ABSTRACT
Phosphatidylinositol transfer proteins (PITPs) are essential regulators of PLC signalling. The PI transfer domain (PITPd) of multi-domain PITPs is reported to be sufficient for in vivo function, questioning the relevance of other domains in the protein. In Drosophila photoreceptors, loss of RDGBα, a multi-domain PITP localized to membrane contact sites (MCSs), results in multiple defects during PLC signalling. Here, we report that the PITPd of RDGBα does not localize to MCSs and fails to support function during strong PLC stimulation. We show that the MCS localization of RDGBα depends on the interaction of its FFAT motif with dVAP-A. Disruption of the FFAT motif (RDGBFF/AA) or downregulation of dVAP-A, both result in mis-localization of RDGBα and are associated with loss of function. Importantly, the ability of the PITPd in full-length RDGBFF/AA to rescue mutant phenotypes was significantly worse than that of the PITPd alone, indicating that an intact FFAT motif is necessary for PITPd activity in vivo. Thus, the interaction between the FFAT motif and dVAP-A confers not only localization but also intramolecular regulation on lipid transfer by the PITPd of RDGBα.
This article has an associated First Person interview with the first author of the paper.
KEY WORDS: Drosophila, FFAT–VAP, Lipid transfer, Membrane contact site, Phosphoinositides
Summary: FFA–VAP interactions regulate lipid transfer activity in Drosophila photoreceptors during PLC signalling.
INTRODUCTION
An essential feature of eukaryotic cells is the presence of membrane-bound organelles. There are several functional consequences of this form of subcellular compartmentalization. One of these is the asymmetric distribution and enrichment of cellular lipids to specific subcellular organelles due to the inability of these hydrophobic molecules to diffuse across the aqueous cytoplasm. The exchange of lipids between organelles occurs through vesicular transport as well as by the activity of lipid transfer proteins (LTPs) that are able to transfer lipids between organelle membranes. Recently, there has been considerable interest in the ability of such LTPs to mediate the localized transfer of lipids at membrane contact sites (MCSs) in cells (Toulmay and Prinz, 2011). MCSs are locations in cells where two organellar membranes are placed in close proximity (<30 nm) to each other without undergoing fusion (Helle et al., 2013).
Among the lipids that show asymmetric distribution in organelles are the phosphoinositides. These seven phosphorylated derivatives of phosphatidylinositol (PI) show a distinctive pattern of distribution in organelles (Idevall-Hagren and De Camilli, 2015). For example, the most abundant phosphoinositide, phosphatidylinositol 4,5 bisphosphate (PIP2) is enriched at the plasma membrane (PM) where it is the substrate for the activity of receptor-activated phospholipase C (PLC). PLC activation has two consequences, depletion of PIP2 and generation of a lipid product diacylglycerol (DAG). Since PIP2 is a low-abundance lipid, it needs to be resynthesized from the precursor PI, which itself is synthesized at the endoplasmic reticulum (ER). Further, DAG generated at the PM is phosphorylated to phosphatidic acid (PA) and this lipid needs to be transferred to the ER for its further metabolism to generate PI. These metabolic reactions, often called the PIP2 cycle, require the transfer of two lipid intermediates, PI from the ER to the PM, and PA from the PM to the ER (Cockcroft and Raghu, 2016). Transfer of PI from the ER to the PM has been ascribed to phosphatidylinositol transfer proteins (PITPs); these are LTPs capable of binding and transferring PI between membranes in vitro (Helmkamp et al., 1974). In vivo, lack of PITPs has been associated with a range of cellular phenotypes including defects in cytokinesis (Giansanti et al., 2006), Ca2+-activated secretion (Hay and Martin, 1993) and neurodegeneration (Alb et al., 2003; Hamilton et al., 1997) reflecting the importance of PITP function. Recently, it has been shown that Class II PITPs are also able to bind and transfer PA (Garner et al., 2012; Yadav et al., 2015) and it has been proposed that Class IIA PITPs might function as PI and PA transfer proteins that mediate the inter-membrane PI and PA transfer required for PIP2 resynthesis (Cockcroft et al., 2016; Kim et al., 2015). A number of studies have looked at the ability of Nir2 (Lev et al., 1999), a mammalian class II PITP in supporting PIP2 resynthesis during PLC signalling (Chang and Liou, 2015; Chang et al., 2013; Kim et al., 2015).
In addition to the biochemical properties of the Class IIA PI/PA transfer proteins, an important issue is the spatial context in which they perform lipid transfer functions. The subcellular localization of the Nir2 (Class IIA PITP in mammals) is debated. In mammalian cells, Nir2 has been reported to localize to the Golgi (Litvak et al., 2004, 2005; Peretti et al., 2008). However, recently, Nir2 has been reported to be distributed throughout the ER in mammalian cells and to translocate to the PM during receptor-mediated PLC activation (Chang and Liou, 2015; Chang et al., 2013; Kim et al., 2015). The precise molecular mechanism underlying this translocation remains unclear. It has previously been reported that an interaction between the FFAT motif of Nir2 and the ER-resident protein VAP (VAMP-associated protein) mediates Nir2 localization and activity (Amarilio et al., 2005; Peretti et al., 2008). However, recent studies have reported that the FFAT motif of overexpressed Nir2 may be dispensable for its translocation to the PM during PLC activation (Chang and Liou, 2015; Chang et al., 2013; Kim et al., 2015) although it may contribute to concentration of the protein at ER to PM contact sites (Chang and Liou, 2015; Kim et al., 2015). It has also been reported that PIP2 resynthesis is unaffected in mammalian cells lacking VAP-A and VAP-B (Dong et al., 2016). In addition, it has been reported that the LNS2 domain may mediate Nir2 localization and support its function during PIP2 resynthesis (Kim et al., 2013). Thus, a critical aspect of the biology of class IIA PITPs, namely their localization in relation to function, remains unresolved.
Drosophila photoreceptors (PRs) are an influential in vivo model to understand PITP function. In these cells, the detection of light is transduced into an electrical response through G-protein-coupled PLC activation (Raghu et al., 2012). PRs experience very bright light (>1×106 effective photons/s); thus a very high level of PLC activity and PIP2 hydrolysis is achieved. These cells express retinal degeneration B (RDGBα, encoded by rdgB), the founding member of the class IIA PITP family (reviewed in Trivedi and Padinjat, 2007). PRs lacking rdgB function show reduced electrical responses to light and undergo light-dependent retinal degeneration that requires ongoing PLC signalling, suggesting a role for RDGBα in light-dependent PIP2 turnover (reviewed in Trivedi and Padinjat, 2007). rdgB mutants also exhibit delayed kinetics of PIP2 turnover following PLC activation (Hardie et al., 2001, 2015; Yadav et al., 2015). RDGBα is a membrane-associated protein (Yadav et al., 2015) with a highly restricted localization within PRs (Suzuki and Hirosawa, 1994; Vihtelic et al., 1993). Endogenous RDGBα is present on sub-microvillar cisternae (SMC), a specialized ER compartment near the base of the rhabdomeral PM, an arrangement reminiscent of an ER–PM membrane contact site (MCS) (Yadav et al., 2016). In a recent study, rdgB mutants were shown to have defects in PI and PA turnover following activation of PLC signalling (Yadav et al., 2015). Thus, both RDGBα and its mammalian orthologue Nir2 (PITPMN1) function by transferring PI and PA at an ER–PM contact site (reviewed in Cockcroft and Raghu, 2016). Despite being a large multi-domain protein that includes several domains other than the PITP domain (PITPd), two independent studies have reported that expression of the PITPd of RDGBα rescues both the electrical response to light as well as retinal degeneration of rdgB mutants (Milligan et al., 1997; Yadav et al., 2015). Thus the significance, if any, of the non-PITPd domains of RDGBα in supporting phototransduction and the underlying light-activated PIP2 cycle remains unclear.
In this study, we report that in photoreceptors, the PITPd of RDGBα is dependent on its C-terminus for SMC localization. We find that the localization of RDGBα requires the presence of an intact FFAT motif and its interacting partner dVAP-A (also known as Vap33), which is enriched at the SMC. Furthermore, in the absence of an intact FFAT–dVAP-A interaction at the SMC in RDGBα, photoreceptors are unable to sustain three key phenotypes: (1) a normal light response, (2) PIP2 resynthesis during PLC activation and (3) photoreceptor structure during illumination. Thus the FFAT–VAP interaction-dependent enrichment of RDGBα at the SMC is necessary and sufficient to mediate a physiological process mediated by G-protein-coupled turnover of PIP2.
RESULTS
Non-PITP domain determinants are required to localize RDGBα to the MCS
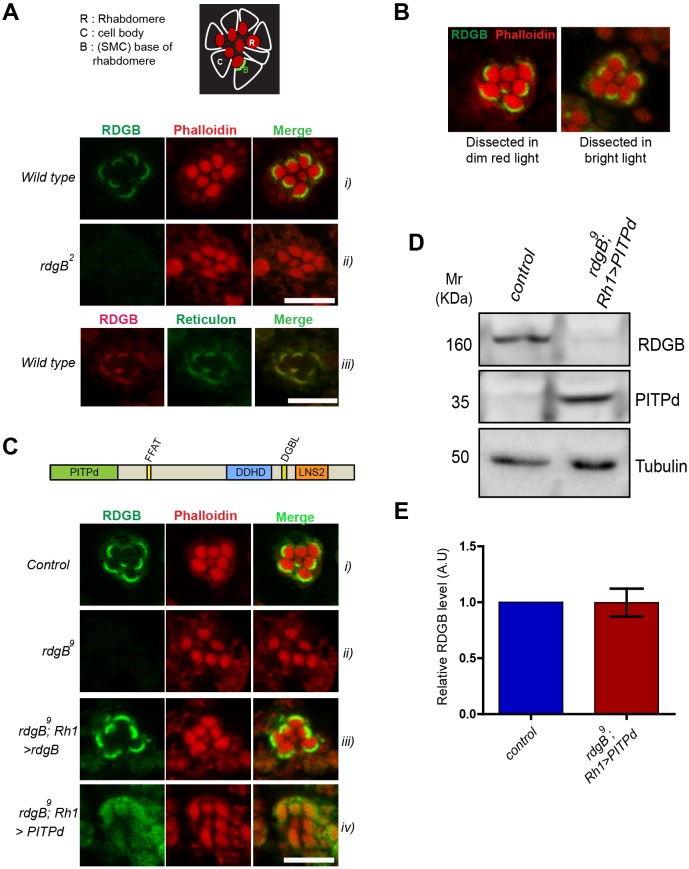
We determined the localization of RDGBα using a polyclonal antibody raised against its PITPd (Yadav et al., 2015). In wild-type PRs this antibody detects RDGBα localization in a crescent-shaped domain located at the base of the rhabdomeres (that are marked by Phalloidin staining) (Fig. 1Ai). This RDGBα staining was not detected in photoreceptors of rdgB2 flies, which are a protein null allele of rdgB, thus validating the antibody (Fig. 1Aii). This localization of RDGBα is consistent with that reported previously by immunoelectron microscopy (Suzuki and Hirosawa, 1994; Vihtelic et al., 1993). In this location, RDGBα co-localized with reticulon, an ER morphology regulator that has been previously reported to localize to the SMC in photoreceptors (Xia and Ready, 2011) (Fig. 1Aiii). Reticulon has been reported to localize to ER tubules in both mammalian (Voeltz et al., 2006) and Drosophila cells (Wakefield and Tear, 2006; Xu et al., 2016).We found that the localization of RDGBα in PRs was not dependent on light exposure (Fig. 1B), suggesting that RDGBα is constitutively present at the MCS at the base of the microvillar PM. Similar to results with rdgB2, no staining was observed in the hypomorph rdgB9. For subsequent experiments, we therefore used rdgB9 because previous characterization has been done with this allele (Yadav et al., 2015).
