Abstract.
Chikungunya fever (CHIK) is an acute viral infection caused by infection with chikungunya virus (CHIKV). The disease affects people in areas where certain Aedes species mosquito vectors are present, especially in tropical and subtropical countries. Indonesia has witnessed CHIK disease since the early 1970s with sporadic outbreaks occurring throughout the year. The CHIK clinical manifestation, characterized by fever, headache, and joint pain, is similar to that of dengue (DEN) disease. During a molecular study of a DEN outbreak in Jambi, Sumatra, in early 2015, DENV-negative samples were evaluated for evidence of CHIKV infection. Among 103 DENV-negative samples, eight samples were confirmed (7.8%) as positive for CHIKV by both molecular detection and virus isolation. The mean age of the CHIK patients was 21.3 ± 9.1 (range 11–35 years). The clinical manifestations of the CHIK patients were mild and mimicked DEN, with fever and headache as the main symptoms. Only three out of eight patients presented with classical joint pain. Sequencing of the envelope glycoprotein E1 gene and phylogenetic analysis identified all CHIKV isolates as belonging to the Asian genotype. Overall, our study confirms sustained endemic CHIKV transmission and the presence of multiple arboviruses circulating during a DEN outbreak in Indonesia. The co-circulation of arboviruses poses a public health threat and is likely to cause misdiagnosis and underreporting of CHIK in DEN-endemic areas such as Indonesia.
INTRODUCTION
Chikungunya (CHIK) is an acute viral disease caused by infection with CHIK virus (CHIKV) which belongs to the genus Alphavirus in the family Togaviridae. The disease is typically characterized by fever, headache, myalgia, rash, and joint pain.1 Known to have originated in Africa, CHIKV has now widely spread in tropical and subtropical regions in the world where the appropriate Aedes species mosquito vectors are prevalent. Three genetically distinct genotypes have been identified, the West African, the East, Central, and South African (ECSA) genotype, and the Asian genotype.2
CHIK has become well established in many Indonesian provinces.3–6 However, the overall burden of disease in the country is still not well determined. During 2001–2003, reemerging CHIK in Indonesia was reported after an absence of about two decades.5 In 2014, the Indonesian Ministry of Health reported the occurrence of CHIK outbreaks in eight districts from three provinces.7 Although most CHIK reports in Indonesia were recognized based on clinical manifestations by epidemic outbreaks, data on sporadic cases are scarce. Diagnosis of CHIK in Indonesia is usually based on clinical symptoms due to limited laboratory resources that prohibit serological and molecular confirmation. Coupled with the clinical similarity to dengue (DEN) and other acute febrile illnesses, the true disease burden of CHIK is likely overshadowed by DEN.
During December 2014–March 2015, DEN cases were found to increase significantly in Jambi, Sumatra. DENV molecular and nonstructural protein-1 (NS1) antigen detection was performed in 210 febrile samples and 107 of them were confirmed as DEN. To determine the etiology of the remaining DENV-negative samples (N = 103), virus isolation and molecular techniques looking for other arboviruses, belonging to the flavivirus and alphavirus families, were performed. These findings of multiple CHIKV-positive cases help to clarify the degree of missed CHIK cases during reported DEN activity.
MATERIALS AND METHODS
Study site, patient recruitment, and sample collection.
