Abstract
Introduction:
High-throughput RNA sequencing (RNA-Seq) studies demonstrate that Alter-native Splicing (AS) is a widespread mechanism that enhances transcriptome diversity, particularly in plants exposed to environmental stress. In an attempt to determine the transcriptome and AS patterns of cabbage inbred line “HO” under Heat Stress (HS), RNA-Seq was carried out using HS-treated and con-trol samples. Genome-wide analysis indicated that AS is differentially regulated in response to HS. The number of AS events markedly increased in HS-treated samples compared to the control.
Conclusion:
We identified 1,864 genes, including Heat shock transcription factor (Hsf) and heat shock protein (Hsp) genes, that exhibited >4-fold changes in expression upon exposure to HS. The enriched Gene Ontology (GO) terms of the 1,864 genes included ‘response to stress/abiotic stimulus/chemical stimulus’, among, which the genes most highly induced by HS encode small Hsps and Hsf proteins. The heat-induced genes also showed an increased number of AS events under HS conditions. In addi-tion, the distribution of AS types was altered under HS conditions, as the level of Intron Retention (IR) decreased, whereas other types of AS increased, under these conditions. Severe HS-induced AS was al-so observed in Hsfs and Hsps, which play crucial roles in regulating heat tolerance. Our results support the notion that AS of HS-related genes, such as HsfA2 and HsfB2a, are important for heat stress adapta-tion in cabbage.
Keywords: Alternative splicing, Cabbage, Heat shock, Heat shock transcription factor, Heat shock protein, Transcriptome
1. INTRODUCTION
Plant growth and development are influenced by various aspects of climate change such as global warming. As heat stress due to high ambient temperatures poses a serious threat to the productivity and quality of various crop plants worldwide, the mechanisms underlying the responses of plants to elevated temperatures are an important focus of study [1-3]. Understanding how plants respond to high temperature at the molecular level will help breeders produce thermo-tolerant crop varieties. Cabbage (Brassica oleracea L.) is a valuable vegetable crop that is sensitive to high temperatures, making heat-tolerant cabbage a strong focus of breeding programs [4].
We previously compared the heat-responsive gene expression levels of four Heat-Tolerant Cabbage Lines (HTCLs) and four heat-sensitive cabbage lines (HSCLs) [4]. We identified differentially expressed heat shock protein (Hsp) and heat shock transcription factor (Hsf) genes under heat stress conditions in inbred lines, ‘HO’ and ‘JK’, using microarray analysis. Hsf and Hsp proteins are central regulatory proteins that function during heat responses and have a significant impact on thermotolerance. The Hsfs comprise a large family consisting of 21 and 25 members in the model species Arabidopsis and rice, respectively [5]. Plant Hsfs can be divided into three classes, A, B, and C, based on variations in their three domains (DNA binding domain, oligomerization domain, and C-terminal activation domain) [6, 7]. Hsf proteins reprogram gene expression to repair protein damage through elevated biosynthesis of Hsp chaperones and proteases [8, 9]. The Hsps are classified into five conserved classes according to their approximate molecular weight: Hsp100, Hsp90, Hsp70, Hsp60, and the small Hsps (sHsps) [10]. The two best-studied families, Hsp60 and Hsp70 recognize and bind to unfolded proteins, thereby preventing their aggregation by functioning as molecular chaperones [11]. Some higher plants have many more sHsps than do other eukaryotes and are characterized by the presence of more than 30 types of sHsps, which are induced upon exposure to stresses, including heat. The diversity of plant sHsps suggests that some aspects of these proteins, possibly including substrate specificity, are quite variable among sHsps in relation to the adaptation of plants to heat stress [12].
AS of pre-mRNA is an important post-transcriptional regulatory mechanism that enhances transcriptome complexity and proteome diversity in metazoan organisms. AS occurs in ~95% of intron-containing genes in human, as revealed by RNA-Seq [13]. Genome-wide studies revealed complex AS patterns in plants [14-16]. In Arabidopsis, 40− 61% of intron-containing genes are alternatively spliced under normal growth conditions. AS is particularly prevalent in plants exposed to environmental stress and is likely important for stress adaptation. Several types of AS events, such as intron retention (IR), alternative 3ʹ or 5ʹ splice site selection, exon skipping (ES), and the production of mutually exclusive exons in plant, can increase protein diversity, cause translational process, and affect mRNA stability in plants under stressful environmental conditions [17, 18].
There are few reports describing how elevated temperature affects pre-mRNA splicing, although several studies demonstrated that temperature changes affect the regulation of splicing [18]. In the current study, based on this reference genome for the mesopolyploid B. oleracea, we characterized the HS transcriptome of cabbage line ‘HO’ under HS. Our genome-wide analysis of AS from whole transcripts helped elucidate which type of AS events affect thermos-tolerance in the heat-tolerant inbred cabbage line, ‘HO’. Our findings about changes in the structures and functions of heat stress proteins due to AS increase our understanding of the complexity of the heat stress response in cabbage. AS represent an important level of regulation of gene expression under heat stress conditions, which should be considered in basic and molecular breeding studies.
2. MATERIALS AND METHODS
2.1. Plant Materials, Growth Conditions and Heat Treatment
Seeds of heat-tolerant inbred line cabbage (Brassica oleracea L.) line ‘HO’ were obtained from the Asia Seed Company (Gyeonggi-Do, Korea). ‘HO’ was selected as an HTCL line, based on its ability to form normal heads after a single summer season in a vinyl greenhouse [4]. The seeds were surface-sterilized with 70% ethanol for 5 min and with 10% chlorox bleach for 30 min, followed by five rinsed with sterilized distilled water for 5 min per rinse. The seeds were directly germinated in sterilized soil and grown for 2 weeks. The plants were maintained under controlled growth conditions of 16 h light/8 h dark cycles with 150 μE m-2s-1 light at 24°C in a growth chamber. For heat treatments, 2-week-old young soil-grown plants were subjected to HS at 42°C for 4 h and sampled for RNA preparation.
