ABSTRACT
Members of the Junctophilin (JPH) protein family have emerged as key actors in all excitable cells, with crucial implications for human pathophysiology. In mammals, this family consists of four members (JPH1-JPH4) that are differentially expressed throughout excitable cells. The analysis of knockout mice lacking JPH subtypes has demonstrated their essential contribution to physiological functions in skeletal and cardiac muscles and in neurons. Moreover, mutations in the human JPH2 gene are associated with hypertrophic and dilated cardiomyopathies; mutations in JPH3 are responsible for the neurodegenerative Huntington's disease-like-2 (HDL2), whereas JPH1 acts as a genetic modifier in Charcot–Marie–Tooth 2K peripheral neuropathy. Drosophila melanogaster has a single junctophilin (jp) gene, as is the case in all invertebrates, which might retain equivalent functions of the four homologous JPH genes present in mammalian genomes. Therefore, owing to the lack of putatively redundant genes, a jp Drosophila model could provide an excellent platform to model the Junctophilin-related diseases, to discover the ancestral functions of the JPH proteins and to reveal new pathways. By up- and downregulation of Jp in a tissue-specific manner in Drosophila, we show that altering its levels of expression produces a phenotypic spectrum characterized by muscular deficits, dilated cardiomyopathy and neuronal alterations. Importantly, our study has demonstrated that Jp modifies the neuronal degeneration in a Drosophila model of Huntington's disease, and it has allowed us to uncover an unsuspected functional relationship with the Notch pathway. Therefore, this Drosophila model has revealed new aspects of Junctophilin function that can be relevant for the disease mechanisms of their human counterparts.
KEY WORDS: Junctophilin, Drosophila, Cardiomyopathy, Huntington's disease, Notch
Summary: This work reveals that the Drosophila Junctophilin protein has similar functions to its mammalian homologues and uncovers new interactions of potential biomedical interest with Huntingtin and Notch signalling.
INTRODUCTION
Junctophilin (JPH) family proteins contribute to the formation and maintenance of junctional membrane complexes (JMCs) by serving as a physical bridge between the plasma membrane (PM) and the endoplasmic reticulum (ER) membrane in excitable cells, allowing the functional crosstalk between ion channels (Takeshima et al., 2015, 2000). Silencing or genetic ablation of the JPH genes produces defects in Ca2+ homeostasis (Hirata et al., 2006; Ito et al., 2001; Li et al., 2010; Moriguchi et al., 2006; Takeshima et al., 2015, 2000). In mammals, this family comprises four members (JPH1-JPH4) that are differentially expressed: JPH1 is predominantly expressed in skeletal muscle, JPH2 in skeletal muscle and heart, and JPH3 and JPH4 genes are coexpressed in neural tissues (Nishi et al., 2000, 2003; Takeshima et al., 2000).
Jph1 knockout (KO) mice exhibit suckling failure and die shortly after birth with morphological and physiological abnormalities in skeletal muscle, including fewer JMCs, failure of normal triad development, abnormal ER features and reduced contractile force (Ito et al., 2001; Komazaki et al., 2002). Jph2 plays a key role in cardiomyocyte development and stability of myocyte ultrastructure (Beavers et al., 2014; Chen et al., 2013; Reynolds et al., 2013; Takeshima et al., 2000). Jph2 null mice die during the early embryonic development, as a result of cardiac failure (Takeshima et al., 2000). Cardiac myocytes from these mutant mice showed deficiency of the JMCs and abnormal structures in ER and mitochondria. Mice with decreased levels of cardiac Jph2 showed defective postnatal T-tubule maturation, whereas mice overexpressing Jph2 had accelerated T-tubule maturation (Reynolds et al., 2013). Inducible cardiac-specific Jph2 knockdown in mice leads to ventricular dilatation, postnatal heart failure and increased mortality (Reynolds et al., 2013).
Jph3 null mice exhibit adult onset, progressive motor dysfunction, whereas Jph3 hemizygous mice have a similar but milder phenotype (Seixas et al., 2012). Knockout mice lacking Jph4 show no obvious abnormalities, suggesting functional redundancy between Jph3 and Jph4 (Moriguchi et al., 2006). Double KO mice lacking both Jph3 and Jph4 genes have severe growth retardation and die within 3-4 weeks after birth, probably as a result of impairment of the neuronal circuit controlling the salivary gland (Moriguchi et al., 2006). In addition, they exhibit impaired motor coordination, learning and memory (Ikeda et al., 2007; Kakizawa et al., 2008; Moriguchi et al., 2006).
JPH genes have been found to play important roles in pathology. In an mdx mouse model of Duchenne muscular dystrophy, aberrant Jph1 proteolysis was detected, providing an association with the development of primary muscle disease (Murphy et al., 2013). JPH2 dysregulation has been associated with variety of heart diseases (Landstrom et al., 2014; Takeshima et al., 2015). Decreased levels of Jph2 expression have been reported in animal models of aortic stenosis (Xu et al., 2007) and hypertrophic and dilated cardiomyopathy (Minamisawa et al., 2004). In addition, in human failing heart samples or in patients with hypertrophic cardiomyopathy, the JPH2 levels are markedly reduced (Landstrom et al., 2011; Zhang et al., 2013). Importantly, dominant mutations in the human JPH2 gene are associated with hypertrophic and dilated cardiomyopathy (Beavers et al., 2013; Landstrom et al., 2007; Sabater-Molina et al., 2016) and constitute a relatively rare cause of congenital cardiomyopathies.
Huntington's disease-like 2 (HDL2) is a rare, autosomal dominant neurodegenerative disorder that is clinically almost indistinguishable from Huntington's disease (HD). HDL2 is caused by a CTG/CAG expansion located within the alternatively spliced exon 2A of the JPH3 gene (Holmes et al., 2001). The pathogenicity of this mutation involves both a toxic gain of function attributable to the expansion and a reduction in the levels of JPH3 protein expression (Seixas et al., 2012). Recently, JPH1 has been described as a genetic modifier for the Charcot–Marie–Tooth 2K (CMT2K) peripheral neuropathy (Pla-Martin et al., 2015).
Drosophila melanogaster has a single junctophilin (jp) gene, as is the case in all invertebrate genomes (Garbino et al., 2009; Takeshima et al., 2015), which can be an advantage for study of the molecular function of this protein family. In addition to the power of genetic analysis in Drosophila, the presence of a single jp gene will prevent masking of the mutant phenotype by other family members, as has been described for murine Jph3 and Jph4. The restricted tissue specificity of the different JPH genes in mammals is not completely clear cut: JPH1 is required in peripheral nerves in addition to its more studied role in muscle (Pla-Martin et al., 2015); and the neural JPH3 and JPH4 proteins are also required in pancreatic β cells and T cells, respectively (Li et al., 2016; Woo et al., 2016). Therefore, an animal model with a single junctophilin gene can help to uncover more ancestral functions. We decided to investigate the phenotypic spectrum of altering jp in Drosophila in order to find out whether it also reproduces histological alterations compatible with those found in KO mice lacking Jph subtypes and in patients with mutations in JPH genes; and to reveal new aspects of Junctophilin function that can be relevant for the disease mechanisms of their human counterparts.
RESULTS
Generation of the overexpression and RNA interference models
We decided to generate models for overexpression (OE) and knockdown (KD) of jp to investigate their effects on the target tissues. Both conditions are based on the Gal4/UAS system for directed expression (Brand and Perrimon, 1993), where a tissue-specific Gal4 construct drives expression of another transgenic construct under the UAS promoter.
The single Drosophila junctophilin gene (jp, initially annotated CG4405) is homologous to the four human JPH genes (JPH1-JPH4), which originated from a single ancestral gene by successive duplications (Garbino et al., 2009). High-throughput studies in Drosophila have shown that jp is expressed in both muscular and neural tissues (Chintapalli et al., 2007; Kumar et al., 2011); data available from http://flybase.org. The jp gene produces five different transcripts that differ in their transcription start sites, four of which produce the same 1054-amino acid-long open reading frame coding for the canonical Jp protein (Fig. 1A). The fifth transcript, coding for a 129-amino acid-long open reading frame, is probably spurious and definitely nonfunctional, because it lacks most functional domains. To generate a UAS-jp, we obtained a stock with an insertion of the P{XP} transgene within the jp locus, P{XP}jpd04563. This P{XP} transgenic construct is inserted upstream of the first coding exon (Fig. 1A) and has two UAS promoters, one at each end and in different orientations, so that the right promoter points towards the jp gene and the left promoter towards the upstream region (Thibault et al., 2004). This second promoter is flanked by FRT recombination sites, so it was removed by crossing to an FLP transgene, leaving only the jp-specific UAS promoter. Details of this process are given in Fig. S1. This insertion will be referred to as UAS-jp. An advantage of this UAS line compared with the ones generated by random insertion is that, as it is already inserted in the target locus, it does not produce insertional mutations in other genes. As the insertion is viable in homozygosis, most probably it does not affect expression of the jp gene substantially, but we cannot discard the possibility that it partly hinders expression and therefore it could be a mild hypomorphic allele. In order to test this possibility, we quantified the jp mRNA levels in individuals heterozygous and homozygous for the insertion. This analysis revealed no statistically significant differences with Oregon-R wild-type flies (Fig. 1B); thus, the insertion does not affect jp transcript expression levels and therefore it is not a hypomorph.
Fig. 1.
Generation and validation of stocks for overexpression and RNAi. (A) Schematic representation of the jp locus, with gene span, structure of the five isoforms identified (coding region light grey), insertion site of the P{XP}jpd04563 construct (UAS, arrow) and target region of the RNAi line P{KK107921}VIE-260B (RNAi, shaded). (B) mRNA levels of the jp gene in jp+ wild-type Oregon-R flies, and in heterozygotes and homozygotes for the UAS-jp insertion do not show any significant differences. (C) Survival of the progeny expressing the jp RNAi driven by each one of the four Gal4 drivers tested. The dashed line indicates the expected proportion of control flies expected (mendelian proportions 1:1 or 1:2) and the white portion of the bar the proportion observed. (D) Relative mRNA levels of jp in the OE and KD genotypes generated by crossing to two different Gal4 lines compared with the control flies bearing only the Gal4 construct. In bar diagrams, data are represented as the mean±s.e.m. One-way ANOVA, *P<0.05, ***P<0.001.
For RNA interference (RNAi), we obtained a stock from the Vienna Drosophila Resource Centre collection (Dietzl et al., 2007) that contains the insertion P{KK107921}VIE-260B, whose target sequence is contained in an exon that is common to the four major isoforms of the jp gene (Fig. 1A). This line is described as having one on-target and no off-target sites, and has the maximum s19 specificity score (the number of 19-mer on-target matches divided by the total number of matches) of one (Dietzl et al., 2007) and only three CAN repeats, which is below the desired threshold of six repeats (Ma et al., 2006). In addition, the KK collection was generated by insertion of the UAS-RNAi constructs into fixed attP sites in order to achieve reproducible expression levels and also avoid position effects and random insertional mutagenesis. This insertion will be referred to as UAS-jpRNAi.
To validate both constructs, we crossed them to different Gal4 lines that drive expression ubiquitously (Ac5C-Gal4/CyO) or specifically in muscular [twi;Mef2-Gal4 (II), early mesoderm and derivatives; Mhc-Gal4 (II), differentiated muscle] or nervous tissues (elav-Gal4/CyO, post-mitotic neurons). As the twi;Mef2-Gal4 and Mhc-Gal4 insertions are homozygous viable, if the cross with one of the UAS lines was lethal we would observe no progeny, making it difficult to assess whether this was true lethality or attributable to a crossing failure. For this reason, we used both UAS lines over a CyO chromosome; even if the cross were lethal, we should observe several CyO flies.
All four gave a viable progeny when crossed to UAS-jp, and so did the combination of UAS-jpRNAi with the late drivers Mhc-Gal4 and elav-Gal4; but the combination of UAS-jpRNAi with early expression drivers Ac5C-Gal4 and twi;Mef2-Gal4 was lethal (Fig. 1C). This suggests that the jp gene is strictly required for development and/or cell survival. We tried other ubiquitous lines and lower culture temperatures, but all attempts resulted in lethality during embryonic or early larval development.
