Abstract
G-protein-coupled receptors (GPCRs) pose challenges for drug discovery efforts because of the high degree of structural homology in the orthosteric pocket, particularly for GPCRs within a single subfamily, such as the nine adrenergic receptors. Allosteric ligands may bind to less-conserved regions of these receptors and therefore are more likely to be selective. Unlike orthosteric ligands, which tonically activate or inhibit signalling, allosteric ligands modulate physiologic responses to hormones and neurotransmitters, and may therefore have fewer adverse effects. The majority of GPCR crystal structures published to date were obtained with receptors bound to orthosteric antagonists, and only a few structures bound to allosteric ligands have been reported. Compound 15 (Cmpd-15) is an allosteric modulator of the β2 adrenergic receptor (β2AR) that was recently isolated from a DNA-encoded small-molecule library1. Orthosteric β-adrenergic receptor antagonists, known as beta-blockers, are amongst the most prescribed drugs in the world and Cmpd-15 is the first allosteric beta-blocker. Cmpd-15 exhibits negative cooperativity with agonists and positive cooperativity with inverse agonists. Here we present the structure of the β2AR bound to a polyethylene glycol-carboxylic acid derivative (Cmpd-15PA) of this modulator. Cmpd-15PA binds to a pocket formed primarily by the cytoplasmic ends of transmembrane segments 1, 2, 6 and 7 as well as intracellular loop 1 and helix 8. A comparison of this structure with inactive- and active-state structures of the β2AR reveals the mechanism by which Cmpd-15 modulates agonist binding affinity and signalling.
There is growing interest in the identification of allosteric ligands for GPCRs owing to their ability to modulate the activity of native hormones and neurotransmitters, and the greater potential for subtype selectivity. We previously reported the structure of the activestate M2 muscarinic receptor with a positive allosteric modulator bound within the extracellular vestibule where it stabilized the active state and slowed agonist dissociation2. Previously, several groups have reported modulators binding within the intracellular surface of GPCRs3,4 and to the outer surface of transmembrane segments5. Cmpd-15, 4-((2S)-3-(((S)-3-(3-bromophenyl)-1-(methylamino)-1-oxopropan-2-yl)amino)-2-(2-cyclohexyl-2-phenylacetamido)-3-oxopropyl)benzamide, is a membrane-permeable negative allosteric modulator of β2AR that was isolated from a DNA-encoded small-molecule library1. Of interest, unlike conventional drug discovery methods, this screen using purified β2AR protein unoccupied by any ligand was unbiased, allowing us to identify molecules capable of binding to any surface of the receptor, not just the orthosteric pocket. Biochemical and pharmacological characterization showed that Cmpd-15 crosses the plasma membrane and binds to the intracellular surface of the β2AR. In an effort to understand the mechanism by which Cmpd-15 stabilizes the inactive state and inhibits coupling to G proteins and β-arrestins, we obtained a crystal structure of the β2AR bound to Cmpd-15.
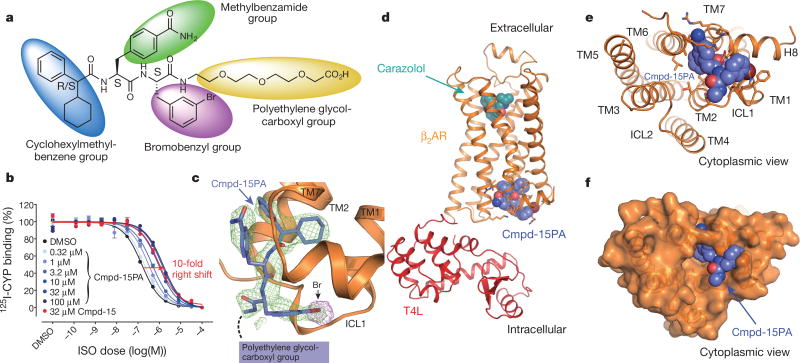
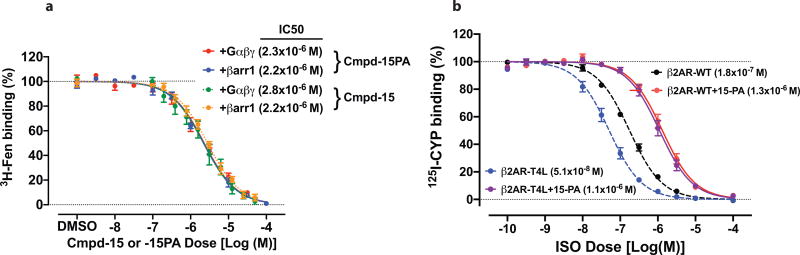
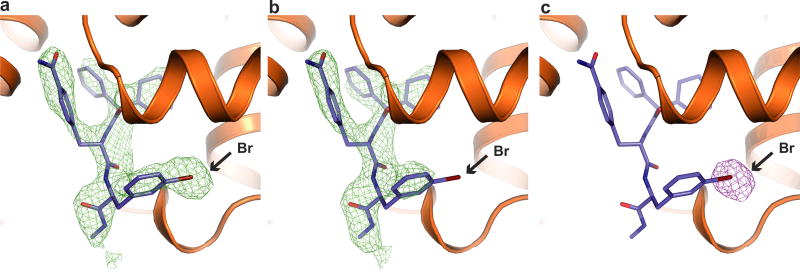
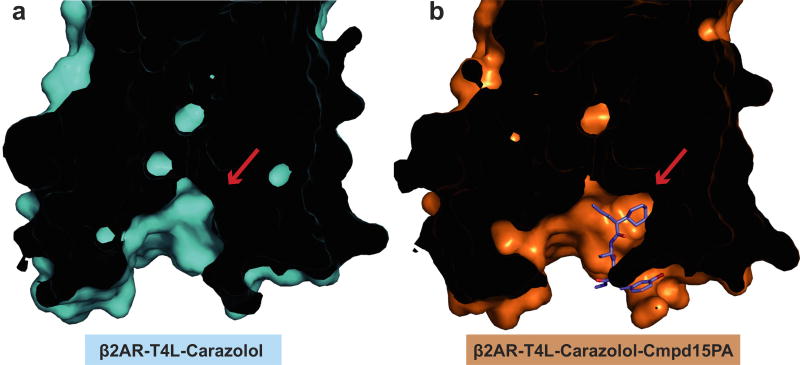
We used a previously described β2AR–T4 lysozyme fusion protein (β2AR–T4L)6 to obtain a crystal structure of the inactive state of the β2AR bound to the orthosteric antagonist carazolol and Cmpd-15. In initial trials, purified β2AR–T4L was crystallized in lipid cubic phase in the presence of 200 µM Cmpd-15 (limit of solubility), and a structure was determined by molecular replacement at 2.5 Å. The structure revealed weak positive electron density near the cytoplasmic ends of transmembrane (TM) segments 1, 2, 6 and 7, but it was not possible to unambiguously dock Cmpd-15 into the density. To improve the occupancy of Cmpd-15 in crystallized β2AR, a molecule of carboxylic acid functionalized polyethylene glycol was added to the position used for coupling of the molecule to its DNA tag (Fig. 1a). The modified Cmpd-15, Cmpd-15 polyethylene glycol-carboxylic acid derivative (denoted as Cmpd-15PA) exhibits comparable effects on orthosteric agonist binding affinity to the parent Cmpd-15 (Fig. 1b, Extended Data Fig. 1a). Furthermore, binding of Cmpd-15PA leads to a comparable decrease in orthosteric agonist binding affinity for both the β2AR–T4L and wild-type β2AR (Extended Data Fig. 1b). As expected, Cmpd-15PA was more soluble than Cmpd-15 and we were able to crystallize β2AR–T4L bound to the orthosteric antagonist carazolol in the presence of 1 mM Cmpd-15PA. The structure was solved by molecular replacement at 2.7 Å (Methods, Extended Data Table 1). We observed a well-defined Fo − Fc simulated annealing omit map for Cmpd-15PA (Fig. 1c, green map). The relatively weak electron density for the bromine substituent is most likely due to its susceptibility to radiation damage. This was confirmed by a ‘radiation damage map’. The diffraction data from each crystal was divided into two sets: data collected early, having the least radiation damage, and the remaining data, having the greatest radiation damage. These early and late data were separately merged, and an isomorphous Foearly − Folate difference electron density map revealed a strong peak around the bromine atom (Fig. 1c, purple map, Extended Data Fig. 2).
