Abstract
Differences in response to medications have a strong genetic component. By leveraging publically available data, the spectrum of such genomic variation can be investigated extensively. Pharmacogenomic variation was extracted from the 1000 Genomes Project Phase 3 data (2504 individuals, 26 global populations). A total of 12 084 genetic variants were found in 120 pharmacogenes, with the majority (90.0%) classified as rare variants (global minor allele frequency <0.5%), with 52.9% being singletons. Common variation clustered individuals into continental super-populations and 23 pharmacogenes contained highly differentiated variants (FST>0.5) for one or more super-population comparison. A median of three clinical variants (PharmGKB level 1A/B) was found per individual, and 55.4% of individuals carried loss-of-function variants, varying by super-population (East Asian 60.9%>African 60.1%>South Asian 60.3%>European 49.3%>Admixed 39.2%). Genome sequencing can therefore identify clinical pharmacogenomic variation, and future studies need to consider rare variation to understand the spectrum of genetic diversity contributing to drug response.
Introduction
Inter-individual differences in response to medications are known to have a strong genetic component and several genes that influence either response to medications or adverse drug reactions (ADRs) have been identified.1, 2 The majority of previous pharmacogenomic studies, however, have either assessed individual candidate genes or analyzed a subset of genetic variation interrogated with genotyping arrays. Current sequencing technologies therefore offer an opportunity to assess the full spectrum of variation present in populations,3 as well as to determine how genes of pharmacogenomic importance are affected by rare genetic variation, the class of genetic variants that are most likely to be deleterious.4 Further, sequencing approaches present a means to investigate understudied populations and identify groups of individuals at risk to certain ADRs on a scale not previously possible.
The 1000 Genomes Project (1000GP) aimed to detect the majority of variants with minor allele frequencies (MAFs) >1% in numerous human populations through the use of current sequencing and array genotyping technologies.5 The final stage consisted of 2504 individuals from 26 populations.6 The current study aimed to leverage these genomic data to determine the spectrum of variation found in pharmacogenes across human populations. A previous study investigated an earlier release of the 1000GP, analyzing 15 populations and a subset of pharmacogenomic variation present on a commercial array (that is, 1156 markers).7 We therefore analyzed the full catalogue of diversity in the protein-coding regions of genes of relevance to pharmacogenomics, incorporating data from across the entire allele frequency spectrum.
The protein-coding regions of these pharmacogenes were the focus of investigation, since these areas were subjected to the most comprehensive sequencing coverage in the 1000GP (mean coverage, complete exome=65.7 × ), compared with the rest of the genome (mean coverage, whole-genome sequencing=7.4 × ).5 Performing pharmacogenomic studies of inclusive population cohorts will lead to a better understanding of the pattern of genetic factors that influence drug safety and effectiveness.
Materials and methods
Selection of pharmacogenes
Pharmacogenes were selected based on curated Pharmacogenomics Knowledgebase (PharmGKB) data and the literature. Autosomal genes annotated as ‘very important pharmacogenes’ and/or containing variants with high to moderate levels of clinical annotation (PharmGKB levels 1 and 2) were prioritized (www.pharmgkb.org/downloads, accessed 26 August 2014). In addition, pharmacogenes with emerging evidence, as highlighted in recent reviews, were included if they had not already fulfilled these criteria.1, 2 Human leukocyte antigen (HLA) and UDP-glucuronosyltransferases (UGT) genes were excluded from analyses due to their complex nomenclature and difficulties associated with current sequencing.8, 9
Study population, genetic data retrieval and functional annotation
The 1000GP Phase 3 consists of 2504 individuals from 26 global populations (Supplementary Table 1), grouped according to five super-populations: African, admixed American, East Asian, European and South Asian. GRCh37 exon locations of pharmacogenes were extracted with the R (R Foundation for Statistical Computing, Vienna, Austria) package, biomaRt, and an intersection between these coordinates and the exome region targeted by the 1000GP was created (ftp://ftp.1000genomes.ebi.ac.uk/vol1/ftp/technical/reference/exome_pull_down_targets, accessed 10 May 2016). The intersection was padded by 25 bp with bedtools (v2.24.0) to capture flanking exon/intron boundaries. Sequencing coverage for the exome capture regions was then calculated with samtools (v0.1.19). Variants were extracted (ftp://ftp-trace.ncbi.nih.gov/1000genomes/ftp/release/20130502, accessed 2 June 2016) using tabix (v1.3) and annotated with the Ensembl Variant Effect Predictor (VEP v83) (per gene, default ranking criteria). Variants were assigned CADD scores and values of ⩾15 were considered deleterious (http://cadd.gs.washington.edu/info, accessed 9 June 2016). The VEP-plugin, Loss-Of-Function Transcript Effect Estimator (LOFTEE), was employed to annotate high-confidence loss-of-function (LOF) variants. LOF variants with a global MAF >30% were manually curated. In order to generate a list of robust LOF variants, we selected variants annotated using GENCODE (v19) transcripts that were annotated as ‘high confidence’ and were not flagged by LOFTEE.
Population genetic analyses
Principal component analysis was performed on a pruned subset of the data with EIGENSOFT (v5.0). Pruning was based on linkage disequillibrium and MAF (parameters: 50 variant window, shifted by a 5-variant interval, r2>0.2, global MAF>0.01) with PLINK (v1.07), and these data were used exclusively for principal component analysis. VCFtools (v0.1.14) was used to calculate global and population-specific allele frequencies, fixation index (FST) statistics and analyze variants in inaccessible regions and/or segmental duplications. Rare variants were defined as those with a MAF <0.5%, while singletons were variants with an allele count of one in all 1000GP individuals. Highly differentiated variants in individual populations were defined as variants that were rare in the global sample (MAF <0.5%), but common in one population (MAF >5.0%).
Clinical pharmacogenomic variants
Clinical variants were defined as variants with a PharmGKB clinical annotation level of evidence of 1A/B with unambiguous allele-defining variants. Level 1A/B variants represent those that are being implemented in clinical practice or have an unequivocal influence on a pharmacogenomic trait, while level 2 variants are ones that are either found in known pharmacogenes or have been replicated with moderate evidence for association (https://www.pharmgkb.org/page/clinAnnLevels, accessed 9 June 2016). Due to pleiotropic effects, the number of minor alleles carried per individual was used to calculate clinical variant statistics. Downstream statistical analyses and plotting were performed in R (dplyr, reshape2 and ggplot2).