2.2. RNA Isolation, Library Construction, and RNA Seq
An equal amount of total RNA from each treated/non-heat-treated sample was pooled for transcriptome sequencing to obtain a comprehensive range of transcripts. Poly(A)+ RNA was purified from the pooled total RNA (20 µg) using oligo(dT) Dynabeads. Impurities were removed from the hybridized sample using a series of low-salt washes. First-strand cDNA was synthesized using oligo(dT) primers. The RNA was then degraded with RNase H (Invitrogen, Carlsbad, CA, USA) and second-strand cDNA was synthesized using DNA polymerase I (New England BioLabs, Ipswich, MA, USA). Double-stranded cDNA was randomly fragmented using a nebulizer. The fragments were then repaired and extended at their 3′ ends by the addition of a single adenine, and different adapters were ligated to the 5′ and 3′ ends. The ligated fragments were separated on a gel, and fragments of ∼200 bp were isolated. After amplification by polymerase chain reaction (PCR), the fragments were separated using electrophoresis, purified, and subjected to Illumina HiSeq2000 sequencing. The forward and reverse paired-end reads of two sequencing data sets were linked, and the indexed adapter sequences were trimmed using the SOLEXA QA package v. 1.13 [19].
2.3. Reads Mapping to the Reference Genome and Splicing Junction Prediction
Reads mapping to the Brassica oleracea genome [20] and splice junction detection were performed using TopHat v. 2.0.6 software [21].
2.4. GO Analysis and Gene Clustering
Among the altered transcripts as a result of HS, genes exhibiting >4-fold changes in expression (up or down-regulated) upon exposure to heat were identified from whole transcripts. To further assess their biological functions, these HS-induced and HS-repressed genes were subjected to functional enrichment analysis using the Agro Gene Ontology (GO) (FDR ≤ 0.05) annotation tool with GI number for Brassica oleracea. All Brassica oleracea genes were annotated by TAIR ID, and analyzed with default criteria (counts ≥ 2 and EASE score ≤ 0.1) to obtain enriched GO terms. In detail, GO terms (biological process) among functionally annotated genes were investigated using the gene GO database. Clustering of Differentially Expressed Genes (DEGs) was performed with Cluster 3.0 software and illustrated by heat map.
2.5. Quantitative Real Time-PCR (qPCR) Analysis and High-Resolution Reverse Transcription-PCR RT-PCR
Total RNA was isolated from B. oleracea tissue using RNAiso Plus (Takara, Tokyo, Japan). The cDNA was synthesized with M-MLV reverse transcriptase and an oligo(dT) primer in a 20 µL volume according to the manufacturer’s instructions (Invitrogen, Carlsbad, CA, USA). Four Hsp and three Hsf genes were selected for RT-PCR. Quantitative real time PCR (q-PCR) was performed in 10 µL reactions containing gene-specific primers, 1 µL of cDNA as template, and SYBR Premix Ex Taq. The reactions were performed using a CFX96 Real-Time PCR system (BioRad, Hercules, CA, USA). The thermal profile for q-PCR was as follows: 3 min at 95°C, followed by 40 cycles consisting of 95°C for 25 s, 60°C for 25 s, and 72°C for 25 s. Primer specificity and the formation of primer-dimers were monitored by dissociation curve analysis. The expression level of B. oleracea Actin2 (BoActin2) was used as an internal standard for normalization of cDNA template quantity. RT-PCR and q-PCR were performed in triplicate.
2.6. Identifications of AS Transcripts Using MultiNA
The PCR mixture (10 μL) contained 100 ng of DNA templates, 2× PCR master Mix (Noblebio, Hwaseong, Korea) and 1 pM of each primer. The following PCR protocol was used: 94°C for 3 min, followed by 32 cycles at 94°C for 30 s, 60°C for 30 s, and 72°C for 30 s and, an extension step at 72°C for 5 min. AS transcripts were separated by electrophoresis in a 1.5% agarose gel using a 1 kb Plus DNA Ladder (BioFact, Daejeon, Korea) to calculate the length of the AS transcripts. The number and abundance of AS transcripts were calculated using MultiNA microchip electrophoresis (Shimadzu Biotech, Kyoto, Japan).
3. RESULTS
3.1. HS Treatment, mRNA Sequencing, and Data Analysis
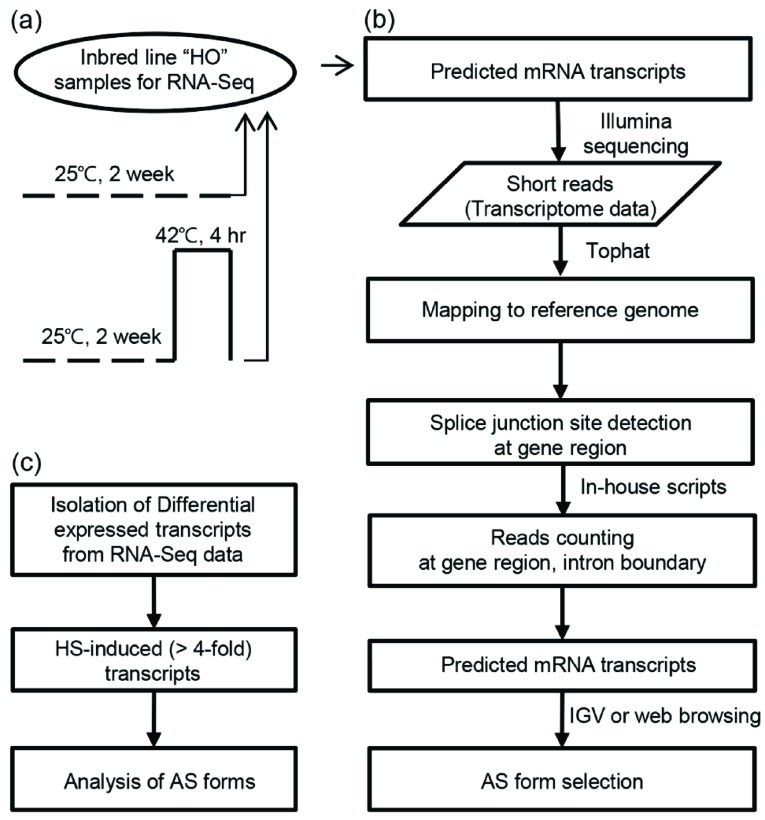
We previously showed that HS at 42°C for 4 h could be used to reveal the obvious heat-tolerant phenotypes of the heat-tolerant inbred cabbage (Brassica oleracea L.) line ‘HO’ compared to heat-sensitive inbred lines [4]. Based on this finding, in the current study, we used ‘HO’ plants grown at 24°C as the control and ‘HO’ plants incubated at 42°C for 4 h as the heat treatment to prepare RNA samples for RNA-Seq (Fig. 1a). The extracted RNA was then used for mRNA isolation, fragmentation, cDNA synthesis, and sequencing. After data trimming and filtering, we performed read mapping to the reference genome and splice junction detection using TopHat and in-house Perl scripts (Fig. 1b). Finally, we counted reads at gene region and intron boundaries, predicted mRNA transcripts, and selected AS forms (Fig. 1b). Nearly 275.4 million reads were generated, including 130.7 and 144.7 million reads for the control and HS samples, respectively (Table S1 (89MB, pdf) ). We mapped the sequence reads to the B. oleracea genome using the TopHat program. In total, 99.1 million reads (75.8%) from the control and 99.6 million reads (68.8%) from the HS samples were mapped to the reference genome. Among the mapped reads, 38.2 million (38.6%) and 30.5 million (30.6%) reads from control and HS samples were alternatively spliced, respectively (Table 1).