Fig. 1.
The PITP domain of RDGBα could not rescue all functions of RDGBα. (A) Localization of endogenous RDGBα in PRs using antibody against the PITP domain. (i) Confocal images showing localization of RDGB in wild-type PRs of 1-day-old flies. Phalloidin stains actin and marks the rhabdomeres. (ii) rdgB2 (null) shows no staining. (iii) RDGBα co-localizes with the reticulon as detected by anti-GFP staining of a reticulon::GFP fusion protein. Top panel is a cartoon representation of an ommatidium showing arrangement of PRs and the base of rhabdomeres. (B) Confocal images showing localization of RDGBα in wild-type PRs when dissected in dim red light versus bright light. (C) Localization of exogenously expressed RDGB and its PITPd. Confocal images showing localization of RDGBα in 1-day-old flies expressing rdgB and PITPd using Rh1Gal4 promoter. (D) Representative western blot image showing level of expression of Rh1>PITPd in rdgB9 head extracts. (E) Quantification of band intensity depicted in D. Error bars represent s.e.m. (n=4 blots). Scale bars: 5 µm.
We studied the localization of RDGBα reconstituted in rdgB9 photoreceptors (rdgB9;Rh1>rdgB) and found that the reconstituted protein localizes to the base of the microvilli, as seen for the endogenous protein (Fig. 1Ci and iii). However, when rdgB9 was reconstituted with the PITPd of RDGBα (rdgB9; Rh1>PITPd), we found that PITPd was diffusely distributed throughout the cell (Fig. 1Civ). The reduced intensity of staining observed at the SMC in flies expressing PITPd was not due to a low level of protein expression, as the level of PITPd expressed was similar to that of RDGBα protein in wild-type flies as quantified in western blots of protein extracts from fly heads (Fig. 1D,E). These results suggest that the PITPd of RDGBα lacks an essential signal responsible for localization of RDGBα to the SMC. This localization signal might reside within the extended C-terminus of RDGBα.
An intact FFAT motif is required to localize RDGBα to the SMC
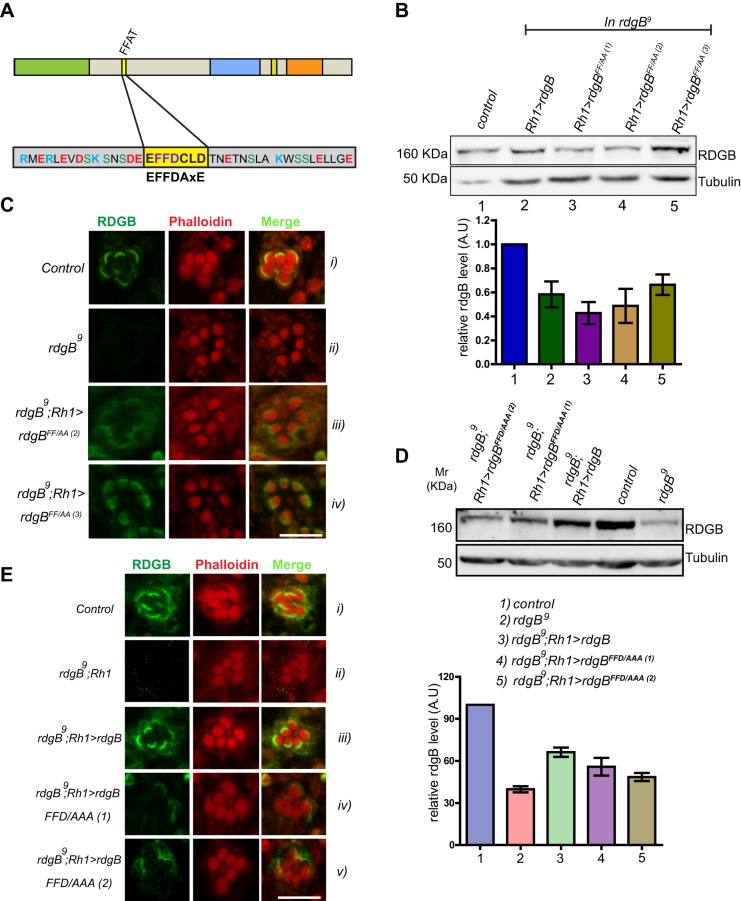
RDGBα is a large protein (1250 amino acids) with the PITPd located at the N-terminus. C-terminal to this are ∼1000 aa within which many domains are present, including an FFAT motif (Fig. 2A). The FFAT motif is implicated in the interaction of many proteins with ER-resident proteins (Loewen et al., 2003). Since RDGBα localizes to a modified ER compartment (i.e. SMC), we investigated the role of its FFAT motif in determining its localization. Structural studies on FFAT-mediated protein interaction suggest that the two phenylalanines of this motif are crucial for its interaction (Loewen et al., 2003). We mutated FF→AA in RDGBα and generated three independent transgenic lines of rdgBFF/AA (rdgBFF/AA(1), rdgBFF/AA(2) and rdgBFF/AA(3)). When reconstituted in rdgB9 photoreceptors (rdgB9; Rh1>rdgBFF/AA), the level of protein expression was found to be equivalent to that of wild-type rdgB (rdgB9; Rh1>rdgB) (Fig. 2B). Immunofluorescence experiments showed that RDGBFF/AA was partially mis-localized (Fig. 2Ci compared with iii,iv) although a significant fraction was still present at or around the SMC.
Fig. 2.
An intact FFAT motif is required to localize RDGBα to the SMC. FFAT mutants were expressed in rdgB9 flies using Rh1-Gal4 and their ability to rescue rdgB9 was tested. (A) Diagrammatic representation of various domains in RDGB (drawn to scale); PITPd, DDHD and LNS2 are three domains whereas DGBL refers to a DAG binding-like region. The core residues of the FFAT motif are highlighted in yellow, along with flanking residues. The FFAT motif was mutated to disable the ability of the FFAT motif to bind to VAP. Two versions were tested: FF→AA (RDGBFF/AA) and FFD→AAA (RDGBFFD/AAA). (B) Representative immunoblot showing expression level of RDGB in different RDGBFF/AA transgenic lines (top panel). Blot was re-probed for tubulin as a loading control. Bar chart shows the quantification of band intensity (n=2 blots). Error bars represent s.e.m. (C) Localization of the RDGBFF/AA protein in control, rdgB9 and two different transgenic lines. (D) Representative immunoblot showing expression level of RDGB in different RDGBFFD/AAA. Quantification of band intensity from these blots is shown in bar chart. Error bars represent s.e.m. (n=3 blots). (E) Confocal images showing localization of RDGBWT and RDGBFFD/AAA (two independent transgenic lines) in the background of rdgB9. Control and rdgB9 photoreceptors are also shown. Scale bars: 5 µm.
The partial mis-localization of RDGBFF/AA raised the following questions i.e. is the FF→AA mutation not sufficient to completely abrogate interaction with the SMC membrane or are there other membrane-targeting determinants? Although the FF→AA mutation is shown to abrogate FFAT-mediated interactions in yeast, it is also reported that making this mutation in a glycolipid transfer protein resulted in only partial loss of interaction and a triple mutation (FFD→AAA) was essential to completely abolish the interaction (Tuuf et al., 2009). To check this possibility, we generated an additional mutant namely rdgBFFD/AAA. When reconstituted in rdgB9(rdgB9;Rh1>rdgBFFD/AAA), this mutant protein was expressed at levels equivalent to that of the rescue construct of RDGB (Fig. 2D,E). Immunolocalization studies showed that rdgBFFD/AAA was completely mis-localized from the base of the rhabdomere and dispersed in the PR cell body (Fig. 2F). Thus, an intact FFAT motif is essential for the localization of RDGBα to the base of the rhabdomere.
