Abstract
Detergents are essential tools for functional and structural studies of membrane proteins. However, conventional detergents are limited in their scope and utility, particularly for eukaryotic membrane proteins. Thus, there are major efforts to develop new amphipathic agents with enhanced properties. Here, a novel class of diastereomeric agents with a preorganized conformation, designated norbornane-based maltosides (NBMs), were prepared and evaluated for their ability to solubilize and stabilize membrane proteins. Representative NBMs displayed enhanced behaviors compared to n-dodecyl-β-d-maltoside (DDM) for all membrane proteins tested. Efficacy of the individual NBMs varied depending on the overall detergent shape and alkyl chain length. Specifically, NBMs with no kink in the lipophilic region conferred greater stability to the proteins than NBMs with a kink. In addition, long alkyl chain NBMs were generally better at stabilizing membrane proteins than short alkyl chain agents. Furthermore, use of one well-behaving NBM enabled us to attain a marked stabilization and clear visualization of a challenging membrane protein complex using electron microscopy. Thus, this study not only describes novel maltoside detergents with enhanced protein-stabilizing properties but also suggests that overall detergent geometry has an important role in determining membrane protein stability. Notably, this is the first systematic study on the effect of detergent kinking on micellar properties and associated membrane protein stability.
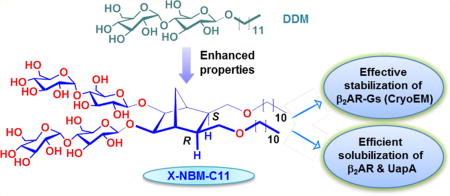
Graphical abstract
INTRODUCTION
Membrane proteins are essential for a number of cellular functions including transport of neutral molecules and ions into and out of the cell and intracellular signal transduction. Additionally, they represent more than one-half of human drug targets.1 Thus, there is a requirement for high-resolution membrane proteins structures in order to gain greater insights into their precise mechanism of action2 and to facilitate rational drug design.3 However, these biomacromolecules tend to undergo rapid protein denaturation and aggregation once extracted from the native lipid bilayers.4 A key requirement of isolation and structural study of membrane proteins is that they must be maintained in a soluble form in aqueous solution. Amphipathic additives play a critical role in this process by shielding the large hydrophobic surface area of proteins from polar aqueous environments.5 Conventional detergents, exemplified by n-octyl-β-d-glucoside (OG), lauryldimethylamine-N-oxide (LDAO), and n-dodecyl-β-d-maltoside (DDM), are widely used for this purpose.6 However, membrane proteins encapsulated even in these popular detergents have the propensity to denature and/or aggregate,7 making it difficult to conduct downstream characterization such as functional studies, spectroscopic analysis, or crystallization trials. The development of new types of amphiphilic molecules or membrane-mimetic systems with enhanced protein stabilization efficacy is therefore of great importance for both functional and structural investigation of these biomacromolecules.8
Over the past two decades, several types of amphiphiles have been invented to solve the issues associated with conventional detergents. Representatives include amphipols,9 peptide-based agents (e.g., lipopeptide detergents (LPDs),10 β-peptides,11 and short peptides),12 nanoassemblies (e.g., nanodiscs (NDs),13 and nanolipodisq).14 These rather large molecules/assemblies proved to be effective at stabilizing the structures of multiple membrane proteins. However, they have a number of disadvantages: they tend to form large protein-detergent complexes (PDCs), are often ineffective at efficiently extracting proteins from the membranes, and are often difficult to synthesize on a bulk scale. In contrast, small amphipathic agents have also been developed as exemplified by tripod amphiphiles (TPAs),15 facial amphiphiles (FAs),16 neopentyl glycol (NG) class amphiphiles (GNGs17 or MNGs),18 mannitol-based amphiphiles (MNAs),19 neopentyl glycol triglucosides (NDTs),20 and penta-saccharide amphiphiles (PSEs).21 These can all be easily synthesized and are largely as effective as DDM at extracting proteins from the membrane. Of these novel small molecule amphiphiles, the NG class agents have contributed to the crystal structures of ~30 new membrane proteins in the last five years, including several G-protein coupled receptors, clearly illustrating the contribution of novel agents to the determination of membrane protein structures.22 In this study we have used a norbornane (i.e., bicyclo[2.2.1]heptane) framework as the core of a group of novel amphiphiles. This core is flanked by two flexible alkyl arms and two maltoside head groups. The two alkyl chains were connected to the norbornane core in a stereospecific manner (endo/endo or exo/exo geometry), designated endo-norbornane-based maltosides (D-NBMs) or exo-norbornane-based maltosides (X-NBMs), respectively. Because of the presence of the bicyclic linker and the well-defined orientation of the alkyl chains, conformational flexibility of the hydrophobic groups is significantly restricted, leading to a unique degree of conformational preorganization in the detergent hydrophobic group (Figure 1). For the same reasons, the alkyl chains of the NBMs were also largely segregated from the hydrophilic groups, thus endowing these agents with a facial property. Thus, they differ in architecture from both conventional detergents which contain either very flexible (e.g., OG and DDM) or very rigid (e.g., 3-[(3-cholamindopropyl) dimethylammonio]-1-propanesulfonate (CHAPS)23) hydrophobic group and other facial agents with very rigid hydrophobic groups (e.g., the steroidal units of FAs and tandem facial amphiphiles (TFAs).16 These two extremes in detergent flexibility contribute to the difficulty in maintaining membrane protein stability in solution and in crystallizing membrane-protein–detergent complexes using conventional detergents. Of the facial agents developed for membrane protein study, the current agents contain systematic chiral variation only in the lipophilic region. The distinct features of the norbornane scaffold are widely exploited where conformational preorganization is paramount: in peptidomimetics24 and medicinal chemistry,25 in supramolecular chemistry as photo-switchable ion carriers,26a,b and as chiral auxiliaries for asymmetric synthesis.26c–g However, they have not so far been applied to membrane protein research. In this study, two sets of stereoisomeric detergents with the norbornane linker were prepared and evaluated for their stabilization efficacy with four different membrane proteins including a G-protein coupled receptor (GPCR) and a membrane protein complex (GPCR:G-protein complex). We found that some of these agents conferred markedly enhanced stability to the target proteins compared to that with DDM, with the X-isomers generally performing better than the D-isomers.
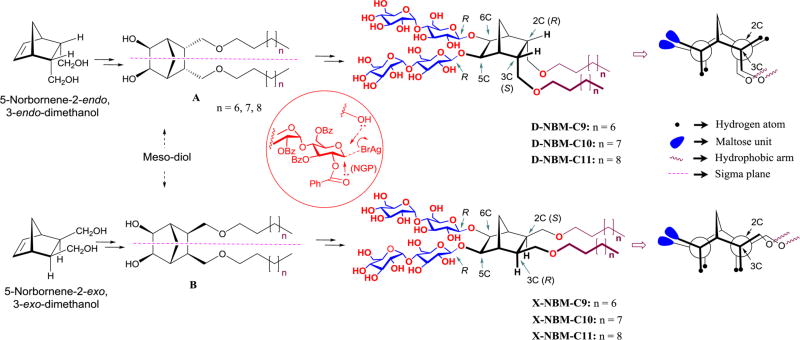
Figure 1.
Chemical structures of the novel NBMs (middle-right) and their Newman projections (extreme right). D-NBMs were derived from 5-norbornene-2-endo,3-endo-dimethanol while X-NBMs were derived from isomeric 2-exo,3-exo-dimethanol (extreme left). Dialkylated norbornane diols (A and B) are meso compounds due to the presence of a symmetry plane passing through the central part of the molecules in a long axis direction, indicated by the purple dotted line (middle left). Newman projections clearly indicate the geometrical difference between these two isomers; the hydrophobic groups of the X-isomers are almost parallel relative to the norbornane linker and the hydrophilic group, while the hydrophobic groups of the D-isomers contain a kink between the norbornane linker and the alkyl chains. Inset within red circle illustrates neighboring group participation (NGP) key for β-selective glycosylation.
RESULTS
Detergent Structures and Physical Characterizations
The NBMs feature two alkyl chains as the hydrophobic group and a branched dimaltoside hydrophilic headgroup, connected via a central norbornane linker (Figure 1). Depending on the spatial orientation of the alkyl chains attached to the linker, these novel agents could be categorized into two groups: D-NBMs, containing two alkyl chains endo-facially (2-endo,3-endo or 2R,3S) connected to the linker, and X-NBMs, with two alkyl chains with exo-facial orientation (2-exo,3-exo or 2S,3R).
