Abstract
Conventional molecular assessment of tissue through histology, if adapted to fresh thicker samples, has the potential to enhance cancer detection in surgical margins and monitoring of 3D cell culture molecular environments. However, in thicker samples, substantial background staining is common despite repeated rinsing, which can significantly reduce image contrast. Recently, “paired-agent” methods—which employ co-administration of a control (untargeted) imaging agent—have been applied to thick-sample staining applications to account for background staining. To date, these methods have included (1) a simple ratiometric method that is relatively insensitive to noise in the data but has accuracy that is dependent on the staining protocol and the characteristics of the sample; and (2) a complex paired-agent kinetic modeling method that is more accurate but is more noise-sensitive and requires a precise serial rinsing protocol. Here, a new simplified mathematical model—the rinsing paired-agent model (RPAM)—is derived and tested that offers a good balance between the previous models, is adaptable to arbitrary rinsing-imaging protocols, and does not require calibration of the imaging system. RPAM is evaluated against previous models and is validated by comparison to estimated concentrations of targeted biomarkers on the surface of 3D cell culture and tumor xenograft models. This work supports the use of RPAM as a preferable model to quantitatively analyze targeted biomarker concentrations in topically stained thick tissues, as it was found to match the accuracy of the complex paired-agent kinetic model while retaining the low noise-sensitivity characteristics of the ratiometric method.
Keywords: breast cancer, quantitative imaging, epidermal growth factor receptor, tracer kinetic modeling
1 INTRODUCTION
Many preclinical and clinical applications in oncology would benefit from a rapid and repeatable method of evaluating cell-surface receptor expression (common targets of drug therapeutics) in thick tissues. For example, such a method could improve cancer resection surgery by providing an intraoperative means of assessing surgical margins for residual cancer (Singletary, 2002; Zhang et al., 2017; Nitin et al., 2009). It could also be used in drug screening to elucidate the molecular mechanisms of targeted therapies and the effects of molecular heterogeneity on drug efficacy in 3-dimensional (3D) cell culture studies (Justice et al., 2009; Haycock, 2011), which are replacing monolayer 2D cell culture studies that fail to mimic the complexity of in vivo tissues (Friedl and Alexander, 2011).
Methods have been developed to assess cell-surface receptor expression in thick fresh tissues via direct topical staining (e.g., with fluorescence (Davis et al., 2013) or surface-enhanced Raman scattering (SERS) agents (W Wang et al., 2016)) without the significant sample processing required in conventional immunohistochemistry, tissue microarrays, or mass spectrometry of non-vital tissues (Dabbs, 2013; Kononen et al., 1998; Lockhart and Winzeler, 2000; Stoeckli et al., 2001; Cukierman et al., 2001). Theoretically, such methods could be carried out with various imaging systems including clinical imaging systems (Keating et al., 2016), confocal fluorescence microscopy (Schiffhauer et al., 2009; Dobbs et al., 2016), spectrally encoded confocal microscopy (Brachtel et al., 2016), multi-photon microscopy (Zipfel et al., 2003), super-resolution microscopy (Huang et al., 2008), wide-area optical-sectioning structured illumination microscopy (SIM) (Fu et al., 2013), and microscopy with UV surface excitation (Levenson et al., 2016). However, most of these approaches require long imaging times, with the exception of wide-field fluorescence, SIM, and UV surface excitation, and all of these approaches are susceptible to high background signals because unbound contrast agents (ie. nonspecific staining) can be difficult or impractical to remove with tissue rinsing. Furthermore, tissue-dependent and wavelength-dependent variations in tissue optical properties also make it difficult to estimate target concentrations based on the detected intensity of the staining agents. These limitations have forced clinicians to rely upon traditional time-consuming histopathology methods to evaluate surgical margins, in which patients are sent home before the results can be attained. Expensive re-excision procedures often result from such post-operative analyses, such as in the case of breast cancer resection, where up to 40% of patients require call-back surgeries (Jacobs, 2008).
Recent work has established the potential for paired-agent methods to overcome the limitations of direct topical staining of thick tissues (Davis et al., 2013; Liu et al., 2009; Sinha et al., 2015; Wang et al., 2014a; Wang et al., 2016; Wang et al., 2015). Such methods utilize the signal from a control (untargeted) imaging agent that is co-administered with one or more targeted agent(s) to account for background staining and variable optical properties (Tichauer et al., 2015). Moreover, a few studies have demonstrated that paired-agent kinetic modeling—an analysis of temporally varying signals (kinetics) of paired-agents—is capable of quantifying targeted receptor concentrations (Blasberg et al., 1987; Huang et al., 1989; Tichauer et al., 2012b). This achievement is essentially the ultimate goal of most molecular imaging protocols and has recently been shown to provide unprecedented levels of sensitivity in cancer detection (Tichauer et al., 2014b). The receptor concentration here is defined as the number of receptors within the volume of entire pixel, which is equivalent to the product of multiplication of the cell density and the average number of receptors per cell (which is different for different cell lines).
To date, most paired-agent kinetic models have been applied to in vivo studies employing systemic administration of imaging agents (Tichauer et al., 2015). Only one model has been applied to thick tissue imaging, a paired-agent model accounting for nonspecific binding (DPM-NS) (Sinha et al., 2015). Though it was the first method capable of quantifying receptor concentration using optical nanoparticles topically applied on fresh tissues, the model requires estimation of 5 kinetic parameters, making it sensitive to experimental noise. Furthermore, it can only analyze serial rinsing data in which each rinse step is identical, a requirement that can be difficult to achieve in certain applications. All other paired-agent applications have employed very simple, quick, and relatively noise-insensitive, “non-kinetic-modeling” ratiometric approaches (Davis et al., 2013; Liu et al., 2009; Wang et al., 2015; Wang et al., 2016): i.e., dividing the targeted agent signal by the control agent signal (ratiometric). These methods have been shown to correlate with receptor concentration under certain conditions (Sinha et al., 2015); however, the linearity of the correlation is highly dependent on the sample type and experimental protocol.
In this paper, an improved simple and adaptable rinsing paired-agent kinetic model (RPAM) is presented that offers an optimal balance between the accuracy of DPM-NS, as well as the noise-insensitivity and simplicity of ratiometric methods. Moreover, RPAM does not require a specific staining and rinsing procedure, and can theoretically be applied to data obtained with arbitrary staining protocols and potentially non-uniform rinsing steps. To evaluate RPAM, data from two distinct experiments were analyzed using RPAM, DPM-NS, and a ratiometric method—one experiment in which 3D cell cultures were stained in vitro, and the other experiment in which tumor xenograft tissues were stained ex vivo. In each case, epidermal growth factor receptor (EGFR) concentrations (a cell surface transmembrane protein upregulated in many cancers (Nicholson et al., 2001)) were targeted and the results from all paired-agent estimates of EGFR concentration were compared against flow cytometry measures.
