Genetic differences between regions are usually studied for individual species. However, many species can reproduce with each other. We studied whether gene flow between two closely related sedge species influences regional differences. Our molecular genetic data support considerable gene flow between the species. Still, we detected clear genetic differences between species and regions, and more pronounced regional differences for the less common one. Thus, gene flow between the species appeared too weak to neutralize differences between the regional genetic structure of our study species. We encourage further regional differentiation studies in groups of cross-compatible species.
Keywords: Carex flava complex, genetic diversity, hybridization, microsatellites, population differentiation
Abstract
Regional genetic differentiation within species is often addressed in evolutionary ecology and conservation biology. Here, we address regional differentiation in two closely related hybridizing taxa, the perennial sedges Carex flava and C. viridula and their hybrid C. × subviridula in 37 populations in the north and centre of their distribution range in Europe (Estonia, Lowland (<1000 m a.s.l.) and Highland Switzerland) using 10 putative microsatellite loci. We ask whether regional differentiation was larger in the less common taxon C. viridula or whether, possibly due to hybridization, it was similar between taxa. Our results showed similar, low to moderate genetic diversity for the three studied taxa. In total, we found 12 regional species-specific alleles. Analysis of molecular variance (AMOVA), STRUCTURE and multidimensional scaling analysis showed regional structure in genetic variation, where intraspecific differentiation between regions was lower for C. flava (AMOVA: 6.84 %) than for C. viridula (20.77 %) or C. × subviridula (18.27 %) populations. Hybrids differed from the parental taxa in the two regions where they occurred, i.e. in Estonia and Lowland Switzerland. We conclude that C. flava and C. viridula clearly differ from each other genetically, that there is pronounced regional differentiation and that, despite hybridization, this regional differentiation is more pronounced in the less common taxon, C. viridula. We encourage future studies on hybridizing taxa to work with plant populations from more than one region.
Introduction
Plants and other organisms differ in their levels of genetic diversity and genetic differentiation (Linhart and Grant 1996). The extent of genetic differentiation among populations and regions depends on the balance of evolutionary forces decreasing and increasing genetic differentiation, that is, gene flow, genetic drift, mutation and selection (Slatkin 1987). The relative importance of these forces may be affected by selection strength, as well as population size, environmental barriers to dispersal and plant life history traits, especially mating system and dispersal mechanism (Loveless and Hamrick 1984). Higher differentiation among populations is generally found for clonally reproducing and selfing species (Loveless and Hamrick 1984; Hamrick and Godt 1996; Nybom 2004; Song et al. 2006) and for species with disjunct distributions or small populations (Ellstrand and Elam 1993), due to effects of genetic drift or reduced gene flow (Slatkin 1987; Schönswetter et al. 2006). Outcrossing and sexually reproducing species, conversely, show less differentiation among populations (Hamrick et al. 1979; Loveless and Hamrick 1984).
While genetic consequences of small population size, i.e. increased inbreeding and genetic drift, are expected to contribute to higher population differentiation and lower genetic diversity within populations (Slatkin 1987; Ellstrand and Elam 1993; Amos and Harwood 1998; Premoli 2003; Leimu et al. 2006; Zhivotovsky et al. 2016), this is not found in all studies (Gitzendanner and Soltis 2000). Loveless and Hamrick (1984) agree that small populations are more susceptible to drift and fixation, but suggest that immigration is more effective in altering gene frequencies in small populations. Thus, gene flow can prevent differentiation and loss of genetic variability, especially in long-lived species. Levels of genetic diversity in populations may also depend on distance from glacial refugia (Schönswetter et al. 2006) and on the landscape (Holderegger and Wagner 2006). For populations at higher altitudes higher radiation intensity may increase the rate of mutations and thus population genetic diversity (Li et al. 1997).
It is generally assumed that species are reproductively isolated without gene flow between them, but in reality hybridization is a widespread phenomenon (Ellstrand et al. 1996; Abbott et al. 2013) and becomes more frequent with continuous climate change and human influence on the environment (Hoffmann and Sgro 2011). Gene flow between taxa can have a profound effect on genetic diversity (Derieg et al. 2008), and due to subsequent genetic drift, may lead to population differentiation (Bain and Golden 2003). Hybridization of closely related species is found to lead to genetic differentiation among regions (Rieseberg et al. 1999; Kane et al. 2009; Lepais et al. 2009; Krebs et al. 2010; Brennan et al. 2016). According to the ‘semipermeable species boundaries’ theory, alleles at some loci can be exchanged between species and species boundaries can vary geographically (Harrison and Larson 2014). Thus, hybridization coupled with backcrossing can be expected to affect population differentiation on a regional scale.
Hybrid individuals are expected to be more heterozygous than their parental taxa due to genetic admixing (Rieseberg and Wendel 1993; Allendorf et al. 2001; Barton 2001; Harrison and Larson 2014; Todesco et al. 2016). Hybridization could result in beneficial evolution by providing additional adaptive genetic variation (Lewontin and Birch 1966; Arnold 2006; Abbott et al. 2013). Conversely, hybridization can reduce biodiversity by causing loss of alleles and genetic diversity by genetic assimilation (Levin et al. 1996). Genetic studies on hybridizing taxa have focused on their genetic variation or the extent of hybridization and introgression (e.g. Friedman et al. 2008; Volkova et al. 2008; Kane et al. 2009; Korpelainen et al. 2010; Krebs et al. 2010). Our study addresses the differentiation of hybrids from parental taxa.
We investigate genetic diversity and regional differentiation between and within Carex flava, C. viridula and their hybrid C. × subviridula for populations from three regions (Estonia in Northern Europe and Lowland and Highland Switzerland in Central Europe). Carex flava and C. viridula var. viridula sensu stricto (s.s.) (henceforth C. viridula) are wind-pollinated, self-compatible, caespitose perennials of the C. flava aggregate (Carex sect. Ceratocystis, Cyperaceae). Although there are no impediments to outcrossing, a large amount of seeds is produced by selfing (Vonk 1979; Schmid 1984a). Both taxa, with circumpolar distribution, occur in the temperate and subarctic Northern hemisphere and also in North Africa (Hultén and Fries 1986; Crins and Ball 1989; Koopman 2011). They often co-occur and hybridize, especially at sites with disturbances (Vonk 1979; Jiménez-Mejías et al. 2012), resulting in C. × subviridula. Carex viridula is considered to be a dispersal generalist, potentially being transported by biotic, e.g. birds, mammals, invertebrates, and abiotic agents, e.g. water and wind (Schmid 1984a; Crins and Ball 1989). Long-distance dispersal is proven experimentally with C. flava var. alpina seeds remaining still partly intact after passing the digestive tract of domesticated ducks within 18 h (Schmid 1984a). Carex viridula is a weak competitor, but able to colonize and survive in fluctuating, relatively unpredictable moist or wet habitats, where it forms small populations (Schmid 1986; Crins and Ball 1989; Kuchel and Bruederle 2000). However, populations of C. viridula are sensitive to anthropogenic influence, such as the drainage of mires, regulation of water levels and eutrophication of shores (Pykälä and Toivonen 1994) and its decrease in southern parts of its distribution could be explained by climate change (Parmesan and Yohe 2003; Chen et al. 2011). Hence, the occurrence of C. viridula has decreased over time and populations have become more fragmented (Davies 1953; Pykälä and Toivonen 1994). According to the latest red list of endangered plants in Switzerland, it is considered near threatened due to loss of habitat (Bornand et al. 2016). Carex flava, on the other hand, is a strong competitor and not as sensitive to environmental changes; its populations are larger and rather constant in time (Schmid 1984a, b, 1986). Intraspecific gene flow is expected to be higher for C. flava and smaller for C. viridula (Schmid 1986).
Previous genetic studies with the C. flava agg. were restricted to small regions, did not consider co-occurrence and hybridization or used allozyme markers of limited variability (Bruederle and Jensen 1991; Kuchel and Bruederle 2000; Hedrén 2002, 2004; Blackstock and Ashton 2010; except Jiménez-Mejías et al. 2012). The novelty of our study originates from considering co-occurrence and hybridization, from comparing geographically and climatically distant regions, and from using contemporary molecular microsatellite (SSR) markers. According to our hypothesis, admixture, i.e. hybridization coupled with backcrossing, can be expected to affect population differentiation on a regional scale. Therefore, species characterized by relatively small populations might be less well differentiated on a regional level than expected without admixing. Thus, we study whether, despite admixing, C. viridula, characterized by relatively small populations, exhibits higher levels of population differentiation than C. flava, which has relatively large populations. In addition, we investigate the variability of hybrid populations and their differentiation from the parental taxa.
Methods
Study taxa and regions
In the studied regions, the C. flava group comprises the four taxa C. flava, C. lepidocarpa, C. demissa and C. viridula var. viridula (Schmid 1981; Toom et al. 2016). In Estonia, the two varieties C. viridula var. pulchella and var. bergrothii are also found (Toom et al. 2016). In this study, we follow Hedrén (2002, 2004) taxonomic treatment, but we use the more common name C. viridula instead of calling it C. oederi.
The studied regions differ in their postglacial history. Swiss populations were established earlier after glaciation, because the territory of Estonia was covered by ice at the end of the last ice age 18000 BP, when the lower parts of Switzerland were ice-free (Hewitt 1999). In addition, the current populations in Switzerland are much closer to possible southern glacial refugia in Iberia, the Apennine or the Balkan Peninsula (Schönswetter et al. 2006) than Estonian populations are. As the populations in Switzerland are older, they may be more amalgamated via interspecific gene flow, hybridization and introgression than the Estonian ones.
