Abstract

Methane-oxidizing bacteria, aerobes that utilize methane as their sole carbon and energy source, are being increasingly studied for their environmentally significant ability to remove methane from the atmosphere. Their genomes indicate that they also have a robust and unusual secondary metabolism. Bioinformatic analysis of the Methylobacter tundripaludum genome identified biosynthetic gene clusters for several intriguing metabolites, and this report discloses the structural and genetic characterization of tundrenone, one of these metabolites. Tundrenone is a highly oxidized metabolite that incorporates both a modified bicyclic chorismate-derived fragment and a modified lipid tail bearing a β,γ-unsaturated α-hydroxy ketone. Tundrenone has been genetically linked to its biosynthetic gene cluster, and quorum sensing activates its production. M. tundripaludum’s genome and tundrenone’s discovery support the idea that additional studies of methane-oxidizing bacteria will reveal new naturally occurring molecular scaffolds and the biosynthetic pathways that produce them.
Research on natural products, genetically encoded members of a small-molecule miscellany, has repeatedly shown that investigating new patches of organismal space can reveal new chemotypes along with their associated biosynthetic machinery and regulatory elements. The increasing availability of genomic sequences has enabled the discovery of previously unreported molecular diversity through bioinformatic analyses—an approach usually called “gene-to-molecule”. In a variation on this approach, we recently characterized a quorum sensing system1 in the aerobic methane-oxidizing bacterium Methylobacter tundripaludum 21/222 that activates the expression of a co-located biosynthetic gene cluster (BGC) detected by the antiSMASH genome mining tool.3 The same quorum sensing system regulated the production of a UV-active molecule observed in the M. tundripaludum supernatant, which was the likely product of the BGC.1 Since quorum sensing systems in bacteria regulate gene expression on the basis of cell density and often control the production of molecules such as siderophores, antibiotics, and electron shuttles, we decided to further investigate the molecule, which we named tundrenone (1) (Figure 1a).4−7
Figure 1.
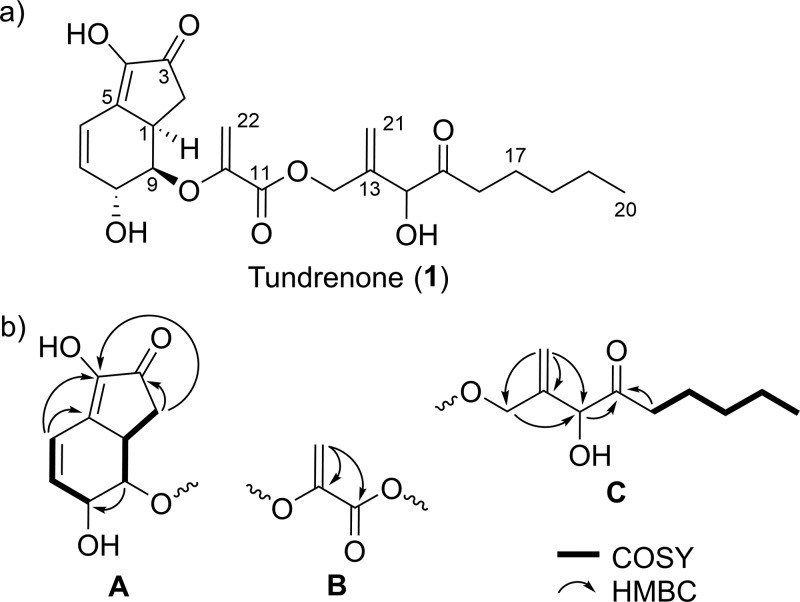
(a) Structure of tundrenone (1). (b) Partial structures assembled by 2D NMR data.
