Abstract
Purpose
MRI-based cell tracking has emerged as a useful tool for identifying the location of transplanted cells, and even their migration. Magnetically labeled cells appear as dark contrast in T2*- weighted MRI, with sensitivities of individual cells. One key hurdle to the widespread use of MRI-based cell tracking is the inability to determine the number of transplanted cells based on this contrast feature. In the case of single cell detection, manual enumeration of spots in 3D MRI in principle is possible; however, it is a tedious and time-consuming task that is prone to subjectivity and inaccuracy on a large scale. This research presents the first comprehensive study on how a computer based intelligent, automatic and accurate cell quantification approach can be designed for spot detection in MRI scans.
Methods
Magnetically labeled mesenchymal stem cells (MSCs) were transplanted into rats using an intracardiac injection, accomplishing single cell seeding in the brain. T2*- weighted MRI of these rat brains were performed where labeled MSCs appeared as spots. Using machine learning and computer vision paradigms, approaches were designed to systematically explore the possibility of automatic detection of these spots in MRI. Experiments were validated against known in vitro scenarios.
Results
Using the proposed deep convolutional neural network (CNN) architecture, an in vivo accuracy up to 97.3% and in vitro accuracy of up to 99.8% was achieved for automated spot detection in MRI data.
Conclusion
The proposed approach for automatic quantification of MRI-based cell tracking will facilitate the use of MRI in large scale cell therapy studies.
Introduction
Cell-based therapies are poised to make a significant impact across a broad spectrum of medical scenarios. In regenerative medicine, stem cell transplants are in various stages of clinical trials for treating or slowing a myriad of diseases, including Parkinson’s disease (1)(2), rheumatoid arthritis (3)(4) and multiple sclerosis (5)(6). Cell-based therapy in the form of cancer immunotherapy is also being tested in clinical trials (7)(8). It is well acknowledged that imaging the location of transplanted cells, both immediately and serially after delivery, will be a crucial component for monitoring the success of the treatment. Two important applications for imaging transplanted cells are:
to non-invasively quantify the number of cells that were delivered or that homed to a particular location, and
to serially determine if there are cells that are leaving desirable or intended locations and entering undesirable locations.
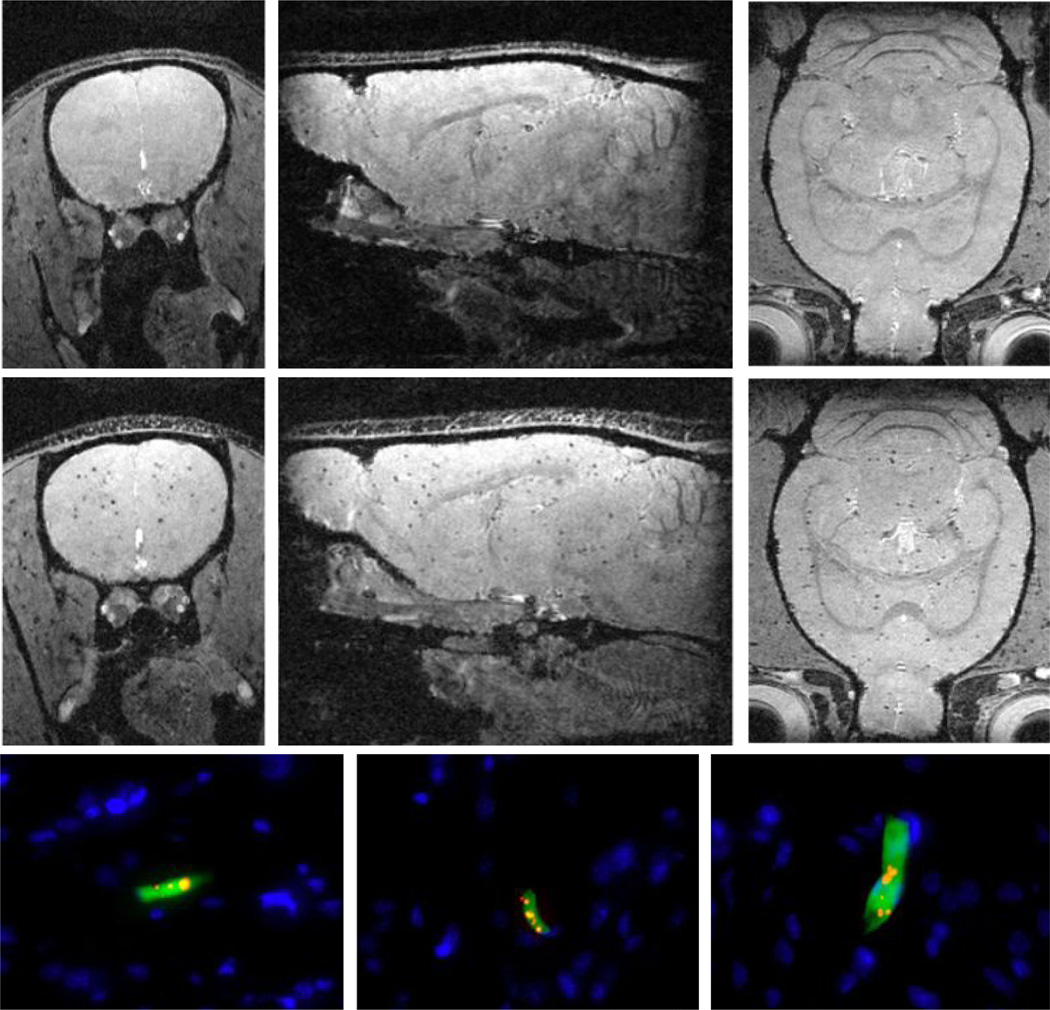
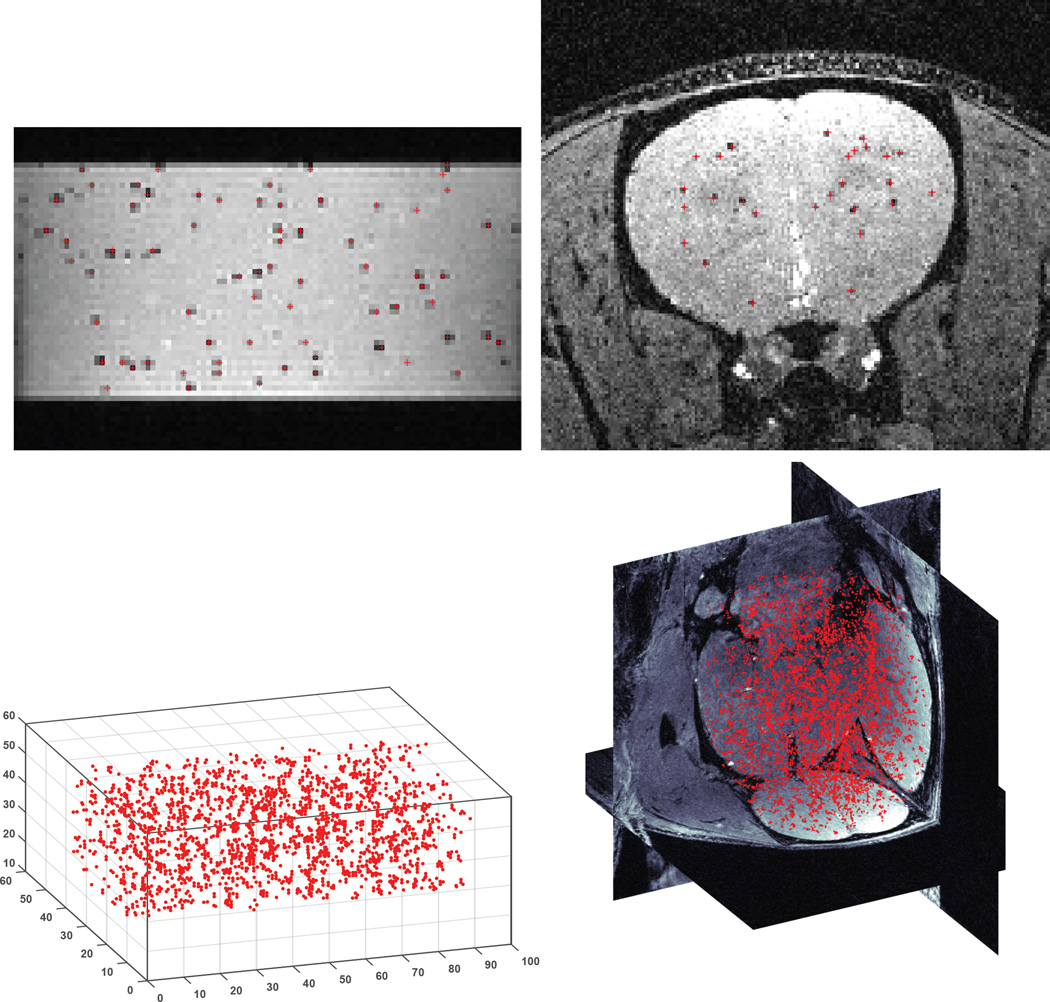
For multiple reasons, including image resolution, lack of radiation, and established safety and imaging versatility, magnetic resonance imaging or MRI has emerged as the most popular and perhaps most promising modality for tracking cells in vivo following transplant or delivery. In general, MRI-based detection of cells is accomplished by first labeling cells with superparamagnetic iron oxide nano- or microparticles, though some cell types can be labeled directly in vivo, such as neural progenitor cells. Following transplant, these labeled cells are then detected in an MRI by using imaging sequences where the signal intensity is sensitive to the local magnetic field inhomogeneity caused by the iron oxide particles. This results in dark contrast in the MRI (9) (10). In the case of a transplant of large numbers of magnetically labeled cells, large areas of dark contrast are formed. In the case of isolated cells, given sufficient magnetic labeling and high image resolution, in vivo single cell detection is possible, indicated by a well-defined and well characterized dark spot in the image (See Fig. 1).
