Abstract
Sarcomas, and the mesenchymal precursor cells from which they arise, express chondroitin sulfate proteoglycan 4 (NG2/CSPG4). However, NG2/CSPG4's function and its capacity to serve as a therapeutic target in this tumor type are unknown. Here, we used cells from human tumors and a genetically engineered autochthonous mouse model of soft-tissue sarcomas (STSs) to determine NG2/CSPG4's role in STS initiation and growth. Inhibiting NG2/CSPG4 expression in established murine and human STSs decreased tumor volume by almost two-thirds and cell proliferation rate by 50%. NG2/CSPG4 antibody immunotherapy in human sarcomas established as xenografts in mice similarly decreased tumor volume, and expression of a lentivirus blocking NG2/CSPG4 expression inhibited tumor cell proliferation and increased the latency of engraftment. Gene profiling showed that Ng2/Cspg4 deletion altered the expression of genes regulating cell proliferation and apoptosis. Surprisingly, Ng2/Cspg4 deletion at the time of tumor initiation resulted in larger tumors. Gene expression profiling indicated substantial down-regulation of insulin-like growth factor binding protein (Igfbp) genes when Ng2/Cspg4 is depleted at tumor initiation, but not when Ng2/Cspg4 is depleted after tumor initiation. Such differences may have clinical significance, as therapeutic targeting of a signaling pathway such as NG2/CSPG4 may have different effects on cell behavior with tumor progression. NG2/CSPG4 depletion has divergent effects, depending on the developmental stage of sarcoma. In established tumors, IGF signaling is active, and NG2 inhibition targets cell proliferation and apoptosis.
Keywords: cell proliferation, chondroitin sulfate, gene expression, insulin-like growth factor (IGF), osteosarcoma, transgenic mice
Introduction
Soft-tissue sarcomas are malignant tumors arising in mesenchymal derived tissues that are classified into subtypes primarily based on histology (1). Undifferentiated pleomorphic sarcoma is the most common and aggressive subtype in adults, with a roughly 50% 5-year survival rate (1–4). This sarcoma type displays a highly heterogeneous and undifferentiated histological phenotype, which suggests that it may represent an end point for other sarcomas that have undergone dedifferentiation from a cell initially of mesenchymal origin (5, 6).
Chondroitin sulfate proteoglycan (CSPG4), also called neural-glia 2 (NG2) is a membrane protein expressed by progenitor mesenchymal cells (e.g. pericytes), immature keratinocytes, melanocytes, and cells in several tumor types (7). As a gene expressed by mesenchymal progenitors, its expression could play a role in sarcoma initiation. It is a transmembrane protein that can potentiate the activities of other signaling-transducing systems, such as integrin and MAPK signaling pathways (8–10). NG2/CSPG4 can bind to and present growth factors (e.g. basic fibroblast growth factor and platelet-derived growth factor) to their cognate receptor tyrosine kinase receptors (11, 12). In human glioblastoma cells, NG2/CSPG4-mediated activation of integrin signaling promotes cell survival through sustained activation of Akt (protein kinase B) (13, 14) and chemoresistance through integrin-dependent PI3K/Akt signaling (8). In human melanomas, NG2/CSPG4 functions to activate the MEK/ERK1/2 pathway by mediating the growth factor-induced activation of receptor tyrosine kinases (15, 16). NG2/CSPG4 can interact with collagen VI, and this NG2/CSPG4-Col VI interplay may regulate interaction between soft-tissue sarcoma cells and the tumor microenvironment (17). Interestingly, driving oncogenic mutations in Ng2/Cspg4-expressing cells results in the formation of sarcomas (18).
Altered NG2/CSPG4 expression and/or distribution may serve as a prognostic factor in various cancer types (19–23). In soft-tissue sarcomas, NG2/CSPG4 expression is correlated with tumor progression (24, 25). Inhibition of NG2/CSPG4 expression or treatment with anti-NG2/CSPG4 antibodies inhibits tumor growth in xenografts from some malignancies (26–28). However, the efficacy of targeted NG2/CSPG4 therapy has not been investigated in sarcomas. Here, we use genetically modified mice, human tumors established as xenografts in mice, and an NG2/CSPG4 antibody-based therapy to study the role of Ng2/Cspg4 in soft-tissue sarcoma initiation and growth in vivo.
Results
Deletion of Ng2/Cspg4 in soft-tissue sarcoma reduces tumor size and cell proliferation
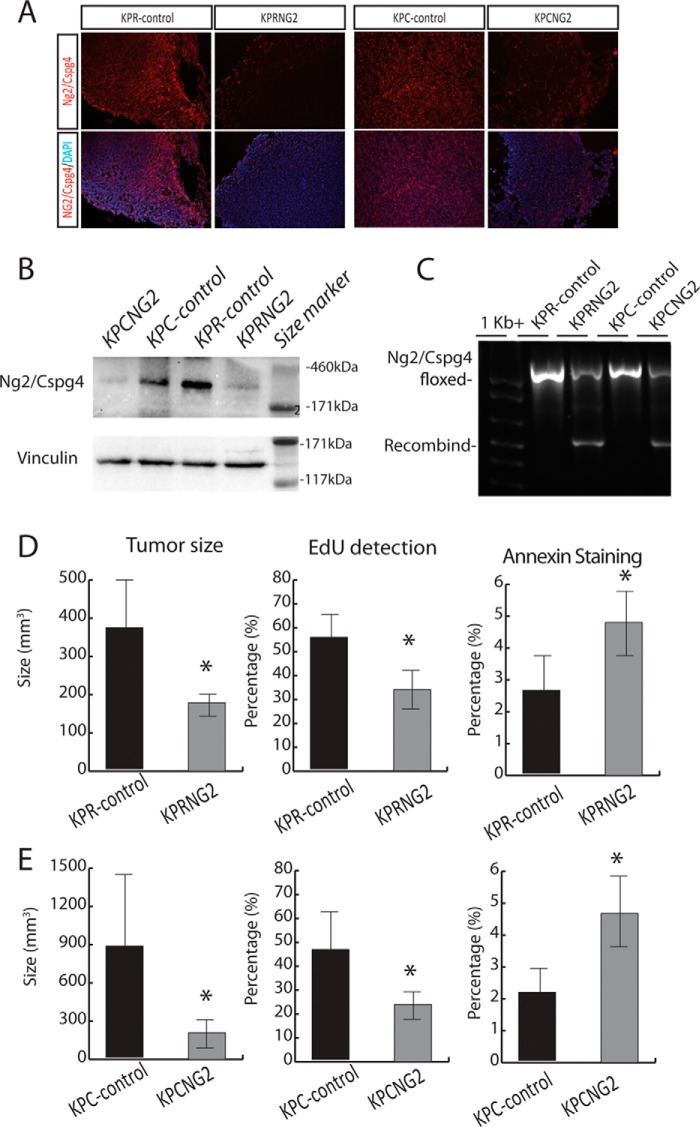
We used an autochthonous mouse model in which undifferentiated soft-tissue sarcoma formation is initiated by deletion of both p53 alleles and expression of an oncogenic mutant Kras driven by either Flp recombinase or Cre recombinase (KP mouse). Previous studies show that when the conditional alleles are activated by local injection of a virus driving expression of Flp or Cre in muscle, this results in tumors with characteristics similar to those of human undifferentiated pleomorphic sarcomas (18, 29, 30). To determine the role Ng2/Cspg4 plays in sarcoma tumor growth and maintenance, we employed a dual recombinase system by crossing Ng2/Cspg4f/f mice with KrasFRT-STOP-FRT-G12D/+; p53FRT/FRT; Rosa26Cre-ER-T2/+ mice to generate KrasFRT-STOP-FRT-G12D/+; p53FRT/FRT; Rosa26Cre-ER-T2/+; Ng2/Cspg4f/f mice (KPRNG2). Primary sarcomas were generated in the hind limbs of these mice by intramuscular injection of adeno-FlpO. After the initial tumor was palpated, tamoxifen (200 μg/g) was delivered via intraperitoneal injection. Tumors from KrasFRT-STOP-FRT-G12D/+; p53FRT/FRT; Rosa26+/+; Ng2/Cspg4f/f (KPR-control) and KrasFRT-STOP-FRT-G12D/+; p53FRT/FRT; Rosa26Cre-ER-T2/+; Ng2/Cspg4f/f mice were collected 12 days after tumor formation, and real-time PCR, immunofluorescence, and Western analysis (Fig. 1 (A and B) and Fig. S1) were used to confirm the deletion of Ng2/Cspg4 and its protein product. Immunofluorescence showed a 65% reduction in the proportion of cells expressing NG2/CSPG4 in KPCNG2 mice and an 80% reduction in KPRNG2 mice. Western analysis showed a relative NG2/CSPG4 protein level of 14% compared with controls in tumors from KPCNG2 mice and 8% compared with controls in tumors from KPRNG2 mice (relative densities are compared using Student's t test, n = 5 in each group, p < 0.01). We also confirmed the recombination at the Ng2/Cspg4 locus in the KrasFRT-STOP-FRT-G12D/+; p53FRT/FRT; Rosa26Cre-ER-T2/+; Ng2/Cspg4f/f tumors by PCR analysis of genomic DNA (Fig. 1C). Deletion of Ng2/Cspg4 in established tumors (KrasFRT-STOP-FRT-G12D/+; p53FRT/FRT; Rosa26Cre-ER-T2/+; Ng2/Cspg4f/f) resulted in a significant reduction in tumor size when compared with the KrasFRT-STOP-FRT-G12D/+; p53FRT/FRT; Rosa26+/+; Ng2/Cspg4f/f tumors (Fig. 1D).
