Abstract
As a membrane-mimetic system, detergent micelles are popularly used to extract membrane proteins from lipid environments and to maintain their solubility and stability in an aqueous medium. However, many membrane proteins encapsulated in conventional detergents tend to undergo structural degradation during extraction and purification, thus necessitating the development of new agents with enhanced properties. In the current study, two classes of new amphiphiles are introduced, resorcinarene-based glucoside and maltoside amphiphiles (designated RGAs and RMAs, respectively), for which the alkyl chains are facially segregated from the carbohydrate head groups. Of these facial amphiphiles, two RGAs (RGA-C11 and RGA-C13) conferred markedly enhanced stability to four tested membrane proteins compared to a gold-standard conventional detergent. The relatively high water solubility and micellar stability of the RGAs compared to the RMAs, along with their generally favourable behaviours for membrane protein stabilisation described here, are likely to be, at least in part, a result of the high conformational flexibility of these glucosides. This study suggests that flexibility could be an important factor in determining the suitability of new detergents for membrane protein studies.
Keywords: facial amphiphiles, membrane proteins, molecular design, protein stability, resorcinarene glycosides
Membrane proteins perform proper functions in their native cellular membrane environments. However, such an environment is incompatible with biophysical methods such as X-ray crystallography and NMR spectroscopy. Thus, membrane proteins need to be extracted from the membrane for structural and many functional analyses. Of the over 120 conventional detergents currently available, n-octyl-β-d-glucoside (OG), n-decyl-β-d-maltoside (DM) and n-dodecyl-β-d-maltoside (DDM) are the most widely used detergents for structural determination of membrane proteins.[1] However, many membrane proteins tend to denature and aggregate when encapsulated in these popular detergents,[2] making it difficult to perform protein structural analysis. This is typically a result of the suboptimal structure of the conventional detergents. Therefore, it is necessary to develop new detergents with distinct architectures to cope with the diverse range of membrane proteins. Recent representative classes include tripod amphiphiles,[2, 3] lipopeptide detergents,[4] hemifluorinated surfactants,[5] glucose or maltose neopentyl glycols (GNGs or MNGs),[6] glyco-diosgenin,[7] facial amphiphiles,[8] neopentyl glycol-derived triglucosides[9] and penta-saccharide-based amphiphiles.[10] Apart from these small amphiphiles, there are polymeric nano-assemblies (polymeric materials or polymer–lipid superassemblies) such as amphipols,[5, 11] nanodiscs[5, 12] and nanolipodisc particles.[13] In terms of membrane-protein stabilisation, most of these new agents were shown to be superior to DDM, the most widely used conventional detergent. However, most are also difficult to synthesise, severely limiting the widespread use of these agents. Furthermore, relatively little effort has been made to develop new facial agents to date. In the current study, we designed and synthesised resorcinarene-based facial glucosides and maltoside amphiphiles, designated RGAs and RMAs, respectively (Scheme 1). These agents contain flexible alkyl chains as a hydrophobic group and are thus distinct from other facial detergents with a very rigid hydrophobic group (e.g., steroidal unit).[8] The glucoside agents (RGAs) could be synthesised through a short synthetic protocol, and several of these agents conferred enhanced stability to all four tested proteins, including a G-protein-coupled receptor, compared to DDM. These glucoside agents also tend to form protein–detergent complexes significantly smaller than those formed by DDM.
Scheme 1.
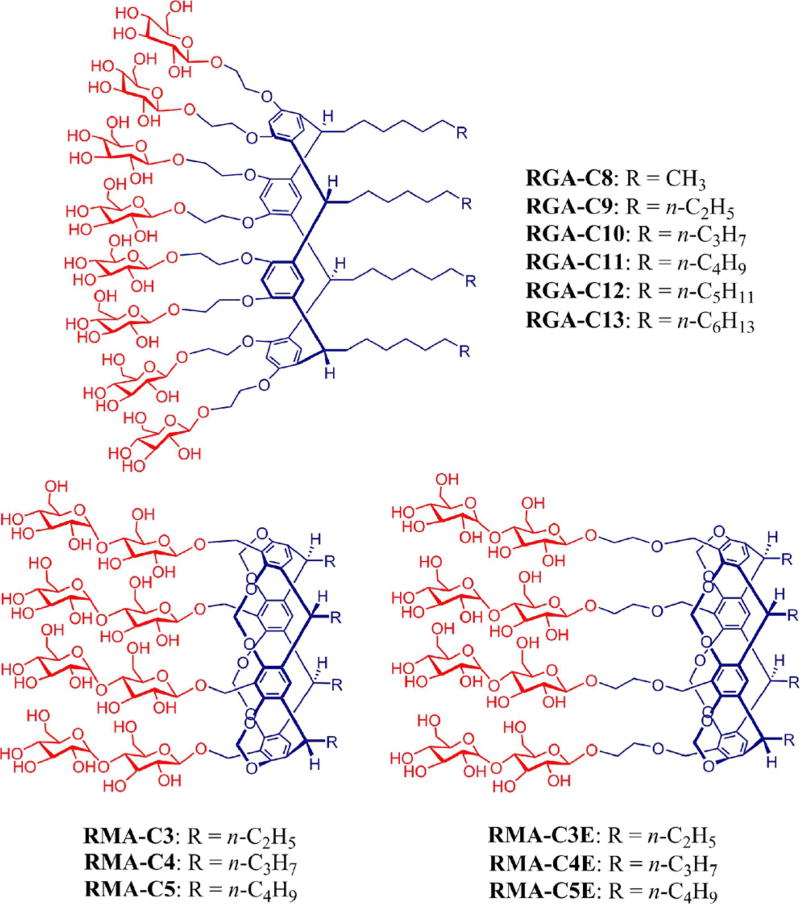
Chemical structures of resorcinarene-based new agents. The agents have glucosides (RGAs) or maltosides (RMAs) as a head group. The alkyl chain length varied from C8 to C13 for the RGAs whereas the RMAs varied in alkyl chain length from C3 to C5. For preparation of the RMAs, the maltoside groups were connected to the resorcinarene core either directly (RMA-Cs) or by an ethylene glycol linker (RMA-CEs).
