Abstract.
In this study, we developed a recombinase polymerase amplification (RPA) assay for specific diagnosis of Plasmodium knowlesi. Genomic DNA was extracted from whole blood samples using a commercial kit. With incubation at 37°C, the samples were successfully amplified within 20 minutes. The end product of RPA was further examined by loading onto agarose gel and a specific band was observed with a size of 128 bp. The RPA assay exhibited high sensitivity with limits of detection down to one copy of the plasmid. From the specificity experiments, it was demonstrated that all P. knowlesi samples (N = 45) were positive while other Plasmodium spp. (N = 42) and negative samples (N = 6) were negative. Therefore, the RPA assay is a highly promising approach with the potential to be used in resource-limited settings. This assay can be further optimized for bedside and on field application.
Among all the parasitic infections, malaria causes the most deaths (range 235,000–639,000) worldwide according to the World Malaria Report, 2016.1 African regions feature the most prevalence (90%) followed by Southeast Asia (7%) and the Eastern Mediterranean (2%). The malaria parasites that commonly infect humans are Plasmodium falciparum, Plasmodium ovale, Plasmodium vivax, and Plasmodium malariae. However, Plasmodium knowlesi, which was only found in monkeys previously, has recently emerged as a new human malaria parasite. The natural hosts for P. knowlesi are long-tailed macaques (Macaca fascicularis). This zoonotic malaria began attracting attention in 2004 during the epidemic of P. knowlesi infections in the Kapit Division of Sarawak, Malaysian Borneo, and there have been a plethora of reports detailing the P. knowlesi infection in humans since then. At this point, P. knowlesi is now the fifth known human malaria parasite and their epidemiology, diagnosis, clinical features, and treatment have been studied broadly.2
Microscopic examination has been the mainstay of malaria diagnosis. It is an inexpensive, rapid, and relatively sensitive procedure. However, interpretation of smears requires considerable expertise, particularly at low-level parasitemia, potentially leading to false-negative results. The P. knowlesi parasite is morphologically similar to that P. falciparum and P. malariae under the microscope.3 To date, nested PCR targeting Plasmodium ssrRNA genes is considered the most sensitive and specific method for Plasmodium detection. Species-specific primers have been used to amplify ssrRNA genes to detect P. knowlesi infection, as well; however, this technique has been described to result in cross-reactions with P. vivax because of the close phylogenetic relationship between the two species.4 On top of that, this methodology is costly and requires trained personnel for its performance.5
The recombinase polymerase amplification (RPA) assay is an isothermal DNA amplification method that does not require any cycling parameters. With this, the RPA assay has emerged as a novel alternative to isothermal amplification for the detection of nucleic acids.6 The RPA assay has been previously documented to take < 20 minutes to perform and requires a constant temperature of just 37–42°C. RPA assays have been applied to the detection of various other infectious agents, including dengue viruses and P. falciparum.7–10 Notably, RPA can be carried out without the need for very high-end machinery as it can be performed using a simple shaking incubator, which is very favorable news for resource-limited laboratories. Herein, this is the first report of using RPA for rapid diagnosis of P. knowlesi.
