenbach K. W., Skaer H., Dow J. A. (2010). The developmental, molecular, and transport biology of Malpighian tubules. Annu. Rev. Entomol. 55, 351–374. 10.1146/annurev-ento-112408-085512 [DOI] [PubMed] [Google Scholar]
- Bianco L. E., Unger E. L., Earley C. J., Beard J. L. (2009). Iron deficiency alters the day-night variation in monoamine levels in mice. Chronobiol. Int. 26, 447–463. 10.1080/07420520902820905 [DOI] [PubMed] [Google Scholar]
- Biteau B., Hochmuth C. E., Jasper H. (2008). JNK activity in somatic stem cells causes loss of tissue homeostasis in the aging Drosophila gut. Cell Stem Cell 3, 442–455. 10.1016/j.stem.2008.07.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc B., Gerez C., Ollagnier de Choudens S. (2015). Assembly of Fe/S proteins in bacterial systems: biochemistry of the bacterial ISC system. Biochim. Biophys. Acta 1853, 1436–1447. 10.1016/j.bbamcr.2014.12.009 [DOI] [PubMed] [Google Scholar]
- Bogaart A. M., Bernini L. F. (1981). The molybdoenzyme system of Drosophila melanogaster. I. Sulfite oxidase: identification and properties. Expression of the enzyme in maroon-like (mal), low-xanthine dehydrogenase (lxd), and cinnamon (cin) flies. Biochem. Genet. 19, 929–946. 10.1007/BF00504258 [DOI] [PubMed] [Google Scholar]
- Bosco G., Clamer M., Messulam E., Dare C., Yang Z., Zordan M., et al. (2015). Effects of oxygen concentration and pressure on Drosophila melanogaster: oxidative stress, mitochondrial activity, and survivorship. Arch. Insect Biochem. Physiol. 88, 222–234. 10.1002/arch.21217 [DOI] [PubMed] [Google Scholar]
- Botebol H., Lesuisse E., Šuták R., Six C., Lozano J. C., Schatt P., et al. (2015). Central role for ferritin in the day/night regulation of iron homeostasis in marine phytoplankton. Proc. Natl. Acad. Sci. U.S.A. 112, 14652–14657. 10.1073/pnas.1506074112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braaten A. C., Bentley M. M. (1993). Sulfite sensitivity and sulfite oxidase activity in Drosophila melanogaster. Biochem. Genet. 31, 375–391. 10.1007/BF02396224 [DOI] [PubMed] [Google Scholar]
- Brand M. D., Goncalves R. L., Orr A. L., Vargas L., Gerencser A. A., Borch Jensen M., et al. (2016). Suppressors of superoxide-H2O2 production at site IQ of mitochondrial complex I protect against stem cell hyperplasia and ischemia-reperfusion injury. Cell Metab. 24, 582–592. 10.1016/j.cmet.2016.08.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridges C. B. (1918). Maroon: a recurrent mutation in Drosophila. Proc. Natl. Acad. Sci. U.S.A. 4, 316–318. 10.1073/pnas.4.10.316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridwell-Rabb J., Fox N. G., Tsai C. L., Winn A. M., Barondeau D. P. (2014). Human frataxin activates Fe-S cluster biosynthesis by facilitating sulfur transfer chemistry. Biochemistry 53, 4904–4913. 10.1021/bi500532e [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browder L. W., Williamson J. H. (1976). The effects of cinnamon on xanthine dehydrogenase, aldehyde oxidase, and pyridoxal oxidase activity during development in Drosophila melanogaster. Dev. Biol. 53, 241–249. 10.1016/0012-1606(76)90226-8 [DOI] [PubMed] [Google Scholar]
- Buchon N., Broderick N. A., Lemaitre B. (2013). Gut homeostasis in a microbial world: insights from Drosophila melanogaster. Nat. Rev. Microbiol. 11, 615–626. 10.1038/nrmicro3074 [DOI] [PubMed] [Google Scholar]
- Buhl E., Bradlaugh A., Ogueta M., Chen K. F., Stanewsky R., Hodge J. J. (2016). Quasimodo mediates daily and acute light effects on Drosophila clock neuron excitability. Proc. Natl. Acad. Sci. U.S.A. 113, 13486–13491. 10.1073/pnas.1606547113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bühning M., Friemel M., Leimkühler S. (2017). Functional complementation studies reveal different interaction partners of Escherichia coli IscS and human NFS1. Biochemistry 56, 4592–4605. 10.1021/acs.biochem.7b00627 [DOI] [PubMed] [Google Scholar]
- Burke R., Commons E., Camakaris J. (2008). Expression and localisation of the essential copper transporter DmATP7 in Drosophila neuronal and intestinal tissues. Int. J. Biochem. Cell Biol. 40, 1850–1860. 10.1016/j.biocel.2008.01.021 [DOI] [PubMed] [Google Scholar]
- Burrows R. (2012). Living with the h-index? Metric assemblages in the contemporary academy. Sociol. Rev. 60, 355–372. 10.1111/j.1467-954X.2012.02077.x [DOI] [Google Scholar]
- Cai K., Tonelli M., Frederick R. O., Markley J. L. (2017). Human mitochondrial ferredoxin 1 (FDX1) and ferredoxin 2 (FDX2) both bind cysteine desulfurase and donate electrons for iron-sulfur cluster biosynthesis. Biochemistry 56, 487–499. 10.1021/acs.biochem.6b00447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calap-Quintana P., González-Fernández J., Sebastiá-Ortega N., Llorens J. V., Moltó M. D. (2017). Drosophila melanogaster models of metal-related human diseases and metal toxicity. Int. J. Mol. Sci. 18:E1456. 10.3390/ijms18071456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calap-Quintana P., Soriano S., Llorens J. V., Al-Ramahi I., Botas J., Moltó M. D., et al. (2015). TORC1 inhibition by rapamycin promotes antioxidant defences in a Drosophila model of Friedreich's Ataxia. PLoS ONE 10:e0132376. 10.1371/journal.pone.0132376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldwell R. B., Toque H. A., Narayanan S. P., Caldwell R. W. (2015). Arginase: an old enzyme with new tricks. Trends Pharmacol. Sci. 36, 395–405. 10.1016/j.tips.2015.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cammack R. (1982). Iron-sulphur cluster interconversions and the activation of aconitase. Nature 298, 792–793. 10.1038/298792a0 [DOI] [PubMed] [Google Scholar]
- Campuzano V., Montermini L., Moltó M. D., Pianese L., Cossée M., Cavalcanti F., et al. (1996). Friedreich's ataxia: autosomal recessive disease caused by an intronic GAA triplet repeat expansion. Science 271, 1423–1427. 10.1126/science.271.5254.1423 [DOI] [PubMed] [Google Scholar]
- Camus M. F., Clancy D. J., Dowling D. K. (2012). Mitochondria, maternal inheritance, and male aging. Curr. Biol. 22, 1717–1721. 10.1016/j.cub.2012.07.018 [DOI] [PubMed] [Google Scholar]
- Camus M. F., Wolf J. B., Morrow E. H., Dowling D. K. (2015). Single nucleotides in the mtDNA sequence modify mitochondrial molecular function and are associated with sex-specific effects on fertility and aging. Curr. Biol. 25, 2717–2722. 10.1016/j.cub.2015.09.012 [DOI] [PubMed] [Google Scholar]
- Cañizares J., Blanca J. M., Navarro J. A., Monrós E., Palau F., Moltó M. D. (2000). DFH is a Drosophila homolog of the Friedreich's ataxia disease gene. Gene 256, 35–42. 10.1016/S0378-1119(00)00343-7 [DOI] [PubMed] [Google Scholar]
- Chandel N. S. (2015). Evolution of mitochondria as signaling organelles. Cell Metab. 22, 204–206. 10.1016/j.cmet.2015.05.013 [DOI] [PubMed] [Google Scholar]
- Chen C., Buhl E., Xu M., Croset V., Rees J. S., Lilley K. S., et al. (2015). Drosophila ionotropic receptor 25a mediates circadian clock resetting by temperature. Nature 527, 516–520. 10.1038/nature16148 [DOI] [PubMed] [Google Scholar]
- Chen J., Larochelle S., Li X., Suter B. (2003). Xpd/Ercc2 regulates CAK activity and mitotic progression. Nature 424, 228–232. 10.1038/nature01746 [DOI] [PubMed] [Google Scholar]
- Chen K., Ho T. S., Lin G., Tan K. L., Rasband M. N., Bellen H. J. (2016a). Loss of Frataxin activates the iron/sphingolipid/PDK1/Mef2 pathway in mammals. eLife 5:e20732. 10.7554/eLife.20732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K., Lin G., Haelterman N. A., Ho T. S., Li T., Li Z., et al. (2016b). Loss of Frataxin induces iron toxicity, sphingolipid synthesis, and Pdk1/Mef2 activation, leading to neurodegeneration. elife 5:e16043. 10.7554/eLife.16043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y. Y., Wang Y., Shin L. J., Wu J. F., Shanmugam V., Tsednee M., et al. (2013). Iron is involved in the maintenance of circadian period length in Arabidopsis. Plant Physiol. 161, 1409–1420. 10.1104/pp.112.212068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z., Qi Y., French S., Zhang G., Covian Garcia R., Balaban R., et al. (2015). Genetic mosaic analysis of a deleterious mitochondrial DNA mutation in Drosophila reveals novel aspects of mitochondrial regulation and function. Mol. Biol. Cell 26, 674–684. 10.1091/mbc.E14-11-1513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Z., Tsuda M., Kishita Y., Sato Y., Aigaki T. (2013). Impaired energy metabolism in a Drosophila model of mitochondrial aconitase deficiency. Biochem. Biophys. Res. Commun. 433, 145–150. 10.1016/j.bbrc.2013.02.040 [DOI] [PubMed] [Google Scholar]
- Chi T., Kim M. S., Lang S., Bose N., Kahn A., Flechner L., et al. (2015). A Drosophila model identifies a critical role for zinc in mineralization for kidney stone disease. PLoS ONE 10:e0124150. 10.1371/journal.pone.0124150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe H. W., Kim Y. J., Park J. H., Morizumi T., Pai E. F., Krauss N., et al. (2011). Crystal structure of metarhodopsin II. Nature 471, 651–655. 10.1038/nature09789 [DOI] [PubMed] [Google Scholar]
- Chouchani E. T., Kazak L., Spiegelman B. M. (2017). Mitochondrial reactive oxygen species and adipose tissue thermogenesis: bridging physiology and mechanisms. J. Biol. Chem. 292, 16810–16816. 10.1074/jbc.R117.789628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chovnick A., Gelbart W., McCarron M., Osmond B. (1976). Organization of the rosy locus in Drosophila melanogaster: evidence for a control element adjacent to the xanthine dehydrogenase structural element. Genetics 84, 233–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhary S., Tomer D., Dubal D., Sambre D., Rikhy R. (2017). Analysis of mitochondrial organization and function in the Drosophila blastoderm embryo. Sci. Rep. 7, 5502. 10.1038/s41598-017-05679-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark D. V. (1994). Molecular and genetic analyses of Drosophila Prat, which encodes the first enzyme of de novo purine biosynthesis. Genetics 136, 547–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobb M. (2017). 60 years ago, Francis Crick changed the logic of biology. PLoS Biol. 15:e2003243. 10.1371/journal.pbio.2003243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins J. F., Glassman E. (1969). A third locus (ipo) affecting pyridoxal oxidase in Drosophila melanogaster. Genetics 61, 833–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Correa C. C., Aw W. C., Melvin R. G., Pichaud N., Ballard J. W. (2012). Mitochondrial DNA variants influence mitochondrial bioenergetics in Drosophila melanogaster. Mitochondrion 12, 459–464. 10.1016/j.mito.2012.06.005 [DOI] [PubMed] [Google Scholar]
- Courtright J. B. (1967). Polygenic control of aldehyde oxidase in Drosophila. Genetics 57, 25–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courtright J. B. (1975). Evidence for a new type of complementation among the cin, lxd and ma-l loci in Drosophila melanogaster. Mol. Gen. Genet. 142, 231–238. 10.1007/BF00425648 [DOI] [PubMed] [Google Scholar]
- Cox R. T., Spradling A. C. (2003). A Balbiani body and the fusome mediate mitochondrial inheritance during Drosophila oogenesis. Development 130, 1579–1590. 10.1242/dev.00365 [DOI] [PubMed] [Google Scholar]
- Cox R. T., Spradling A. C. (2006). Milton controls the early acquisition of mitochondria by Drosophila oocytes. Development 133, 3371–3377. 10.1242/dev.02514 [DOI] [PubMed] [Google Scholar]
- Crick F. H. (1958). On protein synthesis. Symp. Soc. Exp. Biol. 12, 138–163. [PubMed] [Google Scholar]
- Cronan J. E. (2016). Assembly of lipoic acid on its cognate enzymes: an extraordinary and essential biosynthetic pathway. Microbiol. Mol. Biol. Rev. 80, 429–450. 10.1128/MMBR.00073-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cupp-Vickery J. R., Urbina H., Vickery L. E. (2003). Crystal structure of IscS, a cysteine desulfurase from Escherichia coli. J. Mol. Biol. 330, 1049–1059. 10.1016/S0022-2836(03)00690-9 [DOI] [PubMed] [Google Scholar]
- Cypher J. J., Tedesco J. L., Courtright J. B., Kumaran A. K. (1982). Tissue-specific and substrate-specific detection of aldehyde and pyridoxal oxidase in larval and imaginal tissues of Drosophila melanogaster. Biochem. Genet. 20, 315–332. 10.1007/BF00484427 [DOI] [PubMed] [Google Scholar]
- Cyranoski D. (2015). Long-sought biological compass discovered. Nature 527, 283–284. 10.1038/527283a [DOI] [PubMed] [Google Scholar]
- Da-Ré C., De Pittà C., Zordan M. A., Teza G., Nestola F., Zeviani M., et al. (2014). UCP4C mediates uncoupled respiration in larvae of Drosophila melanogaster. EMBO Rep. 15, 586–591. 10.1002/embr.201337972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Paula W. B., Agip A. N., Missirlis F., Ashworth R., Vizcay-Barrena G., Lucas C. H., et al. (2013a). Female and male gamete mitochondria are distinct and complementary in transcription, structure, and genome function. Genome Biol. Evol. 5, 1969–1977. 10.1093/gbe/evt147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Paula W. B., Lucas C. H., Agip A. N., Vizcay-Barrena G., Allen J. F. (2013b). Energy, ageing, fidelity and sex: oocyte mitochondrial DNA as a protected genetic template. Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci. 368:20120263. 10.1098/rstb.2012.0263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLuca S. Z., O'Farrell P. H. (2012). Barriers to male transmission of mitochondrial DNA in sperm development. Dev. Cell 22, 660–668. 10.1016/j.devcel.2011.12.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demerec M. (1950). Biology of Drosophila. New York, NY: John Wiley and Sons. [Google Scholar]
- Dickinson M. H. (2014). Death valley, Drosophila, and the devonian toolkit. Annu. Rev. Entomol. 59, 51–72. 10.1146/annurev-ento-011613-162041 [DOI] [PubMed] [Google Scholar]
- Dickinson W. J. (1970). The genetics of aldehyde oxidase in Drosophila melanogaster. Genetics 66, 487–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson W. J., Gaughan S. (1981). Aldehyde oxidases of Drosophila: contributions of several enzymes to observed activity patterns. Biochem. Genet. 19, 567–583. 10.1007/BF00484627 [DOI] [PubMed] [Google Scholar]
- Dietzl G., Chen D., Schnorrer F., Su K. C., Barinova Y., Fellner M., et al. (2007). A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature 448, 151–156. 10.1038/nature05954 [DOI] [PubMed] [Google Scholar]
- Doran M. L., Knee J. M., Wang N., Rzezniczak T. Z., Parkes T. L., Li L., et al. (2017). Metabolomic analysis of oxidative stress: superoxide dismutase mutation and paraquat induced stress in Drosophila melanogaster. Free Radic. Biol. Med. 113, 323–334. 10.1016/j.freeradbiomed.2017.10.011 [DOI] [PubMed] [Google Scholar]
- Dow J. A. (2017). The essential roles of metal ions in insect homeostasis and physiology. Curr. Opin. Insect Sci. 23, 43–50. 10.1016/j.cois.2017.07.001 [DOI] [PubMed] [Google Scholar]
