Abstract
HOX transcript antisense intergenic RNA has been reported to serve as an important prognostic biomarker in several types of cancers. However, the clinical value of HOX transcript antisense intergenic RNA in digestive cancers remains unclear. Therefore, we tried to investigate the clinical role of expression of HOX transcript antisense intergenic RNA as a prognostic indicator in digestive cancers by a meta-analysis. Literature collection was performed by searching the PubMed, Embase, Web of Science, and Cochrane Library databases (up to October 7, 2017). A quantitative meta-analysis was conducted to assess the eligible articles on the prognostic value of HOX transcript antisense intergenic RNA in digestive cancers. The pooled hazard ratios or odds ratios with 95% confidence intervals were used to evaluate the association between expression of HOX transcript antisense intergenic RNA and clinical outcomes. A total of 1844 patients from 22 studies were included in this meta-analysis. The results found a significant association between expression of HOX transcript antisense intergenic RNA and poor overall survival in digestive cancers (pooled hazard ratio = 2.19, 95% confidence interval, 1.86-2.57, P < .001). Furthermore, subgroup analysis showed that tumor type, region, Newcastle-Ottawa scale, and sample size did not alter the predictive value of HOX transcript antisense intergenic RNA as an independent factor for patients’ survival. In addition, we also revealed that the clinicopathological characteristics such as differentiation, lymph node metastasis, tumor node metastasis (TNM) stage, and distant metastasis were positively related to expression of HOX transcript antisense intergenic RNA digestive cancers. In conclusion, our results suggested high expression of HOX transcript antisense intergenic RNA was correlated with poor clinical outcomes and may serve as a novel prognostic biomarker for patients with digestive cancers.
Keywords: long noncoding RNA, HOTAIR, digestive cancer, biomarker, meta-analysis
Introduction
Digestive cancers are one of the most prevalent cancers and a leading cause of cancer-related deaths worldwild.1 Although there are various effective techniques for cancer diagnosis and treatment, the prognosis of patients with digestive cancers still remains poor.2–5 Therefore, it is necessary to urgently identify some applicable biomarkers for digestive cancers. Recently, long noncoding RNAs (lncRNAs) were reported to play an important role in numerous human diseases, including cancer.
The LncRNAs are evolutionarily conserved nonprotein coding RNAs that are longer than 200 nucleotides in length.6 Although previous opinions agreed that lncRNAs were one kind of transcriptional noise, in recent years, several studies have reported that lncRNAs could regulate gene expression via different molecular mechanisms, including transcriptional and posttranscriptional processing, chromatin modification and epigenetics, genomic imprinting, protein activity modulation, and protein localization.7,8 An emerging evidence has suggested that the aberrant expressions of several confirmed cancer-related lncRNAs including HOX transcript antisense intergenic RNA (HOTAIR), MALAT1, CRNDE, and GAS5 are associated with tumorigenesis, metastasis, and prognosis in digestive cancers. However, the role of most lncRNAs in the progression and prognosis of digestive cancers still remains unclear.9–12
HOX transcript antisense intergenic RNA was first identified as a nuclear lncRNA with a length of 2158 bp and expressed from the HOXC locus on chromosome 12q13.13.13 According to a genome-wide analysis, the silencing of HOTAIR expression could regulate the expression of various cancer-related genes that are associated with tumor growth, apoptosis, cell differentiation, invasion, and metastasis.13–15 Recent studies have reported that aberrant expressions of HOTAIR were found in different types of cancers, including breast cancer16, cervical cancer17, colorectal cancer (CRC)18–21, gastric cancer (GC)9,22–29, pancreatic cancer (PC)30, hepatocellular carcinoma (HCC)31–33, and esophageal squamous cell carcinoma (ESCC)34–38. Yang et al indicated that HOTAIR could activate autophagy by increasing expression of autophagy-related 3 (ATG3) and autophagy-related 7 (ATG7) and promoting HCC cell proliferation.39 Kim et al suggested that HOTAIR is a negative prognostic factor for patients with PC, and HOTAIR-mediated suppression of genes in PC is both PRC2 dependent and PRC2 independent.30 Ge et al illuminate that HOTAIR directly decreased WIF-1 expression by promoting its histone H3K27 methylation in the promoter region and then activated the Wnt/β-catenin signaling pathway. This identified HOTAIR/WIF-1 axis clarified the molecular mechanism of ESCC metastasis.36 Thus, HOTAIR might serve as a biomarker for patients’ prognosis in digestive cancers.
Previous Studies
So far, it was not rare to detect the high HOTAIR expression in both primary and metastasized tumors of digestive cancers, and HOTAIR was also reported to be a novel effective prognostic biomarker and therapeutic target for digestive cancers. There were several studies suggesting that HOTAIR was significantly associated with clinicopathological features. However, most studies that reported about the prognostic value of HOTAIR were limited in discrete outcome and sample size. Therefore, we performed this meta-analysis to evaluate the association between HOTAIR expression and clinical outcomes in digestive cancers, and the prognostic value of HOTAIR expression was investigated as a prognostic biomarker in digestive cancers.
Materials and Methods
Search Strategy
We searched the PubMed, Embase, Web of Science, and Cochrane Library databases to identify the relevant articles according to the following terms: “long intergenic noncoding RNA” or “lncRNA,” “HOX transcript antisense intergenic RNA” or “HOTAIR,” “cancer” or “tumor” or “carcinoma” or “neoplasm.” The published language was limited to English, and the literature search was conducted up to October 7, 2017.