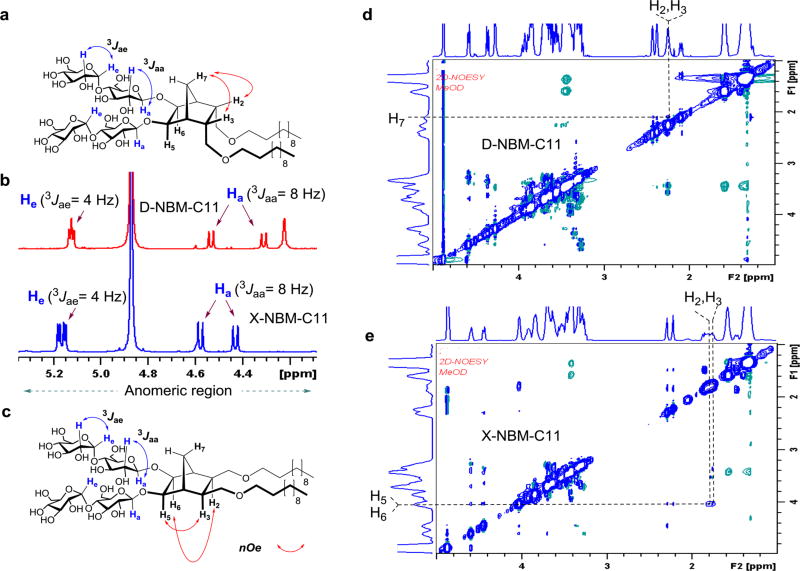
We hypothesized that a difference in the relative orientations of detergent alkyl arms would vary detergent geometry, potentially resulting in substantial changes in amphiphile efficacy for membrane protein stability despite their identical chemical compositions (i.e., identical polar and nonpolar segments). Note that individual hydrophobic groups of the D-/X-NBMs are optically inactive meso compounds due to the internal symmetry plane dissecting the norbornane linker (compounds A and B in Figure 1). As the alkyl chains are connected to the central linker in either endo- or exo-fashion, compounds A and B are diastereomers (i.e., nonmirror-image stereoisomers). The final D-/X-NBMs are also diastereomers of each other, but are optically active because of the lack of a symmetry plane. As hydrophile–lipophile balance (HLB) is a key variable affecting detergent properties, we prepared both sets of NBMs with alkyl chain length variation from C9 to C11, which was used for the designation of the individual detergents. These novel agents were synthesized via straightforward synthetic schemes comprising four high-yielding steps: dialkylation of appropriate norbornene-2,3-dimethanol, alkene syn-dihydroxylation using a typical osmium tetroxide–N-methyl morpholine-N-oxide (OsO4–NMO), glycosylation with perbenzoylated maltosyl bromide, and global deprotection (see the Supporting Information for details). Note that glycosylation was stereospecifically carried out by taking advantage of neighboring group participation (NGP) of the benzoyl group (inset in Figure 1).27 Consequently, this synthetic protocol produced individual NBMs with high diastereomeric purity, confirmed by their individual 1H NMR spectra (Figures 2 and S1). The axial protons of D-NBM-C11 attached to the anomeric carbons, designated Ha (Figure 2a), gave rise to two separate 1H NMR peaks at 4.55 and 4.33 ppm as doublets. The axial protons of the X-isomer also gave two doublet signals, but in different positions, located at 4.57 and 4.42 ppm. In addition, the vicinal coupling constants (3Jaa) for the anomeric protons (Ha) of both isomers were 8.0 Hz, typical of a β-anomer, demonstrating exclusive β-glycosidic bond formation in the glycosylation. Note that the α-anomer contains equatorial anomeric protons, giving a doublet signal in the region of 5.10–5.20 ppm with a smaller coupling constant (3Jae = 4.0 Hz). This spectral feature was identified for another anomeric proton (He) (Figure 2a–c). The exo- or endo-fashioned connection of the alkyl chains to the central linker was confirmed by the through-space interactions seen in 2D NOESY spectra of D-/X-NBM-C11 (Figure 2d,e). Because of the close proximity in space, the strong NOE correlation signals between proton H7 and protons (H2 and H3) were observed in the D-isomer but were not detected for the X-isomer. Instead, the intense NOE correlation signals were obtained between protons [H2 and H3] and protons [H6 and H5] for the X-isomer, indicating their spatial proximity. More detailed analysis on observed NOE correlation signals is given in Figure S2. Because of the high efficiency of each synthetic step, the final amphipathic compounds could be prepared in overall yields of ~75%, making them feasible for preparation in multigram quantities.
Figure 2.
(a, c) Chemical structures of D-/X-NBM-C11 are shown to illustrate anomeric protons (Ha or He), their couplings with the neighboring proton (H) and a set of proton interactions responsible for key NOE correlation signals. (b) Partial 1H NMR spectra in the anomeric region for two NBM isomers showing their high diastereomeric purity (see Figure S1 for the full range of 1H NMR spectra). The spectrum for D-NBM-C11 gave two doublets at 4.55 and 4.33 ppm while that for the X-isomer produced two doublets at 4.57 and 4.42 ppm, along with the same coupling constant (3Jaa) of 8.0 Hz, typical spectral characteristics for a β-anomeric proton (Ha). These NBMs also contain α-anomeric protons (He), giving two doublets in the region of 5.10 to 5.20 ppm with a smaller coupling constant (3Jae = 4.0 Hz). (d, e) Partial 2D NOESY NMR spectra of D-NBM-C11 and X-NBM-C11. Main NOE correlation signals critical for D-/X-isomeric differentiation are assigned (see Figure S2 for detailed analysis for NOE correlation signals and their assignments).
With the exception of X-NBM-C11, all new agents were water-soluble to more than 5 wt %, with no observed precipitation over a month. X-NBM-C11 showed good water-solubility (~5%), but required a brief ultrasonic agitation for complete dissolution. The micelles were characterized in terms of critical micelle concentrations (CMCs) and the hydrodynamic radii (Rh), both of which were estimated at room temperature via fluorophore encapsulation using diphenylhexatriene (DPH)28 and dynamic light scattering (DLS), respectively. The summarized results are presented in Table 1. The CMC values of the NBMs (from 0.012 to 0.006 mM) were much smaller than that of DDM (0.17 mM), indicating their strong tendency to self-assemble. As expected, the CMC values of the new agents decreased with increasing alkyl chain length from C9 to C11 irrespective of isomeric variation. This is likely due to the increased hydrophobicity of the lipophilic groups induced by the alkyl-chain extension.29 In the isomeric comparison, the CMC values of the exo-NBMs were lower than those of the endo-isomers, as exemplified by a comparison of the CMC values of X-NBM-C9 and D-NBM-C9 (~10 vs ~12 µM). This result is indicative of higher tendency of the X-NBMs to self-assemble than the corresponding D-isomers. The sizes of micelles formed by both NBM isomers tend to increase with the alkyl chain length because of the change in molecular geometry from conical to cylindrical shape as the alkyl chain length increases.30 For example, detergent micelle size increased from 3.3 to 3.5 and to 3.7 nm with the variation of alkyl chain length from C9 to C10 and to C11 for the D-isomers. The dependency of micelle size on detergent alkyl chain length became more prominent for the X-isomers. Notably, a substantial difference in micelle size was found between these two isomers. For instance, the micelles formed by D-NBM-C9 had a hydrodynamic radius (Rh) of 3.3 nm, substantially smaller than that of the micelles formed by X-NBM-C9 (3.7 nm). The differences in micelle size were larger with increasing alkyl chain length, reaching a maximum value at C11 alkyl chain length (3.7 vs 17.3 nm). The larger micelle size of the X-isomers compared to that of the D-isomers observed here is likely as a result of the straight architecture of the X-isomers, making their geometry close to cylindrical shape. This result indicates that a small change in detergent alkyl chain orientation could generate a large difference in the properties of their self-assemblies, which could also affect the membrane protein study. When we investigated the size distribution for NBM micelles at room temperature, all isomers showed a single population of micelles, indicative of highly uniform micellar structures (Figure S3).
Table 1.
Molecular Weights, Critical Micelle Concentrations, Water Solubility, and Hydrodynamic Radii of the Micelles of Both the NBMs and a Conventional Detergent, DDM
| detergent | MWa | CMC (mM)b |
water solubility (wt %)b |
Rh (nm)b,c |
|---|---|---|---|---|
| D-NBM-C9 | 1089.3 | ~0.012 | ~20 | 3.3 ± 0.04 |
| X-NBM-C9 | 1089.3 | ~0.010 | ~20 | 3.7 ± 0.03 |
| D-NBM-C10 | 1117.3 | ~0.008 | ~10 | 3.5 ± 0.03 |
| X-NBM-C10 | 1117.3 | ~0.007 | ~10 | 4.0 ± 0.02 |
| D-NBM-C11 | 1145.4 | ~0.007 | ~5 | 3.7 ± 0.05 |
| X-NBM-C11 | 1145.4 | ~0.006 | ~5 | 17.3 ± 0.10 |
| DDM | 510.1 | ~0.17 | ~20 | 3.4 ± 0.03 |
Molecular weight of detergents.