We isolated approximately 3 mg of 1 from the ethyl acetate extract of 12 L of supernatant with subsequent C18 solid-phase extraction and UV-absorbance-guided semi-preparative HPLC. The molecular formula of 1 (C22H28O8), obtained by HRMS (observed [M + H]+m/z 421.1857, theoretical [M + H]+m/z 421.1857, 0 ppm), indicated a highly oxygenated metabolite with no matches in spectral databases.8 The 13C NMR spectrum revealed numerous shielded carbon atoms, including three carbonyls—two ketones (200.9 and 209.9 ppm) and one ester (161.5 ppm)—and eight olefinic carbons between 96.9 and 149.3 ppm that accounted for seven of the nine degrees of unsaturation. There were also four oxygenated sp3-hybridized carbons with resonances between 62.1 and 77.8 ppm.
Further analysis with an array of 2D NMR experiments (gHSQC, gHMBC, gCOSY, and ROESY) led to three partial structures A–C (Figure 1b) that contained all of the carbon atoms in 1. Partial structure A consists of one ketone, two olefins, and two oxygenated sp3-hybridized carbons, all within a [4.3.0] bicyclic system. Partial structure B consists of a dehydroserine hydroxy acid with exo-methylene signals at δC 149.3 and 96.9 and δH 5.29 and 5.03. The last partial structure, C, contains four spin systems and was deduced to be a modified lipid tail functionalized with only an exo-methylene group, an alcohol, and a ketone. Partial structure C is structurally similar to ketalin, a metabolite previously isolated from Streptomyces sp. strain 1668.9 Partial structures A–C were connected to form the planar structure of 1 using key HMBC correlations. A and B were connected by the observation of an HMBC correlation between H9 and C10 to form a vinylic ether linkage. The connection between B and C was established by an HMBC correlation between H12 and C11, thus forming an ester linkage and completing the planar structure elucidation of 1.
The three contiguous stereocenters in the tetrahydroindenone core of 1 were assigned using NMR and chiroptical data and theoretical calculations. The relative configuration was determined by comparing experimental NMR data for 1 with roughly calculated 3JHH coupling constants (Figure 2) and DFT-calculated 13C chemical shifts for i, ii, iii, and iv, where the ester on the acyclic motif was simplified to a carboxylic acid (Table 1).10 The measured 3JHH for H1/H9 in d6-DMSO was 3.3 Hz, which is indicative of a cis H1/H9 configuration and is in good agreement with the calculated113JHH for cis-H1/H9-configured i and iii (1.5 Hz) versus trans-H1/H9-configured ii and iv (10.2–10.4 Hz). However, the calculated 3JHH for trans- and cis-configured H8/H9 in i and iii, respectively, could not be used to confidently resolve their relative configuration. To determine whether the relative stereochemistry of the three contiguous stereocenters in 1 mimics i or iii, the measured 13C chemical shifts for 1 were compared with the DFT-calculated Boltzmann-weighted shifts for i and iii. The latter were obtained through a highly accurate method developed by Tantillo12 that we had previously used.13 Excellent agreement between measured and calculated 13C chemical shifts of 1 and i was observed, with a corrected mean absolute deviation (CMAD) of 1.6 ppm, whereas the CMAD for iii was 5.3 ppm (Table 1). Thus, the chemical shift data strongly support a trans-configured H8/H9 and a cis-configured H1/H9.
Figure 2.
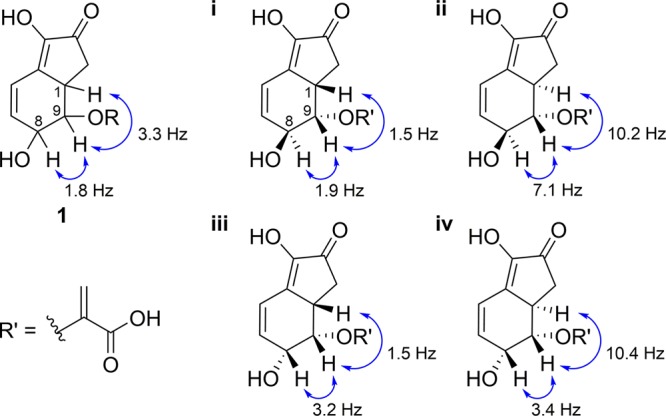
Analysis of 3JHH values [measured vs calculated (i–iv)].
