Abstract
Background
Intra-abdominal abscesses are localized collections of pus, which generally arise from a breach in the normal mucosal defense barrier that allows bacteria from gastrointestinal tract, and less commonly from the gynecologic or urinary tract, to induce inflammation, resulting in an infection. The microbiology of these abscesses is usually polymicrobial, associated with the primary disease process. However, the microbial identity, diversity and richness in intra-abdominal abscesses have not been well characterized, due in part to the difficulty in cultivating commensal organisms using standard culture-based techniques.
Methods
We used culture-independent 16S rRNA Illumina sequencing to characterize bacterial communities in intra-abdominal abscesses collected by percutaneous drainage. A total of 43 abscess samples, including 19 (44.2%) Gram stain and culture-negative specimens, were analyzed and compared with results from conventional microbiologic cultures.
Results
Microbial composition was determined in 8 of 19 culture-negative samples and 18 of 24 culture-positive samples, identifying a total of 221 bacterial taxa or operational taxonomic units (OTUs) and averaging 13.1 OTUs per sample (interquartile range, 8–16.5 OTUs). Microbial richness for monomicrobial and polymicrobial samples was significantly higher than culture-negative samples (17 and 15.2 OTUs vs 8 OTUs, respectively), with a trend toward a higher microbial diversity (Shannon diversity index of 0.87 and 1.18 vs 0.58, respectively).
Conclusions
The bacterial consortia identified by cultures correlated poorly with the microbial composition determined by 16S rRNA sequencing, and in most cases, the cultured isolates were minority constituents of the overall abscess microbiome. Intra-abdominal abscesses were generally polymicrobial with a surprisingly high microbial diversity, but standard culture-based techniques failed to reveal this diversity. These data suggest that molecular-based approaches may be helpful for documenting the presence of bacteria in intra-abdominal abscesses where standard cultures are unrevealing, particularly in the setting of prior antibiotic exposure.
Keywords: Intra-abdominal abscesses, 16S RNA sequencing, Illumina sequencing, Microbial diversity, Bioinformatics
The development of intra-abdominal abscesses is a consequence of inflammatory responses to endogenous microflora that gain access to a normally sterile site, resulting in local inflammation and the formation of pus. Abscesses develop as a result of either direct extension of normal polymicrobial endogenous flora into a normally sterile body site or secondarily through perforation or laceration. If left untreated, abscesses may lead to bacteremia and cause significant morbidity and mortality [1]. Abscesses can arise at any location within the human body, and each abscess collection is associated with unique characteristics. These abscesses are generally not associated with a single organism but reflect diverse ecological niches, whether in chronic wounds [2], periodontal disease [3], or pulmonary infections [4]. Recent studies suggest that molecular interactions within these diverse communities may increase the virulence of known pathogens in a synergistic manner [5] However, standard culture-based studies have shown that only 11%–18% of intracerebral abscesses [6], 11%–40% of liver abscesses [7, 8], and 44% of splenic abscesses [9] are polymicrobial. In some cases, antibiotic exposure prior to drainage of abscesses likely reduces the yield of bacteria recovery and/or alters the microbial profile determined by cultures [10].
The optimal management of large intra-abdominal and pelvic abscesses is drainage followed by adjunctive antimicrobial therapy, which together achieve a success rate of up to 70%–80% [11, 12]. Broad-spectrum antimicrobial therapy with activities against Gram negatives and anaerobes is often adminstered as empiric therapy [13], as the microbiology of intra-abdominal abscesses is thought to largely reflect the endogenous flora at body sites near the location of the abscesses. In the acidic environment of the stomach, which is hostile to most bacteria except for Helicobacter spp., the total bacterial counts are much lower in the stomach compared with the colon [14]. However, the microbial community composition and structure of intra-abdominal abscesses are often not characterized in detail, in part because cultivation and identification of anaerobic organisms are labor-intensive and some organisms may be uncultivable [15]. Thus, the diversity and microbial ecology of intra-abdominal abscesses remain poorly understood and have not been well characterized, and the relative abundance of different microbes in abscesses occurring at various body sites is not known. Furthermore, to what extent the standard microbiologic cultures correlate with microbiome composition determined by culture-independent sequencing is not known.
Recent advances in sequencing technology have greatly contributed to our understanding of the human microbiome and have led to an increasing appreciation of the role of the endogenous microflora in human biology [16–21]. Based on culture-based methods, the number of bacterial species in abscesses has been reported to vary from 2 to 6 [22]. The predominant anaerobes frequently cultured include the Bacteroides fragilis (B. fragilis) group, Prevotella spp., Porphyromonas spp., Peptostreptococcus spp., and Clostridium spp., and the most commonly isolated aerobic and facultative bacteria are the family of Enterobacteriaceae and Enterococci spp. Brook and Frazier examined 52 intra-abdominal abscess specimens retrospectively and found that 34 specimens (65%) were mixed anaerobic and aerobic infections and 47 (90%) were polymicrobial with an average of 3.7 isolates per specimen (on average, 2.1 were anaerobes and 1.6 were facultative anaerobes or aerobes) [23]. The most frequently cultured anaerobes included Peptostreptococcus spp., B. fragilis group, Clostridium spp., and Prevotella spp., and the most commonly isolated aerobic and facultative bacteria included Escherichia coli, Enterococci spp., and Staphlococcus aureus. In a subsequent study, they analyzed 22 intra-abdominal abscesses from diverticulitis and found that 17 (77%) were mixed anaerobic and aerobic infections and 19 (86%) were polymicrobial, with an average of 3.3 isolates per specimen (on average, 1.7 were anaerobes and 1.6 were facultative anaerobes or aerobes) [24]. The most frequently cultured anaerobes were Bacteroides spp., Peptostreptococcus spp., and Clostridium spp., and the most commonly isolated aerobic and facultative bacteria were E. coli and Streptococcus spp. It is now known that more than 5000 bacterial species reside in the gastrointestinal tract [25] and more than 700 reside in the oral cavity [26], which include many previously unidentified or uncultivable organisms. Thus, culture-based methods may underestimate the number of organisms present in abscesses [27], raising the possibility that abscesses of endogenous origin may be more diverse than previously thought.
