Abstract
We recently reported that ZBTB7A is a bona fide transcription repressor of key glycolytic genes and its downregulation in human cancer contributes to tumor metabolism. As reduced expression of ZBTB7A is found only in a subset of human cancers, we explored alternative mechanisms of its inactivation by mining human cancer genome databases. We discovered recurrent somatic mutations of ZBTB7A in multiple types of human cancers with a marked enrichment of mutations within the zinc finger domain. Functional characterization of the mutants demonstrated that mutations within the zinc finger region of ZBTB7A invariably resulted in loss of function. As a consequence, the glycolytic genes were markedly upregulated in cancer cells harboring ZBTB7A zinc finger mutation, leading to increased glycolysis and proliferation. Our study uncovers the loss-of-function mutation in ZBTB7A as a novel mechanism causing elevated glycolysis in human cancer, which carries important therapeutic implication.
INTRODUCTION
ZBTB7A (also known as POKEMON,1 FBI,2 LRF3 and OCZF4) is a member of the POZ/BTB and Krüppel (POK) family transcription factors. Human genome contains 43 genes encoding POK proteins.5 POK proteins harbor C-terminal zinc fingers and an N-terminal BTB domain. Zinc finger domain recognizes and binds specific DNA sequences, whereas the BTB domain mediates homodimerization and/or heterodimerization and interacts with other proteins.6
POK transcription factors have been recognized as critical developmental regulators and have been directly implicated in human cancer.7,8 For instance, BCL6 (B-cell lymphoma 6) and PLZF (promyelocytic leukemia zinc finger) are critical players in the pathogenesis of Non-Hodgkin's lymphoma and acute promyelocytic leukemia, respectively.9,10 ZBTB7A shares structural similarities with BCL6 and PLZF and has a critical role in embryonic development, hematopoiesis and tumorigenesis.1,11–13
Originally, ZBTB7A was found to act as a proto-oncoprotein owing to its ability to repress the transcription of tumor suppressor ARF.1 Recent cancer genomics study indicates the genetic loss of 19p13.3 chromosome region, which contain ZBTB7A, during the progression of multiple human cancers.14,15 Genetic loss of ZBTB7A chromosome region argues against the proto-oncogenic role of ZBTB7A in human cancer. In mouse prostate, loss of Zbtb7a promote the progression of Pten loss driving prostate cancer, suggesting a tumor suppressive function of Zbtb7a in prostate cancer.16 Recently, we have uncovered a novel tumor suppressive function of ZBTB7A that directly represses the expression of key glycolytic genes and its loss results in the induction of glycolysis promoting tumor progression.17
With the finding that reduced ZBTB7A expression owing to chromosome deletion occurs only in a subset of human cancer, we wondered whether there might be alternative mechanisms of ZBTB7A inactivation. We addressed this question by mining human cancer genome databases and discovered recurrent somatic mutations of the ZBTB7A gene with a marked enrichment within the zinc finger domain. We present multiple lines of evidence demonstrating that mutations within the zinc finger domain invariably lead to loss of function. Consistently, cancer cells harboring mutations within ZBTB7A zinc finger domain displayed highly elevated rate of glycolysis and proliferation, and importantly a heightened sensitivity to inhibitors of glycolysis. Our study implicates that ZBTB7A mutations may serve as driving events in promoting tumor progression.
RESULTS
Somatic mutation in the zinc finger domains of ZBTB7A in human cancers
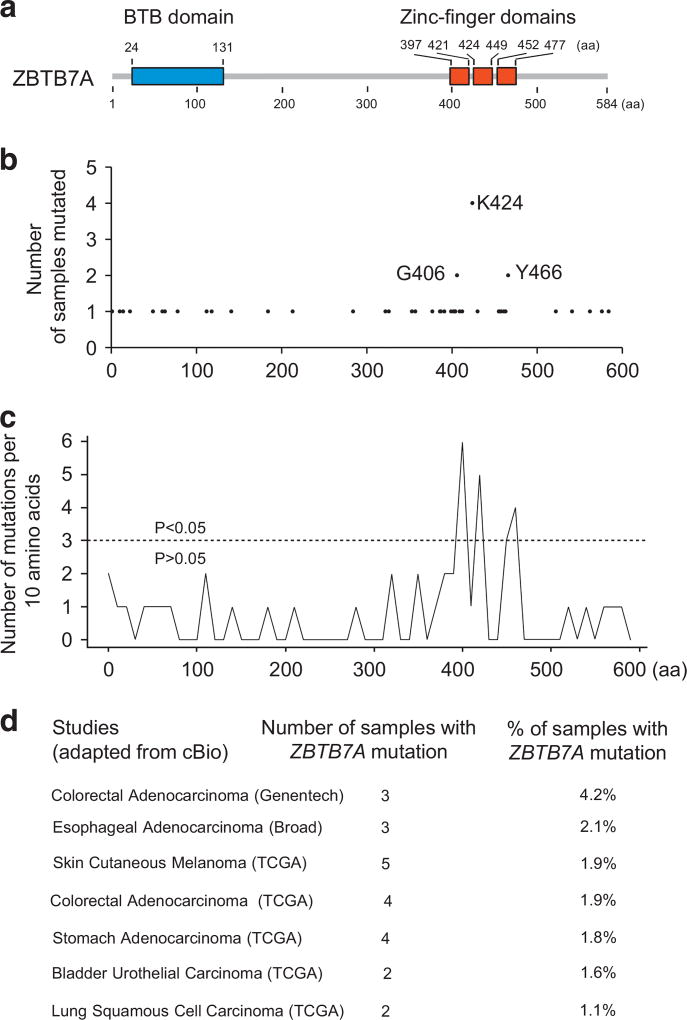
Cancers are caused by genetic alterations of cancer genes with very few (such as P53, PTEN) mutated at a high frequency (>20%), whereas most cancer genes are mutated at a relatively low frequency (2–20%) in a given cancer type.18 The Cancer Genome Atlas and International Cancer Genome Consortium contain information of a vast number of somatic mutations in thousands of human cancers from over 40 cancer types.19,20 With the finding that the decrease in ZBTB7A copy number owing to chromosome loss was detected only in a subset of human cancers, we explored alternative mechanisms that could result in loss of ZBTB7A function. We mined human cancer genomic databases for ZBTB7A somatic mutations and detected a total of 47 missense, and six frame shift and 15 silent mutations (Supplementary Table S1). All these ZBTB7A somatic mutations have not been previously described. Interestingly, of the total 47 missense mutations, 20 (43%) mutations are found in zinc finger domain (Figures 1a and b and Supplementary Table S1). On the basis of the missense mutation density, there is a significant enrichment within the three zinc finger domains when compared with other regions of ZBTB7A (Figure 1c), suggesting a selection of these zinc finger mutants in human cancers. Among several different cancer types, colorectal cancer had the highest ZBTB7A mutation rate, 1.9% and 4.2% based on two different databases (Figure 1d).
