Abstract
Truncated iterative PKS-NRPS megasynthases in which only the C domain is present are widespread in fungi, yet nearly all members have unknown functions. Bioinformatics analysis showed that the C domains of such PKS-C enzymes are noncanonical due to substitution at the second histidine in the active site HHxxxDG motif. Here, we used genome mining strategy to characterize a cryptic PKS-C hybrid from Talaromyces wortmanii and discovered the products are reduced long-chain polyketides amidated with a specific ω-amino acid 5-aminopentanoic acid (5PA). The wortmanamides resemble long-chain N-acyl-amide signaling lipids that target diverse receptors including GPCRs. The noncanonical C domain of this PKS-C hybrid was also demonstrated to be a bona fide condensation domain which specifically selects 5PA and catalyzes amidation to release polyketide chain.
Graphical Abstract
Fungal iterative highly-reducing polyketide synthases (HR-PKSs) are among the most enigmatic enzymatic machineries involved in biosynthesis of diverse polyketide natural products, yet the programming rule remains poorly understood. Elongation and modification of polyketides are achieved by a single set of catalytic domains in the PKSs, which are orchestrated in an iterative and permutative fashion. 1 Additional structural complexity is generated by collaborative work with other biosynthetic modules, notably a C-terminal fused nonribosomal peptide synthetase (NRPS) that introduce nitrogen-containing moieties via the amide bond linkage. Combined functions from PKS-NRPS assembly lines and associated tailoring enzymes in fungi can lead to a variety of natural product scaffolds,. (Figure 1).2
Figure 1.

Representative examples of fungal iterative PKS-NRPS (CcsA) and PKS-C (LovB).x Fungal PKS-NRPS hybrids are featured by the typical bimodular architecture shown here, and are distinct from bacterial multimodular PKS/NRPS hybrid assembly lines.
Interestingly, a large family of fungal iterative PKS-NRPS hybrid enzymes contains a truncated NRPS module with only an intact condensation (C) domain at the C-terminus. Without the adenylation (A) and thiolation (T) domains found in a minimal NRPS module, the functions of the C domains in these PKS-C megasynthases are unclear. To date, the only characterized PKS-C enzymes with known function from this family are LovB and MlcA, which are involved in the biosynthesis of lovastatin and its desmethyl analogue mevastatin, respectively (Figure 1).3a The C domain from LovB has been implicated as a noncanonical condensation domain as the second histidine residue in the active site HHxxxDG motif is substituted by arginine. Indirect biochemical evidences suggest the C domain possibly serves as a Diels-Alderase catalyzing [4+2] cycloaddition to forge the decalin ring in dihydromonacolin L.3
The HHxxxDG motif is a defining signature motif of all catalytic NRPS C domains.4 Except LovB, characterized C domains with mutated HHxxxDG motifs have all been shown to be have noncatalytic roles.5 We conducted bioinformatics analysis and revealed that all C domains from fungal LovB-like PKS-C megasynthases lack the catalytic histidine residue (Fig. S1). Phylogenetic analysis further showed that these C domains from PKS-C are classified into mainly two clades separated from the canonical C domains within intact PKS-NRPS megasynthetases (clade I, Fig. S2). We noted three of the C domains from PKS-C family remained grouped into clade I, suggesting they may be evolutionarily and functionally more closely related to C domains from PKS-NRPSs. The active site histidines in these PKS-C enzymes, however, are mutated to prolines, thereby raising intriguing questions regarding their roles. The gene clusters encoding these three clade I PKS-C megasynthases are highly similar (>50% sequence identity on average) indicating conserved functions.
To determine the role of the C domain and the natural product produced by these clusters, we selected PKS-C TwmB from Talaromyces wortmanii (Figure 2a) as the model pathway. Although RT-PCR analysis showed that genes from twm cluster were transcriptionally active when the host was cultured under laboratory conditions (Fig. S3), no known metabolites are associated with this pathway. Since T. wortmanii is genetically challenging to manipulate, we heterologously overexpressed TwmB along with the auxiliary genes using Aspergillus nidulans as a heterologous host, which led to the production and subsequent isolation of new long-chain N-acyl amides 1–4 (Tables S1–4). Both 1 and 3 contain octaketide (C16) acyl chains, while 2 and 4 contain nonaketide (C18) acyl chains. Whereas the acyl units in 1 and 2 are amidated with 5-aminopentanoic acid (5-PA), 3 and 4 are amidated with β-alanine (Figure 2b). The stereochemistry of the secondary alcohol was determined to be in R-configuration (Tables S7 and S8).