To our knowledge, the microbial composition of intra-abdominal abscesses has not been examined using culture-independent methods and directly compared with microbiologic cultures obtained for clinical indications. Here, we used 16S rRNA deep sequencing to determine the microbial composition of intra-abdominal abscesses drained percutaneously by interventional radiology and compared with Gram stain and culture reported by clinical labs. We show that intra-abdominal abscesses are generally polymicrobial in nature, dominated by anaerobic organisms, and that standard clinical cultures significantly underestimate their overall microbial diversity. Culture-based microbial composition correlated poorly with the abscess microbiome determined by 16S rRNA sequencing, which may be helpful to confirm the presence of bacteria in intra-abdominal abscesses where standard cultures are unrevealing, especially in the setting of prior antibiotic administration.
METHODS
Ethics Statement
De-identified clinical specimens from the Clinical Microbiology Laboratory at University of Florida Health (Gainesville, FL) were used for this study. The study was approved by University of Florida Institutional Review Board.
Samples and DNA Purification
As part of routine clinical care for abscess samples received in anaerobe transport tubes, blood agar plate (BAP), chocolate (CHOC), MacConkey (MAC), bacteriodes bile esculin (BBE)/kanamycin-vancomycin laked blood (KVLB), Brucella, and anaerobic Thiglycolate broth were inoculated in the anaerobic chamber and incubated at 37°C under anaerobic condition. Two microscopic slides were made from the abscess samples inside the anaerobic chamber, Gram stained, and microscopically examined to report the presence of polymorphonuclear leukocytes and bacteria. The plates were examined daily for the presence of colonies over the course of 5 days. Facultative anaerobe and strict anaerobe were determined by comparing the growth of specific bacteria under both aerobic and anaerobic conditions. Gram stains were carried out for each specific bacteria to determine bacterial morphology, color, and spore-forming ability. Distinct colonies were subcultured to obtain pure colonies for further anaerobe identification. Anaerobe identification was performed using RapID ANA and/or mass spectrometry.
For each sample, results of Gram stain and bacterial culture and the body location of percutaneous drainage by the Interventional Radiology Service were recorded. Genomic DNA (gDNA) was extracted using the PSP Spin Stool DNA Kit according to the manufacturer’s instructions (STRATEC Biomedical, Berlin, Germany). The concentration of purified DNA was quantified using a Nanodrop spectrophotometer (ThermoScientific, Carlsbad, CA).
16S rRNA Illumina Sequencing and Bioinformatics Analysis
The V1-V3 hypervariable region (~500 bp) of 16S rRNA gene segment was amplified using barcoded polymerase chain reaction (PCR), and PCR products were gel purified, pooled, and paired-end sequenced at 2 × 300 bp using the MiSeq Reagent Kit v3 on an Illumina MiSeq instrument. Methods for PCR, Illumina sequencing, and bioinformatic analysis are described in detail in the Supplementary Data.
RESULTS
Sample Characteristics and 16S rRNA Sequence Analysis
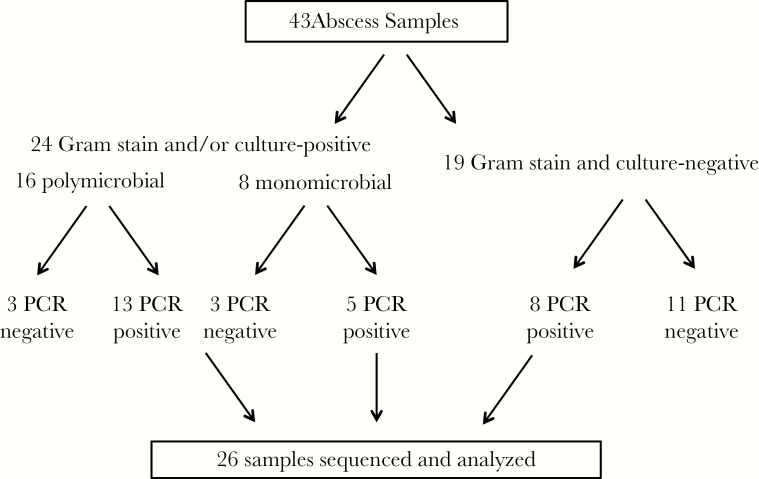
A total of 113 clinical samples drained by interventional radiologists at UF Health were available from the Clinical Microbiology Laboratory. We selected 43 samples that met the following criteria: (1) results of Gram stain and microbiologic culture were available, (2) the sample was from an intra-abdominal location, and (3) the duration from the time of sample collection to storage at −70°C was less than 72 hours (all samples were stored at 4°C while undergoing culture and sensitivity testing per clinical routine). A 72-hour threshold was selected based on an analysis that demonstrated minimal variations in the relative proportions of major operational taxonomic units (OTUs) at 4°C over 72 hours (Supplementary Figure 1). Of the 43 samples, 24 (55.8%) were positive by Gram stain and/or culture and 19 (44.2%) were negative by both Gram stain and cultures (Figure 1). All 43 samples were subjected to V1-V3 16S rRNA gene amplification by barcoded PCR. Of these, 17 (40%) failed repeatedly to yield an amplification product—these included 6 Gram stain/culture-positive samples and 11 Gram stain/culture-negative samples. Thus, these 17 samples were excluded from subsequent analysis. As samples have been previously de-identified, patient-level data including antibiotic history were not available for analysis. Culture results for the 6 Gram stain/culture-positive samples that failed PCR amplification are shown in Supplementary Table 1.
