Abstract
Background:
The worldwide use of glyphosate has dramatically increased, but also has been raising concern over its impact on mineral nutrition, plant pathogen, and soil microbiota. To date, the bulk of previous studies still have shown different results on the effect of glyphosate application on soil rhizosphere microbial communities.
Objective:
This study aimed to clarify whether glyphosate has impact on nitrogen-fixation, pathogen or disease suppression, and rhizosphere microbial community of a soybean EPSPS-transgenic line ZUTS31 in one growth season.
Method:
Comparative analysis of the soil rhizosphere microbial communities was performed by 16S rRNA gene amplicons sequencing and shotgun metagenome sequencing analysis between the soybean line ZUTS31 foliar sprayed with diluted glyphosate solution and those sprayed with water only in seed-filling stage.
Results:
There were no significant differences of alpha diversity but with small and insignificant difference of beta diversity of soybean rhizosphere bacteria after glyphosate treatment. The significantly enriched Gene Ontology (GO) terms were cellular, metabolic, and single-organism of biological process together with binding, catalytic activity of molecular function. The hits and gene abundances of some functional genes being involved in Plant Growth-Promoting Traits (PGPT), especially most of nitrogen fixation genes, significantly decreased in the rhizosphere after glyphosate treatment.
Conclusion:
Our present study indicated that the formulation of glyphosate-isopropylamine salt did not significantly affect the alpha and beta diversity of the rhizobacterial community of the soybean line ZUTS31, whereas it significantly influenced some functional genes involved in PGPT in the rhizosphere during the single growth season.
Keywords: Glyphosate, EPSPS-transgenic soybean line, Soil, Rhizosphere bacterial community, 16S rRNA gene amplicons sequencing, Shotgun metagenome sequencing
1. INTRODUCTION
Glyphosate (N-phosphonomethyl-glycine) was widely but modestly used in the 1980s, because it is a nonselective and broad-spectrum herbicide applied via foliar spray before crop seeding and eradicated almost all herbaceous plants including 90 kinds of emerged grasses, brush and broad-leaf weeds [1].
Glyphosate acts as a herbicide by inhibiting the 5-enolpyruvyl-shikimate-3-phosphatase synthase (EPSPS) and then by blocking the synthesis of necessary aromatic amino acids in the shikimate pathway [2, 3] via translocation within plants [4]. Since transgenic glyphosate-resistant (GR) crops, such as Roundup Ready soybean, became commercially available in 1996 for agricultural planting, the use of glyphosate has dramatically increased [5, 6] and now has been the most widely consumed herbicides in the global market [6].
Although glyphosate has become the dominant herbicide worldwide and has been usually described as environmentally and toxicologically safe [2, 7], it still has raised some concern over the potential impact on plant mineral nutrition, plant pathogen and soil microbial community including rhizosphere microorganisms [8-10] besides glyphosate resistant weeds. Duke et al. intensively reviewed the main concerns and demonstrated that most of available previous studies supported the view that mineral nutrition and plant disease were unaffected by glyphosate although some contradictory studies indicated that glyphosate had such impacts on GR crops [11]. However, the impacts of glyphosate may be covered by functional redundancy of soil microbiota in which overall functions seems not to be affected whereas the composition of microbial community has been changed and some key functions mediated by specific microbial populations have been affected [5]. Actually, Duke et al. also agreed that glyphosate influenced mineral nutrition, disease, and the diversity or richness of rhizosphere microbial community of glyphosate-sensitive plants via its herbicidal effects on roots and other parts of those plants [11]. Furthermore, some intensive studies discovered that glyphosate was released from root into rhizosphere after it translocated within plants [4, 12] and that it was also toxic to some bacteria and fungi [13]. Moreover, the unsafety or toxicity of glyphosate-based herbicide also may result from the additives or surfactants in the commercial formulations [14, 15].
Due to the crucial roles of rhizosphere microbiota affecting plant health and growth [16-19] while plants shape or determine the composition, structure and activity of rhizosphere microbiome via root exudates [20-23], previous studies have investigated the impact of glyphosate on rhizosphere microbiota by using different cultivation-dependent and/or cultivation-independent methods, which were reviewed by Bünemann et al. [24] and Imfeld et al. [5] Newly, deep sequencing of 16S rRNA gene (16S rDNA) amplicons, via next generation sequencing (NGS) platform, has been used to examine the effects of glyphosate on rhizosphere microbiota [25-27].
Recently, the shotgun metagenome sequencing combined with bioinformatics analysis via a NGS platform has been applied to investigate the composition, structure, and function of microbial communities in activated sludge [28], different soil types [29], and other samples [30-33]. However, to the best of published knowledge at the web of science via searching with the combined key words of “glyphosate, metagenome / metagenomic, soil” from all Databases, the effects of glyphosate on rhizosphere microbiota have been rarely investigated by shotgun metagenome sequencing.
In this study, we performed shotgun metagenomic sequencing together with 16S rDNA-based Illumina MiSeq to clarify whether the use of glyphosate affects nitrogen-fixation, pathogen or disease suppression, and rhizosphere microbial community associated with soybean roots during the single growth season.
2. MATERIALS AND METHODs
2.1. Plant Materials
Transgenic soybean line ZUTS31 (or simply Z31), which was same as line L1 generated by Lu et al. [34], contains the g10-epsps gene that was cloned from glyphosate-resistant Deinococcus radiodurans R1 and had been transferred into the soybean cultivar HuaChun3 to produce a glyphosate-resistant 5-enolpyruvylshikimate-3-phosphate synthase (EPSPS).
2.2. Field Design and Sampling
The experimental field (N 31° 53′ 28′′-29′′, E 117° 14 ′ 22′′ -23′′) was located in the Anhui Academy of Agricultural Sciences, Hefei City, Anhui Province, China. The soil type of local area was clay with pH 4.0 to 4.5, which is similar to stagnosol [35]. The experimental field was an area of 576 m2 and was divided into 48 plots (6 m × 2 m per plot) in June 2014. Three replicate plots were used for each treatment of soybean cultivar or line, which were randomly distributed over the field. Soybean seeds of Z31 line were sowed on June 18, 2014. Emerging weeds were manually removed from three plots for planting Z31 line which were foliar sprayed with water as control. Glyphosate solution (Monsanto Company, Malaysia), which contained 41% active ingredient of isopropylamine salt of glyphosate (also named as glyphosate-isopropyl ammonium salt), was foliar sprayed at field rate (3000 ml · ha-1) on July 7, 2014. GR line Z31 plants (samples) treated by glyphosate were named as Z31J1.
The samples of rhizosphere soil were collected as described by Inceoglu et al. [36]. Briefly, two sampling points were in each of three plots, and two soybean plants at seed-filling stage with its surrounding soil were dug out from each sampling point and collected as one biological replicate on September 7, 2014, then placed in a plastic bag, and taken to the laboratory immediately. The soil loosely adhering to the roots were shaken off, and stored at 4°C for enzyme activity analysis or at -70°C freezer for DNA extraction. Then the samples of rhizosphere soil were collected by brushing off the soil that was tightly adhering to the root surface, and then were stored at -80°C freezer for DNA extraction.
2.3. Metagenomic DNA Extraction
In this study, the metagenomic DNA was extracted in duplicate from approximately 2 × 0.60 g soil of every biological replicate using the PowerSoil DNA Isolation Kit (MoBio Laboratories Inc., Carlsbad, CA, USA) as recommended by the manufacturer's instructions with minor modification, which means that soil was homogenized in lysis buffer using Corning LSE vortex mixer (LSE vortex mixer 230V, Coring Inc., Lowell, MA, USA) at 2850 rpm for 10 mins. After mixing well, the concentrations of metagenomic DNA of every biological replicate were checked by a Qubit Fluorometer (Qubit 2.0, Invitrogen, USA), and were more than 10 ng/µl that may minimize the variability in microbial community surveys [37]. DNA integrality was then checked by 1% agarose gel electrophoresis. The DNA samples were stored in a −20°C freezer before using.
2.4. Analyses of 16S rRNA Genes via Amplicon Sequencing
2.4.1. PCR Amplification of 16S rDNA and Illumina Miseq Sequencing
Our strategy is a dual-index sequencing approach [38], which is an improved dual-index paired-end 250 nt approach [39]. In brief, fusion primers were designed to include the appropriate P5 or P7 Illumina adapter sequences, an 8-nt index sequence, and a gene-specific primer for amplifying the V4 region of 16S rDNA, which were 515F (5”- GTGCCAGCMGCCGCGGTAA - 3”) and 806R (5”-GGACTACHVGGGTWTCTAAT - 3”). The primer pair was selected because the error rates decreased to 0.01% for every cluster density within the V4 region data by Illumina Miseq [38], and produced the greatest diversity at the bacterial phylum and domain levels compared with V1-V2, V5-V8 and V6-V8 regions [40]. The 515F plus 806R Dual-index Fusion PCR Primer Cocktail was then added to the PCR Master Mix (NEB Phusion High-Fidelity PCR Master Mix) to amplify the V4 region. Qualified metagenomic DNA was normalized to 30 ng per PCR reaction using 50 µl volume, and its final concentration was higher than 0.4 ng/µl, because the template concentration had a significant effect on the sample profile variability for most samples [37]. The melting temperature was 56°C and PCR cycle is 30. The PCR products were purified using Ampure XP beads (AGENCOURT). High-throughput sequencing of the qualified libraries was conducted by BGI Tech Solutions Co., Ltd (Wuhan, China) by using the Illumina MiSeq NGS platform (Illumina) and MiSeq Reagent Kit with the sequencing strategy paired-end 2 × 250 bp (PE250).
2.4.2. Operational Taxonomic Unit (OTU) Selection
Clean reads were obtained when the raw data were filtered to eliminate the reads with sequencing adapters, ambiguous N base, poly base, or average base quality score less than 20. Then paired-end clean reads with overlap were merged to tags by using Fast Length Adjustment of Short reads (FLASH, v1.2.11) [41]. The tags were then clustered to OTU at 97% sequence similarity by scripts of software USEARCH(v7.0.1090) [42]. OTU representative sequences were taxonomically classified using the Ribosomal Database Project (RDP, Release9, 201203) [43] Classifier v.2.2 trained on the Greengenes database (default: V201305) [44]. Based on the OTU abundance information, principal component analysis (PCA) of OTU was drawn by package “ade4” of software R (v3.0.3).
