Abstract
Acephalic spermatozoa syndrome has been reported for many decades; it is characterized by very few intact spermatozoa and tailless sperm heads in the semen and causes severe male infertility. The only gene in which mutations have been found to be associated with this syndrome encodes Sad1 and UNC84 domain–containing 5 (SUN5), a testis-specific nuclear envelope protein. The functional role of SUN5 has been well-studied in mouse models, but the molecular basis for the pathogenic effects of mutations in the human SUN5 gene remains elusive. Here, we report a new SUN5 mutation (c.475C→T; p.Arg159*), and explore the pathogenic effects of all known SUN5 mutations on acephalic spermatozoa syndrome. Using an artificial splicing system, we found that the intronic mutation affects the splicing of SUN5 mRNA, yielding a premature stop codon that results in a truncated SUN5 protein. We also found that SUN5 interacts with the coupling apparatus protein DnaJ heat shock protein family (Hsp40) member B13 (DNAJB13) during spermatogenesis, and the substitutions in the SUN5 SUN domain impair its interaction with DNAJB13. Furthermore, we observed that many SUN5 mutations affect the secondary structure of the protein and influence its folding and cellular localization. In summary, our findings indicate an interaction of SUN5 with DNAJB13 during spermatogenesis, provide mechanistic insights into the functional role of this interaction in sperm head–tail integration, and elucidate the molecular etiology of acephalic spermatozoa syndrome–associated SUN5 mutations.
Keywords: spermatogenesis, spermatozoa, fertilization, protein folding, pathogenesis, pathology, mutant, alternative splicing, acephalic spermatozoa syndrome, DNAJB13, splicing variant, SUN5 mutation
Introduction
Acephalic spermatozoa syndrome (MIM 617187) is characterized by decapitated flagella, very few intact spermatozoa, and tailless sperm heads in the semen, which leads to severe male infertility (1–5). Acephalic spermatozoa result from impaired spermiogenesis in the testis. The sperm head–tail coupling apparatus (HTCA)4 is disrupted or detached from the sperm head, leading to the separation of the sperm head and the flagellum (6). In the epididymis and semen, more than 90% of the spermatozoa are actually nonfunctional, decapitated flagella, making acephalic spermatozoa syndrome one of the most severe forms of teratozoospermia.
Data from mouse models and acephalic spermatozoa syndrome patients indicate that acephalic spermatozoa syndrome is a result of a genetic disorder. Investigations in mice have found several genes involved in the production of acephalic spermatozoa, including Odf1 (outer dense fiber protein 1) (7), Hook1 (8), Oaz3 (ornithine decarboxylase antizyme 3) (9), and Spata6 (spermatogenesis-associated 6) (10), but none of these mutations were found in acephalic spermatozoa syndrome patients. Sun5 (Sad1 and UNC84 domain-containing 5) is the only gene for which the presence and function were confirmed in a mouse model and also identified as pathogenic mutations in patients. Currently, ten point mutations of SUN5 have been reported, including three non-sense mutations, six missense mutations, and one intron mutation, which may lead to splicing alterations (11). Recently, a homozygous deletion of 5090 bp (including exon 8) in SUN5 was found in three acephalic spermatozoa syndrome patients, predicting the frameshift p.(Leu143Serfs*30) and the inactivation of SUN5 (12). p.(Leu143Serfs*30) denotes a frame shifting change with Leu143 as the first affected amino acid, the new reading frame encoding another 30 alternative amino acids, and then s the stop code.
Although the potential pathogenic mutations have been identified, their effects on SUN5 function remain unknown, and only one mutation, c.340G→A, has been tested experimentally. The 340G→A mutation localizes to the last base on exon 5, predicted to abolish the donor splice site in intron 5. A study reports that the SUN5 exonic variant c.340G→A (p.Gly114Arg) inhibits the splicing of exon 5, causing a frameshift in the SUN5 protein (12).
The discovery of new SUN5 mutations is still under way in the current study, we sequenced SUN5 exons from three unrelated acephalic spermatozoa syndrome patients and found that two patients carried biallelic SUN5 mutations, including an unreported variant. The new SUN5 mutation, identified as c.475C→T (p.Arg159*), resulted in a premature termination codon (PTC) in SUN5 mRNA and increased the number of acephalic spermatozoa syndrome–associated SUN5 mutations to 12.
Few studies have investigated the mechanism by which acephalic spermatozoa are produced, but the Spata6 knockout mouse model provides some clues (10). SPATA6 localizes on HTCA, and the absence of SPATA6 results in the failure of HTCA assembly, which in turn destroys the tight junction between the sperm head and tail. But no SPATA6 mutations are found in patients with acephalic spermatozoa syndrome, so the abovementioned details might not be the actual mechanism that underlies human acephalic spermatozoa syndrome.
To clarify the mechanism by which SUN5 participates in acephalic spermatozoa syndrome, we identified DNAJB13 (DnaJ heat shock protein family (Hsp40) member B13) as a SUN5–interacting protein during spermiogenesis and found that their dynamic interactions are required for the tight junction of the sperm head and tail. Based on these results, we systematically investigated the effects of SUN5 mutations on its function, including the localization, protein secondary structure, and interaction with DNAJB13. Because all of the non-sense mutations resulted in truncated SUN5 lacking the SUN domain, which is required for SUN5–DNAJB13 interaction, we focused our analyses on the remaining six missense mutations, including the one intron mutation. We found that c.425 + 1G→A leads to a partially retained intron 7 in SUN5 mRNA, which introduces a stop codon into the protein translation. Mutations on the coiled-coil domain and SUN domain altered the protein secondary structure and the protein conformation to different extents. Most of the mutations impaired the dynamic interactions between SUN5 and DNAJB13. Taken together, our study reveals mechanistic details of SUN5 mutations in acephalic spermatozoa and provides new insights into the syndrome.
Results
The phenotype and pedigree of the new SUN5 mutation c.475C→T (p.Arg159*)
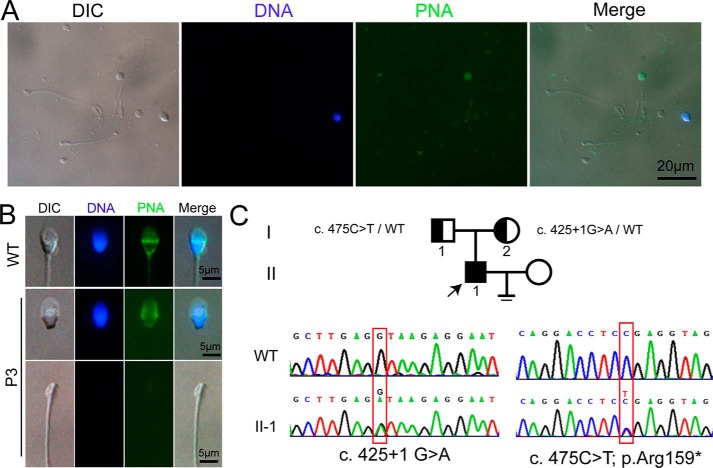
Three infertile men were diagnosed with acephalic spermatozoa syndrome in the Center for Reproductive Medicine in Peking University Third Hospital (Table 1, P1–P3). By sequencing all of the SUN5 exons in the three patients and their parents, we found that two individuals had compound heterozygous SUN5 mutations ((P2: c.781G→A (p.Val261Met), c.1043A→T (p.Asn348Ile); P3: c.425 + 1G→A, c.475C→T (p.Arg159*)). The c.475C→T mutation is an unreported mutation that localizes to exon 8 and leads to a premature stop codon and a truncated form of the SUN5 protein. The spermatozoa in the semen of P3 are typically acephalic spermatozoa (Fig. 1A), and the tailless heads contain nuclei and intact acrosomes (Fig. 1B); no normal spermatozoa were found (Table 1). By analyzing the mutations in the family, we found that the c.425 + 1G→A allele is inherited maternally and the c.475C→T allele is inherited paternally.
Table 1.