Figure 1.
Samples analyzed in this study. A total of 43 clinical samples were identified that met the inclusion criteria. These included 19 samples (44%) that were negative by both Gram stain and culture and 24 samples (56%) that were positive by Gram stain and/or culture. All 43 samples were subjected to V1-V3 16S rRNA gene amplification. Of the 19 Gram stain/culture-negative samples, 8 samples (42%) were successfully amplified and sequenced using Illumina. Of the 24 Gram stain/culture-positive samples, 18 samples (75%) were amplified and sequenced.
Comparison of 16S rRNA Gene Sequencing and Bacterial Culture
To compare 16S rRNA gene sequencing with conventional cultures, we first defined a Gram stain/culture-positive sample as monomicrobial if (1) culture yielded a single microorganism or (2) Gram stain demonstrated a single morphology but culture was negative. We defined a sample as polymicrobial if (1) culture grew more than 1 microorganism or (2) Gram stain demonstrated multiple distinct morphologies but culture was negative. Thus, 16 of 24 (66.7%) were classified as polymicrobial and 8 (33.3%) were monomicrobial. A total of 18 Gram stain/culture-positive samples (13 polymicrobial and 5 monomicrobial samples) yielded 16S rRNA gene amplification products (75%).
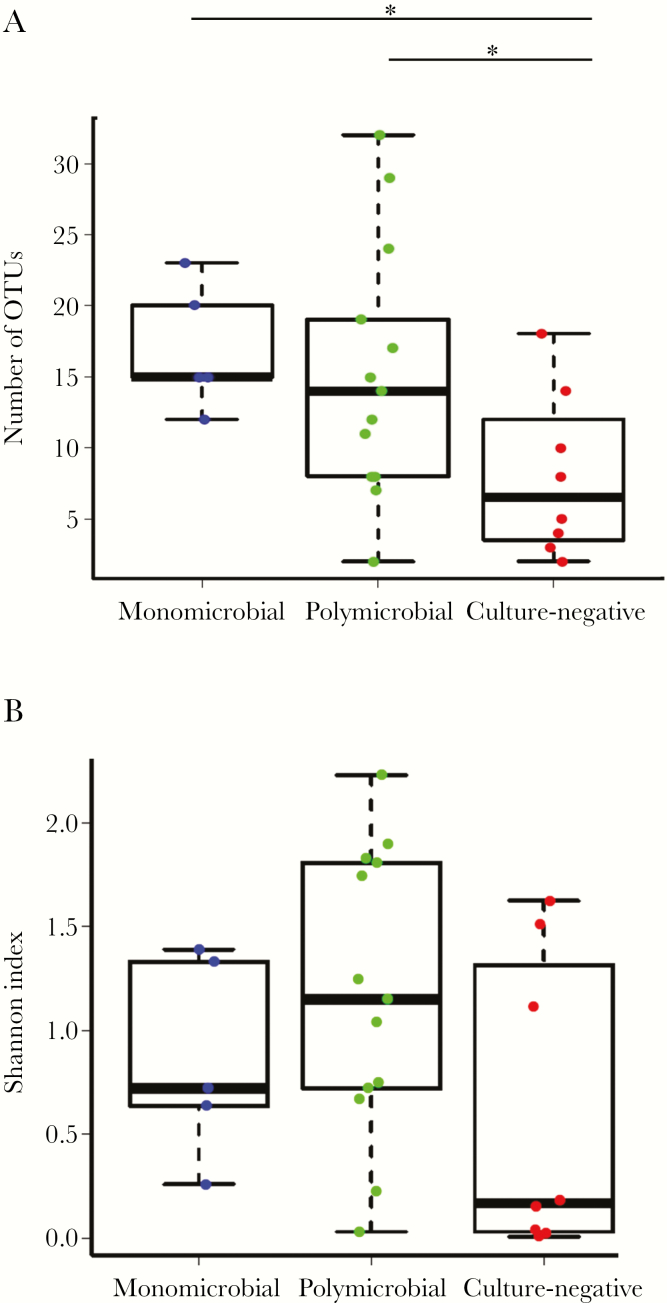
All 26 samples that yielded amplification products were deep sequenced using the Illumina MiSeq platform, yielding 919 669 reads after filtering and quality control, with a mean of 30 656 reads per sample (interquartile range, 26 086–37 051). From 26 samples, we identified a total of 221 distinct operational taxonomic units (mean, 13.1 OTUs per sample; interquartile range, 8–16.5 OTUs). The mean number of OTUs was 17 (range, 12–23) for monomicrobial samples and 15.2 (range, 2–32) for polymicrobial samples. In comparison, the mean number of OTUs for Gram stain/culture-negative samples was 8 (range, 2–18) (Figure 2A). The microbial richness (ie, the number of OTUs) for monomicrobial and polymicrobial samples was significantly higher than culture-negative samples (17 and 15.2, respectively, vs 8; P < .05, unpaired Student t test). Combining monomicrobial and polymicrobial samples, microbial richness for Gram stain/culture-positive samples was significantly higher than for Gram stain/culture-negative samples (15.7 vs 8; P < .05, unpaired Student t test). No significant difference was observed between monomicrobial and polymicrobial samples.