Short-read sequencing accessibility and variant site assessment
Pharmacogenes were assessed for accessibility to short-read sequencing technologies by investigating variants located in (i) potentially inaccessible regions defined by the 1000GP ‘strict mask’ and (ii) segmental duplications (>1000 bp with >90% identity, http://humanparalogy.gs.washington.edu/build37/data/, accessed 23 February 2015). Extreme outlier genes (>3 × interquartile range) with regards to proportion of variants located in either the ‘strict mask’ or segmental duplication regions were flagged as being potentially problematic for short-read technologies. In order to assess the quality of variant calls in the data set, we generated a list of variants that are found in the 1000GP data, but are more likely to be sequencing artefacts. The 1000GP used a support vector machine (SVM) classifier to select high-quality variants and the final call set included single-nucleotide variants with SVM>0. We therefore described marginal quality variants as those close to the SVM separating hyperplane (that is, 0<SVM<0.3), using an upper limit similar to that used by other large-scale sequencing projects.10 Finally, to provide independent in silico verification of the 1000GP variants we compared allele frequency data for overlapping markers found in either the Exome Aggregation Consortium (ExAC)11 and the Human Genome Diversity Project.12
Code availability
The code used to perform these analyses will be made available via GitHub (https://github.com/GalenWright/1000gpPGX).
Results
Summary of pharmacogenomic variation
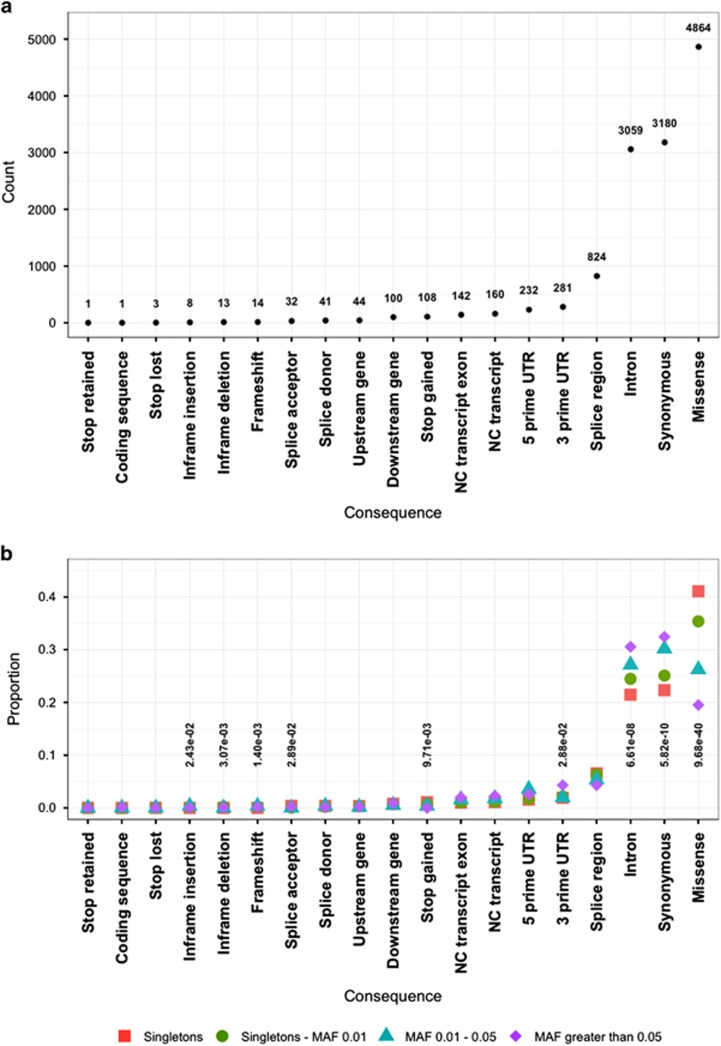
A total of 120 pharmacogenes were included, spanning 369 kb of genomic sequence and containing 12 084 variants, with a mean coverage of 105.2 × for the analyzed pharmacogenomic exome region (Supplementary Table 2). Notably, 6398 (52.9%) of the variants were singletons. Rare variants, with global MAFs <0.5%, made up 90.0% of the data set. Variants that could influence protein function (for example, missense, stop gained, splice acceptor) were enriched in the rare variant classes, while, conversely, those more likely to be benign (for example, synonymous, intronic and 3′UTR) were more frequent in the most common positional annotations (Figure 1). The most significant enrichment was observed for missense variants (corrected P=9.7 × 10−40), where this class was over twice as prevalent in singletons (41.0%) compared with common (global MAF >5.0%) variants (19.5%). Further, rare variants had 50.1% higher mean CADD scores than variants with higher allele frequencies (13.1 versus 8.6 CADD). Supplementary Table 3 presents a per gene summary of the number of variants and select functional annotations.
Figure 1.
Summary of the functional annotation of the pharmacogenomic variants in the 1000 Genomes Project individuals. (a) Counts of the different variant classes according to consequence type. (b) Relative proportion of variants across consequence type stratified by global minor allele frequency (MAF) bins. Consequence types that differ significantly in frequency according to global MAF are annotated with Bonferroni corrected P-values. Missense variants displayed the most significant differences in relative frequencies (P=9.68 × 10−40).
The number of missense variants per coding sequence length ranged from 0.001 (YEATS4, missense variant every ~684 bp) to 0.058 (IFNL3, missense variant every ~17 bp). Seventeen pharmacogenes exclusively carried rare missense variants, while ADRB1 was an outlier with regards to this statistic, with only 66.7% missense variants classified as rare (Supplementary Table 3). Some of the most conserved pharmacogenes were those where somatic mutations are predictive of cancer treatment response (for example, BRAF, KRAS and NRAS), indicating their important role in biological processes. Many of the other conserved pharmacogenes are important for hypertension (NEDD4L, PRKCA and PTGS2), statins (HMGCR) and beta blockers (ADRB1, ADRA2C and PTGS2).