Critical micelle concentration; these data were obtained at 25 °C.
Hydrodynamic radii of the micelles was determined at 1.0 wt % by dynamic light scattering; n = 4.
Detergent Evaluation with Membrane Proteins
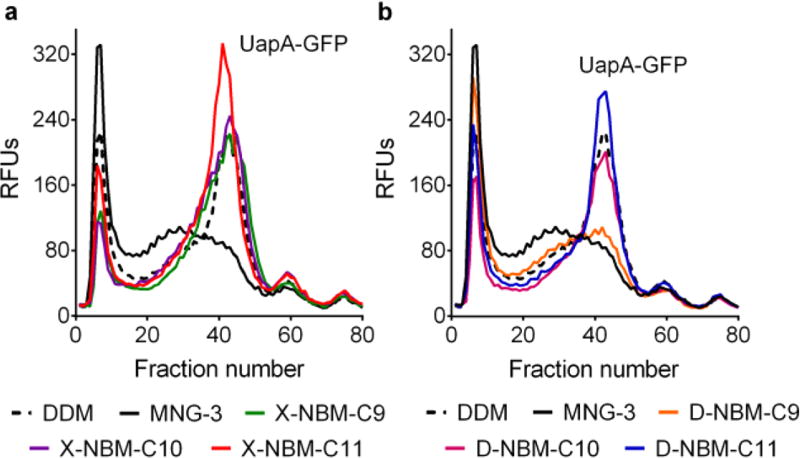
The new agents were first evaluated with a eukaryotic transporter, UapA from Aspergillus nidulans,31 expressed as a fusion protein with a C-terminal GFP in Saccharomyces cerevisiae. The transporter was first extracted from the membranes using 1.0 wt % DDM, MNG-3, or individual NBMs. The protein integrity was then assessed through fluorescent size-exclusion chromatography (FSEC)32 after heat treatment at 40 °C for 10 min. DDM extracted the transporter from the membrane with a solubilization efficiency of ~90%, and the resulting DDM-solubilized UapA-GFP yielded a single monodispersed peak with relatively high intensity (~fraction number 40) following the thermal treatment, implying a good ability to resist heat denaturation (Figure 3). When the X-NBMs were evaluated with the transporter, detergent performance was significantly enhanced with increasing alkyl chain length. Both D-NBM-C9/C10 solubilized the transporter with ~75% yield, but were comparable to DDM in terms of the monodispersed protein peak height (Figure 3a) after thermal treatment. Thus, these agents (D-NBM-C9/C10) were less efficient than DDM at extracting the transporter (~75 vs ~90%), but were more effective at retaining the protein integrity. The use of X-NBM-C11 yielded quantitative extraction of the transporter (~100% yield) and produced a larger monodispersed protein peak compared to that of DDM (Figure 3a). This result indicates that X-NBM-C11 is superior to DDM in terms of both solubilization efficiency and stabilization efficacy for the transporter. Similar to that of the X-NBMs, detergent performance was enhanced with increasing alkyl chain length for the D-NBMs, although these D-isomers were slightly inferior to the corresponding X-isomers in their overall performance. D-NBM-C9/C10 and D-NBM-C11 gave ~60 and ~80% transporter extraction efficiency, respectively; thus, these agents were less efficient at the extraction than the corresponding X-isomers as well as DDM. D-NBM-C9 with the shortest alkyl chain showed a low recovery of monodispersed protein, indicating that significant protein aggregation/denaturation had occurred during heating, while that of D-NBM-C10, with the intermediate chain length, was only slightly worse than that of DDM (Figure 3b). The D-isomer with the longest alkyl chain (i.e., D-NBM-C11) was a little better than DDM. Taking into account the relatively low protein solubilization efficiency, D-NBM-C10/C11 appeared to be better at maintaining protein integrity than DDM during the thermal treatment. It is notable that MNG-3, one of the most successful novel agents for membrane protein structural studies, was ineffective at preventing protein denaturation/aggregation under the same assay conditions (Figure 3a,b).
Figure 3.
UapA stabilization efficacy of DDM, MNG-3, X-NBMs (a), and D-NBMs (b). Fluorescence size-exclusion chromatography (FSEC) was carried out with UapA-GFP fusion protein. UapA-GFP was first extracted from the membrane by DDM, MNG-3, or a NBM at 1.0 wt %, and the detergent-solubilized UapA-GFP fusion protein heated for 10 min at 40 °C. The thermally treated protein samples were loaded onto the SEC column, and the individual relative fluorescent units (RFU) of the fractions assessed. The data is representative of two independent experiments.
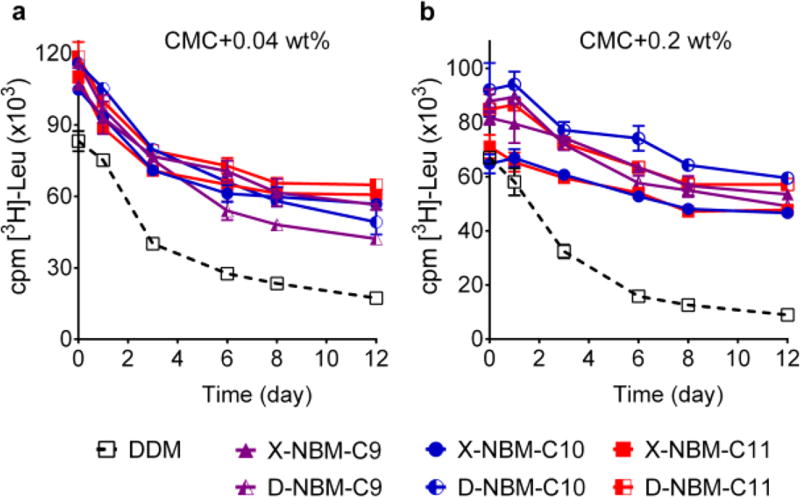
The NBM amphiphiles were further evaluated with the leucine transporter (LeuT), the prokaryotic homologue of the mammalian neurotransmitter/sodium symporter (NSSs family) from Aquifex aeolicus.33 This transporter was initially extracted with 1.0 wt % DDM and purified in 0.05 wt % of the same detergent. The DDM-purified LeuT was diluted into buffer solutions containing individual NBMs or DDM to reach a final detergent concentration of CMC + 0.04 wt %. We assessed protein stability by measuring the ability of the transporter to bind radiolabeled leucine [3H-Leu] via a scintillation proximity assay (SPA).34 The substrate binding activity of the transporter was monitored at regular intervals over the course of a 12 day incubation at room temperature. Upon dilution, LeuT in all the NBMs gave substantially higher transporter activity than in DDM. The enhanced transporter activity relative to that of DDM was well-maintained over 12 days for all the NBMs. When solubilized in X-/D-NBM-C11, transporter activity at the end of incubation (t = 12 day) was only a little less than the initial activity of the protein solubilized in DDM (Figure 4a). However, no clear difference between the X- and D-isomers was observed in this regard. It was also difficult to identify a clear differentiation in transporter activity depending on alkyl chain length variation. When detergent concentration was increased to CMC + 0.2 wt %, a similar trend was observed; the variation in either detergent stereochemistry or alkyl chain length did not give any substantial change in the ability of a detergent to maintain transporter activity. With increasing detergent concentration, however, the increase in detergent efficacy of the NBMs compared to DDM was more evident (Figure 4b). At this concentration after 12 days, D-NBM-C10-solubilized transporter had an activity higher than the activity of the DDM-solubilized protein at the start of the experiment. Overall, all NBMs were effective at preserving the substrate binding ability of the transporter with no noteworthy variation depending on alkyl chain orientation and length of the detergents.
Figure 4.
Long-term substrate binding activity of purified LeuT in the presence of individual NBM isomers or DDM at two detergent concentrations: (a) CMC + 0.04 wt % and (b) CMC + 0.2 wt %. The substrate binding activity of the transporter was measured at regular intervals during a 12 day incubation at room temperature. LeuT activity was measured using a radio-labeled substrate ([3H]-Leu) via scintillation proximity assay (SPA). Error bars, SEM; n = 3.
