ABSTRACT
The RET receptor tyrosine kinase is crucial for the development of the enteric nervous system (ENS), acting as a receptor for Glial cell line-derived neurotrophic factor (GDNF) via GFR co-receptors. Drosophila has a well-conserved RET homolog (Ret) that has been proposed to function independently of the Gfr-like co-receptor (Gfrl). We find that Ret is required for development of the stomatogastric (enteric) nervous system in both embryos and larvae, and its loss results in feeding defects. Live imaging analysis suggests that peristaltic waves are initiated but not propagated in mutant midguts. Examination of axons innervating the midgut reveals increased branching but the area covered by the branches is decreased. This phenotype can be rescued by Ret expression. Additionally, Gfrl shares the same ENS and feeding defects, suggesting that Ret and Gfrl might function together via a common ligand. We identified the TGFβ family member Maverick (Mav) as a ligand for Gfrl and a Mav chromosomal deficiency displayed similar embryonic ENS defects. Our results suggest that the Ret and Gfrl families co-evolved before the separation of invertebrate and vertebrate lineages.
KEY WORDS: Neural development, Enteric nervous system, Ret signaling, GDNF signaling, Axon branching, Drosophila
Summary: Loss of the Ret receptor tyrosine kinase causes larval feeding defects associated with abnormal axon innervation and defective peristalsis of the midgut.
INTRODUCTION
The RET (rearranged during transfection) receptor tyrosine kinase is the leading susceptibility locus for Hirschsprung's disease (HSCR), a congenital lack of neurons in the distal regions of the digestive tract (McKeown et al., 2013; Romeo et al., 1994). HSCR arises due to the abnormal migration and survival of enteric neuron precursors derived from the neural crest, which has been classified as a neurocristopathy (Zhang et al., 2014a). RET is also found to have a role in kidney development and in a subset of neuroendocrine cancers (Davis et al., 2014; Romei et al., 2016; Schuchardt et al., 1994). The ligands for RET are members of the Glial cell line-derived neurotrophic factor (GDNF) family, which act by binding to a GDNF family receptor (GFR) to activate intracellular RET signaling, or the Neural cell adhesion molecule (NCAM) (Jing et al., 1996; Paratcha et al., 2003; Treanor et al., 1996). GDNF is an important component of vertebrate brain development and maintenance, with clinical relevance to Parkinson's disease (Ibáñez and Andressoo, 2017).
GDNF ligands appeared with the emergence of jawed fish and GFRs underwent a gene expansion at the same time (Airaksinen et al., 2006; Hätinen et al., 2007; Kallijärvi et al., 2012). This expansion coincides with the appearance of the neural crest, a distinguishing structure for vertebrates. Homologs of the RET and GFR receptors are present in invertebrates but are thought to function independently of each other, with GFRs operating in conjunction with Fas2/NCAM rather than with a soluble ligand (Kallijärvi et al., 2012) and RET operating with integrins (Soba et al., 2015). In Drosophila, the RET gene (Ret) is expressed by enteric neurons and epithelial progenitor cells of the adult midgut and is required for homeostasis of these populations (Perea et al., 2017). In the Drosophila embryo, Ret is expressed in the developing stomatogastric nervous system (SNS), a population of cells that delaminate and migrate along the developing gut to form the enteric nervous system (ENS), and Ret is also expressed in the Malpighian tubules, the fly equivalent of the kidney (Hahn and Bishop, 2001; Copenhaver, 2007; Hartenstein, 1997). We previously observed expression of Gfrl promoter fragments in the developing SNS, suggesting that Ret and Gfrl might function together in this tissue (Hernández et al., 2015). Here, using CRISPR, we generated Drosophila Ret alleles and found defects in embryonic SNS formation and larval SNS function. These phenotypes led us to identify the novel TGFβ family member Maverick (Mav) as an invertebrate GFR/Ret ligand and a candidate for the ancestor of GDNF. Our results reveal remarkable similarities in the signaling mechanisms used to generate the insect SNS and the vertebrate ENS.
RESULTS
Generation and characterization of Ret alleles
The role of the Drosophila Ret gene in dendrites has previously been analyzed using a transposon insertion in the 3′ UTR of Ret and the adjacent Mcm10 gene (Soba et al., 2015). We sought to generate Ret alleles that disrupted the coding region of the gene using the CRISPR/Cas9 system. We designed guide RNAs (gRNAs) to a site immediately downstream of the signal sequence and also to the cadherin-like domain (CLD). The gRNAs were introduced as transgenic constructs and crossed to sources of Cas9 (Port et al., 2014). The frequency of induced mutations was >90% and three alleles were selected for further analysis: two separate deletions that lead to truncated proteins (LM1, LM3), and an in-frame deletion that removes a tyrosine conserved with the human RET protein (LM2) (Fig. 1A).
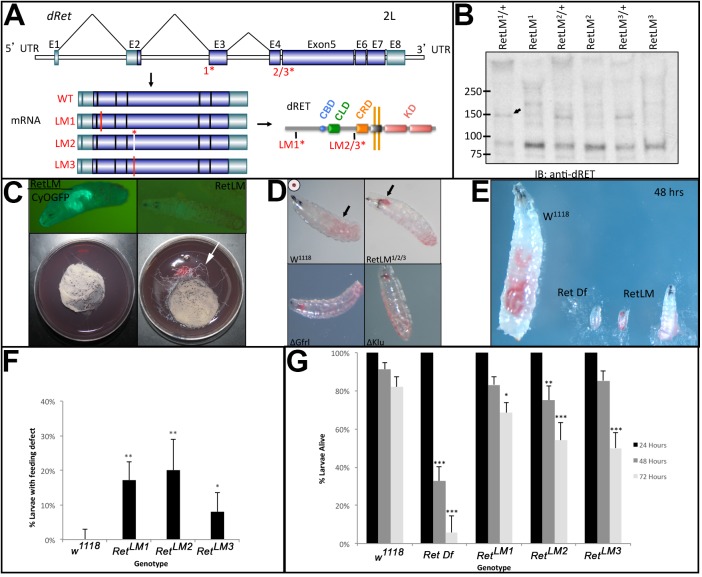
Fig. 1.
Characterization of CRISPR-induced Ret mutations. Mutations in the Drosophila Ret gene were induced using the CRISPR/Cas9 system and phenotypically characterized. (A) Ret gene with the genomic locations of the gRNAs (1*, 2/3*) indicated. Exons (E) are shown as boxes, with dark blue representing protein coding regions and light blue untranslated regions. Three mutations were characterized: two out-of-frame deletions that are predicted to truncate the Ret protein (Δ1, Δ3), and one in-frame deletion that removes six amino acids including a conserved tyrosine residue (Δ2). The positions of these mutations within the mRNAs and protein are shown. Active domains of the protein are labeled to show the calcium-binding domain (CBD), cadherin-like domain (CLD), the cysteine-rich domain (CRD), and the tyrosine kinase domains (KD). (B) Western blot of total protein from late stage (15-17) embryos from each ΔRet allele probed with an anti-Drosophila Ret antibody shows an ∼150 kDa band (arrow) corresponding to full-length Ret in controls, and lack thereof in homozygous mutants. (C) Homozygous larvae for each Ret allele appeared normal on hatching, relative to heterozygous controls (marked by GFP). On grape agar plates with yeast paste in the center, larval tracks can be seen frequently leaving the food (arrow) for the homozygotes. (D) Larvae fed for 48 h on yeast food paste stained with Carmine Red revealed a buildup of food in the midgut (arrow) in w1118 controls. Ret mutants frequently have food stuck in the pharynx and esophagus (arrow) of first and second instar larvae. Gfrl mutants display the same phenotype as a klumpfuss (klu) mutant positive control. (E) Representative image showing feeding defects in Ret mutants, which failed to grow at the same rate as w1118 over a 48 h period. A chromosomal deficiency for the Ret region [Df(2L)BSC312] also failed to grow. (F) Quantification of larval feeding defects in each of the Ret alleles revealed a statistical difference relative to w1118 control (Fisher's exact test with two tails and Bonferroni correction). w1118, n=150; RetLM1, n=207; RetLM2, n=100; RetLM3, n=150. (G) Quantification of mortality at 24 h time points after hatching (n=150, 156, 250, 100, 150 in the order of the columns from left to right). A significant drop off in survival rate was seen in Ret mutants compared with w1118 after 72 h. As expected, a deficiency for the Ret region had greater mortality. Error bars represent a 95% confidence interval and statistical significance relative to the w1118 control was assessed using the Fisher's exact test with two tails and Bonferroni correction. *P<0.05, **P<0.01, ***P<0.001.
We generated an antibody against the Ret ectodomain that recognizes a band of ∼150 kDa when Ret is expressed in cell culture. Immunoblot analysis of embryonic extracts reveals an absence of this band in all three Ret alleles (Fig. 1B), suggesting they are likely to be null alleles. Homozygous larvae were found to hatch and display a foraging phenotype previously seen in mutants with feeding defects (Fig. 1C) (de Belle et al., 1989; Zinke et al., 1999). To evaluate this phenotype, we added Carmine Red dye to yeast paste to examine feeding behavior (Melcher and Pankratz, 2005), and found Ret mutant larvae frequently had food stuck in their esophagus (Fig. 1D). In addition to this striking phenotype, foraging larvae were often found with reduced or absent food in their midguts and without food in their esophagus. These larvae were frequently immobile or sluggish in response to touch. We tested combinations of the three Ret alleles and observed feeding defects in all of them (LM1/LM2, n=55; LM1/LM3, n=40; LM2/LM3, n=46 homozygous larvae examined; Fig. S1). Ret mutant larvae displayed an eating defect 2-4 days after hatching, and failed to grow at a discernible rate (Fig. 1E).
In parallel, we tested a null allele of Gfrl, the fly GFR co-receptor homolog, and found the same phenotype (Fig. 1D). The Gfrl mutant larvae displayed the same range of feeding defects as Ret mutants, with food levels frequently reduced or absent in larvae displaying foraging and sluggish phenotypes. When larvae were separated from their heterozygous siblings on grape agar plates, 82% made it to the pupal stage (n=109), indicating a mortality rate of 18%. Under crowded conditions on grape agar plates in which both heterozygotes and homozygotes were present, homozygous Gfrl larvae displayed a developmental delay of >2 days and decreased viability (3 homozygotes out of a total of 66 larvae scored after 2 days). This suggests that the homozygotes are at a competitive disadvantage. The similarity of the feeding defects suggests that Gfrl functions with Ret for normal larval feeding, in contrast to the proposed independence of GFR in other tissues (Kallijärvi et al., 2012).
