Abstract
Mitochondria are incredibly dynamic organelles that undergo continuous fission and fusion events to control morphology, which profoundly impacts cell physiology including cell cycle progression (Mitra, 2013; Mitra et al., 2009). This is highlighted by the fact that most major human neurodegenerative diseases are due to specific disruptions in mitochondrial fission or fusion machinery and null alleles of these genes result in embryonic lethality (Chan, 2006; Chen et al., 2017; Flippo and Strack, 2017). To gain a better understanding of the pathophysiology of such disorders, tools for the in vivo assessment of mitochondrial dynamics are required. It would be particularly advantageous to simultaneously image mitochondrial fission-fusion coincident with cell cycle progression. To that end, we have generated a new transgenic reporter mouse, called mito::mKate2 that ubiquitously expresses a mitochondria localized far-red mKate2 fluorescent protein. Here we show that mito::mKate2 mice are viable and fertile and that mKate2 fluorescence can be spectrally separated from the previously developed Fucci cell cycle reporters (Sakaue-Sawano et al., 2008). By crossing mito::mKate2 mice to the ROSA26R-mTmG dual fluorescent Cre reporter line (Muzumdar et al., 2007), we also demonstrate the potential utility of mito::mKate2 for genetic mosaic analysis of mitochondrial phenotypes.
INTRODUCTION
Mitochondria are highly specialized organelles that, according to the endosymbiotic theory, are descendants of ancient bacteria that established an interdependent relationship within ancestral eukaryotic cells (Gray, 2017; Sagan, 1967). The most widely known role for mitochondria is as the “powerhouse of the cell” critical for energy (ATP) production by oxidative phosphorylation as well as other types of metabolism (McBride et al., 2006; Siekevitz, 1957). More recent decades of research have uncovered a multitude of roles beyond metabolism such as in calcium signaling, innate immunity, assembly of iron-sulfur clusters, autophagy, and apoptosis (Cloonan and Choi, 2013; Hailey et al., 2010; Stehling et al., 2014; Tait and Green, 2013). Additionally, mitochondrial fission-fusion and electron transport chain activity are known to profoundly influence progenitor cell proliferation and differentiation during development as well as in tumorigenesis (Chen et al., 2017; Flippo and Strack, 2017; Mandal et al., 2010; Mitra, 2013; Owusu-Ansah et al., 2008; Poche et al., 2016). However, the exact functions mitochondria serve during cell cycle progression are not fully understood.
Over the last several years, research has shown that, as cells cycle from G2/M to the G1-S phase transition, mitochondria shift from a mostly fragmented to a mostly fused state (Margineantu et al., 2002; Mishra and Chan, 2014; Mitra et al., 2009). It is possible that fusion at S phase occurs in order to mix mitochondrial protein and DNA content. This might ensure that a more homogenous mitochondrial network will be inherited by the two daughter cells. In subsequent M phase, fission may occur to promote equal distribution of mitochondria during cytokinesis. A previous study suggested that mitochondrial fusion may be instructive for entry into S phase by, at least in part, promoting the buildup of the S phase-promoting protein Cyclin E (Mitra et al., 2009). It is currently unclear how this occurs, but a recent report suggests that a pool of Cyclin E protein is directly recruited to mitochondria (Parker et al., 2015). While these observations and others imply a direct mechanistic link between the cell cycle machinery and mitochondrial fission-fusion proteins (Horn et al., 2011; Mitra et al., 2009; Qiao et al., 2010; Taguchi et al., 2007), there is still much to understand regarding precise cellular and molecular interplay controlling proliferation.
Our present knowledge regarding mitochondrial dynamics predominantly comes from in vitro and non-mammalian model systems, and certain aspects of mammalian mitochondrial function are still unclear. For example, mitochondrial morphology and dynamics can be dramatically different depending on tissue type, cell type, cell identity, and even sub-cellular localization (Collins et al., 2002; Kuznetsov and Margreiter, 2009; Kuznetsov et al., 2006; Mitra et al., 2009). However, in many cases, the significance of this heterogeneity is still largely unknown. Therefore, there is a need for genetic tools to closely monitor dynamic fission-fusion events within different tissues, developmental stages, and disease states such as cancer.
The currently available transgenic mouse strains that ubiquitously express mitochondrial-targeted fluorescent reporters (Mito-EGFP, mtGFP-tg, mtDsRed2-Tg, Acr3-EGFP-CAG-su9-DsRed2, PhAMexcised, Mito-QC, and Mt-Keima) emit within the green and red spectra (Abe et al., 2011; Hasuwa et al., 2010; McWilliams et al., 2016; Pham et al., 2012; Shitara et al., 2001; Sun et al., 2015; Yamaguchi et al., 2012). The same green/red spectral limitation applies to tissue-specific and inducible reporters, such as Mito-Timer, PhAMfloxed, Thy1-mitoDsRed, Thy1-mito-TagRFP, Thy1.2-mitoDendra, Nse-mitoYFP, CaMKIIα-mitoYFP, Hb9-MitoEGFP, and Endo-mitoEGFP (Breckwoldt et al., 2014; Chandrasekaran et al., 2006; Magrane et al., 2014; Misgeld et al., 2007; Pham et al., 2012; Pickles et al., 2013; Stotland and Gottlieb, 2016; Vande Velde et al., 2011; Wang et al., 2011). The only described Cyan Fluorescent Protein mitochondrial reporter mouse (Thy1-mitoCFP) is restricted to neurons (Misgeld et al., 2007). Additionally, a variety of mitochondrial biosensors have been developed to monitor energy production, reactive oxygen species, redox state, secondary messengers (such as Ca2+), Zn2+ homeostasis, and other processes (De Michele et al., 2014; Zhang and Avalos, 2017). The vast majority of these markers fluoresce in the blue and green color range. Thus, the lack of ubiquitous mitochondrial reporters outside of the blue/green/red color range limits the ability of researchers to track mitochondrial dynamics coincident with other processes visualized with biosensors.
Here, we report the generation of a transgenic mouse line, called mito::mKate2, in which the mitochondria are ubiquitously labeled with the mKate2 fluorescent protein. Since mKate2 is a far-red fluorescent protein, it can be visualized coincident with other traditional fluorescent reporters (such as GFP, YFP, CFP, and DsRed) and mitochondrial-specific biosensors. We demonstrate that mito::mKate2 is useful for monitoring mitochondrial dynamics coincident with cell cycle kinetics during mouse embryonic development and in adult tissues. Given that bright, photostable, far-red fluorescent proteins provide greater imaging depth than traditional green and red reporters, the mito::mKate2 reporter is also better-suited for in vivo and ex vivo imaging of mitochondrial dynamics within living tissues (Shcherbo et al., 2007; Shcherbo et al., 2009).
