Abstract
The ability to design and fine-tune non-covalent interactions between organic ligands and proteins is indispensable to rational drug development. Aromatic stacking has long been recognized as one of the key constituents of ligand-protein interfaces. In this communication, we employ a two-parameter geometric model to conduct a large-scale statistical analysis of aromatic contacts in the experimental and computer-generated structures of ligand-protein complexes, considering various combinations of aromatic amino acid residues and ligand rings. The geometry of interfacial π-π stacking in crystal structures accords with experimental and theoretical data collected for simple systems, such as the benzene dimer. Many contemporary ligand docking programs implicitly treat aromatic stacking with van der Waals and Coulombic potentials. Although this approach generally provides a sufficient specificity to model aromatic interactions, the geometry of π-π contacts in high-scoring docking conformations could still be improved. The comprehensive analysis of aromatic geometries at ligand-protein interfaces lies the foundation for the development of type-specific statistical potentials to more accurately describe aromatic interactions in molecular docking. A Perl script to detect and calculate the geometric parameters of aromatic interactions in ligand-protein complexes is available at https://github.com/michal-brylinski/earomatic. The dataset comprising experimental complex structures and computer-generated models is available at https://osf.io/rztha/.
Keywords: aromatic interactions, π-π interactions, parallel stacking, perpendicular stacking, protein-ligand complexes, non-covalent interactions, ligand binding, molecular modeling, ligand docking, molecular docking
1. Introduction
Low molecular weight ligands, such as endogenous compounds and synthetic drugs, reversibly bind to proteins by forming multiple non-covalent interactions predominantly with the side chains of binding pocket residues. In contrast to strong covalent bonds, these rather weak intermolecular contacts comprise a variety of interactions that do not involve sharing electrons. Key interactions between ligands and macromolecules include hydrogen bonds [1, 2], π-π aromatic stacking [3, 4], cation-π interactions [5, 6], hydrophobic effects [7, 8], halogen bonds [9, 10], and salt bridges [11, 12]. A significant effort is directed to study the geometrical properties and energetics of these non-covalent bonds because of their paramount importance in molecular recognition and practical applications in drug discovery [8, 13, 14]. Pharmacology exploits the fact that bioactive compounds have a sufficient specificity and potency to bind and modulate the function of macromolecular targets. At the outset of drug development, the selection of a molecular scaffold requisite for binding is often followed by the optimization of the adjoining chemical moieties non-covalently interacting with pocket residues. Here, the goal is to maximize the affinity of a drug candidate toward the target macromolecule and to reduce its dissociation from the functional site. On that account, the ability to fine-tune a network of non-covalent interactions promoting high-affinity binding of small molecules to their targets is critical for the success of rational drug discovery.
Two distinct, yet complementary computational techniques are used to gain insights into the structure and energy landscape of non-covalent interactions in biological systems. The first approach employs quantum mechanical (QM) calculations, sometimes in combination with molecular mechanics (MM). For instance, the geometrical preferences of various types of hydrogen bonds frequently present at ligand-protein interfaces have been investigated with QM [15]. The derived distance and angle values describing the geometry of hydrogen bonds can subsequently be utilized by empirical methods to effectively model hydrogen bonds upon ligand binding. Another study employed a hybrid QM/MM method to analyze polarization effects playing a significant role in the determination of ligand-protein complex structures [16]. In this approach, fixed charges on ligand atoms obtained from force field parameterization are replaced by those calculated by QM/MM in order to improve the accuracy of the modeling of molecular assemblies. Finally, the relative strength of π-π and cation-π interactions was investigated as a function of the geometry and protonation state in histidine-aromatic complexes with quantum chemistry methods [17]. In addition to important differences in the stability of aromatic interactions in the gas phase, water, and protein-like environments, it was found that π-π stacking is essential for the favorable electron correlation, whereas cation-π contacts produce further electrostatic contributions. The advantage of QM methods is that these calculations can be applied to a variety of systems and the results obtained for idealized molecules are usually straightforward to interpret. Nonetheless, because of limits on the system size as well as the fact that only simple molecules in vacuum are subject to QM calculations, the derived geometric and energy parameters may not be suitable to reliably model a biological system with its highly complex and heterogeneous environment.
On that account, another computational approach to explore the geometry and energy landscape of various non-covalent interactions at ligand-protein interfaces builds on the accumulated knowledge of the atomic structures of molecular assemblies. For example, accurate potentials of mean force (PMF) can be derived from a large number of complexes deposited in the Protein Data Bank (PDB) [18]. The Biomolecular Ligand Energy Evaluation Protocol (BLEEP) was developed to estimate the affinity of ligand binding from the complex structure [19]. BLEEP considers 40 different atom types and employs a reverse Boltzmann methodology to convert the distribution of interaction distances into energy-like pair potential functions. As it was anticipated, these potentials promote short-range polar contacts and hydrogen bonding, whereas the range of hydrophobic interactions is distinctively longer.
Another example of an atomic PMF derived from a database of ligand-protein complexes is the Astex Statistical Potential (ASP) [20]. A unique feature of this new potential is that it accounts for differences in the exposure of various types of protein atoms toward ligand-binding sites. Employing ASP in molecular docking considerably improves the accuracy of pose prediction not only across a large validation set of ligand-protein complexes, but also for a small testing set of pharmaceutically relevant targets. Finally, two distance-dependent statistical scoring functions were developed using probability theory, PoseScore to identify native ligand binding geometries and RankScore to distinguish between binding ligands and non-binding molecules [21]. Both potentials were derived from a set of 8,885 crystallographic structures of ligand-protein complexes employing optimized atomic distance thresholds and including non-native ligand geometries. In addition to experimental structures, the performance of PoseScore and RankScore was evaluated against computer-generated protein models with encouraging results. Because of their remarkable accuracy, these tools can support drug development by predicting ligand-protein complex structures and helping identify potentially bioactive compounds.
In addition to parameters for contact-based and distance-dependent pair potentials routinely derived from interaction statistics in the PDB, this large collection of molecular structures can also be used to parameterize other types of potential functions. For instance, a sophisticated descriptor-based scoring function integrating evolutionary constraints with physics-based energy terms implemented in the GeauxDock docking program [22, 23] was parameterized against a representative snapshot of ligand-protein complexes extracted from the PDB. Scoring terms in GeauxDock include electrostatic and van der Waals interactions, hydrogen bonds, hydrophobic interactions, generic and pocket-specific contact potentials, a pseudo-pharmacophore potential, as well as position restraints on family conserved anchor substructures and the binding site center.
As an example, much deeper potential wells representing strong interactions are assigned by GeauxDock to salt bridges between guanidinium groups in arginine residues and ligand carboxyl moieties, compared to those less favorable, e.g. between arginine and amide groups. Moreover, different types of hydrogen bounds have distinct geometries and strengths. Although the mean interaction distance of hydrogen bond between the hydroxyl group on threonine and ligand primary amine is shorter than that for the tyrosine hydroxyl and an amide group, the latter is slightly stronger at the optimal distance. Also force field parameters to model hydrophobic interactions in GeauxDock are in line with the physicochemical properties of ligand atoms, e.g. aromatic carbons and halogens tend toward nonpolar residues, whereas amine nitrogen and carboxylate oxygen atoms clearly prefer a polar microenvironment. Indubitably, these carefully derived parameters are pivotal for the accuracy of subsequent molecular modeling simulations. Other studies systematically explore structural data in the PDB to calculate distance- and angle-dependent statistical potentials for hydrogen bonds [15], investigate the geometrical and energetic features of halogen bonds in biological molecules [9, 24], as well as characterize hydrophobic and aromatic interactions at the ligand-protein interface [8, 25].
