Abstract
Sexually dimorphic behaviors are often regulated by androgens and estrogens. Steroid receptors and metabolism are control points for evolutionary changes in sexual dimorphism. Electric communication signals of South American knifefishes are a model for understanding the evolution and physiology of sexually dimorphic behavior. These signals are regulated by gonadal steroids and controlled by a simple neural circuit. Sexual dimorphism of the signals varies across species. We used transcriptomics to examine mechanisms for sex differences in electric organ discharges (EODs) of two closely-related species, Apteronotus leptorhynchus and Apteronotus albifrons, with reversed sexual dimorphism in their EODs. The pacemaker nucleus (Pn), which controls EOD frequency (EODf), expressed transcripts for steroid receptors and metabolizing enzymes, including androgen receptors, estrogen receptors, aromatase, and 5α-reductase. The Pn expressed mRNA for ion channels likely to regulate the high-frequency activity of Pn neurons and for neuromodulator and neurotransmitter receptors that may regulate EOD modulations used in aggression and courtship. Expression of several ion channel genes, including those for Kir3.1 inward-rectifying potassium channels and sodium channel β1 subunits, was sex-biased or correlated with EODf in ways consistent with EODf sex differences. Our findings provide a basis for future studies to characterize neurogenomic mechanisms by which sex differences evolve.
Keywords: GENE EXPRESSION, GONADAL STEROID HORMONE, SEX DIFFERENCE, ELECTRIC FISH, ION CHANNELS
INTRODUCTION
Sexually dimorphic behaviors can evolve rapidly and are widespread and diverse across species. Behavioral sex differences are often regulated by androgens and estrogens. Males and females usually differ in gonadal production of these steroids, and they can thus serve as endocrine signals of sex and reproductive condition. By altering gene expression in the brain during development or in adulthood, gonadal steroids can establish or activate sex differences in the structure, connectivity, and function of brain circuits that control sexually dimorphic behaviors. Several systems (e.g., vocal control circuits in birds, frogs, and fishes; spinal circuits that control penile reflexes in mammals; forebrain and neuromuscular systems regulating courtship and aggression in lizards) have yielded valuable insights into the neuroendocrine mechanisms of sexually dimorphic behavior (reviewed in Sengelaub and Forger 2008; Wade 2011; Zornik and Kelley 2011; Feng and Bass 2017; Schlinger and Brenowitz 2017).
Although induction of sex-specific gene expression by gonadal steroids underlies many behavioral sex differences, localized metabolism of steroids and/or de novo steroid synthesis in the brain also substantially influences many sexually dimorphic and reproductive behaviors (Saldanha and Schlinger 2008; Remage-Healey et al. 2010). Furthermore, the actions of sex steroids can also be influenced by co-factors (e.g., steroid receptor co-activators, SRCs) that facilitate or inhibit the ability of ligand-bound receptors to induce transcription of downstream target genes (Auger et al. 2000; Charlier et al. 2005). Localized mechanisms of hormone action mediated by expression of steroid receptors, steroid-metabolizing enzymes, and transcriptional regulators in specific brain regions can also provide flexibility for behaviors to be coopted into or escape from hormonal pleiotropy (Canoine et al. 2007; Voigt and Goymann 2007; Burns et al. 2013). Species diversity in behavioral sex differences may arise through evolution of species differences in the sex-specific timing and levels of peripheral steroid production and/or through variation in brain-region specific expression of hormone receptors, steroidogenic enzymes, transcriptional co-regulators, and/or downstream regulation of target genes by steroids (Brenowitz and Arnold 1985; Shaw and Kennedy 2002; Fuxjager et al. 2016).
The use of microarrays and RNAseq, which simultaneously quantify expression of large numbers of genes, has accelerated the study of sex-specific and hormonally regulated gene expression in the brain and other tissues (Schumer et al. 2011; Peterson et al. 2013; Wong et al. 2014; Stiver et al. 2015). Relating sex-biased gene expression to sexually dimorphic behavior, however, can be challenging. Many studies have quantified sex-biased gene expression in whole brains or in gross subdivisions of the brain. Gene expression varies substantially across brain regions, however, and this heterogeneity can mask region-specific and behaviorally relevant sex-biased gene expression. Indeed, many fewer sex-biased genes are often found in the brain than in more homogeneous tissues like gonads or liver (Bohne et al. 2014; Manousaki et al. 2014; Caetano-Anolles et al. 2015; Liu et al. 2015). To address this challenge, one ideally needs to study sex-biased and hormonally-regulated gene expression specifically in brain regions whose function is dedicated to controlling particular sexually dimorphic behaviors.
The electric communication signals of South American ghost knifefishes (Apteronotidae) provide an outstanding opportunity to investigate neuroendocrine regulation and evolution of diversity in sexually dimorphic behaviors. The electric organs of these fish continuously produce high-frequency electric organ discharges (EODs) that are used to identify nearby objects and to communicate with conspecifics. Electric organ discharge frequency (EODf) varies across species, and in many species is sexually dimorphic (reviewed in Smith 2013; Dunlap et al. 2017).
Both the magnitude and direction of sex differences in EODf vary across apteronotid species and populations. EODf does not differ between males and females in some species, but in other species, EODf is highly sexually dimorphic and a reliable signal of sex (Smith 2013). The direction of sex differences in EODf can also reverse across closely-related species. In brown ghost knifefish (Apteornotus leptorhynchus), EODf is lower in males than in females, whereas in black ghost knifefish (Apteronotus albifrons) EODf is higher in males than in females (Zakon and Dunlap 1999; Kolodziejski et al. 2005).
Variation in sexual dimorphism of EODf is linked to variation in the sensitivity of this signal to gonadal steroid hormones. Sex differences in EODf in species with sexually dimorphic EODs are caused by effects of androgens and/or estrogens on this signal, whereas EODf is less responsive to these hormones in species and populations with reduced or no sex differences in EODf (Meyer et al. 1987; Schaefer and Zakon 1996; Dunlap et al. 1998; Ho et al. 2013; Petzold and Smith 2016). Moreover, reversals in the direction of sex differences in EODf are mediated by reversals of androgenic regulation of EODf. In A. leptorhynchus (EODf M>F), androgens increase EODf, whereas in A. albifrons (EODf F>M), androgens decrease EODf (Zakon and Dunlap 1999).
Understanding how species variation in neuroendocrine mechanisms contributes to diversity in sexual dimorphism of EODf is facilitated by the simplicity of the neural circuit controlling EODf. EODf is controlled by the pacemaker nucleus (Pn) in the medulla (reviewed in Smith 1999). Pacemaker and relay neurons in the Pn determine the firing rate of spinal electromotor neurons whose axons comprise the electric organ. Each action potential in these extensively electrically coupled neurons results in a single discharge of the electric organ. Sex differences in EODf result from androgen- and estrogen-mediated changes in the firing rates of Pn neurons (Schaefer and Zakon 1996). Firing rates of Pn neurons are set by interactions of sodium and potassium currents in these neurons; and androgens and estrogens likely regulate sex differences in EODf by changing the expression of ion channels that carry these currents (Smith and Zakon 2000). In apteronotid fishes, the Pn is a discrete protrusion on the ventral surface of the medulla and can be cleanly dissected. The Pn thus provides a straightforward opportunity to study gene expression in a brain region whose only function is to control a sexually dimorphic communication signal.
The present study used a transcriptomic approach to quantify gene expression in the Pn of two species with reversed EOD sexual dimorphism (A. leptorhynchus: EODf M>F and A. albifrons: EODf M<F). By comparing Pn gene expression across sexes in each species, we sought to answer three questions: (1) which hormone-related genes are expressed in the Pn and are likely contribute to sex differences in EODf; (2) which ion channel genes likely to regulate EODf are expressed in the Pn; and (3) what sex differences in ion channel gene expression might underlie sex and species differences in EODf?
METHODS
Research subjects
Adult, wild-caught A. albifrons (sexually dimorphic population, n=4 males, 4 females) and A. leptorhychus (n=3 males, 3 females) were obtained from commercial suppliers (Ruinemans, Miami, FL, and East Coast Transship, Baltimore, MD). A. albifrons were housed individually in 38-L tanks; A. leptorhynchus were housed socially in a 302-L tank in a 2000-L recirculating aquarium system. Fish were maintained on a 12:12 hour light:dark cycle at 25–27°C, pH 5.5–6.5, and conductivity of 150–300 µS/cm. These housing conditions stimulate gonadal recrudescence in captivity (Kirschbaum 1984).
Sample collection
A pair of wires and a reference electrode were placed next to the fish, and electric organ discharges (EODs) were differentially amplified (Grass-Telefactor model P-55, East Warwick, RI, 100× gain). EOD frequency (EODf) was measured with the frequency function on a digital multimeter (Fluke model 187, Everett, WA, USA). EODf measurements were temperature-compensated to that expected at 26°C by using a Q10°C of 1.62 (Dunlap et al. 2000). After EODf was measured, fish were lightly anesthetized by brief immersion in 2-phenoxyethanol in tank water (1mL/L, Sigma-Aldrich), and were weighed. Blood samples (10–50 µL) were collected from the caudal vein with a heparinized 25G needle and 1-cc syringe for measurement of plasma hormone levels. Fish were then placed back in the 2-phenoxyethanol solution for several minutes to anesthetize them deeply. The fish were placed on ice; and brains were removed rapidly and placed in RNAlater. The dura was removed from the medulla. The pacemaker nucleus (Pn) is visible as an obvious protrusion of grey matter on the ventral surface of the medulla. Under a stereomicroscope (20×), the Pn was conservatively dissected so that the collected sample contained only Pn tissue. The Pn samples were stored in RNALater. Gonads were removed and weighed to calculate gonadosomatic index (GSI, 100 * gonad mass/body mass) as an indication of reproductive condition. The remaining brain tissue, skin, and gonads were collected and stored for future studies.
Measurement of plasma hormone levels
Plasma levels of 11-ketotestosterone (11KT) were measured in males, and plasma levels of estradiol (E2) were measured in females by using enzyme immunoassay kits (Cayman Chemical, Ann Arbor, MI) according to the kit instructions. Samples were diluted in assay buffer 1:50 for the 11KT assay and 1:3–1:5 for the E2 assay. The sensitivity thresholds of the assays were 0.78 pg/ml (i.e., 39 pg/ml in the 1:50 diluted samples) for the 11KT assay and 6.6 pg/ml (19.8–33 pg/ml in the 1:3–1:5 diluted samples) for the E2 assay. Intraassay variation was 2.6% for the 11KT assay and 8.2% for the E2 assay. We did not measure 11KT in females or E2 in males because plasma levels were expected to be below the detection limit of the assays. Insufficient plasma was available to measure E2 in two female A. leptorhynchus.
RNA extraction, library construction, and sequencing
Pn samples were lysed in a microcentrifuge tube with a pestle (USA scientific, Ocala, FL); and RNA was extracted by using the Absolutely RNA Nanoprep kit (Agilent, Santa Clara, CA). Genomic DNA contamination was removed by using the DNAseI treatment step in the Nanoprep kit. RNA sample concentration and quality was measured with the high-sensitivity RNA Screentape on an Agilent 2200 Tapestation. RNA was reverse-transcribed with the SmartSeq Ultra-low kit (ClonTech/Takara, Mountain View, CA), and barcoded libraries were constructed using a Nextera XT kit (Illumina, San Diego, CA). Libraries were size-selected by using SPRISelect beads (Beckman Coulter, Brea, CA). Sequencing was performed by using Illumina NextSeq with 75-bp paired-end reads.
Transcriptome assembly, annotation, and analysis
Trimmomatic (version 0.35) was used to trim reads and to remove adapter sequences and low-quality reads. The transcriptomes for both species were assembled from the unstranded Illumina reads by using Trinity (version 2.1.1) with a minimum contig length of 100. Due to the size of the resulting transcriptomes (931,466 and 1,233,299 transcripts in A. albifrons and A. leptorhynchus, respectively), they were mapped to subsets of the NCBI NRAA database to identify functional regions and relationships. Two subsets were identified -- the complete set of human proteins and a set of fish proteins drawn from eight teleost species: Danio rerio, Esox lucius, Poecilia reticulata, Fundulus heteroclitus, Astyanax mexicanus, Scleropages formosus, Tetraodon nigroviridis, and Poecilia formosa. BLASTX was performed with NCBI-blast (version 2.2.30) with the following non-default parameters: -evalue: 1e-4 -num_threads 60 -seg yes -soft_masking true -culling_limit 3. To determine the abundance of the transcripts, the Illumina reads were mapped with bowtie2 (version 2.1.0) and SAMtools (version 0.1.18) back to the transcripts with the –a option to allow reads to map to multiple isoforms. To eliminate spurious alignment with paralogous gene families and among conserved domains, alignments were further filtered so that only those within an edit distance of 2 of the best match were kept. Read counts and reads per kilobase per million (RPKM) were determined at the transcript level and at the gene level for protein coding genes. To determine read counts and RPKMs at the gene level for protein-coding segments, we had to identify the gene segments themselves. This was challenging because ~20% of the gene clusters had 2 or more distinct protein coding segments, likely due to over assembly by Trinity using unstranded data. BLASTX data was used to identify distinct non-overlapping protein coding segments (presumably independent genes) on each isoform. By using custom Perl scripts employing a greedy strategy, the corresponding protein coding segments were identified across a set of related isoforms on the basis of shared protein-coding annotation. Read counts were then linked to corresponding protein coding gene segments in which a read was considered aligned if it overlapped a gene segment by at least 10 bps. All reads aligned to any of the corresponding protein coding gene segments were combined. Redundancy caused by a single read aligning to multiple isoforms of the same gene was removed, and the counts and RPKM (normalized within species) were then calculated. These read counts were then used to compare gene expression between male and female samples in each species through a differential gene expression analysis with DESeq2. Assembly quality was assessed by conducting a BUSCO analysis of completeness and fragmentation of transcripts versus the Actinopterygii dataset (Simao et al. 2015) and by using Trinity to calculate ExN50 curves.