Figure 1.
Somatic mutation spectrum of ZBTB7A in human cancers. (a) Domain structure of ZBTB7A is shown. (b) Numbers of human cancer samples with ZBTB7A missense point mutations at the indicated amino acid (aa) sites are shown. (c) ZBTB7A mutation density plot of the same data set as b are shown. Three distinct clusters of mutations in ZBTB7A zinc finger domains with P < 0.05 are identified. (d) Cancer genomic studies with 1% or more samples containing ZBTB7A mutations are shown.
Of note, the enrichment of mutation within the zinc finger domains appears to be unique to ZBTB7A as there is no similar mutation pattern found in other POK family members BCL6 or PLZF (Supplementary Figure S1 and Supplementary Figure S2).
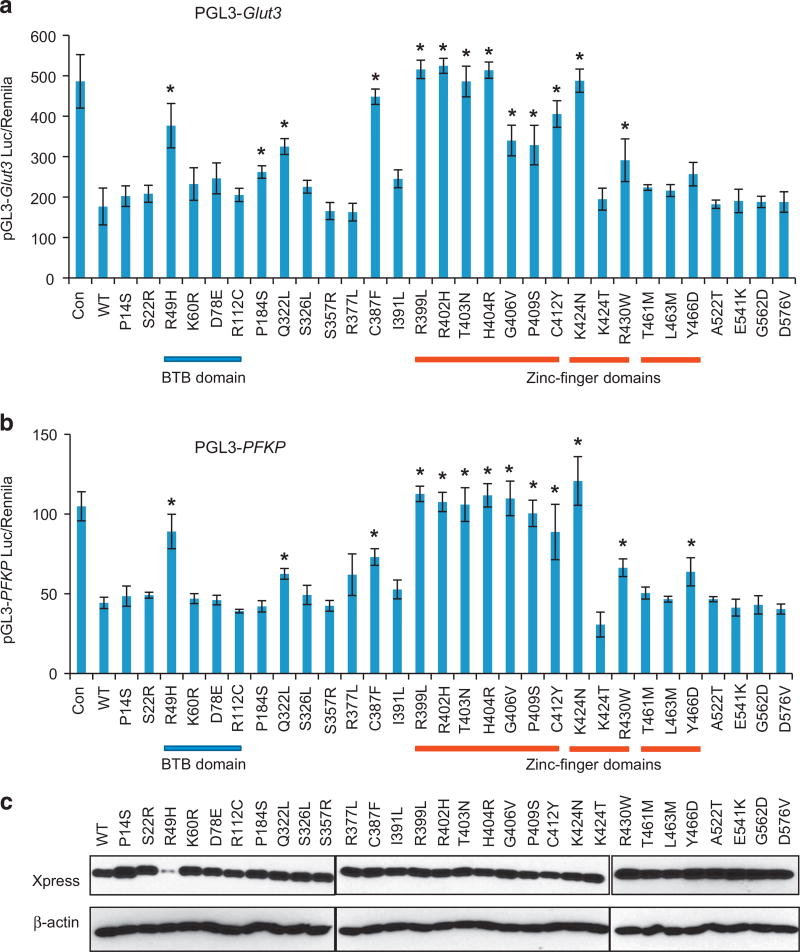
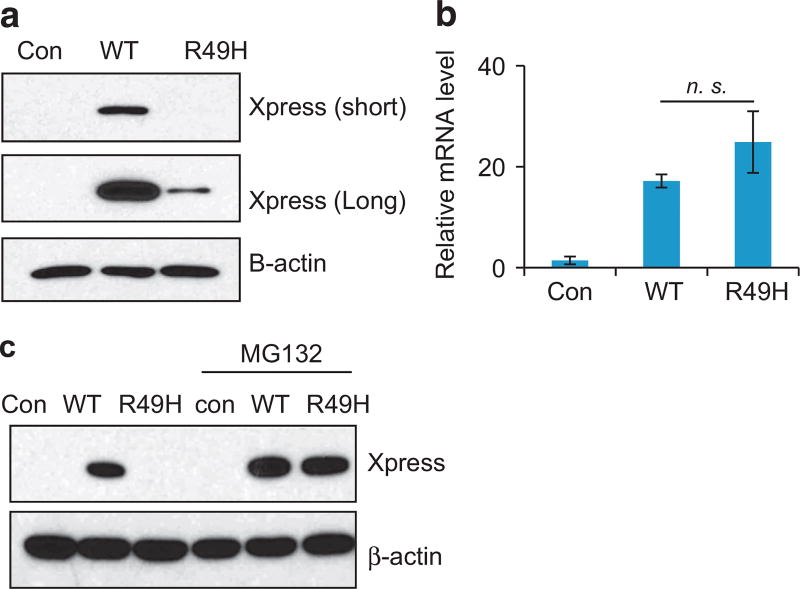
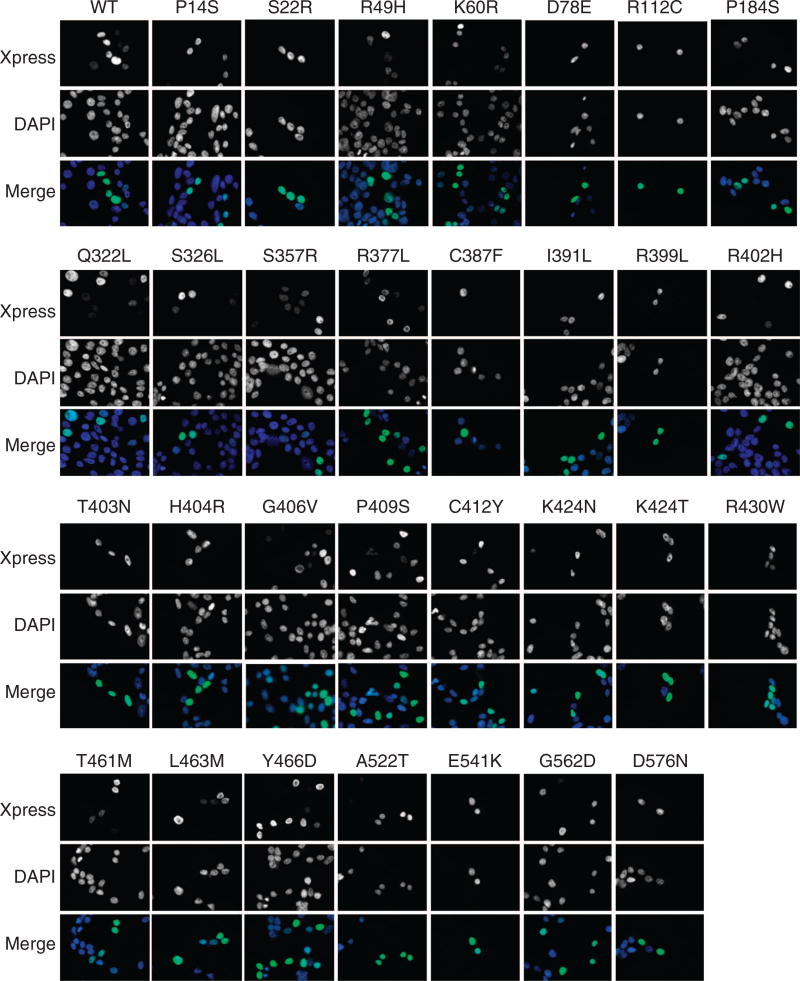
ZBTB7A zinc finger mutants lose transcription repressor activity We went on to characterize the functional consequence of the ZBTB7A mutations found in human cancers. As a POK family of transcription factor, ZBTB7A is known to repress the transcription of target genes by binding to their promoters through zinc finger domain.6 ZBTB7A target genes include ARF,1 GLUT3, PFKP and PKM.17 We used a ZBTB7A target gene promoter-driven luciferase reporter to assess the activity of ZBTB7A mutants. A total of 30 ZBTB7A mutants were generated, confirmed by Sanger sequencing (Supplementary Table S2) and cloned into a pcDNA 3.1/His C vector, with the wild-type human ZBTB7A as a control. As shown previously,17 wild-type ZBTB7A significantly suppressed the activity of GLUT3, PFKP and PKM promoter-driven luciferase (Figure 2 and Supplementary Figure S3). Notably, many ZBTB7A mutants showed a significantly compromised ability to repress the activity of GLUT3, PFKP and PKM promoter-driven luciferase (Figure 2 and Supplementary Figure S3). Remarkably, we found that all the mutations within the first zinc finger domain, R399L, R402H, T403N, H404R, G406V, P409S, C412Y resulted in significantly decreased ability of suppression when compared with wild-type ZBTB7A (Figure 2). The K424N mutation within the second zinc finger domain also significantly diminished ZBTB7A’s transcriptional suppression (Figure 2). Relative to the first