Clinical information of the acephalic spermatozoa syndrome patients
Sperm motility and the percentages of morphologically normal and abnormal spermatozoa were evaluated according to the World Health Organization guidelines (26). Most of the SUN5 mutation–associated spermatozoa were acephalic sperm with low motility. PR, progressive motility; NP, non-progressive motility; IM, immotility.
| Patient | Age | Sperm volume | Concentration | Motility PR/NP/IM | Percentage of spermatozoa morphology |
|||
|---|---|---|---|---|---|---|---|---|
| Normally formed | Abnormal head–tail junction | Decaudated | Acephalic | |||||
| years | ml | 106/ml | % | % | % | % | % | |
| P1 | 29 | 3.40 | 2.22 | 0/11.11/88.89 | 0 | 3.03 | 1.51 | 95.45 |
| P2 | 27 | 3.40 | 1.69 | 8.33/8.33/83.33 | 0 | 2.74 | 17.48 | 79.78 |
| P3 | 34 | 2.0 | 41.3 | 19.93/0.80/79.28 | 0 | 0 | 21.05 | 78.95 |
Figure 1.
Morphology and pedigree of P3 and newly identified SUN5 mutation. A, morphology of the spermatozoa from ejaculated semen smear, showing that most of the sperm are headless tails, nuclear DNA was stained with DAPI (blue), and acrosomes were stained with FITC-labeled peanut agglutinin (FITC-PNA, green). DIC, differential interference contrast. B, single sperm showing that the decapitated tail contains no DNA and acrosome, whereas the decaudated sperm head contains DNA and acrosome. C, pedigree of the family of P3 with new SUN5 mutation (c.475C→T; p.Arg159*) is shown. The individuals I-1, I-2, and II-1 were Sanger-sequenced. The mutation c.425 + G→A was reported previously.
The protein status is altered by mutations at the splicing site and the transmembrane domain
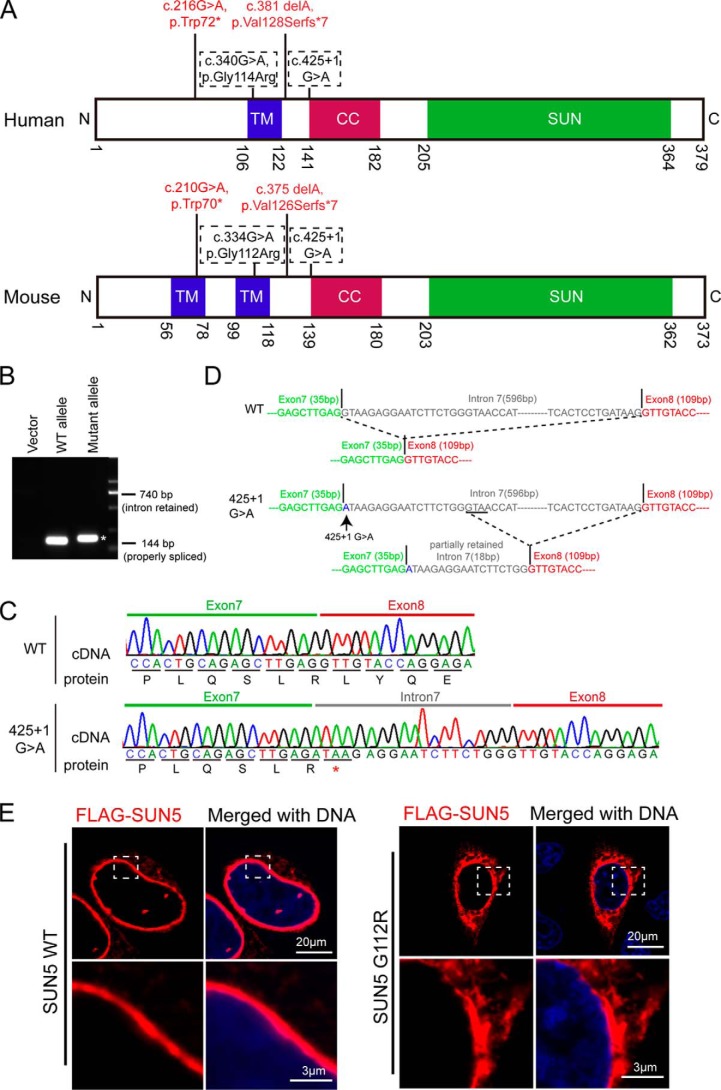
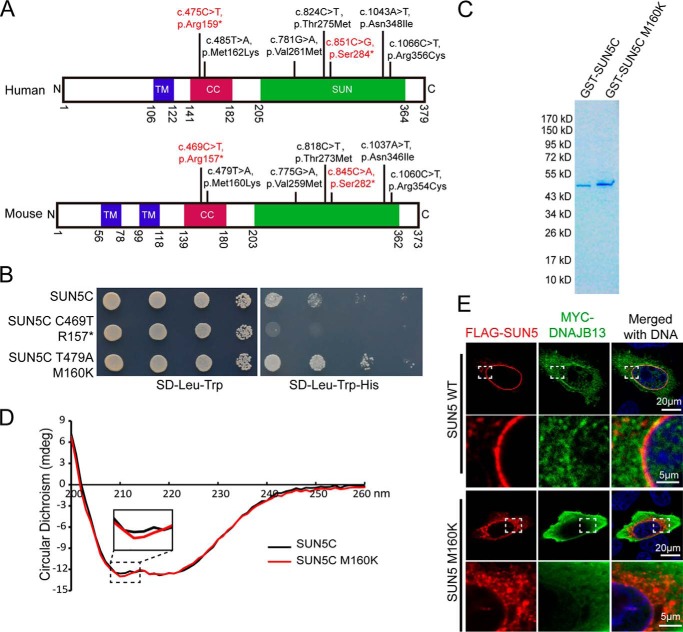
Four mutations of human SUN5 were found in front of the conserved coiled-coil domain and the SUN domain. All of them are evolutionary conserved in mouse, and their corresponding locations in human and mouse SUN5 proteins are shown in Fig. 2A. Among the four mutations, c.210G→A and c.375delA led to truncated SUN5 proteins; c.334G→A was computationally predicted to abolish the donor splice site in intron 5 and confirmed by splicing experiments in cells; and the c.425 + 1G→A splice-site variant was computationally predicted to abolish the donor splice site of intron 7 (11).
Figure 2.
Characteristics of mutations in the splicing site and the transmembrane domain. A, the distribution of SUN5 mutations in the TM domain and splicing sites are shown in the corresponding sites of the human SUN5 and mouse SUN5 proteins. B, the effect of SUN5 425 + 1G→A mutation on mRNA splicing in F9 cells, showing that the SUN5 425 + 1G→A mutation yielded spliced transcripts that were a bit larger than that of the WT. The asterisk indicates the mutant band. C, the sequence of spliced SUN5 mRNA in F9 cells, showing that 18 bp of intron 7 was retained in the 425 + 1G→A mutant transcripts, which introduced a stop codon in SUN5 mRNA. D, the proposed splicing mechanism of the c.425 + 1G→A mutant. When encountering c.425 + 1G→A, the splicing machinery might skip to a similar splicing site near the 425 + 1G site. E, the G112R mutation leads to an altered distribution of full-length SUN5 in mammalian cells. FLAG-tagged full-length WT or G112R SUN5 was transfected into HeLa cells. After a 24-h incubation, immunofluorescence staining was performed using an anti-FLAG antibody (red) to examine the distribution of the overexpressed proteins. The boxed areas have been enlarged in the lower row. DNA was stained with DAPI (blue). WT FLAG–SUN5 was used as a control.