Focal transcript analysis with gene ontology term filtering
We had a priori expectations for which gene classes were likely to be associated with the high-frequency firing that controls the EOD and/or with the hormonal regulation of sex differences in EODf. We therefore focused further analysis of the transcriptomes on genes associated with neuronal excitability (ion channels, gap junction proteins) or hormonal, neurotransmitter, and neuromodulatory regulation (ligands, receptors, metabolizing enzymes). We used a list of gene ontology (GO) terms related to these functions to filter the entire transcriptome for transcripts of interest (Supplemental Table 1). Protein coding gene segments were associated with GO terms of interest based on the human BLASTX output using gene2GO from NCBI (https://ftp.ncbi.nlm.nih.gov/gene/DATA/). Transcripts whose GO annotation matched terms on the list were extracted from the transcriptome. This process included all transcript annotated to any of the GO terms of interest. We further screened the filtered transcripts manually to identify those in specific categories that we most expected to be associated with sex differences in EODf: sodium channels; potassium channels; other voltage-gated ion channels; gap junction-related proteins; steroid hormone receptors and metabolizing enzymes; and neurotransmitters, neuromodulators, and their receptors.
Statistics for sex-biased expression
Sex-biased transcript expression in each species was evaluated with two approaches. At the whole-transcriptome level, we used the more conservative Benjamani-Hochberg false-discovery rate adjustment of p-values from DESeq2 (Benjamini and Hochberg 1995). For the approximately 200 transcripts of interest that were extracted from the entire transcriptome by using the GO-based and manual filtering described above, sex-biased expression was evaluated by correcting p-values for false discovery rate with an independent hypothesis weighting (IHW) approach (Ignatiadis et al. 2016). To assess whether expression of potentially sex-biased ion channel genes was correlated with EOD frequency independently of sex, we first calculated the residuals of the relationship between EODf or gene expression (RPKM) and sex (i.e., the difference between each individual’s EODf or RPKM value and the mean for its sex), and then calculated the Pearson’s correlations between the residuals of EODf vs. the residuals of the RPKM values.
RESULTS
EOD frequency, reproductive condition, plasma steroid hormone levels
As in previous studies, EOD frequency (EODf) was higher in males than in females in A. leptorhynchus, and higher in females than in males in A. albifrons (Table 1). Plasma levels of 11KT were similar in males of the two species, and plasma levels of E2 were similar in the females of the two species (although we only collected sufficient plasma to measure E2 in one of the female A. leptorhynchus; Table 1). 11KT levels were somewhat lower than observed in males in some previous studies (Dunlap et al. 1998). The gonadosomatic indices of males and females in both species, however, were as great or greater than in those studies and indicated that the fish were mature and had fully recrudesced gonads (Table 1; Dunlap et al. 1998; Kolodziejski et al. 2005). Although males tended to be larger than females in both species, body mass varied substantially; and the sex difference was not statistically significant.
Table 1.
Size, EOD Frequency, and Reproductive Condition of Subjects (mean ± SEM)
| A. leptorhynchus | A. albifrons | |||
|---|---|---|---|---|
| Males | Females | Males | Females | |
| Body mass (g) | 14.2 ± 3.5 | 7.7 ± 0.9 | 88.2 ± 38.7 | 34.7 ± 8.0 |
| Gonadosomatic Index (%) | 0.37 ± 0.07 | 7.7 ± 2.6* | 0.47 ± 0.08 | 6.2 ± 1.7* |
| EOD Frequency (Hz) | 847 ± 27 | 689 ± 16** | 940 ± 24 | 1041 ± 16* |
| 11-ketotestosterone (11-KT, ng/ml) | 1.4 ± 0.4 | N/Aa | 1.9 ± 0.8 | N/Aa |
| Estradiol (pg/ml) | N/Aa | 174b | N/Aa | 119 ± 17 |
t-test, Males v. Females,
p<0/05;
p<0.01.
11KT measured only in males; estradiol measured only in females.
Estradiol was measured in only one female A. leptorhynchus because insufficient plasma was obtained from the others.
Transcriptome properties
After trimming, the A. albifrons sequence data contained 3.27×108 reads which assembled into 931,466 transcipts. These transcripts corresponded to 18,694 teleost genes. The A. leptorhynchus sequence data contained 2.3×108 reads which assembled into 1,233,299 transcripts. These transcripts correspond to 21,585 teleost genes. We used the BUSCO (Benchmarking Universal Single-Copy Orthologs) dataset of genes from Actinopterygii (Simao et al. 2015) to assess the completeness and quality of the two transcriptomes. The results of this analysis are shown in Supplemental Table 2. Both transcriptomes contained ~82% complete BUSCO hits. Approximately half of the complete hits were single-copy and the other half were duplicated in the transcriptomes. Because the transcriptomes were derived from multiple individuals from wild populations, the duplicated BUSCOs likely reflect (1) alternative splicing/isoforms; (2) allelic variation (heterozygosity and/or polymorphism); and/or (3) gene duplications. The A. leptorhynchus and A. albifrons transcriptomes contained 7.6% and 7.8% fragmented BUSCO hits, respectively, reflecting a relatively low level of fragmentation in them. The 10.1–10.4% of BUSCOs missing in the two transcriptomes is to be expected, given that the transcriptomes were derived from a single tissue and expression of some genes in the BUSCO database are expected to be tissue-specific. ExN50 curves were also plotted to assess assembly quality (Supplemental Fig. 1). Both ExN50 curves had peaks near the ~90th Expercentile, with Ex90N50 values of 1100 and 1407, respectively for the A. albifrons and A. leptorhynchus transcriptomes, which suggests sufficient read depth was available for the assembly.
Differential expression analysis revealed a relatively small number of transcripts with significant sex-biased expression across the entire transcriptome of each species. In A. leptorhynchus, 47 genes were differentially expressed, with 26 genes upregulated in males and 21 upregulated in females (Supplemental Table 3A). In A. albifrons, 28 genes were differentially expressed between the sexes, with 16 upregulated in males and 12 upregulated in females (Supplemental Table 3B). Only 2 transcripts had significantly sex-biased expression in both species: one upregulated in males of both species (aspartylglucosaminidase, zgc:77327), and one upregulated in females of both species (arf-GAP with SH3 domain, ANK repeat and PH domain-containing protein 2).
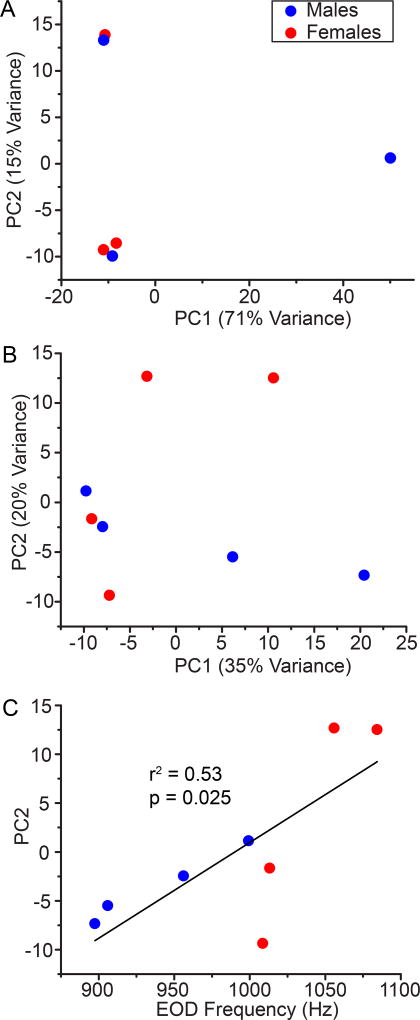
At the whole-transcriptome level, variation in expression across individuals within sex was as great or greater than that between sexes. Two principal components explained 86% and 55% of the individual variation in gene expression in A. leptorhynchus and A. albifrons, respectively (Fig. 1A, B). In neither case, however, did the sexes segregate cleanly based on these principal components (PCs). Interestingly, the second PC in A. albifrons was significantly correlated with EODf (r2 = 0.53, p = 0.025; Fig. 1C). None of the other PCs in either species were correlated with EODf (p>0.15).
Figure 1.
Principal components (PC) analysis of transcript expression in the Pn of A. leptorhynchus (A) and A. albifrons (B). Each data point represents PCs of expression in one individual (males blue circles, females red squares). Variance explained by each PC indicated in the axis title. Male and female PC values did not segregate cleanly in either species. (C) Correlation between EOD frequency (EODf) and PC2 in A. albifrons.
Analysis of GO-filtered transcripts
Because we had a priori hypotheses that certain classes of genes would be related to sex differences in electrocommunication, we focused subsequent analyses in several steps. The first step filtered transcripts to include only those whose annotation matched gene ontology (GO) terms related to hormones, neurotransmission, electrical coupling, and ion channels (see Methods). This filter extracted 3,174 and 3,595 transcripts of potential interest in A. leptorhynchus and A. albifrons, respectively. From these GO-filtered transcripts, we then manually extracted transcripts of particular interest in several categories based on our a priori hypotheses: (1) voltage-gated sodium channels; (2) voltage-gated potassium channels; (3) other ion channels and transporters; (4) connexins; (5) neurotransmitters, neuromodulators and their receptors; (6) sex steroid hormone receptors and metabolizing enzymes; and (7) other hormones and receptors. The transcripts in each of these categories and the abundance of their expression are shown in Tables 2–4 and Supplemental Tables 4A–4D.
Table 2.
Sex Steroid-Hormone Related Transcripts in the Pacemaker Nucleus
| A. leptorhynchus | A. albifrons | ||
|---|---|---|---|
| Short Annotation | RPKM (mean±SEM) |
Short Annotation | RPKM (mean±SEM) |
| Prohibitin 2a | 370 ± 39 | Membrane-associated Progesterone Receptor Component 1 | 251 ± 14 |
| Membrane-associated Progesterone Receptor Component 1 | 315 ± 59 | 17β-hydroxysteroid dehydrogenase 12b | 127 ± 7 |
| Aromatase (cyp19b) | 219 ± 135 | Prohibitin 1 | 113 ± 5 |
| Prohibitin 1 | 143 ± 19 | Prohibitin 2 | 97.6 ± 4.2 |
| 17β-hydroxysteroid dehydrogenase 12b | 114 ± 11 | 3β-hydroxysteroid dehydrogenase 7 (3βHSD 7) | 59.2 ± 8.4 |
| Nuclear receptor coactivator 4 (NCOA4) | 86.0 ± 7.7 | Nuclear receptor coactivator 4 (NCOA4) | 53.7 ± 2.9 |
| 17β-hydroxysteroid dehydrogenase 10 (17βHSD 10) | 79.6 ± 13.8 | 17βHSD 14 | 47.2 ± 3.7 |
| 17β-hydroxysteroid dehydrogenase 14 (17βHSD 14) | 53.2 ± 7.5 | 17βHSD 10 | 30.7 ± 2.0 |
| 3β-hydroxysteroid dehydrogenase 7 (3βHSD 7) | 42.7 ± 4.2 | Androgen receptor | 26.5 ± 3.0 |
| Androgen receptor | 38.4 ± 3.2 | Membrane-associated Progesterone Receptor Component 2 | 21.1 ± 1.5 |
| 11β-hydroxysteroid dehydrogenase 2 (11βHSD 2) | 35.3 ± 9.3 | 17β-hydroxysteroid dehydrogenase 8 (17βHSD 8) | 16.4 ± 1.3 |
| Membrane-associated Progesterone Receptor Component 2 | 28.9 ± 5.1 | 5α-reductase 2 (SRD2) | 10.7 ± 0.9 |
| Membrane-associated Progesterone receptor 2αB | 26.9 ± 2.0 | 5α-reductase 1 (SRD1) | 10.2 ± 0.7 |
| 17β-hydroxysteroid dehydrogenase 8 (17βHSD 8) | 18.6 ± 2.3 | Androgen receptor | 8.7 ± 0.8 |
| Membrane progesterone receptor G3 | 14.4 ± 2.9 | Nuclear receptor interacting protein 1 (NRIP1) | 8.1 ± 0.8 |
| 5α-reductase 1 (SRD1) | 12.1 ± 2.0 | G-Protein coupled estrogen receptor 1 (GPER1) | 6.6 ± 0.6 |
| Nuclear receptor interacting protein 1 (NRIP1) | 8.5 ± 0.7 | 3βHSD δ5 | 5.4 ± 0.6 |
| Estrogen receptor β | 8.1 ± 1.9 | Nuclear receptor coactivator 3 (NCOA3) | 4.0 ± 0.2 |
| 11β-hydroxysteroid dehydrogenase 1 (11βHSD 1) | 7.5 ± 0.6 | Nuclear receptor coactivator 3 (NCOA3) | 3.6 ± 0.2 |
| Membrane progesterone receptor αB | 7.4 ± 1.0 | Aromatase (cyp19b) | 3.0 ± 0.9 |
| 17β-hydroxysteroid dehydrogenase 12a | 7.1 ± 0.8 | 11β-hydroxysteroid dehydrogenase 1 (11βHSD 1) | 2.5 ± 1.0 |
| G-Protein coupled estrogen receptor 1 (GPER1) | 7.0 ± 1.4 | NCOA2 (Nuclear Receptor Co-activator) | 2.5 ± 0.3 |
| Nuclear receptor coactivator 3 (NCOA3) | 7.0 ± 0.9 | 17β-hydroxysteroid dehydrogenase 12 (17βHSD 12) | 2.1 ± 0.6 |
| 17β-hydroxysteroid dehydrogenase 3 (17βHSD 3) | 5.0 ± 37 | 11β-hydroxysteroid dehydrogenase 2 (11βHSD 2) | 2.0 ± 0.2 |
| 3β-hydroxysteroid dehydrogenase 5 (3βHSD 5) | 4.3 ± 1.2 | Estrogen receptor α | 1.7 ± 0.2 |
| 5α-reductase 2 (SRD2) | 3.0 ± 0.9 | ||
| Estrogen receptor α | 2.4 ± 1.3 | ||
Table 4.