Table 1. Comparison of Experimental (1) and Calculated (i, iii) 13C NMR Chemical Shifts.
|
13C NMR chemical shifts (ppm) |
|||
|---|---|---|---|
| atoma | exptl (1)b | calcd (i)c,d | calcd (iii)c,d |
| C1 | 31.0 | 35.2 | 40.2 |
| C2 | 33.7 | 34.8 | 36.4 |
| C3 | 200.9 | 199.9 | 199.0 |
| C4 | 147.5 | 144.8 | 143.7 |
| C5 | 134.6 | 134.5 | 136.5 |
| C6 | 121.7 | 123.6 | 121.1 |
| C7 | 130.0 | 132.6 | 140.5 |
| C8 | 62.1 | 62.6 | 71.3 |
| C9 | 76.2 | 73.1 | 83.3 |
| C10 | 149.3 | 148.6 | 153.2 |
| C11 | 161.5 | 162.0 | 163.7 |
| C22 | 96.9 | 97.1 | 107.1 |
| CMADe | 1.6 | 5.3 | |
| largest Δδ | 4.2 (C1) | 10.5 (C7) | |
Data in d6-DMSO.
Calculated at the SCRF-(IEFPCM/DMSO)-mPW1PW91/6-311+G(2d,p)//B3LYP/6-31+G(d,p) level of theory, scaled, and Boltzmann weighted.
The calculated chemical shift closest in magnitude to the experimental chemical shift for each atom is bolded.
CMAD is the average value of |δcalcd – δexptl|
The absolute stereochemistry of the tetrahydroindenone core was assigned by comparing the specific rotation ([α]D) for 1 with the DFT-calculated Boltzmann-weighted optical rotation for i, where the three stereogenic centers bear the S,S,S configuration. Tundrenone (1) exhibited a specific rotation of −66.4, which has the opposite sign to the calculated [α]D of i (+197).14 Thus, the measured and calculated chiroptical data strongly suggest that the absolute configuration of the three contiguous stereocenters in 1 is R,R,R. Unfortunately, all attempts to determine the configuration of the stereogenic center bearing the hydroxyl group in the fatty acid moiety failed. Attempts to convert the hydroxyl group to a Mosher ester via traditional methods or Mitsunobu conditions yielded no desired product by HRMS.
With the structure of 1 secure, we investigated the connections between 1 and the BGC co-regulated by quorum sensing in M. tundripaludum. While genetic manipulation of this strain is difficult, an in-frame null mutation was made in an annotated acyl-CoA ligase gene (tunJ, T451DRAFT_0812) in the cluster, subsequently named the tun cluster. This mutant did not produce 1 (Figure 3). Additionally, 1 was not detected in the supernatant of the acyl-homoserine lactone synthase mutant ΔmbaI but returned when this strain was grown with the addition of the quorum sensing signal 3-OH-C10-HSL. These experiments confirmed that 1 is produced in a quorum-sensing-dependent manner by the tun cluster,1 with 2.7 ± 0.7 μM detected in the supernatant of stationary-phase wild-type M. tundripaludum cultures. The ΔtunJ mutant still produces the 3-OH-C10-HSL signal (Figure S9), so loss of 1 in this strain is not due to disruption of the quorum sensing system.
Figure 3.
