Abstract
In chronic infections, bacterial pathogens typically grow as small dense cell aggregates embedded in a matrix consisting of, e.g., wound bed sludge or lung mucus. Such biofilm growth mode exhibits extreme tolerance towards antibiotics and the immune defence system. The bacterial aggregates are exposed to physiological heterogeneity and O2 limitation due to steep chemical gradients through the matrix, which is are hypothesised to contribute to antibiotic tolerance. Using a novel combination of microsensor and bioimaging analysis, we investigated growth patterns and chemical dynamics of the pathogen Pseudomonas aeruginosa in an alginate bead model, which mimics growth in chronic infections better than traditional biofilm experiments in flow chambers. Growth patterns were strongly affected by electron acceptor availability and the presence of chemical gradients, where the combined presence of O2 and nitrate yielded highest bacterial growth by combined aerobic respiration and denitrification.
Chronic infection: effects of electron acceptors
Bacterial growth is strongly affected by levels and gradients of electron accepting chemicals such as nitrate ions and oxygen molecules. Thomas Bjarnsholt, Michael Kühl, and colleagues at the University of Copenhagen grew Pseudomonas aeruginosa bacteria in beads composed of a natural carbohydrate called alginate. This method models the growth patterns in chronic infections more effectively than conventional procedures. The model reflects the fact that chronic infections typically involve dense aggregates of cells embedded in materials such as wound debris or mucus. This renders the bacteria resistant to antibiotics and immune defences. The researchers investigated the effect of varying concentration gradients and flow patterns of several electron acceptors. Bacterial growth was strongly influenced by the distribution of the electron acceptors. Adding electron acceptors to treatment regimes may enhance the activity of antibiotics against chronic infections.
Introduction
Bacterial biofilms are ubiquitous in most natural habitats, where they play an integral role in the cycling of elements. However, biofilms are also associated with a wide range of harmful effects ranging from biofouling of ship hulls and drainpipes to the formation of biofilms on medical implants and indwelling devices.1 In chronic infections such as diabetic and venous leg ulcers or in the lungs of patients suffering from the genetic disorder cystic fibrosis (CF),2,3 biofilms grow as small dense cell aggregates devoid of surface association,4,5 in a matrix of exopolymeric substance consisting of polysaccharides, proteins, and eDNA.6–9 Such bacterial aggregates are embedded in a secondary matrix composed of, e.g., wound bed sludge or CF lung mucus. Bacterial aggregates exhibit physiological heterogeneity due to steep chemical gradients forming through the secondary matrix and into the biofilm.10,11 In particular, molecular oxygen (O2) has been shown to reach hypoxic and anoxic levels within the outer 50–100 µm of biofilms and chronic wounds.12–14 Steep O2 gradients are also a recognised feature of chronic infections,13,15–17 where activated polymorphonuclear leucocytes persistently accumulate around the bacterial aggregates4,18 leading to strong depletion of O2 due to formation of reactive oxygen species.19,20 Such complexity of the chemical landscape is believed to result in heterogeneous growth patterns, and the establishment of bacterial subpopulations exhibiting particular metabolic activities.21 This in turn can have an impact on the efficacy of antibiotic treatment as several studies have shown that O2 limitation is correlated to increased antibiotic tolerance of biofilms.10–12,22
The opportunistic pathogenic bacterium Pseudomonas aeruginosa is a key model organism for the study of biofilm infections, and it has been isolated from both chronic wounds and chronically infected lungs of CF patients. In order to gain further insight to the biofilm mode of growth of P. aeruginosa, several optical methods can be applied for visualising growth patterns and biofilm structure. The standard method of confocal laser scanning microscopy (CLSM) of stained or fluorescently tagged P. aeruginosa provides very localised information on biomass distribution (at µm scale). In combination with quantitative peptide nucleic acid fluorescence in situ hybridisation (PNA-FISH)21,23 CLSM can also be applied to quantify the growth potential21 by treating bacterial cells with PNA-FISH probes specific for P. aeruginosa 16S rRNA.24 While providing high-resolution data on bacterial growth, this method is based on fixed samples and is therefore an invasive technique. When aiming to unravel structural biofilm properties at mesoscopic to macroscopic levels (10 µm—mm length scale), optical coherence tomography (OCT) is a suitable alternative imaging technique.25 OCT employs near-infrared radiation (NIR) and provides a non-invasive alternative to light microscopy, enabling high-resolution 3D scanning of larger (mm3 to cm3) biofilm volumes in near-real time.26–28 As previously mentioned, bacterial aggregates are exposed to chemical and physiological heterogeneity due to steep O2 gradients. To further elucidate this aspect, microsensors can be used to investigate the chemical environment in a minimal invasive fashion.29 Microsensors are available for several analytes including O2 and nitrous oxide (N2O), a key intermediate product of denitrification.30 Another approach is to use chemical imaging with optical sensors (either immobilised in sensor films or particles) to visualise the chemical microenvironments in biofilms.31–34
Biofilms are often studied in vitro using continuous flow cell systems, wherein biofilms are grown attached to a surface and can exhibit a variety of structural morphologies including mushroom-shaped structures.35 However, in CF lungs and chronic wounds P. aeruginosa grows in dense suspended aggregates separated by a secondary matrix and with no attachment to a solid substrate or surface.4 The typical growth mode and biofilm shapes observed in flow chambers are thus not representative of the observed in vivo growth patterns of biofilms associated with chronic infections. To better mimic the in vivo conditions of P. aeruginosa in chronic infections, we recently employed an alginate bead model,14 wherein the bacteria form dense, spatially segregated micro colonies similar in size and structure to P. aeruginosa aggregates observed ex vivo in lungs from CF patients21 and chronic wounds.4
P. aeruginosa can grow anaerobically by performing arginine fermentation or using alternative electron acceptors36–38 and there is increasing evidence that P. aeruginosa can utilise the high physiological NO3− and NO2− levels in the CF lungs to grow under O2 limitation by performing denitrification.39,40 In this study, we investigated P. aeruginosa aggregate growth in alginate beads with different O2 and NO3− availability mimicking physiological conditions encountered in the chronic infections.41,42 The bacterial biomass, growth rate, and chemical microenvironment in the beads was characterised using a novel combination of OCT, CLSM, PNA-FISH, viable cell counts, O2 and N2O microsensor analysis, as well as first attempts of chemical O2 imaging. We found significant effects of electron acceptor availability on the growth pattern and metabolic activity of P. aeruginosa forming in vivo-like microcolonies in the alginate bead model, and discuss the implications of our results for better treatment of chronic infections.
Results
Optical coherence tomography
OCT facilitated non-invasive macroscopic imaging of the alginate beads (Supplementary Fig. S1A, B), wherein light scattering bacterial aggregates were identified by their high OCT signal. Due to dense growth of bacterial aggregates in the bead periphery, the vertical OCT signal attenuation was rapid and at about 200 µm below the bead surface the OCT signal approached that of the OCT signal of pure alginate (Supplementary Fig. S2). The OCT dB signal from the outermost peripheral 150 µm of the beads could thus be used as a proxy for bacterial biomass distribution. The OCT dB signal of anoxic beads (without NO3−) was similar to the blank control (without bacteria), thus suggesting no or very minor growth (Fig. 1). In contrast, the OCT signal from anoxic beads with NO3− was significantly higher relative to the control after 48 h (p = 0.035). Normoxic beads and normoxic beads supplemented with NO3− gave rise to the strongest OCT signal, indicative of intense growth and biomass accumulation in the bead periphery (Fig. 1, S1).
Fig. 1.