Fig. (1).
Experimental design and data analysis. (a) HS treatment of heat tolerant inbred cabbage line ‘HO’. Arrows indicate the time of sample collection for total RNA extraction. (b, c) The work flow and algorithm used to detect AS and heat stress-responsive genes.
Table 1.
Statistics of mapping to reference.
| Sample | Total Reads | Mapped Reads | Spliced Reads |
|---|---|---|---|
| Control | 130,704,165 | 99,083,464 (75.81%) | 38,246,833 (38.60%) |
| Heat treatment | 144,692,690 | 99,555,630 (68.80%) | 30,498,399 (30.63%) |
When these reads were located in the 59,225 annotated genes of the cabbage reference (Table S2 (89MB, pdf) ), 88,833 AS events from the control and 118,525 AS events from the HS sample were potentially detected among alternatively spliced transcripts.
3.2. Identification of AS Events in Response to HS
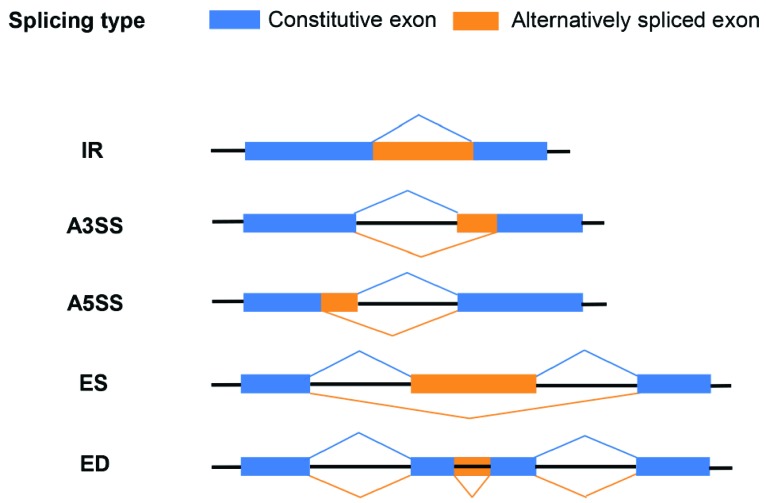
AS generates two or more mRNAs from the same precursor mRNA (pre-mRNA) by using different splicing sites. The different forms of AS events in the transcriptome of inbred line ‘HO’ were divided into five classes: IR, alternative 3′splice site donor (A3SS), alternative 5′ splice site acceptor (A5SS), ES, and exon deletion (ED) (Fig. 2). Of the most common types of AS, IR and ES are the most frequent AS event in plants and human, respectively [14, 22]. In general, ED is not a common type of AS, as shown in previous reports [23-25]. Here, we investigated this type of AS event because a significant number of ED events were identified in the cabbage inbred line regardless of HS condition. The most common AS type was IR, comprising 73-75% of AS events, followed by ES (12%), A5SS and A3SS (6-7%), and ED (1%) (Fig. 2).
Fig. (2).
Schematic representation of alternative splicing. The figure illustrates the different types of alternative splicing: intron retention (IR), alternative 3′ splice site donor (A3SS), alternative 5′ splice site acceptor (A5SS), exon skipping (ES), and exon deletion (ED). Exons, introns, and constitutively/alternatively spliced regions are represented by boxes, black lines, and blue/orange lines, respectively. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this paper.)
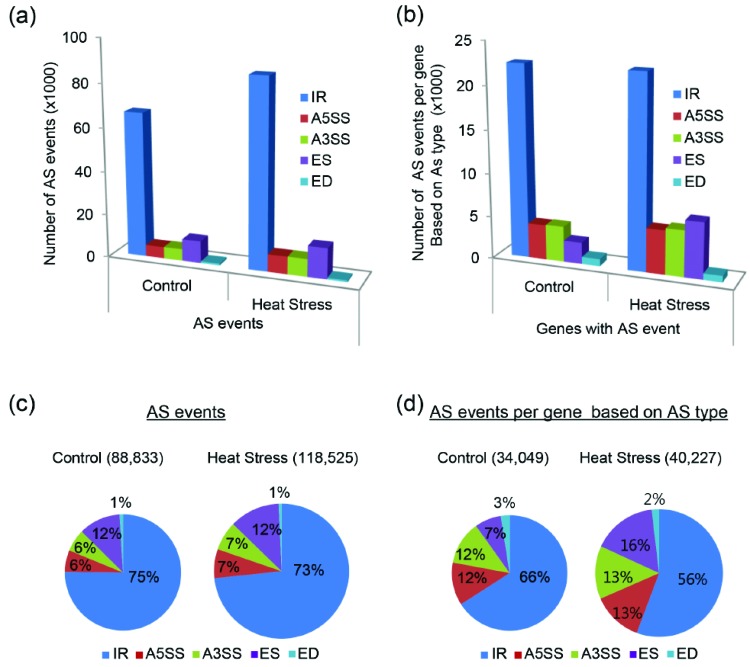
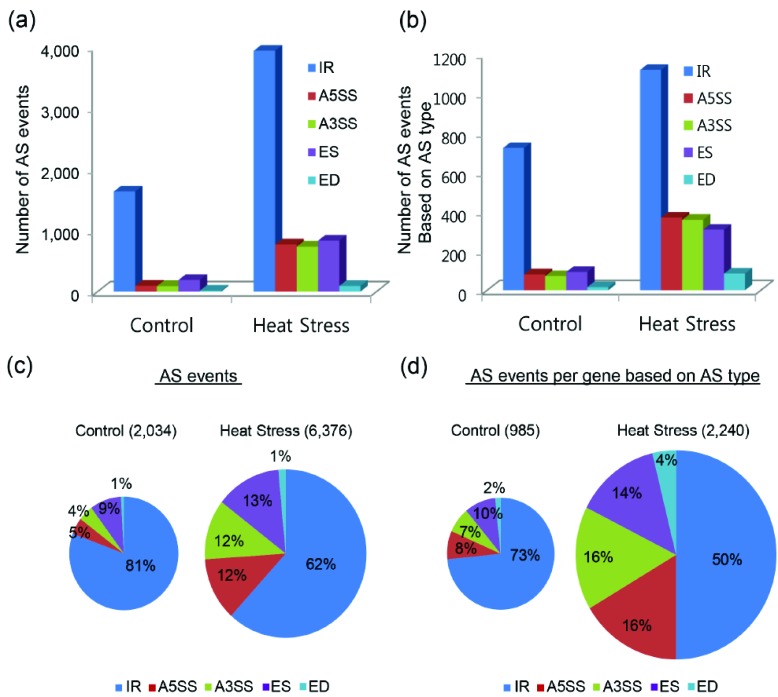
RNA-Seq analysis showed that the number of AS events changed in response to HS (Fig. 3). The total number of predicted AS events in B. oleracea line ‘HO’ with HS increased by more than 30% compared to the control condition (Fig. 3a, c). The distribution of all AS forms in control and HS-treated samples was similar. We analyzed the number of AS events per gene according to type to investigate AS diversity around each gene, finding that AS occurred 30,049 times under control conditions, and 40,227 times under HS-induced conditions (Fig. 3b, d). HS conditions caused a greater number of different AS types than control conditions, with nearly 20% more types of AS per gene in the HS sample versus the control. In particular, the number of ES events was markedly higher in the HS samples, compared to the control (Fig. 3b). The distribution of IR AS forms was markedly reduced under elevated temperatures, but the distribution of A5SS and A3SS AS forms was nearly identical under both conditions. Interestingly, the number of ES events in the HS samples exhibited the greatest increase, suggesting that the AS form of ES may be important in the response to HS (Fig. 3d).