Inclusion and Exclusion Criteria
The studies were considered eligible according to the following criteria: (1) any type of human digestive cancers was studied; (2) studies investigating the prognostic role of HOTAIR in digestive cancers; (3) the expression of HOTAIR in cancerous tissues must be detected; (4) the digestive cancers must be histopathologically confirmed in the study; (5) the sample size was large enough to calculate the hazard ratios (HRs) and 95% confidence intervals (CIs) for survival rates; (6) patients were divided into 2 groups according to the levels of HOTAIR expression; and (7) the language of the study was limited to English. Exclusion criteria were as follows: (1) letters, editorials, expert opinions, case reports, and reviews; (2) duplicate publications; (3) irrelevant, noncomparative, or nonhuman research; and (4) studies without usable data for further studies.
Data Extraction and Quality Assessment
To validate the accuracy of extraction data, 2 authors (YZ and TX) extracted data independently from the eligible studies using standardized data compilation forms, and disagreements were resolved by discussion (SZ). For all included studies, the following information was collected: first author, year of publication, region, type of cancers, tumor node metastasis (TNM) stage, sample size, cutoff values, follow-up, detection method, adjuvant therapy before surgery, survival analysis, and outcome measure. The clinicopathological features including age, gender, tumor size, differentiation, TNM stage, lymph node metastasis, and distant metastasis were also extracted from the studies. Quality assessment was performed independently by 2 investigators (YZ and TX) and reached a consensus on all items through discussion. The quality of included studies was assessed based on the Newcastle-Ottawa quality assessment scale (NOS).40
Statistical Methods
We obtained the HRs and 95% CIs from the studies. The best way to extract the data was by obtaining parameters directly from the studies or calculating the HRs from O-E statistic and variance.41,42 The second way was to estimate the HRs from sample size, survival rate at specified times, log-rank statistic, and P value. Otherwise, Kaplan-Meier curves were analyzed using the Engauge Digitizer version 4.1 (http://digitizer.sourceforge.net/) to retrieve HRs and 95% CIs. A test of heterogeneity of combined odds ratios (ORs) or HRs was conducted by using Cochran Q test and Higgins I 2 statistic. P values <.1 was considered significant. I 2 values >50% indicate heterogeneity among studies. The meta-analysis results were displayed as forest plots. Sensitivity analysis was performed to test the impact of individual study on the pooled data while I 2 values >50%. Begg funnel plots and Egger linear regression test were performed to estimate potential publication bias. This meta-analysis was conducted by software Stata 12.0. All the results were output by Stata 12.0. P values <.05 were considered statistically significant.
Results
Literature Search
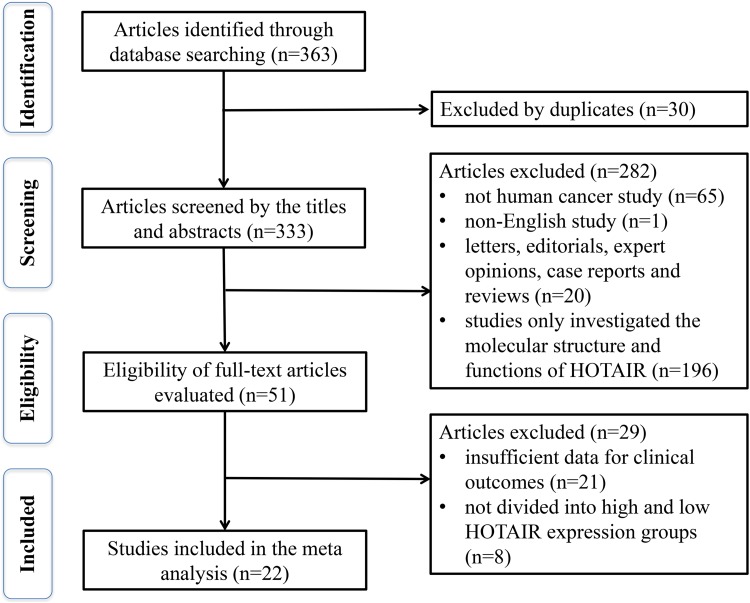
We searched 363 citations from the PubMed, Embase, Web of Science, and Cochrane Library databases using the abovementioned search strategy. After removing duplications, 30 articles were excluded. After detailed screening of the title and abstract, irrelevant and noncomparative articles were excluded, and 51 potential eligible studies were selected. After further evaluation of the full studies, a total of 22 studies with 1844 patients were selected for the further analysis. A flowchart of the study selection process is shown in Figure 1.
Figure 1.
The flow diagram of this meta-analysis.
The characteristics of included studies are shown in Table 1. For all studies, 4 were about CRC, 9 were about GC, 1 was about PC, 3 were about HCC, and 5 were about ESCC. Those 22 studies were reported from around the world: 14 from China, 1 from the USA, 5 from Japan, 1 from Korea, and 1 from Czech, and publication years when these studies were reported ranged from 2011 to 2017. In all 22 studies, patients were divided into high and low HOTAIR expression groups. Some of the studies used receiver–operating characteristic curve (ROC) analysis to define the cutoff value, and some of them used median or mean value of HOTAIR expression, but 3 of them did not report the cutoff value they used in their studies. The follow-up of the 22 studies ranged from 30 to 200 months. The sample size ranged from 39 to 168. Sixteen of the studies reported the overall survival (OS) of patients based on HOTAIR expression levels, 2 of them also reported the metastasis-free survival (MFS), 2 studies reported the disease-free survival (DFS), and 2 studies reported the recurrence-free survival (RFS). We extracted the HRs and 95% CIs from every study. The HRs could be obtained directly from 10 studies, and HRs of 12 of studies were extrapolated by graphical representations of the survival curve.
Table 1.
