Abstract
Focal adhesions (FAs) mechanically couple the extracellular matrix to the dynamic actin cytoskeleton, via transmembrane integrins and actin-binding proteins. The molecular mechanisms by which protein machineries control force transmission along this molecular axis (i.e. modulating integrin activation and controlling actin polymerization) remain largely unknown. Talin is a major actin-binding protein that controls both the inside-out activation of integrins and actin filament anchoring and thus plays a major role in the establishment of the actin-extracellular matrix mechanical coupling. Talin contains three actin-binding domains (ABDs). The N-terminal head domain contains both the F3 integrin-activating domain and ABD1, whereas the C-terminal rod contains the actin-anchoring ABD2 and ABD3. Integrin binding is regulated by an intramolecular interaction between the N-terminal head and a C-terminal five-helix bundle (R9). Whether talin ABDs regulate actin polymerization in a constitutive or regulated manner has not been fully explored. Here, we combine kinetics assays using fluorescence spectroscopy and single actin filament observation in total internal reflection fluorescence microscopy, to examine relevant functions of the three ABDs of talin. We find that the N-terminal ABD1 blocks actin filament barbed-end elongation, whereas ABD2 and ABD3 do not show any activity. By mutating residues in ABD1, we find that this activity is mediated by a positively charged surface that is partially masked by its intramolecular interaction with R9. Our results also demonstrate that, once this intramolecular interaction is released, the integrin-bound talin head retains the ability to inhibit actin assembly.
Keywords: actin, cytoskeleton, fluorescence, focal adhesion, integrin, kinetics, talin, biochemistry
Introduction
Cell adhesion to the extracellular matrix plays a critical role in many physiological functions, such as cell migration, invasion, or epithelial basement membrane attachment. Among the multiple adhesion structures, focal adhesions (FAs)5 play a major role (1, 2). These multiprotein complexes couple various extracellular matrices to the actin cytoskeleton via the transmembrane heterodimeric αβ integrins and actin-binding proteins (ABPs) (3, 4). The control of actin polymerization by ABPs is thought to play an important role to initiate the formation of the actomyosin stress fibers and control their tension by modulating their elongation. We showed previously that vinculin blocks actin filament barbed-end elongation (5), whereas others reported that VASP promotes the elongation of actin filament barbed ends in a processive-like manner (6, 7). Several formins may also play a role in the formation and elongation of stress fibers (8). However, despite these isolated characterizations, the respective roles of the multiple ABPs and their coordination in this process are poorly understood.
The actin-binding protein talin plays a major role in FAs (9) (Fig. S1). First, it acts very early to activate integrins. In this process, the N-terminal PTB (phosphotyrosine-binding) domain, located in the head domain of talin, also known as the F3 subdomain of the FERM (four-point-one, ezrin, radixin, moesin) domain, binds to the cytoplasmic tail of the β subunit of integrins to induce an allosteric conformational change through the membrane, leading to the binding of the extracellular domains to ECM with high affinity (10, 11). Talin head is not constitutively active because F3 is buried by an intramolecular interaction with the R9 five-helix bundle of the rod (12, 13). Several mechanisms have been proposed to explain talin head exposure in FAs. The binding of the N-terminal head of talin to a PIP2-enriched membrane or to the RIAM (Rap1-GTP–interacting adaptor molecule) protein could disrupt the F3-R9 interaction to expose F3, allowing integrin binding (13–16). Talin head is also released by calpain-mediated proteolytic cleavage (17). The isolated head domain of talin, resulting from calpain cleavage, regulates adhesion dynamics (18).
In addition to its role in the inside-out activation of integrins, talin anchors the actin cytoskeleton to FAs (19). Talin contains three actin-binding domains (ABDs), also named actin-binding sites (ABSs) (20). ABD1 spans along the F2 and F3 subdomains of the FERM domain (21). ABD2 is located in the center of the rod (22). ABD3, also known as THATCH (talin/HIP1R/Sla2p actin-tethering C-terminal homology), is located at the C terminus of the protein (23). The nanoscale organization of mature FAs, revealed by superresolution, showed that the N-terminal ABD1 of talin is located proximal to the membrane, whereas ABD3 extends deeper in the cell to anchor actomyosin stress fibers (24). In this conformation, talin behaves as a mechanosensor to transduce the force conveyed by the actin cytoskeleton to FAs (25). These properties allow talin to sense the rigidity of the ECM (26). Single molecule experiments showed that force transduction involves the mechanical stretching of the talin rod domain, which exposes cryptic vinculin-binding sites (27). Our previous work showed that vinculin binding to mechanically stretched talin exposes its ABD, known as vinculin tail, and reinforces actin anchoring (28, 29). In cells, actomyosin force acts on talin via its ABD3 to expose vinculin-binding sites and ABD2, locking talin into an actin-binding configuration that stabilizes FAs (30).
Although actin binding to talin plays a major role in the establishment of the actin-ECM mechanical coupling, the contribution of the three ABDs of talin to the regulation of actin polymerization is poorly understood. Here, we report the detailed characterization of the ABDs of talin. We found that the N-terminal ABD1 blocks actin filament barbed-end elongation, whereas ABD2 and ABD3 do not show any activity. This activity is masked by the intramolecular interaction between the F3 subdomain of ABD1 and R9. When ABD1 is exposed, it associates with the cytoplasmic tail of integrin-β3 and inhibits actin filament barbed-end elongation. Our study reveals a new activity of talin and paves the way to understand the complex regulation of actin assembly in FAs.
Results
Talin ABD1 inhibits actin filament barbed-end elongation
To determine the ability of talin constructs to regulate actin assembly (Fig. S1, A and C), we first monitored barbed-end elongation by measuring the increase in pyrenyl-labeled actin fluorescence upon polymerization in the presence of spectrin-actin seeds (5). In cells, multiple parameters can modify the activity of proteins, such as the charge of membrane lipids, the addition of phosphate groups by kinases, local gradients of pH and cations, crowding effects, or the binding of partners. Hence, in many instances, it proved informative to screen a variety of physico-chemical conditions, including a mild reduction in ionic strength, before establishing the activity of actin-binding proteins, such as formins, VASP, or vinculin (5, 31, 32). Here, varying the ionic strength was sufficient to reveal a new inhibitory activity of ABD1 (Fig. 1A). This activity of ABD1 is higher at low ionic strength, with a half-maximum effect at 50 mm KCl (Fig. S2), which is the KCl concentration used by many groups to establish the activity of actin-binding proteins (33). In conditions where ABD1 inhibited actin polymerization, full-length talin, ABD2, and ABD3 remained inactive (Fig. 1A).
Figure 1.