Next, we tested that the UAS-jp and UAS-jpRNAi lines had the expected effect on the levels of jp transcript, by means of quatitative PCR. With the strongest line, Act5C-Gal4, we obtained a fivefold increase in jp transcript levels with the UAS-jp construct (Fig. 1D), but because the cross to UAS-jpRNAi was lethal, we could not perform the quantification in this condition. To validate the UAS-jpRNAi construct, we dissected thoraxes of Mhc-Gal4/UAS-jpRNAi, where muscle tissue is predominant, and we observed a significant decrease in jp levels (Fig. 1D). This decrease is an underestimation of the real one, because the thorax contains other tissues where Mhc-Gal4 is not expressed, such as the thoracic ganglion of the central nervous system. Further proof of the specificity of this RNAi construct is the fact that it can compensate for overexpression of jp in different tissues (see the results of expression in retina and wing below). We used these Drosophila transgenic lines to analyse the effect of altering the levels of Jp in tissue-specific OE and KD conditions, by comparison with control flies bearing the same Gal4 driver but no UAS construct.
Muscular deficits in the Drosophila Jp models
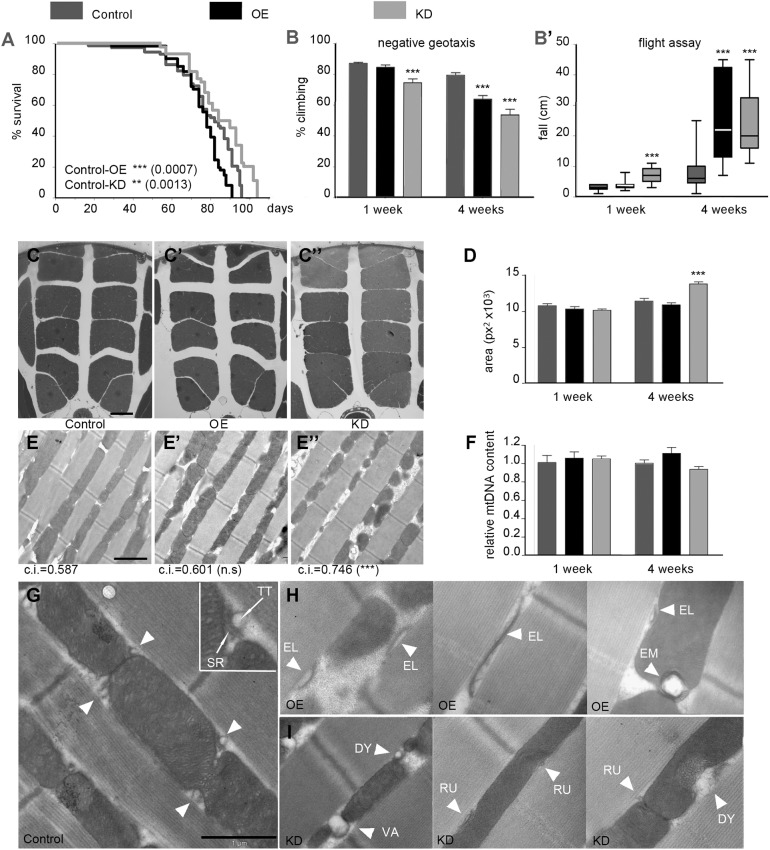
In mammals, JPH1 is the major JPH family member expressed in skeletal muscle (Ito et al., 2001; Komazaki et al., 2002). In Drosophila, as we mentioned above, silencing of jp with the early mesoderm-specific driver twi;Mef2-Gal4 resulted in lethality, so we had to use the muscle-specific Mhc-Gal4 driver that produced viable adults (Fig. 1B). Our control genotype was Mhc-Gal4/+, the OE genotype Mhc-Gal4/UAS-jp, and the KD genotype Mhc-Gal-4/UAS-jpRNAi. Regarding longevity, OE flies had only a slight reduction in viability, whereas KD flies had an extension of the maximal lifespan of ∼10 days (Fig. 2A). The possible role of insulin signalling downregulation in this extended lifespan is explored in the Discussion section. In order to measure the muscular competence, we performed negative geotaxis and flight assays (Fig. 2B,B′). In both tests, the KD flies already showed a markedly decreased performance at 1 week of age; and both OE and KD genotypes had a more severe age-dependent reduction in the ability to climb the vial and to attain stable flight.
Fig. 2.
Muscular deficits in OE and KD flies. (A) Survival curves of the control (Mhc-Gal4), OE and KD flies; log-rank (Mantel–Cox) test shows significant differences from the control genotype. (B,B′) Neuromuscular competence of the three genotypes at 1 and 4 weeks of age estimated in the negative geotaxis assay (B, n=3≥15 flies each) and in the flight assay (B′, n=30). (C-C″) Semi-thin sections of the thorax of control (C), OE (C′) and KD (C″) flies at 4 weeks of age. (D) Area occupied by the IFMs in semi-thin sections at 1 and 4 weeks of age (n≥6, three sections analysed per individual). (E-E″) TEM of ultra-thin longitudinal sections of the IFMs of control (E), OE (E′) and KD (E″) flies at 4 weeks of age; under each panel the mitochondrial circularity index (c.i.) is indicated (n=3, 60 mitochondria per individual). (F) Relative mitochondrial genomic DNA; in each genotype and age the proportion of mitochondrial to nuclear genomic DNAs was calculated, and the results are displayed as mitochondrial genomic DNA abundance relative to the control genotype at each age. (G) Muscle fibre ultrastructure in a 1-week-old control fly. Diads are indicated by white arrowheads, representing the normal morphology of these structures. The inset shows a wild-type diad with indication of the sarcoplasmic reticulum (SR) and T-tubule (TT) components. (H) Examples of aberrant morphology, such as elongated SR cisternae (EL) and diads embedded in the mitochondria (EM), in 1-week-old OE fly muscles. (I) Abnormal rudimentary (RU), dysmorphic (DY) or vacuolated (VA) diads (arrowheads) in 1-week-old KD fly muscles, usually next to disorganised mitochondrial cristae. Scale bars: 100 μm in C-C″; 2 μm in E-E″; 1 μm in G-I. In bar diagrams, data are represented as mean±s.e.m., One-way ANOVA,**P<0.01, ***P<0.001.
To determine whether this motor deficit is attributable to muscular degeneration, we analysed the indirect flight muscles (IFM) in semi-thin sections of the thorax. No significant differences were found at 1 week of age, but at 4 weeks horizontal gaps within each set of muscle packs are evidently reduced in KD flies compared with control and OE flies (Fig. 2C-C″), and this is attributable to an increase in the area of the IFM sections (Fig. 2D). This result suggests an age-dependent muscle hypertrophy in KD flies, which might be related to the defects observed in the climbing assay. To study the ultrastructure of IFM by transmission electron microscopy, we performed longitudinal sections along the muscle fibres. The structure of the myofibrils was not affected in any genotype, but KD flies displayed an aberrant mitochondrial morphology, which was most evident at 4 weeks (Fig. 2E-E″). These mitochondria were smaller and more rounded than in the wild-type control. Mitochondria from control and OE flies are similar, with a circularity index ∼0.6, whereas KD mitochondria are more circular, with an average index of 0.75, and these differences are statistically significant. Given that KD flies have abnormally shaped mitochondria, we estimated mitochondrial biomass by qPCR, to determine the mitochondrial genomic DNA content, using the nuclear genomic DNA for normalization, and we did not find any significant differences between OE or KD and the control.
Given that loss of Jph1 in skeletal muscle results in aberrant triads, we examined the morphology of the equivalent structure in the insect muscle, the diads, formed by a single electron-dense SR cisterna and the adjoining electron-lucent T-tubule (Razzaq et al., 2001). We performed this analysis at 1 week of age, before major morphological changes in mitochondria. Diads are located half-way between the Z and M bands of the myofibril (Fig. 2G). OE muscles have aberrant diads with elongated SR cisternae, and in mitochondria-poor regions unusually elongated SR cisternae are surrounded by electron-lucent material, which might be an expanded T-tubule structure (Fig. 2H). Occasionally, diads also show an abnormal positioning away from the myofibrils and embedded in the mitochondria. Diads in KD muscles have a different phenotype. They are rudimentary or have an aberrant morphology, including a vacuolated SR similar to the one in Jph1 KO mice (Fig. 2I); and the adjoining mitochondrial regions also have abnormal cristae morphology.
Human JPH1 is a modifier of the GDAP1 mutations causing Charcot–Marie–Tooth peripheral neuropathy (Pla-Martin et al., 2015). Previous work from our group found metabolic alterations in Drosophila Gdap1 models (López Del Amo et al., 2017). To determine whether altered levels of Jp result in metabolic changes in the muscle, we carried out a metabolomic study by nuclear magnetic resonance (NMR). In order to detect early and direct metabolic alterations that could contribute to reduced muscular competence, we performed this study in 1-week-old flies. We compared the control genotype with the OE and KD genotypes, and in both cases, we could find a discriminating model by orthogonal projection on latent structure-discriminant analysis (OPLS-DA) (Fig. S2), which means that the compared genotypes are metabolically different. The most marked change in both experimental genotypes is a significant increase in glycogen, the main carbohydrate storage species in the muscle. At the same time, there is a reduction in trehalose, which is the main source of energy for the IFM. Next, we paid attention to metabolites known to reflect the homeostasis of the muscle. The abundance of β-alanine is a marker of muscle degeneration (Sarou-Kanian et al., 2015), and levels of this metabolite had no significant changes. The three branched-chain amino acids (isoleucine, leucine and valine) promote protein synthesis in the muscle (Kimball and Jefferson, 2006), and their abundance was reduced only in KD flies. These results reinforce the notion that imbalanced jp levels affect muscle function but do not result in major muscular degeneration.
Cardiac dysfunction produced by altered Jp expression
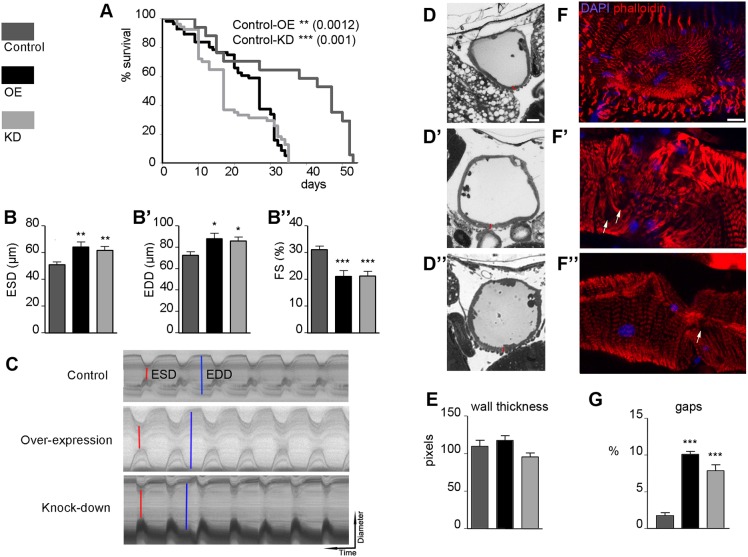
Constitutive or heart-specific loss of Jph2 produces cardiac defects in mice (Takeshima et al., 2000). In order to investigate the relevance of Jp in the Drosophila heart, we used the cardiac-specific driver GMH5-Gal4. All the results we show below have been obtained with a culture temperature of 29°C, at which the cardiac phenotypes were most evident. In this case, our control genotype was GMH5-Gal4/+, the OE genotype GMH5-Gal4/UAS-jp, and the KD genotype GMH5-Gal4/UAS-jpRNAi. Both interference and overexpression of jp caused a reduction of lifespan from 53 days in control flies to only 35 days (Fig. 3A). Mean survival was also reduced from 47 days in control flies to 27 days in OE flies and only 17 days in KD flies. In this survival curve, it is evident that the lifespan of the control flies is very reduced compared with other survival curves in this work (Fig. 2A, Fig. 4A). This reduction is attributable to culture at 29°C, which is a suboptimal temperature but allows a better development of cardiac phenotypes.
