Abstract
Abstract: The phytohormone abscisic acid (ABA) enables plants to adapt to adverse environmental conditions through the modulation of metabolic pathways and of growth and developmental programs. We used comparative microarray analysis to identify genes exhibiting ABA-dependent expression and other hormone-dependent expression among them in Oryza sativa shoot and root. We identified 854 genes as significantly up- or down-regulated in root or shoot under ABA treatment condition. Most of these genes had similar expression profiles in root and shoot under ABA treatment condition, whereas 86 genes displayed opposite expression responses in root and shoot. To examine the crosstalk between ABA and other hormones, we compared the expression profiles of the ABA-dependently regulated genes under several different hormone treatment conditions. Interestingly, around half of the ABA-dependently expressed genes were also regulated by jasmonic acid based on microarray data analysis. We searched the promoter regions of these genes for cis-elements that could be responsible for their responsiveness to both hormones, and found that ABRE and MYC2 elements, among others, were common to the promoters of genes that were regulated by both ABA and JA. These results show that ABA and JA might have common gene expression regulation system and might explain why the JA could function for both abiotic and biotic stress tolerance.
Keywords: Plant stress, ABA signaling, ABA responsive genes, JA responsive genes, Hormone crosstalk, Microarray analysis
1. INTRODUCTION
When plants encounter stress conditions, the stress phytohormone ABA is biosynthesized or converted into an active form from a glucose-conjugated inactive form. As a result, functional ABA contents increase [1], and ABA modulates metabolism, growth and development to enable plants to adapt to adverse environments. ABA has strong effects on gene expression, such that around 10% of protein-encoding genes are transcriptionally regulated by ABA [2]. This transcriptional regulation has been well studied in Arabidopsis thaliana and rice, and the signaling components for ABA have been identified from receptors to transcription factors [3-5]. Typically, PYL/RCARs, ABA receptors sense ABA and bind to subclass A PP2Cs resulting in the activation of SnRK2s, which were suppressed by the subclass A PP2Cs. Activated SnRK2s are able to phosphorylate ABF transcription factors, which in turn promote the expression of ABA-dependent genes [2, 6-8].
To date, studies of ABA signaling pathways have focused on above-ground organs because important agricultural traits such as plant height, tiller number, grain number and weight are determined in those organs. However, recently it has been reported by several research groups that the root plays pivotal roles in plant growth, biomass increase and abiotic stress tolerance, among other characteristics [9-11]. In spite of the importance of the root system in plant growth and abiotic stress tolerance, little is known about any potential differences between the ABA responses of root and shoot.
Recently, cross-talks among several phytohormones have been discovered to lead to a network of complicated interactions during development and abiotic and biotic stress signaling. JA plays roles in biotic stress tolerance, especially against herbivory [12], and recently JA was also found to have functions related to abiotic stress tolerance and stomatal closing [13]. JA signaling from sensing to transcriptional regulation has been characterized in Arabidopsis. The JA receptor is COI1, which has an F-box domain and interacts with JAZ proteins, which repress MYC2 transcription factors. The COI1-JAZ interaction causes the JAZ proteins to be ubiquitinated and subsequently degraded through the 50S proteasome complex pathway. As a result, MYC2 proteins are de-repressed and promote the expression of downstream JA-responsive genes [14].
In this study, we identified ABA-responsive genes in rice root and shoot through microarray analysis and characterized their expression patterns. We compared their gene expression profiles in response to other hormone treatments to identify possible hormone crosstalk mechanisms. Many genes were found to be commonly induced by JA and ABA. These results provide valuable insight into crosstalk between JA and ABA signaling in terms of gene expression regulation.
2. MATERIALS AND METHODS
2.1. Plant Growth and ABA Treatment
Rice (Oryza sativa L. cv. Dongjin) was germinated and grown on 1/2 Murashige and Skoog (MS) medium (Duchefa, Netherland) containing 0.4 g /L MES (Duchefa, Netherland) and 0.4% Phytagel (Sigma, USA) at pH 5.8. The plants were grown for 7 d at 28°C under 16-h liquid in a growth chamber. For hormone treatments, plants were transferred and adapted to 1/2 MS medium for 2 days and JA and ABA were added into medium to final concentration of 50 µM. Root and leaf tissues were harvested 6 h after hormone treatment, frozen in liquid nitrogen and kept at -80°C. light/8-h dark conditions.
2.2. RNA Isolation and cDNA Synthesis
Total RNA was extracted using the RNeasy® Plant Mini Kit (QIAGEN, Germany), according to the manufacturer’s instructions. RNA quality and concentration were determined using a NanoDropTM 1000 Spectrophotometer (Thermo Scientific, USA). cDNAs were prepared using the RNA to cDNA EcoDryTM Premix (Double Primed; Clontech, USA), according to the manufacturer’s instructions [15].
2.3. Microarray Analysis
Expression profiling was conducted with the Oryza sativa 135k v1.0 microarray (Green Gene Bio, Inc., Korea) designed from 31,439 genes deposited in the IRGSP RAP2 database (http://rapdb.lab.nig.ac.jp). The microarray was scanned with a GenePix 4000B (Axon Instruments, Inc., USA) preset with 5-µm resolution for the Cy3 signal. Signals were digitized and analyzed by Nimblescan (Nimblegen, USA). The data were normalized and processed with cubic spline normalization using quantiles to adjust signal variations between chips [16]. Probe-level summarization by Robust Multi-Chip Analysis (RMA) using a median polish algorithm implemented in NimbleScan was used to produce call files.
For the identification of differentially expressed genes at the transcriptional level, statistical analysis was performed using Student’s t-test. The TIGR Multi-Experiment Viewer (MeV, http://www.tm4.org/mev.html) was used to generate heat maps of expression patterns and hierarchical clustering. For analysis of the expression of ABA-responsive genes in response to other hormone treatments, publicly available microarray data for other several hormones were used (the RXP_1000 data set; NCBI GEO accession number GSE39429; stored at RiceXPro, http://ricexpro.dna.affrc.go.jp/).
