Abstract
Preharvest sprouting (PHS), the germination of grain on the mother plant under cool and wet conditions, is a recurring problem for wheat farmers worldwide. α-amylase enzyme produced during PHS degrades starch resulting in baked good with poor end-use quality. The Hagberg-Perten Falling Number (FN) test is used to measure this problem in the wheat industry, and determines how much a farmer's wheat is discounted for PHS damage. PHS tolerance is associated with higher grain dormancy. Thus, breeding programs use germination-based assays such as the spike-wetting test to measure PHS susceptibility. Association mapping identified loci associated with PHS tolerance in U.S. Pacific Northwest germplasm based both on FN and on spike-wetting test data. The study was performed using a panel of 469 white winter wheat cultivars and elite breeding lines grown in six Washington state environments, and genotyped for 15,229 polymorphic markers using the 90k SNP Illumina iSelect array. Marker-trait associations were identified using the FarmCPU R package. Principal component analysis was directly and a kinship matrix was indirectly used to account for population structure. Nine loci were associated with FN and 34 loci associated with PHS based on sprouting scores. None of the QFN.wsu loci were detected in multiple environments, whereas six of the 34 QPHS.wsu loci were detected in two of the five environments. There was no overlap between the QTN detected based on FN and PHS, and there was little correlation between the two traits. However, both traits appear to be PHS-related since 19 of the 34 QPHS.wsu loci and four of the nine QFN.wsu loci co-localized with previously published dormancy and PHS QTL. Identification of these loci will lead to a better understanding of the genetic architecture of PHS and will help with the future development of genomic selection models.
Keywords: preharvest sprouting, falling number, seed dormancy, wheat, association mapping
Introduction
Rainy conditions before harvest can cause mature grain to initiate germination while still on the mother plant (Rodríguez et al., 2015). This problem, called preharvest sprouting (PHS), occurs in many cereal crops such as wheat (Triticum aestivum L.), barley (Hordeum vulgare L.), and sorghum (Sorghum bicolor) (Paterson and Sorrells, 1990; Gualano et al., 2007; Ullrich et al., 2009). Germination is associated with α-amylase enzyme induction in order to mobilize starch reserves for use by the growing embryo (Clarke et al., 1984). This α-amylase induction during PHS in wheat grain leads to problems with poor end-use quality due to starch degradation. Thus, sprouted wheat grain is discounted in the marketplace.
The degree of PHS tolerance is associated with multiple environmental and genetic factors. Grain dormancy can account for up to 60% of variation in PHS tolerance, although spike morphology and epicuticular waxes are also associated with PHS response (King, 1984; King and Richards, 1984; DePauw and McCaig, 1991; King and von Wettstein-Knowles, 2000). Dormant seeds cannot germinate under favorable environmental conditions (light, moisture, and temperature) (Bewley and Black, 1994). Seeds are most dormant at physiological maturity, and then lose dormancy through a period of dry storage called after-ripening (Finkelstein et al., 2008). Dormancy can also be broken by moist chilling (called cold stratification) or seed coat scarification (Paterson et al., 1989; Finkelstein et al., 2008). The degree of wheat grain dormancy and PHS tolerance depends on environmental conditions both before and after the grain reaches physiological maturity. Grain dormancy is higher when the mother plant is exposed to cooler conditions during the maturation phase of grain development (Nakamura et al., 2011). Conversely, cold stratification of mature grain breaks dormancy. If rain and cold temperatures occur after physiological maturity, seed dormancy can be broken through cold stratification and grain is more likely to sprout. Thus, cold temperatures have opposite effects depending on whether they occur prior to or after the maturation date. Because seed dormancy is also broken through dry after-ripening, wheat also becomes more likely to sprout the longer unharvested mature grain stands dry in the field before it rains (Gerjets et al., 2010). Thus, variation in PHS tolerance also depends on when the rain occurred relative to maturation date. Higher PHS tolerance is associated with genetic loci that increase grain dormancy including red kernel color and the synthesis or response to the dormancy-inducing hormone ABA (abscisic acid) (Walker-Simmons, 1987; Flintham, 2000; Warner et al., 2000; Schramm et al., 2010; Himi et al., 2011; Jaiswal et al., 2012; Kulwal et al., 2012; Martinez et al., 2016). Since lack of red kernel color reduces dormancy, other genetic mechanisms supporting PHS tolerance must be identified and selected in wheat with white kernels.
The spike-wetting test is often used to assess PHS tolerance in breeding programs because it examines multiple variables affecting PHS (Paterson et al., 1989). In order to control for variation due to maturity date, intact spikes are harvested from the field at physiological maturity and allowed to dry after-ripen for the same number of days (5–14 days) before placing them under a greenhouse misting system. The use of intact spikes takes into account the effect of awns, erectness of the spike, gloom tightness, and head type (Pool and Patterson, 1958; Ibrahim, 1966; Hong, 1979; King and Richards, 1984). Spikes are assigned a sprouting score using a scale based on visible germination and post-germinative growth throughout the spike (McMaster and Derera, 1976). The sprouting scores of McMaster and Derera (1976) actually reflect three biological stages: (1) initial germination or the first appearance of roots (scores 1-5), (2) coleoptile emergence (scores 6-8), and (3) seedling growth (scores 9-10). Other methods for assessing PHS tolerance include plating assays to assess degree of seed dormancy, the Falling Numbers test (see below), and variations on the spike-wetting test (Paterson et al., 1989; Kumar et al., 2009; Zhang et al., 2014; Jiménez et al., 2016; Zhou et al., 2017).
Damage due to α-amylase induction from PHS is measured in the wheat industry using the Hagberg-Perten Falling Numbers (FN) test (Perten, 1964). During the FN test, a slurry of wheat meal and water is mixed while being heated to 100°C. Then the FN machine measures the time in seconds (sec) that it takes for a stirrer to fall through the slurry. The higher the α-amylase level, the thinner the slurry, allowing the stirrer to fall faster resulting in a lower FN. If the FN is below 300 s, then the farmer receives significantly less money for his/her grain. Because some studies have shown a significant correlation between FN and sprouting scores, one might expect spike-wetting tests and FN to map similar PHS tolerance loci (reviewed by DePauw et al., 2012). However, this has not been directly tested.
Here we present a genome-wide association mapping study (GWAS) for PHS tolerance in white wheat based on sprouting scores and FN. A large panel of 469 white wheat lines representing six northwestern U.S. breeding programs was examined over multiple environments in order to characterize the genetic architecture of PHS tolerance/susceptibility. The goal was to identify quantitative trait nucleotides (QTN) associated with PHS tolerance in white wheat breeding programs, while examining the phenotypic connection between FN and spike-wetting test scores.
Materials and methods
Plant materials
This study used a mapping panel of 469 winter wheat accessions, consisting of advanced soft white breeding lines and cultivars from US Pacific Northwest breeding programs (Supplementary Table 1; Supplementary Figure 1). The accessions included 36% club (T. aestivum ssp compactum) genotypes with compact spike morphology and 64% soft white genotypes with lax spike morphology. The same panel was recently analyzed for soil acidity, aluminum tolerance, Cephalosporium stripe resistance, and stripe rust resistance (Froese and Carter, 2016; Froese et al., 2016; Liu et al., 2017).
Field research environments
The mapping panel was grown as 1.5 m long headrows at Central Ferry, WA in 2014, 2015, and 2016 (C14, C15, C16), or as 8 m2 plots at the Washington State University Spillman Agronomy Farm in Pullman, WA in 2014, 2015, and 2016 (P14, P15, P16) using recommended agronomic practices for those locations. The panel was also grown as headrows in Pullman, WA in 2013 (P13). Heading dates from the Pullman 2014 environment were determined after 50% of the plot reached full spike emergence from the boot. Table 1 lists the planting dates and harvest dates for each environment. Spikes were harvested at physiological maturity, right after the peduncle turned yellow, for the spike-wetting test (see below) only in Pullman 2014, 2015, and 2016 and Central 2014 and 2015.
Table 1.
Environments tested for preharvest sprouting traits. For the (A) Falling Numbers test (seconds) and the (B) spike-wetting test (sprouting score), planting dates, harvest dates, and general statistics are reported.
| Locationa | Year | Planting Date | Harvest dateb | Rain event precipitation | Rain event temperature | nc | t repd | Mean ± SDe | Range (min/max) | |
|---|---|---|---|---|---|---|---|---|---|---|
| PM | HM | cm | °C | |||||||
| (A) | ||||||||||
| Pullman | 2013 | –f | – | Aug 15, 2013 | 0.43 | 22 ± 3 | 459 | 1 | 379 ± 55 | 88/504 |
| Central Ferry | 2014 | Oct 2, 2013 | – | Aug 6, 2014 | 1.91 | 26 ± 5 | 458 | 2 | 331 ± 39 | 202/538 |
| Pullman | 2015 | Oct 8, 2014 | – | Aug 1, 2015 | 0.00 | – | 464 | 2 | 326 ± 31 | 187/410 |
| Central Ferry | 2015 | Oct 1, 2014 | – | Jul 31, 2015 | 3.80 | 31 ± 2 | 397 | 2 | 389 ± 55 | 111/537 |
| Central Ferry | 2016 | Oct 13, 2015 | – | Jul 14, 2016 | 1.80 | 71 ± 2 | 426 | 2 | 347 ± 54 | 154/538 |
| (B) | ||||||||||
| Pullman | 2014 | Oct 10, 2013 | Jul 11–18, 2014 | – | – | – | 427 | 5 | 3.9 ± 1.9 | 1/10 |
| Central Ferry | 2014 | Oct 2, 2013 | Jun 30–Jul 8, 2014 | – | – | – | 230 | 5 | 4.2 ± 1.7 | 1/9 |
| Pullman | 2015 | Oct 8, 2014 | Jul 1–10, 2015 | – | – | – | 416 | 5 | 4.05 ± 2.2 | 1/10 |
| Central Ferry | 2015 | Oct 1, 2014 | Jun 15–25, 2015 | – | – | – | 275 | 5 | 5.8 ± 2.3 | 1/10 |
| Pullman | 2016 | Oct 12, 2015 | Jul 15–22, 2016 | – | – | – | 437 | 5 | 6.5 ± 1.9 | 1/10 |
Pullman, WA and Central Ferry, WA. Bold environments had both the FN and spike-wetting test conducted whereas the other environments had either the FN or spike-wetting test conducted.
FN samples were harvested at harvest maturity (HM) whereas spike-wetting test samples were harvested at physiological maturity (PM).
Number (n) of accessions harvested and conducted in the FN and sprouting tests.
Technical (t) replicates used for each test and environment.
Mean and standard deviation (SD) were calculated; Sprouting scores from Day 5 are reported.
Planting date was not recorded.
Reduced FN was examined in field-grown material after PHS induced either by natural or artificial rain events. PHS-inducing natural rain events occurred after physiological maturity in Pullman 2013 and Central Ferry 2016. Therefore, these two environments were not used for spike-wetting tests, since the material was already sprouted. In Pullman 2013, rain occurred over 3 consecutive days with precipitation amounts of 0.38, 0.025, and 0.025 cm, and high temperatures of 22.8, 18.2, and 23.8°C, respectively (AgWeatherNet, 2016 weather.wsu.edu). Central Ferry 2016 received 1.8 cm of precipitation over 4 days with an average maximum temperature of 17°C. Artificial rain was used to induce PHS in Central Ferry 2014 and 2015 using overhead sprinkler irrigation at 2 weeks past the average physiological maturity date of the trial (precipitation = 1.91, 3.8 cm, average maximum temperature = 26 and 31°C, respectively). Approximately, 0.64 cm was applied daily over 3 days in order to induce mild sprouting. Pullman 2015 was included in the analysis as a “no event” control because there was no natural or artificial rain event after physiological maturity. Grain was harvested from plots 2–3 weeks after physiological maturity for FN tests (see below) when grain moisture was <12%. In Pullman, single plot replicates were harvested using a Wintersteiger Classic small plot combine (Wintersteiger Ag, Ried im Innkreis, Austria). In Central Ferry, one headrow was hand-harvested with a sickle per accession and machine threshed using the Vogel headrow thresher (Bill's Welding, Pullman, WA). All harvested grain was cleaned of chaff using a gravity cleaner.