Figure 1. Crystal structure of β2AR–T4L in complex with carazolol and Cmpd-15PA in inactive conformation.
a, Chemical structure of compound 15 (Cmpd-15) modified with a carboxylic acid functionalized polyethylene glycol (with three monomeric units of ethylene oxide and a carboxylic group, PEG3-COOH), Cmpd-15PA. b, Dose–response curves of isoproterenol (ISO)-competition binding to the β2AR with 125I-CYP were obtained in the presence of various concentrations of Cmpd-15PA as indicated. The condition with Cmpd-15 at 32 µM was included for comparison. Values were expressed as percentages of the maximal 125I-CYP binding level obtained from a one-site competition binding-log IC50 curve fit of the vehicle (0.9% dimethylsulfoxide; DMSO) control data. Points on curves represent mean ± s.e.m. obtained from at least three independent experiments done in duplicate. Dose–response curve fits were obtained using the computer program GraphPad Prism. c, The Cmpd-15PA binding site revealed in an Fo − Fc simulated annealing omit map, contoured at 2.3σ (green). The Br atom is shown by the radiation damage map contoured at 4σ (magenta, see Extended Data Fig. 2). d, Overall structure of β2AR–T4L bound with orthosteric inverse agonist carazolol and intracellular negative allosteric modulator Cmpd-15P. e, f, Cytoplasmic views of Cmpd-15PA bound to β2AR at a pocket formed by TM1, TM2, TM6, TM7, ICL1 and helix 8. The binding site is shown as either cartoon (e) or as surface (f).
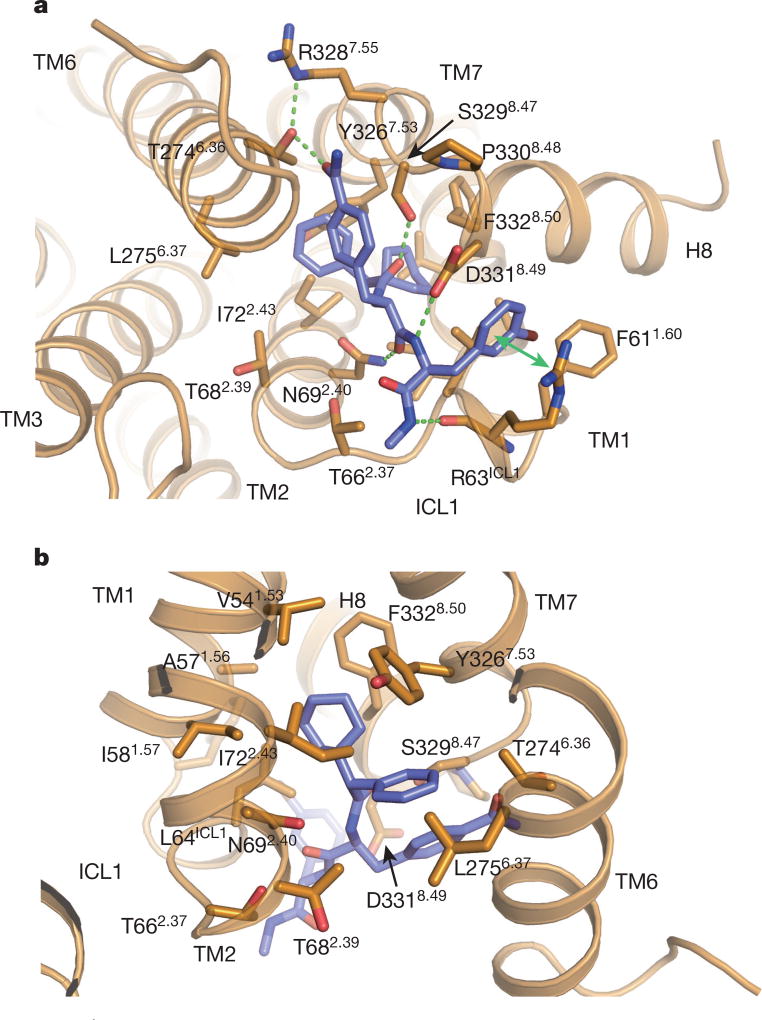
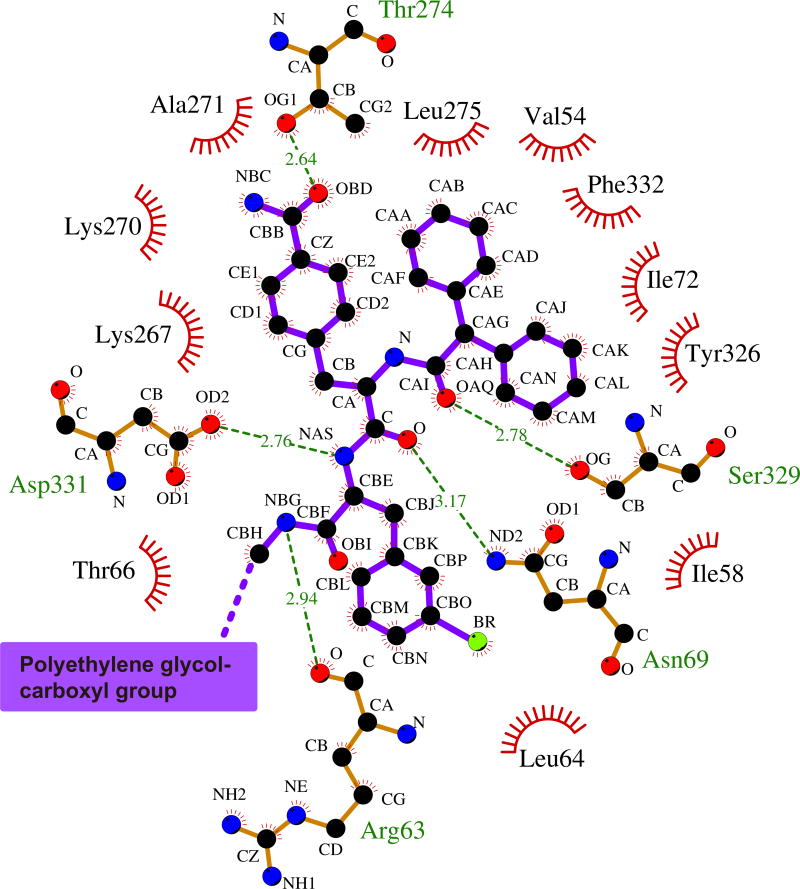
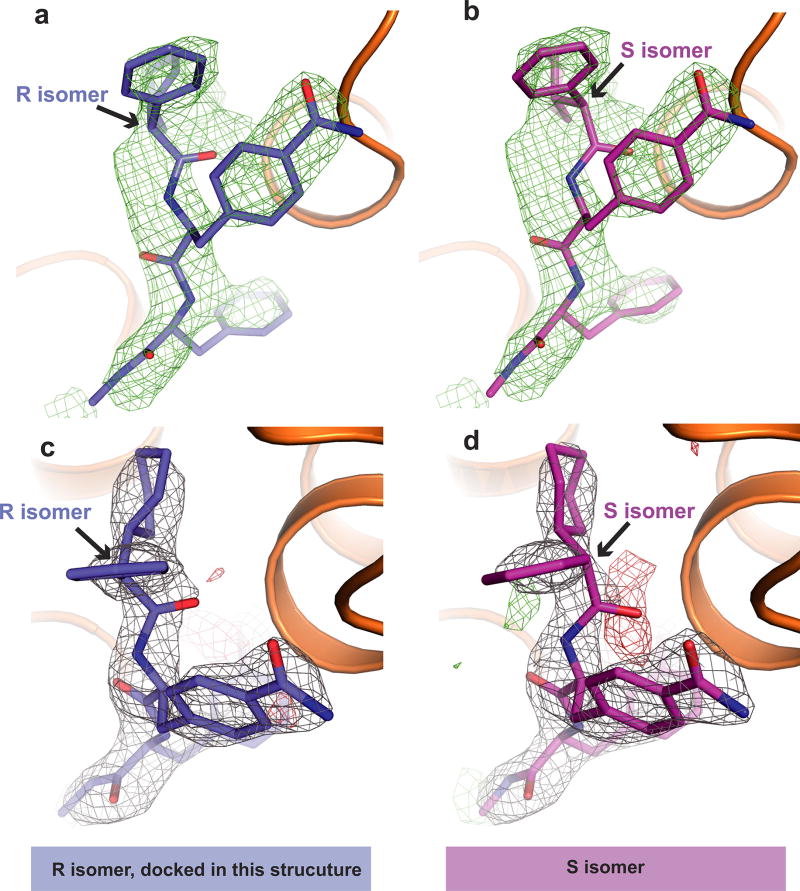
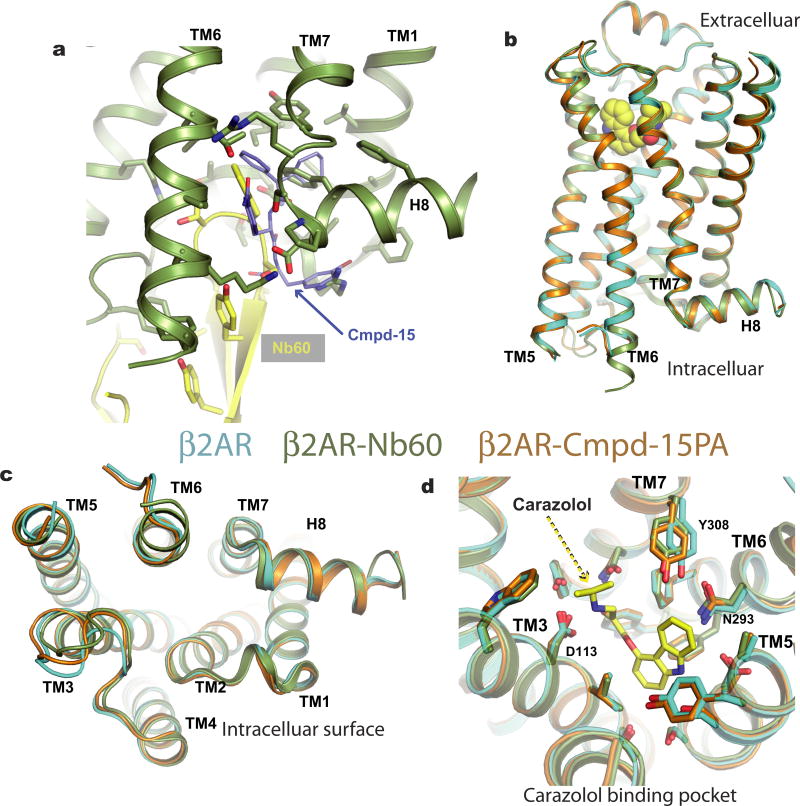
As suggested from previous studies1, Cmpd-15PA binds to the intracellular surface of the β2AR. It lies partially buried in a pocket formed by TMs 1, 2, 6, 7, helix 8 (H8) and intracellular loop 1 (ICL1) (Fig. 1d–f). There is no clear electron density for the polyethylene glycol-carboxylic acid moiety of Cmpd-15PA, suggesting that it is disordered in the crystal structure. Cmpd-15PA forms polar interactions with side chains of Thr2746.36, Ser3298.47, Asp3318.49 and Asn692.40, and with the backbone carbonyl of Arg63ICL1 (Fig. 2a, dashed lines, Extended Data Fig. 3). There is a cation-π interaction between the bromo-benzyl ring and Arg63ICL1 (Fig. 2a, solid green arrow). The cyclohexyl and phenyl rings of Cmpd-15PA are stabilized in a hydrophobic pocket formed by Val541.53 and Ile581.57 at the end of TM1, Leu64ICL1 in ICL1, Ile722.43 in TM2, Leu2756.37 in TM6, Tyr3267.53 from the conserved NPxxY sequence in TM7, and Phe3328.50 in H8 (Fig. 2b, Extended Data Fig. 3). It should be noted that Cmpd-15PA is a racemic mixture with respect to the chiral carbon connecting the cyclohexyl and phenyl rings (Fig. 1a). We docked both isomers and found that the R isomer fits better into the electron density (Extended Data Fig. 4); therefore, we used the R isomer for all figures and for the analysis of interactions between Cmpd-15PA and the β2AR.
Figure 2.
The Cmpd-15PA binding pocket. a, Polar interactions between Cmpd-15PA and β2AR are shown as green dashed lines and a cation-π interaction is shown with a green arrow. b, Hydrophobic interactions are formed between the cyclohexyl and phenyl rings of Cmpd-15PA and hydrophobic residues from β2AR.
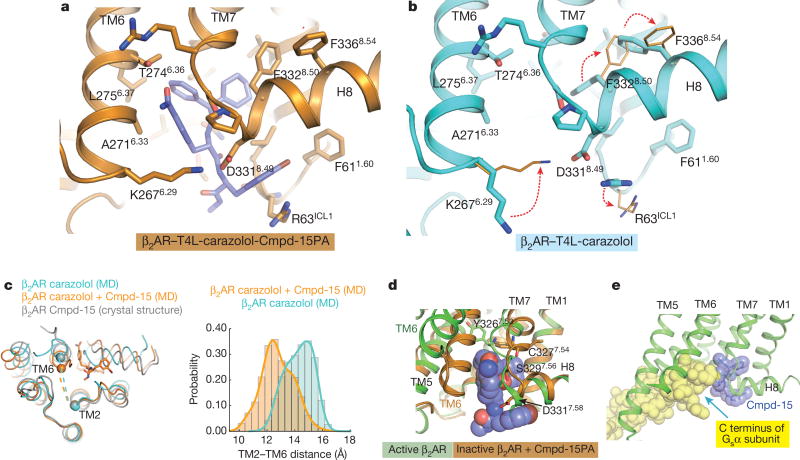
Cmpd-15PA stabilizes TM6 in the inactive state through direct polar interactions with side chain of Thr2746.36 and non-polar interactions with side chains of Ala2716.33 and Leu2756.37 (Fig. 3a). It also indirectly stabilizes interactions between TM6 and H8 through the formation of a new salt bridge. Figure 3 illustrates differences in the crystal structure of β2AR–T4L bound only to carazolol (Fig. 3b) and β2AR–T4L bound to both carazolol and Cmpd-15PA (Fig. 3a). The bromophenyl ring of Cmpd-15PA disrupts a salt bridge between Arg63ICL1 in ICL1 and Asp3318.49 in H8 enabling the formation of a new salt bridge between Lys2676.29 at the end of TM6 and Asp3318.49 in H8. This salt bridge also serves to trap Cmpd-15PA in its binding pocket. In molecular dynamics simulations, this salt bridge exhibited consistent stability in the presence of Cmpd-15. It was present in 86.4 ± 5.1% of simulation