Fig. 1.
Three orthogonal MRI slices extracted from 3D data sets of the brain from animals injected with unlabeled MSCs (top row) and magnetically labeled MSCs (middle row). Note the labeled MSCs appear as distributed dark spots in the brain unlike unlabeled MSCs. The bottom row shows three different fluorescence histology sections from animals injected with magnetically labeled MSCs confirming that these cells were present in the brain mostly as isolated, single cells. Blue indicates cell nuclei, green is the fluorescent label in the cell, red is the fluorescent label of the magnetic particle (See the supporting information Fig S1 for details).
Due to the rather complex relationship between iron content, particle distribution, iron crystal integrity, distribution of magnetic label and cells etc., it is difficult to quantify cell numbers in an MRI-based cell tracking experiment. This is especially the case for a single graft with a large number of cells. There are efficient methods of quantifying iron content, most notably using SWIFT based imaging (11), but the direct correlation to cell number is not straightforward, due to the reasons listed above. MRI-based detection of single cells presents a much more direct way of enumerating cells in certain cell therapy type applications, such as hepatocyte transplant (10), or for immune cells that have homed to an organ or a tumor (12). In this case, the solution is straightforward: if dark spots in the MRI are from single cells, then counting these spots in the MRI should yield cell number. While seemingly straightforward, performing such quantitative analysis on three-dimensional data sets is a difficult task that cannot be accomplished using traditional manual methodologies. Manual analysis and enumeration of cells in MRI is tedious, laborious, and also limited in capturing patterns of cell behavior. In this respect, a manual approach cannot be adopted to analyze large scale datasets comprising dozens of research subjects. Various commercial software that are currently available for MRI can only assist a medical expert in conducting manual analysis. The problem is further compounded in the case of eventual MRI detection of single cells at clinical resolution, which is lower than that achieved on high field small animal systems. At lower image resolution, the well-defined, well-characterized dark spot loses shape and intensity and can be difficult to manually define in a large number of MRI slices.
These hurdles highlight the pressing need to develop an automatic and intelligent approach for detecting and enumerating transplanted cells in MRI, meeting all the aforementioned challenges. An automatic and intelligent approach can allow researchers to efficiently conduct large scale analysis of transplanted cells in MRI, facilitating the exploration of new transplant paradigms and cell sources. Such generalized intelligent tools will find use across a broad spectrum of biomedical pursuits. However, the unique challenges of designing such a tool has not been addressed in any prior literature, especially in the context of detecting cells in MRI.
To design and evaluate an intelligent and automatic approach for cell spot detection in MRI, ground truth definitions, i.e., labels, that annotate spots in MRI images, are required. In (14), authors recognized the need for automation and adopted a threshold based strategy for automatically detecting spots in MRI. However, their approach was not evaluated using a ground truth. Although such threshold reliant approaches are not known to be intelligent for handling variations and diversity in data, their study in fact highlights the need for automation (13) (15). Automatic ML approaches have been successfully used in a wide range of image analysis applications (16)(17)(13). However, it is unexplored how such approaches can be appropriated to the problem of MRI spot detection. Further, state-of-the-art ML approaches rely on a large volume of training data for accurate learning. Unfortunately, due to practical limitations, generating large scale annotated data is challenging in both preclinical and the clinical arenas. Annotation can also be prohibitively time-consuming and can only be performed by a medical expert. Hence, crowd sourcing approaches such as the use of Amazon’s Mechanical Turk (28), cannot be adopted for annotation in such applications. Therefore, the problem of spot detection using a limited amount of annotated training data, is an additional unaddressed challenge. In summary, the problem of 3D spot detection in MRI presents the following key challenges:
Candidate region extraction: Given an MRI scan, how can all the candidate regions that can potentially contain a spot be effectively and efficiently extracted?
Feature design: What will be the best feature representation to accurately capture the inter-class appearance variations of spots in MRI?
Dataset collection: Intelligent ML approaches will require annotated (labeled) MRI datasets for spot detection. Therefore, a diverse set of MRI scans need to be collected and labels must be obtained on them.
Learning with limited data: Further, how can state-of-the-art ML approaches learn to detect spots in MRI despite using very limited MRI data for training?
This paper addresses these challenges of automated spot detection in MRI and presents the first comprehensive study to investigate how different ML approaches encompassing three different paradigms, can be utilized for this purpose. Experimental results of the approaches proposed in this paper show that spots can be automatically detected using ML techniques in unseen in vivo MRI scans with an accuracy of up to 97.3%.
Methods
Datasets
As shown in Tab. 1, a diverse set of 33 in vitro MRI scans of gel samples and 7 in vivo MRI scans of rat brains with transplanted stem cells were utilized in this study. More than 19, 700 manual ground truth labels were collected on 15 of these scans (GA − GD). A flexible software tool (with image pan, zoom, slice advance, contrast manipulation, etc.) was specifically designed to allow our medical expert to put a mouse cursor over a spot in an MRI and click the mouse button to record that spot. These clicked points are taken as our ground truth (labels) on spots in MRI. In addition, theoretically computed cell numbers on 25 scans were also utilized as ground truths during the approach evaluation.
Table 1.
Collection details and characteristics of our MRI database.
| Set | Type | Subject | Labeler | Machine | Labeled Scans | Total Labels | Resolution | Size |
|---|---|---|---|---|---|---|---|---|
| GA | in vivo | Brain | R1 | 11.7T | G1A, G2A, G3A, G4A, G5A | 15442 | 100µm | 256 × 256 × 256 |
| GB | in vivo | Brain | R2 | 7T | G1B, G2B | 2992 | 100µm | 256 × 200 × 256 |
| GC | in vitro | Tube | R2 | 7T | G1C, G2C, G3C, G4C | 814 | 100µm | 128 × 80 × 80 |
| GD | in vitro | Tube | R2 | 7T | G1D, G2D, G3D, G4D | 514 | 200µm | 64 × 40 × 40 |
| GE | in vitro | Tube | t̄ | 7T | G1E, G2E, G3E …, G25E | (2400 × 25) | 100µm | 100 × 64 × 64 |
In vitro
Imaging phantoms were constructed consisting of a known number of 4.5 micron diameter, magnetic microparticles with 10 pg iron per particle, suspended in agarose samples. Each microparticle approximates a single magnetically labeled cell with appropriate iron content for MRI-based single cell detection (18). T2*-weighted gradient echo MRI was then performed on these samples at a field strength of 7T.