Population sampling
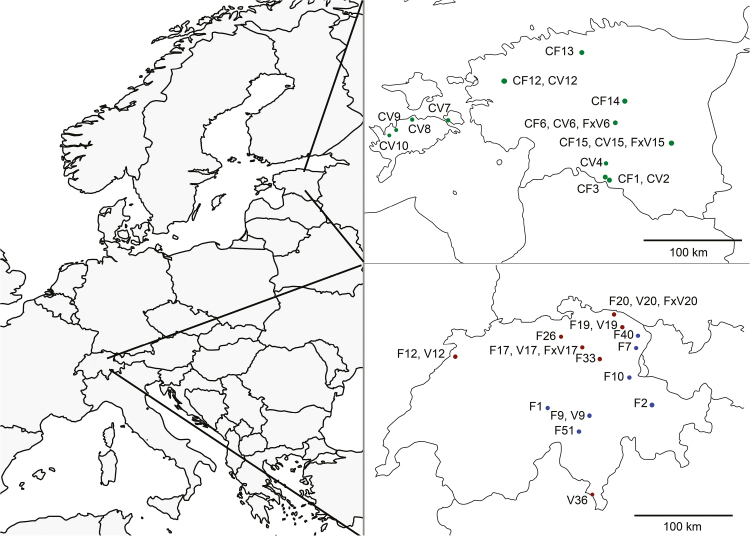
We collected 380 samples of C. flava, C. viridula and C. × subviridula populations from seven sites in Highland Switzerland (>1000 m, in 2012), five sites in Lowland Switzerland (<1000 m, in 2012) and 12 sites in Estonia (2013; Table 1; Fig. 1). At 15 sites we found C. flava and C. viridula growing together or with other sedges, with whom they are able to hybridize, i.e. with other C. flava aggregate members or C. punctata and C. hostiana (Davies 1955; Schmid 1982; Crins and Ball 1989; Więcław and Wilhelm 2014). Samples of the C. flava × C. viridula hybrid (C. × subviridula) were found at two sites in Estonia and two in Lowland Switzerland, while none were found in Highland Switzerland. As hybrids, we classified partly sterile individuals that were morphologically intermediate between the parents, but often were more robust and pale (Schmidt et al. 2017). The sampling populations of C. viridula were smaller, i.e. had fewer individuals, than the ones of C. flava. As we focus on between-region and between-taxa comparisons, for which the population is the unit of replication against which between-region and between-taxa differences are tested, and as the power of such analyses depends on the number of populations, whereas the replication within populations is less decisive (van Kleunen et al. 2014), we sampled a total of 37 populations and on average 10.3 individuals per population.
Table 1.
Populations (Pop) and geographic locations of Carex flava (sect. Ceratocystis, Cyperaceae), Carex viridula var. viridula and their hybrid (C. × subviridula) from three regions: Estonia (EST), Highland (CHH) and Lowland Switzerland (CHL). n, number of analysed individuals; m, altitude in m a.s.l.; N, latitude; E, longitude. The sites where C. flava and C. viridula grew together with other members of the C. flava agg. are termed ‘mixed’, and the sites where only C. flava or only C. viridula occurred ‘pure’. Note: population name codes sharing the same number indicate populations co-occurrence at the same site.
| Species (n) | Pop | State | Location | m | N | E |
|---|---|---|---|---|---|---|
| Highland Switzerland (CHH) | ||||||
| C. flava (12) | F2 | Mixed | Arosa, Peist | 1920 | 46.8013 | 9.6841 |
| C. flava (6) | F1 | Pure | Melchsee-Frutt, Kerns | 1910 | 46.7704 | 8.2809 |
| C. flava (18) | F7 | Mixed | Rüte | 1158 | 47.3221 | 9.4695 |
| C. flava (6) | F9 | |||||
| C. viridula (8) | V9 | Mixed | Fontanivas, Disentis | 1046 | 46.6989 | 8.8565 |
| C. flava (14) | F10 | Mixed | Chapfensee, Mels | 1030 | 47.0483 | 9.3770 |
| C. flava (7) | F12 | |||||
| C. viridula (4) | V12 | Mixed | Etang de Gruere, Saignelegier | 1007 | 47.2381 | 7.0508 |
| C. flava (8) | F40 | Mixed | Gupfloch, Rehetobel | 1015 | 47.4346 | 9.4988 |
| Lowland Switzerland (CHL) | ||||||
| C. flava (14) | F17 | |||||
| C. viridula (11) | V17 | |||||
| Hybrid (3) | FxV17 | Mixed | Robenhuserriet, Wetzikon | 535 | 47.3399 | 8.7811 |
| C. flava (7) | F19 | |||||
| C. viridula (14) | V19 | Mixed | Hudelmoos, Amriswil | 525 | 47.5238 | 9.2869 |
| C. flava (32) | F20 | |||||
| C. viridula (11) | V20 | |||||
| Hybrid (12) | FxV20 | Mixed | Neuweiher, Kreuzlingen | 500 | 47.6311 | 9.1743 |
| C. flava (6) | F33 | Pure | Kaltbrunner Riet, Kaltbrunn | 410 | 47.2152 | 8.9894 |
| C. viridula (20) | V36 | Pure | Luganersee, Caslano | 270 | 45.9613 | 8.8872 |
| Estonia (EST) | ||||||
| C. viridula (9) | CV4 | Pure | Tarvastu, Veisjärv | 97 | 58.1060 | 25.7639 |
| C. flava (9) | CF3 | Pure | Helme, Holdre | 93 | 57.9639 | 25.7434 |
| C. flava (4) | CF1 | |||||
| C. viridula (4) | CV2 | Mixed | Helme, Lagesoo | 87 | 57.9494 | 25.8066 |
| C. flava (9) | CF14 | Pure | Pajusi, Endla | 86 | 58.7603 | 26.1310 |
| C. flava (8) | CF13 | Pure | Anija, Padriku | 64 | 59.2959 | 25.3726 |
| C. flava (14) | CF12 | |||||
| C. viridula (11) | CV12 | Mixed | Risti, Marimetsa bog | 45 | 58.9932 | 24.0743 |
| C. flava (9) | CF6 | |||||
| C. viridula (10) | CV6 | |||||
| Hybrid (10) | FxCV6 | Mixed | Kolga-Jaani, Leie | 37 | 58.4139 | 26.0396 |
| C. flava (13) | CF15 | |||||
| C. viridula (10) | CV15 | |||||
| Hybrid (7) | FxCV15 | Mixed | Luunja, Kabina | 33 | 58.3437 | 26.8252 |
| C. viridula (10) | CV8 | Pure | Leisi, Meiuste | 27 | 58.5860 | 22.5681 |
| C. viridula (10) | CV9 | Pure | Mustjala, Võhma | 19 | 58.5206 | 22.3334 |
| C. viridula (10) | CV10 | Pure | Mustjala, Paatsa | 6 | 58.5053 | 22.3126 |
| C. viridula (10) | CV7 | Pure | Muhu, Nautse | 3 | 58.5774 | 23.1658 |
Figure 1.
Sampling sites in Switzerland and Estonia (top right). For Switzerland blue dots indicate Highland Switzerland populations, dark red dots Lowland Switzerland populations. Sites, where more than one taxon was co-occurring, are marked with one dot. Population codes can be found in Table 1.
As population genetic diversity is expected to be high in the centre and to decline at the margins of distribution ranges (Volis et al. 2016), we studied the taxa neither in the centre nor the margins of theirs. Our southernmost population was at the latitude of 45.96 N in Caslano, Switzerland (V36), and the northernmost one at 59.29 N in Anija, Estonia (CF13). In Switzerland, the population at lowest altitude was in Caslano at 270 m a.s.l. and the one at highest altitude in Arosa at 1920 m a.s.l. Vouchers with samples of all study populations were deposited in the herbarium of the Natural History Museum of the University of Tartu.
Microsatellite analysis
The microsatellite loci tested in this study were developed for other species. Due to good success in cross-amplification among congeners (Rossetto et al. 1999), we chose primer pairs isolated from other Carex species. We chose in total 17 polymorphic microsatellite loci from previous studies that had showed successful cross-species amplification. We screened nine primer pairs developed for Carex scoparia (Hipp et al. 2009), two primers developed for Carex rugulosa (Ohbayashi et al. 2008), four primers developed for Carex kobomugi (Ohsako and Yamane 2007) and two primers developed for Carex limosa (Escudero et al. 2010). Primary microsatellite analysis was performed with few individuals of each taxon and 17 primer pairs. Of the 17 primer pairs tested, 10 aligned successfully with recipient DNA, cross-amplified in C. flava, C. viridula and C. × subviridula, exhibited polymorphism and showed identifiable peaks in fragment analysis. Those 10 primer pairs were used for further analysis with all 380 samples [see Supporting Information—Table S1]. We did not sequence the fragments recovered for the 10 loci nor did we perform progeny analysis. However, the primer pairs used in our study had been successfully used in other population genetic studies (Hipp et al. 2009; Escudero et al. 2010; Korpelainen et al. 2010).
Each primer was optimized for a range of temperatures (Ta: 49–60.1 °C). Magnesium source 1.2 mM MgSO4 was used, except for S177, where 1.6 mM MgCl2 was used. PCR amplifications were performed in 10 µL volumes containing 1–2 µL of genomic DNA, 1.2 µL GoTaq Flexi buffer (1×), 0.6 µL of each dNTP, 0.5 µL of untagged primer, 0.5 µL of fluorescent tag, 0.5 µL of the tagged primer, 0.05 µL of bovine serum albumin (BSA), 0.05 µL GoTaq Flexi DNA polymerase and varying concentrations of MgCl2 or MgSO4. Total genomic DNA was isolated from silica-dried leaves using the CTAB method (Doyle 1987). The extracted DNA was dissolved in 100 µL of TE buffer and diluted to 1:10 for further PCR analyses. DNA from each sample was amplified with the common tag containing one of four fluorescent dyes, 6-FAM, PET, VIC or NED (Applied Biosystems). PCRs were carried out as follows: preliminary denaturation at 95 °C for 5 min, 35 cycles at 95 °C for 1 min, annealing temperature 53.6–60 °C for 1 min, 72 °C for 1 min and a final extension step at 72 °C for 30 min, using a Techne TC-5000 thermocycler (Bibby Scientific). PCR products of different primers, each of 1–2 µL, were mixed together yielding a total of 20-µL mixture. From each mixture 2 µL were pooled with 10 µL buffer (size standard: deionized formamide = 1:25) in the wells of a 96-well plate for fragment analysis on a 3730xl DNA Analyzer (Applied Biosystems), where samples of different plant individuals were randomized. Product sizes were determined using the Peak Scanner Software v1.0 (Applied Biosystems). Scoring errors, e.g. null alleles, were identified and corrected using micro-checker 2.2.3 (van Oosterhout et al. 2004).