frames (mean and standard deviation reported for five independent simulations). Molecular dynamics simulations also suggest that TM6 is stabilized in a more inward conformation when bound to Cmpd-15 (Fig. 3c). The binding of Cmpd-15PA also necessitates concerted rearrangement of the side chains of Phe3328.50 and Phe3368.54, which changes the shape of the binding pocket (Fig. 3a, b and Extended Data Fig. 5). In addition to stabilizing the inactive conformation of TM6, the binding of Cmpd-15PA is incompatible with structural changes associated with activation at the junction of TM7 and H8, where an inward movement of Tyr3267.53, Cys3277.54, Ser3297.56 and Asp3317.58 would sterically clash with Cmpd-15 (Fig. 3d). On the basis of the β2AR–Gs crystal structure7, Cmpd-15 would also be expected to sterically interfere with Gs coupling (Fig. 3e). This will probably be true for β2AR–β-arrestin interactions on the basis of the structure of the rhodopsin–arrestin complex8.
Figure 3. Cmpd-15PA stabilized conformational changes and mechanism of allosteric antagonism.
a, b, Rearrangement of side chains of R63ICL1 from ICL1, K2676.29 from TM6, F3328.50 and F3368.54 from helix 8 are required to accommodate Cmpd-15PA (b, Protein Data Bank, 2RH1). c, Molecular dynamics simulations suggest that binding of Cmpd-15 stabilizes a more inward conformation of TM6. Crystal structures of carazolol-bound β2AR with and without Cmpd-15 bound—both of which are stabilized by the presence of a T4L fusion between TMs 5 and 6—exhibit nearly identical conformations of the entire backbone, including TM6. In simulations with T4L removed, the intracellular end of TM6 moves inward towards TM2 in simulations with Cmpd-15 bound, but not in simulations without Cmpd-15, as seen in simulation snapshots (left) and in histograms of the distance between the α carbons of Glu268 in TM6 and Val67 in TM2 (right). d, Binding of Cmpd-15 is incompatible with conformational changes in TM7 and H8 associated with activation of β2AR, where an inward movement of Tyr3267.53, Cys3277.54, Ser3297.56 and Asp3317.58 would sterically clash with Cmpd-15. Active β2AR structure is shown as green cartoon and inactive β2AR structure is shown as orange cartoon. e, Cmpd-15PA would sterically clash with the C-terminal α5 helix of the G protein Gs (Protein Data Bank, 3SN6).
When bound to Cmpd-15, the β2AR exhibits negative cooperativity for isoproterenol (Fig. 1b) and positive cooperativity for the inverse agonist ICI-118,551 (ref. 1). We previously reported the structure of the β2AR bound to an allosteric nanobody (Nb60) that stabilized the inactive state9. The Nb60 binding pocket partially overlaps with the Cmpd-15PA pocket (Extended Data Fig. 6a), but Nb60 also interacts with TM3 and ICL2. With the exception of side-chains in direct contact with Nb60 or Cmpd-15, the two structures are nearly identical (Extended Data Fig. 6b–d, r.m.s.d. = 0.43 for all Cα atoms). Extended Data Figure 6d shows a comparison of the orthosteric binding pocket of β2AR bound to the inverse agonist carazolol alone (Protein Data Bank, accession number 2RH1), or together with Cmpd-15. Although we observed subtle differences in the position of Y3087.35, this is more likely due to differences in the crystal lattice packing interactions involving extracellular loop 3, and not to allosteric effects of Cmpd-15. The effect of Cmpd-15 and Nb60 on agonist and inverse-agonist binding affinity is probably due to their ability to stabilize TM6 in an inactive conformation. Equilibrium binding affinity represents the average affinity of multiple conformations of the receptor that exchange on time scales that are much faster than the time required for the equilibrium binding study. Therefore, in this experimental setting, as both Cmpd-15 and Nb60 stabilize the inactive state, this results in a lower affinity for agonists and a higher affinity for inverse agonists. Nb60 has a greater effect on agonist binding affinity than Cmpd-15 (70-fold increase in Ki with Nb60 and tenfold increase with Cmpd-15). The greater effect of Nb60 is probably due at least in part to the higher affinity of its interaction with the β2AR (56.8 nM for Nb60 (ref. 9) and 1.7 µM for Cmpd-15 (ref. 1)).