OCT signal intensity (dB) of alginate beads over 48 h of incubation. The OCT signal was averaged for an area covering a vertical depth of 150 μm (from the bead surface into the interior) and a lateral width of 100 μm and was used as a proxy for bacterial growth (see “Methods”). Bars represent average ± standard errors of the means from three replicates. Significant differences as indicated. *p > 0.05; **p ≤ 0.01; ***p ≤ 0.001
Microscopy
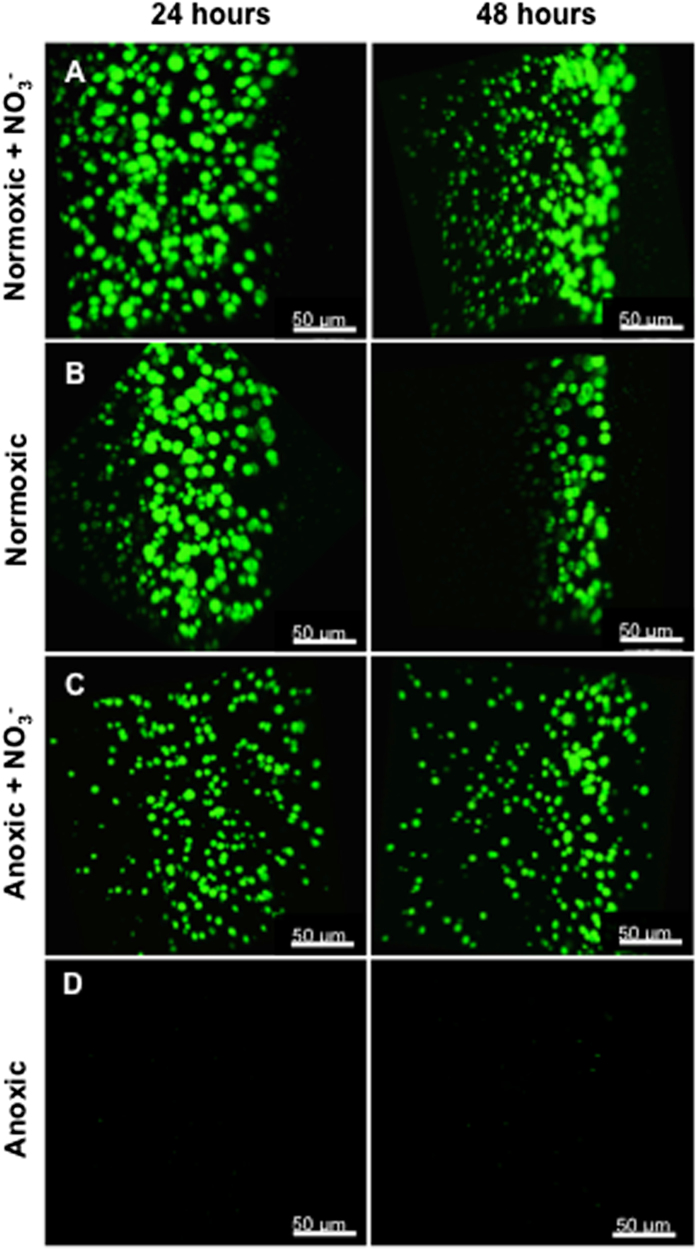
Bacterial growth and organisation in the alginate beads was visualised microscopically by CLSM using green fluorescent protein (GFP)-tagged P. aeruginosa (Fig. 2). In normoxic beads, P. aeruginosa showed peripheral growth (growth in the outermost ~100 µm of the bead) of bacterial microcolonies after 24 and 48 h (Fig. 2b). Normoxic growth in the presence of NO3− resulted in a very intense growth, with larger aggregates in the periphery than observed under anoxic growth of P. aeruginosa in the presence of NO3−. Moreover, 48 h normoxic growth of P. aeruginosa with NO3− resulted in a heterogeneous growth pattern, with bacterial microcolonies decreasing in size with distance from the surface of the alginate bead (Fig. 2a). Anoxic beads with NO3− showed a homogeneous distribution of aggregate size in the bead after 24 h, but also exhibited signs of a more heterogeneous aggregate size distribution after 48 h growth with largest aggregates closer to the bead surface (Fig. 2c). No growth was observed in anoxic beads without alternative electron acceptors at 48 h (Fig. 2d), which was supported by the OCT imaging results.
Fig. 2.
400× CLSM images of the growth dynamics of GFP-tagged P. aeruginosa grown under different conditions after 24 and 48 h. a Normoxic growth in the presence of NO3− resulted in intense and deep growth with a tendency to form smaller aggregates in the deeper parts over time. b Normoxic growth resulted in peripheral growth over time. c Anoxic growth with NO3− also supported growth, but with apparently smaller aggregates. d Anoxic growth without NO3− did not support growth. The edge of the alginate bead is oriented to the right in the images. Size of scale bars: 50 μm
Microscopy combined with quantitative PNA-FISH
The mean fluorescence intensity used here as a proxy for the growth potential (the maximum possible growth rate) of P. aeruginosa21 was quantified and plotted for each of the growth conditions. In normoxic beads, we found that NO3− supplementation induced significantly deeper growth of P. aeruginosa aggregates (Fig. 3a). The median growth depth (interquartile range, IQR) was 51.6 (17.4–92.0) µm vs. 20.0 (2.0–45.0) µm (p < 0.0001) below the alginate bead surface. When NO3− was present, the median growth depth (IQR) was comparable under normoxic vs. anoxic conditions, i.e., 51.6 (17.4–92.0) µm vs. 41.6 (14.2–88.6) µm (p = 0.481).
Fig. 3.

Growth potential of P. aeruginosa grown in alginate beads expressed as mean fluorescence intensity. a Normoxic growth conditions supplemented with NO3−, b normoxic growth conditions without NO3−, and c anoxic growth conditions in the presence of NO3−. P. aeruginosa was fluorescently labelled with Texas-Red-conjugated PNA-FISH probe tagging ribosomes, imaged by CLSM and quantification of intensity was performed by Imaris
In normoxic beads with NO3−, the growth potential of P. aeruginosa was significantly higher after 24 h than after 48 h (10212 FU ± 3298 FU vs. 7141 FU ± 1470 FU; p < 0.001). Furthermore, we found a significantly higher growth depth at 24 h vs. 48 h, with a median growth depth (IQR) of 62.9 (25.9–98.0) µm vs. 38.8 (11.3–70.3) µm (p = 0.023) below the alginate bead surface (Fig. 3a). Growth depth in normoxic beads without NO3− did not differ between the two time points (p = 0.139), but the growth potential, as estimated by mean fluorescence, was significantly higher at 24 h as compared to 48 h (9243 ± 2433 vs. 6586 ± 1614; p < 0.001) (Fig. 3b). In anoxic beads with NO3−, there was no significant difference in neither growth depth (p = 0.761) or in growth potential (p = 0.124) at 24 h vs. 48 h, and the growth potential remained high (9258 FU ± 1865 FU vs. 8750 FU ± 1780 FU) both after 24 and 48 h (Fig. 3c).
When correlating growth potential to growth depth, we found no statistically significant correlation under any conditions. It was not possible to prepare paraffin slices for anoxic beads without NO3−, but as previously shown P. aeruginosa failed to grow under these conditions (Figs. 2, 4).
Fig. 4.

Viable cell counts as colony-forming units (CFU) from dissolved alginate beads after 0, 24, and 48 h. Highest CFU was observed for normoxic beads supplemented with NO3−. No change in CFU over time for the anoxic group was observed. Bars represent average ± standard errors of the means from three replicates. Significant differences as indicated. n.s. not significant; ****p ≤ 0.0001
Colony-forming unit (CFU)
All bead cultures were initiated with a CFU-defined cell density of ~107 cells per bead (mean 1.175 × 107; range 8.67 × 106–1.483 × 107) (Fig. 4). While CFU values remained stable in the anoxic culture without NO3−, the other growth conditions facilitated a significant increase in CFU of 1–2 log units from 0 to 24 h, which then remained constant between 24 and 48 h. The highest CFU was observed for the growth condition with both O2 and NO3− available in the alginate beads (p < 0.0001).
Microsensor measurements
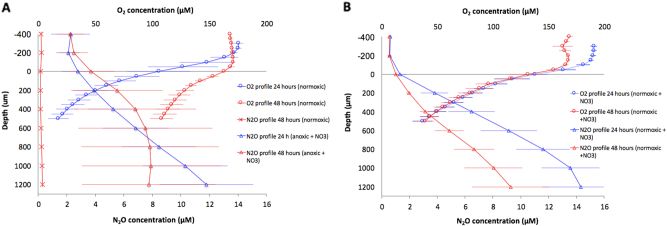
The O2 concentration profiles measured in normoxic alginate beads (without NO3−) after 24 h incubation showed the presence of an oxygenated zone in the peripheral 200 µm with a steep decline in the O2 concentration and an inner hypoxic zone (Fig. 5a). After 48 h, the slope of the profile decreased, indicating an overall decrease in O2 respiration rate from 24 to 48 h (Fig. 5a), leading to more O2 accumulation in the beads over time. No N2O production was detected in normoxic grown alginate beads without NO3− (Fig. 5a). In normoxic alginate beads supplemented with NO3−, the maximal N2O concentration was detected in the lower, hypoxic zone (Fig. 6b). Contrary to normoxic beads (without NO3−), O2 profiles in NO3− supplemented beads were stable between 24 and 48 h, whereas N2O production decreased (Fig. 6b). N2O profiles in anoxic beads with NO3− revealed an increasing N2O concentration throughout the bead, with a slight decrease in production after 48 h (Fig. 6a).
Fig. 5.
