Abstract
Background
Microglia and periphery-derived monocytes infiltrate human and mouse glioblastoma and their density is positively correlated with malignancy. Using microarray and RNA sequencing, we have previously shown that glioblastoma-associated microglia/monocytes (GAMs) express osteopontin/SPP1.
Methods
We used quantitative reverse transcriptase PCR, immunofluorescence stainings, western blot, and flow cytometry to identify the various sources of osteopontin (OPN) expression in human and mouse glioblastoma. We implanted wild type GL261 glioblastoma cells, which do not express significant levels of OPN, into wild type and OPN−/− mice to investigate the role of microenvironment-derived OPN on glioblastoma progression.
Results
Our data indicate that GAMs are the predominant source of OPN in both human and mouse glioblastoma and express only the secreted form of OPN. Loss of microenvironment-derived OPN enhanced tumor progression. Staining by Ki67 and terminal deoxynucleotidyl transferase deoxyuridine triphosphate nick end labeling showed no difference in overall cell proliferation but a decreased apoptosis rate in tumors in OPN−/− mice. CD31 staining showed a significantly decreased number of microvessels in tumors in OPN−/− mice, accompanied by reduced coverage of vessels with platelet derived growth factor receptor β+ pericytes. Flow cytometry analysis revealed a significant increase of CD11b+/CD45low microglia but not of CD11b+/CD45high macrophages/monocytes in tumors in OPN−/− mice. Sorted CD11b+ cells from wild type and OPN−/− naïve brains and tumors did not show a significant difference in the expression pattern of activation marker genes.
Conclusion
Our results show that in tested human and mouse glioblastoma samples, OPN is predominantly expressed and secreted by GAMs and that, in contrast to OPN expression in the tumor cells per se, loss of stroma-derived OPN creates a glioblastoma-promoting microenvironment.
Keywords: glioblastoma, microglia, macrophage, osteopontin, SPP1
Importance of the study
GAMs make up a large percentage of cells in glioblastoma and have previously been shown to support glioblastoma growth. It is necessary to investigate how these cells respond to and interact with the various cells within the tumor microenvironment. We have previously identified a large number of genes that are upregulated in human and mouse GAMs and it is now an important task to determine which of these genes promote or impair glioblastoma progression.
Glioblastoma (GBM; World Health Organization grade IV) is characterized by a high inter- and intratumor heterogeneity, unresponsiveness to multimodal treatment, and a low overall survival rate.1 Microglia and peripheral macrophages/monocytes are part of the glioma microenvironment and might pose attractive targets for therapeutic research; however, the interactions between these cells and the actual tumor cells are still poorly understood.2
Osteopontin (OPN, encoded by the gene SPP1) is an extracellular matrix protein that is present in a number of tissues and has been implicated in the pathogenesis of different cancers, including GBM.3 Glioblastoma cell–derived OPN is known to enhance invasion and promote a stem cell–like character, as well as radiation resistance of GBM cells in vitro and in vivo; and knockdown of OPN in GBM cells reduces tumor growth in vivo.4–7 In addition, OPN secreted by GBM cells has been reported to affect the activation status of microglia/macrophages.8 We have recently determined the expression profile of both human and murine glioblastoma-associated microglia/monocytes (GAMs) and identified several genes that were upregulated compared with naïve microglia. We found that OPN was one of the highest upregulated genes in murine and human GAMs.9,10 Although several studies have investigated the role of GBM cell–derived OPN, only a few studies have focused on the effects of microenvironment-derived OPN on GBM biology.
In this study, we show that OPN is predominantly expressed by GAMs in tested human and mouse GBM. Using the GL261 cell line—which does not produce significant levels of OPN itself—as an in vivo GBM model to study the sole role of loss of microenvironment-derived OPN on GBM growth, we show that OPN−/− mice exhibit a faster disease progression compared with wild type mice.
Materials and Methods
Human Tissue Samples
Human GBM and brain tissue for cell isolation were obtained with patients’ consent from the Departments of Neurosurgery and Neuropathology at Charité University Hospital and the University Medical Center Schleswig-Holstein in strict accordance with the responsible ethics committee (Charité, EA4/098/11; UKSH B307/15). Clinically annotated GBM specimens for immunofluorescence stainings were acquired from the University of Washington Medical Center. Approval for the use of human subject material was granted by the institutional review board of the University of Washington (IRB#44807). Autopsy tissue was used for brain control tissue. Formalin-fixed paraffin-embedded tissue was cut at 5 µm sections and floated onto charged slides for preparation of immunohistochemistry.
Cell Culture
GL261 murine GBM cells were cultured and GL261-conditioned medium was prepared as previously described.11 Mouse primary microglia were prepared from neonatal wild type or OPN−/− mice as previously described.11
Animals
Mice were housed in the animal facilities of the Max Delbrueck Center and handled according to governmental guidelines (LaGeSo G0268/10, G0343/10). We used C57BL/6 wild type, SPP1 knockout (OPN−/−) mice on a C57BL/6 background (The Jackson Laboratory, #004936) and Cx3cr1GFP/wt Ccr2RFP/wt mice. Tumors were implanted as previously described.10 Monitoring of the mice occurred daily; weighing and behavioral assessment reflecting the mice’s health condition started on day 15 post surgery.
Tumor Area Quantification
Wild type and OPN−/− mice (n = 9 per group) were simultaneously inoculated with GL261 cells and sacrificed between days 19 to 21 post-injection in pairs of wild type and knockout on the same day whenever one mouse reached a disease score of 4 or higher. Brains were fixed after perfusion and cut into 40-μm-thick slices. Slices were stained with Hoechst and the tumor area was measured as the percentage of tumor-infiltrated brain area in every sixth slice.
Analysis of Disease Progression
GL261 tumor-bearing wild type and OPN−/− mice (n = 10 per group) were monitored for signs of disease progression using an objective score from 1 (no symptoms) to 5 (severe disease). Scoring of the mice was done in accordance with governmental guidelines, based on weight, posture, and alertness, and integrated into a score from 1, indicating an animal in normal health condition, to 5, representing an animal nonresponsive to acoustic or tactile stimuli or that had lost more than 20% of its starting weight. A mouse was euthanized whenever it reached a score of 4 or higher.
