Abstract
Fluorine has become an important element for the design of synthetic molecules used for medicine, agriculture, and materials. Despite the many advantages provided by fluorine for tuning key molecular properties, it is rarely found in natural metabolism. We seek to expand the molecular space available for discovery through development of new biosynthetic strategies that cross synthetic with natural compounds. Towards this goal, we have engineered a microbial host for organofluorine metabolism and have shown that we can achieve production of a diketide, 2-fluoro-3-hydroxybutyrate, at ~50% yield. This fluorinated diketide can be used as a monomer in vivo to produce fluorinated poly(hydroxyalkanoate) (PHA) bioplastics with fluorine substitutions ranging from ~5–15%. This system provides a platform to produce mM flux through the key fluoromalonyl coenzyme A (CoA) building block, offering the potential to generate a broad range of fluorinated small molecule targets in living cells.
Keywords: Fluorine, polymers, synthetic biology, metabolism
Graphical abstract
In vivo organofluorine metabolism: An engineered microbial host for organofluorine metabolism can produce a fluorinated diketide at ~50% yield. The diketide can be used as a monomer to produce fluorinated poly(hydroxyalkanoate) bioplastics in vivo with fluorine substitution of up to 15%. This system provides a platform to generate a broad range of fluorinated small molecule targets in living cells.
Nature is adept at utilizing organic molecules made mostly of C, H, O, N, S, and P, to carry out a wide range of biological functions, including catalysis, energy generation and storage, self-defense, and construction of cellular architectures. However, the relatively narrow use of the periodic table in living systems can be limiting compared to synthetic chemistry, which takes advantage of a much broader range of elements to achieve the intended goal. One example of this difference is in the use of fluorine, which has become a powerful tool in the design of small molecules, resulting in its inclusion in 20–30% of therapeutics and agrochemicals[1] and its being a key element in organic materials such as liquid crystals[2] and polymers.[3] In contrast, fluorine is quite rare in natural products compared to other halogens (Cl, Br, I: >3,600 known compounds).[4] While there has been significant interest in developing new synthetic methodology to expand the scope of accessible organofluorine structures,[5] the naturally-occurring biosynthetic space is much smaller, consisting of <25 relatively simple structures derived predominantly from a single pathway that produces fluoroacetate and fluorothreonine.[6] Given the potential of living systems to synthesize complex compounds, we have been interested in engineering the biosynthetic machinery of the cell to use simple biogenic fluorinated building blocks to produce new organofluorine targets.[7]
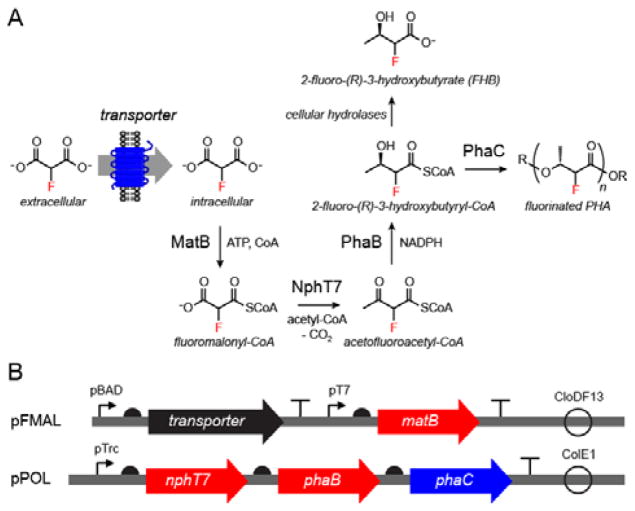
We have previously shown that polyketide synthesis can accommodate the use of fluorinated extender units produced either from fluoroacetate or fluoromalonate, enabling introduction of fluorine into the backbone of polyketides.[8] While it was possible to observe turnover in vivo, the flux was quite low (μM) and we sought to establish this chemistry in living cells in order to take full advantage of the efficiency and scalability of biosynthesis. Since polyketide synthase (PKS) enzymes are typically limited by poor solubility in heterologous hosts, we addressed this problem by designing a high flux organofluorine biosynthetic pathway producing the 2-fluoro-3R-hydroxybutyrate (FHB) diketide through the chain extension of acetyl-CoA with the fluoromalonyl coenzyme A (CoA) extender unit using the acetoacetyl-CoA synthase (NphT7)[9],[10] and an acetoacetyl-CoA reductase (PhaB) (Figure 1A). Furthermore, this fluorinated diketide can be used a monomer to introduce fluorine into bioplastics in the poly(hydroxyalkanoate) (PHA)[11] family (Figure 1A). PHAs are biodegradable polyesters with materials properties similar to poly(propylene) that can accumulate at up to ~80% of dry cell weight in cells from renewable feedstocks.[11b] Although the natural diversity of PHA monomers is high, it draws heavily on variable 3-position substituents because it scavenges monomers from fatty acid catabolism; polymers with 2-substituents are therefore a promising but underexplored class.[12]
Figure 1.