Figure 2.
Characterization of LovB-like cryptic PKS-C hybrid TwmB. a) T. wortmanii gene cluster of interest encoding TwmB. Homologous clusters are shown for comparison. b) Total ion chromatogram (TIC) of the metabolic extract from different fungi transformants. c) Proposed programming of PKS-C TwmB. d) Extracted ion chromatogram (EIC) of compounds 1–4 from different A. nidulans transformants with the overexpressed genes labeled.
The acyl chains of 1–4 are likely synthesized by the same PKS assembly line (Figure 2c). After the octaketide precursor 5 is formed, direct amidation results in release of the shorter N-acyl amides 1 and 3. However, if enoyl-reduction occurs prior to amidation, the PKS-bound 5 can undergo another round of chain elongation, β keto-reduction and dehydration to arrive at the α, β-unsaturated nonaketide 6. This acyl chain is then subjected to chain release via amidation to yield the longer N-acyl amides 2 and 4.
Both octa- and nonaketide chains are amidated by two achiral amino acids: 5PA and β-alanine. The former one is less common and is found in the lysine degradation pathways.6 Consistent with its biogenesis, we showed the nitrogen atoms of 1 and 2 indeed originated from L-lysine by feeding [15Nε]-L-lysine to A. nidulans (Fig. S4). We analyzed the metabolic extracts of T. wortmanii and confirmed that 1 and 2 were produced from the native host culture, but not 3 and 4 (Figure 2b, iv). This suggests that the 5PA-derived 1 and 2 are genuine natural products and are named wortmanamide A and B, respectively; whereas 3 and 4 are heterologous-host dependent products originated from A. nidulans in the absence of TwmA; these compounds are named niduamide A and B, respectively.
To probe the function of the TwmB C domain in forming the N-acyl amides, we expressed the PKS without the C domain (TwmBΔC) and analyzed product profiles from the A. nidulans host coexpressing TwmA and TwmE. As shown in Figure 2d, no amidated polyketides, or any prematurely hydrolyzed polyketide intermediates can be detected in vivo, which indicated that the polyketide chain cannot be released from the PKS in the absence of C domain. Production of nonaketide 2, but not octaketide 1 could be restored in a trans-complementing experiment in which the standalone C domain (TwmB-C) was coexpressed with TwmBΔC. While the overall yield decreased compared to that produced from intact TwmB, this result implicates the essential role of the C domain in formation of the N-acyl amides. The restoration of only nonaketides in trans complementation assay also corroborates with our model that emergence of octaketide may be due to kinetic balance between PKS functions and C domain-catalyzed amidative product release with substrate 5. Since the in trans interactions between the PKS and C domains are expected to be less efficient compared to the in cis assembly line, the PKS can out-compete the C domain for the octaketide substrate 5, leading to exclusive formation of the nonaketide product.