2.4. Real-Time Quantitative RT-PCR Analysis
Real-time quantitative RT-PCR (qRT-PCR) was performed using LightCycler® 480 SYBR Green I Master (Roche, Swiss) and the MyiQ (Bio-Rad, US). Primers used for real-time qRT-PCR experiments were designed with the NCBI primer BLAST program and are listed in (Supplementary Table 1 (1.4MB, pdf) ). Ubi5 (LOC_Os01g22490) was used as an internal control for normalization, LEA3 (LOC_Os05g46480) and TLP (LOC_Os03g46070) were used as ABA- and JA-marker genes, respectively [17]. The qRT-PCR experiments were performed using three biological replicates. The expression ratio was calculated using the ΔΔct method.
2.5. Cis-element Analysis
For cis-element analysis, three genes that were commonly induced by ABA and JA were selected by microarray and qRT-PCR analysis. Ubi5, LEA3, and TLP were also selected for comparison, as ABA and JA marker genes. For the analysis, the 1-kb upstream sequence was obtained for each of the eight rice genes from RAP-DB (http://rapdb.dna.affrc.go.kr/; IRGSP-1.0_1kb-upstream_2013-04-24.fasta) [18, 19]. The cis-elements of those putative promoter regions were analyzed using Signal Scan searches at PLACE, a database of motifs found in plant cis-acting regulatory DNA elements (http://www.dna.affrc.go.jp/PLACE/).
2.6. Gene Ontology Analysis
To get the putative information for molecular and functional role of ABA and JA responsive genes, Gene Ontology analysis was done for all 145 genes. Out of total 145 genes, GO terms for 122 genes were available and retrieved from RGAP database [20, 21]. These terms were sorted on the basis of number of genes categorized under each function. Categories comprising of more than 10 genes were assembled and arranged in graph representation (Supplementary Fig. 1 (1.4MB, pdf) ). Entire gene list with GO term description has been compiled in (Supplementary Table 1 (1.4MB, pdf) ).
2.7. Analysis of Functionally Characterized ABA and JA Responsive Genes from OGRO Database
To further study the role of ABA and JA responsive genes in plant system, previous reported mutant analysis data assembled in public domain of OGRO database (http://qtaro.abr.affrc.go.jp/ogro/table) was analyzed and 11 genes were retrieved, that were studied using several mutants plants under various abiotic stresses. Summary of the 11 genes are listed in (Table 1).
Table 1.
Functionally characterized genes retrieved from OGRO database.
| S. No | ogro_id | Gene | Gene_symbol | Locus_id | Objective |
|---|---|---|---|---|---|
| 1 | 123 | ACC oxidase5 | OsACO5 | Os05g0149400 | Ethylene and Cyanide production. Resistance to blast fungus via restriction of fungal growth. |
| 2 | 1735 | HEAT STRESS-ASSOCIATED 32-KD PROTEIN | HSA32 | Os06g0682900 | Long-term acquired thermotolerance |
| 3 | 1491 | - | Oshox22 | Os04g0541700 | Drought and salinity tolerance. |
| 4 | 1438 | tandem zinc finger 1 | OsTZF1 | Os05g0195200 | Drought and salinity tolerance. |
| 5 | 1051 | Late embryogenesis abundant3-1 | OsLEA3-1 | Os05g0542500 | Drought tolerance. |
| 6 | 509 | - | bel | Os03g0760200 | Herbicide resistance. |
| 7 | 438 | drought sensitive mutant2 | dsm2 | Os03g0125100 | Drought and oxidative stress tolerance. |
| 8 | 1379 | Similar to RCD One 1c | OsSRO1c | Os03g0230300 | Stomatal control. Oxidative stress resistance. |
| 9 | 1931 | stress largely induced 1 | OsSLI1 | Os02g0649300 | Abiotic stress response |
| 10 | 190 | Salicylic acid glucosyltransferase1 | OsSGT1 | Os09g0518200 | Probenazole dependent blast resistance. |
| 11 | 1424 | heat shock transcription factor7 | OsHsfA7 | Os01g0571300 | Drought and salinity tolerance. |
3. RESULTS
3.1. ABA Dependently Regulated Genes in Both Root and Shoot of Rice
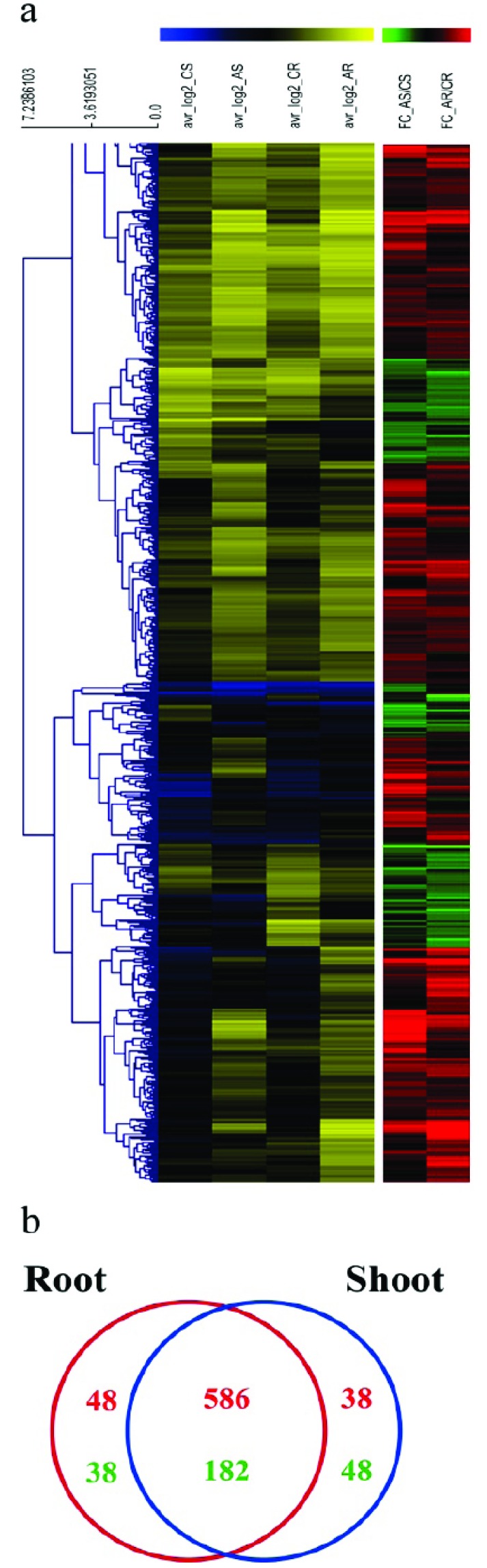
To identify ABA-dependently regulated genes in rice root and shoot, we carried out microarray analysis using total RNA isolated from root and shoot of 10-d-old seedlings treated with or without ABA. A total of 31,588 genes showed detectable signals in at least one microarray experiment. Among them, the expression of 854 genes was significantly changed (with at least two-fold change and p value <0.05) in ABA-treated shoot or root compared with the untreated control (Fig. 1a, Supplementary Table 2 (89MB, pdf) ).
