Graphical Abstract
Introduction
Background and Relevance
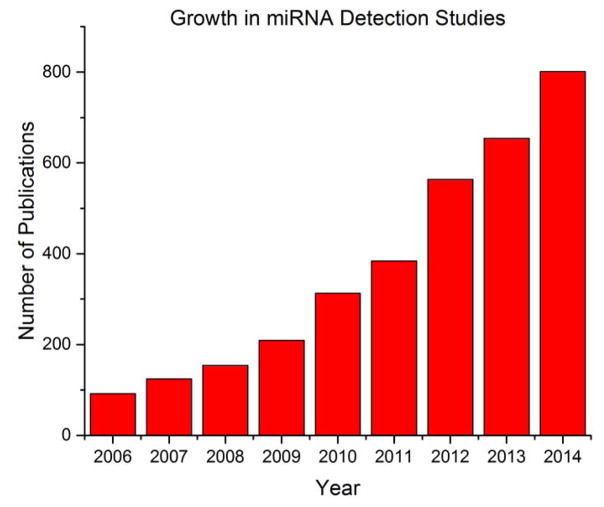
Since their discovery more than two decades ago in C. elegans,1 microRNAs (miRNAs) have emerged as an important class of non-protein coding RNA molecules. miRNAs serve as critical gene expression regulators at the transcriptional and post-transcriptional level and are widely conserved across a broad range of animals, plants, and viruses. Landmark studies have associated miRNAs with key biological events like developmental timing in C. elegans2 and zebrafish,3 and cancer development in humans.4 These studies along with many others that have established miRNA control over numerous biological processes would not have been possible without reliable miRNA detection methods. The impact that these analytical tools have had is reflected in the rapid increase in publications that focus on “miRNA detection”, as shown in Figure 1. The aim of this review is to outline the current state of the art while also highlighting exciting new biosensing approaches to miRNA detection that might help realize the full potential of miRNA expression profiles in both the contexts of fundamental biology studies and clinical diagnostics.
Figure 1.
Growth in publications in microRNA profiling since 2006. These results were obtained from a SciFinder search using the key words “microRNA detection”.
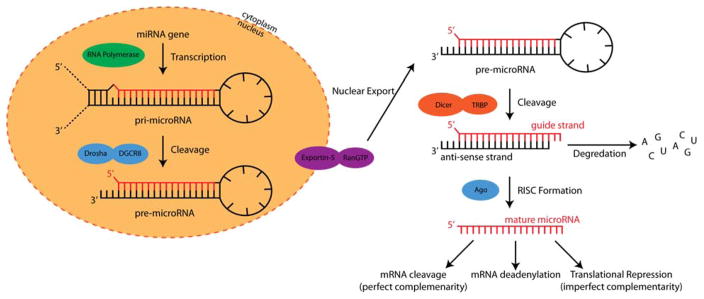
microRNAs are short, non-coding RNAs that are roughly 22 nucleotides in length. miRNA sequences regulate the expression of mRNA targets with either perfect complementarity, which leads to mRNA degradation, or imperfect complementarity, which often results in repression of translation. While the biogenesis of miRNA has been previously reviewed,5–7 the basic process of miRNA expression and maturation is outlined in Figure 2. The genesis of miRNAs is in the nucleus where primary miRNA transcripts (pri-miRNA) are produced. The pri-miRNA sequence is cleaved by the Drosha-DGCR8 complex to produce a pre-miRNA hairpin. This precursor hairpin is then transported to the cytoplasm via Exportin-5-Ran-GTP. In the cytoplasm, the Dicer processing complex cleaves the pre-miRNA hairpin to the mature sequence. One strand of the mature miRNA, the guide strand, is loaded into the miRNA-induced silencing complex (RISC), which contains DICER1 and Argonaute proteins, and directs the RISC to target mRNA sequences. This miRNA directed process affects gene expression through mRNA cleavage, translational repression, or deadenylation.5,6 It is important to note that it is valuable for analytical methodologies to discriminate between pri-, pre-, and mature forms of miRNA sequences.
Figure 2.
Biogenesis of miRNA that starts with transcription in the nucleus and ends with affecting gene translation in the cytoplasm.
miRNA regulation ultimately results in altered protein levels and can have profound consequences on cellular homeostasis. In fact, miRNA expression profiling has identified a host of regulated biological processes, including immune response8, cell differentiation,9,10 and cell proliferation and death.11 Furthermore, if the miRNA regulatory machinery that governs these processes is interrupted, it becomes extremely important to identify these disruptions and understand how they evolve as disease drivers. For example, Rosenfeld and co-workers showed how patterns of aberrantly expressed miRNAs could differentiate between tissues of origin across multiple cancer types.12 Once changes in miRNA expression are better understood in a biological context (i.e. what induces specific upregulation and down regulation patterns), they can then be used as potential therapeutic targets as well as refined into improved diagnostic biomarker panels.13
Recent work aimed at understanding how changes in miRNA expression can lead to specific disease states has shown promise. Initial efforts have implicated aberrant miRNA expression with cancer, 14 neurological disorders,15,16 diabetes,17 and cardiovascular disease,18,19 to name just a few. Equally promising is the fact that the detection of these biopanels is not limited to tissue. A host of studies have shown the ability to detect miRNA biopanels in a variety of biofluids. For example, Weber and co-workers were able to identify meaningful miRNA profiles found in twelve different biofluids, including cerebrospinal fluid, blood, serum, saliva, and urine.20 As increasing attention is focused on non- or minimally-invasive diagnostic methods, the presence of freely circulating miRNA profiles provides a promising approach to disease monitoring. Other studies have again established disease-correlated miRNA levels in blood;21,22 however the tools for detection and interpretation of the biological significance of these alterations in expression must still be refined in order to achieve full clinical adoption and translation.
miRNA Profiling Challenges
While the clinical and biological implications of altered miRNA expression are being elucidated, progress remains to be achieved in the development of robust analytical technologies that facilitate routine miRNA analysis. Improved techniques will help realize the impact that miRNA profiling can have on understanding disease onset and progression, and may play a key role in the realization of personalized medicine. In order to provide improved miRNA detection modalities, one must first understand the analytical challenges that miRNA’s present and the importance of sample processing and miRNA isolation.
Analytical Challenges
There are many unique characteristics of microRNAs that pose analytical challenges for their accurate detection and quantification. The most important of these characteristics are their small size, sequence similarity, wide range in abundance, and their ability to regulate multiple targets.23,24
The small size of miRNAs presents specific thermodynamic considerations and makes their analysis more difficult than the significantly longer mRNAs. Due to their small length, the GC content variation in miRNA sequences lead to a wide range of melting temperatures (Tm). Since the vast majority of miRNA detection methods rely on some sort of hybridization step, this can potentially introduce sequence-specific bias. Additionally, the small size of miRNAs, roughly the same size a traditional primer, complicates the use of the polymerase chain reaction (PCR).
In addition to thermodynamic constraints, there are additional difficulties that complicate the analysis of miRNAs, as compared to mRNAs, due to the very nature of their seuqence. For example, miRNAs lack the poly(A) tail of mRNAs, which is often used as a universal primer for reverse transcription or selective pre-enrichment. This is especially important as miRNAs make up roughly 0.01% of total RNA extracted from a sample of interest.25 This means that any bioanalysis platform must be able to differentiate small amounts of miRNA in the presence of a large abundance of total cell RNA. Moreover, the abundance of particular miRNA sequences can vary from single copies to more than 50,000 copies in a single cell,26 thus requiring exceptional dynamic range. Finally, families of miRNAs are often expressed differing only by single nucleotides and so extremely high sequence selectively is a necessity.
Beyond specificity, selectivity, and large dynamic range, it is also important that analytical methods for miRNA detection allow for multiplexed analysis, whereby levels of more than one miRNA are quantitated simultaneously and from a single sample. This analytical requirement is important on account of the biological mode of action of miRNAs. A single miRNA sequence is capable of regulating up to hundreds of different mRNA sequences and a single mRNA can be targeted by many different miRNAs. Therefore in order to fully comprehend the biological significance of miRNAs in both health and disease, one must be able to analyze the ensemble effects of miRNA expression changes and understand the interrelated consequences on multiparametric regulatory networks that extend across all levels of biomolecular information (i.e. DNA, mRNA, miRNA, and proteins). To this end, it is becoming increasingly clear through both experimental results and computational modeling27,28 that the creation of multiplexed miRNA panels is needed to deconvolute these complex interactions.
Sampling Considerations
It is also worth noting that the reproducibility of experimentally-determined miRNA expression profiles is directly related to the ability to isolate high quality RNA samples. It has been shown that miRNA isolation is possible from cell lines, fresh and formalin fixed paraffin embedded (FFPE) tissue, as well as from various bodily fluids.20,29 It is worthwhile to mention too that while mRNA often suffer from RNase degradation, particularly in FFPE samples, miRNA have been shown to be more stable.30,31 Given some of the aforementioned challenges of miRNA analysis compared to mRNAs, the greater stability of miRNAs in a diverse set of sample matrices provides an opportunity to expand their utility through the study of large libraries of archived tissue and blood samples.
Generally, most workflows to isolate RNA from a sample follow the same procedure,31 using phenol/chloroform purification with chaotrophic salts (i.e. guanidinium thiocynate) to denature RNases and proteins associated with RNAs. After centrifugation, nucleic acids partition into the aqueous and interphase while proteins partition into the organic phase. The RNA from the resulting aqueous phase is then bound to a solid phase silica column and washed. To isolate the small RNA fraction from total RNA, diluted ethanol can be used which will cause large RNAs to dissociate from the silica column and leaves purified small RNAs (<200 nucleotides) in the final elution volume. Finally, the RNA molecules are eluted off the column and analyzed for purity, using UV-Vis spectroscopy, and integrity, using the band intensity ratio of the 28S to 18S rRNA bands measured using gel or capillary electrophoresis. Low integrity samples containing fragmented RNA suffer from a higher background of small RNA sequences, which then leads to lower quality miRNA profiling data with a higher chance of off-target responses.
Scope
It is clear that miRNAs play incredibly important roles in biology; however, many gains remain in the translation of this fundamental insight into the clinical setting. Key to this achievement will be the development of robust and multiplexed analytical technologies that offer strategic advantages over conventional techniques, such as qRT-PCR, microrarrays, and RNA-sequencing. Motivated by the aforementioned analytical challenges, this review focuses on the recent demonstrations of new microRNA detection platforms, with particular emphases placed on reports published since 2013. At the forefront of promising approaches are advanced biosensor technologies. Beyond a discussion of conventional approaches and emerging techniques, we also provide commentary and perspective regarding the role of bioinformatics in constructing multiplexed miRNA panels, as well as how future advances might impact the clinical adoption of panel-based miRNA diagnostics.
Conventional Methods
Three major approaches are used at present to determine levels of miRNA expression: (1) reverse transcription-quantitative polymerase chain reaction (qRT-PCR), (2) hybridization-based microarrays and (3) next generation high-throughput sequencing. This section provides details on each as a way of providing context for the development of emerging biosensing technologies.
Quantitative Reverse Transcription-Polymerase Chain Reaction (qRT-PCR)
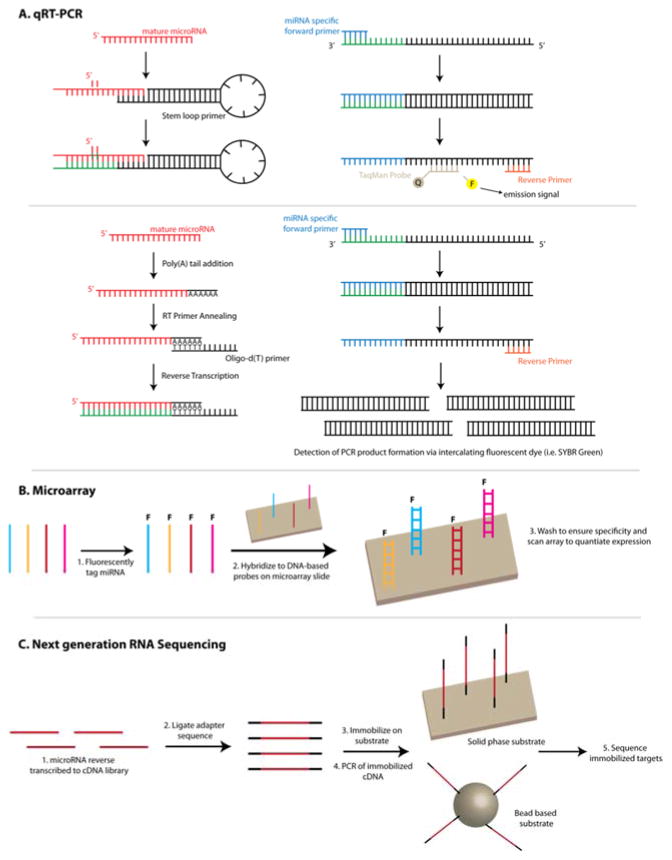
Quantitative reverse transcription polymerase chain reaction (qRT-PCR) is the current gold standard for miRNA analysis. It is commonly used to detect levels of single or small, targeted panels of miRNAs, but also to validate selected results from more global expression studies (i.e. microarrays and next generation sequencing, as described in subsequent sections). qRT-PCR analysis provides a large dynamic range, inherent sensitivity through the ability of PCR to selectively amplify specific target sequences, lower assay costs compared to next generation sequencing, and the ability to measure multiple miRNAs by running reactions in parallel (normally in 96 or 384 well plates). The major downside to running parallel reactions is higher consumption of the sample of interest, qRT-PCR consumables, and enzymes/master mixes. As mentioned earlier, the short size of miRNAs complicates all PCR-based detection schemes due to similarities between the length of the target and primer. However, two of the more common strategies to achieve this goal are: (1) reverse transcription via stem loop primers and the use of TaqMan PCR, and (2) enzymatic addition of a poly(A) tail to RNAs followed by reverse transcription and SYBR Green based qPCR detection.30 The overall workflow for both of these qRT-PCR approaches are illustrated in Figure 3A.
Figure 3.
Overview of conventional techniques: (a) qRT-PCR, (b) microarrays and (c) next generation RNA sequencing. (A) When using TaqMan qRT-PCR, the reverse transcription process utilizes stem-loop primers specific to the miRNA target of interest. During PCR amplification, the DNA polymerase proceeds along the template strands produces by miRNA specific forward and reverse primers and hydrolyses the TaqMan probe bound to the template. This liberates the fluorescent dye from the quencher and results in light emission. In SYBR green-based approaches, miRNAs are typically polyadenylated at the 3′ end and d(T) oligos are used as the reverse transcription primer. PCR amplification is carried out using miRNA specific forward primer and reverse primer. SYBR Green, an intercalating dsDNA dye, is then used to monitor PCR product formation. (B) DNA-based capture probes immobilized on the microarray are used to capture fluorescently tagged miRNAs. The fluorescent signal is then quantitated and the intensity is related to the relative miRNA expression. (C) Most RNA-sequencing workflows begin by reverse transcribing miRNA into a cDNA library. This is followed by adaptor ligation that allows for immobilization on a substrate that are used to obtain sequencing data.