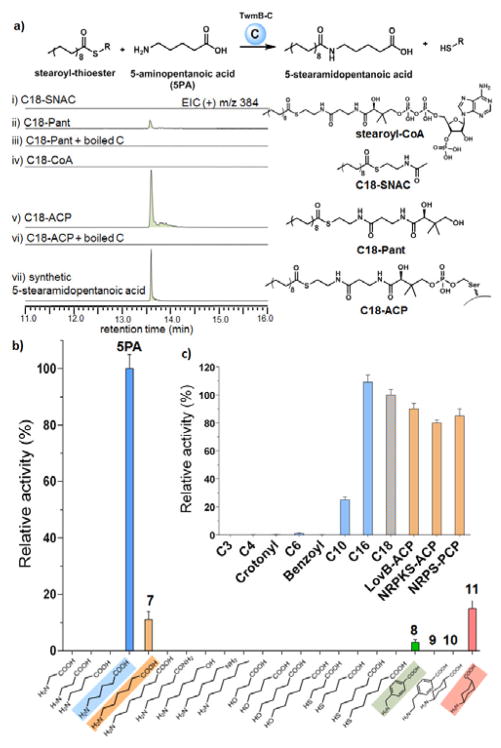
To conclusively establish the role of TwmB-C, we reconstituted its enzymatic activity in vitro using purified enzyme expressed from E. coli (Fig. S5). We used commercial stearoyl-CoA (C18-CoA), as well as synthesized stearoyl-S-N-acetylcysteamine (C18-SNAC), stearoyl-S-pantetheine (C18-Pant) as surrogate substrates to mimic the acyl-carrier protein (ACP) bound nonaketide 6. The C18-thioesters were then incubated with TwmB-C in the presence of 1 mM of 5PA, and the products were analyzed by LC-MS (Figure 3a). Whereas C18-CoA and C18-SNAC did not yield any products, C18-Pant was converted to the anticipated product 5-stearamido-pentanoic acid, confirming the standalone C domain is active. We then prepared stearoyl-S-ACP (C18-ACP) from apo TwmB-ACP and stearoyl-CoA using the phosphopantetheinyl transferase NpgA.7 C18-ACP supported the highest amount of product turnover in the assay (>10-fold than C18-Pant). These results suggest that the pantetheine arm is required for molecular recognition by the C domain, while the extra 3′,5′-diphosphoadenosine moiety from the C18-CoA substrate cannot be accommodated by the enzyme. The protein: protein interactions between the C domain and its ACP substrate provides molecular interactions that enhance catalysis, also observed in other NRPS systems.8
Figure 3.
In vitro characterization of TwmB-C. a) LC-MS analysis of TwmB-C with different thioester substrate mimics. b) Substrate specificity towards amine donor. c) Substrate specificity towards acyl-chain length and carrier protein.
Using C18-ACP as acyl donor substrate, we further characterized the steady-state kinetics of TwmB-C towards the acceptor substrate 5PA (Fig. S6). Notwithstanding that substrate inhibition was observed at high concentration (>20 mM), 9 apparent half maximal-velocity (KM) was reached at 2 mM for 5PA with apparent kcat = 12 min−1. Given that some structural features are missing in the substrate mimic C18-ACP when compared to the native nonaketide substrate 6, the catalytic efficiency (kcat/ KM) measured here could represent a lower limit for TwmB-C. Our data therefore demonstrates that this C domain is a bona fide condensation enzyme despite that the second catalytic histidine in the HHxxxDG motif is substituted by proline.
This result seemingly conflicts with the well-acknowledged importance of HHxxxDG motif found in the NRPS C domains.4 A possible explanation is that instead of using a histidine residue as a general base catalyst,10 or as an electrostatic catalyst stabilizing the tetrahedral anion transition state,11 TwmB-C exerts its catalysis mainly by positioning the approaching nucleophilic amino group of 5PA and aligning its lone electron pair with the Bürgi-Dunitz trajectory for nucleophilic attack at the π* orbital of the thioester carbonyl group, which may take place without a catalytic histidine residue. This hypothesis is in line with the recent proposal for classical condensation domain catalysis.4b Nonetheless, it cannot be excluded that the active site structures of TwmB-C is sufficiently distinct from canonical C domain and an alternative histidine or other polar residue is used for catalysis.
Intrigued by the preference of 5PA as the nucleophile over other more abundant intracellular competitive amine-containing metabolites, we probed the substrate specificity of TwmB-C by assaying different 5PA isosteres and analogues in the presence of C18-ACP (Figure 3b). The terminal carboxylate was found to be essential for molecular recognition, as replacement of the negatively charged carboxylate to neutral (−Me, −OH) or positively charged (−NH3+) failed to yield any product. We also determined the importance of the nucleophile, as substitution of the primary amino head group by either hydroxyl or thiol functional group abolished activity. We next determined the optimal distance between the nucleophile and the carboxylate to be 4 carbon atoms (~10 Å), with 5 carbon atoms to be the only other tolerable length. To reinforce the idea of molecular ruler for specificity, we assayed four more substrate analogues (8 – 11) bearing more rigid linker such as those containing benzene or cyclohexane (Figure 3b). As expected, with benzene ring as linker, only 8 whose length is closer to 10 Å is active (3%). Moreover, comparing 10 and 11 informs that the amino and carboxylate groups must be positioned on the same side of the molecule.