Fig. (1).
Expression profiles of genes regulated by ABA in rice root and shoot. a) Heat map of expression profiles for genes with expression significantly changed by ABA treatment in root or shoot. b) Venn diagram of ABA-dependently regulated genes in rice root (red circle) and shoot (blue circle). Red numbers mean genes up-regulated by ABA. Green numbers mean genes down-regulated by ABA [35]. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this paper.)
Heat map analysis showed that the expression profiles of the 854 genes in root and shoot treated with ABA were quite similar (Fig. 1a). Specifically, 586 genes among 854 were up-regulated and 182 genes were down-regulated in both tissues, whereas the expression of 86 genes was changed in opposite directions in root and shoot (Fig. 1b, Supplementary Table 2 (1.4MB, pdf) ).
3.2. Validation of ABA-responsive Genes Both In Root and Shoot
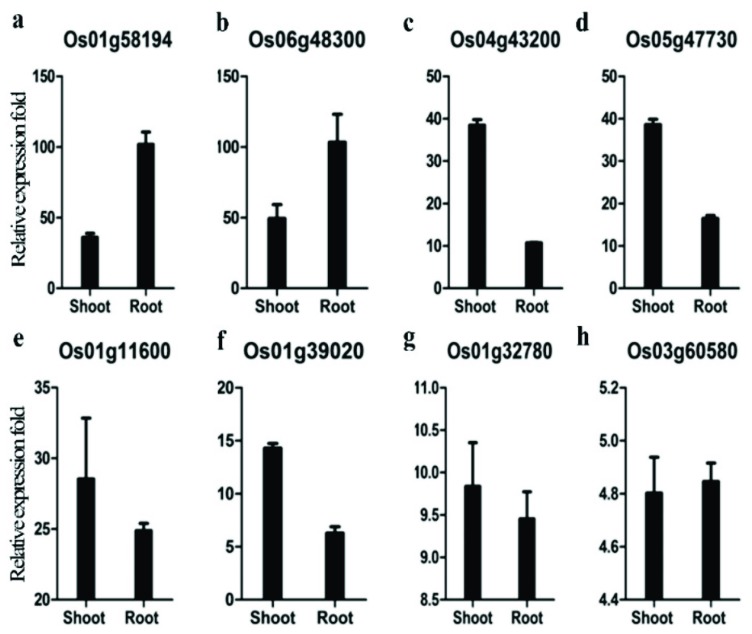
To validate the gene expression patterns derived from microarray analysis of ABA-treated root and shoot, we carried out qRT-PCR analysis of ten genes commonly expressed in root or shoot. The results showed that eight of these genes were clearly induced by ABA treatment both in root and shoot (Fig. 2). All of these genes were induced by ABA in both tissues, although some genes showed differential expression between shoot and root [22-24]. Protease inhibitor and caleosin-related protein were expressed predominantly in shoot, whereas protein disulfide isomerase and phosphatase 2C were much highly expressed in root (Fig. 2).
Fig. (2).
Validation of ABA regulation of genes expressed in root and shoot using qRT-PCR. The ΔΔCT method was used to determine the relative fold change. All data were normalized to the expression level of Ubi5. Error bars represent standard error of three replicates.
3.3. Responsiveness of ABA-dependently Regulated Genes to Other Hormones
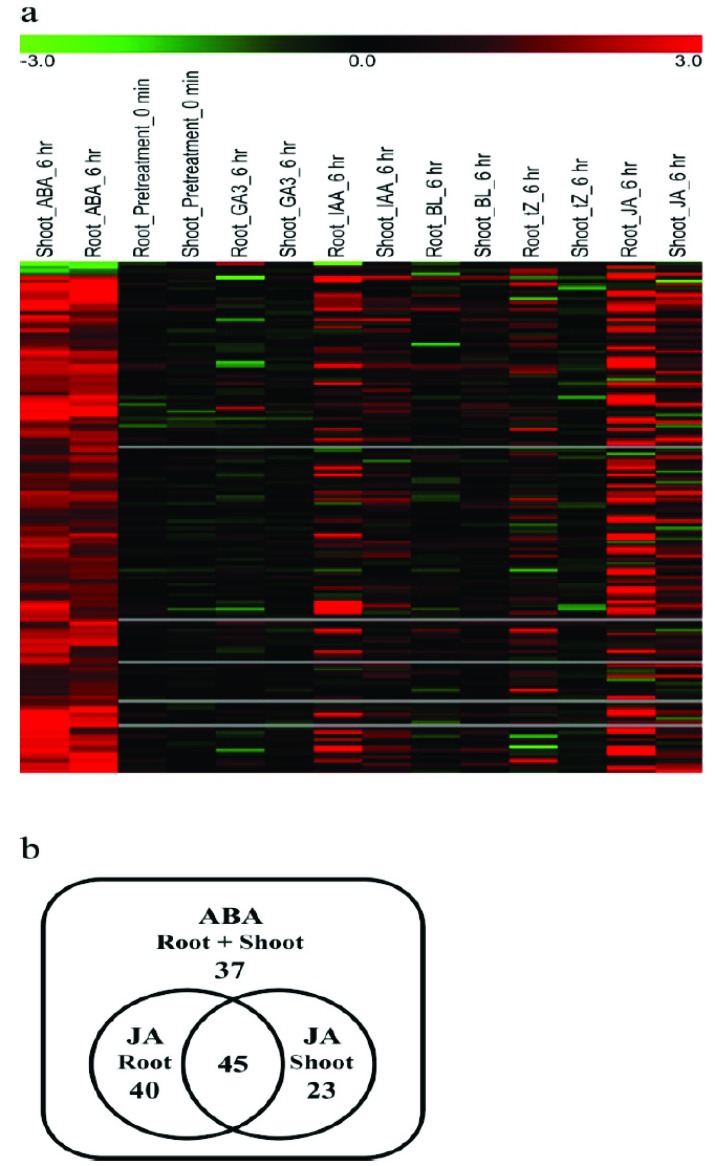
To study the potential cross-talk between ABA and other phytohormones, we selected the 145 genes of which expression was changed significantly more than two folds in the same direction in both root and shoot under ABA treatment [25, 26]. We analyzed the expression profiles of these genes in publicly available rice microarray data for treatment with Gibberellic Acid (GA), Indole Acetic Acid (IAA), brassinolide (BL), transzeatin (tZ), or JA. Very few genes were co-regulated under GA or BL and ABA treatment and IAA and tZ treatment co-regulated the expressions of several genes with ABA treatment. For JA and ABA treatments, the most genes were co-regulated in either root or shoot compared to other hormones (Fig. 3, Supplementary Table 2 (1.4MB, pdf) ). These results suggest that JA and ABA might together modulate the expression of several genes and have a common gene expression system [27-33].
