Abstract
Flow cytometry protocols designed to identify mouse eosinophils typically target Siglec F, an α-2,3-sialic acid binding transmembrane protein expressed universally on cells of this lineage. While a convenient target, antibody-mediated ligation of Siglec F induces eosinophil apoptosis, which limits its usefulness for isolations that are to be followed by functional and/or gene expression studies. We present here a method for FACS isolation which does not target Siglec F and likewise utilizes no antibodies targeting IL5Rα (CD125) or CCR3. Single cell suspensions are prepared from lungs of mice that were sensitized and challenged with Aspergillus fumigatus antigens; eosinophils were identified and isolated by FACS as live SSChi/FSChi CD11c-Gr1-/loMHCII- cells. This strategy was also effective for eosinophil isolation from the lungs of IL5tg mice. Purity by visual inspection of stained cytospin preparations and by Siglec F-diagnostic flow cytometry was 98 − 99% and 97 − 99%, respectively. Eosinophils isolated by this method (yield, ~ 4 × 106 / mouse) generated high-quality RNA suitable for gene expression analysis.
Keywords: Inflammation, Interleukin-5, Allergen
1.0 Introduction
While long-perceived as end-stage cells with limited function, eosinophils are now appreciated as immunomodulatory leukocytes with complex roles in health and disease [1, 2]. Recent studies have underscored several unanticipated features of tissue eosinophils, notably, their heterogeneity and ability to respond to signals from distinct tissue microenvironments [3 - 6].
In order to explore these issues further, it will be critical to have a means to isolate live eosinophils for comparative gene expression and functional studies. There are several protocols available for flow cytometry and FACS isolation of eosinophils generated by our lab and by others (for example, [7 - 10]). These protocols typically target eosinophil-specific or eosinophil-enriched cell surface receptors. Among the most prominent of these receptors is Sialic acid-binding immunoglobulin-type lectin F (Siglec F), a type I transmembrane protein with an immunoreceptor tyrosine-based inhibitory motif (ITIM) that binds α-2,3-linked sialic acids and is the functional paralog of human Siglec 8 (reviewed in [11, 12]). Siglec F is expressed universally on mouse eosinophils [11, 12] and provides reliable quantitative evaluation of eosinophils from multiple tissues [7]. While its biological function has not been fully clarified, antibody-mediated ligation via anti-Siglec F induces eosinophil apoptosis both ex vivo and in vivo [13, 14]. As such, anti-Siglec F may be useful as a means to control eosinophil overabundance in allergic responses and may ultimately be an important clinical tool [11]. Nonetheless, this is of course a negative attribute of this antibody, or any antibody that one would choose to use for FACS-mediated isolation of live, healthy eosinophils for functional studies.
Here, we present a FACS isolation protocol that does not target Siglec F. Eosinophils are isolated at high yield, at 97-99% purity and full viability.
2.0 Methods
2.1 Mice
Wild-type BALB/c mice (8-10 weeks old) were from Charles River Laboratories, Frederick, MD. Interleukin-5 transgenic (IL5tg) mice [15] on the BALB/c background are maintained by the NIAID/Taconic consortium and the 14BS vivarium at NIAID. The National Institute of Allergy and Infectious Diseases Division of Intramural Research Animal Care and Use Program, as part of the National Institutes of Health Intramural Research Program, approved the experimental procedures herein as per protocol LAD 8E.
2.2 Allergen sensitization and challenge
Mice under isoflurane anesthesia were sensitized on days 0 and 7 with intraperitoneal injections of Aspergillus fumigatus extract (Af, 20 μg/mouse, HollisterStier) emulsified with aluminum/magnesium hydroxide (ImjectAlum, ThermoFisher) followed by intranasal inoculation with Af (25 μg/mouse in PBS) on days 12, 13, and 14; mice were euthanized and lungs were removed for preparation of single cell suspensions as described below on day 17.
2.3 Preparation of single-cell lung suspensions
Single cell suspensions were prepared from lungs of IL5tg mice and mice sensitized and challenged with A. fumigatus as described above. After perfusion in situ via the right ventricle with phosphate-buffered saline (PBS) with 500 mM EDTA, the lungs were removed from the body cavity, minced and incubated for 90 min at 37°C with RPMI 1640 and 5% fetal calf serum with DNase I (20 mg/mL, Sigma-Aldrich) and Collagenase D (40 mg/mL, Sigma-Aldrich). After incubation, red blood cells were lysed with sterile dH2O, counted on a hemocytometer with trypan blue exclusion to evaluate viability, and placed in PBS with 0.1% bovine serum albumin (BSA, Sigma-Aldrich) prior to fluorescence-activated cells sorting (FACS). The materials used for preparation of single cell suspensions and for all the methods to follow are listed in Table 1 together with the commercial source and catalog numbers.
