Abstract
Exosomes are small vesicles of endocytic origin, which are released into the extracellular environment and mediate a variety of physiological and pathological conditions. Here we show that Schistosoma mansoni releases exosome-like vesicles in vitro. Vesicles were purified from culture medium by sucrose gradient fractionation and fractions containing vesicles verified by western blot analyses and electron microscopy. Proteomic analyses of exosomal contents unveiled 130 schistosome proteins. Among these proteins are common exosomal markers such as heat shock proteins, energy-generating enzymes, cytoskeletal proteins, and others. In addition, the schistosome extracellular vesicles contain proteins of potential importance for host-parasite interaction, notably peptidases, signaling proteins, cell adhesion proteins (e.g., integrins) and previously described vaccine candidates, including glutathione-S-transferase (GST), tetraspanin (TSP-2) and calpain. S. mansoni exosomes also contain 143 microRNAs (miRNA), of which 25 are present at high levels, including miRNAs detected in sera of infected hosts. Quantitative PCR analysis confirmed the presence of schistosome-derived miRNAs in exosomes purified from infected mouse sera. The results provide evidence of vesicle-mediated secretion in these parasites and suggest that schistosome-derived exosomes could play important roles in host-parasite interactions and could be a useful tool in the development of vaccines and therapeutics.
Introduction
Schistosomiasis is a major parasitic disease, affecting >200 million people in 74 countries, the majority in sub-Saharan Africa1. The disease is caused by blood-dwelling flukes of the genus Schistosoma, primarily S. mansoni, S. haematobium and S. japonicum. Schistosomes have a complex life cycle that requires a snail as an intermediate host. The infective stage of the parasite is a small, short-lived larva (cercaria), which is released by infected snails into fresh water and rapidly invades a human host by penetration through the skin. Shortly after penetration, the cercaria transforms into an immature parasitic larva (schistosomulum), which enters the circulation and migrates towards the hepatic portal system and mesenteries (S. mansoni, S. japonicum) or the venous plexus of the urinary bladder (S. haematobium). Larvae grow during migration, eventually maturing into adult male and female worms, which become tightly coupled and produce large numbers of eggs. The pathology associated with chronic schistosomiasis is due to the eggs. Many eggs leave the body in feces or urine, but some are trapped in tissues and induce a granulomatous immune response, leading to progressive tissue fibrosis and organ damage. Deaths due to schistosomiasis have been estimated at >200,000 per annum in Africa alone. There is no vaccine for schistosomiasis and chemotherapy relies heavily on a single drug (praziquantel), raising concerns about drug resistance2 which highlights the need for more research into new vaccine and drug targets.
In this regard, the secretome is of particular interest. Molecules secreted by parasites play major roles in shaping host interactions and are promising targets for vaccine or chemotherapeutic intervention. Most studies of helminth excretory/secretory products (ESP) have focused on proteins. ESP proteome analyses have been reported for a variety of nematode species3–5 as well as flatworms, including the liver fluke Fasciola hepatica6,7 and several life stages of S. mansoni, S. japonicum and S. bovis8–14. Parasitic worms also secrete microRNAs (miRNA)15–19, which could play important roles in modulating host immune responses. Secreted miRNAs are increasingly recognized as mediators of cell communication20,21 and their regulatory roles in the immune system are well established22.
Despite these advances, and the considerable efforts made to identify schistosome ESPs, very little is known about the mechanisms by which these molecules are released. Many proteins secreted by helminths are unlikely to be released through the classical secretory pathway, for example metabolic enzymes that lack a signal peptide, cytoskeletal proteins, heat shock proteins, and membrane proteins. An alternative export mechanism for these proteins is through membrane-bound vesicles23,24. Extracellular secretory vesicles can be formed by outward budding of the plasma membrane or formed intracellularly and subsequently released to the outside. The best characterized of these vesicles are exosomes, which are small (60–150 nm), typically cup-shaped when observed by transmission electron microscopy (TEM) and are produced from multivesicular intermediates of the endocytic pathway24. There is growing evidence that many proteins secreted by parasites are enclosed within exosome-like vesicles (ELVs)19,25,26. Recent studies of S. japonicum27 and S. mansoni28,29 suggest that schistosomes may use extracellular vesicles as a mechanism of secretion, but the molecular content of these vesicles has only partly been characterized.
Exosomes have attracted considerable interest because of their roles as mediators of intercellular communication, particularly within the immune system and in disease states such as cancer24,30. Exosomes are powerful agents of cell communication because of the many ways they can interact with target cells; they can bind to cell surface receptors, indirectly changing cell behavior, they can fuse with the target cell membrane, releasing their contents into the cytosol, and/or they may be taken up and internalized into phagosomes31. Exosomes also provide an effective mechanism for delivery of parasite molecules to a host cell, either to modulate the immune response or some other host activity that is beneficial to the parasite. Studies primarily in Leishmania25,32,33 and H. polygyrus19 have shown that parasite-derived extracellular vesicles (EVs) can be taken up by host (mammalian) cells in culture; they induce changes in gene expression in host cells and have strong immunomodulatory properties, both in vitro and in infected animals. Leishmania exosomes may be the principal mechanism by which the parasite delivers virulence factors to the host cell25. These studies clearly implicate EVs as important mediators of the host-parasite interaction and underscore the need for further research into their mode of action.
Here we show that S. mansoni release ELVs and describe the proteins and miRNAs contained within these vesicles. The results help to elucidate the mechanism by which proteins and miRNAs are secreted and provide new insight into the role of secreted products in schistosome infections.
Results
Purification of ELVs
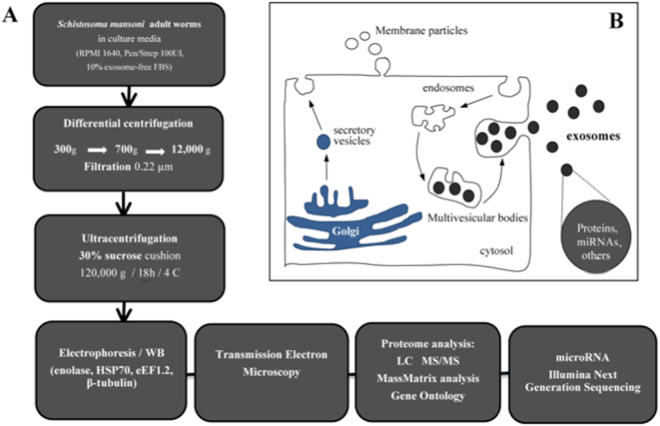
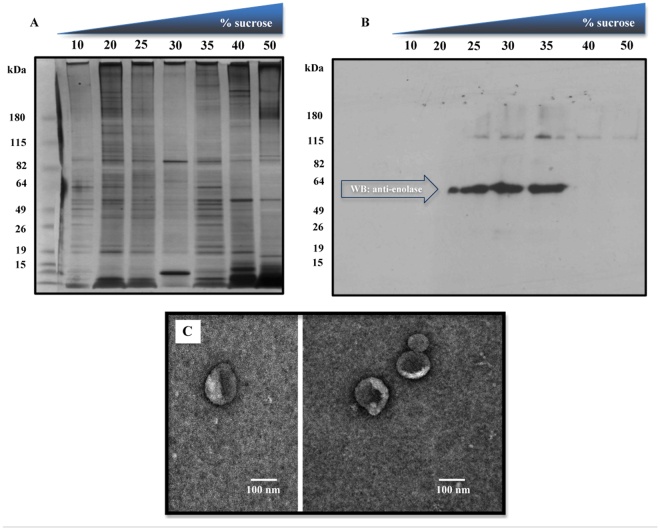
ELVs were initially purified from culture medium by differential centrifugation, followed by filtration and sucrose density ultracentrifugation (Fig. 1) to establish the sucrose concentration required for the sedimentation and enrichment of the S. mansoni ELVs. Purification was monitored by Western blot analysis using an anti-human enolase antibody, a common exosomal marker, which is highly conserved with S. mansoni enolase (75% sequence identity). Fractions exhibited distinct protein profiles, with the 20, 25, 35 and 40% sucrose fractions containing diverse proteins, possibly corresponding to protein aggregates, smaller vesicles or plasma membrane-derived vesicles (Fig. 2A). A predominant anti-enolase reactive band was recovered from the 25%, 30% and 35% sucrose fractions (Fig. 2B), consistent with the presence of exosomes34. Vesicles recovered from the 30% sucrose fraction were tested with antibodies against exosomal markers that are highly conserved in S. mansoni (anti-HSP70, anti-tubulin, anti-elongation factor 1; sequence identities 38–98% compared to mouse homologues) and all showed immunoreactive bands of the expected size (not shown). Subsequent purifications were done by overlaying the 0.2 µm-filtered 100,000 × g pellet directly onto a 30% sucrose cushion followed by ultracentrifugation34. EM analysis showed that the purified cup-shaped vesicles were ~100 nm in diameter (Fig. 2C), consistent with exosomes. Our experimental analysis cannot rule out the possibility that other types of secretory vesicles of similar shape and density may be present.