Fig. (3).
Expression profiles of ABA-regulated genes upon treatment with other hormones. a) Heat map analysis of expression profiles under several hormone treatment conditions for the genes that were significantly regulated by ABA in both root and shoot. The genes for which the p value was below 0.05 and expression was changed two-fold both in root and shoot were selected for comparison. b) Venn diagram of ABA- and JA-regulated genes in rice root and shoot. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this paper.)
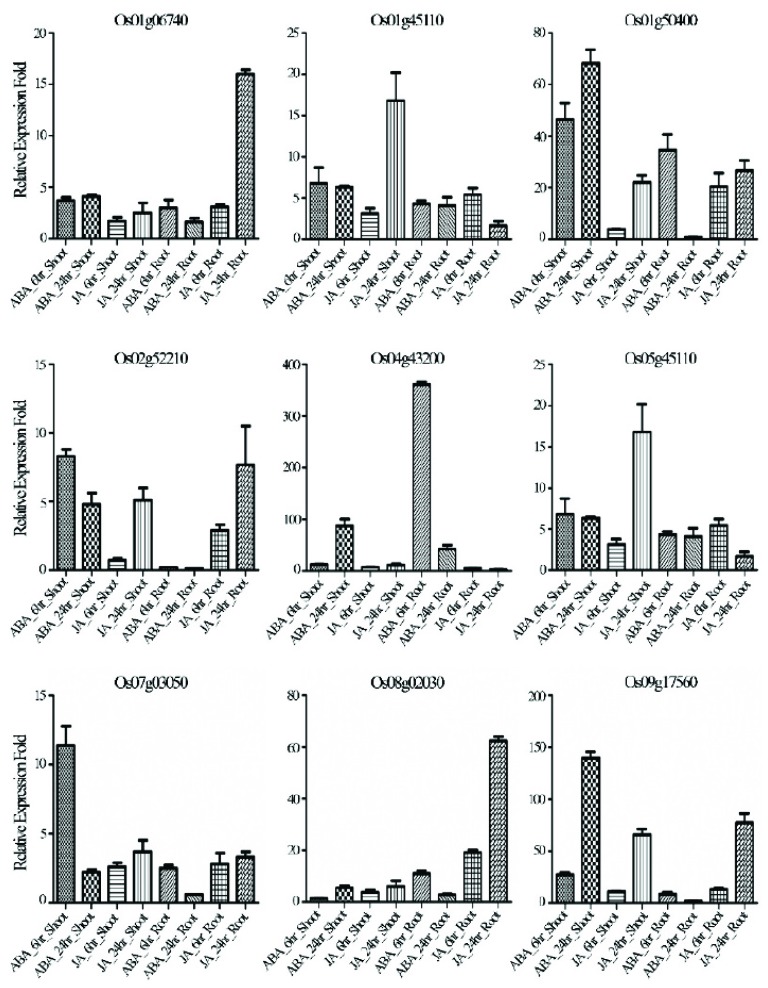
3.4. Validation of Genes Responsive to Both JA and ABA
The above transcriptome analysis revealed that compared to other hormones, JA regulated the most genes commonly with ABA. Those genes could be classified based on tissue specific gene expression pattern. The expressions of forty five genes were up-regulated by ABA and JA in both root and shoot. Although most of the genes were expressed at much higher levels in response to ABA treatment than to JA treatment, Os08g02030 and Os01g06740 were induced more highly in JA than ABA (Fig. 4). For further verification of ABA and JA responsive genes, Gene ontology analysis was done for total 145 genes, and 122 genes were found to be listed with putative functional characteristics. Under GO terms, large number of genes were listed under metabolic process (60 genes) followed by cellular process (38 genes), stress responses (37 genes) and several other biological processes (34) [34]. Detail list of GO terms along with their described functions are listed in (Supplementary Table 4 (1.4MB, pdf) ). Categories comprising of more than 10 genes, were assembled and arranged in graphical representation (Supplementary Fig. 1 (1.4MB, pdf) ). Previously reported studies regarding ABA and JA responsive genes was retrieved from OGRO database (that comprises of total of 1950 genes). Out of 145 genes, 11 genes have been studied under several abiotic stress condition such as drought and salinity suggesting that the remaining genes might as well play a vital role under adverse stress conditions (Table 1).
Fig. (4).
Validation of ABA- and JA-regulated genes using qRT-PCR. The ΔΔCT value method was used to determine the relative fold changes. All data were normalized to the expression level of Ubi5. Error bars represent standard error of three replicates.