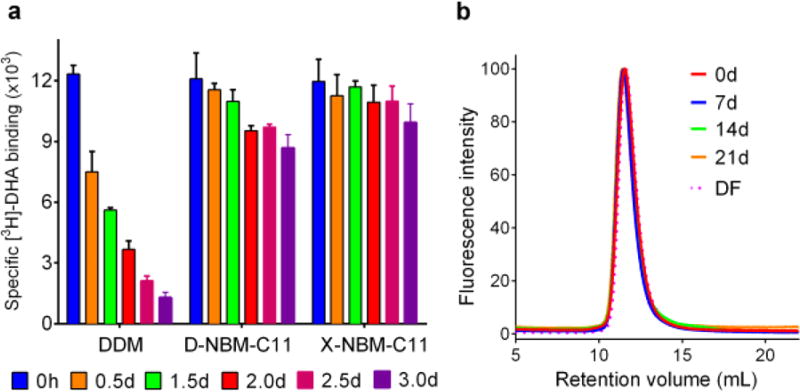
We next moved to the human β2 adrenergic receptor (β2AR), a G-protein-coupled receptor (GPCR),35 for detergent evaluation. For this experiment, the receptor was extracted from the membranes by DDM and purified in the same detergent. The DDM-purified receptor was diluted into individual NBM- or DDM-containing buffers giving a final detergent concentration of CMC + 0.2 wt %. The receptor stability was assessed by measuring ligand binding activity using the antagonist ([3H]-dihydroalprenolol (DHA)).36 As a preliminary study, the activity of the receptor solubilized in the individual NBMs or DDM was measured after a 30 min dilution to allow detergent reconstitution. All C9 and C10 versions of the NBMs showed receptor activity lower than that of DDM (Figure S4). However, receptor activity in the presence of the NBM-C11s was equivalent to that of DDM. In this evaluation, the individual exo-isomers (X-NBM-C9/C10/C11) were a little better than the endo counterparts (D-NBM-C9/C10/C11) regardless of chain length variation (Figure S4). In order to investigate detergent ability to stabilize the receptor long-term, we selected three novel detergents (D-NBM-C11, X-NBM-C11, and DDM), which were well-behaved in the initial study. The receptor solubilized in either of these agents was assessed for radiolabeled ligand binding activity at regular intervals over a 3 day incubation at room temperature (Figure 5a). The DDM-solubilized receptor showed high initial activity, but rapidly lost activity over time, resulting in only ~10% retention of initial receptor activity at the end of the 3 day incubation. In contrast, the D-NBM-C11 or X-NBM-C11 receptor samples retained 70–85% of the initial receptor activity at the end of the incubation period, with X-NBM-C11 performing slightly better than D-NBM-C11. Combined together, overall detergent efficacy order for the receptor stability is NBM-C11s > NBM-C10s > NBM-C9s, with the X-isomers performing better than the D-isomers. This result is more or less in agreement with that obtained for the UapA-GFP fusion protein. The encouraging result of X-NBM-C11 for receptor stability prompted us to compare this agent with MNG-3, a novel detergent which has been extremely successful in GPCR structural studies. As expected, MNG-3 was superior to DDM at maintaining long-term receptor stability at room temperature (Figure S5). DDM-solubilized receptor had almost completely lost ligand binding activity by day 3, while MNG-3 retained ~50% activity even at day 7. In contrast, the receptor in X-NBM-C11 retained ~85% at day 7. After a 10 day incubation at room temperature, MNG-3-solubilized receptor retained ~40% of ligand binding activity, while X-NBM-C11-solubilized receptor retained ~80% activity. This agent was even better than PSE-C11 introduced in our previous study, which retained only ~40% ligand binding activity by day 4.21 Note that this marked behavior of X-NBM-C11 is unprecedented. In addition, in order to exclude the potential effect of residual DDM remaining after dilution, the receptor was directly extracted from the membrane by treatment with 1.0 wt % DDM or X-NBM-C11. Upon protein extraction, the ligand binding ability of the receptor was measured. X-NBM-C11 was comparable to DDM in terms of retaining receptor activity (Figure S6a). These receptor-detergent complexes were further incubated for 7 days at room temperature. During the incubation, receptor stability was monitored at regular intervals by measuring ligand binding ability. Consistent with the previous result described above, the DDM-solubilized receptor lost activity rapidly over time, whereas the X-NBM-C11-protein effectively preserved long-term activity under the same conditions (Figure S6b). This result indicates that X-NBM-C11 alone can be used effectively for both solubilization and purification of the target proteins. Some novel agents are known to be favorable toward membrane protein stability only in the presence of a conventional detergent.37 When we performed SEC with the receptor/DDM or X-NBM-C11 after detergent exchange, we found that X-NBM-C11 formed homogeneous PDCs, with their size being comparable to those formed by DDM (Figure S7). In order to investigate protein functionality, the receptor was conjugated with a fluorophore (monobromobimane; mBBr).38 mBBr-β2AR was used to monitor the conformational changes of the receptor in the presence of binding partners (isopreoterenol (ISO) and Gs-protein) via fluorescence measurement (Figure S8). In the absence of the full agonist (ISO), the DDM- or X-NBM-C11-solubilized receptor gave fluorescence emission spectra corresponding to an inactive receptor. Upon addition of ISO, the fluorescence emission spectrum changed in terms of both the emission intensity and maximum wavelength (λmax) reflecting partial receptor activation in both detergents. A further spectral change corresponding to full receptor activation was observed when Gs-protein and ISO were simultaneously added to the receptor, in accordance with previous results.39,40 These findings indicate that the receptor solubilized in X-NBM-C11 undergoes conformational changes into the partially active (with ISO alone) or fully active states (with ISO+Gs) as occurs in DDM. Next, we focused on β2AR coupled with Gs-protein rather than the receptor alone for detergent evaluation. For this study, DDM-purified receptor and Gs-protein were individually prepared and mixed together to prepare the β2AR–Gs complex in DDM.39 The DDM-purified GPCR–Gs complex was exchanged into X-NBM-C11, and the complex stability was measured at regular intervals over the course of a 21 day incubation at 4 °C. The time-course SEC profiles revealed that X-NBM-C11 maintained perfect complex integrity under these conditions (Figure 5b). This is in stark contrast to the substantial dissociation of the complex into their individual components (the receptor and Gs-protein) after a 2 day incubation reported in the presence of DDM.39 Furthermore, the integrity of the complex was fully maintained even after elution from the gel filtration column using a detergent-free buffer. This result suggests that the NBM molecules associate strongly with the receptor–Gs complex.
Figure 5.
(a) Long-term stability of β2AR solubilized in DDM or a representative NBM (X-NBM-C11 or D-NBM-C11) and (b) time course SEC profiles of β2AR–Gs complex purified in X-NBM-C11. For the long-term stability experiments, DDM-purified receptor was diluted into buffer solution containing individual NBMs to reach a final concentration of CMC + 0.2 wt %. The specific ligand binding activity of the receptor was measured using the antagonist [3H]-dihydroalprenolol (DHA) 30 min after dilution. The activity of the receptor was further measured at regular intervals during a 3 day incubation at room temperature. Error bars, SEM; n = 3. For SEC analysis, the β2AR–Gs complex was prepared from the receptor and the Gs heterotrimer purified in DDM. After detergent exchange with X-NBM-C11, the stability of the complex was assessed over 21 days via analytical gel filtration. The SEC profiles were obtained using detergent-containing or -free running buffer (DF) at designated time points.
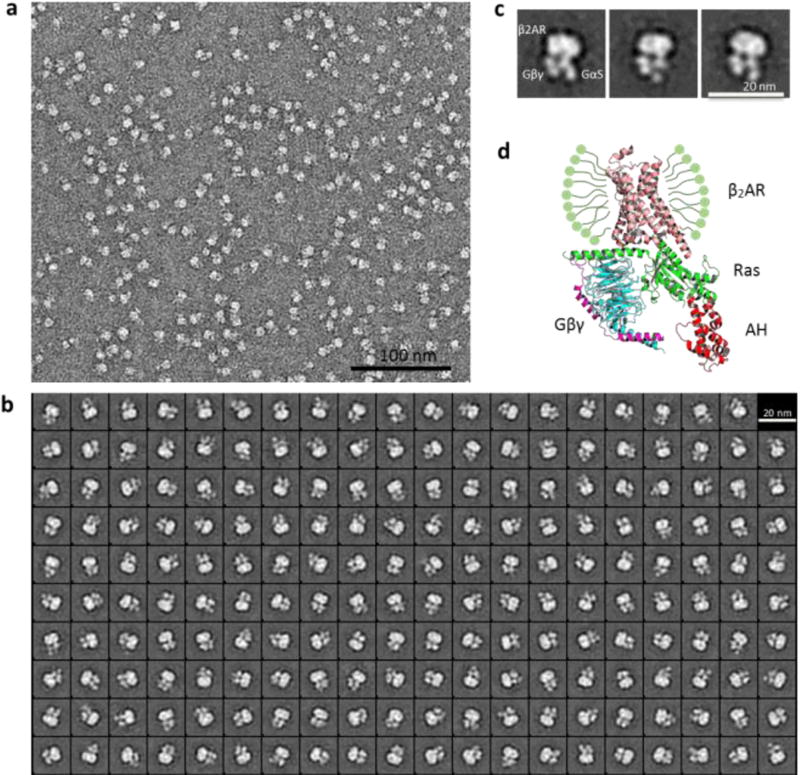
The favorable behavior of the receptor–Gs complex in X-NBM-C11 prompted us to further evaluate this agent for its potential utility in an electron microscopy (EM) study.41,42 β2AR–Gs complex isolated in X-NBM-C11 produced highly monodisperse particles in negative stain EM analysis (Figure 6a) in contrast to the substantial particle aggregation previously observed for DDM-isolated complex.39 Furthermore, the individual components of the complex (β2AR, Gαs, and Gβγ) were clearly distinguished by 2D classification and class averaging of the particles from a single preparation (Figure 6b,c). Even the subdomains of the Gα (the Ras and α-helical (AH) domains) and the individual Gβ and Gγ subunits were discernible in X-NBM-C11. The level of complex visualization achieved here is at least comparable to those obtained from a couple of novel detergents (MNG-3 and PSE-C11),43,21 indicating that the novel agent holds significant potential for the elucidation of dynamic conformational changes of membrane protein complexes via EM study. Overall, X-NBM-C11 was remarkably effective at stabilizing β2AR and its complex with Gs-protein, a feature likely to significantly contribute to GPCR structural and functional analysis.