Principal component analysis and FST analyses (Supplementary Figures 1 and 2) revealed that pharmacogenomic variation tends to separate continental super-populations into different clusters (that is, African, European, South Asian and East Asian). African populations had the highest number of polymorphic sites in their pharmacogenes (Supplementary Figure 3). The average number of singletons per individual per population ranged from 1.2 to 3.6, with the Finnish population displaying the least number of singletons per individual (Supplementary Figure 4). There were 23 pharmacogenes (19.2%) that contained highly differentiated pharmacogenomic variants (pairwise FST>0.5 for one or more continental comparison, Supplementary Figure 5 and Table 1) and 17 (14.1%) possessed a rare variant that was common in one population (Supplementary Table 4 and Supplementary Figure 6).
Table 1. Pharmacogenes containing highly differentiated genetic variants. Twenty-three genes showed at least one variant that had FST values of greater than or equal to 0.5 for one or more super-population comparison (bolded values). These genes are important for various drug classes, with the table presenting the highest mean FST variant for each of these genes.
| Gene | ID | Annotation | CADD | AFR EAS | AFR EUR | AFR SAS | EUR EAS | EUR SAS | SAS EAS | AMR AFR | AMR EAS | AMR EUR | AMR SAS | Important drugs |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ABCG2 | rs2231153 | Intron | 3.91 | 0.43 | 0.66 | 0.51 | 0.12 | 0.06 | 0.01 | 0.38 | 0.00 | 0.17 | 0.03 | Various |
| ADCY9 | rs2230738 | Synonymous | 10.52 | 0.60 | 0.22 | 0.49 | 0.22 | 0.12 | 0.02 | 0.33 | 0.15 | 0.01 | 0.07 | Dalcetrapib |
| ADH1B | rs1229984 | Missense | 8.98 | 0.72 | 0.03 | 0.02 | 0.65 | 0.00 | 0.66 | 0.07 | 0.58 | 0.01 | 0.02 | Alcohol |
| ADH1C | rs2241894 | Synonymous | 0.05 | 0.14 | 0.14 | 0.02 | 0.44 | 0.24 | 0.06 | 0.19 | 0.51 | 0.01 | 0.31 | Anticancer |
| ALK | rs2246745 | Synonymous | 0.09 | 0.58 | 0.59 | 0.48 | 0.00 | 0.02 | 0.02 | 0.48 | 0.02 | 0.03 | 0.00 | Anticancer |
| ANKK1 | rs11214596 | Intron | 3.67 | 0.01 | 0.50 | 0.28 | 0.41 | 0.09 | 0.19 | 0.30 | 0.19 | 0.09 | 0.00 | Antipsychotics, antidepressants |
| BRAF | rs3789806 | Intron | 6.63 | 0.47 | 0.52 | 0.30 | 0.00 | 0.08 | 0.05 | 0.50 | 0.00 | 0.00 | 0.08 | Anticancer |
| CFTR | rs213950 | Missense | 13.09 | 0.52 | 0.47 | 0.40 | 0.00 | 0.01 | 0.03 | 0.41 | 0.03 | 0.01 | 0.00 | Ivacaftor |
| COL22A1 | rs3935045 | Splice region | 2.93 | 0.47 | 0.54 | 0.33 | 0.01 | 0.07 | 0.03 | 0.32 | 0.04 | 0.08 | 0.00 | Salbutamol |
| CYP1A2 | rs2470890 | Synonymous | 2.09 | 0.11 | 0.57 | 0.10 | 0.33 | 0.34 | 0.00 | 0.32 | 0.07 | 0.13 | 0.08 | Antipsychotics, caffeine |
| CYP2D6 | rs1081003 | Synonymous | 4.54 | 0.43 | 0.04 | 0.00 | 0.53 | 0.03 | 0.42 | 0.01 | 0.45 | 0.01 | 0.01 | Various |
| CYP2E1 | rs2480257 | 3′UTR | 7.13 | 0.31 | 0.59 | 0.42 | 0.12 | 0.05 | 0.02 | 0.46 | 0.02 | 0.04 | 0.00 | Analgesics, antituberculosis |
| CYP3A4 | rs2242480 | Intron | 3.78 | 0.52 | 0.74 | 0.40 | 0.11 | 0.21 | 0.02 | 0.39 | 0.03 | 0.25 | 0.00 | Various |
| CYP3A5 | rs15524 | 3′UTR | 2.60 | 0.27 | 0.58 | 0.23 | 0.18 | 0.22 | 0.00 | 0.40 | 0.03 | 0.08 | 0.05 | Immunosuppressives, anticancer |
| ERCC1 | rs11615 | Synonymous | 0.30 | 0.20 | 0.58 | 0.41 | 0.23 | 0.05 | 0.08 | 0.37 | 0.03 | 0.10 | 0.01 | Anticancer |
| F5 | rs13306334 | Missense | 1.68 | 0.63 | 0.02 | 0.19 | 0.56 | 0.12 | 0.33 | 0.12 | 0.43 | 0.04 | 0.03 | Contraceptives, anticancer |
| GRIK4 | rs644057 | Synonymous | 3.51 | 0.54 | 0.13 | 0.30 | 0.30 | 0.06 | 0.15 | 0.22 | 0.24 | 0.02 | 0.01 | Antidepressants |
| GSTP1 | rs4147581 | Intron | 8.01 | 0.58 | 0.37 | 0.53 | 0.07 | 0.04 | 0.00 | 0.21 | 0.21 | 0.05 | 0.17 | Anticancer |
| IFNL3 | rs8103142 | Missense | 0.00 | 0.55 | 0.26 | 0.34 | 0.15 | 0.01 | 0.08 | 0.16 | 0.26 | 0.02 | 0.06 | Interferon |
| P2RY1 | rs701265 | Synonymous | 0.64 | 0.45 | 0.56 | 0.51 | 0.02 | 0.00 | 0.01 | 0.43 | 0.00 | 0.03 | 0.01 | Antiplatelet |
| PTGS1 | rs5788 | Synonymous | 14.54 | 0.60 | 0.46 | 0.52 | 0.06 | 0.01 | 0.03 | 0.33 | 0.17 | 0.03 | 0.07 | Antiplatelet |
| TCF7L2 | rs1056877 | 3′UTR | 0.00 | 0.58 | 0.58 | 0.58 | 0.00 | 0.00 | 0.00 | 0.47 | 0.07 | 0.05 | 0.07 | Sulfonamides |
| TXNRD2 | rs5748469 | Missense | 28.60 | 0.59 | 0.05 | 0.20 | 0.43 | 0.07 | 0.23 | 0.20 | 0.25 | 0.06 | 0.00 | Antidepressants |
Abbreviations: AFR, African; AMR, admixed American; EAS, East Asian; EUR, European; SAS, South Asian; UTR, untranslated region.