RESULTS AND DISCUSSION
Generation of CAG-mito::mKate2 fluorescent reporter mice
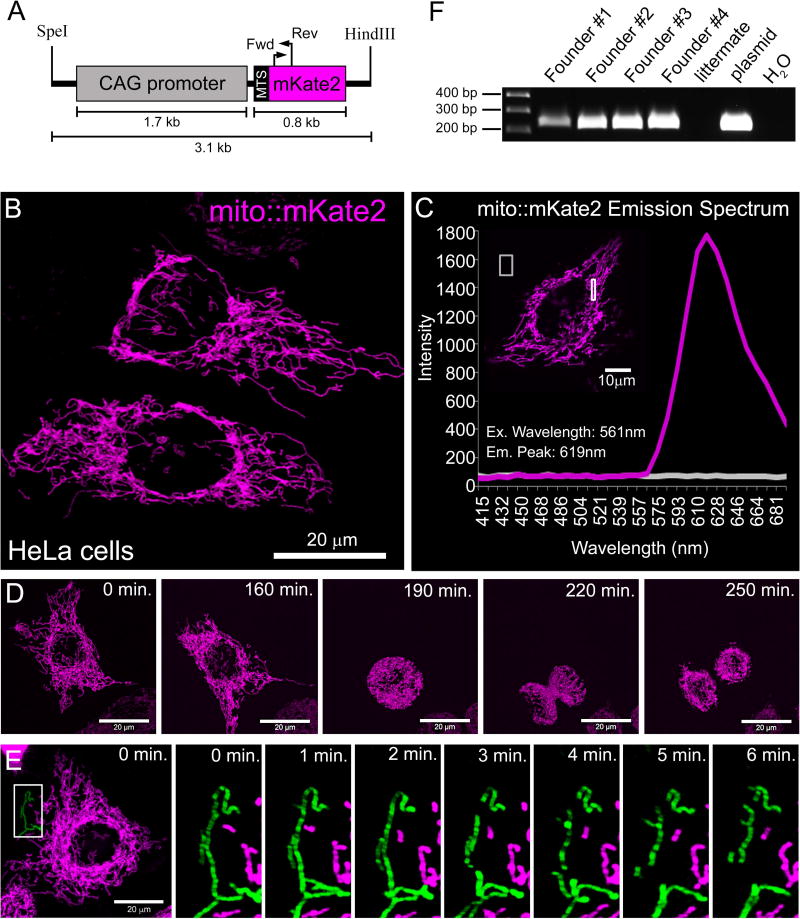
We generated a DNA construct containing a CAG (cytomegalovirus/β-actin) enhancer-promoter (Miyazaki et al., 1989) upstream of a cassette encoding mKate2 (Shcherbo et al., 2009) that is fused (N-terminal) to a mitochondrial targeting sequence (Evrogen) derived from subunit VIII of Cytochrome C Oxidase (Rizzuto et al., 1995; Rizzuto et al., 1989) (Figure 1A). This plasmid was transfected into HeLa cells and mito::mKate2 protein localization was determined by confocal microscopy. The mKate2 signal was detected specifically within the mitochondrial network and provided excellent structural resolution (Figure 1B). Emission fingerprinting revealed a far-red emission peak of 619 nm when excited by a 561 nm laser (Figure 1C). Live imaging of the transfected cells indicated that the mito::mKate2+ mitochondria could be detected over the course of cell division (Figure 1D and Supplemental Movie 1). By acquiring time-lapse movies of the entire cell 3D volume, we were able to resolve discrete fission-fusion events (Supplemental Movies 2 and 3). In order to highlight a fission event that took place over 6 minutes, still images were taken from the 30 minute time-lapse (Supplemental Movie 2) and a segment of the mitochondrial network was pseudo-colored green. During this period, a single mitochondrial segment was observed breaking into three pieces (Figure 1E). Once the transgenic construct was validated in vitro, we generated transgenic mice by pronuclear injection of single-cell C57B6/J mouse zygotes. PCR genotyping revealed that 4 out of 29 weanlings carried the mito::mKate2 transgene (Figure 1F).
Figure 1. In vitro validation of the CAG-mito::mKate2 transgene and generation of mice.
Map of the CAG-driven mito::mKate2 transgene (A). HeLa cell transiently transfected with the CAG-mito::mKate2 transgene-containing plasmid (B). Emission spectrum fingerprinting of mito::mKate2 fluorescence in transfected HeLa cells (C). Stills from a time-lapse movie of a mito::mKate2 transfected HeLa cell (D). Stills from a movie highlighting a mitochondrial fission event (pseudo-colored in green) occurring in a mito::mKate2 transfected HeLa cell (E). PCR genotyping of 4 recovered CAG-mito::mKate2 transgenic founder mice (F). MTS = mitochondrial targeting signal.
Widespread expression of mito::mKate2 in vivo
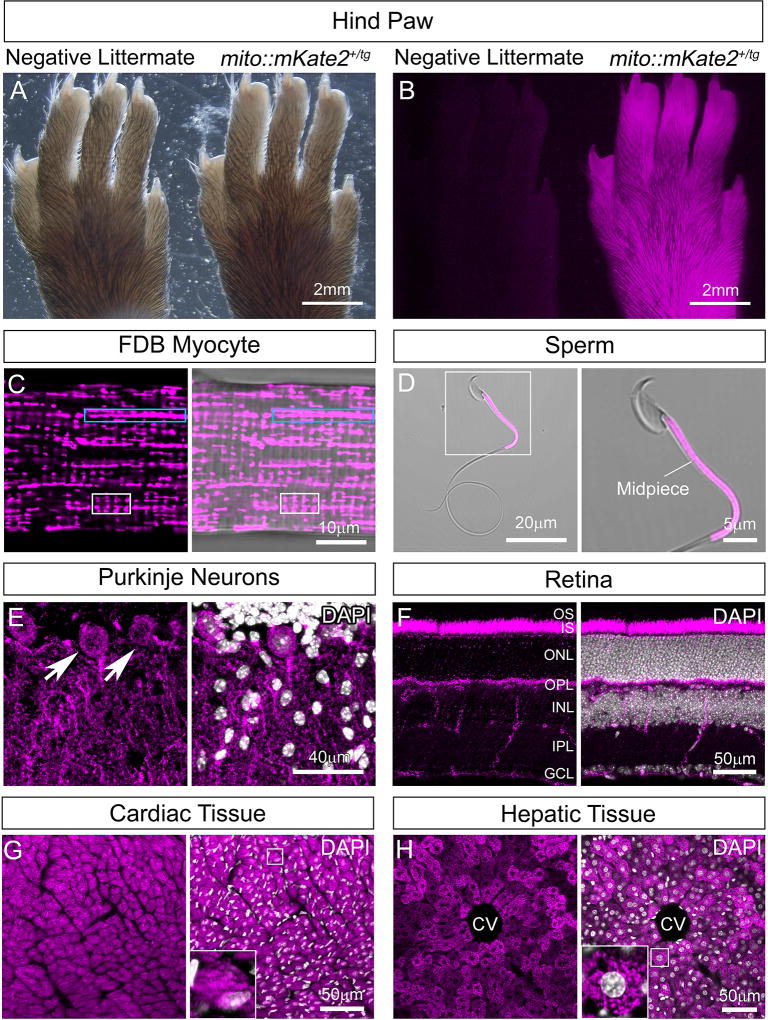
We crossed the four mito::mKate2 founder mice to CD1 mice (Charles River) to determine which founder gave rise to progeny with the brightest mKate2 signal indicated by hind paw fluorescence (Figure 2A–B). We subsequently chose one founder to backcross to C57B6/J mice thereby establishing a CAG-mito::mKate2+/tg line for additional characterization. All CAG-mito::mKate2+/tg progeny from this founder were viable and fertile and transmitted the transgene at the expected ~50% frequency to both male and female offspring. From this established transgenic line, we next harvested several different adult tissues (3–6 weeks) with well-described mitochondrial localization and morphology and performed confocal microscopy.
Figure 2. Widespread mitochondrial labeling in the CAG-mito::mKate2 transgenic line.
Hind paw of a CAG-mito::mKate2+ transgenic founder progeny showing bright mKate2 fluorescence as compared to a negative littermate (A–B). Expression of mito::mKate2 and resolution of tissue-specific mitochondrial morphologies and distribution in the adult flexor digitorum brevis (FDB) muscle (C), sperm midpiece (D), Purkinje neurons of the cerebellum (E), retina (F), heart (G) and liver (H). GCL=Ganglion Cell Layer, IPL=Inner Plexiform Layer, INL=Inner Nuclear Layer, OPL=Outer Plexiform Layer, ONL=Outer Nuclear Layer, IS=Inner Segments, OS=Outer Segments, CV=Central Vein. White nuclear stain is DAPI. n = 3.