Preharvest sprouting evaluation
Spike-wetting tests were used to evaluate preharvest sprouting tolerance of field samples (Anderson et al., 1993). Intact spikes were hand-harvested from the field at physiological maturity, and allowed to dry after-ripen for the 5 days before storing at −15°C to maintain dormancy until tests were conducted (within 2–4 months). A representative set of spikes were tested for moisture content at physiological maturity, following 5 d of after-ripening, and after storage at −15°C, and found to be an average of 31, 14, and 10%, respectively. Spike-wetting tests were conducted in a greenhouse with a 16 h day/8 h night photoperiod and 22–25°C day and 16°C night temperature. Supplemental lighting was used to maintain the photoperiod with a light intensity of 300–400 μmol/m2/s. Spikes were misted for 6 s every minute. The rare moldy spikes were thrown out of the experiment. Sprouting scores based on the McMaster and Derera (1976) 1–10 scale were determined every 24 h for 7 days, except that the “11” value was not used. Note that no sprouting was observed until day 3 of each experiment. Since the greenhouse misting system could test a maximum of 194 genotypes at a time, each environment had to be tested over multiple weeks. Two PHS tolerant controls, “Brevor” and “Clark's Cream,” and two PHS susceptible controls, “Greer” and “Bruneau,” were included in every experiment as a check for consistency (Walker-Simmons, 1987; Tuttle et al., 2015). Spikes were arranged in a randomized order, including five technical replicates (i.e., five spikes) for each genotype.
Analyses of the spike-wetting tests were performed using sprouting scores and a sprouting index designed to give more weight to earlier than later sprouting. Sprouting index (SI) was calculated as (7 × sday1 + 6 × sday2 … + 1 × sday7)/(7 × n) where s is the sprouting score on each day and n is the maximum sprouting score. SI ranged from 0 to 1, where an SI of 1 indicated that the spike reached 100% highly sprouted by day 1 of misting. For the day 3, 4, 5, 6, and 7 sprouting scores and for the SI of each accession, best linear unbiased predictors (BLUPs) were calculated within each environment over the five technical replicates using the MIXED procedure in SAS/STAT v9.4 (Piepho et al., 2008). Furthermore, the week the accession was tested and the tray location in the misting system were used as covariates in the model and accessions were treated as random effects.
Falling number evaluation
The Hagberg-Perten FN test was conducted using cleaned machine-threshed grain from Central Ferry, WA in 2014, 2015, and 2016 and from Pullman, WA in 2013 and 2015 (Table 1). FN can gradually increase during storage at higher temperatures (Ji and Baik, 2016). Grain was stored in sealed containers at −15°C to reduce problems with increasing FN. The FN test was conducted according to the ICC standard No. 107/1 (1995) and the AACC Method 56-81.03A (1999) expect that a 25 g sample was used to represent a plot or headrow rather than a 250 g sample used to represent a field. Twenty-five grams of grain was ground to meal using a Udy Cyclone Sample Mill with a 0.5 mm screen, and stored in air-tight 2 oz jars. Meal moisture content was averaged over four random samples, and applied to a subset of 48 samples. The sample weight used for the test was adjusted for moisture in order to be equivalent to 7 g of meal at 14% moisture. After 25 mL of distilled water was added to a sample, it was placed in a shaker for 5 s, then placed in a Perten Falling Number machine (Model 1600 or 1700). The FN machine determines the time needed for a stirrer to fall to the bottom of the tube after stirring and heating the samples for 60 s (minimum FN is 60 s). A lower FN is indicative of more α-amylase digestion, leading to lower gelling capacity. The FN was corrected for an altitude of 2500 ft (762 m) using FGIS Directive 9180.3 (2009). The material was examined using two technical replicates per accession with the exception of Pullman 2013, which only had one technical replicate. Each technical replicate was run on different days in 2014, within 5 min of one another in 2015, and side-by-side in 2016.
BLUPs for FN were also calculated over the artificial rain environments, the natural rain environments, and the no-rain event environment. Due to each year and environment having a different rain or no-rain event, we did not analyze BLUPs over all years or all environments (Supplementary Figure 2). An analysis of variance between accessions was performed using the MIXED procedure in SAS/STAT v9.4. Covariates were added to the analysis when relevant and included the individual who ran the test, the FN Machine used, and the seed-moisture sample subset (sets of 48 milled and tested together).
Spearman's significant rank correlations between the FN and the sprouting scores were conducted using the CORR procedure in SAS/STAT v9.4 (Supplementary Table 2). Since this is an association panel, the genotypic repeatability (R2), rather than the heritability (H2), was calculated using the lme4 package v1.1-13 in R (Campbell and Lipps, 1998; Bates et al., 2015). For the repeatability calculations, genotypes and covariates were considered to be random effects, whereas FN, sprouting score, or SI was used as the dependent variable.
Genotyping
DNA was harvested and extracted as described in Froese and Carter (2016). Extracted DNA was genotyped using the Illumina Infinium iSelect 90K SNP array, and polymorphic markers were identified and curated using GenomeStudio v2011.1 (Illumina) (Wang et al., 2014). Monomorphic markers were filtered out based on the criteria of only having 0 or 1 accession with an alternate allele (out of 469). Markers with 20% or more missing data and minor allele frequency (MAF) <5% were excluded from the analysis. A consensus map consisting of SNP markers were used to align chromosome locations of polymorphic markers (Wang et al., 2014). Genetic locations of unmapped (unk for unknown) markers were cross referenced with the GrainGenes database and are reported, without reference to a cM position on a chromosome (www.graingenes.org). Missing values for markers with published locations were imputed using default parameters in BEAGLE v3.3.2 (Browning and Browning, 2016). This resulted in a total of 15,229 polymorphic markers, of which 12,681 had known locations covering all chromosomes.
Genome-wide association study
The GAPIT R package identified three principal component sub-groups associated with market class and breeding program of origin in the mapping panel (Tang et al., 2016). Variances captured by the first three principal components (PCs) accounted for 29.6% of the total variance among the genotypes (Liu et al., 2017). In order to account for the presence of population structure, the top three PCs were fitted into the model as fixed effects (Supplementary Figure 1).
A portion of the data were analyzed using multiple statistical models, and the best statistical method selected based on how the observed p-values exceeded the null expectation on the Q-Q plot from GAPIT and FarmCPU (Lipka et al., 2012; Liu et al., 2016; Tang et al., 2016). Using the Pullman 2014 FN and sprouting score phenotypic data, a general linear model (GLM), mixed linear model (MLM), compressed mixed linear model (CMLM), SUPER model, and FarmCPU model were compared (Zhang et al., 2010; Wang et al., 2014; Liu et al., 2016; Supplementary Figure 3). The results indicated that the FarmCPU model performed better than the other models. All subsequent genome-wide association analyses used only the FarmCPU model.
FarmCPU default parameters were used except that the “optimum” bin method with default range and interval parameters was used instead of the “static” method. The FN trait least squares means (LSMeans) and the sprouting score BLUPs were used as dependent variables in this GWAS (Supplementary Figure 3). Markers were identified as significantly associated with the trait after a 1% Bonferroni multiple test correction (p < 2.85E-07; −log10(p) > 6.55).
The proportion of explained phenotypic variance was calculated as follows:
where yi is observed phenotype value, is the estimated phenotype value from a multiple linear regression model that was fitted to all significant SNPs as an independent variable with fixed effect.
Linkage disequilibrium (LD) was calculated using JMP software v6.0 (SAS, Cary, NC). LD of significant markers were used to estimate boundaries of potential quantitative trait loci (QTL) using the criteria of LD (R2 > 0.2), chromosome location (cM) based on Wang et al. (2014), correlation between markers, and marker-trait information among the significant markers. For each designated QTL, the marker with the strongest association with either FN or sprouting scores was reported. Criteria for a strong association include: (a) phenotypic variation explained by the marker (r2); (b) allelic effect; and (c) marker p value.
Tolerant nucleotides of each significant marker were used to determine the pyramiding effect of PHS tolerant loci. A linear model regression was applied to the phenotypic estimates and number of favorable loci per accession. “Favorable” loci nucleotides were those that lowered the sprouting score or increased the FN. Pearson's correlation coefficients were calculated between the trait and number of favorable loci using the “cor” function in R v.3.2.5.
Comparison of QTN locations with previously reported PHS genes and QTL
A comparison of identified QTN with previous studies was performed using the integrated map of Maccaferri et al. (2015) that includes SSR markers, 9k SNP markers, 90k SNP markers, Synthetic × Opata DH GBS markers, and the Diversity Array Technology markers (Supplementary Table 3; Akbari et al., 2006; Cavanagh et al., 2013; Saintenac et al., 2013; Wang et al., 2014). Maccaferri et al. (2015) converted distances in cM into relative % length distances by dividing them by total chromosome length. The approximate relative positions of QTN were estimated based on known marker positions. Note that the LD of QTL from other studies were not recalculated.
Results
Environmental response of PHS-related traits in soft white wheat
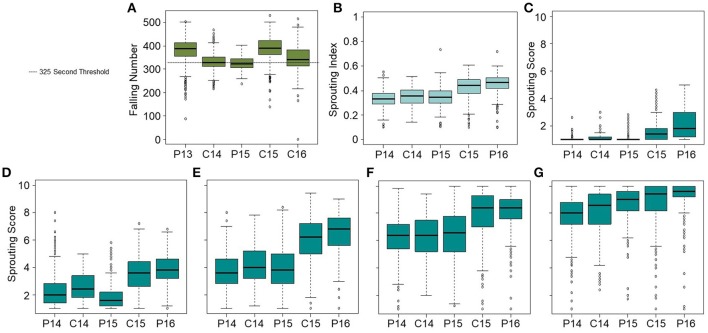
A panel of 469 soft white wheat accessions was evaluated for PHS tolerance based on spike-wetting tests and FN following rain in the field. FN was examined in five environments over 4 years and at two locations. The mean FN ranged from 326 to 389 s for all environments (Table 1). Increasing α-amylase activity tends to correlate with decreasing FN below 325 s (Perten, 1964; Yu et al., 2015). All environments had multiple accessions below the 325 s threshold (ranging from 36 to 234 accessions; Figure 1; Supplementary Figure 2). Natural sprout-inducing rain events occurred in Pullman 2013 and Central Ferry 2016. Artificial rain was applied to induce sprouting in Central Ferry in 2014 and 2015. Pullman 2013, and Central Ferry 2015 and 2016 had wide variation for FN, but Central Ferry 2015 had very few accessions below 325 s. The Pullman 2015 data was included in the analysis as a “no-rain-event” control in an attempt to differentiate PHS-induced differences in FN from differences in FN due to starch or protein composition (AgWeatherNet, 2016 weather.wsu.edu). Pullman 2015 had 234 accessions below 325 s but had lower variation than other environments. This is likely due to poor grain filling since that season had unusually high summer temperatures. QTN mapped based on FN data will be referred to as QFN.wsu.
Figure 1.