dVAP-A is enriched at the SMC and is essential for localization of RDGBα
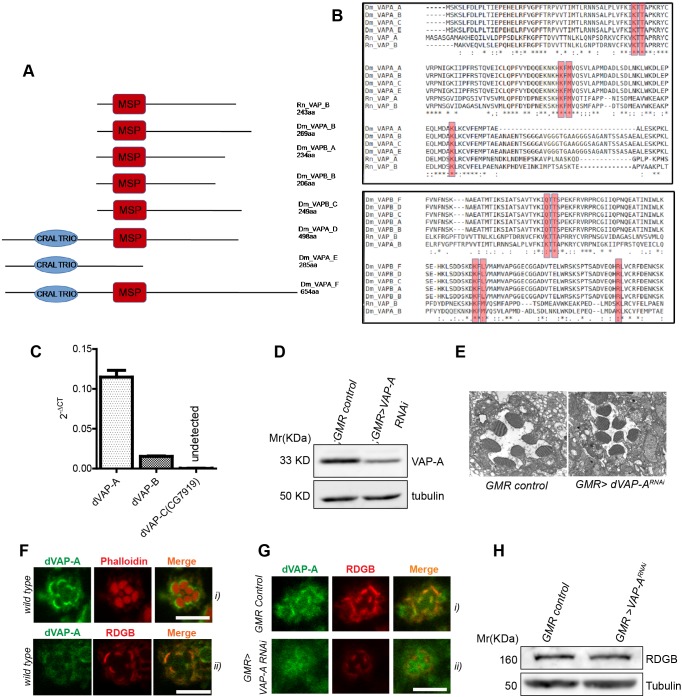
FFAT motif-containing proteins are known to interact with ER membrane VAP proteins (Loewen et al., 2003; Nishimura et al., 1999). The requirement of an intact FFAT motif for SMC localization of RDGBα raised the possibility of its interaction with VAP. However, since RDGBα was exclusively concentrated at the SMC and not on the rest of the ER, it was necessary to ascertain the nature of its interacting partner. The Drosophila genome has three genes that encode VAP, out of which two are ubiquitously expressed, namely dVAP-A (FlyBase annotation: CG5014) and dVAP-B (CG33523). The third VAP gene in Drosophila is called fan (CG7919) and is reported to be expressed exclusively in the testes. Both VAP-A and VAP-B have been reported to interact with the FFAT motif in mammalian systems (Murphy and Levine, 2016) and structural studies have identified five residues in VAPs that are crucial for interaction with an FFAT motif (Kaiser et al., 2005), namely K45, T47, K87, M89 and K118. We analysed the protein sequence of both dVAP-A and dVAP-B for conservation of these residues. dVAP-A has four predicted isoforms, all of which contain the MSP domain. dVAP-B has six isoforms, of which all except isoform E contain the MSP domain (Fig. 3A); in addition, isoforms D, E and F also contain a CRAL-TRIO domain. We aligned all isoforms of dVAP-A and dVAP-B that contain an MSP domain with rat VAP-A and VAP-B. This multiple alignment revealed that all of the five residues implicated in interaction with the FFAT motif were conserved in both dVAP-A and dVAP-B (Fig. 3B).
Fig. 3.
dVAP-A is enriched at the SMC and is essential for RDGBα localization. (A) Protein structure of Drosophila VAP proteins (dVAP-A and dVAP-B) and R. norvegicus VAP proteins (Rn VAP_A and Rn VAP_B). The structure of alternative splice variants of dVAP-A and dVAP-B are shown, as are the major sperm protein (MSP) domain and the CRAIL-TRIO domain. (B) Multiple sequence alignment of the Drosophila and mammalian VAP protein isoforms. Asterisk indicates conservation, dot marks lack of conservation. The conserved residues that coordinate with the FFAT motif of interacting proteins is shown within the red boxes. See Materials and Methods for protein ID. (C) Expression levels of dVAP mRNA in wild-type Drosophila retinae. RP49 was used as housekeeping gene and transcript levels for dVAP isoforms are shown normalized to RP49. (D) Representative immunoblot and quantification showing expression level of Dm-VAP-A in retinae extract of flies of mentioned genotypes (n=2 blots). (E) Representative scanning electron micrograph showing a single ommatidium of 1-day-old flies of indicated genotypes. (F) Confocal images showing localization of endogenously expressed dVAP-A in photoreceptors (i) and co-localization with RDGB (ii). (G) Confocal images showing mis-localization of RDGB in GMR>VAP-ARNAi. (H) Representative immunoblot showing expression level of RDGB in GMR>VAP-ARNAi compared with GMRGal4/+ flies. Scale bar: 5 µm.
We established expression of each of the three VAP genes in Drosophila retinae. Although we found that dVAP-A was highly expressed, dVAP-B mRNA was present at much lower levels; fan expression could not be detected in the retina (Fig. 3C). To test the importance of dVAP-A and dVAP-B in photoreceptors, we downregulated these genes in the Drosophila eye. Downregulation of dVAP-B (GMR>dVAP-BRNAi) using four independent RNAi lines resulted in a rough eye phenotype (data not shown), implying structural defects in the eye during development and precluding any further analysis of structure or function in the adult eye. RNAi of dVAP-A (GMR>dVAP-ARNAi) resulted in a 30% reduction of protein in retinae compared with levels in controls (Fig. 3D). GMR>dVAP-ARNAi flies showed normal eye development and the ultrastructure of the photoreceptors seemed largely undisturbed (Fig. 3E). Therefore, we focused on the role of dVAP-A in regulating RDGBα localization and function.
To determine the distribution of dVAP-A within PRs, we performed immunostaining with an antibody generated against the coiled-coil region of dVAP-A (validation of antibody in Fig. S1). We found reticular staining throughout the PR cell body; however, the staining was enriched at the base of rhabdomeres (Fig. 3Fi); in this location dVAP-A co-localized with RDGBα (Fig. 3Fii). The specificity of dVAP-A staining at the rhabdomere base was established by observing the reduction in staining when dVAP-A was downregulated using GMR>dVAP-ARNAi (Fig. 3G). Importantly, the intensity of RDGBα staining at the rhabdomere base was also reduced in GMR>dVAP-ARNAi compared with that in controls (Fig. 3G). This reduced staining intensity was not due to a reduction in the level of RDGBα protein expression (Fig. 3H). These observations suggest that dVAP-A is enriched at the SMC and is required for the localization of RDGBα at the base of the rhabdomere.
The FFAT–dVAP-A interaction is essential for normal RDGBα function in vivo
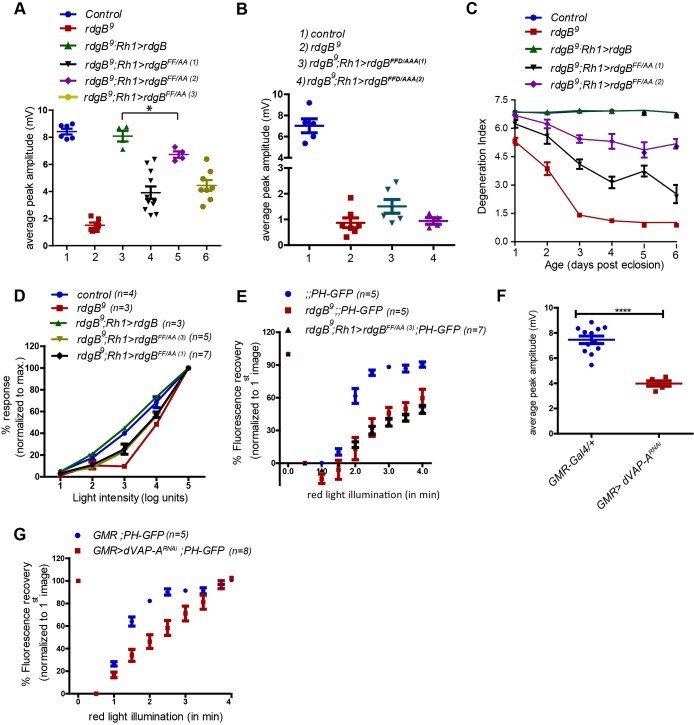
We tested the impact of a mutant FFAT motif on RDGBα function in vivo. When rdgB9 was reconstituted with rdgBFF/AA, its ability to rescue the reduced peak ERG amplitude was partial (Fig. 4A) and the rdgBFFD/AAA transgene was not able to rescue the ERG phenotype of rdgB9 at all (Fig. 4B). Likewise, the ability of rdgBFF/AA to rescue the retinal degeneration and reduced light sensitivity phenotype of rdgB9 was also partial (Fig. 4C,D) despite equivalent levels of protein being expressed (Fig. 2B). Like PITPd, expression of rdgBFF/AA failed to rescue PIP2 recovery kinetics (Fig. 4E). We also tested the physiological consequences of dVAP-A knockdown and found that this resulted in a reduced ERG amplitude (Fig. 4F) although photoreceptor ultrastructure at eclosion was normal (Fig. 3E). We measured the rate of PIP2 turnover and found that similar to results seen in rdgB9, knockdown of dVAP-A resulted in delayed recovery of PIP2 levels (Fig. 4G). Thus, disruption of the FFAT motif of RDGBα or depletion of its interacting partner dVAP-A impairs the ability of RDGBα to function in vivo.
Fig. 4.
FFAT–dVAP-A interaction is essential for normal RDGBα function in vivo. (A,B) Quantification of average ERG response of flies of indicated genotypes. Each data point represents a single fly tested. Error bars represent s.e.m. P-values were calculated using Student's t-test. (C) Comparison of retinal degeneration in flies exposed to 12 h:12 h light-dark regime (light intensity: 1100 lux). Degeneration index represents average number of intact rhabdomeres present per ommatidium. (D) Intensity response functions of 1-day-old flies of indicated genotypes. Average ERG response is depicted over a range of light intensities separated by 1 log unit each. Average ERG response at any given intensity of light is shown as a percentage of the response recorded at maximum light intensity. (E) Recovery kinetics of fluorescent pseudo pupil with time. (F) Quantification of average ERG response. Each data point represents a single fly tested. Error bars represent s.e.m. P-values were calculated using Student's t-test. (G) Recovery kinetics of fluorescent pseudo pupil with time. P-values were calculated using two way ANOVA with Bonferroni post-test corrections. Data points in A-G represent mean±s.e.m. *P<0.05, ****P<0.0001.