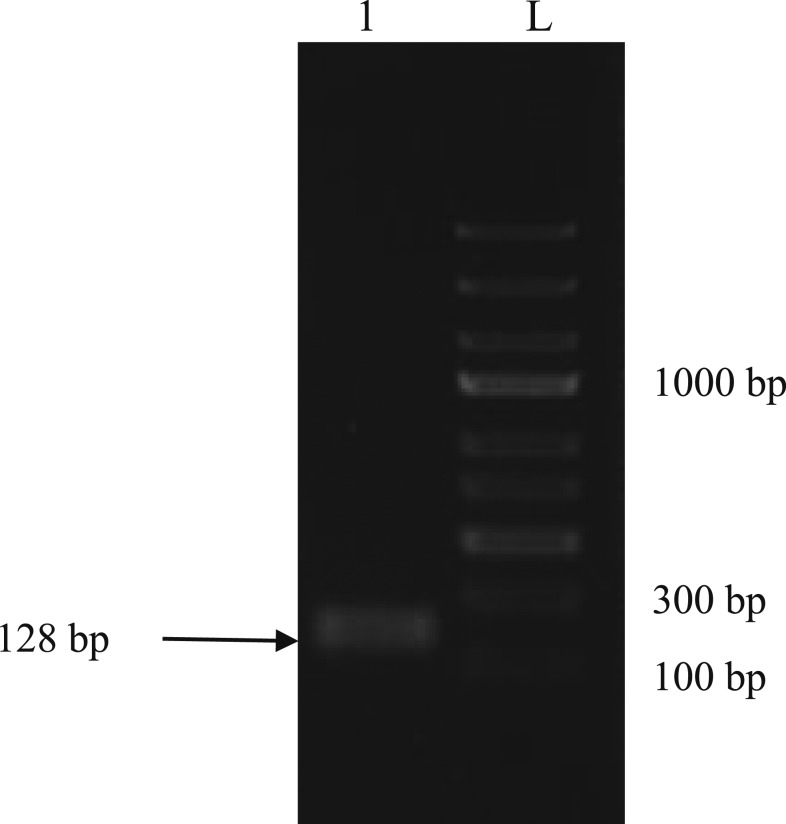
To conduct the RPA assay, blood samples were obtained from the University of Malaya Medical Center and Sarawak State Health Department. DNA was extracted from whole blood using the DNeasy® Blood and Tissue Kit (Qiagen, Hilden, Germany). The samples were previously examined under microscope, diagnosed with the BinaxNOW Malaria Kit (Alere, Scarborough, ME), and nested PCR before RPA assay. This study was approved by the Medical Research and Ethics Committee of the Ministry of Health Malaysia (NMRR-15-672-23975) and the Medical Ethics Committee of the University of Malaya Medical Center (MEC Ref. No. 817.1). The RPA assay was conducted using the primers and probes listed in Table 1. The P. knowlesi small subunit ribosomal RNA (18S rRNA) gene was used as the target gene, and the primers and probes were designed based on the manufacturer’s recommendations (TwistDx, Cambridge, United Kingdom). The probe used in this study was particularly created with an insertion of an abasic nucleotide analogue (a tetrahydrofuran residue, frequently referred to as a “dSpacer”) flanked by a dT-fluorophore (dT-FAM), a corresponding dT-quencher group (BHQ1) and 3′ carbon blocker (C3 spacer). As advised by the manufacturer, the primer and probe were diluted to 10 µM as the working solution throughout the entire study. The RPA assay was performed with a total 50 μL reaction mixture that consisted of 11.2 μL of water, 29.5 μL of 1X rehydration buffer with the addition of 400 nM each of the forward and reverse primers, and 120 nM of the FAM-tagged probe was added to the RPA pellet. After mixing all these components, 2 μL of DNA was added followed by 2.5 μL of 280 mM magnesium acetate, which was pipetted into the tube lids instead of the reaction mixture directly. The lid was closed during the spinning process, and the reaction was initiated as soon as magnesium acetate was added. The reaction tube was then incubated in a real-time PCR machine at 37°C for approximately 20 minutes with 40 reaction cycles. RPA assay was also performed using a shaking incubator at the same setting. The end product of RPA was further investigated by loading onto agarose gel and a specific band was observed with a size of 128 bp (Figure 1).
Table 1.
Primer and probe sequences used in this study
| Name | Sequence (5′-3′) |
|---|---|
| Pkr140 RPA exo | F: CCGTTCTCATGATTTCCATGGTCCAGGGTT |
| Pkr140 RPA exo | R: CCTGAACACCTCATGTCGTGGTAGAAATAG |
| Pkr140 RPA exo Probe | CCGTTCTCATGATTTCCATGGTCCAGGGFTHAGQTTTTTCGGTCCC[3] |
F = FAM-dT; H = Tetrahydrofuran; Q = BHQ1-dT; 3 = C3-spacer.
Figure 1.