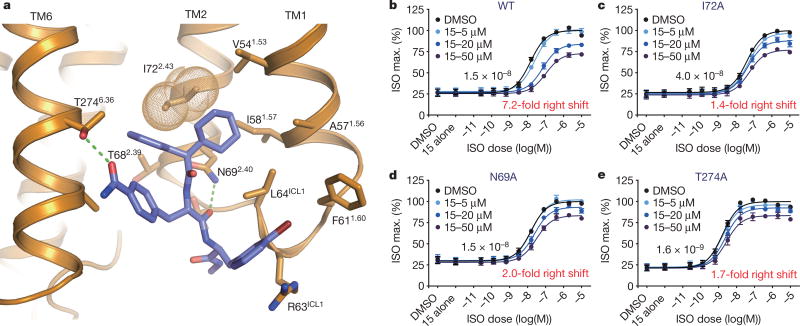
To validate the binding pocket of Cmpd-15, we examined the effect of mutations of amino acid side chains that interact with different regions of the ligand. As can be seen in Fig. 4, mutation of Thr2746.36 to Ala disrupts polar interactions with the formamide functionality of the methylbenzamide group and mutation of Asn692.40 to Ala disrupts polar interactions with a backbone carbonyl. Mutation of Ile722.43 to Ala results in the loss of non-polar van der Waals interactions with the cyclohexylmethyl-benzene group. All three mutations result in a diminution of inhibitory activity of Cmpd-15, both the maximal effect on arrestin recruitment and the shift in half maximal effective concentration (EC50) for isoproterenol (Fig. 4b–e). It is interesting to note that mutations of amino acids that form the Cmpd-15 binding pocket have a greater effect on the fold change in EC50 for isoproterenol than on the maximal inhibition of arrestin recruitment. These results suggest that the ability of Cmpd-15 to sterically disrupt interactions between the β2AR and β-arrestin can be separated from the effects of Cmpd-15 on stabilization of TM6 in an inactive conformation with associated allosteric effects on agonist affinity.
Figure 4. Mutagenesis of Cmpd-15PA binding site.
a, Gln692.40 and Thr2746.36 form polar interactions with Cmpd-15PA (green dashed line) and Ile722.43 (orange dots) forms van der Waals contacts with Cmpd-15PA. b–e, U2OS cells transiently expressing the wild-type or indicated mutant β2AR were pretreated with Cmpd-15 at the indicated concentrations for 20 min. The extent of β-arrestin recruitment to the transiently expressed receptor was measured following stimulation with isoproterenol (ISO) in a dose-dependent manner. Values were expressed as percentages of the maximal level of the isoproterenol-induced activity in the vehicle (0.5% DMSO) control and represent mean ± s.e.m. obtained from four independent experiments done in duplicate. Dose–response curve fits were obtained using the computer program GraphPad Prism.
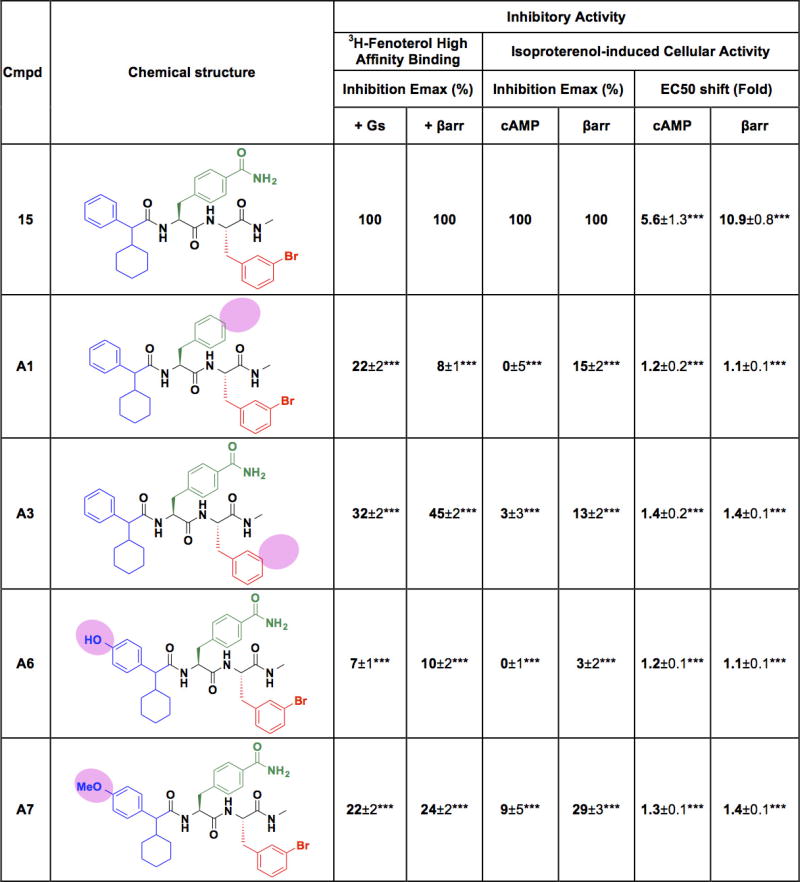
The functional effects of different chemical modifications to Cmpd-15 are shown in Extended Data Fig. 7. The crystal structure helps to explain the observed activity of different analogues on orthosteric agonist binding affinity, and cell-based assays for cAMP production and β-arrestin recruitment. The greatest loss of activity was observed for compounds A1, A6 and A7. Compound A1 lacks the formamide functionality on the methylbenzamide group that forms a hydrogen bond with Thr2746.36 in TM6 (Fig. 4a), whereas either hydroxyl or methoxy substitution at the para-position on the phenyl ring of cyclohexylmethylbenzene group (compounds A6 and A7) would sterically clash with TM6 (Fig. 2b). Loss of the bromine at the meta-position of the bromo-benzyl group (compound A4) also had significant effects in cell-based signalling assays, which may be due to loss of van der Waals interactions of the Br with the side chains of Phe611.60 and Leu64ICL1 in ICL1 (Fig. 4a).
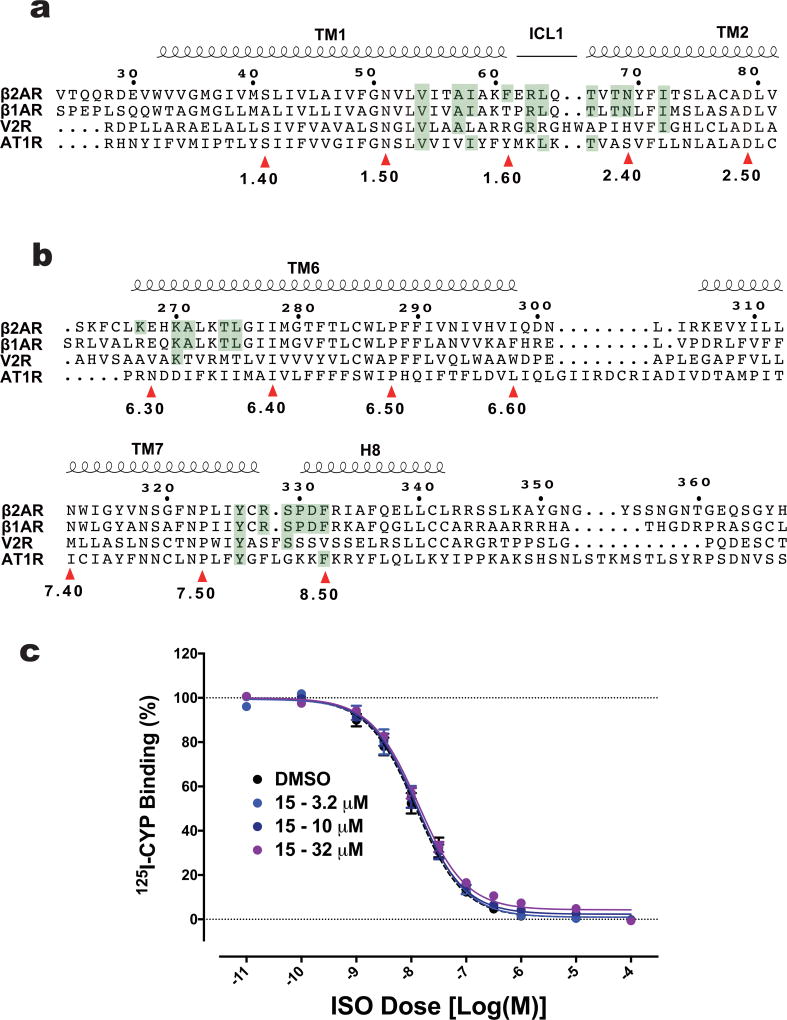
Allosteric ligands may exhibit greater subtype selectivity than orthosteric compounds. Cmpd-15 is more efficacious at inhibiting β2AR activities than β1AR1, although all of the 21 amino acids that form the Cmpd-15 binding pocket are conserved except for Phe61 at the cytoplasmic end of TM1 and Lys2676.29 in TM6, which are Thr and Arg in the homologous position of the β1AR (Extended Data Fig. 8a, b). As noted above, Phe61 may have aromatic interactions with the bromo-benzyl group. Notably, unlike its pronounced effect on agonist binding to β2AR, Cmpd-15 has no effect on orthosteric agonist binding affinity to the β1AR (Extended Data Fig. 8c). This result, together with the previously reported cellular activity data1 and mutagenesis data discussed above, further supports the dual mechanisms by which Cmpd-15 exerts its negative effects on receptor activity. Cmpd-15 specifically reduces the binding affinity of the β2AR orthosteric agonists by stabilizing its inactive conformation (allosteric effect), but it may sterically interfere with transducer coupling to a broader range of receptors such as the β1AR (Fig. 3e). Cmpd-15 was found to have much weaker activity at the Gs-coupled vasopressin receptor type 2, where only 7 of the 21 binding-pocket residues are identical, and almost no activity at the Gq-coupled angiotensin type 1 receptor where only 6 of the 21 residues are identical1 (Extended Data Fig. 8a, b).