As can be seen in Tab. 1, these scans have variation in resolution, matrix sizes, and amount of spots (labels). GE has 25 data sets, collected from 5 samples under 5 different MRI conditions. These conditions were variations in TE from 10 – 30 ms (signal to noise > 30:1), and images with low signal to noise ratio (~ 8:1) at TE = 10 and 20. The effect of increasing TE is to enhance the size of the spots. The higher the TE, the larger the spot (18). The downside of higher TE is that the physics which governs enlargement of the spot, the difference in magnetic susceptibility between the location in and around the magnetic particles and the surrounding tissue, also causes the background tissue to darken. The rationale to collect images with both high and low signal to noise ratio is to test the robustness of our spot detection procedure in two potential in vivo scenarios. Manual ground truths were collected from experts on 8 in vitro MRI scans of GC and GD. These sets were used for training and evaluating ML approaches. For GE, the theoretically computed ground truth was known. This set was used for a direct comparison between the automatically detected spots and the theoretically expected.
In vivo
Two different sets of in vivo MRI were collected from two different machines having different field strengths. Using one machine with a field strength of 11.7T, 5 MRI scans of rats were collected, which are represented by GA in Tab. 1. Three of them were injected intracardiac 1 – 1.5 hours prior to the scan with rat mesenchymal stem cells (MSCs) that had been labeled with micron sized iron oxide particles (MPIOs) to a level of ~14 pg iron per cell. This transplantation scheme delivers cells to the brain - an intravenous injection would deliver cells only to the liver and lungs. Two additional rats were not injected at all. Using another machine with 7T, 2 further brain MRI scans of rats were collected, similarly previously transplanted with MPIO labeled MSCs. GB represents these 2 scans in Tab. 1. The rationale behind collecting these two different in vivo sets was to be able to validate the generalization and robustness of our learned algorithm against potential variations arising from different imaging systems. Note that a different amount of MSCs were injected in different rats to achieve further variations in the data. All MRI were 3D T2*-weighted gradient echo. Further details regarding cell labeling, cell transplant, in vivo MRI and histology are in Supplemental Information.
Machine learning methods
In machine learning, a classification approach maps a real world problem into a classification task where two or more entities (classes) are to be intelligently distinguished from each other (see (19) for basic details). For example, classifying potential candidate regions in MRI as spots or non-spots will also be a classification task.
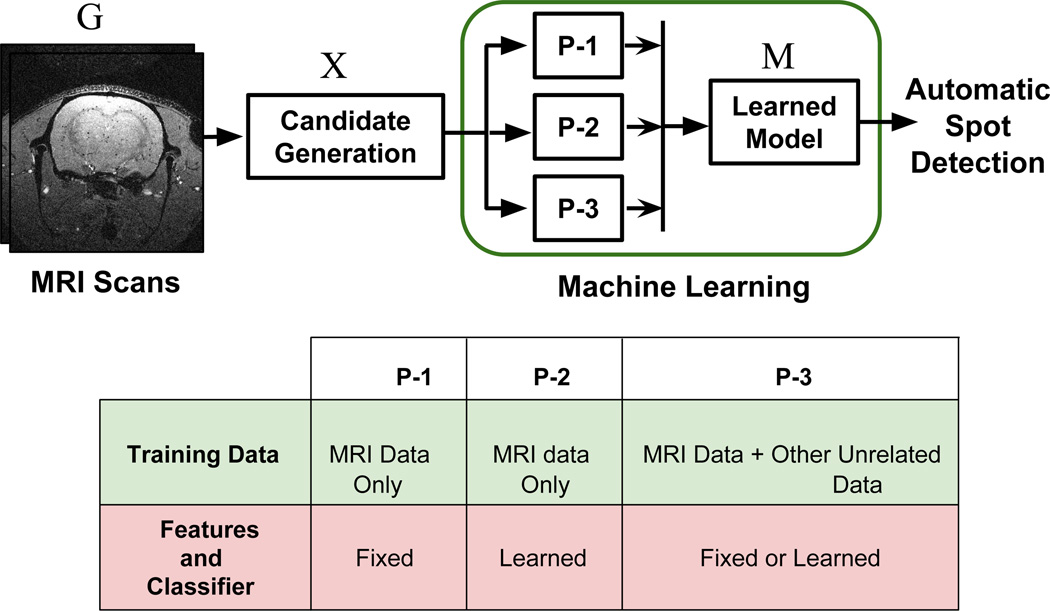
In the context of this work, classification paradigms can be categorized into three fundamental paradigms. In the first paradigm (P-1), discriminating information is extracted from the images using a pre-defined approach that is designed by an expert based on intuition and experience. This information may be in the form of a numeric array of values known as features. For each image such features along with their ground truth classification labels are then forwarded to another algorithm called classifier or classification technique which learns to distinguish between the classes. A classifier can be learned mathematical functions, set of if-then rules etc.
In the second paradigm (P-2), the feature representations are not manually designed by an expert but rather automatically learned from the data. Generally, both, feature representations and the classifiers are learned automatically in a single unified framework. Many neural network based approaches fall into this category which can take image datasets directly as input, along with the labels, and learn a classification model.
In a third classification paradigm (P-3), the model can be learned in the same manner as in P-1 or P-2. The difference here is that learning the model requires more than just the given task’s data (MRI data in this case). Available labeled data for other tasks such as face recognition, that may not be directly related to the given task, is exploited using a transfer learning approach. This approach is useful when collecting large scale annotated data is challenging.
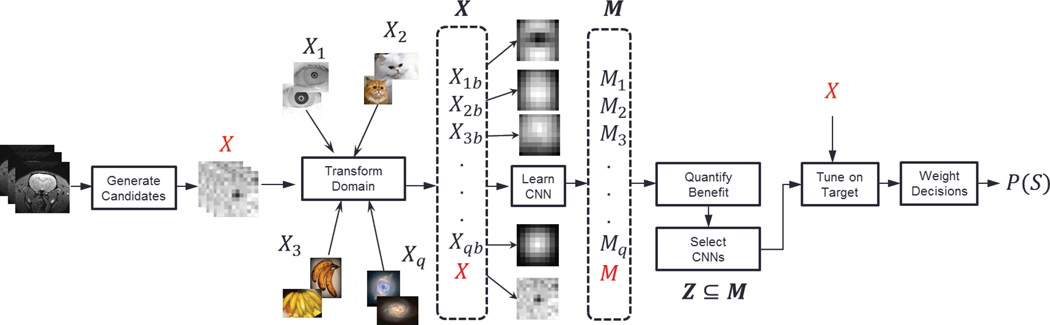
The general architecture of this study and the differences between the ML paradigms are summarized in Fig. 2. Candidate regions are located and extracted from an MRI scan G using the approach proposed in this study. Each candidate region xi may or may not contain a spot. Therefore, all candidate regions in X, along with their manual ground truth labels , yi ∈ {1, 0}, are then forwarded to each of the 3 machine learning paradigms for learning a model M. Depending on the paradigm, this model may be based on a set of if-then rules, mapping functions, a sequence of convolutional filters, etc. For example, in the context of CNN, the model M can take a candidate region as an input and apply a sequence of learned convolutional filters and transformation functions to finally output a value that either describes the candidate region as a spot or a non-spot. Thus, once M is learned, the proposed approach can automatically locate, extract and detect spots in any unseen MRI.
Fig. 2.
(Top) Basic architecture: Three different ML paradigms P-1, P-2 and P-3 are explored to learn a spot detection model. (Bottom) Fundamental design phase differences between the three ML paradigms.
