Abstract
Background
MicroRNAs (miRs) are promising new therapeutics for glioblastoma. However, which miRs are most effective against glioblastomas and how these miRs should be delivered are major unanswered problems.
Methods
To identify potent antiglioma miRs, we selected 8 miRs based on a literature search and screened them against a panel of glioma stem cell (GSC) lines, representing all of the glioblastoma subtypes defined by The Cancer Genome Atlas. To address delivery, we tested the hypothesis that ex vivo cultured bone marrow–derived mesenchymal stem cells (MSCs) can package miRs into exosomes and that these engineered exosomes can systemically deliver antiglioma miRs to glioblastomas.
Results
Of the screened miRs, we identified miR-124a as the most effective antiglioma agent against GSCs. We then transduced MSCs with lentivirus vectors containing miR-124a and isolated vesicles from the medium. Electron microscopy, western blotting, and Nanosight proved that the isolated vesicles were exosomes. Quantitative PCR documented that these exosomes contained high levels of miR-124a, which was not present in control exosomes. In vitro treatment of GSCs with exosomes containing miR-124a (Exo-miR124) resulted in a significant reduction in viability and clonogenicity of GSCs compared with controls. In vivo treatment of mice harboring intracranial GSC267 with systemically delivered Exo-miR124 resulted in 50% of animals living long term. No evidence of tumor was present on histological analysis of the survivors. Mechanistic studies showed that miR-124a acts by silencing Forkhead box (FOX)A2, resulting in aberrant intracellular lipid accumulation.
Conclusion
MSCs can be used as natural biofactories to produce Exo-miR124, which is an effective antiglioma agent worthy of further clinical evaluation.
Keywords: exosomes, glioblastoma, glioma stem cells, mesenchymal stem cells, miR-124a
Importance of the study
This study identifies miR-124a as an effective pan-GSC antiglioma miR and elucidates exosomes derived from MSCs as a novel delivery strategy. Using lentivirus vectors, we engineer MSCs to produce exosomes carrying supraphysiological levels of miR-124a (Exo-miR124a) and showed that Exo-miR124a can inhibit the growth and clonogenicity of patient-derived GSCs. We show for the first time that treatment with systemically delivered Exo-miR124 is capable of curing mice harboring intracranial GSC xenografts. Mechanistic studies showed that miR-124a acts by downregulating FOXA2, a known target of miR-124a, and that apoptotic cell death correlates with FOXA2-mediated aberrant intracellular lipid accumulation. These studies provide some of the first preclinical data that MSCs can be used as biofactories for the ex vivo production of antiglioma miR-carrying exosomes, which can be systemically delivered to patients for the purpose of eradicating malignant gliomas, a disease for which there is no effective treatment.
With a median survival of less than 15 months, there is currently no effective treatment for glioblastoma (GBM). This therapeutic failure is due to the complex biology of GBMs, including the presence of treatment-resistant sphere-forming glioma stem cells (GSCs), as well as delivery challenges imposed by the blood–brain/tumor barrier.1,2 Identifying effective GBM-specific therapeutics and elucidating novel strategies for delivering these new agents would greatly advance the treatment of GBMs.
MicroRNAs (miRs) may be a new class of anticancer agents because of their ability to modulate posttranscriptional gene expression.3,4 Restoration of several downregulated tumor-suppressor miRs has been shown to inhibit the growth of GBMs, suggesting that certain miRs may be effective antiglioma therapeutics.5 But despite their therapeutic potential, it remains unclear which miRs will be most effective against GBMs. Additionally, delivering miRs to tumors has been challenging due to degradation after systemic delivery, poor cellular uptake, and lack of tumor targeting.6,7 Although viral-based delivery systems, liposomes, and artificial nanoparticles have been assessed, all of these approaches exhibit low efficiency.8 Thus, how miRs should be delivered to GBMs is an unresolved problem in their therapeutic application to GBMs.
Exosomes are physiological nanovesicles (40–150 nm diameter) with a lipid bilayer membrane of endocytic origin that are released by cells and can be taken up by neighboring and distant cells because they are stable in blood. As with other cancers, exosomes are an important source of intercellular communication in GBMs, and tumor cells of GBMs release and take up exosomes.9 Several studies indicate that exosomes may be used as delivery vehicles for cancer therapeutics.10 Most recently, Kamerkar et al showed that fibroblast-derived exosomes that were loaded ex vivo by electroporation with anti-KRAS small interfering RNA could be used in the treatment of KRAS mutant pancreatic ductal adenocarcinoma.11
Here we investigate whether exosomes derived from cultured bone marrow–derived human mesenchymal stem cells (MSCs) can be used to systemically deliver antiglioma miRs to GBMs. MSCs are stromal cells that can be acquired from the bone marrow and are readily cultured and engineered ex vivo. Our group was the first to show that MSCs can be used to deliver therapeutic biological agents to GBMs due to their ability to home to gliomas, including GSC xenografts, after systemic (intracarotid) administration.12–16 Gatti et al showed that MSCs are a rich source of exosomes and that MSC-derived exosomes have a tendency to home to wounded tissue, supporting the idea that these exosomes may also home to brain tumors, much like the MSCs from which they are derived.17,18 Recently, several investigators have shown that MSCs naturally package miRs into exosomes, suggesting that MSCs may be exploited to package exogenous therapeutic miRs.19,20 Based on these data, we reasoned that after forced transduction with lentiviruses containing specific antiglioma miRs, ex vivo cultured MSCs would package these miRs into exosomes and release the miR-containing exosomes into the medium. We hypothesized that we could isolate these exosomes and then use them to systemically treat GBMs. Although Lee et al21 and Katakowski et al20 used related strategies, Lee et al only performed intratumoral injections of MSCs loaded with miR-124 and did not isolate exosomes, nor did they explore efficacy in their animal studies; Katakowski et al isolated exosomes (albeit containing miR-146) but also performed intratumoral injections and did not demonstrate a survival advantage in their in vivo studies, which relied only on rat glioma cells, and not human GSCs.20,21 No study has demonstrated the efficacy of systemically delivered MSC-derived exosomes carrying antiglioma miRs in the treatment of human GBM.