During characterization of the CRISPR Ret alleles, a recessive lethal mutation mapping 22 cM from the Ret locus was detected and removed by recombination. This allowed homozygous adults to be recovered. Larvae still displayed eating defects (Fig. 1F) and significant rates of larval mortality (Fig. 1G). Previous work had shown that Ret is strongly expressed in the developing SNS (Hahn and Bishop, 2001), and our phenotypic findings are consistent with a developmental role in this system. Similarly, the expression of Gfrl promoter fragments in the embryonic SNS (Hernández et al., 2015) and the observed feeding defects suggest that Gfrl could function with Ret in the SNS.
Embryonic developmental defects in Ret mutants
To determine the origins of the larval feeding defect, we first examined the development of the embryonic SNS. Drosophila Ret is expressed in three migrating cell clusters that give rise to the SNS (Hahn and Bishop, 2001). The principal features of the embryonic SNS are the frontal ganglion, which lies anterior to the brain commissure; the frontal nerve, which projects anteriorly from the frontal ganglion along the dorsal surface of the pharynx; the recurrent nerve that projects posteriorly to the two esophageal ganglia; and the ventricular ganglion (Fig. 2A,E,I).
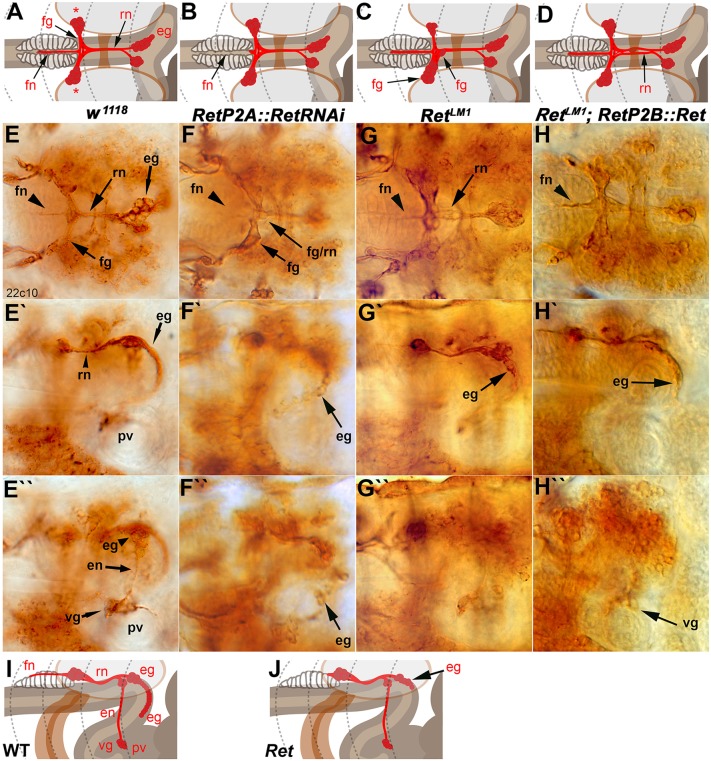
Fig. 2.
Embryonic SNS development is dependent on Ret activity. The embryonic SNS in stage 17 embryos was visualized with monoclonal antibody (mAb) 22c10 (anti-Futsch). (A) Schematic of a dorsal view of the embryonic SNS. The recurrent nerve (rn) runs from the esophageal ganglion (eg) along the esophagus underneath the supraesophageal commissure to the frontal ganglion (fg). The frontal nerve (fn) projects anteriorly from the frontal ganglion. The cell bodies of the frontal ganglion occupy lateral positions (asterisks). (B-D) The embryonic phenotypes scored in this analysis were absence or shortening of the frontal nerve (B), mispositioning of the cells of the frontal ganglion such that the two sides of the frontal ganglion were asymmetric or cells inappropriately occupied midline positions (C), and defasciculation of the recurrent nerve (D). (E) Dorsal view of a w1118 embryo labeled with 22c10 with elements of the SNS annotated as in A. (E′) Lateral view of the same embryo. The esophageal ganglia (eg) can be seen descending along the esophagus as an orderly array of cells. (E″) Alternative focal plane of the lateral embryo view. The esophageal nerve (en), which links the esophageal ganglia to the ventricular ganglion (vg), can be seen. The vg lies on top of the proventriculus (pv), which lies at the start of the anterior midgut, the most posterior portion of the SNS. Compare these images with the schematic in I. (F-F″) RetP2A-GAL4 driving a RetRNAi transgene. The frontal nerve is often shortened or absent or misguided away from the center of the pharyngeal muscles (F, arrowhead), and the frontal ganglion cells often asymmetrically misposition themselves with incomplete fasciculation into the recurrent nerve (F, arrows). Cell bodies of the esophageal ganglia cluster irregularly and more densely than in wild type (F′,F″). (G-G″) RetLM1 CRISPR allele, with SNS defects that were observed in all three alleles. The SNS phenotypic defects are comparable to those seen in the SNS-driven Ret RNAi. (H-H″) Rescue of the RetLM1 mutant by expression of a UAS-Ret transgene driven by RetP2A-GAL4. The frontal nerve visibly rescues to wild-type length, defasciculation in the recurrent nerve is diminished, and esophageal ganglia cells align and descend down the esophagus. (I) The main features of the SNS when viewed from the side of the animal. The esophageal nerve connects the smaller esophageal ganglion to the ventricular ganglion, which lies on the anterior end of the proventriculus. (J) The main phenotype observed in lateral views, namely clustering or mispositioning of the neurons of the esophageal ganglia. Absence or reduction of the frontal nerve is easily observed from the side but can also appear absent in many focal planes.
We used the w1118 stock as a control for all experiments (Fig. 2E-E″). Expression of a transgenic RNA interference (RNAi) hairpin for Ret in the developing SNS frequently led to missing or shortened frontal nerves and disrupted frontal ganglia (Fig. 2B,C,F), and to esophageal ganglia that had not migrated as far as in the control (Fig. 2F′). The same defects were seen in the CRISPR-induced Ret mutants (Fig. 2G,G′), confirming a role for Ret in embryonic SNS development.
To prove that the defects were due to a loss of Ret activity, we expressed a Ret transgene in the developing SNS of RetLM1 mutants and observed rescue of the frontal and recurrent nerve phenotypes, but not those of the frontal or esophageal ganglia (Fig. 2H,H′). We scored the defects shown in Fig. 2B-D,J and quantified them (Fig. 3A-D), confirming that the Ret CRISPR mutants and Ret RNAi have very similar phenotypic defects. None of the phenotypes documented is 100% penetrant, so variability in larval feeding might be due in part to variation in embryonic development. Among the foraging first instar larvae, we observed midgut axon morphologies strikingly similar to those of older animals (Fig. S2A). Differences between alleles might be due to background effects of particular chromosomes or to natural variability in the structures examined. Similarly, for the rescue experiments, lack of statistically significant rescue in the frontal or esophageal ganglia might be due to the variability of these structures or the promoter used might lack adequate expression. In adult flies, Ret function is required in epithelial cells of the intestine (Perea et al., 2017). The Ret-GAL4 lines used express in the epithelial lining of the embryonic midgut, particularly in the earlier stages (Hernández et al., 2015), so we cannot rule out Ret function in the gut as a cause for the SNS patterning defects. However, we used the lines (P2A and P2B) that had the lowest gut expression and that display prominent SNS neuron expression. Gut expression is minimal in the later stages of embryogenesis and undetectable in the first instar larva (Fig. S2B). Overall, the results indicate a role of Ret in the normal patterning of the embryonic SNS.
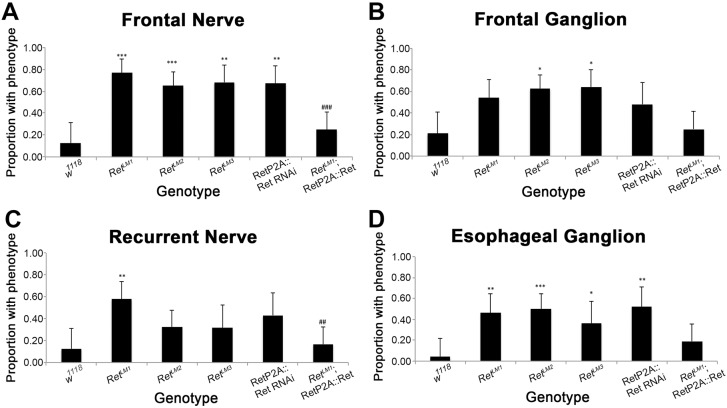
Fig. 3.
Quantification of embryonic SNS defects. (A) Absent or reduced frontal nerves were scored as phenotypes (see Fig. 2B). All mutants showed a statistically significant frequency of phenotypes. Expression of Ret rescued the phenotype, as the proportion of embryos with phenotypes was statistically different from RetLM1 mutants alone (###P=0.0001). (B) Frontal ganglion phenotypes were scored (asymmetric or misplaced cell bodies, see Fig. 2C) and tested for statistical significance. Rescue was not observed. (C) Recurrent nerve phenotypes were quantified (defasciculation, see Fig. 2D) and RetLM1 was rescued by Ret expression (##P=0.0011). (D) Esophageal ganglion (clumped or misplaced, see Fig. 2J) defects in the genotypes examined. Rescue was not observed. Error bars represent a 95% confidence interval and statistical significance relative to the w1118 control was assessed using Fisher's exact test with two tails and Bonferroni correction. Comparison of RetΔ1 and rescue of the mutation were assessed independently with the same test (#). For each panel, n=24, 26, 40, 22, 21, 36 in the order of the columns from left to right. *P<0.05, **P<0.01, ***P<0.001.
Disruption of larval midgut axons in Ret mutants
The relatively subtle nature of the embryonic SNS defects in Ret mutants led us to examine the anatomy of the larval SNS. Second and third instar (2-5 days after hatching at 25°C) Ret mutants had food stuck in their esophagus immediately anterior to the proventriculus, a food-grinding organ and valve (Fig. 1D, Fig. 4A,B). The proventriculus is an infolding of the gut at the junction of the foregut and midgut that regulates food entry into the midgut (Spieß et al., 2008). The proventricular ganglion constitutes the posteriormost cell bodies of the SNS and is located at the anterior end of the proventriculus. The proventricular ganglion forms the midgut nerves, with axons projecting to the anteriormost end of the midgut (Gonzalez-Gaitan and Jackle, 1995).