Figure 1.
Ng2/Cspg4 regulates soft-tissue sarcoma tumor growth. A, immunofluorescence staining for Ng2/Cspg4 in KPR-control, KPRNG2, KPC-control, and KPCNG2 tumor samples showing >80% deletion of Ng2/Cspg4 in KPRNG2 tumors and ∼65% deletion of Ng2/Cspg4 in KPCNG2 tumors. B, Ng2/Cspg4 protein levels in KPR-control, KPRNG2, KPC-control, and KPCNG2 tumors were assessed by Western analysis, and vinculin was used as loading control. C, genotyping of tumors derived from KPR-control, KPRNG2, KPC-control, and KPCNG2 tumors showing recombination at the Ng2/Cspg4 locus in KPRNG2 and KPCNG2 tumors. A representative blot is shown. D and E, tumor size and proliferation was significantly decreased, and apoptosis increased in KPRNG2 and KPCNG2 tumors compared with KPR-control and KPC-control tumors, respectively. Data are shown with 95% confidence intervals indicated (error bars). n = 14 in the KPRNG2 and control groups and 15 in the KPCNG2 and control group. *, p < 0.05. The percentage of EdU-positive cells was found in KPRNG2 tumors compared with KPR-control tumors (n = 6 in each group), and percentage of annexin V–stained cells (n + 6 in each group). Data are shown as means with 95% confidence intervals indicated. *, p < 0.05.
Because the Rosa26Cre-ER-T2/+ allele is expressed in tumor cells and non-tumor cells, the results using the Rosa26Cre-ER-T2/+ mice may be due to a non-sarcoma cell effect of NG2/CSPG4 regulating the niche to alter tumor behavior. Therefore, we generated mice in which Ng2/Cspg4 would be deleted only in the tumor cells. To achieve this, we crossed Ng2/Cspg4f/f mice with KrasFRT-STOP-FRT-G12D; p53FRT/FRT; Col1a1FRT-STOP-FRT-Cre-ER-T2/+ mice to generate KrasFRT-STOP-FRT-G12D; p53FRT/FRT; Col1a1FRT-STOP-FRT-Cre-ER-T2/+; Ng2/Cspg4f/f (KPCNG2) mice. In KrasFRT-STOP-FRT-G12D; p53FRT/FRT; Col1a1FRT-STOP-FRT-Cre-ER-T2 mice, in which Cre-ERT2 is downstream from a FRT-STOP-FRT cassette, cells will only express Cre-ERT2 and have the capacity for tamoxifen-mediated recombination of loxP sites after FlpO-mediated removal of the STOP cassette. Therefore, we utilized Col1a1FRT-STOP-FRT-Cre-ER-T2/+ mice for sequential mutagenesis restricted to the sarcoma cells to investigate the role of tumor-specific Ng2/Cspg4 in tumor maintenance. Sarcomas were generated in the hind limbs of these mice by intramuscular injection of adeno-FlpO. After the initial tumor was palpated, a single dose of 0.75 mg of 4-hydroxytomaxifen (4-OHT)2 in DMSO was delivered via intratumoral injection. Tumors were collected 12 days after the first day of tumor detection. Because complex genetic mice do not always exhibit the expected degree of recombination, we confirmed that Cre-ERT2 was expressed in sarcomas, but not control tissues, using real-time PCR. We then investigated the degree of recombination at the Ng2/Cspg4 locus in the KrasFRT-STOP-FRT-G12D; p53FRT/FRT; Col1a1FRT-STOP-FRT-Cre-ER-T2/+; Ng2/Cspg4f/f tumors by PCR analysis of genomic DNA (Fig. 1C) and used immunofluorescence and Western analysis to assess deletion at the protein level (Fig. 1B). Tumors from KrasFRT-STOP-FRT-G12D; p53FRT/FRT; Col1a1FRT-STOP-FRT-Cre-ER-T2/+; Ng2/Cspg4f/f mice showed partial deletion of Ng2/Cspg4 expression compared with KrasFRT-STOP-FRT-G12D/+; p53FRT/FRT; Rosa26Cre-ER-T2/+; Ng2/Cspg4f/f tumors. This degree of deletion resulted in a significant reduction in tumor size when compared with tumors from KrasFRT-STOP-FRT-G12D; p53FRT/FRT; Col1a1+/+; Ng2/Cspg4f/f mice (Fig. 1E). Thus, deletion of Ng2/Cspg4 in the neoplastic cells themselves reduced the soft-tissue sarcoma tumor size.
Cell proliferation was analyzed by culturing cells derived from tumors with medium containing 5-ethynyl-2′-deoxyuridine (Edu) for 4 h to label cells in S phase. Tumor cells derived from both sets of mice in which Ng2/Cspg4 were deleted (KPRNG2 and KPCNG2) and their respective control tumors, in which Ng2/Cspg4 was expressed, were studied. There was a lower percentage of Edu-positive cells in tumors lacking Ng2/Cspg4 expression than in controls in both genotypes (Fig. 1, D and E), showing that Ng2/Cspg4 positively regulates tumor cell proliferation. Apoptosis was determined using annexin V staining, as detected using flow cytometry. There was an increase the proportion of annexin V staining in the absence of Ng2/Cspg4 expression (Fig. 1, D and E) in both genotypes as well. Thus, Ng2/Cspg4 regulates both cell proliferation and apoptosis in soft-tissue sarcomas.