2.4.3. Analysis of Species Composition and Abundances
The tag number of each taxonomic rank or OTU in different samples was summarized in a profiling histogram which was drawn using software R (v3.0.3). A representative OTU phylogenetic tree was constructed using the QIIME v1.8.0 built-in scripts including the fast tree method for tree construction [45].
2.4.4. Alpha Diversity Analysis
Alpha diversity was applied for analyzing complexity of species diversity for a sample through several indices, including observed OTU number, Chao 1, abundance coverage-based estimator (ACE), Shannon, and Simpson indices, which were calculated by Mothur (v1.31.2) [46]. The corresponding rarefaction curves were drawn by software R (v3.0.3) as follows: calculating OTU numbers based on extracted tags (in multiples of 500); and rarefaction curve was drawn using the indices calculated with extracted tags.
2.4.5. Beta Diversity Analysis
Sequences of each sample were extracted randomly according to the minimum sequence number among the same group to rule out the effects of sequencing depth on beta diversity analyses, which include Bray-Curtis, weighted UniFrac, and unweighted UniFrac, and were then calculated by using software QIIME (v1.80) [45] based on the “OTU table biom” file. Principal coordinate analysis (PCoA) was used to exhibit the differences between the samples according to the matrices of beta diversity distances.
2.5. Shotgun Metagenomic Analyses
2.5.1. Metagenomic DNA Library Construction
0.2 μg DNA was pipetted from each of Z31 or Z31J1 rhizosphere metagenomic DNA, and then the six DNA samples were pooled as one qualified metagenomic DNA, named MGZ31DRh or MGZ31J1DRh, respectively.
Shotgun metagenomic DNA library was constructed according to the manufacturer’s instructions (Illumina) [47] with minor modifications. In brief, a total of 1.2μg qualified DNA of each sample in 80 μl TE was sheared into smaller fragments less than 600 bp by nebulization firstly, fragments were blunted secondly, and were then ligated with Illumina adapter oligo mix after an A(adenine) base was added to the 3' end of the blunt phosphorylated DNA fragments, respectively. Fourthly, the adapter-modified DNA fragments were enriched by NEB Phusion high-fidelity PCR master mix with 65°C melting temperature and 12 cycles. Furthermore, adapted products of 400-600 bp were purified by QIAquick PCR purification kit (QIAGEN), and then were qualified and quantified by Agilent 2100 Bioanaylzer and ABI StepOnePlus Real-Time PCR System. The paired-end (PE) libraries were constructed with insert sizes of 468 bp for MGZ31DRh and 461 bp for MGZ31J1DRh, respectively.
2.5.2. Shotgun Metagenomic Sequencing
High-throughput sequencing of the qualified metagenome libraries was conducted by BGI Tech Solutions Co., Ltd (Shenzhen, China) using the Illumina HiSeq2500 NGS platform (Illumina) and HiSeq PE Cluster Kit v4 (Illumina), with the sequencing strategy PE125.
2.5.3. Quality Control of Raw Data and de novo Metagenome Assembly
Clean reads were obtained after the raw data were filtered to remove the reads with ambiguous N base, sequencing adapters, and average base quality score less than 15. De novo metagenome assembly was firstly performed with SOAPdenovo2 [48] and further assembled with Rabbit [49]; for each sample, reads were assembled with a series of different k-mer size in parallel, and then were mapped back to each assembly result with SOAP2 [50], and the optimal k-mer size and assembly result were chosen depending on both contig N50 and mapping rate. De novo metagenome assembly was reperformed with IDBA-UD (v1.1.1) [51]. Only those contigs with more than 500 bp were kept for further analysis.
2.5.4. Gene Prediction, Catalog Construction and Mapping with Bowtie2
MetaGeneMark [52] (version 2.10, default parameters) was used to predict open reading frames (ORFs) based on assembly results. Genes from different samples were combined and clustered using CD-Hit [53] (sequence identity threshold 95% and alignment coverage threshold 90%). The high quality reads from each sample were aligned against the gene catalogue by Bowtie2 [54] using a sensitive parameter.
2.5.5. Functional Annotation of Predicted Genes and Taxonomic Assignment
All predicted genes were blasted against public databases including databases eggNOG, CAZy, GO, COG, Swiss-Prot, KEGG, ARDB, and NR (blast, e-value < 0.00001), to retrieve proteins with the highest sequence similarity with the given genes along with their protein functional annotations.
Analysis of NR BLAST output files was performed using the MEGAN (version 4.6) [55]. The NCBI taxonomy was displayed as a tree and the size of each node was scaled to indicate number of reads assigned to the corresponding taxonomy. Afterwards, the relative abundance of each taxonomy level was summed from the same taxonomy, and the gross relative abundance was taken as the content of this taxonomy in a sample to generate the taxonomy relative abundance profile of the samples.
Based on the known sequence database of bacteria, fungi and archaeobacteria from the nucleotide database of NCBI, clean reads of each sample were aligned by SOAPaligner (version 2.21) [50], and then mapped clean reads were assigned to the corresponding taxonomy and summed.
2.5.6. Alpha Diversity Analysis
Based on the species profile, the alpha diversity within each sample was calculated to estimate the species richness of a sample using Shannon index, as described previously by Qin et al. [47].
2.5.7. Computation of Relative Gene Abundance
Reads mapping to multiple genes were then reassigned to a “most likely” gene using Pathoscope (version 1.0) [56], which uses a Bayesian framework to examine each read sequence and mapping quality within the context of a global reassignment. Then, for any sample “S”, the hits (number of mapped reads), abundances (copy number of gene with specific length), relative abundances of different genes in single sample were calculated using the formulas described by Qin et al. [47].
2.5.8. Differential Analysis of Gene Abundance Between Two Samples
The number of unambiguous clean reads was denoted as “x” from gene A, given that every gene abundance occupies only a small part of the library, where “x” yielded to the Poisson distribution [57]:
Then, a strict algorithm was developed to identify genes with different abundance between two samples based on the formula described by Audic et al. [57]. N1 and N2 represented the total number of clean reads of samples 1 and 2, respectively. Gene A holds “x” reads in sample 1 and “y” reads in sample 2. The probability of abundance of gene A equally between two samples was calculated with:
Correction was performed on p-value that corresponded to genes with different abundance tests by using Bonferonni method [58]. Correction for false positive (type I) errors and false negative (type II) errors was performed using false discovery rate (FDR) method [59]. We used “FDR ≤ 0.001” and the “absolute value of log2 Ratio ≥ 1” as the default threshold to judge the significance.
2.5.9. Cluster Analysis of Genes
Genes with similar abundance patterns usually have same functional correlations. Therefore, we performed clustering analysis of gene abundance patterns with cluster [60, 61] and java Tree view software [62] according to the provided cluster plans.
2.5.10. Gene Ontology Enrichment
Enrichment analysis of Gene Ontology (GO) provided all GO terms that were significantly enriched in a list of genes with different abundances, compared with a genome background, and filtered the genes that corresponded to specific biological functions. This method firstly mapped all genes with different abundances to GO terms in the database (http://www.geneontology.org/), calculating gene numbers for every term, then used the hypergeometric test to find significantly enriched GO terms in the input list of genes, based on 'GO::TermFinder' (http://www.yeastgenome.org/help/analyze/go-term-finder). A strict algorithm was developed to do the analysis, and the method used is described as follows:
where “N” was the number of all genes with GO annotation; “n” was the number of genes with different abundances in “N”; “M” was the number of all genes that are annotated to certain GO terms; “m” was the number of genes with different abundances in “M”. The calculated p-value went through Bonferonni Correction [58], taking corrected p-value ≤ 0.05 as a threshold. GO terms fulfilling this condition were defined as significantly enriched GO terms in genes with different abundances.
2.5.11. KEGG Pathway Enrichment
Pathway-based analysis was used to further understand genes biological functions. KEGG, the major public pathway-related database, has been used to perform pathway enrichment analysis of genes with different abundances [63]. This analysis identified significantly enriched metabolic pathways or signal transduction pathways in genes with different abundances compared with the whole genome background. The calculating formula was the same with GO enrichment analysis except that “N” was the number of all genes that with KEGG annotation, “n” was the number of gene with different abundances in “N”, “M” was the number of all genes annotated to specific pathways, and “m” was the number of genes with different abundances in “M”.
2.6. Statistical Analyses
Metastats [64] was used to obtain the abundance differences of microbial communities between samples (groups = 2, samples per group ≥ 3). The obtained p-value was adjusted by Benjamini-Hochberg FDR [65] correction (function “p.adjust” in the stats package of R (v3.0.3)). The significance test method for alpha diversity is Wilcoxon Rank-Sum Test.
3. RESULTS
3.1. Composition and Structure of Bacterial Community Revealed by 16S rDNA Amplicon Sequencing
3.1.1. Overall Analysis of 16S rDNA (V4 region) Amplicons Sequencing Data Based Illumina MiSeq
A total of 893,865 qualified pairs of clean reads were obtained with an average of 74,489 (250 bp average) per rhizosphere replicate; and then a total of 29,148 OTUs was identified, except singletons, with an average of 2429 ± 248 OTUs per rhizosphere replicate (Table S1 (9.3MB, pdf) online), and all OTU sequences of rhizosphere soil samples were shown in (File S1 (9.3MB, pdf) ). Moreover, as a systematic contrast study, a total of 684,351 qualified pairs of clean reads were obtained with an average of 57,029 (250 bp average) per surrounding soil replicate and a total of 26,367 OTUs were identified, with an average of 2197 ± 173 OTUs per surrounding soil replicate with the exception of singletons (Table S2 (9.3MB, pdf) online), and all OTU sequences of surrounding soil samples were shown in (File S2 (9.3MB, pdf) ). Based on the OTU abundance of rhizosphere soil samples (Table S3 (9.3MB, pdf) online) and surrounding soil samples (Table S4 (9.3MB, pdf) online), the OTUs of each group together with the specific and common OTU ID were summarized in sheet 1 of Table S3 (9.3MB, pdf) or S4 (9.3MB, pdf) online, respectively, and were also shown in Venn picture (Fig. S1A (9.3MB, pdf) , B (9.3MB, pdf) ).