FN and PHS distributions. (A) FN distributions across environments including the natural rain events in Pullman 2013 (P13) and Central Ferry 2016 (C16), an artificial rain event in Central Ferry 2014 (C14) and 2015 (C15), and an environment without rain in Pullman 2015 (P15). Spike-wetting tests were performed for Pullman 2014 (P14) as well as C14, P15, C15, and Pullman 2016 (P16). (B) Sprouting index was calculated over all 7 days of misting. Sprouting scores after (C) 3, (D) 4, (E) 5, (F) 6, and (G) 7 days of misting were based on a 1-10 scale.
In spike-wetting tests, environments had similar effects on results regardless of whether we examined sprouting scores or the sprouting index. For example, Central Ferry 2015 and Pullman 2016 consistently showed the highest level of sprouting (Figure 1; Table 1). BLUP analysis was performed in order to reduce environmental variation in sprouting scores prior to mapping (Supplementary Figure 2). BLUPs calculated over all environments were highly correlated over days of sprouting and with the sprouting index (SI) (Supplementary Table 2A). Based on the range of values, we expected to have enough variation to map QTN associated with PHS tolerance due to reduced germination or slower post-germinative growth. For ease of communication, QTN identified based on sprouting scores or SI will be referred to collectively as PHS loci or QPHS.wsu.
PHS trait correlations
Correlations were used to examine whether genotypic differences in FN or sprouting scores were consistent between environments, and to examine whether FN and sprouting scores were related. The FN values showed a weak but significant positive correlation between environments, ranging from 0.23 to 0.46 (p < 0.001; Table 2). Sprouting scores were positively correlated between environments, but not always significantly (Supplementary Tables 2B–G). Day 3 sprouting scores showed the least significant correlation between environments, whereas day 5 and day 6 sprouting scores showed significant positive correlations between environments. SI had the strongest and most significant correlation between environments. The genotypic repeatability of FN (R2 = 0.50) was greater than sprouting scores (R2 < 0.31), especially when covariates like operator, machine, subset, and replicate, were taken into account (Table 3). When repeatability was calculated on a line mean basis, both traits had similar repeatability in our experiments (FN R2 = 0.67; SI R2 = 0.70). The genotypic repeatability of sprouting was highest when spikes were misted longer or when all measurements were integrated in the SI (Table 3). Because seed germination induces α-amylase, which in turn lowers FN, we expected higher sprouting scores to negatively correlate with lower FN, but this was not the case (Table 2). BLUPs for SI generated over all days scored showed the strongest and most significant negative correlation with FN in years when natural sprouting events occurred (Table 2B). In Central Ferry 2015, the FN did not correlate to the sprouting scores and SI. Overall, 4 days of misting had the highest correlation across all FN environments.
Table 2.
Rank correlation coefficients for (A) FN LSMeans across environments and (B) PHS score BLUPs compared to FN environments.
| (A) | FN: | P13a | C14 | P15 | C15 | ||
|---|---|---|---|---|---|---|---|
| FN | C14 | 0.29** | |||||
| P15 | 0.23** | 0.42** | |||||
| C15 | 0.23** | 0.29** | 0.29** | ||||
| C16 | 0.33** | 0.46** | 0.30** | 0.34** | |||
| (B) | PHS: | 3 days | 4 days | 5 days | 6 days | 7 days | SI |
| FN | P13 | −0.16** | −0.24** | −0.17** | −0.18** | −0.20** | −0.21** |
| C14 | −0.07 | −0.09* | −0.06 | −0.09 | −0.10* | −0.10* | |
| P15 | −0.07 | −0.13* | −0.12* | −0.12* | −0.17** | −0.15** | |
| C15 | −0.09 | −0.04 | 0.00 | 0.01 | 0.00 | −0.01 | |
| C16 | −0.17** | −0.19** | −0.18** | −0.17** | −0.17** | −0.19** |
Represents a p-value ≤ 0.05 and
represents a p-value ≤ 0.001.
Environments Pullman (P) and Central Ferry (C).
Table 3.
Genotypic repeatability (R2) of FN, spouting scores, and SI across all environments.
| Trait | Simple R2a | Covariate R2b | Line mean basis R2c |
|---|---|---|---|
| FN | 0.197 | 0.500 | 0.667 |
| PHS d3 d | 0.109 | 0.145 | 0.459 |
| PHS d4 | 0.163 | 0.240 | 0.612 |
| PHS d5 | 0.151 | 0.214 | 0.577 |
| PHS d6 | 0.229 | 0.276 | 0.656 |
| PHS d7 | 0.218 | 0.230 | 0.599 |
| SI | 0.228 | 0.315 | 0.697 |
Genetic and environmental variances were calculated using a simple y ~ x model with x (genotypes) as a fixed effect and repeatability R 2 = Vg / (Vg –Ve) was calculated.
Genetic and environmental variances were calculated using a simple y ~ x + covariates model with x (genotypes) and covariates (time, machine, operator, etc.) as a fixed effect.
Repeatability R 2 = Vg/(Vg –(Ve /n)) was expressed by a line mean basis by dividing the environmental variance (Ve, residuals) with the number of technical reps (n; FN = 2 and spike-wetting test = 5).
Sprouting score (PHS) on days (d) 3 through 7.
Association analysis for falling numbers
GWAS for FN was performed using FarmCPU within each environment because environments varied drastically. All environments fit the FarmCPU association mapping model (Supplementary Figure 4). However, Central Ferry 2015 showed no significant marker-trait associations, regardless of which model was used for GWAS (Supplementary Figure 3D). Nine significant QTN associated with FN were mapped on chromosomes 4A, 5A, 5D, 7A, and 7B (Table 4). The three QTN identified without rain in Pullman 2015 likely represent grain characteristics that are independent of PHS. The remaining QTN were identified in natural rain events. The QTN detected on chromosomes 6D (–log10(p) = 5.88) and 7A (–log10(p) = 6. 17) in Central Ferry 2014 were just below the stringent threshold for significance (–log10(p) = 6. 55). QFN.wsu-7A.1 and QFN.wsu-7B.2 had the largest effects, increasing FN by 26 and 27 s, respectively. QFN.wsu-7A.2 had the highest significance (–log10(p) = 12.36) and had an 8 s effect. In order to reduce the effect of loci unrelated to PHS tolerance on the GWAS, the data were re-analyzed with 400 s set as the maximum possible FN. When this was done, none of the nine QFN.wsu were detected and two unique significant QTN, QFN.wsu-6A and QFN.wsu-7A.3 were identified in Central Ferry 2016 (Supplementary Table 4). These two QTN had large effects, increasing FN by 17 and 13 s, respectively, but only explained 1% of the phenotypic variation. A GWAS was also conducted for heading date from Pullman 2014 and there were no heading date QTN discovered that overlapped with QTNs for either FN or PHS (Supplementary Table 5).
Table 4.
Loci significantly associated with Falling Numbers (FN), early preharvest sprouting (PHS) scores (days 3–4), 5 days of misting, late PHS scores (days 6–7), and PHS sprouting index (SI).
| QTLa | Marker | Chrb | cMb | –log10(p) | maf | Effectc | r2 | Environment | Favorable Alleled |
|---|---|---|---|---|---|---|---|---|---|
| QFN.wsu-4A* | IWB1884 | 4A | 152 | 6.63 | 0.48 | 10.28 | 0.00 | C16 FN | A/C |
| QFN.wsu-5A.1* | IWB60191 | 5A | 23 | 7.27 | 0.27 | 7.53 | 0.00 | P15 FN | A/G |
| QFN.wsu-5A.2 | IWB9800 | 5A | 141 | 7.77 | 0.20 | 7.43 | 0.00 | P15 FN | A/G |
| QFN.wsu-5D | IWB36060 | 5D | 202 | 6.11 | 0.35 | 11.70 | 0.08 | P13 FN | A/C |
| QFN.wsu-7A.1 | IWB22966 | 7A | 35 | 8.34 | 0.06 | 26.09 | 000 | P13 FN | A/G |
| QFN.wsu-7A.2 | IWA334 | 7A | 126 | 12.36 | 0.41 | 7.99 | 0.01 | P15 FN | A/C |
| QFN.wsu-7B.1 | IWB39063 | 7B | 162 | 7.91 | 0.48 | 10.88 | 0.01 | C16 FN | A/G |
| QFN.wsu-7B.2 | IWB75387 | 7B | – | 6.15 | 0.09 | 27.35 | 0.00 | P13 FN | A/C |
| QFN.wsu-unk | IWB37658 | unk | – | 6.82 | 0.09 | 15.49 | 0.00 | C16 FN | T/C |
| QPHS.wsu-1A.1 | IWB2320 | 1A | 82 | 6.73 | 0.15 | −0.04 | 0.00 | P14 d3 | T/C |
| QPHS.wsu-1A.2 | IWB6759 | 1A | 155 | 11.70 | 0.47 | −0.31 | 0.15 | P14 d3 | A/G |
| IWB77968 | 1A | 155 | 12.72 | 0.47 | −0.02 | 0.10 | P14 d3 | A/G | |