2 METHODS
2.1 3D cell culture experiment
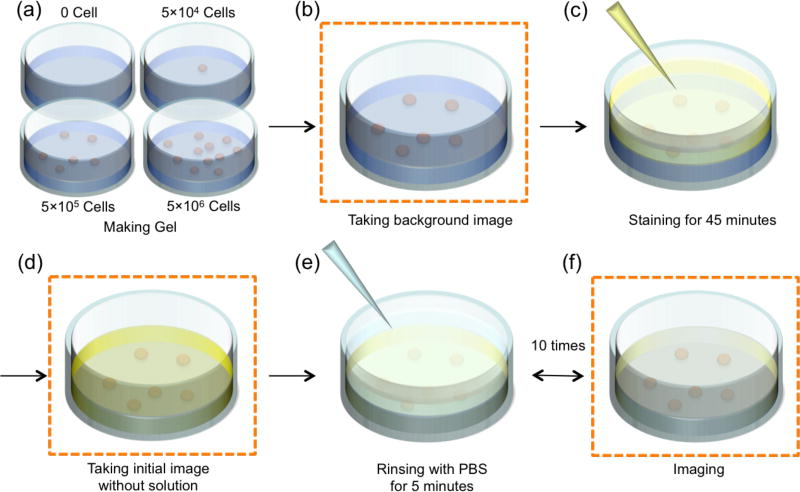
Two cancer cell lines, human epidermoid carcinoma (A431; from Dr. Pogue at Dartmouth College) and human neuronal glioblastoma (U251; from Dr. Pogue at Dartmouth College) were used to test the accuracy of receptor concentration estimation for the various paired-agent methods. Both cell lines were cultured in Dulbecco’s Modified Eagle Medium (DMEM) with 10% fetal bovine serum (FBS) and 1% penicillin-streptomycin (PS). As a simple approximation of standard 3D cell culture, cells were seeded homogeneously in a single layer of 0.3% agarose without adding growth medium in four different concentration groups. This involved resuspension of cells in 500 µL of warm Phosphate Buffered Saline (PBS) solution followed by mixing with 500 µL of 0.6% warm agarose gel. The solutions were immediately seeded into a transparent 6-well plate at various concentrations, including a blank (Control; 0 cells/mL), low (L; 5×104 cells/mL), medium (M; 5×105 cells/mL) and high (H; 5×106 cells/mL) cell concentration (Fig. 1a). These cell densities were chosen to mimic the cell concentration in 3D cell culture (Holy et al., 2000). The depth of all cell culture models was approximately 1 mm.
Figure 1.
Stepwise illustration of staining and rinsing procedures for 3D cell cultures in a well plate. Imaging steps are indicated in panels b, d, and e. All images are obtained with supernatant removed. These procedures were applied to all wells.
After creating 3D cell culture models with various cell concentrations, all samples were stained, followed by repeated rinsing-and-imaging steps (Fig. 1). A planar fluorescence imaging system (Pearl® Impulse, LICOR Biosciences, Lincoln, NE) was used to image white light reflectance and fluorescence emission at both 700–740 nm (“700 channel”) and 800–840 nm (“800 channel”) arising from excitation at 685 nm and 785 nm excitation, respectively. All imaging steps were performed in rapid succession within a 30-s time window. Before adding imaging agents to the four 3D cell culture suspensions, images were first collected from all channels (Fig. 1b) in order to measure the background autofluorescence. A 1-mL mixed solution of 44-nM IRDye® 800CW EGF and a 4-nM IRDye® 700DX NHS ester solution (LICOR Biosciences) was added to the surface of each sample as the “staining” step in Fig. 1c. The targeted agent (IRDye® 800CW EGF) was mixed with the control agent (IRDye® 700DX) only after the control agent was reacted with distilled water for 1 h at room temperature to hydrolyze and deactivate the reactive NHS ester. The 3D cell cultures were allowed to soak in the mixed imaging agent mixture for 45 min to achieve an even distribution of imaging agent within the agarose gel (determined by modeling of agent diffusion in the cellular matrix – results not shown). “Zero-rinse” images, Fig. 1d, were acquired after carefully removing all of the remaining solution on the surface of the cellular matrix. These images were used as initial values for the signal intensity of the imaging agent. Then, ten subsequent rinses were carried out on all wells by incubating the cell cultures in 1 mL of PBS for 5 minutes per rinse, Fig. 1e, 1f. Imaging was carried out after carefully pipetting away the supernatant after each rinse.
2.2 Ex-vivo tumor xenograft experiment
To evaluate the accuracy of receptor concentration estimation on ex vivo tissue samples, data from this study were compared with data collected in a previous study (Sinha et al., 2015). Briefly, in the previous study, excised A431 and U251 tumor xenografts were developed subcutaneously in male nude mice (n = 8 for U251; n = 9 for A431). All animal experiments were carried out under the approval of the Institutional Animal Care and Use Committee (IACUC) at Stony Brook University. Animals were euthanized when the tumor was greater than 5 mm in diameter and then the tumor, and a control muscle tissue of equivalent size, were excised. A tissue-background spectrum was acquired before and after paired-agent staining. The staining procedure involved adding 15 µL of a mixture of targeted and control surface-enhanced Raman scattering nanoparticles (SERS NPs) of different “flavours” (150 pM per NP flavour). The targeted NPs were coated with EGFR-targeted anti-EGFR monoclonal antibodies and the control NPs were coated with isotype control monoclonal antibodies. Spectral signals from the NPs were measured periodically between each of the 10 serial rinses (spray with 0.1 mL of PBS solution followed by gentle removal). Concentrations of the two NPs were then extracted from the raw signals by spectral decomposition (Wang et al., 2014a).
2.3 Image pre-processing
Each pair of fluorescent images (for the targeted and control agents) required correction for 1) autofluorescence; 2) differences in detection efficiency between different imaging channels (only required for ratiometric and DPM-NS methods); and 3) differences in targeted and control imaging agent diffusion into (for staining) and out of (for rinsing) the cellular matrix (only needed for the 3D cell culture data). Autofluorescence was removed through subtraction of pre-staining images (for the respective imaging channels) from all subsequent post-staining images. The relative signal intensities of the targeted and control imaging agents were calibrated by imaging a stock staining solution of a known concentration and concentration ratio (for additional details, see refs (Liu et al., 2009) and (Wang et al., 2014b)). A deconvolution algorithm (Tichauer et al., 2014a) was used to correct for differences in the diffusion of various fluorophores within tissues. Numbers shown were averaged signal intensity over the entire well or tissue.
2.4 Image analysis
Four image-analysis methods were employed to estimate EGFR concentrations in 3D cell cultures and ex vivo experiments.
2.4.1 Conventional fluorescence
The raw signal from targeted imaging agents, after staining and sufficient rinsing (3 rinses), were assessed as a possible correlate of receptor concentration (the vast majority of tissue staining studies assume this to be the case).
2.4.2 Ratiometric
This method involved dividing the pre-processed control imaging agent signal/map (SC) from the targeted agent signal/map (ST) and then subtracting by one (Tichauer et al., 2012a):
| (1) |
where ti is an arbitrary imaging time point after rinsing (evaluated at all points in this study). The binding potential (BP) parameter is often used in quantitative molecular imaging studies as it is equivalent to the product of the affinity of the targeted imaging agent (Ka) for its receptor and the concentration of the receptor (Innis et al., 2007). While used for decades, the ratiometric method was only recently demonstrated to be an approximation of BP, defined here as BPRatio (Tichauer et al., 2012a).