Figure 6.
Single-particle EM of negative-stained β2AR–Gs complex solubilized in X-NBM-C11. Raw image (a), 2D classification (b), representative class averages in the same orientation (c), and crystal structure of the complex with the designations of individual domains (β2AR, GαS (Ras and AH), and Gβγ) (Protein Databank ID: 3SN6) (d). Ras and AH denote the Ras-like domain and α-helical domain of GαS, respectively.
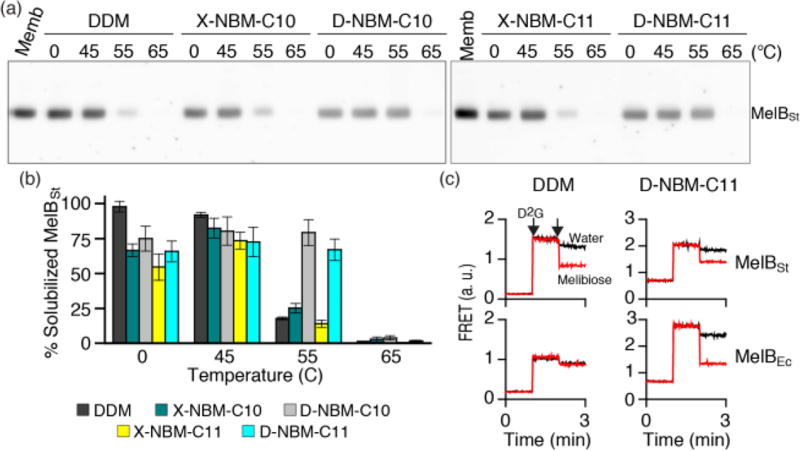
Next, we turned to the melibiose permease of Salmonella typhimurium (MelBSt)44 for further analysis of the stabilization efficacy of the NBMs. Four NBMs (D-NBM-C10, X-NBM-C10, D-NBM-C11, and X-NBM-C11) were selected for this purpose as these agents were effective at stabilizing UapA-GFP, LeuT, and β2AR. Escherichia coli membranes expressing MelBSt at 10 mg/mL were treated with 1.5 wt % DDM or individual NBMs, incubated for 90 min at four different temperatures (0, 45, 55, and 65 °C) and then subjected to ultracentrifugation to remove insoluble material. The amount of soluble MelBSt in the supernatant from each condition was estimated by SDS-PAGE and Western blotting analysis. As shown in Figure 7a,b, DDM quantitatively extracted MelBSt from the membranes at both 0 and 45 °C. However, little or no protein was observed following incubation at 55 or 65 °C, indicating that the DDM-solubilized MelBSt underwent significant aggregation/denaturation at these elevated temperatures. When we used the NBMs for protein extraction at 0 °C, all the NBMs were substantially worse than DDM at efficiently extracting the protein. However, the amounts of soluble MelBSt in the individual NBMs were significantly increased by the extraction at 45 °C; the amounts of soluble MelBSt at this temperature reached a level almost comparable to DDM (Figure 7b). At both incubation temperatures (0 and 45 °C), there was little difference in detergent efficacy between the X- and D-NBM isomers. In contrast, a large difference was found when the incubation temperature was further increased to 55 °C. At this elevated temperature, both X-isomers (X-NBM-C10/C11) yielded only a small amount of soluble MelBSt (~25%), similar to that achieved with DDM. In contrast, the two endo variants of the NBMs (D-NBM-C10 and D-NBM-C11) were highly effective at maintaining MelBSt solubility. At 65 °C, no soluble MelBSt was detectable in any of the tested detergents (Figure 7a,b). The outperformance of D-isomers over X-isomers observed here was opposite to the detergent efficacy order obtained for UapA and β2AR. In order to further assess the advantage of the NBMs over DDM, we performed MelB functional analysis (i.e., melibiose reversal of Förster resonance energy transfer (FRET)). MelB function can be assessed by measuring FRET from tryptophan residues (Trp) to a fluorescent ligand, 2′-(N-dansyl) aminoalkyl-1-thio-β-d-galactopyranoside (D2G).44a,d,e If MelB is well-folded in a given detergent, then the protein is capable of binding both galactosides (D2G and melibiose); thus, FRET signal from D2G bound in the sugar binding site of the protein can be reversed by adding melibiose at a saturation concentration. Of the NBMs, D-NBM-C11 was selected for this assay as this agent was the best at retaining MelBSt stability in solution. MelBSt solubilized in DDM showed an effective melibiose reversal of FRET, indicative of its good ability to bind both galactosides (Figure 7c). However, this ability was significantly impaired for a far less stable MelB homologue from E. coli (MelBEc) solubilized in DDM.44d In contrast, both MelB proteins gave remarkable levels of galactoside binding in D-NBM-C11. Therefore, the overall performance of D-NBM-C11 is superior to that of DDM for maintaining two MelB proteins (MelBSt and MelBEc) in both soluble and functional forms.
Figure 7.
Thermostability of MelBSt solubilized in DDM or a selected novel amphiphile (X-NBM-C10, D-NBM-C10, X-NBM-C11, or D-NBM-C11). Membranes containing MelBSt were treated with the indicated detergent and incubated for 90 min at 0 °C or an elevated temperature (45, 55, or 65 °C). (a) Western blot: the amounts of MelBSt solubilized by detergent treatment were analyzed by SDS-15% PAGE and Western blotting. The untreated membrane sample (Memb) represents the total amount of MelBSt originally present in the membrane. (b) Histogram: the density representing the soluble MelBSt in individual detergents detected in panel (a) was measured by ImageQuant software and expressed as percentages of the total protein amount in the untreated membrane sample. Error bars, SEM, n = 3. (c) FRET reversal by galactoside binding. Right-side-out (RSO) membrane vesicles containing MelBSt or MelBEc were solubilized with DDM or D-NBM-C11. The detergent extracts were used to measure melibiose reversal of FRET from Trp to dansyl-2-galacotside (D2G). D2G at 10 µM and melibiose at a saturating concentration were added at 1 min and 2 min time points, respectively. Control data (black lines) were obtained by addition of water instead of melibiose.
DISCUSSION
Here we describe the development and characterization of a class of novel stereoisomeric amphiphiles with a conformationally preorganized norbornane as a linker. The well-behaved NBMs, particularly X-NBM-C11 and D-NBM-C11, proved to be superior to DDM at stabilizing a range of membrane proteins and protein complex (UapA, LeuT, β2AR, and β2AR–Gs complex). In the comparison of the X- versus D-NBM isomers, the X-isomers yielded significantly enhanced solubilization and stabilization of UapA and β2AR compared to that of the D-isomers. This result indicates the favorable architecture of the X-isomers relative to the D-isomers. In order to explain the different behaviors of the X- and D-NBMs, we considered the detergent–detergent interactions in micelles surrounding a target membrane protein. Favorable detergent–detergent interaction will generate tightly packed detergent micelles, positively associated with micellar stability as well as membrane protein stability. Because of the exo-facial connection of the alkyl chains with the norbornane linker, the molecular shape of the X-isomers is more flat and straight than that of the D-isomers, allowing greater interactions between the individual detergents in the micelles (Figure S9). In contrast, the D-isomers have a bent shape owing to the presence of a central kink, thereby limiting both detergent–detergent interactions and micellar stability. Thus, we suggest that the favorable detergent–detergent interactions possible for the X-isomers relative to the D-isomers are likely to be responsible for the enhanced stability of both the detergent micelles and any associated target protein. Such favorable micellar packing (i.e., detergent–detergent interactions) could also be reflected by the relatively small CMC values of the X-isomers compared to those of the D-isomers. Currently, it is still unclear why there was little difference in detergent efficacy between the stereoisomers in terms of LeuT stability.