Data analysis
For each population the effective number of alleles (Ne), percentage of polymorphic loci (PL), expected heterozygosity (He, also called gene diversity) and observed heterozygosity (Ho) were estimated across all loci using GENALEX 6.5 (Peakall and Smouse 2006). Allelic richness (Ar) and compliance with Hardy–Weinberg expectations were calculated in FSTAT v 2.9.3 (Goudet 2002). The inbreeding coefficient (FIS) shows the probability to observe alleles of an individual (I) that are identical by descent (IBD) in a subpopulation (S). FIS, calculated as (He − Ho)/He, allowed to estimate the prevailing mating systems by region (i.e. Estonia, Lowland and Highland Switzerland) and taxon. Variation in these measures of population diversity was tested with ANOVA, using taxon, region of origin and the interaction of taxon and region of origin as fixed effects, implemented in the software R v 3.1.2 (R Development Core Team 2016).
Population genetic structure and hybrid identification.
We used a Bayesian clustering approach as implemented in STRUCTURE v 2.3.4 (Pritchard et al. 2000) (i) to estimate the number of genetic clusters (K) without a priori knowledge of taxonomy or population, and (ii) to identify the hybrid individuals with admixture analysis. The clustering was conducted with the admixture model and the correlated-allele-frequencies option using a burn-in of 10000 steps and 100000 replications, the remaining parameters were set to the default values. Five independent runs were done for the set of K = {1:10}. We used Structure Harvester v 0.6.92 (Earl 2012) to visualize the optimal number of clusters (K) by using firstly the ΔK method of Evanno et al. (2005) and secondly by examining the distribution of the log-likelihoods for the value with the highest probability and lowest variance using Markov chain Monte Carlo simulations. With K = 2 we detected the posterior probability (q-values), which describes the proportion of an individual genotype originating from each of K categories. We used K = 2 as we expected two taxa contributing to the gene pool of hybrids. We chose a threshold value of 0.9, which was found efficient to distinguish pure individuals (q > 0.9 or q < 0.1) from hybrids and backcrosses (0.1 < q < 0.9) (Vähä and Primmer 2006; Burgarella et al. 2009).
Genetic differentiation.
To compare the degree of differentiation among groups of populations categorized by taxa and region of origin, between-group FST values were calculated, using Arlequin v 3.5.2.2 (Excoffier et al. 2005). The significance of differences in the resulting values was tested with 1000 permutations. To illustrate the dissimilarities among groups of populations categorized by taxa and by region of origin, multidimensional scaling (MDS) analysis was performed in software R v 3.1.2 (R Development Core Team 2016) based on Reynolds distances obtained with the software Arlequin, which estimates the co-ancestry of different samples (Reynolds et al. 1983).
Analysis of molecular variance (AMOVA) enabled us to determine the distribution of microsatellite variation among groups of populations, among populations within groups and among individuals within populations, using Arlequin. We grouped the populations according to the tested hypotheses per taxa and regions. We tested the significance of the variance components by calculating their probabilities based on 9999 permutations of individual samples.
Results
The total number of alleles observed per locus in the overall sample of 380 individuals from 37 populations and 24 sites of three regions ranged from 4 to 11, with overall 64 alleles scored over the 10 loci. Private alleles were detected at seven loci, four of which were found in C. flava in Highland Switzerland [see Supporting Information—Table S2].
Total genetic diversity varied little between the three studied taxa, and the overall absolute values of genetic diversity statistics, which comprise variation within and between regions, were similar in the more common C. flava (Ne = 1.42, PL = 56.11 %, Ar = 1.56, He = 0.21) than in the less common C. viridula (Ne = 1.54, PL = 54.0 %, Ar = 1.64, He = 0.25) (Table 2). The percentage of polymorphic loci was significantly different between two regions (F = 10.2, P = 0.01) and among three regions (F = 5.85, P = 0.01; Table 3). Significant taxon-by-region interactions for allelic richness (F = 3.58, P = 0.04), percentage of polymorphic loci (F = 5.30, P = 0.01) and expected heterozygosity (F = 3.89, P = 0.03) indicate that differences between the two taxa in their levels of genetic diversity depended on the region (Table 3a).
Table 2.
Genetic diversity of (a) Carex flava (sect. Ceratocystis, Cyperaceae), (b) C. viridula var. viridula and (c) hybrid C. × subviridula by region of origin. n, sample size; Ne, effective number of alleles; Ar, allelic richness; PL %, percentage of polymorphic loci; Ho, observed heterozygosity; He, expected heterozygosity (gene diversity); FIS, inbreeding coefficient. For population codes follow Table 1.
| (a) C. flava | ||||||||
|---|---|---|---|---|---|---|---|---|
| Region | Pop | n | N e | Ar | PL % | H o | H e | F IS |
| Overall means | 1.42 | 1.56 | 56.11 | 0.16 | 0.21 | 0.18 | ||
| CHH | F1 | 6 | 1.40 | 1.60 | 70.0 | 0.27 | 0.24 | −0.12 |
| F10 | 14 | 1.52 | 1.71 | 80.0 | 0.16 | 0.29 | 0.43 | |
| F12 | 7 | 1.28 | 1.34 | 40.0 | 0.19 | 0.15 | −0.21 | |
| F2 | 12 | 1.52 | 1.76 | 70.0 | 0.23 | 0.28 | 0.18 | |
| F40 | 8 | 1.33 | 1.54 | 70.0 | 0.16 | 0.21 | 0.23 | |
| F7 | 18 | 1.54 | 1.79 | 90.0 | 0.21 | 0.30 | 0.32 | |
| F9 | 6 | 1.75 | 1.92 | 80.0 | 0.17 | 0.38 | 0.56 | |
| Mean | 1.48 | 1.67 | 71.4 | 0.20 | 0.26 | 0.20 | ||
| CHL | F17 | 14 | 1.32 | 1.40 | 50.0 | 0.16 | 0.15 | −0.04 |
| F19 | 7 | 1.18 | 1.20 | 20.0 | 0.17 | 0.10 | −0.79 | |
| F20 | 32 | 1.43 | 1.71 | 100.0 | 0.12 | 0.27 | 0.54 | |
| F33 | 6 | 1.70 | 1.89 | 80.0 | 0.18 | 0.35 | 0.47 | |
| Mean | 1.41 | 1.55 | 62.5 | 0.16 | 0.22 | 0.05 | ||
| EST | CF1 | 4 | 1.31 | 1.44 | 40.0 | 0.15 | 0.15 | −0.02 |
| CF12 | 14 | 1.54 | 1.56 | 40.0 | 0.18 | 0.22 | 0.20 | |
| CF13 | 8 | 1.10 | 1.16 | 20.0 | 0.06 | 0.06 | 0.04 | |
| CF14 | 9 | 1.23 | 1.35 | 40.0 | 0.17 | 0.14 | −0.18 | |
| CF15 | 13 | 1.26 | 1.37 | 40.0 | 0.09 | 0.14 | 0.33 | |
| CF3 | 9 | 1.71 | 1.71 | 40.0 | 0.09 | 0.24 | 0.63 | |
| CF6 | 9 | 1.44 | 1.54 | 40.0 | 0.07 | 0.19 | 0.65 | |
| Mean | 1.37 | 1.45 | 37.1 | 0.12 | 0.16 | 0.24 | ||
| (b) C. viridula | ||||||||
| Region | Pop | n | N e | Ar | PL % | H o | H e | F IS |
| Overall means | 1.54 | 1.64 | 54.0 | 0.23 | 0.25 | 0.05 | ||
| CHH | V12 | 4 | 1.24 | 1.30 | 30.0 | 0.15 | 0.13 | −0.14 |
| V9 | 8 | 1.43 | 1.49 | 40.0 | 0.20 | 0.19 | −0.04 | |
| Mean | 1.33 | 1.40 | 35.0 | 0.18 | 0.16 | −0.09 | ||
| CHL | V17 | 11 | 1.99 | 2.03 | 70.0 | 0.22 | 0.39 | 0.44 |
| V19 | 14 | 1.51 | 1.67 | 60.0 | 0.18 | 0.24 | 0.25 | |
| V20 | 11 | 2.14 | 2.17 | 90.0 | 0.37 | 0.43 | 0.14 | |
| V36 | 20 | 1.44 | 1.58 | 60.0 | 0.19 | 0.24 | 0.20 | |
| Mean | 1.77 | 1.86 | 70.0 | 0.24 | 0.32 | 0.26 | ||
| (b) C. viridula | ||||||||
| Region | Pop | n | N e | Ar | PL % | H o | H e | F IS |
| EST | CV10 | 10 | 1.42 | 1.53 | 50.0 | 0.17 | 0.21 | 0.20 |
| CV12 | 11 | 1.45 | 1.50 | 40.0 | 0.24 | 0.21 | −0.12 | |
| CV15 | 10 | 1.52 | 1.60 | 60.0 | 0.32 | 0.26 | −0.23 | |
| CV2 | 4 | 1.18 | 1.27 | 30.0 | 0.15 | 0.11 | −0.41 | |
| CV4 | 9 | 1.52 | 1.63 | 40.0 | 0.12 | 0.22 | 0.44 | |
| CV6 | 10 | 1.52 | 1.64 | 60.0 | 0.10 | 0.24 | 0.59 | |
| CV7 | 10 | 1.43 | 1.51 | 50.0 | 0.26 | 0.21 | −0.27 | |
| CV8 | 10 | 1.61 | 1.82 | 70.0 | 0.28 | 0.30 | 0.05 | |
| CV9 | 10 | 1.79 | 1.80 | 60.0 | 0.46 | 0.33 | −0.40 | |
| Mean | 1.49 | 1.59 | 51.1 | 0.23 | 0.23 | −0.02 | ||
| (c) C. × subviridula | ||||||||
| Region | Pop | n | N e | Ar | PL % | H o | H e | F IS |
| Overall means | 1.52 | 1.68 | 57.0 | 0.27 | 0.24 | −0.09 | ||
| CHL | FxV17 | 3 | 1.64 | 1.70 | 50.0 | 0.30 | 0.27 | −0.10 |
| FxV20 | 12 | 1.34 | 1.65 | 80.0 | 0.21 | 0.22 | 0.07 | |
| Mean | 1.49 | 1.68 | 65.0 | 0.25 | 0.25 | −0.02 | ||
| EST | FxCV15 | 7 | 1.55 | 1.67 | 40.0 | 0.29 | 0.23 | −0.26 |
| FxCV6 | 10 | 1.61 | 1.71 | 50.0 | 0.28 | 0.25 | −0.13 | |
| Mean | 1.58 | 1.69 | 45.0 | 0.28 | 0.24 | −0.20 | ||
Overall means per taxon and mean values per taxon and region of origin are in bold.
Table 3.