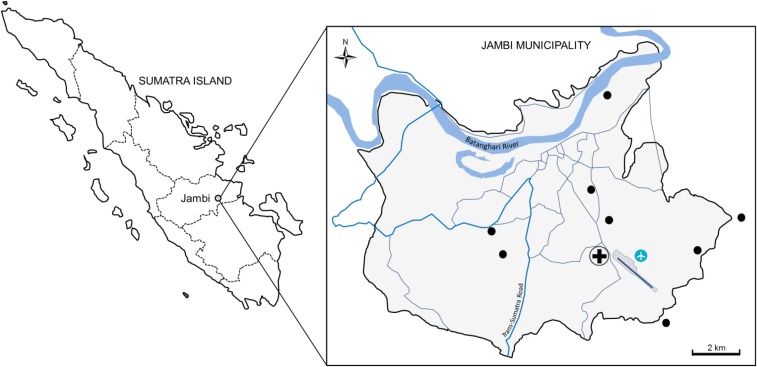
This study was part of a DEN molecular study that was performed during a DEN-like febrile outbreak in Jambi, Sumatra, from December 2014–March 2015.8 Jambi is located in the southern part of Sumatra Island (Figure 1) and populated by approximately 531,857 people (Indonesia Central Statistical Bureau, 2010) with a total area of about 205 km2. Patients presenting at Siloam Hospital, Jambi, with DEN-like illness were recruited after informed consent was signed. The study was approved by the Eijkman Institute Research Ethic Commission (Ethical Approval No. 78). Patients with fever > 38°C accompanied by at least one of the clinical signs of DEN, such as malaise, arthralgia, rash, and retro-orbital pain, within 5 days of post-illness onset were enrolled. Blood samples of 3–5 mL were obtained from a total of 210 patients. Demographic data of the patients including age, gender, geographic location, and clinical information were recorded.
Figure 1.
Study site in Jambi, Sumatra, Indonesia. The patients’ residential areas are shown as black dots. This figure appears in color at www.ajtmh.org.
DENV and CHIKV diagnosis.
DENV infection was detected by using the NS1 Ag Rapid Test (SD Bioline, Korea), PanBio DEN Duo IgM & IgG ELISA (Alere, Waltham, MA), and Simplexa™ real-time reverse transcription polymerase chain reaction (RT-PCR) (Focus Diagnostics, Cypress, CA). Virus RNA was extracted from 200 μL serum samples using the MagNA Pure LC Total NA Isolation Kit (Roche, Mannheim, Germany) in the MagNA Pure LC 2.0 extraction system (Roche). Of 210 patients, we found that 103 cases were clinically diagnosed as DEN but were negative for the presence of DEN virus either by RT-PCR or NS1 antigen. To determine the etiology of the DENV-negative samples, we performed RT-PCR screening for other viruses, including pan-flavivirus and pan-alphavirus,9,10 by conventional RT-PCR following our laboratory algorithm. Because the family consensus primers could amplify other alphaviruses, samples that were positive using the pan-alphavirus RT-PCR were confirmed using CHIKV-specific RT-PCR (targeting the E1 gene) and virus isolation using Vero CCL-81 cells, as described previously.11 Flasks were incubated in a humidified incubator at 37°C, supplemented with 5% CO2 until the detection of cytopathic effect (CPE). Cells that were positive for CPE were confirmed for CHIKV using both an immunofluorescent assay and an RT-PCR assay on the extracted RNA.
CHIKV RT-PCR and complete genome sequencing.
After RNA extraction, a one-step RT-PCR targeting the CHIKV E1 gene was performed using an established protocol.12 A primer pair was used to amplify the sequences from nt 9,870–11,359, using the SuperScript III RT/Platinum Taq High Fidelity Enzyme Mix (Thermo Fisher Scientific, Waltham, MA). The primer sequences used in the templates generation are ChikE1/9870F (5′-ACAAGCCCTTATTCCGCTG) and ChikE1/11359R (5′-GTGTGTCTCTTAGGGGACACATA). Templates were purified using the QIAquick PCR purification kit (Qiagen, Hilden, Germany) and used in cycle sequencing reactions performed using overlapping primers from both strands and BigDye Dideoxy Terminator sequencing kits v3.1 (Thermo Fisher Scientific), following manufacturer’s instructions. Purified DNA was subjected to capillary sequencing performed on a 3130xl genetic analyzer (Thermo Fisher Scientific). Sequence reads were assembled using SeqScape v.2.5 software (Thermo Fisher Scientific) with additional manual adjustments in case of discrepancies. The resulting E1 gene sequences were deposited in GenBank and were granted accession numbers KX097981, KX097983, KX097984, KX097985, and KX097987.