Cell Isolation
Isolation of cluster of differentiation (CD)11b+ cells from naïve and tumor brains and flow cytometry analysis of GL261-bearing brains was carried out as previously described.10 Antibodies for flow cytometry analysis can be found in Supplementary Table S1. Flow cytometry data were analyzed using FlowJo 10.2.
Western Blot
Protein isolation, sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE), and western blotting were performed as previously described.11 Immunoblotting was performed using goat anti-mouse OPN antibody (R&D Systems, #AF808).
Quantitative Reverse Transcriptase PCR
RNA protocols and quantitative reverse transcriptase (qRT)-PCR were carried out as previously described.9 Sequences of primers used can be found in Supplementary Table S2.
Immunofluorescence
Tissue preparation for immunofluorescence stainings of mouse and human tissue was performed as previously described.12 Antibodies used can be found in Supplementary Table S3.
Images were acquired using a Zeiss LSM 780 NLO confocal microscope. All images were acquired with a 20x objective if not otherwise stated.
For quantification of Ki67 positivity, 5–6 randomly distributed fields of view were acquired per tumor, and the percentage of Ki67+ cells was determined and normalized to the total Hoechst+ cell population. Ki67+ cells were defined by a staining intensity of higher than 20 and Ki67high cells by an intensity higher than 55 using TissueQuest software (TissueGnostics). For the quantification of CD31+ blood vessels, 5–6 randomly distributed fields of view were acquired and the number and diameter were determined using ImageJ. For 3D reconstruction, z-stacks were acquired using a 63x objective; 3D reconstructions were made using Volocity (Improvision).
TUNEL Assay
For analysis by terminal deoxynucleotidyl transferase deoxyuridine triphosphate nick end labeling (TUNEL), the Click-iT Plus TUNEL Assay (Thermo Fisher Scientific, #C10618) was used according to the manufacturer’s instructions. Z-stacks were acquired using a Zeiss Axio Imager Z2 upright microscope, and the number of TUNEL+ cells was normalized to the total tumor area.
Statistical Analyses
Graphs were created using GraphPad Prism 7 and were analyzed using an unpaired parametric 2-tailed t-test, assuming equal standard deviations. One-way ANOVA was used in experiments having more than one group to compare with controls. Linear regression was used to compare curves of disease and weight progression. Test details are included in appropriate figure legends.
The cBioPortal (http://www.cbioportal.org/public-portal/) was used to access tumor gene expression data of GBM patients for OPN/SPP1 and macrophage marker genes (Study: Glioblastoma, TCGA Provisional, mRNA Expression z-scores [microarray], accessed March 2017).13,14 Pearson correlation was calculated using GraphPad Prism 7.
Results
Glioblastoma-Associated Microglia/Monocytes Express OPN in Human Glioblastoma
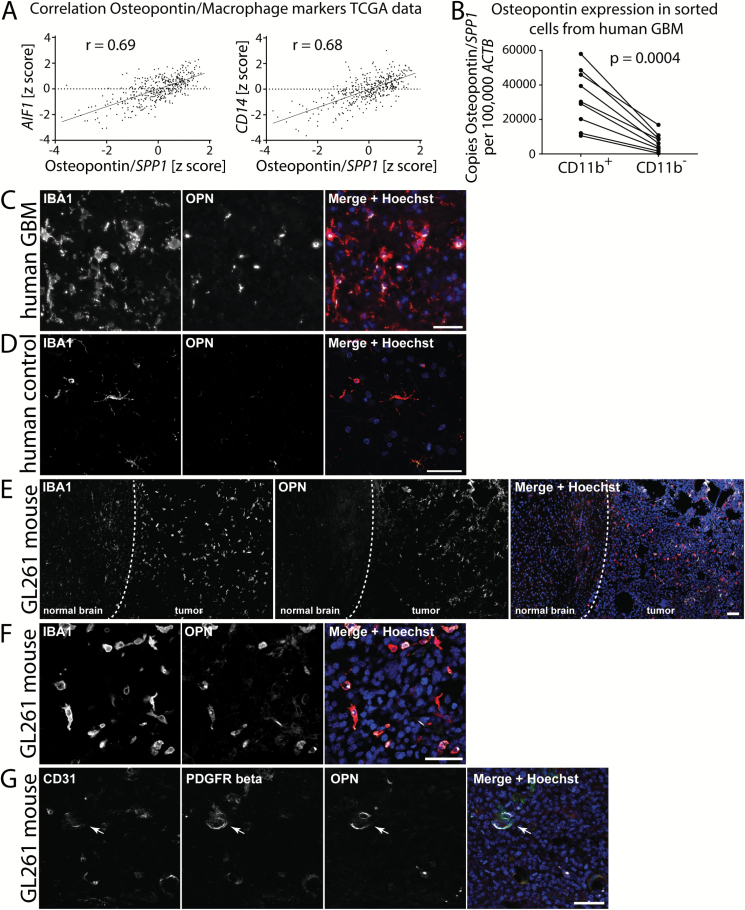
We queried the database of The Cancer Genome Atlas (TCGA) for OPN/SPP1 to identify coexpressed markers. Among the positively correlated genes were several markers for microglia and macrophages/monocytes, such as apoptosis inducing factor 1 (AIF1)/Iba1 (Pearson’s correlation r = 0.69), protein tyrosine phosphatase receptor type C (PTPRC)/CD45 (r = 0.69), CD14 (r = 0.68), CD68 (r = 0.66), integrin subunit alpha M (ITGAM)/CD11b (r = 0.47), and hepatitis A virus cellular receptor 2 (HAVCR2) (r = 0.76), indicating that there is a positive correlation between expression of GAM-specific genes and OPN/SPP1 in the tumor tissue (Fig. 1A, Supplementary Figure S1). In contrast, OPN/SPP1 expression did not significantly correlate with tumor cell markers (Supplementary Fig. S1).
Fig. 1.