Figure 1.
Overview of the procedure used for isolation and characterization of secreted exosome-like vesicles from Schistosoma mansoni. (A) Adult male and female worms were cultured 48-72 h in media containing exosome-depleted serum. Vesicles were purified from the culture media by differential centrifugation, followed by filtration through a 0.2 µm membrane and ultracentrifugation on a discontinuous 25, 30, 35% sucrose gradient, as described72. The purification was monitored by western blot (WB) analysis, using antibodies against known exosomal markers (e.g. enolase) and electron microscopy (EM), prior to analyses of protein and miRNA content. (B) Schematic of the two major types of secretory extracellular vesicles. Membrane particles (or ectovesicles) are formed by outward budding of the plasma membrane. Exosomes are derived from the endocytic pathway via the formation of large multivesicular body (MVB) intermediates, which fuse with the plasma membrane releasing the vesicular cargo, exosomes, as well as other contents into an extracellular environment. Alternatively, the MVBs can be directed to lysosomes and degraded [see 24 for further details].
Figure 2.
Purification of exosome-like vesicles from S. mansoni. Vesicles were collected from worm culture media and partially purified through differential centrifugation as shown in Fig. 1. The resulting crude vesicular pellet was resuspended in PBS, filter sterilized (0.2 µm filter) and subsequently fractionated on a discontinuous 10–50% sucrose gradient. Gradient fractions were tested for total protein content by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) (A) and then western blotting (WB) with an antibody against enolase, a common exosomal marker (B). The results show enolase immunoreactivity between the 25% and 35% sucrose fractions. All subsequent purifications were performed by applying the filter-sterilized, crude vesicular pellets directly onto a discontinuous 25, 30, 35% sucrose gradient followed by ultracentrifugation, as described72 (C) Transmission electron microscopy analysis of purified exosome-like vesicles from S. mansoni. The scale bar indicates 100 nm.
Proteomics studies
LC-MS/MS analysis was repeated on three separate preparations of purified vesicles and candidate proteins identified by searching against the S. mansoni NCBI genome database. These analyses identified 130 S. mansoni proteins, including 125 known proteins and 5 annotated as hypothetical (Table 1). Matching peptides are shown in Table S2. The detected proteins include many homologues of common exosomal markers35 (Table 1). We note, in particular, energy-generating enzymes often associated with exosomes (e.g., enolase, pyruvate kinase, GAPDH, phosphoglycerate kinase 1), heat shock proteins (HSP70), cytoskeletal proteins (actin, tubulin, fimbrin), 14-3-3 proteins, tetraspanin, histones, vesicular traffic proteins (e.g., Rab proteins, dynein) and translational elongation factor eEF1. Their presence in the schistosome vesicles gives further confidence that these are exosomes. Importantly, proteomics data confirmed the presence of enolase.
Table 1.
Protein content of S. mansoni exosome-like vesicles.
| Accession# | Annotationa | peptidesb | ES proteinc references |
|---|---|---|---|
| Metabolic Enzymes | |||
| CCD82906.1 | Glycogen phosphorylase [Sm] | 50 | 8 |
| P16641.3 | Taurocyamine kinase [Sm] | 30 | 8, 13 |
| ABU49845.1 | Creatine kinase [Sm] | 28 | 9 |
| Q27877.1 | Enolase [Sm] d | 25 | 8–13 |
| CCD76480.1 | Pyruvate kinase [Sm] | 22 | 8, 9 |
| CCD81281.1 | Glucose-6-phosphate isomerase [Sm] | 22 | 9 |
| CCD82636.1 | Lactate dehydrogenase [Sm] | 18 | 8, 9, 11 |
| CCD75004.1 | Malate dehydrogenase [Sm] | 17 | 8, 9, 11, 12 |
| CCD75874.1 | Phosphoenolpyruvate carboxykinase [Sm] | 17 | 8, 9, 11, 12 |
| XP_002571535.1 | Ornithine-oxo-acid transaminase [Sm] | 9 | 9 |
| P41759.1 | Phosphoglycerate kinase [Sm] | 8 | 8, 9, 11–13 |
| CCD76263.1 | Transketolase [Sm] | 8 | 9, 12 |
| XP_002581246.1 | Phosphoglucomutase [Sm] | 7 | 8, 9 |
| CCD79691.1 | Fructose 1,6-bisphosphate aldolase [Sm] | 7 | 8, 9, 11–13 |
| CCD59265.1 | Aldehyde dehydrogenase, putative [Sm] | 4 | 9, 11 |
| CCD59437.1 | Adenosylhomocysteinase, putative [Sm] | 4 | |
| CCD75628.1 | Glyceraldehyde-3-phosphate dehydrogenase [Sm] | 3 | 8, 9, 11–13 |
| P09383.1 | Hypoxanthine-guanine phosphoribosyltransferase [Sm] | 3 | |
| AFH56663.1 | Methylthioadenosine phosphorylase [Sm] | 3 | 9 |
| XP_002577577.1 | Aconitate hydratase [Sm] | 3 | 11 |
| Q27778.1 | 6-phosphofructokinase [Sm] | 3 | 8, 9 |
| CCD81640.1 | Long-chain-fatty-acid-CoA ligase [Sm] | 2 | |
| CCD75611.1 | Glycogenin-related [Sm] | 2 | |
| CCD58348.1 | 6-phosphogluconate dehydrogenase,putative [Sm] | 2 | |
| Xenobiotic/Redox Metabolism | |||
| P09792.1 | Glutathione S-transferase 28 kDa isozyme [Sm] | 14 | 8, 9, 11–13 |
| XP_002582203.1 | Glutathione S-transferase 26 kDa [Sm] | 10 | 9, 11–13 |
| CCD59704.1 | Aldo-keto reductase, putative [Sm] | 6 | 11 |
| AAA29889.1 | Glutathione S-transferase, partial [Sm] | 5 | 9, 11 |
| CCD77979.1 | Glyoxalase I [Sm] | 3 | |
| Proteases | |||
| CCD59179.1 | Thimet oligopeptidase (M03 family) [Sm] | 19 | |
| CCD78710.1 | Leucine aminopeptidase (M17 family) [Sm] | 14 | 8, 9 |
| CCD76462.1 | Calpain (C02 family) [Sm] | 11 | 9 |
| P09841.3 | Hemoglobinase (Antigen SM32) [Sm] | 7 | |
| CCD74613.1 | Cathepsin B-like peptidase (C01 family) [Sm] | 6 | 9, 10 |
| XP_002572619.1 | Prolyl oligopeptidase (S09 family) [Sm] | 5 | |
| CCD77256.1 | SpAN g.p. (M12 family) [Sm] | 5 | |
| AHB79081.1 | Serine protease 2 precursor [Sm] | 3 | 10 |
| XP_002578277.1 | Subfamily M12B unassigned peptidase (M12 family)[Sm] | 3 | |
| CCD80658.1 | Family S9 non-peptidase homologue (S09 family) [Sm] | 3 | |
| CCD79473.1 | Xaa-Pro dipeptidase (M24 family) [Sm] | 2 | |
| Fatty acid binding | |||
| 1VYG | Fatty Acid Binding Protein [Sm] | 3 | 8–11, 13 |
| Transporters/Channels | |||
| CCD60986.1 | Plasma membrane calcium-transporting atpase [Sm] | 14 | |
| CCD78964.1 | Sodium potassium transport ATPase alpha subunit [Sm] | 8 | |
| CCD77470.1 | Glucose transport protein [Sm] | 7 | |
| CCD80392.1 | Cation-transporting ATPase [Sm] | 5 | |
| XP_002578298 | Choline transporter-like protein 2 (Ctl2) [Sm] | 4 | |
| CCD75891.1 | Aquaporin-3 [Sm] | 3 | |