- Doyle W. A., Burke J. F., Chovnick A., Dutton F. L., Whittle J. R., Bray R. C. (1996). Properties of xanthine dehydrogenase variants from rosy mutant strains of Drosophila melanogaster and their relevance to the enzyme's structure and mechanism. Eur. J. Biochem. 239, 782–795. 10.1111/j.1432-1033.1996.0782u.x [DOI] [PubMed] [Google Scholar]
- Dubessay P., Garreau-Balandier I., Jarrousse A. S., Fleuriet A., Sion B., Debise R., et al. (2007). Aging impact on biochemical activities and gene expression of Drosophila melanogaster mitochondria. Biochimie 89, 988–1001. 10.1016/j.biochi.2007.03.018 [DOI] [PubMed] [Google Scholar]
- Dubowy C., Sehgal A. (2017). Circadian Rhythms and Sleep in Drosophila melanogaster. Genetics 205, 1373–1397. 10.1534/genetics.115.185157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubreuil R. R. (2004). Copper cells and stomach acid secretion in the Drosophila midgut. Int. J. Biochem. Cell Biol. 36, 745–752. 10.1016/j.biocel.2003.07.004 [DOI] [PubMed] [Google Scholar]
- Duke E. J., Rushing D. R., Glassman E. (1975). Nutritional control of xanthine dehydrogenase. II. Effects on xanthine dehydrogenase and aldehyde oxidase of culturing wild-type and mutant Drosophila on different levels of molybdenum. Biochem. Genet. 13, 53–64. 10.1007/BF00486006 [DOI] [PubMed] [Google Scholar]
- Duke P. S. (1968). An EPR study of live and dead Drosophila adults under aerobic conditions. Exp. Gerontol. 3, 335–340. 10.1016/0531-5565(68)90046-6 [DOI] [PubMed] [Google Scholar]
- Duttaroy A., Paul A., Kundu M., Belton A. (2003). A Sod2 null mutation confers severely reduced adult life span in Drosophila. Genetics 165, 2295–2299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dye T. J., Jain S. V., Simakajornboon N. (2016). Outcomes of long-term iron supplementation in pediatric restless legs syndrome/periodic limb movement disorder (RLS/PLMD). Sleep Med. 32, 213–219. 10.1016/j.sleep.2016.01.008 [DOI] [PubMed] [Google Scholar]
- Dzul S. P., Rocha A. G., Rawat S., Kandegedara A., Kusowski A., Pain J., et al. (2017). In vitro characterization of a novel Isu homologue from Drosophila melanogaster for de novo FeS-cluster formation. Metallomics 9, 48–60. 10.1039/C6MT00163G [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earley C. J., Connor J., Garcia-Borreguero D., Jenner P., Winkelman J., Zee P. C., et al. (2014). Altered brain iron homeostasis and dopaminergic function in Restless Legs Syndrome (Willis-Ekbom Disease). Sleep Med. 15, 1288–1301. 10.1016/j.sleep.2014.05.009 [DOI] [PubMed] [Google Scholar]
- Eck R. V., Dayhoff M. O. (1966). Evolution of the structure of ferredoxin based on living relics of primitive amino Acid sequences. Science 152, 363–366. 10.1126/science.152.3720.363 [DOI] [PubMed] [Google Scholar]
- Edenharter O., Clement J., Schneuwly S., Navarro J. A. (2017). Overexpression of Drosophila frataxin triggers cell death in an iron-dependent manner. J. Neurogenet. 31, 189–202. 10.1080/01677063.2017.1363200 [DOI] [PubMed] [Google Scholar]
- Edwards M. A., Roy S. (2017). Academic research in the 21st century: maintaining scientific integrity in a climate of perverse incentives and hypercompetition (vol 34, pg 51, 2017). Environ. Eng. Sci. 34, 616–616. 10.1089/ees.2016.0223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emery P., Stanewsky R., Hall J. C., Rosbash M. (2000). A unique circadian-rhythm photoreceptor. Nature 404, 456–457. 10.1038/35006558 [DOI] [PubMed] [Google Scholar]
- Enya S., Yamamoto C., Mizuno H., Esaki T., Lin H. K., Iga M., et al. (2017). Dual roles of glutathione in ecdysone biosynthesis and antioxidant function during the larval development in Drosophila. Genetics 207, 1519–1532. 10.1534/genetics.117.300391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esposito G., Vos M., Vilain S., Swerts J., De Sousa Valadas J., Van Meensel S., et al. (2013). Aconitase causes iron toxicity in Drosophila pink1 mutants. PLoS Genet. 9:e1003478. 10.1371/journal.pgen.1003478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farhan S. M., Wang J., Robinson J. F., Lahiry P., Siu V. M., Prasad C., et al. (2014). Exome sequencing identifies NFS1 deficiency in a novel Fe-S cluster disease, infantile mitochondrial complex II/III deficiency. Mol. Genet. Genomic. Med. 2, 73–80. 10.1002/mgg3.46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng C., Tollin G., Enemark J. H. (2007). Sulfite oxidizing enzymes. Biochim. Biophys. Acta 1774, 527–539. 10.1016/j.bbapap.2007.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng G., Tintrup H., Kirsch J., Nichol M. C., Kuhse J., Betz H., et al. (1998). Dual requirement for gephyrin in glycine receptor clustering and molybdoenzyme activity. Science 282, 1321–1324. 10.1126/science.282.5392.1321 [DOI] [PubMed] [Google Scholar]
- Ferguson M., Mockett R. J., Shen Y., Orr W. C., Sohal R. S. (2005). Age-associated decline in mitochondrial respiration and electron transport in Drosophila melanogaster. Biochem. J. 390, 501–511. 10.1042/BJ20042130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferre J., Silva F. J., Real M. D., Mensua J. L. (1986). Pigment patterns in mutants affecting the biosynthesis of pteridines and xanthommatin in Drosophila melanogaster. Biochem. Genet. 24, 545–569. 10.1007/BF00504334 [DOI] [PubMed] [Google Scholar]
- Fiedorczuk K., Letts J. A., Degliesposti G., Kaszuba K., Skehel M., Sazanov L. A. (2016). Atomic structure of the entire mammalian mitochondrial complex I. Nature 538, 406–410. 10.1038/nature19794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filshie B. K., Poulson D. F., Waterhouse D. F. (1971). Ultrastructure of the copper-accumulating region of the Drosophila larval midgut. Tissue Cell 3, 77–102. 10.1016/S0040-8166(71)80033-2 [DOI] [PubMed] [Google Scholar]
- Finnerty V. G., Duck P., Chovnick A. (1970). Studies on genetic organization in higher organisms. IComplementation, I. and fine structure of the maroon-like locus of Drosophila melanogaster. Proc. Natl. Acad. Sci.U.S.A. 65, 939–946. 10.1073/pnas.65.4.939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer C. N., Trautman E. P., Crawford J. M., Stabb E. V., Handelsman J., Broderick N. A. (2017). Metabolite exchange between microbiome members produces compounds that influence Drosophila behavior. eLife 6:e18855. 10.7554/eLife.18855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fogo J. K., Popowsky M. (1949). Spectrophotometric determination of hydrogen sulfide - methylene blue method. Anal. Chem. 21, 732–734. 10.1021/ac60030a028 [DOI] [Google Scholar]
- Forrest H. S., Hanly E. W., Lagowski J. M. (1961). Biochemical differences between the mutants Rosy-2 and maroon-like of Drosophila melanogaster. Genetics 46, 1455–1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox D. J., Conscience-Egli M., Abächerli E. (1972). The soluble citric acid cycle enzymes of Drosophila melanogaster. II. Tissue and intracellular distribution of aconitase and NADP-dependent isocitrate dehydrogenase. Biochem. Genet. 7, 163–175. 10.1007/BF00486086 [DOI] [PubMed] [Google Scholar]
- Fox N. G., Chakrabarti M., McCormick S. P., Lindahl P. A., Barondeau D. P. (2015). The human iron-sulfur assembly complex catalyzes the synthesis of [2Fe-2S] clusters on ISCU2 that can be transferred to acceptor molecules. Biochemistry 54, 3871–3879. 10.1021/bi5014485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox P. L. (2003). The copper-iron chronicles: the story of an intimate relationship. Biometals 16, 9–40. 10.1023/A:1020799512190 [DOI] [PubMed] [Google Scholar]
- Frausto da Silva J. J. R., Williams R. J. P. (2001). The Biological Chemistry of the Elements. New York, NY: Oxford University Press. [Google Scholar]
- Freeman A., Pranski E., Miller R. D., Radmard S., Bernhard D., Jinnah H. A., et al. (2012). Sleep fragmentation and motor restlessness in a Drosophila model of Restless Legs Syndrome. Curr. Biol. 22, 1142–1148. 10.1016/j.cub.2012.04.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman A. A., Mandilaras K., Missirlis F., Sanyal S. (2013). An emerging role for Cullin-3 mediated ubiquitination in sleep and circadian rhythm: insights from Drosophila. Fly 7, 39–43. 10.4161/fly.23506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman M. R., Dobritsa A., Gaines P., Segraves W. A., Carlson J. R. (1999). The dare gene: steroid hormone production, olfactory behavior, and neural degeneration in Drosophila. Development 126, 4591–4602. [DOI] [PubMed] [Google Scholar]
- Frey A. G., Palenchar D. J., Wildemann J. D., Philpott C. C. (2016). A glutaredoxin.bola complex serves as an iron-sulfur cluster chaperone for the cytosolic cluster assembly machinery. J. Biol. Chem. 291, 22344–22356. 10.1074/jbc.M116.744946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friemel M., Marelja Z., Li K., Leimkühler S. (2017). The N-terminus of iron-sulfur cluster assembly factor ISD11 is crucial for subcellular targeting and interaction with L-cysteine desulfurase NFS1. Biochemistry 56, 1797–1808. 10.1021/acs.biochem.6b01239 [DOI] [PubMed] [Google Scholar]
- Fukagawa N. K. (1999). Aging: is oxidative stress a marker or is it causal? Proc. Soc. Exp. Biol. Med. 222, 293–298. 10.1046/j.1525-1373.1999.d01-146.x [DOI] [PubMed] [Google Scholar]
- Furudate N., Komada Y., Kobayashi M., Nakajima S., Inoue Y. (2014). Daytime dysfunction in children with restless legs syndrome. J. Neurol. Sci. 336, 232–236. 10.1016/j.jns.2013.11.001 [DOI] [PubMed] [Google Scholar]
- Garattini E., Fratelli M., Terao M. (2008). Mammalian aldehyde oxidases: genetics, evolution and biochemistry. Cell. Mol. Life Sci. 65, 1019–1048. 10.1007/s00018-007-7398-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia C. J., Khajeh J., Coulanges E., Chen E. I., Owusu-Ansah E. (2017). Regulation of mitochondrial complex I biogenesis in Drosophila flight muscles. Cell Rep. 20, 264–278. 10.1016/j.celrep.2017.06.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner P. R., Fridovich I. (1991). Superoxide sensitivity of the Escherichia coli aconitase. J. Biol. Chem. 266, 19328–19333. [PubMed] [Google Scholar]
- Gardner P. R., Raineri I., Epstein L. B., White C. W. (1995). Superoxide radical and iron modulate aconitase activity in mammalian cells. J. Biol. Chem. 270, 13399–13405. 10.1074/jbc.270.22.13399 [DOI] [PubMed] [Google Scholar]
- Gari K., León Ortiz A. M., Borel V., Flynn H., Skehel J. M., Boulton S. J. (2012). MMS19 links cytoplasmic iron-sulfur cluster assembly to DNA metabolism. Science 337, 243–245. 10.1126/science.1219664 [DOI] [PubMed] [Google Scholar]
- Gentile C., Sehadova H., Simoni A., Chen C., Stanewsky R. (2013). Cryptochrome antagonizes synchronization of Drosophila's circadian clock to temperature cycles. Curr. Biol. 23, 185–195. 10.1016/j.cub.2012.12.023 [DOI] [PubMed] [Google Scholar]
- Georgieva T., Dunkov B. C., Harizanova N., Ralchev K., Law J. H. (1999). Iron availability dramatically alters the distribution of ferritin subunit messages in Drosophila melanogaster. Proc. Natl. Acad. Sci. U.S.A. 96, 2716–2721. 10.1073/pnas.96.6.2716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerber J., Mühlenhoff U., Lill R. (2003). An interaction between frataxin and Isu1/Nfs1 that is crucial for Fe/S cluster synthesis on Isu1. EMBO Rep. 4, 906–911. 10.1038/sj.embor.embor918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glassman E., Mitchell H. K. (1959). Mutants of Drosophila Melanogaster deficient in xanthine dehydrogenase. Genetics 44, 153–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godenschwege T., Forde R., Davis C. P., Paul A., Beckwith K., Duttaroy A. (2009). Mitochondrial superoxide radicals differentially affect muscle activity and neural function. Genetics 183, 175–184. 10.1534/genetics.109.103515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gontijo A. M., Miguela V., Whiting M. F., Woodruff R. C., Dominguez M. (2011). Intron retention in the Drosophila melanogaster rieske iron sulphur protein gene generated a new protein. Nat. Commun. 2, 323. 10.1038/ncomms1328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Morales N., Mendoza-Ortíz M., Á. Blowes L. M., Missirlis F., Riesgo-Escovar J. R. (2015). Ferritin is required in multiple tissues during Drosophila melanogaster development. PLoS ONE 10:e0133499. 10.1371/journal.pone.0133499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon D. M., Lyver E. R., Lesuisse E., Dancis A., Pain D. (2006). GTP in the mitochondrial matrix plays a crucial role in organellar iron homoeostasis. Biochem. J. 400, 163–168. 10.1042/BJ20060904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gradilla A. C., González E., Seijo I., Andrés G., Bischoff M., González-Mendez L., et al. (2014). Exosomes as Hedgehog carriers in cytoneme-mediated transport and secretion. Nat. Commun. 5:5649. 10.1038/ncomms6649 [DOI] [PubMed] [Google Scholar]
- Grant P., Maga T., Loshakov A., Singhal R., Wali A., Nwankwo J., et al. (2016). An eye on trafficking genes: identification of four eye color mutations in Drosophila. G3 6, 3185–3196. 10.1534/g3.116.032508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray N. K., Pantopoulos K., Dandekar T., Ackrell B. A., Hentze M. W. (1996). Translational regulation of mammalian Drosophila citric acid cycle enzymes via iron-responsive elements. Proc. Natl. Acad. Sci. U.S.A. 93, 4925–4930. 10.1073/pnas.93.10.4925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray T. A., Nicholls R. D. (2000). Diverse splicing mechanisms fuse the evolutionarily conserved bicistronic MOCS1A and MOCS1B open reading frames. RNA 6, 928–936. 10.1017/S1355838200000182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross J. C., Chaudhary V., Bartscherer K., Boutros M. (2012). Active Wnt proteins are secreted on exosomes. Nat. Cell Biol. 14, 1036–1045. 10.1038/ncb2574 [DOI] [PubMed] [Google Scholar]
- Gruenewald S., Wahl B., Bittner F., Hungeling H., Kanzow S., Kotthaus J., et al. (2008). The fourth molybdenum containing enzyme mARC: cloning and involvement in the activation of N-hydroxylated prodrugs. J. Med. Chem. 51, 8173–8177. 10.1021/jm8010417 [DOI] [PubMed] [Google Scholar]
- Guarani V., McNeill E. M., Paulo J. A., Huttlin E. L., Fröhlich F., Gygi S. P., et al. (2015). QIL1 is a novel mitochondrial protein required for MICOS complex stability and cristae morphology. eLife 4:e6265. 10.7554/eLife.06265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutiérrez L., Zubow K., Nield J., Gambis A., Mollereau B., Lázaro F. J., et al. (2013). Biophysical and genetic analysis of iron partitioning and ferritin function in Drosophila melanogaster. Metallomics 5, 997–1005. 10.1039/c3mt00118k [DOI] [PubMed] [Google Scholar]