Fig. 3.
Cardiac dysfunction in OE and KD flies. (A) Survival curves of the control (GMH5-Gal4), OE and KD flies; log-rank (Mantel–Cox) test shows significant differences from the control genotype. (B-B″) Cardiac function parameters altered in the experimental genotypes at 1 week of age (n≥15 per genotype): end-systolic diameter (B), end-diastolic diameter (B′) and fractional shortening (B″). (C) Representative M-mode traces (vertical movement of the heart walls in 14 s) of semi-intact Drosophila hearts from the different genotypes; ESD are indicated in red and EDD in blue. (D-D″) Semi-thin sections of adult hearts of 1-week-old flies of the control (D), OE (D′) and KD (D″) genotypes. Red bars represent heart wall thickness measurements like the ones that have been used for the quantification in E. (E) Quantification of heart wall thickness in the three genotypes as mean pixels (n≥10, three measurements per sample). (F-F″) Phalloidin staining of dissected hearts with DAPI staining of nuclei reveals the normal myofibril structure in control 1-week-old flies (F) and abnormal morphologies in same age OE (F′) and KD (D″) hearts; white arrows point to gaps, areas devoid of myofibrils. (G) Quantification of the proportion of surface with gaps in the myofibrillar staining of five hearts of different genotypes (n=5). Scale bars: 10 μm in D-D″ and F-F″. In bar diagrams, data are represented as mean±s.e.m. One-way ANOVA, *P<0.05, **P<0.01, ***P<0.001.
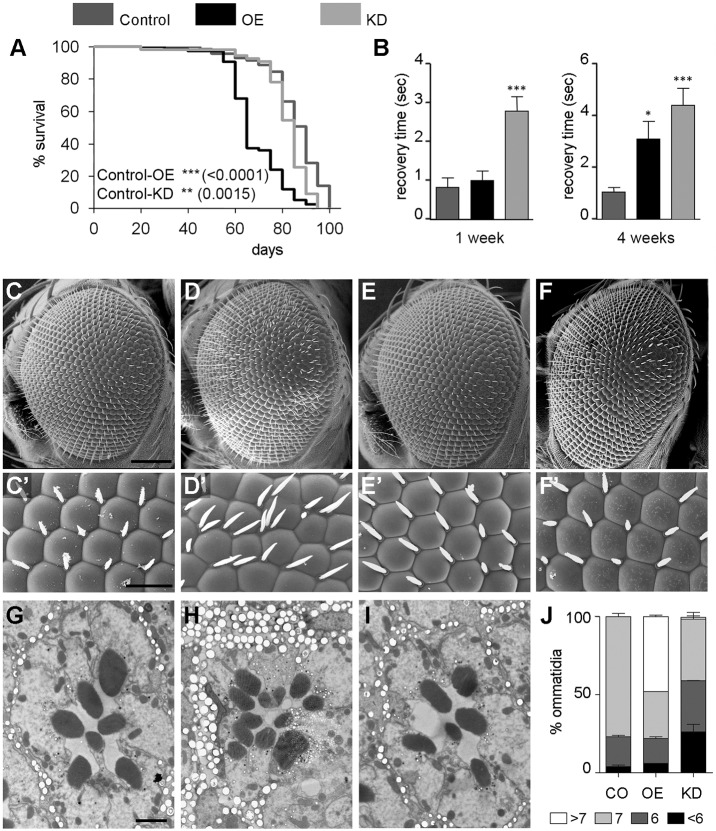
Fig. 4.
Neuronal alterations in the OE and KD genotypes. (A) Survival curves of the control (elav-Gal4), OE and KD flies; log-rank (Mantel–Cox) test shows significant differences with the control genotype. (B) Bang-sensitivity analyses of the three genotypes at 1 and 4 weeks of age, represented as recovery time after mechanical stress-induced paralysis. (n=4, ≥10 flies per experiment). (C,C′) SEM image of a wild-type control eye (GMR-Gal4, C) and higher magnification showing the stereotypical hexagonal arrangement of the ommatidia and the inter-ommatidial bristles (C′). (D,D′) In an OE fly, the eye has a rough aspect (D), and the ommatidia have lost the regular pattern and have supernumerary bristles (D′). (E,E′) KD fly eyes have a wild-type aspect (E) and structural arrangement (E′). (F,F′) The defects in UAS-jp eyes are corrected by coexpression of UAS-jpRNAi. (G) SEM of an ultra-thin section of a control fly ommatidium, showing the wild-type trapezoidal arrangement with seven rhabdomeres. (H) Structure of an ommatidial section of an OE fly with extra rhabdomeres, as a result of the recruitment of extra photoreceptor neurons. (I) Loss of rhabdomeres, hence photoreceptor neurons, in a KD ommatidial section. (J) Distribution of the number photoreceptor neurons in ommatidia of the three genotypes at 1 week of age (n=4, ≥60 ommatidia per section). Scale bars: 100 μm in C-F; 20 μm in C′-F′; 2 μm in G-I. In bar diagrams, data are represented as mean±s.e.m. One-way ANOVA, *P<0.05.
To study heart function, cardiac contractions were analysed in 1-week-old adult fly hearts. There was no alteration of the diastolic interval (DI), the systolic interval (SI) or heart period length (HP, defined as DI+SI) (Fig. S3). The arrhythmia index (AI), an indicator of the variability calculated by dividing the standard deviation of the heart period by its median, was also unaltered (Fig. S3).
By contrast, we found changes in cardiac chamber parameters, including increased end-systolic diameter (ESD) and end-diastolic diameter (EDD) and decreased fractional shortening (FS) percentage, which provides an indication of the cardiac output (Fig. 3B-B″). These changes are evident in the M-mode traces of the three genotypes (Fig. 3C). These data revealed that in flies with altered levels of Jp expression, the heart tube is dilated and there is a dysfunction of the contractile properties that reduces cardiac output. For a more detailed observation of heart morphology in the jp mutants, we examined transverse semi-thin sections of the heart tube (Fig. 3D-D″). We observed an enlargement of the cardiac chamber, which we have previously quantified as an increased EDD (Fig. 3B′). Notably, these sections show that the thickness of the heart wall in the mutant flies does not show any statistically significant difference from control flies, discounting hypertrophy of cardiomyocytes (Fig. 3E).
Drosophila heart tubes have two types of muscle fibres, each with a distinct myofibrillar structure (Mery et al., 2008; Taghli-Lamallem et al., 2008); spirally or transversely oriented myofibrils that represent the contractile ‘working’ myocardium, and longitudinally oriented myofibrils that are found along the ventral surface of the tube (Molina and Cripps, 2001). In young flies, both types of myofibrils exhibit a tight and well-aligned arrangement. Cardiac myofibrils stain uniformly along the entire length of the thin filament with phalloidin (Ao and Lehrer, 1995), which can therefore be used to visualize both types of myofibrils. Phalloidin staining of actin in mutant flies with altered expression of jp revealed structural abnormalities in the parallel alignment of transverse myofibrils in the heart tube in the areas surrounding the ostia (Fig. 3D-D″). The cardiac fibres in the mutant flies were clearly more disorganized and less compact than in control flies. The gaps in myofibrillar staining were quantified by measuring the size of these areas in confocal stacks of five hearts of each genotype (Fig. 3G). The percentage area devoid of myofibrils was significantly smaller in control hearts compared with that of KD or OE (2, 10 and 7.8%, respectively).
Modification of the Jp levels produces neurological abnormalities and affects the number of photoreceptor neurons
To evaluate the neuronal relevance of Jp in Drosophila, we used the pan-neuronal elav-Gal4 driver. The control genotype was elav-Gal4/+, the OE genotype elav-Gal4/UAS-jp, and the KD genotype elav-Gal4/UAS-jpRNAi. Mean survival was strongly reduced in OE flies, whereas a slight reduction was observed in the KD flies (Fig. 4A). The bang-sensitive phenotype, a temporary paralysis when exposed to mechanical stress, has been associated with mutations in genes involved in neuronal function (Graham et al., 2010; Kuebler and Tanouye, 2000; Lee and Wu, 2002; Pavlidis et al., 1994; Schubiger et al., 1994; Trotta et al., 2004). Bang-sensitivity analyses revealed that KD flies exhibit a two- to threefold increase in recovery time at 1 week and at least a fivefold increase at 4 weeks compared with control flies (Fig. 4B). The recovery time of the OE flies was also increased, and the difference was statistically significant at 4 weeks.
The fly retina is a tissue widely used to study neurodegeneration. To drive expression in the retina, we used the GMR-Gal4 (I) construct (control genotype GMR-Gal4/+, OE genotype GMR-Gal4/+; UAS-jp/+, and the KD genotype GMR-Gal4/+; UAS-jpRNAi/+). In the control eyes, we can observe the wild-type external morphology; the ommatidial lenses are dome shaped and arranged in a hexagonal tiling pattern with inter-ommatidial bristles (Fig. 4C,C′). Overexpression of Jp produces a mildly disrupted arrangement, with the presence of nonhexagonal ommatidia and supernumerary bristles (Fig. 4D,D′). By contrast, KD resulted in no observable abnormalities in the external morphology (Fig. 4E,E′). Although GMR-Gal4; UAS-jpRNAi on its own has no phenotype, it is able to correct the phenotype of the eye external morphology caused by overexpression of UAS-jp under the control of GMR-Gal4, which demonstrates the specificity of the abnormal phenotype attributable to the overexpression of jp (Fig. 4F,F′).
A cross-section of a normal ommatidium always cuts through seven rhabdomeres, the light-collecting organs of the neuron, in a stereotypical trapezoidal arrangement (Fig. 4G). Transmission electron microscopy analyses of OE and KD retinas revealed an abnormal number of photoreceptors in both genotypes (Fig. 4G-J). Almost 50% of the OE ommatidia had extra photoreceptor cells, whereas in KD flies the effect was the opposite, with several ommatidia containing fewer photoreceptor cells (Fig. 4I). The presence of extra photoreceptor cells has been extensively described as a consequence of signalling defects during the development in the eye, whereas loss of photoreceptor cells could be a result of defects in such signalling pathways but also neurodegenerative processes, even at early post-eclosion stages, or a mixture of them. To discriminate between these situations, we performed the following experiment. To reduce the expression of the jp RNAi during development, flies were crossed and reared at 18°C until eclosion. At this point, the flies were divided into two groups; one was kept for 1 day at 18°C the other cultured for 7 days at 25°C after eclosion to allow stronger expression of the RNAi. In KD flies that were kept at 18°C, the morphology is slightly compromised, probably as a result of incipient degeneration, because RNAi expression is damped, not abolished, but at this point most ommatidia have seven photoreceptors. KD flies that were moved to 25°C have an enhanced loss of photoreceptors and a more degenerative morphology, with many vacuoles (Fig. S4). Therefore, whereas overexpression of jp results in recruitment of extra photoreceptor neurons, jp RNAi results mainly in neurodegeneration rather than photoreceptor specification.
The Htt-related neurodegeneration is modified by altering the Jp levels
Dominant mutations in the JPH3 gene caused by an expanded CAG/CTG repeat in its alternatively spliced exon 2A are responsible for Huntington's disease-like 2 (HDL2), a phenocopy clinically indistinguishable from Huntington's disease (HD) (Holmes et al., 2001). HD is a fatal neurodegenerative condition caused by expansion of the polyglutamine tract in the Huntingtin (Htt) protein, and the precise disease manifestations and their timing are affected by modifier genes (Gusella and MacDonald, 2009). Despite the phenotypical similarities, a possible role of JPHs as genetic modifiers in HD has not been investigated.