zinc finger domain, the mutations within the third zinc finger domain also compromised ZBTB7A transcription repression ability, albeit to a lesser extent (Figure 2). Interestingly, the extent of decrease in transcriptional repression correlated with the mutation frequency of ZBTB7A in human cancers, which is the highest within the first zinc finger domain than other domains (Figure 1c). Western analysis was performed to ensure a comparable protein expression level of the ZBTB7A mutants. The result indicated that except R49H mutant, all others expressed at a level comparable to wild-type ZBTB7A (Figure 2c). The much lower abundance of the R49H mutant correlated with the significantly compromised transcriptional repression ability. This reduced protein level appeared to result from a posttranscriptional mechanism as there was no decrease in the R49H messenger RNA level (Figure 3). Indeed, treatment of cells with a proteasome inhibitor MG132 (5 µM, 24 h) completely rescued the defect in protein expression, indicating that the R49H mutation was associated with increased protein instability (Figure 3). As a transcriptional repressor, ZBTB7A is primarily localized in the nucleus. We asked whether the compromised ability of the ZBTB7A mutants was caused by the alteration of protein distribution. We addressed this question by examining the subcellular localization of the ZBTB7A mutants by immunofluorescence staining. The results revealed that like the wild-type protein, all ZBTB7A mutants, including zinc finger mutants localized in the nucleus (Figure 4).
Figure 2.
Loss-of-function mutation of ZBTB7A zinc finger mutants. (a and b) Luciferase reporter assay using GLUT3 promoter (a), PFKP promoter (b) driving luciferase reporter in the presence of control, wild-type ZBTB7A and various indicated ZBTB7A mutants are shown. Average values of three independent experiments are shown, error bar indicates ± s.d. * indicates P < 0.05 compared with wild-type ZBTB7A. (c) Expression of wild-type and various mutated ZBTB7A are detected by western blot analysis using anti-Xpress tag antibody. β-Actin serves as loading control. The protein expression of R49H is significantly less than wild type and all other mutants, and is further studied in Figure 3
Figure 3.