In vivo production of fluorinated biopolymers in engineered E. coli. (A) Pathway for the production of FHB and fluorinated PHA synthesis from fluoromalonate. (B) Plasmid design for the synthetic organofluorine pathways.
We constructed a four-gene, two-plasmid system for biosynthesis of 2-fluoro-3-hydroxybutyryl-CoA using fluoromalonate as the initial fluorine source (Table S1). Fluoromalonate is both relatively inexpensive and capable of being produced through enzymatic pathways while avoiding the acute organofluorine poisoning arising from the direct application of fluoroacetate to cells.[13] Plasmid pFMAL1 was designed to encode enzymes for generation of fluoromalonyl-CoA and contains a malonate:CoA ligase (MatB) from Rhodopseudomonas palustris,[14] which showed improved kinetic properties with respect to fluoromalonate activation compared to a previously used homolog (Figure 1B, Table S1 and Figure S1).[8a] A second plasmid, pPOL2, encoding NphT7 and PhaB was also constructed (Figure 1B). We assayed the ability of E. coli BL21(de3) cells harboring both pFMAL1 and pPOL2 to convert fed fluoromalonate (5 mM) to other fluorinated species by 19F NMR analysis of culture supernatant. However, these conditions lead to no new organofluorine products within the limit of detection (50 μM or 1%, Figures S2 and S3) of our assay (Figure 2).
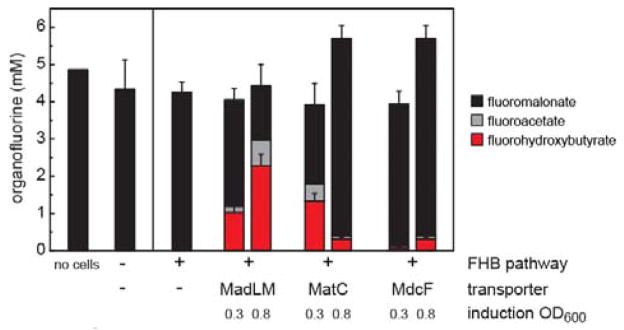
Figure 2.
Organofluorine levels observed from culture of pathway variants. Cultures were incubated for 48 h at 30°C with 5 mM fluoromalonate (no malonate was added). Strains containing pPOL2 with pFMAL plasmids bearing a transporter (pFMAL2, MadLM; pFMAL3, MatC; pFMAL4, MdcF) were induced at different cell densities (OD600 = 0.3 or 0.8). Data are mean ± SD (n = 3) except for the no cell condition where n = 1.
We then reasoned that fluoromalonate transport could be limiting and therefore screened representatives of each of three classes of prokaryotic malonate transporters for their ability to amplify organofluorine synthesis. Using the Transporter Classification Database,[15] we first identified MadLM,[16] a two-component sodium:malonate symporter from Pseudomonas fluorescens. In addition, we identified two putative malonate symporters found in operons with malonate:CoA ligase and malonyl-CoA decarboxylase genes involved in use of malonate as a carbon source (MatC from Streptomyces coelicolor and MdcF from Klebsiella pneumonia). Notably, none of these transporters share significant sequence identity. These transporters were then inserted into the pFMAL1 plasmid to generate plasmids pFMAL2-4 (Table S1). We found that all three transporters were able to accept fluoromalonate, leading to formation of FHB (Figure 2 and Figure S4). Both of these products likely arise from hydrolysis of their respective acyl-CoA precursors by endogenous thioesterases as no exogenous thioesterase was included. Introduction of the transporter enabled mM levels of flux to FHB with both MatC (1.3 ± 0.2 mM) and MadLM (2.3 ± 0.3 mM) (Figure 2). Overall, MadLM was found to achieve titer saturation at lower fluoromalonate levels and was also more versatile with regard to early or late induction compared to MatC (Figure S5). We therefore selected the MadLM-containing plasmid pFMAL2 for further study and BL21(de3) as the production strain after further screening (Figure S6).