HD has been modelled several times in Drosophila by overexpression of pathological expansions under neuronal drivers (Lewis and Smith, 2016), and these models have similar phenotypes to ours, having reduced lifespan when expressed under elav-Gal4 and retinal neuron degeneration under GMR-Gal4 (Fig. 4); and in addition, they also show features typical of HD, such as protein aggregates. The abnormal phenotypes in these models are repeat-length dependent. To model HD in Drosophila, we used a construct for the expression of human HTT exon 1 containing expanded polyglutamine repeats (Htt-Ex1-pQ93) that has been demonstrated to induce neurodegeneration (Steffan et al., 2001). These authors describe that shorter repeats (1Q or 20Q) have no deleterious effect. In order to investigate whether Jp could modify the HD pathogenesis in flies, UAS-jp or UAS-jpRNAi was coexpressed with Htt-Ex1-pQ93. Flies expressing Htt-Ex1-pQ93 have a normal external morphology of the eye at day 1 post-eclosion, but suffer progressive degeneration of the underlying retina, resulting in patchy depigmentation after 4 weeks of age (Fig. 5A-A″). When jp is coexpressed with Htt-Ex1-pQ93, the development of this depigmentation is delayed (Fig. 5B-B″), which means that Jp might act as a partial suppressor of the Htt-Ex1-pQ93-induced degeneration. Conversely, coexpression of Htt-Ex1-pQ93 with the jp RNAi led to enhancement of eye phenotype, because the loss of pigmentation is observed from day 1, and the depigmentation progresses much faster (Fig. 5C-C″). As a control experiment, we aged flies of the GMR-Gal4/+, GMR-Gal4; UAS-jp and GMR-Gal4; UAS-jpRNAi and observed no changes in pigmentation; therefore, the results observed were bona fide genetic modifications.
Fig. 5.
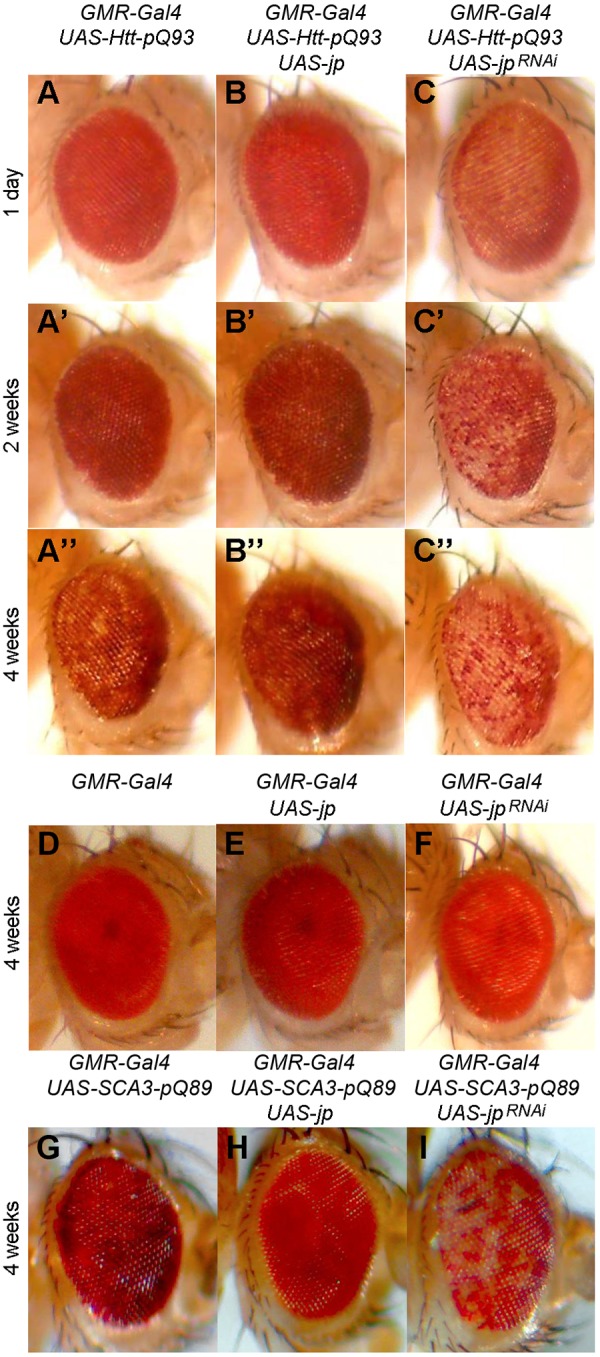
Jp levels modify the phenotype of pathological Htt and SCA3 poly-Q expansions. (A-A″) Progressive degeneration and depigmentation over 4 weeks upon expression of Htt-pQ93 in the eye driven by GMR-Gal4. (B-B″) Coexpression of Jp ameliorates the depigmentation. (C-C″) Coexpression of jp RNAi induces earlier, faster progressing depigmentation. GMR-Gal4 alone (D), GMR-Gal4-driven UAS-jp (E) or UAS-jpRNAi (F) does not produce any depigmentation at 4 weeks. (G) Expression of SCA-Q89 produces eye depigmentation that is evident at 4 weeks. (H) Coexpression of Jp reduces depigmentation. (I) Coexpression of jp RNAi increases depigmentation.
The Htt-Ex1-pQ93 corresponds to a truncated version of the gene coding for only a few endogenous amino acids, so it is possible that jp functions as a modifier for other types of pathogenic poly-Q expansions. To test this possibility, we investigated whether UAS-jp or UAS-jpRNAi was able to modify the phenotype caused by expression of another dominant mutation caused by a poly-Q expansion in the human SCA3 gene causative of spinocerebelar ataxia type 3 (Stochmanski et al., 2012). UAS-SCA3-Q89 contains an expanded tract of glutamines within a full-length SCA3 cDNA, and it also causes depigmentation of the Drosophila retina when expressed under GMR-Gal4 (Fig. 5G). Again, coexpression of UAS-jp slows down depigmentation at 4 weeks (Fig. 5H), whereas coexpression of UAS-jpRNAi enhances it (Fig. 5I). The modification of the phenotype is clear, suggesting that mechanistically Jp can be a modifier of poly-Q expansions in general, but this does not necessarily mean that the modification is clinically relevant, because SCA3 and HD/HDL2 affect different regions of the encephalon.
Jp has functional interactions with the Notch pathway
A closer examination of the normally viable Act5C-Gal4/UAS-jp individuals revealed abnormal phenotypes usually linked to a downregulation of the Notch signalling pathway, such as supernumerary vibrissae under the eye, microchaetae in the notum and sternopleural bristles (Fig. 6A,B) (Schweisguth and Posakony, 1994). Further Notch phenocopies are the duplication of the macrochaetae in the scutellum (Fig. 6C) and delta-shaped wing veins (Fig. 6D). All these phenotypes were 100% penetrant. The phenotype we observed under OE of jp in the retina also points to a downregulation of Notch signalling, because the recruitment of extra cells as photoreceptor neurons is typical of mutants for the Notch ligand Delta (Parks et al., 1995). If this was a result of a functional interaction between Jp and the Notch pathway, either direct or indirect, the prediction would be that jp OE should enhance Notch phenotypes and jp KD should suppress them. To carry out these tests, we could not use the Act5C-Gal4 driver with which we detected the phenotypes, because jp KD with this driver is lethal before the adult age. We also discarded the retina as an experimental tissue, because Notch signalling has successive and complex roles during eye development, including growth, planar cell polarity and several rounds of cell-type specification (Cagan and Ready, 1989). The wing blade is a more suitable model to investigate modification of the Notch/Delta phenotypes; therefore, we used two Gal4 lines that drive expression in the whole wing blade, rn-Gal4 (III) and nub-Gal4 (II).
Fig. 6.
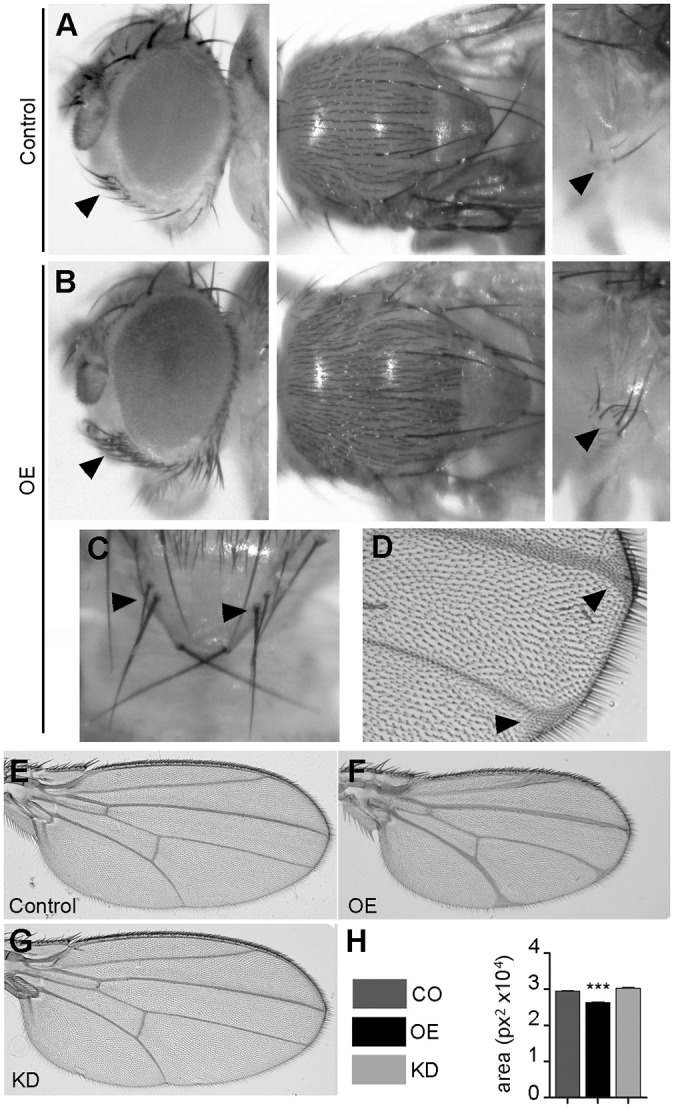
Notch-like phenotypes in OE flies. (A) Detail of the normal number and distribution of head vibrissae, notum microchaetae and sternopleural bristles in a control Act-Gal4/+ fly. (B) Bristles of these types are increased in number in flies of the Ac-Gal4, UAS-jp genotype (OE). (C) Duplicated scutellar macrochaetae in an OE fly. (D) Delta-shaped contacts of the wing veins with the wing margin in an OE fly. (E) Morphology of a wild-type control fly wing (rn-Gal4/+). (F) rn-Gal4, UAS-jp fly wing with expanded wing veins, delta-shaped contacts with the wing margin and decreased wing blade size. (G) rn-Gal4, UAS-jpRNAi wings have wild-type morphology. (H) Quantification of the blade area in wings of the genotypes shown in E-G shows that only OE wings are significantly different from the control (n≥11). In bar diagrams, data are represented as mean±s.e.m. One way-ANOVA, ***P<0.001.
Initially, we tested the effect of jp OE and KD on their own. OE of jp driven by rn-Gal4 driver again mimics a Notch phenotype, with thickening of the longitudinal wing veins and delta-shaped contacts of the veins and the wing margin (De Celis, 2003), and also produces a reduction of the wing area (Fig. 6E,F,H). By contrast, jp KD did not produce any evident phenotypes in the wing morphology (Fig. 6E,G,H). Similar experiments with the other wing driver, nub-Gal4, produced equivalent results and also showed that jp KD can correct jp OE despite not having an effect on its own (Fig. S5). To test whether the extra vein tissue was produced by a reduction in the activation of the Notch pathway, we attempted the modification of the phenotype of a dominant temperature-sensitive allele of the gene encoding the Notch ligand Delta, Dl6B37. These experiments were performed at 29°C, the temperature at which this Dl allele shows a more pronounced phenotype than in the normal rearing conditions at 25°C. In these conditions, again OE wings have a Dl-like phenotype and KD wings are normal (Fig. 7A,B,C). Dl6B37 flies displayed the expected wing vein defects, which are most evident in the L2 vein, and delta-shaped contacts with the wing margin (Fig. 7D). Expression of Jp in the wing blade enhanced these phenotypes (Fig. 7E), whereas expression of the jp RNAi corrected them (Fig. 7F). In order to have a parameter that allowed for a statistical treatment, we measured the length of the contact of L2 with the wing margin, which is wider when it adopts the delta shape. These analyses confirmed that jp behaves as it would be expected from a typical modifier: OE of jp enhances the Dl phenotype and KD partly suppresses it (Fig. 7G).