We first tested a splicing variant with an unknown function, c.425 + 1G→A, which localized to the first base of intron 7 next to exon 7 and was identified in P3 of our cohort. We amplified a fragment from exon 7 to exon 8 from the genomic DNA of P3 and subcloned the fragment into a pRK vector. The forward primer is designed on exon 7 and the reverse on exon 8. Because P3 contains a heterozygous mutation of 425 + 1G→A, we obtained both the WT and mutant allele with the cloning. We then transfected the vectors carrying the WT or mutant allele to a mouse testicular teratoma cell line (F9 cells; CRL-1720, ATCC). mRNA was extracted, reverse-transcribed to cDNA, and amplified using the primers mentioned above. If intron 7 was properly spliced, the amplified fragment would be 144 bp (exon 7 + exon 8), but if intron 7 was fully retained during the splicing, the amplified fragment would be 740 bp (exon 7 + intron 7 + exon 8). Interestingly, we did not get the 740-bp fragment with the mutant allele transcripts but instead obtained a fragment that was a bit larger than the WT allele (Fig. 2B). By sequencing the amplified fragment, we detected that intron 7 was not completely spliced and retained an 18-bp fragment within the mutant transcript. The inserted 18-bp fragment introduced a premature stop codon in SUN5 mRNA and resulted in a truncated SUN5 protein ending at Arg-142 (Fig. 2C). Mechanistically, we propose that when encountering the 425 + 1G→A mutation, the splicing machinery might skip to a similar splicing site near the 425 + 1G site (Fig. 2D).
With c.334G localized on the transmembrane domain (TM) of SUN5, if c.340G→A does not cause a splicing problem; the resulting missense change, p.Gly112Arg, is computationally predicted to severely impair the transmembrane helix of SUN5 (11). To test this possibility, we cloned mouse SUN5 from testis tissues and fused a FLAG tag at the N terminus. We then introduced the mutation Gly112Arg (SUN5 G112R) and overexpressed the recombinant protein in HeLa cells, because this kind of cell spreads well with a flattened morphology, making the protein more easily observed. Using immunostaining of the FLAG tag, we found that WT SUN5 was restricted to the nuclear envelope (NE), whereas a majority of SUN5 G112R “escaped” to the cytosol with some evidence of protein aggregation near the NE (Fig. 2E). These results indicate that both the truncated protein and mutations in the TM domain leads to a dysfunctional SUN5 protein.
SUN5 interacts with DNAJB13 at the coupling apparatus during spermiogenesis
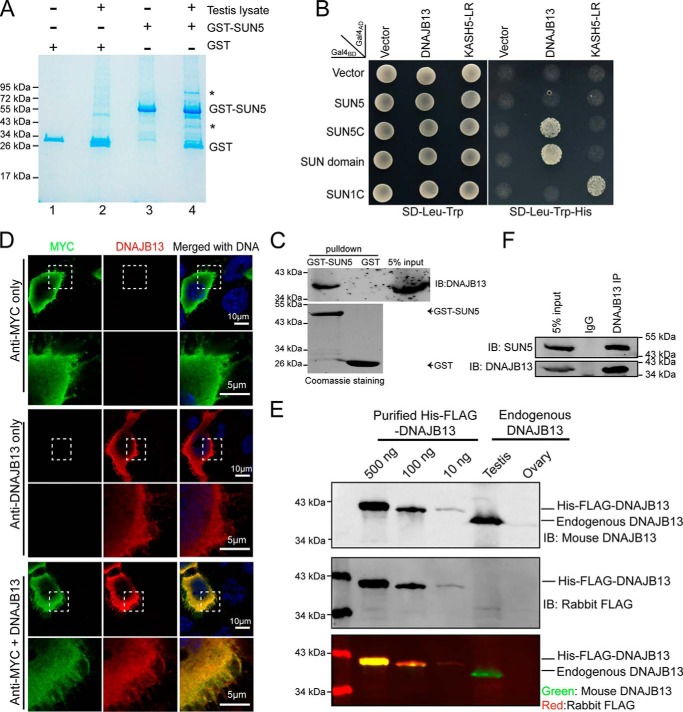
Because the absence of SUN5 leads to a separation of the coupling apparatus from the sperm nucleus, we considered that an interactor or a partner of SUN5 might exist in the coupling apparatus. Using a GST-fused mouse SUN5 protein (glutathione S-transferase fused to the SUN domain of the SUN5 protein hereafter refers to GST–SUN5), we employed a GST pulldown assay to identify SUN5 interactor(s) from mouse testis lysate. Two protein bands were resolved on gel electrophoresis (Fig. 3A, lane 4) and subjected to LC-MS analysis, and 15 and 37 proteins, respectively, were identified by MS (Table 2 and File S1). Gene ontology analysis showed that only one protein, DNAJB13 (from the bottom band in Fig. 3A), was annotated as related to “sperm flagellum,” “axoneme,” or “motile cilium” (File S1). DNAJB13 was also the most abundantly identified testis-specific protein according to the absolute quantitation value obtained by intensity-based absolute quantification (iBAQ) (blank gels at the corresponding molecular weight (Fig. 3A, lane 2) were used as control). We then chose to analyze DNAJB13 using the yeast two-hybrid system (Y2H) to validate the proteomic findings. As reported previously, we used SUN1C and KASH5 LR as positive controls (13) and found that both SUN5C (coiled-coil domain and the SUN domain of SUN5) and the SUN domain of SUN5 interacted with DNAJB13 but not the other candidates (Fig. 3B). Full-length SUN5 did not interact with DNAJB13 in the yeast two-hybrid assay, most likely because it contains a transmembrane domain. The domain may influence the binding capability of the protein, similar to the situation with SUN1 and SUN2 (13).
Figure 3.
SUN5 directly interacts with DNAJB13. A, GST pulldown of SUN5-interacting proteins in testes. The SUN domain of mouse SUN5 was purified and immobilized on GST–beads and then incubated with testis lysate. The eluates were resolved by SDS-PAGE and stained using a Colloidal Blue staining kit. The asterisks indicate the bands that were specifically pulled down by GST–SUN5 beads and retrieved for analysis by mass spectrometry. GST protein was used as a negative control. B, SUN5 interacted with DNAJB13 in the yeast two-hybrid assay. SUN5C (coiled-coil domain and the SUN domain of SUN5) and the SUN domain, instead of full-length SUN5, interacted with DNAJB13 in the yeast two-hybrid assay. SUN1C and KASH5 LR were used as a positive control. Empty vector was used as a negative control. C, GST–SUN5 pulldown results were confirmed with immunoblotting of endogenous DNAJB13. GST pulldown was repeated as in A, but the eluates were analyzed with immunoblotting against DNAJB13 following SDS-PAGE. The analysis showed that DNAJB13 could be detected in the GST–SUN5 pulldown lane but not in the GST alone lane. D, mouse polyclonal DNAJB13 antibody detected overexpressed MYC–DNAJB13 and co-localized with MYC signals in HeLa cells. MYC–DNAJB13 was overexpressed in HeLa cells, and immunofluorescence staining was performed using rabbit anti-MYC antibody (green), mouse anti-DNAJB13 antibody (red), or a combination of the two antibodies, DNA was stained with DAPI (blue). The DNAJB13 signal was detected only in the overexpressed cells (middle panel) and overlapped with the MYC signal, indicating the specificity of murine polyclonal DNAJB13 antibody. The boxed areas have been enlarged in the lower row. E, murine polyclonal DNAJB13 antibody detected purified His—FLAG–DNAJB13 and endogenous DNAJB13 in the testis. 500, 100, and 10 ng of purified His—FLAG–DNAJB13 and 20 μg of adult testis and ovary extracts were loaded and detected by murine DNAJB13 antibody and rabbit FLAG antibody. Ovary extract was used as a negative control. FLAG antibody was used as a positive control for immunoblotting. F, SUN5 bound DNAJB13 in the testis. Endogenous co-immunoprecipitation of SUN5 and DNAJB13 from testis lysate was performed with murine DNAJB13 antibody, and SUN5 was detected with rabbit anti-SUN5 antibody.
Table 2.