Figure 2.
Microbial richness and diversity by subgroups. (A) The number of operational taxonomic units (microbial richness), determined by 16S rRNA sequencing for each of the 3 groups (classified according to culture results), is shown on the y-axis. (B) Shannon index (microbial diversity) is shown on the y-axis. Mean Shannon indices were compared. The asterisk indicates P < .05 (unpaired Student t test).
Microbial diversity for both monomicrobial and polymicrobial samples was higher than for culture-negative samples (0.87 and 1.18, respectively, vs 0.58). However, these differences were not statistically significant. Combining monomicrobial and polymicrobial samples, microbial diversity for Gram stain/culture-positive samples was higher than for Gram stain/culture-negative samples (1.09 vs 0.58; P = .08) but was not statistically significant (Figure 2B).
Monomicrobial Specimens Were Polymicrobial
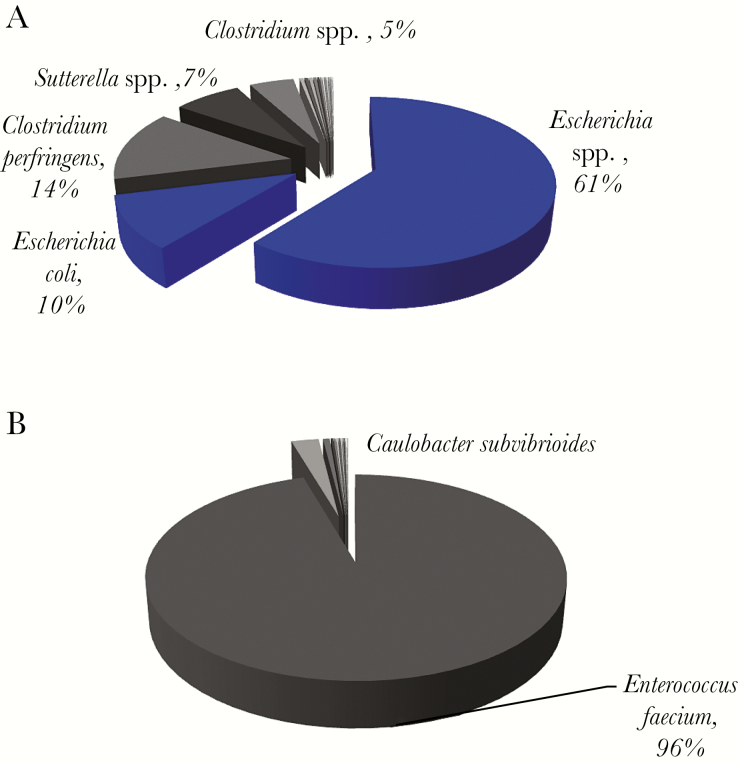
16S rRNA gene sequencing identified a diverse community profile in all 5 monomicrobial samples (Table S2), harboring many OTUs in each sample (Figure 3; Supplementary Figure 2). The organisms identified by microbiologic culture were the dominant OTUs in 3 of the 5 samples (Figure 3A; Supplementary Figure 2A and B). For example, E. coli identified by culture in an intra-abdominal abscess (Figure 3A) was the dominant OTU (71.1%). Interestingly, Clostridium perfringens (13.7%) and other Clostridium spp. were also detected in high abundance, and several organisms with ≥1% in read abundance were mapped to anaerobes or facultative anaerobes. In contrast, for 2 of the 5 monomicrobial samples (Figure 3B; Supplementary Figure 2C), the cultured isolates were not the most abundant organisms in the abscess collections. For example, in a perihepatic abscess (Figure 3B), coagulase-negative staphylococci (CoNS) was identified by culture (which was likely a skin contaminant). However, 16S rRNA gene sequencing revealed a total of 15 OTUs including Enterococcus faecium (95.5%) and Caulobacter subvibrioides (2.6%) as the dominant OTUs, with Staphylococcus epidermidis, a CoNS, as a minority species, constituting 0.08% of total sequence reads.
Figure 3.
Microbial composition and read abundance, determined by 16S rRNA sequencing in representative monomicrobial samples. (A) Gram stain of this sample showed 4+ polymorphonuclear leukocytes (PMNs) and 2+ Gram (+/-) rods, and Escherichia coli was isolated by culture. The dominant operational taxonomic units (OTUs) mapped to Escherichia spp. and E. coli. All organisms with ≥1% frequency were anaerobes or facultative anaerobes. (B) Gram stain of this sample was negative, but subculture of the specimen grew coagulase-negative Staphlococci. The dominant OTU was Enterococcus faecium. Blue pie slices denote concordance between 16S rRNA sequencing data and microbiologic cultures, and the pie slices in shades of gray indicate organisms with their read abundance not identified by culture.