Fig. (3).
Distribution and number of predicted alternative splicing events in heat tolerant Brassica oleracea line ‘HO’ under control and heat stress conditions. (a, b) Total number of AS events and number of AS events based on AS type per a gene. (c, d) Pie charts showing the percentage of each splicing type per total number of AS events and the number of AS each type of AS events per gene. IR (intron retention), A5SS (alternative 5′splice site donor), A3SS (alternative 3′splice site acceptor), ES (exon skipping), and ED (exon deletion). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this paper.)
3.3. Identification of DEGs in Inbred Line ‘HO’ Under HS
In a previous study, we compared the expression levels of HS-related genes between the HTCL, ‘HO’ and the HSCL, ‘JK’ through GeneChip assays [4]. Since the inbred line ‘HO’ used in the current study is the heat tolerant, we expected that genes identified as induced under HS would be involved in the heat tolerance of this line. When the cabbage plants were treated with HS, a complex network of heat-responsive B. oleracea genes appeared. The RNA-Seq data show that HS regulates a number of transcription factor genes and genes associated with the heat stress response. The normalized read count processed by TopHatII was used to quantify transcript abundance [21]. Among the transcripts with altered levels as a result of HS, 1,864 genes/3,355 genes exhibited >4-fold changes in up/downregulated expression upon exposure to HS.
To further assess the biological functions of these genes, 1,864 HS-induced and 3,355 HS-repressed genes were subjected to functional enrichment analysis. (Fig. S1 (89MB, pdf) ) shows the percentage of annotated genes classified in the biological process (BP) category using the Agro GO annotation tool with GI number for Brassica oleracea.
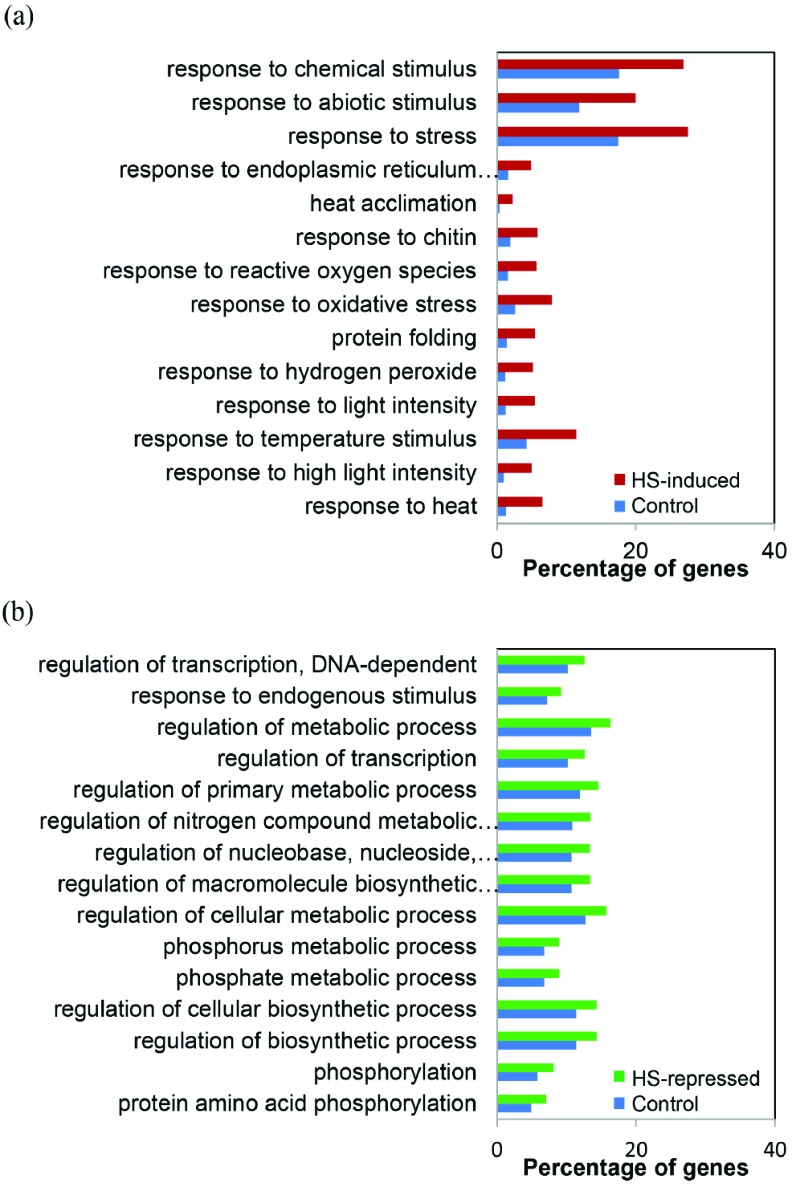
When the GO categories from the 1,864 HS-induced genes were compared to those of the total cabbage reference genes [26], the dominant subcategories were as follows: ‘response to stimulus’, ‘immune system process’, ‘signaling’, and ‘death’ (Fig. S1a (89MB, pdf) ). On the other hand, 3,355 HS-repressed genes were highly enriched in GO terms such as ‘developmental process’, ‘regulation of biological process’, ‘biological regulation’, and ‘multicellular organismal process’ (Fig. S1a (89MB, pdf) ). We also identified over-represented GO terms in the BP category among genes that were functionally annotated in detail (Fig. 4). GO classification of HS-induced genes at the level 2 revealed that ‘response to stress/abiotic stimulus/chemical stimulus/ reactive oxygen species’, and ‘protein folding’ (Fig. 4a), as well as GO terms related to transcription, primary metabolic process, macromolecule biosynthesis, and protein phosphorylation were the most highly represented GO terms among HS-repressed genes (Fig. 4b). These results indicate that specific subsets of genes are differentially regulated by HS.