Potassium Channel Transcripts in the Pacemaker Nucleus
| A. leptorhynchus | A. albifrons | ||
|---|---|---|---|
| Short Annotation | RPKM (mean±SEM) |
Short Annotation | RPKM (mean±SEM) |
| Kv1.8 (KCNA10) | 384 ± 18 | Kv1.8 (KCNA10) | 296 ± 29 |
| Kvβ2 (KCNAB2) | 190 ± 14 | Kir1.2 (KCNJ10) | 207 ± 22 |
| Kir1.2 (KCNJ10) | 84.6 ± 13.9 | Kvβ2 (KCNAB2) | 189 ± 20 |
| Kv3.4 (KCNC4) | 27.1±3.2 | K2P1.1 (KCNK1, TWIK-1) | 66.4 ± 4.9 |
| MinK related peptide 4 (KCNE4) | 26.9±7.0 | KCa2.3 (KCNN3, SK3, Small conductance KCa) | 62.0 ± 6.5 |
| Kv3.2 (KCNC2) | 25.2±5.1 | Kv3.4 (KCNC4) | 28.6 ± 1.9 |
| K2p13.1 (KCNK13, THIK-1) | 23.6 ±13.0 | Kv3.3 (KCNC3) | 25.2 ± 1.3 |
| KCa2.3 (KCNN3, SK3, Small conductance KCa) | 21.6 ±2.8 | K2p1.1b (KCNK1b, TWIK-1b) | 22.0 ± 2.4 |
| K2P1.1 (KCNK1, TWIK-1) | 18.9 ±5.2 | MinK related peptide 4 (KCNE4) | 20.3 ± 1.6 |
| Kv7.5 (KCNQ1) | 12.4 ±1.2 | K2P18.1 (KCNK18, TWIK -18) | 20.0 ± 2.0 |
| Kv11.3 (KCNH7, ether-a-gogo related channel 3) | 11.5 ± 1.4 | Kv7.5 (KCNQ1) | 16.6 ± 1.0 |
| Kv3.3 (KCNC3) | 9.8 ± 2.0 | KChIP1 (KCNIP1) | 16.4 ± 2.4 |
| Kv11.1 (KCNH2, ether-a-gogo 1) | 9.7 ± 1.7 | Kv11.1 (KCNH2, ether-a-gogo 1) | 14.4 ± 1.3 |
| BKβ4 (KCNMB4, Large conductance KCa β4) | 9.3 ± 3.6 | K2P12.1 (KCNK12, THIK-2) | 12.3 ± 1.0 |
| Kvβ1 (KCNAB1) | 8.6 ± 1.4 | Kv6.1 (KCNG1) | 9.3 ± 1.2 |
| KChIP4 (KCNIP4, K+ channel interacting protein 4) | 8.2 ± 3.4 | Kir2.1 (KCNJ2) | 9.0 ± 1.1 |
| Kir2.1 (KCNJ2) | 7.9 ± 1.2 | KChIP2 (KCNIP2) | 7.6 ± 1.4 |
| K2P12.1 (KCNK12, THIK-2) | 7.9 ± 1.5 | K2p13.1 (KCNK13, THIK-1) | 7.2 ± 1.1 |
| KChIP2 (KCNIP2) | 7.4 ± 1.2 | Kir3.3 (KCNJ9, GIRK-3) | 6.4 ± 0.7 |
| Kir3.1 (KCNJ3, GIRK1) | 5.9 ± 1.9 | Kv8.1 (KCNV1) | 6.1 ± 0.9 |
| Kv6.3/Kv10.1 (KCNG3) | 5.3 ± 1.1 | Kir4.1 (KCNJ10) | 6.0 ± 0.5 |
| K2P2.1 (KCNK2, TREK-1) | 5.3 ± 2.1 | K2P5.1 (KCNK5, TASK-2) | 5.8 ± 0.5 |
| Kv8.1 (KCNV1) | 5.3 ± 0.7 | Kv6.3 (KCNG3) | 5.1 ± 0.8 |
| Kv1.4 (KCNA4) | 4.9 ± 0.5 | Kvβ1 (KCNAB1) | 5.0 ± 0.6 |
| Kir4.1 (KCNJ10) | 4.7 ± 0.5 | Kv11.3 (KCNH7, ether-a-gogo related channel 3) | 4.1 ± 0.6 |
| BKβ2 (KCNMB2) | 4.6 ± 0.8 | KChIP4 (KCNIP4) | 4.0 ± 0.8 |
| K2P5.1 (KCNK5, TASK-2) | 4.6 ± 0.8 | BKβ4 (KCNMB4, Large conductance KCa β4) | 3.8 ± 0.7 |
| Kv6.1 (KCNG1) | 4.3 ± 1.2 | Kir3.1 (KCNJ3, GIRK1) | 3.6 ± 0.6 |
| KCHIP1 (KCNIP1) | 3.8 ± 1.2 | BKβ2 (KCNMB2) | 3.5 ± 0.5 |
| Kir6.2 (KCNJ11) | 3.7 ± 0.7 | KCa2.2 (KCNN2, SK2) | 3.3 ± 0.3 |
| Kv5.1 (KCNF2) | 3.5 ± 3.2 | Kv4.2 (KCND2) | 3.2 ± 0.3 |
| Kir7.1 (KCNJ13) | 3.5 ± 1.6 | Kv6.4 (KCNG4) | 3.1 ± 0.5 |
| K2P12.1 (KCNK15, TASK-5) | 3.4 ± 0.9 | Kir6.2 (KCNJ11) | 3.1 ± 0.4 |
| KCa2.2 (KCNN2, SK2) | 3.3 ± 0.2 | MinK1 (KCNE1) | 3.0 ± 0.3 |
| Kv4.3 (KCND3) | 3.1 ± 0.4 | Kv1.4 (KCNA4) | 2.8 ± 0.3 |
| KCa1.1 (KCNMA1, BKα1) | 3.0 ± 1.5 | KCa2.1 (KCNN1, SK1) | 2.5 ± 0.5 |
| SLACK (KCNT1) | 2.5 ± 1.5 | Kvβ2 (KCNAB2) | 2.1 ± 0.8 |
| Mink1 (KCNE1) | 2.4 ± 0.7 | Kv4.1 (KCND1) | 1.9 ± 0.2 |
| Kir3.3 (KCNJ9, GIRK-3) | 2.4 ± 0.6 | Kv4.3 (KCND3) | 1.6 ± 0.2 |
| Kv4.1 (KCND1) | 2.0 ± 0.5 | K2P2.1 (KCNK2, TREK-1) | 1.1 ± 0.1 |
| Kv4.2 (KCND2) | 1.9 ± 0.5 | Kv1.2 (KCNA2) | 1.0 ± 0.3 |
| KCa2.1 (KCNN1, SK1) | 1.4 ± 0.7 | KCa1.1 (KCNMA1, BKα1) | 1.3 ± 0.3 |
| KCa1.1 (KCNMA1, BKα1) | 1.1 ± 0.7 | Kv2.1 (KCNB1) | 0.9 ± 0.4 |
| Kv1.2 (KCNA2) | 1.1 ± 0.1 | ||
Hormone-related genes
Sex differences in EODf are mediated by effects of androgens and/or estrogens on the firing rates of neurons in the Pn (Schaefer and Zakon 1996; Zakon and Dunlap 1999). We therefore hypothesized that the Pn would express genes whose products mediate sex steroid hormone action and that species differences in sexual dimorphism and hormonal regulation of EODf might be associated with differences in the expression of these genes.
Transcripts for numerous sex steroid receptors, metabolizing enzymes, and co-factors were expressed in the Pn of both A. leptorhynchus and A. albifrons (Table 2). The Pn of both species robustly expressed mRNA for androgen receptors (AR). Estrogen receptor β (ERβ) was expressed in the Pn in A. leptorhynchus, but not in A. albifrons; and estrogen receptor α (ERα) was expressed at low levels in the Pn of both species. In addition, the Pn of both species expressed mRNA for the membrane-bound G-protein coupled estrogen receptor (GPER1) and several membrane-associated progesterone receptors. The Pn also expressed transcripts encoding receptors for other hormones, including receptors for glucocorticoids/mineralocorticoids, thyroid hormone, parathyroid hormone, gonadotropin releasing hormone and growth hormone releasing hormone (Supplemental Table 4A).
Transcripts for numerous steroidogenic enzymes were expressed in the Pn of both species (Table 2): aromatase (cyp19b), which converts androgens to estrogens; 5α-reductase (SRD1 and SRD2), which converts testosterone into the nonaromatizable androgen 5α-dihydrotestosterone (5α-DHT); 11β-hydroxysteroid dehydrogenases (11βHSD1 and 11βHSD2), which convert 11β-hydroxytestosterone into the fish androgen 11-ketotestosterone (11KT, Kusakabe et al. 2003); and several different 17β-hydroxysteroid dehydrogenases (17βHSD8, 17βHSD10, 17βHSD12, and 17βHSD14), which oxidize and/or reduce androgens and estrogens to more inactive/active metabolites (Labrie et al. 2000). The Pn of both species also expressed transcripts for co-factors that modulate the transcriptional activity of androgen and/or estrogen receptors: nuclear receptor coactivators/steroid receptor coactivators 2, 3, and 4 (NCOA2, NCOA3, NCOA4); nuclear receptor interacting protein 1 (NRIP1); and Prohibitin 1 and 2.
Although most hormone-related genes were expressed at similar levels in the Pn of both species, the relative expression of some hormone-related genes differed substantially between the two species. The transcript for estrogen receptor β (ESR2) was moderately expressed in the A. leptorhynchus Pn, but was absent in the A. albifrons Pn (Table 2). Similarly, transcripts for aromatase and 11β-HSD 2 were expressed at substantially higher levels in the Pn of A. leptorhynchus than in A. albifrons. Pn expression of most sex steroid-related genes did not differ between the sexes. The only hormone-related genes with any indication of sex-biased expression in the Pn were 17β-HSD 12a (slightly greater expression in females in A. leptorhynchus), ERα (slightly greater expression in males in A. leptorhynchus), Prohibitin-1 (slightly greater expression in females in A. albifrons), and NRIP-1 (slightly greater expression in males in A. albifrons; Tables 5A, 5B). The Pn in female A. albifrons also had greater expression of thyroid hormone receptor α mRNA than male Pns (Table 5B).
Table 5.