| XP_002578385.1 | Chloride channel protein [Sm] | 3 | |
| CCD77770.1 | Anion exchange protein [Sm] | 2 | |
| Signal Transduction and Biological Regulation | |||
| Q26540.1 | 14-3-3 protein homolog 1 [Sm] | 10 | 8, 9, 12, 13 |
| CCD75054.1 | 14-3-3 epsilon [Sm] | 9 | 9, 12, 13 |
| XP_002578585.1 | Annexin [Sm] | 8 | 9, 10 |
| CCD74824.1 | sh3 domain grb2-like protein B1 (endophilin B1) [Sm] | 6 | |
| CCD74840.1 | Kynurenine aminotransferase [Sm] | 6 | |
| CCD77507.1 | Talin [Sm] | 6 | |
| XP_002577660.1 | Integrin alpha-ps [Sm] | 6 | |
| CCD74661.1 | Integrin beta subunit [Sm] | 6 | |
| CCD79674.1 | Atp-diphosphohydrolase 1 [Sm] | 5 | 8 |
| XP_002578604.1 | rpgr-interacting protein 1 related [Sm] | 4 | |
| CCD58613.1 | Hyperpolarization activated cyclic nucleotide-gated potassium channel, putative [Sm] | 4 | |
| XP_002576557.1 | N, N-dimethylarginine dimethylaminohydrolase [Sm] | 3 | |
| XP_002569666.1 | ral guanine nucleotide dissociation stimulator ralgds[Sm] | 3 | |
| CCD75701.1 | Proline-serine-threonine phosphatase interact protein [Sm] | 3 | |
| CCD78505.1 | Lip-related protein (liprin) alpha [Sm] | 3 | |
| XP_002581393.1 | Tetraspanin [Sm] | 3 | |
| XP_002580804.1 | Serine/threonine protein kinase [Sm] | 3 | |
| CCD78813.1 | rap1 [Sm] | 2 | |
| CCD58662.1 | Calponin homolog, putative [Sm] | 2 | 8, 13 |
| CCD78057.1 | Voltage-gated potassium channel [Sm] | 2 | |
| CCD80562.1 | Syntenin [Sm] | 2 | |
| Cytoskeletal/Structural | |||
| CCD58796.1 | Fer-1-related [Sm] | 40 | |
| P53471.1 | Actin-2 [Sm] | 19 | 9, 12 |
| CCD79944.1 | Alpha tubulin [Sm] | 13 | 9, 12 |
| CCD79871.1 | Tubulin beta chain [Sm] | 12 | 8, 9 |
| XP_002574516.1 | Tubulin subunit beta [Sm] | 12 | 8, 9, 12 |
| CCD76120.1 | Collagen alpha-1(V) chain [Sm] | 11 | |
| CCD60034.1 | Prominin (prom) protein, putative [Sm] | 10 | |
| CCD77450.1 | Rab GDP-dissociation inhibitor [Sm] | 7 | |
| CCD82967.1 | Actin [Sm] | 6 | 8, 9, 12, 13 |
| CCD60380.1 | Synaptotagmin, putative [Sm] | 6 | |
| AAA29882.1 | Fimbrin [Sm] | 5 | 8, 9 |
| CCD82452.1 | Signal recognition particle 68 kD protein [Sm] | 5 | |
| XP_002572341.1 | Gelsolin [Sm] | 4 | |
| CCD77020.1 | Intermediate filament proteins [Sm] | 4 | |
| CCD76586.1 | Cytoplasmic dynein light chain [Sm] | 3 | 8, 13 |
| XP_002572850.1 | Collagen alpha chain type IV [Sm] | 2 | |
| CCD79854.1 | Microtubule-associated protein 9 [Sm] | 2 | |
| CCD82782.1 | Rab-2,4,14 [Sm] | 2 | |
| CCD60258.1 | Rab11, putative [Sm] | 2 | |
| CCD74879.1 | Ran [Sm] | 2 | |
| Tegumental antigens | |||
| CCD81232.1 | 200-kDa GPI-anchored surface glycoprotein [Sm] | 24 | |
| CCD76403.1 | Tegumental protein Sm 20.8 [Sm] | 9 | 8, 13 |
| P14202.1 | Tegument antigen SmA 22.6 [Sm] | 7 | 10 |
| CCE94318.1 | Tegumental antigen [Sm] | 3 | |
| CCD59158.1 | Sm23, putative [Sm] | 2 | |
| CCD76286.1 | Sm29 [Sm] | 2 | |
| Histones | |||
| CCD77737.1 | Histone H3 [Sm] | 4 | 8 |
| AAG25601.1 | Histone H4 [Sm] | 4 | 8, 12 |
| CCD75757.1 | Histone H2B [Sm] | 3 | 8, 12 |
| Chaperones | |||
| CAZ34365.1 | Heat shock protein 70 (hsp70)−4, putative [Sm] | 7 | 8, 9, 12, 13 |
| Q26565.1 | Peptidyl-prolyl cis-trans isomerase [Sm] | 5 | 8, 11 |
| CCD76203.1 | Heat shock protein-HSP20/alpha crystallin family [Sm] | 5 | |
| XP_002577613.1 | Chaperonin containing t-complex protein 1 epsilon subunit tcpe [Sm] | 3 | |
| Translation | |||
| CCD76432.1 | Elongation factor 1-alpha (ef-1-alpha) [Sm] | 7 | 8, 9, 11, 13 |
| CCD79146.1 | Eukaryotic translation elongation factor [Sm] | 5 | 9, 12 |
| Others | |||
| XP_002576729.1 | SPRY domain containing protein [Sm] | 10 | |
| CCD60716.1 | Cell division control protein 48 aaa family protein (transitional Endoplasmic reticulum atpase), putative [Sm] | 7 | |
| CCD82557.1 | Band 4.1-like protein [Sm] | 6 | |
| CCD58670.1 | Centrosomal protein of 135 kDa (Cep135 protein) [Sm] | 6 | |
| CCD82376.1 | Excision repair helicase ercc-6-related [Sm] | 5 | |
| XP_002575991.1 | Ubiquitin (ribosomal protein L40) [Sm] | 4 | 8, 9, 11, 13 |
| CCD76953.1 | Ubiquitin-protein ligase BRE1 [Sm] | 4 | |
| XP_002578337.1 | Mixed-lineage leukemia 5 mll5 [Sm] | 4 | |
| CCD58616.1 | Basic helix-loop-helix transcription factor, putative [Sm] | 3 | |
| CCD77867.1 | Late embryogenesis abundant protein [Sm] | 3 | |
| CCD80225.1 | Zinc finger protein [Sm] | 2 | |
| Hypothetical/unnamed | |||
| CCD75352.1 | hypothetical protein Smp_140590 [Sm] (similar to Galectin family) | 9 | |
| XP_002570696.1 | hypothetical protein [Sm] (enolase, fragment) | 7 | |
| CCD81381.1 | hypothetical protein Smp_007640 [Sm] (1,6-glucosidase) | 7 | |
| CCD80386.1 | unnamed protein product [Sm] (actin-like protein) | 6 | |
| CCD78834.1 | hypothetical protein Smp_134750 [Sm] | 5 | |
| CCD82007.1 | hypothetical protein Smp_080920.3 [Sm] | 3 | |
| CCD77946.1 | hypothetical protein Smp_133590 [Sm] membrane, Ca binding vesicle fusion, neurotransmitter exocytosis | 3 | |
| CCD82509.1 | hypothetical protein Smp_024220 [Sm] | 2 | |
| CCD58986.1 | hypothetical protein Smp_155620 [Sm] | 2 | |
| CCD79363.1 | hypothetical protein Smp_145450 [Sm] | 2 | |
| XP_002575612.1 | hypothetical protein [Sm] | 2 | |
| CCD75804.1 | hypothetical protein Smp_159020 [Sm] | 2 | |
| CCD76102.1 | hypothetical protein Smp_006830.1 [Sm] | 2 | |
| Host proteins ( Mus musculus ) | |||
| XP_006536151 | importin-8 isoform X3 [Mm] | 4 | |
| XP_006521203.1 | keratin, type II cytoskeletal 1b isoform X1 [Mm] | 3 | |
| XP_006520948.1 | keratin, type II cytoskeletal 79 isoform X1 [Mm] | 3 | |
| XP_006521177.1 | keratin Kb40 isoform X2 [Mm] | 2 | |
| XP_006520575.1 | keratin, type I cytoskeletal 18 isoform X1 [Mm] | 2 | |
| XP_006523578.1 | axin-1 isoform X1 [Mm] | 2 | |
| XP_006531840.1 | 39 S ribosomal protein L21 [Mm] | 2 | |
aMS data were used to search the S. mansoni and mouse (Mus musculus) genome datasets available at NCBI; Sm, S. mansoni protein; Mm, mouse protein. bNumber of unique (non-overlapping) peptides matching the designated protein across three independent experiments. cProteins previously identified in proteomics analyses of schistosome excretory/secretory (ES) products. Relevant references are provided (refer to reference list for full citations). dProteins in boldface print are featured in ExoCarta’s “top 25” list of the most common exosomal markers from all different species and tissues.