2.4.3 Paired-agent model with nonspecific binding (DPM-NS)
This method is mathematically complex and a full description of the method can be found in a previous publication (Sinha et al., 2015). Briefly, it requires signals from kinetic measurements of contrast agent intensities (both targeted and control) from multi-rinse imaging data in which the rinsing steps are carried out identically at each step to be measured and quantified (i.e., converted to imaging agent concentration). Factors that obfuscate the relationship between the signal from a targeted agent and receptor concentration (specifically, the rinsing and nonspecific binding parameters) are estimated from the kinetics of the control agent and then used in the analysis of the targeted agent kinetics to accurately measure receptor concentration (BP).
2.4.4 Rinsing paired-agent model (RPAM)
This novel method is derived and explained in detail in the Supplementary Section. Briefly, this method takes in any sequence of targeted and control imaging agent signals in the process of repeated staining (ST(t) and SC(t), respectively) and normalizes them to the signals measured at an arbitrary time point, ti (i.e., any one time point in the sequence), to attain the normalized targeted agent signal, ST(t)/ST(ti), and the normalized control agent signal, SC(t)/SC(ti). If the properties of the targeted and control imaging agents are very similar (save the fact that the targeted agent will stick to the biomarker of interest while the control agent will not), then it is possible to mathematically relate the normalized targeted and control imaging agent signals in a region of interest as a function of the binding potential (BP), a parameter directly proportional to the concentration of the biomarker in that region of interest. Assuming the rinsing protocol acts equally on the targeted and control agents, this mathematical relationship can be simplified to obtain an estimate of BP (BPRPAM) as:
| (2) |
where ti is any single arbitrary imaging time point after rinsing, and t denotes a time vector (i.e., ST(ti) is the targeted agent signal at an arbitrary time point; whereas, ST(t) is the signal at multiple time points, which could include ti).
2.5 Simulation experiment
All receptor concentration estimation methods were tested on computer-simulated targeted and control agent kinetic curves in which random Gaussian-distributed noise was added (where “true” values of all fitting parameters, e.g. BP, are known). A detailed description of the simulations can be found in the Supplementary Section. The testing was carried out over a range of rinse durations and BP levels. For every case, receptor concentration estimation methods were applied to 1000 iterations of simulated data (with random noise that approximated experimental noise levels). This allowed us to evaluate the precision and accuracy of each method under the various conditions.
2.6 Statistical analyses
SPSS 23 (IBM) was used for all statistical analyses. A repeated-measures MANOVA with Bonferroni correction to correct for multiple comparisons and Tukey’s-B post hoc tests were used to identify statistically significant effects amongst the different cell concentration and cell type groups, with rinse number as the within-subject variable, and cell concentration and cell type as the between-subjects variables. A univariate ANOVA test was used to compare BP estimates as a function of different concentrations, cell types, and methods for estimating receptor concentration. Statistical significance was based on p < 0.05. All data are presented as mean ± SD.
3 Results
3.1 In vitro 3D cell culture
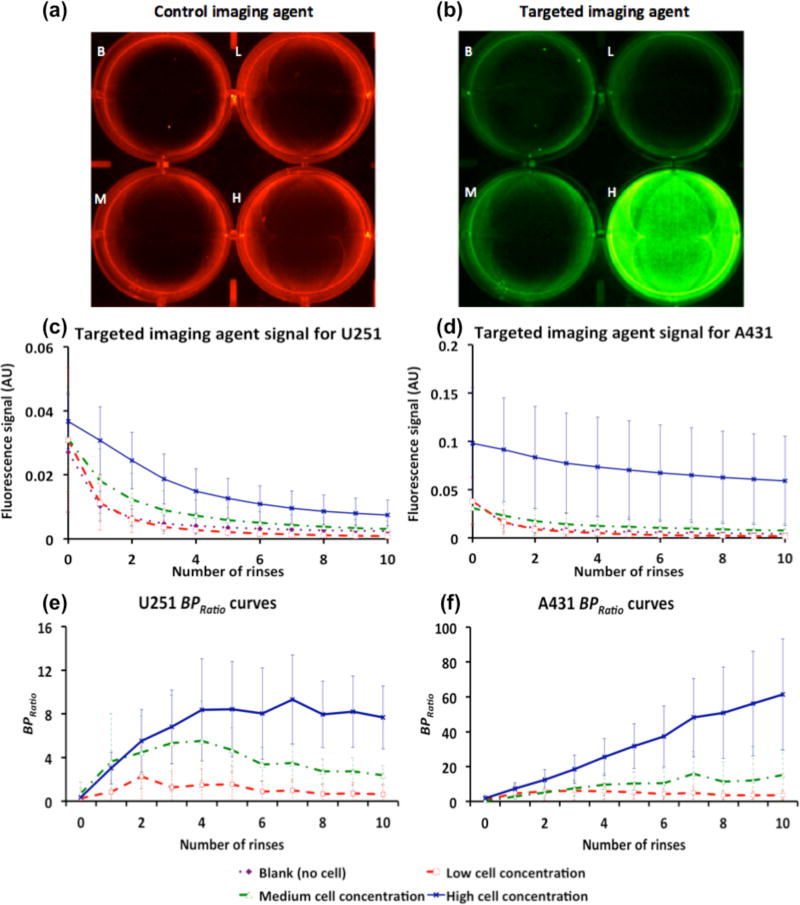
The retention of control and EGFR-targeted fluorescent imaging agents in a U251 3D cell culture (after ten rinses) is shown in Figs. 2a and 2b, respectively. Signal intensity from targeted and control imaging agents averaged over all repeated datasets for each well as a function of the number of rinses are presented in Fig. 2c for the U251 cell line group and in Fig. 2d for the A431 cell line group. Using a repeated measures MANOVA (see Methods), the only statistically significant effects identified were that the fluorescence measured from the high-cell-concentration wells in both U251 and A431 groups were significantly higher than all other wells within the same group (p < 0.001). There were no other statistically significant effects between cell concentration wells within the same cell type groups (p > 0.05, NS). Figures 2e and 2f present the BPRatio from the cell culture studies as a function of rinse number for the U251 and A431 cell type groups, respectively. Paired samples t-tests with Bonferroni corrections indicated significant changes in BPRatio within the first 4 rinses, but no statistically significant differences amongst data collected after rinse number 4. The point with the highest contrast-to-noise between cell concentration groups was rinse 8.
Figure 2.
(a) and (b), Fluorescence images of a 6-well plate after 10 rinses for (a) control imaging agent (700-nm fluorescence channel) and (b) targeted imaging agent (800-nm fluorescence channel), with 0 (Blank, B), 5×104 (Low, L), 5×105 (Medium, M) and 5×106 (High, H) cells. Targeted imaging agent signal as a function of number of rinses for (c) U251 and (d) A431 cell cultures. Calculated BPRatio as a function of number of rinses for (e) U251 and (f) A431 cell cultures. Blank (purple dot diamond marker), Low (red dash square marker), Medium (green dash-dot triangle marker) and High (blue line cross marker). Error bars represent standard deviation.