Candidate Generation using Superpixels
The first challenge in this research is to define a candidate region. Processing each pixel as a candidate region can result in a huge computational burden. We addressed this issue by extracting superpixels as classification units from each MRI scan (15). A superpixel technique groups locally close pixels with similar intensities into a single unit. Since spots are usually darker than their surrounding, they are characterized as superpixels with lower average intensity than the surrounding superpixels. Based on this idea, a novel set of features based on the superpixel intensities, was designed. Experimental results show that these features provide superior performance for spot detection compared to the approach in (13). However, this approach has the following limitations: (1) The accuracy of the approach was dependent on the preciseness of the superpixel algorithms. (2) The approach assumes a superpixel based model for a spot in terms of its depth across consecutive MRI slices. This does not hold true for all spots in different MRI settings.
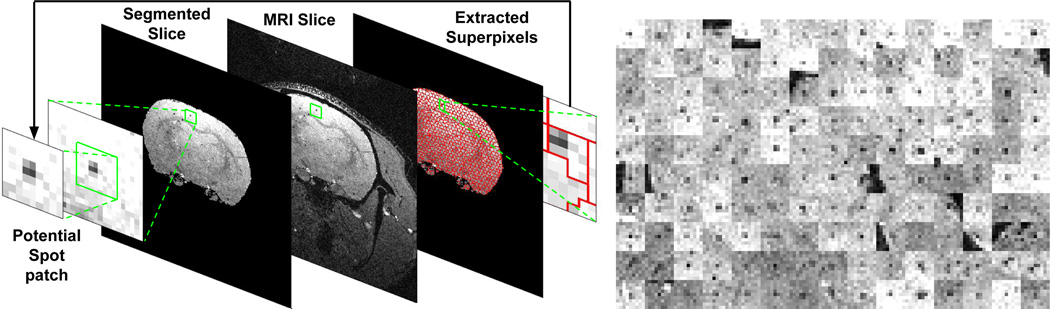
The strategy adopted in this paper is resilient to imprecisions in the superpixel extraction algorithms. Based on each superpixel unit, a representative patch is extracted from the MRI scan as explained in Fig. 3. Each patch is then taken as a candidate region and undergoes a feature extraction process. The approach is model-free and imitates the strategy adopted by a human labeler. All candidate patches are first detected in 2D MRI slices and then neighboring patches detected in consecutive slices are connected without imposing any restriction on their depth in 3D.
Fig. 3.
(Left) Illustrating the generation of candidate regions. For each superpixel a candidate patch is extracted. The darkest pixel in the superpixel acts as the center of the patch. (Right) A mosaic of several 9 × 9 patches extracted from an MRI slice. It can be seen that all patches have a dark region in the center representing a spot in a 2D slice.
The spatial location of each patch in MRI is also recorded. Consequently, these extracted patches are forwarded to the machine learning algorithms as input data.
In summary, the first two paradigms focus on how to accurately and automatically distinguish spot patches from non-spot patches. Then using the 3rd ML paradigm, i.e., transfer learning, we investigated how the best approach out of the first two paradigms could be adapted to achieve better results despite using very limited training data.
Results
Spot detection with fixed designs (P-1)
This is the traditional and most widely adopted paradigm in computer vision and pattern recognition based studies (15)(13). In this paper, an elaborate set of feature extraction methods are utilized that extract shape, intensity, texture and context information about the entities in the candidate patches. In the attached supporting material, Fig. S2, Fig. S3 and Fig. S4 present a brief explanation on how hand-designed features can be extracted specifically for the task of capturing spot appearance in MRI.
Extracted features are finally concatenated to form a feature vector for each candidate patch xi. From this feature vector, the most useful features are selected and the irrelevant features are eliminated using a feature selection module that employs a correlation based feature selection algorithm (22). These feature vectors along with their corresponding labels are then forwarded to tune a classifier. In this study, a diverse group of classifiers such as probabilistic (Naive bayes), functional (Multi-layer perceptron(MLP)), and decision tree (Random Forest), are utilized (see (23) for details).
Spot detection with learned designs (P-2)
Based on expert intuition and experience, features extracted in P-1 can be subjective. Therefore, the key goal of P-2 approaches is to automatically learn the most optimal spot feature representation from the data. Neural networks are a well-known example of P-2 approaches.
Deep convolutional neural network (CNN) (26) (16) have been highly successful in many image based ML studies. Unlike P-1, these features are hierarchically learned in multiple layers in an automatic fashion and not hand-crafted by experts. Consider, M = f() as an overall classification model learned by a P-2 approach. In deep neural networks, f can be decomposed into multiple functional layers:
| [1] |
Each function, fj, j ∈ [1, u], can represent a (a) convolutional layer, (b) non-linear gating layer, (c) pooling layer, (d) full-connected layer (see (26, 27, 16) for more details). For a given task, weights for these convolutional filters are learned automatically using the training data. Different architectures of a CNN are created by utilizing different number of layers and also by sequencing these layers differently. CNN architectures also vary depending on the choice of the non-linear gating function. Filter sizes for convolutional layers are also determined depending on the application at hand. Well-known CNN architectures such as AlexNet (26) or GoogLeNet (32) cannot be utilized for spot detection in MRI. Therefore, a new CNN architecture, specifically designed for spot detection in MRI, is proposed here. The proposed CNN architecture has 3 composite layers and 1 fully connected layer (see Fig S5 in the supporting material). Each composite layer consists of a convolutional layer and a gating function. Note that in a conventional CNN architecture, a pooling layer is also used which reduces the dimensionality of the input data. However, a pooling layer is not utilized in this architecture due to the small size of the input patches (9 × 9). Using a pooling layer, in this context, may result in the loss of valuable information which may be essential to be utilized by the next layers. Further, a gating function is usually added for introducing non-linearity into a CNN. Without non-linear gating, a CNN can be seen as a sequence of linear operations which can hinder its ability to learn the inherent non-linearities in the training data. In conventional neural networks, a sigmoid function or a hyperbolic tangent function was generally utilized for this purpose. However, in recent studies, utilizing ReLU (Rectified Linear Units) has shown significantly superior results for this role (26). Therefore, the proposed architecture uses ReLU as a non-linear gating function.
Further customizing to the task at hand, the sizes of all the convolutional filters were kept small. However, their numbers were kept high. The goal was to provide a higher capacity to the CNN architecture for capturing a diverse set of local features of a patch. Filter sizes and dimensions of resulting feature maps can be seen in Fig. S5 (of supporting material). For any task i, the proposed model (CNN architecture) can be written as
| [2] |
where γ represents a standard softmax function that can be applied to the output of the fully connected layer Lfc. β denotes the non-linear gating function and Cik represents the convolutional layer in the composite layer k.
Performance of the two paradigms
Experiments were performed to answer the following main questions: (1) Which of the two ML technique results in the best detection accuracy for in vivo spots in MRI? (2) How does the best approach perform on in vitro evaluation studies? (4) Can a ML approach learned on in vivo data be tested for spot detection on in vitro data? (5) How is the performance affected if the MRI is conducted at low resolution? (6) Is the proposed approach robust to the differences in MRI machines in terms of field strength, make and model etc.? Importantly, it is also of interest to investigate how the theoretically computed number of spots for in vitro MRI scans compares with the automatically detected spot numbers.
In vivo evaluation studies
In this study, the spot classification performance of a diverse set of approaches was evaluated using the two sets of in vivo MRI scans i.e GA and GB. First, experiments and results are discussed for GA that has 5 different MRI scans obtained from one MRI machine and labeled by one expert. Three of these in vivo scans contain spots that were manually labeled by experts whereas the remaining two were naive. Six combinations of testing and training pairs are created such that two scans are always present in the testing set of each pair, where one of the scans is a naive and the other contains spots. The remaining 3 out of the 5 scans are used for training the ML algorithms. Area Under the Curve (AUC) is utilized as a standard measure for classification accuracy. Experimental results for all the algorithms are listed in Table 2.
Table 2.