Fig. (4).
GO biological process categories of the 1,864 heat-induced genes (a) and 3,555 heat-repressed genes (b). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this paper.)
3.4. HS-Induced Genes Among DEGs and q-PCR Validation
HS-induced genes are enriched in several important functional pathways, such as ‘response to stimulus’ and ‘response to abiotic/temperature stimulus’ (Fig. S1a (89MB, pdf) and Fig. 4a). Among these, the genes most highly induced by HS are sHsps and Hsf proteins, indicating that a subset of genes with critical heat stress response functions are transcriptionally regulated. To investigate trends in gene expression profiles, DEGs that exhibited genes >4-fold upregulated expression between the control and HS transcriptomes were clustered using Cluster 3.0 software. The heat map shows three distinct gene clusters of highly expressed genes (Fig. S2 (89MB, pdf) , arrow-head), including three families of Hsps, sHsps and Hsfs such as the genes encoding Hsp90, H70B, Hsp101, Hsp21, Hsp20-like protein, Hsp17.6, Hsp22, HsfA2, HsfA7A, and HsfB2A (Table S3 (89MB, pdf) ).
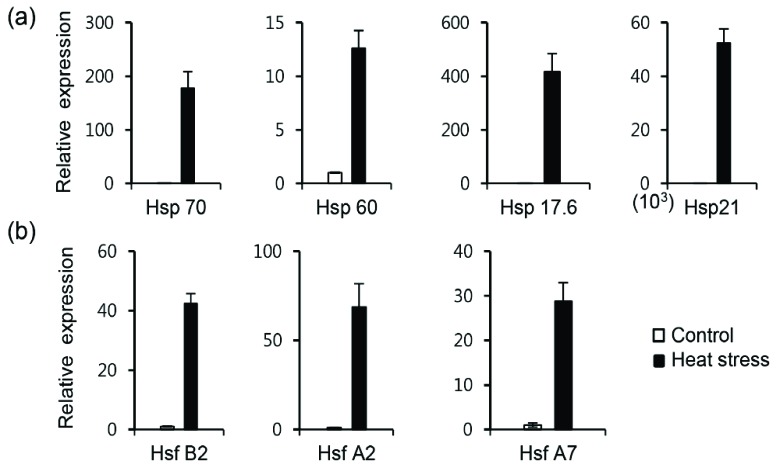
The expression levels of the Hsps, sHsps and Hsfs were further validated by q-PCR using specific primer pairs designed to detect the transcripts of the corresponding genes (Table S4 (89MB, pdf) ). The q-PCR results correlated with the RNA-Seq results (data not shown); the results of q-PCR analysis of seven genes that were highly differentially expressed between the HS and control samples are showed in (Fig. 5). The greatest difference in expression was observed for Hsp21, which exhibited a nearly 50,000-fold increase in expression in response to HS. The others genes exhibited a 10-400 fold increase in expression in response to HS.
Fig. (5).
Validation of RNA-Seq data by q-PCR. (a) Expression levels of Hsps, small Hsps and (b) Hsfs genes. Pooled RNA from control and heat stress-treated samples was analyzed in triplicate for q-PCR. All data points mean ± SE of three independent experiments with different plants.
3.5. AS of HS-Induced Genes
To analyze the post-transcriptional regulation of the 1,864 highly HS-induced genes, we classified the AS events (Fig. 6). The total number of predicted AS events for these genes was over 300% greater under HS than under control conditions (Fig. 6a, c). The distribution of AS types differed between HS-treated sample and the control. The most common AS type was IR, comprising 81% and 62% in the control and HS samples, respectively. While there was an approximately 20% reduction in IR-type events in the HS samples compared to the control, the frequency of ES events
Fig. (6).
Distributions of predicted alternative splicing events of 1,864 heat-induced genes in heat tolerant Brassica oleracea line ‘HO’ under control and heat stress. (a & b) Number of AS events (left panel) and number of AS events based on AS type per gene (right panel). (c & d) Pie charts showing the percentage of each splicing type among total AS events (left panel) and the number of AS events based on AS type per gene (right panel). IR (intron retention), A5SS (alternative 5′ splice site donor), A3SS (alternative 3′ splice site acceptor), ES (exon skipping), and ED (exon deletion). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this paper.)
increased from 9% to 13% in the HS samples versus the control, and the frequency of A5SS and A3SS increased from 4-5% to 12% in the HS samples compared to the control, whereas both the control and HS samples had an ED rate of 1%. We analyzed the number of different types of AS events per gene to explore AS diversity around each HS-induced gene, finding that, among the 1,864 genes, AS occurred 985 times under control conditions, and 2,240 times under HS-induced conditions (Fig. 6b, d). Therefore, HS conditions led to more than twice as many AS events were detected under control conditions. Interestingly, the number of ES, A5SS, and A3SS events increased approximately 2-fold under HS, whereas the number of IR type AS events was reduced by HS. These results indicate that HS-induced genes exhibited more ES-, A3SS-, and A5SS-type AS under HS compared to the control, suggesting that HS-induced genes are regulated at the post-transcriptional level as well as the transcriptional level upon HS.