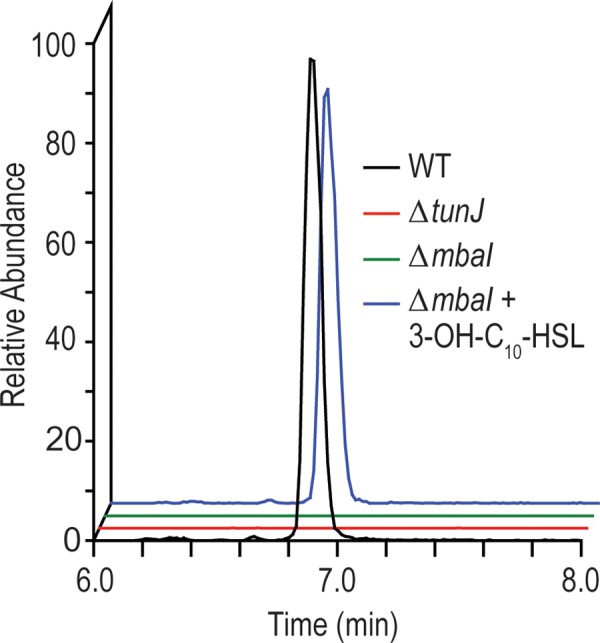
Extracted ion chromatogram (m/z 421.18–421.20) of 1 from supernatant extracts of M. tundripaludum strains, including the wild type (WT), the acyl-CoA ligase mutant (ΔtunJ), and the acyl-homoserine lactone synthase mutant (ΔmbaI) in the absence and presence of 1 μM 3-OH-C10-HSL.
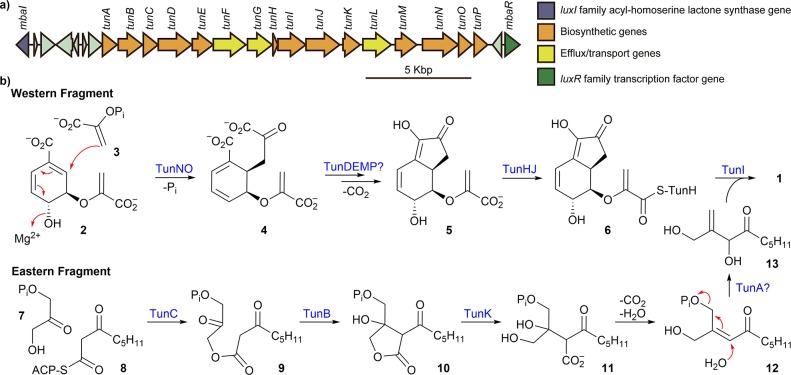
A plausible biosynthesis of 1 that largely reconciles most of the genes in the tun cluster was formulated (Figure 4). The bicyclic western fragment in 1 clearly resembles chorismate (2). Subsequent evaluation of the tun cluster revealed that TunN and TunO resemble—both in sequence and putative function—components of 2-amino-2-deoxyisochorismate synthase (ADS), which utilizes 2 and an ammonia nucleophile, and isochorismate synthase (ICS), which uses 2 and a water nucleophile.15 It is important to note that TunN and TunO are also homologous to anthranilate synthase, which is not found elsewhere in the genome. Therefore, these enzymes may also be necessary for the synthesis of aromatic amino acids in M. tundripaludum, as these bacteria are prototrophic for all amino acids. In this regard, it is important to note that tunN and tunO are constitutively expressed,1 which is consistent with an essential role. In support of this, multiple attempts to knock out either tunN or tunO were unsuccessful. On the basis of the propensity of TunN and TunO to utilize 2 as a coupling partner, a possible route to the tetrahydroindenone core in 1 may initially involve a TunNO-catalyzed SN2′ (or SN1′) transformation with 2 and phosphoenolpyruvate (3), or a related nucleophile, to generate tricarboxylate cyclohexadiene adduct 4. Following adduct formation, cyclization of 4 to give tetrahydroindenone intermediate 5 could occur via catalysis involving the redox enzymes TunDEM and the glutathione S-transferase-like enzyme TunP. Alternatively, cyclization of 4 may be facilitated by a thiamine pyrophosphate-dependent enzyme encoded elsewhere in the genome through a hypothetical formal α-keto acid decarboxylation/5-exo-trig ring closure/1,6-conjugate hydroxyl addition sequence. Activation of 5 for coupling with the eastern fragment (13) (vide infra) may involve TunJ-activated TunH–thioester adduct 6 formed via TunHJ. The supernatant of the ΔtunJ mutant strain contains both 5 and 13, thus supporting this hypothesis (Figures S10 and S11).