We have prepared two kinds of resorcinarene-based amphiphiles with different hydrophilic groups: glucosides (RGAs) and maltosides (RMAs) (Scheme 1). The glucoside agents contain eight glucose units as the terminal hydrophilic groups, whereas the maltoside agents contain four maltose units in the same position. Both classes share four alkyl chains as the hydrophobic group, but alkyl chain length varies from C8 to C13 for the glucosides and from C3 to C5 for the maltosides. The alkyl chain length has been used in the detergent designation. It is noteworthy that there is another structural difference between these two classes: the two hydroxyl groups of each resorcinol unit are connected by a methylene spacer for the maltoside amphiphiles, but there is no such connection in the case of the glucoside agents. Instead, the two corresponding hydroxyl groups are individually conjugated to the glucose groups through an ethylene glycol linker. Accordingly, the glucoside amphiphiles have increased flexibility in the central scaffold relative to the maltoside agents, which could play a favourable role in enhancing membrane-protein stability. In the case of the maltoside amphiphiles, the maltoside head group is attached to the central resorcinarene moiety either directly (RMA-Cs) or by an ethylene glycol linker (RMA-CEs). Irrespective of the presence or absence of the linker, both sets of the RMAs were prepared according to a protocol comprising six synthetic steps: acid-catalysed aldehyde condensation, methylene bridging of two hydroxyls, mono-bromination at the benzylic position, basic hydrolysis/ethylene glycol linkage, β-selective glycosylation using anchimeric assistance and global deprotection (see the Supporting Information for details). The number of synthetic steps in this protocol is in the typical range (five to eight) for those of other new amphiphiles. In contrast, all RGAs were synthesised through three synthetic steps: acid-catalysed aldehyde condensation, β-selective glycosylation using anchimeric assistance and global deprotection, making these agents synthetically more accessible than the RMAs and most other new detergents. Note that perbenzoylated glucosyl (or maltosyl) bromide was used as the glycosyl donor in the glycosylation step. Because the neighbouring group participation of benzoyl group invokes anchimeric assistance via a five-membered cyclic intermediate, a carbohydrate (glucose or maltose) could be connected to the resorcinarene scaffold by a β-glycosidic linkage. The formation of the β-glycosidic bond was confirmed by the 1H NMR spectrum of RMA-C3 (Figure S1 in the Supporting Information). Different behaviour of the RMAs and RGAs was first observed in terms of water solubility. All the maltosides except for RMA-C5 showed a medium range of water solubility [1–5% (w/v)] and tended to form precipitates over time, indicative of the limited water solubility and stability of their micelles. RMA-C5 was poorly water-soluble [<1% (w/v)] and was not studied further. The RMAs with the ethylene glycol linker (RMA-CEs) were slightly more water-soluble than the RMAs with no linker (RMA-Cs). In contrast, all RGAs were highly water soluble [>10% (w/v)], and their micelles were stable enough to afford clear detergent solutions over several months. Thus, the RGAs have three advantages over the RMA agents: synthetic convenience, water solubility and micellar stability.
The new agents were characterised in terms of critical micelle concentration (CMC) and hydrodynamic radius (Rh) of their micelles, estimated by the use of a hydrophobic fluorophore (diphenylhexatriene)[14] and dynamic light-scattering experiments, respectively. The summarised results for the new agents along with a conventional detergent (DDM) are presented in Table 1. Detergent CMC values varied from 2 to 30 µm, at least five times lower than DDM (170 µm), indicating that the new agents have an increased tendency to self-aggregate compared to DDM. The CMC values of the new agents decreased with increasing alkyl chain length, consistent with the general notion that detergent hydrophobicity is the main factor in determining detergent CMC values. The micelle sizes formed by the new agents tended to increase with increasing alkyl chain length. For example, RMA-C3E and RMA-C5E, with the shortest and longest alkyl chain length of the RMA-CEs, produced the smallest (3.6 nm) and largest micelles (5.4 nm), respectively. A similar trend was observed for the RGAs, but there was one clear difference. The micelle size of the glucoside agents gradually increased when the alkyl chains were elongated from C8 to C11 but decreased abruptly at C12 and C13 alkyl chain length. Except for RGA-C11, all RGAs tended to form smaller micelles than the RMAs, indicative of more conical molecular geometry.[15] The micellar volumes formed by all glucoside agents (2.8–3.7 nm) except for RGA-C11 were comparable to that formed by DDM (3.4 nm), whereas the micellar volumes of most maltoside agents (3.6–7.5 nm) were larger than that of DDM. The new detergents were further characterised in terms of their micelle-size distribution. When dynamic light-scattering data were analysed by a number-averaged size distribution, all new agents displayed a single set of micelle populations (Figure S2 in the Supporting Information).