Isolated myocytes from the flexor digitorum brevis (FDB) muscle exhibited the expected mitochondrial morphologies typical of development with some arranged into longitudinal clusters that intersect the striations and others that are shorter, vertical structures running along the striations (Figure 2C, blue box and white box, respectively). These short, vertical structures comprise the early forming T-tubule systems, which are critical for muscle contraction (Boncompagni et al., 2009). Spermatocytes isolated from mito::mKate2+/tg mice revealed fluorescence specifically in the mid-piece region consistent with previous transmission electron microscopy analysis indicating that mitochondria in this region are cylindrically packed (Figure 2D). This is the so-called “mitochondrial sheath”, which surrounds the outer dense fibers and is an important driver of sperm motility (Fawcett, 1975).
We also generated cryosections and determined that mito::mKate2 was widely expressed throughout the animals. In the case of Purkinje neurons of the cerebellum, mito::mKate2+ mitochondria were observed in both the soma and the dendritic arbors; consistent with previous reports (Figure 2E) (Herndon, 1963; Pham et al., 2012). Analysis of the retina showed a bright signal in the photoreceptor inner segments (IS), the outer nuclear layer (ONL), the outer plexiform layer (OPL), inner plexiform layer (IPL), and in the axons of the ganglion cell layer (GCL) (Figure 2F). The photoreceptor inner segments and synaptic terminal regions have previously been identified as sites densely populated by mitochondria (Cohen, 1961; Olney, 1968; Rueda et al., 2016). As previously described, cardiac tissue exhibited densely packed, stacked mitochondria whereas hepatic tissue mitochondria showed a more fragmented morphology (Figure 2G–H, insets) (Claude and Fullam, 1946; Kisch, 1956; Pham et al., 2012).
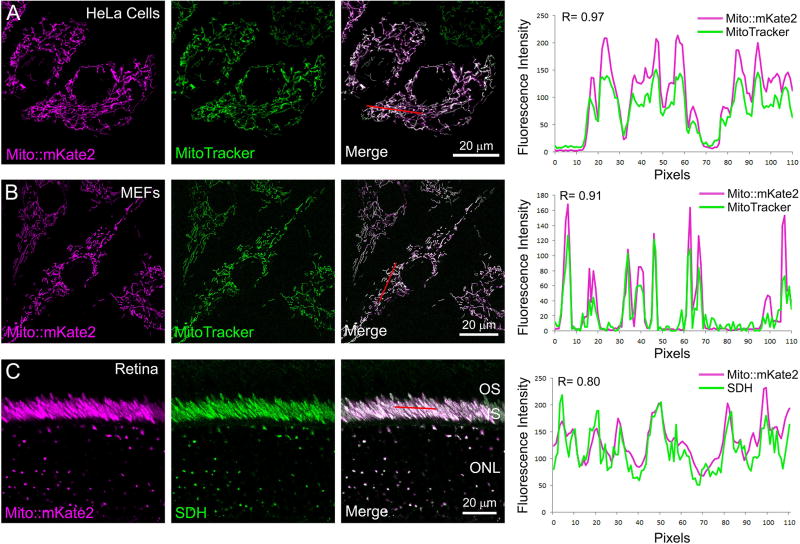
To verify that mito::mKate2 specifically labels mitochondria, we performed co-localization analyses with established mitochondrial markers. Mito::mKate2 transfected HeLa cells were labeled with MitoTracker Green fluorescent dye (Invitrogen), which passively diffuses across the plasma membrane and accumulates in active mitochondria. As expected, confocal imaging confirmed co-localization of the mito::mKate2 and MitoTracker Green fluorescence indicating that mito::mKate2 is specifically targeted to the mitochondria. This was further quantified by measuring the fluorescence intensity of a subset of mito::mKate2+ and MitoTracker Green+ pixels (red line), which showed a positive correlation (R= 0.97) (Figure 3A). The same MitoTracker Green labeling and analysis was performed on mouse embryonic fibroblasts (MEFs) derived from E12.5–E13.5 mito::mKate2+/tg transgenic embryos, and a positive correlation was again observed (R= 0.91) (Figure 3B). To confirm mito::mKate2 mitochondrial fluorescence in vivo, we performed retinal immunofluorescence labeling with an antibody against succinate dehydrogenase (SDH), an electron transport chain protein that is localized to the inner mitochondrial membrane. Both SDH and mito::mKate2 labeled the densely packed mitochondria within the inner segments of the photoreceptors and the mitochondria in the ONL. Co-localization quantification showed a positive correlation between pixel intensities (R= 0.80) (Figure 3C), confirming mitochondrial localization of mito::mKate2 in vivo.
Figure 3. Co-localization of mito::mKate2 fluorescence and mitochondrial-specific markers.
HeLa cells transfected with the CAG-mito::mKate2 plasmid and labeled with MitoTracker Green (A). Mito::mKate2 transgenic MEFs co-labeled with MitoTracker Green (B). Mito::mKate2 transgenic retinal cryosections stained with anti-SDH antibodies (C). Red lines demarcate pixels used for co-localization analyses. n = 3. See text for details.
Mito::mKate2 is useful for genetic mosaic analysis of genes impacting mitochondrial dynamics and structure
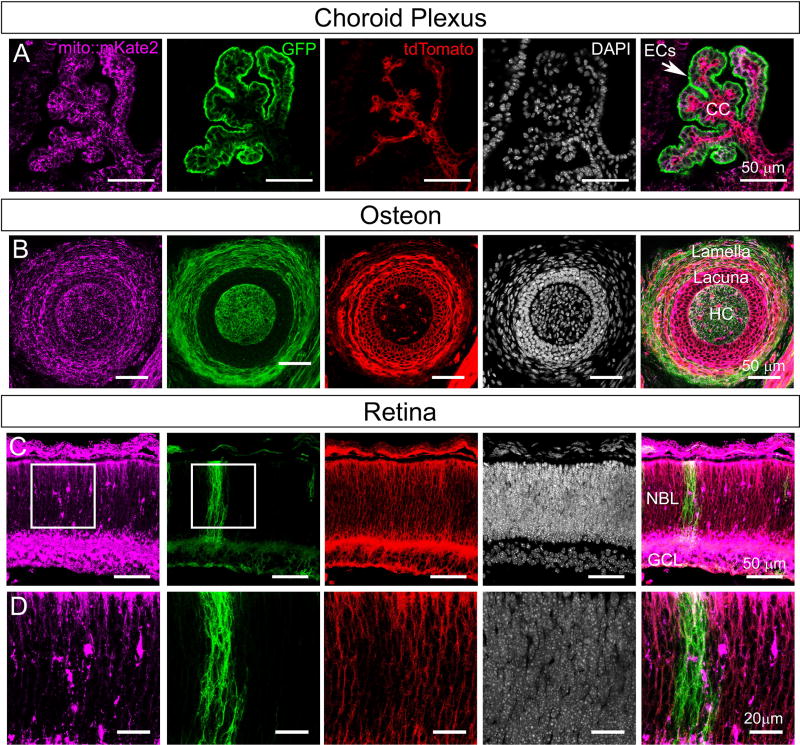
As a proof-of-concept experiment to show that mito::mKate2 can be integrated into genetic mosaic analyses of mitochondrial phenotypes, we crossed mito::mKate2 mice to the ROSA26R-mTmG dual fluorescent Cre reporter line. These mice ubiquitously express a membrane-tethered tandem dimer Tomato protein (tdTomato) prior to Cre recombination and a membrane-tethered GFP upon Cre-mediated recombination of the floxed tdTomato-stop cassette (Muzumdar et al., 2007). We crossed these mice to the Platelet-derived growth factor receptor α Cre line (Pdgfrα-Cre), which mosaically expresses Cre in a variety of tissues (Roesch et al., 2008). Whole heads from postnatal day 0 (P0) Pdgfrα-Cre+/tg; ROSA26R-mTmG+/tg; mito::mKate2+/tg mice were cryosectioned and imaged with confocal microscopy. We focused on tissues with previously reported Pdgfrα expression (Betsholtz, 1995; Orr-Urtreger et al., 1992; Orr-Urtreger and Lonai, 1992). Distinct mito::mKate2, GFP and tdTomato fluorescent signals were detected in the choroid plexus of the developing hindbrain (Figure 4A) and the osteon of the facial bones (Figure 4B). In the case of the retina, a discrete clone of GFP+ cells was observed in the neuroblastic layer (NBL) (Figure 4C–D) and we were able to visualize mitochondria in both the GFP+ and adjacent tdTomato+ regions. In total, these data suggest that the mito::mKate2 mouse will be useful for in vivo mosaic analysis utilizing the ROSA26R-mTmG Cre reporter. By using this reporter system in conditional knockout experiments, researchers will be able to simultaneously analyze both mutant and wild type cells, in the same tissue, for changes in mitochondrial localization and morphology. This approach can also extend to phenotyping of mitochondrial dynamics by live imaging of tissue explant and slice cultures.