3.1.2. Alpha-diversity of Bacterial Community in Rhizosphere and Surrounding Soil
According to alpha diversity of 12 replicates of rhizosphere soil (File S3 (9.3MB, pdf) ) and surrounding soil (File S4 (9.3MB, pdf) ) in detail, the rarefaction curve of the normalized observed OTU number, Chao 1, and ACE of rhizosphere soil samples (File S5 (9.3MB, pdf) ) and surrounding soils (File S6 (9.3MB, pdf) ) almost reached the saturation plateau, indicating that the OTU coverage was sufficient to cover enough detectable species in the bacterial community and to capture the diversity of the bacterial communities in those samples. The mean and standard deviation (SD) of five alpha diversity indices of rhizosphere soil groups (Table S5 (9.3MB, pdf) online) or surrounding soil groups (Table S6 (9.3MB, pdf) online) were then calculated. Wilcoxon test p-values of five indices between two groups were higher than 0.05, which indicated there were no statistically significant difference in the overall indices of alpha diversity either between the rhizosphere soil of Z31J1 and that of Z31 or between the surrounding soil of Z31J1 and that of Z31.
3.1.3. Beta-diversity of Bacterial Community in Rhizosphere and Surrounding Soil
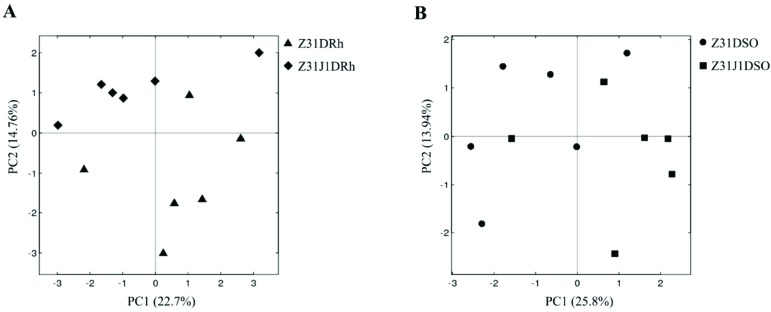
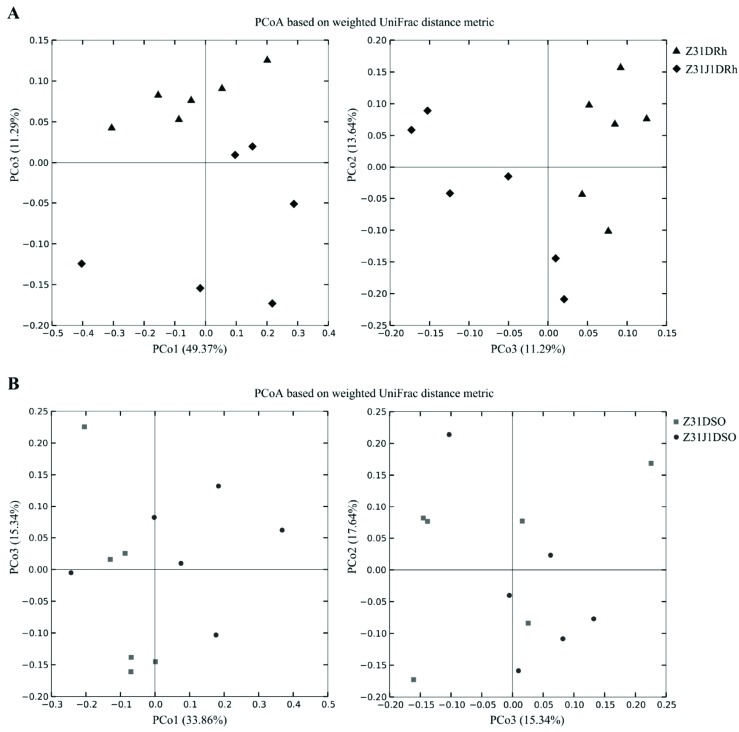
The differences in the OTU composition were firstly examined by using PCA. The rhizosphere soil replicates of Z31J1 seemed to be separated from those of control Z31 (Fig. 1A) whereas the surrounding soil replicates of Z31J1 were not separated from those of Z31 (Fig. 1B). Furthermore, phylogenetic beta diversity analyses were performed to rhizosphere and surrounding soil replicates by PCoA based on weighted UniFrac distance metric. The rhizosphere soil replicates of Z31J1 were separated from those of Z31 using PCoA (Fig. 2A), and the third principal coordinate (PCo3) axis of two dimensions explained 11.29% of the total variance. By comparison, the surrounding soil replicates of Z31J1 were not separated from those of Z31 using PCoA (Fig. 2B).
Fig. (1).
Principal Component Analysis (PCA) based on OTU abundances of bacterial communities. X-axis was 1st principal component and Y-axis was 2nd principal component. Numbers in brackets represented contributions of principal components to the total variance. A) The black rhombuses and black triangles represented rhizosphere soil replicates of the glyphosate treated Z31J1 and its control Z31, respectively. B) The black squares and the black solid circles represented surrounding soil replicates of glyphosate treated Z31J1 and its control Z31, respectively.
Fig. (2).
Principal Coordinate Analysis (PCoA) based on weighted UniFrac distance. The variance explained by each principal coordinate axis was shown in PCo1 vs. PCo3 and PCo3 vs. PCo2. A) The black rhombuses and black triangles represented rhizosphere soil replicates of the glyphosate treated Z31J1 and its control Z31, respectively. B) The black dots and the dark gray squares represented surrounding soil replicates of glyphosate treated Z31J1 and its control Z31, respectively.
Based on the Bray-Curtis distance metric, taxonomic beta diversity analysis was also performed to replicates of rhizosphere and surrounding soil by PCoA. The rhizosphere soil replicates of Z31J1 were not separated from those of Z31 (Fig. S2A (9.3MB, pdf) online), and those surrounding soils replicates of Z31J1 also were not separated from those of Z31 (Fig. S2B (9.3MB, pdf) online).
3.1.4. Comparison of the Major Bacterial Phyla in the Rhizosphere and Surrounding Soil
The taxonomic composition in the rhizosphere or surrounding soil of Z31J1 and its control Z31 at the phylum level were shown in (Table S7 (9.3MB, pdf) online), and the most abundant phylum was Proteobacteria in both the rhizosphere and surrounding soil, which was followed by Bacteroidetes or Acidobacteria and so on. Among these major phyla, only the relative abundance of Gemmatimonadetes was significantly lower in the rhizosphere soil of Z31 compared with that of Z31J1 (Table S7 (9.3MB, pdf) online) based on the systematic contrast analysis of the surrounding soil of Z31J1 compared with that of Z31, which suggested that the relative abundances of Gemmatimonadetes were less decreased in the rhizosphere soil of Z31 after glyphosate treatment. Additionally, both the relative abundances of Proteobacteria and Bacteroidetes increased whereas the relative abundance of Acidobacteria decreased in the rhizosphere soils of Z31J1 and Z31 compared with surrounding soils of Z31J1 and Z31.
3.1.5. Comparison of Differentially Relative Abundance of Bacterial Genera in the Rhizosphere Soil
A total of 559 genera were detected in the rhizosphere soil (File S7 (9.3MB, pdf) ), and only the relative abundances of 17 among 219 characterized genera were significantly different between the rhizosphere of glyphosate-treated Z31J1 and its control Z31 (Sheet 2 of Table S8 (9.3MB, pdf) online). Additionally, only 9 among 192 characterized genera were significantly different between the surrounding soil of glyphosate-treated Z31J1 and its control Z31 (Sheet 3 of Table S8 (9.3MB, pdf) online) whereas 517 genera were detected in the surrounding soils (File S8 (9.3MB, pdf) ). Under the comparative analysis of surrounding soils as a systematic contrast study, the relative abundances of only 3 genera, such as Opitutus, Comamonas, and Dyella, significantly increased in the rhizosphere soils of control Z31 (Z31DRh) compared with Z31J1 (Z31J1DRh), and the relative abundances of 2 genera, Burkholderia and Ralstonia,
increased whereas the relative abundance of Candidatus_Koribacter decreased in the rhizosphere soils of both control Z31 and glyphosate-treated Z31J1 compared with the surrounding soils of both Z31 and Z31J1 (Sheet 1 of Table S8 (9.3MB, pdf) online).
3.1.6. Comparison of Composition of Main Nitrogen-fixing Bacterial Genera in the Rhizosphere Soil
At the genus level, the relative abundance of Burkholderia was significantly higher in the rhizosphere soil of glyphosate-treated Z31J1 compared with its control Z31 (Table S9 (9.3MB, pdf) online), whereas the relative abundances of Bradyrhizobium and Rhizobium were much lower in the rhizosphere soil of glyphosate-treated Z31J1 compared with its control Z31, although no statistically significant difference existed between Z31J1DRh and its control Z31DRh (Table S9 (9.3MB, pdf) online).
Furthermore, the summary of relative abundances of these 9 main symbiotic nitrogen-fixing genera in the rhizosphere of glyphosate-treated Z31J1 (2.116% ±0.404%) were less than those in the rhizosphere of control Z31 (2.513% ±0.546%), which were consistent with the absolute abundances of these main symbiotic nitrogen-fixing genera in the rhizosphere of glyphosate-treated Z31J1 compared with those of control Z31 (Table S10 (9.3MB, pdf) online).
3.2. Metagenomic Analysis of the Effect of Glyphosate on Rhizosphere Microbial Community
3.2.1. Statistical Summary of Assembled Metagenome Data
On average 73,970,948 clean reads and 9.25 Gbp clean data per sample were generated from shotgun metagenomic sequencing (Table S11 (9.3MB, pdf) online). The total clean reads of MGZ31DRh and MGZ31J1DRh were firstly de novo assembled by SOAPdenovo2, respectively, and the mapping rates of both samples were lower than 0.44%, although more than 272,000 reads per sample were mapped (Table S11 (9.3MB, pdf) online). Thus, de novo metagenome assembly of the total clean reads of two samples was reperformed with IDBA-UD (v1.1.1), and the mapping rates of both samples obviously increased to more than 2.61% of MGZ31DRh or 4.91% of MGZ31J1DRh (Table S11 (9.3MB, pdf) online).