Table 1. Reagents used in experimental work in this manuscript.
Shown here are reagents as listed in the Methods Section, together with commercial source and current catalog number.
| Application | Reagent | Source | Catalog No. |
|---|---|---|---|
| Animal study | A. fumigatus extract | HollisterStier | 5021JF |
| ImjectAlum | ThermoFisher Scientific | 77161 | |
| Single cell suspension | DNase I | Sigma-Aldrich | 10104159001 |
| Collagenase D | Sigma-Aldrich | 11088882001 | |
| Flow cytometry | anti-CD16/CD32 | BD Biosciences | 553142 |
| near infrared live-dead | ThermoFisher Scientific | L10119 | |
| anti-CD11c alexaFluor 700 | ThermoFisher Scientific | 56-0114-82 | |
| anti-Gr1 APC | BD Biosciences | 553129 | |
| anti-MHCII PE-cyanine 7 | ThermoFisher Scientific | 25-5321-82 | |
| anti-Siglec F PE | BD Biosciences | 552126 | |
| RNA preparation | RNA-protect | Qiagen | 76526 |
| Trizol | Invitrogen | 15596026 | |
| Direct-zol RNA MiniPrep | Zymo Research | R2050 | |
| RNeasy Mini Kit | Qiagen | 74104 | |
| Agilent RNA 6000 Pico Kit | Agilent | 5067-1513 | |
| cDNA synthesis | First-strand cDNA synthesis kit for RT-PCR (AMV) | Roche | 11 483 188 001 |
| qPCR | RT2-preAMP Master Mix (2X) | ThermoFisher Scientific | 4391128 |
| TaqMan Gene expression assay (FAM) Gusb | ThermoFisher Scientific | ID: Mm01197698_m1 Cat. no. 4453320 | |
| TaqMan Gene Expression Master Mix (2X) | ThermoFisher Scientific | 4369016 |
2.4 FACS, flow cytometry, and cellular visualization
Immediately following preparation of single cell suspensions, cells were incubated with Near-Infrared Live-Dead (ThermoFisher) followed by anti-CD16/CD32 (BD Biosciences), anti-CD11c (ThermoFisher), anti-Gr1 (BD Biosciences), and anti-MHCII (ThermoFisher) at 1 μL per 106 cells. Cells categorized as CD11c-Gr1-/loMHCII- were isolated via fluorescence-activated cell sorting (FACS) on a FACSAria II (BD Biosciences) and sorted into PBS with 0.1% bovine serum albumin (BSA). Purity was assessed by visualization via cytospin preparation stained with modified Giemsa (Diff-Quik; ThermoFisher). Other samples were incubated with anti-Siglec F and evaluated by flow cytometry.
2.5 RNA isolation from FACS-derived eosinophils
Eosinophils isolated via FACS were sorted directly into RNAprotect (Qiagen) at 750 μL per 1.5 × 105 cells eluted. Cells were then lysed with cold Trizol (Invitrogen) and 100% ethanol. RNA was then isolated with the Direct-zol RNA Miniprep Kit (Zymo Research) and purified with the RNeasy Mini Kit (Qiagen) as previously described [3]. RNA quality was determined by using the Agilent RNA 6000 Pico Kit (Agilent) on the Bioanalyzer 2100 Electrophoresis System (Agilent).
2.6 Quantitative PCR amplification of beta-glucuronidase (GusB) from mouse eosinophil RNA
Two ng of RNA per sample was converted into cDNA using the First Strand cDNA Synthesis Kit for RT-PCR (AMV) (Roche) and then pre-amplified with the TaqMan PreAMP Master Mix (ThermoFisher) and beta-glucuronidase (GusB-FAM) TaqMan Gene Expression Assay (ThermoFisher) for 10 cycles as per manufacturers’ instructions. Amplified cDNA products were then amplified further with TaqMan Gene Expression Master Mix (ThermoFisher) on an Applied Biosystems 7500 Real-Time PCR Cycler (Applied Biosystems).