O2 and N2O profiles from alginate-encapsulated P. aeruginosa. a P. aeruginosa cultured under normoxic conditions (circles) and anoxic conditions with 10 mM NO3− (triangles). Furthermore, a control measurement of N2O on alginate-encapsulated P. aeruginosa grown without NO3− (asterisk). b P. aeruginosa cultured under normoxic conditions supplemented with 10 mM NO3−. O2 profiles (circles) and N2O profiles (triangles). Bars represent average ± standard deviation from 3 to 4 replicates. All measurements were performed at 37 °C in growth medium
Fig. 6.
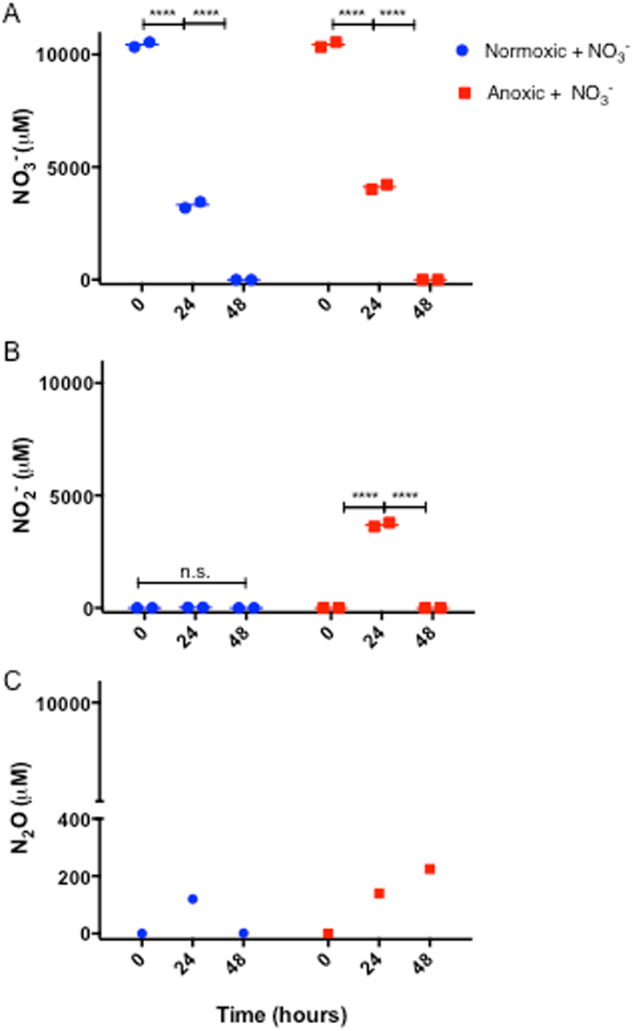
Dynamics of a NO3−, b NO2−, and c N2O (μM) in growth medium quantified by Griess colometric reaction and microsensor measurements directly in growth medium. NO3− and NO2− measurements are based on duplicate sampling from medium (error bars too small to be visualised), and N2O concentration represents on a single measurement directly in medium. Significant differences as indicated. n.s. not significant; ****p ≤ 0.0001
Fluxes of O2 and N2O
We found a stable O2 flux into the normoxic beads supplemented with NO3 (3.18 ± 0.66–3.19 ± 0.84 nmol O2 cm−2 min−1), and a tendency towards a decreasing O2 flux into normoxic beads (without NO3−) between 24 and 48 h (3.07 ± 1.33–1.84 ± 0.32 nmol O2 cm−2 min−1). Calculations of the efflux of N2O from the alginate beads revealed a stable N2O efflux in anoxic beads with NO3− between 24 and 48 h (0.10 ± 0.03–0.13 ± 0.1 nmol N2O cm−2 min−1), whereas the normoxic beads with NO3− showed a tendency towards a decrease in N2O efflux between 24 and 48 h (0.21 ± 0.06–0.09 ± 0.03 nmol N2O cm−2 min−1). None of the changes in flux over time were statistically significant.
NO3−, NO2−, and N2O dynamics in growth medium
Measurements of NO3− and NO2− concentrations in the R2A growth medium supplemented with NO3− showed that the initial concentration (t = 0) of NO3− in the medium was 10.450 µM ± 148 µM (Fig. 6a), while no NO2− could be detected (Fig. 6b). After 24 h, the NO3− concentration had declined significantly to 4.119 ± 0.149 and 3.319 ± 0.191 µM (p < 0.0001) in the anoxic and normoxic cultures, respectively, and was completely depleted within 48 h (Fig. 6a). No NO2− could be detected in the normoxic cultures, but in the anoxic cultures there was a significant accumulation of NO2− after 24 h (p < 0.0001), which was depleted again after 48 h (Fig. 6b). When measuring the N2O concentrations directly in the growth medium with electrochemical sensors, we found no N2O in the growth medium without added NO3− (data not shown). In the anoxic culture flasks with NO3− addition (sealed airtight), denitrification led to an accumulation of N2O over time (Fig. 6c). After 24 h of incubation, a N2O concentration of 140 µM was measured in the medium, increasing to 225 µM after 48 h. In the normoxic culture with NO3−, N2O was measured at a concentration of 120 µM after 24 h, but only 1.5 µM after 48 h.
O2 imaging
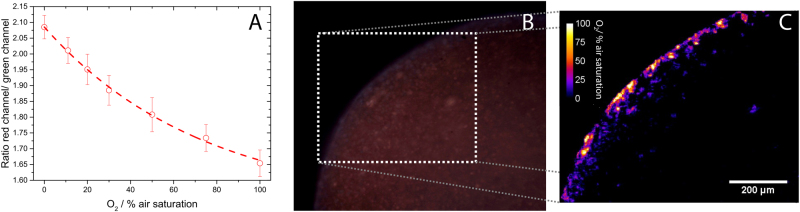
The incorporation of O2-sensitive nanoparticles in the alginate beads enabled visualisation of the O2 distribution relative to the bacterial aggregates. The O2 images revealed steep O2 gradients in the periphery of the beads (Fig. 7c) forming a heterogeneous landscape of O2 concentration within the O2 provisioned part of the beads that roughly followed the aggregate distribution, where aggregates exhibited complete O2 depletion and hypoxia in the surrounding alginate matrix. Atmospheric O2 saturation was only observed at the immediate bead surface.
Fig. 7.
Ratiometric imaging of O2 with oxygen-sensitive nanoparticles incorporated in alginate beads with P. aeruginosa. a Calibration of nanoparticles in alginate beads submerged in water of increasing O2 concentrations. The O2 was quantified as the ratio of the Red and Green channels extracted from RGB colour images obtained with a simple USB microscope. b Alginate bead with sensor nanoparticles, c zoom-in and visualisation of O2 distribution in alginate bead after 24 h of growth with P. aeruginosa. Size of bar: 200 µm
Discussion
Cultures of P. aeruginosa in alginate beads exhibit several characteristics of aggregated bacteria found in the lungs of CF patients and in chronic wounds, in regard to aggregate size, growth, O2 limitation, physiological heterogeneity, and antibiotic tolerance.14 This enabled our study of in vivo-like growth patterns and the chemical microenvironment of P. aeruginosa with and without electron acceptors via a variety of invasive and non-invasive methods. The most intense growth of P. aeruginosa was observed in the presence of the two electron acceptors O2 and NO3−, enabling both aerobic respiration and denitrification in the alginate beads, while complete absence of electron acceptors resulted in an arrested growth state.
We studied the growth patterns of P. aeruginosa in the presence and absence of O2 and NO3− at different spatial scales using OCT for mesoscopic and macroscopic scale, CLSM to unravel structures at a microscopic scale, and quantitative PNA-FISH for information on the growth potential. OCT has previously been used to study the formation and growth dynamics of biofilms in flow chambers and various carrier materials43,44 and of clinical biofilms in nasal polyps,28 but to our knowledge OCT has not previously been used to investigate bacteria growing in alginate beads. The presence of bacteria increased local backscatter in the alginate matrix resulting in a stronger OCT signal.44 However, the alginate matrix also scattered light, and a quantification of bacterial growth thus relies on a clear separation of the OCT signals originating from the bacteria and the alginate. We found that bacterial growth led to an enhancement of the OCT signal relative to the alginate bead, but only within the first 200 µm from the bead surface (see Supplementary Fig. S2). While OCT allows for a potential operational depth of view of several mm’s,45 the high density of bacteria in the outermost bead layers leads to intense multiple scattering of the NIR probing light causing a rapid vertical attenuation and thus loss of image contrast of the collected OCT signal46 resulting in an operational depth of view that was comparable to CLSM.47 Evidently, such application of OCT can be further optimised, e.g., by decreasing scatter in the alginate matrix using washed less optically opaque alginate, but such technical optimisation was beyond the scope of this study. Still, the OCT data in our study complemented higher-resolution microscopy techniques and provided a non-invasive macroscopic overview of the growth dynamics and growth zones in intact alginate beads. With OCT, it was thus possible to monitor the overall growth zone of P. aeruginosa in alginate beads (Fig. S1), while high-resolution CLSM enabled (i) visualisation and quantification of microcolony size and distribution in the alginate beads (Fig. 2) and (ii) local growth potential measurements when used in combination with a quantitative PNA-FISH protocol (Fig. 3).