Sequencing of near-complete genomes of three CHIKV isolates was performed using the Ion Torrent PGM platform on the supernatant of the first passage. CHIKV RNA was treated first with DNAse I (Thermo Scientific) followed by cDNA library construction using the Ovation RNA-Seq System V2 (NuGEN) and the Ion Xpress Plus™ gDNA and Amplicon Library Preparation kit (Thermo Scientific). The sheared and purified cDNA library was analyzed using Bioanalyzer (Agilent Technologies). The cDNA template (100 pM) was then prepared and enriched using Ion PGM Hi-Q OT2 kit (Thermo Scientific). To reach a targeted depth coverage, the enriched ion spheres were then sequenced using the Ion Torrent Personal Genome Machine Sequencer and the Ion PGM Hi-Q Sequencing kit, with a 316 Chip (Thermo Scientific). SMALT (http://www.sanger.ac.uk/science/tools/smalt-0) was used to map the PGM reads to CHIKV genome reference (GenBank accession number KM673291). All mapped reads were then filtered using PRINSEQ (http://prinseq.sourceforge.net) for only high-quality reads (mean Q > 30) with length range of 100–250 bp, and subjected to 5′/3′ dereplication. The filtered reads were assembled using SPAdes (http://bioinf.spbau.ru/spades) in metagenome mode, and the resulting contig was used as an untrusted contig for the next consecutive SPAdes assembling in single cell and careful mode, which used mismatch correction using Burrows-Wheeler aligner internally. The near-complete genomes have been deposited in GenBank repository with accession numbers KX097982, KX097986, and KX097988.
CHIKV phylogenetic and evolutionary analyses.
CHIKV nucleotide sequences from Jambi were grouped with other representative sequences downloaded from GenBank to generate a dataset, aligned using the MUSCLE program. Analysis was conducted based on a 1,050 nt sequence of the E1 gene including amino acid residues 91–439. Sequence identities and similarities were calculated using the SIAS calculator, available online at http://imed.med.ucm.es/Tools/sias.html, with multiple sequence alignment as the input file and BLOSUM62 matrix for global similarity.
A total dataset of 70 CHIKV E1 sequences including eight Jambi isolates and 62 reference sequences from three genotypes of CHIKV retrieved from GenBank was analyzed for their evolutionary rate and the time to most recent common ancestors (TMRCA) using the Bayesian Markov chain Monte Carlo (MCMC) method as implemented in BEAST v.1.8.4.13 The high-passage strains Ross and S27 were excluded to avoid artifacts because of laboratory adaptation.14 Data were prepared using the BEAUti graphical interface and each isolate was calibrated using the year of isolation as the calibration point. Phylogenetic tree was inferred based on the selection of a statistical model for likelihood calculation, optimized for maximum likelihood (ML) tree using jModelTest v.2.1.4.15 Runs were performed using a general time reversible (GTR) model with four gamma parameters and invariant sites (GTR + Γ4 + I) and relaxed uncorrelated lognormal molecular clock using the initial estimated evolutionary rate of 4.33 × 10−4 substitutions per site per year, as previously described.14 Tree prior was set as a coalescent Bayesian skyline plot to facilitate the fewest demographic assumptions. One hundred million chains were run and sampled for every 1,000th iteration, with 10% burn-in used. The convergence of parameters was analyzed using Tracer v.1.6.0 to ensure adequate effective sampling size (ESS) for all parameters. A maximum clade credibility (MCC) tree was created using TreeAnnotator v.1.8.4 and visualized in FigTree v.1.4.3. The TMRCA in each node was estimated as median year with 95% highest posterior density (HPD).
RESULTS
CHIK cases among DEN-negative samples.