3.0 Results and Discussion
3.1 Fluorescence-activated cell sorting (FACS) without targeting Siglec F for isolation of eosinophils from mouse lung tissue
FACS is a useful, rapid method for isolating pure populations of cells (reviewed in [16]). We have used this method to identify subpopulations of eosinophils in the lungs of allergen-challenged mice [4], as have Mesnil and colleagues [3] who recently defined resident populations of regulatory eosinophils in mouse lung tissue. While useful and effective for identification purposes, antibody-mediated ligation of the cell surface antigen, Siglec F, initiates eosinophil apoptosis [14], which precludes use of this reagent for eosinophil isolation for functional or gene expression studies.
A FACS strategy and appropriate gating for isolating eosinophils from single cell suspensions prepared from lung tissue of mice sensitized and challenged with A. fumigatus is shown in Fig. 1. With this strategy, eosinophils are isolated largely by negative selection, as live CD11c-Gr1-/loMHCII- cells, with a yield of ~4 × 106 eosinophils per mouse. This strategy was also used to isolate eosinophils from of lungs IL5tg mice.
Fig. 1. Fluorescence-activated cell sorting (FACS) protocol without anti-Siglec F used to isolate eosinophils from mouse lung tissue.
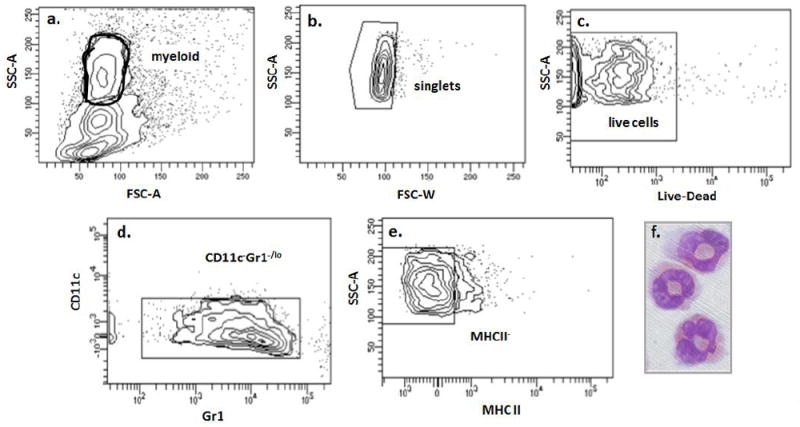
The protocol featured here does not require anti-Siglec F, which induces eosinophil apoptosis. Gating parameters (a. through e.) used to isolate eosinophils from single cell lung suspensions prepared from lungs of BALB/c mice sensitized and challenged with a filtrate from Aspergillus fumigatus. f. Cytospin preparation of cells identified by modified Giemsa-staining as live CD11c-Gr1-/loMHCII- cells are morphologically eosinophils. Image photographed with a Leica DMI4000 microscope with camera; photographs taken at 100X magnification.
As shown, we found that it was not necessary to include anti-CD45 (typically utilized as a pan-leukocyte positive selection marker) as this had no impact on eosinophil purity or yield (see below). Similarly, while the Gr1 gate has been set to be fully inclusive, it can be limited to include only Gr1- eosinophils, but this will reduce yield.
3.2 Evaluation of eosinophil purity
As shown in Fig. 2A, eosinophils isolated from IL5tg mice using this protocol also have normal staining properties, and have typical ring shaped or bow-tie nuclei [17]. The FACS method shown here results in purification of eosinophils from 43% in suspension to ~99% in isolate, as determined by visual inspection of stained cells by light microscopy [Fig. 2B].
Figure 2. Evaluation of live CD11c-Gr1-/loMHCII- eosinophils by light microscopy.