Experimental comparison of in vivo spot detection performance using P-1 and P-2.
| Algorithms | J1 | J2 | J3 | J4 | J5 | J6 | means | |
|---|---|---|---|---|---|---|---|---|
| P-1 | Random Forest | 94.0 | 86.9 | 95.3 | 94.1 | 86.0 | 94.7 | 91.8 ± 4.2 |
| Naive Bayes | 82.9 | 81.8 | 84.3 | 84.1 | 80.1 | 83.7 | 82.8 ± 1.6 | |
| P-2 | CNN | 96.4 | 92.3 | 96.1 | 96.4 | 91.2 | 95.0 | 94.6 ± 2.3 |
| MLP | 91.1 | 85.2 | 90.9 | 91.4 | 84.2 | 90.3 | 88.9 ± 3.3 | |
| MLP (P-1/2) | 93.9 | 89.4 | 95.8 | 95.4 | 90.0 | 95.7 | 93.4 ± 2.9 | |
| means | 91.7 ± 5.2 | 87.1 ± 4.0 | 92.5 ± 5.0 | 92.3 ± 4.9 | 86.3 ± 4.5 | 91.9 ± 5.0 | ||
It was observed that the best results were achieved by a CNN, with a mean accuracy of 94.6%. The superior performance of CNN can be mainly attributed to its ability to automatically explore the most optimal features using training data rather than relying on hand-crafted features utilized in traditional machine learning. Second, CNN learn features in a deep hierarchy across multiple layers. Recent research shows that such a hierarchy provides a superior framework to CNN for learning more complex concepts, unlike traditional machine learning approaches which learns in a shallow manner (26)(32)(27).
The second best results were observed with the simple MLP approach when it takes the carefully designed, handcrafted features as an input, rather than the raw data X. This MLP can be viewed as a mixed paradigm approach (P-1/2). However, the deep learning CNN that inherently extracts hierarchical features without using any hand crafted features resulted in the overall best performance. CNN detected a total of 5246, 5719 and 16048 spots in the 3 labeled rats of GA.
Probabilistic Naive bayes, using P-1, shows the worst detection performance with an average accuracy of 82.8%. This can be because naive bayes assumes complete independence between the features which in many practical problems may not be true. Further, it can be seen in Tab. 2 that J2 and J5 testing sets proved to be the most challenging with low mean accuracies of 87.1% and 86.3%, respectively, from all algorithms. Dataset J4 resulted in the overall best performance with mean accuracy of 92.5%. When investigating this, it was found that both J2 and J5 contained MRI scan GA1 in their test set accompanied with a different naive scan. It was seen that the labeled patches in GA1 were significantly more challenging in terms of morphology and intensity than those extracted from other scans.
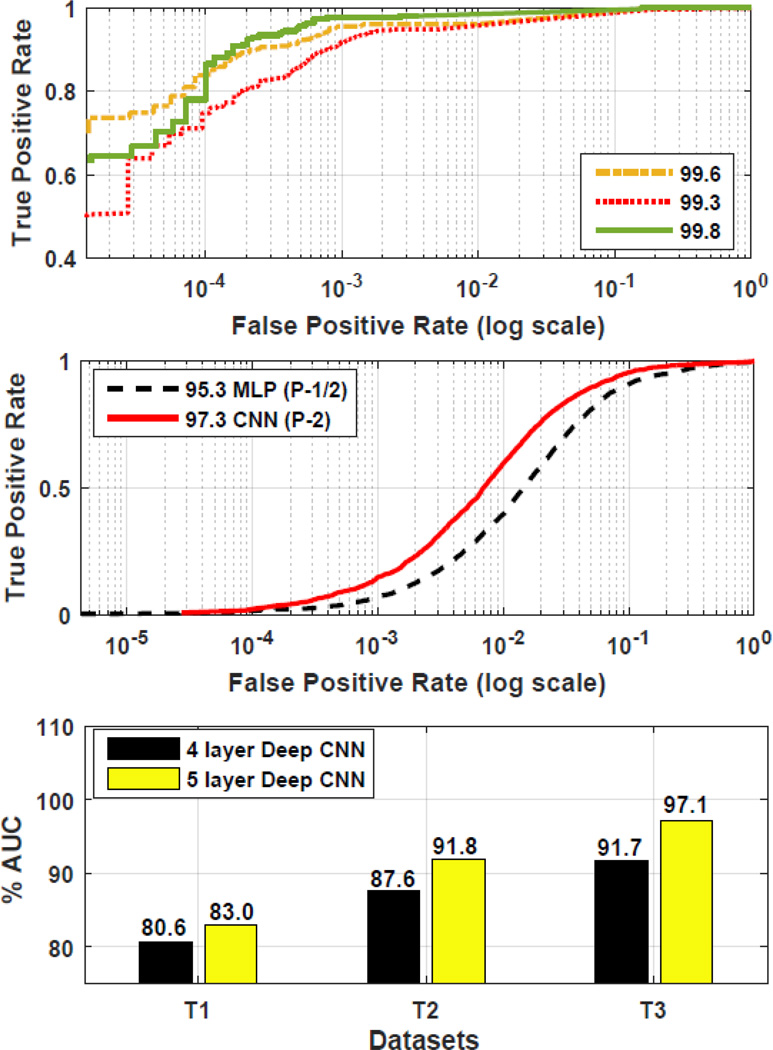
The best two approaches, i.e., MLP(P-1/2) and deep CNN(P-2), were then further compared using another set of in vivo scans i.e GB. This data was collected from a different machine having a different field strength and was also labeled by a different expert. In this study, all the previous 5 scans of GA were used for training both approaches (creating a larger training set), and then the learned spot detection models were tested on the in vivo scans in GB = {GB1, GB2}. Note that despite the differences in machine, its field strength, and also the labeling expert, CNN performed best with an accuracy of 97.3% whereas the mixed paradigm MLP (P-1/2) achieved 95.3%. We show the ROC curves for this test in Fig. 4. In GB, the total number of spots detected by CNN was 4930.
Fig. 4.
Comparison and results: (Top) in vitro results 100 micron, (Middle) generalization test using in vivo scans, (Bottom) in vitro results 200 micron.
In vitro evaluation studies
It can be observed that CNN yields the best result on the in-vivo datasets despite the simplicity of its approach. In this study, its performance is evaluated on the in vitro data in set GC and GD. Its performance is first tested on GC that has 4 in vitro MRI scans each with a 100µm resolution creating a 3D matrix of (128 × 80 × 80). Using these 4 scans, 3 different testing and training pairs were developed. Each testing and training pair has 2 MRI scans. The naive MRI scan was always kept in the test set, thereby generating 3 combinations with the remaining other sets. It was observed that CNN performed with a mean accuracy of 99.6% on in vitro scans. The individual ROC plots for these tests are shown in Fig 4.
A different study was then conducted to see the degradation in performance when each of the 4 in vitro scans are obtained with a much lower resolution of 200µm creating a matrix of (64 × 40 × 40). Such a study is desirable since in some practical applications it may be more convenient to rapidly obtain an MRI at a lower resolution, particularly in human examinations. Using the same procedure as before, three different testing and training pairs were created. It was noted that the mean performance decreased to 86.6% ± 5.6. However, it was also seen that when the number of learning layers for CNN was increased to 5 (4 composite and 1 fully connected) the performance improves to 90.6% ± 7.1. The individual improvements on all the three sets are shown in Fig. 4.
Comparison with theoretically computed spot numbers
A comparison between the automatically detected number of spots with the theoretically computed number of spots was conducted using 25 in vitro MRI scans of set GE. This is an important experiment as it allows a direct comparison with the actual number of injected spots. All the available data from GA to GD was used for training a CNN and then the trained CNN model was used for testing on these 25 scans in set GE. Each scan is expected to contain about 2400 spots. However, it is important to understand that due to the use of manual procedures, the actual number of spots may vary about 2400. The results of automatic spot detection are tabulated in Tab. 3 under different MRI conditions.
Table 3.