Summary of ANOVAs testing the effects of taxon, region of origin and the interaction between taxon and region of origin on genetic diversity statistics. Ne, effective number of alleles; Ar, allelic richness; PL %, percentage of polymorphic loci; Ho, observed heterozygosity; He, expected heterozygosity (gene diversity); FIS, inbreeding coefficient. (a) With Carex flava (sect. Ceratocystis, Cyperaceae) and C. viridula var. viridula from three study regions; (b) C. flava, C. viridula and hybrid C. × subviridula from two study regions. Table reports F and P-values.
| (a) C. flava, C. viridula from three regions | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| df | N e | Ar | PL % | H o | H e | F IS | |||||||
| F | P | F | P | F | P | F | P | F | P | F | P | ||
| Taxon | 1 | 3.06 | 0.09 | 1.24 | 0.28 | 0.13 | 0.72 | 7.01 | 0.01 | 1.54 | 0.23 | 1.16 | 0.29 |
| Region | 2 | 1.56 | 0.23 | 2.28 | 0.12 | 5.85 | 0.01 | 0.72 | 0.50 | 2.84 | 0.08 | 0.06 | 0.94 |
| Taxa × region | 2 | 2.64 | 0.09 | 3.58 | 0.04 | 5.30 | 0.01 | 1.94 | 0.16 | 3.89 | 0.03 | 1.35 | 0.28 |
| (b) C. flava, C. viridula and C. × subviridula from two regions | |||||||||||||
| df | N e | Ar | PL % | H o | H e | F IS | |||||||
| F | P | F | P | F | P | F | P | F | P | F | P | ||
| Taxon | 2 | 2.50 | 0.10 | 2.85 | 0.08 | 1.19 | 0.32 | 6.58 | 0.01 | 3.34 | 0.05 | 0.85 | 0.44 |
| Region | 1 | 2.02 | 0.17 | 3.75 | 0.07 | 10.2 | 0.00 | 0.21 | 0.65 | 4.55 | 0.04 | 0.25 | 0.62 |
| Taxa × region | 2 | 1.39 | 0.27 | 0.88 | 0.43 | 0.10 | 0.90 | 0.30 | 0.74 | 0.54 | 0.59 | 1.15 | 0.33 |
After correcting P-values for multiple comparisons, 10 out of 370 tests (10 loci × 37 populations) showed significant deviations from Hardy–Weinberg expectations (Appendix 1). Large positive inbreeding coefficients (FIS) were generally correlated with significant deviations from Hardy–Weinberg equilibrium (HWE). No locus in any population had a negative inbreeding coefficient that differed significantly from HWE. Locus S180 showed deviations from HWE in three populations (F20, V19, V36), locus Cko2-135 in three populations (F20, F7, V36), locus S177 in two populations (F20, V36) and locus CL101 and locus Cko2-112 in one populations (V36 or F20, respectively). Some pairs (loci × population) could not be tested because loci were not polymorphic.
Population diversity and inbreeding
The genetic diversity in C. viridula populations was highest in Lowland Switzerland (Ne = 1.77, PL = 70.0 %, Ar = 1.86, He = 0.32), followed by Estonia (Ne = 1.49, PL = 51.1 %, Ar = 1.59, He = 0.23). In Highland Switzerland only few populations of C. viridula were found and their genetic diversity was much lower than in the other two study regions (Ne = 1.33, PL = 35.0 %, Ar = 1.40, He = 0.16). We expected C. viridula with its smaller populations to be more prone to inbreeding. Accordingly, C. viridula showed a deficit of heterozygotes in Lowland Switzerland populations and in some populations of Estonia. However, other Estonian populations and the ones in Highland Switzerland showed an excess of heterozygotes (Table 2b).
Mean genetic diversity per population in C. flava was highest in Highland Switzerland (Ne = 1.48, PL = 71.4 %, Ar = 1.67, He = 0.26), intermediate in Lowland Switzerland (Ne = 1.41, PL = 62.5 %, Ar = 1.55, He = 0.22) and lowest in Estonia (Ne = 1.37, PL = 37.1 %, Ar = 1.45, He = 0.16). Independent of the region, C. flava populations varied in their inbreeding coefficients between −0.79 and 0.65 (Table 2a), the heterozygote excess was not significant, however. These results could be explained first with proximity to several glacial refugia and putative mutagenic influence of higher radiation in mountains.
Genetic diversity was similar for hybrids in Lowland Switzerland (Ne = 1.49, PL = 65.0 %, Ar = 1.68, He = 0.25) and in Estonia (Ne = 1.58, PL = 45.0 %, Ar = 1.69, He = 0.24). In both regions hybrid populations showed an excess of heterozygotes (Table 2c), and were more variable than either parental taxon, in line with expectations for populations with genetic admixing.
Nuclear admixture analysis
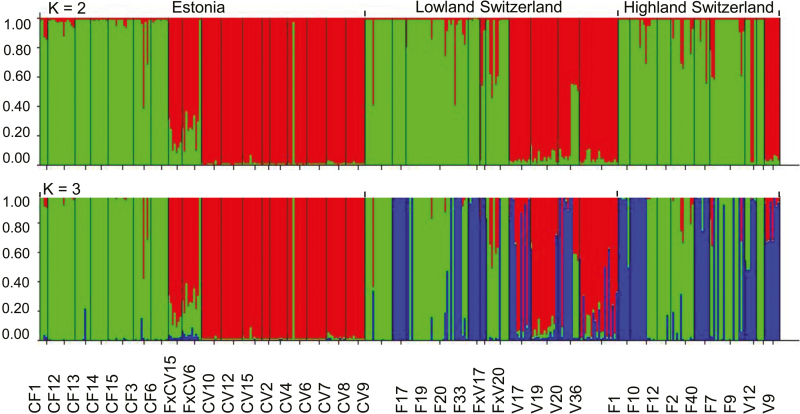
The ΔK method in the Bayesian program STRUCTURE classified all individuals into two clusters [see Supporting Information—Fig. S1], illustrating a clear distinction between taxa, but not among regions within the taxa (K = 2; Fig. 2A). We used the admixture coefficients from the analysis with K = 2 to determine the proportions of admixed individuals. The majority of C. flava and C. viridula individuals from ‘pure’ populations were indeed classified with high admixture coefficients (q > 0.90), i.e. as pure individuals, only two putative C. flava and two C. viridula individuals had q < 0.90, suggesting that they rather were mixed genotypes.
Figure 2.
Bar graph illustrating STRUCTURE analysis of C. flava (CF/F), C. viridula (CV/V) and C. × subviridula (FxCV/FxV) populations from three studied regions Estonia, Highland and Lowland Switzerland. According to the ΔK method, the Bayesian analysis identified two genetic clusters (top, A), while three clusters were found based on observed likelihood values (bottom, B). Each vertical bar represents an individual with coloured partitioning according to genetic clusters. Black vertical lines divide populations. Population names are as in Table 1.
The situation was more complex at ‘mixed’ sites, where two or more taxa of the C. flava complex co-occurred. For the C. flava morphotypes at the mixed sites 136 individuals of 159 putative C. flava were indeed pure C. flava (85.5 %; q > 0.90), 21 of 159 had mixed genotypes (13.3 %; q < 0.90) and two individuals (1.2 %) were even C. viridula-like. For the C. viridula morphotypes at the mixed sites, 102 individuals of 110 putative C. viridula were pure C. viridula (92.7 %; q > 0.90), three had mixed genotype (2.7 %; q < 0.90) and five (4.5 %) were C. flava-like. For the C. × subviridula hybrid morphotypes a wide range of admixture proportions were found (q ranged from 0.103 to 0.897), suggesting the presence of a broad range of hybrid generations and backcrossing to both parental taxa.
With the log-likelihood distribution method, the value where the rate of increase in likelihood reaches a plateau without increase in variance corresponds to three clusters [see Supporting Information—Fig. S1]. K = 3 yielded different results than K = 2, also revealing differences between regions, namely admixture between plants of C. flava and C. viridula in Highland and Lowland Switzerland (Fig. 2B). The third cluster (blue cluster with q > 0.1 in Fig. 2B) occurred mostly for individuals at mixed sites of C. flava and C. viridula (in seven of nine of the mixed sites, and also in Kaltbrunn (F33) and in Melchsee-Frutt (F1), where we exclusively found C. flava).
Overall, the STRUCTURE results indicate clear differentiation between the taxa and in addition further variation between regions. This is in line with the results of genetic diversity, which showed significant differences between regions (for the percentage of polymorphic loci) and significant region-by-taxon interactions (for three measures of genetic diversity; Table 3).
Interspecific differentiation
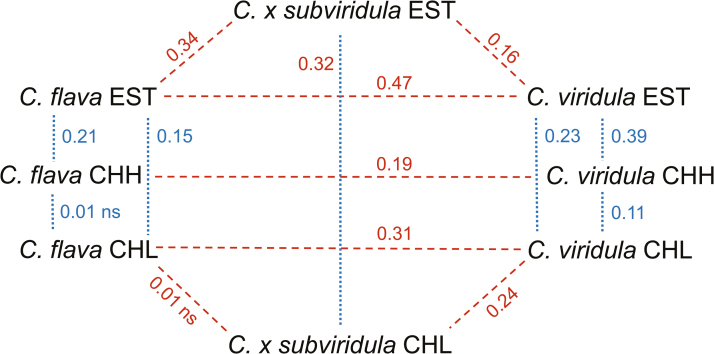
According to the hierarchical AMOVA, the proportion of genetic variance within regions between the studied three taxa was highest in Estonia (39.16 %), followed by Highland (28.27 %) and Lowland Switzerland (17.51 %), i.e. between-taxa differentiation was highest in Estonia and lowest in Lowland Switzerland, where three of the five studied sites were mixed (Table 4). AMOVA results corresponded with the FST values between C. flava and C. viridula, which indicated highest between-taxa differentiation for C. flava and C. viridula populations in Estonia (FST = 0.47), intermediate in Lowland Switzerland (FST = 0.31) and lowest in Highland Switzerland (FST = 0.19; Fig. 3). In Estonia the hybrid C. × subviridula was more differentiated from C. flava (FST = 0.34) than from C. viridula (FST = 0.16), while in Lowland Switzerland the hybrids were more differentiated from C. viridula (FST = 0.24) than from C. flava (FST = 0.01 ns; Figs 3 and 4).
Table 4.