Supplementing P. aeruginosa with two electron acceptors (O2 and NO3−) resulted in higher growth potentials (Figs. 2, 3) at 24 h as compared to 48 h incubation. Moreover, a heterogeneous growth pattern of P. aeruginosa was observed in the alginate beads after 48 h incubation with larger aggregates situated near the oxygenated bead surface. Physiological heterogeneity within biofilms is largely driven by the activity and biomass distribution of the bacteria in combination with mass transfer limitation by diffusion of electron acceptors, substrate and products of bacterial metabolism in the biofilm matrix and in the diffusive boundary layer between the mixed water phase and the biofilm.11 Typically, bacterial depletion of O2 in the biofilm periphery limits diffusion to deeper parts,48 and we found that such development of O2 concentration gradients in the alginate beads lead to a heterogeneous growth pattern over time, where only the bacteria positioned near the bead surface had sufficient access to the preferred electron acceptor O2. While anoxic grown P. aeruginosa with access to NO3− displayed a very homogeneous growth pattern after 24 h, these beads also showed signs of a heterogeneous growth pattern after 48 h (Fig. 2). This shift was probably due to depletion of bead-incorporated NO3−, where after NO3− was only available by diffusion from the growth medium. These results are in accordance with previous studies showing a heterogeneous distribution of bacterial aggregate sizes, with significantly larger aggregates in the periphery as compared to deeper in the beads after 48 h in normoxic grown alginate-encapsulated P. aeruginosa with and without 100 mM NO3− supplement.14
The results from OCT and CLSM corresponded well with the quantitative PNA-FISH results in terms of showing increased growth depth in the presence of NO3−, while the availability of O2 did apparently not affect growth depth into the alginate beads. The latter finding confirms that O2 is strongly affected by diffusion limitation.49 In addition, quantitative PNA-FISH provided an estimate on the growth potential, expressed as fluorescence intensity, which has been shown to correlate with growth rate in P. aeruginosa.21 Contrary to growth depth, the growth potential was affected by the presence of O2, and we found a significantly higher growth potential with O2 at 24 h as compared to 48 h. This suggests that the contribution of O2 to the overall growth may be limited after 48 h, and that the preferred electron acceptor O2 initially facilitates a more intense growth burst than with NO3−, which correlates well with the low growth rate of P. aeruginosa observed in vivo in the mucus of chronically infected CF lungs.21 Furthermore, this is in accordance with a higher energy yield by O2 respiration as compared to denitrification.50,51 Cell counts further confirmed a more intense growth in the presence of both O2 and NO3− (Fig. 4).
Interestingly, bacterial growth in beads with no electron acceptors did not result in a decrease in CFU but rather lead to a steady state in cell counts throughout the study, which is in support of P. aeruginosa’s remarkable ability for long-time survival.52 Previous studies have thus demonstrated that P. aeruginosa is capable of long-term anaerobic survival via fermentation of amino acids,52–54 which were present in the growth medium.
To gain further insight to factors governing the growth dynamics of P. aeruginosa in the beads as observed with OCT, CLSM, and quantitative PNA-FISH, we used microsensors to quantify chemical gradients in the alginate beads. Based on such measurements, we estimated oxic respiration from the net consumption of O2 and denitrification from the net production of N2O in the beads, respectively. The observed decline in O2 in the water just above the alginate bead surface showed the presence of a diffusive boundary layer around the bead,55 and microsensor measurements demonstrated the presence of steep O2 concentration gradients and thus high O2 consumption in beads incubated under normoxic conditions. However, the O2 uptake decreased with time (Fig. 5a) in line with our observation of a decreasing growth rate over time as determined by quantitative PNA-FISH (Fig. 3). This may indicate onset of substrate limitation for bacterial growth in the alginate beads.
Beads incubated in normoxic medium with NO3− showed steep O2 concentration gradients, but contrary to beads grown without NO3−, the O2 uptake did not decrease with time (Fig. 5b). However, such sustained high O2 consumption was not reflected in higher growth potentials, which showed a decrease between 24 and 48 h of incubation (Fig. 3a). Some bacteria can perform so-called aerobic denitrification during hypoxic conditions in the presence of NO3−. Initially described by Robertson and Kuenen,56 aerobic denitrification can explain how NO3− respiration may proceed in the presence of O2.56 In P. aeruginosa this ability is evidenced by the expression of overlapping gene sets depending on O2 concentration, where hypoxia triggers denitrification genes.57 Moreover, the ability to perform intra-aerobic respiration has been proposed, where NO3− is reduced to NO, which is dismutated into N (nitrogen) and O2.58 Such formed O2 would in theory proceed in the aerobic respiration chain, which would then support high O2 consumption rates. However, we note that the presence of aerobic denitrification or intra-aerobic respiration remains to be demonstrated in P. aeruginosa. Maximal N2O concentration was detected in the hypoxic zone of the alginate beads, indicating that denitrification mainly occurred here (Fig. 5). Production of N2O decreased with time (Fig. 5b), which may reflect NO3− depletion in the beads and the surrounding medium. This was supported by the complete exhaustion of NO3− after 48 h (Fig. 6a).
In beads incubated in anoxic medium with NO3−, P. aeruginosa solely relied on denitrification for anaerobic respiration as indicated by the observed N2O production, which decreased over time (Fig. 5b), leading to the appearance of a more heterogeneous size distribution of bacterial aggregates in the beads (Fig. 2) resembling the growth pattern observed in normoxic beads. As N2O is a key intermediate in the denitrification metabolic pathway,36 a decreasing N2O flux could indicate the onset of NO3− limitation within the beads after 48 h of growth, which was supported by our finding of complete consumption of NO3− after 48 h (Fig. 6a). We also followed the fate of NO2− in the growth medium (Fig. 6b), which showed a peak in NO2− for anoxic grown beads after 24 h, and complete depletion after 48 h. In normoxic incubations (with NO3−), no increase in NO2− was observed, but we speculate that such a peak may have occurred before the 24 h measurement, as NO3− in the presence of O2 was shown to facilitate a more intense growth initially. The N2O measurements in the growth medium showed that N2O accumulated to very high levels in the sealed system after 48 h (Fig. 6c) further supporting a strong NO3− consumption during the incubations.