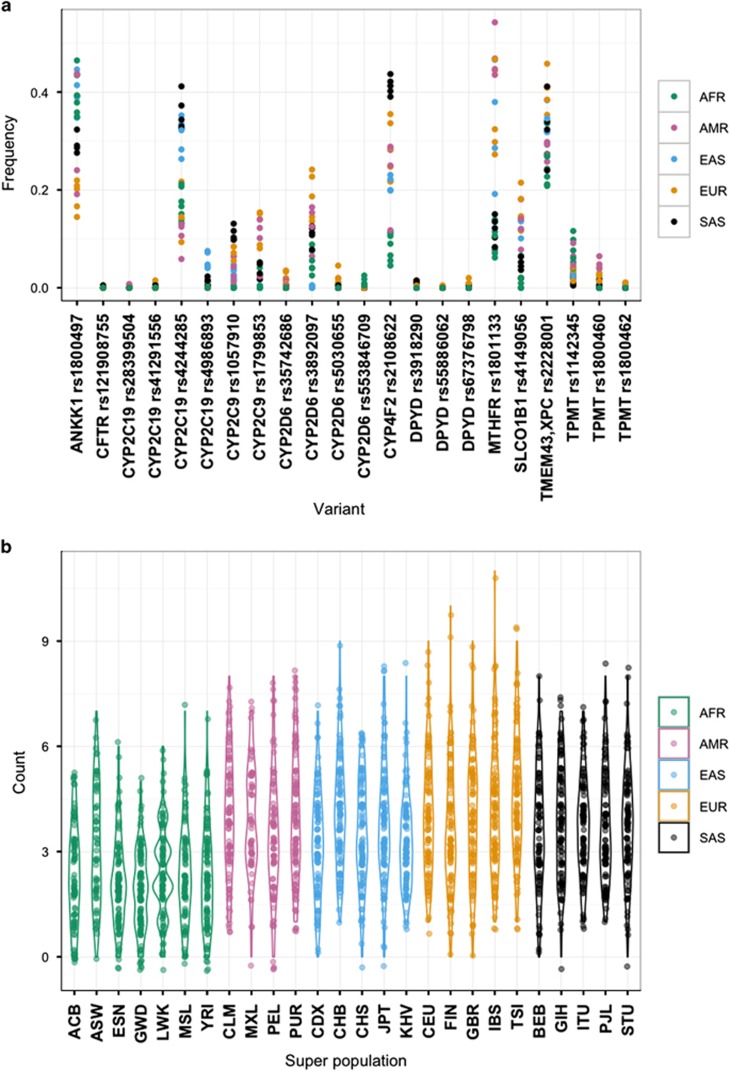
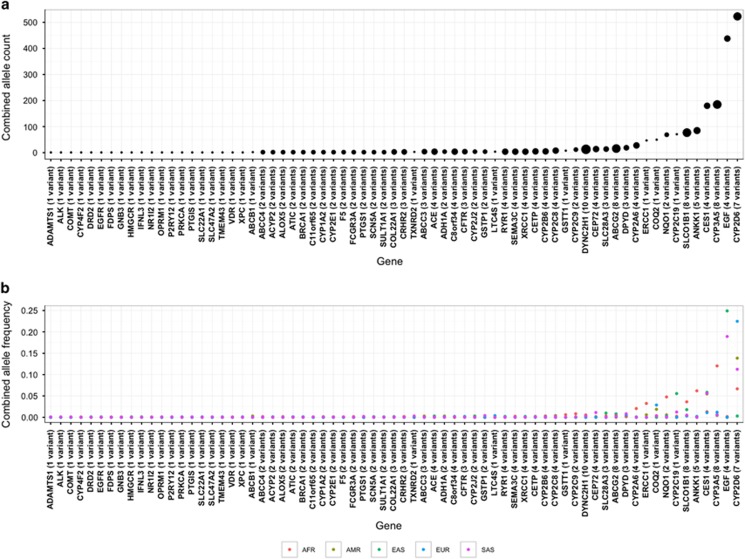
A total of 22 clinical variants were found in 11 pharmacogenes with 7 of these variants displaying global MAF ⩾5.0%. The number of clinical variants per individual varied between 0 and 11 (median 3), with 97% of individuals being carriers (Figure 2). Apart from ANKK1, the coverage of clinical pharmacogenes did not vary substantially between populations (Supplementary Figure 7). High-confidence LOF variants were found in 69 pharmacogenes (57.5%) and we detected 175 unique variants, comprising 1968 alleles (Figure 3). Individuals carried 0–5 of such LOF variants, with 55.4% of individuals being carriers, but this varied by super-population (East Asian 60.9%>African 60.1%>South Asian 60.3%>European 49.3%>Admixed 39.2%). Apart from 12 variants (6.9%), all high-confidence LOF variants were rare (global MAF<0.5%) and many of the higher frequency LOF variants allele frequencies were driven by one super-population. CYP2D6 provided the largest contribution to the LOF allele count and CYP2D6*4 (rs3892097, splice acceptor) displayed the highest global MAF (9.3%).
Figure 2.
Pharmacogenomic variants with a high level of clinical annotation (that is, PharmGKB Level 1A/B). (a) Scatterplot of allele frequencies of clinically relevant variants in the different population groups. Variants in certain genes, such as CY2C19 and CYP4F2, displayed differences in allele frequencies between super-populations. (b) Violin plot of the number of clinically relevant pharmacogenomic variants carried per individual, grouped by population, and coloured by super-population. Ninety-seven percent of the individuals in the 1000 Genomes Project carried at least one such variant (median of 3).
Figure 3.