OPN expression in human and mouse glioblastoma. (A) TCGA gene expression data show that OPN expression in human GBM whole tumor samples strongly correlates with the expression of AIF1 (Iba1) and CD14. Pearson correlation was used to calculate correlation coefficients. (B) CD11b+ sorted cells from human GBM samples express higher levels of OPN than the CD11b− cell fraction from the same tumors. Analysis was done using Student’s t-test. (C) OPN staining in human GBM samples predominantly co-localizes with Iba1+ staining; 40x objective. (D) OPN is sparsely expressed in Iba1+ microglia in human autopsy control brain tissue. (E) OPN is strongly expressed in the tumor tissue of GL261 mouse glioblastoma. Dashed line represents the tumor border. (F) OPN staining in mouse GL261 tumors predominantly co-localizes with Iba1+ staining. (G) PDGFRβ+ pericytes lining CD31+ blood vessels also partially express OPN in GL261 glioblastoma (arrow). Bars represent 50 µm.
We sorted CD11b+ and CD11b− cells from 9 human GBM samples and performed qRT-PCR to measure the overall OPN/SPP1 expression in both cell fractions. We found that OPN/SPP1 was expressed at higher levels in the CD11b+ fractions compared with the CD11b− fraction in all tumors examined (P = 0.0004; Fig. 1B). Using isoform-specific primers we next analyzed which OPN/SPP1 transcript variants were expressed. We found that transcript variant 1 was expressed at the highest levels, followed by variants 2, 5, and 4. We did not detect any expression of transcript variant 3 (Supplementary Figure S2A–D). We used primers for AIF1/Iba1 and glial fibrillary acidic protein (GFAP) to assess the purity of our cell fractions (Supplementary Figure S2E, F).
To characterize the presence of OPN in GBM in more detail, we co-stained sections from 9 human GBM samples for Iba1 and OPN. OPN staining could be observed in all 9 GBM samples. Diffuse OPN staining was detected throughout the tumor tissue. In addition, we found highly localized OPN staining that was predominantly co-localized to Iba1+ cells in all GBM sections stained (Fig. 1C, Supplementary Figure S3). To a lesser degree, we also detected regional OPN staining that co-localized with Iba1− cells in several tumors (Supplementary Figure S4). OPN staining in Iba1+ cells was not uniformly distributed across the tumor and did not correlate with a specific amoeboid or ramified morphology of Iba1+ cells.
We stained 3 human nontumor brain samples for Iba1 and OPN and detected considerably less OPN staining compared with human GBM sections. However, we also detected OPN+ Iba1+ cells in normal tissue samples, although the abundance was low (Fig. 1D).
Glioblastoma-Associated Microglia/Monocytes Express OPN in Murine Glioblastoma
To characterize murine GL261 GBMs we stained sections of GL261-implanted mouse brains for Iba1 and OPN. The highest abundance of OPN was detected in the tumor core regions, but a robust OPN staining could be detected also in the tumor border regions. We observed only weak staining for OPN in the tissue directly adjacent to the actual tumor or in the cortex of the contralateral site (Fig. 1E). In the tumor, we observed a weak diffuse OPN staining as well as highly localized staining that co-localized to Iba1+ cells. We found that in the tumor core regions, 33% of all Iba1+ cells stained positive for OPN and 62% of the cellular OPN was localized to Iba1+ cells. OPN-producing Iba1+ cells were usually found in clusters and were of amoeboid shape (Fig. 1F). Since OPN expression was previously described in pericytes, we next co-stained GL261 tumor sections for CD31, platelet derived growth factor receptor beta (PDGFRβ), and OPN and found that few vessel-lining PDGFRβ+ pericytes also expressed OPN (Fig. 1G).
OPN Is Expressed by Brain-Resident Microglia and Periphery-Derived Macrophages/Monocytes
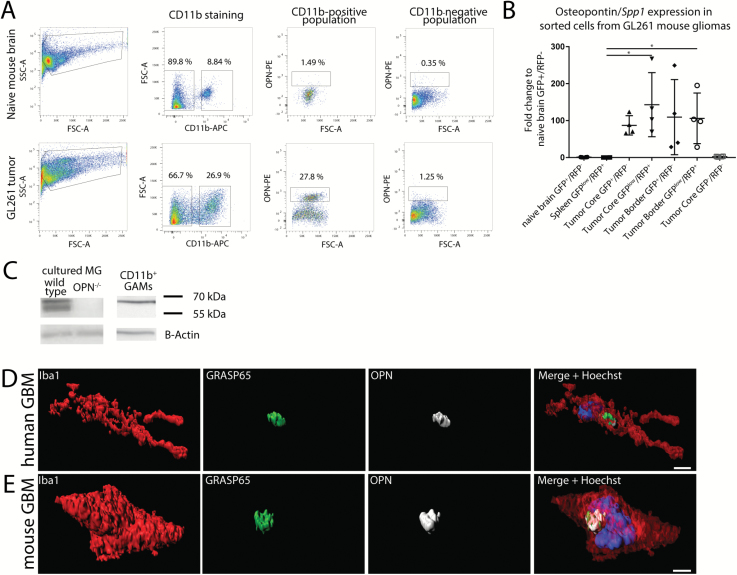
We dissociated naïve mouse brains (n = 2) and tissue from GL261 tumors (n = 4) into single-cell suspensions, then permeabilized and stained the cells for CD11b and OPN. We found that in naïve brain 1.04% (±0.69 SEM) of the CD11b+ cells and 0.89% (±0.57) of the CD11b− cells stained positive for OPN. In contrast, in GL261 tumors a significantly higher percentage of CD11b+ cells compared with CD11b− cells stained positive for OPN (23.49% [±6.39] vs 1.21% [±0.22], P = 0.0162) (Fig. 2A, Supplementary Figure S5A–B).
Fig. 2.
OPN expression in GL261 mouse glioblastoma. (A) Flow cytometry analysis shows that in naïve mouse brains (n = 2) only a few cells stain positive for OPN, while in GL261 tumors (n = 4) predominantly CD11b+ GAMs produce OPN. Percentages of cell populations shown are from one individual tumor and naïve mouse, respectively. (B) Quantitative RT-PCR from sorted cell populations from GL261 tumors shows that glioblastoma-associated GFP+/RFP− brain-resident microglia, as well as GFPlow/RFP+ periphery-derived macrophages/monocytes express OPN, while microglia from naïve brain, spleen monocytes, or GFP−/RFP− cells in the tumor express only little OPN. One-way ANOVA was used to calculate significances. Error bars represent SD. (C) Cultured wild type microglia express both the secreted (60 kDa) and the intracellular forms of OPN (56 kDa), whereas CD11b+ GAMs from GL261 tumors express only the secreted form. (D, E) OPN staining in Iba1+ cells co-localizes with the Golgi marker GRASP65 in human GBM samples (D) and mouse GL261 tumors (E). Bar represents 3 µm.