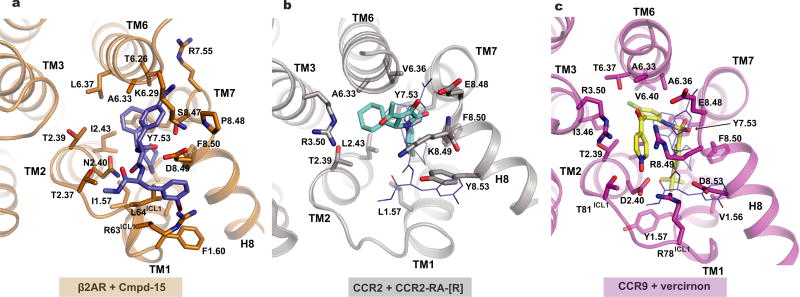
Recently, crystal structures of two chemokine receptors with negative allosteric modulators bound to the intracellular surface were reported. CC chemokine receptor 2 (CCR2) was crystallized with T4 lysozyme inserted between TMs 5 and 6 and bound to the orthosteric antagonist BMS-681 and an allosteric antagonist (CCR2-RA-(R))3. CCR2-RA-(R) binds cooperatively with BMS-681, and the two ligands have an additive effect on the thermostability of CCR2. As shown in Extended Data Fig. 9a, b, the binding pocket for CCR2-RA-(R) is similar to that for Cmpd-15PA involving the cytoplasmic ends of TMs 1, 2, 6 and 7, but the ligand also makes contact with R3.50 in TM3. CCR9 with eight thermostabilizing mutations was crystallized bound to the antagonist vercirnon4. Vercirnon was thought to be an orthosteric antagonist, and it was therefore surprising that they found that it bound to an intracellular pocket with no ligand occupying the orthosteric pocket. No information was provided regarding the effect of vercirnon on the binding of orthosteric ligands. Like CCR2-RA-(R), vercirnon engages more TM segments than Cmpd-15PA and binds to a pocket formed by the intracellular ends of TMs 1, 2, 3, 6 and 7 (Extended Data Fig. 9c). It is interesting to note that the location of this binding site is very similar for all three receptors (a pocket formed by the cytoplasmic ends of TMs 1, 2, 6, 7), yet the structure of the allosteric ligands and the chemical interactions between the receptors and ligands are different. These observations suggest that this site may represent a druggable target for other GPCRs.
The structure of the β2AR bound to Cmpd-15PA in comparison with inactive- and active-state structures of the β2AR provides detailed mechanistic insights into its ability to modulate both orthosteric ligand binding and signalling. Like Nb60, Cmpd-15 sterically prevents coupling to Gs, and probably blocks interactions with GPCR kinases and arrestins as well. The effect of Cmpd-15 on agonist binding affinity is probably the result of stabilizing the inactive conformation of TM6. We previously showed that the G protein Gs and positive allosteric nanobodies for the β2AR7,10,11, M2 muscarinic receptor2 and the μ-opioid receptor12 all stabilize the outward displacement of the cytoplasmic end of TM6, and also stabilize a high-affinity state for agonists. Cmpd-15, like Nb60, stabilizes the inactive conformation of TM6 associated with reduced affinity for agonists1. These results, together with the recent structures of CCR2 and CCR9 bound to intracellular modulators, suggest that the location of this intracellular allosteric pocket may be widely conserved amongst diverse GPCRs and that there may be potential therapeutic value in targeting this surface for drug discovery.
Methods
Data reporting
No statistical methods were used to predetermine sample size. The experiments were not randomized and the investigators were not blinded to allocation during experiments and outcome assessment.
Synthesis of Cmpd-15PA
The detailed methods for the synthesis of Cmpd-15PA are provided in the Supplementary Information.
Protein expression and purification
The same β2AR–T4L construct that yielded the first high-resolution β2AR structure was used in this study6. Protein expression and purification was performed as previously described13 with minor modifications. In brief, the receptor was expressed in Sf9 cells (obtained from Expressions Systems, LLC). Cell membranes were solubilized with solubilization buffer (20 mM HEPES, pH 7.5, 100 mM NaCl, 1% DDM). 30 µM atenolol (Sigma) was added during solubilization to stabilize the receptor. Solubilized receptors were isolate on M1-Flag affinity resin (Sigma). Atenolol was then washed away and target protein was eluted by HMS-CHS buffer (20 mM HEPES, pH 7.5, 350 mM NaCl, 0.1% DDM, 0.02% CHS) plus 0.2 mg ml−1 Flag peptide and 5 mM EDTA. The M1-Flag-purified β2AR–T4L was treated with TCEP/iodoacetamide and applied to alprenolol-Sepharose affinity resin13 to isolate functional protein. The eluted alprenolol bound receptor was then loaded onto a second M1-Flag column, on which the detergent was changed from 0.1% DDM to 0.01% MNG and ligand was changed from alprenolol to 20 µM carazolol (Sigma) and 50 µM Cmpd-15PA. The eluted β2AR–T4L was dialysed against dialysis buffer (20 mM HEPES, pH 7.5, 100 mM NaCl, 0.003% MNG, 0.0003% CHS, 10 µM carazolol, 10 µM Cmpd-15PA) overnight at 4 °C to remove excess EDTA and Flag peptide. PNGase F (New England Biolabs) was added to the protein during dialysis to remove N-linked sugars and purified β2AR–T4L was concentrated to ~50 mg ml−1 with a 50 kDa cutoff Amicon centrifugal filters (Millipore).
Crystallization
Concentrated protein was mixed with monoolein (Sigma) containing 10% (w/w) cholesterol (Sigma) to form lipidic cubic phase (LCP) using the previously reported two-syringe method14. The protein solution to lipid ratio was 2:3 (w/w). Once clear LCP was formed, crystal trials were set up by filling 96-well glass sandwich plates with 30 nl of LCP overlaid with 1 µl precipitant solution using an LCP crystallization robot (Gryphon, Art Robbins Instruments). The best diffracting crystals were grown in 0.1 M Tris-HCl, pH 7.5, 30–37.5% PEG400, 400 mM NH4F, 6% 1,4-butanediol, 1 mM Cmpd-15PA and 1% DMSO. Crystals appeared after 1 day and grew to full size within 1 week.
Data collection and structure determination
Crystals were collected in 50 µm MicroMeso loops (MiTeGen) and flash-frozen in liquid nitrogen. Diffraction data were collected at beamline BL32XU at SPring-8, Japan. Typically, 10 to 20 degrees of data were collected for each crystal. Diffraction data were processed with XDS15. Analysis with UCLA Diffraction Anisotropy Server16 suggested that the diffraction data were anisotropic. An anisotropy-truncated dataset was generated with resolution limits set to 3.1 Å, 2.7 Å and 2.8 Å along the a*, b* and c* axis, respectively, and we report this structure at a maximum resolution of 2.7 Å. The CC1/2 of 0.746 indicates that the high-resolution shell contains useful information, even though the completeness is low owing to anisotropic truncation (Extended Data Table 1). The structure was solved by molecular replacement using phenix.automr17, with the previously reported β2AR–T4L structure (PDB, 2RH1) as a search model. The allosteric ligand and residues that undergo large conformational changes were manually fit into Fo − Fc electron density maps in Coot18. Structure refinement was performed with phenix.refine17, with individual B factors refined for protein and ligands. The occupancy of Cmpd-15PA was refined as a single group, with a final value of 82%. The final model was validated using Molprobity19. Data collection statistics and structure refinement statistics are summarized in Extended Data Table 1. All structure figures were prepared using PyMOL (The PyMOL Molecular Graphics System, Schrödinger, LLC.).