The ZBTB7A R49H mutant is very unstable. (a) Wild-type, R49H ZBTB7A or control vector are transiently expressed, the expression of ZBTB7A (wild type or R49H) are detected by western blot analysis with Xpress antibody. β-Actin serves as loading control. (b) The mRNA levels of wild-type or R49H ZBTB7A are quantified by real-time PCR. Error bars indicate ± s.d. of three experiments. (c) Transiently expressed wild-type, R49H ZBTB7A or control vector are treated with or without proteasome inhibitor MG132 (5 µm, 24 h). The expression of ZBTB7A (wild type or R49H) is detected by western blot analysis with Xpress antibody. β-Actin serves as loading control. mRNA, messenger RNA.
Figure 4.
Subcellular localization of wild-type and various mutated ZBTB7A. Wild-type or various mutated ZBTB7A are transiently expressed in 293 cells, the subcellular localization of Xpress-tagged ZBTB7A are detected by immunofluorescent staining with anti-Xpress antibody.
ZBTB7A zinc finger mutants lost the ability to bind to the target DNA sequence
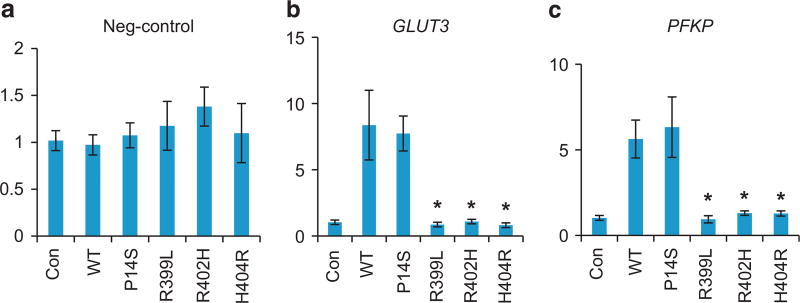
Knowing that the zinc finger domain of ZBTB7A is required for DNA binding, we assessed the ability of the zinc finger mutants to bind the promoter of target gene by chromatin immunoprecipitation. The control vector, Xpress-tagged ZBTB7A, wild type, P14S, R399L, R402H or H404R was expressed in cells and chromatin IP was performed using an anti-Xpress antibody. Quantitative PCR results confirmed that wild-type ZBTB7A could specifically bind to GLUT3 and PFKP promoter but not random negative control genomic sequence (Figures 5a–c). In contrast with wild-type ZBTB7A, the zinc finger mutants R399L, R402H and H404R lost the ability of binding to the GLUT3 and PKM promoters (Figures 5b and c). This effect appeared to be specific to the zinc finger mutation because the P14S mutant retained the promoter binding activity. Importantly, the loss of promoter binding corresponded to the loss of transcriptional repressor activity of these zinc finger mutants, consistent with the requirement of zinc finger domain-mediated DNA binding for the ability of transcriptional suppression.
Figure 5.
Compromised DNA binding ability of ZBTB7A zinc finger mutants. Chromatin immune-precipitation was performed with Xpress antibody in cells transfected with empty control vector, wild-type and P14S, R399L, R402H, H404R ZBTB7A. The associated DNA was quantified by real-time PCR with primers specific to negative control (a), GLUT3 (b) and PFKP promoter (c) genomic DNA. Average values of three independent experiments are shown, error bars indicate ± s.d. * indicates P < 0.05 compared with wild-type ZBTB7A.
Dominant negative activities of ZBTB7A zinc finger mutants
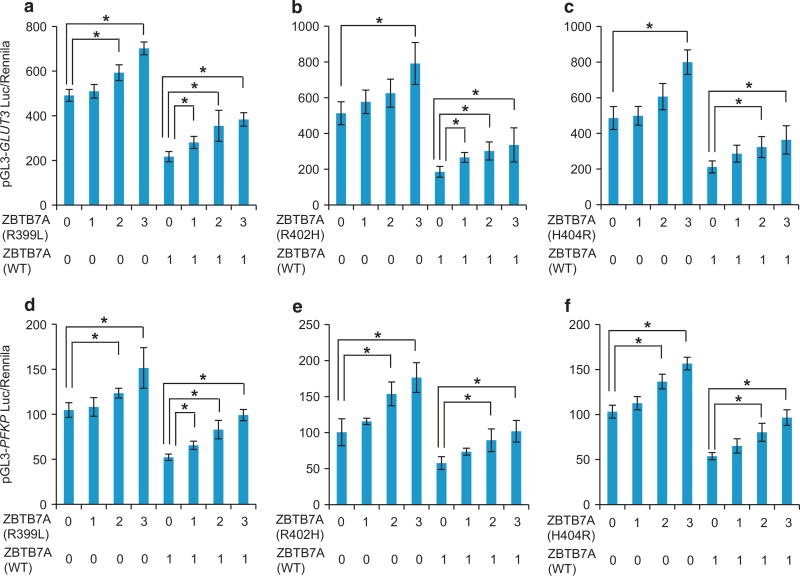
The frequency of gene mutation at both alleles is usually much lower than at single allele. We asked how the mutants and wild-type ZBTB7A might interact with each other when both were present. We transfected increasing doses of zinc finger mutants R399L, R402H or H404R in the presence or absence of wild-type ZBTB7A. With both pGL3-GLUT3 (Figure 6a) and pGL3-PFKP (Figure 6b) promoter-driven luciferase, the expression of zinc finger mutants R399L, R402H or H404R impeded the transcriptional suppressor activity of wild-type ZBTB7A in a dose-dependent manner. The results suggest a dominant negative function of R399L, R402H or H404R zinc finger mutants against wild-type ZBTB7A in transcriptional repression of glycolysis genes.
Figure 6.
Dominant negative activity of ZBTB7A zinc finger mutants. (a–c) Luciferase assay was performed using GLUT3 promoter-driving luciferase reporter. Increasing dose of R399L (a), R402H (b) and H404R (c) ZBTB7A were combined with or without wild-type ZBTB7A. (d–f) Luciferase assay was performed using PFKP promoter-driving luciferase reporter. Increasing dose of R399L (d), R402H (e) and H404R (f) ZBTB7A was combined with or without wild-type ZBTB7A. Average values of three independent experiments are shown, error bars indicate ± s.d. * indicates P < 0.05.