Figure 4.
(a) Annotated tun biosynthetic gene cluster. (b) Proposed biogenesis of 1.
The biogenesis of the acyclic eastern fragment of 1 plausibly begins through formation of 9 via condensation of dihydroxyacetone phosphate (7) with β-keto thioester 8 by TunC, which is similar to AfsA and related enzymes that link 7 and an activated β-keto acid en route to bacterial butyrolactone signals, such as A factor.16 An intramolecular aldol-like cyclization of 9 catalyzed by the δ-aminolevulinic acid dehydratase homologue TunB generates lactone 10. The tun cluster contains a gene encoding TunK, a homologue of the quorum-quenching enzyme AidH, a lactonase that hydrolyzes acyl-homoserine γ-lactones.17 Thus, it is possible that TunK hydrolyzes the lactone in 10 to form β-keto acid 11. Subsequent decarboxylation and β-hydroxyl elimination on 11 followed by SN2′ displacement of the phosphate group with water on 12, possibly catalyzed by haloacid dehydratase homologue TunA, would afford β,γ-unsaturated ketone 13. The final step involves coupling of 13 with TunJ-activated and ACP-bound 6, presumably through the action of condensation enzyme TunI, to generate 1. Further empirical investigation will be needed to elaborate the postulated biosynthesis of the western and eastern fragments.
This initial foray into a gene-to-molecule study on a methane-oxidizing bacterium that relies on methane as the sole carbon and energy source turned up both significant results and promising avenues for future research. In the gene-to-molecule realm, it led to a highly unusual molecule and associated biosynthetic puzzles. Tundrenone (1) incorporates a highly modified chorismate moiety that has not been observed previously. Chorismate serves as the starting material for metabolites ranging from the aromatic amino acids (phenylalanine, tyrosine, and tryptophan) to the plant hormone salicylic acid, large numbers of plant alkaloids, and many metabolites. However, none feature the bicyclic modification seen in 1. The formation is putatively attributed to the action of TunNO, an enzyme incorporating features of known enzymes along chorismate-containing biosynthetic pathways but with a carbon-centered nucleophile. In addition to these structural and mechanistic surprises, the quorum-sensing-regulated production of 1 suggests an as yet unknown functional role.4,18 This project has created both molecular and genetic tools that will be essential in defining the unknown function(s). Finally, further mechanistic understanding will allow the possibility to create a synthetic biology platform leading to microbial factories that utilize methane, the major component of natural gas, as a feedstock for the synthesis of high-value small molecules.19
Acknowledgments
This work was supported by the U.S. Department of Energy, Office of Science, Office of Biological and Environmental Research (Award DE-SC-0010556 to M.E.L.) and the U.S. National Institutes of Health (K99 GM118762 to A.W.P., P41 GM103484 and S10 RR029121 to P.C.D., R01 GM059026 to E.P.G., and R01 AT009874 to J.C.). D.P. was supported by the Deutsche Forschungsgemeinschaft (DFG) (Grant PE 2600/1). Quantum-chemical calculations were run on the Odyssey cluster supported by the FAS Division of Science, Research Computing Group at Harvard University. We thank Amy Schaefer at the University of Washington for helpful discussions and Marina Kalyuzhnaya at San Diego State University for providing resources for strain growth in San Diego.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/jacs.7b12240.
Experimental and computational procedures, NMR spectra, CD spectrum, chromatograms, Cartesian coordinates, and additional computational data (PDF)
Author Contributions
# A.W.P., E.M., and T.R.R. contributed equally to this work.
The authors declare no competing financial interest.