| A. Transcripts of Interest with Potentially Sex-Biased Expression in the A. leptorhynchus Pacemaker Nucleus | |||
|---|---|---|---|
| Short Annotation | Male RPKM | Female RPKM | IHW p-valuea |
| Hormone-Related Genes | |||
| 17β-HSD 12a | 6.0 ± 1.0 | 8.1 ± 0.8 | 0.024 |
| Estrogen Receptor α | 3.8 ± 2.5 | 1.0 ± 0.3 | 0.036 |
| Na+/K+ Channels | |||
| Kvβ2 (KCNAB2) | 170 ± 19 | 210 ±15 | 0.040 |
| K2p13.1 (KCNK13, THIK-1) | 37.3 ± 25.4 | 10.0 ±3.6 | 0.019 |
| BKβ4 (KCNMB4, Large conductance KCa β4) | 14.4 ± 5.9 | 4.1 ± 1.8 | 0.030 |
| KChIP4 (KCNIP4, K+ channel interacting protein 4) | 4.8 ± 2.3 | 11.5 ± 6.6 | 0.018 |
| Kir3.1 (KCNJ3, GIRK1) | 3.8 ± 1.1 | 8.1 ± 3.6 | 0.022 |
| Other Ion Channels/Transporters | |||
| TRPM2 (Transient receptor potential channel M2) | 10.0 ± 1.0 | 12.9 ± 0.8 | 0.014 |
| TRPM6 | 2.0 ± 0.0 | 3.6 ± 0.5 | 0.028 |
| Na+/K+ ATPase β3b | 3.9 ± 1.2 | 1.3 ± 0.3 | 0.033 |
| Neurotransmitters/Neuromodulators and Receptors | |||
| TARPγ2 (CACNG2) | 40.4 ± 3.8 | 49.9 ±2.5 | 0.019 |
| Neuropeptide Y Receptor Y8 | 10.1 ± 1.6 | 16.3 ±0.7 | 0.022 |
| GluR5 | 9.0 ± 0.6 | 13.1 ± 1.6 | 0.014 |
| Prolactin Releasing Peptide Receptor | 14.7 ± 1.4 | 5.8 ± 0.5 | <0.01 |
| GluR2 | 6.9 ± 0.4 | 9.2 ± 0.3 | 0.027 |
| Dopamine D2 Receptor | 10.4 ± 5.7 | 3.3 ± 1.5 | 0.04 |
| Nicotinic acetylcholine receptor α4 (nAChRα4) | 9.5 ± 4.2 | 2.2 ± 0.5 | <0.01 |
| Dopamine β-hydroxylase | 0.2 ± 0.1 | 1.3 ± 0.4 | 0.032 |
| B. Transcripts of Interest with Potentially Sex-Biased Expression in the A. albifrons Pacemaker Nucleus | |||
|---|---|---|---|
| Short Annotation | Male RPKM | Female RPKM | IHW p-valuea |
| Hormone-Related Genes | |||
| Prohibitin-1 | 108.5 ± 6.1 | 116.8 ± 9.2 | 0.029 |
| Nuclear Receptor Interacting Protein 1 (NRIP1) | 9.3 ± 0.9 | 6.9 ± 1.0 | 0.011 |
| Thyroid Hormone Receptor α | 16.1 ± 1.5 | 23.4 ± 2.4 | <0.001 |
| Na+/K+ Channels | |||
| Navβ1 (Danio rerio splice variant D, zβ1D, SCN1B) | 6.4 ± 0.9 | 3.6 ± 0.3 | 0.045 |
| K+ Channnel-Interacting Protein 2 (KChIP2, KCNIP2) | 10.4 ± 1.7 | 4.7 ± 1.0 | <0.001 |
| Kv6.3 (KCNG3) | 3.6 ± 0.5 | 6.6 ± 1.3 | <0.01 |
| Kv6.4 (KCNG4) | 2.3 ± .05 | 3.9 ± 0.5 | <0.01 |
| Kir3.1 (KCNJ3, GIRK1) | 3.0 ± 2.0 | 0.5 ± 0.2 | 0.022 |
| Kv1.2 (KCNA2) | 1.6 ± 0.3 | 0.5 ± 0.1 | <0.01 |
| Kv2.1 (KCNB1) | 1.5 ± 0.7 | 0.2 ± 0.2 | 0.032 |
| Kv1.1 (KCNA1) | 1.2 ± 0.7 | 0.1 ± 0.1 | 0.019 |
| Other Ion Channels/Transporters | |||
| Na+/K+ ATPase α3 | 22.3 ± 3.5 | 10.1 ± 0.7 | <0.001 |
| Acid-Sensing Ion Channel 1 (ASIC-1) | 9.9 ± 2.2 | 4.8 ± 0.5 | 0.011 |
| Cavβ2 (CACNB2, L-type Ca2+ channel β2 subunit) | 5.1 ±1.0 | 2.0 + 0.1 | <0.001 |
| Na+/K+ ATPase β2a | 4.9 ± 1.6 | 1.6 ± 0.3 | 0.030 |
| Na+/K+ ATPase α1a | 2.9 ± 0.7 | 0.9 ± 0.2 | 0.018 |
| Connexins | |||
| Connexin 32.2 (Astyanax mexicanus) | 140 ± 20 | 94 ± 8 | 0.011 |
| Gap Junction Protein β1 (Connexin 29/32) | 72.0 ± 4.6 | 83.5 ± 6.0 | <0.01 |
| Neurotransmitters/Neuromodulators and Receptors | |||
| Prepro-Neuropeptide Y | 27.2 ± 3.5 | 42.4 ± 7.9 | 0.011 |
| GABA Receptor β2 | 39.2 ± 3.2 | 27.2 ± 3.5 | 0.021 |
| Proenkephalin | 15.9 ± 5.1 | 4.9 ± 1.3 | 0.015 |
| Adenosine Receptor A1 | 23.6 ± 2.1 | 27.0 ± 3.6 | 0.047 |
| GABA Receptor ρ2 | 10.6 ± 3.4 | 3.6 ± 0.6 | <0.01 |
| Neuropeptide FF | 1.6 ± 1.2 | 11.0 ± 1.6 | <0.001 |
| Dopamine D2 Receptor | 9.1 ± 3.4 | 3.3 ± 1.3 | 0.031 |
| 5HT1B Receptor | 2.5 ± 0.5 | 4.8 ± 1.2 | 0.022 |
| 5HT1A Receptor | 3.4 ± 0.6 | 1.6 ± 0.4 | 0.017 |
| Substance P Receptor | 3.6 ±0.4 | 1.2 ± 0.3 | <0.001 |
| Dopamine D1B Receptor | 2.7 ± 1.1 | 0.4 ± 0.1 | 0.022 |
| VIP Receptor | 2.3 ± 1.3 | 0.2 ± 0.1 | 0.024 |
Independent Hypothesis-Weighting-corrected p-value with mean normalized read counts as a covariate (Ignatiadis et al., 2016).
Independent Hypothesis-Weighting-corrected p-value (Ignatiadis et al., 2016).
Ion channel and gap junction genes
Sexually dimorphic EOD frequency is controlled directly by the firing rates of pacemaker and relay cells in the Pn, and the firing rates of these neurons are strongly influenced by sodium and potassium currents (Smith and Zakon 2000). The most abundantly expressed sodium channel α-subunit mRNA in both species was SCN8A, which codes for the Nav1.6 Na+ channel (Table 3). In addition, the Pn of A. leptorhynchus, but not A. albifrons, expressed low levels of transcripts for the skeletal muscle sodium channel SCN4AB (Nav1.4b) and for the SCN2A (Nav1.2) α-subunit. The Pn of both species expressed high levels of transcripts for sodium channel beta subunits (SCN1B-4B = Navβ1–4), whose products modify biophysical properties of sodium channels. Additionally, the Pn in both species expressed non-selective Na+ leak channel transcripts (NALCN).
Table 3.
Sodium Channel Transcripts and Related Transcripts in the Pacemaker Nucleus
| A. leptorhynchus | A. albifrons | ||
|---|---|---|---|
| Short Annotation | RPKM (mean±SEM) |
Short Annotation | RPKM (mean±SEM) |
| Navβ4b (SCN4BB) | 498 ± 50 | Navβ4b (SCN4BB) | 287 ± 24 |
| Nav1.6 (SCN8A) | 49.0 ± 2.7 | Navβ3 (SCN3B) | 129 ± 10 |
| Navβ3 (SCN3B) | 41.2 ± 4.8 | Navβ1b (SCN1BB) | 78.2 ± 8.0 |
| Navβ1b (SCN1BB) | 9.0 ± 0.2 | Nav1.6 (SCN8A) | 22.4 ± 2.7 |
| Na+ non-selective leak channel (NALCN) | 8.6 ± 1.0 | Na+ non-selective leak channel (NALCN) | 15.1 ± 1.7 |
| Navβ2 (SCN2B) | 7.4 ± 1.0 | Navβ2 (SCN2B) | 9.9 ± 1.1 |
| Navβ4a (SCN4BA) | 3.7 ± 1.7 | Navβ1 (Danio rerio splice variant D, zβ1D, SCN1B) | 5.0 ± 0.7 |
| Nav1.4b (SCN4AB) | 3.1 ± 0.8 | ||
| Nav1.2 (SCN2A) | 2.9 ± 2.0 | ||
Potassium channels are highly diverse, and numerous K+ channel transcripts were expressed in the Pn of both species (Table 4). The suite of K+ channel genes expressed was well-conserved between A. albifrons and A. leptorhynchus. Thirty-eight K+ channel genes were expressed in the Pn of both species; and most of the transcripts expressed in only one of the two species (3 in A. albifrons and 5 in A. leptorhynchus) were expressed at low levels. The most abundant K+ channel transcript in both species was KCNA10, which codes for the Kv1.8 voltage-gated K+ channel. Transcripts for the β2 K+ channel regulatory subunit (KCNAB2) and the Kir1.2 inward rectifier K+ channel (KCNJ10) were also abundant in the Pn both species. Additionally, the Pn in both species substantially expressed transcripts for Kv3 voltage-gated channels (KCNC2, KCNC3, and/or KCNC4), which are associated with high-frequency firing (Gan and Kaczmarek 1998), and the small conductance calcium-dependent K+ channel SK3 (KCNN3).
The Pn of both species expressed transcripts for voltage-gated Ca2+ channels, and the transcript profile was well-conserved between the two species (Supplemental Table 4B). Ten transcripts for Ca2+ channel α, α/δ, and β subunits were found in the Pn of both species. Only one Ca2+ channel transcript (CACNA1H, which forms T-type Ca2+ channels) was expressed in only one of the species (A. albifrons), and its abundance was low.
The Pn of both species expressed low to moderate levels of transcripts for several other ion channel types, including chloride channel 2 (CLCN2); acid-sensing ion channels (ASIC1); hyperpolarization-activated cation channels (HCN1 and HCN2); and transient receptor potential channels (TRPM2, TRPM4, TRPM6, and TRPN1, Supplemental Table 4B). The expression of these channel types was largely conserved, with eight of the transcripts expressed in the Pn of both species, one (ASIC3) expressed only in A. leptorhynchus, and one (TRPA1) expressed only in A. albifrons.
To achieve the highly synchronous firing rates of over 1 kHz needed to drive the EOD, the pacemaker and relay neurons in the Pn are extensively electrically coupled (Elekes and Szabo 1985). Not surprisingly, the Pn in both species expressed high levels of numerous connexin genes, which encode the proteins that form gap junctions (Supplemental Table 4C). The connexin transcripts expressed in the Pn was largely conserved across species. The two most abundant connexin transcripts in both species were gap junction proteins β1 and γ1, which code 32 kD and 45 kD connexins, respectively. Nine gap junction transcripts were expressed in the Pn of both species, one (δ1) was expressed only in A. leptorhynchus, and two (α1 and α5) were expressed only in A. albifrons.
The high frequency firing of Pn neurons also requires that the Na+ and K+ gradients be continuously maintained. It is thus not surprising that the Pn of both species expressed high levels of transcripts for ion channel transporters (Supplemental Table 4B). Both α- and β-subunits of Na+/K+ ATPases were expressed in the Pn of both species, with α1 and α3 as the most abundant α subunits, and the teleost-specific β233 as the most abundant β subunit. In addition, the Pn of both species robustly expressed transcripts for the electrogenic sodium-bicarbonate co-transporter.
Potentially sex-biased expression of ion channel and transporter genes
Sex differences in EOD frequency are likely caused by sex differences in expression or function of ion channels or other proteins regulating the firing rates of neurons in the Pn (Smith and Zakon 2000). We therefore used independent hypothesis weighting of the differential expression data to ask whether any ion channel, ion transporter, or connexin genes might have sex-biased expression. No sodium channel transcripts differed in Pn expression between males and females in A. leptorhynchus (Table 5A), but one sodium channel beta subunit (annotated to the D-splice variant of the Danio rerio Nav1β subunit, SCNB1), was expressed at higher levels in the Pn of males vs. females in A. albifrons (Table 5B).
Several K+ channel genes had sex-biased expression in each species (Table 5A,5B). In A. leptorhynchus, two genes that form functional voltage-dependent K+ channels were sex-biased. The two-pore K+ channel K2p13.1, which contributes to an inward-rectifying background K+ current (Kang et al. 2014), was expressed at higher levels in the Pn of males than females; and the G-protein modulated inward rectifier Kir3.1 was expressed more in the Pn of females than males. Three genes whose products modify K+ function were sex biased in the Pn of A. leptorhynchus. Kvβ2, which affects the inactivation kinetics of Kv1 channels (Torres et al. 2007), and KChIP4, which affects current density and inactivation of Kv4 channels (Norris et al. 2010), were expressed at higher levels in female Pns than in male Pns. Similarly, in A. albifrons, sex-biased genes included both those whose products form functional channels as well as genes whose products modify K+ channel function. Transcripts for Kir3.1, Kv1.2, Kv2.1, and Kv1.1 channels were all expressed at higher levels in male Pns than in female Pns. Transcripts for KChIP2, which modifies Kv4 channel kinetics, were expressed more in the Pn of males than of females. Transcripts for Kv6.3 and Kv6.4, which slow deactivation kinetics of Kv2 channels (Sano et al. 2002), were expressed at higher levels in female Pns than male Pns.
Other ion channel and transporter transcripts also had sex-biased expression in the Pn. In A. leptorhynchus, transcripts for two transient receptor potential channels (TRPM2 and TRPM6), which are cation channels modulated by temperature and/or nucleotides, were expressed at higher levels in the Pn of females vs. males (Table 5A). In A. albifrons, transcripts for the acid sensing ion channel, ASIC-1, and the Cavβ2 subunit, which modulates inactivation of L-type Ca2+ channels, were expressed at higher levels in male Pns than in female Pns (Table 5B). Transcripts for some sodium-potassium ATPase subunits were expressed at higher levels in the Pn of males vs. females (Na+/K+ ATPase β3b in A. leptorhynchus and Na+/K+ ATPases α3, α1a, and β2a in A. albifrons). Two transcripts for gap junction proteins were sex-biased in the A. albifrons Pn: a transcript annotating to connexin 32.2 of Astyanax mexicanus (greater expression in males) and gap junction protein β1 (Connexin 32/29, greater expression in females).