Stem loop primers are designed to contain a 6–8 nucleotide overhang on the 3′ end that is complimentary to a region of the targeted miRNA and can differentiate between closely related sequences as well as different miRNA forms (i.e. pri-, pre-, and mature).32 Upon hybridization between the stem loop primer and the target miRNA, reverse transcription extends the DNA compliment of the hybridized miRNA from its 3′ end. The use of stem loop primers facilitates better specificity by optimizing the melting temperature and effectively lengthens the miRNA target through ligation to the stem loop, which can then be recognized by a standard PCR primer. The RT-extension step is typically performed at temperatures <16°C to preserve the secondary structure of precursor miRNA sequences. PCR amplicons are then generated using a miRNA-specific forward primer that binds to the 3′ end of the reverse transcription (RT) product and a universal reverse primer that binds to the conserved stem loop region of all RT products. Additionally, a molecular beacon, or TaqMan, probe is present in the PCR reaction solution and is designed to hybridize in between the forward and reverse primers. As the DNA polymerase proceeds along the template and reaches the TaqMan probe, the probe is hydrolyzed and the fluorescent dye is freed from the quencher, resulting in an emission signal proportional to the total amount of PCR product produced. This signal, measured as a function of cycle number, is then used to determine the overall level of a specific miRNA in a sample.
The poly(A) method involves the 3′ polyadenylation of all RNA in a sample normally using either polyadenylate polymerase or T4 ligase. When T4 ligase is used, an additional sequence can be installed following the poly(A) tail that further lengthens the downstream RT product to enable binding of the two PCR primers. After the poly(A) tail addition, binding of a poly(dT) DNA primer, which serves as the reverse transcription primer, initiates the RT reaction and production of cDNA. Conventional PCR primers are then added to initiate amplification, and PCR product formation is measured using a dsDNA-intercalating SYBR green dye.
The main drawback of qRT-PCR is complex primer design requirements and the inability to analyze multiple targets per single sample volume. The design of both RT and PCR primers vary substantially between miRNA targets due to differences in Tm between the resulting primer-target duplexes. However, this problem is partially alleviated through the use of Tm-matched locked nucleic acid primer sequences.33 It also remains difficult to measure multiple miRNAs from within a single qRT-PCR reaction volume, as qPCR is limited by its reliance on spectral multiplexing. Run-to-run inconsistencies due to variability in PCR amplification efficiencies is also a complication that requires careful design of internal controls, which often require the use of global mean averaging,34 referencing common housekeeping genes,35 the spiking in of non-natural miRNA probes that are added before the RNA extraction step,36 or some combination of these control methods.37
Microarrays
Originally developed in the early 1990s for genomic-scale analysis of DNA, microarrays were redeployed as one of the first methods applied to the global analysis of miRNA expression.30,38 Typically, miRNAs are first labeled with a fluorescent reporter. This is accomplished by dephosphorylating the 5′ end of the miRNA followed by ligation of a fluorescently tagged oligonucleotide or short oligonucleotide strand using T4 ligase. The dephosphorylation of the 5′ end is critical to prevent self-circularization of the miRNA and adapter sequences.39 The functionalized miRNA is then introduced to the array surface where they hybridize to complimentary DNA (cDNA) capture probes immobilized on a glass slide, followed by two channel fluorescent imaging, which can provide expression levels. As a result of being a surface-bound hybridization based assay, microarrays require complimentary base-paring between the cDNA:miRNA. The overall workflow for microarray-based analysis of miRNAs is shown in Figure 3B.
Despite the relative ease and historical utility of DNA microarrays, there are some limitations in their application to miRNAs, particularly in light of competitive methods.31 For instance, they are only semi-quantitative due to the absence of a calibration curve from the experimental workflow. As a result, microarrays are best used when comparing miRNA expression levels between multiple states (i.e. heathy vs. disease). To ensure specificity, microarrays also often require additional validation, which often is achieved via qRT-PCR for select targets of interest. Microarrays also suffer have a smaller dynamic range than both qRT-PCR and next generation sequencing.
Despite these drawbacks, microarrays do offer the advantage of being cheaper than global profiling via next generation sequencing. Additionally, substantial effort has already been invested in the development of Tm-normalized cDNA capture probes that incorporate peptide nucleic acids40 and locked nucleic acids.41 The thermal stability of hybridization duplexes across the array can lead to reproducible results and assays with high sensitivity. Other studies have aimed to improve the fluorescent labeling step of the microarray workflow to reduce non-specific background signal. These improvements have focused on the use of labeled binding proteins that only binds to miRNA molecules hybridized at the surface,42 a hybridization based labeling technique termed stacking-hybridized-universal-tagging (SHUT) that allows for the addition of one universal tag,43 and a ligase-assisted sandwich hybridization based approach that eliminates the need for miRNA labeling by ligating a signal probe that binds to capture probe:miRNA hybrids at the array surface.44 The sandwich hybridization based assay improved hybridization efficiency 50,000-fold and allowed quantitation of a synthetic miRNA sequence down to 30 fM. Beyond this limited discussion, other developments in miRNA-detecting microarrays have recently been reviewed.31,45
Next-Generation Sequencing
The advent of next generation sequencing (NGS) platforms has enabled a third major approach for miRNA expression profiling, and with continuing decreases in sequencing cost, this is quickly becoming a dominant method applied to the analysis of miRNAs. The technologies driving NGS has been reviewed.46,47 While the procedures vary depending on specific platform, the first step involves the preparation of a small cDNA library from the RNA-containing sample of interest using a reverse transcription process similar to qRT-PCR. Adaptors are ligated to both the 5′ and 3′ ends of the cDNA products, and the resulting products are attached to either a planar or bead based substrate. This is followed by the massively parallel sequencing of millions of individual cDNA molecules from the library. Bioinformatic analysis of the sequence reads trims the adaptor sequences off of the miRNA sequences. The trimmed sequences are then aligned against a miRNA sequence data base (ex. miRBase) to identify the known miRNAs present in the sample. This sequence data also provides quantification by identifying the number of sequence reads present. Unique to miRNA analysis is the ability for bioinformatic approaches to identify novel miRNA sequences that are not already annotated in miRNA databases by attempting to alight to precursor miRNA sequences. This presents a unique set of advantages as well as roadblocks that must be solved to continue to expand the use of next generation sequencing for miRNA profiling. The overall work for the NGS workflow applied to miRNA analysis is outlined in Figure 3C.
The major advantage of NGS for miRNA analysis is the ability to obtain a truly global expression profile. In addition to known sequences, which could be detected using a pre-synthesized cDNA microarray, previously unknown short RNA sequences can be discovered de novo. Sequencing also obviates the concerns with specificity faced by hybridization-based methods, including qRT-PCR, microarrays, and the majority of the biosensor-based approaches described below. To gain clinical traction, overall sequencing costs, including reagents, still need to be driven down further. Also, streamlined informatics techniques are needed to simplify and/or automate data analysis for use in clinical settings.
The quality of using NGS for miRNA expression profiling in clinical samples has been analyzed in recent publications that compare the results obtained using RNA-sequencing to both qRT-PCR and microarrays.48,49 One study found discrepancies between platforms that were attributed to differences in normalization protocols as well as potential sampling biases.48 The other, though, revealed strong correlation between RNA-seq and qRT-PCR.49 This report also showed good correlation between expression levels from flash-frozen and FFPE samples, which is important for analysis of current and archived clinical samples. Additional studies validating RNA-seq and other conventional analysis methods are important and needed to help put into perspective the vast literature reporting miRNA expression results obtained by different platforms, and ideally may result in the identification of clinically-useful miRNA based tests.
Given the breadth of expression data provided by NGS-based miRNA analysis it is reasonable to consider that most of the resulting information will not be informative in the context of human health and disease. Moreover, an infomatically-robust panel of miRNAs might be extracted from global data sets and correlated with different diagnostic or prognostic outcomes.50,51 Therefore, the workflow by which RNA-seq can be used to identify and translate multiplexed miRNA panels to cost effective biosensing technologies remains an important goal. Once promising panels of miRNAs are proposed, cheaper and less time consuming technologies might be a better fit for high throughput analyses, as well as eventual use in the clinical setting. Going forward, NGS will, if it has not already, become the preferred technique for global miRNA expression profiling and novel sequence identification that establishes correlations with disease. Subpanels may then be informatically-selected and then validated and translated to the clinic using the emerging technologies described in the following section.
Summary of conventional miRNA analysis methods
qRT-PCR, microarrays, and next generation sequencing have all played key roles in advancing our knowledge as to how miRNAs play key roles in regulating gene expression, and in beginning to translate this fundamental insight to application in clinical diagnostics. A high level comparison of the attributes of each of these general classes of techniques is presented in Table 1. However, all face critical hurdles to widespread clinical adoption that portend a role for emerging biosensing technologies. qRT-PCR methods are incredibly sensitive, relatively rapid, and cost effective; however they can only measure levels of one miRNA per assay, thus requiring multiple sample aliquots to profile a panel of targets, which is prohibitive for sample-limited specimens. Microarrays are exceptionally well-suited to multiplexed analyses, but are typically slow, less sensitive, and minimally-quantitative. Next generation sequencing technologies are also well-suited to give a global analysis of all miRNAs present in a sample, but require complex processing steps, an even longer time-to-result, and can present challenges with back end informatics. Therefore, there exists a pressing need for the development of multiplex diagnostic capabilities whereby focused panels of 10s of miRNAs can be simultaneously interrogated using rapid, cost effective, and highly scalable technologies. Such technologies would have broad-reaching utility in both basic and clinical research and be applicable to both tissue and biofluid-based diagnostic applications.
Table 1.
Comparison of conventional miRNA detection platforms
| qRT-PCR | Microarrays | Next Gen. Sequencing | |
|---|---|---|---|
| Time to result | Hours | Days | Weeks |
| Cost | $$ | $ | $$$ |
| Biochemical processing | Ligation with T4 (SYBR Green) Annealing of primers |
No ligation steps | Ligate barcode |
| Input | Low (ng) | Large (ng-μg) | Large (ng-μg) |
| Drawbacks | Results need validation Long time to result |
Single plex | Don’t always need global view |
Biosensor Methods
miRNAs have been a focus of biosensor development over the past decade, with many micro- and nanoscale sensing technologies being applied to this class of analytes. Benefits of many of these schemes lie in their potential for diagnostics at the point of care. However, it is important to point out that for many diagnostic needs, analyses performed in a central laboratory over the multi-hour-to-day timeframe are completely acceptable for many applications, such as in the diagnostic or longitudinal monitoring of relatively slowly progressing diseases, such as cancer. Coupled with these devices have been a number of novel signal transducers, incorporation of modified nucleic acid capture sequences, and the development of new signal amplification strategies. The past few years have shown a movement away from proof-of-principle technology demonstrations and an increasing emphasis on obtaining the sensitivity and sequence selectivity required for clinical applications. A number of reports have also explored low levels of multiplexing; however, most of these studies only analyze 2–3 sequences at a time. Here, we focus broadly on some of the most recent reports considering promising optical, electrochemical/electrical, and magnetoresistive technologies for miRNA detection, emphasizing improvements to analytical sensitivity and selectivity and highlighting some of the more promising demonstrations of multiplexing.
Optical Detection
A wide range of optical detection methods have been applied to miRNA detection. Fluorescent dyes and quantum dots directly conjugated to nucleic acids detection probes have been widely explored in FRET-based analyses and also coupled with enzymatic methods for enhanced performance. In these examples, multiplexing must typically be achieved spectrally, which places some constraints on the number of sequences that can be simultaneously assayed. Electrochemiluminescent assays have also been developed that offer promising enzyme- and nucleic acid-based signal enhancement strategies. Surface-tethered optical methods based on plasmons or waveguide properties promise higher levels of multiplexing through the creation of spatially-resolved sensor arrays.
FRET and related approaches
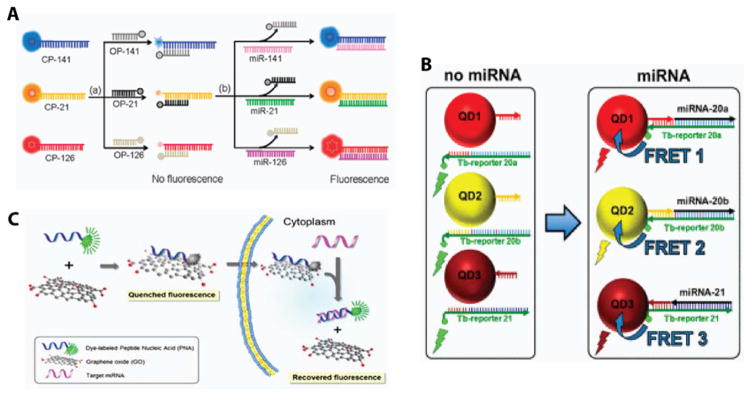
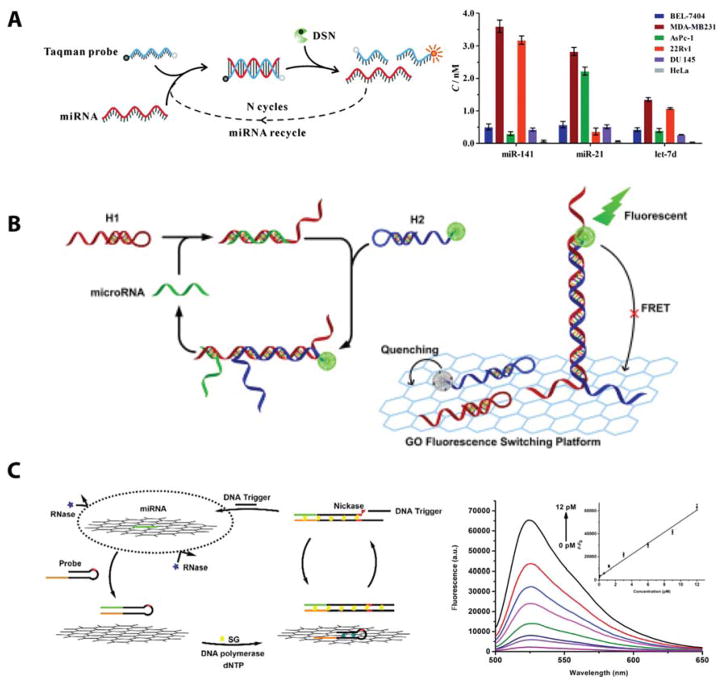
To improve upon microarrays, which most commonly require labeling of the miRNA before detection, fluorescent assays today aim to use fluorescent reporters that bind to the miRNA target via a detection probe. To provide the appropriate specificity, strategies have been designed to ensure that the fluorescent signal is “off” with no miRNA signal is present and “on” when the target is present. Forster resonance energy transfer (FRET) is a common approach to facilitate “on/off” detection schemes. An overview of different FRET-based miRNA detection methods is provided in Figure 5.