Knockdown of Ng2/Cspg4 delays xenograft growth
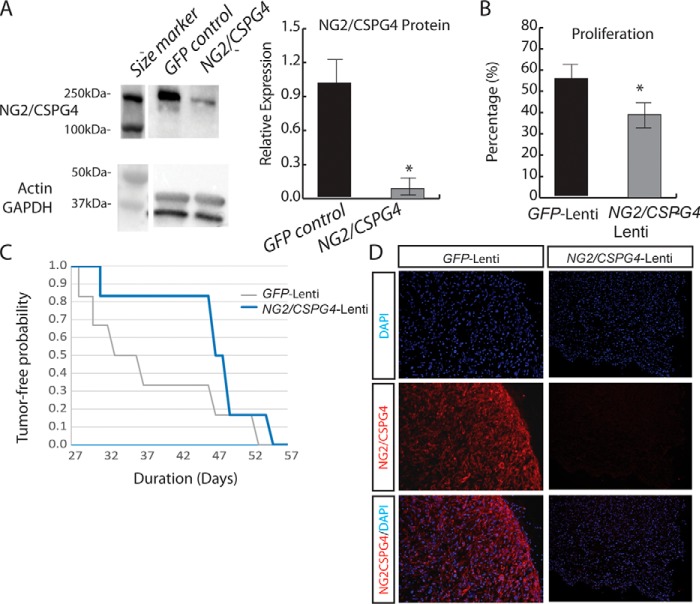
To determine whether NG2/CSPG4 in human soft-tissue tumors plays a similar role, we knocked down NG2/CSPG4 expression in xenografted human tumor cells using a lentivirus expressing shRNA to NG2/CSPG4. Infection of human primary undifferentiated pleomorphic sarcoma cells with a lentivirus expressing shRNA to NG2/CSPG4 resulted in substantial down-regulation of Ng2/CSPG4 expression, as compared with cells transfected with a control GFP-lentivirus (Fig. 2A). In cell cultures, there was a decrease in cell proliferation, as measured by the percentage of EdU-positive cells in NG2/CSPG4-lentivirus–infected cells, compared with that of the same cells transfected with a control lentivirus (Fig. 2B). Lentivirus-infected human undifferentiated pleomorphic sarcoma cells were established as xenografts in NSG mice at serial dilutions ranging from 1 × 105 to 1 × 102 cells/injection. Within this range of number of cells injected, knockdown of NG2/CSPG4 did not affect the tumor-initiating potential of the sarcoma cells (Table S1). However, we found that cells infected with the lentivirus expressing shRNA to NG2/CSPG4 lentivirus showed a delay in engraftment at all four dilutions tested (Fig. 2C). Histological analyses of explanted tumors in which NG2/CSPG4 was knocked down showed an indistinguishable morphology from controls (Fig. S2). To verify NG2/CSPG4 knockdown in the xenografts, an antibody specific for human NG2/CSPG4 was used for immunofluorescent staining analysis in xenograft tumors derived from NG2/CSPG4-lentivirus–infected cells and GFP-lentivirus–infected cells. Xenografts of human tumor cells that had been infected with NG2/CSPG4-lentivirus showed much less NG2/CSPG4 expression (Fig. 2D). Because of the difference in time to engraftment, at any given time, the tumors derived from GFP-lentivirus–infected cells were smaller in size. However, the cause for this change in tumor size cannot be distinguished between a delay in engraftment and changes in cell proliferation, as the lentiviral construct was not inducible and, as such, could not be activated after engraftment.
Figure 2.
NG2/CSPG4 regulates human soft-tissue sarcoma cell proliferation and tumor engraftment. A, protein levels of NG2/CSPG4 in GFP-lentivirus or NG2/CSPG4-lentivirus–infected human soft-tissue sarcoma cells were assessed by Western analysis. Actin and GAPDH were used as loading controls. A significant reduction in the level of NG2/CSPG4 expression was found in cells infected with NG2/CSPG4-lentivirus compared with the GFP-lentivirus–infected control cells. n = 6 in each group. The Western image is from a single blot with additional lanes not relevant to this figure replaced by a space in this image. B, knockdown of NG2/CSPG4 resulted in a decrease in the percentage of EdU-positive cells. n = 6 in each group. Data are shown as means with 95% confidence intervals indicated (error bars). *, p < 0.05. C, representative Kaplan–Meier curve demonstrated delayed tumor engraftments with cells (1 × 104 cells) infected with lentivirus expressing siRNA to NG2/CSPG4 when compared with the ones with GFP-lentivirus–infected control cells. n = 15 in each group. D, immunofluorescence staining of human NG2/CSPG4 in xenografted tumors showing 90% deletion of NG2/CSPG4 in NG2/CSPG4-lentivirus–infected tumors compared with GFP-lentivirus–infected tumors.
NG2/CSPG4 antibody treatment inhibits the growth of human undifferentiated pleomorphic sarcoma xenografts in SCID mice
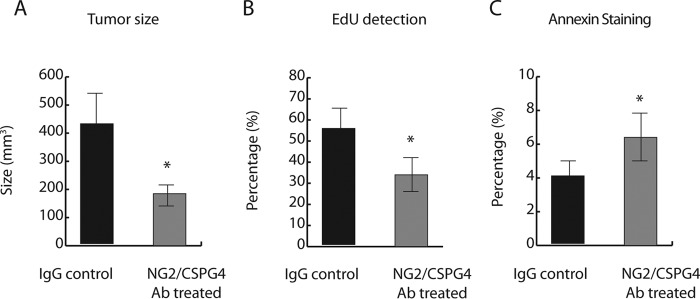
NG2/CSPG4 has been used as an immunotherapy target in xenografts in melanoma, triple-negative breast cancer, and malignant mesothelioma (27, 59, 60). We thus explored targeting NG2/CSPG4 protein with mAb-based immunotherapy in soft-tissue sarcomas. Human undifferentiated pleomorphic sarcomas xenografted into NSG mice were treated with 50 μg/ml/mouse of either mAb 9.2.27 (Abcam) or isotype control IgG every other day for 2 weeks via intraperitoneal injection. Treatment with mAb 9.2.27 significantly reduced the average tumor volume as compared with IgG controls (Fig. 3A). No signs of toxicity were detected in mice treated with mAb 9.2.27. In addition, treatment with mAb 9.2.27 resulted in a decrease in cell proliferation and increase in cell apoptosis (Fig. 3, B and C). Data here suggest that NG2/CSPG4 mAb–based immunotherapy could be developed into an approach for the treatment of NG2/CSPG4-expressing soft-tissue sarcomas.
Figure 3.
NG2/CSPG4-antibody immunotherapy inhibits tumor growth in human undifferentiated pleomorphic sarcoma xenografts. A, NG2/CSPG4-antibody treatments resulted in a decrease in tumor weight when compared with tumors treated with IgG; n = 10 in each group. B, antibody treatment decreased tumor cell proliferation; n = 6 in each group. C, antibody treatment increased tumor cell apoptosis; n = 6 in each group. Data are shown as means with 95% confidence intervals indicated (error bars). *, p < 0.05.
Deletion of Ng2/Cspg4 at the time of tumor initiation resulted in an increase in tumor size
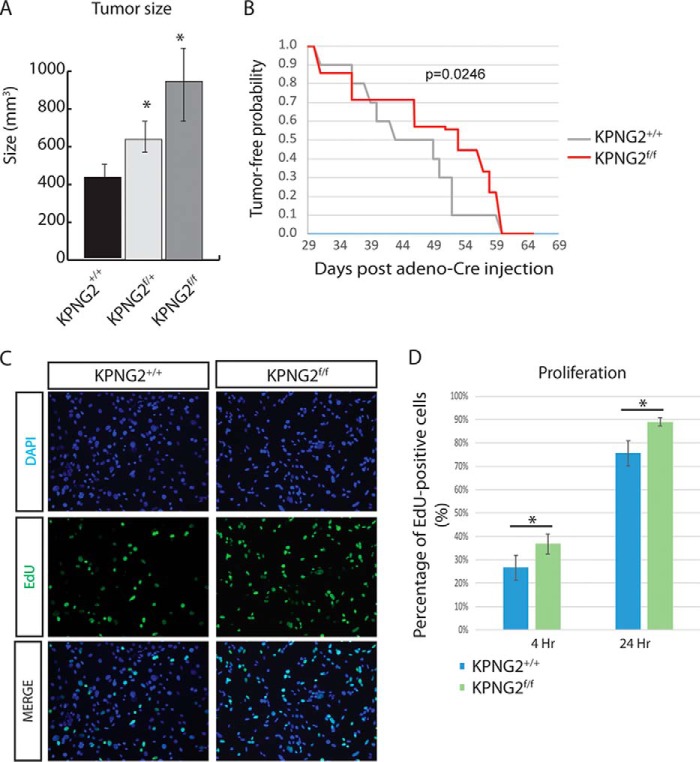
Lentiviral knockdown of NG2/CSPG4 expression in human tumors established as xenografts delayed engraftment. Because Ng2/Cspg4 is expressed in mesenchymal precursor cells and could play a role in regulating stem cell-like properties, and driving oncogenic mutations in Ng2/Cspg4 expressing cells results in sarcoma formation (18), we investigated the role of Ng2/Cspg4 in tumor initiation in the mouse. Ng2/Cspg4f/f mice (32) were crossed with KrasG12D/+; p53f/f mice to generate - KrasG12D; p53f/f; Ng2/Cspg4f/f (KPNG2f/f) mice. Primary tumors were induced with adenovirus expressing Cre-recombinases and harvested 12 days after the initial detection of the tumors. Deletion efficiency was confirmed using real-time PCR, Western blot analysis, and immunofluorescence staining (Fig. S3). Nine of 11 of the KrasG12D/+; p53f/f; Ng2/Cspg4f/f mice developed tumors after the adeno-Cre injection. Whereas this proportion is lower than observed in tumors expressing wildtype Ng2/Cspg4, because these sarcomas lacked Ng2/Cspg4 expression, we know that Ng2/Cspg4 is not required for tumor initiation.