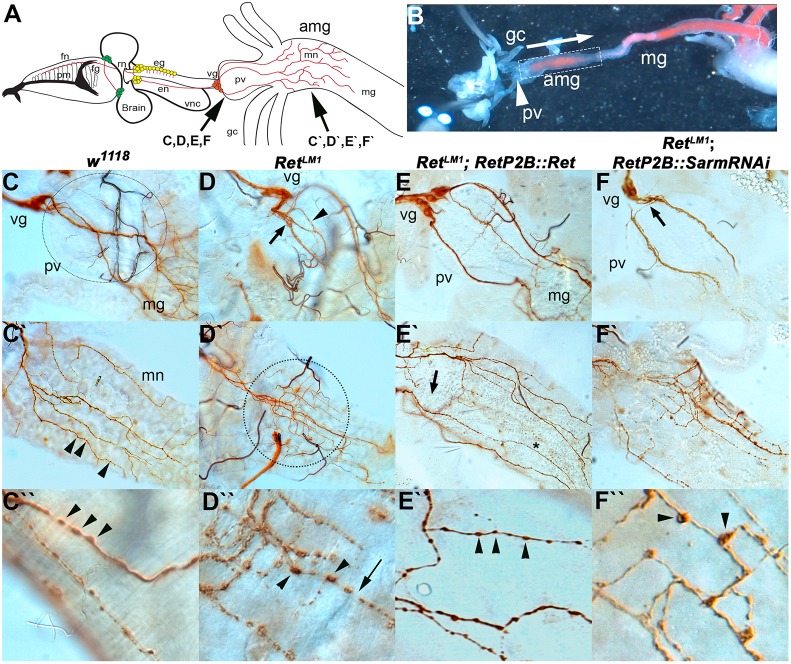
Fig. 4.
Larval midgut SNS architecture is dependent on Ret. SNS of second instar larvae was dissected and visualized with anti-Futsch (mAb 22c10) staining. (A) Larval SNS showing, from left, that the frontal nerve (fn) lies on top of the pharyngeal muscles (pm) and connects to the frontal ganglion (fg). The recurrent nerve (rn) projects anteriorly between the brain lobes to the esophageal ganglia (eg). The esophageal nerve (en) projects to the ventricular ganglion (vg) that lies at the anterior end of the proventriculus (pv). The midgut neuron (mn) axons project from the ventricular ganglion, over the proventriculus to innervate the anterior midgut (amg). The remainder of the midgut (mg) is not innervated. The ventral nerve cord (vnc) is also shown. (B) Dissected third instar larvae pinned in Drosophila S2 cell medium showing the relative positions of the proventriculus, anterior midgut and midgut. This preparation was used to observe spontaneous contractile activity in w1118 and RetLM1. Yeast paste dyed pink can be seen in the midgut. (C) In w1118 animals, the proventriculus (unstained, dotted circle indicates extent) forms at the junction of the foregut and midgut and acts as a valve regulating entry of food into the midgut. The proventriculus and midgut are innervated by cells of the ventricular ganglion, which are located on the anteriormost point of the proventriculus. Axons of the midgut neurons (brown) traverse the proventriculus as two or three tightly fasciculated bundles and only start branching on reaching the midgut. (C′) On encountering the midgut, the midgut neuron axons (brown) branch in a regular fashion to spread out over the anterior end of the midgut. (C″) Multiple small varicosities can be seen throughout the midgut axons as punctate increases in 22c10 staining (C′,C″, arrowheads). (D) RetLM1 allele with midgut SNS defects typically seen in Ret mutants. The midgut nerve axons defasciculate (arrow) a very short distance from the ventricular ganglion and show additional defasciculation (arrowhead) as they traverse the proventriculus. (D′) Upon encountering the midgut, they fail to spread out and remain confined to a much smaller area than in w1118 (dotted circle). Within this area, the midgut neuron axons display a much higher level of branching. (D″) Axonal varicosities appear larger and the distribution of the 22c10 antigen is reduced between varicosities. The appearance initially suggested that axon fragmentation is occurring. (E) Rescue of Ret mutant phenotypes using UAS-Ret driven by RetP2B-GAL4 in a RetLM1 genetic background. Axons from the ventricular ganglion crossing the proventriculus more closely resemble the axon bundles in w1118, remaining tightly fasciculated until the midgut is reached. (E′) Midgut of RetLM1 RetP2B-GAL4::UAS-Ret larva in which the axon pattern resembles w1118, with the axons spreading out to cover a large proportion of the anterior midgut. The branching frequency also resembles that in w1118. (E″) Axonal varicosities resemble w1118, being smaller with a more even distribution of 22c10 antigen between the varicosities. (F) RetP2B-GAL4 driving UAS-SarmRNAi does not rescue the aberrant branching caused by RetLM1, as the proventriculus nerve bundles defasciculate rapidly after leaving the cell bodies of the ventricular ganglion (arrow) and continue to show defasciculation as the axons traverse the proventriculus. (F′) The mn axons also branch more frequently and do not cover the same area of midgut as in w1118. (F″) The increased prominence of axonal varicosities observed in Ret mutants remains.
We examined the midgut nerves of second and third instar larvae with feeding defects by staining for Futsch (mAb 22c10) (Fig. 4). Axons normally project from the proventriculus (ventricular ganglion) neurons posteriorly in three to five well-defined fascicles (Fig. 4C) (Spieß et al., 2008) and begin branching upon encountering the midgut (Fig. 4C′). In Ret mutants, the axon bundles often defasciculate a short distance into their trajectories (Fig. 4D). The most obvious phenotype in Ret mutants is increased axon branching upon encountering the midgut, although the axons cover a smaller area than in w1118 (Fig. 4D′). We quantified this phenotype using ImageJ and the Simple Neurite Tracer plugin (Longair et al., 2011) and found that Ret mutants showed a 2.25-fold increase in the frequency of axon branching that is highly statistically significant [Fig. 5A; P<0.001, Tukey honestly significant difference (HSD) test]. To confirm that the defect was due to lack of Ret, we expressed a Ret transgene in the SNS neurons and found that the proventricular defasciculation (Fig. 4E) and the midgut axon branching phenotypes (Fig. 4E′) were no longer present.
Fig. 5.
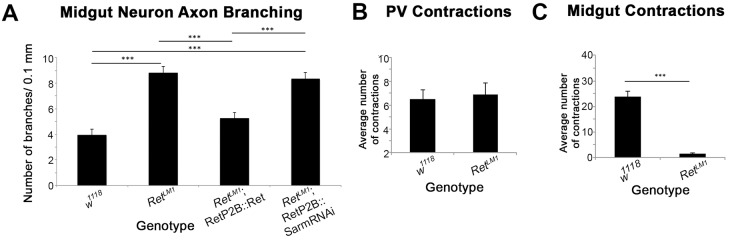
Quantification of larval midgut axon and peristaltic defects. (A) Quantification of the defects seen in the architecture of larval midgut neurons. RetLM1 causes a ∼2-fold increase in the number of branches per unit length of midgut axons measured. This phenotype is rescued by driving expression of Ret with an SNS-specific promoter, but is not rescued by knocking down dSarm. Analysis of individual animals was n=11 for w1118, RetLM1 and Ret transgene rescue; n=10 for the dSarm experiment. Error bars represent s.e.m. and statistical significance (***P<0.001) relative to the w1118 control was assessed using a one-way ANOVA followed by Tukey HSD. (B) Larvae carrying the RetLM1 mutation do not show a change in contractile activity of the proventriculus. Analysis of individual animals was n=16 for w1118 and n=13 for RetLM1. (C) The frequency of peristaltic waves was dramatically reduced in the anterior midgut of RetLM1 mutants. Analysis of individual animals was n=16 for w1118 and n=13 for RetLM1. Error bars represent s.e.m. and statistical significance (***P<0.001) relative to the w1118 control was assessed using an unpaired t-test.
As noted above, the Ret-GAL4 lines express predominantly in SNS neurons, and this pattern of Ret expression continues into the second instar (Fig. S2C). In late second instar and third instar larvae, strong midgut expression is observed, but this is in the middle of the midgut at a considerable distance from the midgut axons. We therefore expect Ret to play a cell-autonomous role in the midgut axons but cannot rule out functions in other tissues. Quantification of the axon branching confirmed the rescue (Fig. 5A).
The midgut neurons are characterized by varicosities, which are likely to be en passant synapses signaling to the underlying gut tissue (Fig. 4C″) (Budnik et al., 1989; Neckameyer and Bhatt, 2012). The Ret mutants frequently display larger varicosities with decreased 22c10 antigen [a microtubule-associated protein (Hummel et al., 2000)] in the axons between the varicosities (Fig. 4D″). The morphology of the axons is reminiscent of the axon fragmentation observed during Wallerian degeneration. Wallerian degeneration can be blocked by reducing the activity of dSarm (Ect4) (Osterloh et al., 2012). However, reducing the activity of dSarm via RNAi failed to suppress the axon branching phenotype or the enlarged varicosities (Fig. 4F,F′, Fig. 5A). To further understand the SNS defect, we dissected live larval guts and observed peristalsis of the midgut. In w1118 larvae, contraction of the proventriculus is followed by a wave of peristalsis that propagates along the midgut (Fig. 5B). In Ret mutants, a similar frequency of proventricular contractions was observed (Fig. 5B), but the wave of peristalsis was mostly absent (Fig. 5C). These observations suggest that the altered neuroanatomy of the midgut neurons reflects an inability to properly signal to the midgut muscles and this results in a midgut contraction but no propagation of the peristaltic wave.