Proteins identified by MS analysis in GST–SUN5 pulldown assay
iBAQ and gene ontology analysis of the listed proteins are available in File S1.
| Top band | UBXN4, DLD, PTPN9, NCL, PDHA2, NPLOC4, STXBP3, STIP1, CLPX, CDKN2AIP, PDHX, CKAP4, NARS, KLHL10, VNN1, |
| Bottom band | NDUFA9, SYNCRIP, 4931406C07RIK, RALY, HNRPDL, COPS6, CLU, GM5428, CSNK1A1, AKR1CL, DLD, DSCR3, SRSF5, COPS5, HMOX2, MDH2, PDHA1, DBT, RPS6, RPS3, FRZB, PPP4C, NUDT18, CRYZL2, EIF2S1, DNAJB13, PDHX, DLAT, TXNL1, GTF2H3, PTGR1, PDK3, MARC2, DHRS1, PRPS2, BLVRA, AGPAT3 |
The interaction between SUN5 and DNAJB13 was further confirmed with immunoblotting following GST pulldown (Fig. 3C). To investigate the physiological interaction between SUN5 and DNAJB13, we generated a murine DNAJB13–specific polyclonal antibody and validated its specificity (Fig. 3, D and E). Next, the interaction between SUN5 and DNAJB13 was confirmed with immunoprecipitation of endogenous SUN5 by DNAJB13 antibody in mouse testis lysate (Fig. 3F).
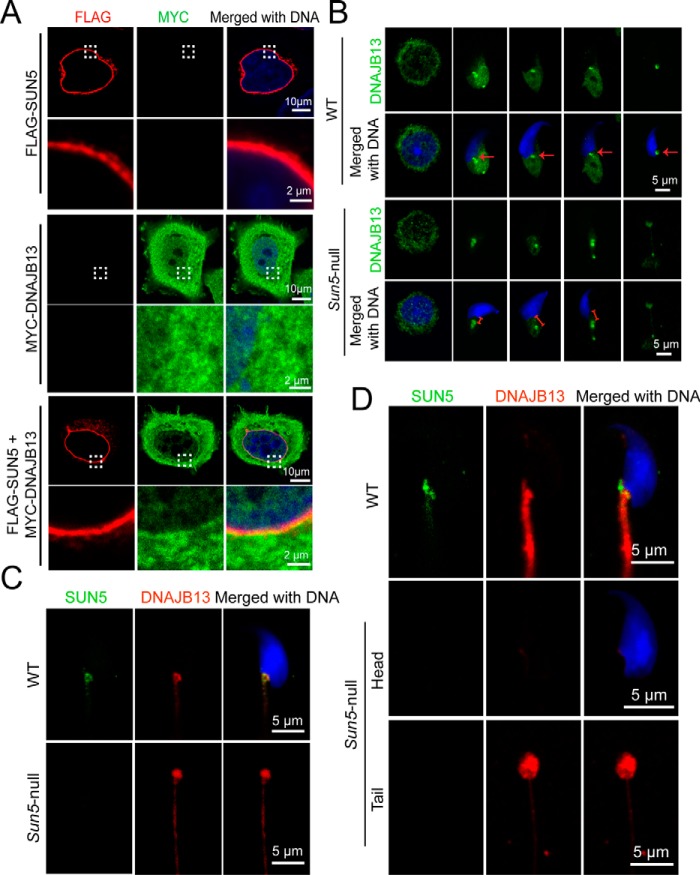
Using immunofluorescence assays, we determined that overexpressed FLAG–SUN5 could recruit MYC–DNAJB13 to the nuclear envelope, whereas MYC–DNAJB13 itself could not be recruited to the nuclear envelope (Fig. 4A). Next, we set out to characterize the dynamic interaction between SUN5 and DNAJB13 during spermiogenesis using immunofluorescence. DNAJB13, a member of the type II HSP40 (heat shock protein 40) family, is defined as an axoneme-associated component in spermatozoa, and subsequent studies found that DNAJB13 is localized on the sperm head–tail coupling apparatus during spermiogenesis (14–16). Additionally, biallelic Dnajb13 mutant mice have been shown to produce abnormal spermatozoa, including headless tails and tailless heads (17). These data support the idea that the interaction between SUN5 and DNAJB13 might be required for the sperm head-to-tail tight junction. We further examined the dynamic distribution of DNAJB13 in developing spermatids. In WT spermatids, DNAJB13 was rapidly enriched in the coupling apparatus with the elongation of the spermatid, and the protein was tightly attached to the implantation fossa during the maturation of the spermatid (Fig. 4B, second row, arrows), which is similar to the distribution of SUN5. In addition, in Sun5-null spermatids, although DNAJB13 was enriched to the coupling apparatus, its tight association with the nucleus was never observed. Instead, there was a gap between the nucleus and DNAJB13 (indicated by the red segment in Sun5-null spermatid in Fig. 4B, bottom row). Finally, in the late-stage spermatid, DNAJB13 was found only in the headless tail spermatozoa (Fig. 4B, bottom row). In WT spermatids, co-localization of SUN5 and DNAJB13 in the sperm head-to-tail coupling apparatus was observed, but in Sun5-null late spermatids (6), the DNAJB13 signal could only be found at the top of headless tails (Fig. 4C). A previous investigation reported that DNAJB13 is localized to the neck region of developing spermatids in testis but the protein migrates to the flagellum in the mature spermatozoa (16). To test this idea, we performed similar immunofluorescence staining of DNAJB13 together with SUN5 in mature spermatozoa from epididymis. We found that in WT sperm, SUN5 was localized to the neck region and DNAJB13 was evenly distributed along the flagellum; the DNAJB13 signals were not enriched in the neck region. In Sun5-null sperms, SUN5 was absent from both the head and the tail, and DNAJB13 was found only in the headless tails (Fig. 4D). These observations suggested that although SUN5 is associated with DNAJB13 during the development of spermatids inside the testis, DNAJB13 might not be the last partner of SUN5 to anchor the sperm head to the tail, because DNAJB13 migrates from the neck region to the tail in mature sperms. Considering that DNAJB13 is a chaperone for protein folding, the dynamic association between SUN5 and DNAJB13 might help to facilitate the proper folding of the SUN5 protein and promote its binding to the right partner in the HTCA in mature sperms.
Figure 4.
Dynamic interaction between SUN5 and DNAJB13 during spermiogenesis. A, FLAG–SUN5 partially recruited MYC–DNAJB13 to the nuclear envelope when overexpressed in HeLa cells. FLAG–SUN5 localized to the nuclear envelope (red, upper panel). MYC–DNAJB13 randomly distributed itself throughout the cell when overexpressed in HeLa cells (green, middle panel). When FLAG–SUN5 and MYC–DNAJB13 were co-transfected in HeLa cells, MYC–DNAJB13 was partially recruited to the nuclear envelope (bottom panel). DNA in the cells was stained with DAPI (blue). B, dynamic distribution of DNAJB13 in developing WT and Sun5-null spermatids was revealed by IF. In WT spermatids, DNAJB13 signals (green) were tightly attached to the implantation fossa during the maturation of the spermatid. The red arrow in the WT spermatids indicate the implantation fossa (upper two panels). In Sun5-null spermatids, although DNAJB13 (green) was enriched to the coupling apparatus, DNAJB13 signals were absent from the spermatid nucleus (lower two panels). The red segment in the Sun5-null spermatid indicates a gap between nucleus and DNAJB13. The spermatid nuclear DNA was stained with DAPI (blue). C, co-localization of SUN5 and DNAJB13 in WT and Sun5-null spermatids from testis. In WT spermatids, co-localization of SUN5 (green) and DNAJB13 (red) in the sperm head-to-tail coupling apparatus was observed (upper panel), but in Sun5-null late spermatids, DNAJB13 signal could be found only at the top of headless tails (lower panel). The spermatid nuclear DNA was stained with DAPI (blue). D, distribution of SUN5 and DNAJB13 in mature spermatozoa from epididymis. In WT mature spermatozoa, DNAJB13 (red) migrated to the flagella, and SUN5 (green) stayed in the HTCA (upper panel). In Sun5-null mature spermatozoa, DNAJB13 could be found only in the tip of the acephalic flagella (lower two panels). The sperm nuclear DNA was stained with DAPI (blue).