- Habarou F., Hamel Y., Haack T. B., Feichtinger R. G., Lebigot E., Marquardt I., et al. (2017). Biallelic mutations in LIPT2 cause a mitochondrial lipoylation defect associated with severe neonatal encephalopathy. Am. J. Hum. Genet. 101, 283–290. 10.1016/j.ajhg.2017.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadorn E., Mitchell H. K. (1951). Properties of mutants of drosophila melanogaster and changes during development as revealed by paper chromatography. Proc. Natl. Acad. Sci. U.S.A. 37, 650–665. 10.1073/pnas.37.10.65.0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadorn E., Schwinck I. (1956). A Mutant of Drosophila without Isoxanthopterine which is non-autonomous for the red eye pigments. Nature 177, 940–941. 10.1038/177940a0 [DOI] [PubMed] [Google Scholar]
- Hall D. O., Cammack R., Rao K. K. (1971). Role for ferredoxins in the origin of life and biological evolution. Nature 233, 136–138. 10.1038/233136a0 [DOI] [PubMed] [Google Scholar]
- Halperin M. L., Cheema-Dhadli S., Taylor W. M., Fritz I. B. (1975). Role of the citrate transporter in the control of fatty acid synthesis. Adv. Enzyme Regul. 13, 435–445. 10.1016/0065-2571(75)90029-1 [DOI] [PubMed] [Google Scholar]
- Hang S., Purdy A. E., Robins W. P., Wang Z., Mandal M., Chang S., et al. (2014). The acetate switch of an intestinal pathogen disrupts host insulin signaling and lipid metabolism. Cell Host Microbe 16, 592–604. 10.1016/j.chom.2014.10.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hänzelmann P., Schindelin H. (2004). Crystal structure of the S-adenosylmethionine-dependent enzyme MoaA and its implications for molybdenum cofactor deficiency in humans. Proc. Natl. Acad. Sci. U.S.A. 101, 12870–12875. 10.1073/pnas.0404624101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hänzelmann P., Schindelin H. (2006). Binding of 5'-GTP to the C-terminal FeS cluster of the radical S-adenosylmethionine enzyme MoaA provides insights into its mechanism. Proc. Natl. Acad. Sci. U.S.A. 103, 6829–6834. 10.1073/pnas.0510711103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmer J. E., Hiscox M. J., Dinis P. C., Fox S. J., Iliopoulos A., Hussey J. E., et al. (2014). Structures of lipoyl synthase reveal a compact active site for controlling sequential sulfur insertion reactions. Biochem. J. 464, 123–133. 10.1042/BJ20140895 [DOI] [PubMed] [Google Scholar]
- Hausmann A., Aguilar Netz D. J., Balk J., Pierik A. J., Mühlenhoff U., Lill R. (2005). The eukaryotic P loop NTPase Nbp35: an essential component of the cytosolic and nuclear iron-sulfur protein assembly machinery. Proc. Natl. Acad. Sci. U.S.A. 102, 3266–3271. 10.1073/pnas.0406447102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haydon M. J., Hearn T. J., Bell L. J., Hannah M. A., Webb A. A. (2013). Metabolic regulation of circadian clocks. Semin. Cell Dev. Biol. 24, 414–421. 10.1016/j.semcdb.2013.03.007 [DOI] [PubMed] [Google Scholar]
- Hegde R. S., Bernstein H. D. (2006). The surprising complexity of signal sequences. Trends Biochem. Sci. 31, 563–571. 10.1016/j.tibs.2006.08.004 [DOI] [PubMed] [Google Scholar]
- Heinrich E. C., Farzin M., Klok C. J., Harrison J. F. (2011). The effect of developmental stage on the sensitivity of cell and body size to hypoxia in Drosophila melanogaster. J. Exp. Biol. 214, 1419–1427. 10.1242/jeb.051904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemler M. E. (2003). Tetraspanin proteins mediate cellular penetration, invasion, and fusion events and define a novel type of membrane microdomain. Annu. Rev. Cell Dev. Biol. 19, 397–422. 10.1146/annurev.cellbio.19.111301.153609 [DOI] [PubMed] [Google Scholar]
- Hermann C., Saccon R., Senthilan P. R., Domnik L., Dircksen H., Yoshii T., et al. (2013). The circadian clock network in the brain of different Drosophila species. J. Comp. Neurol. 521, 367–388. 10.1002/cne.23178 [DOI] [PubMed] [Google Scholar]
- Hill J. H., Chen Z., Xu H. (2014). Selective propagation of functional mitochondrial DNA during oogenesis restricts the transmission of a deleterious mitochondrial variant. Nat. Genet. 46, 389–392. 10.1038/ng.2920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hille R. (1996). The mononuclear molybdenum enzymes. Chem. Rev. 96, 2757–2816. 10.1021/cr950061t [DOI] [PubMed] [Google Scholar]
- Hille R. (2002). Molybdenum enzymes containing the pyranopterin cofactor: an overview. Met. Ions Biol. Syst. 39, 187–226. 10.1201/9780203909331.ch6 [DOI] [PubMed] [Google Scholar]
- Hille R., Hall J., Basu P. (2014). The mononuclear molybdenum enzymes. Chem. Rev. 114, 3963–4038. 10.1021/cr400443z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hille R., Nishino T., Bittner F. (2011). Molybdenum enzymes in higher organisms. Coord. Chem. Rev. 255, 1179–1205. 10.1016/j.ccr.2010.11.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilliker A. J., Duyf B., Evans D., Phillips J. P. (1992). Urate-null rosy mutants of Drosophila melanogaster are hypersensitive to oxygen stress. Proc. Natl. Acad. Sci. U.S.A. 89, 4343–4347. 10.1073/pnas.89.10.4343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho Y. K., Guthrie M. J., Clifford A. J., Ho C. C. (1992). Effect of adenine metabolites on survival of Drosophila melanogaster of low xanthine dehydrogenase activity. Comp. Biochem. Physiol. B 103, 413–417. 10.1016/0305-0491(92)90313-G [DOI] [PubMed] [Google Scholar]
- Homem C. C. F., Steinmann V., Burkard T. R., Jais A., Esterbauer H., Knoblich J. A. (2014). Ecdysone and mediator change energy metabolism to terminate proliferation in Drosophila neural stem cells. Cell 158, 874–888. 10.1016/j.cell.2014.06.024 [DOI] [PubMed] [Google Scholar]
- Hong S., Kim S. A., Guerinot M. L., McClung C. R. (2013). Reciprocal interaction of the circadian clock with the iron homeostasis network in Arabidopsis. Plant Physiol. 161, 893–903. 10.1104/pp.112.208603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton R. (2012). Offline: be careful what you wish for. Lancet 380, 13–13. 10.1016/S0140-6736(12)61078-2 [DOI] [Google Scholar]
- Hover B. M., Yokoyama K. (2015). C-Terminal glycine-gated radical initiation by GTP 3',8-cyclase in the molybdenum cofactor biosynthesis. J. Am. Chem. Soc. 137, 3352–3359. 10.1021/ja512997j [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J., Song D., Flores A., Zhao Q., Mooney S. M., Shaw L. M., et al. (2007). IOP1, a novel hydrogenase-like protein that modulates hypoxia-inducible factor-1alpha activity. Biochem. J. 401, 341–352. 10.1042/BJ20060635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubby J. L., Forrest H. S. (1960). Studies on the Mutant Maroon-like in Drosophila Melanogaster. Genetics 45, 211–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudry B., Khadayate S., Miguel-Aliaga I. (2016). The sexual identity of adult intestinal stem cells controls organ size and plasticity. Nature 530, 344–348. 10.1038/nature16953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes R. K. (1992). Xanthine dehydrogenase from Drosophila melanogaster: purification and properties of the wild-type enzyme and of a variant lacking iron-sulfur centers. Biochemistry 31, 3073–3083. 10.1021/bi00127a007 [DOI] [PubMed] [Google Scholar]
- Hughes R. K., Doyle W. A., Chovnick A., Whittle J. R., Burke J. F., Bray R. C. (1992). Use of rosy mutant strains of Drosophila melanogaster to probe the structure and function of xanthine dehydrogenase. Biochem. J. 285(Pt 2), 507–513. 10.1042/bj2850507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibrahim I. M., Wang L., Puthiyaveetil S., Krauß N., Nield J., Allen J. F. (2017). Oligomeric states in sodium ion-dependent regulation of cyanobacterial histidine kinase-2. Protoplasma. [Epub ahead of print]. 10.1007/s00709-017-1196-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichida K., Matsumura T., Sakuma R., Hosoya T., Nishino T. (2001). Mutation of human molybdenum cofactor sulfurase gene is responsible for classical xanthinuria type IBiochemical, I., and biophysical research Communications 282, 1194–1200. 10.1006/bbrc.2001.4719 [DOI] [PubMed] [Google Scholar]
- Isaya G. (2014). Mitochondrial iron-sulfur cluster dysfunction in neurodegenerative disease. Front. Pharmacol. 5:29. 10.3389/fphar.2014.00029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwamoto H. (2011). Structure, function and evolution of insect flight muscle. Biophysics 7, 21–28. 10.2142/biophysics.7.21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janich P., Arpat A. B., Castelo-Szekely V., Gatfield D. (2016). Analyzing the temporal regulation of translation efficiency in mouse liver. Genomics Data 8, 41–44. 10.1016/j.gdata.2016.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaszczak J. S., Wolpe J. B., Dao A. Q., Halme A. (2015). Nitric oxide synthase regulates growth coordination during Drosophila melanogaster imaginal disc regeneration. Genetics 200, 1219–1228. 10.1534/genetics.115.178053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen L. T., Culotta V. C. (2000). Role of Saccharomyces cerevisiae ISA1 and ISA2 in iron homeostasis. Mol. Cell. Biol. 20, 3918–3927. 10.1128/MCB.20.11.3918-3927.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen L. T., Sanchez R. J., Srinivasan C., Valentine J. S., Culotta V. C. (2004). Mutations in Saccharomyces cerevisiae iron-sulfur cluster assembly genes and oxidative stress relevant to Cu,Zn superoxide dismutase. J. Biol. Chem. 279, 29938–29943. 10.1074/jbc.M402795200 [DOI] [PubMed] [Google Scholar]
- Jiang H., Patel P. H., Kohlmaier A., Grenley M. O., McEwen D. G., Edgar B. A. (2009). Cytokine/Jak/Stat signaling mediates regeneration and homeostasis in the Drosophila midgut. Cell 137, 1343–1355. 10.1016/j.cell.2009.05.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson C., Roos A. K., Montano S. J., Sengupta R., Filippakopoulos P., Guo K., et al. (2011). The crystal structure of human GLRX5: iron-sulfur cluster co-ordination, tetrameric assembly and monomer activity. Biochem. J. 433, 303–311. 10.1042/BJ20101286 [DOI] [PubMed] [Google Scholar]
- Johnstone R. W., Wang J., Tommerup N., Vissing H., Roberts T., Shi Y. (1998). Ciao 1 is a novel WD40 protein that interacts with the tumor suppressor protein WT1. J. Biol. Chem. 273, 10880–10887. 10.1074/jbc.273.18.10880 [DOI] [PubMed] [Google Scholar]
- Jones M. A., Amr S., Ferebee A., Huynh P., Rosenfeld J. A., Miles M. F., et al. (2014). Genetic studies in Drosophila and humans support a model for the concerted function of CISD2, PPT1 and CLN3 in disease. Biol. Open 3, 342–352. 10.1242/bio.20147559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan P., Fromme P., Witt H. T., Klukas O., Saenger W., Krauss N. (2001). Three-dimensional structure of cyanobacterial photosystem I at 2.5 A resolution. Nature 411, 909–917. 10.1038/35082000 [DOI] [PubMed] [Google Scholar]
- Jumbo-Lucioni P., Bu S., Harbison S. T., Slaughter J. C., Mackay T. F., Moellering D. R., et al. (2012). Nuclear genomic control of naturally occurring variation in mitochondrial function in Drosophila melanogaster. BMC Genomics 13:659. 10.1186/1471-2164-13-659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaguni L. S., Rossignol J. M., Conaway R. C., Lehman I. R. (1983). Isolation of an intact DNA polymerase-primase from embryos of Drosophila melanogaster. Proc. Natl. Acad. Sci. U.S.A. 80, 2221–2225. 10.1073/pnas.80.8.2221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamdar K. P., Primus J. P., Shelton M. E., Archangeli L. L., Wittle A. E., Finnerty V. (1997). Structure of the molybdenum cofactor genes in Drosophila. Biochem. Soc. Trans. 25, 778–783. 10.1042/bst0250778 [DOI] [PubMed] [Google Scholar]
- Kamdar K. P., Shelton M. E., Finnerty V. (1994). The Drosophila molybdenum cofactor gene cinnamon is homologous to three Escherichia coli cofactor proteins and to the rat protein gephyrin. Genetics 137, 791–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamleh M. A., Hobani Y., Dow J. A., Watson D. G. (2008). Metabolomic profiling of Drosophila using liquid chromatography fourier transform mass spectrometry. FEBS Lett. 582, 2916–2922. 10.1016/j.febslet.2008.07.029 [DOI] [PubMed] [Google Scholar]
- Kamleh M. A., Hobani Y., Dow J. A., Zheng L., Watson D. G. (2009). Towards a platform for the metabonomic profiling of different strains of Drosophila melanogaster using liquid chromatography-Fourier transform mass spectrometry. FEBS J. 276, 6798–6809. 10.1111/j.1742-4658.2009.07397.x [DOI] [PubMed] [Google Scholar]
- Kanzok S. M., Fechner A., Bauer H., Ulschmid J. K., Müller H. M., Botella-Munoz J., et al. (2001). Substitution of the thioredoxin system for glutathione reductase in Drosophila melanogaster. Science 291, 643–646. 10.1126/science.291.5504.643 [DOI] [PubMed] [Google Scholar]
- Kashima I., Takahashi M., Hashimoto Y., Sakota E., Nakamura Y., Inada T. (2014). A functional involvement of ABCE1, eukaryotic ribosome recycling factor, in nonstop mRNA decay in Drosophila melanogaster cells. Biochimie 106, 10–16. 10.1016/j.biochi.2014.08.001 [DOI] [PubMed] [Google Scholar]
- Katewa S. D., Akagi K., Bose N., Rakshit K., Camarella T., Zheng X., et al. (2016). Peripheral circadian clocks mediate dietary restriction-dependent changes in lifespan and fat metabolism in Drosophila. Cell Metab. 23, 143–154. 10.1016/j.cmet.2015.10.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaut A., Lange H., Diekert K., Kispal G., Lill R. (2000). Isa1p is a component of the mitochondrial machinery for maturation of cellular iron-sulfur proteins and requires conserved cysteine residues for function. J. Biol. Chem. 275, 15955–15961. 10.1074/jbc.M909502199 [DOI] [PubMed] [Google Scholar]
- Keith T. P., Riley M. A., Kreitman M., Lewontin R. C., Curtis D., Chambers G. (1987). Sequence of the structural gene for xanthine dehydrogenase (rosy locus) in Drosophila melanogaster. Genetics 116, 67–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller E. C., Jr., Glassman E. (1964). A third locus (Lxd) affecting xanthine dehydrogenase in Drosophila Melanogaster. Genetics 49, 663–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kijak E., Pyza E. (2017). TOR signaling pathway and autophagy are involved in the regulation of circadian rhythms in behavior and plasticity of L2 interneurons in the brain of Drosophila melanogaster. PLoS ONE 12:e0171848. 10.1371/journal.pone.0171848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K. S., Maio N., Singh A., Rouault T. A. (2018). Cytosolic HSC20 integrates de novo iron-sulfur cluster biogenesis with the CIAO1-mediated transfer to recipients. Hum. Mol. Genet. [Epub ahead of print]. 10.1093/hmg/ddy004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirby K., Hu J., Hilliker A. J., Phillips J. P. (2002). RNA interference-mediated silencing of Sod2 in Drosophila leads to early adult-onset mortality and elevated endogenous oxidative stress. Proc. Natl. Acad. Sci. U.S.A. 99, 16162–16167. 10.1073/pnas.252342899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klemz R., Reischl S., Wallach T., Witte N., Jürchott K., Klemz S., et al. (2017). Reciprocal regulation of carbon monoxide metabolism and the circadian clock. Nat. Struct. Mol. Biol. 24, 15–22. 10.1038/nsmb.3331 [DOI] [PubMed] [Google Scholar]