In this context, we first screened a panel of 8 miRs against 5 GSCs and identified miR-124a as an effective pan-GSC antiglioma miR. Using lentivirus vectors, we then engineered MSCs to produce exosomes carrying miR-124a and showed that these exosomes could be used to inhibit the growth of GSCs, including after systemic administration in vivo. Mechanistic studies showed that miR-124a acts by downregulating Forkhead box (FOX)A2, a known target of miR-124a and a mediator of lipid metabolism. These studies show for the first time that MSCs can be used as natural biofactories for the ex vivo production of anticancer miR-carrying exosomes that can be isolated and delivered to patients for the purpose of eradicating malignant gliomas.
Materials and Methods
Cell Lines
Five GSC lines (GSC267, GSC20, GSC6-27, GSC8-11, and GSC2-14) were used in these studies (Table 1 and Supplementary Figure S1). All GSCs were isolated from surgical specimens of patients according to the methods of Singh et al1 and as we previously published22 (see Supplementary Methods for details of GSCs). Expression profiling and RNA sequencing analyses showed that GSCs represented The Cancer Genome Atlas type of GBMs and expressed mutations commonly found in GBMs (Table 1). MSCs were obtained from Lonza and cultured in Minimum Essential Medium Alpha plus 10% serum. MSCs were used at passage 3–4.
Table 1.
Characteristics of GSCs
| GSC | TCGA Subtype | MGMT Methylation | Radiation Sensitivity | Mouse Survival (days) |
7p11.2-EGFR Amp. |
10q23.31-PTEN Deletion |
17p13.1-TP53 Deletion |
PTEN Mut. |
EGFR Mut. | TP53 Mut. | IDH1 Mut. | CD133 Express, % | CD15 Express, % | CD44 Express, % |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GSC6-27 | Classical | Unmethylated | Resistant | >90 | 0 | Deleted | 0 | Wild | Wild | Mutated | Wild | 0.93 | 86.05 | 62.26 |
| GSC2-14 | Classical | Methylated | Sensitive | >90 | Amplified | 0 | 0 | Mutated | Wild | Mutated | Wild | 0.06 | 52.43 | 0.52 |
| GSC20 | Mesenchymal | Methylated | Sensitive | >90 | 0 | Deleted | 0 | Wild | ND | ND | ND | 0.05 | 91.44 | 0.82 |
| GSC267 | Mesenchymal | Unmethylated | Sensitive | <90 | 0 | Deleted | Deleted | Wild | ND | ND | ND | 2.6 | 0.72 | 99.25 |
| GSC8-11 | Proneural | Unmethylated | ND | <90 | Amplified | 0 | 0 | Mutated | Wild | Mutated | ND | 0.29 | 47.17 | 99.95 |
Abbreviations: TCGA, The Cancer Genome Atlas; MGMT, O6-DNA methylguanine-methyltransferase; EGFR, epidermal growth factor receptor; PTEN, phosphatase and tensin homolog; IDH1, isocitrate dehydrogenase 1; Express, expression; ND = not done.
Lentivirus Constructs and Transduction
Lentiviruses containing miRs were generated and used to overexpress miRs in GSCs and MSCs as outlined in the Supplementary Methods.
Exosome Isolation
Exosomes were isolated from the medium of cultured MSCs using differential centrifugation or sucrose gradients as previously described23 (Supplementary Methods). Methods for analyses of isolated particles, including antibodies, are outlined in the Supplementary Methods.
Cre-loxP Reporter System
GSCs were transduced with reporter lentiviruses (pLV-CMV-LoxP-DsRed-LoxP-eGFP; Addgene, #65726). MSCs were transduced with a lentivirus vector containing Cre (Puro.Cre empty vector) (Addgene #17408). See Supplementary Methods for experimental details.
Animal Studies
Male athymic nude mice (nu/nu) were purchased from the Department of Experimental Radiation Oncology (Houston, Texas) and anesthetized using intraperitoneal injections of ketamine (100 mg/kg)/xylazine (10 mg/kg). Intracranial xenografts were implanted using a guide screw and a multiport micro-infusion syringe pump (Harvard Apparatus) as previously described.24 All animal manipulations were performed in accordance with ethics guidelines under an approved protocol.
FOXA2 Mechanistic Studies
Knockdown of FOXA2 was achieved using short hairpin (sh)RNA technology as described in Supplementary Methods. See the Supplementary Methods for constructs of lentiviruses overexpressing FOXA2 in rescue studies and for treatment protocols.
Statistical Analysis
Statistical differences were assessed by the 2-tailed, unpaired Student’s t-test with significance if P < 0.05. The data are represented as mean ± standard error. For animal studies, statistical significance was assessed by the log-rank test.
Results
MiR-124a Is an Effective Antiglioma miR
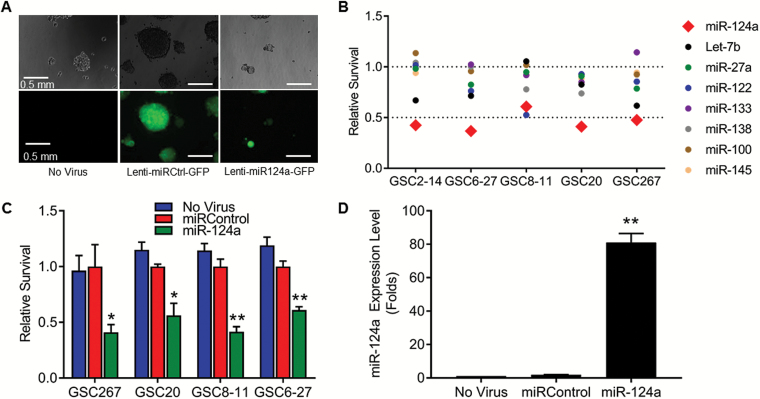
Based on a review of the literature, we identified 8 miRs purported to have antiglioma properties (miR-124a, miR-27a, miR-100, miR-122, miR-133, miR-138, miR-145, Let-7b), and we generated lentiviruses containing the precursor cDNA of each of these miRs under cytomegalovirus (CMV) promoter and green fluorescent protein (GFP) under EF1a promoter (called lenti-miR124a-GFP, lenti-miR27a-GFP, etc). Lentivirus containing a scrambled miR precursor was generated as a control (lenti-miRControl-GFP). To determine the extent to which each of these miRs was capable of inhibiting the growth of gliomas, we used a panel of 5 GSCs, each of which was extensively characterized (see Table 1, Supplementary Figure S1, and Supplementary Methods).25,26 Each of the GSCs was transduced with each of the 8 lentivirus-miR constructs (multiplicity of infection [MOI] 3). Lenti-miRControl-GFP (MOI 3) and medium alone (no virus) were used as negative controls. Visual inspection for green fluorescent cells showed 90%–95% transduction efficiency of all GSCs (Fig. 1A). One week after plating, a colorimetric assay (water-soluble tetrazolium salt 1 [WST1]) showed that miR-124a resulted in the most significant decrease in GSC viability in all GSC lines tested, identifying miR-124 as the most effective antiglioma miR (P < 0.01 vs miRControl in all GSCs, Student’s t-test) (Fig. 1B and Supplementary Figure S2).