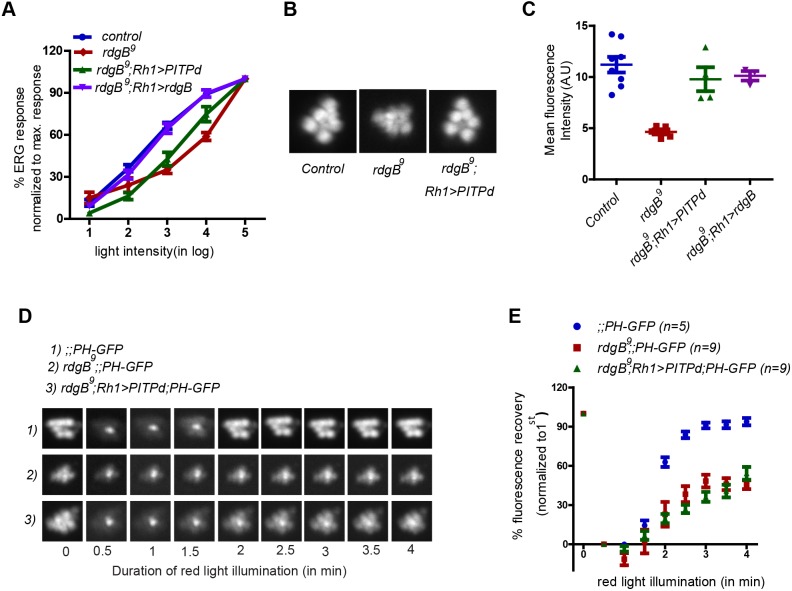
The PITPd alone is insufficient to rescue rdgB9 phenotypes during strong PLC stimulation
It has previously been reported that expression of the PITP domain of RDGBα alone is sufficient to rescue the reduced ERG amplitude of rdgB9. Therefore, it was surprising that rdgBFF/AA and rdgB FFD/AAA, both of which have an intact PITPd were unable to rescue the reduced ERG amplitude of rdgB. To explain this contradiction, we performed a detailed comparison of the functional efficiency of RDGBα and PITPd when reconstituted in rdgB9. As previously reported, in response to a single flash of light, both rdgB9;Rh1>PITPd and rdgB9;Rh1>RDGB showed equivalent rescue of rdgB9. We have previously shown (Yadav et al., 2015) that when rdgB9 flies are stimulated with logarithmically increasing light intensities, their average ERG response does not increase proportionately compared with wild-type controls (i.e. they have reduced light sensitivity). We tested the ability of RDGBα and PITPd to rescue the reduced light-sensitivity defect of rdgB9. Reconstitution with RDGBα (rdgB9;Rh1>rdgB) completely rescued the reduced light sensitivity of rdgB9, whereas the PITPd (rdgB9;Rh1>PITPd) showed only a partial rescue (Fig. 5A). rdgB9 flies are also characterized by reduced basal levels of PM PIP2 as reported by the fluorescent probe PH-PLCδ::GFP and delayed kinetics of PIP2 turnover; these phenotypes are rescued by expression of wild-type RDGBα (Yadav et al., 2015). We tested the ability of PITPd to rescue the delayed PIP2 turnover kinetics in rdgB9. Reconstitution with PITPd restored the basal PIP2 level in rdgB9 to wild-type levels (Fig. 5B,C); however, it did not rescue the delayed turnover kinetics (Fig. 5D,E). Thus, although PITPd is able to rescue some phenotypes of rdgB9, it fails to function as well as the full-length protein does under some physiological conditions. Given that the phenotypes that PITPd failed to rescue were correlated to the use of higher intensities of light for stimulation (see Materials and Methods), it is possible that the presence of the other, non-PITP domains is important to support the function of PITPd during high rates of PLC stimulation.
Fig. 5.
PITPd fails to rescue rdgB9 phenotypes during strong PLC stimulation. (A) Intensity response functions of 1-day-old flies of indicated genotypes. Average ERG response is depicted over a range of light intensities separated by 1 log unit each. Average ERG response at any given intensity of light is shown as a percentage of the response recorded at maximum light intensity. Each data point represents mean±s.e.m. N=5 for each genotype. (B) Representative pseudo pupil image of flies captured after 6 min of dark adaptation. (C) Quantification of mean intensity associated with the pseudo pupil. (D) Representative fluorescent pseudo pupil images acquired at different time points during pseudo pupil imaging of indicated genotypes. (E) Recovery kinetics of fluorescent pseudo pupil over time. Average fluorescence intensity of each pseudo pupil image is expressed as a percentage of the intensity of first image acquired. Each data point represents mean±s.e.m.
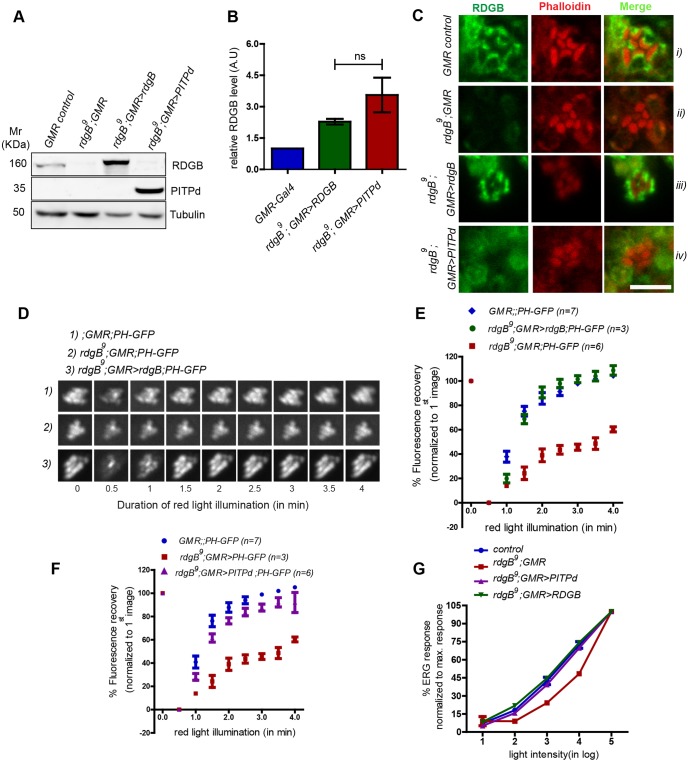
Enrichment of RDGBα at SMC–PM contact sites is necessary for complete rescue of rdgB9
One obvious difference between RDGBα and PITPd is their differential localization within PRs. In contrast to RDGBα, which is enriched at the SMC–PM contact site, PITPd is dispersed throughout the cell body. Therefore, we hypothesized that the lack of complete rescue by PITPd could be due to lower availability of the protein at the SMC (evident as diffuse staining at the SMC in Fig. 1Civ). To test this, we used a strong enhancer (GMR-Gal4) that expresses higher levels of PITPd protein, thus increasing its effective concentration throughout the PRs. As shown in Fig. 6A,B, high levels of PITPd are expressed and this was accompanied by increased staining in PRs (Fig. 6C). Since we were expressing PITPd at twice the wild-type level of RDGBα (Fig. 6B), to exclude any possible effect of such overexpression on PLC signalling, we also expressed full-length RDGBα using the same enhancer (GMR-Gal4). Despite having twice as much RDGBα protein as control cells, in GMR>rdgB, the protein was strictly localized to and around the SMC (Fig. 6Ciii).
Fig. 6.
Enrichment of RDGBα at SMC–PM contact sites is necessary for complete rescue of rdgB9 phenotypes. (A) Representative immunoblot showing expression level of RDGB and PITPd in flies of indicated genotypes. (B) Quantification of band intensity in A (n=3). Error bars represent s.e.m. P-values were calculated using Student's t-test. (C) Confocal images showing localization of PITPd. (D) Representative fluorescent pseudo pupil images acquired at different time points. (E,F) Recovery kinetics of fluorescent pseudo pupil with time. Average fluorescence intensity of each pseudo pupil image is expressed as percentage of the intensity of first image acquired. Each data point represents mean±s.e.m. (G) Intensity response functions of 1-day-old flies of mentioned genotypes. Average ERG response is depicted over a range of light intensities separated by 1 log unit each. Average ERG response at any given intensity of light is shown as a percentage of the response recorded at maximum light intensity. Each data point represents mean±s.e.m. Control, n=8; rdgB9, n=3; rdgB9;Rh1>PITPd, n=6; rdgB9;Rh1>RDGB, n=6.
We studied the ability of GMR>rdgB to rescue the delayed PIP2 turnover defect of rdgB9 and found that it was able to rescue this phenotype in rdgB9 (Fig. 6D,E). Furthermore, and in contrast to Rh1>PITPd, GMR>PITPd was also able to almost completely rescue the PIP2 turnover defect in rdgB9 (Fig. 6F). With high levels of PITPd present around SMC (GMR>PITPd), the light sensitivity defect was also completely rescued (Fig. 6G). Thus, an increase in the effective concentration of PITPd at the base of microvilli was able to rescue all phenotypes of rdgB9.
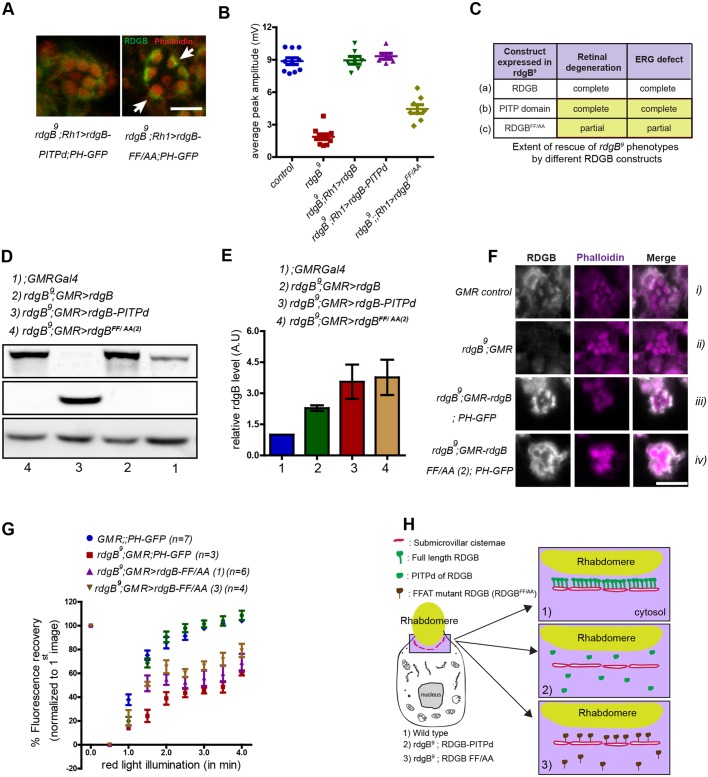
An intact FFAT motif is essential for RDGBα function independent of SMC localization
During these studies, we noted that although the RDGBFF/AA mutant protein retained substantial SMC localization, this was not reflected in its ability to rescue rdgB phenotypes; i.e. we noticed a lack of correlation between the SMC localization and functional rescue shown by RDGBFF/AA. Although, the effective availability of RDGBFF/AA at the SMC in PRs expressing rdgBFF/AA was much higher compared with that in PRs expressing PITPd (see arrows in Fig. 7A), RDGBFF/AA did not completely rescue the reduced ERG response (comparison shown in Fig. 7B) or the retinal degeneration defect (summarized in Fig. 7C). To understand the basis of these differences, we quantified and compared the total amount of protein expressed between flies expressing PITPd and rdgBFF/AA using GMRGal4 and found it to be similar (Fig. 7D,E). Besides an equivalent level of protein expression, the relative intensity of staining at SMC was higher for RDGBFF/AA compared with PITPd (compare Fig. 7Fiv with Fig. 6Civ). This observation suggests that the PITPd of the RDGBFF/AA protein has a reduced function (as measured by its ability to rescue phenotypes) compared with that of the PITPd when expressed alone. If this is true, then increasing the expression of RDGBFF/AA might not confer rescue similar to that shown by overexpression of PITPd (see Fig. 6F,G). We tested this by overexpressing rdgBFF/AA using GMRGal4 (Fig. 7D,E). As shown in Fig. 7F (summarised in Fig. 7H), a significantly higher level (twice that of the wild type) of RDGBFF/AA was expressed and the staining observed at the SMC was similar to that expressed in wild-type flies. Under these conditions we tested its ability to rescue the delayed PIP2 turnover kinetics of rdgB9. Even with an equivalent level of protein present at SMC (evident as similar staining intensities) compared with wild-type, RDGBFF/AA was unable to rescue PIP2 turnover kinetics of rdgB9 (Fig. 7G). These results suggest that the fraction of RDGBFF/AA protein that was still localized to the SMC failed to support PIP2 turnover and suggests that the PITPd associated with the RDGBFF/AA mutant protein was functionally less competent than the PITPd expressed alone.