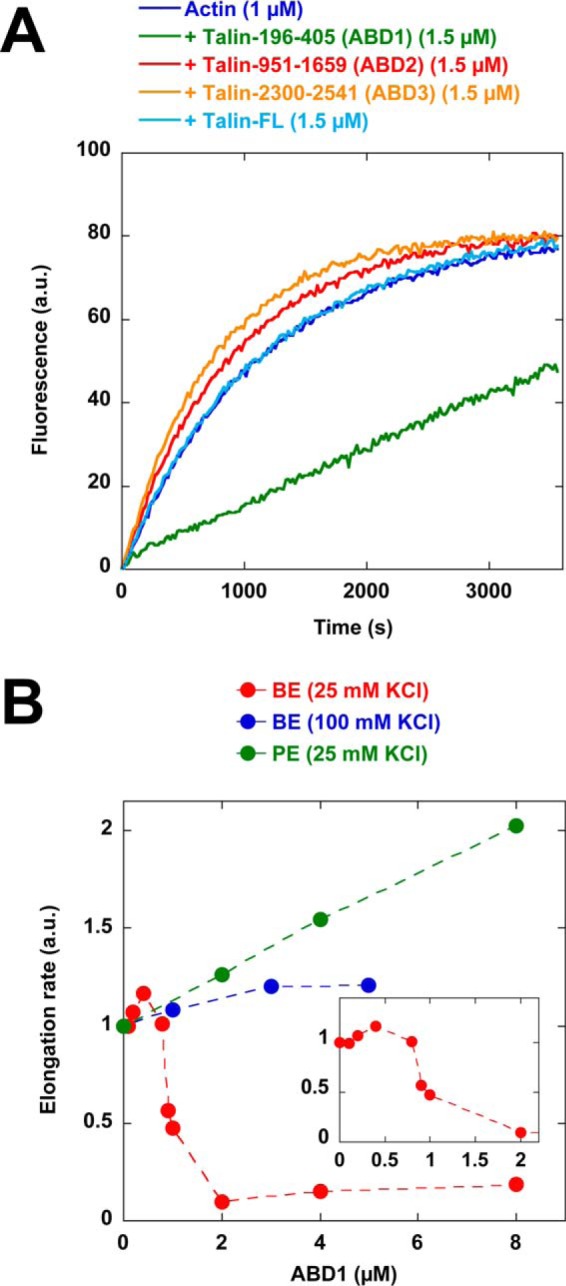
Talin ABD1 inhibits actin filament barbed-end elongation. A, barbed-end elongation was measured in the presence of 100 pm spectrin-actin seeds, 1 μm Mg-ATP–G-actin (10% pyrenyl-labeled) in the absence of talin and in the presence of 1.5 μm full-length talin, 1.5 μm ABD1, 1.5 μm ABD2, and 1.5 μm ABD3. This assay was performed at 25 mm KCl. B, barbed-end (BE) elongation was measured in the presence of 100 pm spectrin-actin seeds, 1 μm Mg-ATP–G-actin (10% pyrenyl-labeled), and increasing concentrations of ABD1 in the presence of 25 mm KCl (red points) and 100 mm KCl (blue points). Pointed end (PE) elongation was measured in the presence of 25 nm gelsolin-actin complex, 2 μm Mg-ATP–G-actin (10% pyrenyl-labeled), and increasing concentrations of ABD1 (green points). The initial rates of pointed-end and barbed-end elongation were plotted versus the concentration of ABD1. The inset shows the detail of the red curve labeled BE (25 mm KCl) between 0 and 2 μm ABD1.
The inhibition curve displays a sigmoidal shape (Fig. 1B), suggesting a cooperative mode of binding. Alternatively, at low concentration, ABD1 could be sequestered along the side of the filaments, preventing it from altering barbed-end dynamics. ABD1 abolished the elongation of spectrin-actin seeds but did not inhibit, or slightly enhanced, the pointed-end elongation of barbed-end capped gelsolin-actin complexes, indicating that ABD1 blocks the elongation of actin filament barbed ends specifically (Fig. 1B).
For the activity of isolated ABD1 to play a physiological role, a significant amount of ABD1-containing talin head, produced by calpain-mediated proteolysis, must exist in cells. To verify this point, we performed the biochemical fractionation of chicken gizzard tissue, extensively used in studies on FA proteins, followed by Western blotting with an antibody directed against talin head (Fig. S3A). We detected talin head in the cytosolic (S1) and membrane (P1) fractions, whereas full-length talin was found associated with the membrane fraction (P1) only. In mild extraction conditions, full-length talin dissociated from the membrane fraction (S2), whereas a large amount of talin head remained associated with the membrane (P2) (Fig. S3, B and C). These observations indicate that ABD1 probably plays a role independent of the context of the full-length protein.
The inhibition of barbed-end elongation is usually the signature of actin filament barbed-end capping proteins. However, our kinetic assays did not rule out the possibility that talin inhibitory activity results from the sequestration of actin monomers. Our results, showing that the saturation of the actin filaments by phalloidin completely abolished ABD1 activity, indicate that ABD1 acts on the filaments and not on the monomers (Fig. S4A). The effect of ABD1 on the linear relationship between the elongation rate and the concentration of polymerizing actin can also provide valuable information about the mechanism. In this assay, a capping protein is expected to decrease the slope of the curve, whereas a monomer-sequestering protein is expected to shift the curve without affecting the slope. We observed that, at nearly saturating concentrations of ABD1, the elongation rate is dramatically reduced (Fig. S4B). The abscissa intercept, which gives the critical concentration for actin assembly, is slightly shifted toward the critical concentration of the free pointed end, which is the usual signature of a barbed-end capping protein, but not further, ruling out an actin-monomer sequestering activity.
In cells, profilin-actin is the main polymerizable actin species, and profilin can participate in several mechanisms as a co-factor in an unpredictable manner. It was therefore important to determine whether ABD1 inhibits the elongation of actin filament barbed ends in the presence of a saturating amount of profilin. We found that profilin did not affect ABD1 activity or slightly enhanced its activity (Fig. S4A).
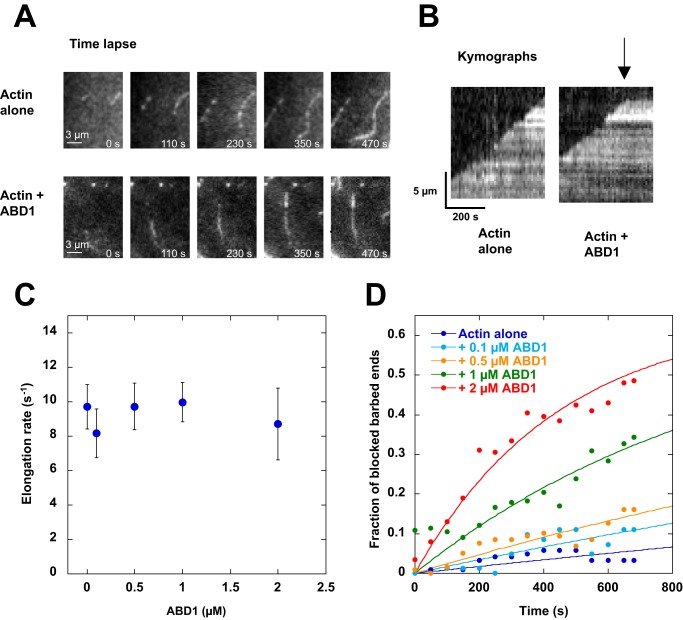
Finally, we directly observed the effect of ABD1 on the elongation of single actin filaments in total internal reflection fluorescence (TIRF) microscopy. In this assay, 5% biotin-labeled actin filaments are trapped on a streptavidin-biotin-PEG–coated surface to facilitate the quantification of the barbed-end elongation rate. In these conditions, we also observed that ABD1 inhibits actin assembly. At the single filament level, the details of the elongation clearly show that pauses occur in the presence of ABD1, resulting in the presence of blocked small filaments, whereas filaments elongate at the rate of the control between pauses (Fig. 2 (A–C) and Movies S1 and S2). In the presence of ABD1, the fraction of blocked filaments increases with time to reach 50% after 600 s at 2 μm ABD1 (Fig. 2D). Altogether, these observations clearly show that ABD1 blocks actin filament barbed-end elongation in a capping-like manner.
Figure 2.