3.6. HsfA2 and HsfB2 Generated Alternatively Spliced Variants by HS in Cabbage
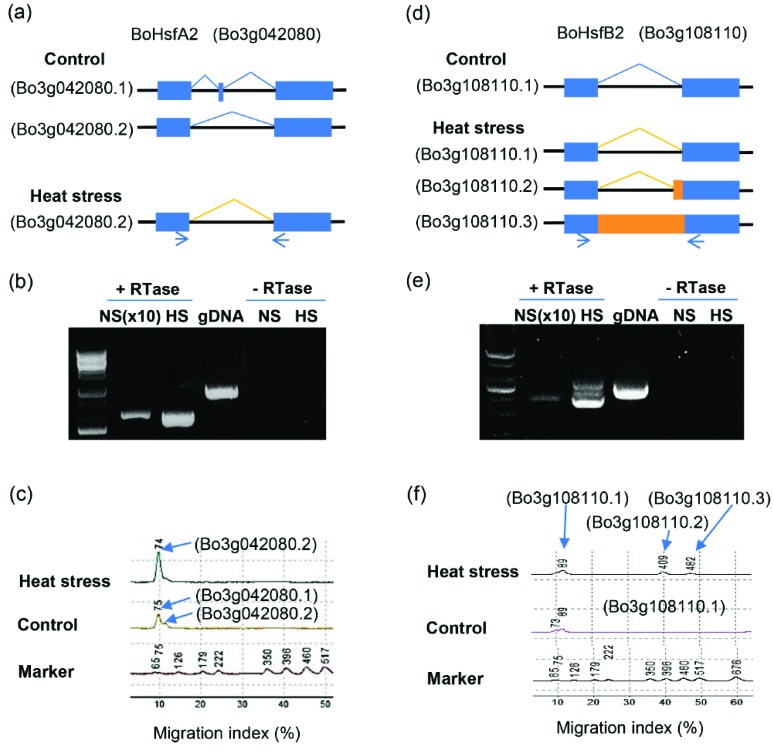
In light of the different splicing types of HS-induced genes in ‘HO’ genotype, we investigated the splicing patterns of Hsps and Hsfs. Severe HS-induced AS also occurred in Hsfs and Hsps, which play crucial roles regulating the heat tolerance response. Specially, we investigated the AS forms of representative Hsfs, HsfA2, and HsfB2a transcription factor genes to determine whether their transcripts exhibited AS under HS. RT-PCR analysis was conducted using specific primers (Table S4 (89MB, pdf) ). The RT-PCR products were cloned and sequenced, revealing altered transcript structures between control and HS conditions (Fig. 7). Upon HS conditions, BoHsfA2 was mainly expressed with ES in the second exon, which was not the major form detected under control conditions. Unlike in Arabidopsis, BoHsfA2 was properly spliced with a mini exon or ES of the mini exon under normal conditions in cabbage. Therefore, BoHsfA2 exhibited the contrary trend in AS, i.e., ES, compared to its homolog in Arabidopsis (Fig. 7a, b, c). BoHsfB2a contained a single intron that was completely removed to generate proper BoHsfB2a transcript under control conditions, whereas upon HS conditions, two new introns retained in AS variants were identified (Fig. 7d, e, f), indicating that HS induces the production of new splice variants post-transcriptionally. Our results support the notion that AS of specific genes, such as HsfA2 and HsfB2, is likely important for the heat stress tolerance of ‘HO’.
Fig. (7).
Validation of alternative splicing events at the BoHsfA2 and BoHsfB2 loci. (a, d) Gene structures and transcripts of BoHsfA2 and BoHsfB2. Exons, introns, and constitutively (with no stress)/alternatively (with heat stress) spliced regions are represented by boxes, black lines, and blue/orange lines, respectively. (b, e) RT-PCR of BoHsfA2 and BoHsfB2. NS/HS indicates no stress/heat stress. PCR of samples under NS used 10-fold (×10) amounts of template compared with those under HS. (c, f) AS transcript patterns determined using MultiNA. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this paper.)
4. DISCUSSION
Heat stress responses are highly sophisticated events that alter the biochemical composition of plant cells to protect them from damage caused by high temperatures. Plants have evolved a variety of strategies for coping with environmental stresses such as HS. One such strategy is AS. A plethora of alternatively spliced isoforms have tissue-preferential expression patterns [27]. In addition, increasing evidence indicates that AS is a critical post-transcriptional event that plays an important role in plant stress responses [28]. Our effort to acquire the transcriptomes of cabbage in response to high-temperature stress will be helpful for developing heat-tolerant cabbage cultivars. In the current study, we used a heat tolerant genotype, ‘HO’, which we previously identified from among eight cabbage genotypes [4] to investigate the responses of cabbage to high temperatures at the global transcriptional level using high-throughput RNA-Seq approach.
CONCLUSION
We detected an increase in the number of AS events in HS-treated samples compared to the control more than 50% of which comprised IR-type AS (Fig. 3). The presence of retained introns is not surprising; in fact, a number of retained introns have been reported in various cell types across many species [29-31]. In the non-vascular plant Physcomitrella patens, the number of IR events is markedly reduced under HS conditions, which helps the plant overcome this stress; IR is repressed in transcripts of genes involved in photosynthesis and protein production but is induced genes for directly involved in protein folding and degradation [32]. Our genome-wide analysis yielded similar results for cabbage: the number of IR-type events was reduced by HS, whereas the number of ES, A3SS, and A5SS-type events was induced by as much as 200% under HS (Fig. 3). Genes undergoing HS-induced AS are mostly involved in specific functions, which suggests that plants regulates AS with transcript specificity under elevated temperatures. Furthermore, the 1,864 HS-induced genes exhibited sensitivity to HS in terms of the regulation of AS. Upon exposure to HS conditions, the relative frequency of IR-type AS events was reduced by approximately 20%, a level greater than the reduction for cabbage genes genome-wide (Fig. 6). A mRNA sequencing study of P. patens under HS revealed that transcripts exhibiting HS-induced IR contain a purine-rich, GAG repetitive motif in their exonic regions, which may serve as a regulatory cis element during HS-mediated AS regulation [32]. This finding led to the speculation that cabbage genes undergoing HS-induced IR also possess this motif; further investigations are needed to elucidate the mechanism underlying HS-induced AS in cabbage.