Requirement for Gfrl, maverick and Pink1 in embryonic SNS development
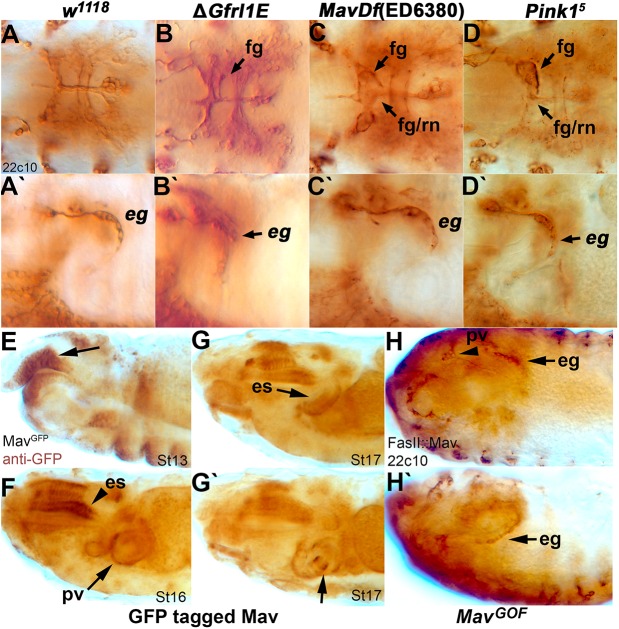
Vertebrate RET is known to function as a complex, and the Drosophila GFR homolog Gfrl is required for larval feeding (Fig. 1D). We therefore analyzed the embryonic SNS in Gfrl mutants and found striking phenotypic similarities to Ret mutants, with asymmetry of the frontal ganglion and disruption to the esophageal ganglia (Fig. 6B,B′, compare with Fig. 6A and Fig. 2C). GDNF is a member of the TGFβ superfamily and in the fly there are seven TGFβ family members. We examined the expression patterns of the seven TGFβ genes and aligned the amino acid sequences to vertebrate RET ligands. None of the proteins contains the distinguishing motifs of the vertebrate ligands, but the Maverick (Mav) protein showed the best, albeit weak, sequence similarities (Fig. S3). The mav gene is expressed in the developing esophagus and midgut at the right time to act as a ligand (Nguyen et al., 2000). Two independent chromosomal deficiencies for the mav region, Df(4)C1-7a and Df(4)ED6380, displayed phenotypic similarities to Ret mutants (Fig. 6C,C′) (Sousa-Neves et al., 2005). These results suggested that a Ret-Gfrl-Mav complex might be functioning in fly SNS development.
Fig. 6.
Gfrl, mav and Pink1 are required for SNS development. Anti-Futsch (mAb 22c10) staining reveals SNS defects in animals carrying mutations in Gfrl, mav and Pink1. (A-D′) Defects are observed in the mature embryonic SNS of ΔGfrl1E (B,B′), Df(ED6380)mav (C,C′) and the Pink15 allele (D,D′), matching defects seen in RetLM alleles. See Fig. 2A-D for schematics of the phenotypes. Defects are seen dorsally in asymmetry of the frontal ganglion and the frontal ganglion/recurrent nerve connection (B,C,D, arrows) and laterally in the esophageal ganglia (B′,C′,D′, arrow), matching those previously observed in animals carrying a Ret mutation. Images are representative of the phenotype seen in randomly selected embryos. (E-G′) A transgenic stock with a GFP-tagged copy of Mav labeled by anti-GFP staining shows expression of the protein in tissues where SNS innervation occurs throughout embryogenesis. Mav protein is observed in the foregut anlage (E, arrow), along the developing esophagus (es) (F,G), and at the proventriculus (G′, arrow). (H,H′) Overexpression (gain of function, GOF) of Mav in developing gut tissues using a Fas2 promoter drives overgrowth of the SNS (arrowhead, arrow) and changes to gut orientation, with 22c10 labeling of the SNS. The proventricular ganglion and proventriculus are displaced to the anterior end of the embryo (H, arrowhead) and projections from the esophageal ganglia cells show increased projections and innervation into the esophagus (H,H′, arrows).
Ret signals through classical receptor tyrosine kinase pathways and genetically interacts with the Pink1 mitochondrial kinase (Ibáñez, 2013; Klein et al., 2014), which has been proposed as a susceptibility locus for HSCR (Meka et al., 2015). Pink1 mutants displayed asymmetric frontal ganglia and altered esophageal ganglia (Fig. 6D,D′), suggesting that Pink1 is functioning in the same pathway.
mav mRNA is expressed in the foregut and esophagus (Nguyen et al., 2000). We looked for dose-sensitive genetic interactions by examining Ret/+; Gfrl/+ transheterozygotes but did not observe any feeding defects or decreases in viability relative to sibling flies. As both mutations are homozygous viable, albeit with decreased viability, we believe sensitized backgrounds will have to be used to demonstrate an interaction.
To examine Mav protein expression, we stained a fosmid-based GFP-tagged Mav with anti-GFP (Sarov et al., 2016). Mav expression is observed in the roof of the stomodeum (mouth) when SNS precursors are invaginating from the epithelium to form migrating clusters (Fig. 6E). Expression continues and broadens within the mouth/pharynx to encompass the esophagus and the proventriculus (Fig. 6F-G′). Based on the migratory behavior of the SNS clusters, mav is expressed at the right time and place to function as a Ret ligand. Overexpression of mav in the digestive tract has a strong effect on embryonic morphology as a whole, possibly affecting midgut differentiation, but SNS neurons also appear to increase in number (Fig. 6H,H′), suggesting that they respond to the Mav ligand.
Mav, Gfrl and Ret form a complex in cell culture
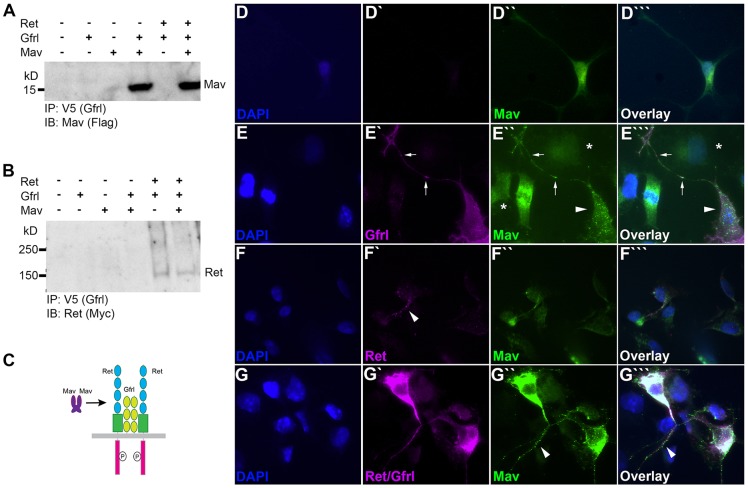
To test whether Drosophila has an equivalent to the RET-GFR-GDNF complex, we expressed epitope-tagged versions of Mav, Gfrl and Ret in COS7 cells and attempted to co-immunoprecipitate the proteins. In vertebrates, GDNF does not bind directly to RET but acts through GFR (Treanor et al., 1996), so we tested for binding between Mav and Gfrl. Co-expression followed by precipitation with anti-Gfrl (V5) and probing with anti-Mav (Flag) demonstrated a strong interaction between Mav and Gfrl (Fig. 7A). The presence of Ret did not alter the interaction. Immunoprecipitation with anti-Gfrl (V5) and probing with Ret revealed that Gfrl and Ret closely interact, both in the presence and absence of Mav (Fig. 7B). Although the interaction appears weak, expression levels of Ret in cell culture were always lower than for Mav or Gfrl (Fig. S4). We also observed a low level of Mav dimers even under the reducing conditions of the SDS sample buffer (Fig. S4). By analogy to GDNF, it seems likely that Mav dimerizes before binding to Gfrl, and Gfrl may already be bound to Ret (Fig. 7C).
Fig. 7.
Ret, Gfrl and Mav form a complex in vitro. Ret, Gfrl and Mav proteins were expressed in HEK 293 and COS7 cells. Protein-protein interactions were assessed by co-immunoprecipitation from HEK 293 (A,B) and via immunofluorescence analysis in COS7 (D-G‴). (A) Gfrl IP in the presence of coexpressed Mav probed with anti-Flag shows a ∼14 kDa band for monomeric Mav. A ∼28 kDa band was also visible (not shown), representing a Mav dimer. The presence of Ret protein does not alter the binding of Mav to Gfrl in the Gfrl immunoprecipitation (IP). IB, immunoblot. (B) A ∼140 kDa band for the Ret monomer is present after Gfrl IP when coexpressed with Gfrl and in a coexpression condition of all three proteins. Mav protein does not interact with the IP primary antibodies alone. Ret does show an interaction with Gfrl in the absence of Mav. (C) Model depicting the presumed Ret-Gfrl-Mav binding complex. (D-G‴) COS7 expression of Ret (magenta), Gfrl (magenta) and Mav (green) 48 h post transfection shows interaction between proteins at the cell membrane. DAPI, blue. (D-D‴) Mav expressed alone is seen in the cytoplasm as well as in the Golgi. (E-E‴) Coexpression of Gfrl and Mav shows puncta of colabeling on the cell membrane of filopodia and at the cell body. Arrows indicate colabeled puncta in a neurite-like extension, arrowheads indicate the changed distribution of Mav staining on a cell expressing Gfrl, and asterisks indicate the weak and diffuse Mav staining on cells not expressing Gfrl. (F-F‴) Coexpression of Ret and Mav shows minimal colabeling of Ret with Mav protein on the cell membrane. Arrowhead indicates puncta of Ret protein that show minimal overlap with Mav staining. Mav protein is seen mostly in the cytoplasm and within the Golgi, as in the condition with Mav expressed alone. (G-G‴) Coexpression of Ret, Gfrl and Mav shows a significant increase in colabeling of puncta on the cell membrane and within the cytoplasm. Arrowhead indicates colabeled puncta in a neurite-like extension.
To verify the results, we performed cell overlay assays in which expression of Mav in COS7 cells was detected by immunohistochemistry. Cells expressing Mav were clearly detected but weak cell surface staining suggesting diffusion into the medium (Fig. 7D″). When Gfrl was co-transfected, a dramatic increase in cell surface levels of Mav was observed and the Mav staining was highly punctate (Fig. 7E″). Colocalization of Mav and Gfrl was also observed, although the proteins have distinct patterns, with Gfrl being more diffuse than the highly punctate Mav (Fig. 7E‴). Expression of Ret alone led to a slight increase in cell surface levels of Mav, with a few puncta present (Fig. 7F″), suggesting a possible weak interaction. Co-transfection of Ret and Gfrl produces Mav binding equivalent to Gfrl alone (Fig. 7G-G′″). The strong alterations in the localization patterns combined with the immunoprecipitation results suggests that Mav, Gfrl and Ret are present in a physiologically relevant complex.