Real-time observation and quantification of the inhibition of barbed-end elongation by talin ABD1 in TIRF microscopy. Conditions were as follows: 1.5 μm Mg-G-actin (5% Alexa 488- and 5% biotin-labeled) in 5 mm Tris, pH 7.8, 200 μm ATP, 0.8% methylcellulose, 1 mm 1,4-diazabicyclo(2,2,2)-octane, 25 mm KCl, 1 mm MgCl2, 200 μm EGTA, 10 mm DTT supplemented with 0, 0.1, 0.5, 1, and 2 μm of talin ABD1. A, time lapse of the elongation of single actin filaments in the absence (top) or presence (bottom) of 1 μm ABD1. Scale bar, 3 μm. See the corresponding Movie S1 and Movie S2. B, kymographs of the filaments shown in A. The fluorescence intensity was measured along the length of a single filament (vertical axis) for each frame of the time lapse (horizontal axis). In contrast with the linear elongation of the control filaments (left), pauses (arrow) were observed in the presence of talin ABD1, indicating barbed-end capping-like events (right). C, elongation rate (s−1) of the filaments during the growing periods (between pauses), in the presence of the indicated concentrations of talin ABD1. Data are mean ± S.D. (error bars). D, quantification of the data shown in A and B. The fraction of blocked filaments is plotted as a function of time in the absence or presence of the indicated concentrations of talin ABD1. For C and D, the number of filaments that we analyzed are n = 121 (0 μm ABD1), n = 82 (0.1 μm ABD1), n = 118 (0.5 μm ABD1), n = 324 (1 μm ABD1), n = 200 (2 μm ABD1).
ABD1 activity is masked by an intramolecular interaction with R9
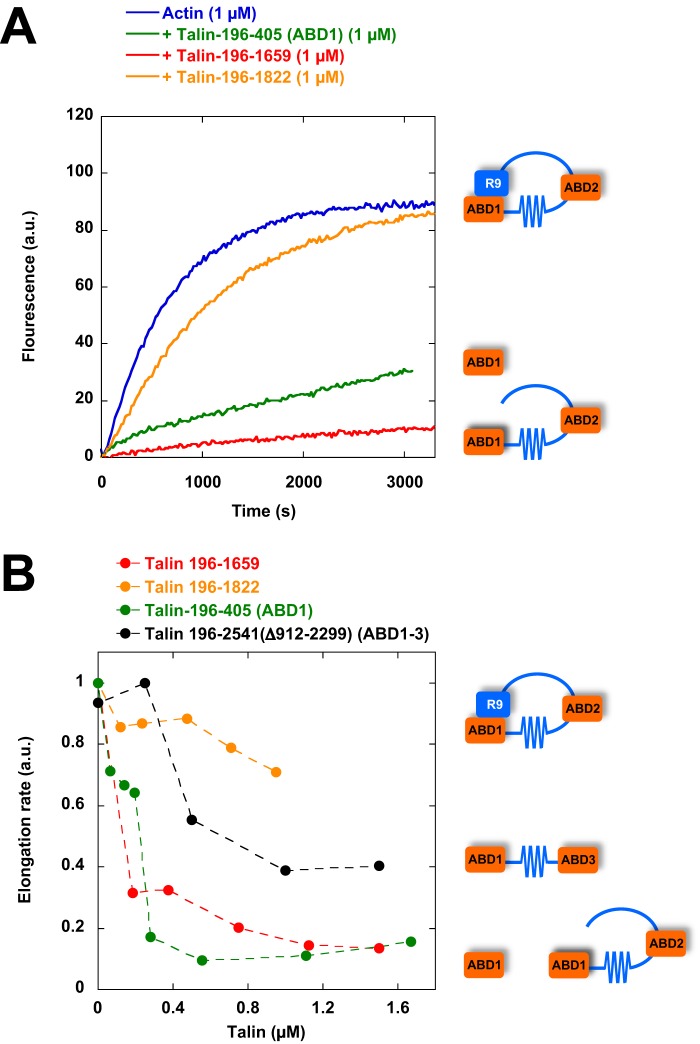
Interestingly, full-length talin, in contrast with isolated ABD1, remained inactive (Fig. 1A), suggesting that ABD1 activity is masked by an intramolecular interaction. To identify the domain that inhibits ABD1 activity, we compared the activity of talin constructs encompassing the N-terminal ABD1 domain and various deletions of the rod domain. We found that a talin construct (talin 196–1822) ending after the R9 five-helix bundle was almost inactive, whereas a talin construct (talin 196–1659) ending before R9 was fully active (Fig. 3, A and B). These observations indicate that R9 plays a critical role to keep ABD1 inactive. Talin constructs, containing combinations of exposed ABD1 and ABD2 (talin 196–1659) or exposed ABD1 and ABD3 (196–2541-Δ912–2299), inhibit actin assembly, indicating that ABD1 activity does not combine with ABD2 or ABD3 to create a different activity (Fig. 3B).
Figure 3.
The activity of talin ABD1 is masked by R9. A, barbed-end elongation was measured in the presence of 100 pm spectrin-actin seeds, 1 μm Mg-ATP–G-actin (10% pyrenyl-labeled) in the absence and in the presence of 1 μm talin ABD1 196–405, 196–1659, and 196–1822. B, the initial rates of barbed-end elongation were plotted versus the concentration of talin-ABD1 196–405, 196–1659, and 196–1822 and talin–ABD1–3 (196–2541Δ912–2299). The conditions used in B are described in A.
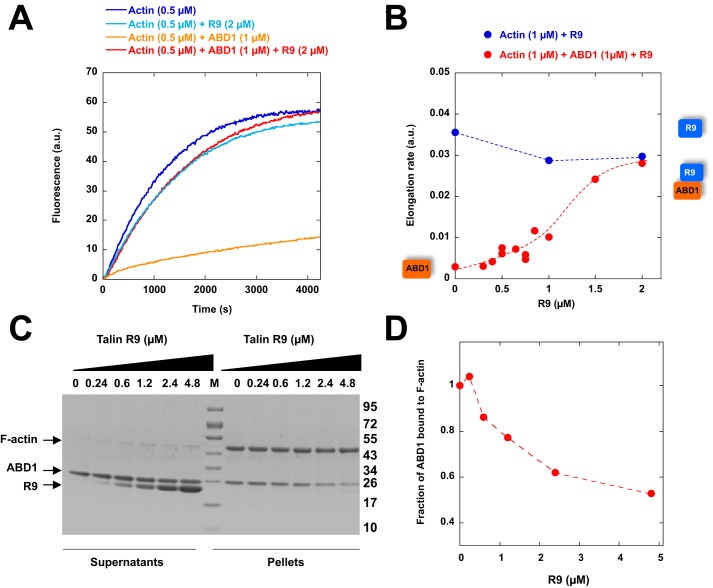
To determine whether R9 was sufficient to inhibit ABD1, we expressed R9 as an isolated protein and tested its role on the inhibitory activity of isolated ABD1. Our results demonstrated that R9 is sufficient to reverse the activity of ABD1, whereas R9 alone did not affect actin assembly (Fig. 4, A and B). This result, indicating that actin and R9 binding to ABD1 are mutually exclusive, was confirmed by a co-sedimentation experiment in which increasing amounts of R9 efficiently displaced ABD1 from actin filaments (Fig. 4, C and D), without binding to actin (Fig. S5).
Figure 4.
The ability of talin ABD1 to bind to actin filaments and inhibit polymerization is regulated by the ABD1-R9 intramolecular interaction. A, barbed-end elongation was measured in the presence of 100 pm spectrin-actin seeds, 0.5 μm Mg-ATP–G-actin (10% pyrenyl-labeled) alone (dark blue) and in the presence of 2 μm R9 alone (light blue), 1 μm ABD1 alone (orange), and 1 μm ABD1 + 2 μm R9 (red). B, quantification of the elongation rate as a function of R9 concentration. C, co-sedimentation of ABD1 (2.3 μm) with actin filaments (5 μm) in the presence of increasing concentrations of R9 as indicated. Supernatant and pellet fractions were resolved by SDS-PAGE and stained by Coomassie Blue. M, molecular weight markers. D, quantification of the fraction of ABD1 bound to actin filaments as a function of R9 concentration.