Organisms Isolated by Culture Were Generally Minority Populations in Polymicrobial Samples
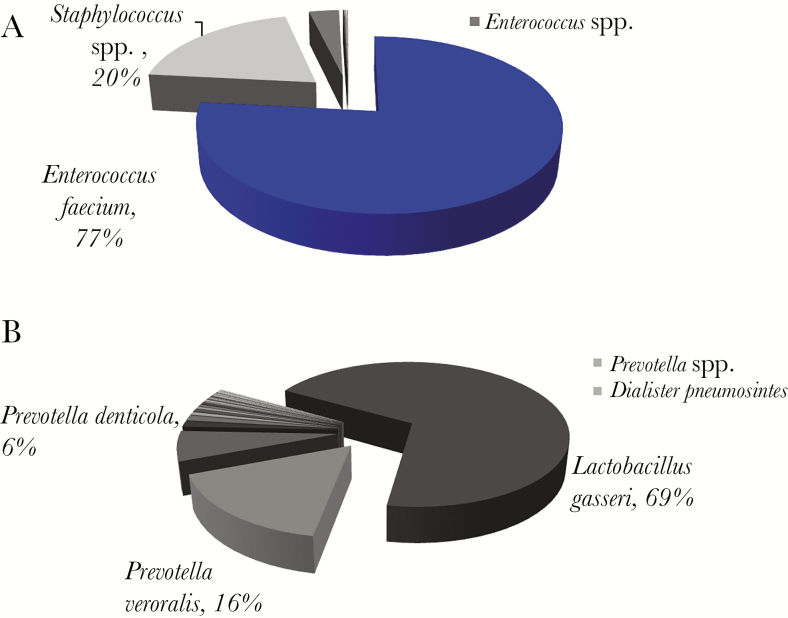
Overall, the number of OTUs in polymicrobial samples (Supplementary Table 3) was not significantly different from that of monomicrobial samples (mean, 15.23 vs 17). However, the range of OTUs varied widely (range, 2–32) (Figure 2A). In contrast to monomicrobial samples, the organism identified by culture was the dominant OTU in only 1 of 13 polymicrobial samples (Figure 4A). For the remaining 12 samples, the cultured organisms were minority populations in the abscess collections, and the dominant OTUs often mapped to Lactobacillus spp. or anaerobes such as Prevotella spp., Fusobacterium spp., and Bacteroides spp. (eg, Figure 4B; Supplementary Figure 3A–K). Sequences belonging to the genera Staphylococcus spp., Streptococcus spp., the family Enterobacteriaceae, and other aerobic Gram-negatives were generally a minority population. For example, E. coli or Enterococcus spp. were identified by culture in 11 of 13 samples, but either constituted a very small minority of the overall microbial population or were not detected at all by deep sequencing (Supplementary Figure 3).
Figure 4.
Microbial composition and read abundance, determined by 16S rRNA sequencing in polymicrobial samples. (A) Gram stain showed 4+ Gram-positive cocci in pairs and chains, 2+ Gram-negative rods, and few Gram-positive rods. Enterococcus spp. and Candida albicans were isolated by cultures. The dominant operational taxonomic units (OTUs) mapped to Enterococcus faecium. (B) Gram stain showed 4+ polymorphonuclear cells and 2+ Gram-variable rods. Escherichia coli was isolated by culture. The dominant OTUs were Lactobaccilus gasseri and Prevotella spp. Blue pie slices denote concordance between 16S rRNA sequencing data and microbiologic cultures, and pie slices in shades of gray indicate organisms not identified by culture. OTUs with ≥5% abundance are labeled on the pie chart. OTUs with <5% but ≥1% abundance are shown in the side bar.
Lower Microbial Richness and Diversity in Gram Stain/Culture-Negative Samples
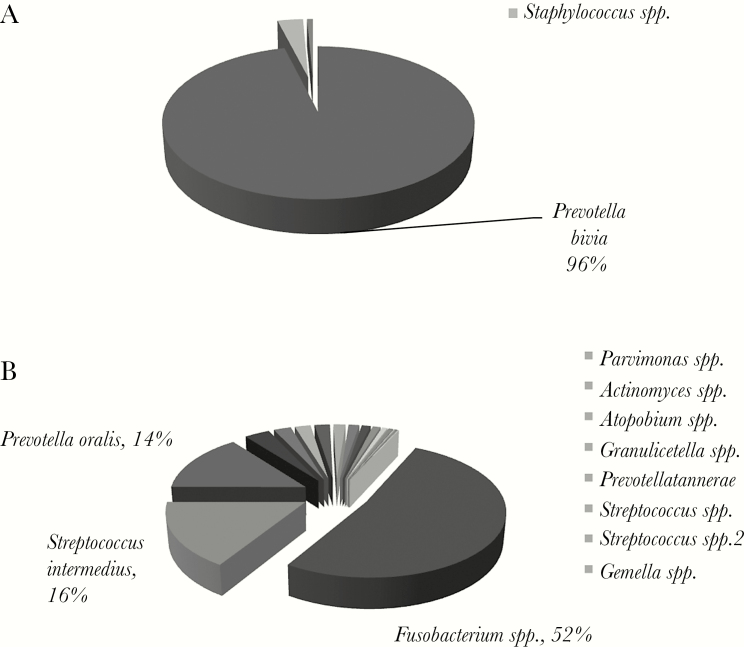
Compared with Gram stain/culture-positive samples, Gram stain/culture-negative samples (Table S4) had a lower number of OTUs (Figure 2). Of the 8 Gram stain/culture-negative samples, 5 had a dominant OTU that represented >95% of all 16S rRNA sequences—these were Streptococcus spp., B. fragilis, Parvimonas spp., Prevotella bivia, and Peptoniphilus spp. (Figure 5A; Supplementary Figure 4A–D). The remaining 3 samples harbored a single dominant OTU with an abundance ranging from 47% to 69%—1 was Fusobacterium (Figure 5B), a Gram-negative anaerobe, and 2 belonged to Streptococcus spp. (Supplementary Figure 4E and F). Overall, aerobic Gram-negative bacilli such as E. coli and Enterobacter comprised a small proportion of all 16S rRNA gene sequences in Gram stain/culture-negative samples (Figures 5; Supplementary Figure 4).
Figure 5.