DISCUSSION
Ret function in the SNS
We have described the effects of mutating the Ret gene in Drosophila and uncovered an evolutionarily conserved role in the development of the ENS. The incorrect positioning of SNS cells in the Drosophila embryo resembles hypoganglionic ENS phenotypes seen when RET is mutated in vertebrates (Lake and Heuckeroth, 2013). In HSCR, the most distal nerves of the digestive tract are affected. Likewise, in Ret mutant larvae we find that the most distal nerves of the SNS, located on the midgut, have an altered anatomy and the larvae show defects in food ingestion. The phenotype resembles the neurotrophic effects of decreased serotonin or CNS dopamine signaling during midgut nerve formation, which also leads to increased axon branching and decreased feeding (Budnik et al., 1989; Neckameyer and Bhatt, 2012).
Although defects are visible in the embryonic SNS, there appear to be two separate lethal phases. Some first instar larvae display feeding defects and die. This is particularly evident in the original alleles that carry the background recessive lethal mutation, and we are investigating the possibility that the background lethal mutation specifically enhances the Ret mutations. Subsequent larval feeding defects often do not emerge until 2-4 days after hatching. Larvae with food in their guts can be observed foraging, suggesting that the larvae have problems with food ingestion. This is supported by observations of mutant larvae with food throughout their midguts, but with peristaltic defects in the anterior midgut (see Movie 2, compare with wild-type peristalsis in Movie 1). We initially suspected a neurodegenerative defect similar to Wallerian degeneration, but failed to suppress the axon defect by reducing dSarm activity. We currently favor a model in which initial SNS defects are amplified as the larva dramatically increases its mass several hundred fold (Ghosh et al., 2013). To keep pace with the expanding midgut, Ret may be required to promote axon growth, guidance, or be fulfilling a pro-synaptic role. These functions have been observed for RET and GDNF (Dudanova and Klein, 2013; Paratcha and Ledda, 2008), including in the human ENS (Böttner et al., 2013).
The midgut axon phenotype resembles defasciculation of the nerves and Gfrl genetically interacts with the fasciculation molecule Fas2 (Kallijärvi et al., 2012), so Ret/Gfrl could potentially be modulating fasciculation as has been observed for other signaling systems (Yu et al., 2000). Alternatively, defasciculation may be a consequence of growth cones searching for sources of ligand, as proposed for Netrin and Bolwig's nerve (Andrews et al., 2008). Decreased midgut innervation and function may provide negative feedback to upstream gut signaling, decreasing the ability to pass food through the pharynx and esophagus (Melcher and Pankratz, 2005; Zhang et al., 2014b). The midgut axons may also be required to maintain communication with downstream enteroendocrine cells (LaJeunesse et al., 2010). An alternative hypothesis raised by the similarity of the Ret and Pink1 phenotypes is that the midgut neurons are running out of energy due to mitochondrial dysfunction (Klein et al., 2014).
GDNF signaling in invertebrates
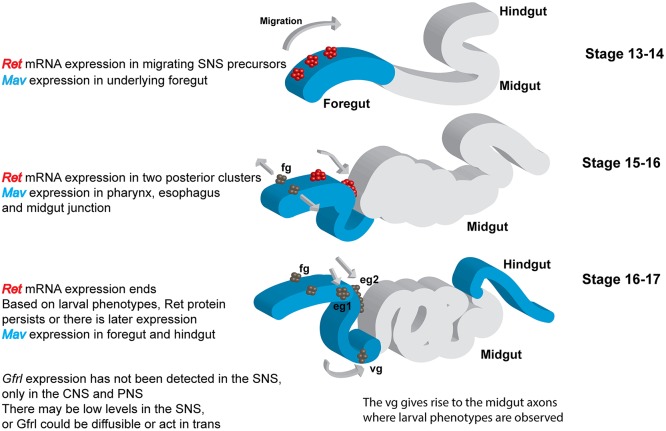
Our analysis enabled us to identify the divergent TGFβ Mav as the elusive ligand for Drosophila Ret (Kallijärvi et al., 2012; Saarenpää et al., 2017). The expression pattern of mav is consistent with a role in embryonic SNS development (Fig 6E-G, Fig. 8). Although the Mav ligand is concentrated in certain regions of the foregut and may create localized gradients, the broad expression pattern suggests that the Ret/Gfrl signaling pathway could be permissive rather than instructive during SNS precursor migration. Embryonic Ret signaling could primarily transduce a neurotrophic signal, and apoptosis has been observed in the migrating SNS precursors (Hartenstein et al., 1994). In vertebrates, models in which GDNF/Ret signaling promotes proliferation rather than cell migration have been proposed to explain development of the nervous system (Newgreen et al., 2013). Experiments are underway to distinguish between these models in the fly. Although Gfrl expression has not yet been observed in the SNS, Gfrl could be acting in a soluble form or in trans (Paratcha et al., 2001). Gfrl promoter fragments continue to drive expression in the anterior midgut of the larvae in support of the trans model (Hernández et al., 2015). Despite extensive sequence divergence in the extracellular domain of Ret (Hahn and Bishop, 2001), domain differences in GFRs (Airaksinen et al., 2006) and low homology of Mav to the GDNF family (Nguyen et al., 2000), the molecular logic of the protein complex appears preserved. In vertebrates, RET and GFR form a preassembled complex (Eketjall et al., 1999), and GDNF binds GFR to activate RET (Treanor et al., 1996). Our molecular data are strikingly similar, as we find that Drosophila Ret and Gfrl can functionally interact in the absence of Mav, and that Mav interacts strongly with Gfrl, but only very weakly with Ret. In flies, Mav modulates synapse formation at the neuromuscular junction of body wall muscles (Fuentes-Medel et al., 2012; Sulkowski et al., 2016). Ret is not expressed in body wall muscles (Hahn and Bishop, 2001), and Mav is likely to be signaling through activin/BMP type 1 receptors (Fuentes-Medel et al., 2012). A Mav homolog, Panda, has been found in the sea urchin Paracentrotus lividus, where it plays a role in dorsoventral axis formation and is also likely to be signaling through type 1 receptors (Haillot et al., 2015). Mav and Panda both lack a key leucine residue, so their binding to type 1 receptors might be weaker than other ligands (Haillot et al., 2015). Candidate Ret and Mav homologs have been found in Strongylocentrotus purpuratus (Lapraz et al., 2006), suggesting that Mav homologs might interact with both type 1 and Ret receptors in sea urchins.
Fig. 8.
Summary of Ret and mav expression during development of the stomatogastric nervous system. Ret exhibits highly dynamic mRNA expression in the embryo (Hahn and Bishop, 2001; Hernández et al., 2015). Ret is also expressed in adult midgut precursors at an earlier stage in development, as well as in discrete cells in the CNS, PNS and Malpighian tubules (not shown). mav mRNA is expressed weakly in the foregut primordium and at later stages in the pharynx, esophagus and proventriculus (Nguyen et al., 2000). Our analysis of an epitope-tagged Mav expressed at endogenous levels indicates strong expression in the epithelial region from which the SNS precursor clusters delaminate (Fig. 6E) and expansion to match the pattern of the mRNA, becoming concentrated near the sites at which the SNS neurons stop migrating (junction of the pharynx and esophagus, proventriculus; Fig. 6F,G). mav is also expressed in the epidermis and visceral mesoderm. Apart from promoter fragments driving reporters, Gfrl expression has not been observed in the SNS (Kallijärvi et al., 2012). Gfrl could therefore be expressed at low levels, or the protein might be acting in trans or in a soluble form (Paratcha et al., 2001). Gfrl promoter fragments continue to drive expression in the anterior midgut of the larvae (Hernández et al., 2015). eg1 and eg2, esophageal ganglia; fg, frontal ganglion; vg, ventricular ganglion.
Despite promiscuity in binding between TGFβ and their receptors in vertebrates, GDNF family members have not been reported to bind BMP/TGFβ receptors (Mueller and Nickel, 2012), suggesting that the ability to interact with more than one receptor was lost during evolution. The GDNF family of ligands, including GDNF, Neurturin, Artemin and Persephin, all appeared when fish gained jaws, as homologs cannot be identified in the published Agnatha sequences. GDNF ligands are distinguished by a highly conserved DLGLGY motif, part of one of two fingers that mediate binding to GFRα (Fig. S3) (Eketjall et al., 1999). This motif is not present in Mav or Panda. The change may have increased affinity or specificity for GFRs and additional changes might have prevented crosstalk with Activin/BMP type 1 receptors. Mav and Panda are similar to GDF-15 (Haillot et al., 2015), a TGFβ placed in the subfamily containing GDNF (Mueller and Nickel, 2012). GDF-15 is an inflammatory cytokine, and although it activates SMAD signaling, GDF-15 does not have an identified receptor (Yadin et al., 2016). GDF-15 has GDNF-like neurotrophic activity for dopaminergic neurons (Strelau et al., 2003), so it would be interesting to test GDF-15 for binding to GFRs.
The limited sequence data available suggest a model in which a divergent TGFβ acquired an ability to bind GFRs and activate Ret, which was followed by extensive co-evolution of the extracellular components. However, the downstream signaling pathways appear to be conserved (Abrescia et al., 2005), so the Ret SNS phenotypes open the door to invertebrate genetic analysis of this clinically important signaling pathway. Particularly exciting is the possibility of functional suppressor screens to identify mutations that could compensate for a lack of Ret signaling. Drosophila has already been used to identify genetic modifiers and a candidate drug to counteract oncogenic Ret signaling (Das and Cagan, 2013; Read et al., 2005).
Conclusions
Ret has an evolutionarily conserved role in the formation and function of the ENS. The GDNF signaling pathway has its origins in TGFβ signaling.
MATERIALS AND METHODS
Drosophila genetics
The w1118 stock was used as a control as it had the most reliable neuroanatomy relative to that described in previous publications. Bloomington Drosophila Stock Center (BDSC) supplied w1118, Act5C-Cas9, Vasa-Cas9, Df(2L)BSC312[Ret]/CyO, Df(4)ED6380[mav]/l(4)102EFf1 (Ryder et al., 2007), FasII-GAL4, Pink15/FM6 and Klu09036 (stock numbers: #6326, #54950, #52669, #24338, #9579, #46123, #51649 and #11733). BDSC also provides the nos-phiC31 attP2 stock #25710 used for embryo injections by Rainbow Transgenic Flies (Camarillo, CA, USA). The Vienna Drosophila Resource Center (VDRC) provided UAS-RetRNAi and UAS-SarmRNAi, #107658 and #22612 (Dietzl et al., 2007). Balancing of stocks was done using w−; KrIf-1/CyOwgβ, w−; KrIf-1/CyOKrGFP, v−; Dr/TM3 and w−; UASτlacZ/CyOwgβ; TM2/TM6 (T.K. laboratory stocks). The Ret-GAL4 (RetP2A and RetP2B) lines used in this work were produced and characterized by our laboratory (Hernández et al., 2015). Df(4)C1-7a was supplied by R. Sousa-Neves (Case Western Reserve University). The ΔGfrl1E allele and UAS-Retflag were provided by J. Kallijärvi (University of Helsinki). UAS-mav and UAS-mavGFP lines were provided by V. Budnik (University of Massachusetts Medical School).