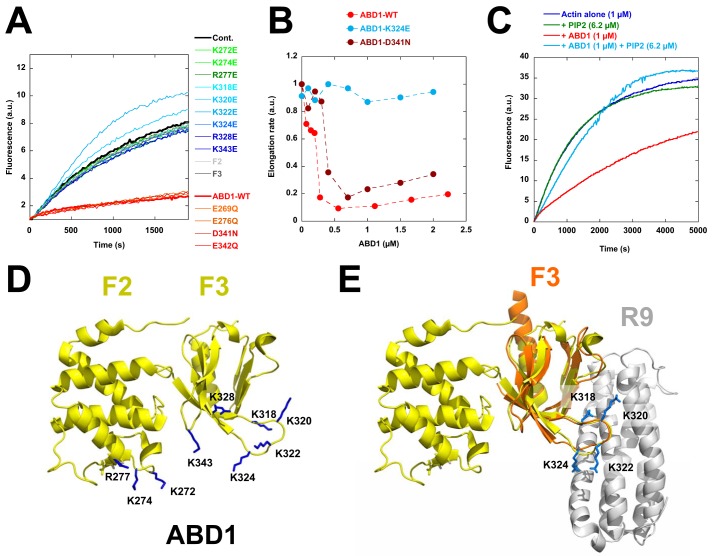
To define the ABD1-actin binding interface involved in the barbed-end capping-like activity, we performed a systematic mutagenesis analysis of ABD1. Because the sensitivity of ABD1 activity to ionic strength indicates that the ABD1-actin interaction is mainly electrostatic, we restricted this analysis to acidic and basic amino acids. We verified that our mutants were not unfolded by showing that they all retained the ability to interact with the side of actin filaments in a co-sedimentation assay (Fig. S6). Interestingly, we found several point mutations in basic residues that abolish ABD1 activity (Fig. 5, A and B). We also showed that the ABD1 K324E mutant, which is defective for barbed-end inhibition but not for side binding (Fig. 5 (A and B) and Fig. S6), reverses the inhibitory activity of the wildtype form of ABD1 in a kinetic assay (Fig. S7). This result indicates that actin filament side binding is necessary but not sufficient for the inhibitory activity of ABD1, which requires an additional set of basic residues to alter barbed-end dynamics.
Figure 5.
Systematic mutagenesis of charged amino acids in ABD1 reveals the ABD1-actin binding interface and explains the mechanism by which R9 masks ABD1 activity. A, barbed-end elongation was measured in the presence of 100 pm spectrin-actin seeds, 1 μm Mg-ATP–G-actin (10% pyrenyl-labeled) in the absence (cont.) and in the presence of 1.5 μm talin-ABD1-WT; the point mutants E269Q, K272E, K274E, E276Q, R277E, K318E, K320E, K322E, K324E, R328E, D341N, E342Q, and K343E; and the F2 and F3 domains. B, the initial rates of barbed-end elongation were plotted versus the concentration of talin-ABD1 WT, K324E, and D341N. The conditions used in B are described in A. C, the inhibition of barbed-end elongation observed in the presence of ABD1 (1 μm) is abrogated by PIP2 (6.2 μm). D, the amino acids whose mutation affects the activity of ABD1 are represented on the known structure (side chains in blue) (43). E, overlay of ABD1 (F2F3) (yellow) and the complex formed by F3 (orange) and R9 (gray) (12, 43). The four basic amino acids (Lys-318, -320, -322, and -324) in F3 (side chains in blue) involved in the inhibition of actin polymerization are also involved in the autoinhibitory contact between ABD1 and R9. The representations of the known structures in D and E have been generated by PyMOL (Schroedinger, LLC, New York).
Because most of these basic residues have been found by previous studies (14) to be involved in PIP2 binding, we tested the effect of PIP2 micelles and found that they reverse the activity of ABD1 (Fig. 5C). When placed on the known crystal structure of ABD1 (talin F2F3), these residues form a clear binding surface that spans along the F2 and F3 subdomains of ABD1 (Fig. 5D). Interestingly, several of these basic residues of F3 were also found to interact with R9 in the crystal structure of the F3-R9 complex (12) (Fig. 5E), explaining why R9 binding to ABD1 interferes with the activity of ABD1 on actin assembly (Fig. 4).
Evidence for an integrin-talin complex that inhibits actin assembly
Previous reports also showed that R9 and the cytoplasmic tail of integrin-β compete for ABD1 binding (12). Whether the binding of integrin-β to ABD1 can release the R9-ABD1 autoinhibitory contact or keep it dissociated after talin activation to form an integrin-talin complex that inhibits barbed-end elongation is not known. Studying the binding of the cytoplasmic tail of integrin-β to talin head in our assays is difficult because the affinity is very low (Kd = 273 μm for talin-1–integrin-β3, (34)). In cells, integrin clustering could favor the binding of integrin to talin through an avidity effect. To determine the activity of an integrin-talin-actin complex, we designed a synthetic peptide of integrin-β3 carrying mutations (DTAN)/(VE) between the membrane-distal (MD) and membrane-proximal (MP) regions that are expected to increase the integrin-talin affinity by a factor of 1000, according to similar mutations performed in integrin-β1D (34) (Fig. S1B). We tested the ability of this integrin peptide to reverse the ABD1-R9 interaction, using the inhibition of actin assembly as a readout of ABD1 exposure. First, we showed that integrin restores the ability of ABD1 to inhibit actin assembly in the presence of a saturating concentration of R9, indicating the existence of an active integrin-ABD1-actin ternary complex that is protected from R9 inhibition (Fig. 6A). We also tested the ability of this integrin peptide to activate longer autoinhibited constructs, such as full-length talin and talin 196–1822. This peptide induced the inhibition of actin assembly by talin 196–1822 (Fig. 6, B and C). However, this peptide could activate neither full-length talin nor full-length talin carrying the E1770A mutation, previously described to break the ABD1-R9 interaction (12), suggesting that the dimerization of full-length talin, together with additional autoinhibitory contacts, stabilizes the inactive conformation of talin (Fig. S8). We also found that the integrin-β3 peptide phosphorylated on tyrosine 747 of the NPLY motif was slightly less efficient at activating talin 196–1822 than the non-phosphorylated one (Fig. 6C), in agreement with previous findings showing that this phosphorylation reduces talin-integrin binding (35).
Figure 6.