The proteins in Table 1 were compared to previously reported schistosome ES proteins. S. mansoni secretome studies have been done mainly on larval stages or eggs8,11,12,14 but a comprehensive dataset is available for adult S. japonicum9, which was used for this comparative analysis. The S. japonicum secretome lists approximately 100 proteins, of which about half are “atypical” (or non-secretory) proteins that lack a signal peptide9. We found ~50% of these atypical secretory proteins (Table 1), in particular heat shock proteins, enolase, GAPDH, GST, 14-3-3 proteins and a fatty acid binding protein. These are all present in the purified vesicles and were previously described as some of the most abundant secreted proteins in S. japonicum and S. mansoni larvae. These results suggest that a significant proportion of protein secretion in schistosomes occurs through vesicles.
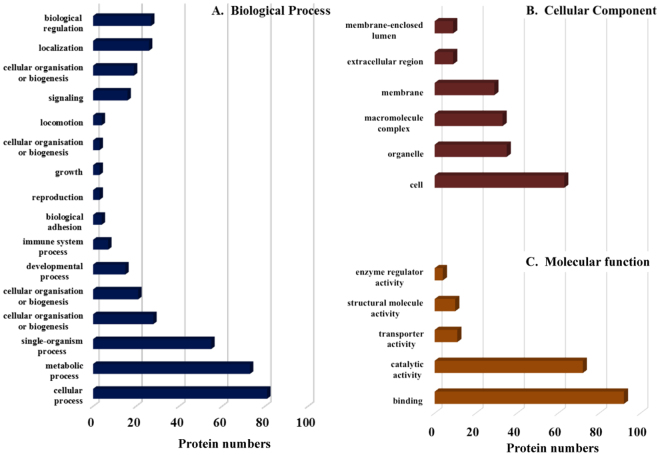
Exosomal proteins were classified by GO annotation according to molecular function, biological process and predicted cellular compartment (Fig. 3). The results show a high prevalence of proteins with catalytic and/or binding activity (“molecular function”) and proteins involved in metabolic and cellular processes (“biological process”). Enzymes are the most common type, representing nearly half of the proteins. The majority of these enzymes are involved in glucose metabolism. Enzymes from other pathways, for example taurine metabolism (taurocyamine kinase), amino acid metabolism (ornithine-oxo-acid transaminase), purine metabolism (hypoxanthine-guanine phosphoribosyltransferase, HGRPT) and detoxification (redox) metabolism (e.g. glutathione-S-transferase (GST), glyoxalase) were detected. The schistosome vesicles are rich in proteases, including metallopeptidases, cysteine peptidase and serine peptidases. The most abundant of these enzymes, based on the number of spectral matches, are thimet oligopeptidase, leucine aminopeptidase (LAP), calpain, hemoglobinase and cathepsin B.
Figure 3.
Gene Ontology (GO) analysis of proteins recovered from S. mansoni exosome-like vesicles. The identified proteins analyzed with Blast2GO83 and were classified according to Biological Process (A), Cellular Component (B) and Molecular Function (D), as defined by the GO consortium.
Many proteins were assigned to GO biological processes terms that relate to signal transduction (“signalling”, “response to stimulus”, “immune processes”) and biological regulation (Table 1). Some of these proteins are common exosomal markers that have broad spectrum signalling activities, for example 14-3-3 proteins36. We also identified proteins associated with calcium-dependent signalling (e.g., calponin), GTP-mediated signalling (rpgr interacting protein 1), a serine/threonine protein kinase, phosphatase-associated proteins, a grb2-like protein normally involved in receptor tyrosine kinase signalling, and two putative potassium channels, one of which is activated by cAMP. A few proteins listed in this category have activities in immune processes and blood coagulation pathways, which could be important for the host-parasite interaction. One example is annexin, a calcium-dependent phospholipid-binding protein implicated in a wide range of cellular processes. Some types of annexins have powerful anti-inflammatory activity37; others function as plasminogen receptors and stimulate fibrinolysis, thus reducing the formation of blood clots38. Also noteworthy are schistosome homologues of ATP-diphosphohydrolase 1, N.N-dimethylarginine dimethylaminohydrolase (DDAH), kynurenine aminotransferase and integrins. In mammals, these proteins have important immune and/or anti-clotting effects. ATP-diphosphohydrolase 1 suppresses inflammation, inhibits platelet aggregation and prevents blood clots by breaking down extracellular pro-inflammatory and prothrombotic ATP and ADP39. DDAH and kynurenine aminotransferase control the levels of important regulators of immune function by indirectly controlling nitric oxide production40 and the levels of kynurenine, a tryptophan metabolite that regulates vasodilation and immune responses41. Finally, integrins mediate cell adhesion and are implicated in a wide range of signalling mechanisms associated with immunity42. We found alpha and beta integrin subunits, as well as talin, which anchors integrins to the cytoskeleton and mediates subsequent signalling, in schistosome exosomes.
“Cellular compartment” analysis shows a high proportion of proteins associated with membrane-bound compartments (vesicles, organelles) as well as the plasma membrane. Membrane proteins constitute roughly 30% of identified proteins (Fig. 3). Some of the most abundant proteins in the dataset are membrane proteins, notably fer-1, a homologue of vertebrate dysferlin, which has been implicated in membrane structure, fusion, and repair associated with wound healing43. Other membrane proteins include transport proteins (e.g., aquaporin, glucose transporter), structural proteins (e.g., prominin), signalling proteins (e.g., tetraspanin, integrins, ion channels), membrane-associated enzymes (e.g., calpain) and a variety of schistosome tegumental antigens (e.g., 200 kDa GPI-anchored glycoprotein) (Table 1).
MS spectra were also searched against the Mus musculus database to test if the schistosome vesicles contained host proteins, which are known to be present in parasite-derived exosomes19,26. Only 7 mouse proteins were reliably identified (Table 1), of which 4 were keratins, a common contaminant in proteomics studies. Three additional mouse proteins (enolase, actin and plasma membrane calcium ATPase) were disregarded because the peptides matched the schistosome homologues with higher scores. Aside from keratins, the most significant hit was an isoform of importin, a protein typically involved in nuclear transport.
Analysis of miRNA content
The majority of miRNA reads mapped to non-miRNA sequences (mRNAs, other noncoding RNAs), repetitive sequences, or identified as “no hits” (Table S3). These sequences were removed from the datasets and not analyzed further. Of the remaining reads, we focused on those that aligned to S. mansoni or Platyhelminth (flatworm) miRNAs in miRbase. Reads that did not match known miRNAs were considered only if they mapped to the genome of S. mansoni and the extended sequences at the mapped genome positions were predicted to form hairpins.
The analysis identified 158 known and predicted miRNAs in whole worm extracts, 143 were also detected in exosomes but their relative abundances differed. This includes only sequences represented by >10 in one of the samples; miRNAs present at <10 reads in both samples were omitted from the analysis. The most abundant exosomal miRNAs (>100 reads) are described in Table 2 and a complete list is provided in Table S4. Among the top hits, roughly 70% are available in miRBase, including known S. mansoni (sma) sequences and conserved homologues from S. japonicum (sja) and the planarian Schmidtea mediterranea (sme). The remaining are novel, putative miRNAs that mapped to the S. mansoni genome within predicted hairpins. Most miRNAs are underrepresented in vesicles, in some instances by more than 100-fold (Fig. 4). Some, however, are present at the same or even higher levels; two of the more abundant miRNAs, sma-miR-71a and sma-miR-125b, were both present at about the same level in the two samples, and other miRNAs, such as sma-bantam and sma-miR-36-3P, were moderately enriched in vesicles. Bantam is an invertebrate-specific miRNA previously detected in serum of helminth-infected hosts and was reported to be secreted by the parasite16,17,44. Our results support these earlier studies and further suggest that sma-bantam is released, at least in part, in secretory vesicles.
Table 2.