Fig. 7.
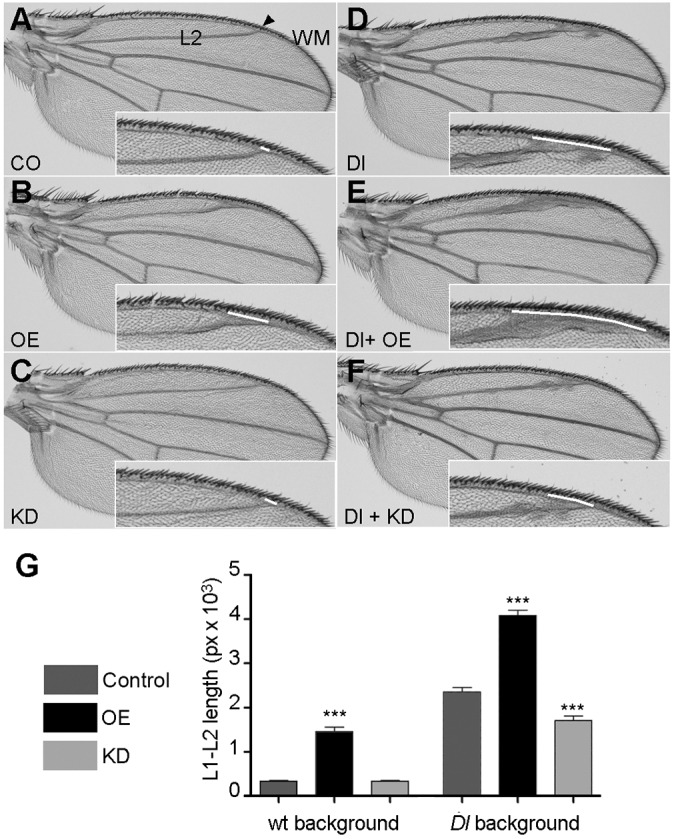
Levels of Jp expression modify the phenotype of a Dl mutant allele. (A-C) At 29°C, wing blades of control nub-Gal4/+ (A), OE (B) and KD (C) genotypes have the same phenotypes as flies of the same genotypes cultured at the standard temperature. (D) Wing of a Dl6B37 fly displaying the typical Dl phenotype of engrossed veins and delta-shaped contacts with the wing margin. (E) Overexpression of Jp in a Dl− background enhances the phenotype. (F) Expression of the jp RNAi in a Dl− background strongly suppresses the wing vein phenotypes. In each panel, the inset shows a higher magnification of the area where measurements were performed, and the white line indicates the length of the contact of vein L2 with the wing margin used in G. (G) Quantification of the length of the contact of vein L2 with the wing margin in flies of the genotypes represented in A-F (n≥17 for each genotype). In bar diagrams, data are represented as mean±s.e.m. One-way ANOVA, ***P<0.001.
DISCUSSION
Drosophila as a model to study Junctophilin function
In the present work, we present data indicating that Drosophila is a good model to study pathologies resulting from mutations in human JPH genes. Our two experimental genotypes are based on a gain and a loss of function, OE and KD, respectively. In general, the specific phenotypes we observe in a particular tissue are also opposite (i.e. the effect on Htt expansions or Notch wing phenotypes), although some of the nonspecific phenotypes, such as overall viability or motility, can be altered in the same sense in both. This reduced fitness indicates that a proper balance in the expression levels of jp is required for normal calcium homeostasis cell functions. The phenotypes we have described in muscle, heart and neurons are compatible with what has been described in patients with pathological mutations in JPH genes or in KO mouse models of these genes. In addition, we describe two new functional relationships that might be relevant in the pathogenesis of junctophilins with pathological poly-Q expansions in Huntington's disease and with Notch signalling.
Junctophilin function in the heart and muscle
Down- and upregulation of jp expression mainly affects the structure of the diads in the IFMs. Elevated levels induce the formation of elongated SR cisternae and, less often, incorrect localization of the diad next to the myofibrils. This effect is similar to the one observed upon elevated expression of Jhp1 in mouse heart (Komazaki et al., 2003). In these mice, cardiac diads had extended SR–T-tubule contacts. Overexpression of Jph2 in heart also increases the SR–T-tubule contacts (Guo et al., 2014). KD of Drosophila jp also produced phenotypes comparable to those observed in Jph1 KO mice: incorrect formation of diads and vacuolated SR and T-tubules. The phenotypes in KD flies are dramatic, with rudimentary or nearly absent diads at 1 week and a fragmentation of the mitochondrial network at 4 weeks. This fragmentation in older flies could be related to the incipient mitochondrial damage observed in younger ones. In murine muscle, there is coexpression of Jph2, so our KD phenotype probably represents a more severe loss of function, thus unveiling that junctophilins are required for the proper formation of the SR–T-tubule contacts, not only their maturation.
In any case, neither our experimental genotypes nor the published KO or overexpression murine models show any structural degeneration of the myofibrils. This is confirmed in the metabolomic profile, because there were no alterations linked to muscular degeneration, only an imbalance in the carbohydrate levels, with increased glycogen and decreased trehalose. This result is confirmed by an independent work, in which a screening for genetic determinants of metabolic traits in whole flies revealed a genetic linkage of jp to energy metabolism traits and increased glycogen levels (Jumbo-Lucioni et al., 2010). A possible explanation for this deregulation of carbohydrate metabolism is an alteration of the insulin signalling pathway.
In fact, downregulation of the insulin pathway could also explain two unexpected phenotypes in the muscle KD flies: the extended lifespan and the muscle hypertrophy. Mitohormesis, the adaptive response of mitochondria to mild stress, has already been reported to produce an extended lifespan in Drosophila (Owusu-Ansah et al., 2013). In this case, mitohormesis triggered by mild muscular stress and mitochondrial fragmentation resulted in lifespan extension, and this effect was mediated by the insulin pathway and the mitochondrial unfolded protein response. There are further examples of extended longevity caused by impairment of insulin signalling and/or mitochondrial stress (Wang and Hekimi, 2015; Zarse et al., 2012). As for muscle hypertrophy, it is usually assumed that low levels of branched-chain amino-acids (BCAA) are indicative of muscle deficiency, but there is a great heterogeneity and many factors impinging at different levels (Tom and Nair, 2006). One of the pathways involved is insulin signalling (Glass, 2005), and in this context, we observe significant alterations of glycogen and trehalose levels. This result could be because of the involvement of different pathways and deserves a deeper study.
Muscle hypetrophy was also observed in mouse skeletal muscle expressing Jph2 with the dominant negative mutations S165F and Y141H, associated with hypertrophic cardiomyopathy in patients (Woo et al., 2012, 2010). Several mutations in human JPH2 have been associated with hypertrophic cardiomyopathy (Beavers et al., 2014), and can cause a hypertrophyc phenotype when downregulated in mouse cardiac muscle cell lines (Landstrom et al., 2011). By contrast, the phenotype of the heart KD flies is more similar to a dilated cardiomyopathy, with increased ESD and EDD and reduced cardiac output. Recently, a new mutation in JPH2 has been found to be associated with dilated cardiomhyopathy (Sabater-Molina et al., 2016). Therefore, mutations in JPH2 could cause hypertrophic or dilated cardiomiopathy depending on factors such as the genetic background, degree of functionality of the mutant protein, or location of the mutation in a particular domain.
A difference between KD in muscle and heart is that in the latter, we could find structural alterations in the cardiac myofibrils. A possible explanation is that the muscular driver we used, Mhc-Gal4, is expressed in differentiated muscle, whereas the cardiac driver, GMH5-Gal4, contains a promoter region from the tinman gene, which drives expression throughout heart development from the early embryo. Alternatively, this difference could be attributable to intrinsic physiological differences between skeletal and cardiac muscle.
Junctophilin function in neurons
A Jph3 KO produces motor deficits in mouse models, and the double KO Jph3/4 has a more severe phenotype, including the impairment of motor, learning and memory abilities (Moriguchi et al., 2006; Nishi et al., 2002; Seixas et al., 2012). Likewise, KD of Jp in Drosophila neurons also affects neuronal function as reflected in the bang-sensitivity test. Alteration of Jp in the retina revealed a mixture of neurodevelopmental defects and degeneration of the retinal neurons. Although neurodevelopmental defects probably involve the mis-regulation of the Notch signalling pathway during cell fate determination and differentiation stages, the neurodegeneration happens in fully differentiated neurons and is dependent on neural function or survival rather than development.
The modification of the Htt-Ex1-pQ93 phenotype by Jp is clear cut; OE of Jp is a suppressor and KD an enhancer of the neurodegeneration. This modification is most probably functionally relevant, because ablation of Drosophila Htt exacerbates the neural toxicity elicited by the same construct we have used in the present work (Zhang et al., 2009). Also, neuronal store-operated calcium entry is a new therapeutic target for HD (Wu et al., 2011). Given that triplet expansions of JPH3 are causative of HDL2 (Holmes et al., 2001), the fact that altering Jp levels modifies the phenotype of flies expressing human Htt-Ex1-pQ93 suggests that both proteins participate in at least one common cellular pathway. In addition to causing HDL2, it is possible that genetic variation in JPH3/4 participates in the clinical variability of HD patients.
A Drosophila model of HDL2 based on the expression of the human JPH3 mutant protein HDL2-Q138 showed that toxicity is attributable to the accumulation of the poly-Q-expanded protein in the nucleus, and this toxicity was alleviated by redirecting it to the cytoplasm (Krench et al., 2016). By contrast, HTT-Q138 aggregates remain cytoplasmic, which suggests that they do not share toxicity mechanisms. Although this finding might seem to contradict ours, it has been demonstrated that the pathogenicity of JPH3 mutations is multifactorial and involves at least two effects, a toxic gain of function of the aggregates and a deficit in JPH3 function attributable to reduced expression levels (Seixas et al., 2012). Therefore, overexpression of the human HTT-Q138 protein and modulation of the endogenous Jp levels could be affecting different cellular mechanisms in the nucleus and the cytoplasm, respectively.
Junctophilin antagonizes Notch signalling
Overexpression of Jp phenocopies the loss of function of the Notch ligand Dl: recruitment of supernumerary photoreceptor neurons, lateral inhibition in sensory organ determination and expansion of the wing veins. A role of Jp as a modifier of Notch is strongly reinforced by the fact that loss of function of Jp can suppress the mutant phenotype of a dominant loss of function allele of Dl. Although this was an unexpected result, there are three previous high-throughput screenings for modifiers of Notch phenotypes in the wing (Cruz et al., 2009; Molnar et al., 2006) or in the eye (Shalaby et al., 2009), in which Drosophila jp was among the identified candidates. All of them were based on overexpression of genes adjacent to insertions with bi-directional UAS promoters, either the same original insertion we have used in our work (P{XP}jpd04563) or a similar construct (P-GS). In consequence, the authors could not discern which of the two genes flanking the insertion, jp or CG3838, was the modifier. Given that we have reproduced the modification after removing the promoter pointing to CG3838, we can single out jp as the Notch-interacting gene.
Our work is the first report of a functional relationship of junctophilins with Notch signalling. The effect of Jp on Notch is most probably through its effect on calcium trafficking. The ER calcium sensor STIM1 has been shown to co-localize with JPH1 during store-operated calcium entry (Pla-Martin et al., 2015) and interacts physically with JPH4 (Woo et al., 2016). Drosophila Stim has also been demonstrated to be a modifier of Notch phenotypes (Eid et al., 2008), although in this case Stim expression is synergistic rather than antagonistic to the pathway. Another screening in Drosophila has unveiled other calcium signalling proteins, such as calmodulin or ryanodine receptor, as modifiers of presenilin-dependent Notch signalling (van de Hoef et al., 2009).