The position of the ligand was found by isomorphous difference Fourier analysis. Diffraction data from crystals without Cmpd-15PA were scaled to the Cmpd-15PA data, and an Fo − Fo map between these two datasets was computed using FFT20 in CCP421. A similar strategy was used to calculate the radiation damage map: diffraction data from the first 5 degrees and the second 5 degrees of each crystal were merged into two different datasets. After scaling these two datasets together, an Fo − Fo difference map was calculated by subtracting the second half of data from the first half of data. The model was confirmed using an Fo − Fc simulated annealing omit map calculated with Phenix17, using a starting temperature of 3,000 K and a starting model in which Cmpd-15PA was omitted. Contact surface between ligand and receptor was calculated with PISA22 in CCP421.
Radio-ligand binding
Assays were performed as previously described1. Competition binding was performed with approximately 0.7 ng of βARs in HDL particles diluted in an assay buffer (20 mM HEPES, pH7.4, 100 mM NaCl, 0.1% BSA and 1 mM ascorbic acid). Receptors reconstituted in HDL particles were prepared as previously described23. βARs in HDL particles were mixed with Cmpd-15PA or Cmpd-15 at different concentrations and a quarter or half log serial dilution of isoproterenol with the constant concentration of 125I-cyanopindolol (CYP) (2,200 Ci mmol−1; PerkinElmer) at 60 pM. Propranolol at 20 µM was used to determine non-specific binding. Following 90-min incubation at room temperature, binding assays were terminated by rapid filtration onto GF/B glass-fibre filters (Brandel) treated with 0.3% polyethyleneimine (PEI). The filters were then washed with 8 ml of a cold binding buffer (20 mM HEPES, pH7.4, 100 mM NaCl) using a harvester (Brandel). Bound [125I] was quantified using a Packard Cobra Quantum gamma counter (Packard) and expressed as specific binding.
3H-methoxyfenoterol (3H-Fen) binding was carried out with isolated membranes from Sf9 cells exogenously expressing the phosphorylated forms of the β2V2R. Membranes with phosphorylated forms of the receptor were prepared essentially following the procedures described previously24. Approximately 40 µg of the isolated membranes in binding buffer (50 mM Tris, pH 7.4, 2 mM EDTA, 12.5 mM MgCl2, 0.05% BSA and 1 mM ascorbic acid) were mixed either with a serial dilution of Cmpd-15 or Cmpd-15PA, or with a single dose of Cmpd-15 analogues at 32 µM. Then 4.3 nM 3H-Fen and transducers (100 nM heterotrimeric Gs or 250 nM β-arrestin1 together with 0.5 µM Fab30) were added to the mixture. Following incubation for 90 min at room temperature, binding assays were terminated by collecting the reaction mixture on to PEI-soaked GF/B filters. Bound 3H was extracted over night with 5 ml scintillation fluid and quantified using a Tri-Carb 2800TR liquid scintillation counter (PerkinElmer).
Measurements of β-arrestin recruitment
The extent of β-arrestin recruitment to the β2AR was monitored using PathHunter, a chemiluminescence-based enzyme fragment complementation assay (DiscoveRx) as previously described1. For the mutagenesis study, U2OS cells stably expressing enzyme acceptor-tagged β-arrestin2 (DiscoveRx) were transiently transfected with ProLink-tagged β2AR mutant constructs using FuGENE 6 (Promega). These mutants were created on the wild-type ProLink-tagged β2AR provided by DiscoveRx using a quick-change site-directed mutagenesis kit (Agilent). On the following day after transfection, cells were plated in 96-well white, clear-bottomed plates at a density of 25,000 cells per well. On another following day, cells were pre-treated with 0.5% dimethylsulfoxide (DMSO) or Cmpd-15 at different concentrations in Hank’s balanced solution (Sigma), supplemented with 20 mM HEPES, pH 7.4 and 0.05% BSA and further incubated for 20 min. Subsequently, cells were stimulated with a serial dilution of isoproterenol for 1 h at 37 °C, which was terminated by adding PathHunter detection reagents (DiscoveRx). After further incubation for 1 h at 27 °C, luminescence signals were read using a NOVOstar microplate reader (BMG Labtech). For the study with Cmpd-15 analogues, U2OS cells stably expressing enzyme acceptor-tagged β-arrestin 2 and ProLink-tagged β2V2R were used.
Measurements of cAMP production
The level of cAMP was monitored using Glosensor, a chemiluminescence-based cAMP biosensor (Promega) as previously described1. HEK-293 cells (ATCC) stably expressing Glosensor were plated in 96-well white, clear-bottomed plates at a density of 80,000 cells per well. On the following day, Glosensor reagents (Promega) were added, and cells were incubated in a 27 °C humidifying incubator for ~1 h. Cells were then further incubated with either the vehicle (DMSO) control or each of the Cmpd-15 analogues at 50 µM in Hank’s balanced solution (Sigma), supplemented with 20 mM HEPES, pH 7.4 and 0.05% BSA, together with 100 µM 3-isobutyl-1-methylxanthine (IBMX, Sigma) for 20 min. At the end of incubation, cells were stimulated with a serial dilution of isoproterenol for 5 min at room temperature. Changes in luminescence were read using a NOVOstar microplate reader (BMG Labtech).
Molecular dynamics simulations setup
Molecular dynamics simulations of β2AR were initiated from the carazolol- and Cmpd-15PA-bound crystal structure described in this manuscript. The receptor was simulated in two distinct conditions: (1) β2AR bound to carazolol and Cmpd-15 and (2) β2AR bound to carazolol only.
The structures were prepared by first removing the crystallized T4 lysozyme fragment and non-receptor molecules except for the ligands, water, and cholesterol. Prime (Schrödinger, Inc.) was used to model in missing side-chains. Hydrogen atoms were added, and protein chain termini were capped with the neutral groups acetyl and methylamide. In the simulations, titratable residues were left in their dominant protonation state at pH 7.0. Previous studies have suggested that residues D792.50 and D1303.49 of β2AR may be deprotonated in the inactive state and protonated in the active state25,26. As the crystal structures represent the inactive state, D792.50 and D1303.49 were deprotonated in our simulations. Histidines were represented with hydrogen on the epsilon nitrogen.
The carazolol tertiary amine nitrogen was protonated, corresponding to the dominant protonation state at pH 7.0 and enabling formation of the conserved salt bridge with neighbouring D1133.32. The prepared protein structures were aligned on the transmembrane helices to the orientation of proteins in membranes (OPM)27 structure of PDB 2RH1. The structures were then inserted into a pre-equilibrated palmitoyl-oleoyl-phosphatidylcholine (POPC) bilayer. Final system dimensions were approximately 75 × 75 × 95 Å3, including approximately 141 lipids, 10,840 water molecules, 23 sodium ions, 29 chloride ions, and 2 cholesterol molecules.
Molecular dynamics simulation force field parameters
We used the CHARMM36 parameter set for protein molecules, lipid molecules, and salt ions, and the CHARMM TIP3P model for water; protein parameters incorporated CMAP terms28–32. Parameters for carazolol and Cmpd-15 were generated using the CHARMM General Force Field (CGenFF)33–35 with the ParamChem server (https://www.paramchem.org/), version 1.0.0. Full parameter sets are available upon request.
Molecular dynamics simulation protocol
Simulations were performed on GPUs using the CUDA version of PMEMD (Particle Mesh Ewald Molecular Dynamics) in Amber1636,37. Prepared systems were minimized, then equilibrated as follows: the system was heated using the Langevin thermostat from 0 to 100 K in the NVT ensemble over 12.5 ps with harmonic restraints of 10.0 kcal mol−1 Å−2 on the non-hydrogen atoms of lipid, protein, and ligand, and initial velocities sampled from the Boltzman distribution. The system was then heated to 310 K over 125 ps in the NPT ensemble with semi-isotropic pressure coupling and a pressure of one bar. Further equilibration was performed at 310 K with harmonic restraints on the protein and ligand starting at 5.0 kcal mol−1 Å−2 and reduced by 1.0 kcal mol−1 Å−2 stepwise every 2 ns for 10 ns and then by 0.1 kcal mol−1 Å−2 stepwise every 2 ns for 20 ns, for a total of 30 ns of additional restrained equilibration. Equilibration was followed by production simulations lasting between 4.0 and 5.0 µs each.
Five independent simulations were performed in the presence of Cmpd-15, for a total of 23.1 µs. Three independent simulations were performed in the absence of Cmpd-15, for a total of 15.1 µs. These simulations were conducted in the NPT ensemble at 310 K and 1 bar, using a Langevin thermostat and Monte Carlo barostat.