Interestingly, tumors from the KrasG12D/+; p53f/f; Ng2/Cspg4f/f mice were significantly larger than the ones from either KrasG12D/+; p53f/f; Ng2/Cspg4+/+ or KrasG12D/+; p53f/f; Ng2/Cspg4f/+ mice (Fig. 4A), and they formed at a similar time following injection as tumors in control mice expressing Ng2/Cspg4 (Fig. 4B). When cells derived from these sarcomas were cultured in Edu-containing medium, there was increased proliferation in the KrasG12D/+; p53f/f; Ng2/Cspg4f/f tumor cells compared with KrasG12D/+; p53f/f; Ng2/Cspg4+/+ cells (Fig. 4, C and D) as detected by EdU staining.
Figure 4.
Deletion of Ng2/Cspg4 at the time of tumor initiation increases tumor size. A, tumor size was significantly larger in KPNG2f/f tumors compared with KPNG2f/+ or KPNG2+/+ tumors. Data are shown as means with 95% confidence intervals (KPNG2+/+, n = 8; KPNG2f/+, n = 17; KPNG2f/f, n = 9. B, KPNG2f/f mice showed a delay in tumor formation compared with KPNG2+/+ animals. p = 0.0246. C, representative images of immunofluorescence staining of EdU showing increased percentage of Edu-positive cells (green channel) in the KPNG2f/f tumor cells compared with KPNG2+/+ tumor cells after 4 h of culturing with medium containing 10 μm Edu. Nuclei were labeled with 4′,6-diamidino-2-phenylindole (DAPI) staining (blue channel). D, percentage of EdU-positive cells were analyzed after 4 and 24 h of culturing with EdU-containing medium. Increased percentages of EdU-positive cells were found at both time points in the KPNG2f/f cells compared with KPNG2+/+ cells. Data are shown as means with 95% confidence intervals indicated (error bars). *, p < 0.05.
Ng2/Cspg4 has divergent effects on gene expression, depending on the developmental stage of the tumor when it is depleted
To investigate an explanation behind the divergent effect of Ng2/Cspg4 deletion on tumor behavior at different stages, we compared RNA expression profiles between tumors expressing Ng2/Cspg4 and those in which Ng2/Cspg4 was deleted either at the time of tumor initiation or after a tumor formed. RNA sequencing was compared between tumors from KrasG12D/+; p53f/f; Ng/Cspg4+/+, KrasG12D/+; p53f/f; Ng/Cspg4f/f, KrasFRT-STOP-FRT-G12D/+; p53FRT/FRT; Rosa26+/+; Ng2/Cspg4f/f and KrasFRT-STOP-FRT-G12D/+; p53FRT/FRT; Rosa26Cre-ER-T2/+; Ng2/Cspg4f/f mice. The data have been deposited in the GEO database (accession number GSE97489).
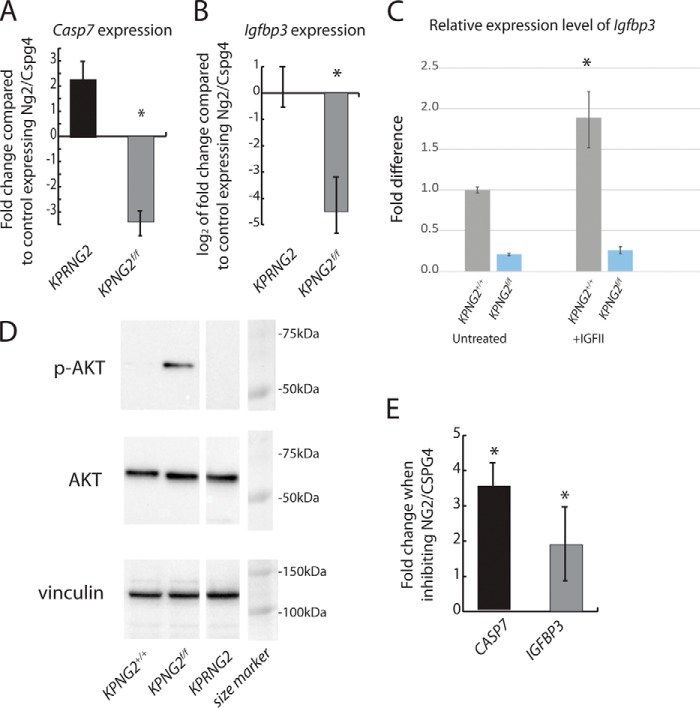
When Ng2/Cspg4 is depleted after a tumor has formed, there is increased expression of genes implicated in the regulation of apoptosis (e.g. Casp7), consistent with our findings of increased annexin V staining in these lesions. This was confirmed by real-time PCR (Table S2 and Fig. 5A). When Ng2/Cspg4 is depleted at the time of tumor initiation, many of these genes are down-regulated (Table S3 and Fig. 5A). Interestingly, there was a significant down-regulation of several Igfbp genes in the KrasG12D; p53f/f; Ng2/Cspg4f/f tumors when compared with either KrasG12D; p53f/f; Ng2/Cspg4+/+ or KrasFRT-STOP-FRT-G12D/+; p53FRT/FRT; Rosa26Cre-ER-T2/+; Ng2/Cspg4f/f tumors (Tables S3 and S4). The down-regulation of Igfbp3 was validated using quantitative real-time PCR (Fig. 5B).
Figure 5.
Divergent effect of Ng2/Cspg4 deletion on gene expression, depending on the developmental stage of the tumor when it is depleted. A, -fold change of Casp7 expression, showing an increase when Ng2/Cspg4 is deleted after the tumor forms but a decrease when deleted at the time of tumor initiation compared with control tumors expressing Ng2/Cspg4. n = 6 for each condition. B, -fold change of Igfbp3 expression, showing a decrease when Ng2/Cspg4 is deleted after the tumor forms but an increase when deleted at the time of tumor initiation compared with control tumors expressing Ng2/Cspg4. n = 6 for each condition. Expression is reported as log2. C, KPNG2+/+ and KPNG2f/f tumor cells were cultured with IGFII (100 ng/ml) for 12 h followed by real-time PCR analysis for Igfbp3 expression. Data are shown as means with 95% confidence intervals indicated. n = 6 for each group. *, p < 0.05. D, increased expression levels of phospho-AKT were found in the KPNG2f/f tumors when compared with tumors derived from KPNG2+/+ or KPRNG2 animals. Vinculin was used as loading control. The Western image is from a single blot with additional lanes not relevant to this figure replaced by a space in this image. E, change in expression of CASP7 and IGFBP3 in a human tumor treated with an NG2/CSPG4 compared with controls (control = 1). There is a significant increase in expression in both genes. Data are shown as means with 95% confidence intervals indicated (error bars). *, p < 0.05.
Because Igfbp3 was substantially down-regulated in the KrasG12D/+; p53f/f; Ng2/Cspg4f/f tumors but not in KrasFRT-STOP-FRT-G12D/+; p53FRT/FRT; Rosa26Cre-ER-T2/+; Ng2/Cspg4f/f tumors, its regulation of expression could not be due to changes in Ng2/Cspg4 alone. Because IGFII-mediated IGF signaling activation up-regulates Igfbp3 expression (33), its down-regulation could be due to the loss of sensitivity to IGFII. We thus examined Ng2/Cspg4 regulation of Igfbp3 regulation upon IGFII stimulation in vitro. Tumor cells derived from KrasG12D/+; p53f/f; Ng2/Cspg4+/+, and KrasG12D/+; p53f/f; Ng2/Cspg4f/f mice were treated with IGFII followed by real-time PCR analysis for Igfbp3 expression. Treatment of IGFII up-regulates Igfbp3 expression in KrasG12D/+; p53f/f; Ng2/Cspg4+/+ tumor cells, but this stimulatory effect is lost in the KrasG12D/+; p53f/f; Ng2/Cspg4f/f tumor cells (Fig. 5C).