Figure 4. Mito::mKate2 fluorescence can be spectrally separated from the ROSA26R-mTmG dual fluorescent Cre reporter.
Postnatal day 0 head cryosections from Pdgfrα-Cre+/tg; ROSA26R-mTmG+/tg; mito::mKate2+/tg mice showing distinct mKate2, GFP and tdTomato fluorescent signals in the choroid plexus (A), osteon (B), and retina (C–D). EC= Ependymal Cells, CC= Choroid Capillaries, HC= Haversian Canal, NBL= Neuroblastic Layer, and GCL= Ganglion Cell Layer. n = 3. See text for details.
Mito::mKate2 mice can be used to monitor coupled mitochondrial dynamics and cell cycle kinetics
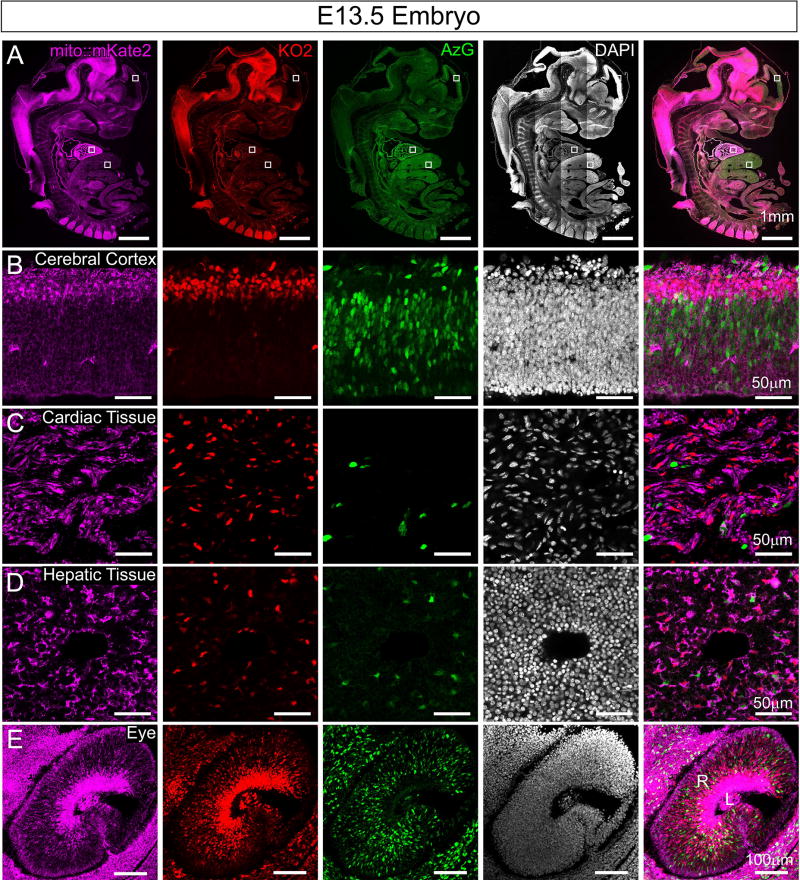
To determine whether we can detect mito::mKate2 expression coincident with cell cycle stage, we generated Fucci-AzG+/tg; Fucci-KO2+/tg; mito::mKate2+/tg triple transgenic mice. Fucci is a previously developed cell cycle reporter system that utilizes two fluorescent proteins - a variant of monomeric Kusabira Orange (KO2) to label nuclei of cells during G1 phase and monomeric Azami Green (AzG) to label nuclei of cells during late S and G2 phase. Cells in early S phase express both proteins (Sakaue-Sawano et al., 2008). Confocal imaging of cryosections from E13.5 embryos showed clear spectral separation of mito::mKate2, Fucci-KO2 and Fucci-AzG fluorescent signals (Figure 5A). Higher magnification images of the cortex, heart, liver and eye show distinct mitochondrial morphologies and localization (Figure 5B–E). In the developing cortex and retina, expression of the Fucci-KO2 reporter is also expressed in cells that have exited the cell cycle (Sakaue-Sawano et al., 2008). In these cells, the mito::mKate2+ mitochondria appear more densely packed as compared to the Fucci-AzG+ regions that are comprised mostly of proliferating progenitor cells (Figure 5B and E).
Figure 5. Mito::mKate2 fluorescence can be spectrally separated from the Fucci cell cycle reporter.
E13.5 embryo cryosections from Fucci-AzG+/tg; Fucci-KO2+/tg; mito::mKate2+/tg mice highlighting distinct fluorescence signals in the mKate2, KO2 and AzG channels throughout the embryo (A). High magnification images of the cerebral cortex (B), heart (C), liver (D) and eye (E). R= Retina and L = Lens. n = 3. See text for details.
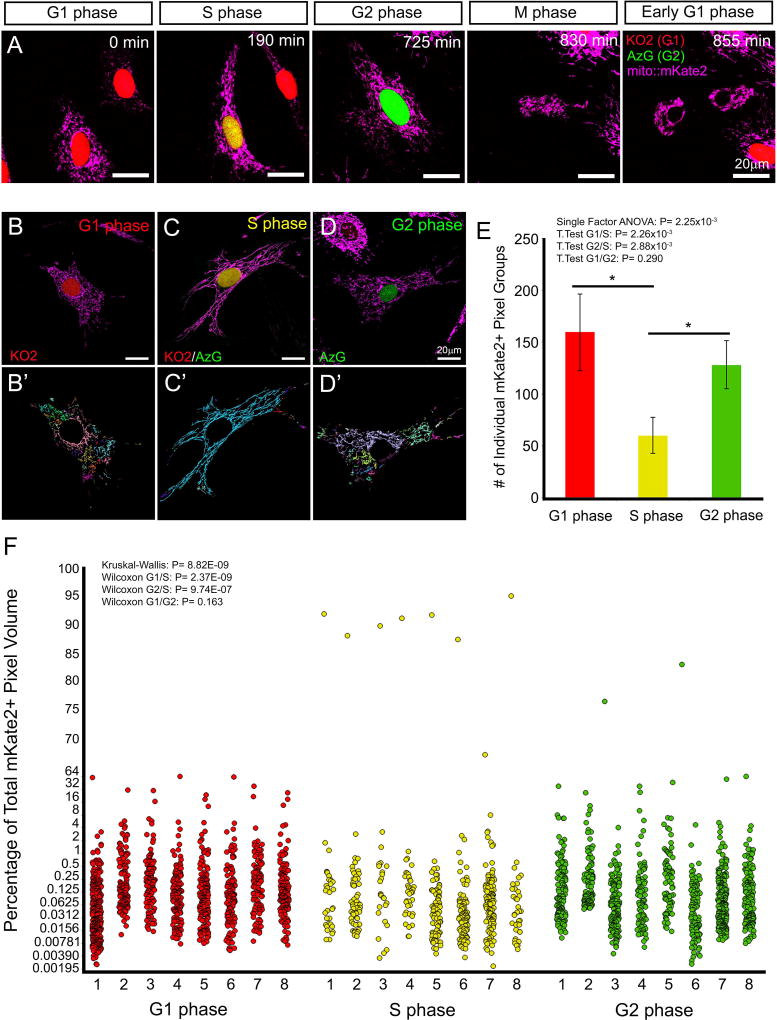
To illustrate the utility of the mito::mKate2 reporter for tracking mitochondrial dynamics at different cell cycle stages, we next isolated MEFs from Fucci-AzG+/tg; Fucci-KO2+/tg; mito::mKate2+/tg embryos and performed live confocal imaging (Supplemental Movie 4). We were able to monitor changes in mitochondrial structure throughout the cell cycle. During early S phase, mitochondria appeared to take on a hyperfused state as described previously (Figure 6A) (Mitra et al., 2009). To determine whether we can detect quantifiable, cell cycle-specific changes in mitochondrial morphology, we obtained 3-dimensional Z-stack images of MEFs in G1, early S and G2 phase (Figure 6B–D). As a measure of the fused state of the mitochondria, we then performed a connected component analysis (Imaris) to identify individual groups of mKate2+ pixels. The number of individual mKate2+ pixel groups and volume of each group were quantified and compared among cells in different cell cycle phases. As a visual representation of the quantification, individual connected components were randomly color-coded (Figure 6B’-D’). The early S phase cells exhibited fewer individual mKate2+ pixel groups (Figure 6E) and generally had 1 large pixel group that made up over 85% of the total pixel volume (Figure 6F). These data are consistent with the hyperfused early S phase mitochondrial network as compared to G1 and G2 phase and confirm mito::Kate2 as a faithful reporter to track mitochondrial dynamics during the cell cycle.