Although π-π stacking, defined as an attractive, non-covalent interaction between aromatic rings, is not as widespread as hydrogen bonds and hydrophobic contacts, it plays a vital role in biological recognition and the organization of biomolecular structures. The benzene dimer is a prototypical system to study π-π aromatic stacking. However, investigating this simple system poses significant practical challenges due to its relatively small binding energy of about 2–3 kcal/mol and the fact that the dimer is stable only at low temperatures. An experimental evidence of the perpendicular conformation in crystal and liquid benzene was obtained by molecular beam electric deflection study [26]. A subsequent theoretical research on π-π interactions indicated that favorable perpendicular and offset-parallel configurations correspond to energy minima of comparable depth, whereas the less stable eclipsed geometry represents an energetic saddle point [27, 28]. Indeed, perpendicular and offset-parallel configurations are dominant in the crystal structures of simple aromatic compounds [29] and proteins [30], in contrast to infrequently observed eclipsed stacking.
Since the major energetic contributors to π-π interactions are London dispersion forces and electrostatics, many molecular force fields and scoring functions simulate aromatic stacking implicitly with van der Waals and Coulomb potentials rather than employing explicit terms. On that account, a systematic evaluation of the geometry of aromatic interactions in computer-generated models compared to that in the experimental structures of organic ligand-protein complexes can cast light on the accuracy of the modeling of π-π stacking in pharmaceutical design. In this communication, we first conduct a large-scale statistical analysis or interfacial aromatic contacts in the crystal structures of organic ligand-protein complexes employing a two-parameter geometric model. Our study considers various combinations of aromatic protein residues and ring structures in ligand molecules. Subsequently, complex models constructed by contemporary docking software are carefully assessed in terms of the predicted geometry of aromatic contacts. The results have important ramifications for the development of molecular force fields and scoring functions for structure-based drug discovery.
2. Methods and Materials
2.1. Geometry of aromatic interactions
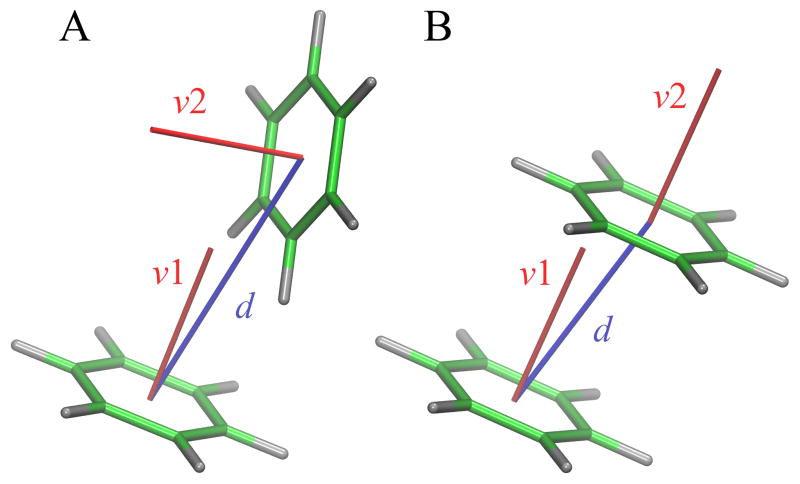
Aromatic rings in ligand molecules are identified with the Chemistry::Ring::Find module available in PerlMol [31]. This module implements a breadth-first ring finding algorithm to identify the Smallest Set of Smallest Rings [32, 33]. Based on atomic contacts detected with Ligand-Protein Contact (LPC) software [34], we subsequently select those interactions involving aromatic residues, phenylalanine (F), tyrosine (Y), tryptophan (W) and histidine (H), and ligand aromatic atoms. This procedure produces a complete list of interacting aromatic rings in a given structure of a ligand-protein complex. Next, we employ a two-parameter model, presented in Figure 1, to describe the geometry of each pair of interacting rings. The first parameter in this model is the Cartesian distance between the geometric centers of two rings, referred to as the distance. The second parameter is an angle between normal vectors of two aromatic rings, referred to as the angle. Further, the interaction type is specified using a notation AP:L, where A is an amino acid, P is the number of amino acid ring atoms, and L is the number of ligand ring atoms. For instance, W6:5 denotes an interaction between a 6-member benzene ring of tryptophan and a 5-member aromatic ring of the ligand.
Figure 1.
Two-parameter model describing the geometry of aromatic interactions. Parameters are the Cartesian distance d between the geometric centers of aromatic rings (blue) and an angle between the normal vectors v1 and v2 of ring planes (red). Two distinct low-energy configurations of the benzene dimer are shown, (A) perpendicular and (B) parallel.
2.2. Dataset of ligand-protein complexes
The protocol employed in this work to compile the dataset of ligand-protein complexes is similar to that previously developed to generate representative and non-redundant sets of ligand-protein complexes for benchmarking of eFindSite [35] and other binding site prediction algorithms. First, we identified in the PDB protein chains composed of 50–999 amino acids that non-covalently bind small organic molecules. Next, we retained those proteins binding a single ligand whose Tanimoto coefficient (TC) to at least one FDA-approved drug is ≥0.5. The TC is calculated for 1024-bit molecular fingerprints with OpenBabel [36] against FDA-approved drugs in the DrugBank database [37]. Subsequently, protein sequences were clustered with CD-HIT [38] at 40% sequence similarity, and a representative set of proteins binding chemically dissimilar ligands (a pairwise TC of <0.5) at different locations (at least 8 Å apart) were selected from each homologous cluster. Finally, we kept only those complexes stabilized by at least one aromatic interaction between the ligand and the receptor protein identified with LPC [34]. This procedure resulted in a non-redundant and representative dataset of 3,079 proteins bound to ligands containing aromatic moieties, referred to as the daTaset tO evalUate alGoritHms for ligand Docking (TOUGH-D1) dataset.
2.3. Molecular docking
In addition to experimental structures acquired from the PDB [18], a series of docking models were constructed for TOUGH-D1 complexes with two academic programs, AutoDock Vina [39] and rDock [40]. Vina has an excellent scoring power according to a recent benchmarking study [41] and it is the most widely used molecular docking tool [42]. rDock is a newly released program that is more accurate and faster than other academic codes. For Vina, MGL tools 1.5.6 [43] and Open Babel 2.4.1 [44] were used to add polar hydrogens and partial charges, as well as to convert target proteins and library compounds to the PDBQT format. The optimal search space centered on the binding site was defined for each docking ligand from its radius of gyration as described previously [45]. Molecular docking was carried out by AutoDock Vina 1.1.2 with the exhaustiveness parameter set to 1000. For rDock, Open Babel 2.4.1 [44] was used to convert receptor proteins and ligands to the required Tripos MOL2 and SDFile formats. The docking box was defined by the rcavity program employing the reference ligand method. Simulations were conducted by rDock 2013.1 with the default scoring function and 100 docking runs per ligand. Finally, we executed Vina for each ligand-protein system with the --local_only option to generate near-native conformations and the --randomize_only option to generate 100 random configurations avoiding atomic clashes.
Docking models constructed for TOUGH-D1 complexes were evaluated with the Contact Mode Score (CMS) [46] against experimental binding poses. The CMS is a new metric assessing the conformational similarity based on intermolecular ligand-protein contacts, which is less dependent on the ligand size compared to the widely used root-mean-square deviation. It ranges from about 0 for random binding poses to 1 for identical configurations. Two sets of conformations were compiled from docking models generated by Vina, a native-like set comprising those configurations having a CMS of ≥0.5 and a random set of models whose CMS to the experimental structure is <0.3. In addition, we prepared a set of high-scoring models for each docking program based on the predicted binding affinity.
2.4. Data analysis and visualization
Two-dimensional histograms of the distribution of geometric parameters describing aromatic interactions, the distance and the angle, were smoothed with the kernel density estimation (KDE) technique [47, 48]. KDE is a non-parametric method estimating the probability density function of a set of variables based on a finite data sample. The parameter space was first discretized to a 100×100 matrix and then populated with observations. i.e. distance and angle values computed for each aromatic interaction in a given dataset. In order to smooth the data with KDE, each observation was represented by a Gaussian kernel, which is a non-negative function integrating to 1 and with a mean of 0. Matrix2png 1.2.2 [49] was then used to generate heat maps showing the correlative distribution of geometric parameters for aromatic interactions. In addition to the visual analysis of heat maps, the overlap between two probability distributions is measured with the G-test [50, 51]:
| Eq. 1 |
where Oi and Ei are the observed and expected counts in the i-th cell and the sum is taken over all non-empty cells in the 100×100 matrix. Finally, pKa values are assigned to histidine residues with PROPKA 3.1 [52, 53] and the molecular structures of ligand-protein complexes are visualized with Visual Molecular Dynamics 1.9.3 [54].