Table 1.
Molecular weights, CMCs of newly synthesised amphiphiles (RGAs/RMAs) and a conventional detergent (DDM) and hydrodynamic radii of their micelles.
| Detergent | M[a] [g mol−1] | CMC [µm] | CMC [wt%] | Rh[b] [nm] |
|---|---|---|---|---|
| RGA-C8 | 2530.8 | ≈ 10 | ≈ 0.0025 | 2.8 ± 0.1 |
| RGA-C9 | 2586.9 | ≈ 7 | ≈ 0.0018 | 2.9 ± 0.1 |
| RGA-C10 | 2643.0 | ≈ 5 | ≈ 0.0013 | 3.5 ± 0.2 |
| RGA-C11 | 2699.1 | ≈ 3 | ≈ 0.0008 | 6.8 ± 0.2 |
| RGA-C12 | 2755.2 | ≈ 2.5 | ≈ 0.0007 | 3.0 ± 0.2 |
| RGA-C13 | 2811.3 | ≈ 2 | ≈ 0.0006 | 3.7 ± 0.1 |
| RMA-C3 | 2066.0 | ≈ 30 | ≈ 0.0006 | 5.5 ± 0.2 |
| RMA-C4 | 2122.1 | ≈ 15 | ≈ 0.0003 | 7.5 ± 0.2 |
| RMA-C3E | 2242.2 | ≈ 15 | ≈ 0.0003 | 3.6 ± 0.1 |
| RMA-C4E | 2298.3 | ≈ 10 | ≈ 0.0002 | 4.6 ± 0.1 |
| RMA-C5E | 2354.4 | ≈ 8 | ≈ 0.0002 | 5.4 ± 0.1 |
| DDM | 510.1 | 170 | 0.0087 | 3.4 ± 0.0 |
Molecular weight of detergents;
hydrodynamic radii of detergents measured at 0.5 wt% by dynamic light scattering, mean ± S.D., n = 5.
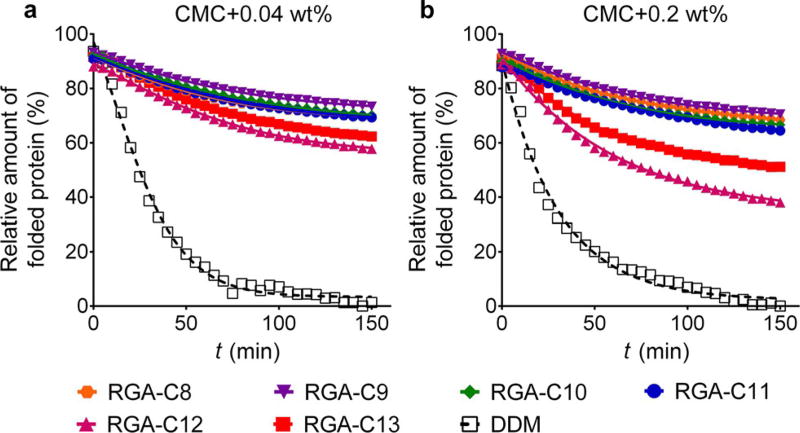
The new agents were first evaluated with the uric acid-xanthine/H+ symporter (UapA) from Aspergillus nidulans.[16] Protein stability was assessed by a thermal denaturation assay using a sulfhydryl-specific fluorophore, N-[4-(7-diethylamino-4-methyl-3-coumarinyl)phenyl]maleimide (CPM).[17] Upon protein unfolding, sulfhydryl groups present in the protein interior become solvent-accessible and thus rapidly react with the fluorophore molecules, leading to an increase in fluorescence emission intensity. Thus, CPM serves as a protein-unfolding sensor in this assay. For the assay, the transporter was first solubilised and purified in a conventional detergent (DDM). The DDM-purified transporter was diluted into buffer solutions including individual new amphiphiles to obtain final detergent concentrations of CMC + 0.04 wt%. The folded state of the transporter was monitored by measuring fluorescence emission intensity during a 150 min incubation at 40°C. Of the tested detergents, DDM was the least effective at maintaining the folded protein, and thus all the data were normalised to the DDM data, thereby providing the relative detergent efficacy toward protein stabilisation under this condition. When we evaluated the RGAs with this protein, all new agents showed significantly better performance than DDM for preserving the folded state of the protein (Figure 1a). RGA-C9 appeared to be best whereas RGA-C12 was the worst, but the differences between the new detergents were marginal. The RMAs were also better than DDM in this regard, but they were less stabilising than the RGAs. Of the maltoside agents, RMA-C3/C4 appeared to be the best (Figure S3a in the Supporting Information). When the detergent concentration was increased to CMC + 0.2 wt%, a similar trend was observed for both classes of amphiphiles. All glucoside agents were significantly better than DDM at this high detergent concentration, with the best stability obtained with RGA-C9 (Figure 1b). RGA-C12, however, was much less effective at stabilising the protein at this concentration. In the case of the maltoside agents, only RMA-C3 was as effective as the glucoside agents at stabilising the protein (Figure S3b in the Supporting Information). The other RMAs were less effective than RMA-C3 but still more effective than DDM. For a more quantitative comparison of detergent efficacy, the half-lives of the transporter solubilised in the individual detergents were calculated and summarised in Table S1 in the Supporting Information by assuming that the CPM data follow a one phase-decay curve. These half-life data more or less correlate with the detergent-efficacy order described above. Therefore, we concluded that the overall detergent efficacy order is RGAs > RMAs > DDM.