| QPHS.wsu-1B.2 | IWB64868 | 1B | 135 | 9.00 | 0.15 | −0.40 | 0.17 | C14 d4 | A/G |
| IWB31676 | 1B | 137 | 8.18 | 0.08 | −0.37 | 0.00 | P16 d3 | A/G | |
| QPHS.wsu-2A.1 | IWB42693 | 2A | 25 | 6.87 | 0.22 | −0.16 | 0.02 | C15 d3 | T/G |
| QPHS.wsu-2D | IWB7652 | 2D | 52 | 10.03 | 0.37 | −0.46 | 0.00 | C14 d4 | T/C |
| IWA8544 | 2D | 50 | 8.73 | 0.46 | −0.28 | 0.01 | P16 d3 | A/G | |
| IWA8544 | 2D | 50 | 9.12 | 0.46 | −0.32 | 0.07 | P16 d4 | A/G | |
| QPHS.wsu-3A.2 | IWB50719 | 3A | 68 | 6.71 | 0.14 | −0.29 | 0.04 | C14 d4 | A/G |
| QPHS.wsu-4A.1 | IWA7535 | 4A | 58 | 8.57 | 0.05 | −0.07 | 0.03 | P14 d3 | A/G |
| QPHS.wsu-4B.2 | IWB21707 | 4B | 75 | 8.93 | 0.10 | −0.42 | 0.07 | P14 d4 | A/G |
| QPHS.wsu-4B.3* | IWB22055 | 4B | 101 | 6.57 | 0.08 | −0.37 | 0.00 | P16 d3 | A/G |
| QPHS.wsu-5B.1 | IWB31067 | 5B | 26 | 8.75 | 0.08 | −0.06 | 0.01 | P14 d3 | T/G |
| QPHS.wsu-7B.1 | IWB54418 | 7B | 3 | 7.46 | 0.03 | −0.26 | 0.01 | P16 d3 | A/G |
| QPHS.wsu-1B.1* | IWB22868 | 1B | 31 | 7.88 | 0.18 | −0.30 | 0.01 | P14 d5 | T/C |
| QPHS.wsu-2D | IWB46396 | 2D | 54 | 9.63 | 0.39 | −0.49 | 0.02 | C14 d5 | A/G |
| QPHS.wsu-3B.2 | IWA6185 | 3B | 62 | 6.55 | 0.44 | −0.23 | 0.01 | P14 d5 | A/G |
| QPHS.wsu-4A.2 | IWB54609 | 4A | 66 | 7.30 | 0.17 | −0.35 | 0.01 | P16 d5 | A/G |
| QPHS.wsu-5A.2* | IWB10250 | 5A | 70 | 9.13 | 0.32 | −0.44 | 0.03 | P15 d5 | T/C |
| QPHS.wsu-5B.3 | IWB73511 | 5B | 129 | 6.73 | 0.30 | −0.28 | 0.01 | P16 d5 | A/G |
| QPHS.wsu-6B* | IWA1838 | 6B | 65 | 10.53 | 0.07 | −0.29 | 0.05 | P14 d5 | A/G |
| QPHS.wsu-7B.2* | IWB7099 | 7B | 133 | 9.00 | 0.00 | −0.34 | 0.00 | C14 d5 | A/G |
| QPHS.wsu-1D* | IWB71680 | 1D | 163 | 7.61 | 0.06 | −0.60 | 0.09 | P14, P16 d6 | A/G |
| QPHS.wsu-2A.2* | IWB17580 | 2A | 53 | 9.02 | 0.07 | −0.69 | 0.02 | C15 d7 | T/C |
| QPHS.wsu-2A.3 | IWB79387 | 2A | – | 6.88 | 0.01 | −0.31 | 0.00 | C14 d6 | A/G |
| QPHS.wsu-2D | IWB7652 | 2D | 52 | 12.69 | 0.37 | −0.85 | 0.12 | C14 d6, d7 | T/C |
| QPHS.wsu-3A.1 | IWB32631 | 3A | 15 | 6.63 | 0.26 | −0.31 | 0.02 | C14 d7 | A/G |
| QPHS.wsu-3B.1* | IWB6430 | 3B | 11 | 8.92 | 0.08 | −0.38 | 0.01 | P14 d7 | T/C |
| QPHS.wsu-3B.3 | IWB9902 | 3B | – | 7.76 | 0.07 | −0.59 | 0.06 | P14 d7 | T/C |
| QPHS.wsu-4A.2 | IWB46089 | 4A | 73 | 6.83 | 0.16 | −0.33 | 0.04 | P16 d6 | A/G |
| QPHS.wsu-4A.3 | IWB1389 | 4A | 151 | 7.51 | 0.23 | −0.45 | 0.02 | C14 d7 | T/G |
| QPHS.wsu-4B.1 | IWB72936 | 4B | 60 | 7.92 | 0.25 | −0.46 | 0.02 | C14 d7 | A/G |
| QPHS.wsu-4B.2 | IWA1382 | 4B | 73 | 8.04 | 0.06 | −0.53 | 0.00 | P15 d7 | A/G |
| QPHS.wsu-5A.2 | IWB60303 | 5A | 70 | 7.56 | 0.34 | −0.39 | 0.01 | C15 d7 | A/G |
| QPHS.wsu-5A.3* | IWB6049 | 5A | 84 | 9.92 | 0.19 | −0.31 | 0.00 | P16 d6 | A/G |
| QPHS.wsu-6A | IWB6726 | 6A | 77 | 7.52 | 0.07 | −0.48 | 0.03 | P14 d7 | T/G |
| QPHS.wsu-6B | IWB76583 | 6B | 65 | 9.76 | 0.05 | −0.33 | 0.03 | P14 d6 | A/G |
| QPHS.wsu-6D* | IWB49280 | 6D | 153 | 7.17 | 0.03 | −0.39 | 0.00 | P15 d7 | A/G |
| QPHS.wsu-7A* | IWB51129 | 7A | 152 | 6.64 | 0.00 | −0.27 | 0.06 | P16 d6 | A/G |
| QPHS.wsu-7B.3 | IWB10815 | 7B | 171 | 10.62 | 0.05 | −0.41 | 0.00 | P14 d7 | T/C |
| QPHS.wsu-1D* | IWB71680 | 1D | 163 | 7.22 | 0.06 | −0.03 | 0.10 | P16 SI | A/G |
| QPHS.wsu-2B | IWB30853 | 2B | 87 | 7.59 | 0.21 | −0.02 | 0.00 | C14 SI | A/G |
| QPHS.wsu-2D | IWB46396 | 2D | 54 | 11.97 | 0.39 | −0.03 | 0.07 | C14 SI | A/G |
| QPHS.wsu-3B.3 | IWB9902 | 3B | – | 7.31 | 0.07 | −0.03 | 0.00 | P14 SI | T/C |
| QPHS.wsu-5A.1 | IWB10998 | 5A | 53 | 8.70 | 0.41 | −0.02 | 0.25 | C14 SI | T/C |
| QPHS.wsu-6B | IWB57747 | 6B | 64 | 6.85 | 0.07 | −0.02 | 0.07 | P16 SI | A/G |
| IWB76583 | 6B | 65 | 6.57 | 0.02 | −0.01 | 0.00 | P14 SI | A/G | |
| QPHS.wsu-7B.2* | IWB7099 | 7B | 133 | 8.63 | 0.00 | −0.02 | 0.01 | C14 SI | A/G |
| IWB7099 | 7B | 133 | 7.58 | 0.01 | −0.02 | 0.00 | P16 SI | A/G |
QTL in bold explained 10% (r2 > 0.1) or more of the phenotypic variation. QTL underlined were significant in 2 environments. Loci more than 10 cM away from previously published QTL were considered to be novel and are indicated with an*.
Chromosome and position according to Wang et al. (2014). Positions are not reported if the location was identified on the GrainGenes database.
The allelic effect is shown in FN seconds or sprouting score BLUPs.
The significant allele is favorable (in bold) if it decreases sprouting scores in the spike-wetting tests or increases Falling Numbers.
Association analysis for sprouting score and sprouting index
For the spike-wetting tests, 34 significant QPHS.wsu were detected based on sprouting scores or on SI (Table 4). There were 12 QPHS.wsu associated with early germination and root emergence (3–4 days of misting) (Figure 1; Table 4; McMaster and Derera, 1976). Seven QPHS.wsu were identified after 5 days of misting, only one of which was also seen with 3–4 days of misting. There were 16 significant QPHS.wsu associated with coleoptile emergence and elongation at 6–7 days of misting, 13 of which were unique to 6–7 days of misting. There were 3 additional QPHS.wsu uniquely detected by SI (Figure 2B). An association at the QPHS.wsu-2D locus was detected over all days of scoring, suggesting that it is not unique to any one sprouting stage (Figure 2A). The QPHS.wsu-1B.2 and QPHS.wsu-6B were detected on two of the scoring days. Of the 34 total QTN found in the spike-wetting tests, only 6 QPHS.wsu on chromosomes 1B, 1D, 2D, 5A, 6B, and 7B were significant in two environments (Figure 2C). No QPHS.wsu were detected in more than 2 environments. Five QPHS.wsu on chromosomes 1A, 1B, 1D, 2D, and 5A explained more than 10% of the phenotypic variation. In Central Ferry 2015, only sprouting scores on days 3 and 7 fit the expected −log(p), whereas days 4, 5, 6, and SI did not (Supplementary Figure 4). Therefore, very few significant QPHS.wsu were identified in this environment.
Figure 2.
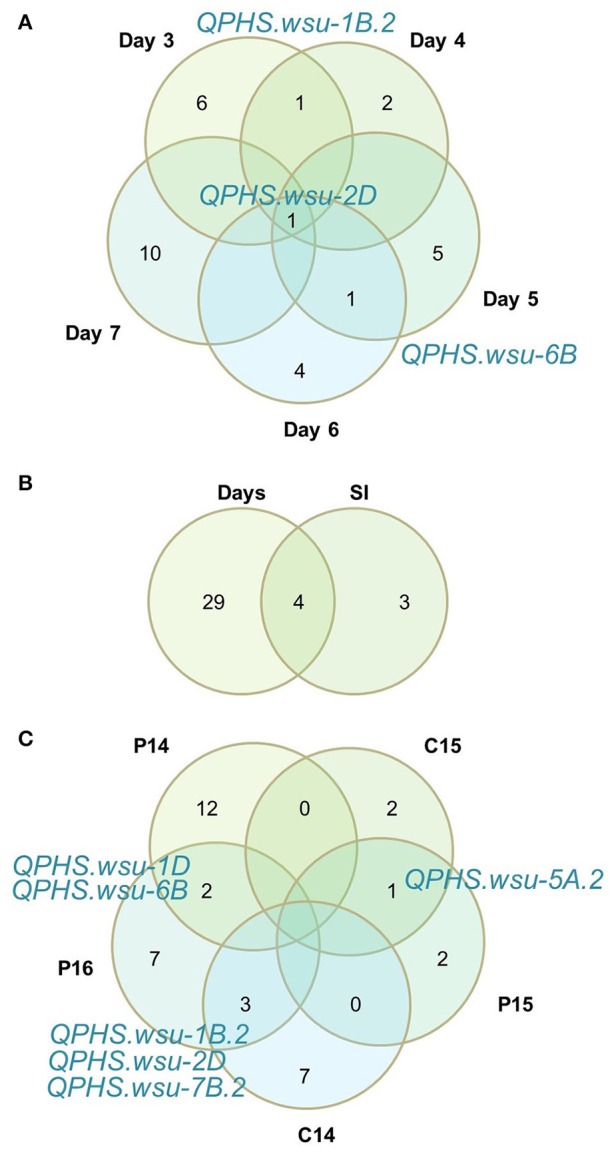
Venn diagrams comparing sprouting score QTN across (A) days misted, (B) all days misted and sprouting index, and (C) the five environments. Shared QTN are printed in blue.
In order to reduce the effect of post-germinative growth versus germination per se, the GWAS was repeated with the maximum sprouting score set at 5 and referred to as QPHSg.wsu (Supplementary Table 4). Out of 46 QTN detected, 32 QTN were unique to this analysis and 21 were no longer significant (Supplementary Figure 5). The major QTN, QPHS.wsu-1D, QPHS.wsu-2D, QPHS.wsu-5A.2, and QPHS.wsu-7B.2 were also detected in this germination-based analysis. The QPHSg.wsu-2D locus was highly significant in this GWAS (7.64 < –log10(p) < 37.13), and had strong effects (0.02 to 0.72).
Association analysis without using the club wheat breeding program as a covariate
Based on the fact that the strong QPHS.wsu-2D locus was near the C locus region for compact spike morphology, we examined whether the club wheat breeding program contributed more PHS tolerance to the GWAS than the lax wheat breeding programs (Figure 3). In Central Ferry 2014, when the tolerant and susceptible loci from the strongest marker within QPHS.wsu-2D were compared, 98% of the tolerant loci were club (Figure 3A). When the BLUPs were compared across all environments, the same trend was observed (Figure 3B). In fact, the club wheat breeding program generally contributed more PHS tolerance in the GWAS than the other programs (Figure 3C). Thus, it is possible that using the principle components as a covariate in the GWAS may artifactually remove some of the PHS loci contributed by the club wheat breeding program. To test this hypothesis, the GWAS was repeated without incorporating the principle components into the model (Supplementary Table 6). The QPHS.wsu-2D QTN became stronger in this analysis, increasing to a –log10(p) of up to 30.03 and an effect of 1.08. An additional 32 QPHSnPC.wsu and 2 QFNnPC.wsu were detected, whereas 15 QPHS.wsu and 2 QFN.wsu were found in common (Supplementary Figure 5).
Figure 3.