ZBTB7A zinc finger mutations stimulate cell proliferation by promoting glycolysis
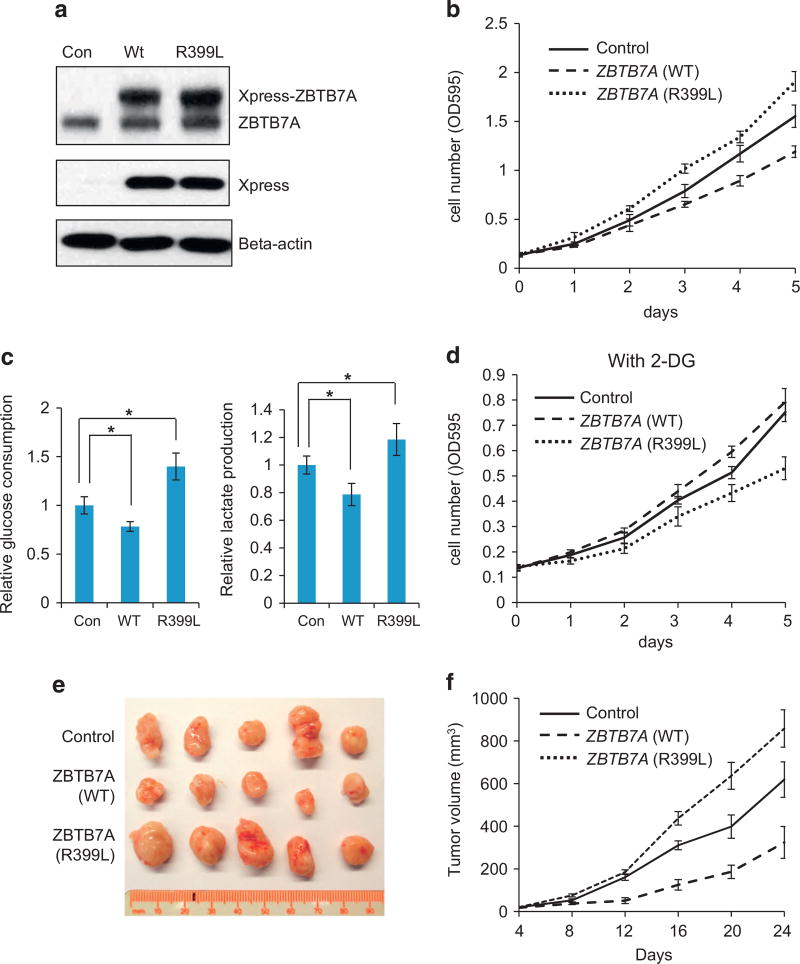
We next investigated the biological consequence of the zinc finger mutants of ZBTB7A with the R399L mutant as the representative. SW48 colon carcinoma cells stably expressing ZBTB7A (R399L) were established with wild-type ZBTB7A included as a control. Western analysis showed that wild-type and R399L mutant of ZBTB7A expressed at a comparable level, which was slightly higher than that of the endogenous counterpart (Figure 7a). Remarkably, the expression of exogenous ZBTB7A proteins had significant impact on the rate of cell proliferation. Additional expression of wild-type ZBTB7A was associated with a decrease in cell proliferation when compared with the vector control cells, suggesting a gene dosage effect of ZBTB7A. On the other hand, the ZBTB7A mutant expressing SW48 cells grew faster than the control cells, consistent with the ability of the mutant to interfere with wild-type ZBTB7A (Figure 7b). In agreement with the ability to regulate glycolytic genes, wild-type ZBTB7A inhibited, whereas the mutant stimulated glycolysis, as reflected by the measurements of glucose consumption and lactate production (Figure 7c). To examine whether the altered metabolism was responsible for the difference in cell proliferation, we treated cells with a glycolysis inhibitor, 2-deoxyglucose. Interestingly, the three cell lines exhibited a different sensitivity to glycolysis inhibition that correlated with the difference in glycolytic metabolism. Relative to the vector control cell line that showed a modest sensitivity to 2-deoxyglucose, wild-type ZBTB7A expressing line was resistant, whereas the R399L expressing line exhibited a heightened sensitivity to inhibition of glycolysis (Figure 7d). To determine the relevance of this observation for tumorigenesis, we generated xenograft tumors with cell lines expressing control vector, wild-type or R399L ZBTB7A in mice. In agreement with the in vitro data, the expression of wild-type ZBTB7A suppressed the tumor growth, whereas the R399L counterpart lost the ability to suppress and instead promoted tumor progression (Figures 7e and f).
Figure 7.
ZBTB7A zinc finger mutant R399L stimulate glycolysis metabolism and promote cell proliferation. (a) Wild-type, R399L ZBTB7A and control vector were stably expressed in SW48 cells, the growth curve of these three cell lines are shown. Error bars indicate ± s.d. of three independent experiments. (b) Western blot analysis of the same cell lines as a, endogenous ZBTB7A and exogenous expressed Xpress-tagged ZBTB7A are separated and detected with anti-ZBTB7A antibody. β-Actin serves as loading control. (c) Glucose consumption and lactate production rates of cells lines stably express control vector, WT or R399L ZBTB7A. Average values of three independent experiments are shown, error bars indicate ± s.d. * indicates P < 0.05. (d) Growth curve of cell lines stably express control vector, WT or R399L ZBTB7A in the presence of glycolysis inhibitor 2-DG. Average values of three independent experiments are shown, error bars indicate ± s.d. (e) Nude mice tumor assay with SW48 cells stably express wild-type, R399L ZBTB7A or control vector, tumors of 24 days are shown. (f) Tumor volume was measured in subcutaneous tumors formed by wild-type, R399L ZBTB7A or control vector expressing SW48 cells. Data shown are mean ± s.d. of five tumors. 2-DG, 2-deoxyglucose.