The Main Characteristics of the Included Studies in the Meta-Analysis.
| First author | Year | Region | Tumor Type | TNM Stage | Sample Size | Cutoff Value | Follow-up (months) | Detection Method | Adjuvant Therapy Before Surgery | Survival Analysis | Outcome Measure | NOS | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Yang | 2011 | China | HCC | N/A | 60 | ROC | 36 | qRT-PCR | N/A | Univariate | Multivariate | RFS | 7 |
| Geng | 2011 | China | HCC | N/A | 63 | ROC | 36 | qRT-PCR | None | Univariate | Multivariate | RFS | 7 |
| Kogo | 2011 | Japan | CRC | N/A | 100 | Mean | 60 | qRT-PCR | None | Univariate | Multivariate | OS | 7 |
| Niinuma | 2012 | Japan | GC | N/A | 39 | Mean | 200 | qRT-PCR | N/A | N/A | N/A | OS | 7 |
| Ishibashi | 2012 | Japan | HCC | N/A | 64 | T/N>1 | 36 | qRT-PCR | N/A | Univariate | Multivariate | OS | 5 |
| Kim | 2012 | USA | PC | I–IV | 102 | N/A | 60 | qRT-PCR | N/A | N/A | N/A | OS | 5 |
| Chen | 2013 | China | ESCC | I–IV | 78 | Mean | 60 | qRT-PCR | None | N/A | Multivariate | OS | 7 |
| Ge | 2013 | China | ESCC | N/A | 137 | T/N>1.5 | 80 | qRT-PCR | None | N/A | Multivariate | OS, MFS | 7 |
| Lv | 2013 | China | ESCC | I-IV | 93 | N/A | 70 | qRT-PCR | N/A | Univariate | Multivariate | OS | 7 |
| Li | 2013 | China | ESCC | I-IV | 100 | T/N>125 | 60 | qRT-PCR | None | N/A | Multivariate | OS | 7 |
| Xu | 2013 | China | GC | I–IV | 83 | ROC | 72 | qRT-PCR | N/A | N/A | N/A | OS | 7 |
| Endo | 2013 | Japan | GC | N/A | 68 | Mean | 60 | qRT-PCR | None | N/A | N/A | OS | 7 |
| WU | 2014 | China | CRC | I–IV | 120 | T/N>5 | 72 | qRT-PCR | None | Univariate | Multivariate | OS, MFS | 7 |
| Svoboda | 2014 | Czech | CRC | I–IV | 73 | ROC | 54 | qRT-PCR | N/A | Univariate | Multivariate | OS | 7 |
| Okugawa | 2014 | Japan | GC | III-IV | 150 | ROC | 60 | qRT-PCR | N/A | Univariate | Multivariate | OS | 7 |
| Lee | 2014 | Korea | GC | I-IV | 50 | Median | 48 | qRT-PCR | N/A | N/A | N/A | DFS | 7 |
| Zhao | 2015 | China | GC | III–IV | 168 | Median | 60 | qRT-PCR | Chemotherapy | N/A | N/A | OS | 5 |
| Zhang | 2015 | China | GC | II-IV | 50 | Median | 36 | qRT-PCR | None | N/A | N/A | OS | 5 |
| Liu | 2015 | China | GC | I-IV | 61 | N/A | 30 | qRT-PCR | None | N/A | N/A | DFS | 5 |
| Luo | 2016 | China | CRC | I-IV | 80 | Mean | 70 | qRT-PCR | None | N/A | N/A | OS | 5 |
| Chen | 2017 | China | GC | I-IV | 65 | Median | 60 | qRT-PCR | None | Univariate | Multivariate | OS | 7 |
| Xu | 2017 | China | ESCC | N/A | 40 | Median | 36 | qRT-PCR | N/A | N/A | N/A | OS | 5 |
Abbreviations: CRC, colorectal cancer; DFS, disease-free survival; ESCC, esophageal squamous cell carcinoma; GC, gastric cancer; HCC, hepatocellular carcinoma; MFS: metastasis-free survival; N, normal; N/A, not available; OS, overall survival; qRT-PCR quantitative real-time PCR; RFS, recurrence-free survival; ROC, receiver–operating characteristic curve; T, tumor.
Association Between HOTAIR and Patients’ Survival in Digestive Cancers
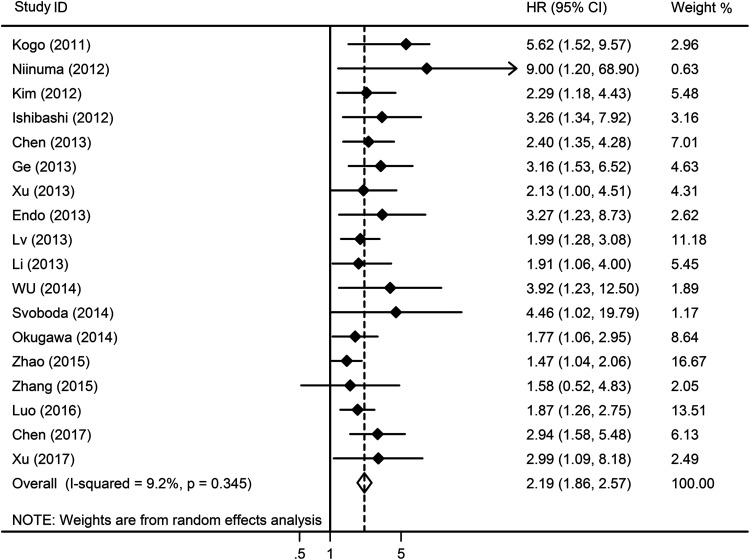
To study the association between expression of HOTAIR and OS in digestive cancers, 16 studies reporting a total of 1505 patients were included. The results showed that high HOTAIR expression was significantly associated with poor OS (pooled HR = 2.19, 95% CI, 1.86-2.57, P < .001) with no heterogeneity (I2 = 9.2%, P = .345; Figure 2). Furthermore, subgroups analysis was performed based on tumor type, region, sample size, and NOS.