Microbial composition and read abundance determined by 16S rRNA sequencing in Gram stain and culture-negative samples. (A) The sample was negative by Gram stain and culture, but 16S rRNA sequencing analysis revealed a single dominant operational taxonomic unit (OTU). (B) The sample was negative by Gram stain and culture but sequencing analysis identified 11 OTUs with ≥1% read abundance. All 11 OTUs corresponded to organisms that were anaerobes or facultative anaerobes. OTUs with ≥5% abundance are labeled on the pie chart, and OTUs with <5% but ≥1% abundance are shown in the side bar.
DISCUSSION
The present study reports the microbial composition (determined by 16S rRNA sequencing) in abscess samples drained percutaneously and compares with microbiologic cultures performed for clinical indications. We found that culture-positive samples had a significantly higher number of OTUs than culture-negative samples, with a trend toward a higher microbial diversity. Of the 5 samples that grew a single organism by culture, sequencing analysis yielded the same organism as the dominant population in 3 samples, but other bacteria in lower abundance were also detected. In contrast, in the majority (12 of 13) of samples that were polymicrobial by culture, the cultured organisms were not the dominant OTUs. These results suggest that microbial richness and diversity of intra-abdominal abscesses are higher than previously thought. Using culture-independent 16S rRNA gene sequencing, many taxa that were not cultivated were identified, revealing the overall abscess microbiome not appreciated using culture-based techniques. In most cases, the consortia of cultured bacteria were not the most abundant bacteria identified by 16S rRNA gene sequencing. However, the data should be interpreted with caution given the lack of patient-level data and antibiotic history.
In samples with positive Gram stain and/or culture, especially those that grew multiple organisms in culture, we found that cultured isolates were generally minority constituents of the overall community that is dominated by anaerobes and facultative anaerobes. The cultured isolates were frequently aerobic Gram-negatives such as E. coli and Klebsiella spp., but other organisms such as Pseudomonas spp., Enterobacter spp., and group B Streptococcus were also observed. This is not surprising as these organisms generally grow well in cultures and likely outcompete strict anaerobes that may be difficult to preserve during transport and require special care to cultivate in the laboratory, which are often not achievable in standard clinical microbiology labs. Interestingly, sequences corresponding to these cultured organisms were often detected at very low frequencies or not detected at all, suggesting that Enterobacteriaceae and Streptococci commonly isolated from intra-abdominal abscesses may constitute a very small minority of the overall bacterial population.
Interestingly, samples that were negative by Gram stain and culture showed significant biodiversity. However, the total number of bacterial taxa was generally lower compared with Gram stain/culture-positive samples. Notably, the prevalence and abundance of members of the phylum Proteobacteria, including E. coli, which is known to co-enrich other known pathogens [28], were low. It’s likely that these samples were obtained from patients who were receiving antibiotics at the time of abscess collection. Unfortunately, the associated clinical data and antibiotic exposure history were not available. Sample 94 was of particular interest (Supplementary Table 4 and Supplementary Figure 4F). This specimen was a liver abscess and was Gram stain/culture-negative. 16S rRNA sequencing analysis revealed a polymicrobial composition dominated by S. intermedius, in addition to E. coli and Enterobacter spp. While organisms in the S. anginosus group (eg, S. intermedius) are often associated with intra-abdominal abscesses including the liver and could be responsible for a monomicrobial infection, our sequence data indicate a polymicrobial composition. Thus, our results suggest that culture-independent methods may be useful to confirm the presence of bacteria in select clinical settings where prior antibiotic usage precludes successful culturing of organisms from clinical samples.
Our data suggest that culture-based approaches may overestimate the prevalence of monomicrobial infections, missing much of the hidden microbial diversity in abscess collections [29]. Similarly, previous studies have suggested the presence of polymicrobial composition in samples with negative culture results [30]. Here we have demonstrated the ability of 16S rRNA sequencing to characterize microbial compositions in both culture-positive and culture-negative samples. All 26 samples that were sequenced had more than 1 distinct taxa (mean, 13.1 OTUs per sample), and the overall composition likely more closely resembled the underlying microbial diversity present in these abscesses. An appreciation of this biodiversity and how the organisms interact with each other, or the “superorganism” [31], is essential for understanding the development of abscess formation and ultimately how these organisms interact with the host to contribute to the pathophysiology of disease. Interspecies communication has been shown to serve as a prerequisite for certain bacterial infections, and pathogen growth in the presence of specific bacterial niches could modulate gene expression to a more virulent phenotype [32]. Additionally, minor communities undetected in traditional cultures could contribute to pathogen persistence after clinical treatment by releasing antibiotic-inactivating proteins, conferring protection to the community as a whole [33, 34].
The standard of care for patients with large intra-abdominal abscesses is percutaneous drainage followed by targeted antibiotic therapy guided by culture and susceptibilty. However, if standard cultures are unrevealing, treatment with broad-spectrum antimicrobials is often implemented, which could lead to undesirable consequences such as antibiotic resistance and complications such as Clostridium difficile infection. Knowledge of the genus or species of bacteria most likely responsible for clinical disease or virulence of the “superorganism” may allow for selection of narrower-spectrum antimicrobial agents. Similarly, targeted treatment against an organism isolated by culture may not be appropriate, as the isolated organisms may not be the primary pathogen. Animal studies of intra-abdominal sepsis have suggested that coliforms contribute to early sepsis while anaerobes are implicated in later stages of abscess formation [35]. This is consistent with our sequencing results, which showed a dominance of a variety of uncultured anaerobic organisms.