Figure 1.
Thermal denaturation profiles of UapA in DDM and the RGAs (RGA-C8, RGA-C9, RGA-C10, RGA-C11, RGA-C12 and RGA-C13) at detergent concentrations of CMC + 0.04 wt% (a) and CMC + 0.2 wt% (b). Protein stability was assessed through a CPM assay for which the protein solubilised in individual detergents was incubated at 40°C for 150 min. The data is representative of three independent experiments.
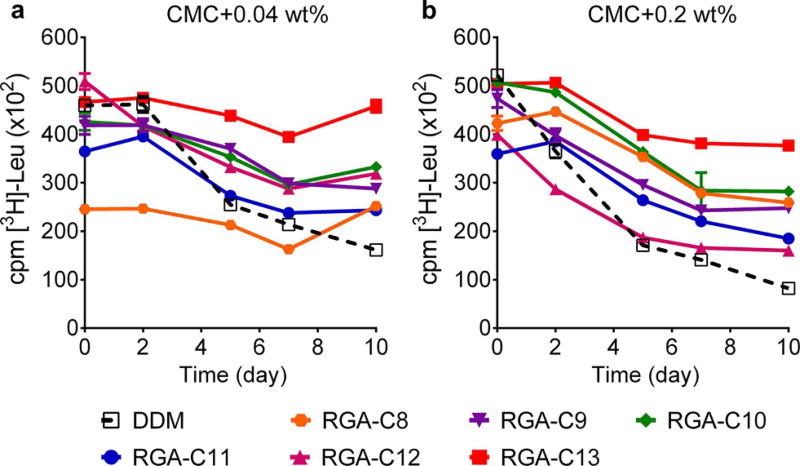
The newly synthesised amphiphiles were further evaluated with the bacterial leucine transporter (LeuT) from Aquifex aeolicus.[18] Initially, the transporter was solubilised and purified in DDM. The DDM-purified protein was diluted into individual detergent-containing solutions to obtain a final concentration of CMC + 0.04 wt%. Protein stability was assessed by measuring the ability of the protein to bind to a radio-labelled substrate ([3H]-leucine) through a scintillation-proximity assay.[19] The substrate-binding activity of the transporter was monitored at regular intervals over the course of a 10 day incubation at room temperature. At this low detergent concentration, all glucoside agents with the exception of RGA-8 were more effective than or comparable to DDM (Figure 2a); of these new glucosides, RGA-C13 with the longest alkyl chain was remarkably effective at retaining the transporter activity. When detergent concentration was increased to CMC + 0.2 wt%, the best performance was also observed for RGA-C13 (Figure 2b). The lowest efficacy was detected for RGA-C12 rather than RGA-C8 at this high detergent concentration. Interestingly, the efficacy of the shortest-chain glucoside agent (RGA-C8) improved significantly at increased detergent concentration from CMC + 0.04 wt% to CMC + 0.2 wt%, whereas the comparative efficacies of the other RGAs were similar at both concentrations. When the RMAs were tested with the transporter, a difference between the RMA-Cs and RMA-CEs was observed. All RMA-CEs were superior to DDM at both detergent concentrations, CMC + 0.04 wt% (Figure S4a in the Supporting Information) and CMC + 0.2 wt% (Figure S4b in the Supporting Information). However, the RMA-Cs with no ethylene glycol linker were generally ineffective at retaining the transporter stability, particularly when evaluated at CMC + 0.2 wt%. The short overall length of these detergents may be responsible for such poor behaviour because detergent dimensions need to match the dimensions of the transporter for effective protein protection. Overall, the long-term functional stability (substrate-binding activity) of the transporter was markedly enhanced when encapsulated by some glucoside agents (e.g., RGA-C13) and the RMA-CEs.
Figure 2.
Long-term stability of LeuT solubilised in DDM or new RGAs (RGA-C8, RGA-C9, RGA-C10, RGA-C11, RGA-C12 and RGA-C13) at concentrations of CMC + 0.04 wt% (a) and CMC + 0.2 wt% (b). DDM-purified transporter was mixed with individual detergent-containing solutions and then incubated for 10 days at room temperature. The substrate-binding activity of LeuT was measured at regular intervals during incubation, using radio-labelled substrate ([3H]-Leu) by a scintillation-proximity assay. Error bars, SEM, n = 2.