3.2.2. Gene Catalogue and Functional Annotation of Predicted Genes
Based on the assembled data by SOAPdenovo2, a total of 54,776 genes with detailed sequence were obtained (File S9 (9.3MB, pdf) ) after ORFs were predicated by MetaGeneMark, while 47,619 and 52,694 genes were identified from MGZ31DRh and MGZ31J1DRh samples, respectively. All predicted 54,776 genes were blasted against public databases including KEGG, and NR etc., and all functional annotations of those genes were summarized in File S10 (9.3MB, pdf) .
Moreover, a total of 523,955 genes with detailed sequence were obtained (File S11 (9.3MB, pdf) ) from the assembled data by IDBA-UD, and 381,428 and 437,494 genes were identified from MGZ31DRh and MGZ31J1DRh samples, respectively. All predicted 523,955 genes were blasted against public databases including KEGG, and NR etc., and the functional annotations of those genes were summarized in File S12 (9.3MB, pdf) .
3.2.3. Computation of Gene Abundances and Taxonomic Assignment of Major Taxons
Based on the assembled data by SOAPdenovo2, the length, the hits (mapped reads), abundances (copy number of gene with specific length), and the relative abundances of 47,619 genes in MGZ31DRh sample and of 52,694 genes in MGZ31J1DRh sample were calculated and summarized in Files S13 (9.3MB, pdf) and S14 (9.3MB, pdf) , respectively. Correspondingly, the length, hits, abundances, and relative abundances of 381,428 genes in MGZ31DRh sample and of 437,494 genes in MGZ31J1DRh sample were calculated and summarized in Files S15 (9.3MB, pdf) and S16 (9.3MB, pdf) , respectively, based on the assembled data by IDBA-UD.
The taxonomic assignment was performed by MEGAN according to predicted genes based on the assembled data by SOAPdenovo2, and the annotated genes were 37,536 and 40,579 among 47,619 genes of MGZ31DRh and 52,694 genes of MGZ31J1DRh, respectively (File S17 (9.3MB, pdf) ). The absolute and relative abundances of annotated taxons at different classification level in detail were summarized in (Table S12 (9.3MB, pdf) online). However, those relative abundances of annotated taxons were different from the relative abundances of taxons at different classification levels that were calculated based on species abundances (Table S13 (9.3MB, pdf) online), although the Wilcoxon test p-values of the Shannon index between MGZ31DRh and MGZ31J1DRh was 1.00 and much higher than 0.05 (Table S14 (9.3MB, pdf) online).
Hence, the taxonomic assignment was further performed by SOAPaligner by aligning clean reads directly to the known sequence database of bacteria, fungi and archaeobacteria from the nucleotide database of NCBI, and then mapped clean reads were assigned to the corresponding taxonomy and summed (File S18 (9.3MB, pdf) ). After comparative analysis, we found that taxonomic assignment results based on directly aligned clean reads seemed consistent with those based on species abundances (Table S15 (9.3MB, pdf) online).
3.2.4. Comparison of Main Nitrogen-fixing Rhizobacterial Genera Based on Metagenome Taxonomic Assignment
According to results of taxonomic assignment based on genes abundances, species abundances, and clean reads alignments, main nitrogen-fixing rhizobacterial genera were collected and compared (Table S16 (9.3MB, pdf) online). The relative abundance of Bradyrhizobium was the richest genus in rhizosphere soil, followed by Cupriavidus or Burkholderia, Rhizobium or Pseudomonas, and so on. Additionally, the relative abundance of Bradyrhizobium was much lower in the rhizosphere soil of Z31J1 compared with control Z31, whereas the relative abundances of Burkholderia and Cupriavidus were higher in the rhizosphere soil of Z31J1.
3.2.5. Differential Analysis of Gene Abundance and Enrichment of GO and KEGG
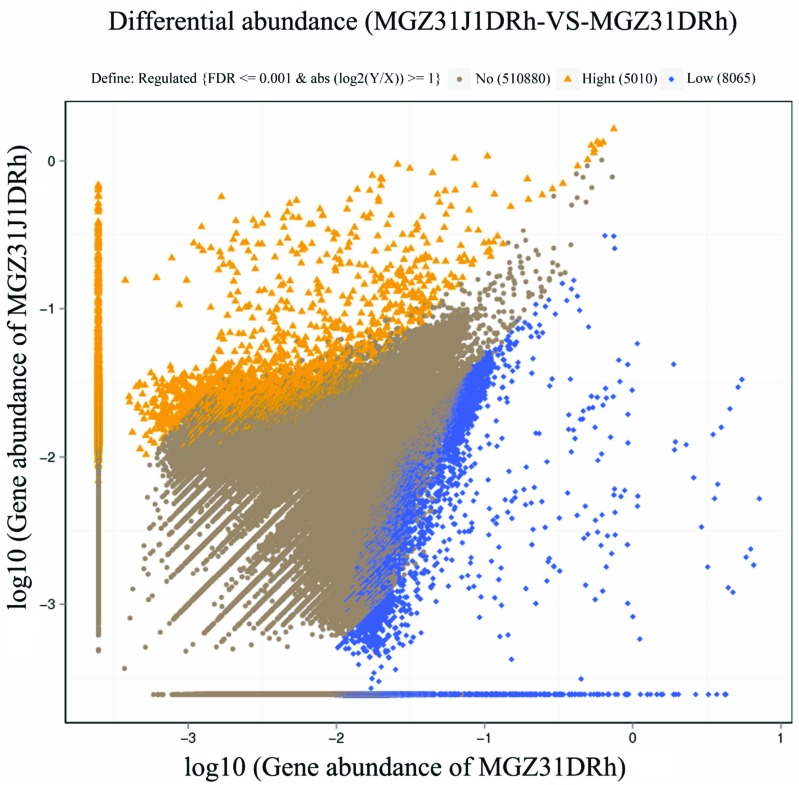
Based on data assembled by SOAPdenovo2, the abundances of 1766 genes were significantly higher, whereas the abundances of 1939 genes were significantly lower in the rhizosphere soil of glyphosate-treated Z31J1 compared with control Z31 among the total of 54,776 genes (File S19 (9.3MB, pdf) ; Files S20 (9.3MB, pdf) , and S21 (9.3MB, pdf) in detail). Correspondingly, the abundances of 5010 genes were significantly higher, whereas the abundances of 8065 genes were significantly lower in the rhizosphere soil of glyphosate-treated Z31J1 compared with control Z31 among the total of 523,955 genes (Fig. 3; Files S22 (9.3MB, pdf) , and S23 (9.3MB, pdf) in detail) based on data assembled by IDBA-UD.
Fig. (3).
Scatter plots of genes with differential abundance. Y and X axis presented value of genes abundance of MGZ31J1DRh and of MGZ31DRh, respectively, based on the assembled data by IDBA-UD. Orange triangles and blue rhombuses indicated genes with significantly higher and lower relative abundance in MGZ31J1DRh, respectively. Brown circles indicate those genes with no significant difference between MGZ31J1DRh and MGZ31DRh. The criterion of screening is on top of the plot. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this paper.)
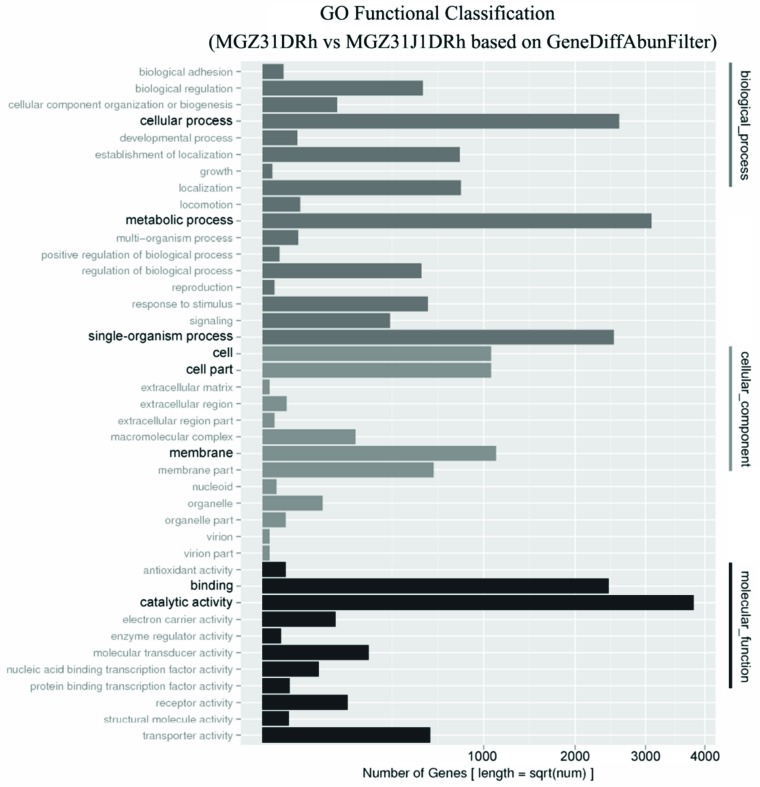
Among those significantly enriched GO terms in genes with different abundances, those remarkable terms were cellular, metabolic, and single-organism of biological process together with binding, catalytic activity of molecular function, and cell, cell part, membrane of cellular component, based on data assembled by IDBA-UD (Fig. 4) as well as those based on data assembled by SOAPdenovo2 (Fig. S3 (9.3MB, pdf) online).
Fig. (4).
GO functional classification for the pairwise of MGZ31DRh VS MGZ31J1DRh. X-axis means number of genes with different abundance based on data assembled by IDBA-UD. Y-axis represents different GO terms. All GO terms were grouped into three ontologies: the dark gray, gray and black color indicated biological process, cellular component, and molecular function, respectively. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this paper.)