In these studies, fluoromalonate was found to be an excellent building block as it was stable in culture medium at 37°C for >2 weeks and was also not transformed by cells carrying various control plasmids with no transporter, MatB, or exogenous pathway genes (Figure 2 and Figure S7). In all of the variations characterized, fluoroacetate was the only off-pathway organofluorine observed either in the growth medium or in cell lysates after 48 h of growth (Figure 2). The fluoroacetate appears to arise from the nonproductive decarboxylation of fluoromalonyl-CoA, which has been observed in other related ketosynthases[9] and is supported by in vitro biochemical experiments (Figures S7 and S8). Given the potential for toxicity from fluoroacetyl-CoA,[13] which is a mechanism-based inhibitor of aconitase that can disable the citric acid cycle, growth of pathway variants was compared in the presence or absence of 5 mM fluoromalonate (Figure S9). The growth inhibition from fluoromalonate-dependent toxicity was modest and could further be alleviated by the co-expression of a fluoroacetyl-CoA specific thioesterase (Figure S9).[17]
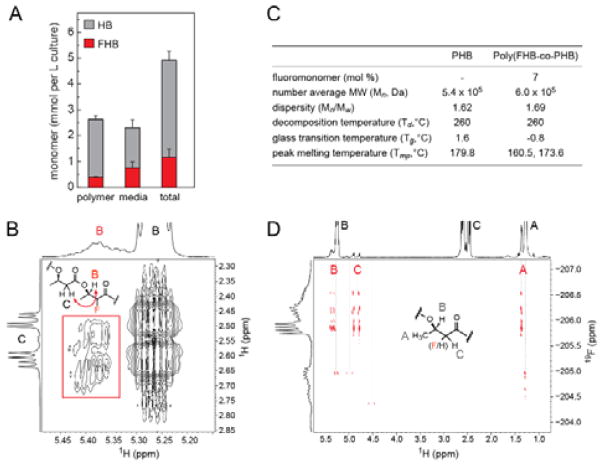
We next investigated if the FHB-CoA monomer could be polymerized to form novel 2-fluorinated polymers with expression of a downstream polymerase. We constructed plasmids containing a PhaC polymerase from Ralstonia eutropha (pPOL1) for co-expression with genes for FHB production. These strains demonstrated production of poly(2-fluoro-3-hydroxybutyrate-co-3-hydroxybutyrate) (poly(FHB-co-HB)) polymers ranging from ~5–15% incorporation (Figure 3A, Figure S10). The nonfluorinated comonomer arises from processing of malonyl-CoA, the first committed metabolite in fatty acid biosynthesis, by the synthetic pathway. The incorporation ratio approximately reflects the relative activity of NphT7 on the two substrates (5-fold difference in kcat/KM).[8a] To further confirm the presence of fluorinated monomers within the polymer, poly(FHB-co-HB) was isolated directly from cells and analyzed by 1H, 19F and 13C NMR, which all contain peaks corresponding to fluorinated monomers with characteristically large JH-F and JC-F coupling constants (Figure S11). The COSY spectrum also contains a crosspeak arising from fluorinated monomers adjacent to their nonfluorinated congeners, indicating the fluorinated monomers are integrated within the polymer (Figure 3B, Figure S12). A polymer with 7% fluorine substitution was characterized using gel-permeation chromatography, which shows that the molecular weight and dispersity index are essentially identical to the non-fluorinated polymer (Figure 3C) and implies that fluorinated monomers do not lead to early chain termination.