Fig. 7.
An intact FFAT motif is essential for RDGBα function independent of SMC localization. (A) High-magnification confocal images of a single ommatidium from rdgB9;Rh1>PITPd and rdgB9; Rh1>rdgBFF/AA. Note the relatively strong and concentrated staining near the base of rhabdomeres in the case of RDGBFF/AA compared with uniformly dispersed staining in case of PITPd. (B) Average ERG response of 1-day-old flies plotted from data pooled across experiments performed under the same conditions (for comparison). Error bars represent s.e.m. (C) Summary of rescue experiments using different RDGB constructs in rdgB9 flies. Compare the level of functional rescue achieved by expressing wild-type RDGB (a), only the PITPd of RDGB (b) and RDGB with mutated FFAT motif but intact wild-type PITP domain (c). Results are shown for equivalent level of protein expression. (D) Representative immunoblot image showing increase in expression level of RDGB and PITPd with GMR-Gal4 promoter. Tubulin was used as a loading control and rgdB9 is a negative control for RDGB. (E) Quantification of band intensity of RDGB normalized to tubulin as depicted in D (n=3 blots). Error bars represent s.e.m. (F) Confocal images showing localization of RDGBFF/AA in PRs using GMR-Gal4 promoter. Appropriate genetic controls were stained simultaneously. (G) Recovery kinetics of fluorescent pseudo pupil with time. Average fluorescence intensity of each pseudo pupil image is expressed as a percentage of the intensity of first image acquired. Each data point represents mean±s.e.m. (H) Cartoon representation (not to scale) of the localization and relative number of PITPd expressed in photoreceptors in case of wild-type flies (1), rdgB9 flies expressing PITPd (2) or RDGBFF/AA (3). Scale bars: 5 µm.
DISCUSSION
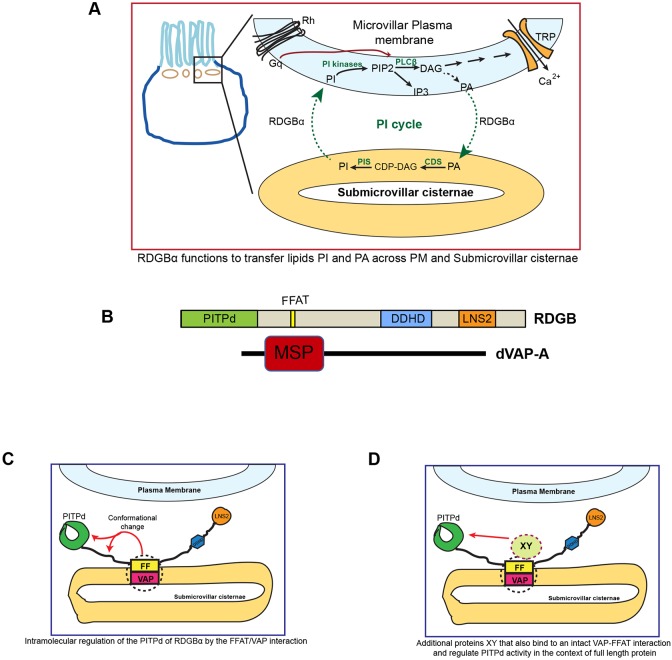
Many examples of receptor-triggered signalling involve PLC activation and the hydrolysis of PIP2. PLC activity triggers a series of biochemical reactions, each of which involves the generation of a lipid product. These biochemical reactions are distributed between two compartments, the PM and the ER, and since lipids cannot diffuse across the aqueous cytosol, it has long been recognized that cells need a mechanism to overcome this topological constraint and transfer lipid intermediates of the PIP2 cycle between the PM and the ER (Michell, 1975). The recent description of class IIA PITPs as proteins whose PITP domain can mediate transfer of PI and PA offers an elegant molecular solution for the transfer of both these key lipid intermediates during PLC signalling (Kim et al., 2015; Yadav et al., 2015). Although the class IIA PITPs possess the relevant biochemical activity, a cell biological mechanism is needed for them to access both the ER and PM to effect lipid transfer. Class IIA PITPs are membrane-associated proteins precluding the diffusion of lipid-bound protein between the ER and PM across aqueous cytosol. This cell biological problem is even more pronounced in cell types or signalling contexts where high rates of PLC activation occur on rapid (millisecond) time scales (e.g. mGluR activation in neurons or PLC activation in photoreceptors). The description of MCSs as cellular locations where ER and PM are relatively closely localized offers an elegant solution to the lipid transfer requirements of the PIP2 cycle but it requires class IIA PITPs to be localized at MCSs in cells. In this study, we have tested the importance of localizing the class IIA PITP, RDGBα to the SMC, part of a stable PM–ER contact site in Drosophila photoreceptors. In contrast to mammalian cells studied thus far, in Drosophila photoreceptors, endogenous RDGBα is constitutively localized to the SMC, an ER–PM contact site. Drosophila photoreceptors show high rates of basal PLC activity (Hardie et al., 2004) and this increases dramatically, several-fold on a millisecond time scale during illumination (Hardie and Raghu, 2001). To prevent PIP2 depletion during this signalling process, a rapid increase of lipid transfer reactions will be required on a rapid time scale and this is probably the reason for the constitutive localization of RDGBα at the photoreceptor MCSs. We observed that molecular changes that mis-localize RDGBα away from the SMC, thereby reducing its concentration at this location, result in an inability to support PIP2 turnover and photoreceptor physiology, particularly during high rates of PLC signalling. During this study, we also found that expressing only the PITPd of RDGBα, but at high levels, was able to rescue all aspects of rdgB mutant phenotypes. This finding implies that a principal function of localizing RDGBα to the SMC is to ensure that an adequate concentration of the PITPd is present at this location (Fig. 7H) to mediate lipid transfer during high rates of PLC stimulation. Thus, our study underscores the importance of localizing Class IIA PITPs to the ER–PM MCSs to support a PIP2-dependent physiological process in vivo. At MCSs, class IIA PITPs are likely to mediate the transfer of lipids through their PITPd, thus supporting in vivo physiology during high rates of PLC signalling (Fig. 8A). In mammalian cells in culture, a distinctive behaviour of overexpressed class IIA PITPs has been reported (e.g. Chang and Liou, 2015; Kim et al., 2013, 2015), namely translocation of the protein from the cell interior to the PM during PLC signalling. The localization and behaviour of endogenous Nir2 protein during PLC signalling has not been studied. It is likely that in mammalian cells, such as neurons, that experience high rates of PLC stimulation, endogenous class IIA PITPs will be localized at equivalent MCS locations to those seen in Drosophila photoreceptors, supporting PIP2 signalling and in vivo physiology. Identification and analysis of such cell types will be of value in the future.
Fig. 8.
RDGBα function at the PM–ER membrane contact site. (A) Schematic representation of the PIP2 cycle, which runs between the plasma membrane and the underlying sub-microvillar cisternae (SMC) in photoreceptors. RDGBα transfers the lipid PI from SMC to PM and transfers PA back from PM to SMC. CDS, CDP diacylglycerol synthase; CDP, cytidine diphosphate; DAG, diacylglcerol; PIS, PI synthase; TRP, transient receptor potential channel. (B) Representation (drawn to scale) of various domains and motifs in RDGBα and dVAP-A. (C) Hypothesized topology of RDGBα and VAP depicting the binding of its FFAT motif to VAP on the SMC. The FFAT–VAP interaction results in an intramolecular conformational change which is important for activity of the PITP domain. (D) Alternative molecular mechanism for the ability of the FFAT–VAP interaction to regulate the function of the PITP domain. The FFAT–VAP interaction recruits and activates a presently undefined effector ‘XY’ which is able to regulate the activity of PITP domain.