Pharmacogenes that carried high-confidence loss-of-function (LOF) variants as designated by LOFTEE. (a) The size of the points is proportional to the number of unique LOF variants in the gene, with the cumulative allele count per gene indicated. (b) Combined allele frequencies of LOF variants per gene in each of the global super-populations. Common LOF variants were frequently driven by one super-population.
Sequencing performance and variant assessment
As our assessment of sequencing performance criteria is stringent8, no pharmacogenes were removed from subsequent analyses, and these metrics should be considered as a reflection of pharmacogenes that should be treated with caution when short-read sequencing technologies are applied. Of 120 pharmacogenes, 16 had variants located within segmental duplications, of which 50% were cytochrome P450 (CYP) genes (Supplementary Table 2). This overrepresentation of CYP genes in the segmental gene list was statistically significant (P=0.001) as CYPs only comprise 10% of the complete set.
Ten pharmacogenes (CES1, CYP2A6, CYP2B6, CYP2D6, CYP3A4, CYP4F2, FCGR3A, GSTT1, IFNL3 and SULT1A1) were extreme outliers with regards to the proportion of variants located in either the 1000GP ‘strict mask’ regions or segmental duplications (that is, >64%). These 10 pharmacogenes had a higher proportion of variants that failed the filtering steps performed by the 1000GP quality control (14.3% versus 1.9%) and more variants that were classified in this study as marginal quality variants (3.4% versus 0.1% Supplementary Figure 8 and Supplementary Table 2). Of note, none of the clinical variants (that is, PharmGKB level 1A/B) failed the 1000GP filtering or fell into our marginal variant category. Further, only four high-confidence LOF variants (SCN5A rs202196386, ABCG2 rs573803020, C8orf34 rs554409474 and SLC28A3 rs548288413) were in the marginal quality variant category. A complete list of the 110 marginal quality variants can be found in Supplementary Table 5.
Validation of our results using genomic data from external projects showed a strong correlation between the 1000GP pharmacogenomic data and results that were generated either by genotyping arrays or exome sequencing (Supplementary Figure 9). Comparison with the ExAC data showed that the allele frequencies for 10 871 variants were comparable, even though different bioinformatic analyses were employed. Previously identified array-genotyped markers (n=136) from the Human Genome Diversity Project correlated well between super-population group (R2⩾0.95) for all populations except the admixed American populations, indicating the difficulty of predicting allele frequencies in highly admixed populations.
Discussion
This study presents an extensive surveillance of pharmacogenomic variation in global populations. Analysis of these regions with current sequencing technologies was shown to be feasible in genes of relevance to drug safety and effectiveness. By assessing the full spectrum of genetic variation, the importance of rare variation in influencing the protein function of pharmacogenes was highlighted. Future pharmacogenomic approaches in clinical practice will need to develop methods to address this class of variation to ensure the maximum predictive value for diagnostic tests. Furthermore, 97% of individuals carried at least one well-established variant of pharmacogenomic relevance, indicating the valuable clinical information related to drug response and/or ADRs that can be obtained through genomic sequencing.
Summary of pharmacogenomic variation
Sequence analysis facilitated the identification of protein-coding pharmacogenomic variation across a globally representative cohort at a scope not previously feasible. The majority of the variation was made up of rare variants and singletons (~90%). Further, the relative frequency of deleterious variants is inversely correlated with allele frequency (Figure 1b), since deleterious variants are more likely to be rare.13 This was demonstrated by the high prevalence of rare missense variants in the pharmacogenes examined in this study and is in line with research involving re-sequencing of drug target genes.14 This is of particular importance to pharmacogenomics, as rare variants are an understudied class of pharmacogenomic variation15 and such low-frequency functional variants are unlikely to be adequately covered on conventional genotyping arrays. One of the pharmacogenes with a highest proportion of missense variants, SLC22A1, encodes the major hepatic uptake transporter of the antidiabetic drug, metformin.16 Over 20 SLC22A1 variants have been associated with either changes in protein function in vitro or clinical traits, such as treatment response.17 Future studies should ensure that variation in highly polymorphic pharmacogenes is adequately genotyped to ensure robust findings. Variation in conserved germline pharmacogenes may be easier to capture through conventional genotyping, although regulatory genetic variants may still have an important role.
Population genetics
The inclusion of diverse populations in genomic studies ensures that the benefits of precision medicine can be applied globally, in accordance with the ethical principle of justice.18 Common pharmacogenomic variants stratified individuals into continental super-populations, with the admixed individuals separating along clines between these clusters (Supplementary Figure 1). This was also observed for the FST analyses of synonymous variants (Supplementary Figure 2). Rare variants have been shown to be geographically localized14 and this clustering makes the design of arrays that adequately capture global variants difficult. This indicates that sequencing is the most appropriate way to assess pharmacogenomic variation across the frequency spectrum.
The pharmacogenes that displayed highly differentiated variants are important for a variety of drug classes (Table 1, Supplementary Figure 5). Consistent with the history of modern humans, most differences were observed between African populations in relation to the other super-populations (91% of such variants displayed differences involving an African population) and there were no highly differentiated variants for the European-South Asian comparisons. The most differentiated polymorphism was a missense ADH1B variant (rs1229984), which is involved in alcohol metabolism. This variant has been linked to an increased oesophageal cancer risk,19 and could contribute towards the elevated prevalence of this cancer in certain Asian populations,20 although this phenotype is multifactorial and the effect size of the variant is modest. The CYP3A4*1G allele (rs2242480), which has been associated with increased tacrolimus metabolism,21 displayed the greatest individual FST statistic (0.74 between Africans-Europeans). Unique patterns of genetic diversity for CYP3A4 in African populations have been documented,22 and this, combined with the fact that African populations have higher frequencies of active CYP3A5,23 indicate that these individuals would require higher dosages of immunosuppressive drug on average.
The angiotensin converting enzyme (ACE) gene, contained the most variants that were globally rare, yet common in one population, with four independent signals (three African and one admixed, Supplementary Table 4). ACE inhibitors display differences response profiles, with African patients displaying less effective blood pressure reduction from these medications than Europeans24 and higher risk for the ADR, angio-oedema.25 Genetic variants identified through these analyses are therefore good candidates for future pharmacogenomic research.