Ion channel gene expression correlated with EOD frequency
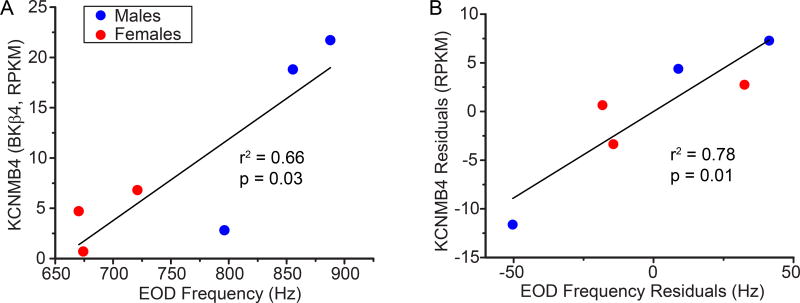
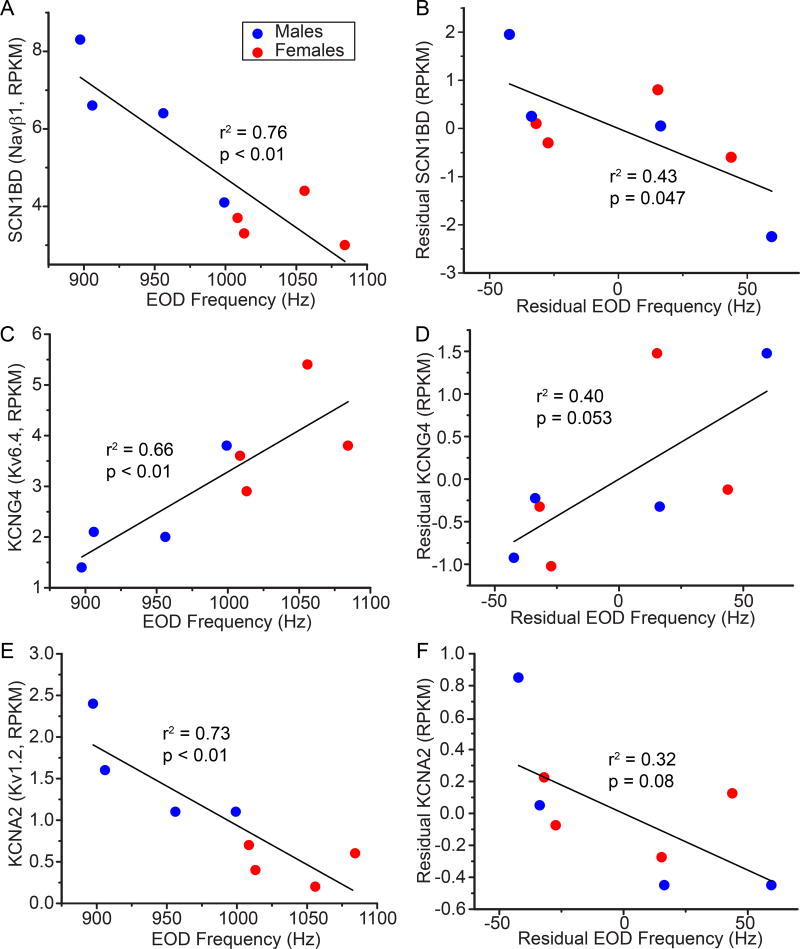
To further test whether ion channel genes with potentially sex-biased expression are linked to sex and species differences in EOD frequency, we asked whether expression of these genes was correlated with EODf. Potential correlations between EODf and sex-biased gene expression are confounded by sex differences in both EODf and gene expression. To assess whether expression of these transcripts was correlated with EODf independently of sex (i.e., whether variation in gene expression was correlated with EODf within sexes), we calculated the residuals of EODf and transcript expression vs. sex, and then tested for correlations between these residuals. In A. leptorhynchus, the only ion channel gene whose expression was correlated with EODf after sex differences were factored out was KCNMB4, which codes for the β4 subunit of the large-conductance Ca2+-dependent K+ channel. KCNMB4 was expressed more in the Pn of A. leptorhynchus with higher EODf (Fig. 2). In A. albifrons, SCN1B(D), which codes for a teleost-specific isoform of the β1 sodium channel subunit, was expressed at significantly higher levels in the Pn of fish with lower EODf (Fig. 3A,B). The KCNG4 transcript, which codes for the Kv6.4 K+ channel subunit, tended to be expressed more in the Pn of A. albifrons with higher EODf (Fig. 3C,D); and KCNA2 transcript, which codes for Kv1.2 K+ channels, tended to be expressed more in the Pn of A. albifrons with low EODf (Fig. 3E,F). Neither of these latter two relationships reached significance, however.
Figure 2.
Expression of transcripts for the large-conductance Ca2+-dependent K+ channels beta subunit (BKβ4, KCNMB4) in the Pn is related to EOD frequency (EODf) in A. leptorhynchus. (A) Fish with higher EODf had greater expression of KCNMB4 in the Pn (r2 = 0.66, p = 0.03). Because males (blue circles) have higher EODf than females (red squares), this relationship is confounded by sex differences in both EODf and KCNMB4 expression. (B) Relationship between the sex-specific residuals of EODf and KCNMB4 expression to factor out the effects of sex differences. Within each sex, individuals with higher EODf had greater KCNMB4 expression than individuals with lower EODf (r2 = 0.78, p = 0.01).
Figure 3.
Expression of sex-biased Na+ and K+ channel genes correlated with EODf in A. albifrons. (A, C, E) Expression of transcripts (RPKM) vs. EODf; (B,D,F) Relationships between sex-specific residuals of gene expression and EODf (sex differences factored out). Fish with lower EODf (males, blue circles) had greater expression of Na+ channel β1 subunit D-isoform (A, SCN1BD, Navβ1) and Kv1.2 K+ channels (E, KCNA2) than fish with higher EODf (females, red squares). (C) The K+ channel modifier KCNG4 (Kv6.4) was expressed at higher levels in the Pn of fish with higher EODf (females). After factoring sex out, the relationship between EODf and channel gene expression remained significant for SCN1BD (B), but was only marginally significant for KCNG4 (D) and KCNA2 (F).
Expression of neurotransmitter and neuromodulator-related genes
Numerous transcripts associated with neurotransmission and neuromodulation were expressed in the Pn of both species (Supplemental Table 4D). The Pn of both species expressed a diversity of transcripts for glutamate receptors, including AMPA, kainate, NMDA, and metabotropic receptors. Sixteen glutamate receptor transcripts were expressed in the Pn of both species, with two ionotropic glutamate receptors (GluR2b and GluR4) expressed only in A. leptorhynchus, and NMDA receptor 3 expressed only in A. albifrons. Similarly, the Pn in both species expressed transcripts for several different ionotropic GABA-A and glycine receptors as well as metabotropic GABA-B receptors (Supplemental Table 4D).
The Pn of both species expressed mRNA for several nicotinic acetylcholine receptors (Supplemental Table 4D). Transcripts for the metabotropic acetylcholine receptor M5 was moderately abundant in the Pn of A. leptorhynchus, but was not found in the Pn of A. albifrons. The Pn of both species expressed transcripts for β2 adrenergic receptors and D2 dopamine receptors, but differed in the expression of serotonin receptor genes. 5HT4 receptor mRNA was present in the Pn of A. albifrons, but not A. leptorhynchus; and the 5HT3 and 5HT1B receptor genes were expressed in the Pn of A. leptorhynchus, but not A. albifrons. Numerous purine receptors, including ionotropic P2X receptors and metabotropic P1/adenosine receptors and P2Y receptors, were expressed in the Pn of both species.
Transcripts for several neuropeptides and their receptors were also expressed in the Pn, though they were less well-conserved across species than other neurotransmitter/neuromodulator receptors (Supplemental Table 4D). Twelve neuropeptide/receptor transcripts were expressed in the Pn of both species; six transcripts (prolactin releasing peptide receptor, neuropeptide Y receptor 2, cholecystokinin, μ-opioid receptor, relaxin 3, and arginine vasopressin receptor 1) were expressed only in A. leptorhynchus; and four transcripts (cholecystokinin receptor, corticotropin releasing factor receptor, tachykinin receptor, and relaxin 3 receptor) were expressed only in A. albifrons. Moreover, the relative abundance of some transcripts differed substantially between species. The transcript for Prepro-neuropeptide Y was the most abundant neuropeptide transcript in A. albifrons, but was expressed at very low levels in the Pn of A. leptorhynchus. Similarly, neuropeptide FF receptor 2 transcripts were moderately abundant in A. albifrons, but were expressed at very low levels in A. leptorhynchus.
DISCUSSION
The goal of this study was to use transcriptomics on the Pn of two electric fish species to identify hormone-related genes that might regulate sex differences in EODf, to identify genes involved in neuronal excitability that might underlie the high frequency rhythm that drives the EOD, and to identify putatively sex-biased ion channel genes whose differential expression might underlie sex differences in EODf. The data in this study provided substantial insights towards these goals. Transcripts for numerous sex steroid hormone receptors and metabolizing enzymes were identified in the Pn. Similarly, we found expression of Na+ and K+ channel genes whose products are likely to contribute to high frequency firing of Pn neurons. In addition, some of these ion channel genes had expression that was potentially sex-biased and/or correlated with EODf.
At the level of the entire Pn transcriptome, we identified relatively few transcripts with strongly sex-biased expression, and the gene expression patterns in the Pn did not cleanly segregate by sex. The paucity of highly sex-biased gene expression in the Pn might be explained by the highly conserved and specialized function of the Pn in generating the command signal for the EOD. Apteronotid Pn neurons are unique in that they fire continuously and highly synchronously at rates that can exceed 1500 Hz. The suite of genes needed to produce this exceptional feat may be constrained, and relatively small changes in gene expression may mediate the 15–30% difference in firing rate between males and females. Interestingly, in A. albifrons, one of the PCs for transcript expression in the Pn was significantly correlated with EODf, which suggests that at least some of the large-scale variation in Pn gene expression may be related to variation in Pn firing rates.
Hormone-related transcripts and hormonal regulation of EOD sex differences
The expression of transcripts for sex steroid hormone receptors and metabolizing enzymes in the Pn is consistent with the role of these hormones in regulating sex differences in EODf. 11-ketotestosterone robustly masculinizes EODf in both A. leptorhynchus and A. albifrons, and does so by changing the firing rates of neurons in the Pn (Schaefer and Zakon 1996; Dunlap et al. 1998). Consistent with this, the Pn of both species robustly expressed transcripts for androgen receptors.
Differences between these species in hormone-related gene expression are also consistent with differences in hormonal regulation of EODs. Testosterone levels are similar in males and females, and in A. leptorhynchus, testosterone feminizes, rather than masculinizes, EODf (Dulka and Maler 1994; Dunlap et al. 1998; Zakon and Dunlap 1999). The feminizing effect of testosterone is thought to be mediated via aromatization to estradiol, because estradiol also feminizes EODf, and the feminizing effects of testosterone are blocked by fadrazole (Schaefer and Zakon 1996; Zucker 1998). In contrast, in A. albifrons, testosterone does not affect EODf (Dunlap et al. 1998). Species differences in the feminizing effects of testosterone might be explained by the differences in the expression of aromatase and estrogen receptor genes reported here. The Pn of A. leptorynchus expressed high levels of aromatase mRNA (mean RPKM > 200) and moderate levels of estrogen receptor β, whereas aromatase was expressed at much lower levels in A. albifrons (mean RPKM of 3.0), and ERβ was not expressed at all. Thus, the feminizing effects of testosterone in A. leptorhynchus may result from robust local aromatization that does not occur in A. albifrons.
The expression of moderate levels of 5α-reductase in the Pn of both species was somewhat surprising. 11-ketotestosterone (11KT) is the primary nonaromatizable masculinizing androgen in teleosts (Fostier et al. 1983). 5α-dihydrotestosterone (5αDHT) has often been used as a nonaromatizable androgen to experimentally to test for androgenic effects on behavior in electric fish (Meyer and Zakon 1982; Meyer 1983, 1984; Bass and Hopkins 1985; Mills and Zakon 1987; Dunlap et al. 1998); the rationale has been that 5αDHT is a potent but less expensive and more readily available androgen than 11KT. The expression of 5α-reductase in the Pn raises the possibility that testosterone may be locally metabolized into 5αDHT in the Pn and that 5αDHT may be an endogenously masculinizing androgen in these fish. 5α-reductase activity has been previously reported in the brains of goldfish and toadfish (Diotel et al. 2011). Interestingly, although 5αDHT masculinizes EOD frequency in many species of electric fish, including A. albifrons (Dunlap et al. 1998), it reportedly does not affect EODf in A. leptorhynchus (Meyer et al. 1987; Dulka 1997; but see Zucker 1998). If true, these findings suggest that 5αDHT may either be an ineffective ligand of the androgen receptor in A. leptorhynchus or that it might be metabolized before reaching the Pn in this species.
In addition to nuclear receptors for androgens and estrogens, the Pn of both species expressed moderate levels of mRNA for G-protein coupled estrogen receptor 1 (GPER1), a membrane-associated receptor that mediates many of the non-genomic actions of estrogens (Evans et al. 2016). The potential role of GPER1 in regulating EODf is unknown. The effects of gonadal steroids on EODf typically take weeks, which implies a traditional, genomic mode of action involving transcriptional regulation (Schaefer and Zakon 1996; Dunlap et al. 1998). In Apteronotus rostratus, rapid (< 1 hour) feminizing effects of estrogen injections have been reported on EODf and the firing rates of Pn neurons recorded in vitro (Meyer 1984). However, saline or 5αDHT injections had similar (though smaller) effects (Meyer 1984), and these findings were not replicated in A. leptorhynchus (G.T.S., unpub. obs.).
The Pn of both species strongly expressed transcripts for several 17β hydroxysteroid dehydrogenases. These enzymes can catalyze the oxidation and/or reduction of androgens and estrogens to more inactive or active forms, and thus might have influence local availability of active ligands for androgen and estrogen receptors in the Pn. 17β-HSDs also have numerous other functions, however, including fatty acid/lipid metabolism (Moeller and Adamski 2009). It is thus unclear whether they play a specific role in steroidal regulation of sex differences in EODf.
The expression of mRNA for steroid receptor coactivators or corepressors in the Pn, (e.g., NCOA, prohibitin, NRIP) raises the possibility that the products of these genes modulate androgenic and estrogenic regulation of sex differences in EODf. Steroid receptor coactivator 1 (SRC1 = NCOA1) facilitates the neonatal defeminization of lordosis and sexually dimorphic nucleus volume by testosterone in rats (Auger et al. 2000) and is necessary for normal masculinization of copulatory behavior and preoptic area volume by gonadal steroids in quail (Charlier et al. 2005). Future studies are needed to determine whether these cofactors in the Pn play a role in the hormonal regulation of EODf.