List of most abundant miRNAs in purified S. mansoni exosomes.
| miR namea | miR sequence | Norm readsb exosome | Norm reads whole worm |
|---|---|---|---|
| sma-miR-125b_R-1c | TCCCTGAGACTGATAATTGCT | 43,394 | 49,431 |
| sma-bantam d | TGAGATCGCGATTAAAGCTGGT | 13,148 | 7,495 |
| sma-miR-71a_R + 1 | TGAAAGACGATGGTAGTGAGAT | 12,322 | 12,408 |
| sma-miR-125a | TCCCTGAGACCCTTTGATTGCC | 12,075 | 33,572 |
| sma-miR-36-3p | CCACCGGGTAGACATTCATTCGC | 8,977 | 4,146 |
| sma-miR-10-5p | AACCCTGTAGACCCGAGTTTGG | 3,281 | 19,497 |
| sma-miR-61_R + 1 | TGACTAGAAAGTGCACTCACTTC | 3,145 | 7,471 |
| sma-miR-2a-3p_R-1 | TCACAGCCAGTATTGATGAAC | 2,198 | 6,603 |
| sme-lin-4-5p_ | TCCCTGAGACCTTAGAGTTGT | 1,893 | 5,902 |
| sja-miR-2162-3p | TATTATGCAACGTTTCACTCT | 1,794 | 3,246 |
| sja-miR-277_R + 2 | TAAATGCATTTTCTGGCCCGTT | 653 | 1,354 |
| PC-5p-12974_124 e | GAGAGATTAAGACTGAACGCC | 580 | 298 |
| sja-miR-277_R + 1 | TAAATGCATTTTCTGGCCCGT | 532 | 1,717 |
| PC-3p-8606_176 | ACGGGCTTGGCAGAATTAGCGGGG | 304 | 167 |
| PC-5p-1634_720 | TCCCTGAGACCTTAGAGTTGTCT | 275 | 595 |
| sme-mir-749-p5 | GTCCGGGGTGCAGGCTTC | 275 | 188 |
| sma-let-7 | GGAGGTAGTTCGTTGTGTGGT | 254 | 3,535 |
| sma-miR-71b-5p | TGAAAGACTTGAGTAGTGAGACT | 243 | 254 |
| PC-5p-15294_107 | ACACTGCGAGGCATTGAAT | 207 | 563 |
| PC-3p-8590_176 | GAGATGGATAGTGGCTAGCATTT | 190 | 39 |
| PC-5p-10382_150 | CCTCCGGAATCCCATAGTACT | 160 | 156 |
| sma-miR-3479-3p | TATTGCACTAACCTTCGCCTTG | 140 | 1,545 |
| PC-5p-14055_115 | TGGCGCTTAGTAGAATGTCACCG | 121 | 200 |
| PC-5p-1776_672 | TGATGGATGTAGTATAGG | 117 | 436 |
| sja-miR-3492 | ATCCGTGCTGAGATTTCGTCT | 111 | 42 |
aOnly the most abundant exosomal miRNAs (>100 reads) are shown. A complete list of all the miRNAs that were detected is provided in Table S2. bNormalized (“norm”) reads were calculated after adjustment for the size of the library, as described in the Methods; cThe suffix R followed by a negative number indicates that the read sequence is shorter than the annotated miRNA in miRbase by one (R-1) or two nucleotides (R-2) at the 3′ end. Conversely, R + 1, R + 2 show that the read sequence is longer than the annotate miRNA by one or two nucleotides; dmiRNAs indicated by bold-face printare present at the same or higher levels in exosomes compared to whole worms. eThe prefix PC (potential candidate) is used to describe novel miRNA sequences.
Figure 4.
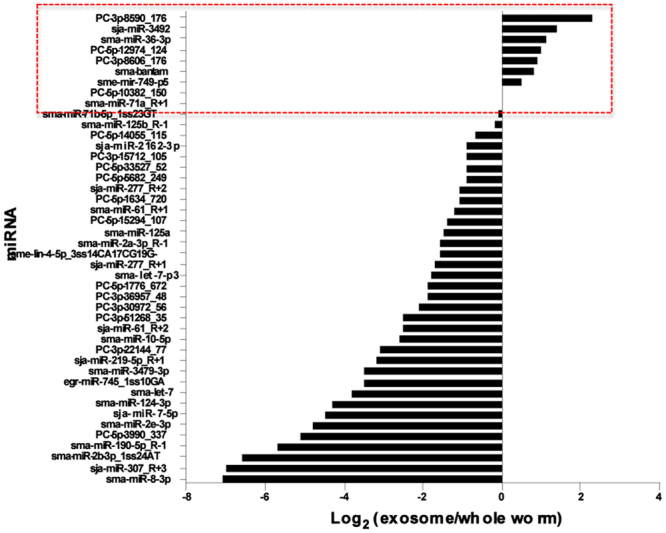
Comparative analysis of S. mansoni microRNAs (miRNA) obtained from whole worms and purified exosome-like vesicles. The data are shown as the Log2 ratio of normalized reads in the exosomal sample relative to the whole worm sample. Only the most abundant miRNAs are shown. Those miRNAs that are present at about the same level in the two samples, or are enriched in exosomes (Log2 ≥ 0) are marked.
Detection of exosomal miRNAs in infected mice
To test if S. mansoni releases exosome-like vesicles in vivo, we purified exosomes from serum of infected mice and screened for schistosome-derived miRNAs by qRT-PCR. The average worm burden was 30–40 worm pairs per mouse. As controls, we used uninfected mice maintained for the same length of time. Although most vesicles isolated using ExoQuick kit are likely of host origin, we hypothesized that even a small proportion of parasite-derived exosomes would be sufficient to detect specific Schistosoma miRNAs by qPCR. To facilitate detection, we focused on the abundant S. mansoni exosomal miRNAs from the RNA seq analysis and used a stem-loop RT method45 combined with TaqMan probe-based qPCR to improve specificity. Attempts to amplify miRNA using a polyA tailing RT method46 and SYBR green for qPCR produced non-specific results (not shown). In contrast, the stem-loop TaqMan probe method was able to specifically detect sma-miR-125a, sma-miR-125b, sma-bantam, sma- miR-71a (Fig. 5). The uninfected sample provides a measure of background amplification, presumably due to non-specific amplification of homologous sequences present in mouse-derived exosomes. The four miRNAs tested showed significant (p < 0.05) amplification compared to background levels and their median signal-to-noise ratios ranged from ~300- to 12,000-fold (Fig. 5). It was surprizing that ELVs isolated from infected mice showed notable difference in the various miRNA level. This was particularly notable for sma-miRNA-125b (Fig. 5).
Figure 5.
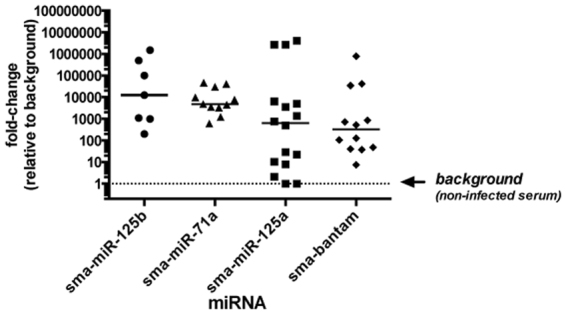
Quantitative qRT-PCR analysis of S. mansoni exosomal miRNAs in sera of infected mice. Circulating exosomes were purified from sera of S. mansoni –infected mice at 6–7 weeks post-infection or uninfected controls of the same age, using ExoQuick. RNA was extracted from the purified exosomes and then used for amplification of four S. mansoni miRNAs (Sma-mir-125a, Sma-mir-125b, Sma-mir-71a, Sma-bantam) by qRT-PCR. Data were median normalized relative to a “spike-in” synthetic miRNA17,80 and are shown as the fold-change relative to the uninfected control sample (background).
Discussion
Exosomes are important agents of intercellular communication in mammals, invertebrates and even prokaryotes23,24. For pathogens, exosomes constitute a powerful mechanism by which virulence factors and other bioactive molecules can be delivered to a host cell to promote infection. This has been well documented for microbial pathogens, in particular fungi, Leishmania and T. cruzi25,47. Evidence for exosome-mediated secretion in helminth infections is more limited19,26,27 and little is known about their functional roles. We further show that S. mansoni releases ELVs and characterize adult schistosome exosomal proteins and miRNAs.
Purified S. mansoni-secreted exosomes and biophysical characterization demonstrated that these vesicles were bona fide exosomes rather than crude vesicular pellets or large protein aggregates. Our approach selects for exosomes based on their distinctive size and floatation properties on a sucrose gradient34. Previous studies have shown that exosomes sediment at density corresponding to ~30% sucrose34, a finding confirmed here. This protocol minimizes contamination by larger membrane vesicles that typically have higher densities34. EM analysis confirmed that the ELVs used in the proteomic studies were ~60–150 nm in diameter and contained most of the “signature” (top 25) exosomal markers described in ExoCarta35. We recognize there may be other secretory vesicles (e.g., plasma membrane-derived ectovesicles) that have similar size and density. Thus, we refer to the material under study as exosome-like vesicles (ELVs).