This intimate relationship of calcium and Notch signalling suggests that defects in Notch could contribute to the pathogenicity of Jp mutations. Disruption of Drosophila Notch pathway members results in a dilated cardiomyopathy similar to the one we describe in our models (Kim et al., 2010). As for the neural function of junctophilins, it is remarkable that the phenotypes of two mouse models, the Jph3/4 double KO and a Notch antisense RNA, have a similar phenotype with impaired synaptic plasticity and long-term potentiation in hippocampal CA1 synapses (Moriguchi et al., 2006; Wang et al., 2004). In the light of all this evidence, the interplay of junctophilins and Notch mediated by calcium could prove to be a relevant disease mechanism in muscular and neural pathologies and deserves further attention.
MATERIALS AND METHODS
Drosophila stocks, maintenance and genetics
The following fly stocks were obtained from the Bloomington Drosophila Stock Center: Oregon-R, w1118, Dl6B37, Act5C-Gal4, GMR-GAL4, Mhc-Gal4, Elav-Gal4, nub-GAL4, UAS-Ser, UAS-Dl, UAS-GFP, P{XP}Jpd04563 and hs-FLP; the RNAi line v100555 expressing a dsRNA for RNAi of jp (CG4405) and UAS-Dcr2 were obtained from the Vienna Drosophila Resource Centre. Other drivers used were GMH5-GAL4 (Wessells and Bodmer, 2004), rn-GAL4 (St Pierre et al., 2002), UAS-Htt-ex1-pQ93 (Steffan et al., 2001). twi;Mef-GAL4 is a recombinant carrying both twi-Gal4 and Mef2-Gal4. For the modification of SCA3 expansions, we used UAS-SCA3-Q89, expressing a full-length cDNA (Stochmanski et al., 2012). Flies were maintained on standard cornmeal medium at 25°C unless stated otherwise in the Results section.
The UAS-jp line was obtained by removing one of the two UAS promoters pointing in opposite directions in the P{XP} transposon in the P{XP}jpd04563 insertion, leaving only the UAS promoter pointing towards the jp gene. For these, we crossed to flies with that express the Flipase protein under the control of a heat shock promoter, hs-FLP. The removal of the UAS was confirmed by PCR and sequencing of the amplified fragment. The following oligonucleotides were used for the PCR: JP-CG4405-FLPout-F (TGCTGTGGTCCGTTCTCTTGGC) and JP-CG4405-FLPout-R (TCGGCTGCTGCTCTAAACGACG).
Nucleic acid isolation and qPCR
Quick Fly Genomic DNA Prep protocol (Berkeley Drosophila Genome Project resources) was used to isolate gDNA for genotyping. The methods for the RNA isolation and the cDNA synthesis were previously described (López Del Amo et al., 2015). The qPCRs were performed with SYBR Green SuperMix (Quanta BioSciences, Beverly, MA, USA) in a LightCycler LC480 real-time PCR instrument (Roche, Basel, Switzerland). Each qPCR was performed in triplicate for all genotypes, and each individual sample was obtained by pooling 10 individual flies in the RNA extraction. The relative mRNA levels were calculated according to the 2−ΔΔCt method. Results were normalized to the expression of the Gapdh or Rpl49 housekeeping genes.
For mitochondrial DNA copy number, total DNA was isolated as previously described (Scialò et al., 2015) and analysed by qPCR using primers against mt:CoxI (for mtDNA) and Rpl32 (nuclear DNA, single-copy, for normalization).
Lifespan and behavioural assays
For lifespan experiments, flies were collected using CO2 anaesthesia within 24-48 h of eclosion and then kept at a density of 20-25 flies per vial at 25°C (29°C in the case of the GMH5-GAL4 driver). Flies were transferred to new vials every 2-3 days, and the number of dead flies was recorded. Lifespan studies were performed with a minimum of 50 flies from three independent experiments.
To examine locomotor ability, the flies were knocked down to the bottom of the vial by quick, firm tapping, and the proportion of flies that had climbed over the 9 cm mark within 10 s was determined. This assay was performed in triplicate for each genotype; at least 15 flies were used per genotype. For the flight assay, individual flies were transferred to a Petri dish, then the lid was removed and the dish inverted over a 45-cm-long cylinder and gently tapped to loosen the fly. Flies were either able to stabilize their flight and stay at the wall of the vessel (this position was scored in centimetres) or fell at the base and were scored as 45 cm. Thirty flies were scored for each genotype. The bang-sensitivity assay was performed as previously described (Graham et al., 2010). A minimum of 10 flies from four independent experiments were tested for any particular genotype.
Cardiac physiological analysis
For the physiological analysis, female flies were collected immediately after eclosion and maintained for 7 days at 29°C. For the heart beat recordings, semi-intact heart preparations and semi-automated optical heartbeat analysis were carried out as previously described (Chakraborty et al., 2015; Ocorr et al., 2007). A minimum of 15 hearts were analysed per genotype.
Histology and microscopy
Flies were examined under an Olympus SZ60 stereomicroscope (Olympus, Tokyo, Japan) equipped with a Scopetek MDC200 Digital Camera (Hangzhou Scopetek Opto-Electric Co., Hangzhou, China). Areas (retina, IFM and wings) and lengths (L1-L2 wing veins) were measured using the ImageJ software (version 1.47; National Institutes of Health, Bethesda, MA, USA). Adult cuticles and wings were mounted in Hoyer's medium and analysed with a Leica DM6000 microscope.
Scanning electron microscopy (SEM) analysis of adult eyes was performed as previously described (Calpena et al., 2015), following the critical point drying method. Images were taken with a Philips XL-30 ESEM scanning electron microscope.
For light and transmission electron microscopy, eyes, thoraxes and abdomens were dissected and fixed overnight with 2% paraformaldehyde and 2% glutaraldehyde in 0.1 M phosphate buffer as previously described (López Del Amo et al., 2015). All samples were post-fixed in OsO4 for 2 h and dehydrated through an ethanol series; thereafter, samples were embedded in Durcupan epoxy resin (Sigma-Aldrich, St Louis, MO; USA). For transmission electron microscopy, 80-nm-thick sections were stained with uranyl acetate and examined with an FEI Tecnai Spirit G2 microscope. For bright field microscopy, 1.5 µm sections of thoraxes and abdomens were stained with Toluidine Blue, and images were examined with a Leica DM6000 microscope. Thorax sections were used for assessment of muscle defects; for the muscle section area, a minimum of six flies per genotype were analysed. Abdomen sections were used to evaluate heart wall thickness. A minimum of 10 sections from different individuals were analysed, and three different measurements of wall thickness were performed on each section. The number of photoreceptor neurons per ommatidium was determined by analysing transmission electron microscopy images of sections of retina from 1-week-old flies. For each genotype, sections from three different individuals were studied, and at least 60 ommatidia were scored in each section.
For confocal microscopy, fly hearts were dissected from 7-day-old females, fixed for 20 min in 4% paraformaldehyde, washed in PBT (PBS containing 0.3% Triton X-100), and stained with phalloidin for 20 min as previously described (Chakraborty et al., 2015). All confocal images were obtained with an Olympus FV1000 microscope. The gaps in myofibrillar staining were quantified by measuring the size of these areas from confocal stacks of five hearts of each genotype using the ImageJ software (version 1.47). The percentage area devoid of myofibrils was calculated, and comparisons were made between the control hearts and the OE or KD genotypes.
Mitochondrial circularity index
For the calculation of the circularity index of the mitochondria, the outline of the mitochondria was manually traced and then analysed with the ImageJ software (version 1.47). Sample size was three individuals per genotype; from each individual, two different muscle electron micrographs were analysed by scoring the index for 30 mitochondria per micrograph (a total of 180 mitochondria per genotype).
NMR spectroscopy
For NMR spectroscopy, six samples were analysed for each one of the three genotypes, each one of them containing 15 flies. Sample preparation, NMR spectroscopy and data analysis were performed as described (López Del Amo et al., 2015).
Statistical analysis
Data were analysed with Prism 5 (GraphPad). In the lifespan experiments, log-rank (Mantel–Cox) test was performed, and each one of the experimental genotypes was compared with the control. In the comparisons between the three genotypes at a single time point, we performed one-way ANOVA with Dunnett's multiple comparison test. Values shown represent means±s.e.m. In all figures, *P<0.05, **P<0.01 and ***P<0.001.
Supplementary Material
Acknowledgements
We thank Mario Soriano for his advice with the histology for electronic microscopy and Martina Palomino for her help with the metabolomics analysis; we also thank Alberto Sanz for the mitochondrial DNA quantification protocol and Hervé Tricoire for sharing fly stocks.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: E.C., V.L.A., M.C., B.L., R.A., C.E., M.I.G.; Formal analysis: E.C., V.L.A., M.C., B.L.; Investigation: E.C., V.L.A., M.C., B.L.; Writing - original draft: E.C., M.I.G.; Writing - review & editing: E.C., V.L.A., M.C., B.L., R.A., C.E., M.I.G.; Supervision: R.A., C.E., M.G.; Funding acquisition: R.A., C.E., M.I.G.
Funding
This work was supported by project grants from Association Française contre les Myopathies [AFM 18540 to M.I.G.], Instituto de Salud Carlos III (ISCIII) [PI12/000453 and PI15/000187 to C.E.], Generalitat Valenciana [PROMETEOII/2014/067 to R.A. as partner], and a collaborative grant from the International Rare Diseases Research Consortium (IRDiRC) and ISCIII [IR11/TREAT-CMT to M.I.G. (partner 12) and C.E. (partner 6)]. C.E. has a ‘Miguel Servet’ contract funded by the ISCIII and Centro de Investigación Príncipe Felipe [CPII14/00002]; M.C. was the recipient of a Santiago Grisolía award from Generalitat Valenciana [GrisoliaP/2013/A/044].