Simulations used periodic boundary conditions, and a time step of 4.0 fs, with hydrogen mass repartitioning38. Bond lengths to hydrogen atoms were constrained using SHAKE. Non-bonded interactions were cut off at 9.0 Å, and long-range electrostatic interactions were computed using the particle mesh Ewald (PME) method with an Ewald coefficient β of approximately 0.31 Å and B-spline interpolation of order 4. The FFT grid size was chosen such that the width of a grid cell was approximately 1 Å.
Analysis protocols for molecular dynamics simulations
Trajectory snapshots were saved every 200 ps during production simulations. Trajectory analysis and visualization were performed using VMD39. For all analysis, we removed frames in the first 1.0 µs of each simulation (to allow receptor conformation to equilibrate). In the presence of compound 15, the mean TM2–TM6 distances for each independent simulation were 13.18 Å, 11.85 Å, 12.75 Å, 13.55 Å and 12.69 Å. In the absence of compound 15, the mean TM2–TM6 distances for each simulation were 14.75 Å, 14.48 Å and 13.55 Å.
Data availability
Atomic coordinates and structure factors have been deposited in the Protein Data Bank (PDB) under accession code 5X7D.
Extended Data
Extended Data Figure 1. Effects of Cmpd-15PA on agonist binding to the β2AR.
a, The level of 3H-Fenoterol (Fen) high-affinity binding to the β2AR was measured after pretreatment with the vehicle control (0.5% DMSO), Cmpd-15 or Cmpd-15PA at various concentrations as indicated in the presence of transducers, either trimeric Gαβγ protein or β-arrestin 1 (β-arr1) together with Fab30. Values were expressed as percentages of the maximal 3H-Fen binding level promoted by each transducer in the vehicle control (0.5% DMSO) and represent mean ± s.e.m. obtained from four independent experiments done in duplicate. b, Isoproterenol (ISO)-125I-CYP competition binding curves were obtained using wild-type (WT) β2AR or β2AR–T4L (T4L) in the absence (vehicle alone) or the presence of Cmpd-15PA at 32 µM. Values are expressed as percentages of the maximal 125I-CYP binding level obtained from a one-site competition binding-log IC50 curve fit and represent mean ± s.e.m. obtained from four independent experiments done in duplicate.
Extended Data Figure 2. Bromine atom of Cmpd-15PA was sensitive to radiation damage.
a–c, Clear differences were observed around the bromine atom of Cmpd-15PA between simulated annealing omit maps of the first half of diffraction data (a, green density, 2.3σ) and maps of the second half of diffraction data (b, green density, 2.3σ), resulting in a strong difference electron density centred around the bromine atom (c, purple density, 4σ).
Extended Data Figure 3. Two-dimensional schematic depiction of the Cmpd-15PA binding pocket.
Carbon atoms are coloured in black, oxygen atoms in red, nitrogen atoms in blue, bromine atom in green. Hydrogen bonds are represented as green dashed lines with distances labelled. The figure is generated using LIGPLOT40.
Extended Data Figure 4. Crystallographic evidence support modelling of R isomer of Cmpd-15PA.
a, b, The R isomer fits better into the simulated annealing omit map (green density map, 2.3σ) than S isomer (b). c, When refined with R isomer, the Cmpd-15PA model fits well into the 2Fo − Fc density (grey map, 1.5σ) with only very weak negative Fo − Fc density (red map, 3.0σ). d, When refined with S isomer, the Cmpd-15PA model does not fit as well to the 2Fo − Fc density (grey map, 1.5σ). The negative density Fo − Fc map (red map, 3.0σ) and positive density Fo − Fc map (green map, 3.0σ) suggest that the S isomer is not supported by crystallographic data.
Extended Data Figure 5. Comparison of modulator binding pocket before and after Cmpd-15PA binding.
a, b, The binding pocket is occluded before Cmpd-15PA binding (a), which would be an obstacle for in silico docking because the shape of the pocket is markedly different after Cmpd-15 binding (b).
Extended Data Figure 6. Comparison of the structures of the β2AR (cyan, PDB, 2RH1), β2AR–Nb60 (green, PDB, 5JQH) and β2AR–Cmpd-15PA (orange).
a, Cmpd-15PA binding pocket overlaps with Nb60 binding pocket. b–d, Different views of superimposed structures of the β2AR, the β2AR–Nb60 and the β2AR–Cmpd-15PA revealing very little structural difference associated with binding of Nb60 or Cmpd-15PA.
Extended Data Figure 7. Relative activities of Cmpd-15 analogues in β2AR-mediated responses.
For each analogue, only the modified region relative to the parent Cmpd-15 is indicated. Values for ‘inhibition Emax (%)’ are expressed as percentages of analogue-induced inhibition relative the Cmpd-15-induced level. Values for ‘EC50 shift (fold)’ are expressed as rightward fold-shifts compared to the EC50 value obtained in the vehicle (DMSO)-treated control curve. All values represent mean ± s.e.m. obtained from at least four independent experiments done in duplicate. Statistical analyses were performed using ‘one-way ANOVA’ with ‘Dunnetts’ post-tests for comparison to the control. ***P < 0.001. cAMP, G protein-mediated cAMP accumulation; βarr, β-arrestin recruitment to the β2AR.
Extended Data Figure 8. Alignment of residues that form the Cmpd-15PA binding pocket in β2AR with those from β1AR, V2R and AT1R.
There are 21 residues from β2AR that form the Cmpd-15 binding pocket (highlighted green). The identical residues from β1AR, V2R and AT1R are also highlighted in green. 10 out of the 21 residues are located at TM1, ICL1 and TM2 (a), while 11 of the 21 residues are located at TM6, TM7 and helix 8 (b). The top numbering refers to protein sequences in β2AR. The bottom numbering refers to Ballesteros–Weinstein numbering. c, Cmpd-15 has no effect on orthosteric agonist binding to the β1AR. Dose–response curves of isoproterenol (ISO)-competition binding to the β1AR with 125I-CYP were obtained in the presence of various concentrations of Cmpd-15 as indicated. Values were expressed as percentages of the maximal 125I-CYP binding level obtained from a one-site competition binding-log IC50 curve fit and represent mean ± s.e.m. obtained from five independent experiments done in duplicate.
Extended Data Figure 9. Comparison of Cmpd-15PA pocket with intracellular allosteric antagonist pockets of CCR2 and CCR9.
a, Cmpd-15PA pocket is formed by residues from TM1, TM2, TM6, TM7, ICL1 and helix 8 in β2AR. b, c, The binding pocket of CCR2-RA-(R) in CCR2 (b) and the pocket of vercirnon in CCR9 (c) are similar to Cmpd-15PA (blue lines) pocket but involve more interactions with TM3. (Protein Data Bank accession numbers, 5T1A for CCR2/CCE2-RA-(R) and 5LWE for CCR9/vercirnon.)
Extended Data Table 1.
Crystallography data collection and refinement statistics
| Data Collection‡ | |
| Number of crystals | 43 |
| Space group | P212121 |
| Cell dimensions | |
| a,b,c (Å) | 40.5, 75.7, 173.4 |
| α,β,γ (°) | 90, 90, 90 |
| Number of reflections | 158506 |
| Number of unique reflections | 12966 |
| Resolution (Å) | 50 – 2.7 (2.77 – 2.7) |
| Rmerge | 0.179 (0.762) |
| CC1/2 (%) | 99.4 (74.6) |
| <I/σI> | 9.10 (1.84) |
| Completeness (%) | 85.0 (13.8) |
| Redundancy | 12.2 (3.8) |
|
| |
| Refinement | |
| Resolution (Å) | 20 – 2.7 |
| Number of reflections (test set) | 11632 (1291) |
| Rwork/Rfree | 0.221/0.268 |
| Number of atoms | |
| Protein | 3523 |
| Carazolol | 22 |
| Cmpd-15 | 43 |
| Others (Lipids, ions, waters) | 83 |
| Average B factors (Å2) | |
| Receptor | 66.1 |
| T4 Lysozyme | 75.9 |
| Carazolol | 61.9 |
| Cmpd-15 | 62.3 |
| Others (Lipids, ions, waters) | 81.2 |
| RMS deviation from ideality | |
| Bond length (Å) | 0.002 |
| Bond angles (°) | 0.579 |
| Ramachandran statistics§ | |
| Favored regions (%) | 98.85 |
| Allowed regions (%) | 1.15 |
| Outliers (%) | 0 |
Highest resolution shell statistics are shown in parentheses.
As defined by MolProbity18.
Supplementary Material
Acknowledgments
We thank K. Hirata at Beamline BL32XU of Spring-8 for assistance in data collection. A. Wall and T. Xu provided technical assistance. NuEvolution for constructive discussions in the course of the work. We acknowledge support from the National Institute of Health grants NS028471 and GM106990 (B.K.K.), HL16037 (R.J.L.) and T32HL007101 (A.W.K. and A.M.), Amgen-China Postdoc fellowship (X.L.) and the Mathers Foundation (B.K.K. and W.I.W.). R.J.L. is an investigator with the Howard Hughes Medical Institute.