Figure 6. Live imaging and structural quantification of mKate2+ mitochondrial dynamics coupled to cell cycle kinetics.
Stills from a time-lapse movie of MEFs, derived from Fucci-AzG+/tg; Fucci-KO2+/tg; mito::mKate2+/tg embryos, showing changes in mitochondrial fission and fusion coincident with specific cell cycle stages (A). Connected component analysis (Imaris) of mitochondrial networks, during G1, S and G2 phase (B–D’). The number of individual mKate2+ pixel groups (E) and volume of each group (F) were quantified. As a visual representation of the quantification, individual connected components were randomly color-coded (B’–D’). n = 8 per cell cycle stage. See text for details.
CONCLUSIONS
We have introduced and validated the mito::mKate2 mouse as a new, far-red fluorescent reporter line that allows for simultaneous monitoring of mitochondrial dynamics and additional reporters in the blue/green/red spectrum. To illustrate this, we crossed mito::mKate2+/tg mice to the Fucci-AzG+/tg; Fucci-KO2+/tg cell cycle reporter line and demonstrated that mito::mKate2 can be used to monitor changes in mitochondrial fission and fusion as cells transition through different stages of the cell cycle. This line will also be useful for genetic mosaic analyses utilizing the ROSA26R-mTmG+/tg Cre reporter by providing visualization of discrete clonal defects impacting mitochondrial structure, distribution and dynamics. Therefore, the mito::mKate2 mouse significantly advances upon the current repertoire of genetic tools for imaging of mitochondrial activity in living tissues.
METHODS
Generation of CAG-mito::mKate2+/tg mice
A cassette encoding a mitochondrial targeting sequence, derived from subunit VIII of cytochrome c oxidase, fused to the N-terminal of mKate2 was released from the pmKate2-mito plasmid (Evrogen) by digestion with NheI followed by blunting with T4 DNA polymerase and subsequent digestion with NotI. This ~800 bp fragment was then ligated to the pCAGEN plasmid (Addgene plasmid #11160, a gift from Connie Cepko) that was digested with EcoRV and NotI. The resulting plasmid containing the transgenic construct was tested by transient transfection of HeLa cells, which resulted in bright and specific labeling of mitochondria. The plasmid was then digested with SalI, HindIII and ScaI and the ~3.1 kb CAG-mito::mKate2 transgene fragment, containing the CAG promoter, mito::mKate2 fusion protein, and a polyadenylation tail was purified and microinjected into single-cell C57BL/6J mouse zygotes by the Baylor College of Medicine Genetically Engineered Mouse Core. PCR genotyping of tail DNA revealed 4 positive founder mice out of 29 weanlings total. The following primers were used and generated a 224 bp product: mKate2F 5’-ACCGTGAACAACCACCACTT-3’ and mKate2R 5’-CTCTCCCATGTGAAGCCCTC-3’. The founder mice were bred to CD1 mice (Charles River Lab) for assessment of mito::mKate2 fluorescence. The line exhibiting the brightest fluorescence was also bred to C57BL/6J mice (Jackson Lab) to establish a inbred CAG-mito::mKate2+/tg line for subsequent experiments. CAG-mito::mKate2+/tg mice will be made available to the research community.
CAG-mito::mKate2+/tg mouse tissue collection and preparation
The FDB muscle was collected from 6-week-old mice and digested in Dulbecco’s Modified Eagle’s Medium (DMEM) (Sigma-Aldrich D6429) with 1% Penicillin-Streptomycin (Pen-Strep) (GenDEPOT CA005-010) and Collagenase (5mg/ml) (Sigma-Aldrich C0130) for 2.5 hours. FDB myocyte bundles were transferred to DMEM media with 10% FBS (Sigma-Aldrich F2442) and 1% Pen-Strep, triturated to create single fibers, and incubated overnight at 37°C, 5% CO2. The next day, single fibers were transferred to 8-well Nunc™ Lab-Tek™ Chambers (Thermo Scientific 155411) coated with ECM gel (Sigma-Aldrich E1270). After 30 minutes, myocytes attached to the ECM and media was aspirated and replaced with DMEM (Life Technologies, 21063-029) with 10% FBS and 1% Pen-Strep. To facilitate imaging of fluorescence, phenol red was omitted from the DMEM.
Using Dumont #5 forceps (FST), sperm were squeezed out of the vas deferens isolated from 6-week-old mice and into 1× PBS. Aliquots of sperm (in 1× PBS) were mounted on a coverslip and immediately imaged with confocal microscopy.
Brain, eyes, heart and liver were harvested from 3 week old mice and fixed in 4% paraformaldehyde at 4°C for 1 hour. The tissues were washed with 1× PBS (pH 7.4), incubated overnight in 15% sucrose solution (Sigma-Aldrich S9378) in 1× PBS at 4° and then transferred to 30% sucrose at 4°C for an additional night. The tissue was then submerged in OCT compound (Sakura 4583), frozen over liquid NO2, and cryosectioned into 20µm sections. Tissue sections were washed with 1× PBS and coverslips mounted with Fluoromount-G (Southern Biotech, #0100-01) prior to confocal microscopy. Whole heads from P0 Pdgfrα-Cre+/tg; ROSA26R-mTmG+/tg; mito::mKate2+/tg mice and E13.5 Fucci-AzG+/tg; Fucci-KO2+/tg; mito::mKate2+/tg embryos were prepared and imaged similarly.
CAG-mito::mKate2+/tg Mouse Embryonic Fibroblast Isolation and Culture
Embryos were collected at E11.5–E15.5 and fluorescent embryos were identified with a stereo microscope (Zeiss Stemi 2000) equipped with a Nightsea Fluorescence Adapter. Fucci-AzG+/tg; Fucci-KO2+/tg; mito::mKate2+/tg embryos were decapitated and stripped of internal organs. The remaining tissue was dissociated in 0.25% trypsin-EDTA (GenDEPOT CA014-010) to isolate single cells. DMEM with 20% FBS and 1% Antibiotic-Antimycotic (GenDEPOT CA002-010) or Pen-Strep was added to the dissociated tissue and the combined media and the dissociated tissue was transferred to 15 ml Falcon tube. Once the larger tissue pieces settled to the bottom, the top layer of cells in solution was transferred to a cell culture dish (Corning 353003) and maintained at 37°C and 5% CO2. To prepare for imaging, MEFs were washed with sterile 1× PBS, incubated in trypsin-EDTA, then plated on an 8-well chambered cover glass and cultured to ~ 60% confluency.