CRISPR generation of mutant Ret alleles
A CRISPR strategy to target Drosophila Ret was organized and carried out as described by Port et al. (2014). We received the pCFD3-dU6:3gRNA plasmid courtesy of P. Miura (University of Nevada, Reno; #49410, Addgene). Three individual gRNA target sites within Ret were chosen using the Drosophila CRISPR/Cas9 gRNA target track in the UCSC Genome Browser (https://genome.ucsc.edu) (Speir et al., 2016). Target sites were checked for specificity using the flyCRISPR Optimal Target Finder (http://tools.flycrispr.molbio.wisc.edu/targetFinder/) (Gratz et al., 2014). Single-stranded oligos corresponding to these target sequences were synthesized by Integrated DNA Technologies (San Diego, CA, USA). Sense and antisense oligos for targeting exons 3 and 5 of the Ret locus are listed in Table S1. Three pCFD3-dU6:3-gRNA:Ret constructs (one targeting exon 3 and two targeting exon 5) were produced and screened using PCR primers listed in Table S2. Rainbow Transgenic Flies produced transgenic stocks expressing Ret gRNAs via a phiC31 injection protocol. Recovered flies were balanced with a TM3 balancer and crossed to Act5C/Vas-Cas9 lines. Recovered progeny from the crossing scheme were balanced with CyOwgB or CyoKrGFP. For candidate alleles, the region of interest was amplified by PCR and sequenced. Sequence analysis was conducted using 4 Peaks (https://nucleobytes.com). Three independent lines were isolated after analysis (RetLM1-3 alleles). Homozygous embryos for each allele were assayed for protein expression using a polyclonal antibody directed against the Ret ectodomain (residues 24-401; Genscript). Embryos were crushed in 50 µl cell lysis buffer (50 mM Tris-HCl pH 7.6, 1 mM EDTA, 150 mM NaCl, 1% Triton X-100) containing protease inhibitor cocktail (Sigma-Aldrich), incubated on ice for 15 min, cleared by centrifugation and analyzed via SDS-PAGE.
Immunohistochemistry
Antibody staining on embryos was performed as described by Patel (1994) with reagents listed in Table S1. 22c10 was used to visualize the developing SNS. Balancer markers and GAL4/UAS expression were visualized with anti-β-galactosidase and anti-GFP antibodies. Typical incubations with primary antibodies were overnight at 4°C and 1 h at room temperature with secondary antibodies. Biotinylated secondary antibodies with VectaStain Elite ABC enhancement were frequently used for stronger labeling of SNS projections in intact embryos. Stage 17 embryos of each genotype were collected at random and scored for the presence, absence or thinning of the frontal nerve, and for defasciculation defects in the recurrent nerve. The arrangement of cell bodies at the frontal ganglion, esophageal ganglion, and proventricular ganglion were also scored. At least 20 embryos were collected for each genotype. Gut preparations of second instar larvae were performed as described (Bhatt and Neckameyer, 2013; Neckameyer and Bhatt, 2012). Larval SNS was visualized using 22c10 and scoring of the SNS in larval preparations was performed in at least ten individual larvae for each genotype. Neural innervation of the midgut was assessed by measuring axon coverage of the innervated midgut tissue and the degree of branching by axons using ImageJ/Simple Neurite Tracer. RapiClear (Cedar Lane Laboratories) was used as a whole-mount medium for imaging late stage 17 embryos and larval gut tissues (Hernández et al., 2015).
Larval behavior
Behavioral and developmental effects in larvae were assayed via an egg lay on grape juice agar with access to a yeast food paste mixture containing Carmine powder as a coloring agent (Sigma-Aldrich; 1.5 mg Carmine per 1 g yeast paste) as described (Melcher and Pankratz, 2005; Zinke et al., 1999). Sets of 50 homozygous mutant larvae from overnight egg collections were allowed to develop for a 72 h period at 25°C and monitored at 24 h intervals for feeding and wandering phenotypes under a dissection microscope. Animals were assayed for each genotype to assess mortality rate, approximate growth rates, and observable feeding defects. At least 100 newly hatched first instar larvae per genotype were assayed for mortality and behavioral phenotypes. Movements of the proventriculus and the midgut were observed in semi-intact third instar larvae (Schoofs et al., 2014). The preparation consisted of CNS, SNS, foregut and midgut dissected from exterior cuticle in a bath of Schneider's Drosophila medium at room temperature. Preparations were allowed to equilibrate in medium to allow for spontaneous contractile activity to occur. At least ten animals of each genotype were assessed in independent preparations. Videos of gut motility were recorded using a Leica MZFLIII with a Jenoptik ProgRes C14 plus camera for periods of ∼10 min. Measurement was by counting peristaltic events at the proventriculus and in the midgut over 1.5 min from the initiation of spontaneous contractile activity.
Statistics
For larval feeding phenotypes, mortality, and embryonic SNS analysis, Fisher's exact test with two tails and 95% confidence intervals was calculated using the GraphPad website (www.graphpad.com/quickcalcs). Statistical significance was tested using Bonferroni correction. For larval proventriculus/midgut contractile activity, a t-test and the Bonferroni correction were performed to determine statistical significance and s.e.m. using the GraphPad website. Statistical analysis of larval midgut neuron axon branching used a one-way ANOVA followed by a Tukey HSD using Statistica (Dell). Sample size was determined using the resource equation method: E=total number of animals−total number of groups. For each experiment a value of E>10 was required. No samples were excluded from the analysis and samples were allocated to experimental groups on the basis of genotype. For the data in Figs 2-5, images or movies of embryos or larvae were given codenames and scored by an experimenter blind to the genotype. All data were tested for fit to a normal distribution. Raw data are available in Table S3.
Cell culture
pcDNA3-FLdRetcmyc was a gift from C. Abrescia and C. Ibáñez (Abrescia et al., 2005). pcDNA3.1-MavcDYK was synthesized from the sequence for the final 112 amino acid residues of the fly Mav protein consisting of the proteolytically cleaved active form of the ligand along with an Ig κ-chain secretion signal added to the N-terminus of the protein for secretion (Genscript). The pSecTag-V5GfrlA was subcloned from pMT-V5GfrlA that was a gift from J. Kallijärvi (Kallijärvi et al., 2012). We PCR amplified the N-terminally tagged GfrlA construct from pMT-V5GfrlA (using forward primer 5′-GGTAAGCCTATCCCTAACCC-3′ and reverse primer 5′-TCATGTCGCCACATCACTC-3′) then subcloned the fragment into pSecTag/FRT/V5-His-TOPO (Invitrogen). Expression of Ret, Gfrl and Mav protein in animal cell lines was performed using COS7 and HEK 293 cells. COS7 or HEK 293 cells at 80% confluence were transfected with DNA expression constructs using Lipofectamine 3000 (Invitrogen) according to the manufacturer's instructions. Ret, Gfrl and mav constructs were transfected individually as well as in combination to assess the ability of each protein to bind in pairs and as a complex.
Immunoprecipitation
Immunoprecipitation (IP) assays from cell culture followed a published protocol (Alavi et al., 2016; Banerjee et al., 2010). In brief, HEK 293 cells were transfected, grown for 48 h, lysed in IP buffer (50 mM HEPES pH 7.2, 100 mM NaCl, 1 mM MgCl2, 1 mM CaCl2, 1% NP-40) and incubated overnight at 4°C with 1 µg antibody. Proteins were immunoprecipitated with 30 µl GammaBind G Sepharose beads (GE Healthcare) for 2 h at room temperature, run on 4-20% Mini-Protean TGX gels (Bio-Rad), immunoblotted and detected with Clarity western ECL substrate (Bio-Rad) and imaged on a ChemiDoc Touch system (Bio-Rad).
Cell overlay assay
Constructs were transfected into COS7 cells for cell immunofluorescent labeling experiments. Chamber slides were coated with rat tail collagen solution (Thermo Fisher Scientific) diluted 1:4 with PBS and allowed to dry. At 48 h post-transfection, medium was removed from the co-transfected receptor (Ret/Gfrl)-expressing cells and replaced with medium containing Mav. All cells were incubated at 37°C for 1-2 h before rinsing three times in 1× PBS and proceeding with fixation and antibody labeling. Alternatively, all three constructs were co-transfected (1:1) into the COS7 cells. Approximately 48 h post-transfection, medium was removed and the cells were washed with 1× PBS. After rinsing, cells were fixed for 15 min in 4% paraformaldehyde at room temperature. Cells were washed once in cold PBS after fix then once in PBST (PBS with 0.1% Triton X-100). Cells were blocked for 30 min at room temperature with 5% normal goat serum (NGS) in PBST. Primary antibodies were added in 5% NGS in PBST and incubated overnight at 4°C. Cells were washed three times in PBST then incubated for 1 h in fluorescent secondary antibody in 5% NGS in PBST. Secondary antibodies were from the AlexaFluor collection (Table S1http://dev.biologists.org/lookup/doi/10.1242/dev.157446.supplemental). Cells were washed five times in cold PBS following labeling. Cells were mounted in FluorSave containing NucBlue DAPI stain (Millipore) to label nuclei.
Supplementary Material
Acknowledgements
We thank P. Miura for advice and reagents for CRISPR; J. Kallijärvi for Gfrl and UAS-Ret reagents; P. Soba for UAS-Ret-mCherry; V. Budnik for mav reagents; C. Ibáñez for Ret reagents; R. Sousa-Neves for Df(4)C1-7a; M. Freeman for Sarm reagents; A. DiAntonio for advice on axon regeneration; G. Hennig for advice on video analysis; S. Ward for advice on the enteric nervous system; L. Heydman and N. Yokdang for help with cell culture experiments; R. Marshall, K. Hernández and other members of the T.K. laboratory for assistance with genetics and imaging; P. Soba and N. Hoyer for communicating results prior to publication; and R. Kellermeyer and T. Gillis for comments on the manuscript. Antibodies were obtained from the Developmental Studies Hybridoma Bank (DHSB) developed under the auspices of the NICHD and maintained by the University of Iowa. Drosophila stocks were obtained from the Bloomington Drosophila Stock Center (NIH P40OD018537). Transgenic fly stocks were obtained from the Vienna Drosophila Resource Center. Funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: L.M., T.K.; Methodology: L.M., H.P.; Formal analysis: L.M.; Investigation: L.M., H.P., M.G.A., T.K.; Writing - original draft: L.M., T.K.; Writing - review & editing: L.M., T.K.