To further understand biological functions of those genes with different abundances, KEGG pathway enrichment analyses were performed between MGZ31DRh and MGZ31J1DRh. Among 32,949 genes with KEGG pathway annotation based on data assembled by SOAPdenovo2, the top 18 pathways were summarized in (Table S17 (9.3MB, pdf) online) after much less related 9 pathways were removed. Moreover, among 39,776 genes with KEGG pathway annotation based on data assembled by IDBA-UD, the top 18 significantly enriched pathways were summarized in (Table S18 (9.3MB, pdf) online) after seven pathways were deleted because of less relation with soil microbes. Those common significantly enriched pathways were purine metabolism, pyrimidine metabolism, ABC transporters, fatty acid metabolism, DNA replication, nitrogen metabolism, and legionellosis between MGZ31DRh versus MGZ31J1DRh. We were more interested in the enriched KEGG pathways of nitrogen metabolism and ABC transporters.
3.2.6. Detection of Functional Genes with Significantly Differential Abundance Involved in PGPT
Based on the functional annotation of predicted genes and differential analysis of genes abundance, together with enrichments of GO and KEGG, we further detected those genes involved in Plant Growth Promoting Traits (PGPT), such as ACC deaminase, nitrogen fixation related genes, plant disease suppression, phosphate solubilization, and iron carriers, based on data assembled by SOAPdenovo2 (Table S19 (9.3MB, pdf) online) and by IDBA-UD (Table 1). Additionally, other nitrogen metabolism related genes also were detected in this study (Tables 1 and S19 (9.3MB, pdf) online).
Table 1.
Detection of functional genes involved in PGPT plus N2-metabolim based on data assembled by IDBA-UD.
| Gene Name | ID of KEGG | ID of [denovogenes] |
MGZ31DRh
(Assembled by IDBA-UD) |
MGZ31J1DRh
(Assembled by IDBA-UD) |
Gene length2
(bp) |
||||
|---|---|---|---|---|---|---|---|---|---|
| Hits1 | Gene Abundance | Hits1 | Gene Abundance | p-value | FDR3 | ||||
| ACC deaminase | K01505 | _73123 | 83 | 0.08185 | 33 | 0.03254 | 1014 | 9.35E-07 | 6.32E-05 |
| Nitrogen fixation | |||||||||
| nifH | K02588 | _103473 | 63 | 0.07023 | 24 | 0.02676 | 897 | 1.04E-05 | 0.0004979 |
| nifD | K02586 | _19302 | 135 | 0.08893 | 59 | 0.03887 | 1518 | 7.25E-09 | 8.97E-07 |
| nifK | K02591 | _17563 | 108 | 0.06936 | 40 | 0.02569 | 1557 | 3.44E-09 | 4.61E-07 |
| nifA | K02584 | _9784 | 146 | 0.08004 | 57 | 0.03125 | 1824 | 3.79E-11 | 7.81E-09 |
| nifB | K02585 | _17564 | 150 | 0.09634 | 47 | 0.03019 | 1557 | 7.76E-15 | 2.89E-12 |
| nifE | K02587 | _12565 | 121 | 0.07101 | 50 | 0.02934 | 1704 | 9.02E-09 | 1.08E-06 |
| nifN | K02592 | _25697 | 97 | 0.06923 | 51 | 0.03640 | 1401 | 5.43E-05 | 0.0018917 |
| nifQ | K15790 | _182142 | 39 | 0.05462 | 12 | 0.01681 | 714 | 7.17E-05 | 0.0024326 |
| nifV | K02594 | _45566 | 91 | 0.07660 | 30 | 0.02525 | 1188 | 4.66E-09 | 6.03E-07 |
| nodB | K14659 | _219374 | 32 | 0.04848 | 8 | 0.01212 | 660 | 6.73E-05 | 0.0023008 |
| nodC | K14666 | _27548 | 112 | 0.08151 | 46 | 0.03348 | 1374 | 2.75E-08 | 2.92E-06 |
| fixA | K03521 | _121736 | 43 | 0.05101 | 11 | 0.01305 | 843 | 4.44E-06 | 0.0002400 |
| fixB | K03522 | _56616 | 85 | 0.07678 | 34 | 0.03071 | 1107 | 7.80E-07 | 5.34E-05 |
| fixC | K00313 | _32844 | 89 | 0.06804 | 28 | 0.02905 | 1308 | 1.68E-06 | 0.0001030 |
| fixJ | K14987 | _242433 | 40 | 0.06319 | 13 | 0.02054 | 633 | 9.83E-05 | 0.0030541 |
| fixL | K14986 | _23774 | 85 | 0.05940 | 41 | 0.02865 | 1431 | 3.14E-05 | 0.0012269 |
| Plant disease suppression | |||||||||
| β-1,3-glucanase | K01210 | _13389 | 26 | 0.01548 | ND 4 | 0.00025 | 1680 | 8.71E-09 | 1.06E-06 |
| _14945 | 125 | 0.07673 | 37 | 0.02271 | 1629 | 2.43E-13 | 7.38E-11 | ||
| Phosphate solubilization | |||||||||
| GDH | K00117 | _3387 | 227 | 0.09554 | 69 | 0.02904 | 2376 | 2.06E-22 | 1.54E-19 |
| Siderophore (iron carrier) | |||||||||
| dhbF | K04780 | _9 | 67 | 0.00774 | 202 | 0.02332 | 8661 | 6.20E-16 | 2.62E-13 |
| _2733 | ND 4 | 0.00025 | 44 | 0.01769 | 2487 | 1.37E-13 | 4.27E-11 | ||
| Nitrogen metabolism other related genes | |||||||||
| cynT/can | K01673 | _18558 | 43 | 0.02799 | 7 | 0.00456 | 1536 | 5.75E-08 | 5.62E-06 |
| ncd2/npd | K00459 | _25423 | 28 | 0.01990 | 6 | 0.00426 | 1407 | 7.31E-05 | 0.002472 |
| gltD | K00266 | _22286 | 105 | 0.07202 | 42 | 0.02881 | 1458 | 3.89E-08 | 3.97E-06 |
| gltB | K00265 | _122 | 160 | 0.03439 | 47 | 0.01010 | 4653 | 8.38E-17 | 3.85E-14 |
| _106 | 461 | 0.09701 | 205 | 0.04314 | 4752 | 5.49E-26 | 5.06E-23 | ||
| _2287 | 68 | 0.02623 | 24 | 0.00926 | 2592 | 1.31E-06 | 8.42E-05 | ||
| nirK | K00368 | _31799 | 1 | 0.00076 | 54 | 0.04091 | 1320 | 4.54E-15 | 1.73E-12 |
| nirB | K00362 | _2403 | 77 | 0.03013 | 41 | 0.01604 | 2460 | 0.000404 | 0.009216 |
| _2606 | 74 | 0.02947 | 24 | 0.00956 | 2511 | 9.77E-08 | 8.86E-06 | ||
| _2886 | 214 | 0.08699 | 85 | 0.03455 | 2460 | 2.68E-15 | 1.05E-12 | ||
1 The number of hits represented the number of mapped reads of single denovogene detected in the sample.
2 The gene length (bp) was listed according to those denovogenes assembled by IDBA-UD.
3 The value in the table cell was underlined when FDR was less than 0.01 but more than 0.001.
4 ND = Not Detected.
Compared with MGZ31DRh, the hits and abundances of nitrogen fixation genes, ACC deaminase, β-1, 3-glucanase and GDH were significantly lower, whereas the hits and abundances of dhbF were significantly higher in the MGZ31J1DRh sample. The present results suggested that the abundance of those PGPT genes except dhbF in rhizosphere soil decreased after glyphosate treatment. As for other nitrogen metabolism related genes detected in this study, the hits and abundances of 5 genes were significantly lower whereas the hits and abundances of nirK were significantly higher in the MGZ31J1DRh sample compared with MGZ31DRh.
One gene of IAA metabolism involved in PGPT, iaaM encoding tryptophan 2-monooxygenase, was not found in either File S10 or File S12, which was the annotation table based on data assembled by SOAPdenovo2 or by IDBA-UD. The other gene of IAA metabolism involved in PGPT, namely, ipdC that encodes indolepyruvate decarboxylase, was not
detected in File S10, but was found in File S12, although its hits and abundances in the MGZ31J1DRh did not significantly differ from those in MGZ31DRh (Table 1).
4. discussion
We collected samples of surrounding and rhizosphere soils from GR transgenic line Z31 plants after glyphosate treatment in seedling, flowering, and seed-filling stages. We then analyzed the effect of glyphosate on the rhizosphere microbes in seed-filling stage because we aimed to determine the effect of glyphosate treatment during a single growth season. In addition, 8% to 12% of the applied glyphosate was still detected in the soil samples incubated with roots one and a half months later [12]. Compared with rhizosphere soil being sampled 3 days later [26] or more than one year [27] after the glyphosate treatment, in this study, the sampling time of two months after the glyphosate treatment was a mid-term period.
Before shotgun metagenome sequencing and analysis were performed, we comparatively analyzed the bacterial communities in the rhizosphere and surrounding soils of the GR transgenic soybean line Z31 treated with glyphosate (or simply Z31J1) versus Z31 treated with water in the seed-filling stage by V4 region of 16S rDNA amplicon based Illumina MiSeq sequencing to clarify whether glyphosate treatment affects rhizosphere bacterial community associated with soybean roots.
It is also important to conduct the analysis of surrounding soil as a systematic contrast study that not only overcomes some of soil heterogeneity but also distinguish those significant differences in some rhizosphere bacterial relative abundances from edaphic factors instead of host plants already in the surrounding and bulk soils [18], especially to distinguish the effect of glyphosate being penetrated from field surface.