Three variants of potential relevance to CYP-related drug metabolism—CYP2B6 (rs28399501, 3′UTR), CYP2C8 (rs11572079, splice region) and CYP2C19 (rs181297724, missense/splice region)—were common in the Finnish, but rare in the global population. Allele frequency differences between the Finnish and other European populations have been documented for other CYP2 polymorphisms.26 Pharmacogenomic studies of related medications and cohorts should include these variants to determine clinical relevance. For example, 27% of patients in Finland were found to discontinue statins (CYP2C8 substrate) during the first year of treatment, and ADRs potentially contributed towards this statistic.27 Another notable finding in the Finnish population was the depletion of singletons in this bottlenecked population (Supplementary Figure 4), which is in line with previous genomic research in these individuals,28 and provides the opportunity to study the effect of rare pharmacogenomic variants in these individuals.
Clinical pharmacogenomics and high-confidence LOF variants
Almost every 1000GP individual (97%) carried a high evidence clinical variant (Figure 2), indicating the clinical utility of current sequencing technologies. In addition, if a patient presents with the absence of pharmacogenomic risk variants for a particular drug, the treating physician can have more confidence prescribing that medication.
Pharmacogenes relevant for anticancer agents featured prominently on this clinical list (DPYD–fluorouracil, MTFHR–methotrexate, TMEM43/XPC–cisplatin, TPMT–mercaptopurine), reflecting an active research field, with several biomarkers available for clinical intervention. This was followed by pharmacogenes involved in warfarin-related traits (CYP2C9 and CYP4F2), with CYP2C9*3 (rs1057910) also having relevance for severe skin reactions from phenytoin.29 The highly polymorphic pharmacogene, CYP2D6, along with CYP2C19, each contributed four clinical variants. CYP2D6 is important in the metabolism of many drugs, including antidepressants as well as analgesics (for example, codeine and tramadol), indicating that carriers of these clinical variants are likely to benefit from receiving these pharmacogenomic results.
The European super-population had the highest mean number of clinical variants (4.1), while the African populations had the lowest number of such variants (2.3) (Figure 2), which is similar to the findings for disease-related variants in different populations.5 This most likely represents database bias, as the clinical pharmacogenomic variants assessed in this study rely on previously published evidence. African populations have been underrepresented in past pharmacogenomic research,6, 30, 31 therefore reiterating the importance of performing research in diverse populations. These genetically diverse individuals are likely to harbour pharmacogenomic variants that are common in African populations, with similar effect sizes, but remain to be identified as being clinically relevant. As only the coding regions were assessed, these clinical carrier counts are underestimated in all populations. For example, increasing the capture region to include a more comprehensive set of transcripts incorporating untranslated regions would allow for the inclusion of additional clinical variants (for example, CYP3A5*3 and VKORC1 rs7294/rs9934438). With the addition of these variants, every individual in the 1000GP would carry a clinical variant, providing support for the use of augmented exome approaches.32
A recent study also highlighted the importance of rare variation in a predominantly European-descent cohort of patients from the eMERGE Network analyzed with the PGRNseq platform.33 This represents a significant advance in incorporating sequencing-based pharmacogenomic approaches into the clinic. Our study adds important additional support for these findings through capturing the diversity of pharmacogenomic alleles observed across the globe, surveying population genetic differences and annotating high-confidence pharmacogenomic LOF variants.
LOF variants have a marked impact on protein function, and consequently, pharmacogenomic traits. We generated a list of pharmacogenes that are impaired by LOF variants that have been annotated with a high degree of confidence, minimizing potential false positives. Of possible clinical relevance, 50% of the top 10 pharmacogenes contributing towards the high-confidence LOF allele count also contain variants with PharmGKB level 1A/B evidence (CYP2C19, SLCO1B1, ANKK1, CYP3A5 and CYP2D6). The high number of CYP2D6 poor metabolizer (PM) alleles is of relevance since poor metabolizers will not receive therapeutic benefit from pro-drugs such as codeine,34 while being placed at risk for ADRs from other medications (for example, tricyclic antidepressants).35 Although >50% of pharmacogenes possessed high-confidence LOF variants, the majority of genes were only affected by rare LOF variation. Sequencing should therefore be considered the best strategy to capture the variation in drug response phenotypes. Finally, the true number of LOF variants is also likely to be higher due to our stringent annotation strategy and because the 1000GP did not report singleton indels,6 a type of variation likely to cause frameshift mutations.
Limitations of current sequencing technologies
This study highlighted pharmacogenes that could be problematic with regards to short-read technologies (Supplementary Figure 8). Our results reiterate the difficulties associated with analyzing the CYP genes with such technologies,8, 36 although our criteria used to flag potential problematic genes could be overly strict for research purposes.8 For clinical sequencing applications, however, variation in these pharmacogenes should be confirmed via alternative methods. The inadequacy of sequencing for highly complex HLA and UGT genes also needs to be addressed since this group represents many important clinical pharmacogenes. The UGT loci play a major role in phase II drug metabolism,9 while the HLA region is important for drug hypersensitivity reactions.37 1000GP Phase 3 employed 76-101bp paired-end sequencing and many limitations will be prevented with longer read technologies. There have been attempts to address some of these issues through novel bioinformatic pipelines and individuals that have been genotyped with numerous platforms.38, 39, 40 Despite these limitations, the overall concordance between the 1000GP and external data with regards to allele frequency patterns was strong (Supplementary Figure 9).
Reference transcripts can have a substantial influence on the annotation of variants, with LOF variants being particularly difficult to assess.41, 42 For example, the important PM allele, CYP2C19*2, was not annotated as a high-confidence LOF variant in our analyses. It has also recently been shown that tools to infer pharmacogenomic alleles are currently inadequate when being used on current sequencing data and need to be improved.43 An additional limitation of only assessing the exome is that non-coding regulatory variation, which is not captured with this approach, can have an important role in pharmacogenomic phenotypes,44 highlighting one of the advantages of performing whole-genome sequencing. Finally, as this was beyond the scope of this study, a dedicated analysis of copy number of pharmacogenes is still required.