Figure 5.
Promising multiplexed detection schemes. (A) Fluorescent FRET pairs were used to probe miRNAs-141, 21, and 126 in multiple cancer cell lines. Reproduced from Wu, P.; Tu, Y.; Qian, Y.; Zhang, H.; Cai, C. Chemical Communications 2014, 50, 1012–1014 (ref 52), with permission of The Royal Society of Chemistry. (B) Quantum dot FRET pairs were also used to profile three miRNAs from diluted serum. Reproduced from Qiu, X.; Hildebrandt, N. ACS Nano 2015, 9, 8449–8457 (ref 55). Copyright 2015 American Chemical Society. (C) FRET-based detection was achieved using a fluorescent peptide nucleic acid detection probe adsorbed onto a graphene oxide nanosheet as a quencher. miRNA-21, 125b, and 96 were quantitated in living cells and the relative expression levels were shown to correlate well with Northern blotting. Reproduced from Ryoo, S.-R.; Lee, J.; Yeo, J.; Na, H.-K.; Kim, Y.-K.; Jang, H.; Lee, J. H.; Han, S. W.; Lee, Y.; Kim, V. N.; Min, D.-H. ACS Nano 2013, 7, 5882–5891 (ref 61). Copyright 2013 American Chemical Society.
A simple, but effective, FRET approach used a DNA strand-displacement scheme, where fluorescently tagged DNA capture probes were initially hybridized with a quencher functionalized compliment. When present, the target miRNA displaced the quenching strand and turned “on” the fluorescent signal. This approach was used to detect the presence of 3 miRNA sequences across multiple cell lines with an LOD of 1 fM and a dynamic range of 4 orders of magnitude.52 Despite its simplicity, the main drawback of this approach is the false positive rate, whereby closely related species might displace the quenching strand giving an erroneous result.
Molecular beacons aim to improve specificity by incorporating a fluorescent tag on one end of the sequence and a fluorescent quencher on the other, thus creating a FRET pair on a single capture sequence. With no miRNA present, the strand adopts a thermodynamically stable hairpin geometry with the fluorophore in close proximity to the quenching molecule. Upon hybridization of a target miRNA, the beacon linearizes, causing separation between the fluorophore and quencher and turning “on” the fluorescence.53 The opposite scheme has also been developed where a molecular beacon was created that was initially held “open” via an interaction with a reporter sequence that featured multiple base pair mismatches. In this case, the fluorophore and quencher started off spatially separate, and the presence of the miRNA of interest displaced, allowing the beacon to fold on itself turning “off” the fluorescent signal.54 This simplified assay took only could be run in as little as 10 minutes and achieved better specificity by using locked nucleic acid detection strands. However, the limit of detection (LOD) was limited to 10 nM. Greater multiplexing capabilities of this molecular beacon-FRET approach were achieved with quantum dots, due to their increased brightness and inherent color tunability. Here, three miRNAs were detected in diluted serum without the need for any washing steps, and a LOD of 1 nM was reported.55
While the aforementioned molecular beacon approaches achieved nanomolar LODs, different FRET pairs have shown improved assay performance. Graphene oxide was used as a quencher, where fluorescently tagged miRNA compliments were absorbed onto the surface.56 In the presence of the miRNA target, the capture probe was released and hybridized with the target, generating a fluorescent signal. Further improvements to specificity were made by incorporating peptide nucleic acids into the molecular beacon sequence absorbed onto the graphene oxide surface.57 Improve performance was also achieved by using DNA probes that were approximately three times larger than the miRNA target, thus offering a higher affinity for adsorption onto the substrate and effectively eliminating non-specific desorption of the fluorescent nucleic acid sequence.58 The target binds a region of the capture probe and an introduced exonuclease cleaved the DNA:miRNA hybrid from the quenching substrate turning “on” the fluorescent signal. LODs as low as 3 fM were reported; however, multiplexing capabilities were not investigated.
While the simplicity of FRET based assays provide many advantages for in vitro diagnostics, they also can facilitate the quantitative profiling of miRNA expression in vivo. For example, a duplex DNA FRET probe was used to detect the intracellular presence of miRNA-294, a marker for neuronal cell differentiation,59 and a molecular beacon approach allowed for imaging of miRNA-126 as a general marker for ischemia.53 Another interesting development involved a single stranded FRET probe that was delivered into cells, where it bound to the complimentary miRNA, and was loaded into the RISC complex. The FRET probe was then hydrolyzed and liberated the fluorescent reporter, allowing the detection of miR-10b, a metastatic marker associated with breast cancer.60 Ryoo, et.al. described an approach with graphene oxide as the quenching substrate used to simultaneously detect the presence of 3 miRNA in living cells.61 A similar approach employing carbon nitride nanosheets as a quencher was also demonstrated.62 While these devices are able to identify the presence of miRNA targets in living cells, these workflows ultimately suffer from poor sensitivity and a limited dynamic range.
The aforementioned FRET-based strategies were stoichiometric, that is a single miRNA molecule led to a single fluorophore being turned “on” or “off”. In order to achieve better gain, several target recycling strategies have been reported by which a single miRNA can lead to multiple fluorophore signatures. Several promising target recycling strategies are highlighted schematically in Figure 6.
Figure 6.
Target recycling approaches. (A) Duplex specific nuclease recycling was used to study the differential expression of three miRNAs in six cancer cell lines. Reproduced from Yin, B.-C.; Liu, Y.-Q.; Ye, B.-C. Journal of the American Chemical Society 2012, 134, 5064–5067 (ref 70). Copyright 2012 American Chemical Society. (B) Toehold-mediated amplification facilitated effective target recycling and detection of miRNA-21 in four cell lines. However, improvements to this assay could focus on quantitating multiple targets per sample. Reprinted from Analytica Chemica Acta, Vol. 888, Huang, R.; Liao, Y.; Zhou, X.; Xing, D. Toehold-mediated nonenzymatic amplification circuit on graphene oxide fluorescence switching platform for sensitive and homogeneous microRNA detection, pp. 162–172 (ref 71). Copyright 2015, with permission from Elsevier. (C) A nickase based recycling strategy combined with the use of a DNA polymerase exponentially amplified nucleic acid sequences related to the target of interest. These dsDNA produces were stained with an intercalating fluorescent dye, and the signal intensity was proportional to the initialtarget concentration. Reproduced from Liu, H.; Li, L.; Wang, Q.; Duan, L.; Tang, B. Analytical Chemistry 2014, 86, 5487–5493 (ref 73). Copyright 2014 American Chemical Society.
Duplex-specific nuclease (DSN), and enzyme that recognizes and selectively cleaves the DNA strand from a DNA:miRNA duplex was employed to detect miRNA, resulting in the release of either a fluorophore or a DNA strand to induce a detectable signal. After cleavage of the DNA, the miRNA is free to bind to another DNA capture sequence and the DSN process is repeated. In this way, a single miRNA strand can interact with thousands of reporter sequences giving high levels of signal gain. This general strategy has been broadly incorporated into a wide variety of optical detection assays, including a fluorescently-tagged molecular beacons,63 DNAzyme capture probes,64 graphene oxide quenching assays,65 WS2 quenching assays,66 magnetic beads,67 gold nanoparticle quenching assays,68 and gold nanoparticle aggregation assays.69 A great example showing the potential of the DSN assay was shown by Yin and co-workers, who designed a simple FRET-based strategy that allowed three miRNA sequences to be simultaneously detected down to 1 fM across a dynamic range close to 5 orders of magnitude.70
Other target regeneration strategies utilized toehold-mediated recycling and nickase based recycling. In both of these approaches, after target recycling the capture probes are introduced to the sample of interest and when the miRNA is present it either linearizes upon hybridization71 or dissociates from a quenching substrate.72 These techniques eliminate the need for fluorescent dye conjugated directly capture probes by staining the hybrid product with an intercalating fluorescent dye. This strategy achieved picomolar detection limits and a dynamic range of 3–4 orders of magnitude. The nickase based strategy immobilized the target miRNA on a graphene oxide substrate to protect from RNases. A stem loop primer was then introduced and hybridized with the miRNA target, which caused desorption of the miRNA from the graphene oxide. Exponential amplification was then achieved in the presence of a DNA polymerase through target recycling using a nicking enzyme. The resulting dsDNA products were stained with an intercalating fluorescent dye. The intercalating dye approach has the advantage of having multiple fluorescent dyes per target rather than only a single fluorophore conjugated to a single capture probe, giving a LOD of 11 fM and 3 order of magnitude dynamic range.73 Of course the limitation of the intercalating dye approach is that the dye will stain any nucleic acid duplex without the sequence specificity that can be engineered using covalently attached fluorophores conjugated to specific strands. Another nickase based recycling approach relied on the formation of a three way junction consisting of the target miRNA sequence, an assistant DNA probe, and an Hg2+ intercalated molecular beacon. Upon complex formation, the intercalated Hg2+ was liberated and able to quench the fluorescent signal from the linearized molecular beacon.74 This approach gave a detection limit of detection of 0.16 nM and a dynamic range of 3 orders of magnitude.
Electrochemiluminescence
Like fluorescence, electorchemiluminescence (ECL) can be used to produce a detectable optical signal that is proportional to the miRNA concentration in a sample. ECL is an alternative approach to lamp or laser based excitation, where an electrochemical excitation can create a luminescent response in the presence of an ECL reporter molecule. This flexible excitation approach, which generally is a turn “on” response, does not require the use of specific wavelength lasers to selectively excite a fluorescent dye. Additionally, these approaches often show exquisite sensitivity due to the elimination of background fluorescent interfering species in solution. As an example of an ECL-based approach to detect miRNAs, a sandwich hybridization approach was successfully used to profile a single miRNA in three cell lines.75 Without any sample recycling or signal amplification, this strategy produced an LOD of 100 pM and a dynamic range of approximately 3 orders of magnitude. While these numbers are respectable, signal amplification strategies can be invoked to further increase limits of detection, improve specificity, and extend dynamic range.
Variations of the hybridization chain reaction and enzymatic amplification can both be utilized for eletrochemiluminescent signal amplification. The hybridization chain reaction allows for a dramatic increase in dsDNA length that is catalyzed in the presence of a target miRNA. This allows for various ECL co-reactants, such as [Ru(phen)3]2+-76 and hemin-conjugated DNAzymes,77 to intercalate into the resulting duplex. The LODs for these techniques were 1 and 1.7 fM, respectively, with linear dynamic range of 4 orders of magnitude.
Enzymatic amplification strategies have been deployed using rolling circle amplification to form DNAzymes,78 cyclic exponential amplification recycling,79 doxorubicin conjugated quantum dots,80 T7 exonuclease recycling and downstream silver deposition,81 ECL quenching via Phi29 DNA polymerase mediated strand displacement,82 and a dual target amplification strategy with combined ECL and fluorescence detection.83 With one exception, all of these strategies report LODs ranging between 10 fM and 100s of aM with dyamic ranges varying 2–5 orders of magnitude. However, the stand displacement system reported a remarkable detection limit of 3.3 aM and a dynamic range of 5 order of magnitude.82 Importantly, none of these examples demonstrated multiplexing capacity, likely in part due to the fact that ECL reporter molecules are not specific to an individual miRNA sequence, similar to that described above for the intercalating dye systems.
Plasmonic and Photonic Approaches
While fluorescence based detection has the benefit of enabling detection in free-solution, it is ultimately limited by the fact that a label is required for detection. Likewise, there are a fixed number of labels (i.e. fluorescent dyes or FRET pairs) that are easily detected with unique, non-overlapping signatures, which limits multiplexing capabilities. Additionally, it is often difficult remove excess fluorescent reporter sequences from solution if a FRET pair is not used, creating a large amount of background signal. In the case of electrochemiluminescence, multiplexing capabilities are difficult due to the need for multi-electrode arrays that can require unique potentiostatic control, and the fact that many ECL reporter molecules are not specific to an individual miRNA sequence. As alternatives that capture the spatial multiplexing features of traditional microarrays, methods that detect surface-hybridization through changes in optical properties, such as refractive index in the case of surface plasmon resonance (SPR) and other approaches, have grown in popularity. These techniques can often be operated in label-free sensing modes or combined with different enhancement strategies to further improve analytical performance metrics.
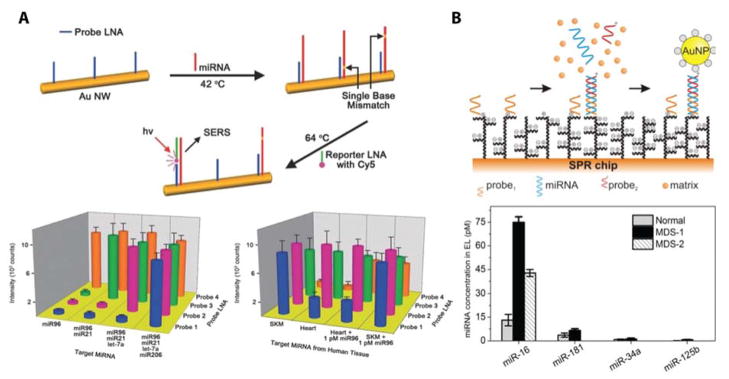
In addition to established label free detection modalities, such as conventional Kretschmann geometry surface plasmon resonance84, other sensing mechanisms have been explored for applications in miRNA analysis, including nano-particle scattering on flexible silicon substrates,85 surface enhanced Raman spectroscopy,86 functionalized gold nanoprisims attached to silanized glass,87 Mach-Zehender interferometers,88 and total internal fluorescence microscopy.89 An impressive gold nanowire plasmonic based detection mechanism was developed as a microfluidic lateral flow assay. A detection limit of 100 aM was achieved for a bi-temperature sandwich based hybridization scheme. The cDNA-modified gold nanowire was incubated at a low temperature to ensure specific base pairing with a target miRNA in solution, followed by an elevated temperature exposure to a LNA functionalized presenting a Cy5 that was detected via its surface-enhanced Raman scattering signal.90 This strategy showed the multiplexing capacity to detect 4 miRNAs simultaneously over a dynamic range of six orders of magnitude. A schematic representation of this detection method is shown in in Figure 7A.
Figure 7.