Detection of recombinase polymerase amplification (RPA) end product after purification. The size of the Plasmodium knowlesi was correctly detected at 128 bp. L: 1 kb ladder; Lane 1: Purified RPA product.
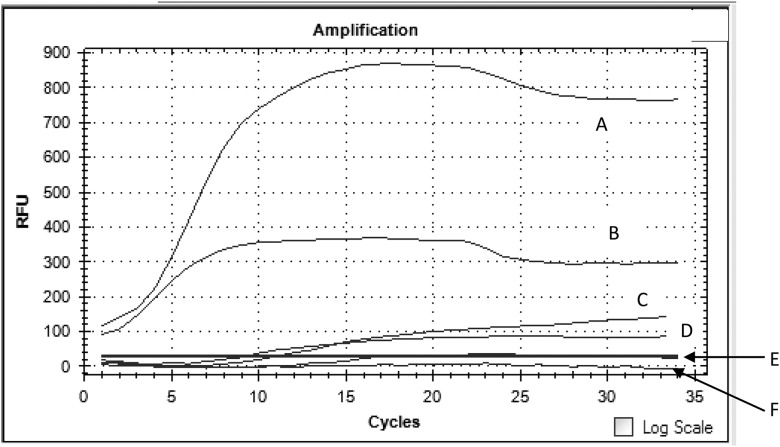
To determine the limit of detection of P. knowlesi RPA, a positive control containing the P. knowlesi 18S rRNA gene was constructed. The copy number of the plasmid was calculated based on the following formula: number of copies = (amount of plasmid (ng) × 6.022 × 1023)/(length of plasmid (bp) × 1 × 109 × 650). A 10-fold serial dilution of the plasmid (106 copies to one copy) was performed with sterile distilled water. To detect the sensitivity of RPA, 2 μL of each of the diluted plasmid DNA was used as the template. In the work presented here, the detection limit of P. knowlesi RPA was as low as one copy (Figure 2). The plasmids from each of the serial dilutions were tested in duplicate and repeated twice to ensure the accuracy of the result. Both sets of results confirmed the lowest limit of detection as one copy. In the pilot study, RPA reactions were incubated at 15, 18, 20, and 23. We found that the RPA reactions were successfully amplified for any given copy number (above one copy) at each of the time points except incubation at 15 and 18 minutes. Meanwhile, to examine the clinical sensitivity and specificity of the P. knowlesi RPA primers, the RPA assay managed to amplify all 45 samples of the P. knowlesi sample whereas the rest of the non-P. knowlesi samples (42 other Plasmodium spp. and six non-Plasmodium spp.) were negative. Both the sensitivity and specificity test for the RPA primers were performed in duplicate and repeated twice. True positive samples were referred to the samples that were tested positive for microscopy, BinaxNow Malaria Kit, and nested PCR.
Figure 2.
Endpoint assessment of recombinase polymerase amplification assay using real-time PCR machine. Panel A, B, C, D, and F: Dilution of plasmids from 103, 102, 101, 100, and 10-1; Panel E, Negative control.
In comparison to the other isothermal detection method, the RPA assay was observed to be more sensitive. Based on our previous report for loop-mediated isothermal amplification (LAMP) detection of P. knowlesi, the limit of detection of LAMP was 10 copies.11 In 2016, using AMA-1 as the target gene, Lau et al.12 furthered their investigation of LAMP detection with five human malaria parasites (including P. knowlesi, P. ovale, P. vivax, P. falciparum, and P. malariae), finding that the limit of detection for P. knowlesi still remained as 10 copies. However, both of the LAMP detection assays of P. knowlesi presented 100% specificity, which was same as our findings in the present study.
Here, we were able to amplify P. knowlesi samples with incubation at both temperatures of 37°C and 30°C using either a real-time PCR machine or a shaking incubator. Besides this, the technology is able to amplify RNA in addition to DNA.8,10,13 RPA is quick—an RPA product was amplified in this study within 20 minutes. This is more advantageous than conventional methods, such as standard PCR and microscopy. As it is understood that both PCR and RPA methods are able to determine a very low concentration of template, RPA is still considered the ideal approach as PCR amplification requires more time. Moreover, even though microscopy has always been the gold standard for malaria diagnosis, this technique requires experienced personnel to examine the slides. Thus, the RPA method is able to facilitate malaria diagnosis without a microscope or veteran microscopist.