Figure 3.
In vivo production of poly(2-fluoro-3-hydroxybutyrate-co-3-hydroxybutyrate) by engineered E. coli. (A) Monomer content of polymer and growth medium for full pathway containing the PhaC polymerase (pPOL1, pFMAL2) in the presence of 5 mM fluoromalonate. Data are mean ± SD (n = 3). (B) Expansion of the COSY spectrum of poly(FHB-co-HB). Crosspeak between HB and HC between a fluorinated and non-fluorinated monomer is highlighted in red. (C) Characterization of poly(FHB-co-HB) compared to PHB. (D) 1H-19F HMBC NMR spectrum shows numerous chemically distinct types of FHB monomers (red) as indicated by the number of fluorine crosspeaks between HA, HB, and HC.
Interestingly, NMR analysis also shows that both diastereomers of 2-fluoro-3R-hydroxybutyryl-CoA appear to be incorporated into polymer, with a ratio similar to that observed in culture medium and in the in vitro reactions of NphT7 and PhaB (Figure S13), suggesting that PhaC does not distinguish the stereochemistry of the 2-fluoro substituent. This lack of stereoregulation could lead to amorphous polymers that avoid the brittleness of bacterial poly(hydroxybutyrates), which are completely crystalline and isotactic. Indeed, the 1H-decoupled 19F spectrum shows that fluoromonomers exist in many distinct chemical environments within the polymer, which is consistent with the 2D 1H-19F spectrum (Figure 3D, Figure S14). A more complex melting peak was observed for the poly(FHB-co-HB) that was shifted to lower temperature relative to nonfluorinated polymer, which may also indicate reduced crystallinity (Figure 3C, Figure S15). Intriguingly, these data show that even a low level of fluorination affects the polymer.
Taken together, we have shown that we are able to engineer a microbial host that can support mM flux through engineered organofluorine biosynthesis pathways that utilize fluoromalonyl-CoA, a key building block for introduction of fluorine into ketosynthase- and polyketide synthase-dependent pathways. We identified functional fluoromalonate transporters that enable this high flux and could be used in a broad range of prokaryotic hosts. Using a downstream polymerase, we have demonstrated that the 2-fluoro-3-hydroxybutyryl-CoA monomer can be utilized for the synthesis of poly(FHB-co-HB) to reliably introduce up to ~15% fluorinated monomers into the biopolymer without appearing to cause significant defects in polymerization. Characterization of the environment of the fluorinated monomers within the polymer indicates that they are chemically inequivalent, yielding promise that differences in stereochemistry, sterics, and electronics introduced through fluorine could alter the intra- and inter-polymer interactions to tune the materials properties of bacterial PHAs.
Supplementary Material
Acknowledgments
This work was supported by the NIH (R01 GM123181 to M.C.Y.C.) and by the NSF under the Center for Sustainable Polymers (CHE-1413862). B.W.T. also acknowledges an Amgen Fellowship in Organic Chemistry (UC Berkeley). We thank Ms. Angelika Neitzel and Prof. Marc Hillmyer (University of Minnesota) for assistance with polymer properties analysis, Dr. Christian Canlas (UC Berkeley College of Chemistry NMR Facility) for assistance with 19F spectra, Dr. Mark Walker for initial investigation into fluoropolymer production, and Dr. Michael Blaisse for cloning of NphT7 expression vectors.