- Klichko V., Sohal B. H., Radyuk S. N., Orr W. C., Sohal R. S. (2014). Decrease in cytochrome c oxidase reserve capacity diminishes robustness of Drosophila melanogaster and shortens lifespan. Biochem. J. 459, 127–135. 10.1042/BJ20131370 [DOI] [PubMed] [Google Scholar]
- Kohler S. A., Henderson B. R., Kühn L. C. (1995). Succinate dehydrogenase b mRNA of Drosophila melanogaster has a functional iron-responsive element in its 5'-untranslated region. J. Biol. Chem. 270, 30781–30786. 10.1074/jbc.270.51.30781 [DOI] [PubMed] [Google Scholar]
- Kondapalli K. C., Kok N. M., Dancis A., Stemmler T. L. (2008). Drosophila frataxin: an iron chaperone during cellular Fe-S cluster bioassembly. Biochemistry 47, 6917–6927. 10.1021/bi800366d [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konopka R. J., Benzer S. (1971). Clock mutants of Drosophila melanogaster. Proc. Natl. Acad. Sci. U.S.A. 68, 2112–2116. 10.1073/pnas.68.9.2112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosman D. J. (2010). Redox cycling in iron uptake, efflux, and trafficking. J. Biol. Chem. 285, 26729–26735. 10.1074/jbc.R110.113217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosmidis S., Botella J. A., Mandilaras K., Schneuwly S., Skoulakis E. M., Rouault T. A., et al. (2011). Ferritin overexpression in Drosophila glia leads to iron deposition in the optic lobes and late-onset behavioral defects. Neurobiol. Dis. 43, 213–219. 10.1016/j.nbd.2011.03.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosmidis S., Missirlis F., Botella J. A., Schneuwly S., Rouault T. A., Skoulakis E. M. (2014). Behavioral decline and premature lethality upon pan-neuronal ferritin overexpression in Drosophila infected with a virulent form of Wolbachia. Front. Pharmacol. 5:66. 10.3389/fphar.2014.00066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuper J., Llamas A., Hecht H. J., Mendel R. R., Schwarz G. (2004). Structure of the molybdopterin-bound Cnx1G domain links molybdenum and copper metabolism. Nature 430, 803–806. 10.1038/nature02681 [DOI] [PubMed] [Google Scholar]
- Kuper J., Palmer T., Mendel R. R., Schwarz G. (2000). Mutations in the molybdenum cofactor biosynthetic protein Cnx1G from Arabidopsis thaliana define functions for molybdopterin binding, molybdenum insertion, and molybdenum cofactor stabilization. Proc. Natl. Acad. Sci. U.S.A. 97, 6475–6480. 10.1073/pnas.110568497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzin B., Roberts I., Peunova N., Enikolopov G. (1996). Nitric oxide regulates cell proliferation during Drosophila development. Cell 87, 639–649. 10.1016/S0092-8674(00)81384-7 [DOI] [PubMed] [Google Scholar]
- LaJeunesse D. R., Buckner S. M., Lake J., Na C., Pirt A., Fromson K. (2004). Three new Drosophila markers of intracellular membranes. Biotechniques 36, 784–788, 790. [DOI] [PubMed] [Google Scholar]
- Land T., Rouault T. A. (1998). Targeting of a human iron-sulfur cluster assembly enzyme, nifs, to different subcellular compartments is regulated through alternative AUG utilization. Mol. Cell 2, 807–815. 10.1016/S1097-2765(00)80295-6 [DOI] [PubMed] [Google Scholar]
- Lane N., Martin W. (2010). The energetics of genome complexity. Nature 467, 929–934. 10.1038/nature09486 [DOI] [PubMed] [Google Scholar]
- Lang M., Braun C. L., Kanost M. R., Gorman M. J. (2012). Multicopper oxidase-1 is a ferroxidase essential for iron homeostasis in Drosophila melanogaster. Proc. Natl. Acad. Sci. U.S.A. 109, 13337–13342. 10.1073/pnas.1208703109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauhon C. T., Skovran E., Urbina H. D., Downs D. M., Vickery L. E. (2004). Substitutions in an active site loop of Escherichia coli IscS result in specific defects in Fe-S cluster and thionucleoside biosynthesis in vivo. J. Biol. Chem. 279, 19551–19558. 10.1074/jbc.M401261200 [DOI] [PubMed] [Google Scholar]
- Lawrence P. A. (2016). The last 50 years: mismeasurement and mismanagement are impeding scientific research. Curr. Top. Dev. Biol. 116, 617–631. 10.1016/bs.ctdb.2015.12.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann F. O., Schützner P. (2010). The respiratory basis of locomotion in Drosophila. J. Insect Physiol. 56, 543–550. 10.1016/j.jinsphys.2009.04.019 [DOI] [PubMed] [Google Scholar]
- Leimkühler S. (2017). Shared function and moonlighting proteins in molybdenum cofactor biosynthesis. Biol. Chem. 398, 1009–1026. 10.1515/hsz-2017-0110 [DOI] [PubMed] [Google Scholar]
- Leimkuhler S., Freuer A., Araujo J. A., Rajagopalan K. V., Mendel R. R. (2003). Mechanistic studies of human molybdopterin synthase reaction and characterization of mutants identified in group B patients of molybdenum cofactor deficiency. J. Biol. Chem. 278, 26127–26134. 10.1074/jbc.M303092200 [DOI] [PubMed] [Google Scholar]
- Leitão-Gonçalves R., Carvalho-Santos Z., Francisco A. P., Fioreze G. T., Anjos M., Baltazar C., et al. (2017). Commensal bacteria and essential amino acids control food choice behavior and reproduction. PLoS Biol. 15:e2000862. 10.1371/journal.pbio.2000862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemaitre B., Miguel-Aliaga I. (2013). The digestive tract of Drosophila melanogaster. Annu. Rev. Genet. 47, 377–404. 10.1146/annurev-genet-111212-133343 [DOI] [PubMed] [Google Scholar]
- Leulier F., Royet J. (2009). Maintaining immune homeostasis in fly gut. Nat. Immunol. 10, 936–938. 10.1038/ni0909-936 [DOI] [PubMed] [Google Scholar]
- Levenbook L., Williams C. M. (1956). Mitochondria in the flight muscles of insects. III. Mitochondrial cytochrome c in relation to the aging and wing beat frequency of flies. J. Gen. Physiol. 39, 497–512. 10.1085/jgp.39.4.497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis E. B. (1978). A gene complex controlling segmentation in Drosophila. Nature 276, 565–570. 10.1038/276565a0 [DOI] [PubMed] [Google Scholar]
- Li H., Mapolelo D. T., Dingra N. N., Naik S. G., Lees N. S., Hoffman B. M., et al. (2009). The yeast iron regulatory proteins Grx3/4 and Fra2 form heterodimeric complexes containing a [2Fe-2S] cluster with cysteinyl and histidyl ligation. Biochemistry 48, 9569–9581. 10.1021/bi901182w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Urwyler O., Suter B. (2010). Drosophila Xpd regulates Cdk7 localization, mitotic kinase activity, spindle dynamics, and chromosome segregation. PLoS Genet. 6:e1000876. 10.1371/journal.pgen.1000876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Huang T. T., Carlson E. J., Melov S., Ursell P. C., Olson J. L., et al. (1995). Dilated cardiomyopathy and neonatal lethality in mutant mice lacking manganese superoxide dismutase. Nat. Genet. 11, 376–381. 10.1038/ng1295-376 [DOI] [PubMed] [Google Scholar]
- Lin G., Xu N., Xi R. (2008). Paracrine wingless signalling controls self-renewal of Drosophila intestinal stem cells. Nature 455, 1119–1123. 10.1038/nature07329 [DOI] [PubMed] [Google Scholar]
- Lind M. I., Missirlis F., Melefors O., Uhrigshardt H., Kirby K., Phillips J. P., et al. (2006). Of two cytosolic aconitases expressed in Drosophila, only one functions as an iron-regulatory protein. J. Biol. Chem. 281, 18707–18714. 10.1074/jbc.M603354200 [DOI] [PubMed] [Google Scholar]
- Lints C. V., Lints F. A., Zeuthen E. (1967). Respiration in Drosophila. I. Oxygen consumption during development of the egg in genotypes of Drosophila melanogaster with contributions to the gradient diver technique. C. R. Trav. Lab. Carlsberg 36, 35–66. [PubMed] [Google Scholar]
- Lints F. A., Lints C. V. (1968). Respiration in Drosophila. II. Respiration in relation to age by wild, inbred and hybrid Drosophila melanogaster imagos. Exp. Gerontol. 3, 341–349. 10.1016/0531-5565(68)90047-8 [DOI] [PubMed] [Google Scholar]
- Liu W., Acín-Peréz R., Geghman K. D., Manfredi G., Lu B., Li C. (2011). Pink1 regulates the oxidative phosphorylation machinery via mitochondrial fission. Proc. Natl. Acad. Sci. U.S.A. 108, 12920–12924. 10.1073/pnas.1107332108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llamas A., Chamizo-Ampudia A., Tejada-Jimenez M., Galvan A., Fernandez E. (2017). The molybdenum cofactor enzyme mARC: Moonlighting or promiscuous enzyme? Biofactors 43, 486–494. 10.1002/biof.1362 [DOI] [PubMed] [Google Scholar]
- Llamas A., Mendel R. R., Schwarz G. (2004). Synthesis of adenylated molybdopterin: an essential step for molybdenum insertion. J. Biol. Chem. 279, 55241–55246. 10.1074/jbc.M409862200 [DOI] [PubMed] [Google Scholar]
- Llamas A., Otte T., Multhaup G., Mendel R. R., Schwarz G. (2006). The Mechanism of nucleotide-assisted molybdenum insertion into molybdopterin. a novel route toward metal cofactor assembly. J. Biol. Chem. 281, 18343–18350. 10.1074/jbc.M601415200 [DOI] [PubMed] [Google Scholar]
- Llorens J. V., Metzendorf C., Missirlis F., Lind M. I. (2015). Mitochondrial iron supply is required for the developmental pulse of ecdysone biosynthesis that initiates metamorphosis in Drosophila melanogaster. J. Biol. Inorg. Chem. 20, 1229–1238. 10.1007/s00775-015-1302-2 [DOI] [PubMed] [Google Scholar]
- Llorens J. V., Navarro J. A., Martínez-Sebastián M. J., Baylies M. K., Schneuwly S., Botella J. A., et al. (2007). Causative role of oxidative stress in a Drosophila model of Friedreich ataxia. FASEB J. 21, 333–344. 10.1096/fj.05-5709com [DOI] [PubMed] [Google Scholar]
- Long X., Ye J., Zhao D., Zhang S. J. (2015). Magnetogenetics: remote non-invasive magnetic activation of neuronal activity with a magnetoreceptor. Sci. Bull. 60, 2107–2119. 10.1007/s11434-015-0902-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorincz P., Lakatos Z., Varga A., Maruzs T., Simon-Vecsei Z., Darula Z., et al. (2016). MiniCORVET is a Vps8-containing early endosomal tether in Drosophila. eLife 5:e14226. 10.7554/eLife.14226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H., Xu H., O'Farrell P. H. (2014). Transmission of mitochondrial mutations and action of purifying selection in Drosophila melanogaster. Nat. Genet. 46, 393–397. 10.1038/ng.2919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahul-Mellier A. L., Huc L., Lemarié A., Irshad S., Hwang M., Datler C., et al. (2015). Stefan Grimm, 1963-2014, a tragic loss for the scientific community. Cell Death Differ. 22, 247. 10.1038/cdd.2014.196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandilaras K. (2012). A Screen for Iron Metabolism Genes that Influence Circadian Rhythms Implicates Ferritin 2 Light Chain Homologue in the Drosophila clock. School of Biological and Chemical Sciences; Queen Mary University of London.
- Mandilaras K., Missirlis F. (2012). Genes for iron metabolism influence circadian rhythms in Drosophila melanogaster. Metallomics 4, 928–936. 10.1039/c2mt20065a [DOI] [PubMed] [Google Scholar]
- Mandilaras K., Pathmanathan T., Missirlis F. (2013). Iron absorption in Drosophila melanogaster. Nutrients 5, 1622–1647. 10.3390/nu5051622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marelja Z. (2013). Detection of the L-cysteine Desulfurase NFS1 in the Cytosol and Analysis of Its Role in Molybdenum Cofactor Biosynthesis in Eukaryotes. Potsdam: Institut für Biochemie und Biologie, University of Potzdam. [Google Scholar]
- Marelja Z., Dambowsky M., Bolis M., Georgiou M. L., Garattini E., Missirlis F., et al. (2014). The four aldehyde oxidases of Drosophila melanogaster have different gene expression patterns and enzyme substrate specificities. J. Exp. Biol. 217, 2201–2211. 10.1242/jeb.102129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marelja Z., Mullick Chowdhury M., Dosche C., Hille C., Baumann O., Löhmannsröben H. G., et al. (2013). The L-cysteine desulfurase NFS1 is localized in the cytosol where it provides the sulfur for molybdenum cofactor biosynthesis in humans. PLoS ONE 8:e60869. 10.1371/journal.pone.0060869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marelja Z., Stöcklein W., Nimtz M., Leimkühler S. (2008). A novel role for human Nfs1 in the cytoplasm: Nfs1 acts as a sulfur donor for MOCS3, a protein involved in molybdenum cofactor biosynthesis. J. Biol. Chem. 283, 25178–25185. 10.1074/jbc.M804064200 [DOI] [PubMed] [Google Scholar]
- Markopoulou K., Artavanis-Tsakonas S. (1989). The expression of the neurogenic locus Notch during the postembryonic development of Drosophila melanogaster and its relationship to mitotic activity. J. Neurogenet. 6, 11–26. 10.3109/01677068909107097 [DOI] [PubMed] [Google Scholar]
- Marsh J. L., Wieschaus E. (1977). Germ-line dependence of the maroon-like maternal effect in Drosophila. Dev. Biol. 60, 396–403. 10.1016/0012-1606(77)90137-3 [DOI] [PubMed] [Google Scholar]
- Martin I., Jones M. A., Rhodenizer D., Zheng J., Warrick J. M., Seroude L., et al. (2009). Sod2 knockdown in the musculature has whole-organism consequences in Drosophila. Free Radic. Biol. Med. 47, 803–813. 10.1016/j.freeradbiomed.2009.06.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Morentin L., Martínez L., Piloto S., Yang H., Schon E. A., Garesse R., et al. (2015). Cardiac deficiency of single cytochrome oxidase assembly factor scox induces p53-dependent apoptosis in a Drosophila cardiomyopathy model. Hum. Mol. Genet. 24, 3608–3622. 10.1093/hmg/ddv106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Pastor M. T., Perea-García A., Puig S. (2017). Mechanisms of iron sensing and regulation in the yeast Saccharomyces cerevisiae. World J. Microbiol. Biotechnol. 33, 75. 10.1007/s11274-017-2215-8 [DOI] [PubMed] [Google Scholar]
- Marzuk O., Peretz G., Bakhrat A., Abdu U. (2013). Drosophila CIAPIN1 homologue is required for follicle cell proliferation and survival. Dev. Dyn. 242, 731–737. 10.1002/dvdy.23951 [DOI] [PubMed] [Google Scholar]
- Matthies A., Nimtz M., Leimkühler S. (2005). Molybdenum cofactor biosynthesis in humans: identification of a persulfide group in the rhodanese-like domain of MOCS3 by mass spectrometry. Biochemistry 44, 7912–7920. 10.1021/bi0503448 [DOI] [PubMed] [Google Scholar]
- Matthies A., Rajagopalan K. V., Mendel R. R., Leimkühler S. (2004). Evidence for the physiological role of a rhodanese-like protein for the biosynthesis of the molybdenum cofactor in humans. Proc. Natl. Acad. Sci. U.S.A. 101, 5946–5951. 10.1073/pnas.0308191101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattila J., Hietakangas V. (2017). Regulation of carbohydrate energy metabolism in Drosophila melanogaster. Genetics 207, 1231–1253. 10.1534/genetics.117.199885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr J. A., Feichtinger R. G., Tort F., Ribes A., Sperl W. (2014). Lipoic acid biosynthesis defects. J. Inherit. Metab. Dis. 37, 553–563. 10.1007/s10545-014-9705-8 [DOI] [PubMed] [Google Scholar]