Fig. 1.
Direct effects of selected miRs on GSCs. (A) Representative photomicrographs of GSC267 spheroids 72 h after treatment with indicated lentiviruses. Upper panel shows light microscopy and lower panel shows fluorescent microscopy; 90%–95% transduction is evident by GFP-labeled cells. Spheres of GSCs transduced with lenti-miR124-GFP are smaller because of antiproliferative effects of miR-124. Similar results were obtained for all other GSC lentivirus transductions. Bars = 0.5 mm. (B) Viability of GSCs after treatment with indicated miRs and assayed for proliferation using the WST1 assay. Values normalized to “miRControl.” Only miR-124a resulted in a significant decrease in cell survival. (C) Viability of several GSCs after treatment with miR-124. Values are relative to miRControl. Values normalized to “miRControl.” *P < 0.05, **P < 0.01. (D) Quantitative RT-PCR analysis of expression level of miR-124a in GSC267 after treatment with lentivirus. **P < 0.01.
To verify the antiglioma effects of miR-124a, the viability of GSCs after treatment with lenti-miR124a-GFP was analyzed through direct cell count in a dye exclusion assay. GSC267, GSC6-27, GSC8-11, and GSC20 were treated with lenti-miR124a-GFP, lenti-miRControl-GFP, or medium alone. Treatment with lenti-miR124a-GFP resulted in a reduction in viability of 58.9% for GSC267 (P = 0.015 vs miRControl), 43.8% for GSC20 (P = 0.016), 58.5% for GSC8-11 (P = 0.001), and 39% for GSC6-27 (P = 0.001) (Fig. 1C).
To directly prove that miR-124a was overexpressed in GSCs transduced with lenti-miR124-GFP, GSC267 was treated with lenti-miR124a-GFP (MOI 3), lenti-miRControl-GFP, or medium alone, and 72 hours later cells were lysed, RNA was collected, and miR-124a levels were analyzed by quantitative reverse transcription (qRT) PCR, using primers for miR-124a and for miR-16, a commonly used internal control for quantifying miRs (see Supplementary Methods). GSCs transduced with lenti-miR124-GFP contained 81-fold more miR-124a compared with GSCs treated with medium alone or with lenti-miRControl-GFP (P < 0.0001) (Fig. 1D).
MSCs Are Effective Delivery Vehicles for miR-124a
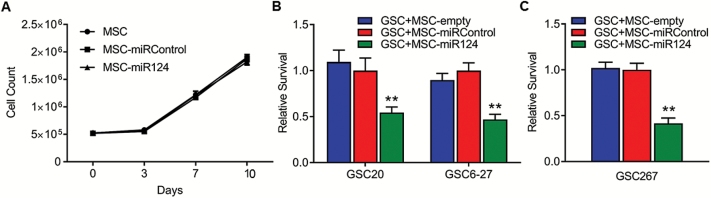
We validated the results of Lee et al,21 showing that MSCs can be used as delivery vehicles for miR-124a in vitro. Specifically, we transduced MSCs with lenti-miR124a-GFP (MOI 3), lenti-miRControl-GFP (MOI 3), or medium alone, generating MSC-miR124, MSC-miRControl, or MSC-empty, respectively. MSCs remained viable after lenti-miR124a-GFP transduction and proliferated similarly to nontransduced MSCs (Fig. 2A), indicating that miR-124a is not deleterious to MSC growth. Forty-eight hours after transduction, we plated MSCs (105 cells/well) in the upper wells of Transwell plates and we plated GSC20 and GSC6-27 (2.5 × 104 cells/well) in the lower wells. The wells were separated by a filter with 0.4-μm pores, which allowed passage of only soluble factors, including nano-sized exosomes, and prevented MSC migration. After 7 days, co-culture with MSCs-miR124 resulted in significant reductions in GSC viability compared with co-culture with control MSCs: 45.6% reduction in viability of GSC20 (P = 0.002 vs MSCs-miRControl), and 52.9% reduction in viability of GSC6-27 (P = 0.0002) (Fig. 2B). Similar results were obtained in an independent Transwell experiment in which MSCs-miR124 were co-cultured with GSC267 and GSC viability was assayed by trypan blue–stained cell count (P = 0.001) (Fig. 2C).
Fig. 2.
Characteristics and effects of MSCs transduced with lenti-miR124-GFP. (A) Viability of MSCs treated with indicated conditions and assayed for proliferation at indicated time points. (B and C) Viability of GSCs after co-culture with MSCs overexpressing miR-124a in Transwell experiment. MSCs were treated with indicated condition and GSCs were assayed for proliferation using WST1 (B) or trypan blue–stained cell count (C). Values normalized to “GSC + MSC-miRControl.” **P < 0.01.