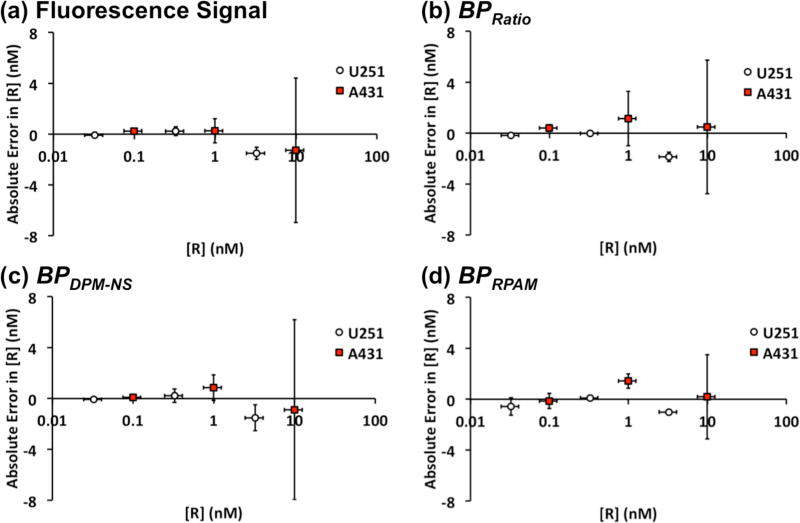
To compare each analysis method with the expected receptor concentration (determined by flow cytometry), root-mean-square error (RMSE) and the sum of absolute error (SAE) were calculated, table 1. RPAM estimation of receptor concentration (BPRPAM) exhibited the smallest error compared with receptor concentration estimates based on raw fluorescence signal, BPDPM-NS, and fluorescence ratios, BPRatio. To visualize the correlation error, absolute errors (fluorescence or BP values) were plotted against the expected receptor concentrations (Fig. 3), which were obtained from flow cytometry experiments carried out in a previous study (Sinha et al., 2015). By the definition of binding potential (BP = KaBavail) (Innis et al., 2007), the slope of the correlation curve between receptor concentration (Bavail) and BP should be Ka (association constant) or 1/KD (dissociation constant). The estimated KD were 0.21 nM, 0.38 nM and 1.77 nM for the ratiometric method, DPM-NS, and RPAM, respectively.
Table 1.
Measurements of correlation error.
| ROOT-MEAN- SQUARE ERROR |
SUM OF ABSOLUTE ERROR (all data points) |
SUM OF ABSOLUTE ERROR (averaged) |
|
|---|---|---|---|
| Fluorescence | 0.343 | 78.7 | 3.61 |
| BPRatio | 0.434 | 75.3 | 4.10 |
| BPDPM-NS | 0.324 | 78.9 | 3.65 |
| BPRPAM | 0.288 | 68.1 | 3.43 |
Figure 3.
Correlation between absolute error in receptor concentration of (a) raw fluorescence intensity, (b) BPRatio, (c) BPDPM-NS, (d) BPRPAM and expected receptor concentration. White circle indicate U251 results and red square indicate A431 results. Error bars indicate standard deviation.
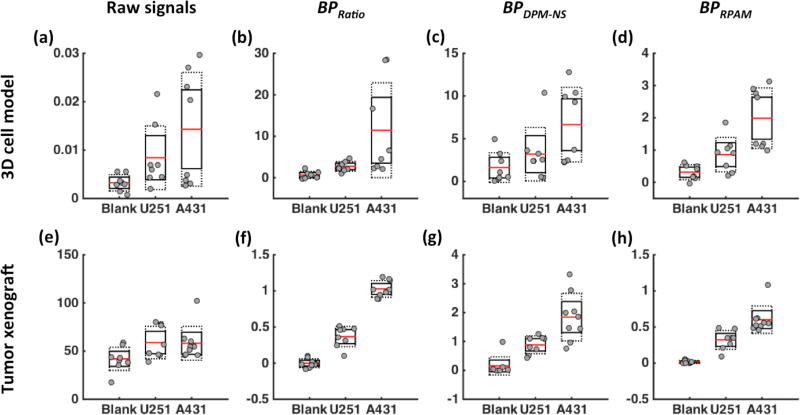
Figs. 4a–d present boxplots of data from 3D cell cultures of blank, U251, and A431 cell lines (medium concentration) 3D cell cultures to further evaluate the characteristics of each method for estimating receptor concentration (raw fluorescence, ratiometric, DPM-NS, and RPAM). No statistically significant differences in cell lines were observed using fluorescence alone (Fig. 4a). BPRatio (Fig. 4b) and BPDPM-NS (Fig. 4c) displayed statistically significant separations between groups (p < 0.05), while RPAM achieved the best separation between the two tumor cell lines as determined by the contrast-to-variance ratio between groups (p < 0.01) (Fig. 4d). Based on flow cytometry validation data, the relative ratio of EGFR expression between A431 cell and U251 cell was used as a method to validate the accuracy of receptor concentration estimations. The expected ratio for EGFR expression was 3.05 ± 0.88 from an in vitro study (Sinha et al., 2015). Based on the experimental result, the estimated EGFR expression ratio (A431 vs. U251) was 1.70 ± 1.93, 4.20 ± 4.56, 2.09 ± 2.47, and 2.31 ± 1.81 for fluorescence, ratiometric, DPM-NS, and RPAM estimations, respectively.
Figure 4.
Estimating receptor expression levels for different tissues (normal tissue, U251 and A431 tumor models (n=8)) based on various methods: (a,e) raw fluorescence signals, (b,f) BPRatio, (c,g) BPDPM-NS, (d,h) BPRPAM. The top row shows data from 3D cell cultures stained with a fluorescent EGFR probe. The bottom row shows data from tumor xenografts in mice stained with EGFR-targeted SERS nanoparticles.
3.2 Ex vivo tumor tissue
Figures 4e–h present similar data to Figs. 4a–d but with the four methods applied to ex vivo tumor tissue data rather than 3D cell culture data. Nanoparticle concentration levels were found to be significantly higher in tumors than in muscle (normal) tissue (p < 0.05); however, no statistically significant difference was observed between the two tumor cell lines, in which the receptor concentration ratio between A431 and U251 xenografts was expected to be 2.1 ± 0.3 (Sinha et al., 2015) (Fig. 4e). Conversely, all other methods, BPRatio (Fig. 4f), BPDPM-NS (Fig. 4g), and BPRPAM (Fig. 4h) were found to exhibit statistically significant differences between normal tissue, U251 xenografts, and A431 xenografts (p < 0.01 for all comparisons with Bonferroni correction). In addition, the relative standard deviation of each group was significantly smaller compared to the raw fluorescence signal data. The EGFR expression ratios between U251 and A431 were 0.99 ± 0.41 based on raw fluorescence signal, 2.80 ± 1.12 for the ratiometric method, 2.09 ± 1.18 for the DPM-NS and 1.87 ± 0.96 for the RPAM.