Previous studies involving the deep sequencing of 16S rDNA amplicon showed that the effects of glyphosate
treatment are the major shift in the relative abundances of Proteobacteria and Acidobacteria at the phylum level for both soybean and corn rhizosphere samples, in which the increase in the relative abundance of Proteobacteria is attributed to the increase in the relative abundance of the class Gammaproteobacteria and the increase in the relative abundance of the family Xanthomonadaceae after glyphosate treatment [27]. However, in our 16S rDNA amplicon sequencing results, the relative abundances of Gemmatimonadetes, Bacteroidetes, and Acidobacteria in the rhizosphere soil were remarkably altered at the phylum level, as indicated by the analysis of surrounding soil as a systematic contrast study (Sheet 1 of Table S7 (9.3MB, pdf) online), and the relative abundances of Gammaproteobacteria and Saprospirae obviously increased under water control treatment compared with glyphosate treatment at the class level in this study (Sheet 2 of Table S7 (9.3MB, pdf) online). Interestingly, our 16S rDNA amplicon deep sequencing results were consistent with those of taxonomic assignment based clean read alignment in our metagenome sequencing data; By contrast, previous results were similar to the taxonomic assignment based on species abundance assembled by SOAPdenovo2 in our metagenome sequencing data (Table S15 (9.3MB, pdf) online, at the class and family level) although the relative abundance of Proteobacteria, which was the major phyla revealed by shotgun metagenome sequencing, decreased in the rhizosphere soil of Z31J1 after glyphosate treatment (Table S15 (9.3MB, pdf) online, at the phylum level).
Previous studies involving the deep sequencing of 16S rDNA amplicon demonstrated that the relative abundance of Acidobacteria, particularly the subgroup Acidobacteria-6, decreases in corn and soybean rhizospheres upon glyphosate treatment [27]. In our results, the relative abundance of Acidobacteria-6 in the rhizosphere soil of soybean Z31 in glyphosate treatment was less than that in water control (Sheet 2 of Table S7 (9.3MB, pdf) online). By comparison, the relative abundances of the phylum Acidobacteria and its major class Acidobacteriia in the rhizosphere soil with glyphosate treatment were much higher than those in the rhizosphere soil with water control, as demonstrated by 16S rDNA amplicon sequencing (Table S7 (9.3MB, pdf) online) and shotgun metagenome sequencing (Table S15 (9.3MB, pdf) online).
The inconsistent results of the deep sequencing of 16S rDNA amplicons might be attributed to different soil types, glyphosate concentrations, sampling times after glyphosate treatment, and PCR programs with different cycles during the amplification of the V4 region of 16S rDNA.
Rhizobium-legume symbioses are essential for land ecosystems by providing ammonia for plant growth via symbiotic nitrogen fixation [23]. Hence, the composition of nitrogen-fixing bacteria was the focus of this study. All 15 main symbiotic nitrogen-fixing bacterial genera with legumes [66] were detected by shotgun metagenome sequencing and analysis of total clean reads direct alignment in the present study (Table S16 (9.3MB, pdf) ). By comparison, only 9 main symbiotic nitrogen-fixing bacterial genera were detected by 16S rDNA amplicons sequencing and analysis in the present study (Table S9 (9.3MB, pdf) ). The relative abundances of several major nitrogen-fixing bacterial genera, such as Bradyrhizobium, and Rhizobium, obviously decreased, whereas the relative abundance of Burkholderia increased in the rhizosphere soil after glyphosate treatment via both shotgun metagenome sequencing and 16S rDNA amplicons sequencing; nevertheless, their relative abundances were different. Moreover, the total relative abundances of main symbiotic nitrogen-fixing bacterial genera obviously decreased in the rhizosphere soil after glyphosate treatment via two methods. This finding was consistent with the significantly decreased hits and gene abundance of most nitrogen-fixation related genes including major nitrogenase genes, such as nifH, nifD, nifK, and nodulation related genes such as nodB, nodC.
With the importance of plant growth promoting rhizobacteria for improving plant growth and health [16, 67-69], in addition to nitrogen-fixation related genes, other functional genes involved in PGPT were also detected in this study. The hits and gene abundances of ACC deaminase, β-1,3-glucanase, and GDH significantly decreased, although a few of them, such as dhbF, and one of nitrogen metabolism related genes, namely, nirK, significantly increased in the rhizosphere soil after glyphosate treatment.
The total clean reads of MGZ31DRh and MGZ31J1DRh were reassembled by IDBA-UD because the metagenome assembly results significantly affected taxonomic assignment, functional gene annotation, and genome reconstruction of different single microbe species.
Shotgun metagenome sequencing combined with bioinformatics analysis is an efficient method to investigate the composition, structure, and function of microbial communities. However, the large datasets generated by current NGS platforms, such as Illumina HiSeq, require massive computational resources and produce relatively short contigs in this study and others studies [70]. In addition to SOAPdenovo2 and IDBA-UD, many bioinformatics tools, such as Kraken [71], GOTTCHA [72], CLARK [73], have been developed to explore the taxonomic assignment and functional composition of metagenomes. Furthermore, the speed and accuracy of 14 metagenome analysis tools have been evaluated [74]; Test datasets have been established with differences in the relative abundance of Bradyrhizobium and Rhizobium for nitrogen fixation functional analysis, and only EBI webserver and MG-RAST tools have predicted the expected shift of nitrogen-fixation [74].
Third-generation sequencing technology, especially PacBio Single Molecule Real Time (SMRT) detection with Circular Consensus Sequencing (CCS), has significantly improved the metagenome assembly when this technology is combined with HiSeq 2000 data. [75] This technology is another important advancement in shotgun metagenome sequencing.
CONCLUSION
Our present study indicated that the formulation of glyphosate-isopropylamine salt did not significantly affect alpha diversity of soybean rhizosphere bacterial community, although it had small but insignificant effect on beta diversity of soybean rhizosphere bacterial community. By contrast, the formulation of glyphosate-isopropylamine salt significantly affected some functional genes involved in PGPT, especially most of nitrogen-fixation genes in rhizosphere soil during the single growth season after glyphosate treatment.
ACKNOWLEDGEMENTS
We were grateful to Jinqun Huang and Jie Li (BGI Tech Solutions Co., Ltd (Shenzhen, China)) for their help in metadata analysis. This research work was supported by grants from the National Important Science & Technology Specific Project (2014ZX08011-003) to Y.-H.Y., the National Natural Science Foundation of China (31372140) to Y.-H.Y, and the Program for Changjiang Scholars and Innovative Research Team in University from the Ministry of Education of China (IRT_14R27) to Y.-H.Y.
LIST OF ABBREVIATIONS
- 16S rDNA
16S ribosomal RNA Gene
- AMPA
Aminomethylphosphonic Acid
- ARDB
Antibiotic Resistance Genes Database
- CAZy
Carbohydrate-Active Enzymes Database
- COG
Cluster of Orthologous Groups of Proteins
- eggNOG
Evolutionary Genealogy of Genes: Non-supervised Orthologous Groups
- EPSPS
5-Enolpyruvylshikimate-3- Phosphate Synthase
- FDR
False Discovery Rate
- GO
Gene Ontology
- GR
Glyphosate Resistant
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- nifA
The Core Gene Encoding Nitrogen Fixation Specific Regulatory Protein
- nifB
The Core Gene Encoding Protein Synthesizes a Fe-S Containing Precursor of FeMo-cofactor
- nifD
The Core Structural Gene Encoding Nitrogenase Molybdenum-iron Protein Alpha Chain
- nifE
The Core Gene Encoding the Molecular Scaffold for Assembly of Mo Cofactor
- nifH
The Core Structural Gene Encoding Nitrogenase Iron Protein
- nifK
The Core Structural Gene Encoding Nitrogenase Molybdenum-iron Protein Beta Chain
- nifN
The Core Gene Encoding the Molecular Scaffold for Assembly of the Fe-Mo Cofactor
- NR
Non-redundant Protein Sequence Database
- ORF
Open Reading Frame
- OTU
Operational Taxonomic Unit
- PCA
Principal Component Analysis
- PCoA
Principal Coordinate Analysis
- PCR
Polymerase Chain Reaction
- PGPR
Plant Growth-promoting Rhizobacteria
- PGPT
Plant Growth-promoting Traits
- Z31
GR Transgenic Soybean Line ZUTS31 Being Foliar Sprayed with Water
- Z31J1
GR Transgenic Line ZUTS31being Foliar Sprayed at Field Rate (3000 ml · ha-1) of Glyphosate
SUPPLEMENTARY MATERIAL
Supplementary material is available on the publisher’s web site along with the published article.
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
Not applicable.
HUMAN AND ANIMAL RIGHTS
No Animals/Humans were used for studies that are base of this research.
CONSENT FOR PUBLICATION
Not applicable.
CONFLICT OF INTEREST
The authors declare no conflict of interest, financial or otherwise.