3. Results
3.1. Geometry of aromatic contacts in experimental complex structures
The TOUGH-D1 dataset of experimental structures compiled in this study comprises 3,079 ligand-protein complexes forming a total number of 8,148 interactions between 4,967 aromatic rings of organic ligands and 5,961 protein residues. The amino acid composition of these interactions is 37.4% F6, 23.5% Y6, 24.7% H5, 5.5% W5, and 8.9% W6. Further, 56.0% and 44.0% of aromatic interactions involve 6- and 5-member rings in ligand molecules, respectively, whose atomic make up is 75.2% carbon, 24.7% nitrogen, 0.1% sulfur, and 0.03% oxygen. Table 1 shows the composition of aromatic interactions in TOUGH-D1 complexes. Phenylalanine, tyrosine, and tryptophan residues form more interactions with 6-member ligand rings, in contrast to histidine residues that prefer to interact with 5-member ligand rings. Aromatic interactions involving a bicyclic side chain of tryptophan more often are formed through its 6-member benzene ring than a 5-member nitrogen-containing pyrrole ring.
Table 1.
Composition of aromatic interactions across the TOUGH-D1 dataset. Interactions between phenylalanine (F6), tyrosine (Y6), tryptophan (W5 and W6) and histidine (H5) residues, and 6- and 5-member ligand aromatic rings are considered.
| Amino acid ring | Ligand ring
|
|
|---|---|---|
| 6-member | 5-member | |
| F6 | 21.8% | 15.6% |
| Y6 | 15.4% | 8.1% |
| W5 | 3.4% | 2.1% |
| W6 | 5.8% | 3.1% |
| H5 | 9.6% | 15.1% |
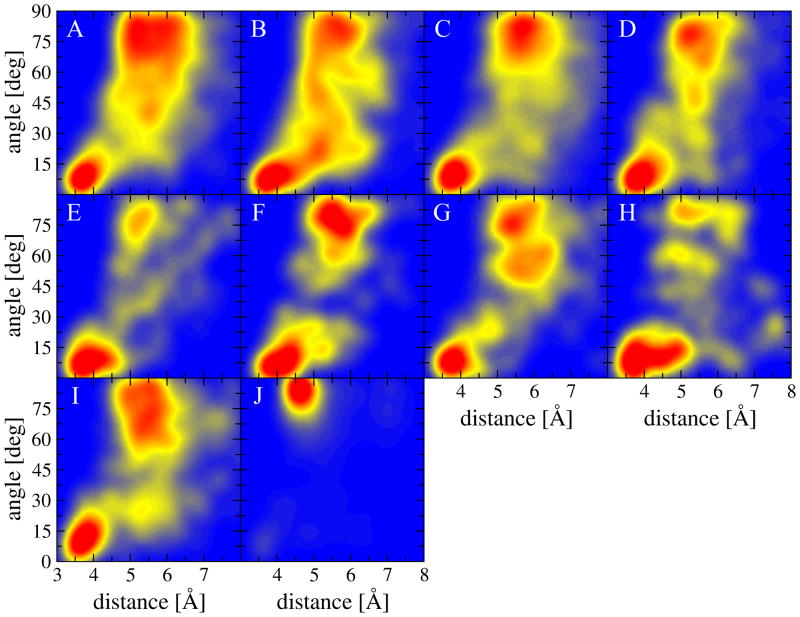
Figure 2 shows the correlative distribution of distance and angle values for all interaction types calculated for the experimental structures of ligand-protein complexes included in the TOUGH-D1 dataset. The distance ranges from 3 to 8 Å, and the angle is within the acute range of 0–90 deg. In general, most interactions have two distinct regions of a high probability density. The first recognizable geometry is described by a relatively close distance of about 3.5 to 4.5 Å between the centers of aromatic rings, and a small angle of 0–15 deg between the normal vectors of ring planes. This range of parameters corresponds to a parallel aromatic stacking, shown for the benzene dimer as a prototypical system in Figure 1B. The second densely populated region described by longer distances of 5–6 Å and near-right angles corresponds to a perpendicular aromatic interaction, another typical geometry illustrated for the benzene dimer in Figure 1A.
Figure 2.
Heat maps showing the distribution of the geometrical properties of aromatic interactions across the experimental structures of ligand-protein complexes included in the TOUGH-D1 dataset. The interaction geometry is described by two parameters, an angle between the normal vectors of aromatic rings and a distance between ring centers. The following interaction types are presented: (A) F6:6, (B) F6:5, (C) Y6:6, (D) Y6:5, (E) W5:6, (F) W5:5, (G) W6:6, (H) W6:5, (I) H5:6, and (J) H5:5.
Distance and angle values obviously mutually depend on one another, e.g. rotating one aromatic ring by 90 deg in order to change the parallel stacking to a perpendicular orientation pushes rings farther apart because of the van der Waals repulsion between the clouds of ring atoms. Further, there are certain differences between interactions involving various amino acids and ligand rings. For instance, the presence of a hydroxyl group in tyrosine perceptibly changes the geometry of aromatic interactions with respect to phenylalanine. Notably more intermediate configurations between parallel and perpendicular regions can be observed for F6:6 (Figure 2A) and F6:5 (Figure 2B) compared to Y6:6 (Figure 2C) and Y6:5 (Figure 2D). The 5-member aromatic ring in tryptophan favors a parallel stacking with 6-member ligand rings (Figure 2E), but forms both parallel and perpendicular interactions with 5-member ligand rings (Figure 2F). Because of the fused ring structure of tryptophan side chains, W6:6 has a dual peak within the perpendicular region (Figure 2G), and the majority of W6:5 interactions are parallel with a somewhat broader range of distances between rings of 3–5.5 Å (Figure 2H).
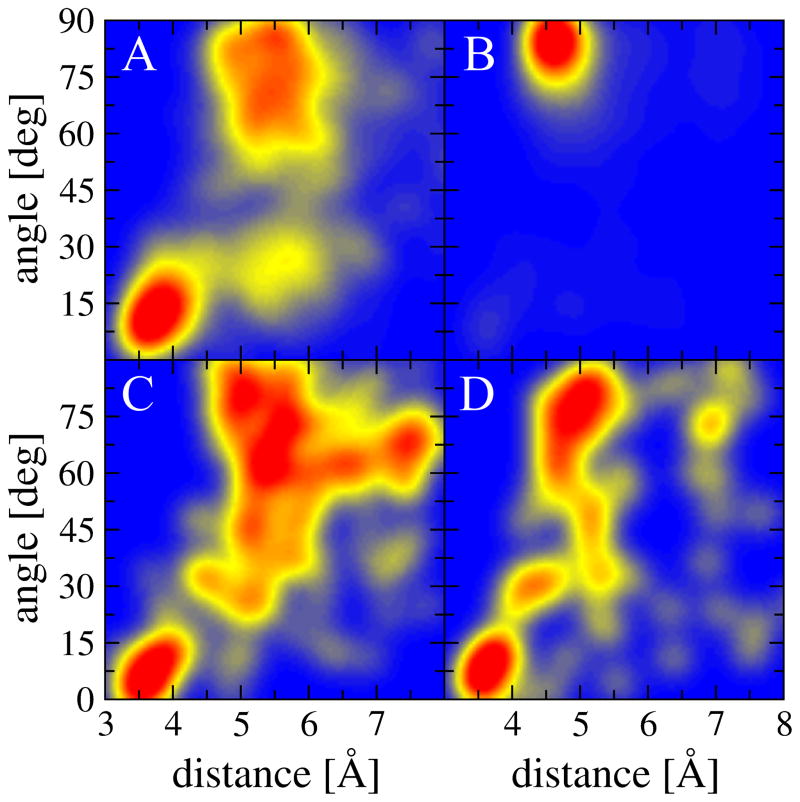
Finally, histidine residues form both parallel and perpendicular interactions with 6-member ligand rings (Figure 2I), whereas perpendicular geometries are the most common for those interactions involving 5-member ligand rings (Figure 2J). Depending on the pH, a histidine switches between the double-protonated form with both δ and ε nitrogen atoms protonated, and the neutral state with either δ or ε nitrogen protonated. On that account, in Figure 3, we show the distribution of geometrical parameters for H5:6 and H5:5 interactions in two groups identified based on the predicted pKa shift from the model value of 6.5 for the imidazole side chain. According to PROPKA3 [52, 53], as many as 84.6% of histidines interacting with ligand aromatic rings are acidic with negative pKa shifts from the model value, most likely because the protonation of histidine residues has a stabilizing effect of about 1–3 kcal/mol [17, 55]. Consequently, interaction geometries shown in Figures 2I and 2J are biased toward protonated histidine residues, for which the distributions of distance and angle values are also independently presented in Figures 3A and 3B. Interestingly, the protonation of the imidazole side chain has a notable effect on the geometry of aromatic interactions. The vast majority of H5:5 interactions involving protonated histidine residues are perpendicular, likely due to the additional favorable contribution from the cation-π electrostatic energy [17]. In contrast, the distributions of geometrical properties of π-π interactions involving neutral histidine residues shown in Figures 3C and 3D are qualitatively similar to those computed for other aromatic amino acids.