Automatically detected number of spots in 5 samples under 5 conditions. The theoretically expected number of spots in each sample is 2400.
| Condition | Tube 1 | Tube 2 | Tube 3 | Tube 4 | Tube 5 |
|---|---|---|---|---|---|
| TE 10 | 2147 | 2272 | 2474 | 2152 | 2270 |
| TE 20 | 2608 | 2750 | 3039 | 2644 | 2660 |
| TE 30 | 2844 | 2993 | 3272 | 2809 | 2909 |
| TE 10 (Low SNR) | 1982 | 2023 | 2247 | 1949 | 2014 |
| TE 20 (Low SNR) | 2419 | 2563 | 2794 | 2401 | 2445 |
Model generalization studies
In this section, the generalization ability of the proposed approach is determined by testing it in different possible practical scenarios. In practice, in vivo scans might be collected with different MRI machines at different laboratories using different field strengths. GA and GB represent two such in vivo datasets. As discussed before in the in vivo evaluation studies, and as shown in Fig. 4, the CNN based approach demonstrates robustness to such variations and achieves 97.3% accuracy despite such differences. Further, it is necessary to know how the performance would be affected if in vivo data is used for training but the in vitro data is used for testing. Therefore, an experiment was conducted where a CNN was trained using GA (in vivo) and then tested it using GC (in vitro). CNN still performed with an accuracy of 96.1%. An in vivo and in vitro MRI slice with automatically detected spots is shown in Fig. 5.
Fig. 5.
Automatic spot detection and visualizations: (left) in vitro, (right) in vivo.
Spot Detection with Transfer Learning (P-3)
The success of deep learning methods for a specific application depends on the availability of large scale annotated datasets. Unfortunately, in many applications, especially those related to medical imaging and radiology, obtaining a large scale annotated (e.g., labeled) dataset can be challenging. Therefore the focus here is devising a strategy to improve the accuracy of a CNN trained only on limited samples.
The concept of transfer learning (29)(27), or inductive transfer, entails the transfer of knowledge from a source task (e.g., document classification) to a target task (e.g., voice recognition). In this paper, transfer learning is exploited in the context of stem cell detection in MRI data (i.e., spot detection), where there is scarcity of labeled training data. Here, transfer learning is implemented via CNNs and involves transplanting network layers from one CNN (derived from the source task) to another (spot detection - known as the target task). The proposed approach is explained below and the basic architecture of this approach is shown in Fig. 6.
Fig. 6.
Architecture of the transfer learning based approach.
First, in addition to the target data, i.e., the given MRI data X, the data of unrelated 20 different real world source entities were collected from publicly available databases (30). These include images of entities such as soccer ball, cherry, egg, cat, bananas etc. (see Fig. S6 for all names in the supporting material). The data of each source i is denoted as Xi where i ∈ {1, 2, …, 20}.
Second, the data from these source tasks were geometrically transformed to ensure compatibility with the target task patches. Therefore, the images in Xi for each source i were transformed to 9 × 9 patches. This transformation is functionally denoted as T(Xi). It was observed that many of the down-sampled images display a spot-like pattern (as shown in Fig. S7 of the supporting material). However, these spots exhibit differences in their shape, size and intensity and, therefore, present completely different distributions.
Third, for each Xi, a two class dataset Xib = {T(Xi), T(Bg)} was developed. T(Xi) consists of samples of one class (positive), while T(Bg) consists of samples of the second class (negative). Images for the negative class, Bg, were selected using a popular search engine by querying the following entries: (a) texture and patterns, (b) sky and ocean, (c) grass. It was observed that the transformed images of these categories show visually rough or uniform characteristics similar to that of the non-spot patches. Collectively, all the obtained datasets can be denoted as X = [X, X1b, X2b, …, X20b].
Fourth, based on each 2 class dataset Xib, a binary classification task was defined. The goal of this task was to learn a CNN Mi that can distinguish between patches in T(Xi) and T(Bg). Generally, to learn a CNN, the weights in all of its layers are first randomly initialized. Let this be denoted as MR. Given a dataset Xib, these weights are iteratively changed. This learning can be functionally denoted as Mi = Θ(MR, Xib).
Consequently, a set of 20 different source CNNs can be learned. Using this approach, a CNN M = Θ(MR, X) can also be learned to differentiate between spot and non-spot patches. Collectively, the set of learned CNNs can be denoted as M = [M, M1, M2, …, M20]. On the other hand, CNNs can also be learned without using a randomly initialized network. For example, a CNN Mxi that can distinguish between spot and non-spot patches can also be learned, i.e., Mxi = Θ(Mi, X). This means, that the weights already learned for a source task i are transferred to initialize another CNN whose goal is to learn to distinguish between spot and non-spot patches (target task). This transfer provides a more useful starting point in learning a target CNN and, thus, results in better generalization of the learned CNN. However, previous research shows that transferring from some source tasks may be significantly more beneficial than that from others (27).
Therefore, as a fifth step, the proposed approach automatically determines which source CNN would be the most beneficial for transfer. There is no previous literature that shows how to automatically rank the available source CNNs based on their predicted benefit to the target task. Note that this is not a learning problem where an objective function can be stated and then optimized using the training data. Instead, it requires a zero-shot prediction which is a challenging task. The approach adopted here automatically measures the potential usefulness of each source CNN, Mi, by measuring its characteristic Ei prior to conducting transfer, where:
| [3] |
Ji measures how different is the learned information (CNN weights) of a source CNN Mi from the target CNN M whereas Ui measures how discriminating is a source CNN Mi for the target task. For details on computing Ji and Ui, see the supporting material on source CNN selection.
To the best of our knowledge, this is the first attempt at ranking source CNNs for a given target task. Note that λ is simply a weighting parameter. In this study, λ = 0.8 was used in all the experiments. Finding an optimal value of λ is not the focus here; however, a more optimal value may result in even better performance.
As a final step, either the top CNN Mi or a group of the top q ∈ {2, …, 20} sources can be selected for transfer. The selected top q sources makes a group Z ⊂ M. When using a group, the predictions of the multiple models are fused using a standard probabilistic approach by utilizing the corresponding Ei for each source CNN Mi as a prior in a bayesian formulation.
Performance of P-3
In this section, experiments were conducted to answer the following questions: (1) Does the spot detection benefit from transfer learning when the annotated training data is very limited? (2) Is the ranking of source CNNs prior to transfer, a beneficial procedure? (3) Why is it useful to combine information from CNNs learned from different sources?