REFERENCES
- 1.Smith E.A., Oehme F.W. The biological-activity of Glyphosate to plants and animals - A literature-review. Vet. Hum. Toxicol. 1992;34(6):531–543. [PubMed] [Google Scholar]
- 2.Duke S.O., Powles S.B. Glyphosate: A once-in-a-century herbicide. Pest Manag. Sci. 2008;64(4):319–325. doi: 10.1002/ps.1518. [DOI] [PubMed] [Google Scholar]
- 3.Pollegioni L., Schonbrunn E., Siehl D. Molecular basis of glyphosate resistance - Different approaches through protein engineering. FEBS J. 2011;278(16):2753–2766. doi: 10.1111/j.1742-4658.2011.08214.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Coupland D., Caseley J.C. Presence of 14c activity in root exudates and guttation fluid from agropyron repens treated with 14c-labelled glyphosate. New Phytol. 1979;83(1):17–22. [Google Scholar]
- 5.Imfeld G., Vuilleumier S. Measuring the effects of pesticides on bacterial communities in soil: A critical review. Eur. J. Soil Biol. 2012;49:22–30. [Google Scholar]
- 6.Benbrook C.M. Trends in glyphosate herbicide use in the United States and globally. Environ. Sci. Eur. 2016;28(3):1–15. doi: 10.1186/s12302-016-0070-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cerdeira A.L., Duke S.O. The current status and environmental impacts of glyphosate-resistant crops: A review. J. Environ. Qual. 2006;35(5):1633–1658. doi: 10.2134/jeq2005.0378. [DOI] [PubMed] [Google Scholar]
- 8.Bott S., Tesfamariam T., Candan H., Cakmak I., Roemheld V., Neumann G. Glyphosate-induced impairment of plant growth and micronutrient status in glyphosate-resistant soybean (Glycine max L.). Plant Soil. 2008;312(1-2):185–194. [Google Scholar]
- 9.Johal G.S., Huber D.M. Glyphosate effects on diseases of plants. Eur. J. Agron. 2009;31(3):144–152. [Google Scholar]
- 10.Yamada T., Kremer R.J., Castro P.R., Wood B.W. Glyphosate interactions with physiology, nutrition, and diseases of plants: Threat to agricultural sustainability? Preface. Eur. J. Agron. 2009;31(3):111–113. [Google Scholar]
- 11.Duke S.O., Lydon J., Koskinen W.C., Moorman T.B., Chaney R.L., Hammerschmidt R. Glyphosate effects on plant mineral nutrition, crop rhizosphere microbiota, and plant disease in glyphosate-resistant crops. J. Agric. Food Chem. 2012;60(42):10375–10397. doi: 10.1021/jf302436u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Laitinen P., Ramo S., Siimes K. Glyphosate translocation from plants to soil - Does this constitute a significant proportion of residues in soil? Plant Soil. 2007;300(1-2):51–60. [Google Scholar]
- 13.Busse M.D., Ratcliff A.W., Shestak C.J., Powers R.F. Glyphosate toxicity and the effects of long-term vegetation control on soil microbial communities. Soil Biol. Biochem. 2001;33(12-13):1777–1789. [Google Scholar]
- 14.Tsui M.T., Chu L.M. Aquatic toxicity of glyphosate-based formulations: comparison between different organisms and the effects of environmental factors. Chemosphere. 2003;52(7):1189–1197. doi: 10.1016/S0045-6535(03)00306-0. [DOI] [PubMed] [Google Scholar]
- 15.Sihtmae M., Blinova I., Kunnis-Beres K., Kanarbik L., Heinlaan M., Kahru A. Ecotoxicological effects of different glyphosate formulations. Appl. Soil Ecol. 2013;72:215–224. [Google Scholar]
- 16.Lugtenberg B., Kamilova F. Plant-growth-promoting rhizobacteria. Annu. Rev. Microbiol. 2009;63:541–556. doi: 10.1146/annurev.micro.62.081307.162918. [DOI] [PubMed] [Google Scholar]
- 17.Berendsen R.L., Pieterse C.M., Bakker P.A. The rhizosphere microbiome and plant health. Trends Plant Sci. 2012;17(8):478–486. doi: 10.1016/j.tplants.2012.04.001. [DOI] [PubMed] [Google Scholar]
- 18.Bulgarelli D., Schlaeppi K., Spaepen S., van Themaat E.V., Schulze-Lefert P. Structure and functions of the bacterial microbiota of plants. Annu. Rev. Plant Biol. 2013;64:807–838. doi: 10.1146/annurev-arplant-050312-120106. [DOI] [PubMed] [Google Scholar]
- 19.Berg G., Grube M., Schloter M., Smalla K. Unraveling the plant microbiome: Looking back and future perspectives. Front. Microbiol. 2014;5:148. doi: 10.3389/fmicb.2014.00148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bais H.P., Weir T.L., Perry L.G., Gilroy S., Vivanco J.M. The role of root exudates in rhizosphere interactions with plants and other organisms. Annu. Rev. Plant Biol. 2006;57:233–266. doi: 10.1146/annurev.arplant.57.032905.105159. [DOI] [PubMed] [Google Scholar]
- 21.Paterson E., Gebbing T., Abel C., Sim A., Telfer G. Rhizodeposition shapes rhizosphere microbial community structure in organic soil. New Phytol. 2007;173(3):600–610. doi: 10.1111/j.1469-8137.2006.01931.x. [DOI] [PubMed] [Google Scholar]
- 22.Aira M., Gomez-Brandon M., Lazcano C., Baath E., Dominguez J. Plant genotype strongly modifies the structure and growth of maize rhizosphere microbial communities. Soil Biol. Biochem. 2010;42(12):2276–2281. [Google Scholar]
- 23.Kondorosi E., Mergaert P., Kereszt A. A paradigm for endosymbiotic life: Cell differentiation of rhizobium bacteria provoked by host plant factors. Annu. Rev. Microbiol. 2013;67:611–628. doi: 10.1146/annurev-micro-092412-155630. [DOI] [PubMed] [Google Scholar]
- 24.Bünemann E.K., Schwenke G.D., Van Zwieten L. Impact of agricultural inputs on soil organisms - A review. Aust. J. Soil Res. 2006;44(4):379–406. [Google Scholar]
- 25.Valverde J.R., Marin S., Mellado R.P. Effect of herbicide combinations on Bt-maize rhizobacterial diversity. J. Microbiol. Biotechnol. 2014;24(11):1473–1483. doi: 10.4014/jmb.1405.05054. [DOI] [PubMed] [Google Scholar]
- 26.Schafer J.R., Hallett S.G., Johnson W.G. Rhizosphere microbial community dynamics in glyphosate-treated susceptible and resistant biotypes of giant ragweed (Ambrosia trifida). Weed Sci. 2014;62(2):370–381. [Google Scholar]
- 27.Newman M.M., Hoilett N., Lorenz N., Dick R.P., Liles M.R., Ramsier C., Kloepper J.W. Glyphosate effects on soil rhizosphere-associated bacterial communities. Sci. Total Environ. 2016;543:155–160. doi: 10.1016/j.scitotenv.2015.11.008. [DOI] [PubMed] [Google Scholar]
- 28.Yu K., Zhang T. Metagenomic and metatranscriptomic analysis of microbial community structure and gene expression of activated sludge. PLoS One. 2012;7(5):e38183. doi: 10.1371/journal.pone.0038183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fierer N., Leff J.W., Adams B.J., Nielsen U.N., Bates S.T., Lauber C.L., Owens S., Gilbert J.A., Wall D.H., Caporaso J.G. Cross-biome metagenomic analyses of soil microbial communities and their functional attributes. Proc. Natl. Acad. Sci. USA. 2012;109(52):21390–21395. doi: 10.1073/pnas.1215210110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fierer N., Lauber C.L., Ramirez K.S., Zaneveld J., Bradford M.A., Knight R. Comparative metagenomic, phylogenetic and physiological analyses of soil microbial communities across nitrogen gradients. ISME J. 2012;6(5):1007–1017. doi: 10.1038/ismej.2011.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bremges A., Maus I., Belmann P., Eikmeyer F., Winkler A., Albersmeier A., Puhler A., Schluter A., Sczyrba A. Deeply sequenced metagenome and metatranscriptome of a biogas-producing microbial community from an agricultural production-scale biogas plant. 2015. [DOI] [PMC free article] [PubMed]
- 32.Mangrola A., Dudhagara P., Koringa P., Joshi C.G., Parmar M., Patel R. Deciphering the microbiota of Tuwa hot spring, India using shotgun metagenomic sequencing approach. Genom. Data. 2015;4:153–155. doi: 10.1016/j.gdata.2015.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tsurumaru H., Okubo T., Okazaki K., Hashimoto M., Kakizaki K., Hanzawa E., Takahashi H., Asanome N., Tanaka F., Sekiyama Y., Ikeda S., Minamisawa K. Metagenomic analysis of the bacterial community associated with the taproot of sugar beet. Microbes Environ. 2015;30(1):63–69. doi: 10.1264/jsme2.ME14109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lu L., Han Q., Li L., Zhou L., Liu R., Song Z., Shen Z., Shou H. Establishment of an efficient transformation protocol for soybean using glyphosate as selective agent and the development of glyphosate-tolerant transgenic soybean lines. Scientia Sinica Vitae. 2014;44(4):406–415. [Google Scholar]
- 35.Rampazzo N., Todorovic G.R., Mentler A., Blum W.E. Adsorption of glyphosate and aminomethylphosphonic acid in soils. Int. Agrophys. 2013;27(2):203–209. [Google Scholar]
- 36.Inceoglu O., Salles J.F., van Overbeek L., van Elsas J.D. Effects of plant genotype and growth stage on the betaproteobacterial communities associated with different potato cultivars in two fields. Appl. Environ. Microbiol. 2010;76(11):3675–3684. doi: 10.1128/AEM.00040-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kennedy K., Hall M.W., Lynch M.D., Moreno-Hagelsieb G., Neufeld J.D. Evaluating bias of illumina-based bacterial 16S rRNA gene profiles. Appl. Environ. Microbiol. 2014;80(18):5717–5722. doi: 10.1128/AEM.01451-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kozich J.J., Westcott S.L., Baxter N.T., Highlander S.K., Schloss P.D. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq illumina sequencing platform. Appl. Environ. Microbiol. 2013;79(17):5112–5120. doi: 10.1128/AEM.01043-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fadrosh D.W., Ma B., Gajer P., Sengamalay N., Ott S., Brotman R.M., Ravel J. An improved dual-indexing approach for multiplexed 16S rRNA gene sequencing on the Illumina MiSeq platform. Microbiome. 2014;2:6. doi: 10.1186/2049-2618-2-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Peiffer J.A., Spor A., Koren O., Jin Z., Tringe S.G., Dangl J.L., Buckler E.S., Ley R.E. Diversity and heritability of the maize rhizosphere microbiome under field conditions. Proc. Natl. Acad. Sci. USA. 2013;110(16):6548–6553. doi: 10.1073/pnas.1302837110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Magoc T., Salzberg S.L. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics. 2011;27(21):2957–2963. doi: 10.1093/bioinformatics/btr507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Edgar R.C. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods. 2013;10(10):996–998. doi: 10.1038/nmeth.2604. [DOI] [PubMed] [Google Scholar]
- 43.Cole J.R., Wang Q., Fish J.A., Chai B.L., McGarrell D.M., Sun Y.N., Brown C.T., Porras-Alfaro A., Kuske C.R., Tiedje J.M. Ribosomal Database Project: Data and tools for high throughput rRNA analysis. Nucleic Acids Res. 2014;42(D1):D633–D642. doi: 10.1093/nar/gkt1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.DeSantis T.Z., Hugenholtz P., Larsen N., Rojas M., Brodie E.L., Keller K., Huber T., Dalevi D., Hu P., Andersen G.L. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 2006;72(7):5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Caporaso J.G., Kuczynski J., Stombaugh J., Bittinger K., Bushman F.D., Costello E.K., Fierer N., Pena A.G., Goodrich J.K., Gordon J.I., Huttley G.A., Kelley S.T., Knights D., Koenig J.E., Ley R.E., Lozupone C.A., McDonald D., Muegge B.D., Pirrung M., Reeder J., Sevinsky J.R., Tumbaugh P.J., Walters W.A., Widmann J., Yatsunenko T., Zaneveld J., Knight R. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods. 2010;7(5):335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schloss P.D., Westcott S.L., Ryabin T., Hall J.R., Hartmann M., Hollister E.B., Lesniewski R.A., Oakley B.B., Parks D.H., Robinson C.J., Sahl J.W., Stres B., Thallinger G.G., Van Horn D.J., Weber C.F. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 2009;75(23):7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Qin J.J., Li Y.R., Cai Z.M., Li S.H., Zhu J.F., Zhang F., Liang S.S., Zhang W.W., Guan Y.L., Shen D.Q., Peng Y.Q., Zhang D.Y., Jie Z.Y., Wu W.X., Qin Y.W., Xue W.B., Li J.H., Han L.C., Lu D.H., Wu P.X., Dai Y.L., Sun X.J., Li Z.S., Tang A.F., Zhong S.L., Li X.P., Chen W.N., Xu R., Wang M.B., Feng Q., Gong M.H., Yu J., Zhang Y.Y., Zhang M., Hansen T., Sanchez G., Raes J., Falony G., Okuda S., Almeida M., LeChatelier E., Renault P., Pons N., Batto J.M., Zhang Z.X., Chen H., Yang R.F., Zheng W.M., Li S.G., Yang H.M., Wang J., Ehrlich S.D., Nielsen R., Pedersen O., Kristiansen K., Wang J. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature. 2012;490(7418):55–60. doi: 10.1038/nature11450. [DOI] [PubMed] [Google Scholar]
- 48.Luo R.B., Liu B.H., Xie Y.L., Li Z.Y., Huang W.H., Yuan J.Y., He G.Z., Chen Y.X., Pan Q., Liu Y.J., Tang J.B., Wu G.X., Zhang H., Shi Y.J., Liu Y., Yu C., Wang B., Lu Y., Han C.L., Cheung D.W., Yiu S.M., Peng S.L., Zhu X.Q., Liu G.M., Liao X.K., Li Y.R., Yang H.M., Wang J., Lam T.W., Wang J. 2012.