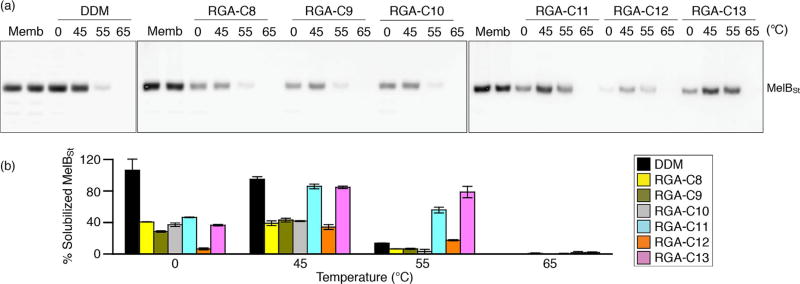
The intriguing results obtained with the two transporters prompted us to test these agents for the solubilisation and stabilisation of another membrane protein, the melibiose permease of Salmonella typhimurium (MelBSt).[20] MelBSt is a major facilitator superfamily permease catalysing cotransport of galactosides with protons, sodium or lithium ion.[21] Initially, membrane fractions prepared from E. coli cells overexpressing MelBSt were treated with 1.5% (w/v) individual new detergent or DDM for 90 min at 0°C. Note that only the RGA agents were tested in this study because the solubility of the RMAs is too low. The amounts of this transporter solubilised by individual detergents were analysed by SDS-PAGE and Western blotting and expressed as percentages of the total amount of the transporter initially present in the untreated membrane sample (Figure 3a, b). As can be seen in Figure 3b, most of the new agents yielded small amounts of the soluble protein (30–50%) relative to DDM (≈100 %), indicating that these new agents were relatively ineffective at extracting the transporter from the membranes at low temperature (0 °C). A similar result was obtained when incubation temperature was increased to 45°C, except for RGA-C11 and RGA-C13. These two new agents produced amounts of soluble transporter comparable to DDM, indicating that these agents were not only efficient at extracting the protein, but were also effective at maintaining it in a soluble state at this elevated temperature. When the incubation temperature was further increased to 55°C, DDM maintained only a small amount of the soluble transporter (≈10%). In contrast, the new agents (RGA-C11 and RGA-C13) still yielded substantial amounts of soluble protein, with RGA-C13 maintaining the same amount of soluble protein at 55 as at 45°C. However, detergent treatment at 65 °C resulted in complete loss of the transporter from the solutions for all detergents. Taken together, the results show that the RGAs were inferior to DDM at extracting the transporter efficiently from the membrane at low temperatures but -more efficient than DDM at an elevated temperature (55 °C). Furthermore, the transporter encapsulated by RGA-C11 or RGA-C13 displayed significantly enhanced thermostability compared to the protein solubilised in DDM.
Figure 3.
Thermo-stability of MelBSt extracted by DDM or an RGA (RGA-C8, RGA-C9, RGA-C10, RGA-C11, RGA-C12 or RGA-C13). (a) Western blot analysis. Membranes containing MelBSt mixed with the indicated detergent were incubated at 0°C or an elevated temperature (45, 55 or 65°C) and subjected to ultracentrifugation. The amounts of the transporter in the solutions were analysed by SDS-15% PAGE and Western blotting. The band from the untreated membrane sample (Memb) represents the total amount of the transporter present in the membranes before detergent treatment. (b) Histogram. The amount of soluble protein in a detergent detected in panel (a) was expressed as a percentage of the total amount of the transporter in the untreated membrane sample. Error bars, SEM, n = 3.