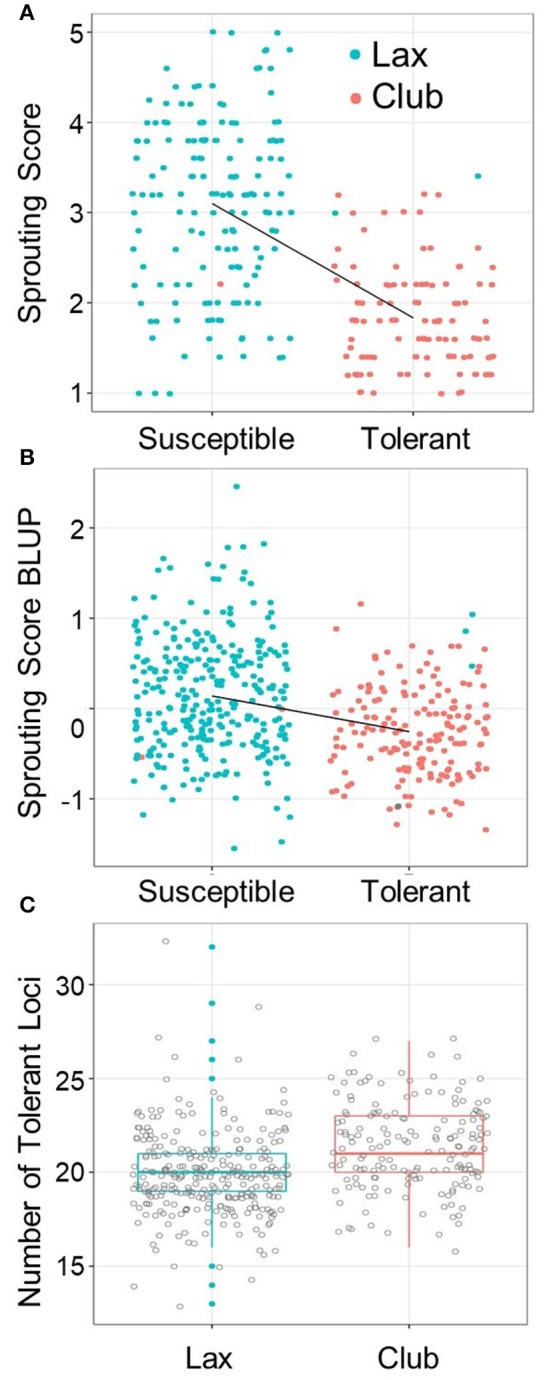
QPHS.wsu-2D in club versus lax mapping lines. Scatter plot of sprouting score (A) raw means over the environment, Central Ferry 2014, that QPHS.wsu-2D had the strongest effect and (B) Sprouting score BLUPs across all five environments after 4 days of misting versus the presence of the QPHS.wsu-2D susceptible (S) or tolerant (T) allele. Club genotypes are in red and lax in blue. (C) Total number of favorable loci are compared to club versus lax spike morphology.
Pyramiding effects of FN and PHS QTN
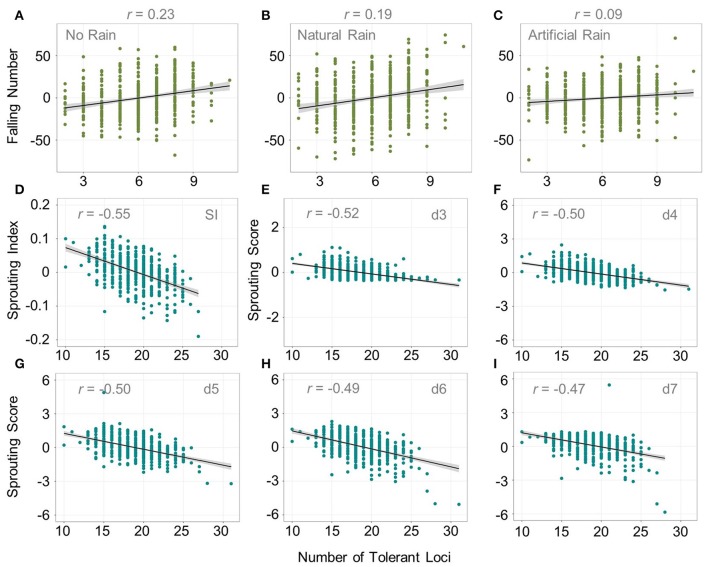
Next, we examined whether an increasing number of favorable QTN were associated with increasing FN or PHS tolerance. The number of favorable QFN.wsu loci within accessions ranged from 2 to 9 (Figures 4A–C). An increasing number of QFN.wsu loci was only weakly correlated to higher FN. The correlation was actually stronger when there was no rain event than when there was a natural or artificial rain event (r = 0.23, 0.19, 0.09, respectively). The number of QPHS.wsu loci varied more widely within the accessions (8 to 26), making it easier to assess the effects of pyramiding multiple tolerance loci (Figures 4D–I). An increasing number of tolerance loci was negatively correlated with sprouting scores ranging from r = −0.47 to r = −0.55 (Supplementary Figure 6). Thus, having more QPHS.wsu loci was associated with more PHS tolerance. Tolerant FN loci only slightly correlated with the increasing sprouting scores, and vice versa (r < 0.20; Supplementary Figure 6).
Figure 4.
The effect of pyramiding multiple QFN.wsu and QPHS.wsu loci. Scatter plots of the number of favorable QFN.wsu loci versus FN BLUPs across: (A) in the absence of rain, (B) both natural rain environments combined, and (C) both artificial rain environments combined. Scatter plots of the number of favorable QPHS.wsu loci versus BLUPs calculated across all environments for (D) sprouting index, and sprouting scores on days (E) 3, (F) 4, (G) 5, (H) 6, and (I) 7 of misting. r is the Pearson correlation coefficient between the trait and number of tolerant loci.
Within the mapping panel, there was only one accession, “A00154,” with all 9 QFN.wsu that had an average FN of 461 s across natural rain events. The accession “6J020288-1” had the highest number of favorable alleles, including 26 out of 34 QPHS.wsu and 6 of 9 QFN.wsu. This was reflected in the phenotype since 6J020288-1 had an average FN of 380 s across natural rain events and a sprouting score of 1 after 5 days of misting from all environments. In contrast, the accession “J950409-10-2” had only 9 out of 34 QPHS.wsu and 5 of 9 QFN.wsu, associated with an average FN of 272 s and a high sprouting score of 7 after 5 days of misting. Interestingly, “Lewjain” had only 8 of the 34 QPHS.wsu and 5 of the 9 QFN.wsu but had an average FN of 401 s and a sprouting score of 4 after 5 days. Thus, these QTN do not always behave in an additive fashion, suggesting that there are epistatic effects. For example, some of the few favorable QTN in Lewjain may have a stronger effect on the phenotype than the unfavorable alleles.
Comparative mapping for PHS
The location of QTN for FN and sprouting scores were compared to locations of PHS-related loci identified in 54 previous studies (Figure 5). This was done using the comparative map of commonly used wheat markers created by Maccaferri et al. (2015). The studies used in the comparison are shown in Supplementary Table 3. Mapped traits that are related to FN and end-use quality included FN, starch content, protein content, and α-amylase activity. Mapped traits related to sprouting and dormancy included germination assays, kernel color, and sprouting scores from spike-wetting tests. There is currently no experimental standard for the spike-wetting test, and methods vary from misting of intact spike to spike immersion to the use of wet sand (McMaster and Derera, 1976; Paterson et al., 1989; Anderson et al., 1993; Humphreys and Noll, 2002; Rehman Arif et al., 2012). Even within each method, the number of days of after-ripening, the duration of spikes wetting, and the day scored vary between studies. Only a few studies score PHS after 4–6 days of misting (Anderson et al., 1993; Munkvold et al., 2009; Kulwal et al., 2012; Somyong et al., 2014). This inconsistency across studies led us to ask whether or not our assay could map previously published cloned genes and QTL. Nineteen of the 34 sprouting QTN detected in this study co-localized with known major PHS QTL and cloned genes such as TaMFT on chromosome 3A (Figure 5; Nakamura et al., 2011). Interestingly, 14 of the 34 sprouting QTN were identified near known FN or quality QTL. Three of 9 FN QTN were identified near known FN and quality QTL, whereas 4 of 9 FN QTN were identified near dormancy or PHS QTL. Of these FN QTN, 2 co-localized with both PHS and FN/quality QTL. A total of 2 QTN for FN and 10 QTN for sprouting score appeared to be unique to this study (marked with a star in Table 4).
Figure 5.
Chromosome positions of PHS-related loci. Comparative mapping of quantitative trait nucleotides identified in this study (black), and previously published quantitative trait loci (QTL) for PHS tolerance (blue), dormancy (purple), Falling Number (green), grain color (red), and quality (orange). Potentially novel QPHS.wsu or QFN.wsu QTN are in bold. Published QTL were aligned to the Maccaferri et al. (2015) comparative map using the flanking markers or significant associated markers. Chromosomes are presented as a standardized relative length. An arrow along a QTL indicates the QTL direction when only one flanking marker was found in the comparative map. The centromeric region is a dark gray oval. ID numbers to the right of the QTL correspond to the references found in Supplementary Table 3. Illumina 9 or 90 k SNP markers were not reported on chromosome 3D, resulting in an incomplete map.
Discussion
This genome-wide association study was, to our knowledge, the first to map preharvest sprouting loci based both on sprouting scores from the spike-wetting test (QPHS.wsu) and FN (QFN.wsu). FN is an important and complex trait that determines the value of the grain in the wheat industry. While there were 34 significant QPHS.wsu loci across the five different sprouting time points and sprouting index, only nine significant QFN.wsu loci were identified. We expected FN and sprouting scores to identify some of the same QTN because the α-amylase expression that lowers FN is a consequence of germination. However, none of the identified significant FN and PHS loci were linked (Table 4). While QFN.wsu-4A and QPHS.wsu-4A.3 appeared to be close to one another (1 cM apart), they were not in the same linkage group (p between 0.05 and 0.01), and had only a −0.13 correlation to one another. Moreover, we failed to find a strong correlation between FN and sprouting scores. This result contrasts with previous studies showing a correlation of up to −0.8 between spike-wetting tests and FN in Canadian and Chilean breeding lines (Rasul et al., 2009; Jiménez et al., 2016).
The lack of a strong correlation between FN and sprouting score in our study has multiple likely causes: our association panel was larger and sampled more variability for FN related traits; our environments were more variable; or the causes of low FN were not solely due to PHS. Eastern Washington is a semi-arid environment where grain is planted deeply to reach moisture and selection for emergence may have led to early and strong induction of α-amylase to fuel seedling growth. This, in turn, may have resulted in lower FN than would be expected for a given sprouting score. Future work may investigate this by determining if a propensity for low FN is associated with earlier expression of α-amylase during germination. The QFN.wsu loci identified, however, did appear to be related to preharvest sprouting because they co-localized with PHS-related loci identified in other studies. Thus, while sprouting scores were not predictive of FN in this study, both traits appeared to be useful for identifying PHS-related loci.
Comparison to previously published PHS QTL and genes
Based on comparative mapping, 12 potentially novel PHS tolerance loci were identified (Figure 5). With the exception of QPHS.wsu-4B.3, the QTN identified during early sprouting (3–4 days misted) were near published QTL (Table 4). In contrast, six novel loci were identified during late sprouting (6–7 days misted). Previous PHS mapping studies mostly used spike-wetting test data collected after 4–6 days of misting (Anderson et al., 1993; Munkvold et al., 2009; Kulwal et al., 2012; Somyong et al., 2014).
Multiple QFN.wsu were located near published QTL or cloned genes governing PHS-related traits (Figure 5; Supplementary Table 3). Three of the six QFN.wsu identified in the presence of natural rain were near known preharvest sprouting and dormancy QTL (Fofana et al., 2009; Kumar et al., 2009, 2015; Kulwal et al., 2012; Albrecht et al., 2015). Previous work has shown FN samples after a rainfall negatively correlate with dormancy which may be another explanation as to why we see FN QTN near dormancy QTL (Biddulph et al., 2008). QFN.wsu-7A.1 had the largest effect on FN, and was near PHS and dormancy QTN found in a European winter wheat GWAS (Albrecht et al., 2015). QFN.wsu-7B.1 localized in a region containing known QTL mapped based on spike-wetting tests, spike immersion, dormancy, and FN (Kulwal et al., 2012; Mohler et al., 2014; Albrecht et al., 2015; Kumar et al., 2015). Given that these QTN were detected after a natural rainfall event, it is curious that they were not detected in any of our spike-wetting test environments. This suggests that either these QFN.wsu loci resulted in higher FN due to PHS tolerance, or that the co-localization of these FN QTN with sprouting QTLs was a coincidence. FN is also controlled by grain starch characteristics. Consistent with this, the QFN.wsu-5A.2 and QFN.wsu-7A.2 loci identified in the absence of rain were located near starch content and starch granule size QTL (Reif et al., 2011; Li et al., 2017).