3.5. Cis-element Analysis
To learn more about the potential mechanism of gene expression regulation by JA and ABA, we searched for shared cis-regulatory elements [36]. We compared the predicted cis-elements of three genes which were regulated by both JA and ABA (LOC_Os01g50400, LOC_Os01g45110, LOC_Os 04g43200) and two genes which are ABA-inducible marker gene LEA3 (Os05g46480) JA-inducible marker genes TLP (LOC_Os03g46070) and a constitutively expressed control gene, Ubi5.
According to PLACE database analysis, there were 196 cis-elements present at least once in those promoters [37]. Among them, we selected the cis-elements which were not present in Ubi5 promoter but present in other promoters and then analyzed the common cis-regulatory elements. Four cis-regulatory elements (ABRE; ACGTG, PRECONSCRHSP70 A; SCGAYNRNNNNNNNNNNNNNNNHD, ASF1MOTIF CAMV; TGACG, MYCATERD1; CATGTG and MYCATRD22; CACATG) were common to the promoter regions of all five selected genes except for Ubi5 (Table 1).
4. DISCUSSION
The root senses the water status in the soil, and ABA is biosynthesized in the root phloem under abiotic stress conditions. The resulting ABA is transported into the shoot and other tissues, where ABA signaling induces downstream responses to adapt to the adverse conditions [1]. Thus, the root plays pivotal roles in sensing environmental stresses from the soil and ABA signal transduction; in fact, the root could be considered a sensing and regulatory organ for other organs [38, 39]. This would suggest that root and shoot could have fairly different gene expression profiles under stress conditions or ABA treatment. However, our microarray analysis showed that the ABA-responsive gene expression patterns in the two tissues are similar and very few genes were differentially expressed in root and shoot under ABA treatment conditions. Intriguingly, among the 86 genes that showed opposite expression responses in root and shoot under ABA treatment, there were five cyclin related F-box proteins. The F-box proteins function to modulate protein degradation through Skp1p-cullin-F-box complex. 687 F-box proteins were annotated in rice genome and COI1, JA receptor and Arabidopsis F-box protein, is studied well in Jasmonic Acid (JA)-regulated defense responses. COI1 is predicted to target repressors of JA signaling to the proteasome for degradation [40, 41].
Previously, antagonistic regulation between ABA and JA has been studied based on differential responses to abiotic and biotic stresses because JA is biotic-stress related and ABA is abiotic-stress related. However, ABA and JA also have common physiological functions in growth inhibition, stomatal closing and tolerance to some stresses [42, 43]. We found that, compared to the other hormones, JA produced to the most similar gene expression patterns to those induced by ABA. This suggests that JA and ABA might share some common gene expression modules or signaling components and points to the importance of studying crosstalk between JA and ABA.
Among several signaling components, wound and JA induced Arabidopsis AtMPK1/AtMPK2, is also activated by ABA. JA- activated MAPKs can regulate several transcription factors and modulate the JA dependent gene expression such as atVSP1 and AtLOX2 [44, 45]. Thus JA/ABA dual activated MAPK might can regulate gene expression which can be regulated commonly in JA and ABA. In terms of transcriptional regulator, the AtMYC2 transcription factor is a candidate for regulating shared JA- and ABA-responsive gene expression in Arabidopsis. AtMYC2, which is a typical JA-responsive gene expression regulator, has been reported to be induced by ABA and can bind MYC2 cis-elements of the abiotic stress-responsive marker gene RD22. In this study we found out two different MYC-binding cis-elements in the promoter regions of the dually regulated genes. Thus AtMYC2 would be one of the responsible transcription factors for ABA/JA dual responsive gene expression [44, 46].
ABRE cis-elements were abundant in the promoter regions of the analyzed dual-regulated genes, as well as in them of the ABA-responsive marker genes. Intriguingly, the JA-responsive marker gene TLP also had ABRE cis-elements. It is possible the common expression of ABA- and JA-responsive genes might be caused by ABRE cis-elements placed in the context of JA-responsive promoters.
JA can regulate physiological responses in biotic stress tolerance as well as in abiotic stress tolerance. Gene Ontology analysis showed that ABA and JA common responsive genes are not only categorized under stress response but also assembled under several cellular and metabolic procedures. Few ABA and JA responsive genes obtained from mutant analysis database provided major information for their specific role under abiotic stress conditions. However, it is not yet clear how JA and ABA under abiotic stress signaling, converge and diverge and what the crosstalk points are. This study provides several candidate genes for further study of the crosstalk between JA and ABA signaling in rice.
CONCLUSION
Hormones regulate several different aspects of plant development and stress tolerance. JA was known that one of the hormones to regulate biotic stress tolerance of plants. However, recently it was known that it also plays important roles in abiotic stress tolerance of plants. JA might have some cross-talk mechanisms with stress phytohormone ABA. Intriguingly the transcriptome analysis of ABA and JA treatment showed that many genes are commonly regulated by JA and ABA. These results were confirmed by q-RT-PCR. We clearly showed that JA and ABA have the mechanisms of common gene expression regulation. And we analyzed the promoter of commonly expressed genes. ABRE and MYC related cis-element are the commonly present in those promoter. Thus we suggest that MYC and bZIPs might be related for co-regulation of JA and ABA. These results might provide substantial contributions toward understanding the crosstalk between ABA and JA signal transduction pathway in rice.
Table 2.
Cis elements in the promoters of genes induced by both ABA and JA.
| Cis-element Sequence | Cis-element Name | Ubi5 | LEA3 | Os01g50400 | Os04g43200 | Os01g45110 | TLP |
|---|---|---|---|---|---|---|---|
| ACGTG | ABRELATERD1 | 0 | 7 | 2 | 5 | 1 | 3 |
| TGACG | ASF1MOTIFCAMV | 0 | 2 | 1 | 1 | 4 | 3 |
| CATGTG | MYCATERD1 | 0 | 2 | 1 | 1 | 2 | 1 |
| CACATG | MYCATRD22 | 0 | 2 | 1 | 1 | 2 | 1 |
| SCGAYNRNNNNNN NNNNNNNNNHD |
PRECONSCRHSP70A | 0 | 3 | 4 | 2 | 1 | 2 |
ACKNOWLEDGEMENTS
This work was supported by the Woo Jang Chun Special Project (project no. PJ009106) and Next-Generation Bio Green21 Program (PJ01187002) of RDA.