Successful multiplexing strategies using plasmonic based biosensors. (A) Plasmonic nanowires detected the presence of four miRNA targets using locked nucleic acid capture probes and Cy5 functionalized reporter locked nucleic acid sequences. These optimized nanosensors were used to profile the expression of four miRNAs from RNA isolated from two tissue types. Reproduced from Ultra-Specific Zeptmole microRNA Detection by Plasmonic Nanowire Interstice Sensor with Bi-Temperature Hybridization, Kang, T.; Kim, H.; Lee, J. M.; Lee, H.; Choi, Y.-S.; Kang, G.; Seo, M.-K.; Chung, B. H.; Jung, Y.; Kim, B. Small, Vol. 10, Issue 20 (ref 91). Copyright 2014 Wiley. (B) A novel ultra-low fouling surface plasmon resonance imaging biosensor detected four miRNAs from erythrocyte lysate. A gold nanoparticle signal enhancement strategy was used to improve limits of detection. Clinical utility was shown by analyzing chances in expression profiles of miR-16, 181, 34a, and 125b in ‘normal’ clinical samples and ones with myelodysplastic syndrome. Reprinted from Biosensors and Bioelectronics, Vol. 70, Vaisocherová, H.; Šípová, H.; Víšová, I.; Bocková, M.; Špringer, T.; Laura Ermini, M.; Song, X.; Krejčík, Z.; Chrastinová, L.; Pastva, O.; Pimková, K.; Dostálová Merkerová, M.; Dyr, J. E.; Homola, J., Rapid and sensitive detection of multiple microRNAs in cell lysate by low-fouling surface plasmon resonance biosensor, pp. 226–231 (ref 95). Copyright 2015, with permission from Elsevier.
Additional label free miRNA sensors aimed to improve upon previously outlined weaknesses. For instance, to avoid diffusion limitations experienced by surface based measurement modalities, a solution-based stem loop primer-functionalized plasmonic nanoparticle aggregation assay was developed. Upon hybridization of the target, the plasmon resonance shifts due to a change in distance between nanoparticles in the aggregate. The magnitude of this shift can then be related to the solution phase concentration.91 The LOD of this assay is 10s of fM; however, the dynamic range was limited to a single order of magnitude.
In an effort to achieve point-of-care miRNA detection, Gao and co-authors developed a lateral flow nanoparticle aggregation assay. Hybridization of a miRNA to cDNA-modified particles produced a visual colorimetric change on account of nanoparticle aggregation in less than 20 minutes.92 To increase sensitivity, a next generation sensor was developed that used nanoparticles conjugated to horseradish peroxidase that, in a subsequent step, catalyzed the oxidation of TMB to produce a blue product. This amplification step improved the limit of detection from 60 pM to 7.5 pM.93 A potential drawback is the lack of multiplexing capabilities as different targets would require parallel detection using separate devices.
Techniques like traditional Kretschmann geometry SPR and SPR imaging have previously been applied to the detection of miRNAs; however, when operated in label-free assays, limits of detection are only modest, as the small size of a captured miRNA does not cause a large change in refractive index at the sensor surface. To improve upon these detection limits, several label-included signal amplification strategies have been developed. Many of these approaches involve a high mass recognition element that can recognize and bind directly to the cDNA:miRNA duplex at the sensor surface. A simple example is that of Vaisocherova and coworkers, where a DNA functionalized nanoparticle recognized half of a miRNA sequence where the other half will be bound to an SPR sensor, forming a conventional sandwich complex. Using this approach, illustrated in Figure 7B, they demonstrated a 4-plex assay for miRNA detection in cell lysate with a LOD of 0.5 pM.94 Another approach used biotin-streptavidin interactions to engender a 24-fold increase in detection sensitivity.95 Lastly, an alternative method used DSN and monitored the decrease in signal over time as the cDNA strand was displaced from the surface. Importantly, the miRNA released from the SPR chip surface can rebind to other cDNAs thereby effectively recycling the target. This assay gave a detection limit of 3 fM and could differentiate miR-21 concentrations in total RNA solutions extracted from the blood of a variety of cancer patients.96
Two notable non-plasmonic, photonic detection technologies have also recently shown promise for miRNA analysis. Cunningham and co-workers have pioneered the development photonic crystals for biosensing applications and have demonstrated their applicability to the detection of miRNAs. Photonic crystals were engineered to enhance the fluorescence signal of tagged miRNA sequences hybridized to a cDNA capture probes on the sensor surface.97 Due to the localization of the electric field the fluorophores experience during excitation and enhanced signal extraction of the fluorophore emission through coupling to modes of the photonic crystal, signal gains of more than 8,000 were reported. This sensor was used to quantitate the expression of miRNA-21 and achieved a 0.1 pM LOD. It is also interesting that the ability to quantitate both miRNA and proteins using the same sensing modality. In attempts to cut down the volume needed to complete analysis, the fluorescence enhanced photonic crystal approach was adapted to a submicron fluid channel.98 This hybridization based assay was dependent on miRNAs binding with molecular beacons functionalized at the photonic crystal surface. Using the narrower channel geometry, only 20 nL of sample was needed to complete analysis.
Silicon photonic microring resonators have also been demonstrated for the detection of miRNAs, 99,100 as well as full length mRNAs.101 A simple detection scheme was developed by monitoring direct hybridization of miRNA targets to DNA capture probes on the photonic ring resonator surface.99 The binding event caused a change in refractive index, which, in turn, created a detectable change in resonant wavelength that can be related to the concentration of the target of interest. Using this approach, it took ten minutes to simultaneously profile four miRNAs, and an LOD of 2 nM was reached. To improve the sensing attributes of the platform, a RNA:DNA heteroduplex specific antibody, S9.6, was used.100 After miRNA:DNA capture probe hybridization, the antibody was introduce to the sensor surface and allowed to bind. The added mass of the antibody at the sensor surface caused a larger shift in the resonance wavelength due to a larger change in the refractive index close to the sensor surface. This resulted in improvements to the LOD of over 2 orders of magnitude (10 pM) and a dynamic range over 4 orders of magnitude. Impressively, this assay was able to quantitate four miRNA simultaneously from total RNA isolated from mouse brain tissue.
Electrochemical Detection
Electrochemical sensors are well-established for a number of classes of target analytes, and obvious successes, such as the portable glucose meter, have established their capabilities for simple, rapid, and low cost bioanalysis. Not surprisingly, there has been considerable growth in the area of electrochemical biosensors for miRNA. Given the requirement for measuring current or voltages at a solid electrode, electrochemical miRNA biosensors are typically based on hybridization at the electrode surface, and similar to plasmonic and photonic optical sensors, can be operated in label free and label-enhanced assay formats. As discussed below, detection is commonly achieved through the measurement of a redox signal or a change in capacitance or impedance.
Label-free approaches to electrochemical detection
Label free detection assays are the simplistic electrochemical measurement schemes, relying only upon target hybridization to a surface-bound capture probe. An example of this strategy involved a novel carbon nanofiber functionalized screen printed electrode functionalized with a capture probe having electrochemically-inactive inosine in place of guanine. Hybridization of the miRNA target then generated a detectable guanine oxidation peak that was measured using differential pulse voltammetry with a detection limit of 1.5 μM.102 However, in comparison with other techniques, the sensitivity of this assay can be greatly improved.
Alternative electrode materials have resulted in significantly improved sensitivity. For instance, a RNA duplex specific binding protein, p19, was immobilized on an electrode surface and only when an RNA duplex was present was a signal due to tryptophan oxidation detected. The LOD of this assay was reported at 160 nM.103 Carbon nanotube functionalized glassy carbon electrodes were used to increase guanine oxidation current density by a factor of ~3 over conventional glassy carbon electrodes, achieving a LOD of 1 pM.104 Another method involved using a quinone based conducting polymer as a redox transducer on a functionalized electrode surface. Differential currents were detected based on the orientation of the DNA capture probe—collapsed on the surface in the absence of the target, yet linear and lifted off the electrode when hybridized to the target miRNA, allowing for more efficient diffusion of counter-ions to the surface. Here, the electrode material had a large influence on the sensitivity, where an LOD of 650 fM was obtained using a glassy carbon electrode105 and 8 fM for a carbon nanotube functionalized electrode.106 Using a similar diffusion based mechanism, a Pd-nanoparticle functionalized electrode was used and changes in the ability of H2O2 to diffuse to the electrode surface was measured. When the miRNA was present, it effectively prohibited diffusion in a concentration dependent manner. A detection limit of 1.7 pM can be achieved using this method.107 Additionally, these techniques show an extended dynamic ranging from 3–5 orders of magnitude.
Labeling with electroactive tags
Another approach to detect hybridization events is to label the miRNA:capture probe duplex with an electroactive species to yield a detectable signal. This was achieved by introducing copper ions to the hybrid which electrostatically interact with the negative nucleic acid backbone and catalyze the turnover of ascorbate.108 Similar approaches used methylene blue as a label.109 Methylene blue has a higher affinity for ssDNA versus miRNA hybrids. miRNA binding therefore decreased the electrochemical for methylene blue, as detected via voltammetry. The limits of detection for these approaches were reported as 8.2 fM and 0.5 fM, respectively.
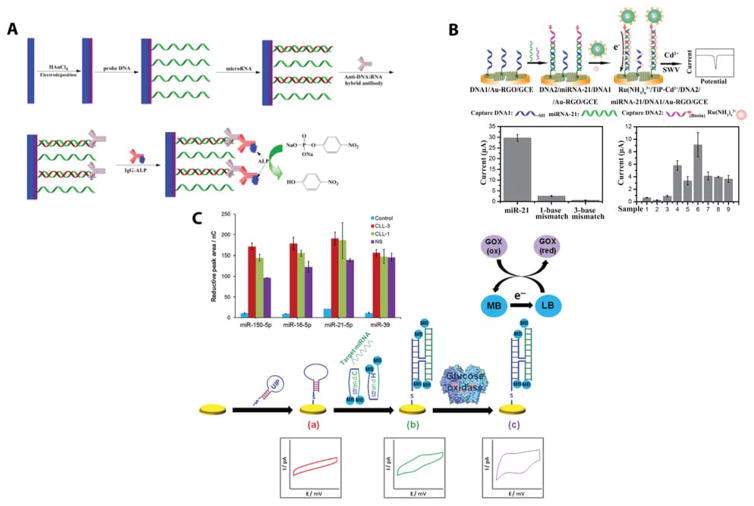
A further step is to directly label the miRNA:capture probe duplexes with either electroactive or catalytic labels. For example, aptamer based capture probes have been designed to bind HRP only when the miRNA is present. This approach offered picomolar LODs, but suffered from the poor specificity of the aptamer sequence.110 Specificity was improved by using a miRNA:DNA specific antibody, S9.6, which, as illustrated in Figure 8A, could then be bound by an ALP-modified IgG antibody and used to achieve a LOD of 0.4 fM.111 Another approach involved ferrocene-boronic acid modified gold nanoparticles, where the boronic acid could form a covalent bond with the ribose sugar of the miRNA bound at the electrode surface. The electrochemical signal from the ferrocene group was then detected using differential pulse voltammetry to give a miRNA detection limit of 1 nM.112
Figure 8.
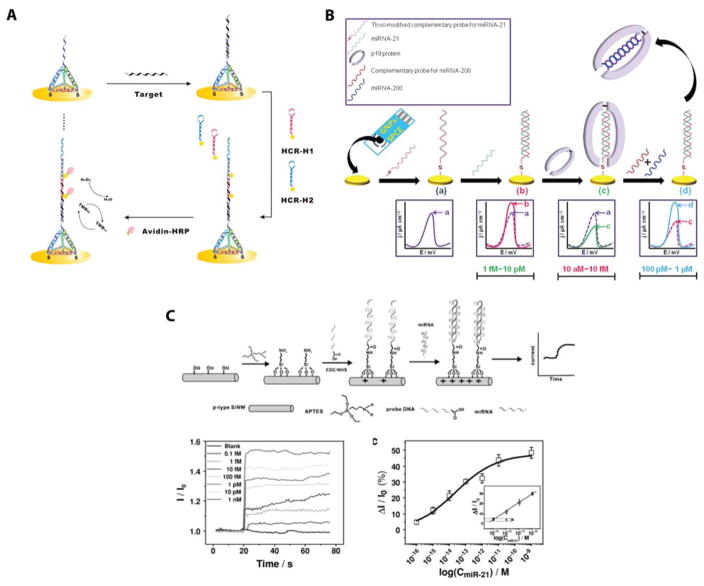
Promising electrochemical signal enhancement approaches by adding an electroactive label to miRNA:DNA hybrids. (A) Labeling was achieved using the S9.6 antibody that binds specifically to RNA:DNA heteroduplexes. After antibody binding, ALP was then conjugated to the surface and provided an electrochemical signal. Reprinted from Electrochemica Acta, Vol. 165, Wang, M.; Li, B.; Zhou, Q.; Yin, H.; Zhou, Y.; Ai, S. Label-free, Ultrasensitive and Electrochemical Immunosensing Platform for microRNA Detection Using Anti-DNA:RNA Hybrid Antibody and Enzymatic Signal Amplification, pp. 130–135 (ref 111). Copyright 2015, with permission from Elsevier. (B) Nanoparticle labeling was achieved using biotin-streptavidin attachement chemistry. This approach was used to identify changes in miRNA-21 expression in serum between healthy patients and patients with various cancer types. Reproduced from Cheng, F.-F.; He, T.-T.; Miao, H.-T.; Shi, J.-J.; Jiang, L.-P.; Zhu, J.-J. ACS Applied Materials & Interfaces 2015, 7, 2979–2985 (ref 118). Copyright 2015 American Chemical Society. (C) A sandwich hybridization based labeling approach was developed using methylene blue conjugated reporter probes that bound to the surface only when the miRNA was present. This strategy effectively placed four methylene blue molecules near the electrode for every one target miRNA, allowing for effective signal amplification. The power of this strategy was sown by detecting three miRNAs in parallel. Reproduced from Labib, M.; Khan, N.; Berezovski, M. V. Analytical Chemistry 2015, 87, 1395–1403 (ref 113). Copyright 2015 American Chemical Society.
Sandwich hybridization based assays have also been shown to facilitate the addition of electroactive species, where a functionalized reporter DNA sequence hybridizes to a DNA capture probe only in the presence of the miRNA target. This approach had the benefit of facilitating the use of a wide variety of reporter tags and electroactive labels. Methylene blue (MB) labeled reporter sequences were designed to bind with the capture probe as well as additional helper sequences to allow the binding of 4 MB molecules per miRNA sequence. An LOD of 100 fM was reported using this assay, coined SensQ, and multiplexing capabilities were demonstrated by the simultaneous quantitation of 3 miRNAs.113 The mechanism of this promising detection platform is shown in Figure 8B.