Fluorescence co-localization analyses
Mito::mKate2 transfected HeLa cells or transgenic MEFs were incubated (30 minutes at 37°C and 5% CO2) in the presence of 100nM MitoTracker Green FM dye (Invitrogen M7514) diluted in serum-free DMEM. After incubation, the cells were washed, placed in normal DMEM, and immediately imaged on a confocal microscope equipped with an environmental chamber maintained at 37°C and 5% CO2.
Immunofluorescence with anti-SDH antibodies (Invitrogen #459230) was performed on retinal cryosections. Tissue was washed in 1× PBS (pH 7.4) and blocked for 1 hour at room temperature in 10% goat serum with 0.3% Triton X. This was followed by incubation (48 hours at 4°C) with anti-SDH antibodies diluted in blocking buffer (1:200). The tissue was again washed with 1× PBS and incubated with goat, anti-mouse IgG Alexafluor 488 secondary antibodies (Invitrogen A11029) diluted in blocking buffer (1:400). Coverslips were mounted over tissue with Fluoromount-G (Southern Biotech #0100-01).
Quantification of co-localization was performed on 2D images using the Profile function in ZEN software (Zeiss Inc.). Specifically, correlation between the changes in pixel intensity of the two fluorescent markers was quantified for all pixels along a line drawn through the fluorescent structures. The Pearson’s Correlation Coefficient (R) was calculated using Microsoft Excel.
Image acquisition and quantification of mitochondrial structure
Imaging of fluorescent reporters was performed using a Zeiss LSM 780 inverted confocal microscope equipped with a 34 channel spectral array and 405, 458, 488, 514, 561, 594, and 633 nm laser lines. For high magnification views of mitochondrial structure in tissues and cells, images were acquired using a C-Apochromat 40×/1.20 W Korr. or a Plan-Apochromat 63×/1.40 Oil DIC objective. For spectral fingerprinting, transfected HeLa cells were excited with a 561 nm laser and imaged with a Plan-Apochromat 63×/1.40 Oil DIC objective. Using the Lambda stack function in ZEN software, emission signal intensities were measured between 415–690 nm at 9 nm intervals, and emission spectra from mito::mKate2-positive and mito::mKate2-negative regions were compared. For low magnification views of whole embryos or tissue sections, an EC Plan-Neofluar 10×/0.30 or Plan-Apochromat 20×/0.8 was used.
For time-lapse imaging, the Zeiss LSM 780 was outfitted with an environmental chamber maintained at 37°C and 5% CO2. Using the C-Apochromat 40×/1.20 W Korr. objective, image Z-stacks were collected every 5 minutes for up to 24 hours with the following laser powers: 1% 561 nm (mito::mKate2), 2% 488 nm (Fucci-AzG), 2% 514 nm (Fucci-KO2).
For quantification of mitochondrial structure within triple transgenic MEFs, image Z stacks, with a step size of 0.28 µm, were acquired for the entire 3D volume of each individual cell. The Z-stack images were subsequently processed using adaptive thresholding. Then, using the surface creation function in Imaris (Bitplane), a connected component analysis of 3D cell reconstructions was performed. This allowed for identification of groups of individual mKate2+ pixels and the measurement of pixel group volume. Individual connected components were randomly color coded to provide a visual representation of the quantification.
To determine whether there was a statistically significant difference in the number of mKate2+ pixel groups among the three cell cycle phases analyzed, we performed a single factor ANOVA (P= 2.25×10−3) and subsequent T Tests (G1/S: P= 2.26×10−3; G2/S: P= 2.88×10−3; G1/G2: P= 0.290). To determine whether mKate2+ pixel group volume data were normally distributed we performed a Kolmogorov-Smirnov test for each distribution (G1: P= 2.89×10−280; S: P= 3.39×10−106; G2: P= 1.13×10−225). Since the data were non-parametric, we performed a Kruskal-Wallis test (P= 8.82×10−9) to determine whether there was a difference among the distributions and subsequent Wilcoxon tests (G1/S: P= 2.37×10−9; G2/S: P= 9.74×10−7; G1/G2: P= 0.163) to determine which distributions were significantly different from each other.
For time-lapse imaging of HeLa cell mitochondrial fission and fusion, cells were cultured in DMEM with no phenol red, 10% FBS, and 1% Pen-Strep. Imaging was performed on a Zeiss LSM 780 outfitted with an environmental chamber maintained at 37°C and 5% CO2. To capture discrete fission events in transfected HeLa cells, a C-Apochromat 40×/1.20 W Korr. objective was used. Z-stack images were collected every minute with the 561 nm laser at a power of 1%. Z-stacks encompassed the entire cell volume and mitochondrial network with a step size of 0.53 um.
Supplementary Material
A MEF, derived from Fucci-AzG+/tg; Fucci-KO2+/tg; mito::mKate2+/tg embryos, as it transitions from G1 to S to G2 to M phase. Follow the cell marked by a white square.
Acknowledgments
We thank Dr. Shang Wang, Dr. Irina Larina and the labs of Dr. Mary Dickinson, and Dr. George Rodney for technical support and members of the Poché lab for critical reading of this manuscript. We also thank the Optical Imaging and Vital Microscopy (OIVM) and Genetically Engineered Mouse (GEM) Core facilities at Baylor College of Medicine for their services. This work was supported by grants from the National Institutes of Health (NIH): R01 EY024906 to Ross Poché and T32 EY007102 to Graeme Mardon.