MSCs Can Be Engineered to Package miR-124a into Exosomes
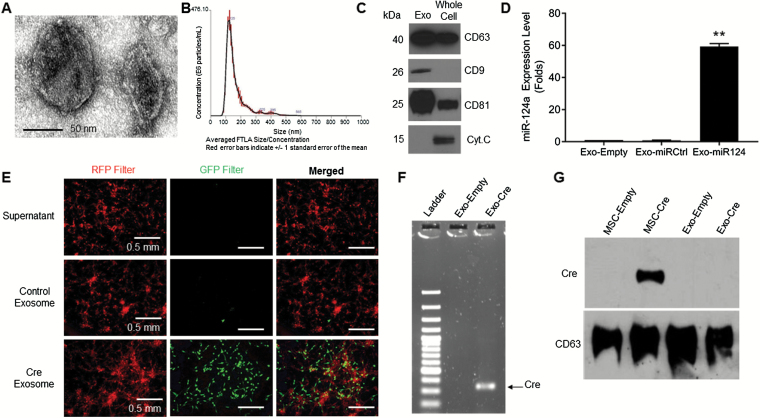
Because MSCs are known to package miRs into exosomes, we reasoned that the effects of MSCs-miR124 in the Transwell experiments were mediated through exosomes, as suggested by Lee et al.21 Therefore, we hypothesized that after transduction with lenti-miR124-GFP, MSCs package miR-124a into exosomes and release these exosomes into the medium. To test this hypothesis, MSCs were transduced (MOI 3) with lenti-miR124-GFP, lenti-miRControl-GFP, or medium, as above, generating MSCs-miR124, MSCs-miRControl, or MSCs-empty. These transduced MSCs were then plated in exosome-free medium and after 48 hour the medium was collected. Vesicles were then isolated from the medium by differential centrifugation. Transmission electron microscopy revealed spherical structures of 50–100 nm diameter with bilayer membranes, consistent with the definition of exosomes (Fig. 3A). Nanoparticle tracking technology (Nanosight) demonstrated a sharp peak of vesicles at 105 nm (Fig. 3B); a similar profile was also shown by highly purified exosomes obtained using the sucrose density gradient method (Supplementary Figure S3), indicating the purity of the isolated particles and a size consistent with previous reports of exosomes using this method.11 Western blotting showed that the isolated vesicles expressed cluster of differentiation (CD)9, CD63, and CD81, which are known exosome markers, whereas mitochondrial protein cytochrome c was not in the isolated vesicles, as expected of exosomes (Fig. 3C). These results verified that the vesicles isolated from the medium had properties consistent with the definition of exosomes.
Fig. 3.
Characteristics of exosomes collected from the medium of MSCs-miR124. (A) Representative photomicrographs of isolated exosomes using electron microscopy. Bar = 50 nm. (B) Nanosight graph showing size (100–125 nm) and concentration of particles isolated using differential centrifugation. (C) Western blot comparing exosomal (CD9, CD63, CD81) and nonexosomal (cytochrome c) markers in isolated particles and whole MSC lysates. (D) Quantitative RT-PCR analyzing expression level of miR-124a in exosomes derived from MSCs transduced with lenti-miR124a-GFP. **P < 0.01. (E) Photomicrographs of GSC267-dsR/GFP treated with MSC derived–exosomes. Only exosomes derived from MSC-Cre induced a color switch from red to green in GSCs. Bars = 0.5 mm. (F) Gel electrophoresis of PCR-amplified Cre mRNA. Only exosomes derived from MSCs transduced with the Cre-LoxP lentivirus construct contained Cre mRNA. (G) Western blot comparing level of Cre protein in MSCs and exosomes. No Cre was present in exosomes, indicating that the color switch was induced by transfer of functional RNA.
To prove that these isolated exosomes contained miR-124a, the exosomes were lysed, and RNA was collected and analyzed by qRT-PCR. The level of miR-124a in the exosomes derived from MSCs-miR124a was nearly 60-fold greater than in exosomes derived from MSCs-miRControl or MSCs-empty, which contained essentially no miR-124a (P = 0.0001) (Fig. 3D). Therefore, forced expression of an antiglioma miR in MSCs by lentiviral transduction results in high expression of that miR in MSC-derived exosomes.
Exosomes Effectively Deliver RNA to GSCs
Having established the ability of MSCs to package miRs into exosomes, we next sought to study the ability of exosomes to deliver genetic material to GSCs. To this end, we utilized the Cre-loxP reporter system. MSCs were plated in exosome-free medium and transduced with lentivirus (MOI 10) containing the cDNA of Cre recombinase (called MSCs-Cre). After 48 hour, exosomes from these MSCs-Cre (Exo-Cre) were isolated from the medium by differential centrifugation and treated with RNase and proteinase K to remove any free-floating RNA and protein. The supernatant obtained after ultracentrifugation (ie, devoid of exosomes) was collected and used as a control. As another control, Exo-empty was generated. Next, GSC267 was stably transduced with a lentivirus containing CMV-LoxP-DsRed-LoxP-eGFP (called GSC-dsRed/GFP), which express red fluorescence at baseline but express green fluorescence when exposed to Cre (see Supplementary Methods). These reporter GSCs were plated and treated with Exo-Cre (106 particles/cell), Exo-empty, or the residual supernatant devoid of exosomes. After 10 days, treatment with Exo-Cre converted GSC267-dsRed/GFP red fluorescent cells to green fluorescent cells, whereas treatment with Exo-empty did not. Importantly, treating GSC267-dsRed/GFP with the residual supernatant collected after exosome isolation also did not result in a color switch, indicating that GSCs do not uptake soluble Cre or free Cre mRNA (Fig. 3E). To prove that these effects were due to the transfer of functional RNA molecules, rather than the Cre protein itself, RNA was isolated from exosomes and RT-PCR was performed using Cre-specific primers. The product was subjected to gel electrophoresis, which demonstrated that Exo-Cre contained high levels of Cre mRNA, whereas controls did not (Fig. 3F). Furthermore, western blotting of protein lysates documented that these Exo-Cre did not contain Cre protein (Fig. 3G). Taken together, these data indicate that exosomes are capable of transferring functional RNA molecules to tumor cells.
MSC-Derived Exosomes Containing miR-124a Are Efficacious Against GSCs In Vitro
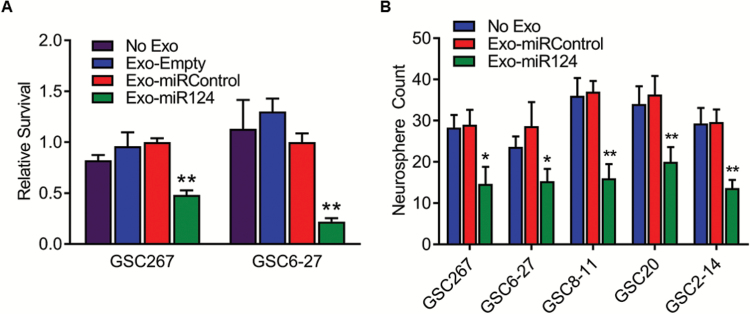
To demonstrate that the miR-124a–carrying MSC-derived exosomes are therapeutically effective, we cultured MSCs-miR124, MSCs-miRControl, or MSCs-empty and isolated the exosomes from the culture medium, as described above (called Exo-miR124, Exo-miRControl, and Exo-empty, respectively). GSC267 or GSC6-27 were cultured (2.5 × 104 cells/well) and treated with Exo-miR124, Exo-miRControl, or Exo-empty (104 particles/cell) daily for 3 days, and cell viability was analyzed after one week. Treatment of GSCs with Exo-miR124 resulted in a significant (>50%) reduction in proliferation compared with treatment with Exo-miRControl (P = 0.0005 for GSC267 and P = 0.001 for GSC6-27) (Fig. 4A).