- 49.You M.S., Yue Z., He W.Y., Yang X.H., Yang G., Xie M., Zhan D.L., Baxter S.W., Vasseur L., Gurr G.M., Douglas C.J., Bai J.L., Wang P., Cui K., Huang S.G., Li X.C., Zhou Q., Wu Z.Y., Chen Q.L., Liu C.H., Wang B., Li X.J., Xu X.F., Lu C.X., Hu M., Davey J.W., Smith S.M., Chen M.S., Xia X.F., Tang W.Q., Ke F.S., Zheng D.D., Hu Y.L., Song F.Q., You Y.C., Ma X.L., Peng L., Zheng Y.K., Liang Y., Chen Y.Q., Yu L.Y., Zhang Y.N., Liu Y.Y., Li G.Q., Fang L., Li J.X., Zhou X., Luo Y.D., Gou C.Y., Wang J.Y., Wang J., Yang H.M., Wang J. A heterozygous moth genome provides insights into herbivory and detoxification. Nat. Genet. 2013;45(2):220–225. doi: 10.1038/ng.2524. [DOI] [PubMed] [Google Scholar]
- 50.Li R.Q., Yu C., Li Y.R., Lam T.W., Yiu S.M., Kristiansen K., Wang J. SOAP2: An improved ultrafast tool for short read alignment. Bioinformatics. 2009;25(15):1966–1967. doi: 10.1093/bioinformatics/btp336. [DOI] [PubMed] [Google Scholar]
- 51.Peng Y., Leung H.C., Yiu S.M., Chin F.Y. IDBA-UD: A de novo assembler for single-cell and metagenomic sequencing data with highly uneven depth. Bioinformatics. 2012;28(11):1420–1428. doi: 10.1093/bioinformatics/bts174. [DOI] [PubMed] [Google Scholar]
- 52.Zhu W.H., Lomsadze A., Borodovsky M. Ab initio gene identification in metagenomic sequences. Nucleic Acids Res. 2010;38(12):e132. doi: 10.1093/nar/gkq275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li W.Z., Godzik A. Cd-hit: A fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 2006;22(13):1658–1659. doi: 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
- 54.Langmead B., Trapnell C., Pop M., Salzberg S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10(3):R25. doi: 10.1186/Gb-2009-10-3-R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Huson D.H., Auch A.F., Qi J., Schuster S.C. MEGAN analysis of metagenomic data. Genome Res. 2007;17(3):377–386. doi: 10.1101/gr.5969107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Francis O.E., Bendall M., Manimaran S., Hong C.J., Clement N.L., Castro-Nallar E., Snell Q., Schaalje G.B., Clement M.J., Crandall K.A., Johnson W.E. Pathoscope: Species identification and strain attribution with unassembled sequencing data. Genome Res. 2013;23(10):1721–1729. doi: 10.1101/gr.150151.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Audic S., Claverie J.M. The significance of digital gene expression profiles. Genome Res. 1997;7(10):986–995. doi: 10.1101/gr.7.10.986. [DOI] [PubMed] [Google Scholar]
- 58.Abdi H. Encyclopedia of Measurement and Statistics. Thousand Oaks, CA: Sage; 2007. The bonferonni and Sidak corrections for multiple comparisons. [Google Scholar]
- 59.Benjamini Y., Yekutieli D. The control of the false discovery rate in multiple testing under dependency. Ann. Stat. 2001;29(4):1165–1188. [Google Scholar]
- 60.Eisen M.B., Spellman P.T., Brown P.O., Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. USA. 1998;95(25):14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.de Hoon M.J., Imoto S., Nolan J., Miyano S. Open source clustering software. Bioinformatics. 2004;20(9):1453–1454. doi: 10.1093/bioinformatics/bth078. [DOI] [PubMed] [Google Scholar]
- 62.Saldanha A.J. Java Treeview-extensible visualization of microarray data. Bioinformatics. 2004;20(17):3246–3248. doi: 10.1093/bioinformatics/bth349. [DOI] [PubMed] [Google Scholar]
- 63.Kanehisa M., Araki M., Goto S., Hattori M., Hirakawa M., Itoh M., Katayama T., Kawashima S., Okuda S., Tokimatsu T., Yamanishi Y. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008;36:D480–D484. doi: 10.1093/nar/gkm882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.White J.R., Nagarajan N., Pop M. Statistical methods for detecting differentially abundant features in clinical metagenomic samples. PLOS Comput. Biol. 2009;5(4):e1000352. doi: 10.1371/journal.pcbi.1000352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Benjamini Y., Hochberg Y. Controlling the false discovery rate - A practical and powerful approach to multiple testing. J. Roy. Stat. Soc. B-Met. 1995;57(1):289–300. [Google Scholar]
- 66.Masson-Boivin C., Giraud E., Perret X., Batut J. Establishing nitrogen-fixing symbiosis with legumes: How many rhizobium recipes? Trends Microbiol. 2009;17(10):458–466. doi: 10.1016/j.tim.2009.07.004. [DOI] [PubMed] [Google Scholar]
- 67.Glick B.R. Bacteria with ACC deaminase can promote plant growth and help to feed the world. Microbiol. Res. 2014;169(1):30–39. doi: 10.1016/j.micres.2013.09.009. [DOI] [PubMed] [Google Scholar]
- 68.Calvo P., Nelson L., Kloepper J.W. Agricultural uses of plant biostimulants. Plant Soil. 2014;383(1-2):3–41. [Google Scholar]
- 69.Pieterse C.M., Zamioudis C., Berendsen R.L., Weller D.M., Van Wees S.C., Bakker P.A. Induced systemic resistance by beneficial microbes. Annu. Rev. Phytopathol. 2014;52:347–375. doi: 10.1146/annurev-phyto-082712-102340. [DOI] [PubMed] [Google Scholar]
- 70.Hess M., Sczyrba A., Egan R., Kim T.W., Chokhawala H., Schroth G., Luo S.J., Clark D.S., Chen F., Zhang T., Mackie R.I., Pennacchio L.A., Tringe S.G., Visel A., Woyke T., Wang Z., Rubin E.M. Metagenomic discovery of biomass-degrading genes and genomes from cow rumen. Science. 2011;331(6016):463–467. doi: 10.1126/science.1200387. [DOI] [PubMed] [Google Scholar]
- 71.Wood D.E., Salzberg S.L. Kraken: Ultrafast metagenomic sequence classification using exact alignments. Genome Biol. 2014;15(3):R46. doi: 10.1186/gb-2014-15-3-r46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Freitas T.A., Li P.E., Scholz M.B., Chain P.S. Accurate read-based metagenome characterization using a hierarchical suite of unique signatures. Nucleic Acids Res. 2015;43(10):e69. doi: 10.1093/nar/gkv180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ounit R., Wanamaker S., Close T.J., Lonardi S. CLARK: Fast and accurate classification of metagenomic and genomic sequences using discriminative k-mers. BMC Genomics. 2015;16:236. doi: 10.1186/s12864-015-1419-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lindgreen S., Adair K.L., Gardner P.P. An evaluation of the accuracy and speed of metagenome analysis tools. Sci. Rep. 2016;6:19233. doi: 10.1038/srep19233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Frank J.A., Pan Y., Tooming-Klunderud A., Eijsink V.G., McHardy A.C., Nederbragt A.J., Pope P.B. Improved metagenome assemblies and taxonomic binning using long-read circular consensus sequence data. 2016. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material is available on the publisher’s web site along with the published article.