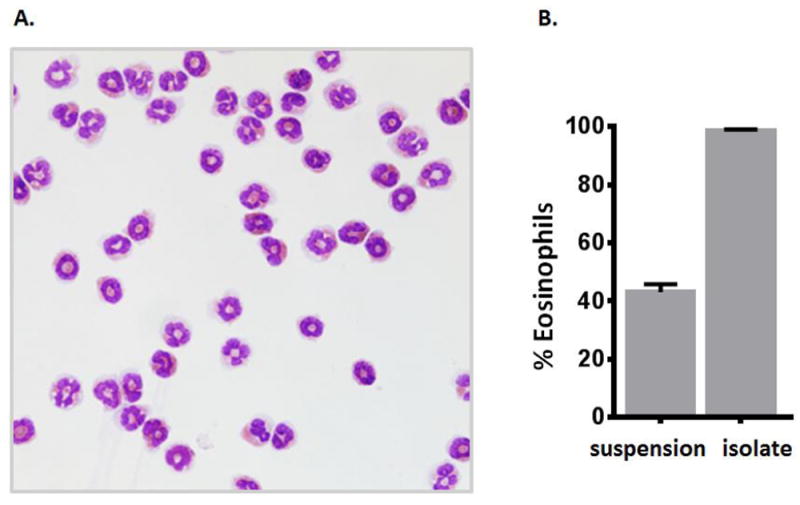
A. Light microscopic image of a larger field of eosinophils isolated from lungs of IL5tg mice by the FACS protocol as shown in Fig. 1. Image photographed with a Leica DMI4000 microscope with camera; photographs taken at 40X magnification. B. Percent eosinophils in the initial single cell suspension and in the post-FACS isolate; > 200 cells counted per sample.
We further evaluated eosinophil purification by flow cytometry, utilizing anti-Siglec F as a diagnostic marker. These results confirmed the light microscopic findings [Fig. 3A]; we find that the FACS isolates from lung tissue of Af sensitized and challenged mice and IL5tg mice included 96.5% and 98.2% Siglec F+ eosinophils, respectively.
Figure 3. Evaluation of live CD11c-Gr1-/loMHCII- eosinophils by flow cytometry and preparation of RNA.
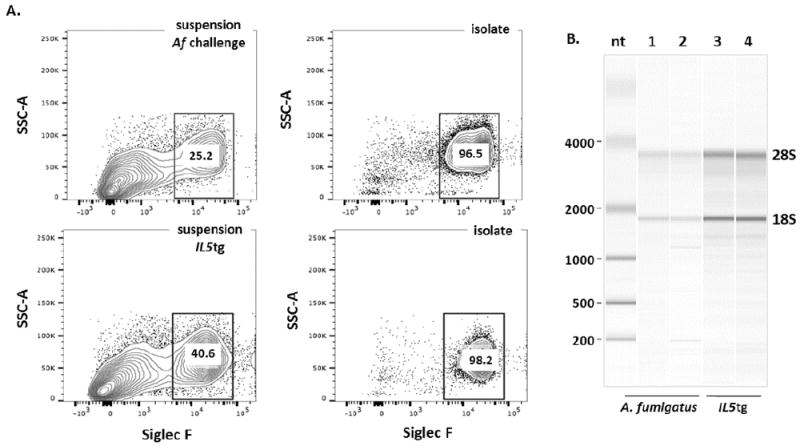
A. Fraction of Siglec F+ cells in the initial single cell suspensions (left-hand panels) and after isolation (FACS sorted cells as in Fig. 1, right-hand panels) from lung tissue from mice sensitized and challenged with a filtrate of A. fumigatus (upper panels) and IL5tg mice (lower panels). B. Independent samples from eosinophils isolated as in Fig. 1 from lungs of A. fumigatus sensitized and challenged mice (lanes 1 and 2) and from IL5tg mice (lanes 3 and 4) and collected into RNA-protect prior to RNA preparation (see Methods). RNA from samples (~3 × 106 cells) were evaluated on an Agilent RNA 6000 PicoChip on a Bioanalyzer 2100 Electrophoresis System and have RNA integrity (RIN) scores of greater than or equal to 7.
3.3 Preparation of RNA from FACS-isolated eosinophils
Preparation of RNA from eosinophils can be challenging, as they are not highly synthetic cells, and cells undergoing activation or perturbation can release enzymatically-active secretory ribonucleases (RNases) from cytoplasmic granules [18].
As such, rapid preparation of intact, viable cells with minimal handling to avoid untoward activation is critical. Eosinophils isolated by the FACS method presented are not only of high purity, RNA samples isolated from these cells remain undegraded [Fig. 3B], and were suitable substrates for quantitative reverse transcriptase-PCR. As shown in Table 2, beta-glucuronidase (gusB) was amplified successfully from RNA isolated from live SSChi/FSChi CD11c-Gr1-/loMHCII- eosinophils from the lungs of IL5tg mice and mice sensitized and challenged with A. fumigatus.