AMOVA of Carex flava (sect. Ceratocystis, Cyperaceae), C. viridula var. viridula and C. × subviridula for SSR data considering the whole data set of all 37 populations with two or three hierarchical levels (a and b) or subsets of populations with two or three hierarchical levels (c–i). P-value = associated significance derived from 16000 permutations. *P < 0.001; **P < 0.05.
| Data set | Source of variation | df | % of variation |
|---|---|---|---|
| (a) 37 populations separated into three groups: Estonia, Highland and Lowland Switzerland | Among groups | 2 | 13.00* |
| Among populations | 34 | 33.09* | |
| Within populations | 723 | 53.90* | |
| (b) 37 populations separated into three groups: C. flava, C. viridula, C. × subviridula | Among groups | 2 | 26.56* |
| Among populations | 34 | 23.27* | |
| Within populations | 723 | 50.18* | |
| (c) 33 populations separated into two groups: C. flava, C. viridula | Among groups | 1 | 32.02* |
| Among populations | 31 | 21.20* | |
| Within populations | 663 | 46.78* | |
| (d) 18 C. flava populations grouped by region of origin | Among groups | 2 | 6.84** |
| Among populations | 15 | 25.76* | |
| Within populations | 374 | 67.40* | |
| (e) 15 C. viridula populations grouped by region of origin | Among groups | 2 | 20.77* |
| Among populations | 12 | 16.07* | |
| Within populations | 289 | 63.16* | |
| (f) 4 C. × subviridula populations grouped by region of origin | Among groups | 1 | 18.27 |
| Among populations | 2 | 23.09* | |
| Within populations | 60 | 58.64* | |
| (g) 18 populations from Estonia grouped by taxa | Among groups | 2 | 39.16* |
| Among populations | 15 | 11.01* | |
| Within populations | 316 | 49.83* | |
| (h) 10 populations from Lowland Switzerland grouped by taxa | Among groups | 2 | 17.51** |
| Among populations | 7 | 24.00* | |
| Within populations | 250 | 58.5* | |
| (i) 9 populations from Highland Switzerland grouped by taxa | Among groups | 1 | 28.27* |
| Among populations | 7 | 14.90* | |
| Within populations | 183 | 56.83* |
Figure 3.
Genetic differentiation (FST values) of C. flava, C. viridula and C. × subviridula from three regions of origin, Estonia (EST), Highland (CHH) and Lowland Switzerland (CHL). Dotted blue lines mark intraspecific differentiation between regions, dashed red lines mark interspecific differentiation within regions. Significances of the pairwise FST values were tested using 1000 permutations; all but two comparisons were significant.
Figure 4.
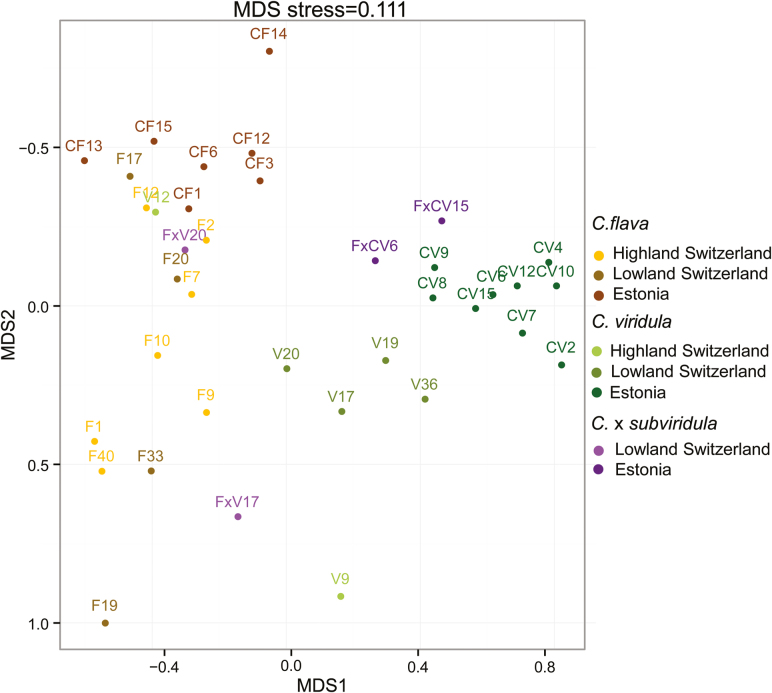
Ordination according to a MDS analysis based on Reynold’s genetic distances between pairs of sampled populations, grouped by the three taxa C. flava (CF/F), C. viridula (CV/V) and C. × subviridula (FxV/FxCV) from the three regions Estonia (EST), Highland (CHH) and Lowland Switzerland (CHL). The stress value of 0.11 indicates a good quality of the graphical representation of the MDS analysis. For population codes see Table 1.
Intraspecific differentiation between regions
Intraspecific differentiation between regions was lower for C. flava (6.84 %) than for C. viridula (20.77 %) or C. × subviridula (18.27 %) populations (AMOVA, Table 4d–f). In accordance, intraspecific between-region pairwise FST values were slightly lower for C. flava (0.01, 0.15 and 0.21) than for C. viridula (0.11, 0.23 and 0.39) or C. × subviridula (0.32; Fig. 3).
Ordination according to the MDS analysis illustrated the genetic distances of populations grouped by taxonomic identity and region of origin (Fig. 4). Estonian C. flava populations formed a distinct cluster, whose difference from clusters of Lowland and Highland Switzerland C. flava populations was smaller than the differences observed among C. viridula populations between the regions (Fig. 4). Estonian C. viridula populations clearly differed from Lowland Switzerland populations and even more from the two populations of C. viridula in Highland Switzerland (V9 and V12), which were also very different from each other (Fig. 4). These findings clearly suggest higher differentiation in C. viridula than in C. flava.
Moreover, in combination, the findings on interspecific and intraspecific differentiation indicate that differentiation between the taxa was stronger than differentiation within the taxa between regions. This was further supported by hierarchical AMOVA, where 26.56 % of the variation resided between the three taxa, while 13.00 % resided between regions (Table 4a and b). As expected, the AMOVA analyses without hybrids C. × subviridula showed higher variance between the parental taxa (32.02 %; Table 4c).
Discussion
Our microsatellite results support growing evidence that interspecific gene flow is more widespread than previously suspected, but that between-species differences are still retained by various mechanisms (Arnold et al. 1990; Friedman et al. 2008; Smith and Waterway 2008; Kane et al. 2009; Nolte et al. 2009; Scascitelli et al. 2010). Microsatellite markers are proven appropriate for population structure and differentiation studies (Pálsson 2000; Song et al. 2006; Tyagi et al. 2016; Volis et al. 2016) and for investigating the relationship among closely related taxa (Korpelainen et al. 2010; Talve et al. 2013, 2014).
We examined whether C. viridula, the taxon with a more disjunct distribution and smaller populations, showed lower genetic diversity and higher inbreeding than the more widespread C. flava, whose populations are larger and more constant in time. However, mean gene diversity (expected heterozygosity) was slightly higher in C. viridula, though not significantly higher from C. flava with He = 0.25 and He = 0.21, respectively. Carex viridula showed highest genetic diversity in Lowland Switzerland, whereas C. flava was most diverse in Highland Switzerland, with He = 0.32 and He = 0.26, respectively. Both taxa had lower diversity in Estonia. Greater allozyme diversity and a lower inbreeding coefficient for C. viridula than for C. flava was also detected in earlier studies using allozymes by Hedrén (2004) and Bruederle and Jensen (1991), where the latter had considered C. viridula s.l., united with C. lepidocarpa, C. demissa and C. viridula s.s., however. Meanwhile, Kuchel and Bruederle (2000) detected low levels of allozyme diversity in North American C. viridula and attributed it to bottlenecks at arrival from Europe and to predominant selfing. Higher diversity in highland populations, as in our study in C. flava, was found in some studies, e.g. for Cystopteris fragilis (Pteridophyta) using isozymes (Gämperle and Schneller 2002), for Primula farinosa (Primulaceae) using RAPD analysis (Reisch et al. 2005) and for Campanula thyrsoides (Campanulaceae) (Frei et al. 2012), and was suggested to be due to higher mutation rates due to elevated radiation (Li et al. 1997). However, others reported higher diversity of lowland populations, as we found for C. viridula (e.g. Premoli 2003; Schönswetter et al. 2006).
In previous studies on sedges C. kobomugi (sect. Macrocephalae), C. macrocephala (sect. Macrocephalae), C. rugulosa (sect. Paludosae) and C. scoparia (sect. Ovales), the mean gene diversities of microsatellites were higher (He = 0.589, 0.523, 0.378 and 0.506, respectively) than in our study (C. flava, He = 0.21; C. viridula, He = 0.25; C. × subviridula, He = 0.24) (Ohbayashi et al. 2008; Hipp et al. 2009; King and Roalson 2009; Ohsako 2010). On the other hand, Escudero et al. (2010) also detected low levels of gene diversity (Hs = 0.10) for C. extensa (sect. Spirostachyae) despite using a wide study area. Possibly our gene diversity values were smaller than the ones detected in other studies, because they generally addressed species with larger and less isolated populations.
Deviation from HWE may indicate inbreeding. In earlier studies, high selfing and evidence for inbreeding has been found in Carex (Arens et al. 2005; King and Roalson 2009; Escudero et al. 2010; Kull and Oja 2010). Inbreeding has also been found to predominate in self-compatible caespitose sedges (Bruederle et al. 2008). We found an excess of heterozygotes in C. flava when growing adjacent to C. viridula (e.g. in populations F12, F19, F17; Table 2a), although this was statistically not significant. The authors who originally published these microsatellites had reported significant excess of heterozygotes in few loci (Ohsako and Yamane 2007; Hipp et al. 2009). Earlier it has been shown that C. flava is the main partner for backcrossing, as it can occasionally be pollinated successfully by F1 hybrids or backcrosses (Schmid 1982; Więcław and Wilhelm 2014). This suggests that C. flava was more prone to between-taxa crosses when growing adjacent to other taxa in section Ceratocystis, e.g. C. viridula, C. lepidocarpa and C. demissa. On the other hand, the direction of hybridization may be affected by the length of the style (Field et al. 2011) which in carices would be determined by the beak length, and hybridization may occur more commonly from long-beaked carices to short-beaked carices due to pollen competition (Derieg et al. 2013), which would suggest higher gene flow from C. flava to C. viridula than vice versa.