3.3 Simulation
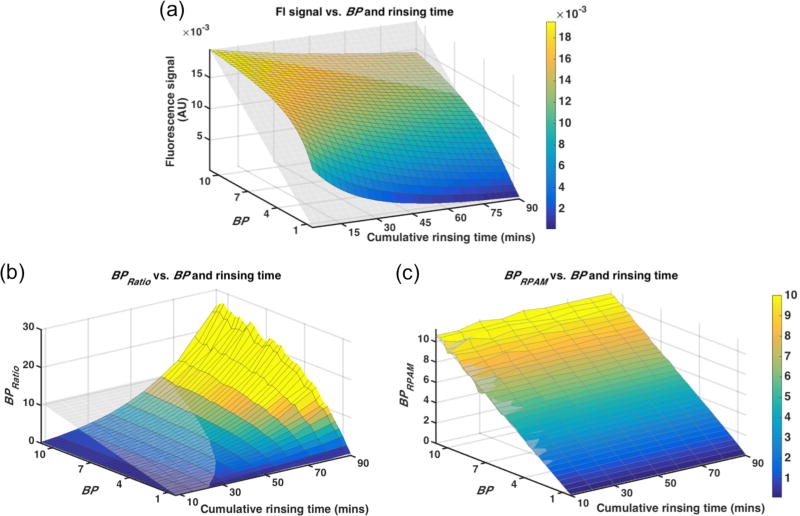
Simulation results are shown in Fig. 5. The fluorescence signal was expected to be linear with the increase of BP (proportional to receptor concentration). However, nonspecific signal from the “free” compartment caused an overestimation in fluorescence signal, particularly at shorter simulated rinsing times and with lower simulated binding potentials. No rinsing protocol was found to correlate linearly with binding potential (Fig. 5a demonstrates the analysis at rinse number 3). BPRatio also failed to correlate linearly with BP using any single rinsing strategy, though longer rinse times resulted in over-enhancement of BP estimation, which could be used to enhance differences between groups (Fig. 5b). On the other hand, BPRPAM demonstrated a strong correlation with BP input for all rinse strategies with the 10-min cumulative rinsing time for 10 rinses resulting in the best correlation (lowest RMSE of the correlation) compared to all other rinsing times.
Figure 5.
Simulation of BP (receptor concentration) estimation using (a) raw fluorescence signal, (b) BPRatio and (c) BPRPAM as a function of different BP inputs and rinsing durations. The coloured surface is the simulated data and the transparent mesh is: (a) the expected signal distribution, (b, c) the BP input map. Colour bar is set to be the same for panel (b) and (c) to compare the estimated BP value.
4 Discussion
Kinetic modeling in tissue fluorescence imaging provides a promising means of improving receptor concentration estimation in a sensitive and linear way for both ex vivo thick-tissue staining applications and in vitro 3D cell culture models. Fluorescence signals from topically applied imaging agents have long been used as a gold standard to approximate and indicate targeted molecule receptor concentrations in thin tissues (after sufficient rinsing). However, this study demonstrated through simulations, 3D cell culture staining experiments, and ex vivo xenograft tumor staining experiments, that this widely used approach can fail to correlate with receptor concentration in many scenarios. Not only did the raw fluorescence signal display the poorest RMSE and SAE in a correlation study analysis (Tab. 1, Fig. 3a), it failed to differentiate between different tissue types in 3D cell culture studies and tumor xenograft studies (Figs. 4a and 4e, respectively) and exhibited a non-linear relationship with receptor concentration in a simulation study (Fig. 5a).
The absence of a correlation between EGFR concentration and measured fluorescence of the EGFR-targeted imaging agents in this study reflects the complications that spatial variability of staining, rinsing, and nonspecific retention (retention of imaging agent in tissue that is not attributable to specific binding to the molecular target) can have on topical molecular imaging of thicker tissue samples. The paired-agent methods presented in this study rely on co-administration of a targeted imaging agent with a second, control imaging-agent to correct for these complications. It should be noted, however, that despite these complications, there is a large body of work demonstrating that topical staining can provide significant improvements in tissue identification (Nitin et al., 2009; DaCosta et al., 2005; Foersch et al., 2010; Hsiung et al., 2008; Nguyen and Tsien, 2013; Shin et al., 2010; Barth and Gibbs, 2017; Park et al., 2014). Other work has demonstrated improvements in signal-to-noise achievable with activatable imaging agents (Cutter et al., 2012; Urano et al., 2011) or induced autofluorescence (Leunig et al., 2001), that mitigate problems of non-specific retention (though can still be affected by uneven staining or rinsing). Paired-agent models do not aim to compete with these approaches, rather, they could be adapted to any of them to improve tissue identification. Three paired-agent molecular imaging analysis methods were tested: a straight forward ratiometric method (Liu et al., 2009), a complex DPM-NS method that aims to correct for nonspecific binding in addition to nonspecific retention of the imaging agents (Sinha et al., 2015), and a new RPAM method that offers a balance between the first two models in terms of the tradeoffs between complexity and sensitivity to noise and experimental error.
Although the measurements of RMSE and SAE for the ratiometric and DPM-NS methods showed that these methods can have a poor correlation with receptor concentration—similar to the raw-fluorescence-based estimates in the correlation analysis (Tab. 1, Figs. 3b and 3c)—both the ratiometric and DPM-NS methods displayed improved separation of the different tissue types (p < 0.05, Fig. 4) by accounting for nonspecific signal retention through paired-agent imaging principles (Tichauer et al., 2015). Thick ex vivo tissue rinsing did show a promising outcome with larger SERS nanoparticles (Figs. 4f and 4g), typically associated with higher levels of nonspecific retention owing to slow diffusion in tissue (Sinha et al., 2015). But there can be multiple problems related to these methods in a situation with lower nonspecific retention, as shown in the in vitro 3D cell culture study.
Specifically, the simulation study (Fig. 5b) demonstrated that non-linearity is the major problem for the ratiometric approach. This is further supported by the overestimation of the ratios observed in BPRatio values between the two tumor models in both ex vivo and in vitro studies (Figs. 4b and 4f). This overestimated ratio, while not accurate, could potentially be used to enhance tumor detection. However, a more rigorous evaluation of contrast-to-noise is warranted as these approaches are expanded to wide-field imaging studies of tissue specimens to ensure that the enhanced differentiation between groups is not overshadowed by an enhancement in the background signal noise and variability. Another limitation of this characteristic is that the pattern of non-linearity will be highly dependent on the conditions of the experiment (e.g., diffusion, tissue thickness, depth sensitivity of imaging system), so there would be a need to calibrate or optimize the method in an application-to-application basis.
Comparatively, DPM-NS estimation of BP was found to have the most linear prediction of receptor concentration in the ex vivo study (Fig. 4g). The limitation of DPM-NS is primarily in the rigidity of the modeling approach, which requires each rinse step to be very similar throughout the study. This is difficult to achieve in practice owing to experimental variability and the fact that rinsing may be driven by the concentration of the imaging agent in the sample (Fick’s Law), which will change over time. Simulations in this work demonstrated that even with equal duration staining and rinsing, the impact of the rinse (what fraction of agent is removed from the sample) can change (diminish) with repeated rinsing (data not shown). For DPM-NS, this change can lead to significant errors in BP estimation, which is why these results were not shown in the simulation studies (Fig. 5) that employed equal rinse durations between imaging.