Figure 2.
Forest plots of the included studies evaluating the HRs for HOTAIR for OS. HRs indicates hazard ratios; HOTAIR, HOX transcript antisense intergenic RNA; OS, overall survival.
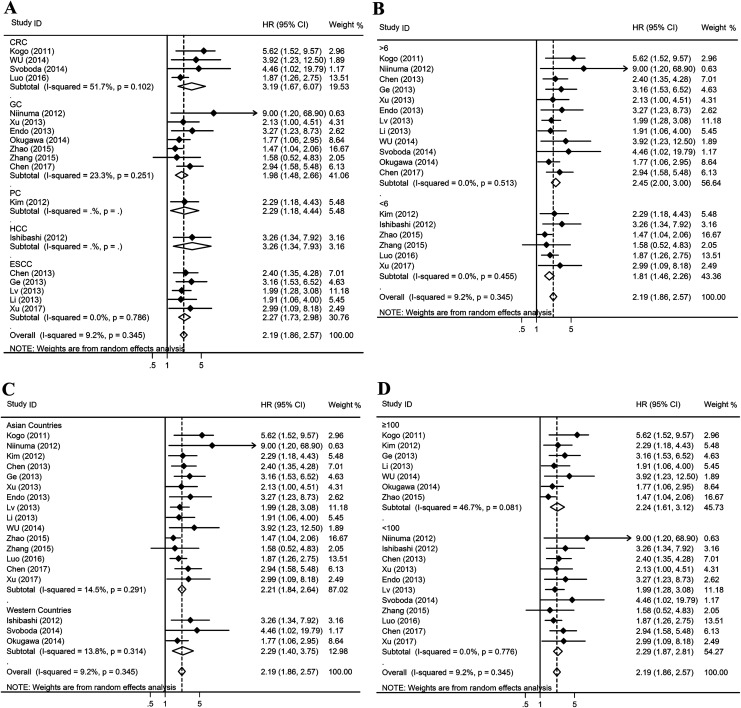
We found that overexpression of HOTAIR was significantly correlated with poor OS in patients with CRC (HR = 3.19, 95% CI, 1.67-6.07, P < .001), GC (HR = 1.98, 95% CI, 1.48-2.66, P < .001), PC (HR = 2.29, 95% CI, 1.18-4.44, P = .014), HCC (HR = 3.26, 95% CI, 1.34-7.93, P = .009), and ESCC (HR = 2.27, 95% CI, 1.73-2.98, P < .001; Figure 3A). Based on NOS, subgroup analysis indicated that there is a significant association between expression of HOTAIR and OS in studies with NOS >6 (HR = 2.45, 95% CI, 2.00-3.00, P < .001) and in studies with NOS <6 (HR = 1.81, 95% CI, 1.46-2.26, P < .001; Figure 3B). We also found that upregulation of HOTAIR expression predicted poor OS in patients with digestive cancers in Asian countries (HR = 2.21, 95% CI, 1.84-2.64, P < .001) and Western countries (HR = 2.29, 95% CI, 1.40-3.75, P = .001; Figure 3C). Besides, our results found that HOTAIR is a vital prognostic marker of OS in studies with sample size >100 (HR = 2.24, 95% CI, 1.61-3.12, P < .001) and the studies with sample size <100 (HR = 2.29, 95% CI, 1.87-2.81, P < .001; Figure 3D).
Figure 3.
Forest plots of the included studies evaluating the HRs for HOTAIR for OS. A, Subgroup analysis of HRs of OS by factor of cancer types. B, Subgroup analysis of HR s of OS by factor of NOS. C, Subgroup analysis of HR s of OS by factor of Region. D, Subgroup analysis of HR s of OS by factor of sample size. HRs indicates hazard ratios; HOTAIR, HOX transcript antisense intergenic RNA; OS, overall survival; NOS, Newcastle-Ottawa scale.
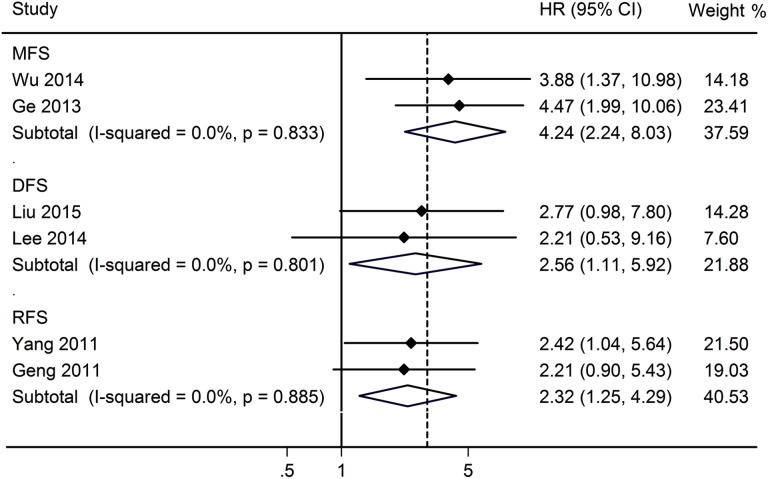
In addition, our analyses showed that there was a significant association between HOTAIR expression and MFS (HR = 4.24, 95% CI, 2.24-8.03, P < .001), RFS (HR = 2.56, 95% CI, 1.11-5.92, P < .001), and DFS (HR = 2.32, 95% CI, 1.25-4.29, P < .001; Figure 4).