Transcripts for glucocorticoid receptors and thyroid hormone receptors were also expressed at moderately high levels in the Pn, though the function of these hormones in the Pn is not well-explored. Cortisol mediates effects of social interactions on chirping behavior in A. leptorhynchus, but this effect is thought to be mediated by actions on the diencephalic prepacemaker nucleus, rather than the Pn (Dunlap et al. 2002; Dunlap et al. 2006). One possibility is that glucocorticoids and/or thyroid hormone regulate the metabolic/energetic demands of extremely high neuronal firing rates in the Pn. Thyroxine shifts the balance of Na+ and K+ currents and excites mammalian cortical neurons (Hoffmann and Dietzel 2004). Thyroxine also increases EODf in A. leptorhynchus (Dunlap and Ragazzi 2015). This hypothesis is also consisted with the greater expression of thyroid receptor transcripts that we found in females than males in A. albifrons, since females have higher EODf than males in this species. Interestingly, thyroid hormone receptor α transcripts are upregulated in the vocal motor nucleus of midshipmen fish (Porichthys notatus), which also contains neurons that fire at relatively high frequencies (Feng et al. 2015).
Transcripts related to high-frequency firing
Neurons in the Pn are the fastest firing neurons known (Moortgat et al. 1998; Smith 1999). The high-frequency activity and precision of these neurons depends on extensive electrical coupling, on transient ionic currents with rapid kinetics to produce short action potentials, and on persistent sodium currents that provide tonic depolarization to support spontaneous firing (Dye 1991; Moortgat et al. 2000; Smith and Zakon 2000). The Pn transcriptomes in this study identified several sodium and potassium channel genes whose products are likely to contribute to these currents. The most abundant sodium channel α subunit gene expressed in the Pn was SCN8A, which encodes for the Nav1.6 sodium channel that is commonly expressed throughout the central nervous system. Interestingly, the Pn of A. leptorhynchus (but not A. albifrons) also expressed an isoform of the SCN4A gene, which encodes the Nav1.4 sodium channel. In other vertebrates, SCN4A is normally expressed in skeletal muscle and is not typically expressed in the CNS. The Pn of both species also expressed numerous transcripts for sodium channel beta subunits, which modify the kinetics of sodium channels, regulate persistent and resurgent sodium currents that support spontaneous firing, and influence subcellular localization of sodium channels (Aman et al. 2009). Indeed, the two most abundant Na+ β subunit transcripts in the Pn of both species were SCN4B, coding for the Navβ4 subunit that strongly promotes resurgent Na+ currents, and SCN3B, coding for the Navβ3 subunit that enhances persistent Na+ currents (Qu et al. 2001; Barbosa et al. 2015). Another possible source of tonic depolarizing current that supports spontaneous activity of Pn neurons is a non-selective Na+ leak conductance carried by NALCN channels. NALCN was expressed in the Pn of both species, and NALCN channels promote spontaneous firing in other neuron types (Lu et al. 2007; Lutas et al. 2016).
Several of K+ channel transcripts expressed in the Pn may also contribute to the high-frequency spontaneous activity of Pn neurons. The effects of channel blockers on the Pn neuronal firing suggested that Kv1 channels are likely to support the spontaneous activity of Pn neurons and regulate their firing rates (Smith and Zakon 2000). Consistent with this previous finding, the most abundantly expressed voltage-gated K+ channel transcript in the Pn of both species was KCNA10, which codes for the Kv1.8 channel. The kinetics of mammalian KCNA10 channels, however, do not seem well-suited to supporting high-frequency firing. These channels typically activate at relatively depolarized potentials (half-activation potential ~ +3.5 mV), activate more slowly than other Kv1 channels (τact ~ 20 ms), and show little or no inactivation. It thus seems likely either that other K+ channels in Pn neurons contribute to their spontaneous activity or that the biophysical properties of KCNA10 channels in apteronotids differ substantially from those in mammals. The Pn also expressed several transcripts for Kv3 channels (KCNC2, 3, and/or 4). Kv3 channels are well-adapted for high-frequency firing. These channels are expressed in phase-locking and time-coding neurons in the mammalian auditory system and in the vestibular nucleus, and their high thresholds and rapid activation and deactivation kinetics allow very brief action potentials that are necessary for neurons to sustain high firing rates (Gan and Kaczmarek 1998; Gittis et al. 2010).
The Pn also expressed transcripts for several accessory subunits that modify K+ channel function. The transcript for the Kvβ2 subunit (KCNAB2) was expressed at high levels in the Pn of both A. albifrons and A. leptorhynchus. Kvβ2 interacts with Kv1 channels to increase current magnitude and accelerate channel activation (Heinemann et al. 1996; Lazaroff et al. 1999). Rapid activation of K+ currents may allow short-duration action potentials and high-frequency firing. The Pn of both species also expressed transcripts for K+ channel interaction proteins (KChIPs) 2 and 4. The KChIPs interact with Kv4 channels to shift inactivation to more depolarized potentials, slow inactivation, and accelerate recovery from inactivation (Pongs and Schwarz 2010). KChIP 2 also enhances Kv4 currents by 10–100 fold. Because Kv4 transcripts are expressed at relatively low levels in the Pn, however, the potential involvement of KChIPs in regulating Pn firing is unclear.
Some of the genes identified here that control the high-frequency firing of neurons in the Pn of apteronotid electric fish are similar to those identified in a transcriptomic study that examined genes responsible for the rapid, synchronous activity of neurons in the vocal motor nucleus (VMN) of midshipmen fish (Feng et al. 2015). Specifically, neurons in the midshipmen VMN, like those in the apteronotid Pn, are electrically coupled, and a connexin transcript expressed in the Pn (gap junction protein β6/connexin 30) is also upregulated in the midshipmen VMN. Similarly, transcripts for Kv3 (KCNC) channels and sodium channel β4 subunits (SCN4B) are expressed in both the apteronotid Pn and midshipmen VMN. Some of the channels hypothesized to support high frequency activity in the VMN (e.g., Kv9.1, Kv2.2, Kv7.2; Feng et al. 2015), however, were not abundantly expressed in the Pn. These differences are not surprising, because the firing rates of Pn neurons (600–1200 Hz) are about an order of magnitude faster than those of VMN neurons (~100 Hz) and thus likely employ different mechanisms.
Gene expression and function of neurotransmitters and neuromodulators in the Pn
Connections between relay and pacemaker neurons in the Pn are mediated by gap junctions, but the Pn also receives descending glutamatergic input from the diencephalic central posterior-prepacemaker nucleus (CP-PPn) and the midbrain sublemniscal prepacemaker nucleus (SPPn) (Kawasaki et al. 1988; Heiligenberg et al. 1996). These inputs transiently excite pacemaker or relay neurons to produce modulations of EODf, including the jamming avoidance response (JAR), chirps (large, abrupt increases in EODf), and gradual frequency rises (GFRs, smaller and slower increases of EODf). In A. leptorhynchus, chirps result when glutamatergic inputs from the chirp subdivision of the CP-PPn (i.e., the PPnC) activate non-NMDA receptors on relay cells in the Pn. GFRs arise when glutamate from neurons in the “gradual rise” subdivision of the CP-PPn (PPnG) activates NMDA receptors on pacemaker cells in the Pn. The JAR results when glutamatergic inputs from the SPPn activate NMDA receptors on the relay cells in the Pn (Kawasaki et al. 1988; Heiligenberg et al. 1996). The Pn expressed mRNA for numerous types of AMPA and kainate receptors that are likely to mediate excitation the leads to chirping, as well as NMDA receptors that could mediate inputs resulting in GFRs and the JAR.
The Pn also expressed ionotropic receptors for the inhibitory neurotransmitters GABA and glycine. The Pn does not receive any known inhibitory inputs from other brain regions, but one possible source of GABA or glycine in the Pn are small, intrinsic interneurons known as parvocells (Smith et al. 2000). Parvocells form chemical synapses on relay and/or pacemaker neurons in the Pn and contain parvalbumin, which is often co-expressed in GABAergic or glycinergic neurons (Aoki et al. 1990; Horn et al. 1994; Laing et al. 1994; Smith et al. 2000). The function of parvocells or of inhibition in the Pn is not known. Similarly, the role of cholinergic neurotransmission, suggested by the robust expression of several nicotinic acetylcholine receptor transcripts in the Pn, remains unexplored.
The expression of numerous transcripts for neuromodulator receptors in the Pn was somewhat unexpected. The CP-PPn, which controls chirping, receives input from numerous neuromodulators, which influence EOD modulations (reviewed in Zupanc and Maler 1997; Smith 2013). For example, sexually dimorphic expression of substance P in the CP-PPn of both A. leptrohynchus and A. albifrons is related to sex differences in the structure of chirps; and serotonergic and catecholaminergic inputs to the CP-PPn regulate the production of chirps and/or the JAR (Maler and Ellis 1987; Weld et al. 1991; Kolodziejski et al. 2005; Smith and Combs 2008). Relatively few studies, however, have investigated the role of neuromodulation in the Pn. The rich expression of transcripts for neuromodulator precursors and receptors in the Pn suggests that these compounds may play a role in setting the ongoing firing rates of pacemaker and relay neurons. Alternatively, these receptors may modulate responses of Pn neurons to descending inputs from the CP-PPn and SPPn, thereby changing the production and/or structure of chirps and JARs in response to environmental or social conditions. In the “pulse type” gymnotiforms Gymnotus omarorum and Brachyhypopomus guaderio, arginine vasotocin (AVT) in the Pn modulates EOD rate in response to the social environment and circadian rhythms (Perrone et al. 2010; Perrone et al. 2014). Investigating the function of neuromodulation in the Pn will be a potentially fruitful line of future inquiry, and the present study has identified several candidate neuromodulators, including catecholeamines, serotonin, and numerous neuropeptides.
Sex-biased gene expression and sex differences in EODf
One of the major goals of this study was to identify candidate genes whose expression in the Pn was sex-biased, and that might thus contribute to sex differences in EODf. We identified several ion channel transcripts whose expression was sexually dimorphic based on a differential expression analysis with independent hypothesis weighted (IHW) false discovery rate correction. An important caveat is that IHW is a relatively non-conservative approach, and type 1 errors are possible. Thus, the putatively sex-biased transcripts identified here should be interpreted cautiously. These transcripts should be viewed as hypothesized candidate genes for mediating sex differences EODf, and those hypotheses should be further tested with future targeted qPCR experiments. Moreover, the sex-biased genes identified here should not be viewed as the only possible mechanisms for hormonal regulation of sex differences in EODf. Sex and species differences in Pn firing rates/EODf could also result from differences in isoform expression or in the synthesis, degradation, or specific localization of proteins in particular cell types or subcellular regions. Such differences would not have been detected in our transcriptomic study.
In A. leptorhynchus, the female Pn expressed more mRNA for the K+ channel β2 subunit (Kvβ2, KCNAB2) than the male Pn. Kvβ2 increases the amplitude of Kv1-mediated K+ currents, and Kv1 channels are likely candidates for influencing Pn firing (Smith and Zakon 2000; Smith et al. 2006). It is possible that increased expression of the Kvβ2 gene in females may lower their EODf by strengthening outward currents during pacemaking ramp potentials, thereby increasing interspike intervals and reducing Pn firing rates. In A. albifrons, expression of several low-abundance K+ channel transcripts was potentially male-biased. Kv1.1 (KCNA1), Kv1.2 (KCNA2) and K2.1 (KCNB1) transcripts were all expressed more in male Pns than in female Pns. These channels have the potential to reduce excitability and neuronal firing rates, and their increased expression in the Pn would be consistent with the lower EODf in male A. albifrons. The very low level of expression of these channel transcripts, however, suggests that they might not play a major role in the excitability of Pn neurons.
The gene for the inward rectifying K+ channel Kir3.1 (KCNJ3) was sex-biased in both A. leptorhynchus and A. albifrons, but was biased in opposite directions in the two species. Consistent with the reversed EOD sexual dimorphism in the two species, Kir3.1 was expressed more in females in A. leptorhynchus, but more in males in A. albifrons. Thus, it was expressed at higher levels in both species in the sex that has lower EODf. Kir3.1 reduces neuronal excitability by hyperpolarizing the resting membrane potential by ~8 mV (Luscher and Slesinger 2010). Elevated expression of Kir3.1 in both species may reduce EODf by hyperpolarizing Pn neurons and slowing their firing rates. If so, this gene may be a target for evolutionary changes in the sexual dimorphism of EODf. Moreover, Kir3.1 is a G-protein coupled channel and is strongly influenced by G-protein coupled receptors (Luscher and Slesinger 2010), so it is also a potential target for the putative neuromodulatory pathways suggested by this study.
Sex differences in Pn expression of mRNA for a sodium channel β1 subunit (Navβ1, SCNAB1) in A. albifrons could also contribute to within- and between sex differences in EODf. Transcripts for an isoform of Navβ1 were expressed at higher levels in males than females in A. albifrons, and within sexes, were expressed at higher levels in fish with lower EODf. Navβ1 interacts with Na+ channel α-subunits to accelerate inactivation and to substantially shift steady-state inactivation voltage-dependence to more hyperpolarized potentials (Liu et al. 2007). This shift in the inactivation voltage-dependence by Navβ1 also decreases persistent sodium currents (Aman et al. 2009). Persistent Na+ currents support high-frequency spontaneous firing of Pn neurons and increase firing rate (Smith and Zakon 2000). Thus, greater expression of Navβ1 in male A. albifrons may lower EODf by reducing persistent Na+ currents and Pn firing rates. Sex differences in the expression of Navβ1 isoforms in the electric organ have also been linked to sex differences in excitability and EOD waveform in the electric fish Sternopygus (Liu et al. 2007).