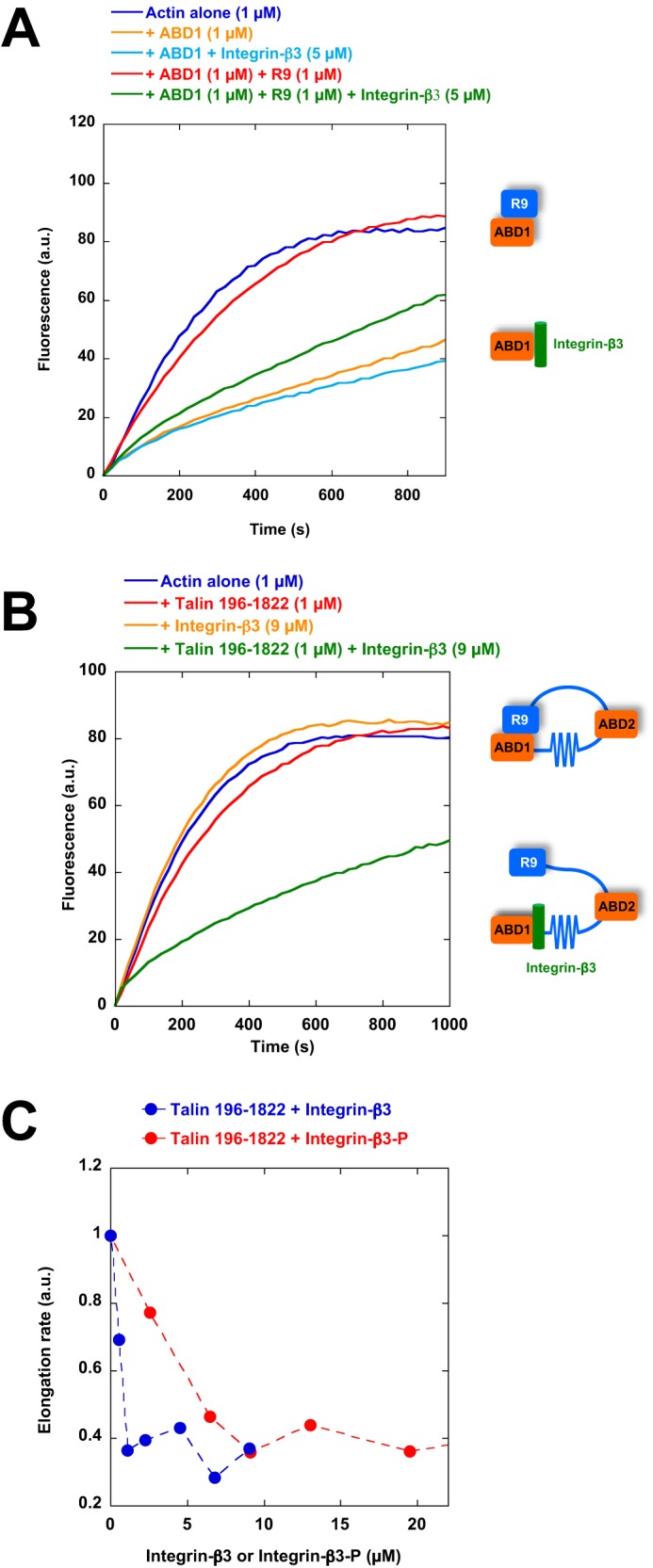
Evidence for an integrin-talin-actin ternary complex that inhibits actin assembly. A, barbed-end elongation was measured in the presence of 100 pm spectrin-actin seeds and 1 μm Mg-ATP–G-actin (10% pyrenyl-labeled) alone (dark blue) and in the presence of 1 μm talin ABD1 alone (orange), 1 μm ABD1 + 5 μm integrin-β3 (DTAN)/(VE) peptide (light blue), 1 μm ABD1 + 1 μm R9 (red), 1 μm ABD1 + 1 μm R9 + 5 μm integrin-β3 (DTAN)/(VE) peptide (green). B, barbed-end elongation was measured in the presence of 100 pm spectrin-actin seeds and 1 μm Mg-ATP–G-actin (10% pyrenyl-labeled) alone (blue) and in the presence of 1 μm talin 196–1822 (red), 9 μm of integrin-β3 (DTAN)/(VE) peptide (orange), and 1 μm talin 196–1822 + 9 μm of integrin-β3 (DTAN)/(VE) peptide (green). C, the initial rates of barbed-end elongation obtained in the conditions described in B were plotted as a function of the concentration of integrin-β3 (DTAN)/(VE) or phosphorylated integrin-β3 (DTAN)/(VE) peptides.
Discussion
Our study revealed that talin ABD1 inhibits actin filament barbed-end elongation in a regulated manner (Fig. 7, A and B). Talin adds to the list of proteins that target actin filaments barbed ends in FAs, including VASP, vinculin, tensin, and formins (5, 7, 8, 36). Like vinculin tail, talin ABD1 binds to the side of actin filaments and inhibits barbed-end elongation. In both mechanisms, barbed-end inhibition requires amino acids that are not necessary for side binding. Determining the result of the combination of these similar activities in the force-dependent talin-vinculin complex will be the subject of future studies.
Figure 7.
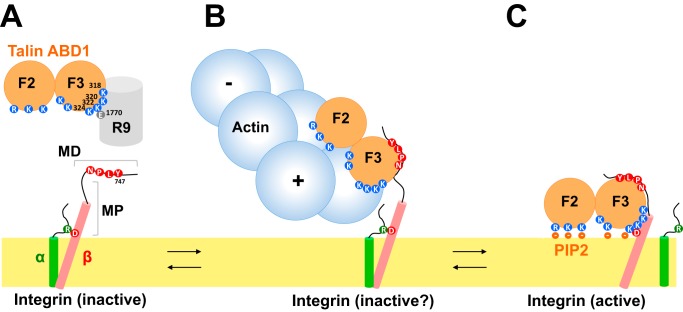
Working model for the activity and regulation of talin. A, schematic representation of talin showing only ABD1, made of F2 and F3, in contact with R9 (top) and integrin (cytoplasmic and transmembrane domains only) in its inactive conformation. The important amino acids are featured. B, the integrin MD site, ABD1, and actin form a ternary complex. In this complex, ABD1 inhibits actin filament barbed-end (+) elongation. In this situation, integrin is inactive, and the α and β subunits remain associated. C, ABD1 binding to a PIP2-containing membrane allows its binding with integrin MD and MP sites. In this situation, integrin is active, and the α and β subunits are dissociated. Actin cannot interact with ABD1.
In FAs, the elongation of actin filaments is expected to release the actomyosin-based tension in stress fibers. Conversely, the stable anchoring provided by actin filament barbed-end capping in FAs could be a means to increase the traction force applied by stress fibers on FAs and ECM. Another interesting possibility is that all the barbed-end capping proteins mentioned above, including talin, anchor the filaments with their barbed ends facing FAs and their pointed ends pointing outward, to create the appropriate orientation for actomyosin contraction.
Interestingly, we identified four lysines in the F3 part of ABD1 (Lys-318, Lys-320, Lys-322, and Lys-324) that are involved in the inhibitory activity and that were previously described in the F3-R9 autoinhibitory interaction (12, 13). This observation explains why an autoinhibited construct of talin (residues 196–1822 or full-length) does not inhibit actin polymerization (Fig. 7A). Although we used a high-affinity mutant of integrin-β3 to disrupt this interaction, as a tool to demonstrate the existence of an integrin-talin-actin complex, the affinity of the wildtype integrin-β3 tail for talin (Kd = 273 μm (34)) is probably not sufficient to break the ABD1-R9 intramolecular interaction. It is more likely that other regulators, such as RIAM, synergize with integrin to disrupt the ABD1-R9 interaction of the autoinhibited conformation (16), allowing ABD1 to bind actin. Alternatively, the calpain-mediated cleavage of the region linking the head and the rod of talin produces enough talin head that constitutively interacts with both integrin and actin (18).
Our findings also shed light on the mechanism by which actin could modulate integrin activation by talin (Fig. 7). Integrin activation occurs through outside-in and inside-out mechanisms. The outside-in mechanism involves the binding of ECM to integrin, inducing integrin extension. The inside-out mechanism involves the binding of talin F3 to the cytoplasmic tail of integrin-β. Talin first interacts with the integrin MD site, inducing its ordering (37). This first step is followed by the binding of the lysine 324 of F3 (Lys-327 in talin-2) to an acidic residue of the integrin MP site (Asp-723 in integrin-β3 or Asp-729 in integrin-β1). This interaction brings the positively charged interface, made of basic residues of F2 and F3, in close proximity to the negative charges of PIP2 heads at the inner face of the plasma membrane (Fig. 7C). The interaction between talin Lys-324 and integrin-β3 Asp-723 also disrupts the salt bridge between this acidic residue and a basic residue of the α subunit (Arg-995 in αIIb), allowing the dissociation of the α and β subunits and their extension in a high-affinity conformation for ECM (38). In this conformation, talin F3 interacts with integrin MD and MP sites, and both F2 and F3 are anchored to the PIP2-containing membrane surface. The actin-binding surface that we identified along F2 and F3 is nearly identical to the one that interacts with PIP2-containing membranes, because all of the mutations that reverse the inhibition of actin assembly by ABD1 have also been described to alter the binding of PIP2 to liposomes in vitro (14). Our results confirm that PIP2 prevents ABD1 to inhibit actin polymerization. Interestingly, our mutagenesis study, showing that the lysine Lys-324 interacts with actin barbed ends, implies that actin prevents lysine Lys-324 from interacting with the MP site, which is normally required for integrin activation. Therefore, in the integrin-ABD1-actin complex, integrin would remain inactive because the F3 part of ABD1 only associates with the integrin MD site, whereas the MP site remains dissociated. In this conformation, ABD1 would not associate with the PIP2-containing membrane leaflet, leaving enough space for an actin filament to bind. In support of this hypothesis, the interaction between talin F3 and the isolated integrin MD site has been reported by others (37, 39). In addition, mutations in the MP region of integrin, including a K324D charge swapping, similar to the K324E we used, abolish integrin activation in cells and abrogate talin binding to the MP site but not to the MD site (37, 38).