Fig. 4.
Effects of Exo-miR124a treatment. (A) Viability (WST assay) of GSCs after treatment with Exo-miR124a. **P < 0.01. (B) Clonogenic neurosphere assay of GSCs after treatment with Exo-miR124a. *P < 0.05, **P < 0.01.
To verify this result, a clonogenic stem cell assay was performed. GSCs (N = 5) were treated with Exo-miRControl, Exo-miR124, or medium alone (no exosomes) for 3 days. Spheroids were dissociated and single cells were placed into each well of a 96-well plate. After one week, clonogenicity was analyzed by counting the number of wells containing a viable spheroid. For each of the GSCs, treatment with Exo-miR124 resulted in a statistically significant reduction in clonogenicity compared with Exo-miRControl or Exo-empty (P < 0.025 vs Exo-miRControl in all GSCs) (Fig. 4B).
Systemic Delivery of miR-124a in Exosomes Increases Mouse Survival In Vivo
We next sought to determine the therapeutic efficacy of Exo-miR124 in vivo. We first explored the antitumorigenic effects of Exo-miR124 on ex vivo treated GSCs that were then implanted into mice. Specifically, GSC267 was treated with Exo-miR124 (104/cell), Exo-miRControl (104/cell), or phosphate buffered saline (PBS) and after 72 hour implanted into the brains of nude mice (5 × 105 cells/animal, N = 10 animals/group).24 All animals treated with Exo-miRControl or Exo-empty died within 48 days, with median survival of 41 days. However, treatment with Exo-miR124 significantly increased animal survival, with a median of 104 days (P < 0.0001; log-rank test) (Fig. 5A).
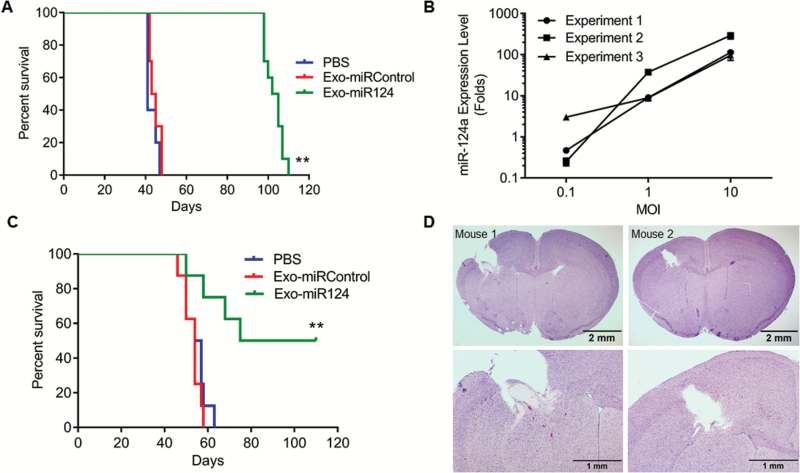
Fig. 5.
Effects of Exo-miR124a on GSCs in vivo. (A) Kaplan–Meier curve showing survival of mice implanted with GSCs that had been treated ex vivo with Exo-miR124a. **P < 0.01. (B) Quantitative RT-PCR analyzing expression level of miR-124a in exosomes derived from MSCs transduced with different MOI (0.1 to 10) of lenti-miR124a-GFP. (C) Kaplan–Meier curve of mice harboring GSC267 intracranial xenografts treated systemically with Exo-miR124a. **P < 0.01. (D) Representative histology from 2 of 4 GSC267-xenografted mice that survived long-term after systemic treatment with Exo-miR124a. Mice were sacrificed after 110 days. Upper figures are low power views of whole brain and bottom figures are high power views around site where tumor was implanted. Bars upper figures = 2 mm; bars lower figures = 1 mm.
Before testing the efficacy of systemic administration of Exo-miR124 in established intracranial GSCs, we sought to maximize the production of Exo-miR124 so that we could reproducibly generate Exo-miR124 needed for 3 day per week dosing in in vivo animal treatments. We plated MSCs-miR124 (5 × 105 cells) in 15-cm2 dishes and collected the exosomes after 48 hour. Using Nanosight, we measured the total number of Exo-miR124 isolated in 3 independent experiments, each performed in triplicate, and calculated that each MSC released, on average, 897 ± 60 exosomes in 3 independent experiments (Supplementary Figure S4). In addition, we transduced MSCs with lenti-miR124a at increasing MOI (0.1, 1.0, 10), isolated 106 exosomes at 48 hour, and determined the amount of miR-124a in the exosomes using qRT-PCR in 3 independent experiments, each performed in triplicate. There was a direct relationship between the MOI of miR lentivirus used to transduce the MSCs and the amount of miRs in the exosomes (Fig. 5B), with MOI 10 resulting in the highest (>95-fold) increase in miR-124a in Exo-miR124 compared with Exo-empty. Based on these data, we transduced MSCs at MOI 10 for our subsequent in vivo experiments in order to maximize the miRs in each exosome. Because miR levels were fairly reproducible with MOI 10 (Fig. 5B), we dosed animals based on exosome concentration (1010 Exos/100 μL) as determined by Nanosight.
We next assessed the therapeutic effect of in vivo systemic administration of Exo-miR124. GSC267 (5 × 105 cells/animal) was implanted into the frontal lobe of nude mice (N = 8/group) and after 7 days animals were treated with Exo-miR124, Exo-miRControl, or PBS given by intraperitoneal injection (1010 Exos/100 μL) every other day. Exosomes were also injected intra-arterially on days 14 and 21. Whereas all controls were dead by 60 days after tumor implantation (median survival of Exo-miRControl = 54 days; PBS = 55 days), 50% of the animals treated with Exo-miR124 were alive at 110 days (median survival = 79 days, P = 0.009) (Fig. 5C). Histological analyses of the 4 surviving mice showed complete regression of tumors, suggesting that the mice were cured of the cancer (Fig. 5D). These data demonstrate that systemically delivered exosomes carrying an antiglioma miR are efficacious against GSCs in vivo. Interestingly, the repeated systemic exosome treatment proved more potent than the single ex vivo treatment, where all mice ultimately died.