A large proportion of the proteins recovered from S. mansoni ELVs are metabolic enzymes, particularly enzymes associated with glycolysis. The prevalence of glycolytic enzymes in exosomes has been reported in other organisms; many of the signature exosomal proteins in ExoCarta are glycolytic enzymes, such as enolase, aldolase, GAPDH, etc. For schistosomes, the discovery of these enzymes in vesicles helps to explain how they are secreted by the parasite, despite lacking classical secretion signal sequences. Why exosomes contain so many glycolytic enzymes is unclear. It is possible that exosomes increase glycolytic activity in a target cell after docking and fusion. Schistosome ELVs contain abundant glycogen phosphorylase and a glucose transporter, which would make more glucose available for metabolism, either through glycogenolysis or increased transport from the outside. Alternatively, these enzymes could have other activities. It is well known that glycolytic enzymes have “moonlighting” functions unrelated to glycolysis. For example, extracellular forms of enolase, aldolase and GAPDH bind to mammalian plasminogen48. If any of these enzymes is present on the surface of the schistosome vesicles, as was recently shown for Leishmania-derived exosomes49, it is tempting to speculate that the plasminogen binding activity could help prevent blood clots and facilitate parasite migration, an important activity for the blood-dwelling schistosome. Other “moonlighting” activities of extracellular glycolytic enzymes include adherence to the extracellular matrix, angiogenesis and the modulation of immune function48, all of which could be important for schistosomes.
Possible roles of exosomes in modulation of clotting pathways deserve consideration. Like other blood parasites, schistosomes must prevent blood clotting and remove clots once formed. This is of particular importance to S. mansoni because of the large size of the worm pair relative to the narrow mesenteric veins in which they reside. The presence of worms is likely to disrupt blood flow and stress the vessel wall, conditions that stimulate coagulation. Schistosomes employ a variety of mechanisms to prevent platelet aggregation and disrupt formation of clots50. Interestingly, some of the proteins found in the S. mansoni exosomes have anti-clotting activities, including ATP-diphosphohydrolase 1 (ATPDase-1), a membrane-associated protein that hydrolyzes extracellular prothrombotic ATP and ADP, inhibiting platelet aggregation and activation39. Previous studies of the schistosome homologue of ATPDase-1 revealed that the enzyme is present on the tegument51 and has ATP and ADP hydrolyzing activity52. Our results show that this enzyme is also secreted in vesicles. Secretion of ATPDase-1 was not reported in vitro52, but the methods employed may not have detected vesicle-associated enzyme. Exosomal ATPDase-1 may represent an important mechanism of haemostatic control. Other exosomal proteins with potential anti-clotting effects include the glycolytic enzymes listed above (enolase, GAPDH, aldolase) due to their plasminogen binding activity48, the schistosomal antigen Sm22.6, which inhibits thrombin53, annexin38 and the calcium-dependent protease calpain. In the host, calpain has a variety of functions, one of which is the regulation of platelet aggregation54. Vesicles carrying parasite calpain could alter normal calpain activity in the host and perhaps disrupt endogenous control of platelet activation. Likewise, if parasite proteins such as integrins and talin function in the same way as the mammalian homologues, vesicle-mediated delivery of these proteins to host cells could help the parasite to modulate the immune response and, importantly, control the formation of blood clots.
S. mansoni exosomes carry a variety of other proteases, including metalloproteases, cysteine and serine proteases that are believed to play important roles in exosome-mediated signaling55, contribute to invasion, migration, nutrient acquisition, immunomodulation and, haemostatic control56. Research on schistosome proteases has focused on secreted enzymes (e.g., cathepsin B) and cercarial elastases that contribute to skin penetration by infective larvae. There is a rich diversity of other proteases in schistosomes, most of which have only recently been identified57 and their functions are poorly understood. Our results show that some of these enzymes are secreted in vesicles and therefore could have important roles in the host-parasite interaction. Examples include a novel type 2 serine protease (SmSP2)58, a prolyl-oligopeptidase of the S9 serine protease family (SmPOP), recently shown to target bradykinin and angiotensin I59 and a homologue of leucine aminopeptidase (LAP). LAP is of considerable interest in flukes due to its involvement in the immune response to schistosome eggs and egg hatching60. LAP is present in the adult worm gut, where it contributes to digestion of blood proteins, in the parasite tegument61, and is secreted8,9, though it lacks a signal peptide. LAP secretion by F. hepatica is vesicle-mediated26 and our results suggest a similar mechanism for S. mansoni.
A proteomic analysis of ELVs derived from adult S. mansoni has been recently reported28. Although we identified some common proteins, notable discrepancies are evident (Table S5). Proteins found in both studies include enolase, GAPDH, glutathione-S-transferase, calpain, LAP, Sm20.8 and Sm22.6. Notable differences include glycogen phosphorylase, taurocyamine kinase and cathepsin B, high abundance proteins detected in this study that were also found in schistosome ESP8–10,13. Differences could be due to variations in protocols.
Proteomic analysis of schistosomulae-derived EVs revealed shared proteins between schistosomulae- and adult worm-derived EVs (e.g., taurocyamine kinase, enolase, glutathione S-transferase, calpain, 14-3-3 epsilon and Sm20.8; Table S5), but also differences. For example, schistosomulae EVs did not contain glycogen phosphorylase or glucose transporters29. LAP, the enzyme responsible for the final stage in catabolism of host hemoglobin61 was also absent from schistosomulae-derived EVs29, probably because the gut is not functional early in schistosomulum development62. Such variations in ESPs between different parasite life stages have previously been reported in helminths63,64 and could be used as specific life-stage markers in diagnosis.
Exosomes carry different types of RNA molecules, including miRNAs21. miRNAs are small molecules of ~23 nucleotides that regulate post-transcriptional silencing of genes. They bind to complementary sequences, typically in the 3′ untranslated region (3′-UTR) of a target transcript, resulting either in translational repression or sequence-specific RNA degradation65. miRNAs can be secreted and much of that secretion is vesicle (exosome)-mediated. Exosomal miRNAs are more stable than extravesicular forms and can be delivered to a target cell when exosomes fuse with the cell membrane21. Helminth parasites have a rich diversity of miRNAs44,65–68, some of which are secreted15–17,19,69, and helminth-secreted miRNAs modulate expression of specific host genes19, suggesting an important role in the host-parasite interaction. Here we provide evidence of vesicle-mediated miRNA secretion in adult schistosomes. Over 140 miRNAs were identified in purified vesicles, some at very high levels, including miRNAs found in sera of schistosome-infected animals16,17. We confirmed the presence of schistosome miRNAs in circulating exosomes from infected mice by qRT-PCR, indicating that exosomal miRNA secretion occurs in the infected host.
The biological relevance of schistosome exosomal miRNA secretion is unknown. Preliminary searches for potential human targets using computational tools (TargetScan Custom (5.2)) detected conserved seed regions in many of the schistosome miRNAs described here, suggesting these could, in principle, recognize human transcripts. For example, sma-miR125b, an abundant miRNAs in schistosome vesicles, has >600 potential human targets based on a conserved 8-mer seed region of the mature miRNA. Similar analysis of Sma-bantam, an invertebrate-specific miRNA, identified 39 potential human targets (not shown). These bioinformatics analyses must be refined and combined with gene expression studies in the context of different host cell environments to elucidate miRNA function. As previously observed in D. immitis64, it is likely that ELVs secreted by male and female schistosomes vary in content. It was also noted that the level of some miRNAs, in particular sma-miR125b, showed a broad variation in the level of miRNA found in the ELVs isolated from the serum of individual mice. These differences could be due to; i) genetic difference in the murine host since CD1 are outbreed mice which may have different responses to S. mansoni infections, ii) difference in worm burdens, or iii) possible differences in the stability of the miRNA transcript. It is also possible that some of the vesicular traffic is directed towards other schistosomes as a mechanism of animal-to-animal communication, as suggested recently for C. elegans70. Future studies will need to consider potential miRNA targets in the parasite as well as the host. miRNAs found in schistosomulae EVs29 were not among the 25 most abundant miRNAs in adult ELVs (Table 2), suggesting that the presence of these miRNA in schistosome ELVs may be stage-specific.