The newly developed RPAM, in general, demonstrated the best performance of all four methods for both in vitro and ex vivo studies. It is mathematically simple and adaptable, and able to account for differences in rinsing efficacy/duration amongst rinse number. Not only did it exhibit the lowest RMSE and SAE in correlation studies (Tab. 1, Fig. 3d), it was also the most effective in differentiating between the different tissue types (Figs. 4d and 4h). Furthermore, in simulation studies, RPAM was the only model that exhibited a statistically significant linear correlation with BP for all rinsing strategies (p < 0.05, Fig. 5). For validation studies with tumor xenografts and 3D cell cultures, the disassociation constant estimated via RPAM (KD, 1.77 nM) were the closest to the expected KD range (2–5 nM) as compared to alternative methods (Klein et al., 2004; Bjorkelund et al., 2011).
It should be noted that all of the paired-agent models evaluated in this study (ratiometric, DPM-NS, and RPAM) are derived from compartment models that assume the free concentration of both targeted and control imaging agents in the tissue can be approximated as a single well-mixed compartment. However, in thick tissues, slower rates of imaging agent diffusion can significantly reduce the potential for even distribution of imaging agents throughout the imaging volume. Preliminary simulations carried out using a numerical solution to a diffusion equation that includes this complexity demonstrated that all models worked best in thick tissue under two conditions: (1) when the staining and total rinsing times are close to the characteristic diffusion time (L2/D; where L is the thickness of the sample and D is the diffusion coefficient), thus allowing time for even mixing and rinsing of the agents through the whole sample; and (2) when staining and rinsing times are much less than the characteristic diffusion time, so that much of the staining happens just at the surface of the tissue (results not shown). Rough estimates of diffusion coefficients of the IRDye-800CW-EGF in 0.6% agarose and SERS nanoparticles in the tissue are on the order of 1 × 10−9 m2/s (Liang et al., 2006) and 1 × 10−15 m2/s (experimentally derived), respectively. For the 3D cell culture, the media was ~1 mm in depth, yielding a characteristic diffusion time of ~ 20 min. With total staining and total rinsing times exceeding 45 min, this experiment matches well with the first condition of single-compartment assumption accuracy. On the other hand, for the ex vivo SERS study, tissue thickness was on the order of 5 mm, yielding a characteristic diffusion time of approximately 800 years. The estimated distance diffused within the staining and rinsing protocol was less than the diameter of a cell, so most of the binding occurred right at the surface of the tissue (which is all we care about for the application of margin assessment), matching well with the second condition of single-compartment assumption accuracy. A more in-depth analysis of the effect of diffusion on paired-agent models and an exploration of alternative models that are less sensitive to these effects are outside of the scope of this article but of key interest going forward.
Other concerns with any targeted molecular imaging study include the influence of non-specific binding and cellular internalization of the targeted agent, each of which can affect the accuracy of the estimation of receptor concentration through binding potential. Significant amounts of non-specific binding or cellular internalization of the targeted imaging agent alone will typically lead to overestimations in binding potential; however, these will be mitigated in all paired-agent methods as long as the level(s) of non-specific binding (and/or internalization) are similar for the control imaging agent. While it is known that the two targeted agents, IRDye800CW-EGF and SERS nanoparticles can be internalized (Felder et al., 1992; Song et al., 2012), little is known about the non-specific binding of these agents. An in-depth analysis of both nonspecific binding and imaging agent internalization are outside of the scope of this article but are also of interest for future work. However, it should be noted that the receptor concentrations estimated using both pairs of imaging agents for the two cell lines in all conditions, matched well with expected levels of EGFR. These agents have also been used extensively and validated in other work, where EGFR concentrations matched well with gold standard and expected levels (Leigh et al., 2013; Wang et al., 2016; Wang et al., 2015; Wang et al., 2014b; Tichauer et al., 2012a; Tichauer et al., 2012b; Samkoe et al., 2014). This body of work suggests that both non-specific binding and internalization are relatively negligible, at least within 1 h of initial administration of the imaging agents.
5 Conclusion
Amongst the three methods compared in this study, RPAM shows the best outcome in receptor concentration prediction and is the only method to provide a linear estimation of receptor concentration. This linearity is crucial for imaging 3D cell cultures and tissues with heterogeneous molecular phenotypes, and can be used to differentiate between different cell lines. Furthermore, this model is not limited to a fixed rinsing protocol and has the flexibility to be applied for the analysis of serial rinsing data with unequal rinsing steps as well. Both in vitro and ex vivo data were tested in this study, demonstrating the strength of the RPAM method compared to conventional detection of a single fluorescent imaging agent, paired-agent ratiometric approximations of binding potential, and a more advanced but rigid DPM-NS model that requires identical rinse steps.
Supplementary Material
Acknowledgments
Work for this study was supported by Illinois Institute of Technology Educational and Research Initiative Fund ERIF awarded to K.T. and the Nayar Prize awarded to K.T. at Illinois Institute of Technology. We thank Shujun Tang for their assistance with 3D cell culture. We thank two “anonymous” reviewers for their insightful comments.
Footnotes
None of the authors had a conflict of interest.
References
- Barth CW, Gibbs SL. Direct Administration of Nerve-Specific Contrast to Improve Nerve Sparing Radical Prostatectomy. Theranostics. 2017;7:573–93. doi: 10.7150/thno.17433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjorkelund H, Gedda L, Andersson K. Comparing the epidermal growth factor interaction with four different cell lines: intriguing effects imply strong dependency of cellular context. PLoS One. 2011;6:e16536. doi: 10.1371/journal.pone.0016536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blasberg RG, Nakagawa H, Bourdon MA, Groothuis DR, Patlak CS, Bigner DD. Regional localization of a glioma-associated antigen defined by monoclonal antibody 81C6 in vivo: kinetics and implications for diagnosis and therapy. Cancer research. 1987;47:4432–43. [PubMed] [Google Scholar]