Another approach utilized aminated reporter detection probes covalently bound to apoferritin-encapsulated copper nanoparticles. Hybridization of the target miRNA led to a pH shift at the electrode, resulting in the release of copper ions from the nanoparticles, which were detected with a LOD of 3.5 fM and a linear range spanning from 0.01 to 10 pM.114 This assay was further modified using streptavidin functionalized reporter sequences to effectively bind multiple apoferritin nanoparticles per reporter strand due to the fact that streptavidin molecules can bind multiple biotins. Trypsase was used to digest the nanoparticles and induce Cu release, and the multivalent streptavidin linkage improved the detection limit by a factor of 10—down to 0.35 fM, with a corresponding improvement in dynamic range as well.115
Biotinylated reporter probes were also used to bind with ALP functionalized streptavidin to provide 0.4 pM LODs and a linear range from 1 pM to 100 nM.116 A similar approach using digoxin functionalized reporter sequences to bind with a HRP-functionalized anti-digoxin antibody claimed a LOD of 0.79 fM with a seven order of magnitude dynamic range. The key innovation of the digoxin based assay that leads to large improvement in sensitivity was the use of a Au and Ag modified dendrimer-chitosan-graphene composite electrode.117 The largest sensitivity enhancements were seen using biotinylated reporter sequences to bind with streptavidin functionalized titanium phosphate nanospheres that have incorporated Cd2+ ions. After binding of the nanospheres to the miRNA:capture probe duplex, [Ru(NH3)6]3+ electrostatically interacted with the nucleic acid backbone and served as an electron carrier between the electrode surface and the nanosphere. The electrochemical response of Cd2+ was then used to quantify the presence of miRNA sequences. Impressively, this workflow reaches an LOD of 0.76 aM with linear range spanning seven orders.118 This impressive detection strategy is shown in Figure 8C.
Redox cycling reactions have also been developed in an attempt to provide high sensitivity miRNA detection. These labeling approaches seek to eliminating diffusion limitations in solution using mercaptophenylboronic acid,119 APBA120 and DNAzyme121 functionalized gold nanoparticles. Despite promise, these early efforts did not show performance metric improvements over the previously discussed electroactive tagging approaches.
Signal amplification via conjugation of multiple electroactive reporters
The small size of miRNA typically means that the sequence can only be covalently tagged with a single label. Similar to that described above for optical methods, target recycling methods can be employed to generate larger per-target electrochemical signals through either the hybridization chain reactions or enzymatic lengthening processing. Particularly effective strategies in this vein are highlighted in Figure 9.
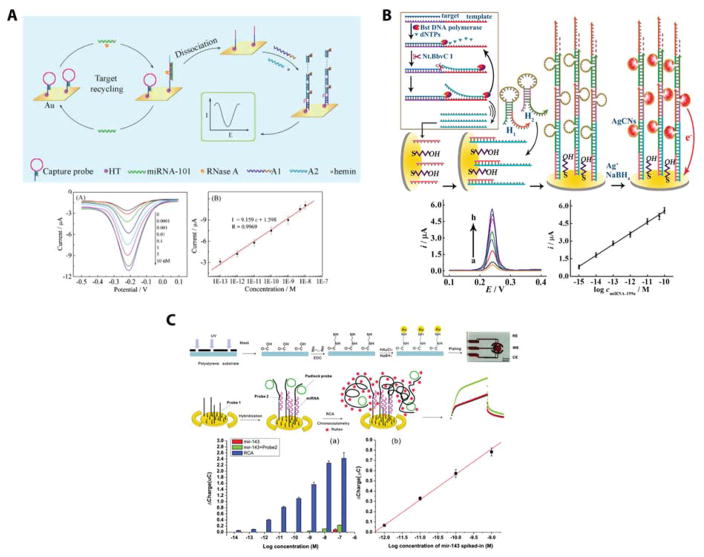
Figure 9.
Electrochemical signal amplification using enzymatic approaches to modify the miRNA:capture probe structure at the sensor surface. (A) Target recycling via RNase A was initially used to increase the number of targets that bound to the surface. The detectable electrochemical signal was then amplified via hybridization chain reaction which formed multiple hemin associated G-quadruplexes and thus an electrochemical signal. Reprinted from Sensors and Actuators B, Vol. 195, Xiang, G.; Jiang, D.; Luo, F.; Liu, F.; Liu, L.; Pu, X. Sensitive detection of microRNAs using hemin/G-quadruplex concatamers as trace labels and RNA endonuclease-aided target recycling for amplification, pp. 515–519 (ref 130). Copyright 2014, with psermission from Elsevier. (B) A nicking enzyme based recycling strategy that relies on DNA polymerase amplification was used to create multiple detection sequences that are proportional to the miRNA target concentration. This is followed by hybridization chain reaction where the primers are designed to associate with silver nanoclusters. Reproduced from Yang, C.; Shi, K.; Dou, B.; Xiang, Y.; Chai, Y.; Yuan, R. ACS Applied Materials & Interfaces 2015, 7, 1188–1193 (ref 132). Copyright 2015 American Chemical Society. (C) Rolling circle amplification was also shown to be an effective way to facilitate binding of multiple electroactive species, in this case Ruhex, per miRNA target. Reproduced from Yao, B.; Liu, Y.; Tabata, M.; Zhu, H.; Miyahara, Y. Chemical Communications 2014, 50, 9704–9706 (ref 135), with permission of The Royal Society of Chemistry.
Enzyme free miRNA recycling techniques, such as mismatch catalytic hairpin assemblies that convert one miRNA molecule into a DNA duplex122 and toehold mediated strand displacement reactions123 have been interfaced with electrochemical detection to reach limits of detection of 0.6 pM and 1.4 fM, respectively. Sensitivities of the catalyzed hairpin assembly were improved 3-fold using TiO2 nanoparticles and redox cycling.124 Enzymatic recycling can also be achieved using double-stranded nuclease125 and T7 exonuclease,126 where LODs were 1 fM and 0.17 fM, respectively. Significant improvements in sensitivity were when LNA G-quaduplex-hemin DNAzymes were located in close proximity to the electrode surface. As the target miRNA target bound, it was degraded by double stranded nuclease making the DNAzyme more accessible to hemin binding and signal amplification. This gave an impressive detection limit of 8 aM; however, the linear range reported was prohibitively narrow.127
Hybridization chain reactions are another amplification technique that do not require enzymatic processing yet can provide “gain” by improving accessibility for signal reporter molecules to intercalate between base pairs. Studies have used this workflow with [Ru(NH3)6]3+ as an intercalating agent to achieve a six order of magnitude dynamic range and an LOD of 100 aM.128 Li and co-workers were able to combine multiple p19 proteins on a magnetic bead and use a hybridization chain reaction mechanism using novel DSA molecules as signal reporters reporting a detection limit of 6 aM.129 Unfortunately, this assay is only linear over one order of magnitude. Target recycling via RNaseA was also combined with the hybridization chain reaction using GC rich sequences that allow for enhanced DNAzyme activity resulting in a LOD of 100 fM with a dynamic range of over 5 orders of magnitude.130
Other methodologies have combined DNA lengthening and target recycling amplification techniques. For example, catalyzed hairpin assembly and hybridization chain reaction were combined with a methylene blue-based read-out,131 and a nicking enzyme and DNA polymerase were utilized to amplify the miRNA target before a hybridization cascade using stem loop primers, which facilitated silver nanocluster association.132 Unfortunately, the limit of detection in both cases was in the femtomolar range, most likely due to the off target response of closely related miRNA species.131 Both assays reported dynamic ranges of 5 orders of magnitude.
Additional enzymatic amplification approaches have also been investigated using a variety of DNA polymerase processing approaches. A simple approach used DNA polymerase to add biotinylated nucleotides as the miRNA strand was elongated to facilitate downstream streptavidin conjugated gold nanoparticle association and signal amplification. This provided a LOD of 99.2 fM and a dynamic range of four orders of magnitude. 133 An alternative approach used strand displacement amplification, enabled by a nicking enzyme and DNA polymerase, followed by sandwich hybridization of the capture probe, the amplification product, and then a biotinylated reporter sequence to bind to streptavidin-HRP conjugates. Using this approach an LOD of 40 pM is achieved with a dynamic range of 2 orders of magnitude.134 A third strategy relied on rolling circle amplification to lengthen a DNA sequence through the production of thousands of repeated sequences. This added sequence was used to conjugate redox probes135 or to prevent diffusion of electroactive species from the electrode surface,136 resulting in LODs of 100 fM and 1.2 fM and dynamic ranges of 4 and 2 order of magnitudes, respectively. Isothermal exponential amplification reaction (EXPAR) has also been explored via sandwich hybridization of the EXPAR product and a biotinylated reporter for ALP amplification137 or DNAzyme formation.138
Lastly, an emerging method, shown in Figure 10A, used design rules from DNA nanotechnology to improve the orientation of capture probes at the surface, thereby mitigating the negative consequences of poor sterics of the electroactive labeling species and reducing and non-specific binding to the electrode surface. This general approach method was utilized in several different assay formats, including a sandwich hybridization assay,139 a target recycling process using a silver nanoparticle functionalized signal probe,140 a rolling circle amplification process,141 and with a hybridization chain reaction scheme.142 The LODs for these respective assays were 1 fM, 0.4 fM, 50 aM, and 10 aM respectively. Additionally, dynamic ranges spanned between 4–6 orders of magnitude. These results underscore how rational surface functionalization can have a profound effect on the ultimate performance of the sensor.
Figure 10.
General electrochemical sensing improvements and additional strategies for generating a detectable signal. (A) DNA nanotechnology has been used to improve surface functionalization by ensuring reproducible capture probe orientation at the electrode surface. Reproduced from Ge, Z.; Lin, M.; Wang, P.; Pei, H.; Yan, J.; Shi, J.; Huang, Q.; He, D.; Fan, C.; Zuo, X. Analytical Chemistry 2014, 86, 2124–2130 (ref 142). Copyright 2014 American Chemical Society. (B) Impedance can be used to detect the presence of miRNAs. Here, three different detection regimes (label free detection, protein binding based signal amplification, and protein dissociation based signal amplification) were identified to extend the dynamic range over 10 orders of magnitude. Reproduced from Labib, M.; Khan, N.; Ghobadloo, S. M.; Cheng, J.; Pezacki, J. P.; Berezovski, M. V. Journal of the American Chemical Society 2013, 135, 3027–3038 ref 146). Copyright 2013 American Chemical Society. (C) Based on impedance, field effect transistors are a simple way to achieve multiplexing capabilities with very low limits of detection. Reproduced from CMOS-Compatible Silicon Nanowire Field-Effect Transistors for Ultrasensitive and Label-Free microRNAs sensing, Lu, N.; Gao, A.; Dai, P.; Song, S.; Fan, C.; Wang, Y.; Li, T. Small, Vol. 10, Issue 10, (ref 154). Copyright 2014, Wiley.
Electrochemical impedance
Electrochemical impedance is another electrochemical property that can be measured in a way to reflect the presence of a targeted miRNA sequence. Strategies to induce a change in charge transfer include the enzymatic turnover of an insulating polymer via DNAzymes,143 hemin conjugated carboxylic graphene144 and DNAzyme functionalized gold nanoparticles145. These assays reported detection limits ranging from 100s of aM to 10 of fM LODs with dynamic ranges of 1.5, 3.5, and 4.5 orders, respectively. The assay that shows the largest gain to sensitivity and specificity of any label addition method is based on a three part impedance system and developed by Labib and coworkers.146 Here, the first step of the assay was to measure the hybridization event of the target miRNA and RNA capture probe. If the signal could not be detected, the p19 RNA binding protein, which is specific to small 21–23 bp RNA duplex via electrostatic and hydrogen bonding interactions between the β-sheet formed by the p19 homodimer and the sugar-phosphate backbone of the dsRNA, was used to amplify the impedance signal. Lastly, a DNA compliment could be added to induce the disassociation of the p19 protein from the electrode signal to further amplify the signal. All three steps form a “three-mode” electrochemical sensor, and each “mode” had a different linear range that together allowed quantitative detection between 10 aM and 1 μM. This multi-step assay is highlighted in Figure 10B. Because of the non-specific nature of the p19 protein, multiplexing must be achieved by splitting the sample into separate volumes; however, the team reported a 3 plex assay in total RNA and validated the results with qPCR.
Magnetic bead-enhanced electrochemical detection
Mass transfer limits are significant hurdles to ultrasensitive target detection. An appealing general approach to circumvent these Langmurian limitations is to use magnetic beads that can diffuse quickly through solution to capture targets of interest, but then be localized onto the surface of a detection element using an external magnetic field. This approach has been widely exploited as a method of sample pre-concentration for a range of analyte classes in both label and label free measurement strategies. Below are several examples where magnetic beads were combined with electrochemical-based read out schemes.
Using a similar inosine-substituted capture probe as described above, arrays of screen printed electrodes were used in combination with magnetic beads to enable multiplexed measurement of miRNAs with increased sensitivity. After pre-concentration with magnetic beads, an alkaline treatment released the duplexes from the beads where they are then absorbed on graphite screen printed electrodes and guanine oxidation is measured. Multiplexing capabilities were shown by running three reactions in parallel with a limit of detection of 143 nM.147 A different iteration of this workflow incubated miRNA solutions with Os(VI)bipy, which electrostatically associates to the miRNA. Magnetic beads with complimentary DNA capture probes were then used to capture target miRNA. The labeled target miRNA were thermally released and quantitated using the peak current detected from the Os label using a mercury drop electrode.148 The main drawback of this strategy this the limited dynamic range that covers approximately 1 order of magnitude. Additionally, redox cycling can improve specificity and extend dynamic range. Workflows have been presented that are dependent on the ligation of a magnetic bead functionalized capture probe and a biotinylated reporter sequence. Conjugation of SA-ALP to the complex catalyzes the production of an electroactive species, 1-naphthol. The supernatant of this process is collected and introduced to a separate electrode, where the concentration of 1-napthol is measured through redox cycling. The LOD was reported to be 3.55 fM and a dynamic range of 4 orders of magnitude.149
Magnetic beads and electrodes have also been employed in tandem. For example, magnetic beads were used to detect biotinylated duplex specific nuclease products. In the presence of the target miRNA, the biotinylated probe was fully digested; however, in the absence of target, it was left intact. These products were absorbed onto a streptavidin coated magnetic bead and pulled down to a magnetic electrode, where an impedance measurement was made. The fully digested probes give lower impedance values compared to the intact capture probes that have a high charge density. This workflow resulted in an LOD of 60 aM and a dynamic range of 2 orders of magnitude.150 An alternative approach uses branching magnetic beads that hybridize to multiple miRNA targets through the formation of Y junctions. As a result of this branching, multiple HRP moieties could bind to the y junctions, and when pulled down to the electrode surface, could give a LOD of 0.22 aM and a four order of magnitude dynamic range.151
An additional approach immobilizes the RNA duplex specific protein, p19, on magnetic beads and uses it as a capture probe for biotinylated RNA capture probe:miRNA hybrids following streptavidin-HRP conjugation. This complex was then pulled down to the electrode where a detectable catalytic current was measured giving a 40 pM LOD.152 However, the approximately 1 order of magnitude is limiting for many applications. The ultimate limit of this p19-based detection approach results from the nanomolar affinity of the RNA duplex:p19 interaction, as a higher affinity would yield lower limits of detection. Lastly, a DNA ligase-dependent sandwich hybridization/redox amplification strategy was reported having a gold nanocluster-ALP complex functionalized to a sequence-specific reporter. After ligation to the MB in the presence of the miRNA, the gold nanocluster-ALP complex catalyzed the production of silver nanoclusters which absorbed onto the bound DNA strand. The resulting product was then brought to the surface via a magnetic electrode and the silver content quantitated. The limit of detection using this approach is 21.5 aM with a dynamic range of 3 orders of magnitude.153
Field Effect Transistors
Field effect transistors (FETs) are an attractive class of sensors from the perspective of potentially low-cost devices that have high sensitivivity to binding-induced changes in charge near the sensor surface. The sensing mechanism of FETs is illustrated in Figure 10C. Recent efforts have aimed at demonstrating the versatility of this transducer through the use of CMOS-compatible silicon nanowire transistors,154 gold nanoparticle functionalized graphene FETs,155 and p19 functionalized FETs,156 with LODs of 0.13 fM, 10 fM, and 1 aM, respectively. The main drawback of this assay is the shallow sensing depth in solution, which is limited by the Debye length. This precludes the use of many labeling or strand extension techniques as they will occur outside the sensing window.