DISCUSSION
ZBTB7A, also known as ‘Pokemon’, was originally identified as a proto-oncogene, owing to its ability to repress the transcription of tumor suppressor ARF.1 Recent studies indicated ZBTB7A as a context-dependent cancer gene. For instance, ZBTB7A was found to be a tumor suppressor in mouse prostate cancer.16 We recently reported a tumor suppressive function of ZBTB7A in multiple human solid tumors and ZBTB7A does so by transcriptional suppression of glycolysis genes.17 Apart from reduction of copy number owing to deletion of 19p13.3 chromosome locus where the ZBTB7A gene resides, we uncovered recurrent somatic mutation within the zinc finger domain of ZBTB7A as an alternative mechanism of inactivation. Of note is the finding that in addition to loss of function, the mutant seemed to be able to interfere with the function of co-expressed wild-type ZBTB7A, consistent with a dominant negative mechanism of action.
Cancer-associated ZBTB7A somatic mutations have not been reported or studied previously, likely owing to its relatively low mutation frequency when compared with frequently mutated cancer genes like P53, PTEN. However, recent cancer genome studies have shown that many cancer-associated gene mutations occur at a relatively low frequency.18 Many of such mutations are quite relevant to the development of human cancer because of the growth advantage associated with the gene mutation or driver mutation.
Although mutations were found in many different regions of the ZBTB7A gene, a marked enrichment of mutations within the zinc finger domain was detected, suggesting a selection event during tumorigenesis. Indeed, cells harboring ZBTB7A (R399L), a mutation within the zinc finger region, exhibited growth advantage over the control cells. Functional characterization of the mutants as a transcription repressor demonstrated that mutations within the zinc finger regions resulted in abrogation of DNA binding, leading to loss of function. Moreover, the zinc finger mutants appeared to have a dominant negative activity against wild-type ZBTB7A. This is significant because the somatic mutations on single allele are usually at a much higher frequency than on both alleles, resulting in co-existence of both wild-type and mutant ZBTB7A proteins in cancer cells. The dominant negative feature will likely provide growth advantage driving cancer progression.
In agreement with the metabolic mechanism of tumor suppression that we recently reported, the loss-of-function mutation in ZBTB7A was associated with upregulation of its target glycolytic genes and consequent glycolysis. Importantly, the oncogenic activity of the ZBTB7A mutant appeared to largely depend on the elevated glycolysis as the growth of cancer cells harboring the mutation were effectively suppressed by inhibition of glycolysis. The heightened sensitivity of the ZBTB7A mutant expressing cancer cells to the glycolytic inhibitor carries important therapeutic implication.
In summary, our study uncovers somatic mutation within the zinc finger domains as a novel mechanism alternative to chromosome deletion in the inactivation of tumor suppressor function of ZBTB7A. The dominant negative function of the zinc finger mutant of ZBTB7A represents an important cancer driver mechanism promoting tumor progression. The dependency of ZBTB7A mutant cancer cells on glycolysis offers an attractive therapeutic opportunity.
MATERIALS AND METHODS
Human cancer genomic analysis
All somatic mutation information of ZBTB7A, BCL6, ZBTB16 (PLZF) are downloaded from cBio,21 COSMIC22 and International Cancer Genome Consortium19 databases. End of data update: 1 December 2014.
To identify regions of ZBTB7A significantly enriched for somatic point mutations, we conducted formal statistical test with the null hypothesis that all 47 ZBTB7A mutations are randomly occurring across the 584 amino acid protein in a 10-amino acid sliding window. We calculated the threshold for significance of at least three mutations per 10 amino acids using 0.05 as the significance level by the formula
which equals 0.01 for k = at least three mutations per 10 amino acids. This means that if there are three or more mutations in a 10-amino acid sliding window, then this region is enriched of mutations.
For BCL6,
There are 706 amino acids and 126 mutations (Supplementary Table S3). Using 10 amino acids as a sliding window, we calculated that the threshold for significant enrichment of mutations is four, and the P-value is 0.036. This means that if there are four or more mutations in a 10-amino acid window, then this region is significantly enriched of mutations.
For PLZF:
PLZF protein has 673 amino acids, in total 117 missense mutations are observed (Supplementary Table S4). Similarly, using 0.05 as the significance level, we calculated the threshold for enrichment of at least four mutations per 10 amino acids using this formula. The right tail probability of observing at least four mutations is equal to 0.03.
Cell culture
Human embryonic kidney 293 cells; human colorectal adenocarcinoma cell SW48 were cultured in DMEM (Corning, Cellgro, Manassas, VA, USA) plus 10% fetal bovine serum (Gibco, Grand Island, NY, USA), 100 U/ml penicillin G and 100 µg/ml streptomycin (Corning, Cellgro).
Antibodies and reagents
Anti-ZBTB7A antibody (13E9, Santa Cruz, Dallas, TX, USA) was used for western blot; Anti-Xpress antibody (Invitrogen, Grand Island, NY, USA) was used for chromatin immunoprecipitation, western blot and immunofluorescence; anti-β-actin antibody (AC-15, Sigma) was used for western blot; anti-Hsp90 antibody (AC-16, Sigma) was used for western blot; MG132 was brought from Sigma.
Plasmid construction and mutation
Human ZBTB7A was cloned into pcDNA 3.1/His C vector (Invitrogen) using KpnI, XbaI restriction enzyme digestion and ligation. The resulting plasmid is termed pcDNA-ZBTB7A. The pcDNA 3.1/His C vector contain Xpress tag for detection by western blot or immunofluorescence. Plasmid pcDNA-ZBTB7A was mutated with QuikChange II XL Site-Directed Mutagenesis Kit (Agilent, Santa Clara, CA, USA) according to the manufacturer’s protocol. Mutation primers are listed in Supplementary Table S2. All the mutations have been confirmed by Sanger sequencing in GENEWIZ.
Promoter activity luciferase assay
Two hundred and ninety-three cells were cultured in 48-well plates, transfection was started when cells reached 50% confluency. Twenty nanograms of pGL3-GLUT3, pGL3-PKM, pGL3-PFKP vectors, 10 ng of pRL-CMV renilla luciferase reporter and 20 ng of pcDNA control or wild-type and various mutated pcDNA-ZBTB7A expression plasmid were co-transfected into 293 cells. To check the dominant negative effects of ZBTB7A mutants, 0, 20 ng, 40 ng and 60 ng of mutated ZBTB7A were co-transfected with 20 ng of wild-type ZBTB7A. After 36 h, the luciferase and renilla activity was measured with the dual luciferase reporter assay system (Promega, Madison, WI, USA) according to manufacturer’s instruction.