Despite the favorable effects on UapA, β2AR, and β2AR–Gs complex, X-NBM-C11 failed to stabilize MelBSt under the conditions tested; the best outcome for this protein was obtained from the D-NBMs rather than the X-isomers, in contrast to the results with the other proteins. Furthermore, this unfavorable behavior of the X-isomers could not be explained by our hypothesis that detergent efficacy is associated with micellar packing propensity as described above. In an attempt to address this paradoxical observation, we measured the micelle size of the detergents at the different temperatures used in the MelBSt assay. We hypothesized that detergent micelles may undergo a size variation with increasing temperature, which may reflect thermal stability of an associated target membrane protein (i.e., PDC). DLS experiments revealed that micelles of the X-isomers (X-NBM-C10 and X-NBM-C11) (Figure S10) underwent a significant increase in size upon incubation at elevated temperature. Specifically, the micelle size (Dh) of X-NBM-C11 doubled (from 27.9 to 55.3 nm) when temperature varied from 5 to 65 °C. As this micellar volume is proportional to R3 where R is the micelle radius, this increase in Dh corresponds to an 8-fold increase in the micellar volume. A similar trend was observed for X-NBM-C10, although the size variation was much less: from 7.8 nm at 5 °C to 10.0 nm at 65 °C. In contrast, all tested D-NBMs (D-NBM-C10 and D-NBM-C11) gave little variation in detergent micelle size under the same conditions. This result indicates that micelles formed by these D-isomers have enhanced thermal stability compared to those formed by the X-isomers, and this in turn is likely to confer enhanced thermal stability to the PDCs formed by the D-isomers with MelBSt at 55 °C. The presence of a kink in the D-isomers, missing in the X-isomers, appeared to play a critical role in maintaining thermal stability of detergent micelles and PDCs at an elevated temperature. It is impossible to know the precise reason for this, but detergent kink would allow less sliding of detergent alkyl chains relative to each other in the thermally agitated micellar assemblies. The restricted movement of detergent molecules would reduce the dynamic nature of micelles, resulting in enhanced micellar stability at a high temperature. Note that absolute micelle size does not seem to be important in this context since large micelles formed by X-NBM-C11 were not substantially different from small micelles formed by X-NBM-C10 in maintaining MelB solubility.
The hydrophobic groups of the diastereomeric NBMs contain structural features distinct from those of conventional or novel detergents. As a result of the chirality variations in C2 and C3, the D-NBMs have a kink in the lipophilic region, lacking in the X-NBMs. There is no systematic study comparing efficacy of detergents with/without a kink in the lipophilic region. Interestingly enough, lipid molecules also contain a variable number of kinks (typically 1–3) in the same region, introduced by the presence of cis-double bond(s). The presence of kink(s) in lipid molecules is known to play a favorable role in maintaining membrane fluidity and permeability relevant for function.45 In contrast to lipid molecules, the current results indicate that the presence of detergent kink in the lipophilic region generally is suboptimal for protein stability as illustrated by the relative poor behaviors of the D-NBMs compared to the X-NBMs. This seemingly contradictory result could be explained by the inherent difference in fluidity between lipid bilayer and detergent micelles. Detergent micelles are highly dynamic, reflected in their small spherical structures; thus, introducing a kink into the detergent structure would further increase the dynamic nature of the micelle.46 Thus, kink-bearing detergents would display unfavorable behaviors toward membrane protein stabilization. However, as described above, the presence of a kink in the NBM architecture appeared favorable at high temperature by reducing the dynamic property of detergent micelles. Thus, the detergent kink appears to have distinct roles in micellar dynamic nature depending on the temperature. In addition, this study suggests that introduction of a kink in detergent architecture could be beneficial for some applications requiring thermally stable micelles or PDCs. It will be interesting to investigate the effect of detergent kink on micellar properties in the context of various detergent architectures.
CONCLUSION
Our exploration of norbornane-based detergents (NBMs) demonstrates their utility as membrane protein solubilizing and stabilizing reagents. Some of the NBMs, particularly for X-NBM-C11 and D-NBM-C11, were superior to or at least comparable to DDM in maintaining the integrity of several membrane proteins. The marked stabilization of β2AR–Gs complex was attained in X-NBM-C11, which enabled us to clearly visualize the individual domains of the complex via EM analysis. As these multiple characteristics, along with synthetic convenience, are often not compatible in a single detergent structure, these NBMs could represent invaluable tools for membrane protein structure study. Stereoisomeric comparison of X- versus D-NBMs strongly indicated that the presence of detergent kink significantly affects micellar properties such as micelle size and protein stabilization efficacy in a temperature-dependent manner. Based on this study, we propose that detergent packing density in micellar environments should be considered as a critical factor when a novel agent is designed for membrane protein study.
Supplementary Material
Acknowledgments
This work was supported by the National Research Foundation of Korea (NRF) funded by the Korean government (MSIP) (grant number 2016R1A2B2011257 to P.S.C. and M.D.). We thank Gérard Leblanc and H. Ronald Kaback for supplying 2′-(N-dansyl)aminoalkyl-1-thio-β-d-galactopyranoside (D2G).
Footnotes
ASSOCIATED CONTENT
- Experimental procedures, characterizations of amphiphiles, and membrane protein stability assays (PDF)
The authors declare the following competing financial interest(s): P.S.C. and M.D. are co-inventors on a patent application that covers the NBM agents.
References
- 1.Overington JP, Al-Lazikani B, Hopkins AL. Nat. Rev. Drug Discovery. 2006;5:993–996. doi: 10.1038/nrd2199. [DOI] [PubMed] [Google Scholar]
- 2.(a) Rosenbaum DM, Rasmussen SGF, Kobilka BK. Nature. 2009;459:356–363. doi: 10.1038/nature08144. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) McCusker EC, Bagneris C, Naylor CE, Cole AR, D’Avanzo N, Nichols CG, Wallace BA. Nat. Commun. 2012;3:1102. doi: 10.1038/ncomms2077. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Ghosh E, Kumari P, Jaiman D, Shukla AK. Nat. Rev. Mol. Cell Biol. 2015;16:69–81. doi: 10.1038/nrm3933. [DOI] [PubMed] [Google Scholar]
- 3.(a) Nam HJ, Jeon J-H, Kim S-U. BMB Rep. 2009;42:697–704. doi: 10.5483/bmbrep.2009.42.11.697. [DOI] [PubMed] [Google Scholar]; (b) Murray CW, Blundell TL. Curr. Opin. Struct. Biol. 2010;20:497–507. doi: 10.1016/j.sbi.2010.04.003. [DOI] [PubMed] [Google Scholar]