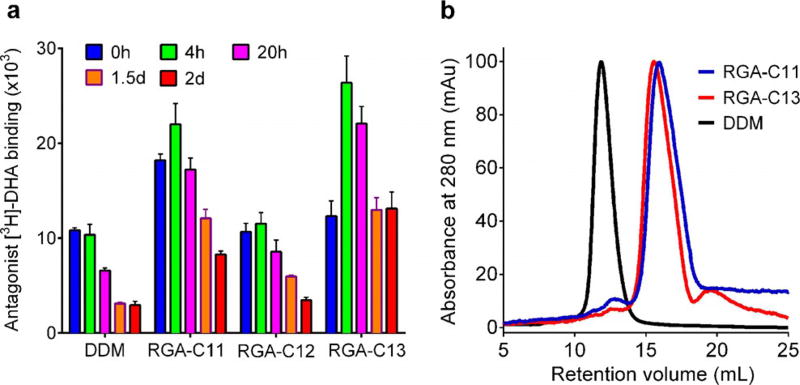
We continued to evaluate the new agents with the human β2 adrenergic receptor (β2AR), a G-protein-coupled receptor.[22] Receptor stability was assessed with a ligand-binding assay using antagonist ([3H]-dihydroalprenolol).[23] To prepare the protein solubilised in individual detergents, the DDM-purified receptor was diluted with a buffer solution containing DDM or a new agent to reach a final detergent concentration of CMC + 0.2 wt%. When ligand binding activity was measured with the receptors solubilised in individual detergents, we found a large variation in detergent efficacy. RGA-C8 and RGA-C9 were inferior to DDM. RGA-C10 was comparable to DDM, whereas RGA-C13 was the best agent followed by RGA-C11 and RGA-C12 (Figure S5 in the Supporting Information). Based on this preliminary result, three agents (RGA-C11/C12/C13) were selected for long-term receptor-stability analysis where the protein–detergent samples were incubated for two days at room temperature (Figure 4a). RGA-C12 was similar to DDM at retaining receptor activity. However, the receptor encapsulated in RGA-C11 or RGA-C13 showed higher initial activity than the protein in DDM and reached maximum values following a 4 h incubation. The maximum activity displayed by the RGA-C13-solubilised receptor was more than 2.5 times higher than that of the DDM-solubilised protein. Over the subsequent additional incubation period, receptor activity gradually decreased over time, but the final receptor activity (t = 2 days) observed for this new agent was even higher than the initial receptor activity (t = 0 h) detected for DDM. It is notable that receptor ligand-binding activity observed at t = 4 h was the highest reported to date,[10, 24] indicating that RGA-C13 has an optimal architecture for receptor stability. The initial increase in receptor activity following the 4 h incubation observed for RGA-C11 and RGA-C13 could be the result of the slow rate of detergent exchange following dilution. Similar behaviours were observed for two maltoside agents; the receptor in RMA-C3E/C5E initially had an enhanced ligand-binding ability relative to the protein in DDM (Figure S6a in the Supporting Information). When RGA-C3E was selected for the long-term experiment, this agent afforded two times higher receptor activity than DDM at t = 0 h, but the final receptor activity (t = 2 days) was just half of the initial receptor activity in DDM (Figure S6b in the Supporting Information). This result suggests that the best RGA (i.e., RGA-C13) is even better than the best RMA (i.e., RMA-C3E) at retaining the ligand binding activity of the receptor. Analysis of the protein–detergent complex size (receptor + RGA-C11/C13) using size-exclusion chromatography revealed that the receptor–detergent complexes formed by these new agents were significantly smaller than those formed by DDM (Figure 4b), a favourable attribute for membrane-protein structure determination by X-ray crystallography and electron microscopy.[25] RGA-C11 appeared to form a protein–detergent complex slightly smaller than RGA-C13, probably owing to the slightly shorter alkyl chain. The large difference in the size of receptor–detergent complexes between RGA-C11/C13 and DDM observed here is unprecedented.[10, 24]
Figure 4.
(a) Long-term stability of β2AR solubilised in an RGA (RGA-C8, RGA-C9, RGA-C10, RGA-C11, RGA-C12 or RGA-C13) or DDM and (b) size-exclusion chromatography profiles of the receptor in selected detergents (RGA-C11, RGA-C13 and DDM). Protein stability was measured by the ligand-binding assay using the antagonist ([3H]-dihydroalprenolol (DHA)) during a two-day incubation at room temperature. For the size-exclusion chromatography analysis, detergent was exchanged from DDM to an RGA (RGA-C11 or RGA-C13) before applying each detergent-solubilised receptor for size-exclusion chromatography. Error bars, SEM, n = 3.
Despite the architectural similarity, the RGAs and RMAs largely differ in water solubility and micellar stability. Both classes contain eight glucose units, but the RGAs showed better water solubility as a function of increasing alkyl chain length than the RMAs. Specifically, RGA-C13 with a long alkyl chain was completely soluble even at 10 %, whereas all RMAs with short alkyl chains (C3/C4/C5) showed poor solubility in aqueous solution. Furthermore, micelles formed by the RGAs were significantly more stable than those formed by the RMAs. We conceive that the relatively flexible RGAs readily adopt a conformation favourable for both micelle formation and water solubility, whereas the RMAs are too rigid to allow similar conformational changes. The relatively small micelle size observed for the RGAs may also be associated with their flexibility. More importantly, the flexibility of the RGAs relative to the RMAs is also associated with the ability of these agents to maintain the structural stability of membrane proteins because the overall performance of the RGAs was superior to that of the RMAs in the current study. Individual membrane proteins have different shapes and dimensions, and thus a detergent needs to be flexible enough to undergo conformational changes to allow favourable interactions with each other and a target membrane protein. However, we cannot rule out that other structural differences between the RGAs and RMAs are responsible for the differences in stability conferred. For instance, they differ in alkyl chain length and head group class (glucose vs. maltose).
All new agents introduced are facial amphiphiles with the hydrophobic group facially separated from the hydrophilic group, but these agents are structurally distinct from previously reported facial agents. For instance, the facial amphiphiles[8, 26] previously developed for membrane protein study contain (a) very rigid cholate unit(s) as the hydrophobic group, whereas the current new agents bear a rigid resorcinarene core along with four flexible alkyl chains. As a result, these new agents are distinct from the previous facial amphiphiles in that the flexible alkyl tips of the new agents rather than rigid cholate units interact with the membrane proteins. In addition, the cholate-based facial agents show little variability in the structure of the hydrophobic group, whereas a variety of hydrophobic groups could be introduced into the RGA architecture using diverse aldehydes in the condensation step. This structural variability is important because it will allow further optimisation of the detergent properties. Furthermore, the preparation of the previously described facial amphiphiles requires five to seven synthetic steps.[8, 26] In contrast, the current RGAs are prepared in three steps. Thus, these agents are significantly more accessible than other carbohydrate-containing facial amphiphiles.