Given the functional importance of localizing RDGBα to the SMC, it is essential to understand the molecular mechanism that underlies localization of this protein. Our finding that the PITPd alone was unable to localize to the SMC implies that the relevant localization sequence/motif exists C-terminal to the PITPd. Although the C-terminal region of the protein contains a number of domains (FFAT motif, DDHD, LNS2 and Ca2+-binding domain), in this study we found that mutation of the FFAT motif (FFD→AAA) resulted in complete mis-localization of RDGBα from the SMC into the cell body. Thus, the FFAT motif is both necessary and sufficient to localize RDGBα to the SMC. FFAT motifs normally interact with VAP on the ER; we found that dVAP-A is enriched at the SMC, and depletion of dVAP-A resulted in mis-localization of RDGBα protein and phenocopied the physiological consequences of RDGBα loss of function. Together, these observations imply that interaction of the FFAT motif with dVAP-A underpins the localization of RDGBα to the SMC. A similar requirement of FFAT–VAP interaction for the localization of Nir2 in the context of ER (Amarilio et al., 2005) and Golgi (Peretti et al., 2008) localization has been described. Interestingly, two previous studies have reported a role for the LNS2 domain (Chang and Liou, 2015; Kim et al., 2013) and a putative DAG binding-like (DGBL) region (Kim et al., 2015) in the translocation of overexpressed Nir2 in cultured mammalian cells. The sequence corresponding to the LNS2 and DGBL are conserved in RDGBα but the significance of these domains for the localization of this protein in Drosophila photoreceptors is not known. Although the LNS2 and DGBL are intact in the RDGBFFD/AAA protein, it remains mis-localized away from the SMC. This finding implies that in Drosophila photoreceptors, these other domains are insufficient to localize RDGBα to the SMC in the absence of an intact FFAT–VAP interaction. It is also possible that while the FFAT–VAP interaction is conserved across different cell types and species, under the conditions of cultured mammalian cells, other domains may also be co-opted and contribute to Nir2 localization.
During our analysis of the importance of the FFAT–VAP interaction in localizing RDGBα, we found that partial disruption of the FFAT motif (RDGBFF/AA) resulted in substantial amounts of residual RDGBα protein at the SMC. Despite this, RDGBFF/AA was less effective in rescuing key phenotypes of rdgB mutants compared with the PITPd alone, when expressed at high levels. Remarkably, this was despite the fact that RDGBFF/AA was present at the SMC at higher concentrations than PITPd in these experiments (Fig. 2Civ). These findings imply that when present in the full-length RDGBFF/AA protein, the PITPd has a reduced function compared with the isolated PITPd (i.e. not in cis with a mutant FFAT motif) as measured by the ability to rescue in vivo phenotypes. Collectively, these observations imply that in RDGBα, the intermolecular interaction between the FFAT motif and dVAP-A confers function on the RDGB PITPd. To date, no study has examined the impact of the FFAT–VAP interaction on the transfer activity of a PITPd in vitro. However, in the case of OSBP which is a sterol/PI4P transfer protein at the Golgi–ER MCSs, the addition of soluble VAP-A appears to regulate the lipid-transfer activity of OSBP in an in vitro reconstitution assay (Mesmin et al., 2013); the relevance of this observation to an in vivo context where VAP-A is membrane-bound and engaged in a range of protein–protein interactions is unclear. Our observations on the impact of FFAT–VAP interaction on the function of RDGBα may represent an in vivo correlate of the in vitro observation of Mesmin et al. (2013).
How might the FFAT–dVAP-A interaction mediate the function of the RDGB PITPd? One possibility is that this could occur at an intramolecular level where conformational changes within RDGBα mediated by the FFAT–dVAP-A interaction could regulate the function of the RDGB PITPd (Fig. 8C). Signals arising from the ER membrane (where the FFAT–dVAP-A interaction would occur), such as changes in the levels of lipid intermediates at the ER membrane that fluctuate during the PIP2 cycle, may trigger this. It is now apparent that VAP interacts with numerous proteins in the cell (Murphy and Levine, 2016). Thus, additional FFAT-containing proteins recruited by dVAP-A to the SMC may themselves contribute to regulation of the lipid-transfer function of RDGBα. In the absence of such additional proteins, the activity of the PITPd in RDGBFF/AA (Fig. 8D) is impaired, resulting in physiological phenotypes in vivo. Proteomic analysis of the VAP-dependent protein complex at the SMC or genetic screens may help uncover the identity of such molecules. Nonetheless, our study provides compelling evidence that the additional domains of a multi-domain PITP are likely to impact its localization and the function of its PITP domain at MCSs in vivo.
MATERIALS AND METHODS
Fly stocks
For all experiments, Red Oregon-R was used as control and is referred to as wild type. All flies (Drosophila melanogaster) were fed on standard corn meal medium with 1.5% yeast and were reared at 25°C in incubators with no internal illumination. UAS-GAL4 binary system was used to drive expression of transgenic constructs. For driving expression in photoreceptors, two Gal4 lines were used, namely Rh1Gal4 (denoted as Rh1>), which drives expression in all peripheral (R1-R6) photoreceptors and GMRGal4 (denoted as GMR>), which carries the transposon P{GAL4-ninaE.GMR} and drives Gal4 expression in all eye cells posterior to the furrow (Ellis et al., 1993).
Transgenic flies
FF→AA mutant was made in full-length RDGB by PCR-based site-directed mutagenesis and cloned in a pUAST insect vector. The complete open reading frame was sequence-verified post-mutagenesis. Multiple lines of transgenic flies were generated by P element-based transgenesis (Rubin and Spradling, 1982). The sequence corresponding to the PITP domain (1-280 aa of clone GH09970) was cloned in pUAST vector. The FFD→AAA mutation was introduced in full-length RDGB using a QuikChange II site-directed mutagenesis kit and was cloned in pUAST-attB vector. Site-specific transgenic flies carrying insertion on the second chromosome were generated. For dVAP-A, the VDRC line v30404 was used. For dVAP-B, all the four VDRC lines (v37237, v37238, v44377 and v110519) were tested and gave a rough eye phenotype when expressed with GMR-Gal4.
Optical neutralization
Imaging was performed as described (Franceschini and Kirschfeld, 1971). Briefly, flies were anesthetized on ice followed by decapitation. The heads were placed on a drop of nail varnish and imaged under an oil-immersion microscope at 40× magnification (Olympus BX43). For all experiments, flies were reared in the dark and transferred to a 12 h:12 h light/dark regime (light intensity: 1500 lux) post eclosion unless otherwise mentioned. A total of 50 ommatidia were scored across at least five different flies. Retinal degeneration was scored by counting the number of intact rhabdomeres per ommatidium. Intact rhabdomeres, which were round in appearance, in well focused ommatidium were given a score of 1, whereas any deformation from the round structure was given a score of 0.5. Missing rhabdomeres were scored 0. Degeneration index was calculated as average number of intact photoreceptors per ommatidium.
Electrophysiology
Flies were anesthetized on ice and immobilized at the end of a 200 µl disposable pipette tip using a drop of nail varnish. The recording electrode was placed on the surface of the eye and the reference electrode was placed on the neck region/thorax. Flies were dark adapted for 5 min followed by 10 repeated green light flashes of 2 s duration, each after an interval of 10 s. Recordings were done using GC 100F-10 borosilicate glass capillaries (1 mm OD and 0.58 mm ID) (Harvard apparatus) filled with 0.8% w/v NaCl solution. Stimulating light was delivered from an LED light source placed within a distance of 5 mm of the fly's eye through a fibre optic guide. Calibrated neutral density filters were used to vary the intensity of the light over 5 log units. Voltage changes were amplified using a DAM50 amplifier (WPI) and recorded using pCLAMP 10.2. Analysis of traces was performed using Clampfit (Axon Laboratories). Green light was used for excitation and the number of photons emitted at five independent ND filters in increasing order was (2×1018)<(2×1019)<(4×1020)<(6×1021)<(7.9×1022).
Live pseudo pupil imaging
Pseudo pupil imaging was done on flies expressing a single copy of PH-PLCδ::GFP (PH domain of PLCδ tagged to GFP) driven by the trp promoter. Briefly, flies were immobilized at the end of a pipette tip and the fluorescent pseudo pupil was focused at 16× magnification using Olympus IX71 inverted microscope. Images were captured (Evolve 512 camera) at nine different time intervals as indicated in respective figures, using Micromanager 1.4 software (Edelstein et al., 2010). Flies were pre-adapted by exposure to red light for 6 min prior to the start of pseudo pupil imaging. Blue light of 90 ms duration was used to image the fluorescent pseudo pupil; this illumination also stimulates phototransduction. In order to terminate the continued stimulation of Gq and PLC by meta-rhodopsin (M*) generated by blue light illumination, the fly eye was illuminated with red light that converts (M*) to rhodopsin. Red light exposure of increasing time duration was given between successive image acquisitions. Changes in fluorescence intensity of the pseudo pupil imaged at each time point reflects diffusion of the probe back into the microvilli and its rebinding to the PM, which presumably depends partly on the rate of PIP2 resynthesis. The fluorescence intensity was measured using ImageJ from NIH. Images were processed equivalently to correct for background intensity. For quantification of percentage recovery, the fluorescence value of the second image was subtracted from all the images and then the fluorescence value of the first image was set as 100. Fluorescence values of the remainder of the images were normalized to the value of the first. For all experiments, 1-day-old flies were used.
RNA extraction and Q-PCR
0- to 12-h-old flies were snap frozen in liquid nitrogen and dehydrated in acetone at −80°C for 48 h. The acetone was then drained off and the heads dried at room temperature. The retinae were cleanly separated from the head at the level of the basement membrane using a scalpel blade. RNA was extracted from 50 retinae in TRIzol (Invitrogen). Purified RNA was treated with amplification-grade DNase I (Invitrogen). cDNA conversion was done using SuperScript II RNaseH reverse transcriptase (Invitrogen) primed with random hexamers (Applied Biosystems). Quantitative PCR (Q-PCR) was performed on an Applied Biosystems 7500 Fast Real-Time PCR instrument. Primers were designed at the exon–exon junction following the parameters recommended for Q-PCR. Transcript levels of ribosomal protein 49 (RP49) were used for normalization across samples. Three separate samples were collected for each genotype. Technical replicates of each sample were performed to ensure consistency of the data. Q-PCR primers used: dVAP-A forward primer (FP): 5′-CAGGTGGAGATCTGCCTTCAGC-3′; reverse primer (RP): 5′-TCGCTTAGATCAGCATCCATGG-3′; dVAP-B FP: 5′-ATCCGAGGGCATGTTGCACAT-3′; RP: 5′-CTTGTACGTCACTGCCGAGGT-3′; dVAP-C FP: 5′-GTGATCGTCTTTGAGGGACCAT-3′; RP: 5′-CGTACATAAAAAAGTTTTGGAG-3′.