References
- Abe T, Kiyonari H, Shioi G, Inoue K, Nakao K, Aizawa S, Fujimori T. Establishment of conditional reporter mouse lines at ROSA26 locus for live cell imaging. Genesis. 2011;49:579–590. doi: 10.1002/dvg.20753. [DOI] [PubMed] [Google Scholar]
- Betsholtz C. Role of platelet-derived growth factors in mouse development. Int J Dev Biol. 1995;39:817–825. [PubMed] [Google Scholar]
- Boncompagni S, Rossi AE, Micaroni M, Beznoussenko GV, Polishchuk RS, Dirksen RT, Protasi F. Mitochondria are linked to calcium stores in striated muscle by developmentally regulated tethering structures. Mol Biol Cell. 2009;20:1058–1067. doi: 10.1091/mbc.E08-07-0783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breckwoldt MO, Pfister FM, Bradley PM, Marinkovic P, Williams PR, Brill MS, Plomer B, Schmalz A, St Clair DK, Naumann R, Griesbeck O, Schwarzlander M, Godinho L, Bareyre FM, Dick TP, Kerschensteiner M, Misgeld T. Multiparametric optical analysis of mitochondrial redox signals during neuronal physiology and pathology in vivo. Nat Med. 2014;20:555–560. doi: 10.1038/nm.3520. [DOI] [PubMed] [Google Scholar]
- Chan DC. Mitochondria: dynamic organelles in disease, aging, and development. Cell. 2006;125:1241–1252. doi: 10.1016/j.cell.2006.06.010. [DOI] [PubMed] [Google Scholar]
- Chandrasekaran K, Hazelton JL, Wang Y, Fiskum G, Kristian T. Neuron-specific conditional expression of a mitochondrially targeted fluorescent protein in mice. J Neurosci. 2006;26:13123–13127. doi: 10.1523/JNEUROSCI.4191-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CC, You JY, Lung J, Huang CE, Chen YY, Leu YW, Ho HY, Li CP, Lu CH, Lee KD, Hsu CC, Gau JP. Aberrant let7a/HMGA2 signaling activity with unique clinical phenotype in JAK2-mutated myeloproliferative neoplasms. Haematologica. 2017;102:509–518. doi: 10.3324/haematol.2016.154385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claude A, Fullam EF. The Preparation of Sections of Guinea Pig Liver for Electron Microscopy. J Exp Med. 1946;83:499–503. [PMC free article] [PubMed] [Google Scholar]
- Cloonan SM, Choi AM. Mitochondria: sensors and mediators of innate immune receptor signaling. Curr Opin Microbiol. 2013;16:327–338. doi: 10.1016/j.mib.2013.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen AI. The fine structure of the extrafoveal receptors of the Rhesus monkey. Exp Eye Res. 1961;1:128–136. doi: 10.1016/s0014-4835(61)80018-3. [DOI] [PubMed] [Google Scholar]
- Collins TJ, Berridge MJ, Lipp P, Bootman MD. Mitochondria are morphologically and functionally heterogeneous within cells. The EMBO Journal. 2002;21:1616–1627. doi: 10.1093/emboj/21.7.1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Michele R, Carimi F, Frommer WB. Mitochondrial biosensors. Int J Biochem Cell Biol. 2014;48:39–44. doi: 10.1016/j.biocel.2013.12.014. [DOI] [PubMed] [Google Scholar]
- Fawcett DW. The mammalian spermatozoon. Dev Biol. 1975;44:394–436. doi: 10.1016/0012-1606(75)90411-x. [DOI] [PubMed] [Google Scholar]
- Flippo KH, Strack S. Mitochondrial dynamics in neuronal injury, development and plasticity. J Cell Sci. 2017;130:671–681. doi: 10.1242/jcs.171017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray MW. Lynn Margulis and the endosymbiont hypothesis: 50 years later. Mol Biol Cell. 2017;28:1285–1287. doi: 10.1091/mbc.E16-07-0509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hailey DW, Rambold AS, Satpute-Krishnan P, Mitra K, Sougrat R, Kim PK, Lippincott-Schwartz J. Mitochondria supply membranes for autophagosome biogenesis during starvation. Cell. 2010;141:656–667. doi: 10.1016/j.cell.2010.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasuwa H, Muro Y, Ikawa M, Kato N, Tsujimoto Y, Okabe M. Transgenic mouse sperm that have green acrosome and red mitochondria allow visualization of sperm and their acrosome reaction in vivo. Exp Anim. 2010;59:105–107. doi: 10.1538/expanim.59.105. [DOI] [PubMed] [Google Scholar]
- Herndon RM. The fine structure of the Purkinje cell. J Cell Biol. 1963;18:167–180. doi: 10.1083/jcb.18.1.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn SR, Thomenius MJ, Johnson ES, Freel CD, Wu JQ, Coloff JL, Yang CS, Tang W, An J, Ilkayeva OR, Rathmell JC, Newgard CB, Kornbluth S. Regulation of mitochondrial morphology by APC/CCdh1-mediated control of Drp1 stability. Mol Biol Cell. 2011;22:1207–1216. doi: 10.1091/mbc.E10-07-0567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kisch B. Electron microscopy of the capillary wall. Exp Med Surg. 1956;14:113–121. [PubMed] [Google Scholar]
- Kuznetsov AV, Margreiter R. Heterogeneity of mitochondria and mitochondrial function within cells as another level of mitochondrial complexity. Int J Mol Sci. 2009;10:1911–1929. doi: 10.3390/ijms10041911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuznetsov AV, Troppmair J, Sucher R, Hermann M, Saks V, Margreiter R. Mitochondrial subpopulations and heterogeneity revealed by confocal imaging: possible physiological role? Biochim Biophys Acta. 2006;1757:686–691. doi: 10.1016/j.bbabio.2006.03.014. [DOI] [PubMed] [Google Scholar]
- Magrane J, Cortez C, Gan WB, Manfredi G. Abnormal mitochondrial transport and morphology are common pathological denominators in SOD1 and TDP43 ALS mouse models. Hum Mol Genet. 2014;23:1413–1424. doi: 10.1093/hmg/ddt528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal S, Freije WA, Guptan P, Banerjee U. Metabolic control of G1-S transition: cyclin E degradation by p53-induced activation of the ubiquitin-proteasome system. J Cell Biol. 2010;188:473–479. doi: 10.1083/jcb.200912024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margineantu DH, Gregory Cox W, Sundell L, Sherwood SW, Beechem JM, Capaldi RA. Cell cycle dependent morphology changes and associated mitochondrial DNA redistribution in mitochondria of human cell lines. Mitochondrion. 2002;1:425–435. doi: 10.1016/s1567-7249(02)00006-5. [DOI] [PubMed] [Google Scholar]
- McBride HM, Neuspiel M, Wasiak S. Mitochondria: more than just a powerhouse. Curr Biol. 2006;16:R551–560. doi: 10.1016/j.cub.2006.06.054. [DOI] [PubMed] [Google Scholar]
- McWilliams TG, Prescott AR, Allen GF, Tamjar J, Munson MJ, Thomson C, Muqit MM, Ganley IG. mito-QC illuminates mitophagy and mitochondrial architecture in vivo. J Cell Biol. 2016;214:333–345. doi: 10.1083/jcb.201603039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misgeld T, Kerschensteiner M, Bareyre FM, Burgess RW, Lichtman JW. Imaging axonal transport of mitochondria in vivo. Nat Methods. 2007;4:559–561. doi: 10.1038/nmeth1055. [DOI] [PubMed] [Google Scholar]
- Mishra P, Chan DC. Mitochondrial dynamics and inheritance during cell division, development and disease. Nat Rev Mol Cell Biol. 2014;15:634–646. doi: 10.1038/nrm3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra K. Mitochondrial fission-fusion as an emerging key regulator of cell proliferation and differentiation. Bioessays. 2013;35:955–964. doi: 10.1002/bies.201300011. [DOI] [PubMed] [Google Scholar]
- Mitra K, Wunder C, Roysam B, Lin G, Lippincott-Schwartz J. A hyperfused mitochondrial state achieved at G1-S regulates cyclin E buildup and entry into S phase. Proc Natl Acad Sci U S A. 2009;106:11960–11965. doi: 10.1073/pnas.0904875106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazaki J, Takaki S, Araki K, Tashiro F, Tominaga A, Takatsu K, Yamamura K. Expression vector system based on the chicken beta-actin promoter directs efficient production of interleukin-5. Gene. 1989;79:269–277. doi: 10.1016/0378-1119(89)90209-6. [DOI] [PubMed] [Google Scholar]