Perspectives
Since their discovery in the 1990s, our understanding of miRNAs has unraveled a new layer of regulatory control over gene expression in organisms. As the importance of miRNAs has become clearer, the number of platforms available to analyze these molecules has grown. Initially efforts focused on applying and modifying traditional techniques from molecular biology to allow for the analysis of miRNAs. For example, the creation of stem loop primers and new enzymatic methods that facilitated qRT-PCR and microarrays to be applied to this class of small RNAs. These techniques were essential to early breakthroughs; however, they are now being replaced with new workflows that offer greater coverage of global expression changes, as evidenced by the rapid gains in next generation sequencing, as applied to small RNA analysis.
While RNA sequencing will continue to be a valuable tool for discovery and fundamental studies, emerging biosensor technologies are posed to play a role in translating basic biomolecular insights into the clinic. The last few years have seen tremendous growth in this area. Optical and electrochemical biosensors have been prominent for decades, but recent improvements in sensitivity, dynamic range, time to result, and surface functionalization have rendered them amenable to miRNA analysis. Novel materials and reagents, such as metallic nanoparticles, semiconductor quantum dots, and intercalating small molecule fluorescent dyes, as well as advantageous electrical and optical properties of innovative micro- and nanostructures continue to open up new opportunities, resulting in detection limits down to aM levels and dynamic ranges spanning seven orders of magnitude. Additionally, these measurements have been made in a wide array of biofluids with minimal, if any, sample pre-treatment, greatly simplifying the analytical workflows compared to existing gold standard techniques from molecular biology. Armed with these and other new detection methodologies, the analytical community is now poised to shift attention from sensor development to deployment where the ultimate successes will be judged by the ability to make meaningful impacts in the clinical space.
Future improvements
While emerging techniques have shown tremendous improvements in sensitivity, specificity, and dynamic range, advances are needed on several fronts in order for the ultimate potential of these technologies to be realized. More attention needs to be given to make these assays capable of making multiplexed measurements. Moreover, the development of disease-relevant diagnostics will only be achieved through coupling with bioinformaticians to parse global expression profiles into clinically-actionable biomarker panels
Challenges with multiplexing
In recent years, it is becoming increasingly clear that miRNA panels can be used in clinical applications, as numerous reports describe the use of miRNA biomarkers for a range of human diseases.157,158 Additionally, systems level studies are revealing the interconnectivity between miRNAs and targets within regulatory networks. To this end, predictive bioinformatic approaches are still needed to both predict miRNA targets and help deconvolve complex miRNA-mRNA regulatory interactions.
Quite possibly due to a paucity of robust technologies able to perform multiplexed analysis on statistically-relevant patient populations, there are a limited number of reports that demonstrate the utility of multiplexed panels. As the push towards multiplexing continues, considerable challenges must be overcome in terms of differentiating between sequences having high levels of similarity, which is difficult to achieve because of subtle thermodynamic differences in hybridization-based assays. A further complication with multiplexing is dynamic range, as miRNAs expression levels can vary by more than 5 orders of magnitude, which would be difficult to span is sequences having highly disparate concentrations were included in a single panel.
Optical detection methods have led the way in terms of the analytical capacity to perform multiplexed detection. Studies featuring both fluorescence and plasmonic sensors were used to demonstrate three-52,55,61,70 and four-plex90,94 assays with a relatively short time to result. However, increased clinical utility will likely be gained beyond proof-of-principle studies as the multiplexing is increased into the 10s of targets. Electrochemical sensors have also been shown to be promising for miRNA detection, with reported demonstrations of up to three-plex assays using a variety of different specific detection mechanisms.113,146,147,151 However, the instrumentation required to facilitate multiplexed detection requires either parallel analysis, which then requires multiple sample aliquots, or more complex instrumentation that involves multi-electrode configurations and multiple potentiostats.
Challenges in informatics and the identification of disease-relevant miRNA panels
While the engineering of multiplexed detection approaches is promising from the perspective of technology alone, a significant challenge remains in the informatics behind panel construction. Next generations sequencing approaches can provide an almost overwhelming amount of expression information that must be informatically-reduced in the appropriate disease and population statistical context. As multiplexed detection approaches continue to mature, progress in informatics will ideally ripen at an equivalent rate.
One difficulty in correlating miRNA expression with their functional effects on disrupting mRNA translation stems from how miRNAs are bioinformatically identified. As opposed to siRNA, miRNAs do not require a perfect antisense match against a potential mRNA target. These so-called “noncanonical” miRNA-mRNA interactions are not confined to translational repression through binding to the 3′ untranslated region (UTR) of the mRNA, as originally suspected, but play multiple complex and not completely understood roles in mRNA regulation.159 To this end, recent studies have aimed to build bioinformatics approaches that broadly consider both canonical and noncanonical miRNA-mRNA targets,160,161 and other promising computational tools applied to miRNAs have recently been reviewed.162
In addition to challenges associated with target prediction, another layer of computational complexity lies in the fact that multiple miRNAs often act on single mRNA targets, and therefore it is difficult to conclude what effect a single miRNA has on a biological state. Despite this multi-factorial regulation, a number of single miRNA knockout studies exist in the literature although their relevance to a broad understanding of miRNAs in disease is not clear. Therefore, incredible opportunities exist for sensor scientists to work together with bioinformaticians to develop multiplexed panels that can together assemble technologies and relevant panels to help elucidate deep and meaningful correlations between multi-node regulatory networks.
Challenges in matching technologies to appropriate clinical needs
In order to achieve widespread use outside academic labs, miRNA assays must be cost-effective and easy to use. This is an important consideration when engineering signal amplification steps that require complicated liquid handling steps. Additionally, researchers must consider the tradeoff between time to result and assay sensitivity. Depending on the application, label free assays that can be completed in 10 minutes might suffice; whereas, signal amplification techniques that provide sensitivity improvements that take 2 hours or more to complete might be required for other applications. Lastly, the pre-analytical requirements for different technologies and applications must be considered. That is, a label free assay might suffice for applications where miRNA has been extracted for a sample of interest, whereas one may have to leverage signal amplification and pre-concentration strategies to make high fidelity measurements directly from highly complex matrices.
In order to optimize assays and determine the appropriate balance between time to result, sensitivity, and multiplexing capabilities, interdisciplinary collaborations between analytical chemists, clinical chemists, clinical practitioners, and statisticians/bioinformaticians will be essential. Appropriate large sample set studies will not only validate emerging sensor technologies, but more importantly also establish the broad utility of panel-based diagnostics that will then have impact beyond any one specific technology platform.
Brief conclusions
Though many of the challenges outlined above are significant, the miRNA sensing field is poised for a bright future. Keeping in mind the great strides that have been made to overall assay attributes (i.e. sensitivity and dynamic range), miRNA detection using emerging biosensing technologies is far beyond the proof-of-principle stage. The next few years will hopefully show a shift away from fundamental sensor development towards the identification and validation of multiplexed panels, and then onto clinical translation. Strategic collaborations should enhance this process as bioinformatic approaches evolve alongside detection technology maturation. These collaborations will help catalyze this shift in focus from further pushing LODs and performing proof-of-concept sensing studies to placing instrumentation in clinical settings to revolutionize diagnostic capabilities by supporting or replacing current gold standard techniques. Although there are sure to be challenges along the way, the ultimate goal of sensitive, multiplexed, and easy to use miRNA detection devices is on the horizon, and will hopefully help miRNAs fill a key role in the realization of informative diagnostics guiding individualized medicine.
Acknowledgments
The authors gratefully acknowledge financial support from the National Cancer Institute of the Institutes of Health through Grant R33CA1774. R.M.G. acknowledges support at UIUC from the NIH National Cancer Institute Alliance for Nanotechnology in Cancer ‘Midwest Cancer Nanotechnology Training Center’ Grant R25CA154015A. The content within is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Biographies
Richard M. Graybill is a NIH-funded Midwest Cancer Nanotechnology Center trainee and Ph.D. candidate in the Department of Chemistry at the University of Illinois at Urbana-Champaign. He received a B.S. in Chemistry from Furman University in 2012. At Furman, he investigated the chemotherapeutic properties of novel Cr(III) tris-diimine complexes using a variety of electrophoretic techniques under the direction of John F. Wheeler. His work in the lab of Ryan C. Bailey focuses on the development of silicon photonic microring resonators for use as multiplexed miRNA biosensors, with specific applications in disease diagnosis using circulating miRNA biomarkers, cancer subclassficiation of tumor tissue using miRNA panels, and discrimination of tissue types based on miRNA signatures.
Ryan C. Bailey Ryan C. Bailey is a Professor in the Department of Chemistry at the University of Illinois at Urbana-Champaign. He received degrees in chemistry from Eastern Illinois University (B.S.) and Northwestern University (Ph.D.), and completed postdoctoral training at Caltech and the Institute for Systems Biology. Prof. Bailey’s research focuses on developing enabling bioanalytical tools for a range of applications, with a focus on translational biomedical microdevices. Prof. Bailey was named a Technology Review Magazine Top Innovator under 35 and has been recognized with a Sloan Foundation Fellowship, the Arthur F. Findeis Award for Achievements by a Young Analytical Scientist, Pittcon Achievement Award, and a NIH Director’s New Innovator Award, among others.
References
- 1.Wightman B, Ha I, Ruvkun G. Cell. 1993;75:855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- 2.Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR, Ruvkun G. Nature. 2000;403:901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- 3.Wienholds E, Kloosterman WP, Miska E, Alvarez-Saavedra E, Berezikov E, Bruijn Ed, Horvitz HR, Kauppinen S, Plasterk RHA. Science. 2005;309:310–311. doi: 10.1126/science.1114519. [DOI] [PubMed] [Google Scholar]