Exo-miR124 Inhibits FOXA2 and Alters GSC Lipid Metabolism
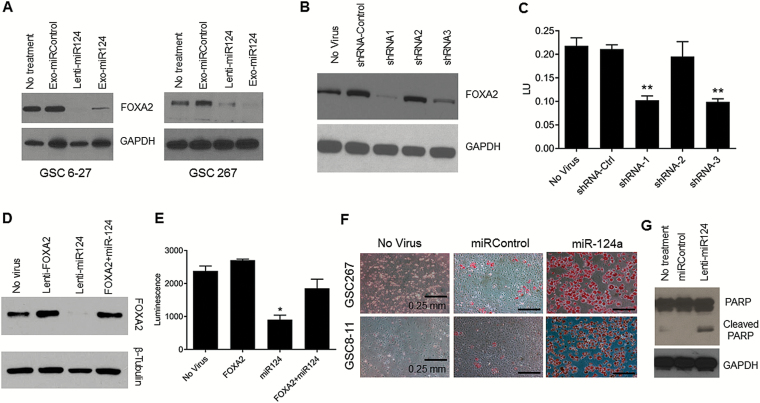
In order to understand the mechanism of action of miR-124a on GSCs and to further show that the miR-124a delivered into GSCs by MSC-derived exosomes is functional, we analyzed a variety of known targets of miR-124a. Specifically, GSCs were treated with Exo-miR124a, Exo-miRControl, or medium alone. GSCs were also treated with lenti-miR124a-GFP to test the effect of direct overexpression of miR-124a on gene expression. Three days after treatment, protein lysates were collected and analyzed by western blotting for the expression of sex determining region Y-box 2 (SOX2), SOX9, signal transducer and activator of transcription 3, polypyrimidine tract binding protein 1, repressor element-1 silencing transcription factor, and FOXA2, all known targets of miR-124a.27 Treatment with lenti-miR124-GFP or Exo-miR124 significantly inhibited the expression of only FOXA2 (Fig. 6A and Supplementary Figure S5).
Fig. 6.
Mechanism of action of miR-124a. (A) Western blot showing FOXA2 expression level in GSCs after indicated treatment. GAPDH used as loading control. (B and C) Western blot showing FOXA2 expression (B) and graph showing GSC267 viability (C) after treatment with anti-FOXA2 shRNAs. FOXA2 knockdown correlated with GSC viability reduction. β-Tubulin used as loading control. **P < 0.01. (D and E) Western blot showing FOXA2 expression (D) and graph showing GSC267 viability (E) after treatment with indicated condition. FOXA2 expression and GSC proliferation were rescued when treating with both lentiviruses. GAPDH used as loading control. (F) Representative photomicrographs of GSCs treated with lenti-miR124a-GFP and stained with Oil Red O solution for lipids. Lipid accumulation (red staining) can be seen in GSCs treated with lenti-miR124a-GFP. Bars = 0.25 mm. (G) Western blot showing induction of apoptotic cell death after treatment of GSC267 with lenti-miR124a-GFP.
Given this result, we next sought to quantitatively determine the percentage knockdown of FOXA2 after in vivo treatment of Exo-miR124. Therefore, we again implanted GSC267 into nude mice. We then injected Exo-miR124 or Exo-miRControl (1010 Exos/5 μL) intratumorally and after 48 hour sacrificed the mice, collected the brain, and isolated the tumor cells by punch biopsy. These cells were lysed and analyzed by qRT-PCR using FOXA2-specific primers and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) as an internal control. Treatment with Exo-miR124 resulted in a 32.3% knockdown of FOXA2 compared with no treatment (P = 0.0008), providing further evidence that FOXA2 is a target of miR-124a in GBM (Supplementary Figure S6).
We next directly linked FOXA2 knockdown to reduced GSC viability. We constructed 3 lentiviruses containing anti-FOXA2 shRNA (shRNA1/2/3-FOXA2) and transduced GSC267. Based on western blotting 3 days after treatment, shRNA1-FOXA2 and shRNA3-FOXA2 resulted in a significant decrease in FOXA2 expression, whereas shRNA2-FOXA2 had minimal effect (Fig. 6B). Consistent with this finding, based on a WST1 assay one week after treatment, shRNA1-FOXA2 and shRNA3-FOXA2 significantly inhibited the viability of GSC267 (P < 0.0001 vs shRNA-Control), whereas shRNA2-FOXA2 had minimal effect compared with shRNA-Control (Fig. 6C). These results indicate a causal and dose-dependent effect of inhibiting FOXA2 on GSC viability.
To further demonstrate a causal relationship between miR-124a expression, FOXA2 inhibition, and GSC proliferation, we performed rescue experiments using a lentivirus vector for a miR-124a insensitive FOXA2 (lenti-FOXA2). This lentiviral construct generates a FOXA2 that lacks the miR-124a binding site, making it resistant to inhibition by miR-124a and permitting phenotypic rescue. Next, GSC267 was treated with medium alone (no lentivirus), lenti-FOXA2 alone, lenti-miR124a-GFP alone, or both lenti-FOXA2 and lenti-miR124a-GFP. One week after culturing, GSC viability was analyzed by WST1 assay, and FOXA2 expression was determined by western blotting. As expected, treatment with lenti-miR124-GFP alone resulted in a significant reduction in the expression of FOXA2 (Fig. 6D) and a concomitant inhibition of GSC viability compared with control (P = 0.028) (Fig. 6E). Also as expected, treatment with lenti-FOXA2 alone resulted in a slight increase in FOXA2 expression and a slight, but not statistically significant, increase in GSC viability compared with control (Fig. 6D and 6E). However, co-treatment with miR-124a and lenti-FOXA2 showed that lenti-FOXA2 was capable of restoring the expression of FOXA2 after treatment with miR-124a as well as rescuing the survival of GSCs (Fig. 6D and 6E). This result shows that the mechanism by which miR-124a inhibits GSC viability is, at least in part, through knockdown of FOXA2.