Mutations in the coiled-coil domain affect the secondary structure of the SUN5 protein
The last five mutations and the newly found c.475C→T were all localized to the conserved coiled-coil domain and the SUN domain. All were evolutionary conserved in the mouse (Fig. 5A). Two mutations were found in the coiled-coil domain of the human SUN5 protein, c.475C→T (p.Arg159*) and c.485T→A (p.Met162Lys), which correspond to mouse c.469C→T (p.Arg157*) and c.479T→A (Met160Lys). The c.469C→T (p.Arg157*) led to a truncated protein and disrupted the coiled-coil SUN domain. We first tested the ability of the mutant proteins to bind DNAJB13 in the Y2H assay and found that the truncation at Arg-157 abolished the interaction of the protein with DNAJB13 (Fig. 5B), as predicted. On the other hand, the M160K mutation did not influence the SUN5C–DNAJB13 interaction. On the contrary, this mutation seemed to enhance the interaction. To reveal the differences in the protein properties between the mutant and WT SUN5C, we purified GST-tagged WT mouse SUN5C (coiled-coil and SUN domain) and the mutant (SUN5C M160K) (Fig. 5C) and then examined their secondary structure by circular dichroism (CD). We found that the secondary structure of SUN5C is altered by the mutation M160K, particularly in the α-helix region (Fig. 5D).
Figure 5.
The secondary structure of the SUN5 protein was altered by mutations in the coiled-coil domain. A, the mutations in the coiled-coil domain and the SUN domain were observed in human and mouse SUN5, showing that these mutations are highly conserved. Due to codon degeneracy, human Ser-284 is coded by TCA, whereas mouse Ser-282 is coded by TCG. Consistent with the human non-sense mutation, 851C→G (TCA-TGA), the corresponding nucleotide (845C) in mouse was mutated to A (TCG-TAG). Other mutations in mouse were consistent with those found in human. B, influence of two mutations in the coiled-coil domain on the SUN5–DNAJB13 interaction as revealed by the Y2H assay. WT SUN5C or its variants were co-transformed with DNAJB13 in the Y2H assay, and yeast cells were diluted by 101, 102, and 103. Aliquots were spotted onto control (SD-Leu-Trp) and selective (SD-Leu-Trp-His) plates. C, protein purification of GST–SUN5C and its variant. The samples were separated by SDS-PAGE and stained with Coomassie Blue. D, the secondary structure of SUN5C was altered by M160K as revealed by CD analysis. E, the NE localization of full-length SUN5 in HeLa cells was impaired by the M160K mutation. WT–FLAG–SUN5 or FLAG–SUN5 M160K, together with MYC–DNAJB13, were co-transfected into HeLa cells. After a 24-h incubation, the transfected cells were immunostained with mouse anti-FLAG (red) and rabbit anti-MYC (green) antibodies. The nuclear DNA was stained with DAPI (blue). WT–FLAG–SUN5 localized to the NE, and DNAJB13 was partially recruited to the NE (upper panels), whereas FLAG–SUN5 M160K showed abnormal localization and could not recruit DNAJB13 to the NE (lower panels). The boxed areas have been enlarged in the lower row.
Finally, we co-expressed the full-length FLAG–SUN5 and MYC–DNAJB13 in mammalian cells to examine their properties in vivo. Our studies revealed that WT SUN5 was localized on the NE and partially recruited DNAJB13 to the NE, but the distribution of SUN5 R160K was not as restricted as WT SUN5. Many of the SUN5 R160K signals were found in the cytosol and localized to a protein aggregate. In addition, the mutant lost its ability to recruit DNAJB13 to the NE (Fig. 5E).
Mutations in the SUN domain impaired SUN5 protein folding and SUN5–DNAJB13 interactions
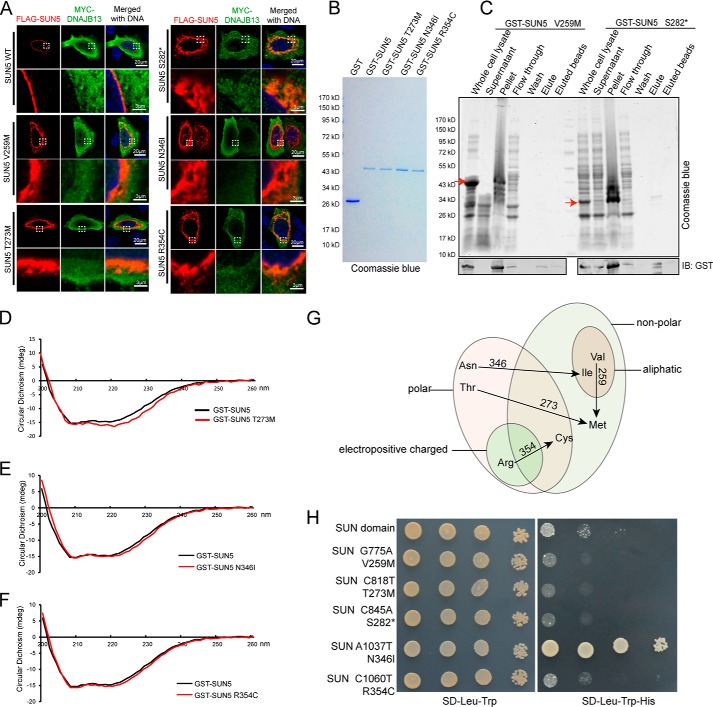
The results of the M160K mutation show that the in vivo status might reflect the protein property faithfully, and so we introduced the four missense and one non-sense mutation identified in the SUN domain into full-length SUN5 and transfected them into HeLa cells with DNAJB13, respectively. We found that each mutation caused an abnormal distribution of SUN5 inside the cells, and their ability to recruit DNAJB13 was also impaired (Fig. 6A). These results suggested that the protein folding of these mutants may also be impaired, and so we attempted to purify Escherichia coli-expressed GST-tagged SUN domain proteins carrying each of the mutations. Most of the mutants could be purified successfully (Fig. 6B), but to our surprise, the Y259M and S282* mutations led to protein precipitation, suggesting that these mutations brought striking changes to protein conformation and yielded misfolded proteins (Fig. 6C).
Figure 6.
Mutations in the SUN domain affected protein secondary structures, localization, and interaction with DNAJB13. A, mutations in the SUN domain affected the distribution of full-length SUN5 in mammalian cells and recruitment of DNAJB13. WT–FLAG–SUN5 or its variants, together with MYC–DNAJB13, were co-transfected into HeLa cells. After a 24-h incubation, the transfected cells were immunostained with anti-FLAG (red) and anti-MYC (green) antibodies. The nuclear DNA was stained with DAPI (blue). WT–FLAG–SUN5 localized to the NE, and DNAJB13 was partially recruited to the NE. FLAG–SUN5 V259M, T273M, S282*, N346I, and R354C showed abnormal localization and could not recruit DNAJB13 to the NE. The boxed areas have been enlarged in the lower row. B, GST–SUN5 and its variants. GST–SUN5 and its variant proteins were expressed in E. coli and purified by GST–beads. They were separated using SDS-PAGE and stained with Coomassie Blue. C, the mutations V259M and S282* led to protein precipitation during in vitro purification. The samples were separated by SDS-PAGE and stained with Coomassie Blue. D–F, the secondary structure of the GST–SUN5 protein was affected by incorporated mutations T273M, N346I, and R354C, as revealed by CD analysis. G, all of the SUN domain mutations caused striking changes to amino acid properties. H, most mutations in the SUN domain impaired SUN5–DNAJB13 interaction as characterized by the Y2H system. WT SUN (SUN domain of SUN5) or its variants were co-transformed with DNAJB13 in the Y2H assay, and yeast cells were diluted by 101, 102, and 103 and then spotted onto control (SD-Leu-Trp) and selective (SD-Leu-Trp-His) plates.
For the purified mutants, we examined their secondary structure by CD and found that each mutation caused changes in the secondary structure at different levels, especially in the β-sheet region compared with the GST–SUN5 WT protein (Fig. 6, D–F). These changes are presumably due to changes in the amino acid properties, such as from nonpolar to polar or from charged to uncharged state (Fig. 6G). We then introduced the mutants into the Y2H system and monitored their interaction with DNAJB13. As expected, most SUN mutants showed reduced interaction with DNAJB13 (Fig. 6H), with only one exception. Importantly, the c.1037A→T, p.Asn346Ile (N346I) mutant showed an enhanced interaction with DNAJB13. These results suggest that the dynamic interaction between SUN5 and DNAJB13 is required for spermiogenesis and that the disruption of any step of the binding–releasing process might be harmful to sperm head–tail tight junction.