Figure 3.
Heat maps showing the distribution of the geometrical properties of aromatic interactions involving histidine residues across the experimental structures of TOUGH-D1 complexes. The interaction geometry is described by two parameters, an angle between the normal vectors of aromatic rings and a distance between ring centers. Interactions are divided into two groups with (A, B) negative and (C, D) positive shifts from the model pKa value for the imidazole side chain. The following interaction types are presented: (A, C) H5:6 and (B, D) H5:5.
3.2. Examples of aromatic interactions stabilizing ligand-protein complexes
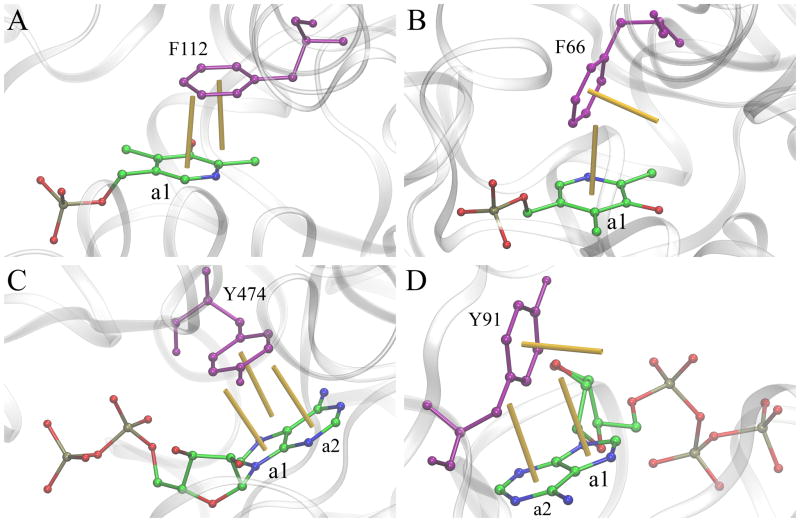
We selected a series of representative examples to examine the molecular structures of aromatic interactions in the context of high probability density regions in heat maps presented in Figure 2. Figure 4 shows four ligand-protein complexes stabilized by various aromatic contacts. Predominant interactions are exemplified by those formed by pyridoxal-5′-phosphate and phenylalanine residues. The offset-parallel aromatic stacking in glutamine aminotransferase from Thermus thermophilus HB8 (Figure 4A, PDB-ID: 1v2d, chain A) [56] has a distance of 3.8 Å and an angle of 8.5 deg, whereas the perpendicular stacking in maize serine racemase (Figure 4B, PDB-ID: 5cvc, chain B) [57] has a distance of 4.9 Å and an angle of 79.5 deg. Further, complexes shown in Figures 4C and 4D typify interactions between nucleotides and tyrosine residues. The offset-parallel aromatic stacking between ADP and Y474 in isocitrate dehydrogenase kinase/phosphatase from Escherichia coli (Figure 4C, PDB-ID: 3lc6, chain A) [58] consists of two interactions, Y6:5 and Y6:6. The former involves a 5-member ring a1 and has a distance (angle) of 4.0 Å (7.4 deg), and the latter involves a 6-member ring a2 and has a distance (angle) of 4.0 Å (6.3 deg). ATP bound to Hmd co-occurring protein HcgE from Methanothermobacter marburgensis forms two perpendicular interactions with Y91 (Figure 4D, PDB-ID: 3wv8, chain B) [59]. In this complex structure, Y6:5 (ring a1) has a distance of 5.1 Å and an angle of 76.9 deg, whereas Y6:6 (ring a2) has a distance of 5.1 Å and an angle of 77.2 deg.
Figure 4.
Examples of aromatic stacking in ligand-protein complexes involving phenylalanine and tyrosine residues. (A, B) pyridoxal-5′-phosphate and phenylalanine residues, and (C, D) ADP/ATP and tyrosine residues. Two distinct stacking geometries are presented: (A, C) offset-parallel and (B, D) perpendicular. Ligands are colored by atom type (C – green, N – blue, O – red, P – ochre), whereas aromatic binding residues are purple. Normal vectors of aromatic rings are shown as yellow sticks.
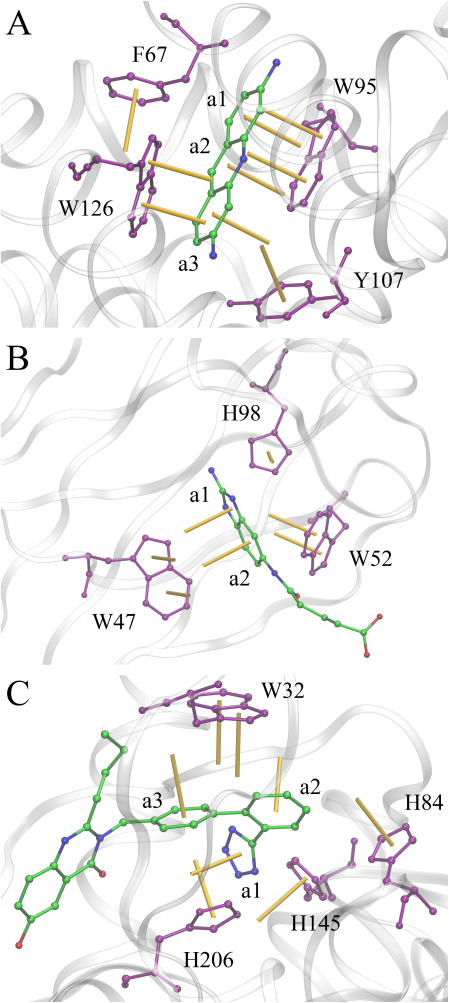
Figure 5 presents three examples of ligand-protein complexes stabilized by multiple aromatic interactions involving tryptophan and histidine residues. Proflavin forms a 3-layer parallel stacking with two tryptophan residues, W95 and W126, when bound to multidrug binding protein EbrR from Streptomyces lividans (Figure 5A, PDB-ID: 3hth, chain A). For example, the middle ring a2 in proflavin forms two offset-parallel interactions with W95, W5:6 with a distance (angle) of 4.6 Å (4.2 deg) and W6:6 with a distance (angle) of 3.7 Å (4.1 deg), as well as a near-parallel interaction with W126, W5:6 whose distance (angle) is 5.9 Å (36.5 deg). In addition, the EbrR/proflavin complex is stabilized by aromatic interactions with F67 and Y107, for instance, proflavin interacts with F67 through a perpendicular stacking F6:6 with a distance (angle) of 5.2 Å (83.9 deg). A series of perpendicular aromatic interactions are formed between tryptophan residues W47 and W52 of the human catalytic elimination antibody 13G5 and a hapten molecule (Figure 5B, PDB-ID: 3fo2, chain B) [60]. Distance (angle) values for selected contacts involving W47, W5:5, W6:6, and W5:6 are 5.7 Å (85.1 deg), 6.3 Å (85.6 deg), and 6.1 Å (85.6 deg), respectively. Also, histidine H98 perpendicularly interacts with the ring a1 of the ligand with a distance of 4.8 Å and an angle of 75.1 deg. The last example is metallo-β-lactamase from Bacteroides fragilis bound to a potent inhibitor (Figure 5C, PDB-ID: 1a8t, chain A) [61]. This complex is stabilized by a variety of aromatic interactions involving three histidine residues, H84, H145 and H206, as well as tryptophan W32. For instance, H206 forms a parallel stacking H5:6 against ring a3 with a distance (angle) of 5.6 Å (22.0 deg), and a perpendicular interaction H5:5 against ring a1 with a distance (angle) of 4.0 Å (72.0 deg). Furthermore, both rings of W32 make parallel contacts with ring a3 of the inhibitor whose distances (angles) are 5.7 Å (11.8 deg) for W5:6 and 5.5 Å (11.4 deg) for W6:6.