- 4.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, Downing JR, Jacks T, Horvitz HR, Golub TR. Nature. 2005;435:4653–4662. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 5.Winter J, Jung S, Keller S, Gregory RI, Diederichs S. Nature Cell Biology. 2009;11:228–234. doi: 10.1038/ncb0309-228. [DOI] [PubMed] [Google Scholar]
- 6.Lin S, Gregory RI. Nat Rev Cancer. 2015;15:321–333. doi: 10.1038/nrc3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ha M, Kim VN. Nat Rev Mol Cell Biol. 2014;15:509–524. doi: 10.1038/nrm3838. [DOI] [PubMed] [Google Scholar]
- 8.O’Connell RM, Rao DS, Baltimore D. Annual Review of Immunology. 2012;30:295–312. doi: 10.1146/annurev-immunol-020711-075013. [DOI] [PubMed] [Google Scholar]
- 9.Johnston RJ, Hobert O. Nature. 2003;426:845–849. doi: 10.1038/nature02255. [DOI] [PubMed] [Google Scholar]
- 10.Shenoy A, Blelloch RH. Nat Rev Mol Cell Biol. 2014;15:565–576. doi: 10.1038/nrm3854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hwang HW, Mendell JT. Br J Cancer. 2006;94:776–780. doi: 10.1038/sj.bjc.6603023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rosenfeld N, Aharonov R, Meiri E, Rosenwald S, Spector Y, Zepeniuk M, Benjamin H, Shabes N, Tabak S, Levy A, Lebanony D, Goren Y, Silberschein E, Targan N, Ben-Ari A, Gilad S, Sion-Vardy N, Tobar A, Feinmesser M, Kharenko O, Nativ O, Nass D, Perelman M, Yosepovich A, Shalmon B, Polak-Charcon S, Fridman E, Avniel A, Bentwich I, Bentwich Z, Cohen D, Chajut A, Barshack I. Nat Biotech. 2008;26:462–469. doi: 10.1038/nbt1392. [DOI] [PubMed] [Google Scholar]
- 13.Keshavarz M, Behpour M, Rafiee-pour H-A. RSC Advances. 2015;5:35651–35660. [Google Scholar]
- 15.Kumar P, Dezso Z, MacKenzie C, Oestreicher J, Agoulnik S, Byrne M, Bernier F, Yanagimachi M, Aoshima K, Oda Y. PLoS ONE. 2013;8:e69807. doi: 10.1371/journal.pone.0069807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shafi G, Aliya N, Munshi A. Canadian Journal of Neurological Sciences/Journal Canadien des Sciences Neurologiques. 2010;37:177–185. doi: 10.1017/s0317167100009902. [DOI] [PubMed] [Google Scholar]
- 17.Guay C, Regazzi R. Nature Reviews Endocrinology. 2013;9:513–521. doi: 10.1038/nrendo.2013.86. [DOI] [PubMed] [Google Scholar]
- 18.Thum T, Mayr M. Cardiovascular Research. 2012;93:543–544. doi: 10.1093/cvr/cvs085. [DOI] [PubMed] [Google Scholar]
- 19.Olson EN. Science Translational Medicine. 2014;6:239ps233–239ps233. doi: 10.1126/scitranslmed.3009008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Weber JA, Baxter DH, Zhang S, Huang DY, How Huang K, Jen Lee M, Galas DJ, Wang K. Clinical Chemistry. 2010;56:1733–1741. doi: 10.1373/clinchem.2010.147405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, Peterson A, Noteboom J, O’Briant KC, Allen A, Lin DW, Urban N, Drescher CW, Knudsen BS, Stirewalt DL, Gentleman R, Vessella RL, Nelson PS, Martin DB, Tewari M. Proceedings of the National Academy of Sciences. 2008;105:10513–10518. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schwarzenbach H, Hoon DSB, Pantel K. Nat Rev Cancer. 2011;11:426–437. doi: 10.1038/nrc3066. [DOI] [PubMed] [Google Scholar]
- 23.Cissell KA, Shrestha S, Deo SK. Analytical Chemistry. 2007;79:4754–4761. [Google Scholar]
- 24.Qavi A, Kindt J, Bailey R. Anal Bioanal Chem. 2010;398:2535–2549. doi: 10.1007/s00216-010-4018-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dong H, Lei J, Ding L, Wen Y, Ju H, Zhang X. Chemical Reviews. 2013;113:6207–6233. doi: 10.1021/cr300362f. [DOI] [PubMed] [Google Scholar]
- 26.Bartel DP. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 27.Rajewsky N. Nat Genet. 2006 [Google Scholar]
- 28.Muniategui A, Nogales-Cadenas R, Vázquez M, Aranguren LX, Agirre X, Luttun A, Prosper F, Pascual-Montano A, Rubio A. PLoS ONE. 2012;7:e30766. doi: 10.1371/journal.pone.0030766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meng W, McElroy JP, Volinia S, Palatini J, Warner S, Ayers LW, Palanichamy K, Chakravarti A, Lautenschlaeger T. PLoS ONE. 2013;8:e64393. doi: 10.1371/journal.pone.0064393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pritchard CC, Cheng HH, Tewari M. Nature Reviews Genetics. 2012;13:358–369. doi: 10.1038/nrg3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hunt EA, Broyles D, Head T, Deo SK. Annual Reviews in Analytical Chemistry. 2015;8:217–237. doi: 10.1146/annurev-anchem-071114-040343. [DOI] [PubMed] [Google Scholar]
- 32.Chen C, Ridzon DA, Broomer AJ, Zhou Z, Lee DH, Nguyen JT, Barbisin M, Xu NL, Mahuvakar VR, Andersen MR, Lao KQ, Livak KJ, Guegler KJ. Nucleic Acids Research. 2005;33:e179–e179. doi: 10.1093/nar/gni178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ballantyne KN, van Oorschot RAH, Mitchell RJ. Genomics. 2008;91:301–305. doi: 10.1016/j.ygeno.2007.10.016. [DOI] [PubMed] [Google Scholar]
- 34.D’haene B, Mestdagh P, Hellemans J, Vandesompele J. In: Next-Generation MicroRNA Expression Profiling Technology. Fan J-B, editor. Humana Press; 2012. pp. 261–272. [Google Scholar]
- 35.Peltier HJ, Latham GJ. RNA. 2008;14:844–852. doi: 10.1261/rna.939908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sarkar D, Parkin R, Wyman S, Bendoraite A, Sather C, Delrow J, Godwin AK, Drescher C, Huber W, Gentleman R, Tewari M. Nucleic Acids Research. 2009;37:e17–e17. doi: 10.1093/nar/gkn932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roberts TC, Coenen-Stass AML, Wood MJA. PLoS ONE. 2014;9:e89237. doi: 10.1371/journal.pone.0089237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thomson JM, Parker J, Perou CM, Hammond SM. Nat Meth. 2004;1:47–53. doi: 10.1038/nmeth704. [DOI] [PubMed] [Google Scholar]
- 39.Berezikov E, Cuppen E, Plasterk RHA. Nat Genet. 2006 doi: 10.1038/ng1794. [DOI] [PubMed] [Google Scholar]
- 40.Weiler J, Gausepohl H, Hauser N, Jensen ON, Hoheisel JD. Nucleic Acids Research. 1997;25:2792–2799. doi: 10.1093/nar/25.14.2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Castoldi M, Schmidt S, Benes V, Noerholm M, Kulozik AE, Hentze MW, Muckenthaler MU. RNA. 2006;12:913–920. doi: 10.1261/rna.2332406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee JM, Cho H, Jung Y. Angewandte Chemie International Edition. 2010;49:8662–8665. doi: 10.1002/anie.201004000. [DOI] [PubMed] [Google Scholar]
- 43.Duan D, Zheng K-x, Shen Y, Cao R, Jiang L, Lu Z, Yan X, Li J. Nucleic Acids Research. 2011 doi: 10.1093/nar/gkr774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ueno T, Funatsu T. PLoS ONE. 2014;9:e90920. doi: 10.1371/journal.pone.0090920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li W, Ruan K. Anal Bioanal Chem. 2009;394:1117–1124. doi: 10.1007/s00216-008-2570-2. [DOI] [PubMed] [Google Scholar]
- 46.Metzker ML. Nat Rev Genet. 2010;11:31–46. doi: 10.1038/nrg2626. [DOI] [PubMed] [Google Scholar]
- 47.Mardis ER. Annual Review of Analytical Chemistry. 2013;6:287–303. doi: 10.1146/annurev-anchem-062012-092628. [DOI] [PubMed] [Google Scholar]
- 48.Git A, Dvinge H, Salmon-Divon M, Osborne M, Kutter C, Hadfield J, Bertone P, Caldas C. RNA. 2010;16:991–1006. doi: 10.1261/rna.1947110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tam S, de Borja R, Tsao M-S, McPherson JD. Lab Invest. 2014;94:350–358. doi: 10.1038/labinvest.2013.157. [DOI] [PubMed] [Google Scholar]
- 50.Calin GA, Ferracin M, Cimmino A, Di Leva G, Shimizu M, Wojcik SE, Iorio MV, Visone R, Sever NI, Fabbri M, Iuliano R, Palumbo T, Pichiorri F, Roldo C, Garzon R, Sevignani C, Rassenti L, Alder H, Volinia S, Liu C-g, Kipps TJ, Negrini M, Croce CM. New England Journal of Medicine. 2005;353:1793–1801. doi: 10.1056/NEJMoa050995. [DOI] [PubMed] [Google Scholar]
- 51.Schultz NA, Dehlendorff C, Jensen BV, et al. JAMA. 2014;311:392–404. doi: 10.1001/jama.2013.284664. [DOI] [PubMed] [Google Scholar]
- 52.Wu P, Tu Y, Qian Y, Zhang H, Cai C. Chemical Communications. 2014;50:1012–1014. doi: 10.1039/c3cc46773b. [DOI] [PubMed] [Google Scholar]
- 53.Lee CH, Chae Ji, Ko HY, Kim S. Journal of Molecular Imaging & Dynamics. 2013:2. [Google Scholar]
- 54.Larkey NE, Almlie CK, Tran V, Egan M, Burrows SM. Analytical Chemistry. 2014;86:1853–1863. doi: 10.1021/ac403866g. [DOI] [PubMed] [Google Scholar]
- 55.Qiu X, Hildebrandt N. ACS Nano. 2015;9:8449–8457. doi: 10.1021/acsnano.5b03364. [DOI] [PubMed] [Google Scholar]
- 56.Zhang H, Wang Y, Zhao D, Zeng D, Xia J, Aldalbahi A, Wang C, San L, Fan C, Zuo X, Mi X. ACS Applied Materials & Interfaces. 2015;7:16152–16156. doi: 10.1021/acsami.5b04773. [DOI] [PubMed] [Google Scholar]
- 57.Lee J, Park G, Min D-H. Chemical Communications. 2015;51:14597–14600. doi: 10.1039/c5cc04706d. [DOI] [PubMed] [Google Scholar]
- 58.Tu Y, Li W, Wu P, Zhang H, Cai C. Analytical Chemistry. 2013;85:2536–2542. doi: 10.1021/ac303772m. [DOI] [PubMed] [Google Scholar]
- 59.Ko HY, Lee J, Lee YS, Gu H-N, Ali BA, Al-Khedhairy AA, Heo H, Cho S, Kim S. Chemical Communications. 2015;51:2159–2161. doi: 10.1039/c4cc08898k. [DOI] [PubMed] [Google Scholar]
- 60.Yoo B, Kavishwar A, Ghosh SK, Barteneva N, Yigit MV, More A, Medarova Z. Chemistry & Biology. 2014;21:199–204. doi: 10.1016/j.chembiol.2013.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ryoo S-R, Lee J, Yeo J, Na H-K, Kim Y-K, Jang H, Lee JH, Han SW, Lee Y, Kim VN, Min D-H. ACS Nano. 2013;7:5882–5891. doi: 10.1021/nn401183s. [DOI] [PubMed] [Google Scholar]
- 62.Liao X, Wang Q, Ju H. Analyst. 2015;140:4245–4252. doi: 10.1039/c5an00128e. [DOI] [PubMed] [Google Scholar]
- 63.Lin X, Zhang C, Huang Y, Zhu Z, Chen X, Yang CJ. Chemical Communications. 2013;65:7243–7245. doi: 10.1039/c3cc43224f. [DOI] [PubMed] [Google Scholar]
- 64.Yan L, Yan Y, Pei L, Wei W, Zhao J. Scientific Reports. 2014;4:7400. doi: 10.1038/srep07400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Guo S, Yang F, Zhang Y, Ning Y, Yao Q, Zhang G-J. Analytical Methods. 2014;6:3598–3603. [Google Scholar]
- 66.Xi Q, Zhou D-M, Kan Y-Y, Ge J, Wu Z-K, Yu R-Q, Jiang J-H. Analytical Chemistry. 2014;86:1361–1365. doi: 10.1021/ac403944c. [DOI] [PubMed] [Google Scholar]
- 67.Shen W, Yeo KH, Gao Z. Analyst. 2015;140:1932–1938. doi: 10.1039/c4an02146k. [DOI] [PubMed] [Google Scholar]
- 68.Degliangeli F, Kshirsagar P, Brunetti V, Pompa PP, Fiammengo R. Journal of the American Chemical Society. 2014;136:2264–2267. doi: 10.1021/ja412152x. [DOI] [PubMed] [Google Scholar]
- 69.Wang Q, Li R-D, Yin B-C, Ye B-C. Analyst. 2015;140:6306–6312. doi: 10.1039/c5an01350j. [DOI] [PubMed] [Google Scholar]
- 70.Yin B-C, Liu Y-Q, Ye B-C. Journal of the American Chemical Society. 2012;134:5064–5067. doi: 10.1021/ja300721s. [DOI] [PubMed] [Google Scholar]
- 71.Huang R, Liao Y, Zhou X, Xing D. Analytica Chimica Acta. 2015;888:162–172. doi: 10.1016/j.aca.2015.07.041. [DOI] [PubMed] [Google Scholar]
- 72.Zhu D, Zhang L, Ma W, Lu S, Xing X. Biosensors and Bioelectronics. 2015;65:152–158. doi: 10.1016/j.bios.2014.10.019. [DOI] [PubMed] [Google Scholar]
- 73.Liu H, Li L, Wang Q, Duan L, Tang B. Analytical Chemistry. 2014;86:5487–5493. doi: 10.1021/ac500752t. [DOI] [PubMed] [Google Scholar]
- 74.Dong H, Hao K, Tian Y, Jin S, Lu H, Zhou S-F, Zhang X. Biosensors and Bioelectronics. 2014;53:377–383. doi: 10.1016/j.bios.2013.09.061. [DOI] [PubMed] [Google Scholar]
- 75.Liu W, Zhou X, Xing D. Biosensors and Bioelectronics. 2014;58:388–394. doi: 10.1016/j.bios.2014.02.082. [DOI] [PubMed] [Google Scholar]
- 76.Liu T, Chen X, Hong C-Y, Xu X-P, Yang H-H. Microchim Acta. 2014;181:731–736. [Google Scholar]
- 77.Zhang P, Wu X, Chai Y, Yuan R. Analyst. 2014;139:2748–2753. doi: 10.1039/c4an00284a. [DOI] [PubMed] [Google Scholar]
- 78.Zhang P, Wu X, Yuan R, Chai Y. Analytical Chemistry. 2015;87:3202–3207. doi: 10.1021/ac504455z. [DOI] [PubMed] [Google Scholar]
- 79.Hao N, Li X-L, Zhang H-R, Xu J-J, Chen H-Y. Chemical Communications. 2014;50:14828–14830. doi: 10.1039/c4cc06801g. [DOI] [PubMed] [Google Scholar]
- 80.Chen Y, Xiang Y, Yuan R, Chai Y. Analytica Chimica Acta. 2015;891:130–135. doi: 10.1016/j.aca.2015.07.059. [DOI] [PubMed] [Google Scholar]
- 81.Zhang P, Zhuo Y, Chang Y, Yuan R, Chai Y. Analytical Chemistry. 2015 doi: 10.1021/acs.analchem.5b02495. [DOI] [PubMed] [Google Scholar]
- 82.Chen A, Gui G-F, Zhuo Y, Chai Y-Q, Xiang Y, Yuan R. Analytical Chemistry. 2015;87:6328–6334. doi: 10.1021/acs.analchem.5b01168. [DOI] [PubMed] [Google Scholar]