It is easy to envision that vesicles secreted within the confines of the mesenteric veins could directly target the endothelial lining of the blood vessels, platelets or cells of the immune system. Most research on parasite-derived vesicles has focused on the immune response. Protozoal pathogen exosomes have either pro- or anti-inflammatory effects, depending on the parasite and type of vesicle25, and modulate expression of a variety of host genes associated with immunity71. Whether these effects extend to schistosomes is unclear. A recent study of S. japonicum reported that treatment of macrophage RAW264.7 cells with parasite-derived vesicles promoted M1-type polarization and increased production of pro-inflammatory cytokines such as TNF-α and IL-1227. We tested S. mansoni exosomes in cultures of bone marrow-derived mouse (C57/BL6) macrophages and detected no change in TNF-α or IL12 production at concentrations of up to 50 µg exosomal protein/ml (data not shown). This discrepancy could be a function of the different target cells (RAW264.7 versus bone-marrow derived primary cells), different purity of vesicle preparations or different species of schistosome.
We have characterized miRNAs present in adult s. mansoni ELVs, which constitute the first report of exosomal miRNA secretion from adult schistosomes. We also provide a molecular profiling of exosomal protein content, offering contrast to a previous report28 (Table S5), which is possibly due to differences between our protocols. Both proteomic and miRNA analysis of schistosomulae-derived EVs29 revealed variations between schistosomulae- and adult worm-derived EVs (Table S5), which could serve as specific life-stage markers in diagnosis. However, further investigation will be required to exploit their potential in the development of vaccines, therapeutics and diagnostic methods.
Methods
Parasites
Schistosoma mansoni adults were obtained from CD1 mice after a six week experimental infection by perfusion72. Mice were maintained in the animal facility at the Small Animal Research Unit (SARU) on the Macdonald Campus of McGill University (Montreal, Canada) according to the McGill University Animal Care Committee (Permit # 2001–3346). Briefly, mice were euthanized by CO2 asphyxiation and cervical dislocation and Schistosoma mansoni adults harvested from mesenteric venules and thoroughly washed with Phosphate-Buffered Saline (PBS), and maintained in RPMI-1640 medium supplemented with 100 U penicillin, 100 mg/mL streptomycin, and 10% exosome-depleted Fetal Bovine Serum. Exosome-depletion was performed in advance by ultracentrifugation of FBS under sterile and apyrogenic conditions for 18 h at 120,000 × g and 4 °C as described73. FBS was of the highest grade and lowest endotoxin level (Life Technologies; Ref: 16000). Adult male and female worm pairs were maintained in 6-well plates for 72 h at a density of 20 worms (10 males and 10 females)/well in 10 ml culture medium at 37 °C. Conditioned medium was collected after 48 and 72 h. Worms remained active during this period of incubation, with no apparent change in morphology or movement. All experiments and protocols were approved by the McGill University Animal Care Committee according to the high standards established by the Canadian Council on Animal Care.
Extracellular vesicle isolation
ELVs were purified by differential centrifugation followed by membrane filtration and sucrose density ultracentrifugation as described34,73. Briefly, after 48 and 72 h in culture, the parasite conditioned culture medium was collected and centrifuged first at low speed (300 × g/ 20 min), then at 3,000 × g (20 min) to remove larger debris, and the supernatant was centrifuged at 12,000 × g for 45 min. Supernatants were filter-sterilized using a 0.22 µm hydrophilic PVDF Durapore membrane (EMD Millipore; SVGV01015) and centrifuged at 100,000 × g for 2 h using a SW-41 rotor on a Beckman-Coulter ultracentrifuge. The pellet was re-suspended in sterile cell culture grade PBS (Life Technologies) and fractionated on a discontinuous 10–50% sucrose gradient (1.03–1.23 g/cm3), using a SW-28 rotor at 120,000 × g for 18 h. Alternatively, exosomes were purified by loading the re-suspended pellet onto a sucrose step gradient (25%, 30% and 35%)34, then centrifuged at 120,000 × g for 18 h as above. Following centrifugation, the 30% sucrose fraction was collected, diluted 4-5 fold with sterile PBS and vesicles were re-pelleted by centrifugation in an SW-28 rotor at 100,000 × g for 2 h. The pellets were washed once by re-suspension in PBS and centrifugation repeated at 100,000 × g. For the last spin, the pellets were re-suspended in 1.0 ml PBS, transferred to 1.5 ml Beckman-Coulter ultracentrifuge tube and centrifuged in an Optima TL100 tabletop ultracentrifuge (Beckman-Coulter) for 1.5 h at 100,000 × g using a TLA-100.3 rotor. Exosomes were quantified based on protein concentration using a Pierce BCA Protein Assay Kit (ThermoFischer Scientific). The pelleted material was snap-frozen and stored at −80 °C.
Western blot assays
Purified ELVs (30 μg total protein) were treated with lysis buffer (Invitrogen) supplemented with 50 mM Tris (2-carboxyethyl)phosphine (TCEP) and a cocktail of protease inhibitors (Sigma-Aldrich) for 15 min at room temperature, followed by heating for 15 min at 65 °C. Proteins were resolved on a 4–20% Tris–glycine gel (Novex, San Diego, CA) and transferred to a nitrocellulose membrane according to standard protocols. Proteins were detected using goat anti-rabbit enolase antibodies (Santa Cruz Biotechnology; sc-7455). Secondary antibodies were a goat anti-rabbit IgG HRP-conjugate from Millipore (EMD Millipore).
Electron microscopy (EM)
EM analyses of S. mansoni ELVs were performed at the Facility for Electron Microscopy Research of McGill University using standard protocols34. Briefly, exosomes were fixed in 2% paraformaldehyde in PBS for 4 h, deposited on Formvar-carbon coated EM grids, washed in PBS, treated with 1% glutaraldehyde and then washed with water. Samples were stained with a solution of 4% uranyl oxalate, pH 7, then contrasted and embedded in a mixture of 2% methyl cellulose/4% uranyl acetate, pH 4 (9:1 v/v, methyl cellulose-UA) and finally placed in methyl cellulose-UA for 10 min on ice to increase the contrast. Grids were observed in a FEI Tecnai 12 transmission electron microscope operated at 80 kV. Images were acquired with a digital AMT XR80C CCD Camera System.
Proteomics analysis
For protein identification purified ELVs (200 μg total protein) were dissolved in a MS-compatible protein solubilizer (Invitrosol LC/MS Protein Solubilizer, Invitrogen) according to the manufacturer’s protocol and diluted with 5 volumes 50 mM ammonium bicarbonate, pH 8.5. Samples were heated to 60 °C for 30 min and sequentially reduced with 5 mM DTT (60 °C/30 min), alkylated with 5 mM iodoacetamide (20 °C in the dark/30 min) and finally treated with a small amount of DTT (10 µl 10 mM DTT) at 20 °C for 30 min to neutralize iodoacetamide. Proteins were quantified with a Pierce BCA Protein Assay Kit and samples were treated with Promega Gold Mass Spectrometry Grade Trypsin (1:50 enzyme:substrate ratio) and incubated for 14–16 h at 37 °C with shaking. Digestions were stopped with 1% trifluoroacetic acid (TFA) (Sigma-Aldrich) and peptides were purified with a C18 Zip-Tip (Millipore) and the peptide eluate was dried under vacuum. Peptides were reconstituted in 10 µl 0.1% formic acid before analyzing by LC-MS/MS. For analysis, peptides were fractionated on a 360 μm ID x 4 mm C18 trap column prior to separation on a 150 µm ID x 10 cm nano-LC column (Jupiter C18, 3 μm, 300 Å, Phenomex) and sequenced using an ESI-LC-MS/MS system, LTQ-Orbitrap Elite hybrid mass spectrometer with a nanoelectrospray ion source (ThermoFisher, San Jose, CA) coupled with an Eksigent nano-LC 2D pump (Dublin, CA). Peptides were separated using a linear gradient of 5–40% acetonitrile (0.2% formic acid) for 53 min and a flow rate of 600 nl/min. MS survey scans were acquired in profile mode at a resolution of 60,000 at m/z 400. MS/MS spectra were acquired in the ion trap using collision-induced dissociation (CID) for multiply charged ions exceeding a threshold of 10,000 counts. Three independent LC-MS/MS experiments were performed.
Protein data analyses
MS data were acquired using Xcalibur (v. 2.0 SR1). Peak lists were then generated with Mascot distiller (v. 2.1.1, Matrix Science) and MS processing was achieved using the LCQ_plus_zoom script. Database searches were performed against the S. mansoni proteome dataset available at the National Center for Biotechnology Information (NCBI) and the mouse proteome (Mus musculus) to test for the presence of host proteins, using MassMatrix (v. 2.4.2). The following search parameters were consistently applied: trypsin digestion with two missed cleavages allowed, cysteine carbamidomethylation as a fixed modification, methionine oxidation as variable modification, 0.8 Da tolerance for fragment ion masses, 10 parts per million mass tolerance for precursor ion masses and a minimum peptide length of 6 amino acids. A decoy reversed sequence database was included in every search. Peptide matches were considered significant if they exceeded the threshold statistical scores defined by MassMatrix74,75 and the false discovery rate was <5% (p < 0.05), as determined by decoy database searching. Functional annotation of protein sequences was done with Blast2GO software76 against the entire non-redundant NCBI nr database using default parameters. Annotation was augmented with Annotation Expander (ANNEX)77 and InterPro database scans were performed within Blast2Go to retrieve additional GO terms associated with functional domains.