Many QPHS.wsu were found near published QTL or cloned genes associated with seed dormancy and PHS tolerance (Figure 5). QPHS.wsu-2A.1 was located near a preharvest sprouting QTL, QPhs.ccsu-2A.5, found in the dormant accession “SPR8198” (Mohan et al., 2009). QPHS.wsu-2B was located near the locus providing dormancy and PHS tolerance in the soft white wheat cultivar “Cayuga” and in a European winter wheat QTL (Somyong et al., 2014; Albrecht et al., 2015). The QPHS.wsu-2D QTN had the strongest effect, and co-localized with the QPhs.spa-2D locus identified in Canadian wheat (Kumar et al., 2015). QPHS.wsu-3A.1 was within 1 cM of the major dormancy and PHS tolerance gene MOTHER OF FT AND TFL1 (TaMFT), identified by map-based cloning in both Japanese and U.S. wheat (Nakamura et al., 2011; Liu et al., 2013). QPHS.wsu-4A.1 and QPHS.wsu-4A.2 are within the Phs-A1 region associated with dormancy and PHS tolerance in mapping studies world-wide (Mares and Mrva, 2014; Barrero et al., 2015; Shorinola et al., 2016, 2017; Torada et al., 2016). A polymorphism in the MITOGEN-ACTIVATED PROTEIN KINASE KINASE 3 (TaMKK3-A) gene likely accounts for the seed dormancy providing PHS tolerance on chromosome 4A. Future work will need to examine if the PHS tolerance loci mapped in the current study are associated with the known dormancy-associated polymorphisms in TaMFT and TaMKK3. If so, then these perfect markers can be used for selecting PHS tolerance within the breeding programs represented in this GWAS. Identifying QTL near regions of known PHS QTL validates the GWAS and suggests that breeding programs in the northwestern U.S. have historically used multiple sources of PHS tolerance.
The strong QPHS.wsu-2D locus co-localized both with a known PHS locus and with the C locus that determines club head type (Supplementary Table 3; Johnson et al., 2008). The strong QPHS.wsu-2D locus associated with the C locus may partly be an artifact because the club wheat breeding program was the dominant source of this PHS tolerance locus in the mapping panel. Since QPHS.wsu-2D is <1 cM from the C locus flanking marker wmc144, this invited the question as to whether the C locus itself provided PHS tolerance or whether there was another PHS-tolerance locus in tight linkage with the C locus. The latter seemed more likely because previous studies found that the club head type took up more water during rain events and was sometimes associated with higher preharvest sprouting in near-isogenic lines (Hong, 1979; King and Richards, 1984; R.E. Allan, personal comm.). Interestingly, there were three PHS tolerant lax wheat lines that carried the QPHS.wsu-2D locus (J950409-10-4, J950409-10-5, and ID581), and there were two PHS susceptible club wheat lines (J970057-5 and ARS00226) that did not carry the QPHS.wsu-2D locus. While these counter-examples suggest that there was a PHS QTL strongly linked to the C locus, they are not proof because these PHS phenotypes may have resulted from variation at other loci. Future work will need to examine this question using near-isogenic lines that differ only for the C locus and for the QPHS.wsu-2D locus.
Breeding for PHS tolerance based on spike-wetting tests and FN
Sprouting scores for this population did not correlate strongly with FN (Table 2). While increasing number of QPHS.wsu loci correlated to increasing sprouting index and sprouting scores, we observed little or no correlation to increasing FN in natural rain, artificial rain, or no-rain environments (Figure 4; Supplementary Figure 6). Moreover, there was no strong association between increasing QFN.wsu loci and sprouting scores. In fact, sprouting scores and FN provided complementary information. It is possible that the tendency toward low FN is dependent more on the timing and strength of α-amylase induction during seed imbibition, then on the timing of visible sprout/germination per se. For example, there may be varieties that induce α-amylase earlier in the germination program, prior to germination per se. Such varieties would be prone to higher α-amylase/lower FN than expected based on the timing of visible sprout.
Breeding for FN is complicated by the fact that it is a complex trait governed by multiple factors. Although the intention was to map preharvest sprouting QTL, our FN field environments may have also experienced conditions that induced late maturity α-amylase (LMA). During LMA, α-amylase is induced in response to large temperature fluctuations during late grain filling (Farrell and Kettlewell, 2008; Mares and Mrva, 2014). While four of the five environments experienced either natural or artificial rain events, we cannot rule out the possibility that the wheat also experienced LMA. Indeed, the QFN.wsu-7B.1 locus co-localized with a large LMA QTL (Figure 5; Mrva and Mares, 2001; McNeil et al., 2009; Emebiri et al., 2010). In the environment without rain, differences in FN likely resulted from differences in properties of grain starch and protein (Graybosch et al., 2000; Guo et al., 2003; Ross et al., 2012). Such properties likely also impact FN over 300 s when there is a rain event. The FN test has a fairly high standard deviation within technical replicates (Supplementary Figure 7). The genotypic repeatability (R2) of FN increased when we took experimental covariates (such as machine and operator) and technical replicates into account (Table 3). The FN test has other limitations for breeding such as the need for an expensive instrument, and the fact that it is more time-consuming to run FN than spike-wetting tests. Future research should examine whether α-amylase enzyme assays (Phadebas™ or Megazyme) or ELISA assays may serve as a faster, cheaper, or less variable proxy to FN (Mares and Mrva, 2008; Barrero et al., 2013). Environmental factors also caused variation in FN, resulting in only moderate correlations between environments in the current study (Table 2A). While the Zhang et al. (2014) study had higher correlations between FN in different environments (0.43 > r > 0.80), it had fewer samples below 300 s suggesting that less sprouting occurred in their environments. Breeders cannot rely on natural rain occurring when they want to screen for low FN due to PHS, making the use of artificial rain necessary. Other differences in the environment, such as temperature during maturation or temperature during the sprout-inducing rainfall, can impact grain dormancy, PHS susceptibility, and FN. Future artificial rain experiments may be improved by applying the artificial rain during lower evening temperatures or letting the wheat after-ripen longer in the field prior to misting.
The spike-wetting test has long been favored for selecting preharvest sprouting tolerance because the experimental design takes into account after-ripening time, spike morphology, and grain dormancy/germinability (Paterson et al., 1989). A limitation of FN testing of field-harvested grain is that differences in maturation date can be a major covariate, since early maturing varieties may have lost more dormancy through after-ripening than late maturing varieties. The spike-wetting test reduces this problem by harvesting spikes at physiological maturity and then after-ripening for the same number of days before conducting the test. When screening large numbers of breeding lines, it would be convenient to avoid scoring daily over 3 to 7 days of misting. Based on the correlations between spike-wetting test and all FN environments (with and without rain), scoring after 4 days of misting should provide breeders with both adequate variation and higher correlation to FN (Table 2B). Scoring after 4 days misting also provides good insight into initial germination capacity (scores 1-5) rather than speed of seedling growth (scores 6-10) (Figure 1). However, it should be noted that the day 6 sprouting score had the highest genotypic repeatability in this study. One drawback of selecting for PHS tolerance based on seed dormancy, is that too much dormancy may result in poor seedling emergence of winter wheat when grain is planted ~8 weeks after harvest (Rodríguez et al., 2015). Future work will need to develop a genomic selection model for breeding wheat with sufficient seed dormancy to prevent preharvest sprouting without compromising seedling emergence.
Seed dormancy and PHS tolerance are stronger if temperatures are cool during grain maturation (Nakamura et al., 2011). One could remove temperature during grain development as a variable in spike-wetting tests by growing plants in a controlled environment instead of in the field. We might have also seen better correlations if spike-wetting tests had been performed for the entire trial in the 2 years with natural rain events. However, when spike-wetting tests were performed on 162 accessions in Pullman 2013, only a −0.26 correlation was observed. A more likely explanation is that FN in these environments was impacted by multiple factors in addition to preharvest sprouting, including LMA, starch, and protein characteristics. Thus, within this study, FN and spike-wetting tests were not two ways to measure the same trait.
PHS tolerance is profoundly impacted by environmental conditions during grain maturation and during the sprout-inducing rain event (Cao et al., 2016; Kashiwakura et al., 2016; Martinez et al., 2016). Thus, it is not uncommon to find lack of agreement between environments in PHS tolerance association studies based on spike-wetting tests (Ogbonnaya et al., 2008; Jaiswal et al., 2012; Kulwal et al., 2012; Zhou et al., 2017). A total of 6 out of 34 QPHS.wsu were identified in at least two of the five environments. In fact, the correlations between our environments were as good as those in other spike-wetting test studies (Jaiswal et al., 2012; Kulwal et al., 2012). The fact that this study identified some QTN in multiple environments and that many of the QTN identified agreed with previous studies, suggests that this association study will lay a strong foundation for future efforts to develop genomic selection for PHS tolerance in northwestern U.S. wheat.
Author contributions
SM, KC, and CS: Designed the experiments; SM: Conducted the experiments, analyzed the phenotypic data, performed the statistical analysis and GWAS, and constructed the visualizations; JG: Curated and filtered the genotypic data and performed the LD analysis; MH: Calculated the phenotypic variation; MH and ZZ: Provided expert guidance on performing the GWAS; KC and AC: Provided field resources; CS, KC, AC, and ZZ: Obtained funding; SM and CS: Wrote the manuscript.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors wish to thank Rehana Parveen, Tracy Harris, Patricia DeMacon, Gary Shelton, and Xavian Thompson for expert technical assistance. The authors sincerely thank Dan Dreesman for constructing the greenhouse misting system. Thanks are due to members of the KG, AC, and CS labs for assistance with planting, sampling, and harvesting. Special thanks go to the KG and CS labs for helpful feedback on the research and manuscript.
Footnotes
Funding. This work was funded by the NIFA projects 2015-05798 and 2016-68004-24770, the USDA-ARS project 424575, and the WA Grain Commission projects 7599, 5389, and 126593.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2018.00141/full#supplementary-material
References
- AgWeatherNet (2016). Station Details: Pullman. Avaliable online at: https://weather.wsu.edu/?p=90150&UNIT_ID=300029 (Accessed October 2, 2017).