In order to distinguish between intrinsic microglia and periphery-derived monocytes we intracranially injected GL261 cells into Cx3cr1GFP/wtCcr2RFP/wt mice (Supplementary Figure S5C–D). OPN/SPP1 was expressed only at very low levels in green flourescent protein–positive/red flourescent protein–negative (GFP+/RFP−) microglia sorted from naïve brains, as well as in GFPlow/RFP+ macrophages/monocytes sorted from naïve spleens. In turn, OPN/SPP1 was expressed at high levels in GFP+/RFP− microglia, as well as in GFPlow/RFP+ macrophages/monocytes in both the tumor core and border region. The GFP−/RFP− cell fraction displayed only very low expression of OPN/SPP1, indicating that GAMs are the main source in this tumor model (Fig. 2B). Using isoform-specific primers, we next analyzed which OPN/SPP1 transcript variants were expressed. We found that the transcript variants 3–4 were expressed at the highest levels, followed by variants 1–2 and 5 (Supplementary Figure S5E, F).
GAMs Express Only the Secreted Form of OPN
We investigated the presence of OPN in primary neonatal microglial cultures, GL261 cells, and magnetic affinity cell sorted CD11b+ GAMs from GL261 tumors by SDS-PAGE and western blot. We could detect 2 OPN bands in lysates from cultured wild type microglia, whereas these bands were absent in cultured OPN−/− microglia and GL261 cells (Fig. 2C, Supplementary Figure S6A). The slightly larger band migrated at around 60 kDa and corresponded to the secreted form of OPN (sOPN). In addition, we could detect a second, slightly smaller band that migrated at around 56 kDa and represented the intracellular form of OPN (iOPN).15,16 Stimulation with GL261-conditioned medium did not change the expression of sOPN or iOPN after 24 h incubation. In contrast, in CD11b+ sorted GAMs we could detect only the 60 kDa large band that corresponds to sOPN, whereas iOPN was completely absent.
In accordance with this, OPN staining in Iba1+ cells in human and GL261 sections was predominantly localized to perinuclear regions and co-localized with Golgi reassembly-stacking protein of 65 kDa (GRASP65), a marker for the Golgi apparatus, indicating that most of the OPN produced by Iba1+ GAMs may be secreted (Fig. 2D, E, Supplementary Figure S6B–E). To exclude the possibility that GAMs phagocytosed OPN, we co-labeled human and mouse GAMs with lysosomal-associated membrane protein 1 (LAMP1), a marker for lysosomes that, however, can also be found in the Golgi apparatus.17 In human GAMs, LAMP1 staining was localized to specific cell compartments, whereas OPN staining was predominantly restricted to the area previously identified as the Golgi apparatus. LAMP1 staining did partially overlap with OPN staining in the Golgi area; however, OPN staining did not co-localize with the other areas that stained positive for LAMP1 (Supplementary Figure S6F). In murine GAMs, LAMP1 staining was also restricted to specific cell compartments and did not overlap with the OPN staining (Supplementary Figure S6G).
Loss of Microenvironment-Derived OPN Enhances Tumor Growth and Disease Progression
To evaluate the role of microenvironment-derived OPN on tumor growth and disease progression, we intracranially implanted GL261 cells into OPN−/− and wild type control mice. We chose the GL261 model as these cells do not express significant levels of OPN, allowing us to investigate the effect of OPN ablation in a system where OPN is predominantly derived from the microenvironment.
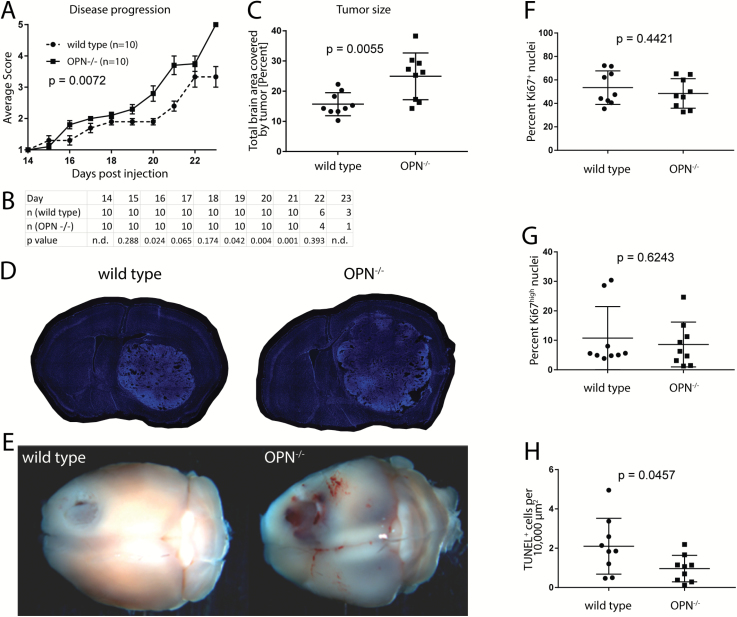
OPN−/− mice displayed a significantly faster progression of tumor-associated symptoms compared with control wild type mice (P = 0.0072; Fig. 3A, B, Supplementary Figure S7). In order to compare the tumor size between the 2 groups, we prepared 40-µm-thick sections from tumor-bearing brains and measured the percentage of brain area that was occupied by the tumor. Consistent with the findings from the disease progression, tumors in OPN−/− mice were significantly larger compared with wild type control mice (P = 0.0055; Fig. 3C–E).
Fig. 3.
Loss of microenvironment-derived OPN expression enhances glioblastoma growth. (A) OPN−/− GL261-implanted animals show a significantly faster disease progression compared with wild type control animals (n = 10 animals per group). Disease scores are defined in Materials and Methods. Significance was calculated using linear regression analysis. (B) Animal numbers in each group and P-values showing significantly different disease scores between groups are listed for each day. (C) Tumors in OPN−/− mice are significantly larger than tumors in wild type control mice (n = 9 animals per group). (D) Representative 40-µm-thick sections of wild type and OPN−/− GL261-bearing mouse brains. Sections were stained with Hoechst. (E) Representative wild type and OPN−/− GL261-implanted brains 21 days post-injection. (F, G) No difference in the percentage of Ki67+ (F) and Ki67high (G) cells could be detected between tumors in OPN−/− and wild type mice. (H) Tumors in OPN−/− mice show less TUNEL+ cells per area than tumors in wild type mice. Analysis was done using Student’s t-test (B, C, F, G, H). Error bars represent SEM (A) or SD (C, F, G, H).