Igfbp3 can function as a pro-apoptotic factor to decrease the survival cancer cells (34–37). IGFBPs also down-regulate IGF signaling (38), and IGF signaling can activate AKT to affect tumor cell viability (39, 40). Thus, deletion of Ng2/Cspg4 at the time of tumor initiation could result in an increase in tumor size by promoting tumor cell proliferation through down-regulation of Igfbp3. In support of this notion, a link between Ng2/Cspg4 and AKT activity was shown in lung cancer cells (41). Thus, we examined the AKT activity in the tumor cells derived from KrasG12D/+; p53f/f; Ng2/Cspg4+/+, and KrasG12D; p53f/f; Ng2/Cspg4f/f mice using Western analysis. We found a significant increase in the level of phospho-AKT in the KrasG12D/+; p53f/f; Ng2/Cspg4f/f cells when compared with the KrasG12D; p53f/f; Ng2/Cspg4+/+ cells (Fig. 5D). To determine whether the findings in the mouse correlated with the situation in human tumors, we examined the expression of CAS7 and IGFBP3 in tumors treated with NG2/CSPG4 antibody and found increased expression in both genes with inhibition (Fig. 5E). Thus, the human tumors behave similarly to the mouse tumors in which Ng2/Cspg4 is depleted in an established tumor. Taken together, these data raise the possibility that deleting Ng2/Cspg4 at tumor initiation activates IGF signaling, a pathway known to positively regulate soft-tissue sarcoma growth (42), whereas in established tumors, there is an opposite effect.
Discussion
Here we show that Ng2/Cspg4 plays an important role in soft-tissue sarcoma growth and maintenance. Inhibition of Ng2/Cspg4 in established sarcomas by gene deletion or NG2/CSPG4 antibody immunotherapy significantly reduced tumor size in murine and human sarcomas. This was associated with a decrease in cell proliferation and increase in apoptosis.
Surprisingly, deleting Ng2/Cspg4 at the time of tumor initiation resulted in the opposite effect on tumor growth. We found that deletion of Ng2/Cspg4 at tumor initiation resulted in activation of IGF signaling and a loss of the normal regulation of insulin growth factor–binding proteins, which are known to inhibit IGF signaling and regulate cell proliferation. It is possible that cells might adapt to loss of Ng2/Cspg4 at tumor initiation in ways that are different from Ng2/Cspg4 after the tumor is already formed. This adaptation can include activation of IGF signaling through the loss of normal regulation of IGFBPs. This might be related to an epigenetic change, allowing activation of IGF signaling through the loss of normal up-regulation of IGFPBs. Another possibility is that because Ng2/Cspg4 is a membrane proteoglycan that interacts with other cells and extracellular matrix, loss of Ng2/Cspg4 at tumor initiation may alter a critical extracellular interaction that promotes tumor growth. Deletion of Ng2/Cspg4 at tumor initiation revealed a pro-apoptotic effect of Ng2/Cspg4, so ablation of Ng2/Cspg4 might account for decreased cell death at the time of tumor initiation. Consistent with that notion, Ng2/Cspg4 has been shown to play a pro-apoptotic role in fibroblasts (43, 44). Another possibility is that by deleting Ng2/Cspg4 at tumor initiation, we are changing the cell type that becomes the tumor or changing the type of tumor that develops; however, in our analysis to date, we have not found differences between the sarcomas to suggest a difference in tumor cell type. Our data on phospho-AKT, and a lack of normal down-regulation of Igfbp3 expression, suggest that there is constitutive IGF activity in the tumors that lost Ng2/Cspg4 at tumor initiation. This adds support to the notion that IGF plays a critical role in tumor cell behavior in sarcomas (45, 46).
Soft-tissue sarcoma comprises a heterogeneous group of tumors; therefore, precise categorization of patient tumor types is important for choosing appropriate and effective treatments. Our data suggest that targeting Ng2/Cspg4 can be developed into a novel therapeutic approach for soft-tissue sarcomas expressing this transmembrane proteoglycan. Furthermore, our data also illustrate the complexity in interpreting results from genetically engineered mice, as the data from deletion of Ng2/Cspg4 at tumor initiation probably do not necessarily pertain to the more clinically relevant experiment of Ng2/Cspg4 inhibition in established tumors. Issues related to experiments studying the role of genes in tumor initiation compared with their role in tumor maintenance should be considered when translating data from mouse genetics to proposed therapies.
Experimental procedures
Mice
All experiments were performed in accordance with National Institutes of Health guidelines and were approved by the Duke University Division of Laboratory Resources. We crossed Ng2/Cspg4−/−, Ng2/Cspg4f/f (32, 47), LSL-KrasG12D; p53f/f (29), KrasFRT-STOP-FRT-G12D; p53FRT/FRT (48, 49), and R26Cre-ER-T2 or Col1a1FRT-STOP-FRT-Cre-ER-T2 transgenic mice. Tumors were generated by intramuscular injection of a calcium phosphate precipitate of adenovirus expression Cre recombinase or flippase (University of Iowa) into the right hind leg of 6–8-week-old mice. Animals were euthanized within 12 days of tumor formation. Volume was calculated by using the formula, (ab2)π/6, where a is the longest measurement and b is the shortest.
Tomoxifen-induced genes were activated using 4-OHT (Sigma-Aldrich) dissolved in 100% ethanol at a concentration of 250 mg/ml and then diluted with DMSO (Sigma-Aldrich) to a final working solution concentration of 25 mg/ml. Soft-tissue sarcomas in the mouse hind limb were infiltrated with a single intramuscular injection of 30 μl of 4-OHT (0.75 mg) working solution via a 27.5-gauge insulin syringe.
NG2/CSPG4 lentiviral particle transfection and human sarcoma xenografts
Human undifferentiated pleomorphic sarcoma cells were dissociated into single cells as described previously (50, 51). Lentiviral particles containing three target-specific constructs that encode 19–25-nucleotide shRNA targeting human NG2/CSPG4 and lentiviral particles expressing copGFP control lentiviral particles were purchased from Santa Cruz Biotechnology. Cells were plated 24 h before viral transfection at 0.6 × 105 cells/well. For cell infection, viral particles (multiplicity of infection = 4) were supplemented with 10 μg/ml Polybrene and incubated with cells for 24 h. Puromycin (2 μg/ml) was used to select stable clones expressing the shRNA. Various numbers of cells (1 × 105 to 1 × 102) were suspended with Matrigel (BD Biosciences) and injected subcutaneously into 6–8-week-old NSG mice (Jackson Laboratory).
Histology and immunofluorescence
Tumor samples were formalin-fixed in 4% paraformaldehyde, embedded in Tissue Tek O.C.T compound (Fisher Scientific), and sectioned. Sections (10 μm) of the tumor samples were stained with hematoxylin and eosin following standard procedures. For immunofluorescence, the sections were stained with antibodies against human NG2/CSPG4 (mouse mAb 9.2.27, 1:100) or mouse Ng2/Cspg4 (rabbit anti-Ng2/EC, 1:100) at 4 °C overnight. Secondary antibodies conjugated with Alexa Fluor 488 (1:1000; Invitrogen) or Alexa Fluor 568 (1:1000; Thermo Fisher Scientific) were incubated for 1 h at room temperature to detect the primary antibody. Finally, the sections were mounted in 4′,6-diamidino-2-phenylindole containing Fluoroshield (Abcam) and imaged.
Western analysis
Western analysis was performed using standard protocols. Immunoblotting was performed overnight at 4 °C with the following primary antibodies: human NG2/CSPG4 mAb 9.2.27 (1:1000), mouse Ng2/Cspg4 (1:2000), phospho-AKT (Ser-473) from Cell Signaling (1:1000), pan-AKT from Cell Signaling (1:1000), actin from Thermo Fisher (1:5000), and vinculin from Millipore (1:2000).