- Akbari M., Wenzl P., Caig V., Carling J., Xia L., Yang S., et al. (2006). Diversity arrays technology (DArT) for high-throughput profiling of the hexaploid wheat genome. Theor. Appl. Genet. 113, 1409–1420. 10.1007/s00122-006-0365-4 [DOI] [PubMed] [Google Scholar]
- Albrecht T., Oberforster M., Kempf H., Ramgraber L., Schacht J., Kazman E., et al. (2015). Genome-wide association mapping of preharvest sprouting resistance in a diversity panel of European winter wheats. J. Appl. Genet. 56, 277–285. 10.1007/s13353-015-0286-5 [DOI] [PubMed] [Google Scholar]
- Anderson J. A., Sorrells M. E., Tanksley S. D. (1993). RFLP analysis of genomic regions associated with resistance to preharvest sprouting in wheat. Crop Sci. 33, 453–459. 10.2135/cropsci1993.0011183X003300030008x [DOI] [Google Scholar]
- Barrero J. M., Cavanagh C., Verbyla K. L., Tibbits J. F. G., Verbyla A. P., Huang B. E., et al. (2015). Transcriptomic analysis of wheat near-isogenic lines identifies PM19-A1 and A2 as candidates for a major dormancy QTL. Genome Biol. 16, 93. 10.1186/s13059-015-0665-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrero J. M., Mrva K., Talbot M. J., White R. G., Taylor J., Gubler F., et al. (2013). Genetic, hormonal, and physiological analysis of late maturity α-amylase in wheat. Plant Physiol. 161, 1265–1277. 10.1104/pp.112.209502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates D., Maechler M., Bolker B., Walker S. (2015). Fitting linear mixed-effects models using lme4. J. Stat. Softw. 67, 1–48. 10.18637/jss.v067.i01 [DOI] [Google Scholar]
- Bewley J. D., Black M. (1994). Seeds: Physiology of Development and Germination, 2nd Edn. New York, NY: Plenum Publishing Corporation. [Google Scholar]
- Biddulph T. B., Plummer J. A., Setter T. L., Mares D. J. (2008). Seasonal conditions influence dormancy and preharvest sprouting tolerance of wheat (Triticum aestivum L.) in the field. Field Crops Res. 107, 116–128. 10.1016/j.fcr.2008.01.003 [DOI] [Google Scholar]
- Browning B. L., Browning S. R. (2016). Genotype imputation with millions of reference samples. Am. J. Hum. Genet. 98, 116–126. 10.1016/j.ajhg.2015.11.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell K. A. G., Lipps P. E. (1998). Allocation of resources: sources of variation in fusarium head blight screening nurseries. Phytopathology 88, 1078–1086. 10.1094/PHYTO.1998.88.10.1078 [DOI] [PubMed] [Google Scholar]
- Cao L., Hayashi K., Tokui M., Mori M., Miura H., Onishi K. (2016). Detection of QTLs for traits associated with pre-harvest sprouting resistance in bread wheat (Triticum aestivum L.). Breed Sci. 66, 260–270. 10.1270/jsbbs.66.260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavanagh C. R., Chao S., Wang S., Huang B. E., Stephen S., Kiani S., et al. (2013). Genome-wide comparative diversity uncovers multiple targets of selection for improvement in hexaploid wheat landraces and cultivars. Proc. Natl. Acad. Sci. U.S.A. 110, 8057–8062. 10.1073/pnas.1217133110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke J. M., Ronald M., De P., Christensen J. V. (1984). Effect of weathering on falling numbers of standing and windrowed wheat. Can. J. Plant Sci. 64, 457–463. 10.4141/cjps84-068 [DOI] [Google Scholar]
- DePauw R. M., McCaig T. N. (1991). Components of variation, heritabilities and correlations for indices of sprouting tolerance and seed dormancy in Triticum spp. Euphytica 52, 221–229. 10.1007/BF00029399 [DOI] [Google Scholar]
- DePauw R. M., Knox R. E., Singh A. K., Fox S. L., Humphreys D. G., Hucl P. (2012). Developing standardized methods for breeding preharvest sprouting resistant wheat, challenges and successes in Canadian wheat. Euphytica 188, 7–14. 10.1007/s10681-011-0611-y [DOI] [Google Scholar]
- Emebiri L. C., Oliver J. R., Mrva K., Mares D. (2010). Association mapping of late maturity α-amylase (LMA) activity in a collection of synthetic hexaploid wheat. Mol. Breed. 26, 39–49. 10.1007/s11032-009-9375-7 [DOI] [Google Scholar]
- Farrell A. D., Kettlewell P. S. (2008). The effect of temperature shock and grain morphology on alpha-amylase in developing wheat grain. Ann. Bot. 102, 287–293. 10.1093/aob/mcn091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein R. R., Reeves W., Ariizumi T., Steber C. M. (2008). Molecular aspects of seed dormancy. Annu. Rev. Plant Biol. 59, 387–415. 10.1146/annurev.arplant.59.032607.092740 [DOI] [PubMed] [Google Scholar]
- Flintham J. E. (2000). Different genetic components control coat-imposed and embryo-imposed dormancy in wheat. Seed Sci. Res. 10, 43–50. 10.1017/S0960258500000052 [DOI] [Google Scholar]
- Fofana B., Humphreys D. G., Rasul G., Cloutier S., Brûlé-Babel A., Woods S., et al. (2009). Mapping quantitative trait loci controlling pre-harvest sprouting resistance in a red × white seeded spring wheat cross. Euphytica 165, 509–521. 10.1007/s10681-008-9766-6 [DOI] [Google Scholar]
- Froese P. S., Carter A. H. (2016). Single nucleotide polymorphisms in the wheat genome associated with tolerance of acidic soils and aluminum toxicity. Crop Sci. 56, 1662–1677. 10.2135/cropsci2015.10.0629 [DOI] [Google Scholar]
- Froese P. S., Murray T. D., Carter A. H. (2016). Quantitative Cephalosporium stripe disease resistance mapped in the wheat genome. Crop Sci. 56, 1586–1601. 10.2135/cropsci2015.09.0568 [DOI] [Google Scholar]
- Gerjets T., Scholefield D., Foulkes M. J., Lenton J. R., Holdsworth M. J. (2010). An analysis of dormancy, ABA responsiveness, after-ripening and pre-harvest sprouting in hexaploid wheat (Triticum aestivum L.) caryopses. J. Exp. Bot. 61, 597–607. 10.1093/jxb/erp329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graybosch R. A., Guo G., Shelton D. R. (2000). Aberrant falling numbers of waxy wheats independent of α-amylase activity. Cereal Chem. J. 77, 1–3. 10.1094/CCHEM.2000.77.1.1 [DOI] [Google Scholar]
- Gualano N., Carrari F., Rodriguez M. V., Perez-Flores L., Sanchez R., Iusem N., et al. (2007). Reduced embryo sensitivity to abscisic acid in a sprouting-susceptible sorghum (Sorghum bicolor) variety is associated with altered ABA signalling. Seed Sci. Res. 17, 81–90. 10.1017/S0960258507708115 [DOI] [Google Scholar]
- Guo G., Jackson D., Graybosch R., Parkhurst A. (2003). Asian salted noodle quality: impact of amylose content adjustments using waxy wheat flour. Faculty Publications Food Sci. Technol. 80, 437–445. 10.1094/CCHEM.2003.80.4.437 [DOI] [Google Scholar]
- Himi E., Maekawa M., Miura H., Noda K. (2011). Development of PCR markers for Tamyb10 related to R-1, red grain color gene in wheat. Theor. Appl. Genet. 122, 1561–1576. 10.1007/s00122-011-1555-2 [DOI] [PubMed] [Google Scholar]
- Hong B. H. (1979). Genetic and Environmental Aspects of Preharvest Sprouting and Related Traits in Triticum aestivum L. em Thell, Ph.D. dissertation, Washington State University, Pullman, WA. [Google Scholar]
- Humphreys D. G., Noll J. (2002). Methods for characterization of preharvest sprouting resistance in a wheat breeding program. Euphytica 126, 61–65. 10.1023/A:1019671622356 [DOI] [Google Scholar]
- Ibrahim H. A. (1966). Studies on the Sprouting Problem in Soft White Wheats. Ph.D. dissertation Cornell University, Ithaca, NY. [Google Scholar]
- Jaiswal V., Mir R. R., Mohan A., Balyan H. S., Gupta P. K. (2012). Association mapping for pre-harvest sprouting tolerance in common wheat (Triticum aestivum L.). Euphytica 188, 89–102. 10.1007/s10681-012-0713-1 [DOI] [Google Scholar]
- Ji T., Baik B.-K. (2016). Storage conditions affecting increase in falling number of soft red winter wheat grain. Cereal Chem. J. 93, 263–267. 10.1094/CCHEM-08-15-0167-R [DOI] [Google Scholar]
- Jiménez N., Mares D., Mrva K., Lizana C., Contreras S., Schwember A. R. (2016). Susceptibility to preharvest sprouting of Chilean and Australian elite cultivars of common wheat. Crop Sci. 57, 462–474. 10.2135/cropsci2016.02.0138 [DOI] [Google Scholar]
- Johnson E. B., Nalam V. J., Zemetra R. S., Riera-Lizarazu O. (2008). Mapping the compactum locus in wheat (Triticum aestivum L.) and its relationship to other spike morphology genes of the Triticeae. Euphytica 163, 193–201. 10.1007/s10681-007-9628-7 [DOI] [Google Scholar]
- Kashiwakura Y., Kobayashi D., Jikumaru Y., Takebayashi Y., Nambara E., Seo M., et al. (2016). Highly sprouting-tolerant wheat grain exhibits extreme dormancy and cold imbibition-resistant accumulation of abscisic acid. Plant Cell Physiol. 57, 715–732. 10.1093/pcp/pcw051 [DOI] [PubMed] [Google Scholar]
- King R. W. (1984). Water uptake in relation to pre-harvest sprouting damage in wheat: grain characteristics. Aust. J. Agric. Res. 35, 337–345. 10.1071/AR9840337 [DOI] [Google Scholar]
- King R. W., Richards R. A. (1984). Water uptake in relation to pre-harvest sprouting damage in wheat: ear characteristics. Aust. J. Agric. Res. 35, 327–336. 10.1071/AR9840327 [DOI] [Google Scholar]
- King R. W., von Wettstein-Knowles P. (2000). Epicuticular waxes and regulation of ear wetting and pre-harvest sprouting in barley and wheat. Euphytica 112, 157–166. 10.1023/A:1003832031695 [DOI] [Google Scholar]
- Kulwal P., Ishikawa G., Benscher D., Feng Z., Yu L.-X., Jadhav A., et al. (2012). Association mapping for pre-harvest sprouting resistance in white winter wheat. Theor. Appl. Genet. 125, 793–805. 10.1007/s00122-012-1872-0 [DOI] [PubMed] [Google Scholar]
- Kumar A., Kumar J., Singh R., Garg T., Chhuneja P., Balyan H. S., et al. (2009). QTL analysis for grain colour and pre-harvest sprouting in bread wheat. Plant Sci. 177, 114–122. 10.1016/j.plantsci.2009.04.004 [DOI] [Google Scholar]
- Kumar S., Knox R. E., Clarke F. R., Pozniak C. J., DePauw R. M., Cuthbert R. D., et al. (2015). Maximizing the identification of QTL for pre-harvest sprouting resistance using seed dormancy measures in a white-grained hexaploid wheat population. Euphytica 205, 287–309. 10.1007/s10681-015-1460-x [DOI] [Google Scholar]
- Li J., Rasheed A., Guo Q., Dong Y., Liu J., Xia X., et al. (2017). Genome-wide association mapping of starch granule size distribution in common wheat. J. Cereal Sci. 77, 211–218. 10.1016/j.jcs.2017.08.016 [DOI] [Google Scholar]
- Lipka A. E., Tian F., Wang Q., Peiffer J., Li M., Bradbury P. J., et al. (2012). GAPIT: Genome Association and Prediction Integrated Tool. Bioinformatics 28, 2397–2399. 10.1093/bioinformatics/bts444 [DOI] [PubMed] [Google Scholar]