Western blotting
Protein extracts from flies were prepared by homogenizing heads in 2× SDS-PAGE sample buffer (with 4% SDS) followed by incubation at 90°C for 5 min. Following separation by SDS-PAGE, proteins were transferred to nitrocellulose membrane (Amersham) using Bio-Rad Trans-Blot SD semi-dry transfer assembly. Dilutions of primary antibodies used are: anti RDGB (1:4000, polyclonal; made in-house against the PITP domain of RDGB), anti-tubulin (1:4000, Clone E7, DSHB), anti dVAP-A (1:2000, generated in house, see below). All secondary antibodies (Jackson Immunochemicals, cat nos. 711-035-152,112-035-003, 115-035-003) were used at 1:10,000 dilution.
Electron microscopy
For EM study, flies were reared in darkness. One-day-old flies were decapitated and the head was bisected in a drop of ice-cold fixative (2.5% glutaraldehyde in 0.1 M PIPES buffer, pH 7.4). The bisected heads were then incubated in fixative for 10-12 h at 4°C. Post primary fixation these were washed with 0.1 M PIPES and placed in 1% OsO4 (30 min) for secondary fixation. The samples were stained en bloc in 2% uranyl acetate for 1 h after which they were dehydrated in a series of ethanol concentrations (50%, 70%, 90% and 100%) and embedded in epoxy. The blocks were heated at 60°C for 2 days and ultrathin sections (50 nm) were cut and picked in grids. Imaging was done using a scanning electron microscope model MERLIN Compact VP.
Antibody generation
For generation of anti-VAP antibody, the coiled-coiled region of VAP (cc-VAP) was chosen as an epitope. The coding region spanning 150 to 243 aa was cloned into the BamHI and SalI site of the p-GEX4T1 vector. The protein was expressed in E. coli (BL21DE3) cells by induction with 1 mM isopropyl-D-thiogalactoside (IPTG). The cells were lysed by sonication in lysis buffer [50 mM Tris-HCl, pH 7.5, 500 mM NaCl, 0.5% Triton X-100] with protease inhibitor cocktail (Roche) and 10 mg of the recombinant protein was purified by affinity chromatography using glutathione beads (GE healthcare). The GST tag was cleaved from the recombinant protein using thrombin and purified protein was used for antibody generation from rabbits (Abexome Biosciences). The antibody (VAP-Rb-446) was validated both in western blots and in tissues. The antibody gave single bands with purified protein (data not shown) and levels monitored by both overexpression and knockdown of VAP in S2 cells (Fig. S1). As expected, the antibody crossreacted with a VAP(P58S) variant that forms cellular inclusions. Reduction of VAP by RNAi led to a decrease of expression of protein and thus the inclusions.
Immunohistochemistry
For immunofluorescence studies, retinae from flies were dissected under low red light in phosphate-buffered saline (PBS). For Fig. 1B, retinae were dissected in bright light. Retinae were fixed in 4% paraformaldehyde in PBS with 1 mg/ml saponin at room temperature for 30 min. Fixed eyes were washed three times in PBST (1× PBS+0.3% Triton X-100) for 10 min. The sample was then blocked in a blocking solution (5% fetal bovine serum in PBST) for 2 h at room temperature, after which the sample was incubated with primary antibody in blocking solution overnight at 4°C on a shaker. The following antibodies were used: anti-RDGB (1:300), anti-VAP (1:3000) and anti-GFP (1:5000, Abcam, ab13970). Appropriate secondary antibodies conjugated with a fluorophore were used at 1:300 dilutions [Alexa Fluor 488 anti-rabbit (A11034), anti-rat (A11006) and Alexa Fluor 633 anti-rabbit (A21070), anti-rat (A21094), anti-chicken (A21103) IgG (Molecular Probes)] and incubated for 4 h at room temperature. When required, during the incubation with secondary antibody, Alexa Fluor 568–Phalloidin (Invitrogen, A12380) was also added to the tissues to stain F-actin. After three washes in PBST, sample was mounted in 70% glycerol in 1× PBS. Whole-mounted preps were imaged on an Olympus FV1000 confocal microscope using a Plan-Apochromat 60×, NA 1.4 objective (Olympus).
Multiple sequence alignment
The sequences of VAP proteins were obtained from FlyBase (http://flybase.org/). The sequence identifiers are as follows: Rn_VAP_A: NP_113819.3; Rn_VAP_B: NP_068619.1; DmVAPA_A: FBpp0089169; DmVAPA_B: FBpp0070589; DmVAPA_C: FBpp0089170; DmVAPA_E: FBpp0309607; DmVAPB_A: FBpp0090957; DmVAPB_Bb: FBpp0090955; DmVAPB_Cc: FBpp0090956; DmVAPB_Dd: FBpp0090954; DmVAPB_F: FBpp0303152. Two separate multiple sequence alignments were obtained using CLUSTALO (Sievers et al., 2011). First alignment includes the VAP-A and VAP-B sequence from Rattus norvegicus and the four isoforms of the D. melanogaster VAP-A sequence. The second one includes the VAP-B sequence from R. norvegicus, VAP-A sequence from D. melanogaster and the five isoforms of the D. melanogaster VAP-B sequence. The Pfam domains and the conserved residues involved in binding to FFAT motif-containing proteins were manually mapped on the sequences and the alignment.
Supplementary Material
Acknowledgements
We thank the National Centre for Biological Sciences Electron Microscopy, Central Imaging and Flow Cytometry Facility and Fly Facility for assistance. We thank colleagues for critical reading of this manuscript and suggestions.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: R.P.; Methodology: S.Y., R.T., P.G., S.D., H.K., G.R.; Formal analysis: S.Y., H.K.; Investigation: R.T., H.K.; Resources: P.G., S.D., G.R.; Writing - original draft: S.Y., R.P.; Writing - review & editing: S.Y., R.T., R.P.; Supervision: G.R., R.P.; Project administration: R.P.; Funding acquisition: R.P.
Funding
This work was supported by the National Centre for Biological Sciences TIFR and a Wellcome Trust/DBT India Alliance Senior Fellowship to R.P. Work in the G.R. lab was supported by intramural grants from the Indian Institute of Science Education and Research (IIISER, Pune to G.R.) and the DST Science and Engineering Research Board (SERB) (EMR/2014/000367). Deposited in PMC for release after 6 months.
Supplementary information
Supplementary information available online at http://jcs.biologists.org/lookup/doi/10.1242/jcs.207985.supplemental
References
- Alb J. G. Jr, Cortese J. D., Phillips S. E., Albin R. L., Nagy T. R., Hamilton B. A. and Bankaitis V. A. (2003). Mice lacking phosphatidylinositol transfer protein-alpha exhibit spinocerebellar degeneration, intestinal and hepatic steatosis, and hypoglycemia. J. Biol. Chem. 278, 33501-33518. 10.1074/jbc.M303591200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amarilio R., Ramachandran S., Sabanay H. and Lev S. (2005). Differential regulation of endoplasmic reticulum structure through VAP-Nir protein interaction. J. Biol. Chem. 280, 5934-5944. 10.1074/jbc.M409566200 [DOI] [PubMed] [Google Scholar]
- Chang C.-L. and Liou J. (2015). Phosphatidylinositol 4,5-bisphosphate homeostasis regulated by Nir2 and Nir3 proteins at endoplasmic reticulum-plasma membrane junctions. J. Biol. Chem. 290, 14289-14301. 10.1074/jbc.M114.621375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C.-L., Hsieh T.-S., Yang T. T., Rothberg K. G., Azizoglu D. B., Volk E., Liao J.-C. and Liou J. (2013). Feedback regulation of receptor-induced Ca2+ signaling mediated by E-Syt1 and Nir2 at endoplasmic reticulum-plasma membrane junctions. Cell Rep. 5, 813-825. 10.1016/j.celrep.2013.09.038 [DOI] [PubMed] [Google Scholar]
- Cockcroft S. and Raghu P. (2016). Topological organisation of the phosphatidylinositol 4,5-bisphosphate-phospholipase C resynthesis cycle: PITPs bridge the ER-PM gap. Biochem. J. 473, 4289-4310. 10.1042/BCJ20160514C [DOI] [PubMed] [Google Scholar]
- Cockcroft S., Garner K., Yadav S., Gomez-Espinoza E. and Raghu P. (2016). RdgBα reciprocally transfers PA and PI at ER-PM contact sites to maintain PI(4,5)P2 homoeostasis during phospholipase C signalling in Drosophila photoreceptors. Biochem. Soc. Trans. 44, 286-292. 10.1042/BST20150228 [DOI] [PubMed] [Google Scholar]
- Dong R., Saheki Y., Swarup S., Lucast L., Harper J. W. and De Camilli P. (2016). Endosome-ER contacts control actin nucleation and retromer function through VAP-dependent regulation of PI4P. Cell 166, 408-423. 10.1016/j.cell.2016.06.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelstein A., Amodaj N., Hoover K., Vale R. and Stuurman N. (2010). Computer control of microscopes using μ manager. Curr. Protoc. Mol. Biol. 14, 1-17. 10.1002/0471142727.mb1420s92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis M. C., O'Neill E. M. and Rubin G. M. (1993). Expression of Drosophila glass protein and evidence for negative regulation of its activity in non-neuronal cells by another DNA-binding protein. Development 119, 855-865. [DOI] [PubMed] [Google Scholar]
- Franceschini N. and Kirschfeld K. (1971). [In vivo optical study of photoreceptor elements in the compound eye of Drosophila]. Kybernetik 8, 1-13. 10.1007/BF00270828 [DOI] [PubMed] [Google Scholar]
- Garner K., Hunt A. N., Koster G., Somerharju P., Groves E., Li M., Raghu P., Holic R. and Cockcroft S. (2012). Phosphatidylinositol transfer protein, cytoplasmic 1 (PITPNC1) binds and transfers phosphatidic acid. J. Biol. Chem. 287, 32263-32276. 10.1074/jbc.M112.375840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giansanti M. G., Bonaccorsi S., Kurek R., Farkas R. M., Dimitri P., Fuller M. T. and Gatti M. (2006). The class I PITP giotto is required for Drosophila cytokinesis. Curr. Biol. 16, 195-201. 10.1016/j.cub.2005.12.011 [DOI] [PubMed] [Google Scholar]