Although it is known that FOXA2 is important for GBM pathogenesis,28 its mechanism of action remains unclear. FOXA2 plays an important role during animal development and in the regulation of cellular metabolism, especially in lipid metabolism.29 The recognition that metabolic reprogramming is a key feature of transformed cells has led to significant interest in targeting metabolism as a cancer therapy. Thus, we hypothesized that miR-124a knockdown of FOXA2 causes cell death by perturbing lipid metabolism in GSCs. To begin to demonstrate a link between treatment with miR-124a and abnormal lipid metabolism, we treated GSCs with medium alone, lenti-miRControl-GFP, and lenti-miR124a-GFP and analyzed lipid production by Oil Red O staining. GSCs treated with miR124a-GFP demonstrated dramatic increases in lipid accumulation in the GSCs compared with controls (Fig. 6F). These data indicate an association between miR-124a treatment and perturbation of lipid metabolism, through downregulation of FOXA2 (Fig. 6F) and ultimately cell death by apoptosis as evidenced by poly(ADP-ribose) polymerase cleavage (Fig. 6G).
Discussion
Our results show that overexpression of miR-124a inhibits the growth of a diverse group of GSCs, identifying miR-124a as an effective antiglioma agent, which is consistent with previously reported results.21,30 We also show that MSCs induced to overexpress miR-124a effectively inhibit the growth of GSCs in Transwell assays, confirming MSCs as potential delivery vehicles of miRs. Most importantly, we demonstrate that MSCs can be engineered to function as natural biofactories for the production of exosomes containing supraphysiological levels of miR-124a. These Exo-miR124 exosomes can inhibit the growth and clonogenicity of GSCs and, when administered systemically, are capable of curing mice harboring intracranial GSC xenografts. Mechanistic studies showed that miR-124a acts by downregulating FOXA2, a known target of miR-124a, and that miR-124a induced apoptotic cell death correlates with FOXA2-mediated aberrant intracellular lipid accumulation.
Although miR-124a inhibited GSC growth better than the other miRs that were tested, there was variability in the ability of miR-124a to inhibit the growth of the individual GSC lines, with growth reductions varying from 39% to 59% among the GSCs. Furthermore, other miRs were effective against several of the GSCs—for instance, miR-122 was particularly effective against GSC8-11, and miR-138 and Let-7 had inhibitory effects on several lines. Hence, the inhibitory effect of miRs should be studied with greater granularity, using many GSC lines, in a large-scale screen. Although this investigation analyzed the effects of delivering only a single miR, this result also suggests that delivering multiple antiglioma miRs simultaneously may result in greater inhibition of GSCs through the downregulation of multiple growth-driving genes. Nevertheless, our studies support the notion that specific miRs, particularly miR-124a, are potentially powerful therapies against a variety of GBMs.
While the use of MSCs as delivery vehicles has been well studied,12,13,21,31 the use of miR-carrying exosomes in GBM therapy is relatively novel and unexplored. In this context, we showed for the first time that treating GSCs with miR-124a–carrying exosomes inhibits GSC survival in both in vitro assays and in vivo gold-standard animal models. Together these results support the notion that MSCs can be used as ex vivo biofactories for the packaging, production, and therapeutic use of exosomes carrying antiglioma miRs. The advantage of delivering exosomes rather than MSCs is that the larger MSCs are trapped in the lungs, precluding intravenous delivery strategies and necessitating intra-arterial (ie, intracarotid) administration. Intravenous administration is more easily achieved and can be conducted frequently, thereby allowing for a wider range of dose schedules. Overall, our result provides strong evidence for the translational feasibility and efficacy of this novel approach, which combines an effective therapy, miR-124, and an effective delivery mechanism, exosomes.
Our results also suggest a possible new mechanism for the effects of miR-124a on GSCs. Recent reports suggest that a variety of cancers, including GBM, are dependent on capturing and metabolizing exogenous lipids for their growth, demonstrating a metabolic adaptation to facilitate tumor growth and survival.32,33 Thus, FOXA2’s regulation of lipid metabolism represents a potential vulnerability in GBMs.29,34,35 In this context, we showed that miR-124a downregulates FOXA2, an oncogenic transcription factor, and that FOXA2 downregulation results in reduced GSC viability. We showed that overexpression of miR-124a results in intracellular accumulation of lipids, indicating that miR-124a renders GSCs unable to efficiently metabolize lipids leading to toxic levels, and supports the notion that downregulation of FOXA2 by miR-124a reduces viability of GSCs due to an induced inability to utilize lipids. Further investigation is required to completely elucidate the role of FOXA2 in glioma lipid metabolism and its link to tumor growth.
Supplementary Material
Supplementary material is available at Neuro-Oncology online.
Acknowledgment
F.M.L was a National Finalist in the 2014 Siemens Foundation Science and Technology Competition.
Funding
This work was supported by NCI P50 CA127001, the Broach Foundation, the Elias Family Fund, the Gene Pennebaker Fund, the Sorenson Foundation, the Gold Fund, and the Brian McCulloch Fund.
Conflict of interest statement. All authors declare no conflict of interest pertaining to this work.