- Mehta A., Deshpande A., Bettedi L., Missirlis F. (2009). Ferritin accumulation under iron scarcity in Drosophila iron cells. Biochimie 91, 1331–1334. 10.1016/j.biochi.2009.05.003 [DOI] [PubMed] [Google Scholar]
- Meister M. (2016). Physical limits to magnetogenetics. eLife 5:e17210 10.7554/eLife.17210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melber A., Na U., Vashisht A., Weiler B. D., Lill R., Wohlschlegel J. A., et al. (2016). Role of Nfu1 and Bol3 in iron-sulfur cluster transfer to mitochondrial clients. Elif e 5:e15991. 10.7554/eLife.15991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellor J. (2016). The molecular basis of metabolic cycles and their relationship to circadian rhythms. Nat. Struct. Mol. Biol. 23, 1035–1044. 10.1038/nsmb.3311 [DOI] [PubMed] [Google Scholar]
- Mendel R. R., Leimkühler S. (2015). The biosynthesis of the molybdenum cofactors. J. Biol. Inorg. Chem. 20, 337–347. 10.1007/s00775-014-1173-y [DOI] [PubMed] [Google Scholar]
- Mercer S. W., Burke R. (2016). Evidence for a role for the putative Drosophila hGRX1 orthologue in copper homeostasis. Biometals 29, 705–713. 10.1007/s10534-016-9946-0 [DOI] [PubMed] [Google Scholar]
- Merkey A. B., Wong C. K., Hoshizaki D. K., Gibbs A. G. (2011). Energetics of metamorphosis in Drosophila melanogaster. J. Insect Physiol. 57, 1437–1445. 10.1016/j.jinsphys.2011.07.013 [DOI] [PubMed] [Google Scholar]
- Merzetti E., Hackett J. M., Clark D. V. (2013). Transcriptional regulation of the purine de novo synthesis gene Prat in Drosophila melanogaster. Gene 518, 280–286. 10.1016/j.gene.2013.01.024 [DOI] [PubMed] [Google Scholar]
- Metzendorf C., Lind M. I. (2010). Drosophila mitoferrin is essential for male fertility: evidence for a role of mitochondrial iron metabolism during spermatogenesis. BMC Dev. Biol. 10:68. 10.1186/1471-213X-10-68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miquel J. (1998). An update on the oxygen stress-mitochondrial mutation theory of aging: genetic and evolutionary implications. Exp. Gerontol. 33, 113–126. 10.1016/S0531-5565(97)00060-0 [DOI] [PubMed] [Google Scholar]
- Miśkiewicz K., Jose L. E., Bento-Abreu A., Fislage M., Taes I., Kasprowicz J., et al. (2011). ELP3 controls active zone morphology by acetylating the ELKS family member Bruchpilot. Neuron 72, 776–788. 10.1016/j.neuron.2011.10.010 [DOI] [PubMed] [Google Scholar]
- Missirlis F. (2003). Understanding the Aging fly Through Physiological Genetics. Advances in Cell Aging and Gerontology. Amsterdam: Elsevier B.V. [Google Scholar]
- Missirlis F., Holmberg S., Georgieva T., Dunkov B. C., Rouault T. A., Law J. H. (2006). Characterization of mitochondrial ferritin in Drosophila. Proc. Natl. Acad. Sci. U.S.A. 103, 5893–5898. 10.1073/pnas.0601471103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Missirlis F., Hu J., Kirby K., Hilliker A. J., Rouault T. A., Phillips J. P. (2003a). Compartment-specific protection of iron-sulfur proteins by superoxide dismutase. J. Biol. Chem. 278, 47365–47369. 10.1074/jbc.M307700200 [DOI] [PubMed] [Google Scholar]
- Missirlis F., Kosmidis S., Brody T., Mavrakis M., Holmberg S., Odenwald W. F., et al. (2007). Homeostatic mechanisms for iron storage revealed by genetic manipulations and live imaging of Drosophila ferritin. Genetics 177, 89–100. 10.1534/genetics.107.075150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Missirlis F., Phillips J. P., Jäckle H. (2001). Cooperative action of antioxidant defense systems in Drosophila. Curr. Biol. 11, 1272–1277. 10.1016/S0960-9822(01)00393-1 [DOI] [PubMed] [Google Scholar]
- Missirlis F., Phillips J. P., Jackle H., Rouault T. A. (2003b). Drosophila and antioxidant therapy design, in Proceedings of the Meeting of the Society for Free Radical Research, European Section. (Ioannina: ). [Google Scholar]
- Missirlis F., Ulschmid J. K., Hirosawa-Takamori M., Grönke S., Schäfer U., Becker K., et al. (2002). Mitochondrial and cytoplasmic thioredoxin reductase variants encoded by a single Drosophila gene are both essential for viability. J. Biol. Chem. 277, 11521–11526. 10.1074/jbc.M111692200 [DOI] [PubMed] [Google Scholar]
- Mistry R., Kounatidis I., Ligoxygakis P. (2017). Interaction between familial transmission and a constitutively active immune system shapes gut microbiota in Drosophila melanogaster. Genetics 206, 889–904. 10.1534/genetics.116.190215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell H. K., Glassman E. (1959). Hypoxanthine in rosy and maroon-like mutants of Drosophila melanogaster. Science 129:268. 10.1126/science.129.3344.268 [DOI] [PubMed] [Google Scholar]
- Mockett R. J., Orr W. C., Rahmandar J. J., Benes J. J., Radyuk S. N., Klichko V. I., et al. (1999). Overexpression of Mn-containing superoxide dismutase in transgenic Drosophila melanogaster. Arch. Biochem. Biophys. 371, 260–269. 10.1006/abbi.1999.1460 [DOI] [PubMed] [Google Scholar]
- Mohr S. E., Hu Y., Kim K., Housden B. E., Perrimon N. (2014). Resources for functional genomics studies in Drosophila melanogaster. Genetics 197, 1–18. 10.1534/genetics.113.154344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan T. H. (1910). Sex limited inheritance in Drosophila. Science 32, 120–122. 10.1126/science.32.812.120 [DOI] [PubMed] [Google Scholar]
- Morgan T. H. (1911). The origin of five mutations in eye color in Drosophila and their modes of inheritance. Science 33, 534–537. 10.1126/science.33.849.534-a [DOI] [PubMed] [Google Scholar]
- Morita T. (1958). Purine catabolism in Drosophila melanogaster. Science 128:1135. 10.1126/science.128.3332.1135 [DOI] [PubMed] [Google Scholar]
- Mortenson L. E. (1964). Ferredoxin and atp, requirements for nitrogen fixation in cell-free extracts of clostridium pasteurianum. Proc. Natl. Acad. Sci. U.S.A. 52, 272–279. 10.1073/pnas.52.2.272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muckenthaler M., Gunkel N., Frishman D., Cyrklaff A., Tomancak P., Hentze M. W. (1998). Iron-regulatory protein-1 (IRP-1) is highly conserved in two invertebrate species–characterization of IRP-1 homologues in Drosophila melanogaster and Caenorhabditis elegans. Eur. J. Biochem. 254, 230–237. 10.1046/j.1432-1327.1998.2540230.x [DOI] [PubMed] [Google Scholar]
- Muckenthaler M. U., Rivella S., Hentze M. W., Galy B. (2017). A red carpet for iron metabolism. Cell 168, 344–361. 10.1016/j.cell.2016.12.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mühlenhoff U., Balk J., Richhardt N., Kaiser J. T., Sipos K., Kispal G., et al. (2004). Functional characterization of the eukaryotic cysteine desulfurase Nfs1p from Saccharomyces cerevisiae. J. Biol. Chem. 279, 36906–36915. 10.1074/jbc.M406516200 [DOI] [PubMed] [Google Scholar]
- Mühlenhoff U., Molik S., Godoy J. R., Uzarska M. A., Richter N., Seubert A., et al. (2010). Cytosolic monothiol glutaredoxins function in intracellular iron sensing and trafficking via their bound iron-sulfur cluster. Cell Metab. 12, 373–385. 10.1016/j.cmet.2010.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhlenhoff U., Richter N., Pines O., Pierik A. J., Lill R. (2011). Specialized function of yeast Isa1 and Isa2 proteins in the maturation of mitochondrial [4Fe-4S] proteins. J. Biol. Chem. 286, 41205–41216. 10.1074/jbc.M111.296152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller F. L., Lustgarten M. S., Jang Y., Richardson A., Van Remmen H. (2007). Trends in oxidative aging theories. Free Radic. Biol. Med. 43, 477–503. 10.1016/j.freeradbiomed.2007.03.034 [DOI] [PubMed] [Google Scholar]
- Munro D., Treberg J. R. (2017). A radical shift in perspective: mitochondria as regulators of reactive oxygen species. J. Exp. Biol. 220, 1170–1180. 10.1242/jeb.132142 [DOI] [PubMed] [Google Scholar]
- Na U., Yu W., Cox J., Bricker D. K., Brockmann K., Rutter J., et al. (2014). The LYR factors SDHAF1 and SDHAF3 mediate maturation of the iron-sulfur subunit of succinate dehydrogenase. Cell Metab. 20, 253–266. 10.1016/j.cmet.2014.05.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nag R. C., Niggli S., Sousa-Guimarães S., Vazquez-Pianzola P., Suter B. (2018). Mms19 is a mitotic gene that permits Cdk7 to be fully active as a Cdk-activating kinase. Development 145:dev156802. 10.1242/dev.156802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagano S., Scheerer P., Zubow K., Michael N., Inomata K., Lamparter T., et al. (2016). The crystal structures of the N-terminal photosensory core module of agrobacterium phytochrome Agp1 as parallel and anti-parallel dimers. J. Biol. Chem. 291, 20674–20691. 10.1074/jbc.M116.739136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naranuntarat A., Jensen L. T., Pazicni S., Penner-Hahn J. E., Culotta V. C. (2009). The interaction of mitochondrial iron with manganese superoxide dismutase. J. Biol. Chem. 284, 22633–22640. 10.1074/jbc.M109.026773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasta V., Giachetti A., Ciofi-Baffoni S., Banci L. (2017). Structural insights into the molecular function of human [2Fe-2S] BOLA1-GRX5 and [2Fe-2S] BOLA3-GRX5 complexes. Biochim. Biophys. Acta 1861, 2119–2131. 10.1016/j.bbagen.2017.05.005 [DOI] [PubMed] [Google Scholar]
- Navarro J. A., Botella J. A., Metzendorf C., Lind M. I., Schneuwly S. (2015). Mitoferrin modulates iron toxicity in a Drosophila model of Friedreich's ataxia. Free Radic. Biol. Med. 85, 71–82. 10.1016/j.freeradbiomed.2015.03.014 [DOI] [PubMed] [Google Scholar]
- Navarro J. A., Llorens J. V., Soriano S., Botella J. A., Schneuwly S., Martínez-Sebastián M. J., et al. (2011). Overexpression of human and fly frataxins in Drosophila provokes deleterious effects at biochemical, physiological and developmental levels. PLoS ONE 6:e21017. 10.1371/journal.pone.0021017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro J. A., Ohmann E., Sanchez D., Botella J. A., Liebisch G., Moltó M. D., et al. (2010). Altered lipid metabolism in a Drosophila model of Friedreich's ataxia. Hum. Mol. Genet. 19, 2828–2840. 10.1093/hmg/ddq183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro J. A., Schneuwly S. (2017). Copper and zinc homeostasis: lessons from Drosophila melanogaster. Front. Genet. 8:223. 10.3389/fgene.2017.00223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson C. R., Szauter P. (1992). Cytogenetic analysis of chromosome region 89A of Drosophila melanogaster: isolation of deficiencies and mapping of Po, Aldox-1 and transposon insertions. Mol. Gen. Genet. 235, 11–21. 10.1007/BF00286176 [DOI] [PubMed] [Google Scholar]
- Netz D. J., Stümpfig M., Doré C., Mühlenhoff U., Pierik A. J., Lill R. (2010). Tah18 transfers electrons to Dre2 in cytosolic iron-sulfur protein biogenesis. Nat. Chem. Biol. 6, 758–765. 10.1038/nchembio.432 [DOI] [PubMed] [Google Scholar]
- Nitabach M. N., Taghert P. H. (2008). Organization of the Drosophila circadian control circuit. Curr. Biol. 18, R84–R93. 10.1016/j.cub.2007.11.061 [DOI] [PubMed] [Google Scholar]
- Nitschke W., Russell M. J. (2009). Hydrothermal focusing of chemical and chemiosmotic energy, supported by delivery of catalytic Fe, Ni, Mo/W, Co, S and Se, forced life to emerge. J. Mol. Evol. 69, 481–496. 10.1007/s00239-009-9289-3 [DOI] [PubMed] [Google Scholar]
- Nolte D. J. (1961). Pigment granules in compound eyes of Drosophila. Heredity 16, 25–38. 10.1038/hdy.1961.2 [DOI] [Google Scholar]
- Nordman J., Li S., Eng T., Macalpine D., Orr-Weaver T. L. (2011). Developmental control of the DNA replication and transcription programs. Genome Res. 21, 175–181. 10.1101/gr.114611.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochiai E. I. (1978). Principles in the selection of inorganic elements by organisms–application to molybdenum enzymes. Biosystems 10, 329–337. 10.1016/0303-2647(78)90016-3 [DOI] [PubMed] [Google Scholar]
- Ohlstein B., Spradling A. (2006). The adult Drosophila posterior midgut is maintained by pluripotent stem cells. Nature 439, 470–474. 10.1038/nature04333 [DOI] [PubMed] [Google Scholar]
- Ohlstein B., Spradling A. (2007). Multipotent Drosophila intestinal stem cells specify daughter cell fates by differential notch signaling. Science 315, 988–992. 10.1126/science.1136606 [DOI] [PubMed] [Google Scholar]
- Oka S., Hirai J., Yasukawa T., Nakahara Y., Inoue Y. H. (2015). A correlation of reactive oxygen species accumulation by depletion of superoxide dismutases with age-dependent impairment in the nervous system and muscles of Drosophila adults. Biogerontology 16, 485–501. 10.1007/s10522-015-9570-3 [DOI] [PubMed] [Google Scholar]
- Okazaki F., Matsunaga N., Okazaki H., Azuma H., Hamamura K., Tsuruta A., et al. (2016). Circadian clock in a mouse colon tumor regulates intracellular iron levels to promote tumor progression. J. Biol. Chem. 291, 7017–7028. 10.1074/jbc.M115.713412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osaki S., Johnson D. A., Frieden E. (1966). The possible significance of the ferrous oxidase activity of ceruloplasmin in normal human serum. J. Biol. Chem. 241, 2746–2751. [PubMed] [Google Scholar]
- Ozer H. K., Dlouhy A. C., Thornton J. D., Hu J., Liu Y., Barycki J. J., et al. (2015). Cytosolic Fe-S cluster protein maturation and iron regulation are independent of the mitochondrial Erv1/Mia40 import system. J. Biol. Chem. 290, 27829–27840. 10.1074/jbc.M115.682179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagliarini D. J., Rutter J. (2013). Hallmarks of a new era in mitochondrial biochemistry. Genes Dev. 27, 2615–2627. 10.1101/gad.229724.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palandri A., L'hôte D., Cohen-Tannoudji J., Tricoire H., Monnier V. (2015). Frataxin inactivation leads to steroid deficiency in flies and human ovarian cells. Hum. Mol. Genet. 24, 2615–2626. 10.1093/hmg/ddv024 [DOI] [PubMed] [Google Scholar]
- Pang K., You H., Chen Y., Chu P., Hu M., Shen J., et al. (2017). MagR alone is insufficient to confer cellular calcium responses to magnetic stimulation. Front. Neural Circuits 11:11. 10.3389/fncir.2017.00011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papanikolaou G., Pantopoulos K. (2017). Systemic iron homeostasis and erythropoiesis. IUBMB Life 69, 399–413. 10.1002/iub.1629 [DOI] [PubMed] [Google Scholar]
- Papatriantafyllou M. (2012). DNA Metabolism: MMS19: CIA agent for DNA-linked affairs. Nat. Rev. Mol. Cell Biol. 13, 538. 10.1038/nrm3411 [DOI] [PubMed] [Google Scholar]
- Parent A., Elduque X., Cornu D., Belot L., Le Caer J. P., Grandas A., et al. (2015). Mammalian frataxin directly enhances sulfur transfer of NFS1 persulfide to both ISCU and free thiols. Nat. Commun. 6, 5686. 10.1038/ncomms6686 [DOI] [PubMed] [Google Scholar]
- Paul V. D., Lill R. (2015). Biogenesis of cytosolic and nuclear iron-sulfur proteins and their role in genome stability. Biochim. Biophys. Acta 1853, 1528–1539. 10.1016/j.bbamcr.2014.12.018 [DOI] [PubMed] [Google Scholar]