Figure 4.
Forest plots of the included studies evaluating the HRs for HOTAIR for MFS, DFS, and RFS. HRs indicates hazard ratios; HOTAIR, HOX transcript antisense intergenic RNA; MFS, metastasis-free survival; DFS, disease-free survival; RFS, recurrence-free survival.
Association Between HOTAIR and Clinicopathological Characteristics of Cancers
The association between HOTAIR expression and clinicopathological characteristics was assessed in 14 studies with 5 types of digestive cancer including 1179 patients. The result revealed that differentiation grade (OR = 1.65, 95% CI, 1.02-2.65, P = .040), TNM stage (OR = 3.58, 95% CI, 2.43-5.30, P < .001), lymph node metastasis (LNM; OR = 2.52, 95% CI, 1.89-3.36, P < .001), and distant metastasis (OR = 4.20, 95% CI, 2.17-8.14, P < .001) were all positively associated with high HOTAIR expression. However, no relevance was expressed between high HOTAIR expression and age, gender, or tumor size (Table 2).
Table 2.
Correlation Between Expression of HOTAIR and Clinicopathological Characteristics of Cancers.
| Clinical Parameters | No. of Studies | No. of Patients | OR (95% CI) | P-Value | Heterogeneity | |
|---|---|---|---|---|---|---|
| I2 (%) | P Value | |||||
| Age (elderly vs young) | 11 | 1045 | 0.81 (0.62-1.05) | .113 | 0 | .915 |
| Gender (male vs female) | 14 | 1277 | 1.12 (0.88-1.43) | .346 | 0 | .857 |
| Tumor size (large vs small) | 9 | 924 | 1.08 (0.80-1.46) | .617 | 44.8 | .080 |
| Differentiation (poor vs well) | 5 | 386 | 1.65 (1.02-2.65) | .040 | 78.5 | .001 |
| TNM stage (III + IV vs I + II) | 6 | 460 | 3.58 (2.43-5.30) | <.001 | 29.2 | .205 |
| Lymph node metastasis (present vs absent) | 11 | 967 | 2.52 (1.89-3.36) | <.001 | 0.0 | .457 |
| Distant metastasis (present vs absent) | 4 | 403 | 4.20 (2.17-8.14) | <.001 | 39.1 | .177 |
Abbreviation: HOTAIR, HOX transcript antisense intergenic RNA.
Publication Bias
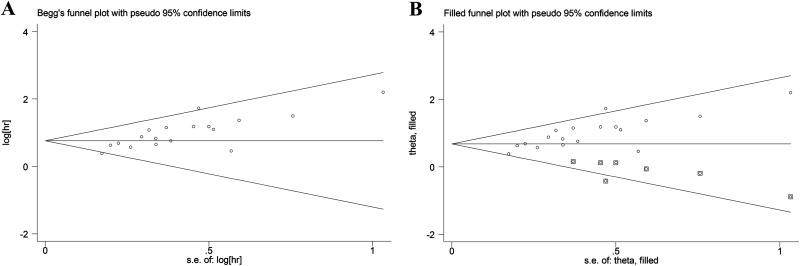
To assess the publication bias, Begg funnel plot and Egger test were performed. The results indicated that the shapes of the funnel plots are asymmetrical, and publication bias was significant by Begg test (z = 2.65, P = .008) and Egger test (t = 5.16, P < .001; Figure 5A). Then, the nonparametric “trim-and-fill” method was used to replace 7 missing studies. After the “trim-and-fill” adjustment, the estimated pooled HR was 1.97, with 95% CI being 1.64-2.37 (P < .001; Figure 5B).
Figure 5.
Publication bias in this meta-analysis. A, Funnel plots of the included studies for overall survival; B, filled funnel plot of meta-analysis using “trim-and-fill” method.
Discussion
HOTAIR is one of the most important lncRNAs that is implicated in various types of cancers. It has been demonstrated that lncRNAs involved in progression and metastasis of cancer through the mechanism of chromosome remodeling, transcription, and posttranscriptional processing. Recent study found that HOTAIR influences progression of PC by regulating the expression of miR-663b via the histone modification.43 In addition, Zhang et al suggested that HOTAIR might regulate cell cycle progression through enhancer of zeste homolog 2 (EZH2) in glioma.44 Meanwhile, HOTAIR adjusts the expression of human epidermal growth factor receptor 2 (HER2), which results in the regulation of GC progression.45 Kim et al showed that HOTAIR could promote tumor aggressiveness through the upregulation of vascular endothelial growth factor (VEGF), matrix metalloproteinase-9 (MMP-9), and epithelial-to-mesenchymal transition (EMT)-related genes in cervical cancer.46 Gao et al illuminate that HOTAIR is important in the progression and recurrence of HCC, partly through the regulation of the Wnt/β-catenin signaling pathway.47 Also, the results of many studies revealed that the activities of several kinds of regulatory factors might be affected by the genetic variants of HOTAIR. And further, it could cause the overexpression of HOTAIR, which might be one of the underlying mechanisms that affect cancer prognosis. Jin et al demonstrated that HOTAIR rs7958904 might influence cervical cancer susceptibility through modulation of cervical cancer cell proliferation and could serve as a diagnostic biomarker.48 It has been reported that the predisposition for cervical cancer might be associated with one kind of single nucleotide polymorphism (SNP) in HOTAIR (rs920778).49 Thus, it is important and necessary to combine these published data through meta-analysis and evaluate the association between expression of HOTAIR and prognosis as well as clinicopathological characteristics in patients with digestive cancer.