LIST OF ABBREVIATIONS
- ABA
Abscisic Acid
- COI1
Coronatine-insensitive Protein 1
- JA
Jasmonic Acid
- JAZ
Jasmonate ZIM-domain
- PP2C
Protein Phosphatase 2C
- PYL/RCAR
Pyrabactin Resistance 1-like/Regulatory Components of ABA Receptor
- SnRK2
SNF1-Related Protein Kinase 2
SUPPLEMENTARY MATERIAL
Supplementary material is available on the publisher’s web site along with the published article.
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
Not applicable.
HUMAN AND ANIMAL RIGHTS
No Animals/Humans were used for studies that are base of this research.
CONSENT FOR PUBLICATION
Not applicable.
CONFLICT OF INTEREST
The authors declare no conflict of interest, financial or otherwise.
REFERENCES
- 1.Xu Z.Y., Kim D.H., Hwang I. ABA homeostasis and signaling involving multiple subcellular compartments and multiple receptors. Plant Cell Rep. 2013;32:807–813. doi: 10.1007/s00299-013-1396-3. [DOI] [PubMed] [Google Scholar]
- 2.Fujita Y., Fujita M., Shinozaki K. Yamaguchi-Shinozaki, K. ABA-mediated transcriptional regulation in response to osmotic stress in plants. J. Plant Res. 2011;124:509–525. doi: 10.1007/s10265-011-0412-3. [DOI] [PubMed] [Google Scholar]
- 3.Yazaki J., Kishimoto N., Nagata Y., Ishikawa M., Fujii F., Hashimoto A., Shimbo K., Shimatani Z., Kojima K., Suzuki K., Yamamoto M., Honda S., Endo A., Yoshida Y., Sato Y., Takeuchi K., Toyoshima K., Miyamoto C., Wu J., Sasaki T., Sakata K., Yamamoto K., Iba K., Oda T., Otomo Y., Murakami K., Matsubara K., Kawai J., Carninci P., Hayashizaki Y., Kikuchi S. Genomics approach to abscisic acid- and gibberellin-responsive genes in rice. DNA Res. 2003;10:249–261. doi: 10.1093/dnares/10.6.249. [DOI] [PubMed] [Google Scholar]
- 4.Kim H., Hwang H., Hong J.W., Lee Y.N., Ahn I.P., Yoon I.S., Yoo S.D., Lee S., Lee S.C., Kim B.G. A rice orthologue of the ABA receptor, OsPYL/RCAR5, is a positive regulator of the ABA signal transduction pathway in seed germination and early seedling growth. J. Exp. Bot. 2012;63:1013–1024. doi: 10.1093/jxb/err338. [DOI] [PubMed] [Google Scholar]
- 5.Kim H., Lee K., Hwang H., Bhatnagar N., Kim D.Y., Yoon I.S., Byun M.O., Kim S.T., Jung K.H., Kim B.G. Overexpression of PYL5 in rice enhances drought tolerance, inhibits growth, and modulates gene expression. J. Exp. Bot. 2014;65:453–464. doi: 10.1093/jxb/ert397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nakashima K., Fujita Y., Kanamori N., Katagiri T., Umezawa T., Kidokoro S., Maruyama K., Yoshida T., Ishiyama K., Kobayashi M., Shinozaki K., Yamaguchi-Shinozaki K. Three Arabidopsis SnRK2 protein kinases, SRK2D/SnRK2.2, SRK2E/ SnRK2.6/OST1 and SRK2I/SnRK2.3, involved in ABA signaling are essential for the control of seed development and dormancy. Plant Cell Physiol. 2009;50:1345–1363. doi: 10.1093/pcp/pcp083. [DOI] [PubMed] [Google Scholar]
- 7.Park S.Y., Fung P., Nishimura N., Jensen D.R., Fujii H., Zhao Y., Lumba S., Santiago J., Rodrigues A., Chow T.F., Alfred S.E., Bonetta D., Finkelstein R., Provart N.J., Desveaux D., Rodriguez P.L., McCourt P., Zhu J.K., Schroeder J.I., Volkman B.F., Cutler S.R. Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science. 2009;324:1068–1071. doi: 10.1126/science.1173041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yoshida T., Fujita Y., Sayama H., Kidokoro S., Maruyama K., Mizoi J., Shinozaki K., Yamaguchi-Shinozaki K. AREB1, AREB2, and ABF3 are master transcription factors that cooperatively regulate ABRE-dependent ABA signaling involved in drought stress tolerance and require ABA for full activation. Plant J. 2010;61:672–685. doi: 10.1111/j.1365-313X.2009.04092.x. [DOI] [PubMed] [Google Scholar]
- 9.Jeong J.S., Kim Y.S., Baek K.H., Jung H., Ha S.H., Do Choi Y., Kim M., Reuzeau C., Kim J.K. Root-specific expression of OsNAC10 improves drought tolerance and grain yield in rice under field drought conditions. Plant Physiol. 2010;153:185–197. doi: 10.1104/pp.110.154773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Redillas M.C., Jeong J.S., Kim Y.S., Jung H., Bang S.W., Choi Y.D., Ha S.H., Reuzeau C., Kim J.K. The overexpression of OsNAC9 alters the root architecture of rice plants enhancing drought resistance and grain yield under field conditions. Plant Biotechnol. J. 2012;10:792–805. doi: 10.1111/j.1467-7652.2012.00697.x. [DOI] [PubMed] [Google Scholar]
- 11.Minh-Thu P.T., Hwang D.J., Jeon J.S., Nahm B.H., Kim Y.K. Transcriptome analysis of leaf and root of rice seedling to acute dehydration. Rice (N. Y.) 2013;6:38. doi: 10.1186/1939-8433-6-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wasternack C., Hause B. Jasmonates: biosynthesis, perception, signal transduction and action in plant stress response, growth and development. An update to the 2007 review in Annals of Botany. Ann. Bot. 2013;111:1021–1058. doi: 10.1093/aob/mct067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Munemasa S., Mori I.C., Murata Y. Methyl jasmonate signaling and signal crosstalk between methyl jasmonate and abscisic acid in guard cells. Plant Signal. Behav. 2011;6:939–941. doi: 10.4161/psb.6.7.15439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fonseca S., Chico J.M., Solano R. The jasmonate pathway: The ligand, the receptor and the core signalling module. Curr. Opin. Plant Biol. 2009;12:539–547. doi: 10.1016/j.pbi.2009.07.013. [DOI] [PubMed] [Google Scholar]