Conclusions and future directions
As is typical of many transcriptomic studies, this study generated more hypotheses than it strongly tested. Consistent with the role of androgens and estrogens in regulating sex differences in EODf, we found robust expression of steroidogenic enzymes and receptors in the Pn. However, these findings raise several new questions: (1) what roles do 5α-reductase and 5αDHT play in masculinizing EODf? (2) do steroid receptor co-factors influence steroidal regulation of sex differences in EODf? (3) do the abundantly-expressed 17βHSDs influence androgenic or estrogenic regulation of the EOD? (4) what roles do membrane bound receptors (GPER1 and membrane-associated progesterone receptors) play in regulating EODf? Similarly, we identified a complex suite of ion channel and connexin genes that are expressed in the Pn, and some of those genes encode channels previously predicted to regulate the rhythm that underlies the EOD. Fully understanding the roles of these ion channel genes in the Pn, however, will require identifying their localization, characterizing their biophysical properties, and modeling their interactions. Finally, we identified several ion channel genes whose expression in the Pn was putatively sexually dimorphic and/or correlated with EODf. Testing the hypothesis that these channel genes underlie hormonally regulated sex differences in EODf will require confirming their sex-biased expression with subsequent qPCR studies and experiments to determine whether their expression is regulated by androgens and/or estrogens. Such experiments have the potential not only to elucidate the genomic mechanisms of sex differences, but also to examine how those mechanisms evolve to produce species diversity in sexual dimorphism.
Supplementary Material
Acknowledgments
The authors thank Jie Huang and James Ford at the Indiana University Center for Genomics and Bioinformatics for constructing and sequencing libraries, and the Indiana University Center for the Integrative Study of Animal Behavior (CISAB) Mechanisms of Behavior Core Laboratory for the use of facilities to isolate RNA from samples. Supported by NSF IOS 0950721 to GTS and NIH T32049336 to ARS. Animal care and experimental procedures were in conducted in accordance with ethical standards in the NIH Guide by using protocols approved by the Bloomington Institutional Animal Care and Use Committee at Indiana University.
List of Abbreviations
- 11KT
11-ketotestosterone
- 11β-HSD
11β hydroxysteroid dehydrogenase
- 17β-HSD
17β hydroxysteroid dehydrogenase
- 5αDHT
5α dihydrotestosterone
- 5HT
serotonin
- AVT
arginine vasotocin
- AR
androgen receptor
- ER
estrogen receptor
- BUSCO
benchmarking universal single-copy orthologs
- CP-PPn
central posterior-prepacemaker nucleus
- E2
estradiol
- EOD
electric organ discharge
- EODf
electric organ discharge frequency
- GFR
gradual frequency rise
- GO
gene ontology
- GPER
G-protein coupled estrogen receptor
- GSI
gonadosomatic index
- IHW
independent hypothesis weighted
- JAR
jamming avoidance response
- KChIP
K+ channel-interacting protein
- NCOA
nuclear receptor coactivator
- NRIP
nuclear receptor interacting protein
- PCs
principal components
- Pn
pacemaker nucleus
- SPPn
sublemniscal prepacemaker nuclueus
- SRC
steroid receptor coactivator
- VMN
vocal motor nucleus
References Cited
- Aman TK, Grieco-Calub TM, Chen C, Rusconi R, Slat EA, Isom LL, Raman IM. Regulation of persistent Na current by interactions between β subunits of voltage-gated Na channels. J Neurosci. 2009;29(7):2027–2042. doi: 10.1523/Jneurosci.4531-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki E, Semba R, Seto-Ohshima A, Heizmann CW, Kashiwamata S. Coexistence of parvalbumin and glycine in the rat brainstem. Brain Res. 1990;525(1):140–143. doi: 10.1016/0006-8993(90)91329-F. [DOI] [PubMed] [Google Scholar]
- Auger AP, Tetel MJ, McCarthy MM. Steroid receptor coactivator-1 (SRC-1) mediates the development of sex-specific brain morphology and behavior. Proc Natl Acad Sci U S A. 2000;97(13):7551–7555. doi: 10.1073/pnas.97.13.7551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbosa C, Tan ZY, Wang RZ, Xie WR, Strong JA, Patel RR, Vasko MR, Zhang JM, Cummins TR. Navβ4 regulates fast resurgent sodium currents and excitability in sensory neurons. Mol Pain. 2015;11 doi: 10.1186/s12990-015-0063-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bass AH, Hopkins CD. Hormonal control of sex differences in the electric organ discharge (EOD) of mormyrid fishes. J Comp Physiol A. 1985;156(5):587–604. doi: 10.1007/BF00619109. [DOI] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate - a practical and powerful approach to multiple testing. J Roy Stat Soc B Met. 1995;57(1):289–300. doi: 10.2307/2346101. [DOI] [Google Scholar]
- Bohne A, Sengstag T, Salzburger W. Comparative transcriptomics in east african cichlids reveals sex- and species-specific expression and new candidates for sex differentiation in fishes. Genome Biol Evol. 2014;6(9):2567–2585. doi: 10.1093/gbe/evu200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenowitz EA, Arnold AP. Lack of sexual dimorphism in steroid accumulation in vocal control brain-regions of duetting song birds. Brain Res. 1985;344(1):172–175. doi: 10.1016/0006-8993(85)91205-3. [DOI] [PubMed] [Google Scholar]
- Burns CMB, Rosvall KA, Ketterson ED. Neural steroid sensitivity and aggression: comparing individuals of two songbird subspecies. J Evol Biol. 2013;26(4):820–831. doi: 10.1111/jeb.12094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caetano-Anolles K, Seo M, Rodriguez-Zas S, Oh JD, Han JY, Lee K, Park TS, Shin S, Jiao ZJ, Ghosh M, Jeong DK, Cho S, Kim H, Song KD, Lee HK. Comprehensive identification of sexual dimorphism-associated differentially expressed genes in two-way factorial designed RNA-seq data on japanese quail (Coturnix coturnix japonica) Plos One. 2015;10(9) doi: 10.1371/journal.pone.0139324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canoine V, Fusani L, Schlinger B, Hau M. Low sex steroids, high steroid receptors: Increasing the sensitivity of the nonreproductive brain. Devel Neurobiol. 2007;67(1):57–67. doi: 10.1002/neu.20296. [DOI] [PubMed] [Google Scholar]
- Charlier TD, Ball GF, Balthazart J. Inhibition of steroid receptor coactivator-1 blocks estrogen and androgen action on male sex behavior and associated brain plasticity. J Neurosci. 2005;25(4):906–913. doi: 10.1523/Jneurosci.3533-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diotel N, Do Rego JL, Anglade I, Vaillant C, Pellegrini E, Vaudry H, Kah O. The brain of teleost fish, a source, and a target of sexual steroids. Front Neurosci-Switz. 2011;5 doi: 10.3389/fnins.2011.00137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dulka JG. Androgen-induced neural plasticity and the regulation of electric-social behavior in the brown ghost knifefish: current status and future directions. 1997;17(1–6):195–202. doi: 10.1023/A:1007778815126. [DOI] [Google Scholar]
- Dulka JG, Maler L. Testosterone modulates female chirping behavior in the weakly electric fish Apteronotus leptorhynchus. J Comp Physiol A. 1994;174:331–343. doi: 10.1007/BF00240215. [DOI] [Google Scholar]
- Dunlap KD, Castellano JF, Prendaj E. Social interaction and cortisol treatment increase cell addition and radial glia fiber density in the diencephalic periventricular zone of adult electric fish, Apteronotus leptorhynchus. Horm Behav. 2006;50:10–17. doi: 10.1016/j.yhbeh.2006.01.003. [DOI] [PubMed] [Google Scholar]
- Dunlap KD, Pelczar PL, Knapp R. Social interactions and cortisol treatment increase the production of aggressive electrocommunication signals in male electric fish, Apteronotus leptorhynchus. Horm Behav. 2002;42:97–108. doi: 10.1006/hbeh.2002.1807. [DOI] [PubMed] [Google Scholar]
- Dunlap KD, Ragazzi MA. Thermal acclimation and thyroxine treatment modify the electric organ discharge frequency in an electric fish, Apteronotus leptorhynchus. Physiol Behav. 2015;151:64–71. doi: 10.1016/j.physbeh.2015.06.036. [DOI] [PubMed] [Google Scholar]
- Dunlap KD, Silva AC, Smith GT, Zakon HH. Weakly electric fish: behavior, neurobiology, and neuroendocrinology. In: Pfaff DW, Joels M, editors. Hormones, brain, and behavior. 3. Vol. 2. Academic Press; New York: 2017. pp. 69–100. [Google Scholar]
- Dunlap KD, Smith GT, Yekta A. Temperature dependence of electrocommunication signals and their underlying neural rhythms in the weakly electric fish, Apteronotus leptorhynchus. Brain Behav Evol. 2000;55:152–162. doi: 10.1159/000006649. [DOI] [PubMed] [Google Scholar]
- Dunlap KD, Thomas P, Zakon HH. Diversity of sexual dimorphism in electrocommunication signals and its androgen regulation in a genus of electric fish, Apteronotus. J Comp Physiol A. 1998;183:77–86. doi: 10.1007/s003590050236. [DOI] [PubMed] [Google Scholar]
- Dye J. Ionic and synaptic mechanisms underlying a brainstem oscillator: an in vitro study of the pacemaker nucleus of Apteronotus. J Comp Physiol A. 1991;168:521–532. doi: 10.1007/BF00215074. [DOI] [PubMed] [Google Scholar]
- Elekes K, Szabo T. Synaptology of the medullary command (pacemaker) nucleus of the weakly electric fish (Apteronotus leptorhynchus) with particular reference to comparative aspects. Exp Brain Res. 1985;60:509–520. doi: 10.1007/BF00236936. [DOI] [PubMed] [Google Scholar]
- Evans NJ, Bayliss AL, Reale V, Evans PD. Characterisation of signalling by the endogenous GPER1 (GPR30) receptor in an embryonic mouse hippocampal cell line (mHippoE-18) Plos One. 2016;11(3) doi: 10.1371/journal.pone.0152138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng NY, Bass AH. Neural, hormonal, and genetic mechanisms of alternative reproductive tactics. In: Pfaff DW, Joels M, editors. Hormones, brain, and behavior. 3. Vol. 2. Academic Press; Oxford, UK: 2017. pp. 47–65. [DOI] [Google Scholar]
- Feng NY, Fergus DJ, Bass AH. Neural transcriptome reveals molecular mechanisms for temporal control of vocalization across multiple timescales. Bmc Genomics. 2015;16 doi: 10.1186/s12864-015-1577-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fostier A, Jalabert B, Billard R, Breton B, Zohar Y. The gonadal steroids. In: Hoar WS, Randall DJ, Donaldson EM, editors. Fish physiology. 9A. Elsevier; New York: 1983. pp. 277–372. [Google Scholar]
- Fuxjager MJ, Schuppe ER, Hoang J, Chew J, Shah M, Schlinger BA. Expression of 5α-and 5β–reductase in spinal cord and muscle of birds with different courtship repertoires. Front Zool. 2016;13 doi: 10.1186/s12983-016-0156-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan L, Kaczmarek LK. When, where, and how much? Expression of the Kv3.1 potassium channel in high-frequency firing neurons. J Neurobiol. 1998;37:69–79. doi: 10.1002/(SICI)1097-4695(199810)37:1<69::AID-NEU6>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- Gittis AH, Moghadam SH, du Lac S. Mechanisms of sustained high firing rates in two classes of vestibular nucleus neurons: Differential contributions of resurgent Na, Kv3, and BK currents. J Neurophysiol. 2010;104(3):1625–1634. doi: 10.1152/jn.00378.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heiligenberg W, Metzner W, Wong CJH, Keller CH. Motor control of the jamming avoidance response of Apteronotus leptorhynchus: evolutionary changes of a behavior and its neural substrate. J Comp Physiol A. 1996;179:653–674. doi: 10.1007/BF00216130. [DOI] [PubMed] [Google Scholar]
- Heinemann SH, Rettig J, Graack HR, Pongs O. Functional characterization of Kv channel beta subunits from rat brain. J Physiol (Lond) 1996;493(3):625–633. doi: 10.1113/jphysiol.1996.sp021409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho WW, Rack JM, Smith GT. Divergence in androgen sensitivity contributes to population differences in sexual dimorphism of electrocommunication behavior. Horm Behav. 2013;63(1):49–53. doi: 10.1016/j.yhbeh.2012.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann G, Dietzel ID. Thyroid hormone regulates excitability in central neurons from postnatal rats. Neuroscience. 2004;125(2):369–379. doi: 10.1016/j.neuroscience.2004.01.047. [DOI] [PubMed] [Google Scholar]
- Horn AK, Buttner-Ennever JA, Wahle P, Reichenberger I. Neurotransmitter profile of saccadic omnipause neurons in nucleus raphe interpositus. J Neurosci. 1994;14(4):2032–2046. doi: 10.1523/JNEUROSCI.14-04-02032.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ignatiadis N, Klaus B, Zaugg JB, Huber W. Data-driven hypothesis weighting increases detection power in genome-scale multiple testing. Nat Methods. 2016;13(7):577-+. doi: 10.1038/Nmeth.3885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang D, Hogan JO, Kim D. THIK-1 (K2P13.1) is a small-conductance background K+ channel in rat trigeminal ganglion neurons. Pflug Arch Eur J Phy. 2014;466(7):1289–1300. doi: 10.1007/s00424-013-1358-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawasaki M, Maler L, Rose GJ, Heiligenberg W. Anatomical and functional organization of the prepacemaker nucleus in gymnotiform electric fish: the accommodation of two behaviors in one nucleus. J Comp Neurol. 1988;276:113–131. doi: 10.1002/cne.902760108. [DOI] [PubMed] [Google Scholar]