Many of Hsfs share a common feature, namely AS. The conserved splicing position in these proteins plays an important role in Hsf gene evolution across organisms [33]. On reason that plants contain many more functional Hsfs than other organisms is that plant Hsfs undergo AS followed by specialization, eventually leading to the formation of different plant Hsfs. The results from our RT-PCR analysis of Hsfs in cabbage (Fig. 7) support the notion that Hsf genes are involved in post-transcriptional thermoregulation. HsfA2 AS has been extensively studied in Arabidopsis, Medicago sativa, rice, and moss [32-35]. These studies indicate that intronic HsfA2 exhibits AS, a post-transcriptional regulatory mechanism that is conserved in plants. In the current study, we found that pattern of AS in cabbage HsfA2 differed from that of Arabidopsis: a mini exon was produce under control conditions and mini ES occurred under HS conditions, a pattern that is the opposite of that of Arabidopsis, despite we verified the AS with three replicated cloning and sequencing of the RT-PCR products. Further investigation is needed to clarify the regulation of BoHsfA2 expression in cabbage. Heat-inducible HsfB2a is involved in the development of the female germ line and slightly represses HsfA2 activity in Arabidopsis [36], however, to date, little information is available about the regulation of HsfB2a by AS so far. In this study, we demonstrated that BoHsfB2a generates two AS variants, which may contribute additional complexity to the transcriptional regulation of Hsf, under HS conditions. Heat-induced transcripts and their alternative spliced forms in inbred line ‘HO’ might also contribute to the heat tolerance of ‘HO’.
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
Not applicable.
HUMAN AND ANIMAL RIGHTS
No Animals/Humans were used for studies that are base of this research.
CONSENT FOR PUBLICATION
Not applicable.
ACKNOWLEDGEMENTS
This work was supported by the Agricultural Biotechnology Developmental Program (no. 114061-3 and no. 115076-2) of the Ministry of Agriculture, Food, and Rural Affairs: and by the KRIBB Initiative Program to HS Cho.
SUPPLEMENTARY MATERIAL
Supplementary material is available on the publisher’s web site along with the published article.
CONFLICT OF INTEREST
The authors declare no conflict of interest, financial or otherwise.
REFERENCES
- 1.Hedhly A., Hormaza J.I., Herrero M. Global warming and sexual plant reproduction. Trends Plant Sci. 2009;14:30–36. doi: 10.1016/j.tplants.2008.11.001. [DOI] [PubMed] [Google Scholar]
- 2.Lobell D.B., Field C.B. Global scale climate-crop yield relationships and the impacts of recent warming. Environ. Res. Lett. 2007;2:1–7. [Google Scholar]
- 3.Lobell D.B., Schlenker W., Costa-Roberts J. Climate trends and global crop production since 1980. Science. 2011;333:616–620. doi: 10.1126/science.1204531. [DOI] [PubMed] [Google Scholar]
- 4.Park H.J., Jung W.Y., Lee S.S., Song J.H., Kwon S.Y., Kim H., Kim C., Ahn J.C., Cho H.S. Use of heat stress responsive gene expression levels for early selection of heat tolerant cabbage (Brassica oleracea L.). Int. J. Mol. Sci. 2013;14:11871–11894. doi: 10.3390/ijms140611871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Scharf K.D., Berberich T., Ebersberger I., Nover L. The plant heat stress transcription factor (Hsf) family, structure, function and evolution. Biochim. Biophys. Acta. 2012;1819:104–119. doi: 10.1016/j.bbagrm.2011.10.002. [DOI] [PubMed] [Google Scholar]
- 6.Nover L., Bharti K., Doring P., Mishra S.K., Ganguli A., Scharf K.D. Arabidopsis and the heat stress transcription factor world, how many heat stress transcription factors do we need? Cell Stress Chaperones. 2001;6:177–189. doi: 10.1379/1466-1268(2001)006<0177:aathst>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bharti K., Von Koskull-Doring P., Bharti S., Kumar P., Tintschl-Korbitzer A., Treuter E., Nover L. Tomato heat stress transcription factor HsfB1 represents a novel type of general transcription coactivator with a histone-like motif interacting with the plant CREB binding protein ortholog HAC1. Plant Cell. 2004;16:1521–1535. doi: 10.1105/tpc.019927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Morimoto R.I. Dynamic remodeling of transcription complexes by molecular chaperones. Cell. 2002;110:281–284. doi: 10.1016/s0092-8674(02)00860-7. [DOI] [PubMed] [Google Scholar]
- 9.Baniwal S.K., Bharti K., Chan K.Y., Fauth M., Ganguli A., Kotak S., Mishra S.K., Nover L., Port M., Scharf K.D., Tripp J., Weber C., Zielinski D., von Koskull-Doring P. Heat stress response in plants, a complex game with chaperones and more than twenty heat stress transcription factors. J. Biosci. 2004;29:471–487. doi: 10.1007/BF02712120. [DOI] [PubMed] [Google Scholar]
- 10.Gupta S.C., Sharma A., Mishra M., Mishra R.K., Chowdhuri D.K. Heat shock proteins in toxicology, how close and how far? Life Sci. 2010;86:377–384. doi: 10.1016/j.lfs.2009.12.015. [DOI] [PubMed] [Google Scholar]
- 11.Wang W., Vinocur B., Shoseyov O., Altman A. Role of plant heat-shock proteins and molecular chaperones in the abiotic stress response. Trends Plant Sci. 2004;9:244–252. doi: 10.1016/j.tplants.2004.03.006. [DOI] [PubMed] [Google Scholar]
- 12.Waters E.R. The evolution, function, structure, and expression of the plant sHSPs. J. Exp. Bot. 2013;64:391–403. doi: 10.1093/jxb/ers355. [DOI] [PubMed] [Google Scholar]
- 13.Pan Q., Shai O., Lee L.J., Frey B.J., Blencowe B.J. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat. Genet. 2008;40:1413–1415. doi: 10.1038/ng.259. [DOI] [PubMed] [Google Scholar]
- 14.Syed N.H., Kalyna M., Marquez Y., Barta A., Brown J.W. Alternative splicing in plants--coming of age. Trends Plant Sci. 2012;17:616–623. doi: 10.1016/j.tplants.2012.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thatcher S.R., Zhou W., Leonard A., Wang B.B., Beatty M., Zastrow-Hayes G., Zhao X., Baumgarten A., Li B. Genome-wide analysis of alternative splicing in Zea mays, landscape and genetic regulation. Plant Cell. 2014;26:3472–3487. doi: 10.1105/tpc.114.130773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reddy A.S., Marquez Y., Kalyna M., Barta A. Complexity of the alternative splicing landscape in plants. Plant Cell. 2013;25:3657–3683. doi: 10.1105/tpc.113.117523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Capovilla G., Pajoro A., Immink R.G., Schmid M. Role of alternative pre-mRNA splicing in temperature signaling. Curr. Opin. Plant Biol. 2015;27:97–103. doi: 10.1016/j.pbi.2015.06.016. [DOI] [PubMed] [Google Scholar]