Culture-independent methods are limited by the inability to determine antimicrobial susceptibility, leaving the clinician to make treatment decisions based on known microbe characteristics and local antibiogram. Additionally, the clinical significance of bacterial taxa identified by sequencing is difficult to deduce. Further studies are necessary to understand the dynamic interplay within bacterial communities during abscess formation. Moreover, sequencing may lend itself to potential artifacts of amplifying contaminating environmental microbes as can occur in clinical cultures following percutaneous access. In our study, we have focused our discussion on taxa that were present at ≥1% relative read abundance.
As this study used universal primers for bacterial small subunit ribosomal RNA, fungal pathogens could not be identified. In 2 cases, conventional culture identified Candida albicans, 1 with concomitant normal gut flora and the other with Enterococcus spp. Sequencing of these samples identified multiple bacterial species. In future studies, fungal rDNA PCR could be used where pyogenic fungal infection is suspected or when both conventional methods and 16S rRNA sequencing fail to identify a putative pathogen.
Illumina sequencing of the bacterial 16S rRNA gene provides a semiquantitative measure of microbial compositions in abscess collections that cannot be deduced using culture-based methods. Conventional diagnostics require the presence of viable organisms, which could be compromised by antibiotic use. Moreover, the drainage and sampling approach, the transit time from the bedside to the laboratory, conditions of specimen storage (ie, anaerobic storage), the presence of fastidious microorganisms (eg, obligate intracellular bacteria or obligate anaerobes), and antibiotic therapy can all affect culture yield and biochemical tests significantly more than culture-independent approaches [36, 37]. In contrast, the 16S rRNA gene acts as a molecular barcode, allowing taxa-specific identification of bacteria without the need for culture. Thus, when culture-based methods fail, metagenomic approaches may be a reasonable alternative for identifying fastidious or uncultivable organisms [38]. On the other hand, we note that 17/43 (40%) specimens failed to yield PCR amplication products. Among them, 11 were Gram stain/culture-negative samples that likely harbored no bacteria at the time the samples were collected. Of the other 6 Gram stain/culture-positive samples, the presence of PCR inhibitors may have contributed to the failure in PCR amplification. Thus, while the sequence-based approach may be useful, technical or sample-specific factors may be a limitation in select samples.
In summary, the current study uncovered a hidden microbial diversity in intra-abdominal abscesses and suggests that conventional culture-based methods may selectively isolate aerobic and/or facultative aerobes, which are minority constituents of the overall microbial community in abscesses in some cases. As the role of a polymicrobial community in abscess formation remains poorly understood, future studies should focus on understanding the interplay between various bacteria in abscess collections and the clinical implications, including antimicrobial management and treatment response.
Supplementary Data
Supplementary materials are available at Open Forum Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Acknowledgements
We thank members of Wang laboratory for helpful discussion.
Financial support. This work was supported by the Gatorade Trust through funds distributed by the University of Florida, Department of Medicine, and in part by the Infectious Diseases Society of America medical scholars program to A.K.
Potential conflicts of interest. All authors: no reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Kaplan GG, Gregson DB, Laupland KB. Population-based study of the epidemiology of and the risk factors for pyogenic liver abscess. Clin Gastroenterol Hepatol 2004; 2:1032–8. [DOI] [PubMed] [Google Scholar]
- 2. Han A, Zenilman JM, Melendez JH et al. The importance of a multifaceted approach to characterizing the microbial flora of chronic wounds. Wound Repair Regen 2011; 19:532–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Mendes L, Azevedo NF, Felino A, Pinto MG. Relationship between invasion of the periodontium by periodontal pathogens and periodontal disease: a systematic review. Virulence 2015; 6:208–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Dickson RP, Erb-Downward JR, Prescott HC et al. Analysis of culture-dependent versus culture-independent techniques for identification of bacteria in clinically obtained bronchoalveolar lavage fluid. J Clin Microbiol 2014; 52:3605–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Murray JL, Connell JL, Stacy A et al. Mechanisms of synergy in polymicrobial infections. J Microbiol 2014; 52:188–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Al Masalma M, Armougom F, Scheld WM et al. The expansion of the microbiological spectrum of brain abscesses with use of multiple 16S ribosomal DNA sequencing. Clin Infect Dis 2009; 48:1169–78. [DOI] [PubMed] [Google Scholar]
- 7. Chen SC, Tsai SJ, Chen CH et al. Predictors of mortality in patients with pyogenic liver abscess. Neth J Med 2008; 66:196–203. [PubMed] [Google Scholar]
- 8. Alvarez JA, González JJ, Baldonedo RF et al. Single and multiple pyogenic liver abscesses: etiology, clinical course, and outcome. Dig Surg 2001; 18:283–8. [DOI] [PubMed] [Google Scholar]
- 9. Ferraioli G, Brunetti E, Gulizia R et al. Management of splenic abscess: report on 16 cases from a single center. Int J Infect Dis 2009; 13:524–30. [DOI] [PubMed] [Google Scholar]