Figure 5.
Examples of aromatic stacking in ligand-protein complexes involving tryptophan and histidine residues. (A) EbrR complexed with proflavin, (B) catalytic antibody 13G5 complexed with a hapten, and (C) metallo-β-lactamase complexed with a biphenyl tetrazole inhibitor. Ligands are colored by atom type (C – green, N – blue, O – red), whereas aromatic binding residues are purple. Normal vectors of aromatic rings are shown as yellow sticks.
3.3. Aromatic contacts in computer-generated complex structures
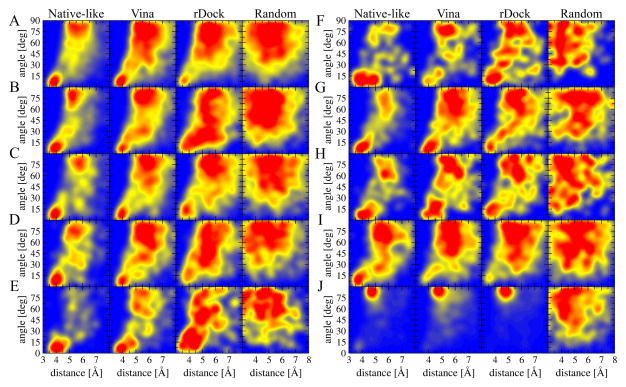
Heat maps presented in Figure 2 clearly demonstrate that aromatic interactions in the experimental structures of ligand-protein complexes tend to adopt certain geometries. An important question from a modeling point of view is whether similar stacking configurations are observed in the theoretical models of complex structures generated by molecular docking. In order to look into this facet of ligand docking, we analyze aromatic interactions in four sets of theoretical models of ligand-protein assemblies constructed for the TOUGH-D1 dataset. In addition to heat maps presented in Figure 6, the deviation from the reference probability distribution obtained for experimental structures is quantified by the G-test. G-values are reported in Table 2 for all interaction types with smaller values indicating a better agreement with the reference distribution. Heat maps generated for native-like configurations (1st column in Figure 6) are very similar to those shown in Figure 2 for experimental structures. Further, the average G-value for this set is as low as 0.184. This can be expected because employing a CMS threshold of ≥0.5 ensures that the modeled ligand-protein contacts are highly correlated with those in experimental structures.
Figure 6.
Heat maps showing the distribution of the geometrical properties of aromatic interactions across the computer-generated models of ligand-protein complexes included in the TOUGH-D1 dataset. The interaction geometry is described by two parameters, an angle between the normal vectors of aromatic rings and a distance between ring centers. The following interaction types are presented: (A) F6:6, (B) F6:5, (C) Y6:6, (D) Y6:5, (E) W5:6, (F) W5:5, (G) W6:6, (H) W6:5, (I) H5:6, and (J) H5:5. Four models are considered for each interaction type: native-like conformations, high-scoring models reported by Vina and rDock, and ligands randomly positioned within binding sites.
Table 2.
Deviation of the geometry of aromatic interactions from experimental structures for computer-generated models of TOUGH-D1 complexes. The deviation is measured with the G-test for native-like conformations, high-scoring models constructed by Vina and rDock, and ligands randomly positioned within binding sites.
| Interaction | Protein-ligand conformations
|
|||
|---|---|---|---|---|
| Native-like | Vina | rDock | Random | |
| F6:6 | 0.045 | 0.122 | 0.179 | 1.622 |
| F6:5 | 0.113 | 0.199 | 0.203 | 2.198 |
| Y6:6 | 0.082 | 0.118 | 0.165 | 1.733 |
| Y6:5 | 0.132 | 0.333 | 0.354 | 2.232 |
| W5:6 | 0.274 | 0.359 | 0.753 | 2.325 |
| W5:5 | 0.296 | 1.065 | 0.581 | 3.479 |
| W6:6 | 0.131 | 0.274 | 0.271 | 1.585 |
| W6:5 | 0.418 | 0.628 | 0.862 | 3.049 |
| H5:6 | 0.130 | 0.312 | 0.340 | 1.530 |
| H5:5 | 0.215 | 0.601 | 0.278 | 2.093 |
|
| ||||
| Average | 0.184 | 0.401 | 0.399 | 2.185 |
Vina and rDock construct ligand conformations in which the geometry of aromatic interaction does not deviate far away from that in experimental complexes (2nd and 3rd columns in Figure 6). Although the average G-values for both docking tools are comparable, there are notable differences with respect to individual interaction types. In general, binding poses generated by Vina have better geometries of aromatic interactions involving 6-member rings in ligand molecules than rDock. For instance, G-values for F6:6, Y6:6, W5:6, and H5:6 modeled by Vina are 0.122, 0.118, 0.359, and 0.312, respectively, compared to 0.179, 0.165, 0.753, and 0.340 for rDock. However, rDock seems to model certain interactions involving 5-member rings more accurately than Vina, e.g. G-value for W5:5 (H5:5) is 0.581 (0.278) for rDock and 1.065 (0.601) for Vina. The quality of other interaction types, such as F6:5, Y6:5, W6:6, and H5:6, in docking models is to a large extent independent of the docking algorithm. For comparison, the last column in Figure 6 shows the distribution of distance and angle values across a dataset of random ligand-protein configurations. The correlative distribution of geometric parameters for aromatic interactions is totally lost when ligands are arbitrarily positioned within their target binding sites only to avoid steric clashes. In addition, G-values for random conformations reported in Table 2 are much higher than those calculated for high-scoring models by Vina and rDock, e.g. the average G-value across the random dataset is as high as 2.185, compared to only 0.401 for Vina and 0.399 for rDock.
4. Discussion
In this communication, we report the results of a statistical analysis of aromatic contacts at the ligand-protein interface. A two-parameter model considering a distance between the geometric centers of aromatic rings and an angle between normal vectors of ring planes is employed to investigate interfacial π-π interactions in experimental as well as computer-generated complex structures. Our analysis of X-ray crystallography data is consistent with the geometrical and energetic properties of aromatic stacking reported previously. Exploring the potential energy surface for the benzene dimer with ab initio methods revealed two nearly isoenergetic structures, perpendicular and offset-parallel configurations [62]. The ideal offset-parallel conformation with an angle between ring planes of 0 deg has the lowest energy of −3.33 kcal/mol at a horizontal displacement of 1.54 Å, whereas perfectly perpendicular structures with an angle of 90 deg have interaction energies of −2.84 kcal/mol (point-face) and −2.51 kcal/mol (edge-face) [27]. Another QM study predicted that perpendicular and offset-parallel configurations are nearly isoenergetic with binding energies of 2.7 and 2.8 kcal/mol, respectively. These two low-energy arrangements of aromatic rings appear as distinct areas of high probability density on the distance-angle maps constructed in this study for different combinations of protein aromatic residues and ligand ring structures present in the TOUGH-D1 dataset.