Discussion
Most of our knowledge about acephalic spermatozoa was obtained from animal models. SUN5 is the first gene for which the function has been validated in both an animal model and acephalic spermatozoa syndrome patients. Because species variation exists for many genes and proteins, we aligned the protein sequences of SUN5 and its homologues in other mammals and found that they are evolutionary conserved from mouse to human (6). In a previous patient cohort, we found that 47.06% of the patients had SUN5 mutations, and we found that two of three patients carried biallelic SUN5 mutations and one patient showed a novel SUN5 mutation. These results suggest that acephalic spermatozoa syndrome is associated with a high frequency of SUN5 mutations. Among the 11 known point mutations, six led to premature stop codons and truncated proteins, among which five localized upstream of the conserved coiled-coil domain and only one mutation was found inside the SUN domain. In summary, these non-sense mutations are highly harmful to protein function. In the current study, we investigated the effects of missense mutations on the function of SUN5 and found that the substituted amino acids changed either the protein conformation or its distribution in cells, influencing the dynamic interactions of the protein with DNAJB13. Because all these mutations in the patients are evolutionarily conserved in the mouse, we believe that the mouse results also apply to human spermatid development.
SUN5 itself cannot bring the sperm head and tail together to form a tight junction; it must interact with another or other partners. We identified the novel SUN5 interactor as DNAJB13. From the beginning of the spermatid elongation, we observed that SUN5 and DNAJB13 were co-localized to the neck region of spermatids. DNAJB13 knockout has been shown to result in acephalic spermatozoa in mice (17), and mutations of DNAJB13 have been implicated in human ciliary dyskinesia and male infertility (18). These findings suggest that DNAJB13 plays a vital role in spermiogenesis, and the direct interaction of the protein with SUN5 may be essential to sperm head–tail integration. We found that when the elongation is completed in mature spermatozoa, DNAJB13 is no longer enriched in the neck region, but the tight junction between the sperm head and tail already has been established based on SUN5. This result suggests that there must be another interactor with SUN5 in the mature spermatozoa, and mutations of this interactor are predicted to be associated with acephalic spermatozoa syndrome.
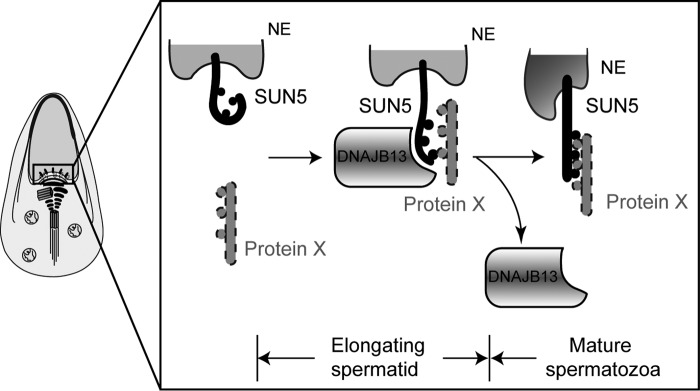
We noticed that most of the mutations reduced the interaction between SUN5 and DNAJB13, but two mutations, M160K and N346I, strengthened the binding of SUN5 to DNAJB13, which also led to acephalic spermatozoa. This implies that both overly weak and overly strong interactions between SUN5 and DNAJB13 are harmful to protein function. A weak interaction between SUN5 and DNAJB13 may not be sufficient to provide enough force to pull the nuclear envelope and HTCA together during elongation, whereas a strong binding between the two proteins might inhibit the release of DNAJB13 after the elongation of the spermatids, thus preventing the continuous functioning of DNAJB13 in mature spermatozoa. We speculated that during the complete process of spermiogenesis, DNAJB13 might serve as a chaperone as well as an interactor in the sperm head-to-tail anchoring mechanism. Considering the dynamic interaction between SUN5 and DNAJB13 during spermiogenesis, we suggest that DNAJB13 might be a protein chaperone that assures SUN5 folds properly and binds to another protein in the HTCA. As a working model for SUN5–DNAJB13 and the potential SUN5 interactor, we propose that SUN5, localized to the nuclear envelope, needs DNAJB13 to help its folding and to facilitate its binding to a “protein X” during spermatid elongation. In mature spermatozoa, the well-folded SUN5 will build a solid interaction with protein X to anchor the sperm head to the tail. Once DNAJB13 finishes its mission, it is released from the neck region (Fig. 7).
Figure 7.
Potential functional roles of DNAJB13 and SUN5 during spermiogenesis. SUN5 interacts with DNAJB13 in the elongating spermatids, and DNAJB13 facilitated SUN5 protein folding to promote its binding to an unknown protein X during spermatid elongation. In mature spermatozoa, the well-folded SUN5 will build a solid interaction with protein X to anchor the sperm head to the tail. Meanwhile, DNAJB13 is released from the neck region.
In summary, our study identified the functional interactor of SUN5 during spermatogenesis, DNAJB13, and explored the pathogenesis of SUN5 mutations recently found in acephalic spermatozoa syndrome patients (11). The splicing variant leads to a premature stop codon, yielding a truncated SUN5 protein that lacks the essential SUN domain. We also characterized the missense mutations that affected SUN5 protein folding, localization, or interaction with DNAJB13. Together, our studies provide mechanistic insights into the functional role of SUN5 in connecting the sperm head to the tail and the etiology of acephalic spermatozoa syndrome–associated SUN5 mutations.
Experimental procedures
Patients
This study conforms to the principles of the “Declaration of Helsinki” and was approved by the Reproductive Study Ethics Committee of Peking University Third Hospital (Reference No. 201552-008). Three patients gave informed consent for their samples to be used for research, to identify the genetic cause of their infertility. The data were treated confidentially and anonymized. Related examinations of these samples are listed in Table 1. Amplification and sequencing of SUN5 exons for the patients were performed as described (11).
Animals
The Sun5-null mice and WT mice were kept as described (6), and all of the animal experiments were performed with the approval of the Institutional Animal Care and Use Committee (IACUC) of the Institute of Zoology, Chinese Academy of Sciences.
Plasmids
Full-length mouse Sun5 was amplified and subcloned into pRK using SalI and NotI. Mouse SUN5 (full-length), SUN5C (residue 133–374), the SUN domain (residue 226–374), and SUNIC (residues 458–913) were subcloned into pGBT9 using EcoRI and SalI. SUN5C and SUN domain were also cloned into pGEX-4T-1 for protein purification using EcoRI and SalI. Mouse Dnajb13 was cloned into pCS2+ using EcoRI and AscI. Mutations of SUN5 were generated by site-directed mutagenesis.
Antibodies
The rabbit DNAJB13 antibody has been described previously (16). The mouse DNAJB13 antibody was generated in our laboratory. Briefly, FLAG-tagged full-length Dnajb13 was inserted into pET28a+, and the His–FLAG-tagged protein was purified using nickel–beads (GE Healthcare) in purification buffer (50 mm Tris-HCl, pH 8.0, 150 mm NaCl, 2 mm MgCl2, and 5% glycerol). The recombinant protein was dialyzed in PBS and used to immunize mice for 6 weeks. The antisera against DNAJB13 was recovered from mouse blood and used in the following experiments. Rabbit anti-MYC antibody (ab1027t) was purchased from Boaoruijing (Beijing, China), mouse anti-FLAG antibody (M20008) was purchased from Abmart (Berkeley Heights, NJ), and rabbit anti-FLAG antibody (PM020) was purchased from MBL International (Aichi, Japan).
Testis smear
The indicated mice (8 weeks old) were euthanized by cervical dislocation. The testes were removed surgically, and the tunica albuginea was removed from the testes. Then, the testes were digested with 1 mg/ml collagenase and 1 mg/ml hyaluronidase. Cells were dissociated by gentle pipetting, filtered through a 70-μm filter, and then pelleted by centrifugation at 500 × g for 10 min. Cells were suspended in 1 ml of PBS (Gibco, C14190500BT), fixed with 4% paraformaldehyde solution, washed with PBS, and finally spread onto polylysine-coated slides for staining.