- Brachtel EF, Johnson NB, Huck AE, Rice-Stitt TL, Vangel MG, Smith BL, Tearney GJ, Kang D. Spectrally encoded confocal microscopy for diagnosing breast cancer in excision and margin specimens. Laboratory Investigation. 2016 doi: 10.1038/labinvest.2015.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cukierman E, Pankov R, Stevens DR, Yamada KM. Taking cell-matrix adhesions to the third dimension. Science. 2001;294:1708–12. doi: 10.1126/science.1064829. [DOI] [PubMed] [Google Scholar]
- Cutter JL, Cohen NT, Wang J, Sloan AE, Cohen AR, Panneerselvam A, Schluchter M, Blum G, Bogyo M, Basilion JP. Topical application of activity-based probes for visualization of brain tumor tissue. PloS one. 2012;7:e33060. doi: 10.1371/journal.pone.0033060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dabbs DJ. Diagnostic immunohistochemistry. Elsevier Health Sciences; 2013. [Google Scholar]
- DaCosta RS, Wilson BC, Marcon NE. Optical techniques for the endoscopic detection of dysplastic colonic lesions. Current opinion in gastroenterology. 2005;21:70–9. [PubMed] [Google Scholar]
- Davis SC, Gibbs SL, Gunn JR, Pogue BW. Topical dual-stain difference imaging for rapid intra-operative tumor identification in fresh specimens. Optics letters. 2013;38:5184–7. doi: 10.1364/OL.38.005184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobbs JL, Shin D, Krishnamurthy S, Kuerer H, Yang W, Richards-Kortum R. Confocal fluorescence microscopy to evaluate changes in adipocytes in the tumor microenvironment associated with invasive ductal carcinoma and ductal carcinoma in situ. International journal of cancer. 2016 doi: 10.1002/ijc.30160. [DOI] [PubMed] [Google Scholar]
- Felder S, LaVin J, Ullrich A, Schlessinger J. Kinetics of binding, endocytosis, and recycling of EGF receptor mutants. The Journal of cell biology. 1992;117:203–12. doi: 10.1083/jcb.117.1.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foersch S, Kiesslich R, Waldner MJ, Delaney P, Galle PR, Neurath MF, Goetz M. Molecular imaging of VEGF in gastrointestinal cancer in vivo using confocal laser endomicroscopy. Gut. 2010;59:1046–55. doi: 10.1136/gut.2009.202986. [DOI] [PubMed] [Google Scholar]
- Friedl P, Alexander S. Cancer invasion and the microenvironment: plasticity and reciprocity. Cell. 2011;147:992–1009. doi: 10.1016/j.cell.2011.11.016. [DOI] [PubMed] [Google Scholar]
- Fu HL, Mueller JL, Javid MP, Mito JK, Kirsch DG, Ramanujam N, Brown JQ. Optimization of a widefield structured illumination microscope for non-destructive assessment and quantification of nuclear features in tumor margins of a primary mouse model of sarcoma. PloS one. 2013;8:e68868. doi: 10.1371/journal.pone.0068868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haycock JW. 3D cell culture: a review of current approaches and techniques. Methods in molecular biology. 2011;695:1–15. doi: 10.1007/978-1-60761-984-0_1. [DOI] [PubMed] [Google Scholar]
- Holy CE, Shoichet MS, Davies JE. Engineering three-dimensional bone tissue in vitro using biodegradable scaffolds: Investigating initial cell-seeding density and culture period. Journal of Biomedical Materials Research. 2000;51:376–82. doi: 10.1002/1097-4636(20000905)51:3<376::aid-jbm11>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- Hsiung PL, Hardy J, Friedland S, Soetikno R, Du CB, Wu AP, Sahbaie P, Crawford JM, Lowe AW, Contag CH, Wang TD. Detection of colonic dysplasia in vivo using a targeted heptapeptide and confocal microendoscopy. Nature medicine. 2008;14:454–8. doi: 10.1038/nm1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang B, Wang W, Bates M, Zhuang X. Three-dimensional super-resolution imaging by stochastic optical reconstruction microscopy. Science. 2008;319:810–3. doi: 10.1126/science.1153529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang SC, Bahn MM, Barrio JR, Hoffman JM, Satyamurthy N, Hawkins RA, Mazziotta JC, Phelps ME. A double-injection technique for in vivo measurement of dopamine D2-receptor density in monkeys with 3-(2'-[18F]fluoroethyl)spiperone and dynamic positron emission tomography. Journal of cerebral blood flow and metabolism : official journal of the International Society of Cerebral Blood Flow and Metabolism. 1989;9:850–8. doi: 10.1038/jcbfm.1989.119. [DOI] [PubMed] [Google Scholar]
- Innis RB, Cunningham VJ, Delforge J, Fujita M, Gjedde A, Gunn RN, Holden J, Houle S, Huang SC, Ichise M, Iida H, Ito H, Kimura Y, Koeppe RA, Knudsen GM, Knuuti J, Lammertsma AA, Laruelle M, Logan J, Maguire RP, Mintun MA, Morris ED, Parsey R, Price JC, Slifstein M, Sossi V, Suhara T, Votaw JR, Wong DF, Carson RE. Consensus nomenclature for in vivo imaging of reversibly binding radioligands. Journal of cerebral blood flow and metabolism : official journal of the International Society of Cerebral Blood Flow and Metabolism. 2007;27:1533–9. doi: 10.1038/sj.jcbfm.9600493. [DOI] [PubMed] [Google Scholar]
- Jacobs L. Positive margins: the challenge continues for breast surgeons. Annals of surgical oncology. 2008;15:1271–2. doi: 10.1245/s10434-007-9766-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Justice BA, Badr NA, Felder RA. 3D cell culture opens new dimensions in cell-based assays. Drug discovery today. 2009;14:102–7. doi: 10.1016/j.drudis.2008.11.006. [DOI] [PubMed] [Google Scholar]
- Keating J, Tchou J, Okusanya O, Fisher C, Batiste R, Jiang J, Kennedy G, Nie S, Singhal S. Identification of breast cancer margins using intraoperative near-infrared imaging. Journal of surgical oncology. 2016;113:508–14. doi: 10.1002/jso.24167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein P, Mattoon D, Lemmon MA, Schlessinger J. A structure-based model for ligand binding and dimerization of EGF receptors. Proc Natl Acad Sci U S A. 2004;101:929–34. doi: 10.1073/pnas.0307285101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kononen J, Bubendorf L, Kallioniemi A, Barlund M, Schraml P, Leighton S, Torhorst J, Mihatsch MJ, Sauter G, Kallioniemi OP. Tissue microarrays for high-throughput molecular profiling of tumor specimens. Nature medicine. 1998;4:844–7. doi: 10.1038/nm0798-844. [DOI] [PubMed] [Google Scholar]
- Leigh SY, Som M, Liu JT. Method for assessing the reliability of molecular diagnostics based on multiplexed SERS-coded nanoparticles. PloS one. 2013;8:e62084. doi: 10.1371/journal.pone.0062084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leunig A, Mehlmann M, Betz C, Stepp H, Arbogast S, Grevers G, Baumgartner R. Fluorescence staining of oral cancer using a topical application of 5-aminolevulinic acid: fluorescence microscopic studies. Journal of photochemistry and photobiology. B, Biology. 2001;60:44–9. doi: 10.1016/s1011-1344(01)00117-8. [DOI] [PubMed] [Google Scholar]
- Levenson R, Harmany ZT, Krueger AR, Fereidouni F. Rapid, Inexpensive Slide-Free Histology via MUSE: Ultraviolet Surface Excitation Microscopy for Imaging Unsectioned Tissue. Modern Pathol. 2016;29:517A-A. [Google Scholar]
- Liang S, Xu J, Weng L, Dai H, Zhang X, Zhang L. Protein diffusion in agarose hydrogel in situ measured by improved refractive index method. Journal of controlled release : official journal of the Controlled Release Society. 2006;115:189–96. doi: 10.1016/j.jconrel.2006.08.006. [DOI] [PubMed] [Google Scholar]