- 15.Nakata M., Mitsuda N., Herde M., Koo A.J., Moreno J.E., Suzuki K., Howe G.A., Ohme-Takagi M. A bHLH-type transcription factor, aba-inducible bhlh-type transcription factor/ja-associated myc2-like1, acts as a repressor to negatively regulate jasmonate signaling in arabidopsis. Plant Cell. 2013;25:1641–1656. doi: 10.1105/tpc.113.111112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Workman C., Jensen L.J., Jarmer H., Berka R., Gautier L., Nielser H.B., Saxild H.H., Nielsen C., Brunak S., Knudsen S. A new non-linear normalization method for reducing variability in DNA microarray experiments. 2002. [DOI] [PMC free article] [PubMed]
- 17.Xu Z.Y., Kim D.H., Hwang I. ABA homeostasis and signaling involving multiple subcellular compartments and multiple receptors. Plant Cell Rep. 2013;32:807–813. doi: 10.1007/s00299-013-1396-3. [DOI] [PubMed] [Google Scholar]
- 18.Fujita Y., Fujita M., Shinozaki K., Yamaguchi-Shinozaki K. ABA-mediated transcriptional regulation in response to osmotic stress in plants. J. Plant Res. 2011;124:509–525. doi: 10.1007/s10265-011-0412-3. [DOI] [PubMed] [Google Scholar]
- 19.Yazaki J., Kishimoto N., Nagata Y., Ishikawa M., Fujii F., Hashimoto A., Shimbo K., Shimatani Z., Kojima K., Suzuki K., Yamamoto M., Honda S., Endo A., Yoshida Y., Sato Y., Takeuchi K., Toyoshima K., Miyamoto C., Wu J., Sasaki T., Sakata K., Yamamoto K., Iba K., Oda T., Otomo Y., Murakami K., Matsubara K., Kawai J., Carninci P., Hayashizaki Y., Kikuchi S. Genomics approach to abscisic acid- and gibberellin-responsive genes in rice. DNA Res. 2003;10:249–261. doi: 10.1093/dnares/10.6.249. [DOI] [PubMed] [Google Scholar]
- 20.Kim H., Hwang H., Hong J.W., Lee Y.N., Ahn I.P., Yoon I.S., Yoo S.D., Lee S., Lee S.C., Kim B.G. A rice orthologue of the ABA receptor, OsPYL/RCAR5, is a positive regulator of the ABA signal transduction pathway in seed germination and early seedling growth. J. Exp. Bot. 2012;63:1013–1024. doi: 10.1093/jxb/err338. [DOI] [PubMed] [Google Scholar]
- 21.Kim H., Lee K., Hwang H., Bhatnagar N., Kim D.Y., Yoon I.S., Byun M.O., Kim S.T., Jung K.H., Kim B.G. Overexpression of PYL5 in rice enhances drought tolerance, inhibits growth, and modulates gene expression. J. Exp. Bot. 2014;65:453–464. doi: 10.1093/jxb/ert397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nakashima K., Fujita Y., Kanamori N., Katagiri T., Umezawa T., Kidokoro S., Maruyama K., Yoshida T., Ishiyama K., Kobayashi M., Shinozaki K., Yamaguchi-Shinozaki K. Three Arabidopsis SnRK2 protein kinases, SRK2D/SnRK2.2, SRK2E/ SnRK2.6/OST1 and SRK2I/SnRK2.3, involved in ABA signaling are essential for the control of seed development and dormancy. Plant Cell Physiol. 2009;50:1345–1563. doi: 10.1093/pcp/pcp083. [DOI] [PubMed] [Google Scholar]
- 23.Park S.Y., Fung P., Nishimura N., Jensen D.R., Fujii H., Zhao Y., Lumba S., Santiago J., Rodrigues A., Chow T.F., Alfred S.E., Bonetta D., Finkelstein R., Provart N.J., Desveaux D., Rodriguez P.L., McCourt P., Zhu J.K., Schroeder J.I., Volkman B.F., Cutler S.R. Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science. 2009;324:1068–1071. doi: 10.1126/science.1173041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yoshida T., Fujita Y., Sayama H., Kidokoro S., Maruyama K., Mizoi J., Shinozaki K., Yamaguchi-Shinozaki K. AREB1, AREB2, and ABF3 are master transcription factors that cooperatively regulate ABRE-dependent ABA signaling involved in drought stress tolerance and require ABA for full activation. Plant J. 2010;61:672–685. doi: 10.1111/j.1365-313X.2009.04092.x. [DOI] [PubMed] [Google Scholar]
- 25.Jeong J.S., Kim Y.S., Baek K.H., Jung H., Ha S.H., Do Choi Y., Kim M., Reuzeau C., Kim J.K. Root-specific expression of OsNAC10 improves drought tolerance and grain yield in rice under field drought conditions. Plant Physiol. 2010;153:185–197. doi: 10.1104/pp.110.154773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Redillas M.C., Jeong J.S., Kim Y.S., Jung H., Bang S.W., Choi Y.D., Ha S.H., Reuzeau C., Kim J.K. The overexpression of OsNAC9 alters the root architecture of rice plants enhancing drought resistance and grain yield under field conditions. Plant Biotechnol. J. 2012;10:792–805. doi: 10.1111/j.1467-7652.2012.00697.x. [DOI] [PubMed] [Google Scholar]
- 27.Minh-Thu P.T., Hwang D.J., Jeon J.S., Nahm B.H., Kim Y.K. Transcriptome analysis of leaf and root of rice seedling to acute dehydration. Rice (N. Y.) 2013;6:38. doi: 10.1186/1939-8433-6-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wasternack C., Hause B. Jasmonates: Biosynthesis, perception, signal transduction and action in plant stress response, growth and development. An update to the 2007 review in Annals of Botany. Ann. Bot. 2013;111:1021–1058. doi: 10.1093/aob/mct067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Munemasa S., Mori I.C., Murata Y. Methyl jasmonate signaling and signal crosstalk between methyl jasmonate and abscisic acid in guard cells. Plant Signal. Behav. 2011;6:939–941. doi: 10.4161/psb.6.7.15439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fonseca S., Chico J.M., Solano R. The jasmonate pathway: the ligand, the receptor and the core signalling module. Curr. Opin. Plant Biol. 2009;12:539–547. doi: 10.1016/j.pbi.2009.07.013. [DOI] [PubMed] [Google Scholar]