To investigate why tumors implanted into OPN−/− mice grew faster compared with tumors in wild type mice, we stained 40-µm-thick sections of tumor brains (n = 9 animals for each genotype) for Ki67, a marker for proliferating cells. We analyzed 5–6 randomly distributed fields of view for each tumor and determined the percentage of Ki67+ cells by dividing the number of Ki67+ nuclei by the total number of Hoechst+ nuclei. We did not see a significant difference in the percentage of Ki67+ cells between both genotypes. Around 53.5% of all cells (±4.8 SEM) were positive for Ki67 in tumors in wild type animals versus 48.5% (±4.2) in tumors in OPN−/− mice (P = 0.44; Fig. 3F). When considering only Ki67high cells, we found that 10.75% (±3.6) of all cells in tumors in wild type animals and 8.57% (±2.5) of all cells in tumors in OPN−/− animals were positive (P = 0.62; Fig. 3G). In turn, TUNEL staining (n = 9 animals for each genotype) revealed a significantly higher percentage of TUNEL+ cells in tumors in wild type animals (2.10% ± 0.47 SEM) compared with tumors in OPN−/− animals (0.96% ± 0.23) (P = 0.0457; Fig. 3H).
Loss of Microenvironment-Derived OPN Alters the Tumor Microvasculature
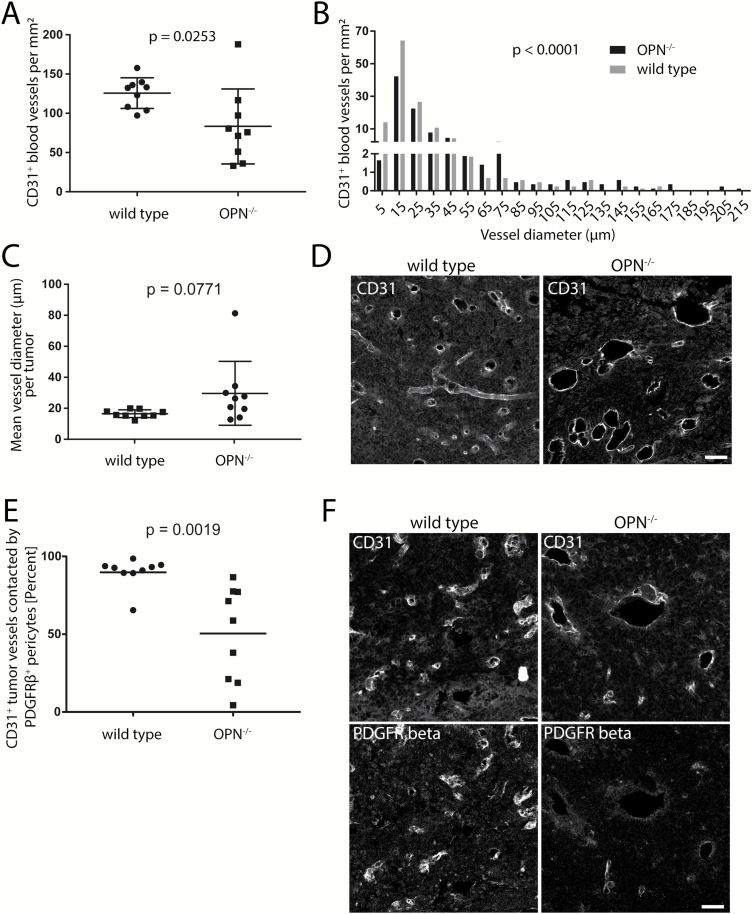
We observed larger macroscopically visible hemorrhage in tumors in OPN−/− mice (8 of 9 tumors) compared with tumors in wild type mice (2 of 9 tumors) (Fig. 3E). To investigate differences in the tumor vasculature, we stained 40-µm-thick sections (n = 9 animals per genotype) for CD31 and measured the diameter of tumor blood vessels. Tumors in OPN−/− mice had a lower number of blood vessels per square millimeter in general (P = 0.0253; Fig. 4A) and reduced numbers specifically of microvessels with a diameter of <5 µm (1.64 vs 13.96 vessels per mm2) and a diameter between 5 and 15 µm (42.28 vs 64.24 vessels per mm2) in OPN−/− mice versus wild type mice, respectively. In turn, tumors in OPN−/− mice had a higher frequency of vessels with a diameter greater than 65 µm (Fig. 4B). Finally, the mean tumor vessel diameter showed little variability between different wild type animals, whereas we observed an increased variability between different OPN−/− mice as well as a trend toward larger vessels (P = 0.0771; Fig. 4C, D).
Fig. 4.
Loss of microenvironment-derived OPN alters the tumor microvasculature. (A) Tumors in OPN−/− mice had a significantly lower number of CD31+ blood vessels per mm2 compared with wild type mice. (B) Tumors in OPN−/− mice had a reduction specifically of microvessels with a diameter of <5 µm and 5–15 µm. (C) A trend toward larger vessel diameter of CD31+ blood vessels was detected in tumors in OPN−/− mice compared with wild type mice. (D) Representative images of CD31+ vessels in tumors in OPN−/− and wild type mice. (E) CD31+ tumor vessels in OPN−/− mice show significantly less co-localization with PDGFRβ+ pericytes. (F) Representative images from tumor sections stained with CD31 and PDGFRβ. Bars represents 50 µm. Analysis was done using Student’s t-test (A, B, C, E). Error bars represent SD.
OPN−/− mice have been shown to exhibit defects in the reestablishment of blood–brain barrier integrity after stroke, which was in part mediated by inhibited polarization of reactive astrocytes.18 We stained tumor slides from wild type and OPN−/− mice for CD31 and GFAP and found that GFAP+ astrocytes were mostly located surrounding the tumor tissue, whereas the actual tumor tissue was mostly devoid of GFAP+ cells (Supplementary Figure S5A). Likewise, aquaporin-4 (AQP4) staining was restricted to the outer tumor border regions, whereas we did not observe any AQP4 staining in the tumor core regions (Supplementary Figure S5B). We did not observe a difference between both genotypes.