We observed hybrids in most of the sites where C. flava and C. viridula grew together sympatrically. Mean gene diversity (expected heterozygosity) was not higher in hybrid populations than in the parental taxa (He = 0.24 vs. 0.21 and 0.25). Korpelainen et al. (2010) studied sedge hybrids in Finland using microsatellite data and found for C. aquatilis × recta (sect. Phacocystis) similar gene diversity than for its parental taxa (He = 0.348 vs. 0.308 and 0.460) and for C. paleacea × recta higher diversity (He = 0.603 vs. 0.185 and 0.460). In contrast, high genetic diversity, using RAPD analysis, was found in Fallopia × bohemica (Polygonaceae) in Germany and Switzerland (Krebs et al. 2010).
We determined hybrids based on partial sterility and morphological differences from the parental taxa. Most hybrid individuals had utricles without fully developed achenes, but some hybrid individuals had circa 5 % of fully developed achenes (own observation). The admixture proportions detected by microsatellites for hybrid individuals were very variable, indicating that these comprised F1 to Fn hybrids and backcrosses. Our genetic data showed that some of the supposed intermediate individuals were not hybrids, but rather backcrosses, or in rare cases even pure parental taxa. Thus, our results imply that hybrids can be identified well based on morphological criteria, but that morphological criteria do not allow for distinguishing backcrosses and that they may even lead to occasional, but very rare, errors in identifying pure individuals.
Interspecific differentiation
The results of our STRUCTURE analysis suggested that we dealt with two species and their hybrids (K = 2) and with some regional differentiation (K = 3; Fig. 2). The presence of separate taxa was further supported by hierarchical AMOVA, where we detected 13.00 % of genetic variation among studied regions, but more than twice this variation between taxa (32.02 %; Table 4). We conclude that there are solid differences between the two species despite evident hybridization. This is in line with earlier studies on the taxonomic relationships of C. flava and C. viridula (Bruederle and Jensen 1991; Hedrén 2002; Jiménez-Mejías et al. 2012). Similarly, Morgan‐Richards and Wolff (1999) found differences between sympatric Plantago major (Plantaginaceae) taxa preserved despite intraspecific gene flow. Kane et al. (2009) also concluded that hybridizing Helianthus taxa (Asteraceae) remained largely reproductively isolated and morphologically and ecologically distinct despite high levels of interspecific gene flow. Brennan et al. (2016) found similar results for hybridizing Senecio taxa (Asteraceae) and explained them with selection against hybrids and locally maladapted hybrid individuals.
Our STRUCTURE analysis with K = 3 revealed a widely present third genetic cluster in Switzerland, which is extremely rare in Estonia (Fig. 2). This third cluster occurred in all three studied taxa C. flava, C. viridula and C. × subviridula, at both low and high altitudes in Switzerland. This suggests high gene flow between taxa within Switzerland and lower between Estonia and Switzerland, as further supported by AMOVA (Table 4). In Estonia, hybridization and introgression occurs, whereas in Lowland Switzerland gene flow between the species seems to be more frequent and to affect the genetic structure of C. flava and C. viridula.
Intraspecific differentiation between regions
As expected for rarer taxa with smaller populations, we found higher among-region differentiation for C. viridula (AMOVA; 20.77 %) than for the more widespread C. flava or hybrid (6.84 and 18.27 %, respectively). In addition, groupwise FST values between regions were higher for C. viridula, especially between Estonia and both altitudes in Switzerland (Fig. 3), indicating that hybridization was not strong enough to prevent stronger regional differentiation in C. viridula. High levels of differentiation might be caused by low levels of wind pollination, which is not very effective for small herbs in closed habitats (Kull and Oja 2010). Another explanation for the higher differentiation of C. viridula is the loss of suitable habitats and subsequent fragmentation of populations, as also shown for other taxa (Pykälä and Toivonen 1994; Reisch et al. 2005; Schönswetter et al. 2006; Kull and Oja 2010; DeWoody et al. 2015). Taking into account that C. viridula had a wider distribution in the past and now shows reduced occurrence and increased fragmentation, our finding of higher differentiation between regions for C. viridula fits theoretical expectations.
We found especially low genetic differentiation among the C. flava groups of populations between Highland and Lowland Switzerland (Fig. 3). This supports the idea of higher gene flow between populations of C. flava, as suggested by Schmid (1984b, 1986). With the use of microsatellite markers, Escudero et al. (2013) have shown considerable mixing among populations in the widespread C. scoparia (sect. Ovales) and explained it with long-distance dispersal. Potential for long-distance pollen and seed dispersal was suggested to contribute to low geographic differentiation of circumpolar C. bigelowii (sect. Phacocystis) (Schönswetter et al. 2008). Theoretical studies have shown that only a small amount of long-distance gene flow is needed to prevent population differentiation for neutral alleles (Loveless and Hamrick 1984). Differences in phenology with altitude are expected to reduce gene flow via pollen and increase differentiation instead (Premoli 2003; Reisch et al. 2005). As flowering times differ notably in Switzerland (Körner 1999), we suggest that this may explain the observed low differentiation among Swiss C. flava populations of similar altitude.
Hybrids showed unexpectedly high differentiation between Estonia and Switzerland (FST = 0.32). This difference could originate from genetic drift in the hybrid populations (Nolte et al. 2009). Moreover, backcross patterns may have differed between the regions. A further explanation could be that in natural populations hybrid swarms involve more than two species (Lepais et al. 2009). At some sites, we found C. lepidocarpa and C. demissa growing beside C. flava and C. viridula, which might increase the number of potential parental taxa and differentiation of hybrids. These mechanisms of hybrid and backcross differentiation between regions are very different from the case of hybrids in the invasive Fallopia species complex, which arise from crosses between different taxa in their home origin, and where different hybrids were introduced to different regions, leading to high regional differentiation in the introduced range (Krebs et al. 2010).
Conclusions
Our in-depth analysis of 380 individuals belonging to two sedge taxa and their hybrids and involving populations from three regions suggest that hybridization and introgression are neither strong enough to prevent clear differentiation between taxa nor to prevent stronger regional differentiation for the less common taxon. We encourage further studies on regional differentiation of hybrids and parental taxa to see whether our findings for the C. flava complex represent a more general pattern. Moreover, we suggest also considering hybrids and closely related taxa when addressing genetic diversity and differentiation for rare and endangered taxa.
Sources of Funding
The Estonian Ministry of Education and Research, institutional research funding (IUT 20-28, 20-29) and the European Union through the European Regional Development Fund (Centre of Excellence EcolChange) supported this study.
Contributions by the Authors
L.S., T.O. and M.F. conceived the study and its design. L.S. conducted field and lab work, analysed data and led the writing of the manuscript. T.O. advised lab work. Both T.O. and M.F. contributed to data analysis, data interpretation and writing of the manuscript. This manuscript forms part of the PhD thesis of the first author (L.S.). All authors have made a substantial contribution to the paper.
Conflict of Interest
None declared.
Supporting Information
The following additional information is available in the online version of this article—
Table S1. Microsatellite loci used to study genetic diversity and species relationships in Carex flava (sect. Ceratocystis, Cyperaceae), C. viridula var. viridula and their hybrid (C. × subviridula). Presented are primer sequences, 5′ tag, repeat motif, size range, annealing temperature Ta (°C), number of alleles detected for each locus. The 5′ tags M13R (AGGAAACAGCTATGACCAT) and CAGT (ACAGTCGGGCGTCATCA) were used for incorporation of the fluorescent tag. S082, S180, S245, S175, S119, S177 are from Hipp et al. (2009), Cr37 is from Ohbayashi et al. (2008), Cko2-112, Cko2-135 are from Ohsako and Yamane (2007), CL101 is from Escudero et al. (2010).
Table S2. Presence and frequency of private alleles in seven (Cko2-112, Cko2-135, S082, S245, CL101, S180, S175) of the 10 studied microsatellite loci in Carex flava (sect. Ceratocystis, Cyperaceae), C. viridula var. viridula and their hybrid (C. × subviridula). The three other analysed loci (S119, S177, Cr37) did not show private alleles.
Figure S1. Estimating the optimal number of clusters with admixture analyses of plants of Carex flava (sect. Ceratocystis, Cyperaceae), C. viridula var. viridula and their hybrid (C. × subviridula) from Estonia, Highland and Lowland Switzerland. (A) The ΔK method indicated two genetic groups. (B) The distribution of log-likelihoods indicated three genetic groups.
Acknowledgements
We acknowledge very helpful information on populations and discussions with B. Schmid, University of Zurich. We are thankful to Ü. Aarna, M. Mürk and T. Talve for guidance and help with the laboratory work, University of Tartu. We thank the DORA programme (Doctoral Studies and Internationalisation Programme) of the Archimedes Foundation for supporting the sampling in Switzerland.