Of 210 DEN-suspected patients, we detected 103 patients that were DEN-negative as confirmed by the absence of DENV specific RNA, NS1 antigen, and IgM/IgG.8 Of 103 DEN-negative samples, we detected eight samples positive by alphavirus-specific PCR. Inoculation of those sera in Vero cell line generated CPE consistent with alphavirus infection (not shown). Sequencing was then performed on RNA from these isolations and we identified CHIKV in all eight samples. In addition to CHIKV, screening of samples using flavivirus-specific primers resulted in one positive case which, after virus isolation and genome sequencing, was identified as Zika virus (ZIKV) which has been reported elsewhere.16
The age of the CHIK patients ranged from 11 to 35 years old (mean = 21.3 ± 9.1). Seven out of eight were male. The clinical presentations of all patients were mild, while the most notable symptoms were headache and fever (Table 1). Arthralgia, the most characteristic symptom of CHIK, was only observed in three of eight patients. Rash was only found in two patients. Half of the patients presented with malaise and loss of appetite. Low thrombocyte count was also seen in some patients (Table 1). No fatalities related to CHIK were reported.
Table 1.
Chikungunya patients characteristics from Jambi, Sumatra
| Patient characteristics | Cases (N = 8) |
|---|---|
| Demographic data | |
| Sex (male) | 7 (87.5%) |
| Age (year)* | 21.3 ± 9.1 |
| Clinical symptoms | |
| Fever | 8 (100%) |
| Duration of fever (days)* | 2.0 ± 0.93 |
| Temperature (°C)* | 38.58 ± 0.80 |
| Headache | 7 (87.5%) |
| Malaise | 4 (50%) |
| Cough | 1 (12.5%) |
| Runny nose | 2 (25%) |
| Nausea | 2 (25%) |
| Vomiting | 2 (25%) |
| Epigastric pain | 2 (25%) |
| Loss of appetite | 4 (50%) |
| Diarrhea | 0 (0%) |
| Arthralgia | 3 (37.5%) |
| Rash | 2 (25%) |
| Itching | 1 (12.5%) |
| Joint swelling | 0 (0%) |
| Hematology data | |
| Hemoglobin (g/dL)* | 14.08 ± 0.84 |
| Leukocyte (103/µL)* | 6.36 ± 2.07 |
| Platelet count (103/µL)* | 196.88 ± 70.71 |
Mean ± STDEV.
The geographical distribution of the CHIK cases is shown in Figure 1. The patients’ residences were scattered throughout the Jambi municipality, with no indication of residential clustering. The nearest cases were separated by about 2 km (Figure 1).
Phylogenetic and evolutionary analyses.
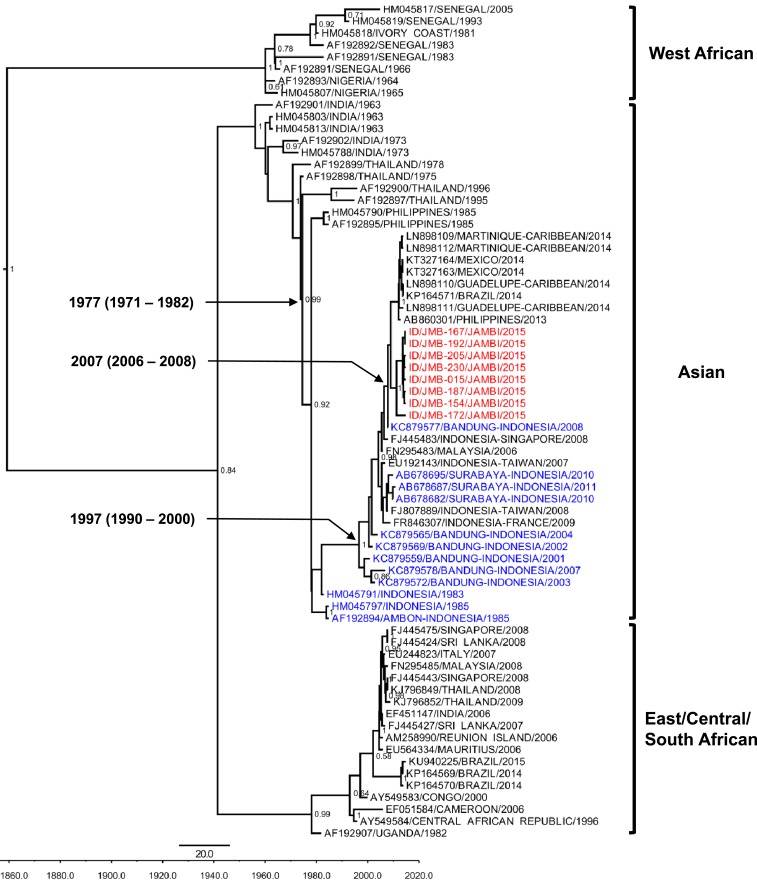