In this meta-analysis, 22 studies including 1844 patients were involved. It assessed the prognostic role of HOTAIR expression in digestive cancers and provided sufficient evidence for the association between HOTAIR expression and clinicopathological characteristics of digestive cancers. In 5 types of digestive cancer (CRC, GC, PC, HCC, and ESCC), overexpression of HOTTAIR could predict a poor outcome for OS. All the results of our article indicated that HOTAIR might be a hopeful prognostic biomarker for patients with digestive cancer. Besides, subgroup analysis was also used in order to investigate the association between HRs and these variables, including type of cancer, region of participants, sample size, and NOS. Then, the association between expression of HOTAIR and clinicopathological characteristics was examined. We found that abnormal expression of HOTAIR in digestive cancer was significantly correlated with clinicopathological variables including differentiation, TNM stage, lymph node metastasis, and distant metastasis. As a result, these findings indicated that HOTAIR may act as a hopeful prognostic biomarker to predict patients’ survival in different types of digestive cancers. We also tested the publication bias of studies, and the result of Begg funnel plot and Egger test showed the shapes of the funnel plots seemed asymmetrical. However, through the trim-and-fill adjustment, we believed this meta-analysis was stable.
In previous studies, Wang et al performed a meta-analysis including 13 studies and indicated that HOTAIR could be exploited as a novel prognostic biomarker for patients with digestive cancer.50 However, our article had larger sample size and updated data. Furthermore, we did a more reasonable subgroup analysis and examined the relation between high HOTAIR expression and clinicopathological variables. In 2015, Tian et al performed a meta-analysis including 8 studies and provided evidence that HOTAIR rs920778 may modify the susceptibility to certain cancer types.51 Meanwhile, this meta-analysis only included 8 studies, and the majority of the participants were Chinese, and the number of caucasian participants included in this study was relatively small. Finally, they did not examine the relation between genetic variants of HOTAIR and clinicopathological variables. Consequently, the results from our article might be more credible and valuable.
Several limitations exist in this meta-analysis. First, we should have made a specific definition of the cutoff value of HOTAIR expression level, while the researchers of all included studies did not use the same cutoff value and some of the studies even did not report it. Second, in our article, some of the studies only showed Kaplan-Meier curves and sample size and the survival rate at specified time without HRs and CIs. Hence, the HRs and CIs were only calculated by the data and figures, and it might cause some imprecision. Third, different kinds of treatment strategies might affect patients’ survival, which cause heterogeneities among studies. However, we did not perform the stratified analysis by the treatment strategies because we did not find any randomized clinical trials, or the data were not available. Fourth, the language of the involved studies was limited to English, and most of the included studies reported positive results; however, the studies that reported negative results were rare. Therefore, the unpublished articles in other languages were missed, which might cause a publication bias. Thus, our results might overstate the prognostic value of HOTAIR in digestive cancers. The above issues which were mentioned should be addressed in the further randomized controlled trial.
In conclusion, this meta-analysis indicated that there was an association between expression of HOTAIR and prognosis in patients with digestive cancer. Meanwhile, the results of our article showed that abnormal HOTAIR expression in digestive cancer was significantly correlated with clinicopathological variables including differentiation, TNM stage, lymph node metastasis, and distant metastasis. Therefore, HOTAIR could sever as a novel prognostic biomarker in patients with digestive cancers. In further studies, the relation between HOTAIR and digestive cancers and even other kinds of tumors may be identified by studies that have larger sample size and updated data.
Abbreviations
- ATG3
autophagy-related 3
- ATG7
autophagy-related 7
- CI
confidence interval
- CRC
colorectal cancer
- DFS
disease-free survival
- EMT
epithelial-to-mesenchymal transition
- ESCC
esophageal squamous cell carcinoma
- EZH2
enhancer of zeste homolog 2
- GC
gastric cancer
- HCC
hepatocellular carcinoma
- HER2
human epidermal growth factor receptor 2
- HOTAIR
HOX transcript antisense intergenic RNA
- HR
hazard ratio
- lncRNAs
long noncoding RNAs
- LNM
lymph node metastasis
- MFS
metastasis-free survival
- MMP-9
matrix metalloproteinase-9
- NOS
Newcastle-Ottawa scale
- OR
odds ratio
- OS
overall survival
- PC
pancreatic cancer
- RCT
randomized controlled trial
- RFS
recurrence-free survival
- ROC
receiver–operating characteristic curve
- VEGF
vascular endothelial growth factor.
Footnotes
Authors’ Note: Yun Zhang and Yu Zhou contributed equally to this work.
Declaration of Conflicting Interests: The author(s) declared no potential conflict of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) received no financial support for the research, authorship, and/or publication of this article.