Literature Cited
- Abbott R, Albach D, Ansell S, Arntzen JW, Baird SJ, Bierne N, Boughman J, Brelsford A, Buerkle CA, Buggs R, Butlin RK, Dieckmann U, Eroukhmanoff F, Grill A, Cahan SH, Hermansen JS, Hewitt G, Hudson AG, Jiggins C, Jones J, Keller B, Marczewski T, Mallet J, Martinez-Rodriguez P, Möst M, Mullen S, Nichols R, Nolte AW, Parisod C, Pfennig K, Rice AM, Ritchie MG, Seifert B, Smadja CM, Stelkens R, Szymura JM, Väinölä R, Wolf JB, Zinner D. 2013. Hybridization and speciation. Journal of Evolutionary Biology 26:229–246. [DOI] [PubMed] [Google Scholar]
- Allendorf FW, Leary RF, Spruell P, Wenburg JK. 2001. The problems with hybrids: setting conservation guidelines. Trends in Ecology and Evolution 16:613–622. [Google Scholar]
- Amos W, Harwood J. 1998. Factors affecting levels of genetic diversity in natural populations. Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences 353:177–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arens P, Bijlsma RJ, van’t Westende W, van Os B, Smulders MJ, Vosman B. 2005. Genetic structure in populations of an ancient woodland sedge, Carex sylvatica Hudson, at a regional and local scale. Plant Biology 7:387–396. [DOI] [PubMed] [Google Scholar]
- Arnold ML. 2006. Evolution through genetic exchange, 1st edn. New York: Oxford University Press Inc. [Google Scholar]
- Arnold ML, Bennett BD, Zimmer EA. 1990. Natural hybridization between Iris fulva and Iris hexagona: pattern of ribosomal DNA variation. Evolution 44:1512–1521. [DOI] [PubMed] [Google Scholar]
- Bain JF, Golden JL. 2003. Phylogeographic relationships within Packera sanguisorboides (Asteraceae), a narrow endemic species that straddles a major biogeographic boundary. American Journal of Botany 90:1087–1094. [DOI] [PubMed] [Google Scholar]
- Barton NH. 2001. The role of hybridization in evolution. Molecular Ecology 10:551–568. [DOI] [PubMed] [Google Scholar]
- Blackstock N., Ashton PA. 2010. Genetic markers and morphometric analysis reveal past hybridization and introgression in putative Carex flava L. s. str. (Cyperaceae) hybrid populations. Plant Systematics and Evolution 287:37–47. [Google Scholar]
- Bornand C, Gygax A, Juillerat P, Jutzi M, Möhl A, Romentsch S, Sage RL, Santiago H, Eggenberg S. 2016. Rote liste gefässpflanzen. Gefährdete arten der schweiz. Switzerland: Bundesamt für Umwelt BAFU, Info Flora. [Google Scholar]
- Brennan AC, Hiscock SJ, Abbott RJ. 2016. Genomic architecture of phenotypic divergence between two hybridizing plant species along an elevational gradient. AoB PLANTS 8:plw022; doi:10.1093/aobpla/plw022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruederle LP, Jensen U. 1991. Genetic differentiation of Carex flava and Carex viridula in West Europe (Cyperaceae). Systematic Botany 16:41–49. [Google Scholar]
- Bruederle LP, Yarbrough SL, Fehlberg SD. 2008. Allozyme variation in the genus Carex … 15 years later: 1986–2001. In: Naczi RFC, Ford BA, eds. Sedges: uses, diversity, and systematics of the Cyperaceae. St. Louis, MO: Missouri Botanical Garden Press, 187–196. [Google Scholar]
- Burgarella C, Lorenzo Z, Jabbour-Zahab R, Lumaret R, Guichoux E, Petit RJ, Soto A, Gil L. 2009. Detection of hybrids in nature: application to oaks (Quercus suber and Q. Ilex). Heredity 102:442–452. [DOI] [PubMed] [Google Scholar]
- Chen IC, Hill JK, Ohlemüller R, Roy DB, Thomas CD. 2011. Rapid range shifts of species associated with high levels of climate warming. Science 333:1024–1026. [DOI] [PubMed] [Google Scholar]
- Crins WJ, Ball PW. 1989. Taxonomy of the Carex flava complex (Cyperaceae) in North America and northern Eurasia. II. Taxonomic treatment. Canadian Journal of Botany 67:1048–1065. [Google Scholar]
- Davies EW. 1953. Notes on Carex flava and its allies. IV. Geographic distribution. Watsonia 3:80–84. [Google Scholar]
- Davies EW. 1955. The cytogenetics of Carex flava and its allies. Watsonia 3:129–137. [Google Scholar]
- Derieg NJ, Sangaumphai A, Bruederle LP. 2008. Genetic diversity and endemism in North American Carex section Ceratocystis (Cyperaceae). American Journal of Botany 95:1287–1296. [DOI] [PubMed] [Google Scholar]
- Derieg NJ, Weil SJ, Reznicek AA, Bruederle LP. 2013. Carex viridistellata sp. nov. (Cyperaceae), a new cryptic species from prairie fens of the eastern United States. Systematic Botany 38:82–91. [Google Scholar]
- DeWoody J, Trewin H, Taylor G. 2015. Genetic and morphological differentiation in Populus nigra L.: isolation by colonization or isolation by adaptation?Molecular Ecology 24:2641–2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle JJ. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochemical Bulletin 19:11–15. [Google Scholar]
- Earl DA. 2012. STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conservation Genetics Resources 4:359–361. [Google Scholar]
- Ellstrand NC, Elam DR. 1993. Population genetic consequences of small population size: implications for plant conservation. Annual Review of Ecology and Systematics 24:217–242. [Google Scholar]
- Ellstrand NC, Whitkus R, Rieseberg LH. 1996. Distribution of spontaneous plant hybrids. Proceedings of the National Academy of Sciences 93:5090–5093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escudero M, Vargas P, Arens P, Ouborg NJ, Luceño M. 2010. The east-west-north colonization history of the Mediterranean and Europe by the coastal plant Carex extensa (Cyperaceae). Molecular Ecology 19:352–370. [DOI] [PubMed] [Google Scholar]
- Escudero M, Weber JA, Hipp AL. 2013. Species coherence in the face of karyotype diversification in holocentric organisms: the case of a cytogenetically variable sedge (Carex scoparia, Cyperaceae). Annals of Botany 112:515–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evanno G, Regnaut S, Goudet J. 2005. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Molecular Ecology 14:2611–2620. [DOI] [PubMed] [Google Scholar]
- Excoffier L, Laval G, Schneider S. 2005. ARLEQUIN (version 3.0): an integrated software package for population genetics data analysis. Evolutionary Bioinformatics Online 1:47–50. [PMC free article] [PubMed] [Google Scholar]
- Field DL, Ayre DJ, Whelan RJ, Young AG. 2011. Patterns of hybridization and asymmetrical gene flow in hybrid zones of the rare Eucalyptus aggregata and common E. Rubida. Heredity 106:841–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frei ES, Scheepens JF, Stöcklin J. 2012. High genetic differentiation in populations of the rare alpine plant species Campanula thyrsoides on a small mountain. Alpine Botany 122:23–34. [Google Scholar]
- Friedman JM, Roelle JE, Gaskin JF, Pepper AE, Manhart JR. 2008. Latitudinal variation in cold hardiness in introduced Tamarix and native Populus. Evolutionary Applications 1:598–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gämperle E, Schneller JJ. 2002. Phenotypic and isozyme variation in Cystopteris fragilis (Pteridophyta) along an altitudinal gradient in Switzerland. Flora-Morphology, Distribution, Functional Ecology of Plants 197:203–213. [Google Scholar]
- Gitzendanner MA, Soltis PS. 2000. Patterns of genetic variation in rare and widespread plant congeners. American Journal of Botany 87:783–792. [PubMed] [Google Scholar]
- Goudet J. 2002. FSTAT, a software to estimate and test gene diversities and differentiation statistics from codominant markers (version 2.9.3.2). http://www2.unil.ch/popgen/softwares/fstat.htm. [Google Scholar]
- Hamrick J, Godt MJW. 1996. Effects of life history traits on genetic diversity in plant species. Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences 351:1291–1298. [Google Scholar]
- Hamrick JL, Linhart YB, Mitton JB. 1979. Relationships between life history characteristics and electrophoretically detectable genetic variation in plants. Annual Review of Ecology and Systematics 10:173–200. [Google Scholar]
- Harrison RG, Larson EL. 2014. Hybridization, introgression, and the nature of species boundaries. The Journal of Heredity 105:795–809. [DOI] [PubMed] [Google Scholar]
- Hedrén M. 2002. Patterns of allozyme and morphological differentiation in the Carex flava complex (Cyperaceae) in Fennoscandia. Nordic Journal of Botany 22:257–301. [Google Scholar]
- Hedrén M. 2004. Species delimitation and the partitioning of genetic diversity‐an example from the Carex flava complex (Cyperaceae). Biodiversity and Conservation 13:293–316. [Google Scholar]
- Hewitt GM. 1999. Post‐glacial re‐colonization of European biota. Biological Journal of the Linnean Society 68:87–112. [Google Scholar]
- Hipp AL, Kettenring KM, Feldheim KA, Weber JA. 2009. Isolation of 11 polymorphic tri- and tetranucleotide microsatellite loci in a North American sedge (Carex scoparia: Cyperaceae) and cross-species amplification in three additional Carex species. Molecular Ecology Resources 9:625–627. [DOI] [PubMed] [Google Scholar]
- Hoffmann AA, Sgro CM. 2011. Climate change and evolutionary adaptation. Nature 470:479–485. [DOI] [PubMed] [Google Scholar]
- Holderegger R, Wagner HH. 2006. A brief guide to landscape genetics. Landscape Ecology 21:793–796. [Google Scholar]
- Hultén E, Fries M. 1986. Atlas of North European vascular plants (North of the Tropic of Cancer). Königstein: Koeltz Scientific Books. [Google Scholar]
- Jiménez-Mejías P, Martín-Bravo S, Luceño M. 2012. Systematics and taxonomy of Carex sect. Ceratocystis (Cyperaceae) in Europe: a molecular and cytogenetic approach. Systematic Botany 37:382–398. [Google Scholar]
- Kane NC, King MG, Barker MS, Raduski A, Karrenberg S, Yatabe Y, Knapp SJ, Rieseberg LH. 2009. Comparative genomic and population genetic analyses indicate highly porous genomes and high levels of gene flow between divergent Helianthus species. Evolution 63:2061–2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King MG, Roalson EH. 2009. Isolation and characterization of 11 microsatellite loci from Carex macrocephala (Cyperaceae). Conservation Genetics 10:531–533. [Google Scholar]
- Koopman J. 2011. Carex Europaea: the genus Carex L. (Cyperaceae) in Europe, 1: accepted names, hybrids, synonyms, distribution, chromosome numbers. Weikersheim: Magraf Press. [Google Scholar]
- Körner C. 1999. Alpine plant life: functional plant ecology of high mountain ecosystems. Berlin: Springer-Verlag. [Google Scholar]
- Korpelainen H, Virtanen V, Kostamo K, Väre H. 2010. Hybridization and introgression in Carex aquatilis and C. paleacea. Plant Systematics and Evolution 287:141–151. [Google Scholar]