Footnotes
Supporting information for this article is given via a link at the end of the document
References
- 1.a) Hagmann WK. J Med Chem. 2008;51:4359–4369. doi: 10.1021/jm800219f. [DOI] [PubMed] [Google Scholar]; b) Purser S, Moore P, Swallow S, Gouverneur V. Chem Soc Rev. 2008;37:320–330. doi: 10.1039/b610213c. [DOI] [PubMed] [Google Scholar]
- 2.Ameduri B, Sawada H. Fluorinated Polymers. The Royal Society of Chemistry; Cambridge: 2017. [Google Scholar]
- 3.Hird M. Chem Soc Rev. 2007;36:2070–2095. doi: 10.1039/b610738a. [DOI] [PubMed] [Google Scholar]
- 4.Gribble GW. In: Organofluorines. Neilson AH, editor. Springer; Berlin Heidelberg, Berlin, Heidelberg: 2002. pp. 121–136. [Google Scholar]
- 5.Furuya T, Kuttruff CA, Ritter T. Curr Opin Drug Discovery Dev. 2008;11:803–819. [PubMed] [Google Scholar]
- 6.Harper DB, O’Hagan D, Murphy CD. In: Natural Production of Organohalogen Compounds. Gribble G, editor. Springer; Berlin Heidelberg, Berlin, Heidelberg: 2003. pp. 141–169. [Google Scholar]
- 7.Thuronyi BW, Chang MCY. Acc Chem Res. 2015;48:584–592. doi: 10.1021/ar500415c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.a) Walker MC, Thuronyi BW, Charkoudian LK, Lowry B, Khosla C, Chang MCY. Science. 2013;341:1089–1094. doi: 10.1126/science.1242345. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Ad O, Thuronyi BW, Chang MCY. Proc Natl Acad Sci U S A. 2017;114:E660–E668. doi: 10.1073/pnas.1614196114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Okamura E, Tomita T, Sawa R, Nishiyama M, Kuzumaya T. Proc Natl Acad Sci U S A. 2010;107:11265–11270. doi: 10.1073/pnas.1000532107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Matsumoto K, Yamada M, Leong CR, Jo SJ, Kuzumaya T, Taguchi S. Biosci, Biotechnol, Biochem. 2011;75:364–366. doi: 10.1271/bbb.100682. [DOI] [PubMed] [Google Scholar]
- 11.a) Brandl H, Gross RA, Lenz RW, Fuller RC. Adv Biochem Eng/Biotechnol. 1990;41:77–93. doi: 10.1007/BFb0010232. [DOI] [PubMed] [Google Scholar]; b) Lu J, Tappel RC, Nomura CT. J Macromol Sci, Polym Rev. 2009;49:226–248. [Google Scholar]; c) Stubbe JJ, Tian J. Natural Product Reports. 2003;20:445–457. doi: 10.1039/b209687k. [DOI] [PubMed] [Google Scholar]
- 12.a) Agnew DE, Pfleger BF. Chem Eng Sci. 2013;103:58–67. doi: 10.1016/j.ces.2012.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Bengtsson S, Pisco AR, Reis MAM, Lemos PC. J Biotechnol. 2010;145:253–263. doi: 10.1016/j.jbiotec.2009.11.016. [DOI] [PubMed] [Google Scholar]; c) Fuchtenbusch B, Fabritius D, Steinbüchel AA. FEMS Microbiol Lett. 1996;138:153–160. [Google Scholar]; d) Pisco AR, Bengtsson S, Werker A, Reis MAM, Lemos PC. Appl Environ Microbiol. 2009;75:4676–4686. doi: 10.1128/AEM.02486-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lauble H, Kennedy MC, Emptage MH, Beinert H, Stout CD. Proc Natl Acad Sci U S A. 1996;93:13699–13703. doi: 10.1073/pnas.93.24.13699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Crosby HA, Rank KC, Rayment I, Escalante-Semerena JC. Appl Environ Microbiol. 2012;78:6619–6629. doi: 10.1128/AEM.01733-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Saier MH, Reddy VS, Tamang DG, Västermark A. Nucleic Acids Res. 2014;42:D251–258. doi: 10.1093/nar/gkt1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schaffitzel C, Berg M, Dimroth P, Pos KM. J Bacteriol. 1998;180:2689–2693. doi: 10.1128/jb.180.10.2689-2693.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.a) Weeks AM, Chang MCY. Proc Natl Acad Sci U S A. 2012;109:19667–19672. doi: 10.1073/pnas.1212591109. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Weeks AM, Coyle SM, Jinek M, Doudna JA, Chang MCY. Biochemistry. 2010;49:9269–9279. doi: 10.1021/bi101102u. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Weeks AM, Keddie NS, Wadoux RDP, O’Hagan D, Chang MCY. Biochemistry. 2014;53:2053–2063. doi: 10.1021/bi4015049. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.