Gene expression analysis
RNA isolated from at least three independent experiments was analyzed by quantitative real-time PCR in triplicate for each treatment condition and primer set. The reactions used TaqMan Universal PCR master mix (Applied Biosystems) with TaqMan gene expression assays for human NG2/CSPG4, mouse Ng2/Cspg4, and mouse Igfbp3 (Applied Biosystems). The gene expression levels between samples were analyzed using the 2ΔΔCt method (52). Gapdh (Applied Biosystems) was used as endogenous control for target gene normalization. 50 ng of total RNA was used for cDNA synthesis using RT2 SYBR Green quantitative PCR master mixes and the RT2 First Strand kit (Qiagen). Gene expression profiling was performed using the RT2 Profiler Mouse Cancer PathwayFinder PCR Array (Qiagen) according to the manufacturer's protocol.
RNA sequencing
RNA-seq data were processed using the TrimGalore toolkit, which employs Cutadapt to trim low-quality bases and Illumina sequencing adapters from the 3′-end of the reads. Only reads that were 20 nucleotides or longer after trimming were kept for further analysis. Reads were mapped to the GRCm38v68 version of the mouse genome and transcriptome (53) using the STAR RNA-seq alignment tool (54). Reads were kept for subsequent analysis if they mapped to a single genomic location. Gene counts were compiled using the HTSeq tool. Only genes that had at least 10 reads in any given library were used in subsequent analysis. Normalization and differential expression was carried out using the DESeq2 (55) Bioconductor (56) package with the R statistical programming environment. The false discovery rate was calculated to control for multiple hypothesis testing. Gene set enrichment analysis (57) was performed to identify differentially regulated pathways and gene ontology terms for each of the comparisons performed.
Proliferation and apoptosis
Cell proliferation was measured using the Click-iT EdU imaging kit (Invitrogen) according to the manufacturer's protocols. Cells isolated from sarcomas were cultured with medium containing 10 μm Edu for 4 or 24 h followed by fixation and EdU detection. Apoptosis was measured using annexin V incorporation in a manner identical to that previously reported (58).
Immunotherapy studies
Human undifferentiated pleomorphic sarcoma cells (1 × 104) were suspended with Matrigel (BD Biosciences) and injected subcutaneously into 6–8-week-old NSG mice. After 7 weeks, the animals were randomly divided into two groups. Animals were treated with mouse monoclonal antibody 9.2.27 to human NG2/CSPG4 or IgG control at 50 μg/ml/mouse (Abcam) via intraperitoneal injection every other day for 2 weeks.
Statistical analyses
For all of the data, the mean, 95% confidence interval, and S.D were calculated for each condition. Statistical significance was calculated using two-tailed, unpaired Student's t test. The threshold for statistical significance was p < 0.05.
Author contributions
S.-C. H., W. B. S., D. G. K., and B. A. A. designed experiments, interpreted results, and wrote the manuscript. S.-C. H. performed genetic and cell culture experiments. P. N. and V. P. performed the xenograft immunotherapy experiment.
Supplementary Material
This work was supported by NCI, National Institutes of Health, Grant R01 CA183811. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Tables S1–S4 and Figs. S1–S3.
RNA sequencing data have been deposited to the GEO database and are available under accession number GSE97489.
- 4-OHT
- 4-hydroxytomaxifen
- Edu
- 5-ethynyl-2′-deoxyuridine
- IGF
- insulin-like growth factor.
References
- 1. Fletcher C. D. (2014) Recently characterized soft tissue tumors that bring biologic insight. Mod. Pathol. 27, S98–S112 10.1038/modpathol.2013.172 [DOI] [PubMed] [Google Scholar]
- 2. King D. M., Hackbarth D. A., Kilian C. M., and Carrera G. F. (2009) Soft-tissue sarcoma metastases identified on abdomen and pelvis CT imaging. Clin. Orthop. Relat. Res. 467, 2838–2844 10.1007/s11999-009-0989-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Borden E. C., Baker L. H., Bell R. S., Bramwell V., Demetri G. D., Eisenberg B. L., Fletcher C. D., Fletcher J. A., Ladanyi M., Meltzer P., O'Sullivan B., Parkinson D. R., Pisters P. W., Saxman S., Singer S., et al. (2003) Soft tissue sarcomas of adults: state of the translational science. Clin. Cancer Res. 9, 1941–1956 [PubMed] [Google Scholar]
- 4. Helman L. J., and Meltzer P. (2003) Mechanisms of sarcoma development. Nat. Rev. Cancer 3, 685–694 10.1038/nrc1168 [DOI] [PubMed] [Google Scholar]
- 5. Lehnhardt M., Daigeler A., Homann H. H., Schwaiberger V., Goertz O., Kuhnen C., and Steinau H. U. (2009) MFH revisited: outcome after surgical treatment of undifferentiated pleomorphic or not otherwise specified (NOS) sarcomas of the extremities: an analysis of 140 patients. Langenbecks Arch. Surg. 394, 313–320 10.1007/s00423-008-0368-5 [DOI] [PubMed] [Google Scholar]
- 6. Matushansky I., Charytonowicz E., Mills J., Siddiqi S., Hricik T., and Cordon-Cardo C. (2009) MFH classification: differentiating undifferentiated pleomorphic sarcoma in the 21st century. Expert Rev. Anticancer Ther. 9, 1135–1144 10.1586/era.09.76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Stallcup W. B. (2002) The NG2 proteoglycan: past insights and future prospects. J. Neurocytol. 31, 423–435 10.1023/A:1025731428581 [DOI] [PubMed] [Google Scholar]
- 8. Chekenya M., Krakstad C., Svendsen A., Netland I. A., Staalesen V., Tysnes B. B., Selheim F., Wang J., Sakariassen P. Ø., Sandal T., Lønning P. E., Flatmark T., Enger P. Ø., Bjerkvig R., Sioud M., and Stallcup W. B. (2008) The progenitor cell marker NG2/MPG promotes chemoresistance by activation of integrin-dependent PI3K/Akt signaling. Oncogene 27, 5182–5194 10.1038/onc.2008.157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Makagiansar I. T., Williams S., Mustelin T., and Stallcup W. B. (2007) Differential phosphorylation of NG2 proteoglycan by ERK and PKCα helps balance cell proliferation and migration. J. Cell Biol. 178, 155–165 10.1083/jcb.200612084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. You W. K., Yotsumoto F., Sakimura K., Adams R. H., and Stallcup W. B. (2014) NG2 proteoglycan promotes tumor vascularization via integrin-dependent effects on pericyte function. Angiogenesis 17, 61–76 10.1007/s10456-013-9378-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Nishiyama A., Lin X. H., Giese N., Heldin C. H., and Stallcup W. B. (1996) Co-localization of NG2 proteoglycan and PDGF α-receptor on O2A progenitor cells in the developing rat brain. J. Neurosci. Res. 43, 299–314 10.1002/(SICI)1097-4547(19960201)43:3%3C299::AID-JNR5%3E3.0.CO%3B2-E [DOI] [PubMed] [Google Scholar]