- Peck V. M., Gerner E. W., Cress A. E. (1992). Delta-type DNA polymerase characterized from Drosophila melanogaster embryos. Nucleic Acids Res. 20, 5779–5784. 10.1093/nar/20.21.5779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Z., Dittmer N. T., Lang M., Brummett L. M., Braun C. L., Davis L. C., et al. (2015). Multicopper oxidase-1 orthologs from diverse insect species have ascorbate oxidase activity. Insect Biochem. Mol. Biol. 59, 58–71. 10.1016/j.ibmb.2015.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkhulyn N. V., Rovenko B. M., Lushchak O. V., Storey J. M., Storey K. B., Lushchak V. I. (2017). Exposure to sodium molybdate results in mild oxidative stress in Drosophila melanogaster. Redox Rep. 22, 137–146. 10.1080/13510002.2017.1295898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfluger P. T., Kabra D. G., Aichler M., Schriever S. C., Pfuhlmann K., García V. C., et al. (2015). Calcineurin links mitochondrial elongation with energy metabolism. Cell Metab. 22, 838–850. 10.1016/j.cmet.2015.08.022 [DOI] [PubMed] [Google Scholar]
- Phillips J. P., Forrest H. S. (1980). Ommochromes and pteridines, in The Genetics and Biology of Drosophila, eds Ashburner M., Wright T. R. F. (London: Academic Press; ), 542–623. [Google Scholar]
- Poetini M. R., Araujo S. M., Trindade de Paula M., Bortolotto V. C., Meichtry L. B., Polet de Almeida F., et al. (2018). Hesperidin attenuates iron-induced oxidative damage and dopamine depletion in Drosophila melanogaster model of Parkinson's disease. Chem. Biol. Interact. 279, 177–186. 10.1016/j.cbi.2017.11.018 [DOI] [PubMed] [Google Scholar]
- Politi Y., Gal L., Kalifa Y., Ravid L., Elazar Z., Arama E. (2014). Paternal mitochondrial destruction after fertilization is mediated by a common endocytic and autophagic pathway in Drosophila. Dev. Cell 29, 305–320. 10.1016/j.devcel.2014.04.005 [DOI] [PubMed] [Google Scholar]
- Pomatto L. C. D., Carney C., Shen B., Wong S., Halaszynski K., Salomon M. P., et al. (2017). The mitochondrial lon protease is required for age-specific and sex-specific adaptation to oxidative stress. Curr. Biol. 27, 1–15. 10.1016/j.cub.2016.10.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponka P., Sheftel A. D., English A. M., Scott Bohle D., Garcia-Santos D. (2017). Do mammalian cells really need to export and import heme? Trends Biochem. Sci. 42, 395–406. 10.1016/j.tibs.2017.01.006 [DOI] [PubMed] [Google Scholar]
- Poulson D. F., Bowen V. T. (1952). Organization and function of the inorganic constituents of nuclei. Exp. Cell Res. 2(Suppl), 161–180. [Google Scholar]
- Pulipparacharuvil S., Akbar M. A., Ray S., Sevrioukov E. A., Haberman A. S., Rohrer J., et al. (2005). Drosophila Vps16A is required for trafficking to lysosomes and biogenesis of pigment granules. J. Cell Sci. 118, 3663–3673. 10.1242/jcs.02502 [DOI] [PubMed] [Google Scholar]
- Puri M., Goyal A., Senutovich N., Dowd S. R., Minden J. S. (2008). Building proteomic pathways using Drosophila ventral furrow formation as a model. Mol. Biosyst. 4, 1126–1135. 10.1039/b812153b [DOI] [PubMed] [Google Scholar]
- Qi W., Li J., Chain C. Y., Pasquevich G. A., Pasquevich A. F., Cowan J. A. (2013). Glutathione-complexed iron-sulfur clusters. Reaction intermediates and evidence for a template effect promoting assembly and stability. Chem. Commun. 49, 6313–6315. 10.1039/c3cc43620a [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin S., Yin H., Yang C., Dou Y., Liu Z., Zhang P., et al. (2016). A magnetic protein biocompass. Nat. Mater. 15, 217–226. 10.1038/nmat4484 [DOI] [PubMed] [Google Scholar]
- Radford S. J., Goley E., Baxter K., McMahan S., Sekelsky J. (2005). Drosophila ERCC1 is required for a subset of MEI-9-dependent meiotic crossovers. Genetics 170, 1737–1745. 10.1534/genetics.104.036178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajagopalan K. V. (1997). Biosynthesis and processing of the molybdenum cofactors. Biochem. Soc. Trans. 25, 757–761. 10.1042/bst0250757 [DOI] [PubMed] [Google Scholar]
- Rajagopalan K. V., Johnson J. L., Hainline B. E. (1982). The pterin of the molybdenum cofactor. Fed. Proc. 41, 2608–2612. [PubMed] [Google Scholar]
- Rawls J. M., Jr. (2006). Analysis of pyrimidine catabolism in Drosophila melanogaster using epistatic interactions with mutations of pyrimidine biosynthesis and beta-alanine metabolism. Genetics 172, 1665–1674. 10.1534/genetics.105.052753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reaume A. G., Clark S. H., Chovnick A. (1989). Xanthine dehydrogenase is transported to the Drosophila eye. Genetics 123, 503–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reaume A. G., Knecht D. A., Chovnick A. (1991). The rosy locus in Drosophila melanogaster: xanthine dehydrogenase and eye pigments. Genetics 129, 1099–1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiff T., Jacobson J., Cognigni P., Antonello Z., Ballesta E., Tan K. J., et al. (2015). Endocrine remodelling of the adult intestine sustains reproduction in Drosophila. elife 4:e06930. 10.7554/eLife.06930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resnik-Docampo M., Koehler C. L., Clark R. I., Schinaman J. M., Sauer V., Wong D. M., et al. (2017). Tricellular junctions regulate intestinal stem cell behaviour to maintain homeostasis. Nat. Cell Biol. 19, 52–59. 10.1038/ncb3454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rey G., Valekunja U. K., Feeney K. A., Wulund L., Milev N. B., Stangherlin A., et al. (2016). The pentose phosphate pathway regulates the circadian clock. Cell Metab. 24, 462–473. 10.1016/j.cmet.2016.07.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynaud E., Lomelí H., Vázquez M., Zurita M. (1999). The Drosophila melanogaster homologue of the Xeroderma pigmentosum D gene product is located in euchromatic regions and has a dynamic response to UV light-induced lesions in polytene chromosomes. Mol. Biol. Cell 10, 1191–1203. 10.1091/mbc.10.4.1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards T. A., van der Giezen M. (2006). Evolution of the Isd11-IscS complex reveals a single alpha-proteobacterial endosymbiosis for all eukaryotes. Mol. Biol. Evol. 23, 1341–1344. 10.1093/molbev/msl001 [DOI] [PubMed] [Google Scholar]
- Roche B., Aussel L., Ezraty B., Mandin P., Py B., Barras F. (2013). Iron/sulfur proteins biogenesis in prokaryotes: formation, regulation and diversity. Biochim. Biophys. Acta 1827, 455–469. 10.1016/j.bbabio.2012.12.010 [DOI] [PubMed] [Google Scholar]
- Rodríguez-Manzaneque M. T., Tamarit J., Bellí G., Ros J., Herrero E. (2002). Grx5 is a mitochondrial glutaredoxin required for the activity of iron/sulfur enzymes. Mol. Biol. Cell 13, 1109–1121. 10.1091/mbc.01-10-0517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosas-Arellano A., Vásquez-Procopio J., Gambis A., Blowes L. M., Steller H., Mollereau B., et al. (2016). Ferritin assembly in enterocytes of Drosophila melanogaster. Int. J. Mol. Sci. 17:27. 10.3390/ijms17020027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross R. E. (2000). Age-specific decrease in aerobic efficiency associated with increase in oxygen free radical production in Drosophila melanogaster. J. Insect Physiol. 46, 1477–1480. 10.1016/S0022-1910(00)00072-X [DOI] [PubMed] [Google Scholar]
- Rouault T. A. (2012). Biogenesis of iron-sulfur clusters in mammalian cells: new insights and relevance to human disease. Dis. Model. Mech. 5, 155–164. 10.1242/dmm.009019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouault T. A., Maio N. (2017). Biogenesis and functions of mammalian iron-sulfur proteins in the regulation of iron homeostasis and pivotal metabolic pathways. J. Biol. Chem. 292, 12744–12753. 10.1074/jbc.R117.789537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rovenko B. M., Perkhulyn N. V., Lushchak O. V., Storey J. M., Storey K. B., Lushchak V. I. (2014). Molybdate partly mimics insulin-promoted metabolic effects in Drosophila melanogaster. Compar. Biochem. Physiol. Toxicol. Pharmacol. 165, 76–82. 10.1016/j.cbpc.2014.06.002 [DOI] [PubMed] [Google Scholar]
- Roy A., Solodovnikova N., Nicholson T., Antholine W., Walden W. E. (2003). A novel eukaryotic factor for cytosolic Fe-S cluster assembly. EMBO J. 22, 4826–4835. 10.1093/emboj/cdg455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudolf J., Makrantoni V., Ingledew W. J., Stark M. J., White M. F. (2006). The DNA repair helicases XPD and FancJ have essential iron-sulfur domains. Mol. Cell 23, 801–808. 10.1016/j.molcel.2006.07.019 [DOI] [PubMed] [Google Scholar]
- Ruiz de Mena I., Fernández-Moreno M. A., Bornstein B., Kaguni L. S., Garesse R. (1999). Structure and regulated expression of the delta-aminolevulinate synthase gene from Drosophila melanogaster. J. Biol. Chem. 274, 37321–37328. 10.1074/jbc.274.52.37321 [DOI] [PubMed] [Google Scholar]
- Ruland C., Berlandi J., Eikmeier K., Weinert T., Lin F. J., Ambree O., et al. (2017). Decreased cerebral Irp-1B limits impact of social isolation in wild type and Alzheimer's disease modeled in Drosophila melanogaster. Genes Brain Behav. [Epub ahead of print]. 10.1111/gbb.12451 [DOI] [PubMed] [Google Scholar]
- Runko A. P., Griswold A. J., Min K. T. (2008). Overexpression of frataxin in the mitochondria increases resistance to oxidative stress and extends lifespan in Drosophila. FEBS Lett. 582, 715–719. 10.1016/j.febslet.2008.01.046 [DOI] [PubMed] [Google Scholar]
- Ruprecht J., Nield J. (2001). Determining the structure of biological macromolecules by transmission electron microscopy, single particle analysis and 3D reconstruction. Prog. Biophys. Mol. Biol. 75, 121–164. 10.1016/S0079-6107(01)00004-9 [DOI] [PubMed] [Google Scholar]
- Russell M. J., Martin W. (2004). The rocky roots of the acetyl-CoA pathway. Trends Biochem. Sci. 29, 358–363. 10.1016/j.tibs.2004.05.007 [DOI] [PubMed] [Google Scholar]
- Ruzicka F. J., Beinert H. (1978). The soluble “high potential” type iron-sulfur protein from mitochondria is aconitase. J. Biol. Chem. 253, 2514–2517. [PubMed] [Google Scholar]
- Sahashi R., Crevel G., Pasko J., Suyari O., Nagai R., Saura M. M., et al. (2014). DNA polymerase alpha interacts with PrSet7 and mediates H4K20 monomethylation in Drosophila. J. Cell Sci. 127, 3066–3078. 10.1242/jcs.144501 [DOI] [PubMed] [Google Scholar]
- Salomé P. A., Oliva M., Weigel D., Krämer U. (2013). Circadian clock adjustment to plant iron status depends on chloroplast and phytochrome function. EMBO J. 32, 511–523. 10.1038/emboj.2012.330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samson M. L. (2000). Drosophila arginase is produced from a nonvital gene that contains the elav locus within its third intron. J. Biol. Chem. 275, 31107–31114. 10.1074/jbc.M001346200 [DOI] [PubMed] [Google Scholar]
- Sandoval H., Yao C. K., Chen K., Jaiswal M., Donti T., Lin Y. Q., et al. (2014). Mitochondrial fusion but not fission regulates larval growth and synaptic development through steroid hormone production. eLife 3:e3558 10.7554/eLife.03558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanz A. (2016). Mitochondrial reactive oxygen species: do they extend or shorten animal lifespan? Biochim. Biophys. Acta 1857, 1116–1126. 10.1016/j.bbabio.2016.03.018 [DOI] [PubMed] [Google Scholar]
- Sardiello M., Tripoli G., Romito A., Minervini C., Viggiano L., Caggese C., et al. (2005). Energy biogenesis: one key for coordinating two genomes. Trends Genet. 21, 12–16. 10.1016/j.tig.2004.11.009 [DOI] [PubMed] [Google Scholar]
- Schatz G. (2013). Getting mitochondria to center stage. Biochem. Biophys. Res. Commun. 434, 407–410. 10.1016/j.bbrc.2013.03.081 [DOI] [PubMed] [Google Scholar]
- Scheerer P., Park J. H., Hildebrand P. W., Kim Y. J., Krauss N., Choe H. W., et al. (2008). Crystal structure of opsin in its G-protein-interacting conformation. Nature 455, 497–502. 10.1038/nature07330 [DOI] [PubMed] [Google Scholar]
- Schoepp-Cothenet B., van Lis R., Philippot P., Magalon A., Russell M. J., Nitschke W. (2012). The ineluctable requirement for the trans-iron elements molybdenum and/or tungsten in the origin of life. Sci. Rep. 2:263. 10.1038/srep00263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schott D. R., Baldwin M. C., Finnerty V. (1986). Molybdenum hydroxylases in Drosophila. III. further characterization of the low xanthine dehydrogenase gene. Biochem. Genet. 24, 509–527. 10.1007/BF00504332 [DOI] [PubMed] [Google Scholar]
- Schwarz G., Schulze J., Bittner F., Eilers T., Kuper J., Bollmann G., et al. (2000). The molybdenum cofactor biosynthetic protein Cnx1 complements molybdate-repairable mutants, transfers molybdenum to the metal binding pterin, and is associated with the cytoskeleton. Plant Cell 12, 2455–2472. 10.1105/tpc.12.12.2455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarzländer M., Murphy M. P., Duchen M. R., Logan D. C., Fricker M. D., Halestrap A. P., et al. (2012). Mitochondrial ‘flashes’: a radical concept repHined. Trends Cell Biol. 22, 503–508. 10.1016/j.tcb.2012.07.007 [DOI] [PubMed] [Google Scholar]
- Schwarzländer M., Wagner S., Ermakova Y. G., Belousov V. V., Radi R., Beckman J. S., et al. (2014). The ‘mitoflash’ probe cpYFP does not respond to superoxide. Nature 514, E12–E14. 10.1038/nature13858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scopelliti A., Cordero J. B., Diao F., Strathdee K., White B. H., Sansom O. J., et al. (2014). Local control of intestinal stem cell homeostasis by enteroendocrine cells in the adult Drosophila midgut. Curr. Biol. 24, 1199–1211. 10.1016/j.cub.2014.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sehadova H., Glaser F. T., Gentile C., Simoni A., Giesecke A., Albert J. T., et al. (2009). Temperature entrainment of Drosophila's circadian clock involves the gene nocte and signaling from peripheral sensory tissues to the brain. Neuron 64, 251–266. 10.1016/j.neuron.2009.08.026 [DOI] [PubMed] [Google Scholar]
- Sellers V. M., Wang K. F., Johnson M. K., Dailey H. A. (1998). Evidence that the fourth ligand to the [2Fe-2S] cluster in animal ferrochelatase is a cysteine. Characterization of the enzyme from Drosophila melanogaster. J. Biol. Chem. 273, 22311–22316. 10.1074/jbc.273.35.22311 [DOI] [PubMed] [Google Scholar]
- Sen A., Damm V. T., Cox R. T. (2013). Drosophila clueless is highly expressed in larval neuroblasts, affects mitochondrial localization and suppresses mitochondrial oxidative damage. PLoS ONE 8:e54283. 10.1371/journal.pone.0054283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanbhag S., Tripathi S. (2009). Epithelial ultrastructure and cellular mechanisms of acid and base transport in the Drosophila midgut. J. Exp. Biol. 212, 1731–1744. 10.1242/jeb.029306 [DOI] [PubMed] [Google Scholar]