References
- 1. Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61(2):69–90. [DOI] [PubMed] [Google Scholar]
- 2. Rullier E, Rouanet P, Tuech JJ, et al. Organ preservation for rectal cancer (GRECCAR 2): a prospective, randomised, open-label, multicentre, phase 3 trial. Lancet. 2017;390(10093):469–479. [DOI] [PubMed] [Google Scholar]
- 3. Rice TW, Ishwaran H, Hofstetter WL, et al. Recommendations for pathologic staging (pTNM) of cancer of the esophagus and esophagogastric junction for the 8th edition AJCC/UICC staging manuals. Dis Esophagus. 2016;29(8):897–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Dumonceau JM, Deprez PH, Jenssen C, et al. Indications, results, and clinical impact of endoscopic ultrasound (eus)-guided sampling in gastroenterology: European society of gastrointestinal endoscopy (esge) clinical guideline—updated January 2017. Endoscopy. 2017;49(7):695–714. [DOI] [PubMed] [Google Scholar]
- 5. Bartley AN, Washington MK, Colasacco C, et al. HER2 testing and clinical decision making in gastroesophageal adenocarcinoma: guideline from the college of American pathologists, American society for clinical pathology, and the American society of clinical oncology. J Clin Oncol. 2017;35(4):446–464. [DOI] [PubMed] [Google Scholar]
- 6. Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell. 2011;43(6):904–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wilusz JE, Sunwoo H, Spector DL. Long noncoding RNAs: functional surprises from the RNA world. Genes Dev. 2009;23(13):1494–1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Malone CD, Hannon GJ. Small RNAs as guardians of the genome. Cell. 2009;136(4):656–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Xu ZY, Yu QM, Du YA, et al. Knockdown of long non-coding RNA HOTAIR suppresses tumor invasion and reverses epithelial-mesenchymal transition in gastric cancer. Int J Biol Sci. 2013;9(6):587–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Yao W, Bai Y, Li Y, et al. Upregulation of MALAT-1 and its association with survival rate and the effect on cell cycle and migration in patients with esophageal squamous cell carcinoma. Tumour Biol. 2016;37(4):4305–4312. [DOI] [PubMed] [Google Scholar]
- 11. Jiang H, Wang Y, Ai M, et al. Long noncoding RNA CRNDE stabilized by hnRNPUL2 accelerates cell proliferation and migration in colorectal carcinoma via activating Ras/MAPK signaling pathways. Cell Death Dis. 2017;8(6):e2862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zheng Y, Song D, Xiao K, et al. LncRNA GAS5 contributes to lymphatic metastasis in colorectal cancer. Oncotarget. 2016;7(50):83727–83734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Rinn JL, Kertesz M, Wang JK, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129(7):1311–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Shi X, Sun M, Liu H, Yao Y, Song Y. Long non-coding RNAs: a new frontier in the study of human diseases. Cancer Lett. 2013;339(2):159–166. [DOI] [PubMed] [Google Scholar]
- 15. Gupta RA, Shah N, Wang KC, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464(7291):1071–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sorensen KP, Thomassen M, Tan Q, et al. Long non-coding RNA HOTAIR is an independent prognostic marker of metastasis in estrogen receptor-positive primary breast cancer. Breast Cancer Res Treat. 2013;142(3):529–536. [DOI] [PubMed] [Google Scholar]
- 17. Lee M, Kim HJ, Kim SW, et al. The long non-coding RNA HOTAIR increases tumour growth and invasion in cervical cancer by targeting the Notch pathway. Oncotarget. 2016;7(28):44558–44571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wu ZH, Wang XL, Tang HM, et al. Long non-coding RNA HOTAIR is a powerful predictor of metastasis and poor prognosis and is associated with epithelial-mesenchymal transition in colon cancer. Oncol Rep. 2014;32(1):395–402. [DOI] [PubMed] [Google Scholar]
- 19. Svoboda M, Slyskova J, Schneiderova M, et al. HOTAIR long non-coding RNA is a negative prognostic factor not only in primary tumors, but also in the blood of colorectal cancer patients. Carcinogenesis. 2014;35(7):1510–1515. [DOI] [PubMed] [Google Scholar]
- 20. Luo ZF, Zhao D, Li XQ, et al. Clinical significance of HOTAIR expression in colon cancer. World J Gastroenterol. 2016;22(22):5254–5259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kogo R, Shimamura T, Mimori K, et al. Long noncoding RNA HOTAIR regulates polycomb-dependent chromatin modification and is associated with poor prognosis in colorectal cancers. Cancer Res. 2011;71(20):6320–6326. [DOI] [PubMed] [Google Scholar]
- 22. Zhao W, Dong S, Duan B, et al. HOTAIR is a predictive and prognostic biomarker for patients with advanced gastric adenocarcinoma receiving fluorouracil and platinum combination chemotherapy. Am J Transl Res. 2015;7(7):1295–1302. [PMC free article] [PubMed] [Google Scholar]
- 23. Zhang ZZ, Shen ZY, Shen YY, et al. HOTAIR long noncoding RNA promotes gastric cancer metastasis through suppression of poly r(c)-binding protein (PCBP) 1. Mol Cancer Ther. 2015;14(5):1162–1170. [DOI] [PubMed] [Google Scholar]
- 24. Okugawa Y, Toiyama Y, Hur K, et al. Metastasis-associated long non-coding RNA drives gastric cancer development and promotes peritoneal metastasis. Carcinogenesis. 2014;35(12):2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Niinuma T, Suzuki H, Nojima M, et al. Upregulation of miR-196a and HOTAIR drive malignant character in gastrointestinal stromal tumors. Cancer Res. 2012;72(5): 1126–1136. [DOI] [PubMed] [Google Scholar]