Our phylogenetic analysis based on partial E1 gene sequences confirmed that all eight CHIKV isolates belonged to the Asian genotype (Figure 2). Although these isolates formed their own clade, they grouped together with CHIKV strains from the Americas, the Philippines, and another city in Indonesia, Bandung, West Java. Aligned with those strains, Jambi isolates showed sequence identity and similarity ranging from 92.8% to 99.9%, with a normalized global amino acid similarity score ranging from 0.91 to 0.99. Jambi CHIKV isolates have 99.16% ± 0.05% sequence identity and similarity with the Bandung strain from 2008.4 The amino acid similarity score of the Jambi isolates compared with Bandung strains was 0.98. To further explore the genetic characteristic of the CHIKV isolates, we performed amino acid analysis of partial E1 genes but did not find mutation in the Alanine residue at position 226 which was implicated in better transmissibility of CHIKV by Aedes albopictus in the ECSA genotype.17 It is important to note, however, that this particular mutation is not associated with differences in transmission in the context of the Asian genotype backbone.18
Figure 2.
Maximum clade credibility (MCC) tree of chikungunya virus (CHIKV) generated by Bayesian inference method as implemented in BEAST using GTR evolution model and gamma parameter rates from the E1 gene sequences. The Jambi CHIKV isolates (red font) grouped with other Asian genotype strains including isolates from other cities in Indonesia (blue font). The posterior probabilities of the clades were indicated as numbers in the node labels, with value of > 0.5 shown. This figure appears in color at www.ajtmh.org.
The coefficient of variation of the 60 CHIKV E1 genes was 1.3 (95% HPD: 0.72–2.28) with the overall evolutionary rate estimated as 9.0 × 10−4 nucleotide substitution per site per year (95% HPD: 5.7–12.6 × 10−4 subs/nt/yr). The Jambi isolates exhibited mean evolutionary rates of 7.3 ± 2.0 × 10−4 subs/nt/yr.
Phylogenetic analysis using representative near-complete genomes of three Jambi isolates (JMB-154, JMB-192, and JMB-230) confirmed the earlier results of E1 gene generating a similar phylogenetic tree (data not shown).
DISCUSSION
The first report of possible CHIK endemicity in Indonesia could be traced back to 1779 when a physician, David Bylon, municipal surgeon for the city of Batavia (now Jakarta), acquired a CHIKV-like disease.19 Recent work including laboratory testing has clearly shown CHIK endemicity in Indonesia for decades; however, only limited data are published regarding its circulation and characterization. In particular, the virological and clinical features of CHIK in Sumatra have never been reported previously. Here, we documented the first detection of endemic CHIKV in Sumatra which occurred during a DEN outbreak. We performed phylogenetic and evolutionary analyses of CHIKV based on the E1 gene to better understand its evolutionary dynamics.