With respect to the geometry of π-π interactions, a distance of 4.96 Å between the centers of mass of individual rings in the perpendicular benzene dimer in the gas phase was measured by rotational experiments [63]. Similar distances of 5.0–5.1 Å were obtained with QM calculations [27, 28, 62, 64]. These values match a lower bound of the inter-monomer distance range in ligand-protein complexes, which in our analysis extends to about 6 Å. This broader distance range is likely caused by divers chemical moieties attached to aromatic rings in ligand molecules. It has been reported that adding a substituent to one of the rings in the benzene dimer impacts the π-π interaction energy and geometry compared to unsubstituted systems [65]. Specifically, Monte Carlo simulations demonstrated that the presence of a substituent tends to increase the inter-monomer separation in perpendicular conformations. For instance, the lowest energy structure of the benzene/phenol dimer has a distance of 5.6 Å, which increases to 6.2 Å as the dimer adopts an edge-to-edge configuration. Furthermore, ab initio calculations together with the analysis of X-ray crystallography data showed that aromatic molecules form stacks with the vertical separation of 3.3–4.1 Å between ring planes in various parallel orientations [27–29], closely matching the distance range for the parallel aromatic stacking in our analysis. Interestingly, a quantum chemistry study of the benzene dimer revealed the presence of a shallow minimum on the path interconverting offset-parallel structures through a perpendicular saddle point [27]. This configuration corresponds to a tilt angle between phenyl ring planes of about 45 deg, which can also be observed as a medium probability density area at a distance of approximately 5.5 Å in our distance-angle maps generated for phenylalanine and tyrosine residues.
Previous studies investigating aromatic stacking at the ligand-protein interface are often focused only on nucleotide binding, for instance, it was reported that 65% of adenylate-protein complexes form π-π interactions between adenine bases and aromatic side chains with two predominant orientations, offset-parallel and perpendicular [66]. A similar model to that used in the present study was employed to survey the structures of complexes between proteins and adenine- and guanine-containing ligands in the PDB [4]. Reported values of the vertical distance between ring planes of less than 4.5 Å for parallel and less than 5.5 Å for perpendicular orientations are in line with our analysis of the X-ray crystallography data. Nonetheless, a somewhat broader range of distances observed for the perpendicular configurations in our distance-angle maps arise from the fact that geometric parameters describing aromatic stacking in ligand-protein complexes are derived in the present study from a more diverse collection of compounds containing aromatic groups.
In addition to the experimental structures of ligand-protein complexes, we examine the geometry of aromatic interactions in theoretical models constructed by molecular docking. To the best of our knowledge, scoring functions implemented in modern docking programs treat π-π stacking as van der Waals and Coulombic interactions. This description of aromatic interactions is sufficient in some cases. For instance, docking of a series of agonists to the binding pocket of the homology model of the serotonin 5-HT2C G protein-coupled receptor with the PatchDock server [67] produced ligand conformations interacting with tryptophan and phenylalanine residues through parallel and perpendicular aromatic stacking [68]. Another example is a molecular modeling study conducted with Glide [69] for subnanomolar affinity antagonists of the cannabinoid receptor CB2 [70]. Docking calculations revealed a number of parallel and perpendicular aromatic contacts between chloromethylphenyl and methylbenzyl rings of the compounds and a cluster of aromatic residues in CB2, suggesting that π-π interactions are critical to the efficacy of these antagonists.
Post-processing of docking models is a common practice to improve the prediction accuracy of molecular docking. For instance, molecular mechanics with continuum solvation offers a rigorous approach to decompose free energy into individual contributions from various interaction groups enhancing the screening and ranking power of AutoDock [71, 72]. In addition, it has been reported that re-ranking guanosine triphosphate (GTP) docking poses generated by the GOLD [73] program by explicitly accounting for the π-π stacking yields a higher accuracy of the modeling of GTP-protein complexes compared to the default goldscore scoring function [4]. Our assessment of docking models constructed by AutoDock Vina [39] and rDock [40] for a diverse collection of ligands and proteins included in the TOUGH-D1 dataset indicate that although state-of-the-art molecular docking force fields provide sufficient specificity to reliably model aromatic interactions, the geometry of π-π contacts in high-scoring conformations could still be improved. The comprehensive analysis of aromatic geometries at ligand-protein interfaces presented in this study lies the foundation for the development of type-specific statistical potentials to more accurately treat π-π interactions in molecular docking. A Perl script to detect and calculate the geometric parameters of aromatic interactions in ligand-protein complexes is available at https://github.com/michal-brylinski/earomatic. The TOUGH-D1 dataset comprising experimental structures and computer-generated models of ligand-protein complexes is available at https://osf.io/rztha/.
Acknowledgments
The author is grateful to Louisiana State University for providing computing resources. Research reported in this publication was supported by the National Institute of General Medical Sciences of the National Institutes of Health under Award Number R35GM119524. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Conflict of Interest: none
References
- 1.Panigrahi SK, Desiraju GR. Strong and weak hydrogen bonds in the protein-ligand interface. Proteins. 2007;67:128–41. doi: 10.1002/prot.21253. [DOI] [PubMed] [Google Scholar]
- 2.Williams MA, Ladbury JE. Hydrogen bonds in protein-ligand complexes. Weinheim, FRG: Wiley-VCH Verlag GmbH & Co. KGaA; 2003. Hydrogen bonds in protein-ligand complexes; pp. 137–61. [Google Scholar]
- 3.Boehr DD, Farley AR, Wright GD, Cox JR. Analysis of the pi-pi stacking interactions between the aminoglycoside antibiotic kinase APH(3′)-IIIa and its nucleotide ligands. Chem Biol. 2002;9:1209–17. doi: 10.1016/s1074-5521(02)00245-4. [DOI] [PubMed] [Google Scholar]
- 4.Pyrkov TV, Pyrkova DV, Balitskaya ED, Efremov RG. The role of stacking interactions in complexes of proteins with adenine and Guanine fragments of ligands. Acta Naturae. 2009;1:124–7. [PMC free article] [PubMed] [Google Scholar]
- 5.Scrutton NS, Raine AR. Cation-pi bonding and amino-aromatic interactions in the biomolecular recognition of substituted ammonium ligands. Biochem J. 1996;319(Pt 1):1–8. doi: 10.1042/bj3190001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu R, McMahon TB. Investigation of cation-pi interactions in biological systems. J Am Chem Soc. 2008;130:12554–5. doi: 10.1021/ja802117s. [DOI] [PubMed] [Google Scholar]