- Krebs C, Mahy G, Matthies D, Schaffner U, Tiébré MS, Bizoux JP. 2010. Taxa distribution and RAPD markers indicate different origin and regional differentiation of hybrids in the invasive Fallopia complex in central-western Europe. Plant Biology 12:215–223. [DOI] [PubMed] [Google Scholar]
- Kuchel SD, Bruederle LP. 2000. Allozyme data support a Eurasian origin for Carex viridula subsp. viridula var. viridula (Cyperaceae). Madroño 47:147–158. [Google Scholar]
- Kull T, Oja T. 2010. Allozyme diversity and geographic variation among populations of the locally endangered taxon Carex magellanica subsp. irrigua (Cyperaceae). Folia Geobotanica 45:323–336. [Google Scholar]
- Leimu R, Mutikainen PIA, Koricheva J, Fischer M. 2006. How general are positive relationships between plant population size, fitness and genetic variation?Journal of Ecology 94:942–952. [Google Scholar]
- Lepais O, Petit RJ, Guichoux E, Lavabre JE, Alberto F, Kremer A, Gerber S. 2009. Species relative abundance and direction of introgression in oaks. Molecular Ecology 18:2228–2242. [DOI] [PubMed] [Google Scholar]
- Levin DA, Francisco‐Ortega J, Jansen RK. 1996. Hybridization and the extinction of rare plant species. Conservation Biology 10:10–16. [Google Scholar]
- Lewontin RC, Birch LC. 1966. Hybridization as a source of variation for adaptation to new environments. Evolution 20:315–336. [DOI] [PubMed] [Google Scholar]
- Li J, Wang P, Han D, Chen F, Deng L, Guo Y. 1997. Mutation effect of high altitude balloon flight on rice and green pepper seeds. Space Medicine & Medical Engineering 10:79–83. [PubMed] [Google Scholar]
- Linhart YB, Grant MC. 1996. Evolutionary significance of local genetic differentiation in plants. Annual Review of Ecology and Systematics 27:237–277. [Google Scholar]
- Loveless MD, Hamrick JL. 1984. Ecological determinants of genetic structure in plant populations. Annual Review of Ecology and Systematics 15:65–95. [Google Scholar]
- Morgan‐Richards M, Wolff K. 1999. Genetic structure and differentiation of Plantago major reveals a pair of sympatric sister species. Molecular Ecology 8:1027–1036. [Google Scholar]
- Nolte AW, Gompert Z, Buerkle CA. 2009. Variable patterns of introgression in two sculpin hybrid zones suggest that genomic isolation differs among populations. Molecular Ecology 18:2615–2627. [DOI] [PubMed] [Google Scholar]
- Nybom H. 2004. Comparison of different nuclear DNA markers for estimating intraspecific genetic diversity in plants. Molecular Ecology 13:1143–1155. [DOI] [PubMed] [Google Scholar]
- Ohbayashi K, Hodoki Y, Nakayama S, Shimada M, Kunii H. 2008. Development of new microsatellite markers from a salt-marsh sedge Carex rugulosa by compound simple sequence repeat-polymerase chain reaction. Molecular Ecology Resources 8:1497–1499. [DOI] [PubMed] [Google Scholar]
- Ohsako T. 2010. Clonal and spatial genetic structure within populations of a coastal plant, Carex kobomugi (Cyperaceae). American Journal of Botany 97:458–470. [DOI] [PubMed] [Google Scholar]
- Ohsako T, Yamane K. 2007. Isolation and characterization of polymorphic microsatellite loci in Asiatic sand sedge, Carex kobomugi Ohwi (Cyperaceae). Molecular Ecology Notes 7:1023–1025. [Google Scholar]
- Pálsson S. 2000. Microsatellite variation in Daphnia pulex from both sides of the Baltic Sea. Molecular Ecology 9:1075–1088. [DOI] [PubMed] [Google Scholar]
- Parmesan C, Yohe G. 2003. A globally coherent fingerprint of climate change impacts across natural systems. Nature 421:37–42. [DOI] [PubMed] [Google Scholar]
- Peakall ROD, Smouse PE. 2006. GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Molecular Ecology Resources 6:288–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Premoli AC. 2003. Isozyme polymorphisms provide evidence of clinal variation with elevation in Nothofagus pumilio. The Journal of Heredity 94:218–226. [DOI] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. 2000. Inference of population structure using multilocus genotype data. Genetics 155:945–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pykälä J, Toivonen H. 1994. Taxonomy of the Carex flava complex (Cyperaceae) in Finland. Nordic Journal of Botany 14:173–191. [Google Scholar]
- R Development Core Team 2016. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing. [Google Scholar]
- Reisch C, Anke A, Röhl M. 2005. Molecular variation within and between ten populations of Primula farinosa (Primulaceae) along an altitudinal gradient in the northern Alps. Basic and Applied Ecology 6:35–45. [Google Scholar]
- Reynolds J, Weir BS, Cockerham CC. 1983. Estimation of the coancestry coefficient: basis for a short-term genetic distance. Genetics 105:767–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieseberg LH, Wendel JF. 1993. Introgression and its consequences in plants. In: Harrison RG, ed. Hybrid zones and the evolutionay process. New York: Oxford University Press, 70–106. [Google Scholar]
- Rieseberg LH, Whitton J, Gardner K. 1999. Hybrid zones and the genetic architecture of a barrier to gene flow between two sunflower species. Genetics 152:713–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossetto M, Shepherd M, Cordeiro GM, Harriss FCL, Lee LS, Henry RJ. 1999. Cross species amplification of microsatellite loci: a valuable tool for genetic studies in plants. Plant and Animal Genome VII Conference, San Diego, USA (abstract). [Google Scholar]
- Scascitelli M, Whitney KD, Randell RA, King M, Buerkle CA, Rieseberg LH. 2010. Genome scan of hybridizing sunflowers from texas (Helianthus annuus and H. debilis) reveals asymmetric patterns of introgression and small islands of genomic differentiation. Molecular Ecology 19:521–541. [DOI] [PubMed] [Google Scholar]
- Schmid BW. 1981. Die Verbreitung der Artengruppe Carex flava L. s.l. in der Schweiz. Botanica Helvetica 91:3–8. [Google Scholar]
- Schmid BW. 1982. Karyology and hydridization in the Carex flava complex in Switzerland. Feddes Repertorium 93:23–59. [Google Scholar]
- Schmid BW. 1984a. Life histories in clonal plants of the Carex flava group. Journal of Ecology 72:93–114. [Google Scholar]
- Schmid BW. 1984b. Niche width and variation within and between populations in colonizing species (Carex flava group). Oecologia 63:1–5. [DOI] [PubMed] [Google Scholar]
- Schmid BW. 1986. Patterns of variation and population structure in the Carex flava group. Symbolae Botanicae Upsalienses 27:113–126. [Google Scholar]
- Schmidt L, Fischer M, Schmid BW, Oja T. 2017. Despite admixing two closely related Carex species differ in their regional morphological differentiation. Plant Systematics and Evolution 7:901–914. [Google Scholar]
- Schönswetter P, Elven R, Brochmann C. 2008. Trans-atlantic dispersal and large-scale lack of genetic structure in the circumpolar, arctic-alpine sedge Carex bigelowii s. l. (Cyperaceae). American Journal of Botany 95:1006–1014. [DOI] [PubMed] [Google Scholar]
- Schönswetter P, Popp M, Brochmann C. 2006. Central Asian origin of and strong genetic differentiation among populations of the rare and disjunct Carex atrofusca (Cyperaceae) in the Alps. Journal of Biogeography 33:948–956. [Google Scholar]
- Slatkin M. 1987. Gene flow and the geographic structure of natural populations. Science 236:787–792. [DOI] [PubMed] [Google Scholar]
- Smith TW, Waterway MJ. 2008. Evaluating species limits and hybridization in the Carex complanata complex using morphology, amplified fragment length polymorphisms, and restriction fragment analysis. Botany 86:809–826. [Google Scholar]
- Song BH, Clauss MJ, Pepper A, Mitchell-Olds T. 2006. Geographic patterns of microsatellite variation in Boechera stricta, a close relative of Arabidopsis. Molecular Ecology 15:357–369. [DOI] [PubMed] [Google Scholar]
- Talve T, McGlaughlin ME, Helenurm K, Wallace LE, Oja T. 2013. Population genetic diversity and species relationships in the genus Rhinanthus L. based on microsatellite markers. Plant Biology 16:495–502. [DOI] [PubMed] [Google Scholar]
- Talve T, Mürk M, Lindell T, Oja T. 2014. Rhinanthus plants found in calcareous fens on Gotland (Sweden): are they related to Rhinanthus osiliensis from Saaremaa (Estonia)?Biochemical Systematics and Ecology 54:113–122. [Google Scholar]
- Todesco M, Pascual MA, Owens GL, Ostevik KL, Moyers BT, Hübner S, Heredia SM, Hahn MA, Caseys C, Bock DG. 2016. Hybridization and extinction. Evolutionary Applications 9:1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toom M, Liira J, Kull T. 2016. Tarnad - the genus Carex in Estonia [in Estonian], 1st edn. Tartu: University of Tartu. [Google Scholar]
- Tyagi A, Singh S, Mishra P, Singh A, Tripathi AM, Jena SN, Roy S. 2016. Genetic diversity and population structure of Arabidopsis thaliana along an altitudinal gradient. AoB Plants 8:1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vähä JP, Primmer CR. 2006. Efficiency of model-based Bayesian methods for detecting hybrid individuals under different hybridization scenarios and with different numbers of loci. Molecular Ecology 15:63–72. [DOI] [PubMed] [Google Scholar]
- van Kleunen M, Dawson W, Bossdorf O, Fischer M. 2014. The more the merrier: multi-species experiments in ecology. Basic and Applied Ecology 15:1–9. [Google Scholar]
- van Oosterhout C, Hutchinson WF, Wills DPM, Shipley P. 2004. MICRO‐CHECKER: software for identifying and correcting genotyping errors in microsatellite data. Molecular Ecology Notes 4:535–538. [Google Scholar]
- Volis S, Ormanbekova D, Shulgina I. 2016. Role of selection and gene flow in population differentiation at the edge vs. interior of the species range differing in climatic conditions. Molecular Ecology 25:1449–1464. [DOI] [PubMed] [Google Scholar]
- Volkova PA, Shipunov AB, Elven R, Brochmann C. 2008. The seashore sedges of the Russian Kola Peninsula: how many species?Flora-Morphology, Distribution, Functional Ecology of Plants 203:523–533. [Google Scholar]
- Vonk DH. 1979. Biosystematic studies of the Carex flava complex I. Flowering. Plant Biology 28:1–20. [Google Scholar]
- Więcław H, Wilhelm M. 2014. Natural hybridization within the Carex flava complex (Cyperaceae) in Poland: morphometric studies. Annales Botanici Fennici 51:129–147. [Google Scholar]
- Zhivotovsky LA, Teterina AA, Mukhina NV, Stroganov AN, Rubtsova GA, Afanasiev KI. 2016. Effects of genetic drift in a small population of Atlantic cod (Gadus morhua kildinensis Derjugin) landlocked in a meromictic lake: genetic variation and conservation measures. Conservation Genetics 17:229–238. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.