The RGAs were superior to DDM despite being glucoside detergents that are generally less effective at stabilising membrane proteins than maltoside agents.[1a, 6b] Notably, the current new agents appeared to be even superior to MNG-3, one of the most successful new detergents for membrane-protein studies.[6a] MNG-3 was roughly as effective as DDM at maintaining UapA in a stable state, whereas the current new agents were significantly better (Figure S3 in the Supporting Information). In the case of LeuT, a previous study has indicated that MNG-3 is slightly superior to DDM at maintaining transporter stability.[27] In contrast, some of the RGAs conferred substantially enhanced stability to the transporter under the same conditions. RGA-C11/C13 produced substantially smaller protein–detergent complex sizes compared to MNG-3, similar to that formed by DDM.[28] Combined together, this study not only introduces new detergent tools with potential for membrane-protein study, but also proposes a detergent flexibility–efficacy relationship important for new amphiphile design.
Experimental Section
Experimental details on the synthesis and characterisation of new amphiphiles as well as membrane protein stability assays can be found in the Supporting Information.
Supplementary Material
Acknowledgments
This work was supported by the National Research Foundation of Korea (NRF) funded by the Korean government (MSIP) (2016R1A2B2011257 to P.S.C., H.H., M.D. and M.E.).
Footnotes
Supporting Information and the ORCID identification numbers for the authors of this article can be found under: https://doi.org/10.1002/chem.201605016.
Conflict of interest
P.S.C. and H.H. are co-inventors on a patent application that covers the RGA/RMA agents.
References
- 1.a) Privé GG. Methods. 2007;41:388–397. doi: 10.1016/j.ymeth.2007.01.007. [DOI] [PubMed] [Google Scholar]; b) Carpenter EP, Beis K, Cameron AD, Iwata S. Curr. Opin. Struct. Biol. 2008;18:581–586. doi: 10.1016/j.sbi.2008.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chae PS, Bae HE, Ehsan M, Hussain H, Kim JW. Org. Biomol. Chem. 2014;12:8480–8487. doi: 10.1039/c4ob01375a. [DOI] [PubMed] [Google Scholar]
- 3.Chae PS, Kruse AC, Gotfryd K, Rana RR, Cho KH, Rasmussen SGF, Bae HE, Chandra R, Gether U, Guan L, Kobilka BK, Loland CJ, Byrne B, Gellman SH. Chem. Eur. J. 2013;19:15645–15651. doi: 10.1002/chem.201301423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Privé GG. Curr. Opin. Struct. Biol. 2009;19:379–385. doi: 10.1016/j.sbi.2009.07.008. [DOI] [PubMed] [Google Scholar]
- 5.a) Breyton C, Chabaud E, Chaudier Y, Pucci B, Popot J-L. FEBS Lett. 2004;564:312–318. doi: 10.1016/S0014-5793(04)00227-3. [DOI] [PubMed] [Google Scholar]; b) Popot J-L. Ann. Rev. Biochem. 2010;79:737–775. doi: 10.1146/annurev.biochem.052208.114057. [DOI] [PubMed] [Google Scholar]; c) Abla M, Unger S, Keller S, Bonneté F, Ebel C, Pucci B, Breyton C, Durand G. J. Colloid Interface Sci. 2015;445:127–136. doi: 10.1016/j.jcis.2014.12.066. [DOI] [PubMed] [Google Scholar]
- 6.a) Chae PS, Rasmussen SGF, Rana RR, Gotfryd K, Chandra R, Goren MA, Kruse AC, Nurva S, Loland CJ, Pierre Y, Drew D, Popot J-L, Picot D, Fox BG, Guan L, Gether U, Byrne B, Kobilka B, Gellman SH. Nat. Methods. 2010;7:1003–1008. doi: 10.1038/nmeth.1526. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Chae PS, Rana RR, Gotfryd K, Rasmussen SGF, Kruse AC, Cho KH, Capaldi S, Carlsson E, Kobilka B, Loland CJ, Gether U, Banerjee S, Byrne B, Lee JK, Gellman SH. Chem. Commun. 