- 12. Cattaruzza S., Ozerdem U., Denzel M., Ranscht B., Bulian P., Cavallaro U., Zanocco D., Colombatti A., Stallcup W. B., and Perris R. (2013) Multivalent proteoglycan modulation of FGF mitogenic responses in perivascular cells. Angiogenesis 16, 309–327 10.1007/s10456-012-9316-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Downward J. (2004) PI 3-kinase, Akt and cell survival. Semin. Cell Dev. Biol. 15, 177–182 10.1016/j.semcdb.2004.01.002 [DOI] [PubMed] [Google Scholar]
- 14. Joy A. M., Beaudry C. E., Tran N. L., Ponce F. A., Holz D. R., Demuth T., and Berens M. E. (2003) Migrating glioma cells activate the PI3-K pathway and display decreased susceptibility to apoptosis. J. Cell Sci. 116, 4409–4417 10.1242/jcs.00712 [DOI] [PubMed] [Google Scholar]
- 15. Yang J., Price M. A., Neudauer C. L., Wilson C., Ferrone S., Xia H., Iida J., Simpson M. A., and McCarthy J. B. (2004) Melanoma chondroitin sulfate proteoglycan enhances FAK and ERK activation by distinct mechanisms. J. Cell Biol. 165, 881–891 10.1083/jcb.200403174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yu L., Favoino E., Wang Y., Ma Y., Deng X., and Wang X. (2011) The CSPG4-specific monoclonal antibody enhances and prolongs the effects of the BRAF inhibitor in melanoma cells. Immunol. Res. 50, 294–302 10.1007/s12026-011-8232-z [DOI] [PubMed] [Google Scholar]
- 17. Cattaruzza S., Nicolosi P. A., Braghetta P., Pazzaglia L., Benassi M. S., Picci P., Lacrima K., Zanocco D., Rizzo E., Stallcup W. B., Colombatti A., and Perris R. (2013) NG2/CSPG4-collagen type VI interplays putatively involved in the microenvironmental control of tumour engraftment and local expansion. J. Mol. Cell. Biol. 5, 176–193 10.1093/jmcb/mjt010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sato S., Tang Y. J., Wei Q., Hirata M., Weng A., Han I., Okawa A., Takeda S., Whetstone H., Nadesan P., Kirsch D. G., Wunder J. S., and Alman B. A. (2016) Mesenchymal tumors can derive from Ng2/Cspg4-expressing pericytes with β-catenin modulating the neoplastic phenotype. Cell Rep. 16, 917–927 10.1016/j.celrep.2016.06.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kageshita T., Yamada M., Arao T., and Ferrone S. (1992) Expression of high molecular weight-melanoma associated antigen (HMW-MAA) in primary ALM lesions is associated with a poor prognosis. Pigment Cell Res. 2, 132–135 [DOI] [PubMed] [Google Scholar]
- 20. Smith F. O., Rauch C., Williams D. E., March C. J., Arthur D., Hilden J., Lampkin B. C., Buckley J. D., Buckley C. V., Woods W. G., Dinndorf P. A., Sorensen P., Kersey J., Hammond D., and Bernstein I. D. (1996) The human homologue of rat NG2, a chondroitin sulfate proteoglycan, is not expressed on the cell surface of normal hematopoietic cells but is expressed by acute myeloid leukemia blasts from poor-prognosis patients with abnormalities of chromosome band 11q23. Blood 87, 1123–1133 [PubMed] [Google Scholar]
- 21. Petrovici K., Graf M., Hecht K., Reif S., Pfister K., and Schmetzer H. (2010) Use of NG2 (7.1) in AML as a tumor marker and its association with a poor prognosis. Cancer Genomics Proteomics 7, 173–180 [PubMed] [Google Scholar]
- 22. Wang J., Svendsen A., Kmiecik J., Immervoll H., Skaftnesmo K. O., Planagumà J., Reed R. K., Bjerkvig R., Miletic H., Enger P. Ø., Rygh C. B., and Chekenya M. (2011) Targeting the NG2/CSPG4 proteoglycan retards tumour growth and angiogenesis in preclinical models of GBM and melanoma. PLoS One 6, e23062 10.1371/journal.pone.0023062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Svendsen A., Verhoeff J. J., Immervoll H., Brøgger J. C., Kmiecik J., Poli A., Netland I. A., Prestegarden L., Planagumà J., Torsvik A., Kjersem A. B., Sakariassen P. Ø., Heggdal J. I., Van Furth W. R., Bjerkvig R., et al. (2011) Expression of the progenitor marker NG2/CSPG4 predicts poor survival and resistance to ionising radiation in glioblastoma. Acta Neuropathol. 122, 495–510 10.1007/s00401-011-0867-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Benassi M. S., Pazzaglia L., Chiechi A., Alberghini M., Conti A., Cattaruzza S., Wassermann B., Picci P., and Perris R. (2009) NG2 expression predicts the metastasis formation in soft-tissue sarcoma patients. J. Orthop. Res. 27, 135–140 10.1002/jor.20694 [DOI] [PubMed] [Google Scholar]
- 25. Nicolosi P. A., Dallatomasina A., and Perris R. (2015) Theranostic impact of NG2/CSPG4 proteoglycan in cancer. Theranostics 5, 530–544 10.7150/thno.10824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wagner S., Krepler C., Allwardt D., Latzka J., Strommer S., Scheiner O., Pehamberger H., Wiedermann U., Hafner C., and Breiteneder H. (2008) Reduction of human melanoma tumor growth in severe combined immunodeficient mice by passive transfer of antibodies induced by a high molecular weight melanoma-associated antigen mimotope vaccine. Clin. Cancer Res. 14, 8178–8183 10.1158/1078-0432.CCR-08-0371 [DOI] [PubMed] [Google Scholar]
- 27. Wang X., Osada T., Wang Y., Yu L., Sakakura K., Katayama A., McCarthy J. B., Brufsky A., Chivukula M., Khoury T., Hsu D. S., Barry W. T., Lyerly H. K., Clay T. M., and Ferrone S. (2010) CSPG4 protein as a new target for the antibody-based immunotherapy of triple-negative breast cancer. J. Natl. Cancer Inst. 102, 1496–1512 10.1093/jnci/djq343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Rivera Z., Ferrone S., Wang X., Jube S., Yang H., Pass H. I., Kanodia S., Gaudino G., and Carbone M. (2012) CSPG4 as a target of antibody-based immunotherapy for malignant mesothelioma. Clin. Cancer Res. 18, 5352–5363 10.1158/1078-0432.CCR-12-0628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kirsch D. G., Dinulescu D. M., Miller J. B., Grimm J., Santiago P. M., Young N. P., Nielsen G. P., Quade B. J., Chaber C. J., Schultz C. P., Takeuchi O., Bronson R. T., Crowley D., Korsmeyer S. J., Yoon S. S., et al. (2007) A spatially and temporally restricted mouse model of soft tissue sarcoma. Nat. Med. 13, 992–997 10.1038/nm1602 [DOI] [PubMed] [Google Scholar]
- 30. Mito J. K., Riedel R. F., Dodd L., Lahat G., Lazar A. J., Dodd R. D., Stangenberg L., Eward W. C., Hornicek F. J., Yoon S. S., Brigman B. E., Jacks T., Lev D., Mukherjee S., and Kirsch D. G. (2009) Cross species genomic analysis identifies a mouse model as undifferentiated pleomorphic sarcoma/malignant fibrous histiocytoma. PLoS One 4, e8075 10.1371/journal.pone.0008075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Deleted in proof.