As we detected OPN expression in tumor vessel–lining pericytes in wild type tumors, we next co-stained tumor sections from wild type and OPN−/− mice for CD31 and PDGFRβ. We found a significant reduction of CD31+ vessels that were lined by PDGFRβ+ pericytes (P = 0.0019). While in wild type animals 89.8% (±3.2 SEM) of CD31+ tumor vessels were lined by PDGFRβ+ pericytes, only 50.4% (±10.2) of vessels were lined by pericytes in OPN−/− mice (Fig. 4E, F).
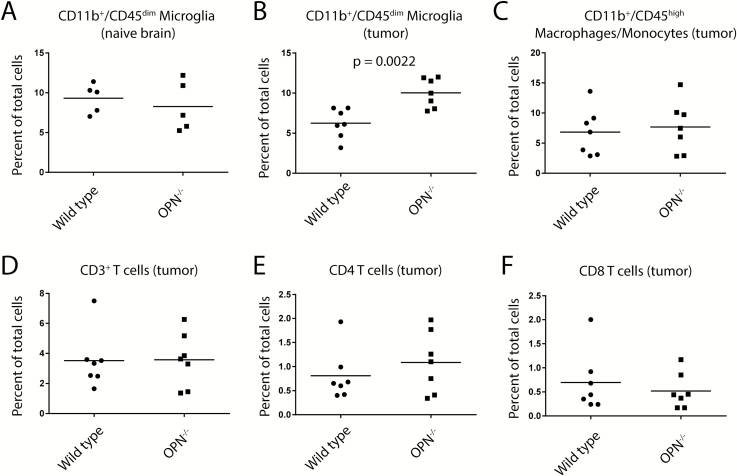
Increased Accumulation of CD11b+/CD45low Microglia in OPN−/− Tumors
We intracranially implanted GL261 cells into OPN−/− and wild type mice, sacrificed the animals between days 19 and 21 post-injection, and analyzed the immune cell infiltrates using flow cytometry. We used CD11b and CD45 as markers to distinguish between brain-resident microglia and periphery-derived macrophages/monocytes, as well as CD3, CD4, and CD8 to analyze the composition of the T-cell populations in these tumors.
We first analyzed the percentage of CD11b+/CD45low brain-resident microglia in naïve brains of OPN−/− and wild type control mice and found no difference between both genotypes (9.3% ± 0.8 SEM versus 8.3% ± 1.4 of all cells, respectively, P = 0.53) (Fig. 5A, Supplementary Figure S6A). In tumor-bearing animals we detected a significantly higher abundance of CD11b+/CD45low brain-resident microglia in OPN−/− mice compared with wild type mice (10.0% ± 0.69 vs 6.2% ± 0.7 of all cells, respectively, P = 0.0022) but no significant change in the abundance of CD11b+/CD45high periphery-derived macrophages/monocytes (7.7% ± 1.6 vs 6.8% ± 1.5 of all cells, respectively, P = 0.70) (Fig. 5B, C, Supplementary Figure S6B). Furthermore, we found no significant difference in the abundance of CD3+ T cells (3.57% ± 0.7 vs 3.52% ± 0.7 of all cells, respectively, P = 0.93), CD3+CD4+ T cells (1.09% ± 0.2 vs 0.81% ± 0.2 of all cells, respectively, P = 0.25), or CD3+CD8+ T cells (0.52% ± 0.14 vs 0.70% ± 0.24 of all cells, P = 0.527) in the tumor tissue between OPN−/− and wild type mice (Fig. 5D–F).
Fig. 5.
Increased accumulation of CD11b+/CD45low microglia in tumors in OPN−/− versus wild type mice. (A, B) We observed no differences in the frequency of CD11b+/CD45low microglia between naïve OPN−/− and wild type brains (A) but an increased frequency of these cells in GL261 tumors (B). (C) We observed no differences in the frequency of CD11b+/CD45+ macrophages/monocytes in GL261 tumors in OPN−/− and wild type mice. (D–F) We observed no differences in the frequency of CD3+ (D), CD3+CD4+ (E), or CD3+CD8+ (F) T cells in GL261 tumors in OPN−/− and wild type mice. Analysis was done using Student’s t-test.
OPN−/− Glioblastoma-Associated Microglia/Monocytes Do Not Exhibit an Altered Expression of Pro- or Anti-Inflammatory Cytokines
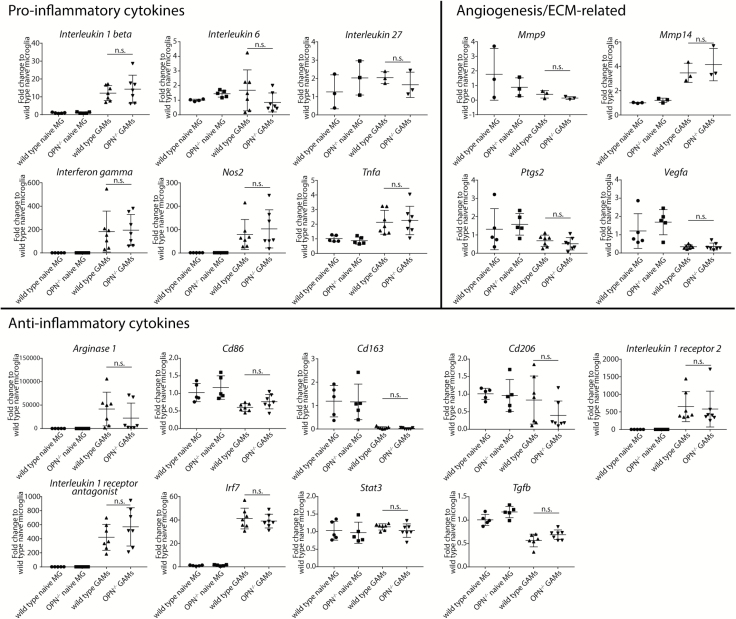
We sorted CD11b+ cells from OPN−/− and wild type naïve and tumor-bearing brains and measured the gene expression of several pro- and anti-inflammatory effector proteins and 4 genes known to play a role in angiogenesis and/or remodeling of extracellular matrix via qRT-PCR.