- 83.Hao N, Dai P-P, Yu T, Xu J-J, Chen H-Y. Chemical Communications. 2015;51:13504–13507. doi: 10.1039/c5cc05350a. [DOI] [PubMed] [Google Scholar]
- 84.Fang S, Lee HJ, Wark AW, Corn RM. Journal of the American Chemical Society. 2006;128:14044–14046. doi: 10.1021/ja065223p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lee J, Park J, Lee J-Y, Yeo J-S. Advanced Science. 2015;2:n/a–n/a. [Google Scholar]
- 86.Guven B, Dudak FC, Boyaci IH, Tamer U, Ozsoz M. Analyst. 2014;139:1141–1147. doi: 10.1039/c3an01600e. [DOI] [PubMed] [Google Scholar]
- 87.Joshi GK, Deitz-McElyea S, Johnson M, Mali S, Korc M, Sardar R. Nano Letters. 2014;14:6955–6963. doi: 10.1021/nl503220s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Liu Q, Shin Y, Kee JS, Kim KW, Rafei SRM, Perera AP, Tu X, Lo G-Q, Ricci E, Colombel M, Chiong E, Thiery JP, Park MK. Biosensors and Bioelectronics. 2015;71:365–372. doi: 10.1016/j.bios.2015.04.052. [DOI] [PubMed] [Google Scholar]
- 89.Ho S-L, Chan H-M, Wong RN-S, Li H-W. Analytica Chimica Acta. 2014;823:61–68. doi: 10.1016/j.aca.2014.03.020. [DOI] [PubMed] [Google Scholar]
- 90.Kang T, Kim H, Lee JM, Lee H, Choi Y-S, Kang G, Seo M-K, Chung BH, Jung Y, Kim B. Small. 2014;10:4200–4206. doi: 10.1002/smll.201400164. [DOI] [PubMed] [Google Scholar]
- 91.Wang Y, MacLachlan E, Nguyen BK, Fu G, Peng C, Chen JIL. Analyst. 2015;140:1140–1148. doi: 10.1039/c4an02189d. [DOI] [PubMed] [Google Scholar]
- 92.Gao X, Xu H, Baloda M, Gurung AS, Xu L-P, Wang T, Zhang X, Liu G. Biosensors and Bioelectronics. 2014;54:578–584. doi: 10.1016/j.bios.2013.10.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Gao X, Xu L-P, Wu T, Wen Y, Ma X, Zhang X. Talanta. 2015 doi: 10.1016/j.talanta.2015.06.060. [DOI] [PubMed] [Google Scholar]
- 94.Vaisocherová H, Šípová H, Víšová I, Bocková M, Špringer T, Laura Ermini M, Song X, Krejčík Z, Chrastinová L, Pastva O, Pimková K, Dostálová Merkerová M, Dyr JE, Homola J. Biosensors and Bioelectronics. 2015;70:226–231. doi: 10.1016/j.bios.2015.03.038. [DOI] [PubMed] [Google Scholar]
- 95.Zhang D, Yan Y, Cheng W, Zhang W, Li Y, Ju H, Ding S. Microchim Acta. 2013;180:397–403. [Google Scholar]
- 96.Qiu X, Liu X, Zhang W, Zhang H, Jiang T, Fan D, Luo Y. Analytical Chemistry. 2015;87:6303–6310. doi: 10.1021/acs.analchem.5b01159. [DOI] [PubMed] [Google Scholar]
- 97.George S, Chaudhery V, Lu M, Takagi M, Amro N, Pokhriyal A, Tan Y, Ferreira P, Cunningham BT. Lab on a Chip. 2013;13:4053–4064. doi: 10.1039/c3lc50579k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Tan Y, Sutanto E, Alleyne AG, Cunningham BT. Journal of Biophotonics. 2014;7:266–275. doi: 10.1002/jbio.201300158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Qavi AJ, Bailey RC. Angewandte Chemie International Edition. 2010;49:4608–4611. doi: 10.1002/anie.201001712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Qavi AJ, Kindt JT, Gleeson MA, Bailey RC. Analytical Chemistry. 2011;83:5949–5956. doi: 10.1021/ac201340s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kindt JT, Bailey RC. Analytical Chemistry. 2012;84:8067–8074. doi: 10.1021/ac3019813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Erdem A, Eksin E, Congur G. Journal of Electroanalytical Chemistry. 2015;755:167–173. [Google Scholar]
- 103.Kilic T, Nur Topkaya S, Ozsoz M. Biosensors and Bioelectronics. 2013;48:165–171. doi: 10.1016/j.bios.2013.04.011. [DOI] [PubMed] [Google Scholar]
- 104.Li F, Peng J, Wang J, Tang H, Tan L, Xie Q, Yao S. Biosensors and Bioelectronics. 2014;54:158–164. doi: 10.1016/j.bios.2013.10.061. [DOI] [PubMed] [Google Scholar]
- 105.Tran HV, Piro B, Reisberg S, Anquetin G, Duc HT, Pham MC. Anal Bioanal Chem. 2014;406:1241–1244. doi: 10.1007/s00216-013-7292-4. [DOI] [PubMed] [Google Scholar]
- 106.Tran HV, Piro B, Reisberg S, Tran LD, Duc HT, Pham MC. Biosensors and Bioelectronics. 2013;49:164–169. doi: 10.1016/j.bios.2013.05.007. [DOI] [PubMed] [Google Scholar]
- 107.Wu X, Chai Y, Yuan R, Su H, Han J. Analyst. 2013;138:1060–1066. doi: 10.1039/c2an36506e. [DOI] [PubMed] [Google Scholar]
- 108.Wang Z, Si L, Bao J, Dai Z. Chemical Communications. 2015;51:6305–6307. doi: 10.1039/c5cc01081k. [DOI] [PubMed] [Google Scholar]
- 109.Li F, Peng J, Zheng Q, Guo X, Tang H, Yao S. Analytical Chemistry. 2015;87:4806–4813. doi: 10.1021/acs.analchem.5b00093. [DOI] [PubMed] [Google Scholar]
- 110.Cai Z, Song Y, Wu Y, Zhu Z, James Yang C, Chen X. Biosensors and Bioelectronics. 2013;41:783–788. doi: 10.1016/j.bios.2012.10.002. [DOI] [PubMed] [Google Scholar]
- 111.Wang M, Li B, Zhou Q, Yin H, Zhou Y, Ai S. Electrochimica Acta. 2015;165:130–135. [Google Scholar]
- 112.Xia N, Wang X, Deng D, Wang G, Zhai H, Li S-J. International journal of Electrochemical Science. 2013;8:9714–9722. [Google Scholar]
- 113.Labib M, Khan N, Berezovski MV. Analytical Chemistry. 2015;87:1395–1403. doi: 10.1021/ac504331c. [DOI] [PubMed] [Google Scholar]
- 114.Wang M, Yin H, Fu Z, Guo Y, Wang X, Zhou Y, Ai S. J Solid State Electrochem. 2014;18:2829–2835. [Google Scholar]
- 115.Yin H, Wang M, Zhou Y, Zhang X, Sun B, Wang G, Ai S. Biosensors and Bioelectronics. 2014;53:175–181. doi: 10.1016/j.bios.2013.09.053. [DOI] [PubMed] [Google Scholar]
- 116.Wu S, Chen H, Zuo Z, Wang M, Luo R, Xu H. International journal of Electrochemical Science. 2015;10:3848–3858. [Google Scholar]
- 117.Liu L, Jiang S, Wang L, Zhang Z, Xie G. Microchim Acta. 2015;182:77–84. [Google Scholar]
- 118.Cheng F-F, He T-T, Miao H-T, Shi J-J, Jiang L-P, Zhu J-J. ACS Applied Materials & Interfaces. 2015;7:2979–2985. doi: 10.1021/am508690x. [DOI] [PubMed] [Google Scholar]
- 119.Xia N, Zhang L, Wang G, Feng Q, Liu L. Biosensors and Bioelectronics. 2013;47:461–466. doi: 10.1016/j.bios.2013.03.074. [DOI] [PubMed] [Google Scholar]
- 120.Liu L, Xia N, Liu H, Kang X, Liu X, Xue C, He X. Biosensors and Bioelectronics. 2014;53:399–405. doi: 10.1016/j.bios.2013.10.026. [DOI] [PubMed] [Google Scholar]
- 121.Xia N, Zhang Y, Wei X, Huang Y, Liu L. Analytica Chimica Acta. 2015;878:95–101. doi: 10.1016/j.aca.2015.04.018. [DOI] [PubMed] [Google Scholar]
- 122.Zhang Y, Yan Y, Chen W, Cheng W, Li S, Ding X, Li D, Wang H, Ju H, Ding S. Biosensors and Bioelectronics. 2015;68:343–349. doi: 10.1016/j.bios.2015.01.026. [DOI] [PubMed] [Google Scholar]
- 123.Shi K, Dou B, Yang C, Chai Y, Yuan R, Xiang Y. Analytical Chemistry. 2015;87:8578–8583. doi: 10.1021/acs.analchem.5b02418. [DOI] [PubMed] [Google Scholar]
- 124.Wu X, Chai Y, Zhang P, Yuan R. ACS Applied Materials & Interfaces. 2015;7:713–720. doi: 10.1021/am507059n. [DOI] [PubMed] [Google Scholar]
- 125.Ren Y, Deng H, Shen W, Gao Z. Analytical Chemistry. 2013;85:4784–4789. doi: 10.1021/ac400583e. [DOI] [PubMed] [Google Scholar]
- 126.Wang M, Fu Z, Li B, Zhou Y, Yin H, Ai S. Analytical Chemistry. 2014;86:5606–5610. doi: 10.1021/ac5010376. [DOI] [PubMed] [Google Scholar]
- 127.Zhang X, Wu D, Liu Z, Cai S, Zhao Y, Chen M, Xia Y, Li C, Zhang J, Chen J. Chemical Communications. 2014;50:12375–12377. doi: 10.1039/c4cc05541a. [DOI] [PubMed] [Google Scholar]
- 128.Hong C-Y, Chen X, Liu T, Li J, Yang H-H, Chen J-H, Chen G-N. Biosensors and Bioelectronics. 2013;50:132–136. doi: 10.1016/j.bios.2013.06.040. [DOI] [PubMed] [Google Scholar]
- 129.Li C, Liu Z, Cai S, Wen F, Wu D, Liu Y, Wu F, Lan J, Han Z, Chen J. Electrochemistry Communications. 2015;60:185–189. [Google Scholar]
- 130.Xiang G, Jiang D, Luo F, Liu F, Liu L, Pu X. Sensors and Actuators B: Chemical. 2014;195:515–519. [Google Scholar]
- 131.Wu X, Chai Y, Yuan R, Zhuo Y, Chen Y. Sensors and Actuators B: Chemical. 2014;203:296–302. [Google Scholar]
- 132.Yang C, Shi K, Dou B, Xiang Y, Chai Y, Yuan R. ACS Applied Materials & Interfaces. 2015;7:1188–1193. doi: 10.1021/am506933r. [DOI] [PubMed] [Google Scholar]
- 133.Peng Y, Jiang J, Yu R. Analytical Methods. 2014;6:2889–2893. [Google Scholar]
- 134.Wang M, Shen B, Yuan R, Cheng W, Xu H, Ding S. Journal of Electroanalytical Chemistry. 2015;756:147–152. [Google Scholar]
- 135.Yao B, Liu Y, Tabata M, Zhu H, Miyahara Y. Chemical Communications. 2014;50:9704–9706. doi: 10.1039/c4cc03330b. [DOI] [PubMed] [Google Scholar]
- 136.Tian Q, Wang Y, Deng R, Lin L, Liu Y, Li J. Nanoscale. 2015;7:987–993. doi: 10.1039/c4nr05243a. [DOI] [PubMed] [Google Scholar]
- 137.Yan Y, Zhao D, Yuan T, Hu J, Zhang D, Cheng W, Zhang W, Ding S. Electroanalysis. 2013;25:2354–2359. [Google Scholar]
- 138.Yu Y, Chen Z, Shi L, Yang F, Pan J, Zhang B, Sun D. Analytical Chemistry. 2014;86:8200–8205. doi: 10.1021/ac501505a. [DOI] [PubMed] [Google Scholar]
- 139.Lin M, Wen Y, Li L, Pei H, Liu G, Song H, Zuo X, Fan C, Huang Q. Analytical Chemistry. 2014;86:2285–2288. doi: 10.1021/ac500251t. [DOI] [PubMed] [Google Scholar]
- 140.Miao P, Wang B, Chen X, Li X, Tang Y. ACS Applied Materials & Interfaces. 2015;7:6238–6243. doi: 10.1021/acsami.5b01508. [DOI] [PubMed] [Google Scholar]
- 141.Miao P, Wang B, Meng F, Yin J, Tang Y. Bioconjugate Chemistry. 2015;26:602–607. doi: 10.1021/acs.bioconjchem.5b00064. [DOI] [PubMed] [Google Scholar]
- 142.Ge Z, Lin M, Wang P, Pei H, Yan J, Shi J, Huang Q, He D, Fan C, Zuo X. Analytical Chemistry. 2014;86:2124–2130. doi: 10.1021/ac4037262. [DOI] [PubMed] [Google Scholar]
- 143.Wan J, Liu X, Zhang Y, Gao Q, Qi H, Zhang C. Sensors and Actuators B: Chemical. 2015;213:409–416. [Google Scholar]
- 144.Zhou Y, Wang M, Xu Z, Ni C, Yin H, Ai S. Biosensors and Bioelectronics. 2014;54:244–250. doi: 10.1016/j.bios.2013.11.026. [DOI] [PubMed] [Google Scholar]
- 145.Meng X, Zhou Y, Liang Q, Qu X, Yang Q, Yin H, Ai S. Analyst. 2013;138:3409–3415. doi: 10.1039/c3an36788f. [DOI] [PubMed] [Google Scholar]
- 146.Labib M, Khan N, Ghobadloo SM, Cheng J, Pezacki JP, Berezovski MV. Journal of the American Chemical Society. 2013;135:3027–3038. doi: 10.1021/ja308216z. [DOI] [PubMed] [Google Scholar]
- 147.Erdem A, Congur G. Talanta. 2014;118:7–13. doi: 10.1016/j.talanta.2013.09.041. [DOI] [PubMed] [Google Scholar]
- 148.Bartosik M, Hrstka R, Palecek E, Vojtesek B. Analytica Chimica Acta. 2014;813:35–40. doi: 10.1016/j.aca.2014.01.023. [DOI] [PubMed] [Google Scholar]
- 149.Cheng F-F, Zhang J-J, He T-T, Shi J-J, Abdel-Halim ES, Zhu J-J. Analyst. 2014;139:3860–3865. doi: 10.1039/c4an00777h. [DOI] [PubMed] [Google Scholar]
- 150.Zhang J, Wu D-Z, Cai S-X, Chen M, Xia Y-K, Wu F, Chen J-H. Biosensors and Bioelectronics. 2016;75:452–457. doi: 10.1016/j.bios.2015.09.006. [DOI] [PubMed] [Google Scholar]
- 151.Wang Z, Zhang J, Guo Y, Wu X, Yang W, Xu L, Chen J, Fu F. Biosensors and Bioelectronics. 2013;45:108–113. doi: 10.1016/j.bios.2013.02.007. [DOI] [PubMed] [Google Scholar]
- 152.Campuzano S, Torrente-Rodríguez RM, López-Hernández E, Conzuelo F, Granados R, Sánchez-Puelles JM, Pingarrón JM. Angewandte Chemie International Edition. 2014;53:6168–6171. doi: 10.1002/anie.201403270. [DOI] [PubMed] [Google Scholar]
- 153.Si Y, Sun Z, Zhang N, Qi W, Li S, Chen L, Wang H. Analytical Chemistry. 2014;86:10406–10414. doi: 10.1021/ac5028885. [DOI] [PubMed] [Google Scholar]
- 154.Lu N, Gao A, Dai P, Song S, Fan C, Wang Y, Li T. Small. 2014;10:2022–2028. doi: 10.1002/smll.201302990. [DOI] [PubMed] [Google Scholar]
- 155.Cai B, Huang L, Zhang H, Sun Z, Zhang Z, Zhang G-J. Biosensors and Bioelectronics. 2015;74:329–334. doi: 10.1016/j.bios.2015.06.068. [DOI] [PubMed] [Google Scholar]
- 156.Ramnani P, Gao Y, Ozsoz M, Mulchandani A. Analytical Chemistry. 2013;85:8061–8064. doi: 10.1021/ac4018346. [DOI] [PubMed] [Google Scholar]
- 157.Lujambio A, Lowe SW. Nature. 2012;482:347–355. doi: 10.1038/nature10888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Li Y, Qiu C, Tu J, Geng B, Yang J, Jiang T, Cui Q. Nucleic Acids Research. 2014;42:D1070–D1074. doi: 10.1093/nar/gkt1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Agarwal V, Bell GW, Nam J-W, Bartel DP. eLife. 2015:4. doi: 10.7554/eLife.05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Khorshid M, Hausser J, Zavolan M, van Nimwegen E. Nat Meth. 2013;10:253–255. doi: 10.1038/nmeth.2341. [DOI] [PubMed] [Google Scholar]
- 161.Marín RM, Šulc M, Vaníček J. RNA. 2013;19:467–474. doi: 10.1261/rna.035634.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Peterson SM, Thompson JA, Ufkin ML, Sathyanarayana P, Liaw L, Congdon CB. Frontiers in Genetics. 2014;5 doi: 10.3389/fgene.2014.00023. [DOI] [PMC free article] [PubMed] [Google Scholar]