- Liu JT, Helms MW, Mandella MJ, Crawford JM, Kino GS, Contag CH. Quantifying cell-surface biomarker expression in thick tissues with ratiometric three-dimensional microscopy. Biophysical journal. 2009;96:2405–14. doi: 10.1016/j.bpj.2008.12.3908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockhart DJ, Winzeler EA. Genomics, gene expression and DNA arrays. Nature. 2000;405:827–36. doi: 10.1038/35015701. [DOI] [PubMed] [Google Scholar]
- Nguyen QT, Tsien RY. Fluorescence-guided surgery with live molecular navigation--a new cutting edge. Nature reviews. Cancer. 2013;13:653–62. doi: 10.1038/nrc3566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson RI, Gee JM, Harper ME. EGFR and cancer prognosis. European journal of cancer. 2001;37(Suppl 4):S9–15. doi: 10.1016/s0959-8049(01)00231-3. [DOI] [PubMed] [Google Scholar]
- Nitin N, Carlson AL, Muldoon T, El-Naggar AK, Gillenwater A, Richards-Kortum R. Molecular imaging of glucose uptake in oral neoplasia following topical application of fluorescently labeled deoxy-glucose. International journal of cancer. Journal international du cancer. 2009;124:2634–42. doi: 10.1002/ijc.24222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park MH, Hyun H, Ashitate Y, Wada H, Park G, Lee JH, Njiojob C, Henary M, Frangioni JV, Choi HS. Prototype nerve-specific near-infrared fluorophores. Theranostics. 2014;4:823–33. doi: 10.7150/thno.8696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samkoe KS, Tichauer KM, Gunn JR, Wells WA, Hasan T, Pogue BW. Quantitative in vivo immunohistochemistry of epidermal growth factor receptor using a receptor concentration imaging approach. Cancer research. 2014;74:7465–74. doi: 10.1158/0008-5472.CAN-14-0141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiffhauer LM, Boger JN, Bonfiglio TA, Zavislan JM, Zuley M, Fox CA. Confocal microscopy of unfixed breast needle core biopsies: a comparison to fixed and stained sections. BMC cancer. 2009;9:1. doi: 10.1186/1471-2407-9-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin D, Vigneswaran N, Gillenwater A, Richards-Kortum R. Advances in fluorescence imaging techniques to detect oral cancer and its precursors. Future oncology. 2010;6:1143–54. doi: 10.2217/fon.10.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singletary SE. Surgical margins in patients with early-stage breast cancer treated with breast conservation therapy. American journal of surgery. 2002;184:383–93. doi: 10.1016/s0002-9610(02)01012-7. [DOI] [PubMed] [Google Scholar]
- Sinha L, Wang Y, Yang C, Khan A, Brankov JG, Liu JTC, Tichauer KM. Quantification of the binding potential of cell-surface receptors in fresh excised specimens via dual-probe modeling of SERS nanoparticles. Scientific reports. 2015;5:8582. doi: 10.1038/srep08582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J, Zhou J, Duan H. Self-assembled plasmonic vesicles of SERS-encoded amphiphilic gold nanoparticles for cancer cell targeting and traceable intracellular drug delivery. Journal of the American Chemical Society. 2012;134:13458–69. doi: 10.1021/ja305154a. [DOI] [PubMed] [Google Scholar]
- Stoeckli M, Chaurand P, Hallahan DE, Caprioli RM. Imaging mass spectrometry: a new technology for the analysis of protein expression in mammalian tissues. Nature medicine. 2001;7:493–6. doi: 10.1038/86573. [DOI] [PubMed] [Google Scholar]
- Tichauer KM, Diop M, Elliott JT, Samkoe KS, Hasan T, Lawrence KS, Pogue BW. Accounting for pharmacokinetic differences in dual-tracer receptor density imaging. Physics in medicine and biology. 2014a;59:2341–51. doi: 10.1088/0031-9155/59/10/2341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tichauer KM, Samkoe KS, Gunn JR, Kanick SC, Hoopes PJ, Barth RJ, Kaufman PA, Hasan T, Pogue BW. Microscopic lymph node tumor burden quantified by macroscopic dual-tracer molecular imaging. Nature medicine. 2014b;20:1348–53. doi: 10.1038/nm.3732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tichauer KM, Samkoe KS, Sexton KJ, Gunn JR, Hasan T, Pogue BW. Improved tumor contrast achieved by single time point dual-reporter fluorescence imaging. Journal of biomedical optics. 2012a;17:066001. doi: 10.1117/1.JBO.17.6.066001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tichauer KM, Samkoe KS, Sexton KJ, Hextrum SK, Yang HH, Klubben WS, Gunn JR, Hasan T, Pogue BW. In vivo quantification of tumor receptor binding potential with dual-reporter molecular imaging. Molecular imaging and biology : MIB : the official publication of the Academy of Molecular Imaging. 2012b;14:584–92. doi: 10.1007/s11307-011-0534-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tichauer KM, Wang Y, Pogue BW, Liu JT. Quantitative in vivo cell-surface receptor imaging in oncology: kinetic modeling and paired-agent principles from nuclear medicine and optical imaging. Physics in medicine and biology. 2015;60:R239–69. doi: 10.1088/0031-9155/60/14/R239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urano Y, Sakabe M, Kosaka N, Ogawa M, Mitsunaga M, Asanuma D, Kamiya M, Young MR, Nagano T, Choyke PL, Kobayashi H. Rapid cancer detection by topically spraying a gamma-glutamyltranspeptidase-activated fluorescent probe. Science translational medicine. 2011;3:110ra9. doi: 10.1126/scitranslmed.3002823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- W Wang Y, Doerksen JD, Kang S, Walsh D, Yang Q, Hong D, Liu JT. Multiplexed Molecular Imaging of Fresh Tissue Surfaces Enabled by Convection-Enhanced Topical Staining with SERS-Coded Nanoparticles. Small. 2016 doi: 10.1002/smll.201601829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Altaz K, Leigh SY, Wang D, Chen Y, Meza D, Liu JTC. Comprehensive spectral endoscopy of topically applied SERS nanoparticles in the rat esophagus. Biomedical optics express. 2014a;5:2883. doi: 10.1364/BOE.5.002883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Kang S, Khan A, Ruttner G, Leigh SY, Murray M, Abeytunge S, Peterson G, Rajadhyaksha M, Dintzis S, Javid S, Liu JT. Quantitative molecular phenotyping with topically applied SERS nanoparticles for intraoperative guidance of breast cancer lumpectomy. Scientific reports. 2016;6:21242. doi: 10.1038/srep21242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YW, Kang S, Khan A, Bao PQ, Liu JT. In vivo multiplexed molecular imaging of esophageal cancer via spectral endoscopy of topically applied SERS nanoparticles. Biomedical optics express. 2015;6:3714–23. doi: 10.1364/BOE.6.003714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YW, Khan A, Som M, Wang D, Chen Y, Leigh SY, Meza D, McVeigh PZ, Wilson BC, Liu JT. Rapid ratiometric biomarker detection with topically applied SERS nanoparticles. Technology. 2014b;2:118–32. doi: 10.1142/S2339547814500125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang RR, Schroeder AB, Grudzinski JJ, Rosenthal EL, Warram JM, Pinchuk AN, Eliceiri KW, Kuo JS, Weichert JP. Beyond the margins: real-time detection of cancer using targeted fluorophores. Nat Rev Clin Oncol. 2017 doi: 10.1038/nrclinonc.2016.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipfel WR, Williams RM, Webb WW. Nonlinear magic: multiphoton microscopy in the biosciences. Nat Biotechnol. 2003;21:1369–77. doi: 10.1038/nbt899. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.