We further studied the substrate specificity of TwmB-C at the condensation donor site. To test the chain-length requirement of the acyl donor, we enzymatically loaded short to long fatty-acyl CoA onto apo TwmB ACP using NpgA, and incubated these acyl-S-ACPs with 5PA and TwmB-C. As shown in Figure 3c, TwmB-C clearly prefers long-chain fatty acyl substrate with no significant discrimination between C16 and C18, which is consistent with simultaneous formation of both octaketide and nonaketide acyl amides. We also tested the acyl-carrier protein substrate specificity by employing three dissected fungal acyl or peptidyl carrier proteins (PCP) from different PKSs and NRPS: PKS-C hybrid (LovB), NRPKS (TlnB), and NRPS (SidC-PCP3). In spite of low sequence identity (<25%) between any two of these acyl carrier proteins (Figure S7), TwmB-C was able to recognize and process all carrier protein-bound stearoyl thioesters,. A proposed scheme of the TwmB-C active site to account for all the above observations is depicted in Figure 4.
Figure 4.

Schematic proposal of substrate binding in the TwmB-C active site.
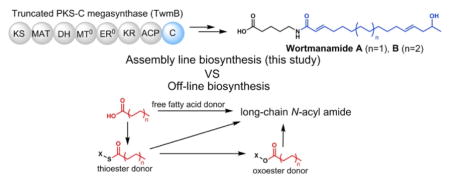
The wortmanamides discovered here resemble long-chain fatty acyl amides which are known to activate diverse channels and receptors, including GPCRs.12 Interestingly, a GPCR-like protein TwmC is encoded in the cluster and is co-transcribed with PKS-C hybrid TwmB (Figs. S3 and S8).. The assembly line biosynthesis of wortmanamides are different from the known mechanisms of fatty acyl amides formation: free fatty acids are either directly transferred to amino acids by ATP-grasp enzyme; or activated as thioesters or oxoesters followed by transfer to amine nucleophiles by N-acyltransferase or N-transacylase, respectively.12a, 13 The use of a dedicated and programmed PKS assembly line enables synthesis of functionalized fatty acyl chain which may play a specific biological role and avoids releasing it into fatty acid pool. Although both are non-canonical based on the active site motif, the function of the C domain from TwmB-C is strikingly different from that of LovB-C, contributing to the dramatic structural difference of final products. Our study offers the prospect that diverse functions of noncanonical C domains and new natural products may be discovered from this family of PKS-C megasynthases.
Supplementary Material
Acknowledgments
This work was supported by the NIH (1R35GM118056 to Y.T.). Y.H. is a Life Sciences Research Foundation Fellow. We thank Drs. S.-S. Gao, M.-C. Tang, and M. Ohashi for helpful discussions. We also thank Dr. M. Chen for providing apo TlnB ACP protein sample.
Footnotes
Notes
The authors declare no competing financial interest.
Experimental details, spectroscopic data. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.(a) Cox RJ. Org Biomol Chem. 2007;5:2010. doi: 10.1039/b704420h. [DOI] [PubMed] [Google Scholar]; (b) Chooi YH, Tang Y. J Org Chem. 2012;77:9933. doi: 10.1021/jo301592k. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Roberts DM, Bartel C, Scott A, Ivison D, Simpson TJ, Cox RJ. Chem Sci. 2017;8:1116. doi: 10.1039/c6sc03496a. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Cacho RA, Thuss J, Xu W, Sanichar R, Gao Z, Nguyen A, Vederas JC, Tang Y. J Am Chem Soc. 2015;137:15688. doi: 10.1021/jacs.5b11814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.(a) Boettger D, Hertweck C. ChemBioChem. 2013;14:28–42. doi: 10.1002/cbic.201200624. [DOI] [PubMed] [Google Scholar]; (b) Fisch KM, Bakeer W, Yakasai AA, Song Z, Pedrick J, Wasil Z, Bailey AM, Lazarus CM, Simpson TJ, Cox RJ. J Am Chem Soc. 2011;133:16635. doi: 10.1021/ja206914q. [DOI] [PubMed] [Google Scholar]; (c) Li XW, Ear A, Nay B. Nat Prod Rep. 2013;30:765. doi: 10.1039/c3np70016j. [DOI] [PubMed] [Google Scholar]
- 3.(a) Ma SM, Li JWH, Choi JW, Zhou H, Lee KKM, Moorthie VA, Xie X, Kealey JT, Da Silva NA, Vederas JC, Tang Y. Science. 2009;326:589. doi: 10.1126/science.1175602. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Kakule TB, Lin Z, Schmidt EW. J Am Chem Soc. 2014;136:17882. doi: 10.1021/ja511087p. [DOI] [PubMed] [Google Scholar]; (c) Boettger D, Bergmann H, Kuehn B, Shelest E, Hertweck C. ChemBioChem. 2012;13:2363. doi: 10.1002/cbic.201200449. [DOI] [PubMed] [Google Scholar]
- 4.(a) Except cyclization domain (Cy) in bacterial NRPSs. Bloudoff K, Schmeing TM. Biochim Biophys Acta. 2017;1865:1587. doi: 10.1016/j.bbapap.2017.05.010.