- 7.Efremov RG, Chugunov AO, Pyrkov TV, Priestle JP, Arseniev AS, Jacoby E. Molecular lipophilicity in protein modeling and drug design. Curr Med Chem. 2007;14:393–415. doi: 10.2174/092986707779941050. [DOI] [PubMed] [Google Scholar]
- 8.Gallina AM, Bork P, Bordo D. Structural analysis of protein-ligand interactions: the binding of endogenous compounds and of synthetic drugs. J Mol Recognit. 2014;27:65–72. doi: 10.1002/jmr.2332. [DOI] [PubMed] [Google Scholar]
- 9.Lu Y, Wang Y, Zhu W. Nonbonding interactions of organic halogens in biological systems: implications for drug discovery and biomolecular design. Phys Chem Chem Phys. 2010;12:4543–51. doi: 10.1039/b926326h. [DOI] [PubMed] [Google Scholar]
- 10.Zhou P, Tian F, Zou J, Shang Z. Rediscovery of halogen bonds in protein-ligand complexes. Mini Rev Med Chem. 2010;10:309–14. doi: 10.2174/138955710791331016. [DOI] [PubMed] [Google Scholar]
- 11.Kukic P, Nielsen JE. Electrostatics in proteins and protein-ligand complexes. Future Med Chem. 2010;2:647–66. doi: 10.4155/fmc.10.6. [DOI] [PubMed] [Google Scholar]
- 12.Neves-Petersen MT, Petersen SB. Protein electrostatics: a review of the equations and methods used to model electrostatic equations in biomolecules--applications in biotechnology. Biotechnol Annu Rev. 2003;9:315–95. doi: 10.1016/s1387-2656(03)09010-0. [DOI] [PubMed] [Google Scholar]
- 13.Zhou P, Huang J, Tian F. Specific noncovalent interactions at protein-ligand interface: implications for rational drug design. Curr Med Chem. 2012;19:226–38. doi: 10.2174/092986712803414150. [DOI] [PubMed] [Google Scholar]
- 14.Chen K, Kurgan L. Investigation of atomic level patterns in protein--small ligand interactions. PLoS One. 2009;4:e4473. doi: 10.1371/journal.pone.0004473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu Z, Wang G, Li Z, Wang R. Geometrical Preferences of the Hydrogen Bonds on Protein-Ligand Binding Interface Derived from Statistical Surveys and Quantum Mechanics Calculations. J Chem Theory Comput. 2008;4:1959–73. doi: 10.1021/ct800267x. [DOI] [PubMed] [Google Scholar]
- 16.Cho AE, Guallar V, Berne BJ, Friesner R. Importance of accurate charges in molecular docking: quantum mechanical/molecular mechanical (QM/MM) approach. J Comput Chem. 2005;26:915–31. doi: 10.1002/jcc.20222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cauet E, Rooman M, Wintjens R, Lievin J, Biot C. Histidine-Aromatic Interactions in Proteins and Protein-Ligand Complexes: Quantum Chemical Study of X-ray and Model Structures. J Chem Theory Comput. 2005;1:472–83. doi: 10.1021/ct049875k. [DOI] [PubMed] [Google Scholar]
- 18.Berman HM, Battistuz T, Bhat TN, Bluhm WF, Bourne PE, Burkhardt K, et al. The Protein Data Bank. Acta Crystallogr D Biol Crystallogr. 2002;58:899–907. doi: 10.1107/s0907444902003451. [DOI] [PubMed] [Google Scholar]
- 19.Mitchell JBO, Laskowski RA, Alex A, Thornton JM. BLEEP - potential of mean force describing protein-ligand interactions: I. Generating potential. J Comput Chem. 1999;20:1165–76. [Google Scholar]
- 20.Mooij WT, Verdonk ML. General and targeted statistical potentials for protein-ligand interactions. Proteins. 2005;61:272–87. doi: 10.1002/prot.20588. [DOI] [PubMed] [Google Scholar]
- 21.Fan H, Schneidman-Duhovny D, Irwin JJ, Dong G, Shoichet BK, Sali A. Statistical potential for modeling and ranking of protein-ligand interactions. J Chem Inf Model. 2011;51:3078–92. doi: 10.1021/ci200377u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fang Y, Ding Y, Feinstein WP, Koppelman DM, Moreno J, Jarrell M, et al. GeauxDock: Accelerating structure-based virtual screening with heterogeneous computing. PLoS One. 2016;11:e0158898. doi: 10.1371/journal.pone.0158898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ding Y, Fang Y, Feinstein WP, Ramanujam J, Koppelman DM, Moreno J, et al. GeauxDock: A novel approach for mixed-resolution ligand docking using a descriptor-based force field. J Comput Chem. 2015;36:2013–26. doi: 10.1002/jcc.24031. [DOI] [PubMed] [Google Scholar]
- 24.Auffinger P, Hays FA, Westhof E, Ho PS. Halogen bonds in biological molecules. Proc Natl Acad Sci U S A. 2004;101:16789–94. doi: 10.1073/pnas.0407607101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Salentin S, Schreiber S, Haupt VJ, Adasme MF, Schroeder M. PLIP: fully automated protein-ligand interaction profiler. Nucleic Acids Res. 2015;43:W443–7. doi: 10.1093/nar/gkv315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Janda KC, Hemminger JC, Winn JS, Novick SE, Harris SJ, Klemperer W. Benzene dimer: A polar molecule. J Chem Phys. 1975;63:1419–21. [Google Scholar]
- 27.Jaffe RL, Smith GD. A quantum chemistry study of benzene dimer. J Chem Phys. 1996;105:2780–8. [Google Scholar]
- 28.Sinnokrot MO, Valeev EF, Sherrill CD. Estimates of the ab initio limit for pi-pi interactions: the benzene dimer. J Am Chem Soc. 2002;124:10887–93. doi: 10.1021/ja025896h. [DOI] [PubMed] [Google Scholar]
- 29.Dahl T. The nature of stacking interactions between organic molecules elucidated by analysis of crystal structures. Acta Chem Scand. 1994;48:95–106. [Google Scholar]
- 30.Hunter CA, Singh J, Thornton JM. Pi-pi interactions: the geometry and energetics of phenylalanine-phenylalanine interactions in proteins. J Mol Biol. 1991;218:837–46. doi: 10.1016/0022-2836(91)90271-7. [DOI] [PubMed] [Google Scholar]
- 31.PerlMol. PerlMol ( http://www.perlmol.org/)
- 32.Leach AR, Dolata DP, Prout K. Automated conformational analysis and structure generation: algorithms for molecular perception. J Chem Inf Comput Sci. 1990;30:316–24. doi: 10.1021/ci00067a017. [DOI] [PubMed] [Google Scholar]
- 33.Figueras J. Ring perception using breadth-first search. J Chem Inf Comput Sci. 1996;36:986–91. [Google Scholar]
- 34.Sobolev V, Sorokine A, Prilusky J, Abola EE, Edelman M. Automated analysis of interatomic contacts in proteins. Bioinformatics. 1999;15:327–32. doi: 10.1093/bioinformatics/15.4.327. [DOI] [PubMed] [Google Scholar]
- 35.Brylinski M, Feinstein WP. eFindSite: Improved prediction of ligand binding sites in protein models using meta-threading, machine learning and auxiliary ligands. J Comput Aided Mol Des. 2013;27:551–67. doi: 10.1007/s10822-013-9663-5. [DOI] [PubMed] [Google Scholar]
- 36.Guha R, Howard MT, Hutchison GR, Murray-Rust P, Rzepa H, Steinbeck C, et al. The Blue Obelisk-interoperability in chemical informatics. J Chem Inf Model. 2006;46:991–8. doi: 10.1021/ci050400b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wishart DS, Knox C, Guo AC, Shrivastava S, Hassanali M, Stothard P, et al. DrugBank: a comprehensive resource for in silico drug discovery and exploration. Nucleic Acids Res. 2006;34:D668–72. doi: 10.1093/nar/gkj067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fu L, Niu B, Zhu Z, Wu S, Li W. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics. 2012;28:3150–2. doi: 10.1093/bioinformatics/bts565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Trott O, Olson AJ. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2010;31:455–61. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ruiz-Carmona S, Alvarez-Garcia D, Foloppe N, Garmendia-Doval AB, Juhos S, Schmidtke P, et al. rDock: a fast, versatile and open source program for docking ligands to proteins and nucleic acids. PLoS Comput Biol. 2014;10:e1003571. doi: 10.1371/journal.pcbi.1003571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang Z, Sun H, Yao X, Li D, Xu L, Li Y, et al. Comprehensive evaluation of ten docking programs on a diverse set of protein-ligand complexes: the prediction accuracy of sampling power and scoring power. Phys Chem Chem Phys. 2016;18:12964–75. doi: 10.1039/c6cp01555g. [DOI] [PubMed] [Google Scholar]