Table 2. Quantitative RT-PCR amplification of cDNA prepared from RNA isolated from live CD11c-Gr1-/loMHCII- eosinophils.
The housekeeping gene, beta-glucuronidase (gusB) was amplified from first-strand cDNA (+RT) generated from RNA samples shown in Fig. 3B prepared from FACS-isolated eosinophils from lungs of IL5tg mice and mice sensitized and challenged with A. fumigatus (Af) shown in Fig. 3B. No amplification product was detected from RNA samples in which reverse transcriptase was omitted (-RT) or in the absence of RNA template (ntc, no template control); undet, undetected, Ct > 40 cycles.
| Well | Detector | cDNA sample | Ct (cycles) |
|---|---|---|---|
| B1 | gusb | IL5tg +RT | 27.00 |
| B2 | gusb | IL5tg +RT | 26.98 |
| B5 | gusb | IL5tg - RT | undet |
| B6 | gusb | IL5tg - RT | undet |
| B10 | gusb | ntc | undet |
| D1 | gusb | Af + RT | 27.09 |
| D2 | gusb | Af + RT | 27.10 |
| D5 | gusb | Af - RT | undet |
| D6 | gusb | Af - RT | undet |
| D10 | gusb | ntc | undet |
Thus far, we have utilized this FACS method to isolate viable eosinophils in substantial purity and yield from lungs of allergen-challenged and IL5tg mice, both conditions in which the non-leukocyte and non-eosinophil populations are relatively limited. This method may require adjustments in other tissues where less favorable conditions prevail. However, given the substantial focus on asthma and allergic inflammation in the the respiratory tract, utilization of this method under these (and related) conditions alone represent a substantial advance in our ability to examine eosinophils and their responses to specific provocation at the molecular level.
4.0 Conclusions
We present a useful and generally applicable method for rapid FACS-mediated isolation of viable mouse eosinophils at high purity (97-99%) from lungs of allergen challenged mice. The method does not utilize anti-CD125, anti-CCR3 or anti-Siglec F, the latter antibody known to induce eosinophil apoptosis. Large numbers eosinophils (~4 × 106/mouse) isolated by this method yield high-quality RNA suitable for gene expression analysis.
Highlights.
Mouse eosinophils are typically identified by flow cytometry by targeting Siglec F.
Ligation with anti-Siglec F induces mouse eosinophil apoptosis.
We present a method for FACS isolation which does not target Siglec F, CCR3 or IL5Rα.
FACS-isolated eosinophils from Af-challenged mice are live CD11c-Gr1-/loMHCII-.
Isolated eosinophils are high purity (98-99%) and generate high quality RNA.
Acknowledgments
We are grateful for assistance from members of the NIAID Flow Cytometry core facility, notably from Dr. Kevin Holmes, Ms. Dareskedar Admassu, and Mr. Calvin Eigsti. We are also grateful for the assistance on the RNA Bioanalyzer from Dr. Qin Su. Work in our lab is funded by NIAID Division of Intramural Research (Z01-AI-000941 to HFR).
Abbreviations
- FACS
fluorescence-activated cell sorter
- IL5tg
interleukin-5 transgenic
- Siglec F
sialic acid-binding immunoglobulin-type lectin F
- ITIM
immunoreceptor tyrosine-based inhibitory motif
- gusB
beta-glucuronidase
- Af
Aspergillus fumigatus
Footnotes
Author Contributions
WEG designed and performed experiments, and reviewed the manuscript.
CMP provided critical assistance to WEG, analyzed data, and reviewed the manuscript.