Immunofluorescence (IF)
The immunofluorescence and immunohistochemical assays were performed as described previously (19). For IF staining of human sperm, semen spread on polylysine-coated slides was air-dried and then fixed with 4% paraformaldehyde and stained with FITC-peanut agglutinin for acrosomes and DAPI for DNA. For cellular immunofluorescence, 1 × 105 HeLa cells were plated on glass coverslips in a 35-mm dish and cultured in DMEM with 10% fetal bovine serum for 24 h. The cells were transfected with the corresponding plasmids for 24 h and then rinsed three times with PBS, fixed, and stained with the corresponding primary and secondary antibodies. To test the functions of SUN5 mutations, WT FLAG–SUN5 was transfected as a control group. For IF staining of the testis smear, the slides was rinsed three times with PBS, treated with 0.5% Triton X-100, blocked with 5% BSA, and incubated with the corresponding antibodies overnight. The signals were detected using the corresponding secondary antibodies. The testis smears from the WT mice were used as a control group in these experiments. The IF images were taken immediately using an LSM 780/710 microscope (Zeiss, Oberkochen, Germany) or SP8 microscope (Leica, Wetzlar, Germany).
mRNA splicing assay
The fragment containing exon 7–intron 7–exon 8 from genomic DNA of P3 was subcloned into a pRK vector. The forward primer was found on exon 7 (5′-GCGTCGACCGATGACAGCATAAATGGT-3′) and the reverse primer on exon 8 (5′-TTGCGGCCGCCTCATCGGACATGGCTTCCA-3′). For the P3 patient with a heterozygous mutation of 425 + 1G→A, we could get both WT and mutant allele during the cloning. The vectors carrying the WT and mutant allele were then transfected to 293T cells, and mRNA was extracted and reverse-transcribed to cDNA. The spliced cDNA was amplified using the primers designated above, and PCR product were analyzed by 2% agarose gel and subsequent sequencing.
GST pulldown assay
The SUN domain (residues 226–374) of the SUN5 protein was cloned into a pET42a vector and expressed in E. coli. A 20-μg quantity of GST–SUN5 or GST protein was immobilized on glutathione–Sepharose 4B (GE Healthcare) in immunoprecipitation buffer (50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 2 mm MgCl2, and 0.1% Triton X-100) with 0.1% BSA and 1 mm phenylmethylsulfonyl fluoride (M145, Amresco, Solon, OH). WT mouse testes were lysed in radioimmune precipitation assay buffer with 1 mm phenylmethylsulphonyl fluoride (Amresco, M145, Solon, OH) and a proteinase inhibitor mixture (No. 04693132001, Roche Diagnostics) for 30 min on ice and then cleared twice at 13,200 × g for 15 min at 4 °C. Then, the supernatant was diluted with an equal volume of IP buffer and then cleaned by incubating with GST–beads to remove endogenous GST proteins. The cleaned testes extract was incubated with immobilized GST or GST–SUN5 overnight, and then the beads were washed with wash buffer (25 mm Tris-HCl, pH 7.5, 2 mm EGTA, and 0.1% Nonidet P-40) three times. After the addition of SDS loading buffer, the samples were separated by SDS-PAGE and stained using a Colloidal Blue staining kit (LC6025, Thermo Fisher Scientific).
Liquid chromatography–mass spectrometry analysis
The specific bands from the Colloidal Blue-stained gel were excised, denatured, alkylated, and trypsin-digested using previously described methods (20). The digested peptides were desalted with StageTip (Thermo Fisher Scientific) and analyzed by LTQ Orbitrap Velos mass spectrometer (Thermo Finnigan). The MS/MS spectra were searched against the UniProt protein database using MaxQuant version 1.3.0.5 for protein identification, with a false discovery rate of 1% for both peptides and proteins (21). Three replicates were performed, and for label-free quantification, the protein expression levels were estimated using the iBAQ algorithm embedded in MaxQuant (22). Any protein with more than two unique peptide reads, uniquely identified in the GST pulldown lane with statistical difference (Student's t test, p < 0.01), was considered for analysis. Gene ontology annotation was performed using DAVID Bioinformatics Resources 6.8 (23). Testis-specific gene annotation was from previously published data (24). The MS proteomics data of GST-pulldown in this study have been deposited in the ProteomeXchange dataset via the PRIDE partner repository under identifier PXD007815.
Yeast two-hybrid screen
Yeast two-hybrid screening was performed as described previously (25). Mouse SUN5 (full-length), SUN5C (residues 133–374), the SUN domain (residues 226–374), and SUNIC (residues 458–913) were subcloned into pGBT9 as bait. Mouse Dnajb13 and KASH LR (residues 627–648) were subcloned into pGAD424 as prey. The bait and prey plasmids were co-transformed into PJ69-4a and selected on an SD-Leu-Trp-His plate.
Immunoprecipitation
Testes were lysed with radioimmune precipitation assay buffer and cleared followed by diluting them with IP buffer as described above. The supernatant was incubated overnight with mouse anti-DNAJB13 serum or normal mouse serum before immunization. Then the antibodies were immobilized on protein A–beads (GE Healthcare) for 4 h. After the washing with the wash buffer mentioned above, SDS loading buffer was added, and then the samples were separated by SDS-PAGE and immunoblotted with rabbit anti-SUN5 and mouse anti-DNAJB13 antibodies.
Circular dichroism spectroscopy
To determine the secondary structure of GST–SUN5C, GST–SUN5, and their variants, the CD spectra of the proteins were taken at 0.1 mg/ml in 50 mm Tris-HCl, pH 8.0, 150 mm NaCl, 2 mm MgCl2, and 5% glycerol using a Chirascan Plus CD spectrometer (Applied Photophysics, Surrey, UK) and a 1-mm quartz sample cell equilibrated at 20 °C. Scans between 200 and 260 nm were performed at a scan rate of 50 nm/min.
Author contributions
Y. S., J. Y., W. T., S. X., L. Y., L. C., H. J., J. Q., and W. L. resources; Y. S., J. Y., W. T., C. L., S. X., and J. Q. data curation; Y. S., C. L., S. X., Y. G., and X. G. software; Y. S., J. Y., W. T., and W. L. formal analysis; Y. S. validation; Y. S., J. Y., W. T., C. L., S. X., Y. G., L. Y., L. C., H. J., X. G., J. Q., and W. L. investigation; Y. S., L. Y., L. C., and H. J. visualization; Y. S., J. Y., W. T., C. L., S. X., Y. G., L. Y., L. C., H. J., X. G., J. Q., and W. L. methodology; Y. S., J. Y., W. T., J. Q., and W. L. writing-original draft; J. Q. and W. L. supervision; J. Q. and W. L. funding acquisition; J. Q. and W. L. writing-review and editing; W. L. project administration.
Supplementary Material
Acknowledgments
We thank Dr. Tracey Baas for critical reading of the manuscript. We are grateful to Prof. Wei Xiao from Capital Normal University (Beijing) for providing the yeast two-hybrid system.
This work was supported by Grant 2016YFA0500901 from the National Key Research and Development Program of China, Grants 31471277 and 91649202 from the National Nature Science Foundation of China (to W. L.), and Grant 2017YFC1002002 from the National Key Research and Development Program (to J. Y.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains File S1.
The MS proteomics data of GST-pulldown in this study have been deposited in the ProteomeXchange dataset via the PRIDE partner repository under identifier PXD007815.
- HTCA
- head–tail coupling apparatus
- PTC
- premature termination codon
- TM
- transmembrane domain
- NE
- nuclear envelope
- iBAQ
- intensity-based absolute quantification
- Y2H
- yeast two-hybrid
- IF
- immunofluorescence
- DAPI
- 4′,6-diamidino-2-phenylindole.