During December 2014–March 2015, we detected 7.8% CHIKV infection (and 0.97% ZIKV infection) among patients presenting with DEN-like illness in Jambi. This study demonstrates the co-circulation of multiple mosquito-borne viruses all with similar clinical symptoms during an outbreak of DEN in Sumatra. Concomitant circulation of CHIKV and DENV has been reported in Indonesia4,20 as both are transmitted by the same vectors. Co-infection of DENV and CHIKV reported in Asia21 was not determined in our study as CHIKV was screened only in those that were negative for DENV. The CHIK incidence in Jambi is a little higher than reported in Bandung, West Java, which showed CHIK accounted for 7.1% of febrile patients.4 These data reflect the significance of CHIK in Jambi and the considerable burden it may pose on the local health and economy. The absence of confirmatory laboratory testing for CHIKV in most hospitals in Indonesia, including Jambi, leaves the true degree of CHIK activity in the community difficult to assess, especially during inter-outbreak periods.
The CHIK patients in this study resided throughout the Jambi municipality with no indication of clustering. This was different from the typical epidemic CHIKV transmissions that have occurred and been reported in Indonesia in which CHIK cases were mostly clustered and affected many people in villages/suburbs.5 Thus, our finding confirms that CHIK in Jambi is sporadic and suggests that sustainable and silent CHIKV transmission might be occurring in Jambi. This would not be unexpected; given the significant presence of Aedes mosquitoes in the municipality, the likelihood of CHIK outbreaks in this area is high.
Unlike previous CHIK outbreaks reported in Indonesia5 which were mostly characterized by severe manifestations, the CHIK cases in this study only manifested with mild clinical symptoms similar to classical DEN fever. Although fever, rash, and arthralgia are the hallmarks of CHIK,22,23 patients in our study mainly exhibited fever and headache (Table 1). Our study demonstrated that CHIKV infection can be mild without the classical symptoms, causing it to be misdiagnosed or overlooked by physicians. Similar clinical manifestations were also observed in other regions of Indonesia.4
Currently, only limited information on the genotype of CHIKV circulating in Indonesia is available. No data on CHIKV genotypes in Sumatra existed before this work. Knowledge of the genotype(s) circulating is valuable as it may be associated with clinical severity, high pathogenicity or virulence, or enhanced transmissibility in mosquito vector species.24 The CHIKV isolates found in Jambi belong to the Asian genotype which is linked with sporadic CHIK cases in several cities in Indonesia, such as Bandung and Surabaya.4,25 Many CHIK outbreaks in Southeast Asian countries have also been associated with the variant Indian Ocean lineage of the ECSA genotype26 which has been linked with enhanced transmissibility of CHIKV by Ae. albopictus mosquitoes.17,27 Although the ECSA genotype has caused outbreaks in Southeast Asia, the Asian genotype has not been supplanted and is still the main cause of CHIK outbreaks in Indonesia and Malaysia.4,6,28 It is interesting to observe that only the Asian genotype has detected in Indonesia with the exception of the single ESCA genotype reported in Kalimantan,6 whereas both Asian and ECSA genotypes are circulating in nearby Singapore.12
Our Bayesian analysis of the partial E1 gene estimated the overall evolutionary rate to be higher than the estimated evolutionary rate previously reported.14 However, the estimated overall evolutionary rate of CHIKV in this study is still within 95% HPD range of the epidemic strains.
Jambi CHIKV isolates have formed a clade with isolates from the Americas, the Philippines, and Bandung (Indonesia), with an estimated TMRCA of 2007 (95% HPD: 2006–2008) (Figure 2). Over the last four decades the Indonesian CHIKV strains were suggested to have originated from Thailand (TMRCA of 1977, 95% HPD: 1971–1982). The recent outbreaks of CHIKV in Indonesia were presumed to have common ancestors for the last two decades (TMRCA of 1997, 95% HPD: 1990–2000) and were closely related to the isolates from the Philippines (Figure 2). One interesting finding was the closeness of Jambi isolates with American strains (from South America and the Caribbean). Our data support the evidence of CHIKV importation to the Americas from Southeast Asia.29
Reports described that in 2001–2003, reemergence of CHIK in Indonesia was detected after an absence of about two decades.5,30 Based on these evolutionary analyses, the absence of CHIK outbreaks during those times was confirmed. The TMRCAs estimated by this phylogenetic analysis are in accordance with those outbreak data, including the gap (Figure 2). However, there are some potential limitations of the evolutionary clock analyses as only those who were acutely ill and not asymptomatic individuals have been sampled and analyzed; thus, minority populations that may exist in these alternative samples are not evaluated in the consensus model. However, analysis using the E1 gene alone might be inadequate to fully resolve the phylogenetic history of CHIKV in Indonesia14 and phylogenetic analysis using representative whole genome ORF from three isolates was performed to confirm the results.