Footnotes
Supplementary Information is available in the online version of the paper.
Author Contributions X.L. expressed and purified β2AR–T4L, performed crystallization, data collection, data processing, structure determination and refinement. S.A. designed and performed in vitro radio ligand binding and mutagenesis studies. A.W.K. designed the chemical synthetic route for Cmpd-15PA. B.P. designed and performed the in vitro radio ligand binding experiments. A.M. assisted in the design of mutagenesis studies and the development of Cmpd-15PA. K.-C.M. synthesized Cmpd-15PA and analysed the spectral data. X.C. designed the chemical structure and synthetic route for Cmpd-15PA, analysed the spectral data and assisted in manuscript preparation. N.R.L. performed and analysed molecular dynamics simulations, with assistance from A.J.V. R.O.D. oversaw molecular dynamics simulations and analysis. W.I.W. oversaw data processing, structure determination and refinement. The manuscript was written by B.K.K. with assistance from X.L. R.J.L. and B.K.K. coordinated the experiments and supervised the overall research. All authors contributed to the editing of the manuscript.
The authors declare no competing financial interests.
References
- 1.Ahn S, et al. Allosteric ‘beta-blocker’ isolated from a DNA-encoded small molecule library. Proc. Natl Acad. Sci. 2017;114:1708–1713. doi: 10.1073/pnas.1620645114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kruse AC, et al. Activation and allosteric modulation of a muscarinic acetylcholine receptor. Nature. 2013;504:101–106. doi: 10.1038/nature12735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zheng Y, et al. Structure of CC chemokine receptor 2 with orthosteric and allosteric antagonists. Nature. 2016;540:458–461. doi: 10.1038/nature20605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Oswald C, et al. Intracellular allosteric antagonism of the CCR9 receptor. Nature. 2016;540:462–465. doi: 10.1038/nature20606. [DOI] [PubMed] [Google Scholar]
- 5.Jazayeri A, et al. Extra-helical binding site of a glucagon receptor antagonist. Nature. 2016;533:274–277. doi: 10.1038/nature17414. [DOI] [PubMed] [Google Scholar]
- 6.Rosenbaum DM, et al. GPCR engineering yields high-resolution structural insights into β2-adrenergic receptor function. Science. 2007;318:1266–1273. doi: 10.1126/science.1150609. [DOI] [PubMed] [Google Scholar]
- 7.Rasmussen SG, et al. Crystal structure of the β2 adrenergic receptor–Gs protein complex. Nature. 2011;477:549–555. doi: 10.1038/nature10361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kang Y, et al. Crystal structure of rhodopsin bound to arrestin by femtosecond X-ray laser. Nature. 2015;523:561–567. doi: 10.1038/nature14656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Staus DP, et al. Allosteric nanobodies reveal the dynamic range and diverse mechanisms of G-protein-coupled receptor activation. Nature. 2016;535:448–452. doi: 10.1038/nature18636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ring AM, et al. Adrenaline-activated structure of β2-adrenoceptor stabilized by an engineered nanobody. Nature. 2013;502:575–579. doi: 10.1038/nature12572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rasmussen SG, et al. Structure of a nanobody-stabilized active state of the β2 adrenoceptor. Nature. 2011;469:175–180. doi: 10.1038/nature09648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang W, et al. Structural insights into μ-opioid receptor activation. Nature. 2015;524:315–321. doi: 10.1038/nature14886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kobilka BK. Amino and carboxyl terminal modifications to facilitate the production and purification of a G protein-coupled receptor. Anal. Biochem. 1995;231:269–271. doi: 10.1006/abio.1995.1533. [DOI] [PubMed] [Google Scholar]
- 14.Caffrey M, Cherezov V. Crystallizing membrane proteins using lipidic mesophases. Nat. Protoc. 2009;4:706–731. doi: 10.1038/nprot.2009.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kabsch W. Xds. Acta Crystallogr. D. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Strong M, et al. Toward the structural genomics of complexes: crystal structure of a PE/PPE protein complex from Mycobacterium tuberculosis. Proc. Natl Acad. Sci. 2006;103:8060–8065. doi: 10.1073/pnas.0602606103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr. D. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen VB, et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ten Eyck LF. Fast Fourier transform calculation of electron density maps. Methods Enzymol. 1985;115:324–337. doi: 10.1016/0076-6879(85)15024-x. [DOI] [PubMed] [Google Scholar]
- 21.Winn MD, et al. Overview of the CCP4 suite and current developments. Acta Crystallogr. D. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krissinel E. Stock-based detection of protein oligomeric states in jsPISA. Nucleic Acids Res. 2015;43(W1):W314–W319. doi: 10.1093/nar/gkv314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Whorton MR, et al. A monomeric G protein-coupled receptor isolated in a high-density lipoprotein particle efficiently activates its G protein. Proc. Natl Acad. Sci. 2007;104:7682–7687. doi: 10.1073/pnas.0611448104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Strachan RT, et al. Divergent transducer-specific molecular efficacies generate biased agonism at a G protein-coupled receptor (GPCR) J. Biol. Chem. 2014;289:14211–14224. doi: 10.1074/jbc.M114.548131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ghanouni P, et al. The effect of pH on β2 adrenoceptor function Evidence for protonation-dependent activation. J. Biol. Chem. 2000;275:3121–3127. doi: 10.1074/jbc.275.5.3121. [DOI] [PubMed] [Google Scholar]
- 26.Ranganathan A, Dror RO, Carlsson J. Insights into the role of Asp792.50 in β2 adrenergic receptor activation from molecular dynamics simulations. Biochemistry. 2014;53:7283–7296. doi: 10.1021/bi5008723. [DOI] [PubMed] [Google Scholar]
- 27.Lomize MA, Lomize AL, Pogozheva ID, Mosberg HI. OPM: orientations of proteins in membranes database. Bioinformatics. 2006;22:623–625. doi: 10.1093/bioinformatics/btk023. [DOI] [PubMed] [Google Scholar]
- 28.Huang J, MacKerell AD., Jr CHARMM36 all-atom additive protein force field: validation based on comparison to NMR data. J. Comput. Chem. 2013;34:2135–2145. doi: 10.1002/jcc.23354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Klauda JB, et al. Update of the CHARMM all-atom additive force field for lipids: validation on six lipid types. J. Phys. Chem. B. 2010;114:7830–7843. doi: 10.1021/jp101759q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.MacKerell AD, et al. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J. Phys. Chem. B. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 31.Best RB, et al. Optimization of the additive CHARMM all-atom protein force field targeting improved sampling of the backbone φ, ψ and side-chain χ1 and χ2 dihedral angles. J. Chem. Theory Comput. 2012;8:3257–3273. doi: 10.1021/ct300400x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Best RB, Mittal J, Feig M, MacKerell AD., Jr Inclusion of many-body effects in the additive CHARMM protein CMAP potential results in enhanced cooperativity of α-helix and β-hairpin formation. Biophys. J. 2012;103:1045–1051. doi: 10.1016/j.bpj.2012.07.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vanommeslaeghe K, et al. CHARMM general force field: a force field for drug-like molecules compatible with the CHARMM all-atom additive biological force fields. J. Comput. Chem. 2010;31:671–690. doi: 10.1002/jcc.21367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vanommeslaeghe K, Raman EP, MacKerell AD., Jr Automation of the CHARMM General Force Field (CGenFF) II: assignment of bonded parameters and partial atomic charges. J. Chem. Inf. Model. 2012;52:3155–3168. doi: 10.1021/ci3003649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vanommeslaeghe K, MacKerell AD., Jr Automation of the CHARMM General Force Field (CGenFF) I: bond perception and atom typing. J. Chem. Inf. Model. 2012;52:3144–3154. doi: 10.1021/ci300363c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Salomon-Ferrer R, Götz AW, Poole D, Le Grand S, Walker RC. Routine microsecond molecular dynamics simulations with AMBER on GPUs. 2. Explicit solvent particle mesh ewald. J. Chem. Theory Comput. 2013;9:3878–3888. doi: 10.1021/ct400314y. [DOI] [PubMed] [Google Scholar]
- 37.Case DA, et al. AMBER 2016. 2016 [Google Scholar]
- 38.Hopkins CW, Le Grand S, Walker RC, Roitberg AE. Long-time-step molecular dynamics through hydrogen mass repartitioning. J. Chem. Theory Comput. 2015;11:1864–1874. doi: 10.1021/ct5010406. [DOI] [PubMed] [Google Scholar]
- 39.Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 40.Wallace AC, Laskowski RA, Thornton JM. LIGPLOT: a program to generate schematic diagrams of protein–ligand interactions. Protein Eng. 1995;8:127–134. doi: 10.1093/protein/8.2.127. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Atomic coordinates and structure factors have been deposited in the Protein Data Bank (PDB) under accession code 5X7D.