- Liu S., Sehgal S. K., Li J., Lin M., Trick H. N., Yu J., et al. (2013). Cloning and characterization of a critical regulator for preharvest sprouting in wheat. Genetics 195, 263–273. 10.1534/genetics.113.152330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W., Naruoka Y., Miller K., Garland-Campbell K. A., Carter A. H. (2017). Characterizing and validating stripe rust resistance loci in US Pacific Northwest winter wheat accessions (Triticum aestivum L.) by genome-wide association and linkage mapping. Plant Genome. 11 10.3835/plantgenome2017.10.0087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Huang M., Fan B., Buckler E. S., Zhang Z. (2016). Iterative usage of fixed and random effect models for powerful and efficient genome-wide association studies. PLoS Genet. 12:e1005767. 10.1371/journal.pgen.1005767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maccaferri M., Zhang J., Bulli P., Abate Z., Chao S., Cantu D., et al. (2015). A genome-wide association study of resistance to stripe rust (Puccinia striiformis f. sp. tritici) in a worldwide collection of hexaploid spring wheat (Triticum aestivum L.). G3 5, 449–465. 10.1534/g3.114.014563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mares D. J., Mrva K. (2014). Wheat grain preharvest sprouting and late maturity alpha-amylase. Planta 240, 1167–1178. 10.1007/s00425-014-2172-5 [DOI] [PubMed] [Google Scholar]
- Mares D., Mrva K. (2008). Late-maturity α-amylase: low falling number in wheat in the absence of preharvest sprouting. J. Cereal Sci. 47, 6–17. 10.1016/j.jcs.2007.01.005 [DOI] [Google Scholar]
- Martinez S. A., Tuttle K. M., Takebayashi Y., Seo M., Campbell K. G., Steber C. M. (2016). The wheat ABA hypersensitive ERA8 mutant is associated with increased preharvest sprouting tolerance and altered hormone accumulation. Euphytica 212, 229–245. 10.1007/s10681-016-1763-6 [DOI] [Google Scholar]
- McMaster G. J., Derera N. F. (1976). Methodology and sample preparation when screening for sprouting damage in cereals. Cereal Res. Commun. 4, 251–254. [Google Scholar]
- McNeil M. D., Diepeveen D., Wilson R., Barclay I., McLean R., Chalhoub B., et al. (2009). Haplotype analyses in wheat for complex traits: tracking the chromosome 3B and 7B regions associated with late maturity alpha amylase (LMA) in breeding programs. Crop Pasture Sci. 60, 463–471. 10.1071/CP08340 [DOI] [Google Scholar]
- Mohan A., Kulwal P., Singh R., Kumar V., Mir R. R., Kumar J., et al. (2009). Genome-wide QTL analysis for pre-harvest sprouting tolerance in bread wheat. Euphytica 168, 319–329. 10.1007/s10681-009-9935-2 [DOI] [Google Scholar]
- Mohler V., Albrecht T., Mrva K., Schweizer G., Hartl L. (2014). Genetic analysis of falling number in three bi-parental common winter wheat populations. Plant Breed. 133, 448–453. 10.1111/pbr.12177 [DOI] [Google Scholar]
- Mrva K., Mares D. J. (2001). Quantitative trait locus analysis of late maturity α-amylase in wheat using the doubled haploid population Cranbrook Halberd. Aust. J. Agric. Res. 52, 1267–1273. 10.1071/AR01047 [DOI] [Google Scholar]
- Munkvold J. D., Tanaka J., Benscher D., Sorrells M. E. (2009). Mapping quantitative trait loci for preharvest sprouting resistance in white wheat. Theor. Appl. Genet. 119, 1223–1235. 10.1007/s00122-009-1123-1 [DOI] [PubMed] [Google Scholar]
- Nakamura S., Abe F., Kawahigashi H., Nakazono K., Tagiri A., Matsumoto T., et al. (2011). A wheat homolog of MOTHER OF FT AND TFL1 acts in the regulation of germination. Plant Cell 23, 3215–3229. 10.1105/tpc.111.088492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogbonnaya F. C., Imtiaz M., Ye G., Hearnden P. R., Hernandez E., Eastwood R. F., et al. (2008). Genetic and QTL analyses of seed dormancy and preharvest sprouting resistance in the wheat germplasm CN10955. Theor. Appl. Genet. 116, 891–902. 10.1007/s00122-008-0712-8 [DOI] [PubMed] [Google Scholar]
- Paterson A. H., Sorrells M. E. (1990). Inheritance of grain dormancy in white-kernelled wheat. Crop Sci. 30, 25–30. 10.2135/cropsci1990.0011183X003000010006x [DOI] [Google Scholar]
- Paterson A. H., Sorrells M. E., Obendorf R. L. (1989). Methods of evaluation for preharvest sprouting resistance in wheat breeding programs. Can. J. Plant Sci. 69, 681–689. 10.4141/cjps89-084 [DOI] [Google Scholar]
- Perten H. (1964). Application of the falling number method for evaluating α-amylase activity. Cereal Chem. 41, 127–140. [Google Scholar]
- Piepho H. P., Möhring J., Melchinger A. E., Büchse A. (2008). BLUP for phenotypic selection in plant breeding and variety testing. Euphytica 161, 209–228. 10.1007/s10681-007-9449-8 [DOI] [Google Scholar]
- Pool M., Patterson F. L. (1958). Moisture relations in soft red winter wheats. II. Awned versus Awnless and Waxy versus Nonwaxy Glumes. Agron. J. 50, 158–160. 10.2134/agronj1958.00021962005000030013x [DOI] [Google Scholar]
- Rasul G., Humphreys D. G., Brûlé-Babel A., McCartney C. A., Knox R. E., DePauw R. M., et al. (2009). Mapping QTLs for pre-harvest sprouting traits in the spring wheat cross 'RL4452/AC Domain.' Euphytica 168, 363–378. 10.1007/s10681-009-9934-3 [DOI] [Google Scholar]
- Rehman Arif M. A., Neumann K., Nagel M., Kobiljski B., Lohwasser U., Börner A. (2012). An association mapping analysis of dormancy and pre-harvest sprouting in wheat. Euphytica 188, 409–417. 10.1007/s10681-012-0705-1 [DOI] [Google Scholar]
- Reif J. C., Gowda M., Maurer H. P., Longin C. F. H., Korzun V., Ebmeyer E., et al. (2011). Association mapping for quality traits in soft winter wheat. Theor. Appl. Genet. 122, 961–970. 10.1007/s00122-010-1502-7 [DOI] [PubMed] [Google Scholar]
- Rodríguez M. V., Barrero J. M., Corbineau F., Gubler F., Benech-Arnold R. L. (2015). Dormancy in cereals (not too much, not so little): about the mechanisms behind this trait. Seed Sci. Res. 25, 99–119. 10.1017/S0960258515000021 [DOI] [Google Scholar]
- Ross A. S., Flowers M. D., Zemetra R. S., Kongraksawech T. (2012). Effect of grain protein concentration on falling number of ungerminated soft white winter wheat. Cereal Chem. J. 89, 307–310. 10.1094/CCHEM-07-12-0085-RC [DOI] [Google Scholar]
- Saintenac C., Jiang D., Wang S., Akhunov E. (2013). Sequence-based mapping of the polyploid wheat genome. G3 3, 1105–1114. 10.1534/g3.113.005819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schramm E. C., Abellera J. C., Strader L. C., Campbell K. G., Steber C. M. (2010). Isolation of ABA-responsive mutants in allohexaploid bread wheat (Triticum aestivum L.): drawing connections to grain dormancy, preharvest sprouting, and drought tolerance. Plant Sci. 179, 620–629. 10.1016/j.plantsci.2010.06.004 [DOI] [Google Scholar]
- Shorinola O., Balcárková B., Hyles J., Tibbits J. F. G., Hayden M. J., Holušova K., et al. (2017). Haplotype ANalysis of the pre-harvest sprouting resistance locus Phs-A1 reveals a causal role of TaMKK3-A in global germplasm. Front. Plant Sci. 8:1555. 10.3389/fpls.2017.01555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shorinola O., Bird N., Simmonds J., Berry S., Henriksson T., Jack P., et al. (2016). The wheat Phs-A1 pre-harvest sprouting resistance locus delays the rate of seed dormancy loss and maps 0.3 cM distal to the PM19 genes in UK germplasm. J. Exp. Bot. 67, 4169–4178. 10.1093/jxb/erw194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somyong S., Ishikawa G., Munkvold J. D., Tanaka J., Benscher D., Cho Y.-G., et al. (2014). Fine mapping of a preharvest sprouting QTL interval on chromosome 2B in white wheat. Theor. Appl. Genet. 127, 1843–1855. 10.1007/s00122-014-2345-4 [DOI] [PubMed] [Google Scholar]
- Tang Y., Liu X., Wang J., Li M., Wang Q., Tian F., et al. (2016). GAPIT Version 2: an enhanced integrated tool for genomic association and prediction. Plant Genome 9, 1–9. 10.3835/plantgenome2015.11.0120 [DOI] [PubMed] [Google Scholar]
- Torada A., Koike M., Ogawa T., Takenouchi Y., Tadamura K., Wu J., et al. (2016). A causal gene for seed dormancy on wheat chromosome 4A encodes a MAP kinase kinase. Curr. Biol. 26, 782–787. 10.1016/j.cub.2016.01.063 [DOI] [PubMed] [Google Scholar]
- Tuttle K. M., Martinez S. A., Schramm E. C., Takebayashi Y., Seo M., Steber C. M. (2015). Grain dormancy loss is associated with changes in ABA and GA sensitivity and hormone accumulation in bread wheat, Triticum aestivum (L.). Seed Sci. Res. 25, 179–193. 10.1017/S0960258515000057 [DOI] [Google Scholar]
- Ullrich S. E., Lee H., Clancy J. A., del Blanco I. A., Jitkov V. A., Kleinhofs A., et al. (2009). Genetic relationships between preharvest sprouting and dormancy in barley. Euphytica 168, 331–345. 10.1007/s10681-009-9936-1 [DOI] [Google Scholar]
- Walker-Simmons M. (1987). ABA Levels and sensitivity in developing wheat embryos of sprouting resistant and susceptible cultivars. Plant Physiol. 84, 61–66. 10.1104/pp.84.1.61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Wong D., Forrest K., Allen A., Chao S., Huang B. E., et al. (2014). Characterization of polyploid wheat genomic diversity using a high-density 90 000 single nucleotide polymorphism array. Plant Biotechnol. J. 12, 787–796. 10.1111/pbi.12183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner R. L., Kudrna D. A., Spaeth S. C., Jones S. S. (2000). Dormancy in white-grain mutants of Chinese Spring wheat (Triticum aestivum L.). Seed Sci. Res. 10, 51–60. 10.1017/S0960258500000064 [DOI] [Google Scholar]
- Yu N., Laurenz R., Siler L., Ng P. K. W., Souza E., Lewis J. M. (2015). Evaluation of α-amylase activity and falling number around maturity for soft white and soft red wheat varieties in Michigan. Cereal Res. Commun. 43, 1–10. 10.1556/0806.43.2015.026 [DOI] [Google Scholar]
- Zhang J., Chen J., Bowman B. C., O'Brien K., Marshall J. M., Bonman J. M. (2014). Association mapping of hagberg falling number in hard white spring wheat. Crop Sci. 54:1243 10.2135/cropsci2013.08.0551 [DOI] [Google Scholar]
- Zhang Z., Ersoz E., Lai C.-Q., Todhunter R. J., Tiwari H. K., Gore M. A., et al. (2010). Mixed linear model approach adapted for genome-wide association studies. Nat. Genet. 42, 355–360. 10.1038/ng.546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Tang H., Cheng M.-P., Dankwa K. O., Chen Z.-X., Li Z.-Y., et al. (2017). Genome-wide association study for pre-harvest sprouting resistance in a large germplasm collection of chinese wheat landraces. Front. Plant Sci. 8:401. 10.3389/fpls.2017.00401 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.