It is generally believed that, in FAs, the ABD1-containing talin head lies on the membrane, where it activates integrin by the inside-out pathway (11), whereas the C-terminal ABD3 domain senses the actomyosin force to induce the formation of the mechanosensitive talin-vinculin complex, which is the first step toward the maturation of FAs (9). More recently, ABD2 has been involved in force transmission too (30). Our findings suggest an alternative mechanism to couple integrin to the actin cytoskeleton through the N-terminal ABD1. In this conformation, the talin rod does not sense force, and the integrin MP region cannot contribute to the inside-out integrin activation. This mechanism would allow the retrograde actin flow to drag inactive integrins inside FAs. The mechanism that we identified could also prevent inactive integrins from diffusing away from FAs between two activation events, which appears necessary for a very dynamic and adaptive process like integrin activation. Finally, it is also possible that the integrin-ABD1-actin complex transmits enough force to induce the mechanically induced transition from the closed to the open high-affinity conformation of integrin (40, 41).
Experimental procedures
Recombinant cDNA constructs and peptides
cDNAs encoding for human talin-1 196–405 (ABD1), 196–309 (F2), 309–405 (F3), 196–1659, 196–1822, 951–1659 (ABD2),1655–1822 (R9), and 2300–2541 (ABD3) were constructed by PCR amplification of the full-length talin cDNAs and subcloning of the resulting DNA in a pGEX6P1 plasmid (GE Healthcare). An N-terminal glutathione S-transferase (GST) tag is present in the pGEX6P1, and a C-terminal His6 tag was introduced by PCR for talin 196–1659, talin 196–1822, and ABD2. Talin 196–2541Δ912–2299 (ABD1–3) was cloned into a pETM plasmid with an N-terminal StrepTagII and a C-terminal His6 tag. Full-length talin carrying E1770A was generated from full-length talin cloned into pET101-TOPO (28, 29). The following integrin-β3 peptides were synthesized by ProteoGenix (France): KLLITIHDRKEFAKFEEERARAKWVENPLYPKEATSTFTNITYRGTC and KLLITIHDRKEFAKFEEERARAKWVENPLpYPKEATSTFTNITYRGTC (where pY represents phosphotyrosine).
Protein purification
All of the recombinant proteins were expressed using a similar protocol. First, the plasmid of interest was transformed in Escherichia coli (BL21 DE3, Invitrogen). Typically, transformed bacteria were grown in 4–8 liter of LB medium containing the appropriate antibiotics at 37 °C until the absorbance measured at 600 nm reached 0.6–0.8. The culture was then incubated on ice until the temperature reached 16 °C precisely. The expression of the recombinant proteins of interest was induced by adding 1 mm isopropyl 1-thio-β-d-galactopyranoside to the medium during 16 h at 16 °C. After centrifugation, the pellet was lysed by sonication in 50 mm Tris, pH 7.8, 500 mm NaCl, 1% Triton X-100, 1 mm β-mercaptoethanol, 10 μg/ml benzamidine, and 1 mm PMSF.
The proteins expressed as N-terminal GST fusions were bound to glutathione-Sepharose (GE Healthcare) during 2 h at 4 °C on a rotating wheel and washed with 50 mm Tris, pH 7.8, 500 mm NaCl, 1 mm β-mercaptoethanol. The protein of interest was cleaved from its GST tag by the PreScission protease (GE Healthcare) during 16 h at 4 °C on a rotating wheel and recovered in the supernatant after a low-speed centrifugation. Proteins containing a C-terminal His6 tag (ABD2, 196–1659, and 196–1822) were further bound to Ni2+-nitrilotriacetic acid–agarose (Macherey-Nalgene); washed with 50 mm Tris, pH 7.8, 500 mm NaCl, 20 mm imidazole, 1 mm β-mercaptoethanol; and eluted with 50 mm Tris, pH 7.8, 500 mm NaCl, 250 mm imidazole, 1 mm β-mercaptoethanol. Talin 196–2541Δ912-2299 (ABD1–3) was bound to Ni2+-nitrilotriacetic acid–agarose (Macherey-Nalgene), washed with 50 mm Tris, pH 7.8, 500 mm NaCl, 20 mm imidazole, 1 mm β-mercaptoethanol and eluted with 50 mm Tris, pH 7.8, 500 mm NaCl, 250 mm imidazole, 1 mm β-mercaptoethanol. Proteins were finally dialyzed in 20 mm Tris, pH 7.8, 100 mm KCl, 1 mm β-mercaptoethanol; frozen in liquid nitrogen; and stored at −80 °C. Full-length talin and the E1770A mutant were expressed and purified as described previously (28, 29).
Chicken gizzard fractionation
To fractionate chicken gizzards, we adapted the beginning of the classic protocol used for talin purification (42). Chicken gizzards (7 g) were mixed in 70 ml of ice-cold buffer (5 mm Tris, pH 7.0, 1 mm PMSF (protease inhibitor), and 5 mm EGTA) and homogenized in a Waring blender with three 10-s bursts at full speed. The suspension was centrifuged at 4 °C for 10 min at 10,000 × g. The supernatant (S1) was kept. The pellet (P1) was resuspended in 70 ml of ice-cold buffer (50 mm Tris, pH 9.0, 1 mm PMSF, and 5 mm EGTA) and homogenized in a Waring blender with three 10-s bursts at full speed. The suspension was then incubated at 37 °C for 30 min and centrifuged at 4 °C for 10 min at 10,000 × g to separate the soluble FA proteins (S2) and the membrane-associated proteins (P2). Comparable samples of S1, P1, S2, and P2 were submitted to Western blot analysis with an antibody directed against talin head (TA205, Bio-Rad). This experiment was reproduced twice with the same results.
F-actin co-sedimentation assay
We performed co-sedimentation assays to measure the competition between actin filaments and R9 for talin ABD1. Increasing concentrations of R9 (0–4.8 μm) were incubated with 2.3 μm ABD1 and 5 μm F-actin in 5 mm Tris, pH 7.8, 25 mm KCl, 1 mm MgCl2, 200 μm ATP, 1 mm DTT for 15 min at room temperature. After centrifugation at 90,000 rpm in a TL100 centrifuge (Beckman), the pellets and supernatants were separated, loaded on SDS-PAGE, and quantified using the ImageJ software. This experiment was reproduced three times in similar conditions with the same results.
Polymerization assay
Actin polymerization was monitored by the increase in fluorescence of 10% pyrenyl-labeled actin. Actin polymerization was induced by the addition of 25 mm KCl (unless specified), 1 mm MgCl2, and 0.2 mm EGTA to a solution of 10% pyrenyl-labeled Ca-ATP–G-actin containing the proteins of interest. Fluorescence measurements were carried out in a Xenius spectrofluorimeter (Safas, Monaco). For kinetic experiments, 100 pm spectrin-actin seeds were added to the reaction for barbed-end elongation measurements, and 25 nm gelsolin-actin (1:2) complexes were added for pointed-end elongation measurements. All of the experiments have been reproduced two or three times with the same conclusions.