While our microsensor measurements revealed insight to the overall dynamics of O2 and N2O gradients in the alginate beads when incubated under different electron acceptor availability, we note that such measurements cannot provide detailed insight to the heterogeneous chemical landscape of individual microcolonies in the alginate beads. In a first attempt to link microcolony heterogeneity to the distribution in O2, we used ratiometric O2 imaging in a 24 h old alginate bead supplemented with O2-sensitive nanoparticles and grown under normoxic conditions (Fig. 7c). Ratiometric O2 imaging showed pronounced local variation in the outermost 100 µm of the alginate bead. However, the used camera system for O2 imaging did not have enough spatial resolution to investigate co-localisation and enable correlations between microcolony size and O2 distribution. However, such combined biomass and O2 imaging34 in the alginate beads has a strong potential to further resolve links between the chemical microenvironment and growth of P. aeruginosa in the alginate bead. Such approach would require more measurements of O2 distribution with nanoparticles in the beads using higher-resolution microscopic imaging.33,59
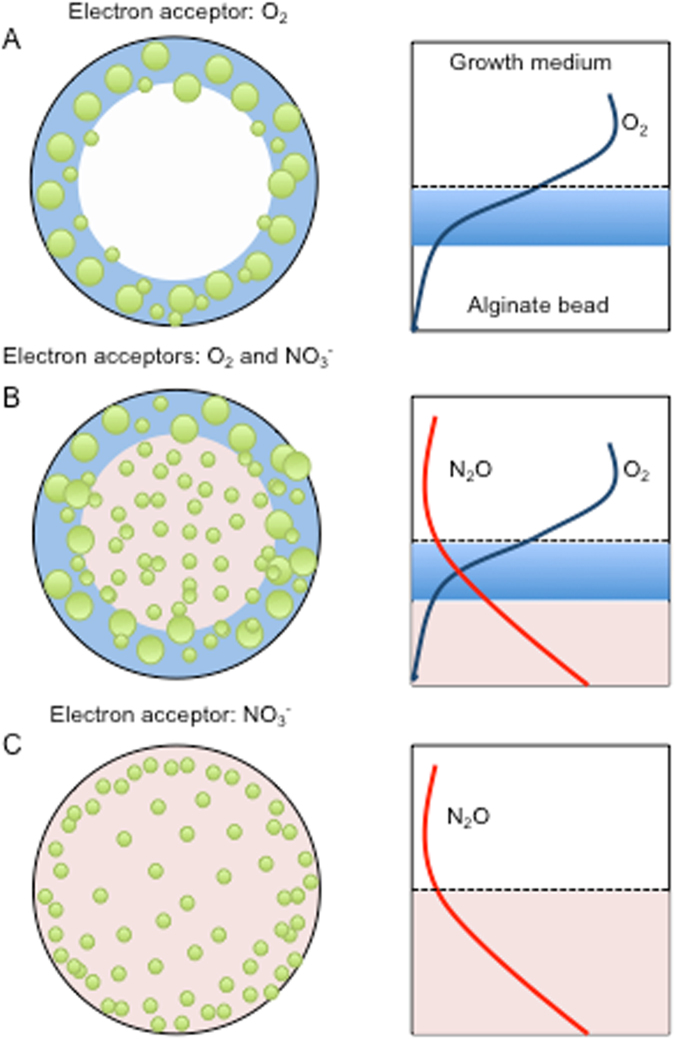
In conclusion, growth in alginate beads represents a useful in vitro model for the in vivo growth of P. aeruginosa in chronic infections. This model system is suitable for testing responses in bacterial metabolism and growth patterns to the availability of different electron acceptors and donors during chronic infections, and the effect of the different treatments are summarised in Fig. 8. The alginate bead model may also prove very suitable for testing antimicrobial susceptibility and tolerance of P. aeruginosa and other pathogens involved in mono-species and multi-species infections. It is well known that O2 limitation contributes to antibiotic tolerance of bacteria in biofilms,22 but addition of electron acceptors such as NO3− and O2 may antagonise this effect and enhance the susceptibility towards antibiotics.60,61
Fig. 8.
Schematic presentation of growth dynamics and chemical dynamics in alginate beads under different growth conditions. a Normoxic growth, b normoxic growth with NO3−, and c anoxic growth with NO3−. Green circles represent aggregates of P. aeruginosa within the alginate bead. O2: blue/light blue. NO3−: red/light red. Sizes not to scale
Methods
Bacterial strains
A wild-type P. aeruginosa strain PAO1 was obtained from the Pseudomonas Genetic Stock Center (http://www.pseudomonas.med.ecu.edu). To enable visualisation of PAO1 cells, GFP constitutively expressed on plasmid pMRP962 was used.
Growth conditions
Cultures were propagated from −80 °C freeze culture stocks and grown overnight (ON) in lysogeny broth (LB) for approximately 18 h at 37 °C under continuous shaking at 180 RPM. Subsequently, the LB ON culture was used for inoculation in low nutritional R2A broth (Lab M Limited, UK) supplemented with 0.05 M Tris-HCl buffer (pH 7.6) and 0.5% glucose (abbreviated R2A), and left to acclimatise ON until further use. The medium to volume ratio was 1:2.5.
Bead preparation
Alginate beads of 2.4 mm ± 0.1 mm in diameter (mean ± standard deviation) were prepared by a previously described method,14 with the exception of adding 10 mM potassium nitrate (KNO3−) (P8394, Sigma-Aldrich, USA) to some of the beads. In all experiments, beads were incubated in R2A medium (with or without 10 mM KNO3−) at 100 RPM at 37 °C for 24 or 48 h, respectively. For normoxic conditions, culture flasks were sealed with cotton. To achieve anoxic growth conditions, the medium was flushed with nitrogen gas (N2) for 5 min, where after the culture flask was immediately sealed airtight.
The bacterial growth of P. aeruginosa in the alginate beads was studied after 24 and 48 h in response to four permutations of the growth conditions: (i) Normoxic culture without NO3−, (ii) normoxic culture with NO3−, (iii) anoxic culture without NO3−, and (iv) anoxic culture with NO3−. Separate culture flasks were prepared for each time point, but all cultures originated from the same two batches of alginate beads (with and without NO3−) inoculated with the same PAO1 ON culture.
Optical coherence tomography
We used a commercially available spectral-domain OCT system (Ganymede II, Thorlabs GmbH, Germany) equipped with an objective lens with an effective focal length of 36 mm, and a working distance of 25.1 mm (LSM03; Thorlabs GmbH, Germany). The operating principle and components of the OCT system are described in detail elsewhere63,64 and in Supplementary. In order to use the OCT signal to compare bacterial growth between experimental treatments, OCT measurements were performed under well-defined optical conditions in terms of OCT system settings. A single alginate bead was placed in a black screw cap filled with 800 µL of distilled water. Using z-axis OCT scans (so-called A-scans), the image was brought into focus via the manual focusing stage and by adjusting the reference light intensity as well as the position of the reference length.64 After optimisation of image acquisition settings, the configuration was not changed, and subsequent measurements on alginate beads were performed under identical conditions. Each alginate bead was first scanned at high resolution in three (technical) replicate cross-sectional scans (so-called B-scans) followed by one full 3D scan (so-called C-scan) and rendering of the entire bead. OCT imaging was done in (biological) triplicates for each treatment after 24 and 48 h of incubation, respectively.
Visualisation of OCT B-scan and C-scan was done with the manufacturers imaging software (ThorImage 4.2; Thorlabs GmbH, Germany) using the built-in brightness and contrast functions. The images were visualised assuming a constant refractive index of water (n = 1.33). The attenuation of the A-scan signal can be used to understand changes in the structure of the alginate bead. We extracted three vertical A-scans from the B-scans over the area surrounding the highest point of the alginate bead (Fig. S1). OCT images were extracted in B-scan mode with manually optimised brightness and contrast adjustment and assuming a refractive index of 1.33 (for water). These adjustments separated the background noise from the OCT signal generated from the alginate beads.
Microscopy
A confocal laser scanning microscope (Zeiss Imager.Z2, LSM710 CLSM; Zeiss) operated with the manufacturers software (Zen2010, version 6.0; Zeiss, Germany) was used for imaging alginate-encapsulated, GFP-tagged PAO1. Samples were prepared by cutting the beads in half with a sterile scalpel. Cut beads were mounted in the dents of a flow-cell with the cut surface facing upwards. 10 µL milliQ water was applied to the cut bead surface, and a cover glass was fixed to the flow-cell with silicone sealant, making sure the cut surface was in close contact with the cover glass. The flow-cell was mounted on the microscope, and images were taken from the cut surface of the bead enabling visualisation of the distribution of bacterial aggregates from the periphery of the beads and towards the bead interior. Imaging of GFP-tagged P. aeruginosa was done with a 40×/NA1.3 oil objective, using laser excitation at 488 nm, and an emission filter range from 495 to 605 nm with a peak at 510 nm. The resulting images were processed with Imaris image processing software (v8.3.1; Bitplane, Switzerland).
Microscopy combined with quantitative PNA-FISH
Alginate beads from the four permutations, sampled after 24 and 48 h, were stored for at least 24 h in 4% formaldehyde (Hounisen, Denmark) kept at 4 °C and supplemented with 0.25 CaCl2 for stabilisation. Subsequently, the beads were embedded in paraffin, cut in 4-µm thick sections with a standard microtome, fixed on cover slides and kept dark at 4 °C until further use; we note that this procedure did not work with the anoxic beads incubated without NO3− due to bead disruption. Staining of bead sections was conducted with a Texas Red-conjugated PNA-FISH probe specific for P. aeruginosa 16S rRNA (AdvanDx, USA) by previously described methods.14,21 A microscope slide with the fixated and PNA-FISH stained 4-µm sections of alginate beads was mounted on the microscope. Imaging of the alginate bead sections for quantitative PNA-FISH was done with identical image acquisition settings for all pictures: Fluorescence images were recorded as 1-μm step size z-stacks at an image resolution of 4096 × 4096 pixels, with an averaging of two scans, and 16-bit colour depth using a 63×/1.4NA oil immersion objective, laser excitation at 594 nm, and emission range from 600 to 695 nm with a peak of 615 nm. Quantification of microcolony fluorescence (mean intensity) was performed using Imaris image processing software (v8.3.1; Bitplane, Switzerland). Image thresholding was applied, which discriminated background and foreground fluorescence with the use of the Measuring Pro expansion pack for the Imaris software (Bitplane, Switzerland). A minimum colony size was set to 10 µm3 to avoid inclusion of planktonic bacteria in the analysis of bacterial aggregates.14 The mean fluorescence intensity of each micro colony was measured on a 16-bit scale from 0–65535 fluorescence intensity units (FU). Kragh et al.21 previously described a linear correlation between growth rate and fluorescence intensity of PNA-FISH stained rRNA molecules in P. aeruginosa. Based on this relationship, the fluorescence intensity of PNA-FISH stained samples could be used as a proxy for apparent growth rate. Colony distance from the discernible periphery of alginate beads was measured manually for individual colonies using the measuring tool in the Imaris software (Bitplane, Switzerland) and was plotted against fluorescence intensity.