Notes
To facilitate future compound dereplication, MS/MS spectra were annotated and included in the Global Natural Product Social Molecular Networking database (GNPS)8 under MassIVE ID MSV000081637 (https://massive.ucsd.edu/).
Supplementary Material
References
- Puri A. W.; Schaefer A. L.; Fu Y.; Beck D. A.; Greenberg E. P.; Lidstrom M. E. J. Bacteriol. 2017, 199, e00773-16. 10.1128/JB.00773-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalyuzhnaya M. G.; Lamb A. E.; McTaggart T. L.; Oshkin I. Y.; Shapiro N.; Woyke T.; Chistoserdova L. Genome Announc. 2015, 3, e00103-15. 10.1128/genomeA.00103-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blin K.; Wolf T.; Chevrette M. G.; Lu X.; Schwalen C. J.; Kautsar S. A.; Suarez Duran H. G.; de Los Santos E. L. C.; Kim H. U.; Nave M.; Dickschat J. S.; Mitchell D. A.; Shelest E.; Breitling R.; Takano E.; Lee S. Y.; Weber T.; Medema M. H. Nucleic Acids Res. 2017, 45, W36. 10.1093/nar/gkx319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandara H. M. H. N.; Lam O. L. T.; Jin L. J.; Samaranayake L. Crit. Rev. Microbiol. 2012, 38, 217. 10.3109/1040841X.2011.652065. [DOI] [PubMed] [Google Scholar]
- Duerkop B. A.; Varga J.; Chandler J. R.; Peterson S. B.; Herman J. P.; Churchill M. E. A.; Parsek M. R.; Nierman W. C.; Greenberg E. P. J. Bacteriol. 2009, 191, 3909. 10.1128/JB.00200-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierson L. S. III; Keppenne V. D.; Wood D. W. J. Bacteriol. 1994, 176, 3966. 10.1128/jb.176.13.3966-3974.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seyedsayamdost M. R.; Chandler J. R.; Blodgett J. A. V.; Lima P. S.; Duerkop B. A.; Oinuma K.-I.; Greenberg E. P.; Clardy J. Org. Lett. 2010, 12, 716. 10.1021/ol902751x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M.; Carver J. J.; Phelan V. V.; Sanchez L. M.; Garg N.; Peng Y.; Nguyen D. D.; Watrous J.; Kapono C. A.; Luzzatto-Knaan T.; Porto C.; Bouslimani A.; Melnik A. V.; Meehan M. J.; Liu W.-T.; Crüsemann; Boudreau P. D.; Esquenazi E.; Sandoval-Calderón; Kersten R. D.; Pace L. A.; Quinn R. A.; Duncan K. R.; Hsu C.-C.; Floros D. J.; Gavilan R. G.; Kleigrewe K.; Northen T.; Dutton R. J.; Parrot D.; Carlson E. E.; Aigle B.; Michelsen C. F.; Jelsbak L.; Sohlenkamp C.; Pevzner P.; Edlund A.; McLean J.; Piel J.; Murphy B. T.; Gerwick L.; Liaw C.-C.; Yang Y.-L.; Humpf H.-U.; Maansson M.; Keyzers R. A.; Sims A. C.; Johnson A. R.; Sidebottom A. M.; Sedio B. E.; Klitgaard A.; Larson C. B.; Boya P C. A.; Torres-Mendoza D.; Gonzalez D. J.; Silva D. B.; Marques L. M.; Demarque D. P.; Pociute E.; O’Neill E. C.; Briand E.; Helfrich E. J. N.; Granatosky E. A.; Glukhov E.; Ryffel F.; Houson H.; Mohimani H.; Kharbush J. J.; Zeng Y.; Vorholt J. A.; Kurita K. L.; Charusanti P.; McPhail K. L.; Nielsen K. F.; Vuong L.; Elfeki M.; Traxler M. F.; Engene N.; Koyama N.; Vining O. B.; Baric R.; Silva R. R.; Mascuch S. J.; Tomasi S.; Jenkins S.; Macherla V.; Hoffman T.; Agarwal V.; Williams P. G.; Dai J.; Neupane R.; Gurr J.; Rodriguez A. M. C.; Lamsa A.; Zhang C.; Dorrestein K.; Duggan B. M.; Almaliti J.; Allard P.