Immunofluorescence
Two hundred and ninety-three cells were seeded in 24-well plates containing round glass coverslips cells, and were transfected with plasmid pcDNA-ZBTB7A (wild-type or various mutated ZBTB7A) using Lipofectamine 2000 reagent (Invitrogen). Forty-eight hours after transfection, cells were fixed with 4% paraformaldehyde, permeabilized in 0.1% Triton-X-100/phosphate-buffered saline. Coverslips were then incubated with primary antibody diluted in 1% BSA/phosphate-buffered saline for 1 h. After washing, coverslips were incubated in secondary antibody diluted in 1% BSA/phosphate-buffered saline for 1 h. Coverslips were washed, stained with DAPI, mounted and analyzed by fluorescent microscopy.
Immunoblot
Cells were lysed in buffer (50 mm Tris, pH8.0; 150 mm NaCl; 1% NP-40 and 0.1% SDS). Protein concentrations of the lysates were measured by Bradford assay. The protein lysates were boiled with Laemmli protein sample buffer, and then resolved by SDS–polyacrylamide gel electrophoresis and immunoblotted with the indicated antibodies.
Cell proliferation assay
Cells were seeded in 12-well plates at a density of 104/well, then left to grow for 4 days in the presence or absence of 2 mm 2-deoxy-d-glucose (2-deoxyglucose; Sigma). Cells were fixed by paraformaldehyde at each time point, and stained with crystal violet. After extensive washing, crystal violet was re-solubilized in 10% acetic acid and quantified at 595 nm as a relative measure of cell number as described previously.23
Glucose uptake and lactate production measurement
A total 3 × 104 cells were seeded into 24-well plates; the next day, cells were washed two times with culture medium and fresh culture medium was added; 5 h later, cell culture medium was collected and frozen in −80 °C, the cell numbers in each well were counted using a hemocytometer. Glucose and lactate concentration in the collected cell culture medium were measured using colorimetric kit (Biovision, Milpitas, CA, USA) according to the attached protocol.
ZBTB7A wild type, R399L, R402H stable expression cell line generation
The pcDNA-ZBTB7A (wt, R399L, R402H) plasmids were transfected into SW48 cells; 2 days later, G418 selection (500 µg/ml) was started for 2 weeks; the polyclonal cells after selection were used for downstream analysis.
Chromatin immunoprecipitation
Chromatin immunoprecipitation was performed as described previously.24 Briefly, protein–DNA complexes were crosslinked for 10 min at room temperature with 1% formaldehyde added directly into the culture medium. Reaction was stopped by addition of glycine (final concentration: 0.125 mol/l) incubated for 5 min through gentle rocking. The cells were washed with phosphate-buffered saline and buffer (10 mm Tris pH 8.0, 10 mm EDTA, 0.5 mm EGTA, 0.25% Triton X-100), then suspended in 200 µl lysis buffer (1.1% Triton X-100, 4 mm EDTA, 40 mm Tris pH 8.1, 300 mm NaCl) and submitted to sonication to produce small DNA fragments (200–1000 bp). Chromatin was pre-cleared and immunoprecipitated with the anti-Xpress antibody (Invitrogen). Precipitated DNA and protein complexes were reverse crosslinked, proteins digested with proteinase K (Fermentas, Grand Island, NY, USA) and DNA fragment were purified with a QIAquick PCR Purification Kit (Qiagen, Hamburg, Germany). The purified DNAs were amplified by real-time quantitative PCR using StepOnePlus (Applied Biosystems, Grand Island, NY, USA) and SYBR Green JumpStart Taq ReadyMix (Sigma). Primers to quantify the abundance of human GLUT3 and PFKP promoter: GLUT3CHIP-5 5′-CCCCTGAAGCAATCTTGTGATC-3′; GLUT3CHIP-3 5′-AAAAACCCAGGGTGGAGAGAG-3′; PFKPCHIP-5 5′-TCATCTCT AGAGCCCCCAAC-3′; PFKPCHIP-3 5′-GTGTGGGCAGGAGCATCTAC-3′. Primers as internal control: NegCHIP-5 5′-ATGGTTGCCACTGGGGATCT-3′; NegCHIP-3 5′-TGCCAAAGCCTAGGGGAAGA-3′.
Nude mice tumor formation assay
A total 2 × 106 SW48 cells stably expressing wild-type, R399L ZBTB7A or control vector were subcutaneously injected into the flank of nude mice (nu/nu, female, 6–8 weeks old, Charlies River Laboratories, Wilmington, MA, USA). The size of the tumor was measured with a caliper every 4 days. Tumor volume was calculated by using the formula V = (π /6) (d1 × d2)3/2. At the end point, mice were euthanized and the tumors from each mouse were excised and total RNA was extracted with TRIzol Reagents, or the tumor was fixed in formaldehyde, embedded in paraffin and sectioned for histological staining. All animal procedures were conducted in accordance with the Guidelines for the Care and Use of Laboratory Animals and were approved by the Institutional Animal Care and Use Committee at the Harvard School of Public Health.
Statistical analysis
Unpaired two-tailed Student’s t-test was used for the comparisons of the means, error bars represent s.d. The statistical significances between data sets were expressed as P-values, and P < 0.05 was considered statistically significant.
Supplementary Material
Acknowledgments
We are grateful to current and former members of the Yuan lab for experimental support, advice and helpful discussions. This work was supported in part by the Morningside Foundation and grants from NIH/NCI (R01CA085679, RO1CA167814 and RO1CA125144).
Footnotes
CONFLICT OF INTEREST
The authors declare no conflict of interest.