2013;49:2287–2289. doi: 10.1039/c2cc36844g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chae PS, Rasmussen SGF, Rana RR, Gotfryd K, Kruse AC, Manglik A, Cho KH, Nurva S, Gether U, Guan L, Loland CJ, Byrne B, Kobilka BK, Gellman SH. Chem. Eur. J. 2012;18:9485–9490. doi: 10.1002/chem.201200069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.a) Zhang Q, Ma X, Ward A, Hong WX, Jaakola VP, Stevens RC, Finn MG, Chang G. Angew. Chem. Int. Ed. 2007;46:7023–7025. doi: 10.1002/anie.200701556. Angew. Chem.2007, 119, 7153–7155. [DOI] [PubMed] [Google Scholar]; b) Lee SC, Bennett BC, Hong W-X, Fu Y, Baker KA, Marcoux J, Robinson CV, Ward AB, Halpert JR, Stevens RC, Stout CD, Yeager MJ, Zhang Q. Proc. Natl. Acad. Sci. USA. 2013;110:E1203–E1211. doi: 10.1073/pnas.1221442110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sadaf A, Mortensen JS, Capaldi S, Tikhonova E, Hariharan P, de Castro Ribeiro O, Loland CJ, Guan L, Byrne B, Chae PS. Chem. Sci. 2016;7:1933–1939. doi: 10.1039/c5sc02900g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ehsan M, Du Y, Scull NJ, Tikhonova E, Tarrasch J, Mortensen JS, Loland CJ, Skiniotis G, Guan L, Byrne B, Kobilka BK, Chae PS. J. Am. Chem. Soc. 2016;138:3789–3796. doi: 10.1021/jacs.5b13233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tribet C, Audebert R, Popot J-L. Proc. Natl. Acad. Sci. USA. 1996;93:15047–15050. doi: 10.1073/pnas.93.26.15047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Denisov IG, Sligar SG. Nat. Struct. Mol. Biol. 2016;23:481–486. doi: 10.1038/nsmb.3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Orwick-Rydmark M, Lovett JE, Graziadei A, Lindholm L, Hicks MR, Watts A. Nano. Lett. 2012;12:4687–4692. doi: 10.1021/nl3020395. [DOI] [PubMed] [Google Scholar]
- 14.De Vendittis E, Palumbo G, Parlato G, Bocchini V. Anal. Biochem. 1981;115:278–286. doi: 10.1016/0003-2697(81)90006-3. [DOI] [PubMed] [Google Scholar]
- 15.Israelachvili JN, Mitchell DJ, Ninham BW. J. Chem. Soc. Faraday Trans. 2. 1976;72:1525–1568. [Google Scholar]
- 16.Pantazopoulou A, Diallinas G. Mol. Membr. Biol. 2006;23:337–348. doi: 10.1080/09687860600738239. [DOI] [PubMed] [Google Scholar]
- 17.Alexandrov AI, Mileni M, Chien EYT, Hanson MA, Stevens RC. Structure. 2008;16:351–359. doi: 10.1016/j.str.2008.02.004. [DOI] [PubMed] [Google Scholar]
- 18.Deckert G, Warren PV, Gaasterland T, Young WG, Lenox AL, Graham DE, Overbeek R, Snead MA, Keller M, Aujay M, Huber R, Feldman RA, Short JM, Olsen GJ, Swanson RV. Nature. 1998;392:353–358. doi: 10.1038/32831. [DOI] [PubMed] [Google Scholar]
- 19.Quick M, Javitch JA. Proc. Natl. Acad. Sci. USA. 2007;104:3603–3608. doi: 10.1073/pnas.0609573104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guan L, Nurva S, Ankeshwarapu SP. J. Biol. Chem. 2011;286:6367–6374. doi: 10.1074/jbc.M110.206227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ethayathulla AS, Yousef MS, Amin A, Leblanc G, Kaback HR, Guan L. Nat. Commun. 2014;5:3009. doi: 10.1038/ncomms4009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kolb P, Rosenbaum DM, Irwin JJ, Fung JJ, Kobilka BK, Shoichet BK. Proc. Natl. Acad. Sci. USA. 2009;106:6843–6848. doi: 10.1073/pnas.0812657106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Riva MA, Creese I. Mol. Pharmacol. 1989;36:211–218. [PubMed] [Google Scholar]
- 24.Hussain H, Du Y, Scull NJ, Mortensen JS, Tarrasch J, Bae HE, Loland CJ, Byrne B, Kobilka BK, Chae PS. Chem. Eur. J. 2016;22:7068–7073. doi: 10.1002/chem.201600533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.a) Moraes I, Evans G, Sanchez-Weatherby J, Newstead S, Stewart PDS. Biochim. Biophys. Acta Biomembr. 2014;1838:78–87. doi: 10.1016/j.bbamem.2013.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Hendrickson WA. Nat. Struct. Mol. Biol. 2016;23:464–467. doi: 10.1038/nsmb.3215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chae PS, Gotfryd K, Pacyna J, Miercke LJW, Rasmussen SGF, Robbins RA, Rana RR, Loland CJ, Kobilka B, Stroud R, Byrne B, Gether U, Gellman SH. J. Am. Chem. Soc. 2010;132:16750–16752. doi: 10.1021/ja1072959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cho KH, Husri M, Amin A, Gotfryd K, Lee HJ, Go J, Kim JW, Loland CJ, Guan L, Byrne B, Chae PS. Analyst. 2015;140:3157–3163. doi: 10.1039/c5an00240k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rasmussen SGF, DeVree BT, Zou Y, Kruse AC, Chung KY, Kobilka TS, Thian FS, Chae PS, Pardon E, Calinski D, Mathiesen JM, Shah STA, Lyons JA, Caffrey M, Gellman SH, Steyaert J, Skiniotis G, Weis WI, Sunnahara RK, Kobilka BK. Nature. 2011;477:549–555. doi: 10.1038/nature10361. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.