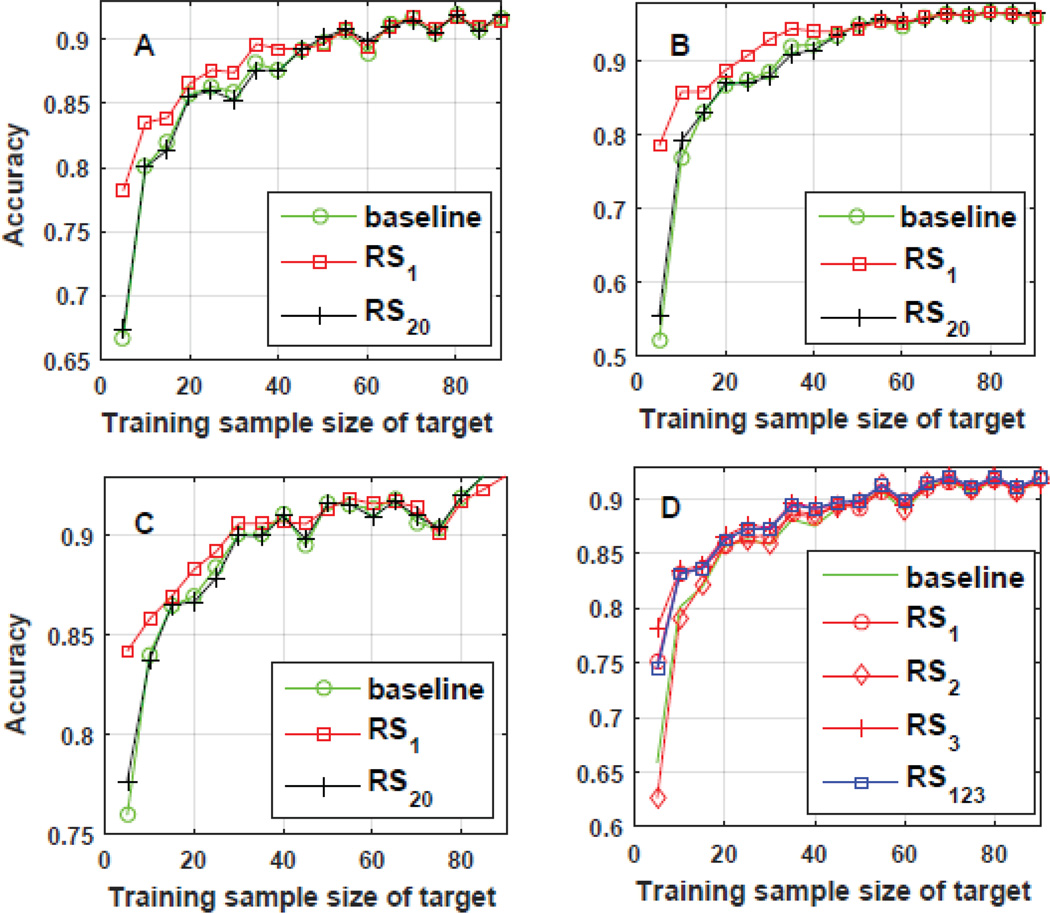
In Fig. 7(A,B,C), using all of the three labeled datasets of GA, three different testing scenarios are shown. On the x-axis, the amount of training data was varied and on the y-axis the performance of differently trained CNNs was observed. The baseline method is a CNN that did not undergo any transfer learning and only used the target data for training. The remaining two CNNs underwent transfer learning with the best ranked (RS1) and worst ranked (RS20) sources, respectively. We observed that the performance gain due to transfer from the best source is significantly higher when the training data from the target dataset is small. For example, in Fig. 7(B), using only 5% of the training data, the accuracy substantially improved from 52% to nearly 78% on the test set. Such an increase is highly encouraging in the clinical arena where there is scarcity of data and, where, the proposed approach would be highly relevant.
Fig. 7.
Results: D represent the case where information fusion provides robustness to ranking mistakes.
Next, we noted that the source which was ranked the “best” significantly outperforms the source which was ranked the “worst”. Thus, our strategy to rank each source prior to invoking the transfer learning paradigm is clearly of importance. Further, we observed that choosing a group of top ranked CNNs can be more useful than simply choosing one. In one scenario, the proposed approach mistakenly ranked one source as the second best as shown in Fig. 7(D). However, it can be seen that the decision weighting of multiple sources resulted in a performance that was significantly better than the worst source when there is limited target training data. In fact, at many points along the x-axis, we find that the resulting performance was higher than the other two sources as well.
Discussion
Automated spot detection presents a set of unique challenges that were carefully considered while designing computer vision and machine learning based approaches.
First, for thorough evaluation and training, an annotated MRI database needed to be developed. Therefore, a diverse database consisting of 40 MRI scans was assembled and more than 19, 700 manual labels were assigned.
Second, given an MRI scan, a set of candidate regions needed to be extracted effectively. Each candidate region must represent a region in MRI that can potentially contain a spot. This paper discussed how a superpixel based strategy can be designed to extract relevant regions. The proposed approach has clear advantages over some traditional alternatives.
Third, spots have high intra-class variation due to their diverse appearances in terms of shape and intensity. Therefore, for machine learning approaches to work effectively, a set of robust feature descriptors needed to be extracted from the candidate regions. A novel CNN architecture was designed to automatically extract the most useful spot features. The performance of these features was systematically compared against those extracted by appropriating hand-crafted feature extraction techniques. Results show that automatically learned features performed better with an accuracy of up to 97.3% in vivo.
Fourth, machine learning approaches typically require a large training dataset for accurate learning. However, in applications in the medical domain, it can be challenging to obtain a large volume of training data. Therefore, this paper explored how automatic spot detection can be performed using a limited amount of training data. A novel transfer learning strategy for CNNs was developed, where the best source task is automatically selected from an ensemble of many tasks.
It is important to note that MRI-based cell tracking has remained largely phenomenological for its history, starting in the late 80s. Moving forward, automated spot detection for MRI-based cell tracking would prove useful across a broad spectrum of research tracks. For example, Walczak et al, infused neural stem cells via the carotid artery in an effort to target stroke lesions (33). High resolution in vivo and in vitro MRI appear to show small clusters of cells, perhaps even single cells, distributed in the brain as a function of the intervention. Only qualitative analysis was performed on this imaging data; automated spot detection would have enabled quantitative metrics of cell numbers. Another application would be for the evaluation of transplanted islets encapsulated with iron oxide nanoparticles within alginate microspheres. These imaging features, typically are individual hypointensities, examples being (34) and (35). In both cases, only qualitative or semi- quantitative data were compiled, without a direct enumeration of transplanted and surviving grafts. A last example would be for enumeration of kidney glomeruli in conjunction with the use of cationized ferritin as a contrast agent (36).
The general use of MRI-based cell tracking and this specific approach to quantifying this data has some limitations. Still, MRI of magnetically labeled cells only detects the iron, not the cell itself, and this method is still unable to distinguish live cells from dead cells. Further, if more than one cell generates a particular spot in the MRI, then the calculated cell number would be inaccurate. In this work, only 67% of spots were resultant from individual cells, the other 33% from 2 or 3 cells. It remains an open question as to how accurate an automated spot detection algorithm for MRI-based cell tracking needs to be in order to provide useful clinical information. However, we do not feel that heterogeneous magnetic cell labeling is a significant problem. Indeed, cells with more internalized iron would have darker and larger spots on MRI, while cells with less internalized iron would have lighter and smaller spots. However, our automated quantification algorithm can account for differences in spot size and intensity to compensate for heterogeneous cell labeling.
Conclusion
In summary, this paper presented a comprehensive study on spot detection in MRI using machine learning (ML) approaches. Challenges unique to spot detection in MRI were highlighted. Novel approaches were designed for spot detection using different ML paradigms and were then experimentally compared. For this study, a new labeled database of MRI scans was developed. Results show that features that are automatically learned using a deep-learning approach outperform hand-crafted features. It was also observed that the transfer learning paradigm can provide significant performance improvement when the training dataset is small. Further, using deep convolutional neural networks, the proposed approach achieved up to 97.3% accuracy in vivo and about 99.8% in vitro.
Supplementary Material
Acknowledgments
We are thankful to the support provided by NIH grants DP2 OD004362 (EMS), R21 CA185163 (EMS), R01 DK107697 (EMS), R21 AG041266 (EMS). We are very grateful to the team of expert trainers, including Matt Latourette, Jason Unold, Michael Baer, Josie Alana and Jacob Nathaniel.
References
- 1.Clinical trial: Outcomes Data of Adipose Stem Cells to Treat Parkinson’s Disease. Website https://clinicaltrials.gov/ct2/show/NCT02184546 (First received: July 3, 2014 Last updated: June 17, 2015 Last verified: June 2015)
- 2.Clinical trial: A Study to Evaluate the Safety of Neural Stem Cells in Patients With Parkinson’s Disease. Website https://clinicaltrials.gov/ct2/show/NCT02452723 (First received: May 18, 2015 Last updated: March 10, 2016 Last verified: February 2016)
- 3.Clinical trial: Umbilical Cord Tissue-derived Mesenchymal Stem Cells for Rheumatoid Arthritis. Website https://clinicaltrials.gov/ct2/show/NCT01985464 (First received: October 31, 2013 Last updated: February 4, 2016 Last verified: February 2016)
- 4.Clinical trial: Cx611-0101, eASCs Intravenous Administration to Refractory Rheumatoid Arthritis Patients. Website https://clinicaltrials.gov/ct2/show/NCT01663116 (First received: August 5, 2011 Last updated: March 5, 2013 Last verified: February 2013)
- 5.Clinical trial: Evaluation of Autologous Mesenchymal Stem Cell Transplantation (Effects and Side Effects) in Multiple Sclerosis. Website https://clinicaltrials.gov/ct2/show/NCT01377870 (First received: June 19, 2011 Last updated: April 24, 2014 Last verified: August 2010)
- 6.Clinical trial: Stem Cell Therapy for Patients With Multiple Sclerosis Failing Alternate Approved Therapy- A Randomized Study. Website https://clinicaltrials.gov/ct2/show/NCT00273364 (First received: January 5, 2006 Last updated: March 21, 2016 Last verified: March 2016)
- 7.Clinical trial: Pilot Study of Redirected Autologous T Cells Engineered to Contain Humanized Anti-CD19 in Patients With Relapsed or Refractory CD19+ Leukemia and Lymphoma Previously Treated With Cell Therapy. Website https://clinicaltrials.gov/ct2/show/NCT02374333 (First received: February 23, 2015 Last updated: February 23, 2016 Last verified: February 2016)
- 8.Clinical trial: Genetically Modified T-cells in Treating Patients With Recurrent or Refractory Malignant Glioma. Website https://clinicaltrials.gov/ct2/show/NCT02208362.