However, there are still certain limitations to this technology. The main issue is cross contamination, which did occur in this study occasionally. Usually, contamination arose when the tube was opened outside of a sterile environment. Hence, strong pipetting practices are the best way to overcome this problem. Having different sets of pipettes can be thought of as another way to reduce the issue of contamination.
In conclusion, as there is an inability of existing diagnosis tools to discern most infectious diseases, nucleic acid amplification methods are always recommended. In this study, we propose RPA assay as an important molecular tool for malaria diagnosis. There is no requirement for expensive equipment or high-temperature incubation. Seeing that the results of RPA can be obtained within 20 minutes, it would be useful for clinicians when treating patients. Ultimately, based on the numerous advantages of the RPA method, RPA is highly advised to be used in a variety of the world’s major infectious diseases. Further, RPA is also a potential technique to be developed as a point-of-care molecular diagnostic tool for P. knowlesi infections as well as other infectious diseases.
REFERENCES
- 1.World Health Organization , 2016. The World Malaria Report 2016. Geneva, Switzerland: World Health Organization. [Google Scholar]
- 2.Singh B, Kim Sung L, Matusop A, Radhakrishnan A, Shamsul SS, Cox-Singh J, Thomas A, Conway DJ, 2004. A large focus of naturally acquired Plasmodium knowlesi infections in human beings. Lancet 363: 1017–1024. [DOI] [PubMed] [Google Scholar]
- 3.Lee KS, Cox-Singh J, Singh B, 2009. Morphological features and differential counts of Plasmodium knowlesi parasites in naturally acquired human infections. Malar J 8: 73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Imwong M, Tanomsing N, Pukrittayakamee S, Day NP, White NJ, Snounou G, 2009. Spurious amplification of a Plasmodium vivax small-subunit RNA gene by use of primers currently used to detect P. knowlesi. J Clin Microbiol 47: 4173–4175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lau YL, Lai MY, Anthony CA, Chang PY, Palaeya V, Fong MY, Mahmud R, 2015. Comparison of three molecular methods for the detection and speciation of five human Plasmodium species. Am J Trop Med Hyg 92: 28–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Piepenburg O, Williams CH, Stemple DL, Armes NA, 2006. DNA detection using recombination protein. PLoS Biol 4: e204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Teoh BT, , et al. 2015. Early detection of dengue virus by use of reverse transcription-recombinase polymerase amplification. J Clin Microbiol 53: 830–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Abd El Wahed A, , et al. 2015. Recombinase polymerase amplification assay for rapid diagnostics of dengue infection. PLoS One 10: e0129682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kersting S, Rausch V, Bier FF, Nickisch-Rosenegk MV, 2014. Rapid detection of Plasmodium falciparum with isothermal recombinase polymerase amplification and lateral flow analysis. Malar J 13: 99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moorea MD, Jaykus LA, 2017. Development of a recombinase polymerase amplification assay for detection of epidemic human noroviruses. Sci Rep 7: 40244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lau YL, Fong MY, Mahmud R, Chang PY, Palaeya V, Cheong FW, Chin LC, Anthony CN, Al-Mekhlafi AM, Chen Y, 2014. Specific, sensitive and rapid detection of human Plasmodium knowlesi infection by loop-mediated isothermal amplification (LAMP) in blood samples. Malar J 10: 197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lau YL, Lai MY, Fong MY, Jelip J, Mahmud R, 2016. Comparison of three molecular methods for the detection and speciation of five human Plasmodium species. Am J Trop Med Hyg 94: 336–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yan L, Zhou J, Zheng Y, Gamson AS, Roembke BT, Nakayama S, Sintim HO, 2014. Isothermal amplified detection of DNA and RNA. Mol Biosyst 10: 970–1003. [DOI] [PubMed] [Google Scholar]