- 5.(a) Hillson NJ, Balibar CJ, Walsh CT. Biochemistry. 2004;43:11344–51. doi: 10.1021/bi0489199. [DOI] [PubMed] [Google Scholar]; (b) Haslinger K, Peschke M, Brieke C, Maximowitsch E, Cryle MJ. Nature. 2015;521:105. doi: 10.1038/nature14141. [DOI] [PubMed] [Google Scholar]
- 6.According to the Metacyc database: superpathway of L-lysine degradation Caspi R, Altman T, Billington R, Dreher K, Foerster H, Fulcher CA, Holland TA, Keseler IM, Kothari A, Kubo A, Krummenacker M, Latendresse M, Mueller LA, Ong Q, Paley S, Subhraveti P, Weaver DS, Weerasinghe D, Zhang P, Karp PD. Nucleic Acids Res. 2014;42:D459. doi: 10.1093/nar/gkt1103.
- 7.Mootz HD, Schörgendorfer K, Marahiel MA. FEMS Microbiol Lett. 2002;213:51. doi: 10.1111/j.1574-6968.2002.tb11285.x. [DOI] [PubMed] [Google Scholar]
- 8.(a) Liu X, Walsh CT. Biochemistry. 2009;48:8746. doi: 10.1021/bi901123r. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Gao X, Haynes SW, Ames BD, Wang P, Vien LP, Walsh CT, Tang Y. Nat Chem Biol. 2012;8:823. doi: 10.1038/nchembio.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Similar behavior was observed for VibH towards norspermidine: Keating TA, Marshall CG, Walsh CT. Biochemistry. 2000;39:15513. doi: 10.1021/bi001651a.
- 10.Bergendahl V, Linne U, Marahiel MA. Eur J Biochem. 2002;269:620. doi: 10.1046/j.0014-2956.2001.02691.x. [DOI] [PubMed] [Google Scholar]
- 11.Samel SA, Schoenafinger G, Knappe TA, Marahiel MA, Essen LO. Structure. 2007;15:781. doi: 10.1016/j.str.2007.05.008. [DOI] [PubMed] [Google Scholar]
- 12.(a) Farrell EK, Merkler DJ. Drug Discov Today. 2008;13:558. doi: 10.1016/j.drudis.2008.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Cohen LJ, Esterhazy D, Kim SH, Lemetre C, Aguilar RR, Gordon EA, Pickard AJ, Cross JR, Emiliano AB, Han SM, Chu J, Vila-Farres X, Kaplitt J, Rogoz A, Calle PY, Hunter C, Bitok JK, Brady SF. Nature. 2017;549:48. doi: 10.1038/nature23874. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Vizcaino MI, Engel P, Trautman E, Crawford JM. J Am Chem Soc. 2014;136:9244. doi: 10.1021/ja503450q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.(a) Torring T, Shames SR, Cho W, Roy CR, Crawford JM. ChemBioChem. 2017;18:638. doi: 10.1002/cbic.201600618. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Van Wagoner RM, Clardy J. Structure. 2006;14:1425. doi: 10.1016/j.str.2006.07.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.