- 9.Slotkin JR, Cahill KS, Tharin SA, Shapiro EM. Cellular magnetic resonance imaging: nanometer and micrometer size particles for noninvasive cell localization. Neurotherapeutics. 2007;4(3):428–433. doi: 10.1016/j.nurt.2007.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shapiro EM, Sharer K, Skrtic S, Koretsky AP. In vivo detection of single cells by MRI. Magnetic Resonance in Medicine. 2006;55(2):242–249. doi: 10.1002/mrm.20718. [DOI] [PubMed] [Google Scholar]
- 11.Zhou R, Djaudat I, Steen M, Curt C, Hualei Z, Hui Q, Jia Z, Michael G. SWIFT detection of SPIO labeled stem cells grafted in the myocardium. Magnetic Resonance in Medicine. 2007;63(5):1154–1161. doi: 10.1002/mrm.22378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu YL, Ye Q, Eytan DF, Liu L, Rosario BL, Hitchens TK, Yeh FC, Ho C. Magnetic resonance imaging investigation of macrophages in acute cardiac allograft rejection after heart transplantation. Circulation: Cardiovascular Imaging. 2013;6(6):965–973. doi: 10.1161/CIRCIMAGING.113.000674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Smal I, Loog M, Niessen W, Meijering E. Quantitative comparison of spot detection methods in fluorescence microscopy. IEEE Transactions on Medical Imaging. 2010;29(2):282–301. doi: 10.1109/TMI.2009.2025127. [DOI] [PubMed] [Google Scholar]
- 14.Mori Y, Chen T, Fujisawa T, Kobashi S, Ohno K, Yoshida S, Tago Y, Komai Y, Hata Y, Yoshioka Y. From cartoon to real time MRI: in vivo monitoring of phagocyte migration in mouse brain. Scientific reports. 2014;4 doi: 10.1038/srep06997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Afridi MJ, Liu X, Shapiro EM, Ross A. Automatic in vivo Cell Detection in MRI. Proc. of 18th International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI); Munich, Germany. 2015. [Google Scholar]
- 16.Taigman Y, Ming Y, Marc’Aurelio R, Lars W. Deepface: Closing the gap to human-level performance in face verification; Proc. of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR); 2014. pp. 1701–1708. [Google Scholar]
- 17.Afridi MJ, Liu X, McGrath JM. An Automated System for Plant-Level Disease Rating in Real Fields; In proc. of 22nd International Conference on Pattern Recognition (ICPR); 2014. pp. 148–153. [Google Scholar]
- 18.Shapiro EM, Skrtic S, Koretsky AP. Sizing it up: Cellular MRI using micronsized iron oxide particles. Magnetic Resonance in Medicine. 2005;53(2):329–338. doi: 10.1002/mrm.20342. [DOI] [PubMed] [Google Scholar]
- 19.Witten IH, Frank E. Data Mining: Practical machine learning tools and techniques. Morgan kaufmann. 2005 [Google Scholar]
- 20.Liu MY, Tuzel O, Ramalingam S, Chellappa R. Entropy rate superpixel segmentation; Proc. of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR); 2011. pp. 2097–2104. [Google Scholar]
- 21.Turk M, Pentland AP. Face recognition using eigenfaces; Proc. of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR); 1991. pp. 586–591. [Google Scholar]
- 22.Hall MA. Doctoral dissertation. The University of Waikato; 1999. Correlation-based feature selection for machine learning. [Google Scholar]
- 23.Bouckaert R, Frank E, Hall M, Kirkby R, Reutemann P, Seewald A, Scuse D. Weka manual for version 3-7-8. 2013 [Google Scholar]
- 24.Dalal N, Triggs B. Histograms of oriented gradients for human detection. Proc. of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR) 2005;1:886–893. [Google Scholar]
- 25.Oliva A, Torralba A. Building the gist of a scene: The role of global image features in recognition. Progress in Brain Research. 2006;155:23–36. doi: 10.1016/S0079-6123(06)55002-2. [DOI] [PubMed] [Google Scholar]
- 26.Krizhevsky A, Ilya S, Geoffrey EH. Imagenet classification with deep convolutional neural networks. Advances in Neural Information Processing Systems. 2012:1097–1105. [Google Scholar]
- 27.Yosinski J, Jeff C, Yoshua B, Hod L. How transferable are features in deep neural networks? Advances in Neural Information Processing Systems. 2014:3320–3328. [Google Scholar]
- 28.Sorokin A, Forsyth D. Utility data annotation with amazon mechanical turk. 2008 [Google Scholar]
- 29.Pan SJ, Qiang Y. A survey on transfer learning. IEEE Transactions on Knowledge and Data Engineering. 2010;22(10):1345–1359. [Google Scholar]
- 30.Deng J, Dong W, Socher R, Li LJ, Li K, Fei-Fei L. Imagenet: A large-scale hierarchical image database; Proc. of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR); 2009. pp. 248–255. [Google Scholar]
- 31.Schmidhuber J. Deep learning in neural networks: An overview. Neural Networks. 2015;61:85–117. doi: 10.1016/j.neunet.2014.09.003. [DOI] [PubMed] [Google Scholar]
- 32.Szegedy C, Liu W, Jia Y, Sermanet P, Reed S, Anguelov D, Erhan D, Vanhoucke V, Rabinovich A. Going deeper with convolutions; Proc. of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR); 2015. pp. 1–9. [Google Scholar]
- 33.Walczak P, Zhang J, Gilad AA, Kedziorek DA, Ruiz-Cabello J, Young RG, Pittenger MF, van Zijl PC, Huang J, Bulte JW. Dual-modality monitoring of targeted intraarterial delivery of mesenchymal stem cells after transient ischemia. Stroke. 2008;39(5):1569–1574. doi: 10.1161/STROKEAHA.107.502047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Arifin DR, Valdeig S, Anders RA, Bulte JW, Weiss CR. Magnetoencapsulated human islets xenotransplanted into swine: a comparison of different transplantation sites. Xenotransplantation. 2016 doi: 10.1111/xen.12235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang P, Schuetz C, Vallabhajosyula P, Medarova Z, Tena A, Wei L, Yamada K, Deng S, Markmann JF, Sachs DH, Moore A. Monitoring of Allogeneic Islet Grafts in Nonhuman Primates Using MRI. Transplantation. 2015;99(8):1574–1581. doi: 10.1097/TP.0000000000000682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Baldelomar EJ, Charlton JR, Beeman SC, Hann BD, Cullen-McEwen L, Pearl VM, Bertram JF, Wu T, Zhang M, Bennett KM. Phenotyping by magnetic resonance imaging nondestructively measures glomerular number and volume distribution in mice with and without nephron reduction. Kidney international. 2015 doi: 10.1038/ki.2015.316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bennewitz MF, Tang KS, Markakis EA, Shapiro EM. Specific chemotaxis of magnetically labeled mesenchymal stem cells: Implications for MRI of glioma. Molecular Imaging and Biology. 2012;14(6):676–687. doi: 10.1007/s11307-012-0553-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.