- 18.Staiger D., Brown J.W. Alternative splicing at the intersection of biological timing, development, and stress responses. Plant Cell. 2013;25:3640–3656. doi: 10.1105/tpc.113.113803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cox M.P., Peterson D.A., Biggs P.J., Solexa Q.A. At-a-glance quality assessment of Illumina second-generation sequencing data. BMC Bioinformatics. 2010;11:485. doi: 10.1186/1471-2105-11-485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Parkin I.A., Koh C., Tang H., Robinson S.J., Kagale S., Clarke W.E., Town C.D., Nixon J., Krishnakumar V., Bidwell S.L., Denoeud F., Belcram H., Links M.G., Just J., Clarke C., Bender T., Huebert T., Mason A.S., Pires J.C., Barker G., Moore J., Walley P.G., Manoli S., Batley J., Edwards D., Nelson M.N., Wang X., Paterson A.H., King G., Bancroft I., Chalhoub B., Sharpe A.G. Transcriptome and methylome profiling reveals relics of genome dominance in the mesopolyploid Brassica oleracea. Genome Biol. 2014;15:R77. doi: 10.1186/gb-2014-15-6-r77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Trapnell C., Pachter L., Salzberg S.L. TopHat, discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang E.T., Sandberg R., Luo S., Khrebtukova I., Zhang L., Mayr C., Kingsmore S.F., Schroth G.P., Burge C.B. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Allen M., Pratscher B., Krepler C., Frei K., Schofer C., Pehamberger H., Muller M., Lucas T. Alternative splicing of IL-24 in melanocytes by deletion of exons 3 and 5. Int. J. Immunogenet. 2005;32:375–378. doi: 10.1111/j.1744-313X.2005.00540.x. [DOI] [PubMed] [Google Scholar]
- 24.Capell B.C., Collins F.S., Nabel E.G. Mechanisms of cardiovascular disease in accelerated aging syndromes. Circ. Res. 2007;101:13–26. doi: 10.1161/CIRCRESAHA.107.153692. [DOI] [PubMed] [Google Scholar]
- 25.Dwianingsih E.K., Malueka R.G., Nishida A., Itoh K., Lee T., Yagi M., Iijima K., Takeshima Y., Matsuo M. A novel splicing silencer generated by DMD exon 45 deletion junction could explain upstream exon 44 skipping that modifies dystrophinopathy. J. Hum. Genet. 2014;59:423–429. doi: 10.1038/jhg.2014.36. [DOI] [PubMed] [Google Scholar]
- 26.Liu S., Liu Y., Yang X., Tong C., Edwards D., Parkin I.A., Zhao M., Ma J., Yu J., Huang S., Wang X., Wang J., Lu K., Fang Z., Bancroft I., Yang T.J., Hu Q., Wang X., Yue Z., Li H., Yang L., Wu J., Zhou Q., Wang W., King G.J., Pires J.C., Lu C., Wu Z., Sampath P., Wang Z., Guo H., Pan S., Yang L., Min J., Zhang D., Jin D., Li W., Belcram H., Tu J., Guan M., Qi C., Du D., Li J., Jiang L., Batley J., Sharpe A.G., Park B.S., Ruperao P., Cheng F., Waminal N.E., Huang Y., Dong C., Wang L., Li J., Hu Z., Zhuang M., Huang Y., Huang J., Shi J., Mei D., Liu J., Lee T.H., Wang J., Jin H., Li Z., Li X., Zhang J., Xiao L., Zhou Y., Liu Z., Liu X., Qin R., Tang X., Liu W., Wang Y., Zhang Y., Lee J., Kim H.H., Denoeud F., Xu X., Liang X., Hua W., Wang X., Wang J., Chalhoub B., Paterson A.H. The Brassica oleracea genome reveals the asymmetrical evolution of polyploid genomes. Nat. Commun. 2014;5:3930. doi: 10.1038/ncomms4930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Emrich S.J., Barbazuk W.B., Li L., Schnable P.S. Gene discovery and annotation using LCM-454 transcriptome sequencing. Genome Res. 2007;17:69–73. doi: 10.1101/gr.5145806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mastrangelo A.M., Marone D., Laido G., De Leonardis A.M., De Vita P. Alternative splicing, enhancing ability to cope with stress via transcriptome plasticity. Plant Sci. 2012;185-186:40–49. doi: 10.1016/j.plantsci.2011.09.006. [DOI] [PubMed] [Google Scholar]
- 29.Sakabe N.J., de Souza S.J. Sequence features responsible for intron retention in human. BMC Genomics. 2007;8:59. doi: 10.1186/1471-2164-8-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marquez Y., Brown J.W., Simpson C., Barta A., Kalyna M. Transcriptome survey reveals increased complexity of the alternative splicing landscape in Arabidopsis. Genome Res. 2012;22:1184–1195. doi: 10.1101/gr.134106.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sebe-Pedros A., Irimia M., Del Campo J., Parra-Acero H., Russ C., Nusbaum C., Blencowe B.J., Ruiz-Trillo I. Regulated aggregative multicellularity in a close unicellular relative of metazoa. eLife. 2013;2:e01287. doi: 10.7554/eLife.01287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chang C.Y., Lin W.D., Tu S.L. Genome-wide analysis of heat-sensitive alternative splicing in physcomitrella patens. Plant Physiol. 2014;165:826–840. doi: 10.1104/pp.113.230540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.He Z.S., Xie R., Zou H.S., Wang Y.Z., Zhu J.B., Yu G.Q. Structure and alternative splicing of a heat shock transcription factor gene, MsHSF1, in Medicago sativa. Biochem. Biophys. Res. Commun. 2007;364:1056–1061. doi: 10.1016/j.bbrc.2007.10.131. [DOI] [PubMed] [Google Scholar]
- 34.Sugio A., Dreos R., Aparicio F., Maule A.J. The cytosolic protein response as a subcomponent of the wider heat shock response in Arabidopsis. Plant Cell. 2009;21:642–654. doi: 10.1105/tpc.108.062596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cheng Q., Zhou Y., Liu Z., Zhang L., Song G., Guo Z., Wang W., Qu X., Zhu Y., Yang D. An alternatively spliced heat shock transcription factor, OsHSFA2dI, functions in the heat stress-induced unfolded protein response in rice. Plant Biol. (Stuttg) 2015;17:419–429. doi: 10.1111/plb.12267. [DOI] [PubMed] [Google Scholar]
- 36.Wunderlich M., Gross-Hardt R., Schoffl F. Heat shock factor HSFB2a involved in gametophyte development of Arabidopsis thaliana and its expression is controlled by a heat-inducible long non-coding antisense RNA. Plant Mol. Biol. 2014;85:541–550. doi: 10.1007/s11103-014-0202-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material is available on the publisher’s web site along with the published article.