Colony-forming units
For quantification of CFU, beads were dissolved using a solution of Na2CO3 (0.05 M) and citric acid (0.02 M).65 The dissolved bead slurry was serially diluted before plating on LB plates for enumeration of cells via colony formation. CFUs were determined in biological triplicates.
Microsensor measurements
Beads were submerged in a Petri dish filled with pre-warmed R2A medium (with or without NO3−) that was kept at 37 °C under gentle ventilation by a fine air or nitrogen stream directed towards the surface via a Pasteur pipette connected to an air pump or N2 gas cylinder. Profiles of O2 concentration vs. depth in the alginate bead were measured with an amperometric O2 microsensor (25 µm tip diameter; OX25, Unisense A/S, Denmark) mounted on a motorised micromanipulator (MU1 Pyro-Science GmbH, Germany). Similarly, N2O concentration profiles were measured with an O2-insensitive amperometric N2O microsensor (25 µm tip diameter; N2O25, Unisense A/S, Denmark). All measurements were performed in 3−4 biological replicates. Both microsensors were connected to a pA metre (Unisense A/S, Denamark) that was interfaced to a PC via an A/D converter (Profix; Pyroscience GmbH, Germany). Linear calibrations of the microsensors were performed as specified by the manufacturer via measurements with the O2 microsensor in air saturated and O2-free water, and measurements with the N2O microsensor in N2O-free water, followed by measurement upon addition of defined aliquots of N2O saturated water.
Concentration profiles were recorded in beads incubated for 24 and 48 h after encapsulation of P. aeruginosa in alginate, respectively. The position, where the microsensor tip touched the bead surface (depth = 0), was determined visually with the aid of a USB microscope (model AM7515MZTL, dino-lite.eu) aiming after the centre of the uppermost bead surface. Profile measurements were conducted in steps of 50 or 200 µm through the bead. Microsensor positioning and data acquisition were done with dedicated profiling software (Profix; Pyro-Science GmbH, Germany).
To assess N2O concentration directly in the growth medium, the sensor tip was submerged in the growth medium and the resulting concentration was recorded as quickly as possible after removal of the cotton or rubber sealing.
Calculations of O2 and N2O flux
Net production of N2O and net consumption of O2 in the alginate beads were estimated as gas fluxes from measured steady-state concentration profiles, by a modified version of Fick’s first law of diffusion,66 where the slope of the profile in the alginate bead was calculated from three consecutive measurements, from the upper quasi-linear parts of the profiles:39
| 1 |
where J is the flux of O2 or N2O (nmol cm−2 min−1), D is the molecular diffusion coefficient of O2 (1.5 × 10−5 cm2 s−1)55 or N2O (2.76 × 10−5 cm2 s−1)67 in water at 37 °C, and C is the concentration of O2 or N2O (µmol L−1) at depth xn (µm), where n = a, b or c denotes three subsequent measurements at increasing depth (Eq. 1).
NO3− and NO2− quantification
The NO3− and NO2− concentration in fresh and spent NO3−-supplemented R2A medium during the time course of the experiments was quantified by the Griess colorimetric reaction (no. 780001, Cayman Chemicals, USA), in technical duplicates, as previously described.39
Imaging of O2
In order to image the O2 distribution within alginate beads harbouring P. aeruginosa, O2-sensitive sensor nanoparticles were incorporated into the beads. The sensor nanoparticles contained an O2-sensitive indicator (PtTFPP) and an insensitive reference dye (MY) and were prepared as described elsewhere.33,68 The sensor nanoparticles were added to the alginate-bacterial solution prior to bead preparation (sensor particle to alginate ratio: 1:50 vol/vol). After 24 h of growth, ratiometric O2 imaging of the bead was performed as described below.
For imaging, a bead was cut in half and placed on a cover glass with the cut surface facing the glass surface. The cut bead was covered with fresh medium and left to acclimate for a few minutes. A USB RGB microscope with built-in UV (405 nm) LED illumination (model AM4113-FVT, dino-lite.eu) was placed below the cover glass and was used to obtain images of the cut bead surface. After image acquisition, the obtained RGB images were split into the three colour channels (red, green, and blue) using the free image analysis software ImageJ (imagej.net). As the O2-sensitive indicator emits in the red channel and the reference indicator in the green channel, the ratio between these two channels could be used to obtain an O2 image. Ratios were calculated using the plugin Ratio Plus (https://imagej.nih.gov/ij/plugins/ratio-plus.html) and were linked to O2 concentration by a previously obtained calibration using the curve fitting function of ImageJ. For the calibration, alginate beads with sensor nanoparticles but without bacteria were submerged and imaged in water containing known O2 levels. The O2 levels were adjusted by flushing the water with an air/N2 mixture using a PC-controlled gas mixer (Sensorsense, The Netherlands). Detailed additional information on ratiometric O2 imaging using sensor nanoparticles can be found in recent publications.32,33,68
Statistics
Data were analysed for statistical significance with SPSS version 24 (IBM, USA) and GraphPad Prism version 6 (GraphPad Software, USA). Data were illustrated with GraphPad Prism. Group comparisons for quantitative PNA-FISH data (Fig. 3) were made using either Mann–Whitney U test (for growth depth calculated as IQR) or independent samples t-test on log transformed values (apparent growth rate, expressed as mean fluorescence intensity). Log growth rate and depth were correlated using Pearson correlation. Flux rates (Fig. 5) were compared by Mann–Whitney U test. Changes in CFU (Fig. 4), concentrations of NO3− and NO2− (Fig. 6), as well as differences in OCT dB signal over time (Fig. 1) were analysed by two-way ANOVA. Based on our experience with the effect of treatment and the standard deviation of the methods rarely exceeding 40% of the effect we expect that three replicates will allow us to detect significant differences with p < 0.05 and a power of 0.80. A two-sided α of 0.05 was considered significant.
Electronic supplementary material
Acknowledgements
We thank Heidi Marie Paulsen for cutting alginate beads for the quantitative PNA-FISH analysis. The project was supported by grants from the Danish Research Council/Natural Sciences (Sapere Aude Advanced grant to M.K.), the Danish Research Council/Technical and Production Sciences (M.Kü., P.Ø.J.), the Carlsberg Foundation (M.Kü., D.W.), and the Lundbeck Foundation (to T.B.).
Author contributions
M.S., M.Kü., K.K., and T.B. conceptualised the study, M.S. and M.Kü. wrote the main manuscript text, K.K. performed data acquisition for Figs. 5 and 7 and prepared Fig. 7, D.W. performed OCT data acquisition and analysis (Fig. 1, and Supplementary Figs. S1, S2), K.N.K. analysed data for Fig. 3, P.Ø.J. analysed the results and revised the manuscript, M.Ko. assisted with data collection for Fig. 6. All authors reviewed the manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies the paper on the npj Biofilms and Microbiomes website (10.1038/s41522-018-0047-4).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Thomas Bjarnsholt, Email: tbjarnsholt@sund.ku.dk.
Michael Kühl, Email: mkuhl@bio.ku.dk.