-M.; Phapale P.; Nothias L.-F.; Alexandrov T.; Litaudon M.; Wolfender J.-L.; Kyle J. E.; Metz T. O.; Peryea T.; Nguyen D.-T.; VanLeer D.; Shinn P.; Jadhav A.; Müller R.; Waters K. M.; Shi W.; Liu X.; Zhang L.; Knight R.; Jensen P. R.; Palsson B. Ø.; Pogliano K.; Linington R. G.; Gutiérrez M.; Lopes N. P.; Gerwick W. H.; Moore B. S.; Dorrestein P. C.; Bandeira N. Nat. Biotechnol. 2016, 34, 828. 10.1038/nbt.3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- König W. A.; Drautz H.; Zähner H. Liebigs Ann. Chem. 1980, 1980, 1384. 10.1002/jlac.198019800906. [DOI] [Google Scholar]
- These truncated diastereomers were modeled in order to mitigate complexity and computational burden arising from the flexible alkyl chain in 1. We do not expect the absence of the pendant chain to exhibit a significant impact on the ability to compare experimental and calculated data.
- Haasnoot C. A. G.; de Leeuw F. A. A. M.; Altona C. Tetrahedron 1980, 36, 2783. 10.1016/0040-4020(80)80155-4. [DOI] [Google Scholar]
- a Lodewyk M. W.; Soldi C.; Jones P. B.; Olmstead M. M.; Rita J.; Shaw J. T.; Tantillo D. J. J. Am. Chem. Soc. 2012, 134, 18550. 10.1021/ja3089394. [DOI] [PubMed] [Google Scholar]; b Lodewyk M. W.; Siebert M. R.; Tantillo D. J. Chem. Rev. 2012, 112, 1839. 10.1021/cr200106v. [DOI] [PubMed] [Google Scholar]; c Scaling factors:Pierens G. K. J. Comput. Chem. 2014, 35, 1388. 10.1002/jcc.23638. [DOI] [PubMed] [Google Scholar]
- Kim K. H.; Ramadhar T. R.; Beemelmanns C.; Cao S.; Poulsen M.; Currie C. R.; Clardy J. Chem. Sci. 2014, 5, 4333. 10.1039/C4SC01136H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The difference in magnitude between the experimental and calculated [α]D is likely attributed to the dilute nature of the experimental sample and the truncation of the tail in i.
- He Z.; Stigers Lavoie K. D.; Bartlett P. A.; Toney M. D. J. Am. Chem. Soc. 2004, 126, 2378. 10.1021/ja0389927. [DOI] [PubMed] [Google Scholar]
- a Ando N.; Matsumori N.; Sakuda S.; Beppu T.; Horinouchi S. J. Antibiot. 1997, 50, 847. 10.7164/antibiotics.50.847. [DOI] [PubMed] [Google Scholar]; b Kato J. Y.; Funa N.; Watanabe H.; Ohnishi Y.; Horinouchi S. Proc. Natl. Acad. Sci. U. S. A. 2007, 104, 2378. 10.1073/pnas.0607472104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mei G.-Y.; Yan X.-X.; Turak A.; Luo Z.-Q.; Zhang L.-Q. Appl. Environ. Microbiol. 2010, 76, 4933. 10.1128/AEM.00477-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster M.; Sexton D. J.; Diggle S. O.; Greenberg E. P. Annu. Rev. Microbiol. 2013, 67, 43. 10.1146/annurev-micro-092412-155635. [DOI] [PubMed] [Google Scholar]
- Kalyuzhnaya M. G.; Puri A. W.; Lidstrom M. E. Metab. Eng. 2015, 29, 142. 10.1016/j.ymben.2015.03.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.