Supplementary information
Supplementary information available online at http://dmm.biologists.org/lookup/doi/10.1242/dmm.029082.supplemental
References
- Ao X. and Lehrer S. S. (1995). Phalloidin unzips nebulin from thin filaments in skeletal myofibrils. J. Cell Sci. 108, 3397-3403. [DOI] [PubMed] [Google Scholar]
- Beavers D. L., Wang W., Ather S., Voigt N., Garbino A., Dixit S. S., Landstrom A. P., Li N., Wang Q., Olivotto I. et al. (2013). Mutation E169K in junctophilin-2 causes atrial fibrillation due to impaired RyR2 stabilization. J. Am. Coll. Cardiol. 62, 2010-2019. 10.1016/j.jacc.2013.06.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beavers D. L., Landstrom A. P., Chiang D. Y. and Wehrens X. H. T. (2014). Emerging roles of junctophilin-2 in the heart and implications for cardiac diseases. Cardiovasc. Res. 103, 198-205. 10.1093/cvr/cvu151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand A. H. and Perrimon N. (1993). Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118, 401-415. [DOI] [PubMed] [Google Scholar]
- Cagan R. L. and Ready D. F. (1989). Notch is required for successive cell decisions in the developing Drosophila retina. Genes Dev. 3, 1099-1112. 10.1101/gad.3.8.1099 [DOI] [PubMed] [Google Scholar]
- Calpena E., Palau F., Espinós C. and Galindo M. I. (2015). Evolutionary history of the Smyd gene family in metazoans: a framework to identify the orthologs of human Smyd genes in Drosophila and other animal species. PLoS ONE 10, e0134106 10.1371/journal.pone.0134106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty M., Selma-Soriano E., Magny E., Couso J. P., Perez-Alonso M., Charlet-Berguerand N., Artero R. and Llamusi B. (2015). Pentamidine rescues contractility and rhythmicity in a Drosophila model of myotonic dystrophy heart dysfunction. Dis. Model. Mech. 8, 1569-1578. 10.1242/dmm.021428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B., Guo A., Zhang C., Chen R., Zhu Y., Hong J., Kutschke W., Zimmerman K., Weiss R. M., Zingman L. et al. (2013). Critical roles of junctophilin-2 in T-tubule and excitation-contraction coupling maturation during postnatal development. Cardiovasc. Res. 100, 54-62. 10.1093/cvr/cvt180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chintapalli V. R., Wang J. and Dow J. A. T. (2007). Using FlyAtlas to identify better Drosophila melanogaster models of human disease. Nat. Genet. 39, 715-720. 10.1038/ng2049 [DOI] [PubMed] [Google Scholar]
- Cruz C., Glavic A., Casado M. and de Celis J. F. (2009). A gain-of-function screen identifying genes required for growth and pattern formation of the Drosophila melanogaster wing. Genetics 183, 1005-1026. 10.1534/genetics.109.107748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Celis J. F. (2003). Pattern formation in the Drosophila wing: the development of the veins. BioEssays 25, 443-451. 10.1002/bies.10258 [DOI] [PubMed] [Google Scholar]
- Dietzl G., Chen D., Schnorrer F., Su K.-C., Barinova Y., Fellner M., Gasser B., Kinsey K., Oppel S., Scheiblauer S. et al. (2007). A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature 448, 151-156. 10.1038/nature05954 [DOI] [PubMed] [Google Scholar]
- Eid J.-P., Arias A. M., Robertson H., Hime G. R. and Dziadek M. (2008). The Drosophila STIM1 orthologue, dSTIM, has roles in cell fate specification and tissue patterning. BMC Dev. Biol. 8, 104 10.1186/1471-213X-8-104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garbino A., van Oort R. J., Dixit S. S., Landstrom A. P., Ackerman M. J. and Wehrens X. H. T. (2009). Molecular evolution of the junctophilin gene family. Physiol. Genomics 37, 175-186. 10.1152/physiolgenomics.00017.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass D. J. (2005). Skeletal muscle hypertrophy and atrophy signaling pathways. Int. J. Biochem. Cell Biol. 37, 1974-1984. 10.1016/j.biocel.2005.04.018 [DOI] [PubMed] [Google Scholar]
- Graham B. H., Li Z., Alesii E. P., Versteken P., Lee C., Wang J. and Craigen W. J. (2010). Neurologic dysfunction and male infertility in Drosophila porin mutants: a new model for mitochondrial dysfunction and disease. J. Biol. Chem. 285, 11143-11153. 10.1074/jbc.M109.080317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo A., Zhang X., Iyer V. R., Chen B., Zhang C., Kutschke W. J., Weiss R. M., Franzini-Armstrong C. and Song L.-S. (2014). Overexpression of junctophilin-2 does not enhance baseline function but attenuates heart failure development after cardiac stress. Proc. Natl. Acad. Sci. USA 111, 12240-12245. 10.1073/pnas.1412729111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gusella J. F. and MacDonald M. E. (2009). Huntington's disease: the case for genetic modifiers. Genome Med. 1, 80 10.1186/gm80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirata Y., Brotto M., Weisleder N., Chu Y., Lin P., Zhao X., Thornton A., Komazaki S., Takeshima H., Ma J. et al. (2006). Uncoupling store-operated Ca2+ entry and altered Ca2+ release from sarcoplasmic reticulum through silencing of junctophilin genes. Biophys. J. 90, 4418-4427. 10.1529/biophysj.105.076570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes S. E., O'Hearn E., Rosenblatt A., Callahan C., Hwang H. S., Ingersoll-Ashworth R. G., Fleisher A., Stevanin G., Brice A., Potter N. T. et al. (2001). A repeat expansion in the gene encoding junctophilin-3 is associated with Huntington disease-like 2. Nat. Genet. 29, 377-378. 10.1038/ng760 [DOI] [PubMed] [Google Scholar]
- Ikeda A., Miyazaki T., Kakizawa S., Okuno Y., Tsuchiya S., Myomoto A., Saito S.-Y., Yamamoto T., Yamazaki T., Iino M. et al. (2007). Abnormal features in mutant cerebellar Purkinje cells lacking junctophilins. Biochem. Biophys. Res. Commun. 363, 835-839. 10.1016/j.bbrc.2007.09.062 [DOI] [PubMed] [Google Scholar]
- Ito K., Komazaki S., Sasamoto K., Yoshida M., Nishi M., Kitamura K. and Takeshima H. (2001). Deficiency of triad junction and contraction in mutant skeletal muscle lacking junctophilin type 1. J. Cell Biol. 154, 1059-1067. 10.1083/jcb.200105040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jumbo-Lucioni P., Ayroles J. F., Chambers M. M., Jordan K. W., Leips J., Mackay T. F. C. and De Luca M. (2010). Systems genetics analysis of body weight and energy metabolism traits in Drosophila melanogaster. BMC Genomics 11, 297 10.1186/1471-2164-11-297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakizawa S., Moriguchi S., Ikeda A., Iino M. and Takeshima H. (2008). Functional crosstalk between cell-surface and intracellular channels mediated by junctophilins essential for neuronal functions. Cerebellum 7, 385-391. 10.1007/s12311-008-0040-1 [DOI] [PubMed] [Google Scholar]
- Kim I. M., Wolf M. J. and Rockman H. A. (2010). Gene deletion screen for cardiomyopathy in adult Drosophila identifies a new notch ligand. Circ. Res. 106, 1233-1243. 10.1161/CIRCRESAHA.109.213785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimball S. R. and Jefferson L. S. (2006). Signaling pathways and molecular mechanisms through which branched-chain amino acids mediate translational control of protein synthesis. J. Nutr. 136, 227S-231S. [DOI] [PubMed] [Google Scholar]
- Komazaki S., Ito K., Takeshima H. and Nakamura H. (2002). Deficiency of triad formation in developing skeletal muscle cells lacking junctophilin type 1. FEBS Lett. 524, 225-229. 10.1016/S0014-5793(02)03042-9 [DOI] [PubMed] [Google Scholar]
- Komazaki S., Nishi M. and Takeshima H. (2003). Abnormal junctional membrane structures in cardiac myocytes expressing ectopic junctophilin type 1. FEBS Lett. 542, 69-73. 10.1016/S0014-5793(03)00340-5 [DOI] [PubMed] [Google Scholar]
- Krench M., Cho R. W. and Littleton J. T. (2016). A Drosophila model of Huntington disease-like 2 exhibits nuclear toxicity and distinct pathogenic mechanisms from Huntington disease. Hum. Mol. Genet. 25, 3164-3177. 10.1093/hmg/ddw166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuebler D. and Tanouye M. A. (2000). Modifications of seizure susceptibility in Drosophila. J. Neurophysiol. 83, 998-1009. [DOI] [PubMed] [Google Scholar]
- Kumar S., Konikoff C., Van Emden B., Busick C., Davis K. T., Ji S., Wu L.-W., Ramos H., Brody T., Panchanathan S. et al. (2011). FlyExpress: visual mining of spatiotemporal patterns for genes and publications in Drosophila embryogenesis. Bioinformatics 27, 3319-3320. 10.1093/bioinformatics/btr567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landstrom A. P., Weisleder N., Batalden K. B., Bos J. M., Tester D. J., Ommen S. R., Wehrens X. H. T., Claycomb W. C., Ko J.-K., Hwang M. et al. (2007). Mutations in JPH2-encoded junctophilin-2 associated with hypertrophic cardiomyopathy in humans. J. Mol. Cell. Cardiol. 42, 1026-1035. 10.1016/j.yjmcc.2007.04.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landstrom A. P., Kellen C. A., Dixit S. S., van Oort R. J., Garbino A., Weisleder N., Ma J., Wehrens X. H. T. and Ackerman M. J. (2011). Junctophilin-2 expression silencing causes cardiocyte hypertrophy and abnormal intracellular calcium-handling. Circ. Heart Fail. 4, 214-223. 10.1161/CIRCHEARTFAILURE.110.958694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landstrom A. P., Beavers D. L. and Wehrens X. H. T. (2014). The junctophilin family of proteins: from bench to bedside. Trends Mol. Med. 20, 353-362. 10.1016/j.molmed.2014.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. and Wu C. F. (2002). Electroconvulsive seizure behavior in Drosophila: analysis of the physiological repertoire underlying a stereotyped action pattern in bang-sensitive mutants. J. Neurosci. 22, 11065-11079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis E. A. and Smith G. A. (2016). Using Drosophila models of Huntington's disease as a translatable tool. J. Neurosci. Methods 265, 89-98. 10.1016/j.jneumeth.2015.07.026 [DOI] [PubMed] [Google Scholar]
- Li H., Ding X., Lopez J. R., Takeshima H., Ma J., Allen P. D. and Eltit J. M. (2010). Impaired Orai1-mediated resting Ca2+ entry reduces the cytosolic [Ca2+] and sarcoplasmic reticulum Ca2+ loading in quiescent junctophilin 1 knock-out myotubes. J. Biol. Chem. 285, 39171-39179. 10.1074/jbc.M110.149690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Pan Z.-F., Huang X., Wu B.-W., Li T., Kang M.-X., Ge R.-S., Hu X.-Y., Zhang Y.-H., Ge L.-J. et al. (2016). Junctophilin 3 expresses in pancreatic beta cells and is required for glucose-stimulated insulin secretion. Cell Death Dis. 7, e2275 10.1038/cddis.2016.179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- López Del Amo V., Seco-Cervera M., García-Giménez J. L., Whitworth A. J., Pallardó F. V. and Galindo M. I. (2015). Mitochondrial defects and neuromuscular degeneration caused by altered expression of Drosophila Gdap1: implications for the Charcot-Marie-Tooth neuropathy. Hum. Mol. Genet. 24, 21-36. 10.1093/hmg/ddu416 [DOI] [PubMed] [Google Scholar]
- López Del Amo V., Palomino-Schätzlein M., Seco-Cervera M., García-Giménez J. L., Pallardó F. V., Pineda-Lucena A. and Galindo M. I. (2017). A Drosophila model of GDAP1 function reveals the involvement of insulin signalling in the mitochondria-dependent neuromuscular degeneration. Biochim. Biophys. Acta 1863, 801-809. 10.1016/j.bbadis.2017.01.003 [DOI] [PubMed] [Google Scholar]
- Ma Y., Creanga A., Lum L. and Beachy P. A. (2006). Prevalence of off-target effects in Drosophila RNA interference screens. Nature 443, 359-363. 10.1038/nature05179 [DOI] [PubMed] [Google Scholar]
- Mery A., Taghli-Lamallem O., Clark K. A., Beckerle M. C., Wu X., Ocorr K. and Bodmer R. (2008). The Drosophila muscle LIM protein, Mlp84B, is essential for cardiac function. J. Exp. Biol. 211, 15-23. 10.1242/jeb.012435 [DOI] [PubMed] [Google Scholar]