- 42.Chen YC. Beware of docking! Trends Pharmacol Sci. 2015;36:78–95. doi: 10.1016/j.tips.2014.12.001. [DOI] [PubMed] [Google Scholar]
- 43.Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, et al. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J Comput Chem. 2009;30:2785–91. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.O’Boyle NM, Banck M, James CA, Morley C, Vandermeersch T, Hutchison GR. Open Babel: An open chemical toolbox. J Cheminform. 2011;3:33. doi: 10.1186/1758-2946-3-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Feinstein WP, Brylinski M. Calculating an optimal box size for ligand docking and virtual screening against experimental and predicted binding pockets. J Cheminform. 2015;7:18. doi: 10.1186/s13321-015-0067-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ding Y, Fang Y, Moreno J, Ramanujam J, Jarrell M, Brylinski M. Assessing the similarity of ligand binding conformations with the Contact Mode Score. Comput Biol Chem. 2016;64:403–13. doi: 10.1016/j.compbiolchem.2016.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Parzen E. On estimation of a probability density function and mode. Ann Math Stat. 1962;33:1065–76. [Google Scholar]
- 48.Rosenblatt M. Remarks on some nonparametric estimates of a density function. Ann Math Stat. 1956;27:832–7. [Google Scholar]
- 49.Pavlidis P, Noble WS. Matrix2png: a utility for visualizing matrix data. Bioinformatics. 2003;19:295–6. doi: 10.1093/bioinformatics/19.2.295. [DOI] [PubMed] [Google Scholar]
- 50.McDonald JH. G-test of goodness-of-fit. Baltimore, Maryland: Sparky House Publishing; 2014. G-test of goodness-of-fit; pp. 53–8. [Google Scholar]
- 51.Sokal RR, Rohlf FJ. Biometry: The Principles and Practice of Statistics in Biological Research. 2. New York: Freeman; 1981. [Google Scholar]
- 52.Olsson MH, Sondergaard CR, Rostkowski M, Jensen JH. PROPKA3: Consistent Treatment of Internal and Surface Residues in Empirical pKa Predictions. J Chem Theory Comput. 2011;7:525–37. doi: 10.1021/ct100578z. [DOI] [PubMed] [Google Scholar]
- 53.Sondergaard CR, Olsson MH, Rostkowski M, Jensen JH. Improved Treatment of Ligands and Coupling Effects in Empirical Calculation and Rationalization of pKa Values. J Chem Theory Comput. 2011;7:2284–95. doi: 10.1021/ct200133y. [DOI] [PubMed] [Google Scholar]
- 54.Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J Mol Graph. 1996;14:33–8. 27–8. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 55.Loewenthal R, Sancho J, Fersht AR. Fluorescence spectrum of barnase: contributions of three tryptophan residues and a histidine-related pH dependence. Biochemistry. 1991;30:6775–9. doi: 10.1021/bi00241a021. [DOI] [PubMed] [Google Scholar]
- 56.Goto M, Omi R, Miyahara I, Hosono A, Mizuguchi H, Hayashi H, et al. Crystal structures of glutamine:phenylpyruvate aminotransferase from Thermus thermophilus HB8: induced fit and substrate recognition. J Biol Chem. 2004;279:16518–25. doi: 10.1074/jbc.M311575200. [DOI] [PubMed] [Google Scholar]
- 57.Zou L, Song Y, Wang C, Sun J, Wang L, Cheng B, et al. Crystal structure of maize serine racemase with pyridoxal 5′-phosphate. Acta Crystallogr F Struct Biol Commun. 2016;72:165–71. doi: 10.1107/S2053230X16000960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zheng J, Jia Z. Structure of the bifunctional isocitrate dehydrogenase kinase/phosphatase. Nature. 2010;465:961–5. doi: 10.1038/nature09088. [DOI] [PubMed] [Google Scholar]
- 59.Fujishiro T, Kahnt J, Ermler U, Shima S. Protein-pyridinol thioester precursor for biosynthesis of the organometallic acyl-iron ligand in [Fe]-hydrogenase cofactor. Nat Commun. 2015;6:6895. doi: 10.1038/ncomms7895. [DOI] [PubMed] [Google Scholar]
- 60.Debler EW, Muller R, Hilvert D, Wilson IA. An aspartate and a water molecule mediate efficient acid-base catalysis in a tailored antibody pocket. Proc Natl Acad Sci U S A. 2009;106:18539–44. doi: 10.1073/pnas.0902700106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Toney JH, Fitzgerald PM, Grover-Sharma N, Olson SH, May WJ, Sundelof JG, et al. Antibiotic sensitization using biphenyl tetrazoles as potent inhibitors of Bacteroides fragilis metallo-beta-lactamase. Chem Biol. 1998;5:185–96. doi: 10.1016/s1074-5521(98)90632-9. [DOI] [PubMed] [Google Scholar]
- 62.Hobza P, Selzle HL, Schlag EW. Potential energy surface for the benzene dimer. Results of ab Initio CCSD(T) calculations show two nearly isoenergetic structures: T-shaped and parallel-displaced. J Phys Chem. 1996;100:18790–4. [Google Scholar]
- 63.Arunan E, Gutowsky HS. The rotational spectrum, structure and dynamics of a benzene dimer. J Chem Phys. 1993;98:4294–6. [Google Scholar]
- 64.Hobza P, Selzle HL, Schlag EW. Potential energy surface of the benzene dimer: Ab initio theoretical study. J Am Chem Soc. 1994;116:3500–6. [Google Scholar]
- 65.Smith T, Slipchenko LV, Gordon MS. Modeling pi-pi interactions with the effective fragment potential method: the benzene dimer and substituents. J Phys Chem A. 2008;112:5286–94. doi: 10.1021/jp800107z. [DOI] [PubMed] [Google Scholar]
- 66.Mao L, Wang Y, Liu Y, Hu X. Molecular determinants for ATP-binding in proteins: a data mining and quantum chemical analysis. J Mol Biol. 2004;336:787–807. doi: 10.1016/j.jmb.2003.12.056. [DOI] [PubMed] [Google Scholar]
- 67.Schneidman-Duhovny D, Inbar Y, Nussinov R, Wolfson HJ. PatchDock and SymmDock: servers for rigid and symmetric docking. Nucleic Acids Res. 2005;33:W363–7. doi: 10.1093/nar/gki481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cordova-Sintjago T, Villa N, Fang L, Booth RG. Aromatic interactions impact ligand binding and function at serotonin 5-HT2C G protein-coupled receptors: Receptor homology modeling, ligand docking, and molecular dynamics results validated by experimental studies. Mol Phys. 2014;112:398–407. doi: 10.1080/00268976.2013.833656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Repasky MP, Shelley M, Friesner RA. Flexible ligand docking with Glide. Curr Protoc Bioinformatics. 2007;Chapter 8(Unit 8):12. doi: 10.1002/0471250953.bi0812s18. [DOI] [PubMed] [Google Scholar]
- 70.Kotsikorou E, Navas F, 3rd, Roche MJ, Gilliam AF, Thomas BF, Seltzman HH, et al. The importance of hydrogen bonding and aromatic stacking to the affinity and efficacy of cannabinoid receptor CB2 antagonist, 5-(4-chloro-3-methylphenyl)-1-[(4-methylphenyl)methyl]-N-[(1S,2S,4R)-1,3,3-trimet hylbicyclo[2.2.1]hept-2-yl]-1H-pyrazole-3-carboxamide (SR144528) J Med Chem. 2013;56:6593–612. doi: 10.1021/jm400070u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sun H, Li Y, Shen M, Tian S, Xu L, Pan P, et al. Assessing the performance of MM/PBSA and MM/GBSA methods. 5. Improved docking performance using high solute dielectric constant MM/GBSA and MM/PBSA rescoring. Phys Chem Chem Phys. 2014;16:22035–45. doi: 10.1039/c4cp03179b. [DOI] [PubMed] [Google Scholar]
- 72.Sun H, Li Y, Tian S, Xu L, Hou T. Assessing the performance of MM/PBSA and MM/GBSA methods. 4. Accuracies of MM/PBSA and MM/GBSA methodologies evaluated by various simulation protocols using PDBbind data set. Phys Chem Chem Phys. 2014;16:16719–29. doi: 10.1039/c4cp01388c. [DOI] [PubMed] [Google Scholar]
- 73.Jones G, Willett P, Glen RC, Leach AR, Taylor R. Development and validation of a genetic algorithm for flexible docking. J Mol Biol. 1997;267:727–48. doi: 10.1006/jmbi.1996.0897. [DOI] [PubMed] [Google Scholar]