Funding
This project was supported by a grant from the National Institutes of Health (NIH) (R15NS075918) to T.K. This study also received funding from NIH National Institute of General Medical Sciences grants P20 GM103554, P20 GM103650, P20 GM103440. Additionally, Michael (Mick) J. M. Hitchcock, PhD Graduate Student Research Fund to L.M. Deposited in PMC for release after 12 months.
Supplementary information
Supplementary information available online at http://dev.biologists.org/lookup/doi/10.1242/dev.157446.supplemental
References
- Abrescia C., Sjöstrand D., Kjaer S. and Ibáñez C. F. (2005). Drosophila RET contains an active tyrosine kinase and elicits neurotrophic activities in mammalian cells. FEBS Lett. 579, 3789-3796. 10.1016/j.febslet.2005.05.075 [DOI] [PubMed] [Google Scholar]
- Airaksinen M. S., Holm L. and Hätinen T. (2006). Evolution of the GDNF family ligands and receptors. Brain Behav. Evol. 68, 181-190. 10.1159/000094087 [DOI] [PubMed] [Google Scholar]
- Alavi M., Song M., King G. L. A., Gillis T., Propst R., Lamanuzzi M., Bousum A., Miller A., Allen R. and Kidd T. (2016). Dscam1 forms a complex with Robo1 and the N-terminal fragment of slit to promote the growth of longitudinal axons. PLoS Biol. 14, e1002560 10.1371/journal.pbio.1002560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews G. L., Tanglao S., Farmer W. T., Morin S., Brotman S., Berberoglu M. A., Price H., Fernandez G. C., Mastick G. S., Charron F. et al. (2008). Dscam guides embryonic axons by Netrin-dependent and -independent functions. Development 135, 3839-3848. 10.1242/dev.023739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee S., Blauth K., Peters K., Rogers S. L., Fanning A. S. and Bhat M. A. (2010). Drosophila neurexin IV interacts with Roundabout and is required for repulsive midline axon guidance. J. Neurosci. 30, 5653-5667. 10.1523/JNEUROSCI.6187-09.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatt P. K. and Neckameyer W. S. (2013). Functional analysis of the larval feeding circuit in Drosophila. J. Vis. Exp. 81, e51062 10.3791/51062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böttner M., Harde J., Barrenschee M., Hellwig I., Vogel I., Ebsen M. and Wedel T. (2013). GDNF induces synaptic vesicle markers in enteric neurons. Neurosci. Res. 77, 128-136. 10.1016/j.neures.2013.08.012 [DOI] [PubMed] [Google Scholar]
- Budnik V., Wu C. F. and White K. (1989). Altered branching of serotonin-containing neurons in Drosophila mutants unable to synthesize serotonin and dopamine. J. Neurosci. 9, 2866-2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copenhaver P. F. (2007). How to innervate a simple gut: familiar themes and unique aspects in the formation of the insect enteric nervous system. Dev. Dyn. 236, 1841-1864. 10.1002/dvdy.21138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das T. K. and Cagan R. L. (2013). A Drosophila approach to thyroid cancer therapeutics. Drug Discov. Today Technol. 10, e65-e71. 10.1016/j.ddtec.2012.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis T. K., Hoshi M. and Jain S. (2014). To bud or not to bud: the RET perspective in CAKUT. Pediatr. Nephrol. 29, 597-608. 10.1007/s00467-013-2606-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Belle J. S., Hilliker A. J. and Sokolowski M. B. (1989). Genetic localization of foraging (for): a major gene for larval behavior in Drosophila melanogaster. Genetics 123, 157-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietzl G., Chen D., Schnorrer F., Su K.-C., Barinova Y., Fellner M., Gasser B., Kinsey K., Oppel S., Scheiblauer S. et al. (2007). A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature 448, 151-156. 10.1038/nature05954 [DOI] [PubMed] [Google Scholar]
- Dudanova I. and Klein R. (2013). Integration of guidance cues: parallel signaling and crosstalk. Trends Neurosci. 36, 295-304. 10.1016/j.tins.2013.01.007 [DOI] [PubMed] [Google Scholar]
- Eketjall S., Fainzilber M., Murray-Rust J. and Ibáñez C. F. (1999). Distinct structural elements in GDNF mediate binding to GFRalpha1 and activation of the GFRalpha1-c-Ret receptor complex. EMBO J. 18, 5901-5910. 10.1093/emboj/18.21.5901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuentes-Medel Y., Ashley J., Barria R., Maloney R., Freeman M. and Budnik V. (2012). Integration of a retrograde signal during synapse formation by glia-secreted TGF-beta ligand. Curr. Biol. 22, 1831-1838. 10.1016/j.cub.2012.07.063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh S. M., Testa N. D. and Shingleton A. W. (2013). Temperature-size rule is mediated by thermal plasticity of critical size in Drosophila melanogaster. Proc. Biol. Sci. 280, 20130174 10.1098/rspb.2013.0174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Gaitan M. and Jackle H. (1995). Invagination centers within the Drosophila stomatogastric nervous system anlage are positioned by Notch-mediated signaling which is spatially controlled through wingless. Development 121, 2313-2325. [DOI] [PubMed] [Google Scholar]
- Gratz S. J., Ukken F. P., Rubinstein C. D., Thiede G., Donohue L. K., Cummings A. M. and O'Connor-Giles K. M. (2014). Highly specific and efficient CRISPR/Cas9-catalyzed homology-directed repair in Drosophila. Genetics 196, 961-971. 10.1534/genetics.113.160713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn M. and Bishop J. M. (2001). Expression pattern of Drosophila ret suggests a common ancestral origin between the metamorphosis precursors in insect endoderm and the vertebrate enteric neurons. Proc. Natl. Acad. Sci. USA 98, 1053-1058. 10.1073/pnas.98.3.1053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haillot E., Molina M. D., Lapraz F. and Lepage T. (2015). The maternal Maverick/GDF15-like TGF-beta ligand panda directs dorsal-ventral axis formation by restricting nodal expression in the sea urchin embryo. PLoS Biol. 13, e1002247 10.1371/journal.pbio.1002247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartenstein V. (1997). Development of the insect stomatogastric nervous system. Trends Neurosci. 20, 421-427. [DOI] [PubMed] [Google Scholar]
- Hartenstein V., Tepass U. and Gruszynski-Defeo E. (1994). Embryonic development of the stomatogastric nervous system in Drosophila. J. Comp. Neurol. 350, 367-381. [DOI] [PubMed] [Google Scholar]
- Hätinen T., Holm L. and Airaksinen M. S. (2007). Loss of neurturin in frog--comparative genomics study of GDNF family ligand-receptor pairs. Mol. Cell. Neurosci. 34, 155-167. 10.1016/j.mcn.2006.10.009 [DOI] [PubMed] [Google Scholar]
- Hernández K., Myers L. G., Bowser M. and Kidd T. (2015). Genetic tools for the analysis of Drosophila stomatogastric nervous system development. PLoS ONE 10, e0128290 10.1371/journal.pone.0128290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hummel T., Krukkert K., Roos J., Davis G. and Klämbt C. (2000). Drosophila Futsch/22C10 is a MAP1B-like protein required for dendritic and axonal development. Neuron 26, 357-370. 10.1016/S0896-6273(00)81169-1 [DOI] [PubMed] [Google Scholar]
- Ibáñez C. F. (2013). Structure and physiology of the RET receptor tyrosine kinase. Cold Spring Harb. Perspect. Biol. 5, a009134 10.1101/cshperspect.a009134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibáñez C. F. and Andressoo J.-O. (2017). Biology of GDNF and its receptors - relevance for disorders of the central nervous system. Neurobiol. Dis. 97, 80-89. 10.1016/j.nbd.2016.01.021 [DOI] [PubMed] [Google Scholar]
- Jing S., Wen D., Yu Y., Holst P. L., Luo Y., Fang M., Tamir R., Antonio L., Hu Z., Cupples R. et al. (1996). GDNF-induced activation of the ret protein tyrosine kinase is mediated by GDNFR-alpha, a novel receptor for GDNF. Cell 85, 1113-1124. 10.1016/S0092-8674(00)81311-2 [DOI] [PubMed] [Google Scholar]
- Kallijärvi J., Stratoulias V., Virtanen K., Hietakangas V., Heino T. I. and Saarma M. (2012). Characterization of Drosophila GDNF receptor-like and evidence for its evolutionarily conserved interaction with neural cell adhesion molecule (NCAM)/FasII. PLoS ONE 7, e51997 10.1371/journal.pone.0051997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein P., Muller-Rischart A. K., Motori E., Schonbauer C., Schnorrer F., Winklhofer K. F. and Klein R. (2014). Ret rescues mitochondrial morphology and muscle degeneration of Drosophila Pink1 mutants. EMBO J. 33, 341-355. 10.1002/embj.201284290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaJeunesse D. R., Johnson B., Presnell J. S., Catignas K. and Zapotoczny G. (2010). Peristalsis in the junction region of the Drosophila larval midgut is modulated by DH31 expressing enteroendocrine cells. BMC Physiol. 10, 14 10.1186/1472-6793-10-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lake J. I. and Heuckeroth R. O. (2013). Enteric nervous system development: migration, differentiation, and disease. Am. J. Physiol. Gastrointest. Liver Physiol. 305, G1-G24. 10.1152/ajpgi.00452.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapraz F., Röttinger E., Duboc V., Range R., Duloquin L., Walton K., Wu S.-Y., Bradham C., Loza M. A., Hibino T. et al. (2006). RTK and TGF-beta signaling pathways genes in the sea urchin genome. Dev. Biol. 300, 132-152. 10.1016/j.ydbio.2006.08.048 [DOI] [PubMed] [Google Scholar]
- Longair M. H., Baker D. A. and Armstrong J. D. (2011). Simple Neurite Tracer: open source software for reconstruction, visualization and analysis of neuronal processes. Bioinformatics 27, 2453-2454. 10.1093/bioinformatics/btr390 [DOI] [PubMed] [Google Scholar]