In summary, we documented the circulation of CHIKV during a DEN outbreak in Jambi, Sumatra. Our finding of concurrent circulation of arboviruses emphasizes the importance of continuous monitoring, surveillance, and simultaneous testing of febrile samples for DENV, CHIKV, and ZIKV in endemic areas. This study has provided information to improve our previously limited knowledge of the clinical, virological, endemicity, evolution, and dynamics of CHIKV circulation in Sumatra.
Acknowledgments:
We would like to thank the patients and physicians involved in this study. We greatly appreciate the help of Drs. Harjanto Soelaiman, Samsirun Halim, Andri Budiman, Lanceria Sijabat, and Ifo Faujiah from Siloam Hospital, Jambi, for their support with patient recruitment and Rama Dhenni for assisting with complete genome sequencing. The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the Centers for Disease Control and Prevention. This study was approved by Eijkman Institute Research Ethic Commission (Ethical Approval No. 78).
REFERENCES
- 1.Petersen LR, Powers AM, 2016. Chikungunya: epidemiology. F1000 Res 5: 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Powers AM, Brault AC, Tesh RB, Weaver SC, 2000. Re-emergence of chikungunya and O’nyong-nyong viruses: evidence for distinct geographical lineages and distant evolutionary relationships. J Gen Virol 81: 471–479. [DOI] [PubMed] [Google Scholar]
- 3.Yoshikawa MJ, Kusriastuti R, 2013. Surge of dengue virus infection and chikungunya fever in Bali in 2010: the burden of mosquito-borne infectious diseases in a tourist destination. Trop Med Health 41: 67–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kosasih H, et al. 2013. Evidence for endemic chikungunya virus infections in Bandung, Indonesia. PLoS Negl Trop Dis 7: e2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Laras K, et al. 2005. Tracking the re-emergence of epidemic chikungunya virus in Indonesia. Trans R Soc Trop Med Hyg 99: 128–141. [DOI] [PubMed] [Google Scholar]
- 6.Maha MS, Susilarini NK, Hariastuti NI, Subangkit, 2015. Chikungunya virus mutation, Indonesia, 2011. Emerg Infect Dis 21: 379–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Indonesia Ministry of Health 2015. Indonesia health profile year 2014. Available at: http://www.depkes.go.id/resources/download/pusdatin/profil-kesehatan-indonesia/Indonesia Health Profile 2014.pdf. Accessed March 31, 2016.