- 32. Chang Y., She Z. G., Sakimura K., Roberts A., Kucharova K., Rowitch D. H., and Stallcup W. B. (2012) Ablation of NG2 proteoglycan leads to deficits in brown fat function and to adult onset obesity. PLoS One 7, e30637 10.1371/journal.pone.0030637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Fottner C., Engelhardt D., and Weber M. M. (1999) Characterization of insulin-like growth factor binding proteins (IGFBPs) secreted by bovine adrenocortical cells in primary culture: regulation by insulin-like growth factors (IGFs) and adrenocorticotropin (ACTH). Horm. Metab. Res. 31, 203–208 10.1055/s-2007-978720 [DOI] [PubMed] [Google Scholar]
- 34. Granata R., Trovato L., Garbarino G., Taliano M., Ponti R., Sala G., Ghidoni R., and Ghigo E. (2004) Dual effects of IGFBP-3 on endothelial cell apoptosis and survival: involvement of the sphingolipid signaling pathways. FASEB J. 18, 1456–1458 [DOI] [PubMed] [Google Scholar]
- 35. Williams A. C., Collard T. J., Perks C. M., Newcomb P., Moorghen M., Holly J. M., and Paraskeva C. (2000) Increased p53-dependent apoptosis by the insulin-like growth factor binding protein IGFBP-3 in human colonic adenoma-derived cells. Cancer Res. 60, 22–27 [PubMed] [Google Scholar]
- 36. Butt A. J., Firth S. M., King M. A., and Baxter R. C. (2000) Insulin-like growth factor-binding protein-3 modulates expression of Bax and Bcl-2 and potentiates p53-independent radiation-induced apoptosis in human breast cancer cells. J. Biol. Chem. 275, 39174–39181 10.1074/jbc.M908888199 [DOI] [PubMed] [Google Scholar]
- 37. Hollowood A. D., Lai T., Perks C. M., Newcomb P. V., Alderson D., and Holly J. M. (2000) IGFBP-3 prolongs the p53 response and enhances apoptosis following UV irradiation. Int. J. Cancer 88, 336–341 10.1002/1097-0215(20001101)88:3%3C336::AID-IJC3%3E3.0.CO%3B2-A [DOI] [PubMed] [Google Scholar]
- 38. Walter H. J., Berry M., Hill D. J., Cwyfan-Hughes S., Holly J. M., and Logan A. (1999) Distinct sites of insulin-like growth factor (IGF)-II expression and localization in lesioned rat brain: possible roles of IGF binding proteins (IGFBPs) in the mediation of IGF-II activity. Endocrinology 140, 520–532 10.1210/endo.140.1.6463 [DOI] [PubMed] [Google Scholar]
- 39. Duan C., Liimatta M. B., and Bottum O. L. (1999) Insulin-like growth factor (IGF)-I regulates IGF-binding protein-5 gene expression through the phosphatidylinositol 3-kinase, protein kinase B/Akt, and p70 S6 kinase signaling pathway. J. Biol. Chem. 274, 37147–37153 10.1074/jbc.274.52.37147 [DOI] [PubMed] [Google Scholar]
- 40. Kulik G., Klippel A., and Weber M. J. (1997) Antiapoptotic signalling by the insulin-like growth factor I receptor, phosphatidylinositol 3-kinase, and Akt. Mol. Cell. Biol. 17, 1595–1606 10.1128/MCB.17.3.1595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Jin Q., Lee H. J., Min H. Y., Smith J. K., Hwang S. J., Whang Y. M., Kim W. Y., Kim Y. H., and Lee H. Y. (2014) Transcriptional and posttranslational regulation of insulin-like growth factor binding protein-3 by Akt3. Carcinogenesis 35, 2232–2243 10.1093/carcin/bgu129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Van der Ven L. T., Roholl P. J., Gloudemans T., Van Buul-Offers S. C., Welters M. J., Bladergroen B. A., Faber J. A., Sussenbach J. S., and Den Otter W. (1997) Expression of insulin-like growth factors (IGFs), their receptors and IGF binding protein-3 in normal, benign and malignant smooth muscle tissues. Br. J. Cancer 75, 1631–1640 10.1038/bjc.1997.278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Joo N. E., Watanabe T., Chen C., Chekenya M., Stallcup W. B., and Kapila Y. L. (2008) NG2, a novel proapoptotic receptor, opposes integrin α4 to mediate anoikis through PKCα-dependent suppression of FAK phosphorylation. Cell Death Differ. 15, 899–907 10.1038/cdd.2008.22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Joo N. E., Miao D., Bermúdez M., Stallcup W. B., and Kapila Y. L. (2014) Shedding of NG2 by MMP-13 attenuates anoikis. DNA Cell Biol. 33, 854–862 10.1089/dna.2014.2399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Scotlandi K., Benini S., Sarti M., Serra M., Lollini P. L., Maurici D., Picci P., Manara M. C., and Baldini N. (1996) Insulin-like growth factor I receptor-mediated circuit in Ewing's sarcoma/peripheral neuroectodermal tumor: a possible therapeutic target. Cancer Res. 56, 4570–4574 [PubMed] [Google Scholar]
- 46. Ho L., Stojanovski A., Whetstone H., Wei Q. X., Mau E., Wunder J. S., and Alman B. (2009) Gli2 and p53 cooperate to regulate IGFBP-3-mediated chondrocyte apoptosis in the progression from benign to malignant cartilage tumors. Cancer Cell 16, 126–136 10.1016/j.ccr.2009.05.013 [DOI] [PubMed] [Google Scholar]
- 47. Grako K. A., Ochiya T., Barritt D., Nishiyama A., and Stallcup W. B. (1999) PDGF α-receptor is unresponsive to PDGF-AA in aortic smooth muscle cells from the NG2 knockout mouse. J. Cell Sci. 112, 905–915 [DOI] [PubMed] [Google Scholar]
- 48. Young N. P., Crowley D., and Jacks T. (2011) Uncoupling cancer mutations reveals critical timing of p53 loss in sarcomagenesis. Cancer Res. 71, 4040–4047 10.1158/0008-5472.CAN-10-4563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ventura A., Kirsch D. G., McLaughlin M. E., Tuveson D. A., Grimm J., Lintault L., Newman J., Reczek E. E., Weissleder R., and Jacks T. (2007) Restoration of p53 function leads to tumour regression in vivo. Nature 445, 661–665 10.1038/nature05541 [DOI] [PubMed] [Google Scholar]
- 50. Wei Q., Tang Y. J., Voisin V., Sato S., Hirata M., Whetstone H., Han I., Ailles L., Bader G. D., Wunder J., and Alman B. A. (2015) Identification of CD146 as a marker enriched for tumor-propagating capacity reveals targetable pathways in primary human sarcoma. Oncotarget 6, 40283–40294 10.18632/oncotarget.5375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wang C. Y., Wei Q., Han I., Sato S., Ghanbari-Azarnier R., Whetstone H., Poon R., Hu J., Zheng F., Zhang P., Wang W., Wunder J. S., and Alman B. A. (2012) Hedgehog and Notch signaling regulate self-renewal of undifferentiated pleomorphic sarcomas. Cancer Res. 72, 1013–1022 10.1158/0008-5472.CAN-11-2531 [DOI] [PubMed] [Google Scholar]
- 52. Livak K. J., and Schmittgen T. D. (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(−ΔΔC(T)) method. Methods 25, 402–408 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- 53. Kersey P. J., Staines D. M., Lawson D., Kulesha E., Derwent P., Humphrey J. C., Hughes D. S., Keenan S., Kerhornou A., Koscielny G., Langridge N., McDowall M. D., Megy K., Maheswari U., Nuhn M., et al. (2012) Ensembl Genomes: an integrative resource for genome-scale data from non-vertebrate species. Nucleic Acids Res. 40, D91–D97 10.1093/nar/gkr895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Dobin A., Davis C. A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., and Gingeras T. R. (2013) STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Love M. I., Huber W., and Anders S. (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Huber W., Carey V. J., Gentleman R., Anders S., Carlson M., Carvalho B. S., Bravo H. C., Davis S., Gatto L., Girke T., Gottardo R., Hahne F., Hansen K. D., Irizarry R. A., Lawrence M., et al. (2015) Orchestrating high-throughput genomic analysis with Bioconductor. Nat. Methods 12, 115–121 10.1038/nmeth.3252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Mootha V. K., Lindgren C. M., Eriksson K. F., Subramanian A., Sihag S., Lehar J., Puigserver P., Carlsson E., Ridderstråle M., Laurila E., Houstis N., Daly M. J., Patterson N., Mesirov J. P., Golub T. R., et al. (2003) PGC-1α-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 34, 267–273 10.1038/ng1180 [DOI] [PubMed] [Google Scholar]
- 58. Poon R., Hong H., Wei X., Pan J., and Alman B. A. (2012) A high throughput screen identifies Nefopam as targeting cell proliferation in β-catenin driven neoplastic and reactive fibroproliferative disorders. PLoS One 7, e37940 10.1371/journal.pone.0037940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Campoli M. R., Chang C. C., Kageshita T., Wang X., McCarthy J. B., and Ferrone S. (2004) Human high molecular weight-melanoma-associated antigen (HMW-MAA): a melanoma cell surface chondroitin sulfate proteoglycan (MSCP) with biological and clinical significance. Crit. Rev. Immunol. 24, 267–296 10.1615/CritRevImmunol.v24.i4.40 [DOI] [PubMed] [Google Scholar]
- 60. Mittelman A., Chen G. Z., Wong G. Y., Liu C., Hirai S., and Ferrone S. (1995) Human high molecular weight-melanoma associated antigen mimicry by mouse anti-idiotypic monoclonal antibody MK2–23: modulation of the immunogenicity in patients with malignant melanoma. Clin. Cancer Res. 1, 705–713 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.