Observation and measurement of single actin filament elongation by TIRF microscopy
Our protocol is a modification of the protocol used to study vinculin activity (5). To force the filaments to grow at the surface of the coverslip, we first irradiated coverslips with deep UV radiation for 1 min and incubated them with 10% biotin-labeled PLL-PEG for 1 h at room temperature. The coverslip was then washed extensively with water and dried with a nitrogen stream. Flow cells containing 40–60 μl of liquid were prepared by sticking the PLL-PEG–coated coverslip to a slide with double-faced adhesive spacers. The chamber was first incubated with 0.1 mg/ml streptavidin for 5 min and washed with washing buffer (5 mm Tris, pH 7.8, 200 μm ATP, 10 mm DTT, 1 mm MgCl2, 25 mm KCl). The chamber was then saturated with 10% BSA for 5 min and washed with washing buffer. The final reaction was then injected into the chamber. A typical reaction was composed of 1.5 μm 5% Alexa 488, 5% biotin-labeled Mg-G-actin in 5 mm Tris, pH 7.8, 200 μm ATP, 0.8% methylcellulose, 1 mm 1,4-diazabicyclo(2,2,2)-octane, 25 mm KCl, 1 mm MgCl2, 200 μm EGTA, 10 mm DTT supplemented with 0, 0.1, 0.5, 1, or 2 μm talin ABD1. Finally, we sealed the flow chamber with VALAP (a mixture of Vaseline, lanolin, and paraffin) and observed the reaction on an Olympus AX71 inverted microscope equipped with a ×60 (numerical aperture 1.45) objective (Olympus) and a Blues 473-nm laser (Cobolt). The time-lapse videos were acquired by Metamorph and subsequently analyzed by the ImageJ software.
Author contributions
C. C. identified the activity of talin, purified proteins, and performed some of the experiments shown in Figs. 1 and 4. H. W. performed the experiments and analyzed the data shown in Fig. 2. V. H. made cDNA constructs, purified proteins, and performed the experiments shown in Fig. 5. C. M. performed some of the experiments shown in Fig. 4. A. F. performed some of the experiments shown in Fig. 3. S. C. performed some of the experiments shown in Fig. 1 and Fig. S2. C. V. performed some of the experiments shown in Fig. 3. B. F. made cDNA constructs and purified proteins (Fig. S1). C. L. C. performed some of the experiments shown in Figs. 3 and 6 and Figs. S3–S8, conceived and coordinated the study, and wrote the paper.
Supplementary Material
Acknowledgments
We thank Pierre Montaville for help with structure representations and all the members of the “Cytokeleton Dynamics and Motility” team for helpful discussions.
This work was supported in part by Agence Nationale pour la Recherche Grants ANR-09-JCJC-0111 ADERACTIN (to C. L. C.) and ANR-16-CE13-0007-02 PHAGOMECANO (to C. L. C.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1–S8 and Movies S1 and S2.
- FA
- focal adhesion
- ABP
- actin-binding protein
- VASP
- vasodilator-stimulated phosphoprotein
- ABD
- actin-binding domain
- ABS
- actin-binding site
- TIRF
- total internal reflection fluorescence
- PMSF
- phenylmethlylsulfonyl fluoride
- a.u.
- arbitrary units
- PLL
- poly-l-lysine.
References
- 1. Wehrle-Haller B. (2012) Structure and function of focal adhesions. Curr. Opin. Cell Biol. 24, 116–124 10.1016/j.ceb.2011.11.001 [DOI] [PubMed] [Google Scholar]
- 2. Wehrle-Haller B. (2012) Assembly and disassembly of cell matrix adhesions. Curr. Opin. Cell Biol. 24, 569–581 10.1016/j.ceb.2012.06.010 [DOI] [PubMed] [Google Scholar]
- 3. Ciobanasu C., Faivre B., and Le Clainche C. (2012) Actin dynamics associated with focal adhesions. Int. J. Cell Biol. 2012, 941292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ciobanasu C., Faivre B., and Le Clainche C. (2013) Integrating actin dynamics, mechanotransduction and integrin activation: the multiple functions of actin binding proteins in focal adhesions. Eur. J. Cell Biol. 92, 339–348 10.1016/j.ejcb.2013.10.009 [DOI] [PubMed] [Google Scholar]
- 5. Le Clainche C., Dwivedi S. P., Didry D., and Carlier M. F. (2010) Vinculin is a dually regulated actin filament barbed end-capping and side-binding protein. J. Biol. Chem. 285, 23420–23432 10.1074/jbc.M110.102830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Breitsprecher D., Kiesewetter A. K., Linkner J., and Faix J. (2009) Analysis of actin assembly by in vitro TIRF microscopy. Methods Mol. Biol. 571, 401–415 10.1007/978-1-60761-198-1_27 [DOI] [PubMed] [Google Scholar]
- 7. Breitsprecher D., Kiesewetter A. K., Linkner J., Urbanke C., Resch G. P., Small J. V., and Faix J. (2008) Clustering of VASP actively drives processive, WH2 domain-mediated actin filament elongation. EMBO J. 27, 2943–2954 10.1038/emboj.2008.211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Skau C. T., Plotnikov S. V., Doyle A. D., and Waterman C. M. (2015) Inverted formin 2 in focal adhesions promotes dorsal stress fiber and fibrillar adhesion formation to drive extracellular matrix assembly. Proc. Natl. Acad. Sci. U.S.A. 112, E2447–E2456 10.1073/pnas.1505035112 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 9. Calderwood D. A., Campbell I. D., and Critchley D. R. (2013) Talins and kindlins: partners in integrin-mediated adhesion. Nat. Rev. Mol. Cell Biol. 14, 503–517 10.1038/nrm3624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Tadokoro S., Shattil S. J., Eto K., Tai V., Liddington R. C., de Pereda J. M., Ginsberg M. H., and Calderwood D. A. (2003) Talin binding to integrin beta tails: a final common step in integrin activation. Science 302, 103–106 10.1126/science.1086652 [DOI] [PubMed] [Google Scholar]
- 11. Calderwood D. A., Zent R., Grant R., Rees D. J., Hynes R. O., and Ginsberg M. H. (1999) The Talin head domain binds to integrin β subunit cytoplasmic tails and regulates integrin activation. J. Biol. Chem. 274, 28071–28074 10.1074/jbc.274.40.28071 [DOI] [PubMed] [Google Scholar]
- 12. Goult B. T., Bate N., Anthis N. J., Wegener K. L., Gingras A. R., Patel B., Barsukov I. L., Campbell I. D., Roberts G. C., and Critchley D. R. (2009) The structure of an interdomain complex that regulates talin activity. J. Biol. Chem. 284, 15097–15106 10.1074/jbc.M900078200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Goksoy E., Ma Y. Q., Wang X., Kong X., Perera D., Plow E. F., and Qin J. (2008) Structural basis for the autoinhibition of talin in regulating integrin activation. Mol. Cell 31, 124–133 10.1016/j.molcel.2008.06.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Saltel F., Mortier E., Hytönen V. P., Jacquier M. C., Zimmermann P., Vogel V., Liu W., and Wehrle-Haller B. (2009) New PI(4,5)P2- and membrane proximal integrin-binding motifs in the talin head control β3-integrin clustering. J. Cell Biol. 187, 715–731 10.1083/jcb.200908134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lee H. S., Lim C. J., Puzon-McLaughlin W., Shattil S. J., and Ginsberg M. H. (2009) RIAM activates integrins by linking talin to ras GTPase membrane-targeting sequences. J. Biol. Chem. 284, 5119–5127 10.1074/jbc.M807117200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yang J., Zhu L., Zhang H., Hirbawi J., Fukuda K., Dwivedi P., Liu J., Byzova T., Plow E. F., Wu J., and Qin J. (2014) Conformational activation of talin by RIAM triggers integrin-mediated cell adhesion. Nat. Commun. 5, 5880 10.1038/ncomms6880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. O'Halloran T., Beckerle M. C., and Burridge K. (1985) Identification of talin as a major cytoplasmic protein implicated in platelet activation. Nature 317, 449–451 10.1038/317449a0 [DOI] [PubMed] [Google Scholar]