- Sharon G., Segal D., Ringo J. M., Hefetz A., Zilber-Rosenberg I., Rosenberg E. (2010). Commensal bacteria play a role in mating preference of Drosophila melanogaster. Proc. Natl. Acad. Sci. U.S.A. 107, 20051–20056. 10.1073/pnas.1009906107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheftel A. D., Wilbrecht C., Stehling O., Niggemeyer B., Elsässer H. P., Mühlenhoff U., et al. (2012). The human mitochondrial ISCA1, ISCA2, and IBA57 proteins are required for [4Fe-4S] protein maturation. Mol. Biol. Cell 23, 1157–1166. 10.1091/mbc.E11-09-0772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen E. Z., Song C. Q., Lin Y., Zhang W. H., Su P. F., Liu W. Y., et al. (2014). Mitoflash frequency in early adulthood predicts lifespan in Caenorhabditis elegans. Nature 508, 128–132. 10.1038/nature13012 [DOI] [PubMed] [Google Scholar]
- Shibata T., Hadano J., Kawasaki D., Dong X., Kawabata S. I. (2017). Drosophila TG-A transglutaminase is secreted via an unconventional Golgi-independent mechanism involving exosomes and two types of fatty acylations. J. Biol. Chem. 292, 10723–10734. 10.1074/jbc.M117.779710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shidara Y., Hollenbeck P. J. (2010). Defects in mitochondrial axonal transport and membrane potential without increased reactive oxygen species production in a Drosophila model of Friedreich ataxia. J. Neurosci. 30, 11369–11378. 10.1523/JNEUROSCI.0529-10.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiehzadegan S., Le Vinh Thuy J., Szabla N., Angilletta M. J., Jr., VandenBrooks J. M. (2017). More oxygen during development enhanced flight performance but not thermal tolerance of Drosophila melanogaster. PLoS ONE 12:e0177827 10.1371/journal.pone.0177827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin S. C., Kim S. H., You H., Kim B., Kim A. C., Lee K. A., et al. (2011). Drosophila microbiome modulates host developmental and metabolic homeostasis via insulin signaling. Science 334, 670–674. 10.1126/science.1212782 [DOI] [PubMed] [Google Scholar]
- Shoup J. R. (1966). The development of pigment granules in the eyes of wild type and mutant Drosophila melanogaster. J. Cell Biol. 29, 223–249. 10.1083/jcb.29.2.223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieber M. H., Thomsen M. B., Spradling A. C. (2016). Electron transport chain remodeling by GSK3 during Oogenesis connects nutrient state to reproduction. Cell 164, 420–432. 10.1016/j.cell.2015.12.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simcox J. A., Mitchell T. C., Gao Y., Just S. F., Cooksey R., Cox J., et al. (2015). Dietary iron controls circadian hepatic glucose metabolism through heme synthesis. Diabetes 64, 1108–1119. 10.2337/db14-0646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simoni A., Wolfgang W., Topping M. P., Kavlie R. G., Stanewsky R., Albert J. T. (2014). A mechanosensory pathway to the Drosophila circadian clock. Science 343, 525–528. 10.1126/science.1245710 [DOI] [PubMed] [Google Scholar]
- Skandalis D. A., Stuart J. A., Tattersall G. J. (2011). Responses of Drosophila melanogaster to atypical oxygen atmospheres. J. Insect Physiol. 57, 444–451. 10.1016/j.jinsphys.2011.01.005 [DOI] [PubMed] [Google Scholar]
- Slaninova V., Krafcikova M., Perez-Gomez R., Steffal P., Trantirek L., Bray S. J., et al. (2016). Notch stimulates growth by direct regulation of genes involved in the control of glycolysis and the tricarboxylic acid cycle. Open Biol. 6:150155. 10.1098/rsob.150155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J. Y., Cha J., Lee J., Roe J. H. (2006). Glutathione reductase and a mitochondrial thioredoxin play overlapping roles in maintaining iron-sulfur enzymes in fission yeast. Eukaryot. Cell 5, 1857–1865. 10.1128/EC.00244-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soriano S., Calap-Quintana P., Llorens J. V., Al-Ramahi I., Gutiérrez L., Martínez-Sebastián M. J., et al. (2016). Metal homeostasis regulators suppress FRDA phenotypes in a Drosophila model of the disease. PLoS ONE 11:e0159209. 10.1371/journal.pone.0159209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soriano S., Llorens J. V., Blanco-Sobero L., Gutiérrez L., Calap-Quintana P., Morales M. P., et al. (2013). Deferiprone and idebenone rescue frataxin depletion phenotypes in a Drosophila model of Friedreich's ataxia. Gene 521, 274–281. 10.1016/j.gene.2013.02.049 [DOI] [PubMed] [Google Scholar]
- Sparacino-Watkins C. E., Tejero J., Sun B., Gauthier M. C., Thomas J., Ragireddy V., et al. (2014). Nitrite reductase and nitric-oxide synthase activity of the mitochondrial molybdopterin enzymes mARC1 and mARC2. J. Biol. Chem. 289, 10345–10358. 10.1074/jbc.M114.555177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan C., Liba A., Imlay J. A., Valentine J. S., Gralla E. B. (2000). Yeast lacking superoxide dismutase(s) show elevated levels of “free iron” as measured by whole cell electron paramagnetic resonance. J. Biol. Chem. 275, 29187–29192. 10.1074/jbc.M004239200 [DOI] [PubMed] [Google Scholar]
- St Johnston D. (2013). Using mutants, knockdowns, and transgenesis to investigate gene function in Drosophila. Wiley Interdisc. Rev. Develop. Biol. 2, 587–613. 10.1002/wdev.101 [DOI] [PubMed] [Google Scholar]
- Stallmeyer B., Drugeon G., Reiss J., Haenni A. L., Mendel R. R. (1999). Human molybdopterin synthase gene: identification of a bicistronic transcript with overlapping reading frames. Am. J. Hum. Genet. 64, 698–705. 10.1086/302295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stallmeyer B., Nerlich A., Schiemann J., Brinkmann H., Mendel R. R. (1995). Molybdenum co-factor biosynthesis: the Arabidopsis thaliana cDNA cnx1 encodes a multifunctional two-domain protein homologous to a mammalian neuroprotein, the insect protein Cinnamon and three Escherichia coli proteins. Plant J. 8, 751–762. 10.1046/j.1365-313X.1995.08050751.x [DOI] [PubMed] [Google Scholar]
- Stanewsky R., Fry T. A., Reim I., Saumweber H., Hall J. C. (1996). Bioassaying putative RNA-binding motifs in a protein encoded by a gene that influences courtship and visually mediated behavior in Drosophila: in vitro mutagenesis of nonA. Genetics 143, 259–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanewsky R., Jamison C. F., Plautz J. D., Kay S. A., Hall J. C. (1997). Multiple circadian-regulated elements contribute to cycling period gene expression in Drosophila. EMBO J. 16, 5006–5018. 10.1093/emboj/16.16.5006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanewsky R., Kaneko M., Emery P., Beretta B., Wager-Smith K., Kay S. A., et al. (1998). The cryb mutation identifies cryptochrome as a circadian photoreceptor in Drosophila. Cell 95, 681–692. 10.1016/S0092-8674(00)81638-4 [DOI] [PubMed] [Google Scholar]
- Stehling O., Vashisht A. A., Mascarenhas J., Jonsson Z. O., Sharma T., Netz D. J., et al. (2012). MMS19 assembles iron-sulfur proteins required for DNA metabolism and genomic integrity. Science 337, 195–199. 10.1126/science.1219723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiban J., Farnum G. A., Hovde S. L., Kaguni L. S. (2014). The N-terminal domain of the Drosophila mitochondrial replicative DNA helicase contains an iron-sulfur cluster and binds DNA. J. Biol. Chem. 289, 24032–24042. 10.1074/jbc.M114.587774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stüeken E. E., Buick R., Guy B. M., Koehler M. C. (2015). Isotopic evidence for biological nitrogen fixation by molybdenum-nitrogenase from 3.2 Gyr. Nature 520, 666–669. 10.1038/nature14180 [DOI] [PubMed] [Google Scholar]
- Suarez R. K. (2000). Energy metabolism during insect flight: biochemical design and physiological performance. Physiol. Biochem. Zool. 73, 765–771. 10.1086/318112 [DOI] [PubMed] [Google Scholar]
- Suganuma T., Mushegian A., Swanson S. K., Abmayr S. M., Florens L., Washburn M. P., et al. (2010). The ATAC acetyltransferase complex coordinates MAP kinases to regulate JNK target genes. Cell 142, 726–736. 10.1016/j.cell.2010.07.045 [DOI] [PubMed] [Google Scholar]
- Suganuma T., Mushegian A., Swanson S. K., Florens L., Washburn M. P., Workman J. L. (2012). A metazoan ATAC acetyltransferase subunit that regulates mitogen-activated protein kinase signaling is related to an ancient molybdopterin synthase component. Mol. Cell. Proteomics 11, 90–99. 10.1074/mcp.M111.015818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suganuma T., Swanson S. K., Florens L., Washburn M. P., Workman J. L. (2016). Moco biosynthesis and the ATAC acetyltransferase engage translation initiation by inhibiting latent PKR activity. J. Mol. Cell Biol. 8, 44–50. 10.1093/jmcb/mjv070 [DOI] [PubMed] [Google Scholar]
- Sullivan D. T., Sullivan M. C. (1975). Transport defects as the physiological basis for eye color mutants of Drosophila melanogaster. Biochem. Genet. 13, 603–613. 10.1007/BF00484918 [DOI] [PubMed] [Google Scholar]
- Surdej P., Richman L., Kühn L. C. (2008). Differential translational regulation of IRE-containing mRNAs in Drosophila melanogaster by endogenous IRP and a constitutive human IRP1 mutant. Insect Biochem. Mol. Biol. 38, 891–894. 10.1016/j.ibmb.2008.05.010 [DOI] [PubMed] [Google Scholar]
- Tahoe N. M., Dean A. M., Curtsinger J. W. (2002). Nucleotide variations in the lxd region of Drosophila melanogaster: characterization of a candidate modifier of lifespan. Gene 297, 221–228. 10.1016/S0378-1119(02)00916-2 [DOI] [PubMed] [Google Scholar]
- Takashima S., Mkrtchyan M., Younossi-Hartenstein A., Merriam J. R., Hartenstein V. (2008). The behaviour of Drosophila adult hindgut stem cells is controlled by Wnt and Hh signalling. Nature 454, 651–655. 10.1038/nature07156 [DOI] [PubMed] [Google Scholar]
- Takeuchi T., Suzuki M., Fujikake N., Popiel H. A., Kikuchi H., Futaki S., et al. (2015). Intercellular chaperone transmission via exosomes contributes to maintenance of protein homeostasis at the organismal level. Proc. Natl. Acad. Sci. U.S.A. 112, E2497–E2506. 10.1073/pnas.1412651112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang X., Roessingh S., Hayley S. E., Chu M. L., Tanaka N. K., Wolfgang W., et al. (2017). The role of PDF neurons in setting the preferred temperature before dawn in Drosophila. eLife 6:e23206. 10.7554/eLife.23206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang X., Zhou B. (2013a). Ferritin is the key to dietary iron absorption and tissue iron detoxification in Drosophila melanogaster. FASEB J. 27, 288–298. 10.1096/fj.12-213595 [DOI] [PubMed] [Google Scholar]
- Tang X., Zhou B. (2013b). Iron homeostasis in insects: insights from Drosophila studies. IUBMB Life 65, 863–872. 10.1002/iub.1211 [DOI] [PubMed] [Google Scholar]
- Tassetto M., Kunitomi M., Andino R. (2017). Circulating immune cells mediate a systemic RNAi-based adaptive antiviral response in Drosophila. Cell 169, 314–325.e13. 10.1016/j.cell.2017.03.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira F. K., Sanchez C. G., Hurd T. R., Seifert J. R., Czech B., Preall J. B., et al. (2015). ATP synthase promotes germ cell differentiation independent of oxidative phosphorylation. Nat. Cell Biol. 17, 689–696. 10.1038/ncb3165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tejeda-Guzmán C., Rosas-Arellano A., Kroll T., Webb S. M., Barajas-Aceves M., Osorio B., et al. (2018). Biogenesis of zinc storage granules in Drosophila melanogaster. J. Exp. Biol. 2018:jeb.168419 10.1242/jeb.168419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tennessen J. M., Baker K. D., Lam G., Evans J., Thummel C. S. (2011). The Drosophila estrogen-related receptor directs a metabolic switch that supports developmental growth. Cell Metab. 13, 139–148. 10.1016/j.cmet.2011.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tennessen J. M., Bertagnolli N. M., Evans J., Sieber M. H., Cox J., Thummel C. S. (2014). Coordinated metabolic transitions during Drosophila embryogenesis and the onset of aerobic glycolysis. G3 4, 839–850. 10.1534/g3.114.010652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thörig G. E., Heinstra P. W., Scharloo W. (1981a). The action of the notch locus in Drosophila melanogaster. I. Effects of the notch8 deficiency on mitochondrial enzymes. Mol. Gen. Genet. 182, 31–38. [DOI] [PubMed] [Google Scholar]
- Thörig G. E., Heinstra P. W., Scharloo W. (1981b). The action of the notchlocus in Drosophila melanogaster. II. Biochemical effects of recessive lethals on mitochondrial enzymes. Genetics 99, 65–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong W. H., Jameson G. N., Huynh B. H., Rouault T. A. (2003). Subcellular compartmentalization of human Nfu, an iron-sulfur cluster scaffold protein, and its ability to assemble a [4Fe-4S] cluster. Proc. Natl. Acad. Sci. U.S.A. 100, 9762–9767. 10.1073/pnas.1732541100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong W. H., Rouault T. (2000). Distinct iron-sulfur cluster assembly complexes exist in the cytosol and mitochondria of human cells. EMBO J. 19, 5692–5700. 10.1093/emboj/19.21.5692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong W. H., Rouault T. A. (2006). Functions of mitochondrial ISCU and cytosolic ISCU in mammalian iron-sulfur cluster biogenesis and iron homeostasis. Cell Metab. 3, 199–210. 10.1016/j.cmet.2006.02.003 [DOI] [PubMed] [Google Scholar]
- Tong W. H., Rouault T. A. (2007). Metabolic regulation of citrate and iron by aconitases: role of iron-sulfur cluster biogenesis. Biometals 20, 549–564. 10.1007/s10534-006-9047-6 [DOI] [PubMed] [Google Scholar]
- Tovar J., León-Avila G., Sánchez L. B., Sutak R., Tachezy J. M., et al. (2003). Mitochondrial remnant organelles of Giardia function in iron-sulphur protein maturation. Nature 426, 172–176. 10.1038/nature01945 [DOI] [PubMed] [Google Scholar]
- Tricoire H., Palandri A., Bourdais A., Camadro J. M., Monnier V. (2014). Methylene blue rescues heart defects in a Drosophila model of Friedreich's ataxia. Hum. Mol. Genet. 23, 968–979. 10.1093/hmg/ddt493 [DOI] [PubMed] [Google Scholar]
- Tsai C. L., Barondeau D. P. (2010). Human frataxin is an allosteric switch that activates the Fe-S cluster biosynthetic complex. Biochemistry 49, 9132–9139. 10.1021/bi1013062 [DOI] [PubMed] [Google Scholar]
- Tsimilli-Michael M., Haldimann P. (2017). Sustainability of photosynthesis research–when research is impeded by the cults of audit and management. Photosynthetica 55, 391–400. 10.1007/s11099-017-0686-3 [DOI] [Google Scholar]
- Uhrigshardt H., Rouault T. A., Missirlis F. (2013). Insertion mutants in Drosophila melanogaster Hsc20 halt larval growth and lead to reduced iron-sulfur cluster enzyme activities and impaired iron homeostasis. J. Biol. Inorg. Chem. 18, 441–449. 10.1007/s00775-013-0988-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhrigshardt H., Singh A., Kovtunovych G., Ghosh M., Rouault T. A. (2010). Characterization of the human HSC20, an unusual DnaJ type III protein, involved in iron-sulfur cluster biogenesis. Hum. Mol. Genet. 19, 3816–3834. 10.1093/hmg/ddq301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbina H. D., Silberg J. J., Hoff K. G., Vickery L. E. (2001). Transfer of sulfur from IscS to IscU during Fe/S cluster assembly. J. Biol. Chem. 276, 44521–44526. 10.1074/jbc.M106907200 [DOI] [PubMed] [Google Scholar]