We did not detect a different expression of the investigated pro- (IL1b, IL6, IL27, Ifng, Nos2, and Tnfα) or anti-inflammatory (Arg1, Cd86, Cd163, Cd206, IL1r2, IL1rn, Irf7, Stat3, Tgfb) transcripts between wild type or OPN−/− cells, indicating that wild type and OPN−/− naïve microglia and GAMs do not exhibit a significantly different activation status. In addition, we could not detect any difference in the expression of Mmp9, Mmp14, Ptgs2, and Vegfa (Fig. 6).
Fig. 6.
OPN−/− and wild type GAMs do not differ in the expression of selected pro- or anti-inflammatory cytokines. We observed no differences in the expression of pro- and anti-inflammatory genes, as well as extracellular matrix- and angiogenesis-related genes in CD11b+ sorted cells from naïve and GL261-bearing wild type and OPN−/− brains. Analysis was done using one-way ANOVA. Error bars represent SD.
Discussion
In the present study we observed that GAMs are an important source for OPN in human and GL261 GBM in vivo. In the naïve brain, microglia, at least in mouse, express only low levels of OPN and the expression levels are upregulated when these cells are confronted with glioma. These results are in line with a study showing that macrophages and neutrophils express OPN in GBM.19 Our data, however, do not exclude that human GBM cells also express OPN. Previous reports have demonstrated that cultured GBM cells express OPN in vitro and that in vivo tumor growth is reduced when OPN-depleted GBM cells are transplanted into mice.5,8,20 In addition, a recent study has shown that OPN is expressed by more stemlike GBM initiating cells.21 In fact, we also found small populations of OPN-expressing Iba1− cells in human GBM sections. We observed small clusters of OPN-expressing cells in GL261 tumors implanted in OPN−/− mice, indicating that also some GL261 cells express OPN. The fact that OPN might be expressed by more stemlike tumor cells might also explain why so many tumor cells express OPN in vitro, since sphere-forming cultures enrich for more stemlike cells.
It is puzzling that the depletion of OPN from the GBM cells per se versus the host microenvironment has such opposing results on GBM growth in vivo. An explanation might be the difference in the cell lines used in different studies. Studies investigating the role of GBM cell–derived OPN used cell lines that depend on OPN for their biology, as seen by the diminished growth and sphere-forming potential in vitro and in vivo, as well as differences in the formation of tumor–microenvironment interactions in vivo, after OPN knockdown.5–8,20,21 In turn, the GL261 cell line and other GBM lines do not express OPN/SPP1 in vitro and do not seem to depend on OPN for their growth or stemness in vitro.7,21 As these cells do not require OPN for their growth, the lack of host-derived OPN does not negatively affect tumor growth in vivo. This allowed us to evaluate the effect of OPN deficiency on the interaction between the tumor and the microenvironment, where other factors, such as the increased abundance of CD11b+/CD45low microglia, and other yet unknown factors, in OPN−/− mice might be beneficial for tumor growth in vivo. In turn, using this model we did not investigate the effect of host-derived OPN on OPN-responsive tumor cells—such as invasiveness/aggressiveness, stemness, and response to treatment. Hence, different results might be expected when using an in vivo model where the GBM cells themselves are dependent on OPN signaling.4
Our results are in line with studies that investigated the role of microenvironment-derived OPN in chemically induced models of hepatocellular carcinoma and squamous cell carcinoma, as well as a virally induced model of prostate cancer, which also found a more severe tumor progression in OPN−/− mice compared with wild type mice.15,22–24 In contrast, 2 studies on hepatocellular carcinoma and melanoma found a decrease in disease burden in OPN−/− compared with wild type mice.25,26 The effect of OPN on tumor–stroma interactions seems to differ between tumors in different tissues. Danzaki et al found an enhanced infiltration of leukocytes into tumors in OPN−/− mice.22 We did not find an altered infiltration of periphery-derived myeloid cells but an increased accumulation of brain-resident microglia. Furthermore, they did not see an altered tumor vasculature, whereas we saw a significant reduction in the numbers of microvessels.22 Additionally, tumor-associated macrophages in different cancer types might produce different forms of OPN. Fan et al found that tumor-associated macrophages in hepatocellular carcinoma predominantly produce the intracellular form of OPN, which interferes with the Toll-like receptor signaling protein MyD88 and strongly suppresses the production of pro-inflammatory cytokines.15 We found that GAMs express only the secreted form of OPN.
Using flow cytometry we show that there is an increased frequency of CD11b+/CD45low microglia but not of CD11b+/CD45+ macrophages/monocytes or CD3+, CD3+CD4+, or CD3+CD8+ T-cell populations in tumor-bearing OPN−/− compared with tumor-bearing wild type mice. By crossing OPN−/− mice with Rag2−/− mice, Danzaki et al found that the adaptive immune response does not play a critical role in the accelerated tumor progression.22 The accelerated disease progression in OPN−/− mice might at least in part be attributed to the enhanced frequency of CD11b+/CD45low microglia, as these cells have previously been shown to support tumor progression. We furthermore analyzed the expression of several activation markers in a CD11b+ sorted microglia/monocyte population from OPN−/− and wild type mice but did not find any significant differences in the expression of these markers. A more in-depth analysis will be necessary to study the differences between knockout and wild type cells.
One of the most prominent differences between tumors in wild type and OPN−/− mice was the altered tumor vasculature, since we found a significant reduction in the number of tumor microvessels in OPN−/− mice but in turn found a higher frequency of vessels with a diameter greater than 65 µm. OPN has previously been reported to promote angiogenesis.27 We tested whether sorted GAMs from wild type and knockout animals express altered levels of Vegfa or Ptgs2 but could not detect a difference between both genotypes. In addition, we detected more hemorrhage in tumors in knockout animals and found that blood vessels in these tumors were significantly less covered with PDGFRβ+ pericytes. Since we also detected that some tumor vessel–lining pericytes express OPN, it might act as an autocrine factor for the pericytes to promote vessel growth. It is also possible that GAM-derived OPN affects the pericytes.
Supplementary Material
Supplementary material is available at Neuro-Oncology online.