- McKeown S. J., Stamp L., Hao M. M. and Young H. M. (2013). Hirschsprung disease: a developmental disorder of the enteric nervous system. Wiley Interdiscip. Rev. Dev. Biol. 2, 113-129. 10.1002/wdev.57 [DOI] [PubMed] [Google Scholar]
- Meka D. P., Müller-Rischart A. K., Nidadavolu P., Mohammadi B., Motori E., Ponna S. K., Aboutalebi H., Bassal M., Annamneedi A., Finckh B. et al. (2015). Parkin cooperates with GDNF/RET signaling to prevent dopaminergic neuron degeneration. J. Clin. Invest. 125, 1873-1885. 10.1172/JCI79300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melcher C. and Pankratz M. J. (2005). Candidate gustatory interneurons modulating feeding behavior in the Drosophila brain. PLoS Biol. 3, e305 10.1371/journal.pbio.0030305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller T. D. and Nickel J. (2012). Promiscuity and specificity in BMP receptor activation. FEBS Lett. 586, 1846-1859. 10.1016/j.febslet.2012.02.043 [DOI] [PubMed] [Google Scholar]
- Neckameyer W. S. and Bhatt P. (2012). Neurotrophic actions of dopamine on the development of a serotonergic feeding circuit in Drosophila melanogaster. BMC Neurosci. 13, 26 10.1186/1471-2202-13-26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newgreen D. F., Dufour S., Howard M. J. and Landman K. A. (2013). Simple rules for a “simple” nervous system? Molecular and biomathematical approaches to enteric nervous system formation and malformation. Dev. Biol. 382, 305-319. 10.1016/j.ydbio.2013.06.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen M., Parker L. and Arora K. (2000). Identification of maverick, a novel member of the TGF-beta superfamily in Drosophila. Mech. Dev. 95, 201-206. 10.1016/S0925-4773(00)00338-5 [DOI] [PubMed] [Google Scholar]
- Osterloh J. M., Yang J., Rooney T. M., Fox A. N., Adalbert R., Powell E. H., Sheehan A. E., Avery M. A., Hackett R., Logan M. A. et al. (2012). dSarm/Sarm1 is required for activation of an injury-induced axon death pathway. Science 337, 481-484. 10.1126/science.1223899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paratcha G. and Ledda F. (2008). GDNF and GFRalpha: a versatile molecular complex for developing neurons. Trends Neurosci. 31, 384-391. 10.1016/j.tins.2008.05.003 [DOI] [PubMed] [Google Scholar]
- Paratcha G., Ledda F., Baars L., Coulpier M., Besset V., Anders J., Scott R. and Ibáñez C. F. (2001). Released GFRalpha1 potentiates downstream signaling, neuronal survival, and differentiation via a novel mechanism of recruitment of c-Ret to lipid rafts. Neuron 29, 171-184. 10.1016/S0896-6273(01)00188-X [DOI] [PubMed] [Google Scholar]
- Paratcha G., Ledda F. and Ibáñez C. F. (2003). The neural cell adhesion molecule NCAM is an alternative signaling receptor for GDNF family ligands. Cell 113, 867-879. 10.1016/S0092-8674(03)00435-5 [DOI] [PubMed] [Google Scholar]
- Patel N. H. (1994). Imaging neuronal subsets and other cell types in whole-mount Drosophila embryos and larvae using antibody probes. Methods Cell Biol. 44, 445-487. 10.1016/S0091-679X(08)60927-9 [DOI] [PubMed] [Google Scholar]
- Perea D., Guiu J., Hudry B., Konstantinidou C., Milona A., Hadjieconomou D., Carroll T., Hoyer N., Natarajan D., Kallijärvi J. et al. (2017). Ret receptor tyrosine kinase sustains proliferation and tissue maturation in intestinal epithelia. EMBO J. 36, 3029-3045. 10.15252/embj.201696247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Port F., Chen H.-M., Lee T. and Bullock S. L. (2014). Optimized CRISPR/Cas tools for efficient germline and somatic genome engineering in Drosophila. Proc. Natl. Acad. Sci. USA 111, E2967-E2976. 10.1073/pnas.1405500111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Read R. D., Goodfellow P. J., Mardis E. R., Novak N., Armstrong J. R. and Cagan R. L. (2005). A Drosophila model of multiple endocrine neoplasia type 2. Genetics 171, 1057-1081. 10.1534/genetics.104.038018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romei C., Ciampi R. and Elisei R. (2016). A comprehensive overview of the role of the RET proto-oncogene in thyroid carcinoma. Nat. Rev. Endocrinol. 12, 192-202. 10.1038/nrendo.2016.11 [DOI] [PubMed] [Google Scholar]
- Romeo G., Ronchetto P., Luo Y., Barone V., Seri M., Ceccherini I., Pasini B., Bocciardi R., Lerone M., Kääriäinen H. et al. (1994). Point mutations affecting the tyrosine kinase domain of the RET proto-oncogene in Hirschsprung's disease. Nature 367, 377-378. 10.1038/367377a0 [DOI] [PubMed] [Google Scholar]
- Ryder E., Ashburner M., Bautista-Llacer R., Drummond J., Webster J., Johnson G., Morley T., Chan Y. S., Blows F., Coulson D. et al. (2007). The DrosDel deletion collection: a Drosophila genomewide chromosomal deficiency resource. Genetics 177, 615-629. 10.1534/genetics.107.076216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saarenpää T., Kogan K., Sidorova Y., Mahato A. K., Tascón I., Kaljunen H., Yu L., Kallijärvi J., Jurvansuu J., Saarma M. et al. (2017). Zebrafish GDNF and its co-receptor GFRalpha1 activate the human RET receptor and promote the survival of dopaminergic neurons in vitro. PLoS ONE 12, e0176166 10.1371/journal.pone.0176166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarov M., Barz C., Jambor H., Hein M. Y., Schmied C., Suchold D., Stender B., Janosch S., KJ V. V., Krishnan R. T. et al. (2016). A genome-wide resource for the analysis of protein localisation in Drosophila. Elife 5, e12068 10.7554/eLife.12068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoofs A., Hückesfeld S., Surendran S. and Pankratz M. J. (2014). Serotonergic pathways in the Drosophila larval enteric nervous system. J. Insect Physiol. 69, 118-125. 10.1016/j.jinsphys.2014.05.022 [DOI] [PubMed] [Google Scholar]
- Schuchardt A., D'Agati V., Larsson-Blomberg L., Costantini F. and Pachnis V. (1994). Defects in the kidney and enteric nervous system of mice lacking the tyrosine kinase receptor Ret. Nature 367, 380-383. 10.1038/367380a0 [DOI] [PubMed] [Google Scholar]
- Soba P., Han C., Zheng Y., Perea D., Miguel-Aliaga I., Jan L. Y. and Jan Y. N. (2015). The Ret receptor regulates sensory neuron dendrite growth and integrin mediated adhesion. Elife 4, e05491 10.7554/eLife.05491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa-Neves R., Lukacsovich T., Mizutani C. M., Locke J., Podemski L. and Marsh J. L. (2005). High-resolution mapping of the Drosophila fourth chromosome using site-directed terminal deficiencies. Genetics 170, 127-138. 10.1534/genetics.104.032516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speir M. L., Zweig A. S., Rosenbloom K. R., Raney B. J., Paten B., Nejad P., Lee B. T., Learned K., Karolchik D., Hinrichs A. S. et al. (2016). The UCSC Genome Browser database: 2016 update. Nucleic Acids Res. 44, D717-D725. 10.1093/nar/gkv1275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spieß R., Schoofs A. and Heinzel H.-G. (2008). Anatomy of the stomatogastric nervous system associated with the foregut in Drosophila melanogaster and Calliphora vicina third instar larvae. J. Morphol. 269, 272-282. 10.1002/jmor.10581 [DOI] [PubMed] [Google Scholar]
- Strelau J., Schober A., Sullivan A., Schilling L. and Unsicker K. (2003). Growth/differentiation factor-15 (GDF-15), a novel member of the TGF-beta superfamily, promotes survival of lesioned mesencephalic dopaminergic neurons in vitro and in vivo and is induced in neurons following cortical lesioning. J. Neural Transm. Suppl. 65, 197-203. 10.1007/978-3-7091-0643-3_12 [DOI] [PubMed] [Google Scholar]
- Sulkowski M. J., Han T. H., Ott C., Wang Q., Verheyen E. M., Lippincott-Schwartz J. and Serpe M. (2016). A novel, noncanonical BMP pathway modulates synapse maturation at the Drosophila neuromuscular junction. PLoS Genet. 12, e1005810 10.1371/journal.pgen.1005810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treanor J. J. S., Goodman L., de Sauvage F., Stone D. M., Poulsen K. T., Beck C. D., Gray C., Armanini M. P., Pollock R. A., Hefti F. et al. (1996). Characterization of a multicomponent receptor for GDNF. Nature 382, 80-83. 10.1038/382080a0 [DOI] [PubMed] [Google Scholar]
- Yadin D., Knaus P. and Mueller T. D. (2016). Structural insights into BMP receptors: specificity, activation and inhibition. Cytokine Growth Factor Rev. 27, 13-34. 10.1016/j.cytogfr.2015.11.005 [DOI] [PubMed] [Google Scholar]
- Yu H. H., Huang A. S. and Kolodkin A. L. (2000). Semaphorin-1a acts in concert with the cell adhesion molecules fasciclin II and connectin to regulate axon fasciculation in Drosophila. Genetics 156, 723-731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D., Ighaniyan S., Stathopoulos L., Rollo B., Landman K., Hutson J. and Newgreen D. (2014a). The neural crest: a versatile organ system. Birth Defects Res. C Embryo Today Rev. 102, 275-298. 10.1002/bdrc.21081 [DOI] [PubMed] [Google Scholar]
- Zhang W., Yan Z., Li B., Jan L. Y. and Jan Y. N. (2014b). Identification of motor neurons and a mechanosensitive sensory neuron in the defecation circuitry of Drosophila larvae. Elife 3, e03293 10.7554/eLife.03293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinke I., Kirchner C., Chao L. C., Tetzlaff M. T. and Pankratz M. J. (1999). Suppression of food intake and growth by amino acids in Drosophila: the role of pumpless, a fat body expressed gene with homology to vertebrate glycine cleavage system. Development 126, 5275-5284. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.