- 4.(a) White SH, Wimley WC. Annu. Rev. Biophys. Biomol. Struct. 1999;28:319–365. doi: 10.1146/annurev.biophys.28.1.319. [DOI] [PubMed] [Google Scholar]; (b) Bowie JU. Curr. Opin. Struct. Biol. 2001;11:397–402. doi: 10.1016/s0959-440x(00)00223-2. [DOI] [PubMed] [Google Scholar]; (c) Lacapere JJ, Pebay-Peyroula E, Neumann JM, Etchebest C. Trends Biochem. Sci. 2007;32:259–327. doi: 10.1016/j.tibs.2007.04.001. [DOI] [PubMed] [Google Scholar]; (d) Phillips R, Ursell T, Wiggins P, Sens P. Nature. 2009;459:379–385. doi: 10.1038/nature08147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang Q, Tao H, Hong W-X. Methods. 2011;55:318–323. doi: 10.1016/j.ymeth.2011.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chae PS, Laible PD, Gellman SH. Mol. BioSyst. 2010;6:89–94. doi: 10.1039/b915162c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Prive GG. Methods. 2007;41:388–397. doi: 10.1016/j.ymeth.2007.01.007. [DOI] [PubMed] [Google Scholar]
- 8.(a) Loll JP. J. Struct. Biol. 2003;142:144–153. doi: 10.1016/s1047-8477(03)00045-5. [DOI] [PubMed] [Google Scholar]; (b) White SH. Protein Sci. 2004;13:1948–1949. doi: 10.1110/ps.04712004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.(a) Tribet C, Audebert R, Popot J-L. Proc. Natl. Acad. Sci. U. S. A. 1996;93:15047–15050. doi: 10.1073/pnas.93.26.15047. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Popot J-L, Althoff T, Bagnard D, Banères JL, Bazzacco P, Billon-Denis E, Catoire LJ, Champeil P, Charvolin D, Cocco MJ, et al. Annu. Rev. Biophys. 2011;40:379–408. doi: 10.1146/annurev-biophys-042910-155219. [DOI] [PubMed] [Google Scholar]
- 10.McGregor C-L, Chen L, Pomroy NC, Hwang P, Go S, Chakrabartty A, Prive GG. Nat. Biotechnol. 2003;21:171–176. doi: 10.1038/nbt776. [DOI] [PubMed] [Google Scholar]
- 11.Tao H, Lee SC, Moeller A, Roy RS, Siu FY, Zimmermann J, Stevens RC, Potter CS, Carragher B, Zhang Q. Nat. Methods. 2013;10:759–761. doi: 10.1038/nmeth.2533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhao X, Nagai Y, Reeves PJ, Kiley P, Khorana HG, Zhang S. Proc. Natl. Acad. Sci. U. S. A. 2006;103:17707–17712. doi: 10.1073/pnas.0607167103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nath A, Atkins WM, Sligar SG. Biochemistry. 2007;46:2059–2069. doi: 10.1021/bi602371n. [DOI] [PubMed] [Google Scholar]
- 14.Orwick-Rydmark M, Lovett JE, Graziadei A, Lindholm L, Hicks MR, Watts A. Nano Lett. 2012;12:4687–4692. doi: 10.1021/nl3020395. [DOI] [PubMed] [Google Scholar]
- 15.(a) McQuade DT, Quinn MA, Yu SM, Polans AS, Krebs MP, Gellman SH. Angew. Chem., Int. Ed. 2000;39:758–761. [PubMed] [Google Scholar]; (b) Chae PS, Wander MJ, Bowling AP, Laible PD, Gellman SH. ChemBioChem. 2008;9:1706–1709. doi: 10.1002/cbic.200800169. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Chae PS, Cho KH, Wander MJ, Bae HE, Gellman SH, Laible PD. Biochim. Biophys. Acta, Biomembr. 2014;1838:278–286. doi: 10.1016/j.bbamem.2013.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.(a) Lee SC, Bennett BC, Hong W-X, Fu Y, Baker KA, Marcoux J, Robinson CV, Ward AB, Halpert JR, Stevens RC, Stout CD, Yeager MJ, Zhang Q. Proc. Natl. Acad. Sci. U. S. A. 2013;110:E1203–E1211. doi: 10.1073/pnas.1221442110. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Chae PS, Gotfryd K, Pacyna J, Miercke LJW, Rasmussen SGF, Robbins RA, Rana RR, Loland CJ, Kobilka B, Stroud R, Byrne B, Gether U, Gellman SH. J. Am. Chem. Soc. 2010;132:16750–16752. doi: 10.1021/ja1072959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.(a) Chae PS, Rana RR, Gotfryd K, Rasmussen SGF, Kruse AC, Cho KH, Capaldi S, Carlsson E, Kobilka BK, Loland CJ, Gether U, Banerjee S, Byrne B, Lee JK, Gellman SH. Chem. Commun. 2013;49:2287–2289. doi: 10.1039/c2cc36844g. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Cho KH, Bae HE, Das M, Gellman SH, Chae PS. Chem. - Asian J. 2014;9:632–638. doi: 10.1002/asia.201301303. [DOI] [PubMed] [Google Scholar]
- 18.(a) Chae PS, Rasmussen SGF, Rana RR, Gotfryd K, Chandra R, Goren MA, Kruse AC, Nurva S, Loland CJ, Pierre Y, Drew D, Popot J-L, Picot D, Fox BG, Guan L, Gether U, Byrne B, Kobilka B, Gellman SH. Nat. Methods. 2010;7:1003–1008. doi: 10.1038/nmeth.1526. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Cho KH, Byrne B, Chae PS. ChemBioChem. 2013;14:452–455. doi: 10.1002/cbic.201200759. [DOI] [PubMed] [Google Scholar]
- 19.Hussain H, Du Y, Scull NJ, Mortensen JS, Tarrasch J, Bae HE, Loland CJ, Byrne B, Kobilka BK, Chae PS. Chem. - Eur. J. 2016;22:7068–7073. doi: 10.1002/chem.201600533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sadaf A, Mortensen JS, Capaldi S, Tikhonova E, Hariharan P, Ribeiro O, Loland CJ, Guan L, Byrne B, Chae PS. Chem. Sci. 2016;7:1933–1939. doi: 10.1039/c5sc02900g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ehsan M, Du Y, Scull NJ, Tikhonova E, Tarrasch J, Mortensen JS, Loland CJ, Skiniotis G, Guan L, Byrne B, Kobilka BK, Chae PS. J. Am. Chem. Soc. 2016;138:3789–3796. doi: 10.1021/jacs.5b13233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.(a) Rosenbaum DM, Zhang C, Lyons J, Holl R, Aragao D, Arlow DH, Rasmussen SGF, Choi H-J, DeVree BT, Sunahara RK, Chae PS, Gellman SH, Dror RO, Shaw DE, Weis WI, Caffrey M, Gmeiner P, Kobilka BK. Nature. 2011;469:236–240. doi: 10.1038/nature09665. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Haga K, Kruse AC, Asada H, Yurugi-Kobayashi T, Shiroishi M, Zhang C, Weis WI, Okada T, Kobilka BK, Haga T, Kobayashi T. Nature. 2012;482:547–551. doi: 10.1038/nature10753. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Granier S, Manglik A, Kruse AC, Kobilka TS, Thian FS, Weis WI, Kobilka BK. Nature. 2012;485:400–404. doi: 10.1038/nature11111. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) White JF, Noinaj N, Shibata Y, Love J, Kloss B, Xu F, Gvozdenovic-Jeremic J, Shah P, Shiloach J, Tate CG, Grisshammer R. Nature. 2012;490:508–513. doi: 10.1038/nature11558. [DOI] [PMC free article] [PubMed] [Google Scholar]; (e) Kruse AC, Ring AM, Manglik A, Hu J, Hu K, Eitel K, Hubner H, Pardon E, Valant C, Sexton PM, Christopoulos A, Felder CC, Gmeiner P, Steyaert J, Weis WI, Garcia KC, Wess J, Kobilka BK. Nature. 2013;504:101–106. doi: 10.1038/nature12735. [DOI] [PMC free article] [PubMed] [Google Scholar]; (f) Ring AM, Manglik A, Kruse AC, Enos MD, Weis WI, Garcia KC, Kobilka BK. Nature. 2013;502:575–579. doi: 10.1038/nature12572. [DOI] [PMC free article] [PubMed] [Google Scholar]; (g) Miller PS, Aricescu AR. Nature. 2014;512:270–275. doi: 10.1038/nature13293. [DOI] [PMC free article] [PubMed] [Google Scholar]; (h) Karakas E, Furukawa H. Science. 2014;344:992–997. doi: 10.1126/science.1251915. [DOI] [PMC free article] [PubMed] [Google Scholar]; (i) Suzuki H, Nishizawa T, Tani K, Yamazaki Y, Tamura A, Ishitani R, Dohmae N, Tsukita S, Nureki O, Fujiyoshi Y. Science. 2014;344:304–307. doi: 10.1126/science.1248571. [DOI] [PubMed] [Google Scholar]; (j) Kane Dickson V, Pedi L, Long SB. Nature. 2014;516:213–218. doi: 10.1038/nature13913. [DOI] [PMC free article] [PubMed] [Google Scholar]; (k) Shukla AK, Westfield GH, Xiao K, Reis RI, Huang L-Y, Tripathi-Shukla P, Qian J, Li S, Blanc A, Oleskie AN, et al. Nature. 2014;512:218–222. doi: 10.1038/nature13430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bae HE, Gotfryd K, Thomas J, Hussain H, Ehsan M, Go J, Loland CJ, Byrne B, Chae PS. ChemBioChem. 2015;16:1454–1459. doi: 10.1002/cbic.201500151. [DOI] [PubMed] [Google Scholar]