- Muzumdar MD, Tasic B, Miyamichi K, Li L, Luo L. A global double-fluorescent Cre reporter mouse. Genesis. 2007;45:593–605. doi: 10.1002/dvg.20335. [DOI] [PubMed] [Google Scholar]
- Olney JW. An electron microscopic study of synapse formation, receptor outer segment development, and other aspects of developing mouse retina. Invest Ophthalmol. 1968;7:250–268. [PubMed] [Google Scholar]
- Orr-Urtreger A, Bedford MT, Do MS, Eisenbach L, Lonai P. Developmental expression of the alpha receptor for platelet-derived growth factor, which is deleted in the embryonic lethal Patch mutation. Development. 1992;115:289–303. doi: 10.1242/dev.115.1.289. [DOI] [PubMed] [Google Scholar]
- Orr-Urtreger A, Lonai P. Platelet-derived growth factor-A and its receptor are expressed in separate, but adjacent cell layers of the mouse embryo. Development. 1992;115:1045–1058. doi: 10.1242/dev.115.4.1045. [DOI] [PubMed] [Google Scholar]
- Owusu-Ansah E, Yavari A, Mandal S, Banerjee U. Distinct mitochondrial retrograde signals control the G1-S cell cycle checkpoint. Nat Genet. 2008;40:356–361. doi: 10.1038/ng.2007.50. [DOI] [PubMed] [Google Scholar]
- Parker DJ, Iyer A, Shah S, Moran A, Hjelmeland AB, Basu MK, Liu R, Mitra K. A new mitochondrial pool of cyclin E, regulated by Drp1, is linked to cell-density-dependent cell proliferation. J Cell Sci. 2015;128:4171–4182. doi: 10.1242/jcs.172429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham AH, McCaffery JM, Chan DC. Mouse lines with photo-activatable mitochondria to study mitochondrial dynamics. Genesis. 2012;50:833–843. doi: 10.1002/dvg.22050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickles S, Cadieux-Dion M, Alvarez JI, Lecuyer MA, Peyrard SL, Destroismaisons L, St-Onge L, Terouz S, Cossette P, Prat A, Vande Velde C. Endo-MitoEGFP mice: a novel transgenic mouse with fluorescently marked mitochondria in microvascular endothelial cells. PLoS One. 2013;8:e74603. doi: 10.1371/journal.pone.0074603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poche RA, Zhang M, Rueda EM, Tong X, McElwee ML, Wong L, Hsu CW, Dejosez M, Burns AR, Fox DA, Martin JF, Zwaka TP, Dickinson ME. RONIN Is an Essential Transcriptional Regulator of Genes Required for Mitochondrial Function in the Developing Retina. Cell Rep. 2016;14:1684–1697. doi: 10.1016/j.celrep.2016.01.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao X, Zhang L, Gamper AM, Fujita T, Wan Y. APC/C-Cdh1: from cell cycle to cellular differentiation and genomic integrity. Cell Cycle. 2010;9:3904–3912. doi: 10.4161/cc.9.19.13585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzuto R, Brini M, Pizzo P, Murgia M, Pozzan T. Chimeric green fluorescent protein as a tool for visualizing subcellular organelles in living cells. Curr Biol. 1995;5:635–642. doi: 10.1016/s0960-9822(95)00128-x. [DOI] [PubMed] [Google Scholar]
- Rizzuto R, Nakase H, Darras B, Francke U, Fabrizi GM, Mengel T, Walsh F, Kadenbach B, DiMauro S, Schon EA. A gene specifying subunit VIII of human cytochrome c oxidase is localized to chromosome 11 and is expressed in both muscle and non-muscle tissues. J Biol Chem. 1989;264:10595–10600. [PubMed] [Google Scholar]
- Roesch K, Jadhav AP, Trimarchi JM, Stadler MB, Roska B, Sun BB, Cepko CL. The transcriptome of retinal Muller glial cells. J Comp Neurol. 2008;509:225–238. doi: 10.1002/cne.21730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rueda EM, Johnson JE, Jr, Giddabasappa A, Swaroop A, Brooks MJ, Sigel I, Chaney SY, Fox DA. The cellular and compartmental profile of mouse retinal glycolysis, tricarboxylic acid cycle, oxidative phosphorylation, and ~P transferring kinases. Mol Vis. 2016;22:847–885. [PMC free article] [PubMed] [Google Scholar]
- Sagan L. On the origin of mitosing cells. J Theor Biol. 1967;14:255–274. doi: 10.1016/0022-5193(67)90079-3. [DOI] [PubMed] [Google Scholar]
- Sakaue-Sawano A, Kurokawa H, Morimura T, Hanyu A, Hama H, Osawa H, Kashiwagi S, Fukami K, Miyata T, Miyoshi H, Imamura T, Ogawa M, Masai H, Miyawaki A. Visualizing spatiotemporal dynamics of multicellular cell-cycle progression. Cell. 2008;132:487–498. doi: 10.1016/j.cell.2007.12.033. [DOI] [PubMed] [Google Scholar]
- Shcherbo D, Merzlyak EM, Chepurnykh TV, Fradkov AF, Ermakova GV, Solovieva EA, Lukyanov KA, Bogdanova EA, Zaraisky AG, Lukyanov S, Chudakov DM. Bright far-red fluorescent protein for whole-body imaging. Nat Methods. 2007;4:741–746. doi: 10.1038/nmeth1083. [DOI] [PubMed] [Google Scholar]
- Shcherbo D, Murphy CS, Ermakova GV, Solovieva EA, Chepurnykh TV, Shcheglov AS, Verkhusha VV, Pletnev VZ, Hazelwood KL, Roche PM, Lukyanov S, Zaraisky AG, Davidson MW, Chudakov DM. Far-red fluorescent tags for protein imaging in living tissues. Biochem J. 2009;418:567–574. doi: 10.1042/BJ20081949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shitara H, Kaneda H, Sato A, Iwasaki K, Hayashi J, Taya C, Yonekawa H. Non-invasive visualization of sperm mitochondria behavior in transgenic mice with introduced green fluorescent protein (GFP) FEBS Lett. 2001;500:7–11. doi: 10.1016/s0014-5793(01)02574-1. [DOI] [PubMed] [Google Scholar]
- Siekevitz P. "Powerhouse of the cell". Scientific American. 1957;197:131–140. [Google Scholar]
- Stehling O, Wilbrecht C, Lill R. Mitochondrial iron-sulfur protein biogenesis and human disease. Biochimie. 2014;100:61–77. doi: 10.1016/j.biochi.2014.01.010. [DOI] [PubMed] [Google Scholar]
- Stotland A, Gottlieb RA. alpha-MHC MitoTimer mouse: In vivo mitochondrial turnover model reveals remarkable mitochondrial heterogeneity in the heart. J Mol Cell Cardiol. 2016;90:53–58. doi: 10.1016/j.yjmcc.2015.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun N, Yun J, Liu J, Malide D, Liu C, Rovira II, Holmstrom KM, Fergusson MM, Yoo YH, Combs CA, Finkel T. Measuring In Vivo Mitophagy. Mol Cell. 2015;60:685–696. doi: 10.1016/j.molcel.2015.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taguchi N, Ishihara N, Jofuku A, Oka T, Mihara K. Mitotic phosphorylation of dynamin-related GTPase Drp1 participates in mitochondrial fission. J Biol Chem. 2007;282:11521–11529. doi: 10.1074/jbc.M607279200. [DOI] [PubMed] [Google Scholar]
- Tait SW, Green DR. Mitochondrial regulation of cell death. Cold Spring Harb Perspect Biol. 2013;5 doi: 10.1101/cshperspect.a008706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vande Velde C, McDonald KK, Boukhedimi Y, McAlonis-Downes M, Lobsiger CS, Bel Hadj S, Zandona A, Julien JP, Shah SB, Cleveland DW. Misfolded SOD1 associated with motor neuron mitochondria alters mitochondrial shape and distribution prior to clinical onset. PLoS One. 2011;6:e22031. doi: 10.1371/journal.pone.0022031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Pan Y, Price A, Martin LJ. Generation and characterization of transgenic mice expressing mitochondrial targeted red fluorescent protein selectively in neurons: modeling mitochondriopathy in excitotoxicity and amyotrophic lateral sclerosis. Mol Neurodegener. 2011;6:75. doi: 10.1186/1750-1326-6-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi J, Nishiyama S, Shimanuki M, Ono T, Sato A, Nakada K, Hayashi J, Yonekawa H, Shitara H. Comprehensive application of an mtDsRed2-Tg mouse strain for mitochondrial imaging. Transgenic Res. 2012;21:439–447. doi: 10.1007/s11248-011-9539-1. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Avalos JL. Traditional and novel tools to probe the mitochondrial metabolism in health and disease. Wiley Interdiscip Rev Syst Biol Med. 2017;9 doi: 10.1002/wsbm.1373. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A MEF, derived from Fucci-AzG+/tg; Fucci-KO2+/tg; mito::mKate2+/tg embryos, as it transitions from G1 to S to G2 to M phase. Follow the cell marked by a white square.