- 10. Mancini N, Carletti S, Ghidoli N et al. The era of molecular and other non-culture-based methods in diagnosis of sepsis. Clin Microbiol Rev 2010; 23:235–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cinat ME, Wilson SE, Din AM. Determinants for successful percutaneous image-guided drainage of intra-abdominal abscess. Arch Surg 2002; 137:845–9. [DOI] [PubMed] [Google Scholar]
- 12. Robert B, Chivot C, Fuks D et al. Percutaneous, computed tomography-guided drainage of deep pelvic abscesses via a transgluteal approach: a report on 30 cases and a review of the literature. Abdom Imaging 2013; 38:285–9. [DOI] [PubMed] [Google Scholar]
- 13. Solomkin JS, Mazuski JE, Bradley JS et al. Diagnosis and management of complicated intra-abdominal infection in adults and children: guidelines by the Surgical Infection Society and the Infectious Diseases Society of America. Clin Infect Dis 2010; 50:133–64. [DOI] [PubMed] [Google Scholar]
- 14. Pei Z, Bini EJ, Yang L et al. Bacterial biota in the human distal esophagus. Proc Natl Acad Sci U S A 2004; 101:4250–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Fenollar F, Raoult D. Molecular diagnosis of bloodstream infections caused by non-cultivable bacteria. Int J Antimicrob Agents 2007; 30(Suppl 1):S7–15. [DOI] [PubMed] [Google Scholar]
- 16. Sibley CD, Church DL, Surette MG et al. Pyrosequencing reveals the complex polymicrobial nature of invasive pyogenic infections: microbial constituents of empyema, liver abscess, and intracerebral abscess. Eur J Clin Microbiol Infect Dis 2012; 31:2679–91. [DOI] [PubMed] [Google Scholar]
- 17. Woo PC, Lau SK, Teng JL et al. Then and now: use of 16S rDNA gene sequencing for bacterial identification and discovery of novel bacteria in clinical microbiology laboratories. Clin Microbiol Infect 2008; 14:908–34. [DOI] [PubMed] [Google Scholar]
- 18. Cai HY, Caswell JL, Prescott JF. Nonculture molecular techniques for diagnosis of bacterial disease in animals: a diagnostic laboratory perspective. Vet Pathol 2014; 51:341–50. [DOI] [PubMed] [Google Scholar]
- 19. Wolcott RD, Gontcharova V, Sun Y et al. Bacterial diversity in surgical site infections: not just Aerobic cocci any more. J Wound Care 2009; 18:317–23. [DOI] [PubMed] [Google Scholar]
- 20. Mishra AK, Dufour H, Roche PH et al. Molecular revolution in the diagnosis of microbial brain abscesses. Eur J Clin Microbiol Infect Dis 2014; 33:2083–93. [DOI] [PubMed] [Google Scholar]
- 21. Salipante SJ, Sengupta DJ, Rosenthal C et al. Rapid 16S rRNA next-generation sequencing of polymicrobial clinical samples for diagnosis of complex bacterial infections. PLoS One 2013; 8:e65226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Brogden KA, Guthmiller JM, Taylor CE. Human polymicrobial infections. Lancet 2005; 365:253–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Brook I, Frazier EH. Microbiology of subphrenic abscesses: a 14-year experience. Am Surg 1999; 65:1049–53. [PubMed] [Google Scholar]
- 24. Brook I, Frazier EH. Aerobic and anaerobic microbiology in intra-abdominal infections associated with diverticulitis. J Med Microbiol 2000; 49:827–30. [DOI] [PubMed] [Google Scholar]
- 25. Dethlefsen L, Huse S, Sogin ML, Relman DA. The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLoS Biol 2008; 6:e280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Dewhirst FE, Chen T, Izard J et al. The human oral microbiome. J Bacteriol 2010; 192:5002–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sibley CD, Church DL, Surette MG et al. Pyrosequencing reveals the complex polymicrobial nature of invasive pyogenic infections: microbial constituents of empyema, liver abscess, and intracerebral abscess. Eur J Clin Microbiol Infect Dis 2012; 31:2679–91. [DOI] [PubMed] [Google Scholar]
- 28. Pettengill JB, McAvoy E, White JR et al. Using metagenomic analyses to estimate the consequences of enrichment bias for pathogen detection. BMC Res Notes 2012; 5:378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Brook I. Microbiology of polymicrobial abscesses and implications for therapy. J Antimicrob Chemother 2002; 50:805–10. [DOI] [PubMed] [Google Scholar]
- 30. Al Masalma M, Armougom F, Scheld WM et al. The expansion of the microbiological spectrum of brain abscesses with use of multiple 16S ribosomal DNA sequencing. Clin Infect Dis 2009; 48:1169–78. [DOI] [PubMed] [Google Scholar]
- 31. Eberl G. A new vision of immunity: homeostasis of the superorganism. Mucosal Immunol 2010; 3:450–60. [DOI] [PubMed] [Google Scholar]
- 32. Duan K, Dammel C, Stein J et al. Modulation of Pseudomonas aeruginosa gene expression by host microflora through interspecies communication. Mol Microbiol 2003; 50:1477–91. [DOI] [PubMed] [Google Scholar]
- 33. Brook I. Beta-lactamase-producing bacteria in mixed infections. Clin Microbiol Infect 2004; 10:777–84. [DOI] [PubMed] [Google Scholar]
- 34. Brook I. Microbiology of polymicrobial abscesses and implications for therapy. J Antimicrob Chemother 2002; 50:805–10. [DOI] [PubMed] [Google Scholar]
- 35. Bartlett JG, Onderdonk AB, Louie T et al. A review. Lessons from an animal model of intra-abdominal sepsis. Arch Surg 1978; 113:853–7. [DOI] [PubMed] [Google Scholar]
- 36. Baron E, Thomson R. 2011. Specimen collection, transport, and processing: bacteriology. In: Versalovic J, Carroll K, Funke G. et al. , eds. Manual of Clinical Microbiology. 10th ed Washington, DC: ASMPress: 228–71. [Google Scholar]
- 37. Peters RP, van Agtmael MA, Danner SA et al. New developments in the diagnosis of bloodstream infections. Lancet Infect Dis 2004; 4:751–60. [DOI] [PubMed] [Google Scholar]
- 38. Pallen MJ. Diagnostic metagenomics: potential applications to bacterial, viral and parasitic infections. Parasitology 2014; 141:1856–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.