Funding
This work was supported by the Deutsche Forschungsge meinschaft (KE 329/30-1, SY 144/4-1, and SZ 350/1-1), NeuroCure, and grants from the National Institutes of Health (U01CA160882-01A1) and the Berlin Institute of Health.
Conflict of interest statement. The authors declare no conflicts of interest.
Supplementary Material
Acknowledgments
We thank Regina Piske, Nadine Scharek, Julio Vazquez Lopez, and David L McDonald for excellent technical assistance.
References
- 1. Reifenberger G, Wirsching HG, Knobbe-Thomsen CB, Weller M. Advances in the molecular genetics of gliomas—implications for classification and therapy. Nat Rev Clin Oncol. 2016;14(7):434–452. [DOI] [PubMed] [Google Scholar]
- 2. Hambardzumyan D, Gutmann DH, Kettenmann H. The role of microglia and macrophages in glioma maintenance and progression. Nat Neurosci. 2016;19(1):20–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Subraman V, Thiyagarajan M, Malathi N, Rajan ST. OPN-revisited. J Clin Diagn Res. 2015;9(6):ZE10–ZE13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Pietras A, Katz AM, Ekström EJ et al. . Osteopontin-CD44 signaling in the glioma perivascular niche enhances cancer stem cell phenotypes and promotes aggressive tumor growth. Cell Stem Cell. 2014;14(3):357–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lamour V, Henry A, Kroonen J et al. . Targeting osteopontin suppresses glioblastoma stem-like cell character and tumorigenicity in vivo. Int J Cancer. 2015;137(5):1047–1057. [DOI] [PubMed] [Google Scholar]
- 6. Henry A, Nokin MJ, Leroi N et al. . New role of osteopontin in DNA repair and impact on human glioblastoma radiosensitivity. Oncotarget. 2016;7(39):63708–63721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lamour V, Le Mercier M, Lefranc F et al. . Selective osteopontin knockdown exerts anti-tumoral activity in a human glioblastoma model. Int J Cancer. 2010;126(8):1797–1805. [DOI] [PubMed] [Google Scholar]
- 8. Ellert-Miklaszewska A, Wisniewski P, Kijewska M et al. . Tumour-processed osteopontin and lactadherin drive the protumorigenic reprogramming of microglia and glioma progression. Oncogene. 2016;35(50):6366–6377. [DOI] [PubMed] [Google Scholar]
- 9. Szulzewsky F, Arora S, de Witte L et al. . Human glioblastoma-associated microglia/monocytes express a distinct RNA profile compared to human control and murine samples. Glia. 2016;64(8):1416–1436. [DOI] [PubMed] [Google Scholar]
- 10. Szulzewsky F, Pelz A, Feng X et al. . Glioma-associated microglia/macrophages display an expression profile different from M1 and M2 polarization and highly express Gpnmb and Spp1. PLoS One. 2015;10(2):e0116644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Vinnakota K, Hu F, Ku MC et al. . Toll-like receptor 2 mediates microglia/brain macrophage MT1-MMP expression and glioma expansion. Neuro Oncol. 2013;15(11):1457–1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kempermann G, Gast D, Kronenberg G, Yamaguchi M, Gage FH. Early determination and long-term persistence of adult-generated new neurons in the hippocampus of mice. Development. 2003;130(2):391–399. [DOI] [PubMed] [Google Scholar]
- 13. Cerami E, Gao J, Dogrusoz U et al. . The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2(5):401–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gao J, Aksoy BA, Dogrusoz U et al. . Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6(269):pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Fan X, He C, Jing W et al. . Intracellular osteopontin inhibits toll-like receptor signaling and impedes liver carcinogenesis. Cancer Res. 2015;75(1):86–97. [DOI] [PubMed] [Google Scholar]
- 16. Zhao K, Zhang M, Zhang L et al. . Intracellular osteopontin stabilizes TRAF3 to positively regulate innate antiviral response. Sci Rep. 2016;6:23771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cook NR, Row PE, Davidson HW. Lysosome associated membrane protein 1 (Lamp1) traffics directly from the TGN to early endosomes. Traffic. 2004;5(9):685–699. [DOI] [PubMed] [Google Scholar]
- 18. Gliem M, Krammes K, Liaw L, van Rooijen N, Hartung HP, Jander S. Macrophage-derived osteopontin induces reactive astrocyte polarization and promotes re-establishment of the blood brain barrier after ischemic stroke. Glia. 2015;63(12):2198–2207. [DOI] [PubMed] [Google Scholar]
- 19. Atai NA, Bansal M, Lo C et al. . Osteopontin is up-regulated and associated with neutrophil and macrophage infiltration in glioblastoma. Immunology. 2011;132(1):39–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jan HJ, Lee CC, Shih YL et al. . Osteopontin regulates human glioma cell invasiveness and tumor growth in mice. Neuro Oncol. 2010;12(1):58–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kijewska M, Kocyk M, Kloss M et al. . The embryonic type of SPP1 transcriptional regulation is re-activated in glioblastoma. Oncotarget. 2017;8(10):16340–16355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Danzaki K, Kanayama M, Alcazar O, Shinohara ML. Osteopontin has a protective role in prostate tumor development in mice. Eur J Immunol. 2016;46(11):2669–2678. [DOI] [PubMed] [Google Scholar]
- 23. Crawford HC, Matrisian LM, Liaw L. Distinct roles of osteopontin in host defense activity and tumor survival during squamous cell carcinoma progression in vivo. Cancer Res. 1998;58(22):5206–5215. [PubMed] [Google Scholar]
- 24. Hsieh YH, Margaret Juliana M, Ho KJ et al. . Host-derived osteopontin maintains an acute inflammatory response to suppress early progression of extrinsic cancer cells. Int J Cancer. 2012;131(2):322–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lee SH, Park JW, Woo SH et al. . Suppression of osteopontin inhibits chemically induced hepatic carcinogenesis by induction of apoptosis in mice. Oncotarget. 2016;7(52):87219–87231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kumar S, Sharma P, Kumar D, Chakraborty G, Gorain M, Kundu GC. Functional characterization of stromal osteopontin in melanoma progression and metastasis. PLoS One. 2013;8(7):e69116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Dai J, Peng L, Fan K et al. . Osteopontin induces angiogenesis through activation of PI3K/AKT and ERK1/2 in endothelial cells. Oncogene. 2009;28(38):3412–3422. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.