References
- 1.Stewart PS. Mechanisms of antibiotic resistance in bacterial biofilms. Int. J. Med. Microbiol. 2002;292:107–113. doi: 10.1078/1438-4221-00196. [DOI] [PubMed] [Google Scholar]
- 2.Costerton JW, Stewart PS, Greenberg EP. Bacterial biofilms: a common cause of persistent infections. Science. 1999;284:1318–1322. doi: 10.1126/science.284.5418.1318. [DOI] [PubMed] [Google Scholar]
- 3.Hall-Stoodley L, Stoodley P. Evolving concepts in biofilm infections. Cell Microbiol. 2009;11:1034–1043. doi: 10.1111/j.1462-5822.2009.01323.x. [DOI] [PubMed] [Google Scholar]
- 4.Bjarnsholt T, et al. The in vivo biofilm. Trends Microbiol. 2013;21:466–474. doi: 10.1016/j.tim.2013.06.002. [DOI] [PubMed] [Google Scholar]
- 5.Kirketerp-Moller K, et al. Distribution, organization, and ecology of bacteria in chronic wounds. J. Clin. Microbiol. 2008;46:2717–2722. doi: 10.1128/JCM.00501-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Flemming HC, Neu TR, Wozniak DJ. The EPS matrix: the “house of biofilm cells”. J. Bacteriol. 2007;189:7945–7947. doi: 10.1128/JB.00858-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Flemming HC, Wingender J. The biofilm matrix. Nat. Rev. Microbiol. 2010;8:623–633. doi: 10.1038/nrmicro2415. [DOI] [PubMed] [Google Scholar]
- 8.Mayer C, et al. The role of intermolecular interactions: studies on model systems for bacterial biofilms. Int. J. Biol. Macromol. 1999;26:3–16. doi: 10.1016/S0141-8130(99)00057-4. [DOI] [PubMed] [Google Scholar]
- 9.Burmolle M, et al. Biofilms in chronic infections—a matter of opportunity—monospecies biofilms in multispecies infections. FEMS Immunol. Med. Microbiol. 2010;59:324–336. doi: 10.1111/j.1574-695X.2010.00714.x. [DOI] [PubMed] [Google Scholar]
- 10.Pabst B, Pitts B, Lauchnor E, Stewart PS. Gel-entrapped Staphylococcus aureus bacteria as models of biofilm infection exhibit growth in dense aggregates, oxygen limitation, antibiotic tolerance, and heterogeneous gene expression. Antimicrob. Agents Chemother. 2016;60:6294–6301. doi: 10.1128/AAC.01336-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stewart PS, Franklin MJ. Physiological heterogeneity in biofilms. Nat. Rev. Microbiol. 2008;6:199–210. doi: 10.1038/nrmicro1838. [DOI] [PubMed] [Google Scholar]
- 12.Walters MC, Roe F, Bugnicourt A, Franklin MJ, Stewart PS. Contributions of antibiotic penetration, oxygen limitation, and low metabolic activity to tolerance of Pseudomonas aeruginosa biofilms to ciprofloxacin and tobramycin. Antimicrob. Agents Chemother. 2003;47:317–323. doi: 10.1128/AAC.47.1.317-323.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.James GA, et al. Microsensor and transcriptomic signatures of oxygen depletion in biofilms associated with chronic wounds. Wound Repair Regen. 2016;24:373–383. doi: 10.1111/wrr.12401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sønderholm, M. et al. Pseudomonas aeruginosa aggregate formation in an alginate bead model system exhibits in vivo-like characteristics. Appl. Environ. Microbiol. 83, 10.1128/AEM.00113-17 (2017). [DOI] [PMC free article] [PubMed]
- 15.Cowley, E.S., Kopf, S.H., LaRiviere, A., Ziebis, W. & Newman, D.K. Pediatric cystic fibrosis sputum can be chemically dynamic, anoxic, and extremely reduced due to hydrogen sulfide formation. Mbio6, 10.1128/mBio.00767-15 (2015). [DOI] [PMC free article] [PubMed]
- 16.Hassett DJ, et al. Pseudomonas aeruginosa hypoxic or anaerobic biofilm infections within cystic fibrosis airways. Trends Microbiol. 2009;17:130–138. doi: 10.1016/j.tim.2008.12.003. [DOI] [PubMed] [Google Scholar]
- 17.Worlitzsch D, et al. Effects of reduced mucus oxygen concentration in airway Pseudomonas infections of cystic fibrosis patients. J. Clin. Invest. 2002;109:317–325. doi: 10.1172/JCI0213870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bjarnsholt T, et al. Pseudomonas aeruginosa biofilms in the respiratory tract of cystic fibrosis patients. Pediatr. Pulm. 2009;44:547–558. doi: 10.1002/ppul.21011. [DOI] [PubMed] [Google Scholar]
- 19.Jensen PO, Kolpen M, Kragh KN, Kühl M. Microenvironmental characteristics and physiology of biofilms in chronic infections of CF patients are strongly affected by the host immune response. Acta Pathol. Microbiol. Immunol. Scand. 2017;125:276–288. doi: 10.1111/apm.12668. [DOI] [PubMed] [Google Scholar]
- 20.Kolpen M, et al. Polymorphonuclear leucocytes consume oxygen in sputum from chronic Pseudomonas aeruginosa pneumonia in cystic fibrosis. Thorax. 2010;65:57–62. doi: 10.1136/thx.2009.114512. [DOI] [PubMed] [Google Scholar]
- 21.Kragh KN, et al. Polymorphonuclear leukocytes restrict growth of Pseudomonas aeruginosa in the lungs of cystic fibrosis patients. Infect. Immun. 2014;82:4477–4486. doi: 10.1128/IAI.01969-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Borriello G, et al. Oxygen limitation contributes to antibiotic tolerance of Pseudomonas aeruginosa in biofilms. Antimicrob. Agents Chemother. 2004;48:2659–2664. doi: 10.1128/AAC.48.7.2659-2664.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bjarnsholt T, Tolker-Nielsen T, Givskov M, Janssen M, Christensen LH. Detection of bacteria by fluorescence in situ hybridization in culture-negative soft tissue filler lesions. Dermatol. Surg. 2009;35:1620–1624. doi: 10.1111/j.1524-4725.2009.01313.x. [DOI] [PubMed] [Google Scholar]
- 24.Perry-O’Keefe H, et al. Identification of indicator microorganisms using a standardized PNA FISH method. J. Microbiol. Methods. 2001;47:281–292. doi: 10.1016/S0167-7012(01)00303-7. [DOI] [PubMed] [Google Scholar]
- 25.Wagner, M. & Horn, H. Optical coherence tomography in biofilm research: a comprehensive review. Biotechnol. Bioeng. 9999, 10.1002/bit.26283 (2017). [DOI] [PubMed]
- 26.Blauert F, Horn H, Wagner M. Time-resolved biofilm deformation measurements using optical coherence tomography. Biotechnol. Bioeng. 2015;112:1893–1905. doi: 10.1002/bit.25590. [DOI] [PubMed] [Google Scholar]
- 27.Haisch C, Niessner R. Visualisation of transient processes in biofilms by optical coherence tomography. Water Res. 2007;41:2467–2472. doi: 10.1016/j.watres.2007.03.017. [DOI] [PubMed] [Google Scholar]
- 28.Toth L, et al. Optical coherence tomography for biofilm detection in chronic rhinosinusitis with nasal polyposis. Eur. Arch. Otorhinolaryngol. 2013;270:555–563. doi: 10.1007/s00405-012-2051-2. [DOI] [PubMed] [Google Scholar]
- 29.Andersen K, Kjaer T, Revsbech NP. An oxygen insensitive microsensor for nitrous oxide. Sens. Actuators B Chem. 2001;81:42–48. doi: 10.1016/S0925-4005(01)00924-8. [DOI] [Google Scholar]
- 30.Revsbech NP. Analysis of microbial communities with electrochemical microsensors and microscale biosensors. Methods Enzymol. 2005;397:147–166. doi: 10.1016/S0076-6879(05)97009-2. [DOI] [PubMed] [Google Scholar]
- 31.Moßhammer, M. et al. Design and application of an optical sensor for simultaneous imaging of pH and dissolved O2 with low cross-talk. ACS Sensors. 6, 681–687 (2016).