- Uzarska M. A., Nasta V., Weiler B. D., Spantgar F., Ciofi-Baffoni S., Saviello M. R., et al. (2016). Mitochondrial Bol1 and Bol3 function as assembly factors for specific iron-sulfur proteins. Elife 5:e16673. 10.7554/eLife.16673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vakifahmetoglu-Norberg H., Ouchida A. T., Norberg E. (2017). The role of mitochondria in metabolism and cell death. Biochem. Biophys. Res. Commun. 482, 426–431. 10.1016/j.bbrc.2016.11.088 [DOI] [PubMed] [Google Scholar]
- Vallières C., Holland S. L., Avery S. V. (2017). Mitochondrial ferredoxin determines vulnerability of cells to copper excess. Cell Chemical Biology. 24, 1228–1237. 10.1016/j.chembiol.2017.08.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Gelder R. N., Bae H., Palazzolo M. J., Krasnow M. A. (1995). Extent and character of circadian gene expression in Drosophila melanogaster: identification of twenty oscillating mRNAs in the fly head. Curr. Biol. 5, 1424–1436. 10.1016/S0960-9822(95)00280-6 [DOI] [PubMed] [Google Scholar]
- Van Vranken J. G., Bricker D. K., Dephoure N., Gygi S. P., Cox J. E., Thummel C. S., et al. (2014). SDHAF4 promotes mitochondrial succinate dehydrogenase activity and prevents neurodegeneration. Cell Metab. 20, 241–252. 10.1016/j.cmet.2014.05.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vann A. C., Webster G. C. (1977). Age-related changes in mitochondrial function in Drosophila melanogaster. Exp. Gerontol. 12, 1–5. 10.1016/0531-5565(77)90025-0 [DOI] [PubMed] [Google Scholar]
- Venton D. (2013). Highlight: on the origin of the sexes. Genome Biol. Evol. 5, 2073–2074. 10.1093/gbe/evt164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villee C. A. (1948). Studies in biochemical genetics in Drosophila. J. Gen. Physiol. 31, 337–345. 10.1085/jgp.31.4.337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent A., Briggs L., Chatwin G. F., Emery E., Tomlins R., Oswald M., et al. (2012). Parkin-induced defects in neurophysiology and locomotion are generated by metabolic dysfunction and not oxidative stress. Hum. Mol. Genet. 21, 1760–1769. 10.1093/hmg/ddr609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vo A. T. V., Fleischman N. M., Froehlich M. J., Lee C. Y., Cosman J. A., Glynn C. A., et al. (2017). Identifying the protein interactions of the cytosolic iron sulfur cluster targeting complex essential for its assembly and recognition of apo-targets. Biochemistry. [Epub ahead of print]. 10.1021/acs.biochem.7b00072 [DOI] [PubMed] [Google Scholar]
- Vrailas-Mortimer A., del Rivero T., Mukherjee S., Nag S., Gaitanidis A., Kadas D., et al. (2011). A muscle-specific p38 MAPK/Mef2/MnSOD pathway regulates stress, motor function, and life span in Drosophila. Dev. Cell 21, 783–795. 10.1016/j.devcel.2011.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wachnowsky C., Fidai I., Cowan J. A. (2016). Iron-sulfur cluster exchange reactions mediated by the human Nfu protein. J. Biol. Inorg. Chem. 21, 825–836. 10.1007/s00775-016-1381-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wächtershäuser G. (1988). Before enzymes and templates: theory of surface metabolism. Microbiol. Rev. 52, 452–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl R. C., Rajagopalan K. V. (1982). Evidence for the inorganic nature of the cyanolyzable sulfur of molybdenum hydroxylases. J. Biol. Chem. 257, 1354–1359. [PubMed] [Google Scholar]
- Wahl R. C., Warner C. K., Finnerty V., Rajagopalan K. V. (1982). Drosophila melanogaster ma-l mutants are defective in the sulfuration of desulfo Mo hydroxylases. J. Biol. Chem. 257, 3958–3962. [PubMed] [Google Scholar]
- Walker D. W., Hájek P., Muffat J., Knoepfle D., Cornelison S., Attardi G., et al. (2006). Hypersensitivity to oxygen and shortened lifespan in a Drosophila mitochondrial complex II mutant. Proc. Natl. Acad. Sci. U.S.A. 103, 16382–16387. 10.1073/pnas.0607918103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Fang H., Groom L., Cheng A., Zhang W., Liu J., et al. (2008). Superoxide flashes in single mitochondria. Cell 134, 279–290. 10.1016/j.cell.2008.06.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warburg O. (1925). Iron, the oxygen-carrier of respiration-ferment. Science 61, 575–582. 10.1126/science.61.1588.575 [DOI] [PubMed] [Google Scholar]
- Warner C. K., Finnerty V. (1981). Molybdenum hydroxylases in Drosophila. II. Molybdenum cofactor in xanthine dehydrogenase, aldehyde oxidase and pyridoxal oxidase. Mol. Gen. Genet. MGG 184, 92–96. 10.1007/BF00271201 [DOI] [PubMed] [Google Scholar]
- Watson J. A., Lowenstein J. M. (1970). Citrate and the conversion of carbohydrate into fat. Fatty acid synthesis by a combination of cytoplasm and mitochondria. J. Biol. Chem. 245, 5993–6002. [PubMed] [Google Scholar]
- Wei-LaPierre L., Gong G., Gerstner B. J., Ducreux S., Yule D. I., Pouvreau S., et al. (2013). Respective contribution of mitochondrial superoxide and pH to mitochondria-targeted circularly permuted yellow fluorescent protein (mt-cpYFP) flash activity. J. Biol. Chem. 288, 10567–10577. 10.1074/jbc.M113.455709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicks S., Bain N., Duttaroy A., Hilliker A. J., Phillips J. P. (2009). Hypoxia rescues early mortality conferred by superoxide dismutase deficiency. Free Radic. Biol. Med. 46, 176–181. 10.1016/j.freeradbiomed.2008.09.036 [DOI] [PubMed] [Google Scholar]
- Wiedemann N., Urzica E., Guiard B., Müller H., Lohaus C., Meyer H. E., et al. (2006). Essential role of Isd11 in mitochondrial iron-sulfur cluster synthesis on Isu scaffold proteins. EMBO J. 25, 184–195. 10.1038/sj.emboj.7600906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiley K., Forrest H. S. (1981). Terminal synthesis of xanthommatin in Drosophila melanogaster. IV Enzymatic and nonenzymatic catalysis. Biochem. Genet. 19, 1211–1221. 10.1007/BF00484574 [DOI] [PubMed] [Google Scholar]
- Wingert R. A., Galloway J. L., Barut B., Foott H., Fraenkel P., Axe J. L., et al. (2005). Deficiency of glutaredoxin 5 reveals Fe-S clusters are required for vertebrate haem synthesis. Nature 436, 1035–1039. 10.1038/nature03887 [DOI] [PubMed] [Google Scholar]
- Wittle A. E., Kamdar K. P., Finnerty V. (1999). The Drosophila cinnamon gene is functionally homologous to Arabidopsis cnx1 and has a similar expression pattern to the mammalian gephyrin gene. Mol. Gen. Genet. 261, 672–680. 10.1007/s004380050010 [DOI] [PubMed] [Google Scholar]
- Wofford J. D., Lindahl P. A. (2015). Mitochondrial iron-sulfur cluster activity and cytosolic iron regulate iron traffic in Saccharomyces cerevisiae. J. Biol. Chem. 290, 26968–26977. 10.1074/jbc.M115.676668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff J. N., Pichaud N., Camus M. F., Côté G., Blier P. U., Dowling D. K. (2016). Evolutionary implications of mitochondrial genetic variation: mitochondrial genetic effects on OXPHOS respiration and mitochondrial quantity change with age and sex in fruit flies. J. Evol. Biol. 29, 736–747. 10.1111/jeb.12822 [DOI] [PubMed] [Google Scholar]
- Wong A. C., Wang Q. P., Morimoto J., Senior A. M., Lihoreau M., Neely G. G., et al. (2017). Gut microbiota modifies olfactory-guided microbial preferences and foraging decisions in Drosophila. Curr. Biol. 27, 2397 e4–2404e4. 10.1016/j.cub.2017.07.022 [DOI] [PubMed] [Google Scholar]
- Wong H. S., Dighe P. A., Mezera V., Monternier P. A., Brand M. D. (2017). Production of superoxide and hydrogen peroxide from specific mitochondrial sites under different bioenergetic conditions. J. Biol. Chem. 292, 16804–16809. 10.1074/jbc.R117.789271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C. K., Dailey H. A., Rose J. P., Burden A., Sellers V. M., Wang B. C. (2001). The 2.0 A structure of human ferrochelatase, the terminal enzyme of heme biosynthesis. Nat. Struct. Biol. 8, 156–160. 10.1038/84152 [DOI] [PubMed] [Google Scholar]
- Wuebbens M. M., Rajagopalan K. V. (1993). Structural characterization of a molybdopterin precursor. J. Biol. Chem. 268, 13493–13498. [PubMed] [Google Scholar]
- Xia H., Cao Y., Dai X., Marelja Z., Zhou D., Mo R., et al. (2012). Novel frataxin isoforms may contribute to the pathological mechanism of Friedreich ataxia. PLoS ONE 7:e47847. 10.1371/journal.pone.0047847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao G., Wan Z., Fan Q., Tang X., Zhou B. (2014). The metal transporter ZIP13 supplies iron into the secretory pathway in Drosophila melanogaster. eLife 3:e03191. 10.7554/eLife.03191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao G., Zhou B. (2018). ZIP13: a study of Drosophila offers an alternative explanation for the corresponding human disease. Front. Genet. 8:234 10.3389/fgene.2017.00234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanaka N., Rewitz K. F., O'Connor M. B. (2013). Ecdysone control of developmental transitions: lessons from Drosophila research. Annu. Rev. Entomol. 58, 497–516. 10.1146/annurev-ento-120811-153608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan L. J., Levine R. L., Sohal R. S. (1997). Oxidative damage during aging targets mitochondrial aconitase. Proc. Natl. Acad. Sci. U.S.A. 94, 11168–11172. 10.1073/pnas.94.21.11168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M., Cobine P. A., Molik S., Naranuntarat A., Lill R., Winge D. R., et al. (2006). The effects of mitochondrial iron homeostasis on cofactor specificity of superoxide dismutase 2. EMBO J. 25, 1775–1783. 10.1038/sj.emboj.7601064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye H., Jeong S. Y., Ghosh M. C., Kovtunovych G., Silvestri L., Ortillo D., et al. (2010). Glutaredoxin 5 deficiency causes sideroblastic anemia by specifically impairing heme biosynthesis and depleting cytosolic iron in human erythroblasts. J. Clin. Invest. 120, 1749–1761. 10.1172/JCI40372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeom E., Hong S. T., Choi K. W. (2015). Crumbs interacts with Xpd for nuclear division control in Drosophila. Oncogene 34, 2777–2789. 10.1038/onc.2014.202 [DOI] [PubMed] [Google Scholar]
- Yin L., Wu N., Curtin J. C., Qatanani M., Szwergold N. R., Reid R. A., et al. (2007). Rev-erbalpha, a heme sensor that coordinates metabolic and circadian pathways. Science 318, 1786–1789. 10.1126/science.1150179 [DOI] [PubMed] [Google Scholar]
- Yin S., Qin Q., Zhou B. (2017). Functional studies of Drosophila zinc transporters reveal the mechanism for zinc excretion in Malpighian tubules. BMC Biol. 15:12. 10.1186/s12915-017-0355-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon S., Cho B., Shin M., Koranteng F., Cha N., Shim J. (2017). Iron homeostasis controls myeloid blood cell differentiation in Drosophila. Mol. Cells 40, 976–985. 10.14348/molcells.2017.0287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshii T., Todo T., Wülbeck C., Stanewsky R., Helfrich-Förster C. (2008). Cryptochrome is present in the compound eyes and a subset of Drosophila's clock neurons. J. Comp. Neurol. 508, 952–966. 10.1002/cne.21702 [DOI] [PubMed] [Google Scholar]
- Yoshiyama T., Namiki T., Mita K., Kataoka H., Niwa R. (2006). Neverland is an evolutionally conserved Rieske-domain protein that is essential for ecdysone synthesis and insect growth. Development 133, 2565–2574. 10.1242/dev.02428 [DOI] [PubMed] [Google Scholar]
- Yu Z., O'Farrell P. H., Yakubovich N., DeLuca S. Z. (2017). The mitochondrial DNA polymerase promotes elimination of paternal mitochondrial genomes. Curr. Biol. 27, 1033–1039. 10.1016/j.cub.2017.02.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D. L., Ghosh M. C., Rouault T. A. (2014). The physiological functions of iron regulatory proteins in iron homeostasis - an update. Front. Pharmacol. 5:124. 10.3389/fphar.2014.00124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F., Scheerer P., Oberpichler I., Lamparter T., Krauß N. (2013). Crystal structure of a prokaryotic (6-4) photolyase with an Fe-S cluster and a 6,7-dimethyl-8-ribityllumazine antenna chromophore. Proc. Natl. Acad. Sci. U.S.A. 110, 7217–7222. 10.1073/pnas.1302377110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K., Li Z., Jaiswal M., Bayat V., Xiong B., Sandoval H., et al. (2013). The C8ORF38 homologue Sicily is a cytosolic chaperone for a mitochondrial complex I subunit. J. Cell Biol. 200, 807–820. 10.1083/jcb.201208033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Gladyshev V. N. (2008). Molybdoproteomes and evolution of molybdenum utilization. J. Mol. Biol. 379, 881–899. 10.1016/j.jmb.2008.03.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Lyver E. R., Nakamaru-Ogiso E., Yoon H., Amutha B., Lee D. W., et al. (2008). Dre2, a conserved eukaryotic Fe/S cluster protein, functions in cytosolic Fe/S protein biogenesis. Mol. Cell. Biol. 28, 5569–5582. 10.1128/MCB.00642-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Wan D., Zhou X., Long C., Wu X., Li L., et al. (2017). Diurnal variations in iron concentrations and expression of genes involved in iron absorption and metabolism in pigs. Biochem. Biophys. Res. Commun. 490, 1210–1214. 10.1016/j.bbrc.2017.06.187 [DOI] [PubMed] [Google Scholar]
- Zhao W., Zheng H. Z., Niu Y. J., Yuan Y., Fang B. X., Liu Y. N., et al. (2015). CIA2 deficiency results in impaired oxidative stress response and enhanced intracellular basal UPR activity in Saccharomyces cerevisiae. FEMS Microbiol. Lett. 362:fnv013. 10.1093/femsle/fnv013 [DOI] [PubMed] [Google Scholar]
- Zheng L., Cash V. L., Flint D. H., Dean D. R. (1998). Assembly of iron-sulfur clusters. Identification of an iscSUA-hscBA-fdx gene cluster from Azotobacter vinelandii. J. Biol. Chem. 273, 13264–13272. 10.1074/jbc.273.21.13264 [DOI] [PubMed] [Google Scholar]
- Zheng L., White R. H., Cash V. L., Dean D. R. (1994). Mechanism for the desulfurization of L-cysteine catalyzed by the nifS gene product. Biochemistry 33, 4714–4720. 10.1021/bi00181a031 [DOI] [PubMed] [Google Scholar]
- Zheng L., White R. H., Cash V. L., Jack R. F., Dean D. R. (1993). Cysteine desulfurase activity indicates a role for NIFS in metallocluster biosynthesis. Proc. Natl. Acad. Sci. U.S.A. 90, 2754–2758. 10.1073/pnas.90.7.2754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z. J., Wu K. C., Qian Z. M., Yung W. H., Ke Y. (2014). Drosophila models for studying iron-related neurodegenerative diseases. Sheng li xue Bao. 66, 47–54. 10.13294/j.aps.2014.0007 [DOI] [PubMed] [Google Scholar]
- Zouni A., Witt H. T., Kern J., Fromme P., Krauss N., Saenger W., et al. (2001). Crystal structure of photosystem II from Synechococcus elongatus at 3.8 A resolution. Nature 409, 739–743. 10.1038/35055589 [DOI] [PubMed] [Google Scholar]