- Hamilton B. A., Smith D. J., Mueller K. L., Kerrebrock A. W., Bronson R. T., van Berkel V., Daly M. J., Kruglyak L., Reeve M. P., Nemhauser J. L. et al. (1997). The vibrator mutation causes neurodegeneration via reduced expression of PITP alpha: positional complementation cloning and extragenic suppression. Neuron 18, 711-722. 10.1016/S0896-6273(00)80312-8 [DOI] [PubMed] [Google Scholar]
- Hardie R. C. and Raghu P. (2001). Visual transduction in Drosophila. Nature 413, 186-193. 10.1038/35093002 [DOI] [PubMed] [Google Scholar]
- Hardie R. C., Raghu P., Moore S., Juusola M., Baines R. A. and Sweeney S. T. (2001). Calcium influx via TRP channels is required to maintain PIP2 levels in Drosophila photoreceptors. Neuron 30, 149-159. 10.1016/S0896-6273(01)00269-0 [DOI] [PubMed] [Google Scholar]
- Hardie R. C., Gu Y., Martin F., Sweeney S. T. and Raghu P. (2004). In vivo light induced and basal phospholipase C activity in Drosophila photoreceptors measured with genetically targeted PIP2 sensitive ion channels (Kir2.1). J. Biol. Chem. 279, 47773-47782. 10.1074/jbc.M407525200 [DOI] [PubMed] [Google Scholar]
- Hardie R. C., Liu C.-H., Randall A. S. and Sengupta S. (2015). In vivo tracking of phosphoinositides in Drosophila photoreceptors. J. Cell Sci. 128, 4328-4340. 10.1242/jcs.180364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay J. C. and Martin T. F. (1993). Phosphatidylinositol transfer protein required for ATP-dependent priming of Ca(2+)-activated secretion. Nature 363, 210-211. [DOI] [PubMed] [Google Scholar]
- Helle S. C. J., Kanfer G., Kolar K., Lang A., Michel A. H. and Kornmann B. (2013). Organization and function of membrane contact sites. Biochim. Biophys. Acta 1833, 2526-2541. 10.1016/j.bbamcr.2013.01.028 [DOI] [PubMed] [Google Scholar]
- Helmkamp G. M., Harvey M. S., Wirtz K. W. and Van Deenen L. L. (1974). Phospholipid exchange between membranes. Purification of bovine brain proteins that preferentially catalyze the transfer of phosphatidylinositol. J. Biol. Chem. 249, 6382-6389. [PubMed] [Google Scholar]
- Idevall-Hagren O. and De Camilli P. (2015). Detection and manipulation of phosphoinositides. Biochim. Biophys. Acta 1851, 736-745. 10.1016/j.bbalip.2014.12.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser S. E., Brickner J. H., Reilein A. R., Fenn T. D., Walter P. and Brunger A. T. (2005). Structural basis of FFAT motif-mediated ER targeting. Structure 13, 1035-1045. 10.1016/j.str.2005.04.010 [DOI] [PubMed] [Google Scholar]
- Kim S. H., Kedan A., Marom M., Gavert N., Keinan O., Selitrennik M., Laufman O. and Lev S. (2013). The phosphatidylinositol-transfer protein Nir2 binds phosphatidic acid and positively regulates phosphoinositide signalling. EMBO Rep. 14, 891-899. 10.1038/embor.2013.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y. J., Guzman-Hernandez M.-L., Wisniewski E. and Balla T. (2015). Phosphatidylinositol-phosphatidic acid exchange by Nir2 at ER-PM contact sites maintains phosphoinositide signaling competence. Dev. Cell 33, 549-561. 10.1016/j.devcel.2015.04.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lev S., Hernandez J., Martinez R., Chen A., Plowman G. and Schlessinger J. (1999). Identification of a novel family of targets of PYK2 related to Drosophila retinal degeneration B (rdgB) protein. Mol. Cell Biol. 19, 2278-2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litvak V., Argov R., Dahan N., Ramachandran S., Amarilio R., Shainskaya A. and Lev S. (2004). Mitotic phosphorylation of the peripheral Golgi protein Nir2 by Cdk1 provides a docking mechanism for Plk1 and affects cytokinesis completion. Mol. Cell 14, 319-330. 10.1016/S1097-2765(04)00214-X [DOI] [PubMed] [Google Scholar]
- Litvak V., Dahan N., Ramachandran S., Sabanay H. and Lev S. (2005). Maintenance of the diacylglycerol level in the Golgi apparatus by the Nir2 protein is critical for Golgi secretory function. Nat. Cell Biol. 7, 225-234. 10.1038/ncb1221 [DOI] [PubMed] [Google Scholar]
- Loewen C. J. R., Roy A. and Levine T. P. (2003). A conserved ER targeting motif in three families of lipid binding proteins and in Opi1p binds VAP. EMBO J. 22, 2025-2035. 10.1093/emboj/cdg201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesmin B., Bigay J., Moser von Filseck J., Lacas-Gervais S., Drin G. and Antonny B. (2013). A four-step cycle driven by PI(4)P hydrolysis directs sterol/PI(4)P exchange by the ER-golgi tether OSBP. Cell 155, 830-843. 10.1016/j.cell.2013.09.056 [DOI] [PubMed] [Google Scholar]
- Michell R. H. (1975). Inositol phospholipids and cell surface receptor function. Biochim. Biophys. Acta 415, 81-147. 10.1016/0304-4157(75)90017-9 [DOI] [PubMed] [Google Scholar]
- Milligan S. C., Alb J. G., Elagina R. B., Bankaitis V. A. and Hyde D. R. (1997). The phosphatidylinositol transfer protein domain of Drosophila retinal degeneration B protein is essential for photoreceptor cell survival and recovery from light stimulation. J. Cell Biol. 139, 351-363. 10.1083/jcb.139.2.351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy S. E. and Levine T. P. (2016). VAP, a versatile access point for the endoplasmic reticulum: review and analysis of FFAT-like motifs in the VAPome. Biochim. Biophys. Acta 1861, 952-961. 10.1016/j.bbalip.2016.02.009 [DOI] [PubMed] [Google Scholar]
- Nishimura Y., Hayashi M., Inada H. and Tanaka T. (1999). Molecular cloning and characterization of mammalian homologues of vesicle-associated membrane protein-associated (VAMP-associated) proteins. Biochem. Biophys. Res. Commun. 254, 21-26. 10.1006/bbrc.1998.9876 [DOI] [PubMed] [Google Scholar]
- Peretti D., Dahan N., Shimoni E., Hirschberg K. and Lev S. (2008). Coordinated lipid transfer between the endoplasmic reticulum and the Golgi complex requires the VAP proteins and is essential for Golgi-mediated transport. Mol. Biol. Cell 19, 3871-3884. 10.1091/mbc.E08-05-0498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghu P., Yadav S. and Mallampati N. B. N. (2012). Lipid signaling in Drosophila photoreceptors. Biochim. Biophys. Acta 1821, 1154-1165. 10.1016/j.bbalip.2012.03.008 [DOI] [PubMed] [Google Scholar]
- Rubin G. M. and Spradling A. C. (1982). Genetic transformation of Drosophila with transposable element vectors. Science 218, 348-353. 10.1126/science.6289436 [DOI] [PubMed] [Google Scholar]
- Sievers F., Wilm A., Dineen D., Gibson T. J., Karplus K., Li W., Lopez R., McWilliam H., Remmert M., Söding J. et al. (2011). Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7, 539 10.1038/msb.2011.75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki E. and Hirosawa K. (1994). Immunolocalization of a drosophila phosphatidylinositol transfer protein (Rdgb) in normal and Rdga mutant photoreceptor cells with special reference to the Subrhabdomeric cisternae. J. Electron. Microscopy 43, 183-189. [PubMed] [Google Scholar]
- Toulmay A. and Prinz W. A. (2011). Lipid transfer and signaling at organelle contact sites: the tip of the iceberg. Curr. Opin. Cell Biol. 23, 458-463. 10.1016/j.ceb.2011.04.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trivedi D. and Padinjat R. (2007). RdgB proteins: functions in lipid homeostasis and signal transduction. Biochim. Biophys. Acta 1771, 692-699. 10.1016/j.bbalip.2007.04.014 [DOI] [PubMed] [Google Scholar]
- Tuuf J., Wistbacka L. and Mattjus P. (2009). The glycolipid transfer protein interacts with the vesicle-associated membrane protein-associated protein VAP-A. Biochem. Biophys. Res. Commun. 388, 395-399. 10.1016/j.bbrc.2009.08.023 [DOI] [PubMed] [Google Scholar]
- Vihtelic T. S., Goebl M., Milligan S., O'Tousa J. E. and Hyde D. R. (1993). Localization of Drosophila retinal degeneration B, a membrane-associated phosphatidylinositol transfer protein. J. Cell Biol. 122, 1013-1022. 10.1083/jcb.122.5.1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voeltz G. K., Prinz W. A., Shibata Y., Rist J. M. and Rapoport T. A. (2006). A class of membrane proteins shaping the tubular endoplasmic reticulum. Cell 124, 573-586. 10.1016/j.cell.2005.11.047 [DOI] [PubMed] [Google Scholar]
- Wakefield S. and Tear G. (2006). The Drosophila reticulon, Rtnl-1, has multiple differentially expressed isoforms that are associated with a sub-compartment of the endoplasmic reticulum. Cell. Mol. Life Sci. 63, 2027-2038. 10.1007/s00018-006-6142-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia H. and Ready D. F. (2011). Ectoplasm, ghost in the R cell machine? Dev. Neurobiol. 71, 1246-1257. 10.1002/dneu.20898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z., Chikka M. R., Xia H. and Ready D. F. (2016). Ire1 supports normal ER differentiation in developing Drosophila photoreceptors. J. Cell Sci. 129, 921-929. 10.1242/jcs.180406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav S., Garner K., Georgiev P., Li M., Gomez-Espinosa E., Panda A., Mathre S., Okkenhaug H., Cockcroft S. and Raghu P. (2015). RDGBα, a PtdIns-PtdOH transfer protein, regulates G-protein-coupled PtdIns(4,5)P2 signalling during Drosophila phototransduction. J. Cell Sci. 128, 3330-3344. 10.1242/jcs.173476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav S., Cockcroft S. and Raghu P. (2016). The Drosophila photoreceptor as a model system for studying signalling at membrane contact sites. Biochem. Soc. Trans. 44, 447-451. 10.1042/BST20150256 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.