Conclusions
Sequencing technologies will continue to be used for pharmacogenomic applications in both research and clinical settings at an increasing rate. This study highlighted that this approach remains the best way to capture rare variants, which although independently rare, make up the bulk of the variation in pharmacogenes.
To facilitate clinical uptake, it will be important to address the analysis burden associated with high-throughput sequencing-related data. Developing variant interpretation systems that include drug response prediction beyond well-characterized clinical factors will help achieve this goal. Rare variants will need to be considered in such approaches, a task that will be assisted by improvements in computational prediction.
Sequencing is a globally inclusive technique, as genotypes are not restricted to a predetermined panel of variants. Our clinical analyses detected variants that were mainly relevant to anticancer agents and warfarin, suggesting literature biases. Additional robust pharmacogenomic studies using globally representative cohorts are therefore essential. Further, once sequenced, a genome can be used throughout a patient’s lifetime and can provide a constant source of medically relevant information that can be used to achieve a balance between mitigating ADRs and achieving drug efficacy.
Acknowledgments
We thank the 1000 Genomes Project Consortium for providing public access to the sequencing data analyzed in this study as well as the individuals who participated in the 1000 Genomes Project. We also thank the Canadian Pharmacogenomics Network for Drug Safety Consortium for providing support, the Canadian Institutes for Health Research for funding and Dr Britt Drögemöller for valuable discussions and scientific input regarding this study.
Footnotes
Supplementary Information accompanies the paper on the The Pharmacogenomics Journal website (http://www.nature.com/tpj)
The authors declare no conflict of interest.
Supplementary Material
References
- Lee JW, Aminkeng F, Bhavsar AP, Shaw K, Carleton BC, Hayden MR et al. The emerging era of pharmacogenomics: current successes, future potential, and challenges. Clin Genet 2014; 86: 21–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yip V, Hawcutt DB, Pirmohamed M. Pharmacogenetic markers of drug efficacy and toxicity. Clin Pharmacol Ther 2015; 98: 61–70. [DOI] [PubMed] [Google Scholar]
- Katsila T, Patrinos GP. Whole genome sequencing in pharmacogenomics. Front Pharmacol 2015; 6: 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacArthur DG, Balasubramanian S, Frankish A, Huang N, Morris J, Walter K et al. A systematic survey of loss-of-function variants in human protein-coding genes. Science 2012; 335: 823–828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genomes Project CGenomes Project CAbecasis GR Genomes Project CAuton A Genomes Project CBrooks LD Genomes Project CDePristo MA Genomes Project CDurbin RM et al. An integrated map of genetic variation from 1,092 human genomes. Nature 2012; 491: 56–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genomes Project CGenomes Project CAuton A Genomes Project CBrooks LD Genomes Project CDurbin RM Genomes Project CGarrison EP Genomes Project CKang HM et al. A global reference for human genetic variation. Nature 2015; 526: 68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos E, Doumatey A, Elkahloun AG, Shriner D, Huang H, Chen G et al. Pharmacogenomics, ancestry and clinical decision making for global populations. Pharmacogenomics J 2014; 14: 217–222. [DOI] [PubMed] [Google Scholar]
- Drogemoller BI, Wright GE, Niehaus DJ, Emsley R, Warnich L. Next-generation sequencing of pharmacogenes: a critical analysis focusing on schizophrenia treatment. Pharmacogenet Genomics 2013; 23: 666–674. [DOI] [PubMed] [Google Scholar]
- Tourancheau A, Margaillan G, Rouleau M, Gilbert I, Villeneuve L, Levesque E et al. Unravelling the transcriptomic landscape of the major phase II UDP-glucuronosyltransferase drug metabolizing pathway using targeted RNA sequencing. Pharmacogenomics J 2016; 16: 60–70. [DOI] [PubMed] [Google Scholar]
- Fu W, O'Connor TD, Jun G, Kang HM, Abecasis G, Leal SM et al. Analysis of 6,515 exomes reveals the recent origin of most human protein-coding variants. Nature 2013; 493: 216–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lek M, Karczewski K, Minikel E, Samocha K, Banks E, Fennell T et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature 2016; 536: 285–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Zhan X, Bragg-Gresham J, Kang HM, Stambolian D, Chew EY et al. Ancestry estimation and control of population stratification for sequence-based association studies. Nat Genet 2014; 46: 409–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein DB, Allen A, Keebler J, Margulies EH, Petrou S, Petrovski S et al. Sequencing studies in human genetics: design and interpretation. Nat Rev Genet 2013; 14: 460–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson MR, Wegmann D, Ehm MG, Kessner DSt, Jean P, Verzilli C et al. An abundance of rare functional variants in 202 drug target genes sequenced in 14,002 people. Science 2012; 337: 100–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drogemoller BI, Wright GE, Warnich L. Considerations for rare variants in drug metabolism genes and the clinical implications. Expert Opin Drug Metab Toxicol 2014; 10: 873–884. [DOI] [PubMed] [Google Scholar]
- Chen L, Shu Y, Liang X, Chen EC, Yee SW, Zur AA et al. OCT1 is a high-capacity thiamine transporter that regulates hepatic steatosis and is a target of metformin. Proc Natl Acad Sci USA 2014; 111: 9983–9988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arimany-Nardi C, Koepsell H, Pastor-Anglada M. Role of SLC22A1 polymorphic variants in drug disposition, therapeutic responses, and drug-drug interactions. Pharmacogenomics J 2015; 15: 473–487. [DOI] [PubMed] [Google Scholar]
- Wright GE, Koornhof PG, Adeyemo AA, Tiffin N. Ethical and legal implications of whole genome and whole exome sequencing in African populations. BMC Med Ethics 2013; 14: 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y, He Y, Xu J, Xu L, Du J, Zhu C et al. Genetic variants at 4q21, 4q23 and 12q24 are associated with esophageal squamous cell carcinoma risk in a Chinese population. Hum Genet 2013; 132: 649–656. [DOI] [PubMed] [Google Scholar]