- Minamisawa S., Oshikawa J., Takeshima H., Hoshijima M., Wang Y., Chien K. R., Ishikawa Y. and Matsuoka R. (2004). Junctophilin type 2 is associated with caveolin-3 and is down-regulated in the hypertrophic and dilated cardiomyopathies. Biochem. Biophys. Res. Commun. 325, 852-856. 10.1016/j.bbrc.2004.10.107 [DOI] [PubMed] [Google Scholar]
- Molina M. R. and Cripps R. M. (2001). Ostia, the inflow tracts of the Drosophila heart, develop from a genetically distinct subset of cardial cells. Mech. Dev. 109, 51-59. 10.1016/S0925-4773(01)00509-3 [DOI] [PubMed] [Google Scholar]
- Molnar C., López-Varea A., Hernández R. and de Celis J. F. (2006). A gain-of-function screen identifying genes required for vein formation in the Drosophila melanogaster wing. Genetics 174, 1635-1659. 10.1534/genetics.106.061283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriguchi S., Nishi M., Komazaki S., Sakagami H., Miyazaki T., Masumiya H., Saito S.-Y., Watanabe M., Kondo H., Yawo H. et al. (2006). Functional uncoupling between Ca2+ release and afterhyperpolarization in mutant hippocampal neurons lacking junctophilins. Proc. Natl. Acad. Sci. USA 103, 10811-10816. 10.1073/pnas.0509863103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy R. M., Dutka T. L., Horvath D., Bell J. R., Delbridge L. M. and Lamb G. D. (2013). Ca2+-dependent proteolysis of junctophilin-1 and junctophilin-2 in skeletal and cardiac muscle. J. Physiol. 591, 719-729. 10.1113/jphysiol.2012.243279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishi M., Mizushima A., Nakagawara K. and Takeshima H. (2000). Characterization of human junctophilin subtype genes. Biochem. Biophys. Res. Commun. 273, 920-927. 10.1006/bbrc.2000.3011 [DOI] [PubMed] [Google Scholar]
- Nishi M., Hashimoto K., Kuriyama K., Komazaki S., Kano M., Shibata S. and Takeshima H. (2002). Motor discoordination in mutant mice lacking junctophilin type 3. Biochem. Biophys. Res. Commun. 292, 318-324. 10.1006/bbrc.2002.6649 [DOI] [PubMed] [Google Scholar]
- Nishi M., Sakagami H., Komazaki S., Kondo H. and Takeshima H. (2003). Coexpression of junctophilin type 3 and type 4 in brain. Brain Res. Mol. Brain Res. 118, 102-110. 10.1016/S0169-328X(03)00341-3 [DOI] [PubMed] [Google Scholar]
- Ocorr K., Perrin L., Lim H. Y., Qian L., Wu X. and Bodmer R. (2007). Genetic control of heart function and aging in Drosophila. Trends Cardiovasc. Med. 17, 177-182. 10.1016/j.tcm.2007.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owusu-Ansah E., Song W. and Perrimon N. (2013). Muscle mitohormesis promotes longevity via systemic repression of insulin signaling. Cell 155, 699-712. 10.1016/j.cell.2013.09.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks A. L., Turner F. R. and Muskavitch M. A. T. (1995). Relationships between complex Delta expression and the specification of retinal cell fates during Drosophila eye development. Mech. Dev. 50, 201-216. 10.1016/0925-4773(94)00336-L [DOI] [PubMed] [Google Scholar]
- Pavlidis P., Ramaswami M. and Tanouye M. A. (1994). The Drosophila easily shocked gene: a mutation in a phospholipid synthetic pathway causes seizure, neuronal failure, and paralysis. Cell 79, 23-33. 10.1016/0092-8674(94)90397-2 [DOI] [PubMed] [Google Scholar]
- Pla-Martin D., Calpena E., Lupo V., Márquez C., Rivas E., Sivera R., Sevilla T., Palau F. and Espinós C. (2015). Junctophilin-1 is a modifier gene of GDAP1-related Charcot-Marie-Tooth disease. Hum. Mol. Genet. 24, 213-229. 10.1093/hmg/ddu440 [DOI] [PubMed] [Google Scholar]
- Razzaq A., Robinson I. M., McMahon H. T., Skepper J. N., Su Y., Zelhof A. C., Jackson A. P., Gay N. J. and O'Kane C. J. (2001). Amphiphysin is necessary for organization of the excitation-contraction coupling machinery of muscles, but not for synaptic vesicle endocytosis in Drosophila. Genes Dev. 15, 2967-2979. 10.1101/gad.207801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds J. O., Chiang D. Y., Wang W., Beavers D. L., Dixit S. S., Skapura D. G., Landstrom A. P., Song L.-S., Ackerman M. J. and Wehrens X. H. T. (2013). Junctophilin-2 is necessary for T-tubule maturation during mouse heart development. Cardiovasc. Res. 100, 44-53. 10.1093/cvr/cvt133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabater-Molina M., Navarro M., Garcia-Molina Saez E., Garrido I., Pascual-Figal D., Gonzalez Carrillo J. and Gimeno Blanes J. R. (2016). Mutation in JPH2 cause dilated cardiomyopathy. Clin. Genet. 90, 468-469. 10.1111/cge.12825 [DOI] [PubMed] [Google Scholar]
- Sarou-Kanian V., Joudiou N., Louat F., Yon M., Szeremeta F., Même S., Massiot D., Decoville M., Fayon F. and Beloeil J.-C. (2015). Metabolite localization in living drosophila using high resolution magic angle spinning NMR. Sci. Rep. 5, 9872 10.1038/srep09872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubiger M., Feng Y., Fambrough D. M. and Palka J. (1994). A mutation of the Drosophila sodium pump alpha subunit gene results in bang-sensitive paralysis. Neuron 12, 373-381. 10.1016/0896-6273(94)90278-X [DOI] [PubMed] [Google Scholar]
- Schweisguth F. and Posakony J. W. (1994). Antagonistic activities of Suppressor of Hairless and Hairless control alternative cell fates in the Drosophila adult epidermis. Development 120, 1433-1441. [DOI] [PubMed] [Google Scholar]
- Scialò F., Sriram A., Naudí A., Ayala V., Jové M., Pamplona R. and Sanz A. (2015). Target of rapamycin activation predicts lifespan in fruit flies. Cell Cycle 14, 2949-2958. 10.1080/15384101.2015.1071745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seixas A. I., Holmes S. E., Takeshima H., Pavlovich A., Sachs N., Pruitt J. L., Silveira I., Ross C. A., Margolis R. L. and Rudnicki D. D. (2012). Loss of junctophilin-3 contributes to Huntington disease-like 2 pathogenesis. Ann. Neurol. 71, 245-257. 10.1002/ana.22598 [DOI] [PubMed] [Google Scholar]
- Shalaby N. A., Parks A. L., Morreale E. J., Osswalt M. C., Pfau K. M., Pierce E. L. and Muskavitch M. A. T. (2009). A screen for modifiers of notch signaling uncovers Amun, a protein with a critical role in sensory organ development. Genetics 182, 1061-1076. 10.1534/genetics.108.099986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- St Pierre S. E., Galindo M. I., Couso J. P. and Thor S. (2002). Control of Drosophila imaginal disc development by rotund and roughened eye: differentially expressed transcripts of the same gene encoding functionally distinct zinc finger proteins. Development 129, 1273-1281. [DOI] [PubMed] [Google Scholar]
- Steffan J. S., Bodai L., Pallos J., Poelman M., McCampbell A., Apostol B. L., Kazantsev A., Schmidt E., Zhu Y.-Z., Greenwald M. et al. (2001). Histone deacetylase inhibitors arrest polyglutamine-dependent neurodegeneration in Drosophila. Nature 413, 739-743. 10.1038/35099568 [DOI] [PubMed] [Google Scholar]
- Stochmanski S. J., Therrien M., Laganière J., Rochefort D., Laurent S., Karemera L., Gaudet R., Vyboh K., Van Meyel D. J., Di Cristo G. et al. (2012). Expanded ATXN3 frameshifting events are toxic in Drosophila and mammalian neuron models. Hum. Mol. Genet. 21, 2211-2218. 10.1093/hmg/dds036 [DOI] [PubMed] [Google Scholar]
- Taghli-Lamallem O., Akasaka T., Hogg G., Nudel U., Yaffe D., Chamberlain J. S., Ocorr K. and Bodmer R. (2008). Dystrophin deficiency in Drosophila reduces lifespan and causes a dilated cardiomyopathy phenotype. Aging Cell 7, 237-249. 10.1111/j.1474-9726.2008.00367.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeshima H., Komazaki S., Nishi M., Iino M. and Kangawa K. (2000). Junctophilins: a novel family of junctional membrane complex proteins. Mol. Cell 6, 11-22. 10.1016/S1097-2765(05)00005-5 [DOI] [PubMed] [Google Scholar]
- Takeshima H., Hoshijima M. and Song L.-S. (2015). Ca2+ microdomains organized by junctophilins. Cell Calcium 58, 349-356. 10.1016/j.ceca.2015.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thibault S. T., Singer M. A., Miyazaki W. Y., Milash B., Dompe N. A., Singh C. M., Buchholz R., Demsky M., Fawcett R., Francis-Lang H. L. et al. (2004). A complementary transposon tool kit for Drosophila melanogaster using P and piggyBac. Nat. Genet. 36, 283-287. 10.1038/ng1314 [DOI] [PubMed] [Google Scholar]
- Tom A. and Nair K. S. (2006). Assessment of branched-chain amino Acid status and potential for biomarkers. J. Nutr. 136, 324S-330S. [DOI] [PubMed] [Google Scholar]
- Trotta N., Rodesch C. K., Fergestad T. and Broadie K. (2004). Cellular bases of activity-dependent paralysis in Drosophila stress-sensitive mutants. J. Neurobiol. 60, 328-347. 10.1002/neu.20017 [DOI] [PubMed] [Google Scholar]
- van de Hoef D. L., Hughes J., Livne-Bar I., Garza D., Konsolaki M. and Boulianne G. L. (2009). Identifying genes that interact with Drosophila presenilin and amyloid precursor protein. Genesis 47, 246-260. 10.1002/dvg.20485 [DOI] [PubMed] [Google Scholar]
- Wang Y. and Hekimi S. (2015). Mitochondrial dysfunction and longevity in animals: Untangling the knot. Science 350, 1204-1207. 10.1126/science.aac4357 [DOI] [PubMed] [Google Scholar]
- Wang Y., Chan S. L., Miele L., Yao P. J., Mackes J., Ingram D. K., Mattson M. P. and Furukawa K. (2004). Involvement of Notch signaling in hippocampal synaptic plasticity. Proc. Natl. Acad. Sci. USA 101, 9458-9462. 10.1073/pnas.0308126101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessells R. J. and Bodmer R. (2004). Screening assays for heart function mutants in Drosophila. BioTechniques 37, 58-60, 62, 64 passim. [DOI] [PubMed] [Google Scholar]
- Woo J. S., Hwang J.-H., Ko J.-K., Weisleder N., Kim D. H., Ma J. and Lee E. H. (2010). S165F mutation of junctophilin 2 affects Ca2+ signalling in skeletal muscle. Biochem. J. 427, 125-134. 10.1042/BJ20091225 [DOI] [PubMed] [Google Scholar]
- Woo J. S., Cho C.-H., Lee K. J., Kim D. H., Ma J. and Lee E. H. (2012). Hypertrophy in skeletal myotubes induced by junctophilin-2 mutant, Y141H, involves an increase in store-operated Ca2+ entry via Orai1. J. Biol. Chem. 287, 14336-14348. 10.1074/jbc.M111.304808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo J. S., Srikanth S., Nishi M., Ping P., Takeshima H. and Gwack Y. (2016). Junctophilin-4, a component of the endoplasmic reticulum-plasma membrane junctions, regulates Ca2+ dynamics in T cells. Proc. Natl. Acad. Sci. USA 113, 2762-2767. 10.1073/pnas.1524229113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J., Shih H.-P., Vigont V., Hrdlicka L., Diggins L., Singh C., Mahoney M., Chesworth R., Shapiro G., Zimina O. et al. (2011). Neuronal store-operated calcium entry pathway as a novel therapeutic target for Huntington's disease treatment. Chem. Biol. 18, 777-793. 10.1016/j.chembiol.2011.04.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M., Zhou P., Xu S.-M., Liu Y., Feng X., Bai S.-H., Bai Y., Hao X.-M., Han Q., Zhang Y. et al. (2007). Intermolecular failure of L-type Ca2+ channel and ryanodine receptor signaling in hypertrophy. PLoS Biol. 5, e21 10.1371/journal.pbio.0050021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarse K., Schmeisser S., Groth M., Priebe S., Beuster G., Kuhlow D., Guthke R., Platzer M., Kahn C. R. and Ristow M. (2012). Impaired insulin/IGF1 signaling extends life span by promoting mitochondrial L-proline catabolism to induce a transient ROS signal. Cell Metab. 15, 451-465. 10.1016/j.cmet.2012.02.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S., Feany M. B., Saraswati S., Littleton J. T. and Perrimon N. (2009). Inactivation of Drosophila Huntingtin affects long-term adult functioning and the pathogenesis of a Huntington's disease model. Dis. Model. Mech. 2, 247-266. 10.1242/dmm.000653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H.-B., Li R.-C., Xu M., Xu S.-M., Lai Y.-S., Wu H.-D., Xie X.-J., Gao W., Ye H., Zhang Y.-Y. et al. (2013). Ultrastructural uncoupling between T-tubules and sarcoplasmic reticulum in human heart failure. Cardiovasc. Res. 98, 269-276. 10.1093/cvr/cvt030 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.