Supplementary Information accompanies this paper on the Oncogene website (http://www.nature.com/onc)
References
- 1.Maeda T, Hobbs RM, Merghoub T, Guernah I, Zelent A, Cordon-Cardo C, et al. Role of the proto-oncogene Pokemon in cellular transformation and ARF repression. Nature. 2005;433:278–285. doi: 10.1038/nature03203. [DOI] [PubMed] [Google Scholar]
- 2.Pessler F, Pendergrast PS, Hernandez N. Purification and characterization of FBI-1, a cellular factor that binds to the human immunodeficiency virus type 1 inducer of short transcripts. Mol Cell Biol. 1997;17:3786–3798. doi: 10.1128/mcb.17.7.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu CJ, Prazak L, Fajardo M, Yu S, Tyagi N, Di Cesare PE. Leukemia/lymphoma-related factor, a POZ domain-containing transcriptional repressor, interacts with histone deacetylase-1 and inhibits cartilage oligomeric matrix protein gene expression and chondrogenesis. J Biol Chem. 2004;279:47081–47091. doi: 10.1074/jbc.M405288200. [DOI] [PubMed] [Google Scholar]
- 4.Kukita A, Kukita T, Ouchida M, Maeda H, Yatsuki H, Kohashi O. Osteoclast-derived zinc finger (OCZF) protein with POZ domain, a possible transcriptional repressor, is involved in osteoclastogenesis. Blood. 1999;94:1987–1997. [PubMed] [Google Scholar]
- 5.Stogios PJ, Downs GS, Jauhal JJ, Nandra SK, Prive GG. Sequence and structural analysis of BTB domain proteins. Genome Biol. 2005;6:R82. doi: 10.1186/gb-2005-6-10-r82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee SU, Maeda T. POK/ZBTB proteins: an emerging family of proteins that regulate lymphoid development and function. Immunol Rev. 2012;247:107–119. doi: 10.1111/j.1600-065X.2012.01116.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kelly KF, Daniel JM. POZ for effect--POZ-ZF transcription factors in cancer and development. Trends Cell Biol. 2006;16:578–587. doi: 10.1016/j.tcb.2006.09.003. [DOI] [PubMed] [Google Scholar]
- 8.Costoya JA. Functional analysis of the role of POK transcriptional repressors. Brief Funct Genomic Proteomic. 2007;6:8–18. doi: 10.1093/bfgp/elm002. [DOI] [PubMed] [Google Scholar]
- 9.Lo Coco F, Ye BH, Lista F, Corradini P, Offit K, Knowles DM, et al. Rearrangements of the BCL6 gene in diffuse large cell non-Hodgkin's lymphoma. Blood. 1994;83:1757–1759. [PubMed] [Google Scholar]
- 10.Chen Z, Brand NJ, Chen A, Chen SJ, Tong JH, Wang ZY, et al. Fusion between a novel Krüppel-like zinc finger gene and the retinoic acid receptor-alpha locus due to a variant t(11;17) translocation associated with acute promyelocytic leukaemia. EMBO J. 1993;12:1161–1167. doi: 10.1002/j.1460-2075.1993.tb05757.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maeda T, Merghoub T, Hobbs RM, Dong L, Maeda M, Zakrzewski J, et al. Regulation of B versus T lymphoid lineage fate decision by the proto-oncogene LRF. Science. 2007;316:860–866. doi: 10.1126/science.1140881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maeda T, Ito K, Merghoub T, Poliseno L, Hobbs RM, Wang G, et al. LRF is an essential downstream target of GATA1 in erythroid development and regulates BIM-dependent apoptosis. Dev Cell. 2009;17:527–540. doi: 10.1016/j.devcel.2009.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sakurai N, Maeda M, Lee SU, Ishikawa Y, Li M, Williams JC, et al. The LRF transcription factor regulates mature B cell development and the germinal center response in mice. J Clin Invest. 2011;121:2583–2598. doi: 10.1172/JCI45682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Beroukhim R, Mermel CH, Porter D, Wei G, Raychaudhuri S, Donovan J, et al. The landscape of somatic copy-number alteration across human cancers. Nature. 2010;463:899–905. doi: 10.1038/nature08822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zack TI, Schumacher SE, Carter SL, Cherniack AD, Saksena G, Tabak B, et al. Pan-cancer patterns of somatic copy number alteration. Nat Genet. 2013;45:1134–1140. doi: 10.1038/ng.2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang G, Lunardi A, Zhang J, Chen Z, Ala U, Webster KA, et al. Zbtb7a suppresses prostate cancer through repression of a Sox9-dependent pathway for cellular senescence bypass and tumor invasion. Nat Genet. 2013;45:739–746. doi: 10.1038/ng.2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu XS, Haines JE, Mehanna EK, Genet MD, Ben-Sahra I, Asara JM, et al. ZBTB7A acts as a tumor suppressor through the transcriptional repression of glycolysis. Genes Dev. 2014;28:1917–1928. doi: 10.1101/gad.245910.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lawrence MS, Stojanov P, Mermel CH, Robinson JT, Garraway LA, Golub TR, et al. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature. 2014;505:495–501. doi: 10.1038/nature12912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.International Cancer Genome Consortium. International network of cancer genome projects. Nature. 2010;464:993–998. doi: 10.1038/nature08987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cancer Genome Atlas Research Network. The Cancer Genome Atlas Pan-Cancer analysis project. Nat Genet. 2013;45:1113–1120. doi: 10.1038/ng.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Forbes SA, Bindal N, Bamford S, Cole C, Kok CY, Beare D, et al. COSMIC: mining complete cancer genomes in the Catalogue of Somatic Mutations in Cancer. Nucleic Acids Res. 2011;39:D945–D950. doi: 10.1093/nar/gkq929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carnero A, Hudson JD, Price CM, Beach DH. p16INK4A and p19ARF act in overlapping pathways in cellular immortalization. Nat Cell Biol. 2000;2:148–155. doi: 10.1038/35004020. [DOI] [PubMed] [Google Scholar]
- 24.Nelson JD, Denisenko O, Bomsztyk K. Protocol for the fast chromatin immunoprecipitation (ChIP) method. Nat Protoc. 2006;1:179–185. doi: 10.1038/nprot.2006.27. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.