- 8.Haryanto S, et al. 2016. The molecular and clinical features of dengue during outbreak in Jambi, Indonesia in 2015. Pathog Glob Health 110: 119–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kuno G, Chang GJ, Tsuchiya KR, Karabatsos N, Cropp CB, 1998. Phylogeny of the genus Flavivirus. J Virol 72: 73–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ledermann JP, Zeidner N, Borland EM, Mutebi J-P, Lanciotti RS, Miller BR, Lutwama JJ, Tendo JM, Andama V, Powers AM, 2014. Sunguru virus: a novel virus in the family Rhabdoviridae isolated from a chicken in north-western Uganda. J Gen Virol 95: 1436–1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Riswari SF, et al. 2016. Study of viremic profile in febrile specimens of chikungunya in Bandung, Indonesia. J Clin Virol 74: 61–65. [DOI] [PubMed] [Google Scholar]
- 12.Ng L-C, et al. 2009. Entomologic and virologic investigation of chikungunya, Singapore. Emerg Infect Dis 15: 1243–1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Drummond AJ, Rambaut A, 2007. BEAST: bayesian evolutionary analysis by sampling trees. BMC Evol Biol 7: 214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Volk SM, et al. 2010. Genome-scale phylogenetic analyses of chikungunya virus reveal independent emergences of recent epidemics and various evolutionary rates. J Virol 84: 6497–6504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Darriba D, Taboada GL, Doallo R, Posada D, 2012. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods 9: 772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Perkasa A, et al. 2016. Isolation of Zika virus from febrile patient, Indonesia. Emerg Infect Dis 22: 924–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tsetsarkin KA, Vanlandingham DL, McGee CE, Higgs S, 2007. A single mutation in chikungunya virus affects vector specificity and epidemic potential. PLoS Pathog 3: e201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tsetsarkin KA, Chen R, Leal G, Forrester N, Higgs S, Huang J, Weaver SC, 2011. Chikungunya virus emergence is constrained in Asia by lineage-specific adaptive landscapes. Proc Natl Acad Sci USA 108: 7872–7877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Halstead SB, 2015. Reappearance of chikungunya, formerly called dengue, in the Americas. Emerg Infect Dis 21: 557–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Capeding MR, et al. 2013. Dengue and other common causes of acute febrile illness in Asia: an active surveillance study in children. PLoS Negl Trop Dis 7: e2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Singh P, Mittal V, Rizvi MMA, Chhabra M, Sharma P, Rawat DS, Bhattacharya D, Chauhan LS, Rai A, 2012. The first dominant co-circulation of both dengue and chikungunya viruses during the post-monsoon period of 2010 in Delhi, India. Epidemiol Infect 140: 1337–1342. [DOI] [PubMed] [Google Scholar]
- 22.Borgherini G, Poubeau P, Staikowsky F, Lory M, Le Moullec N, Becquart JP, Wengling C, Michault A, Paganin F, 2007. Outbreak of chikungunya on Reunion Island: early clinical and laboratory features in 157 adult patients. Clin Infect Dis 44: 1401–1407. [DOI] [PubMed] [Google Scholar]
- 23.Horwood PF, Buchy P, 2015. Chikungunya. Rev Sci Tech 34: 479–489. [DOI] [PubMed] [Google Scholar]
- 24.Tsetsarkin KA, Chen R, Sherman MB, Weaver SC, 2011. Chikungunya virus: evolution and genetic determinants of emergence. Curr Opin Virol 1: 310–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mulyatno KC, Susilowati H, Yamanaka A, Soegijanto S, Konishi E, 2012. Primary isolation and phylogenetic studies of chikungunya virus from Surabaya, Indonesia. Jpn J Infect Dis 65: 92–94. [PubMed] [Google Scholar]
- 26.Pulmanausahakul R, Roytrakul S, Auewarakul P, Smith DR, 2011. Chikungunya in Southeast Asia: understanding the emergence and finding solutions. Int J Infect Dis 15: e671–e676. [DOI] [PubMed] [Google Scholar]
- 27.Sam I-C, Chan YF, Chan SY, Loong SK, Chin HK, Hooi PS, Ganeswrie R, Abubakar S, 2009. Chikungunya virus of Asian and Central/East African genotypes in Malaysia. J Clin Virol 46: 180–183. [DOI] [PubMed] [Google Scholar]
- 28.AbuBakar S, Sam I-C, Wong P-F, MatRahim N, Hooi P-S, Roslan N, 2007. Reemergence of endemic chikungunya, Malaysia. Emerg Infect Dis 13: 147–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weaver SC, Lecuit M, 2015. Chikungunya virus and the global spread of a mosquito-borne disease. N Engl J Med 372: 1231–1239. [DOI] [PubMed] [Google Scholar]
- 30.Mackenzie JS, et al. 2001. Emerging viral diseases of Southeast Asia and the Western Pacific. Emerg Infect Dis 7 (Suppl): 497–504. [DOI] [PMC free article] [PubMed] [Google Scholar]