- 26. Liu YW, Sun M, Xia R, et al. LincHOTAIR epigenetically silences miR34a by binding to PRC2 to promote the epithelial-to-mesenchymal transition in human gastric cancer. Cell Death Dis. 2015;6:e1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lee NK, Lee JH, Park CH, et al. Long non-coding RNA HOTAIR promotes carcinogenesis and invasion of gastric adenocarcinoma. Biochem Biophys Res Commun. 2014;451(2):171–178. [DOI] [PubMed] [Google Scholar]
- 28. Endo H, Shiroki T, Nakagawa T, et al. Enhanced expression of long non-coding RNA HOTAIR is associated with the development of gastric cancer. PLos One. 2013;8(10):e77070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Chen WM, Chen WD, Jiang XM, et al. HOX transcript antisense intergenic RNA represses E-cadherin expression by binding to EZH2 in gastric cancer. World J Gastroenterol. 2017;23(33):6100–6110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kim K, Jutooru I, Chadalapaka G, et al. HOTAIR is a negative prognostic factor and exhibits pro-oncogenic activity in pancreatic cancer. Oncogene. 2013;32(13):1616–1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yang Z, Zhou L, Wu LM, et al. Overexpression of long non-coding RNA HOTAIR predicts tumor recurrence in hepatocellular carcinoma patients following liver transplantation. Ann Surg Oncol. 2011;18(5):1243–1250. [DOI] [PubMed] [Google Scholar]
- 32. Ishibashi M, Kogo R, Shibata K, et al. Clinical significance of the expression of long non-coding RNA HOTAIR in primary hepatocellular carcinoma. Oncol Rep. 2013;29(3): 946–950. [DOI] [PubMed] [Google Scholar]
- 33. Geng YJ, Xie SL, Li Q, Ma J, Wang GY. Large intervening non-coding RNA HOTAIR is associated with hepatocellular carcinoma progression. J Int Med Res. 2011;39(6): 2119–2128. [DOI] [PubMed] [Google Scholar]
- 34. Lv XB, Lian GY, Wang HR, Song E, Yao H, Wang MH. Long noncoding RNA HOTAIR is a prognostic marker for esophageal squamous cell carcinoma progression and survival. PLoS One. 2013;8(5):e63516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Li X, Wu Z, Mei Q, et al. Long non-coding RNA HOTAIR, a driver of malignancy, predicts negative prognosis and exhibits oncogenic activity in oesophageal squamous cell carcinoma. Br J Cancer. 2013;109(8):2266–2278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ge XS, Ma HJ, Zheng XH, et al. HOTAIR, a prognostic factor in esophageal squamous cell carcinoma, inhibits WIF-1 expression and activates Wnt pathway. Cancer Sci. 2013;104(12):1675–1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Chen FJ, Sun M, Li SQ, et al. Upregulation of the long non-coding RNA HOTAIR promotes esophageal squamous cell carcinoma metastasis and poor prognosis. Mol Carcinog. 2013;52(11):908–915. [DOI] [PubMed] [Google Scholar]
- 38. Xu F, Zhang J. Long non-coding RNA HOTAIR functions as miRNA sponge to promote the epithelial to mesenchymal transition in esophageal cancer. Biomed Pharmacother. 2017;90:888–896. [DOI] [PubMed] [Google Scholar]
- 39. Yang L, Zhang X, Li H, Liu J. The long noncoding RNA HOTAIR activates autophagy by upregulating ATG3 and ATG7 in hepatocellular carcinoma. Mol BioSyst. 2016;12(8):2605–2612. [DOI] [PubMed] [Google Scholar]
- 40. Oremus M, Oremus C, Hall GB, McKinnon MC; Ect and Cognition Systematic Review T. Inter-rater and test-retest reliability of quality assessments by novice student raters using the jadad and newcastle-ottawa scales. BMJ Open. 2012;2(4): pii: e001368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Tierney JF, Stewart LA, Ghersi D, Burdett S, Sydes MR. Practical methods for incorporating summary time-to-event data into meta-analysis. Trials. 2007;8:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. McShane LM, Altman DG, Sauerbrei W, et al. REporting recommendations for tumor MARKer prognostic studies (REMARK). Breast Cancer Res Treat. 2006;100(2):229–235. [DOI] [PubMed] [Google Scholar]
- 43. Cai H, An Y, Chen X, et al. Epigenetic inhibition of miR-663b by long non-coding RNA HOTAIR promotes pancreatic cancer cell proliferation via up-regulation of insulin-like growth factor 2. Oncotarget. 2016;7(52):86857–86870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zhang K, Sun X, Zhou X, et al. Long non-coding RNA HOTAIR promotes glioblastoma cell cycle progression in an EZH2 dependent manner. Oncotarget. 2015;6(1):537–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Liu XH, Sun M, Nie FQ, et al. Lnc RNA HOTAIR functions as a competing endogenous RNA to regulate HER2 expression by sponging miR-331-3p in gastric cancer. Mol Cancer. 2014;13:92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kim HJ, Lee DW, Yim GW, et al. Long non-coding RNA HOTAIR is associated with human cervical cancer progression. Int J Oncol. 2015;46(2):521–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Gao JZ, Li J, Du JL, Li XL. Long non-coding RNA HOTAIR is a marker for hepatocellular carcinoma progression and tumor recurrence. Oncol Lett. 2016;11(3):1791–1798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Jin H, Lu X, Ni J, et al. HOTAIR rs7958904 polymorphism is associated with increased cervical cancer risk in a Chinese population. Sci Rep. 2017;7(1):3144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Guo L, Lu X, Zheng L, Liu X, Hu M. Association of long non-coding RNA HOTAIR polymorphisms with cervical cancer risk in a Chinese population. PLoS One. 2016;11(7):e0160039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wang S, Wang Z. Prognostic value of long noncoding RNA HOTAIR in digestive system malignancies. J Gastroenterol Hepatol. 2015;30(7):1123–1133. [DOI] [PubMed] [Google Scholar]
- 51. Tian T, Li C, Xiao J, et al. Quantitative assessment of the polymorphisms in the HOTAIR lncRNA and cancer risk: a meta-analysis of 8 case-control studies. PLoS One. 2016;11(3):e0152296. [DOI] [PMC free article] [PubMed] [Google Scholar]