Supplementary Material
References
- 1. Singh SK, Hawkins C, Clarke ID et al. Identification of human brain tumour initiating cells. Nature. 2004;432(7015):396–401. [DOI] [PubMed] [Google Scholar]
- 2. DeAngelis LM. Brain tumors. N Engl J Med. 2001;344(2):114–123. [DOI] [PubMed] [Google Scholar]
- 3. Ambros V. The functions of animal microRNAs. Nature. 2004;431(7006):350–355. [DOI] [PubMed] [Google Scholar]
- 4. Huntzinger E, Izaurralde E. Gene silencing by microRNAs: contributions of translational repression and mRNA decay. Nat Rev Genet. 2011;12(2):99–110. [DOI] [PubMed] [Google Scholar]
- 5. Silber J, Lim DA, Petritsch C et al. miR-124 and miR-137 inhibit proliferation of glioblastoma multiforme cells and induce differentiation of brain tumor stem cells. BMC Med. 2008;6:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bora RS, Gupta D, Mukkur TK, Saini KS. RNA interference therapeutics for cancer: challenges and opportunities (review). Mol Med Rep. 2012;6(1):9–15. [DOI] [PubMed] [Google Scholar]
- 7. Li Z, Rana TM. Therapeutic targeting of microRNAs: current status and future challenges. Nat Rev Drug Discov. 2014;13(8):622–638. [DOI] [PubMed] [Google Scholar]
- 8. van der Meel R, Fens MH, Vader P, van Solinge WW, Eniola-Adefeso O, Schiffelers RM. Extracellular vesicles as drug delivery systems: lessons from the liposome field. J Control Release. 2014;195:72–85. [DOI] [PubMed] [Google Scholar]
- 9. Skog J, Würdinger T, van Rijn S et al. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol. 2008;10(12):1470–1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Johnsen KB, Gudbergsson JM, Skov MN, Pilgaard L, Moos T, Duroux M. A comprehensive overview of exosomes as drug delivery vehicles—endogenous nanocarriers for targeted cancer therapy. Biochim Biophys Acta. 2014;1846(1):75–87. [DOI] [PubMed] [Google Scholar]
- 11. Kamerkar S, LeBleu VS, Sugimoto H et al. Exosomes facilitate therapeutic targeting of oncogenic KRAS in pancreatic cancer. Nature. 2017;546(7659):498–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Nakamizo A, Marini F, Amano T et al. Human bone marrow-derived mesenchymal stem cells in the treatment of gliomas. Cancer Res. 2005;65(8):3307–3318. [DOI] [PubMed] [Google Scholar]
- 13. Yong RL, Shinojima N, Fueyo J et al. Human bone marrow-derived mesenchymal stem cells for intravascular delivery of oncolytic adenovirus Delta24-RGD to human gliomas. Cancer Res. 2009;69(23):8932–8940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hata N, Shinojima N, Gumin J et al. Platelet-derived growth factor BB mediates the tropism of human mesenchymal stem cells for malignant gliomas. Neurosurgery. 2010;66(1):144–156; discussion 156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Doucette T, Rao G, Yang Y et al. Mesenchymal stem cells display tumor-specific tropism in an RCAS/Ntv-a glioma model. Neoplasia. 2011;13(8):716–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Shinojima N, Hossain A, Takezaki T et al. TGF-β mediates homing of bone marrow-derived human mesenchymal stem cells to glioma stem cells. Cancer Res. 2013;73(7):2333–2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gatti S, Bruno S, Deregibus MC et al. Microvesicles derived from human adult mesenchymal stem cells protect against ischaemia-reperfusion-induced acute and chronic kidney injury. Nephrol Dial Transplant. 2011;26(5):1474–1483. [DOI] [PubMed] [Google Scholar]
- 18. Xin H, Li Y, Cui Y, Yang JJ, Zhang ZG, Chopp M. Systemic administration of exosomes released from mesenchymal stromal cells promote functional recovery and neurovascular plasticity after stroke in rats. J Cereb Blood Flow Metab. 2013;33(11):1711–1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Xin H, Li Y, Buller B et al. Exosome-mediated transfer of miR-133b from multipotent mesenchymal stromal cells to neural cells contributes to neurite outgrowth. Stem Cells. 2012;30(7):1556–1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Katakowski M, Buller B, Zheng X et al. Exosomes from marrow stromal cells expressing miR-146b inhibit glioma growth. Cancer Lett. 2013;335(1):201–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lee HK, Finniss S, Cazacu S et al. Mesenchymal stem cells deliver synthetic microRNA mimics to glioma cells and glioma stem cells and inhibit their cell migration and self-renewal. Oncotarget. 2013;4(2):346–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hossain A, Gumin J, Gao F et al. Mesenchymal stem cells isolated from human gliomas increase proliferation and maintain stemness of glioma stem cells through the IL-6/gp130/STAT3 pathway. Stem Cells. 2015;33(8):2400–2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Théry C, Amigorena S, Raposo G, Clayton A. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr Protoc Cell Biol. 2006;Chapter 3:Unit 3.22. [DOI] [PubMed] [Google Scholar]
- 24. Lal S, Lacroix M, Tofilon P, Fuller GN, Sawaya R, Lang FF. An implantable guide-screw system for brain tumor studies in small animals. J Neurosurg. 2000;92(2):326–333. [DOI] [PubMed] [Google Scholar]
- 25. Bhat KPL, Balasubramaniyan V, Vaillant B et al. Mesenchymal differentiation mediated by NF-κB promotes radiation resistance in glioblastoma. Cancer Cell. 2013;24(3):331–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Jiang H, Gomez-Manzano C, Aoki H et al. Examination of the therapeutic potential of Delta-24-RGD in brain tumor stem cells: role of autophagic cell death. J Natl Cancer Inst. 2007;99(18):1410–1414. [DOI] [PubMed] [Google Scholar]
- 27. Sun Y, Luo ZM, Guo XM, Su DF, Liu X. An updated role of microRNA-124 in central nervous system disorders: a review. Front Cell Neurosci. 2015;9:193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Qi J, Nakayama K, Cardiff RD et al. Siah2-dependent concerted activity of HIF and FoxA2 regulates formation of neuroendocrine phenotype and neuroendocrine prostate tumors. Cancer Cell. 2010;18(1):23–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kurtz CL, Peck BC, Fannin EE et al. MicroRNA-29 fine-tunes the expression of key FOXA2-activated lipid metabolism genes and is dysregulated in animal models of insulin resistance and diabetes. Diabetes. 2014;63(9):3141–3148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wei J, Wang F, Kong LY et al. miR-124 inhibits STAT3 signaling to enhance T cell-mediated immune clearance of glioma. Cancer Res. 2013;73(13):3913–3926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Parker Kerrigan BC, Shimizu Y, Andreeff M, Lang FF. Mesenchymal stromal cells for the delivery of oncolytic viruses in gliomas. Cytotherapy. 2017;19(4):445–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bensaad K, Favaro E, Lewis CA et al. Fatty acid uptake and lipid storage induced by HIF-1α contribute to cell growth and survival after hypoxia-reoxygenation. Cell Rep. 2014;9(1):349–365. [DOI] [PubMed] [Google Scholar]
- 33. Mashimo T, Pichumani K, Vemireddy V et al. Acetate is a bioenergetic substrate for human glioblastoma and brain metastases. Cell. 2014;159(7):1603–1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. von Meyenn F, Porstmann T, Gasser E et al. Glucagon-induced acetylation of Foxa2 regulates hepatic lipid metabolism. Cell Metab. 2013;17(3):436–447. [DOI] [PubMed] [Google Scholar]
- 35. Wan M, Leavens KF, Saleh D et al. Postprandial hepatic lipid metabolism requires signaling through Akt2 independent of the transcription factors FoxA2, FoxO1, and SREBP1c. Cell Metab. 2011;14(4):516–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.