- 18. Franco S. J., Rodgers M. A., Perrin B. J., Han J., Bennin D. A., Critchley D. R., and Huttenlocher A. (2004) Calpain-mediated proteolysis of talin regulates adhesion dynamics. Nat. Cell Biol. 6, 977–983 10.1038/ncb1175 [DOI] [PubMed] [Google Scholar]
- 19. Jiang G., Giannone G., Critchley D. R., Fukumoto E., and Sheetz M. P. (2003) Two-piconewton slip bond between fibronectin and the cytoskeleton depends on talin. Nature 424, 334–337 10.1038/nature01805 [DOI] [PubMed] [Google Scholar]
- 20. Hemmings L., Rees D. J., Ohanian V., Bolton S. J., Gilmore A. P., Patel B., Priddle H., Trevithick J. E., Hynes R. O., and Critchley D. R. (1996) Talin contains three actin-binding sites each of which is adjacent to a vinculin-binding site. J. Cell Sci. 109, 2715–2726 [DOI] [PubMed] [Google Scholar]
- 21. Lee H. S., Bellin R. M., Walker D. L., Patel B., Powers P., Liu H., Garcia-Alvarez B., de Pereda J. M., Liddington R. C., Volkmann N., Hanein D., Critchley D. R., and Robson R. M. (2004) Characterization of an actin-binding site within the talin FERM domain. J. Mol. Biol. 343, 771–784 10.1016/j.jmb.2004.08.069 [DOI] [PubMed] [Google Scholar]
- 22. Gingras A. R., Bate N., Goult B. T., Patel B., Kopp P. M., Emsley J., Barsukov I. L., Roberts G. C., and Critchley D. R. (2010) Central region of talin has a unique fold that binds vinculin and actin. J. Biol. Chem. 285, 29577–29587 10.1074/jbc.M109.095455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gingras A. R., Bate N., Goult B. T., Hazelwood L., Canestrelli I., Grossmann J. G., Liu H., Putz N. S., Roberts G. C., Volkmann N., Hanein D., Barsukov I. L., and Critchley D. R. (2008) The structure of the C-terminal actin-binding domain of talin. EMBO J. 27, 458–469 10.1038/sj.emboj.7601965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kanchanawong P., Shtengel G., Pasapera A. M., Ramko E. B., Davidson M. W., Hess H. F., and Waterman C. M. (2010) Nanoscale architecture of integrin-based cell adhesions. Nature 468, 580–584 10.1038/nature09621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Haining A. W., Lieberthal T. J., and Del Río Hernández A. (2016) Talin: a mechanosensitive molecule in health and disease. FASEB J. 30, 2073–2085 10.1096/fj.201500080R [DOI] [PubMed] [Google Scholar]
- 26. Austen K., Ringer P., Mehlich A., Chrostek-Grashoff A., Kluger C., Klingner C., Sabass B., Zent R., Rief M., and Grashoff C. (2015) Extracellular rigidity sensing by talin isoform-specific mechanical linkages. Nat. Cell Biol. 17, 1597–1606 10.1038/ncb3268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. del Rio A., Perez-Jimenez R., Liu R., Roca-Cusachs P., Fernandez J. M., and Sheetz M. P. (2009) Stretching single talin rod molecules activates vinculin binding. Science 323, 638–641 10.1126/science.1162912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ciobanasu C., Faivre B., and Le Clainche C. (2014) Actomyosin-dependent formation of the mechanosensitive talin-vinculin complex reinforces actin anchoring. Nat. Commun. 5, 3095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ciobanasu C., Faivre B., and Le Clainche C. (2015) Reconstituting actomyosin-dependent mechanosensitive protein complexes in vitro. Nat. Protoc. 10, 75–89 [DOI] [PubMed] [Google Scholar]
- 30. Atherton P., Stutchbury B., Wang D. Y., Jethwa D., Tsang R., Meiler-Rodriguez E., Wang P., Bate N., Zent R., Barsukov I. L., Goult B. T., Critchley D. R., and Ballestrem C. (2015) Vinculin controls talin engagement with the actomyosin machinery. Nat. Commun. 6, 10038 10.1038/ncomms10038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Romero S., Le Clainche C., Didry D., Egile C., Pantaloni D., and Carlier M. F. (2004) Formin is a processive motor that requires profilin to accelerate actin assembly and associated ATP hydrolysis. Cell 119, 419–429 10.1016/j.cell.2004.09.039 [DOI] [PubMed] [Google Scholar]
- 32. Laurent V., Loisel T. P., Harbeck B., Wehman A., Gröbe L., Jockusch B. M., Wehland J., Gertler F. B., and Carlier M. F. (1999) Role of proteins of the Ena/VASP family in actin-based motility of Listeria monocytogenes. J. Cell Biol. 144, 1245–1258 10.1083/jcb.144.6.1245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Doolittle L. K., Rosen M. K., and Padrick S. B. (2013) Measurement and analysis of in vitro actin polymerization. Methods Mol. Biol. 1046, 273–293 10.1007/978-1-62703-538-5_16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Anthis N. J., Wegener K. L., Critchley D. R., and Campbell I. D. (2010) Structural diversity in integrin/talin interactions. Structure 18, 1654–1666 10.1016/j.str.2010.09.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Anthis N. J., Haling J. R., Oxley C. L., Memo M., Wegener K. L., Lim C. J., Ginsberg M. H., and Campbell I. D. (2009) β integrin tyrosine phosphorylation is a conserved mechanism for regulating talin-induced integrin activation. J. Biol. Chem. 284, 36700–36710 10.1074/jbc.M109.061275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Chuang J. Z., Lin D. C., and Lin S. (1995) Molecular cloning, expression, and mapping of the high affinity actin-capping domain of chicken cardiac tensin. J. Cell Biol. 128, 1095–1109 10.1083/jcb.128.6.1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wegener K. L., Partridge A. W., Han J., Pickford A. R., Liddington R. C., Ginsberg M. H., and Campbell I. D. (2007) Structural basis of integrin activation by talin. Cell 128, 171–182 10.1016/j.cell.2006.10.048 [DOI] [PubMed] [Google Scholar]
- 38. Anthis N. J., Wegener K. L., Ye F., Kim C., Goult B. T., Lowe E. D., Vakonakis I., Bate N., Critchley D. R., Ginsberg M. H., and Campbell I. D. (2009) The structure of an integrin/talin complex reveals the basis of inside-out signal transduction. EMBO J. 28, 3623–3632 10.1038/emboj.2009.287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ling K., Doughman R. L., Iyer V. V., Firestone A. J., Bairstow S. F., Mosher D. F., Schaller M. D., and Anderson R. A. (2003) Tyrosine phosphorylation of type Iγ phosphatidylinositol phosphate kinase by Src regulates an integrin-talin switch. J. Cell Biol. 163, 1339–1349 10.1083/jcb.200310067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Friedland J. C., Lee M. H., and Boettiger D. (2009) Mechanically activated integrin switch controls α5β1 function. Science 323, 642–644 10.1126/science.1168441 [DOI] [PubMed] [Google Scholar]
- 41. Zhu J., Luo B. H., Xiao T., Zhang C., Nishida N., and Springer T. A. (2008) Structure of a complete integrin ectodomain in a physiologic resting state and activation and deactivation by applied forces. Mol. Cell 32, 849–861 10.1016/j.molcel.2008.11.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. O'Halloran T., Molony L., and Burridge K. (1986) Purification and assay of vinculin, metavinculin, and talin. Methods Enzymol. 134, 69–77 10.1016/0076-6879(86)34076-X [DOI] [PubMed] [Google Scholar]
- 43. García-Alvarez B., de Pereda J. M., Calderwood D. A., Ulmer T. S., Critchley D., Campbell I. D., Ginsberg M. H., and Liddington R. C. (2003) Structural determinants of integrin recognition by talin. Mol. Cell 11, 49–58 10.1016/S1097-2765(02)00823-7 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.