- Kirschbaum F. Reproduction of weakly electric teleosts - just another example of convergent development. Env Biol Fish. 1984;10(1–2):3–14. [Google Scholar]
- Kolodziejski JA, Nelson BS, Smith GT. Sex and species differences in neuromodulatory input to a premotor nucleus: A comparative study of substance P and communication behavior in weakly electric fish. J Neurobiol. 2005;62:299–315. doi: 10.1002/neu.20095. [DOI] [PubMed] [Google Scholar]
- Kusakabe M, Nakamura I, Young G. 11 β-Hydroxysteroid dehydrogenase complementary deoxyribonucleic acid in rainbow trout: Cloning, sites of expression, and seasonal changes in gonads. Endocrinology. 2003;144(6):2534–2545. doi: 10.1210/en.2002-220446. [DOI] [PubMed] [Google Scholar]
- Labrie F, Luu-The V, Lin SX, Simard J, Labrie C. Role of 17 β-hydroxysteroid dehydrogenases in sex steroid formation in peripheral intracrine tissues. Trends Endocrin Met. 2000;11(10):421–427. doi: 10.1016/S1043-2760(00)00342-8. [DOI] [PubMed] [Google Scholar]
- Laing I, Todd AJ, Heizmann CW, Schmidt HH. Subpopulations of GABAergic neurons in laminae I–III of rat spinal dorsal horn defined by coexistence with classical transmitters, peptides, nitric oxide synthase, or parvalbumin. J Comp Neurol. 1994;384(4):580–596. doi: 10.1016/0306-4522(94)90065-5. [DOI] [PubMed] [Google Scholar]
- Lazaroff MA, Hofmann AD, Ribera AB. Xenopus embryonic spinal neurons express potassium channel Kvβ subunits. J Neurosci. 1999;19(24):10706–10715. doi: 10.1523/JNEUROSCI.19-24-10706.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Lamm MS, Rutherford K, Black MA, Godwin JR, Gemmell NJ. Large-scale transcriptome sequencing reveals novel expression patterns for key sex-related genes in a sex-changing fish. Biol Sex Differ. 2015;6 doi: 10.1186/s13293-015-0044-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Wu MM, Zakon HH. Individual variation and hormonal modulation of a sodium channel beta subunit in the electric organ correlate with variation in a social signal. Devel Neurobiol. 2007;67(10):1289–1304. doi: 10.1002/Dneu.20404. Doi. [DOI] [PubMed] [Google Scholar]
- Lu BX, Su YH, Das S, Liu J, Xia JS, Ren DJ. The neuronal channel NALCN contributes resting sodium permeability and is required for normal respiratory rhythm. Cell. 2007;129(2):371–383. doi: 10.1016/j.cell.2007.02.041. [DOI] [PubMed] [Google Scholar]
- Luscher C, Slesinger PA. Emerging roles for G protein-gated inwardly rectifying potassium (GIRK) channels in health and disease. Nat Rev Neurosci. 2010;11(5):301–315. doi: 10.1038/nrn2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutas A, Lahmann C, Soumillon M, Yellen G. The leak channel NALCN controls tonic firing and glycolytic sensitivity of substantia nigra pars reticulata neurons. Elife. 2016;5 doi: 10.7554/eLife.15271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maler L, Ellis WG. Inter-male aggressive signals in weakly electric fish are modulated by monoamines. Behav Brain Res. 1987;25(1):75–81. doi: 10.1016/0166-4328(87)90046-5. [DOI] [PubMed] [Google Scholar]
- Manousaki T, Tsakogiannis A, Lagnel J, Sarropoulou E, Xiang JZ, Papandroulakis N, Mylonas CC, Tsigenopoulos CS. The sex-specific transcriptome of the hermaphrodite sparid sharpsnout seabream (Diplodus puntazzo) Bmc Genomics. 2014;15 doi: 10.1186/1471-2164-15-655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer JH. Steroid influences upon the discharge frequencies of a weakly electric fish. J Comp Physiol A. 1983;153:29–37. doi: 10.1007/BF00610339. [DOI] [Google Scholar]
- Meyer JH. Steroid influences upon discharge frequencies of intact and isolated pacemakers of weakly electric fish. J Comp Physiol A. 1984;154:659–668. doi: 10.1007/BF01350219. [DOI] [Google Scholar]
- Meyer JH, Leong M, Keller CH. Hormone-induced and maturational changes in electric organ discharges and electroreceptor tuning in the weakly electric fish Apteronotus. J Comp Physiol A. 1987;160:385–394. doi: 10.1007/BF00613028. [DOI] [PubMed] [Google Scholar]
- Meyer JH, Zakon HH. Androgens alter the tuning of electroreceptors. Science. 1982;217(4560):635–637. doi: 10.1126/science.217.4560.635. [DOI] [PubMed] [Google Scholar]
- Mills A, Zakon HH. Coordination of EOD frequency and pulse duration in a weakly electric wave fish: the influence of androgens. J Comp Physiol A. 1987;161:417–430. doi: 10.1007/BF00603967. [DOI] [Google Scholar]
- Moeller G, Adamski J. Integrated view on 17β-hydroxysteroid dehydrogenases. Mol Cell Endocrinol. 2009;301(1–2):7–19. doi: 10.1016/j.mce.2008.10.040. [DOI] [PubMed] [Google Scholar]
- Moortgat KT, Bullock TH, Sejnowski TJ. Precision of the pacemaker nucleus in a weakly electric fish: Network versus cellular influences. J Neurophysiol. 2000;83(2):971–983. doi: 10.1152/jn.2000.83.2.971. doi:10.1.1.156.2618. [DOI] [PubMed] [Google Scholar]
- Moortgat KT, Keller CH, Bullock TH, Sejnowski TJ. Submicrosecond pacemaker precision is behaviorally modulated: the gymnotiform electromotor pathway. Proc Natl Acad Sci U S A. 1998;95:4684–4689. doi: 10.1073/pnas.95.8.4684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norris AJ, Foeger NC, Nerbonne JM. Interdependent roles for accessory KChIP2, KChIP3, and KChIP4 subunits in the generation of Kv4-encoded I-A channels in cortical pyramidal neurons. J Neurosci. 2010;30(41):13644–13655. doi: 10.1523/Jneurosci.2487-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrone R, Batista G, Lorenzo D, Macadar O, Silva A. Vasotocin actions on electric behavior: interspecific, seasonal, and social context-dependent differences. Front Behav Neurosci. 2010;4 doi: 10.3389/fnbeh.2010.00052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrone R, Migliaro A, Comas V, Quintana L, Borde M, Silva A. Local vasotocin modulation of the pacemaker nucleus resembles distinct electric behaviors in two species of weakly electric fish. Journal of Physiology-Paris. 2014;108(2–3):203–212. doi: 10.1016/j.jphysparis.2014.07.007. [DOI] [PubMed] [Google Scholar]
- Peterson MP, Rosvall KA, Choi JH, Ziegenfus C, Tang HX, Colbourne JK, Ketterson ED. Testosterone affects neural gene expression differently in male and female juncos: A role for hormones in mediating sexual dimorphism and conflict. Plos One. 2013;8(4) doi: 10.1371/journal.pone.0061784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petzold JM, Smith GT. Androgens regulate sex differences in signaling but are not associated with male variation in morphology in the weakly electric fish Parapteronotus hasemani. Horm Behav. 2016;78:67–71. doi: 10.1016/j.yhbeh.2015.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pongs O, Schwarz JR. Ancillary subunits associated with voltage-dependent K+ channels. Physiol Rev. 2010;90(2):755–796. doi: 10.1152/physrev.00020.2009. [DOI] [PubMed] [Google Scholar]
- Qu Y, Curtis R, Lawson D, Gilbride K, Ge P, DiStefano PS, Silos-Santiago I, Catterall WA, Scheuer T. Differential modulation of sodium channel gating and persistent sodium currents by the β1, β2, and β3 subunits. Mol Cell Neurosci. 2001;18(5):570–580. doi: 10.1006/mcne.2001.1039. [DOI] [PubMed] [Google Scholar]
- Remage-Healey L, London SE, Schlinger BA. Birdsong and the neural production of steroids. J Chem Neuroanat. 2010;39(2):72–81. doi: 10.1016/j.jchemneu.2009.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saldanha CJ, Schlinger BA. Steroidogenesis and neuroplasticity in the songbird brain. In: Ritsner MS, Weizman A, editors. Neuroactive steroids in brain function, behavior, and neuropsychiatric disorders. Springer; New York: 2008. pp. 201–216. [Google Scholar]
- Sano Y, Mochizuki S, Miyake A, Kitada C, Inamura K, Yokoi H, Nozawa K, Matsushime H, Furuichi K. Molecular cloning and characterization of Kv6.3, a novel modulatory subunit for voltage-gated K+ channel Kv2.1. FEBS Lett. 2002;512(1–3):230–234. doi: 10.1016/S0014-5793(02)02267-6. [DOI] [PubMed] [Google Scholar]
- Schaefer JE, Zakon HH. Opposing actions of androgen and estrogen on in vitro firing frequency of neuronal oscillators in the electromotor system. J Neurosci. 1996;16(8):2860–2868. doi: 10.1523/JNEUROSCI.16-08-02860.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlinger BA, Brenowitz EA. Neural and hormonal control of birdsong. In: Pfaff DW, Joels M, editors. Hormones, brain, and behavior. 3. Vol. 2. Academic Press; Oxford, UK: 2017. pp. 255–282. [DOI] [Google Scholar]
- Schumer M, Krishnakant K, Renn SCP. Comparative gene expression profiles for highly similar aggressive phenotypes in male and female cichlid fishes (Julidochromis) J Exp Biol. 2011;214(19):3269–3278. doi: 10.1242/jeb.055467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengelaub DR, Forger NG. The spinal nucleus of the bulbocavemosus: Firsts in androgen-dependent neural sex differences. Horm Behav. 2008;53(5):596–612. doi: 10.1016/j.yhbeh.2007.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw BK, Kennedy GG. Evidence for species differences in the pattern of androgen receptor distribution in relation to species differences in an androgen-dependent behavior. J Neurobiol. 2002;52(3):203–220. doi: 10.1002/neu.10079. [DOI] [PubMed] [Google Scholar]
- Simao FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics. 2015;31(19):3210–3212. doi: 10.1093/bioinformatics/btv351. [DOI] [PubMed] [Google Scholar]
- Smith GT. Ionic currents that contribute to a sexually dimorphic communication signal in weakly electric fish. J Comp Physiol A. 1999;185:379–387. doi: 10.1007/s003590050398. [DOI] [PubMed] [Google Scholar]
- Smith GT. Evolution and hormonal regulation of sex differences in the electrocommunication behavior of ghost knifefishes (Apteronotidae) J Exp Biol. 2013;216(13):2421–2433. doi: 10.1242/Jeb.082933. [DOI] [PubMed] [Google Scholar]
- Smith GT, Combs N. Serotonergic activation of 5HT1A and 5HT2 receptors modulates sexually dimorphic communication signals in the weakly electric fish Apteronotus leptorhynchus. Horm Behav. 2008;54:69–82. doi: 10.1016/j.yhbeh.2008.01.009. [DOI] [PubMed] [Google Scholar]
- Smith GT, Lu Y, Zakon HH. Parvocells: A novel interneuron type in the pacemaker nucleus of a weakly electric fish. J Comp Neurol. 2000;423:427–439. doi: 10.1002/1096-9861(20000731)423:3<427::AID-CNE6>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- Smith GT, Unguez GA, Weber CM. Distribution of Kv1-like potassium channels in the electromotor and electrosensory systems of the weakly electric fish, Apteronotus leptorhynchus. J Neurobiol. 2006;66(9):1011–1031. doi: 10.1002/neu.20283. [DOI] [PubMed] [Google Scholar]
- Smith GT, Zakon HH. Pharmacological characterization of ionic currents that regulate the pacemaker rhythm in a weakly electric fish. J Neurobiol. 2000;42:270–286. doi: 10.1002/(SICI)1097-4695(20000205)42:2<270::AID-NEU10>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- Stiver KA, Harris RM, Townsend JP, Hofmann HA, Alonzo SH. Neural gene expression profiles and androgen levels underlie alternative reproductive tactics in the ocellated wrasse, Symphodus ocellatus. Ethology. 2015;121(2):152–167. doi: 10.1111/eth.12324. [DOI] [Google Scholar]
- Torres YP, Morera FJ, Carvacho I, Latorre R. A marriage of convenience: β-subunits and voltage-dependent K+ channels. J Biol Chem. 2007;282(34):24485–24489. doi: 10.1074/jbc.R700022200. [DOI] [PubMed] [Google Scholar]
- Voigt C, Goymann W. Sex-role reversal is reflected in the brain of African black coucals (Centropus grillii) Devel Neurobiol. 2007;67(12):1560–1573. doi: 10.1002/dneu.20528. [DOI] [PubMed] [Google Scholar]
- Wade J. Relationships among hormones, brain and motivated behaviors in lizards. Horm Behav. 2011;59(5):637–644. doi: 10.1016/j.yhbeh.2010.08.014. [DOI] [PubMed] [Google Scholar]
- Weld MM, Maler L, Quirion R, Kar S. Sexually dimorphic distribution of substance P and its role in regulation of communication in an electric fish. Abstr Soc Neurosci. 1991;17:1407. [Google Scholar]
- Wong RY, McLeod MM, Godwin J. Limited sex-biased neural gene expression patterns across strains in Zebrafish (Danio rerio) Bmc Genomics. 2014;15 doi: 10.1186/1471-2164-15-905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakon HH, Dunlap KD. Sex steroids and communication signals in electric fish: A tale of two species. Brain Behav Evol. 1999;54(1):61–69. doi: 10.1159/000006612. [DOI] [PubMed] [Google Scholar]
- Zornik E, Kelley DB. A neuroendocrine basis for the hierarchical control of frog courtship vocalizations. Front Neuroendocrinol. 2011;32(3):353–366. doi: 10.1016/j.yfrne.2010.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zucker MS. Doctoral Dissertation. University of Texas; Austin, TX: 1998. Hormonal basis of sexual dimorphism and steroid sensitivity of electric organ discharge frequency of the gymnotiform fish, Apteronotus leptorhynchus. [Google Scholar]
- Zupanc GKH, Maler L. Neuronal control of behavioral plasticity: the prepacemaker nucleus of weakly electric gymnotiform fish. J Comp Physiol A. 1997;180:99–111. doi: 10.1007/s003590050031. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.