References
- 1. Perotti M. E., and Gioria M. (1981) Fine structure and morphogenesis of “headless” human spermatozoa associated with infertility. Cell Biol. Int. Rep. 5, 113 10.1016/0309-1651(81)90018-7 [DOI] [PubMed] [Google Scholar]
- 2. Perotti M. E., Giarola A., and Gioria M. (1981) Ultrastructural study of the decapitated sperm defect in an infertile man. J. Reprod. Fertil. 63, 543–549 10.1530/jrf.0.0630543 [DOI] [PubMed] [Google Scholar]
- 3. Baccetti B., Selmi M. G., and Soldani P. (1984) Morphogenesis of 'decapitated' spermatozoa in a man. J. Reprod. Fertil. 70, 395–397 10.1530/jrf.0.0700395 [DOI] [PubMed] [Google Scholar]
- 4. Chemes H. E., Carizza C., Scarinci F., Brugo S., Neuspiller N., and Schwarsztein L. (1987) Lack of a head in human spermatozoa from sterile patients: A syndrome associated with impaired fertilization. Fertil. Steril. 47, 310–316 10.1016/S0015-0282(16)50011-9 [DOI] [PubMed] [Google Scholar]
- 5. Chemes H. E., and Alvarez Sedo C. (2012) Tales of the tail and sperm head aches: Changing concepts on the prognostic significance of sperm pathologies affecting the head, neck and tail. Asian J. Androl. 14, 14–23 10.1038/aja.2011.168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Shang Y., Zhu F., Wang L., Ouyang Y. C., Dong M. Z., Liu C., Zhao H., Cui X., Ma D., Zhang Z., Yang X., Guo Y., Liu F., Yuan L., Gao F., et al. (2017) Essential role for SUN5 in anchoring sperm head to the tail. Elife 6, e28199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Yang K., Meinhardt A., Zhang B., Grzmil P., Adham I. M., and Hoyer-Fender S. (2012) The small heat shock protein ODF1/HSPB10 is essential for tight linkage of sperm head to tail and male fertility in mice. Mol. Cell. Biol. 32, 216–225 10.1128/MCB.06158-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mendoza-Lujambio I., Burfeind P., Dixkens C., Meinhardt A., Hoyer-Fender S., Engel W., and Neesen J. (2002) The Hook1 gene is non-functional in the abnormal spermatozoon head shape (azh) mutant mouse. Hum. Mol. Genet. 11, 1647–1658 10.1093/hmg/11.14.1647 [DOI] [PubMed] [Google Scholar]
- 9. Tokuhiro K., Isotani A., Yokota S., Yano Y., Oshio S., Hirose M., Wada M., Fujita K., Ogawa Y., Okabe M., Nishimune Y., and Tanaka H. (2009) OAZ-t/OAZ3 is essential for rigid connection of sperm tails to heads in mouse. PLoS Genet. 5, e1000712 10.1371/journal.pgen.1000712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Yuan S., Stratton C. J., Bao J., Zheng H., Bhetwal B. P., Yanagimachi R., and Yan W. (2015) Spata6 is required for normal assembly of the sperm connecting piece and tight head-tail conjunction. Proc. Natl. Acad. Sci. U.S.A. 112, E430–E439 10.1073/pnas.1424648112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhu F., Wang F., Yang X., Zhang J., Wu H., Zhang Z., Zhang Z., He X., Zhou P., Wei Z., Gecz J., and Cao Y. (2016) Biallelic SUN5 mutations cause autosomal-recessive acephalic spermatozoa syndrome. Am. J. Hum. Genet. 99, 942–949 10.1016/j.ajhg.2016.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Elkhatib R. A., Paci M., Longepied G., Saias-Magnan J., Courbière B., Guichaoua M. R., Lévy N., Metzler-Guillemain C., and Mitchell M. J. (2017) Homozygous deletion of SUN5 in three men with decapitated spermatozoa. Hum. Mol. Genet. 26, 3167–3171 [DOI] [PubMed] [Google Scholar]
- 13. Morimoto A., Shibuya H., Zhu X., Kim J., Ishiguro K., Han M., and Watanabe Y. (2012) A conserved KASH domain protein associates with telomeres, SUN1, and dynactin during mammalian meiosis. J. Cell Biol. 198, 165–172 10.1083/jcb.201204085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yang H. M., Liu G., Nie Z. Y., Nie D. S., Deng Y., and Lu G. X. (2005) Molecular cloning of a novel rat gene Tsarg1, a member of the DnaJ/HSP40 protein family. DNA Seq 16, 166–172 10.1080/10425170500129736 [DOI] [PubMed] [Google Scholar]
- 15. Guan J., and Yuan L. (2008) A heat-shock protein 40, DNAJB13, is an axoneme-associated component in mouse spermatozoa. Mol. Reprod. Dev. 75, 1379–1386 10.1002/mrd.20874 [DOI] [PubMed] [Google Scholar]
- 16. Guan J., Kinoshita M., and Yuan L. (2009) Spatiotemporal association of DNAJB13 with the annulus during mouse sperm flagellum development. BMC Dev. Biol. 9, 23 10.1186/1471-213X-9-23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Oji A., Noda T., Fujihara Y., Miyata H., Kim Y. J., Muto M., Nozawa K., Matsumura T., Isotani A., and Ikawa M. (2016) CRISPR/Cas9 mediated genome editing in ES cells and its application for chimeric analysis in mice. Sci. Rep. 6, 31666 10.1038/srep31666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. El Khouri E., Thomas L., Jeanson L., Bequignon E., Vallette B., Duquesnoy P., Montantin G., Copin B., Dastot-Le Moal F., Blanchon S., Papon J. F., Lorès P., Yuan L., Collot N., et al. (2016) Mutations in DNAJB13, encoding an HSP40 family member, cause primary ciliary dyskinesia and male infertility. Am. J. Hum. Genet. 99, 489–500 10.1016/j.ajhg.2016.06.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Liu C., Wang H., Shang Y., Liu W., Song Z., Zhao H., Wang L., Jia P., Gao F., Xu Z., Yang L., Gao F., and Li W. (2016) Autophagy is required for ectoplasmic specialization assembly in Sertoli cells. Autophagy 12, 814–832 10.1080/15548627.2016.1159377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Guo X., Zhao C., Wang F., Zhu Y., Cui Y., Zhou Z., Huo R., and Sha J. (2010) Investigation of human testis protein heterogeneity using 2-dimensional electrophoresis. J. Androl. 31, 419–429 10.2164/jandrol.109.007534 [DOI] [PubMed] [Google Scholar]
- 21. Wang G., Guo Y., Zhou T., Shi X., Yu J., Yang Y., Wu Y., Wang J., Liu M., Chen X., Tu W., Zeng Y., Jiang M., Li S., Zhang P., et al. (2013) In-depth proteomic analysis of the human sperm reveals complex protein compositions. J. Proteomics 79, 114–122 10.1016/j.jprot.2012.12.008 [DOI] [PubMed] [Google Scholar]
- 22. Schwanhäusser B., Busse D., Li N., Dittmar G., Schuchhardt J., Wolf J., Chen W., and Selbach M. (2011) Global quantification of mammalian gene expression control. Nature 473, 337–342 10.1038/nature10098 [DOI] [PubMed] [Google Scholar]
- 23. Huang da W., Sherman B. T., and Lempicki R. A. (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 10.1038/nprot.2008.211 [DOI] [PubMed] [Google Scholar]
- 24. Shen Y., Yue F., McCleary D. F., Ye Z., Edsall L., Kuan S., Wagner U., Dixon J., Lee L., Lobanenkov V. V., and Ren B. (2012) A map of the cis-regulatory sequences in the mouse genome. Nature 488, 116–120 10.1038/nature11243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Guo H., Wen R., Wang Q., Datla R., and Xiao W. (2016) Three Brachypodium distachyon Uev1s promote Ubc13-mediated Lys63-linked polyubiquitination and confer different functions. Front. Plant Sci. 7, 1551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. World Health Organization (2010) WHO Laboratory Manual for the Examination and Processing of Human Semen, 5th Ed., World Health Organisation Press, Geneva [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.