- Cui R, Kamatani Y, Takahashi A, Usami M, Hosono N, Kawaguchi T et al. Functional variants in ADH1B and ALDH2 coupled with alcohol and smoking synergistically enhance esophageal cancer risk. Gastroenterology 2009; 137: 1768–1775. [DOI] [PubMed] [Google Scholar]
- Uesugi M, Hosokawa M, Shinke H, Hashimoto E, Takahashi T, Kawai T et al. Influence of cytochrome P450 (CYP) 3A4*1G polymorphism on the pharmacokinetics of tacrolimus, probability of acute cellular rejection, and mRNA expression level of CYP3A5 rather than CYP3A4 in living-donor liver transplant patients. Biol Pharm Bull 2013; 36: 1814–1821. [DOI] [PubMed] [Google Scholar]
- Drogemoller B, Plummer M, Korkie L, Agenbag G, Dunaiski A, Niehaus D et al. Characterization of the genetic variation present in CYP3A4 in three South African populations. Front Genet 2013; 4: 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oetting WS, Schladt DP, Guan W, Miller MB, Remmel RP, Dorr C et al. Genomewide Association Study of tacrolimus concentrations in African American kidney transplant recipients identifies multiple CYP3A5 alleles. Am J Transplant 2016; 16: 574–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewster LM, Seedat YK. Why do hypertensive patients of African ancestry respond better to calcium blockers and diuretics than to ACE inhibitors and beta-adrenergic blockers? A systematic review. BMC Med 2013; 11: 141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDowell SE, Coleman JJ, Ferner RE. Systematic review and meta-analysis of ethnic differences in risks of adverse reactions to drugs used in cardiovascular medicine. BMJ 2006; 332: 1177–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sistonen J, Fuselli S, Palo JU, Chauhan N, Padh H, Sajantila A. Pharmacogenetic variation at CYP2C9, CYP2C19, and CYP2D6 at global and microgeographic scales. Pharmacogenet Genomics 2009; 19: 170–179. [DOI] [PubMed] [Google Scholar]
- Helin-Salmivaara A, Lavikainen P, Korhonen MJ, Halava H, Junnila SY, Kettunen R et al. Long-term persistence with statin therapy: a nationwide register study in Finland. Clin Ther 2008; 30 (Pt 2): 2228–2240. [DOI] [PubMed] [Google Scholar]
- Lim ET, Wurtz P, Havulinna AS, Palta P, Tukiainen T, Rehnstrom K et al. Distribution and medical impact of loss-of-function variants in the Finnish founder population. PLoS Genet 2014; 10: e1004494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung WH, Chang WC, Lee YS, Wu YY, Yang CH, Ho HC et al. Genetic variants associated with phenytoin-related severe cutaneous adverse reactions. JAMA 2014; 312: 525–534. [DOI] [PubMed] [Google Scholar]
- Drogemoller BI, Wright GE, Niehaus DJ, Emsley RA, Warnich L. Whole-genome resequencing in pharmacogenomics: moving away from past disparities to globally representative applications. Pharmacogenomics 2011; 12: 1717–1728. [DOI] [PubMed] [Google Scholar]
- Aminkeng F, Ross CJ, Rassekh SR, Brunham LR, Sistonen J, Dube MP et al. Higher frequency of genetic variants conferring increased risk for ADRs for commonly used drugs treating cancer, AIDS and tuberculosis in persons of African descent. Pharmacogenomics J 2014; 14: 160–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patwardhan A, Harris J, Leng N, Bartha G, Church DM, Luo S et al. Achieving high-sensitivity for clinical applications using augmented exome sequencing. Genome Med 2015; 7: 71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bush WS, Crosslin DR, Obeng AO, Wallace J, Almoguera B, Basford MA et al. Genetic variation among 82 pharmacogenes: the PGRN-Seq data from the eMERGE network. Clin Pharmacol Ther 2016; 100: 160–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crews KR, Gaedigk A, Dunnenberger HM, Leeder JS, Klein TE, Caudle KE et al. Clinical Pharmacogenetics Implementation Consortium guidelines for cytochrome P450 2D6 genotype and codeine therapy: 2014 update. Clin Pharmacol Ther 2014; 95: 376–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leckband SG, Kelsoe JR, Dunnenberger HM, George ALJr., Tran E, Berger R et al. Clinical Pharmacogenetics Implementation Consortium guidelines for HLA-B genotype and carbamazepine dosing. Clin Pharmacol Ther 2013; 94: 324–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujikura K, Ingelman-Sundberg M, Lauschke VM. Genetic variation in the human cytochrome P450 supergene family. Pharmacogenet Genomics 2015; 25: 584–594. [DOI] [PubMed] [Google Scholar]
- Pirmohamed M, Ostrov DA, Park BK. New genetic findings lead the way to a better understanding of fundamental mechanisms of drug hypersensitivity. J Allergy Clin Immunol 2015; 136: 236–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Numanagic I, Malikic S, Pratt VM, Skaar TC, Flockhart DA, Sahinalp SC. Cypiripi: exact genotyping of CYP2D6 using high-throughput sequencing data. Bioinformatics 2015; 31: i27–i34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang H, Liu X, Ramirez J, Choudhury N, Kubo M, Im HK et al. Establishment of CYP2D6 reference samples by multiple validated genotyping platforms. Pharmacogenomics J 2014; 14: 564–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twist GP, Gaedigk A, Miller NA, Farrow EG, Willig LK, Dinwiddie DL et al. Constellation: a tool for rapid, automated phenotype assignment of a highly polymorphic pharmacogene, CYP2D6, from whole-genome sequences. Npj Genomic Med 2016; 1: 15007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankish A, Uszczynska B, Ritchie GR, Gonzalez JM, Pervouchine D, Petryszak R et al. Comparison of GENCODE and RefSeq gene annotation and the impact of reference geneset on variant effect prediction. BMC Genomics 2015; 16: S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy DJ, Humburg P, Kanapin A, Rivas MA, Gaulton K, Cazier JB et al. Choice of transcripts and software has a large effect on variant annotation. Genome Med 2014; 6: 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samwald M, Blagec K, Hofer S, Freimuth RR. Analyzing the potential for incorrect haplotype calls with different pharmacogenomic assays in different populations: a simulation based on 1000 Genomes data. Pharmacogenomics 2015; 16: 1713–1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson C, Cairns J, Wang L, Sinha S. Computational discovery of transcription factors associated with drug response. Pharmacogenomics J 2015; doi:10.1038/tpj.2015.74. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.