- 32.Koren K, Jakobsen SL, Kühl M. In-vivo imaging of O2 dynamics on coral surfaces spray-painted with sensor nanoparticles. Sens. Actuators B Chem. 2016;237:1095–1101. doi: 10.1016/j.snb.2016.05.147. [DOI] [Google Scholar]
- 33.Koren K, Brodersen KE, Jakobsen SL, Kühl M. Optical sensor nanoparticles in artificial sediments—a new tool to visualize O2 dynamics around the rhizome and roots of seagrasses. Environ. Sci. Technol. 2015;49:2286–2292. doi: 10.1021/es505734b. [DOI] [PubMed] [Google Scholar]
- 34.Kühl M, Rickelt LF, Thar R. Combined imaging of bacteria and oxygen in biofilms. Appl. Environ. Microbiol. 2007;73:6289–6295. doi: 10.1128/AEM.01574-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pamp SJ, Sternberg C, Tolker-Nielsen T. Insight into the microbial multicellular lifestyle via flow-cell technology and confocal microscopy. Cytom. Part A. 2009;75:90–103. doi: 10.1002/cyto.a.20685. [DOI] [PubMed] [Google Scholar]
- 36.Zumft WG. Cell biology and molecular basis of denitrification. Microbiol. Mol. Biol. Rev. 1997;61:533–616. doi: 10.1128/mmbr.61.4.533-616.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hassett DJ, et al. Anaerobic metabolism and quorum sensing by Pseudomonas aeruginosa biofilms in chronically infected cystic fibrosis airways: rethinking antibiotic treatment strategies and drug targets. Adv. Drug Deliv. Rev. 2002;54:1425–1443. doi: 10.1016/S0169-409X(02)00152-7. [DOI] [PubMed] [Google Scholar]
- 38.Thauer RK, Jungermann K, Decker K. Energy conservation in chemotrophic anaerobic bacteria. Bacteriol. Rev. 1977;41:100–180. doi: 10.1128/br.41.1.100-180.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kolpen M, et al. Nitrous oxide production in sputum from cystic fibrosis patients with chronic Pseudomonas aeruginosa lung infection. PLoS ONE. 2014;9:e84353. doi: 10.1371/journal.pone.0084353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Line L, et al. Physiological levels of nitrate support anoxic growth by denitrification of Pseudomonas aeruginosa at growth rates reported in cystic fibrosis lungs and sputum. Front. Microbiol. 2014;5:1–11. doi: 10.3389/fmicb.2014.00554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Grasemann H, et al. Nitric oxide metabolites in cystic fibrosis lung disease. Arch. Dis. Child. 1998;78:49–53. doi: 10.1136/adc.78.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Palmer KL, Mashburn LM, Singh PK, Whiteley M. Cystic fibrosis sputum supports growth and cues key aspects of Pseudomonas aeruginosa physiology. J. Bacteriol. 2005;187:5267–5277. doi: 10.1128/JB.187.15.5267-5277.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nguyen CT, et al. Noninvasive in vivo optical detection of biofilm in the human middle ear. Proc. Natl Acad. Sci. USA. 2012;109:9529–9534. doi: 10.1073/pnas.1201592109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xi, C.W., Marks, D., Schlachter, S., Luo, W. & Boppart, S.A. High-resolution three-dimensional imaging of biofilm development using optical coherence tomography. J. Biomed. Opt. 11, 10.1117/1.2209962 (2006). [DOI] [PubMed]
- 45.Huang D, et al. Optical coherence tomography. Science. 1991;254:1178–1181. doi: 10.1126/science.1957169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Levitz D, et al. Quantitative characterization of developing collagen gels using optical coherence tomography. J. Biomed. Opt. 2010;15:026019. doi: 10.1117/1.3377961. [DOI] [PubMed] [Google Scholar]
- 47.Vroom JM, et al. Depth penetration and detection of pH gradients in biofilms by two-photon excitation microscopy. Appl. Environ. Microbiol. 1999;65:3502–3511. doi: 10.1128/aem.65.8.3502-3511.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Morris RL, Schmidt TM. Shallow breathing: bacterial life at low O(2) Nat. Rev. Microbiol. 2013;11:205–212. doi: 10.1038/nrmicro2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gosmann B, Rehm HJ. Oxygen-uptake of microorganisms entrapped in Ca-alginate. Appl. Microbiol. Biot. 1986;23:163–167. doi: 10.1007/BF00261907. [DOI] [Google Scholar]
- 50.Chen J, Strous M. Denitrification and aerobic respiration, hybrid electron transport chains and co-evolution. Biochim. Biophys. Acta. 2013;1827:136–144. doi: 10.1016/j.bbabio.2012.10.002. [DOI] [PubMed] [Google Scholar]
- 51.Strohm TO, Griffin B, Zumft WG, Schink B. Growth yields in bacterial denitrification and nitrate ammonification. Appl. Environ. Microbiol. 2007;73:1420–1424. doi: 10.1128/AEM.02508-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Eschbach M, et al. Long-term anaerobic survival of the opportunistic pathogen Pseudomonas aeruginosa via pyruvate fermentation. J. Bacteriol. 2004;186:4596–4604. doi: 10.1128/JB.186.14.4596-4604.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schreiber K, et al. Anaerobic survival of Pseudomonas aeruginosa by pyruvate fermentation requires an Usp-type stress protein. J. Bacteriol. 2006;188:659–668. doi: 10.1128/JB.188.2.659-668.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vander Wauven C, Pierard A, Kley-Raymann M, Haas D. Pseudomonas aeruginosa mutants affected in anaerobic growth on arginine: evidence for a four-gene cluster encoding the arginine deiminase pathway. J. Bacteriol. 1984;160:928–934. doi: 10.1128/jb.160.3.928-934.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ploug H, Kuhl M, BuchholzCleven B, Jorgensen BB. Anoxic aggregates—an ephemeral phenomenon in the pelagic environment? Aquat. Microb. Ecol. 1997;13:285–294. doi: 10.3354/ame013285. [DOI] [Google Scholar]
- 56.Robertson LA, Kuenen JG. Aerobic denitrification—old wine in new bottles? Antonie Van Leeuwenhoek. 1984;50:525–544. doi: 10.1007/BF02386224. [DOI] [PubMed] [Google Scholar]
- 57.Alvarez-Ortega C, Harwood CS. Responses of Pseudomonas aeruginosa to low oxygen indicate that growth in the cystic fibrosis lung is by aerobic respiration. Mol. Microbiol. 2007;65:153–165. doi: 10.1111/j.1365-2958.2007.05772.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ettwig KF, et al. Nitrite-driven anaerobic methane oxidation by oxygenic bacteria. Nature. 2010;464:543–548. doi: 10.1038/nature08883. [DOI] [PubMed] [Google Scholar]
- 59.Staal M, Borisov SM, Rickelt LF, Klimant I, Kuhl M. Ultrabright planar optodes for luminescence life-time based microscopic imaging of O2 dynamics in biofilms. J. Microbiol. Methods. 2011;85:67–74. doi: 10.1016/j.mimet.2011.01.021. [DOI] [PubMed] [Google Scholar]
- 60.Borriello G, Richards L, Ehrlich GD, Stewart PS. Arginine or nitrate enhances antibiotic susceptibility of Pseudomonas aeruginosa in biofilms. Antimicrob. Agents Chemother. 2006;50:382–384. doi: 10.1128/AAC.50.1.382-384.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kolpen M, et al. Reinforcement of the bactericidal effect of ciprofloxacin on Pseudomonas aeruginosa biofilm by hyperbaric oxygen treatment. Int. J. Antimicrob. Agents. 2016;47:163–167. doi: 10.1016/j.ijantimicag.2015.12.005. [DOI] [PubMed] [Google Scholar]
- 62.Bjarnsholt T, et al. Pseudomonas aeruginosa tolerance to tobramycin, hydrogen peroxide and polymorphonuclear leukocytes is quorum-sensing dependent. Microbiology. 2005;151:373–383. doi: 10.1099/mic.0.27463-0. [DOI] [PubMed] [Google Scholar]
- 63.Fujimoto JG, Pitris C, Boppart SA, Brezinski ME. Optical coherence tomography: an emerging technology for biomedical imaging and optical biopsy. Neoplasia. 2000;2:9–25. doi: 10.1038/sj.neo.7900071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wangpraseurt, D., Wentzel, C., Jacques, S.L., Wagner, M. & Kühl, M. In vivo imaging of coral tissue and skeleton with optical coherence tomography. J. R. Soc. Interface14, 10.1098/rsif.2016.1003 (2017). [DOI] [PMC free article] [PubMed]
- 65.Mater DDG, Barbotin JN, Saucedo JEN, Truffaut N, Thomas D. Effect of gelation temperature and gel-dissolving solution on cell viability and recovery of 2 Pseudomonas putida strains co-immobilized within calcium alginate or k-carrageenan gel beads. Biotechnol. Tech. 1995;9:747–752. doi: 10.1007/BF00159242. [DOI] [Google Scholar]
- 66.de Beer, D., Stoodley, P. in The Prokaryotes (eds Dworkin, M., Falkow, S., Rosenberg, E., Schleifer, K.H., Stackebrandt, E.) 904–937 (Springer Science, New York, 2006).
- 67.Broecker WS, Peng TH. Gas-exchange rates between air and sea. Tellus. 1974;26:21–35. doi: 10.3402/tellusa.v26i1-2.9733. [DOI] [Google Scholar]
- 68.Elgetti Brodersen K, Koren K, Lichtenberg M, Kuhl M. Nanoparticle-based measurements of pH and O2 dynamics in the rhizosphere of Zostera marina L.: effects of temperature elevation and light-dark transitions. Plant Cell Environ. 2016;39:1619–1630. doi: 10.1111/pce.12740. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.