HFR created the manuscript, and wrote the first and subsequent drafts.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Rosenberg HF, Dyer KD, Foster PS. Eosinophils: changing perspectives in health and disease. Nat Rev Immunol. 2013;13:9–22. doi: 10.1038/nri3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee JJ, Jacobsen EA, McGarry MP, Schleimer RP, Lee NA. Eosinophils in health and disease: the LIAR hypothesis. Clin Exp Allergy. 2010;40:563–575. doi: 10.1111/j.1365-2222.2010.03484.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mesnil C, Raulier S, Paulissen G, Xiao X, Birrell MA, Pirottin D, Janss T, Starkl P, Ramery E, Henket M, Schleich FN, Radermecker M, Thielemans K, Gillet L, Thiry M, Belvisi MG, Louis R, Desmet C, Marichal T, Bureau F. Lung-resident eosinophils represent a distinct regulatory eosinophil subset. J Clin Invest. 2016;126:3279–3295. doi: 10.1172/JCI85664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Percopo CM, Brenner TA, Ma M, Kraemer LS, Hakeem RM, Lee JJ, Rosenberg HF. Siglec F+Gr1hi eosinophils are a distinct subpopulation within the lungs of allergen-challenged mice. J Leukoc Biol. 2017;101:321–328. doi: 10.1189/jlb.3A0416-166R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Geslewitz WE, Percopo CM, Rosenberg HF. Eosinophil persistence in vivo and sustained viability ex vivo in response to respiratory challenge with fungal allergens. Clin Exp Allergy 2017. 2017 doi: 10.1111/cea.13050. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wicher SA, Jacoby DB, Fryer AD. Newly divided eosinophils limit ozone-induced airway hyperreactivity in non-sensitized guinea pigs. Am J Physiol Lung Cell Mol Physiol. 312:L969–L982. doi: 10.1152/ajplung.00530.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dyer KD, Garcia-Crespo KE, Killoran KE, Rosenberg HF. Antigen profiles for the quantitative assessment of eosinophils in mouse tissues by flow cytometry. J Immunol Methods. 2011;369:91–97. doi: 10.1016/j.jim.2011.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stevens WW, Kim TS, Pujanauski LM, Hao X, Braciale TJ. Detection and quantitation of eosinophils in the murine respiratory tract by flow cytometry. J Immunol Methods. 2007;327:63–74. doi: 10.1016/j.jim.2007.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Berek C, Beller A, Chu VT. Isolation of eosinophils from the lamina propria of the murine small intestine. Methods Mol Biol. 2016;1422:213–221. doi: 10.1007/978-1-4939-3603-8_20. [DOI] [PubMed] [Google Scholar]
- 10.Kastenschmidt JM, Avetyan I, Armando Villalta S. Characterization of the inflammatory response in dystrophic muscle using flow cytometry. Methods Mol Biol. 2018;1687:43–56. doi: 10.1007/978-1-4939-7374-3_4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.O’Sullivan JA, Carroll DJ, Bochner BS. Glycobiology of eosinophilic inflammation: contributions of siglecs, glycans and other glycan-binding proteins. Front Med. 2017;4:116. doi: 10.3389/fmed.2017.00116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kiwamoto T, Katoh T, Tiemeyer M, Bochner BS. The role of lung epithelial ligands for Siglec-8 and Siglec-F in eosinophilic inflammation. Curr Opin Allergy Clin Immunol. 2013;13:106–111. doi: 10.1097/ACI.0b013e32835b594a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zimmermann N, McBride ML, Yamada Y, Hudson SA, Jones C, Cromie KD, Crocker PR, Rothenberg ME, Bochner BS. Siglec-F antibody administration to mice selectively reduces blood and tissue eosinophils. Allergy. 2008;63:1156–1163. doi: 10.1111/j.1398-9995.2008.01709.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang M, Angata T, Cho JY, Miller M, Broide DH, Varki A. Defining the in vivo function of Siglec-F, a CD33-related Siglec expressed on mouse eosinophils. Blood. 2007;109:4280–4287. doi: 10.1182/blood-2006-08-039255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dent LA, Strath M, Mellor AL, Sanderson CJ. Eosinophilia in transgenic mice expressing interleukin 5. J Exp Med. 1990;172:1425–1431. doi: 10.1084/jem.172.5.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Herzenberg LA, Parks D, Sahaf B, Perez O, Roederer M, Herzenberg LA. The history and future of the fluorescence activated cell sorter and flow cytometry. Clin Chem. 2002;48:1819–1827. [PubMed] [Google Scholar]
- 17.McGarry MP, Protheroe CA, Lee JJ. A laboratory manual. Cold Spring Harbor Laboratory Press; New York: 2010. Mouse hematology. [Google Scholar]
- 18.Shamri R, Melo RC, Young KM, Bivas-Benita M, Xenakis JJ, Spencer LA, Weller PF. CCL11 elicits secretion of RNases from mouse eosinophils and their cell-free granules. FASEB J. 2012;26:2084–2093. doi: 10.1096/fj.11-200246. [DOI] [PMC free article] [PubMed] [Google Scholar]