- 24.(a) Karle I, Ranganathan D. J. Mol. Struct. 2003;647:85–96. [Google Scholar]; (b) Chakraborty TK, Srinivasu P, Kumar SK, Kunwar AC. J. Org. Chem. 2002;67:2093–2100. doi: 10.1021/jo010937y. [DOI] [PubMed] [Google Scholar]; (c) Hibbs DE, Hursthouse MB, Jones IG, Jones W, Abdul Malik KM, North M. J. Org. Chem. 1998;63:1496–1504. doi: 10.1021/jo990140v. [DOI] [PubMed] [Google Scholar]
- 25.(a) Buchbauer G, Pauzenberger I. Pharmazie. 1999;54:5–18. [PubMed] [Google Scholar]; (b) Buchbauer G, Spreitzer H, Frei H. Pharmazie. 1991;46:161–170. [PubMed] [Google Scholar]; (c) Oliver DW, Malan SF. Med. Chem. Res. 2008;17:137–151. [Google Scholar]; (d) Moreno-Vargas AJ, Schutz C, Scopelliti R, Vogel P. J. Org. Chem. 2003;68:5632–5640. doi: 10.1021/jo0301088. [DOI] [PubMed] [Google Scholar]; (e) Hutchinson SA, Baker SP, Scammells PJ. Bioorg. Med. Chem. 2002;10:1115–1122. doi: 10.1016/s0968-0896(01)00384-4. [DOI] [PubMed] [Google Scholar]
- 26.(a) Winkler T, Bayrhuber M, Sahlmann B, Herges R. DaltonTrans. 2012;41:7037–7040. doi: 10.1039/c2dt30242j. [DOI] [PubMed] [Google Scholar]; (b) Winkler T, Dix I, Jones PG, Herges R. Angew. Chem., Int. Ed. 2003;42:3541–3544. doi: 10.1002/anie.200350959. [DOI] [PubMed] [Google Scholar]; (c) Verdaguer X, Vazquez J, Fuster G, Bernardes-Genisson V, Greene AE, Moyano A, Pericas MA, Riera A. J. Org. Chem. 1998;63:7037–7052. doi: 10.1021/jo9809985. [DOI] [PubMed] [Google Scholar]; (d) Lattanzi A, Iannece P, Vicinanza A, Scettri A. Chem. Commun. 2003:1440–1441. [PubMed] [Google Scholar]; (e) Gnas Y, Glorius F. Synthesis. 2006;2006:1899–1930. [Google Scholar]; (f) Dalko PI, Moisan L. Angew. Chem., Int. Ed. 2001;40:3726–3748. doi: 10.1002/1521-3773(20011015)40:20<3726::aid-anie3726>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]; (g) Piatek A, Chapuis C, Jurczak J. Helv. Chim. Acta. 2002;85:1973–1988. [Google Scholar]
- 27.Veeneman GH, van Leeuwen SH, van Boom JH. Tetrahedron Lett. 1990;31:1331–1334. [Google Scholar]
- 28.Chattopadhyay A, London E. Anal. Biochem. 1984;139:408–412. doi: 10.1016/0003-2697(84)90026-5. [DOI] [PubMed] [Google Scholar]
- 29.Tanford C. Hydrophobic Effect: Formation of Micelles and Biological Membranes. John Wiley & Sons; New York: 1973. [Google Scholar]
- 30.Cho KH, Husri M, Amin A, Gotfryd K, Lee HJ, Go J, Kim JW, Loland CJ, Guan L, Byrne B, Chae PS. Analyst. 2015;140:3157–3163. doi: 10.1039/c5an00240k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Alguel Y, Amillis S, Leung J, Lambrinidis G, Capaldi S, Scull NJ, Craven G, Iwata S, Armstrong A, Mikros E, Diallinas G, Cameron AD, Byrne B. Nat. Commun. 2016;7:11336. doi: 10.1038/ncomms11336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Leung J, Karachaliou M, Alves C, Diallinas G, Byrne B. Protein Expression Purif. 2010;72:139–146. doi: 10.1016/j.pep.2010.02.002. [DOI] [PubMed] [Google Scholar]
- 33.Huber R, Swanson RV, Deckert G, Warren PV, Gaasterland T, Young WG, Lenox AL, Graham DE, Overbeek R, Snead MA, et al. Nature. 1998;392:353–358. doi: 10.1038/32831. [DOI] [PubMed] [Google Scholar]
- 34.(a) Hart HE, Greenwald EB. Mol. Immunol. 1979;16:265–267. doi: 10.1016/0161-5890(79)90065-8. [DOI] [PubMed] [Google Scholar]; (b) Quick M, Javitch JA. Proc. Natl. Acad. Sci. U. S. A. 2007;104:3603–3608. doi: 10.1073/pnas.0609573104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rosenbaum DM, Cherezov V, Hanson MA, Rasmussen SG, Thian FS, Kobilka TS, Choi HJ, Yao XJ, Weis WI, Stevens RC, Kobilka BK. Science. 2007;318:1266–1273. doi: 10.1126/science.1150609. [DOI] [PubMed] [Google Scholar]
- 36.(a) Yao X, Parnot C, Deupi X, Ratnala VRP, Swaminath G, Farrens D, Kobilka B. Nat. Chem. Biol. 2006;2:417–422. doi: 10.1038/nchembio801. [DOI] [PubMed] [Google Scholar]; (b) Swaminath G, Steenhuis J, Kobilka B, Lee TW. Mol. Pharmacol. 2002;61:65–72. doi: 10.1124/mol.61.1.65. [DOI] [PubMed] [Google Scholar]
- 37.Howell SC, Fraser NJ, Mittal R, Huang L, Travis B, Breyer RM, Sanders CR. Biochemistry. 2010;49:9572–9583. doi: 10.1021/bi101334j. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Li Q, Mittal R, Huang L, Travis B, Sanders CR. Biochemistry. 2009;48:11606–11608. doi: 10.1021/bi9018708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yao X, Parnot C, Deupi X, Ratnala VRP, Swaminath G, Farrens D, Kobilka B. Nat. Chem. Biol. 2006;2:417–422. doi: 10.1038/nchembio801. [DOI] [PubMed] [Google Scholar]
- 39.Rasmussen SGF, Choi H-J, Fung JJ, Pardon E, Casarosa P, Chae PS, DeVree BT, Rosenbaum DM, Thian FS, Kobilka TS, Schnapp A, Konetzki I, Sunahara RK, Gellman SH, Pautsch A, Steyaert J, Weis WI, Kobilka BK. Nature. 2011;469:175–180. doi: 10.1038/nature09648.(b) Ref 3a. Murray CW, Blundell TL. Curr. Opin. Struct. Biol. 2010;20:497–507. doi: 10.1016/j.sbi.2010.04.003.
- 40.Ref 36b.
- 41.Paulsen CE, Armache J-P, Gao Y, Cheng Y, Julius D. Nature. 2015;520:511–517. doi: 10.1038/nature14367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bartesaghi A, Merk A, Banerjee S, Matthies D, Wu X, Milne JLS, Subramaniam S. Science. 2015;348:1147–1151. doi: 10.1126/science.aab1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Westfield GH, Rasmussen SGF, Su M, Dutta S, DeVree BT, Chung KY, Calinski D, Velez-Ruiz G, Oleskie AN, Pardon E, Chae PS, Liu T, Li S, Woods VL, Steyaert J, Kobilka BK, Sunahara RK, Skiniotis G. Proc. Natl. Acad. Sci. U. S. A. 2011;108:16086–16091. doi: 10.1073/pnas.1113645108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.(a) Guan L, Nurva S, Ankeshwarapu SP. J. Biol. Chem. 2011;286:6367–6374. doi: 10.1074/jbc.M110.206227. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Ethayathulla AS, Yousef MS, Amin A, Leblanc G, Kaback HR, Guan L. Nat. Commun. 2014;5:3009. doi: 10.1038/ncomms4009. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Amin A, Ethayathulla AS, Guan L. J. Bacteriol. 2014;196:3134–3139. doi: 10.1128/JB.01868-14. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Amin A, Hariharan P, Chae PS, Guan L. Biochemistry. 2015;54:5849–5855. doi: 10.1021/acs.biochem.5b00660. [DOI] [PMC free article] [PubMed] [Google Scholar]; (e) Cordat E, Mus-Veteau I, Leblanc G. J. Biol. Chem. 1998;273:33198–33202. doi: 10.1074/jbc.273.50.33198. [DOI] [PubMed] [Google Scholar]
- 45.(a) Collier JH, Messersmith PB. Annu. Rev. Mater. Res. 2001;31:237–263. [Google Scholar]; (b) Koike T, Ishida G, Taniguchi M, Higaki K, Ayaki Y, Saito M, Sakakihara Y, Iwamori M, Ohno K. Biochim. Biophys. Acta, Mol. Basis Dis. 1998;1406:327–335. doi: 10.1016/s0925-4439(98)00019-2. [DOI] [PubMed] [Google Scholar]
- 46.(a) Garavito RM, Ferguson-Miller S. J. Biol. Chem. 2001;276:32403–32406. doi: 10.1074/jbc.R100031200. [DOI] [PubMed] [Google Scholar]; (b) Tieleman DP, van der Spoel D, Berendsen HJC. J. Phys. Chem. B. 2000;104:6380–6388. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.