- 31.Devoto A., Nieto-Rostro M., Xie D., Ellis C., Harmston R., Patrick E., Davis J., Sherratt L., Coleman M., Turner J.G. COI1 links jasmonate signalling and fertility to the SCF ubiquitin-ligase complex in Arabidopsis. Plant J. 2002;32:457–566. doi: 10.1046/j.1365-313x.2002.01432.x. [DOI] [PubMed] [Google Scholar]
- 32.Xu L., Liu F., Lechner E., Genschik P., Crosby W.L., Ma H., Peng W., Huang D., Xie D. The SCF(COI1) ubiquitin-ligase complexes are required for jasmonate response in Arabidopsis. Plant Cell. 2002;14:1919–1935. doi: 10.1105/tpc.003368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ortiz-Masia D., Perez-Amador M.A., Carbonell J., Marcote M.J. Diverse stress signals activate the C1 subgroup MAP kinases of Arabidopsis. FEBS Lett. 2007;581:1834–1840. doi: 10.1016/j.febslet.2007.03.075. [DOI] [PubMed] [Google Scholar]
- 34.Savchenko T., Kolla V.A., Wang C.Q., Nasafi Z., Hicks D.R., Phadungchob B., Chehab W.E., Brandizzi F., Froehlich J., Dehesh K. Functional convergence of oxylipin and abscisic acid pathways controls stomatal closure in response to drought. Plant Physiol. 2014;164:1151–1160. doi: 10.1104/pp.113.234310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Takahashi F., Yoshida R., Ichimura K., Mizoguchi T., Seo S., Yonezawa M., Maruyama K., Yamaguchi-Shinozaki K., Shinozaki K. The mitogen-activated protein kinase cascade MKK3-MPK6 is an important part of the jasmonate signal transduction pathway in Arabidopsis. Plant Cell. 2007;19:805–818. doi: 10.1105/tpc.106.046581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schweighofer A., Meskiene I. Regulation of stress hormones jasmonates and ethylene by MAPK pathways in plants. Mol. Biosyst. 2008;4:799–803. doi: 10.1039/b718578m. [DOI] [PubMed] [Google Scholar]
- 37.Kazan K., Manners J.M. MYC2: The master in action. Mol. Plant. 2013;6:686–703. doi: 10.1093/mp/sss128. [DOI] [PubMed] [Google Scholar]
- 38.Nakata M., Mitsuda N., Herde M., Koo A.J., Moreno J.E., Suzuki K., Howe G.A., Ohme-Takagi M. A bHLH-type transcription factor, ABA-inducible bhlh-type transcription factor/ja-associated myc2-like1, acts as a repressor to negatively regulate jasmonate signaling in arabidopsis. Plant Cell. 2013;25:1641–1656. doi: 10.1105/tpc.113.111112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Workman C., Jensen L.J., Jarmer H., Berka R., Gautier L., Nielser H.B., Saxild H.H., Nielsen C., Brunak S., Knudsen S. A new non-linear normalization method for reducing variability in DNA microarray experiments. 2002. [DOI] [PMC free article] [PubMed]
- 40.Devoto A., Nieto-Rostro M., Xie D., Ellis C., Harmston R., Patrick E., Davis J., Sherratt L., Coleman M., Turner J.G. COI1 links jasmonate signalling and fertility to the SCF ubiquitin-ligase complex in Arabidopsis. Plant J. 2002;32:457–466. doi: 10.1046/j.1365-313x.2002.01432.x. [DOI] [PubMed] [Google Scholar]
- 41.Xu L., Liu F., Lechner E., Genschik P., Crosby W.L., Ma H., Peng W., Huang D., Xie D. The SCF(COI1) ubiquitin-ligase complexes are required for jasmonate response in Arabidopsis. Plant Cell. 2002;14:1919–1935. doi: 10.1105/tpc.003368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ortiz-Masia D., Perez-Amador M.A., Carbonell J., Marcote M.J. Diverse stress signals activate the C1 subgroup MAP kinases of Arabidopsis. FEBS Lett. 2007;581:1834–1840. doi: 10.1016/j.febslet.2007.03.075. [DOI] [PubMed] [Google Scholar]
- 43.Savchenko T., Kolla V.A., Wang C.Q., Nasafi Z., Hicks D.R., Phadungchob B., Chehab W.E., Brandizzi F., Froehlich J., Dehesh K. Functional convergence of oxylipin and abscisic acid pathways controls stomatal closure in response to drought. Plant Physiol. 2014;164:1151–1160. doi: 10.1104/pp.113.234310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Takahashi F., Yoshida R., Ichimura K., Mizoguchi T., Seo S., Yonezawa M., Maruyama K., Yamaguchi-Shinozaki K., Shinozaki K. The mitogen-activated protein kinase cascade MKK3-MPK6 is an important part of the jasmonate signal transduction pathway in Arabidopsis. Plant Cell. 2007;19:805–818. doi: 10.1105/tpc.106.046581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schweighofer A., Meskiene I. Regulation of stress hormones jasmonates and ethylene by MAPK pathways in plants. Mol. Biosyst. 2008;4:799–803. doi: 10.1039/b718578m. [DOI] [PubMed] [Google Scholar]
- 46.Kazan K., Manners J.M. MYC2: The master in action. Mol. Plant. 2013;6:686–703. doi: 10.1093/mp/sss128. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material is available on the publisher’s web site along with the published article.