Illumina RNA sequencing
S. mansoni-derived exosomes were purified from worm-conditioned culture media as described above and the RNA contained within the vesicles was isolated using a Norgen Total RNA extraction kit (Thorold, Canada), according to the manufacturer’s protocol. Total RNA was extracted from adult male and female pairs using the same kit. All subsequent steps were performed at LC Sciences, LLC (Houston, Texas, USA) using their standard protocols. Briefly, S. mansoni ELV (1000 worm pairs – 20 ng of miRNA) and whole worm (400 worm pairs – 100 ng of miRNA) small RNA libraries were generated, reverse-transcribed (RT) and the resulting cDNA constructs were purified using an Illumina Truseq™ Small RNA Preparation kit. ELV and whole worm cDNA libraries were used for cluster generation on a Cluster Station fluidics device and then sequenced on an Illumina GAIIx instrument, according to vendor’s instructions. Raw sequencing reads (40 nt) were obtained using Illumina’s Sequencing Control Studio software version 2.8 (SCS v2.8), following real-time sequencing image analysis and base-calling by Illumina’s Real-Time Analysis version 1.8.70 (RTA v1.8.70). A proprietary pipeline script, ACGT101-miR v4.2 (LC Sciences), was used to analyze the data. Starting with raw sequencing reads, a series of digital filters (LC Sciences) were employed to remove reads <15 nt and unmappable sequences (low quality reads, adaptors and other “impurities” due to sample preparation and sequencing chemistry). The remaining sequences with lengths between 15 and 32 nt were mapped to S. mansoni and related Platyhelminth (flatworm) miRNAs available in the latest release of miRBase (v.20) (http://mirbase.org) (Accessed September 18, 2015). Mapping was also performed against other small RNAs in the RFam database (http://rfam.janelia.org), against repetitive sequences in RepBase (http://www.girinst.org/repbase) and the latest assemblies of the S. mansoni genome and transcriptome available at the S. mansoni GeneDB78,79 (Accessed September 28, 2015). For comparisons between ELV and whole worm samples, the number of read counts from each sample was tracked during mapping and normalized to account for differences in library size. Normalization was achieved by dividing the counts by a scaling factor that reflected the size of each library. Scaling factors were calculated according to a median count method80 as outlined by LC Sciences.
Isolation of ELV RNA from mouse serum
Serum was collected from individual S. mansoni-infected mice 6–7 weeks post-infection and from age-matched uninfected controls. Exosomes were isolated with an ExoQuick Exosome Precipitation kit (SBI System Biosciences), starting with 500 µl serum per purification according to the recommendations of the manufacturer. The resulting exosomal pellets were resuspended in sterile lysis buffer and the RNA contained within the vesicles was isolated by column purification as specified in the kit protocol. A constant amount (1011 copies) of a spike-in synthetic RNA oligonucleotide (5′- CGUAUCGAGUGAUGUCACGUA- 3′) was added at the time of exosomal lysis, prior to RNA purification, and then carried through all additional steps. The spike-in RNA does not match any known miRNA in miRBase and was used for normalization of qRT-PCR data as described17.
Taqman qRT-PCR
Five highly abundant schistosome-derived miRNA sequences (Sma-Bantam, Sma-miR-71a, Sma-miR-36-3p, Sma-miR-125a and Sma-miR-125b) were selected for PCR amplification. At least 4 mismatches distinguished these miRNAs from the closest vertebrate miRNA homologue identified in miRBase, as determined by BLASTn alignments. A stem-loop method of miRNA amplification81 was used throughout the study. Target-specific stem-loop RT primers were synthesized and re-folded as described45. RT was performed according to standard protocols in a 20 µl reaction containing 0.5 mM dNTPs, 200 units MMLV reverse transcriptase (Life technologies) (or no enzyme in the -RT control tubes), 1 × First strand RT buffer, 10 mM DTT, 20 units RNase out inhibitor, 5 nM RT stem-loop primer and either synthetic miRNA or serum-derived exosomal RNA as the template. qPCR amplifications were performed in a total volume of 20 µl containing Luminaris Probe qPCR colorless master mix (Thermo Scientific), 2 µl fresh RT product, 1.5 µM sequence-specific forward primer, 0.7 µM universal reverse primer and 0.8 µM hydrolysis TaqMan probe. Primer and probe concentrations are considered optimal for stem-loop qRT-PCR of miRNAs45. Reactions were performed in replicates of 2–4 in an Applied Biosystems 7500 Fast thermocycler (Life Technologies) according to the following cycling protocol: 2 min at 50 °C, 10 min at 95 °C, followed by 60 cycles of denaturation at 95 °C/15 s, annealing at 58 °C/45 s and extension at 70 °C/30 s. Quantitative qRT-PCR was first performed with synthetic miRNAs to optimize reaction conditions. Amplification efficiencies were calculated from the slopes of serial dilution curves (where Efficiency (E) = 10 (−1/slope)) and were determined to be approximately 90–100% for all five targets and the spike-in control. For qRT-PCR of serum-derived exosomal samples, we used a fixed volume (5 µl) of RNA corresponding to approximately 100 µl serum and RT was performed with two different stem-loop primers added simultaneously and at the same concentration (5 nM), one targeting the specific miRNA of interest and the other targeting the spike-in RNA used for data normalization. RT reactions were split in half and used for parallel qPCR amplification of the target miRNA and spike-in control, using appropriate primers. A complete list of all the primers and TaqMan probes used in these experiments are described in Table S1. Synthetic miRNA sequences and oligonucleotides (stem-loop RT primers, forward primers, universal reverse primer) and hydrolysis TaqMan probes were purchased from Life Technologies (Foster City, USA).
qPCR data analysis
A relative ΔCt method was used to compare qPCR data from schistosome-infected mice and uninfected controls. The method was adapted for the analysis of circulating miRNAs from serum and other biological fluids, where conventional data normalization based on a housekeeping gene is not feasible. Ct values were first “median normalized” to the spike-in oligo as described17,82 to correct for sample-to-sample variation and differences in RT efficiency. The normalized Ct (Ctn) values were then used to calculate the ΔCtn for each sample. This was done by subtracting a median Ctn of all the uninfected control samples (background amplification) from the Ctn of the test (infected) sample. Fold-change relative to the uninfected control was calculated from the formula 2−∆Ctn. The median Ctn values for the uninfected controls varied between 53 and 60 for the four miRNA targets. Samples where there was no detectable amplification of the target miRNA were arbitrarily assigned a maximum Ct value of 60 for data calculations.
Electronic supplementary material
Acknowledgements
The authors are grateful to the Biomedical Research Institute (Bethesda, MD, USA) and BEI Resources, (Manassas, VA, USA) for providing Biomphalaria glabrata snails infected with S. mansoni. We also thank Louis-Philippe Leroux and Rajesh Valanparambil for their assistance in exosome purification and analysis. This work is dedicated to the memory of our dearly departed friend and colleague, Professor Paula Ribeiro who supervised the majority of the work reported in this manuscript.
Author Contributions
P.R. and A.J. conceived the experiments; V.S., M.D., V.G. and A.G. conducted the experiments; P.R. and V.S. analyzed the data; P.R., A.J. and M.D. wrote the manuscript; M.D., J.A. and A.J. reviewed the manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Paula Ribeiro is Deceased.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-21587-4.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Gryseels B, Polman K, Clerinx J, Kestens L. Human schistosomiasis. Lancet. 2006;368(9541):1106–1118. doi: 10.1016/S0140-6736(06)69440-3. [DOI] [PubMed] [Google Scholar]
- 2.Wang W, Wang L, Liang YS. Susceptibility or resistance of praziquantel in human schistosomiasis: a review. Parasitol Res. 2012;111:1871–1877. doi: 10.1007/s00436-012-3151-z. [DOI] [PubMed] [Google Scholar]
- 3.Moreno Y, et al. Proteomic analysis of excretory-secretory products of Heligmosomoides polygyrus assessed with next-generation sequencing transcriptomic information. PLoS Negl Trop Dis. 2011;5:e1370. doi: 10.1371/journal.pntd.0001370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chehayeb JF, Robertson AP, Martin RJ, Geary TG. Proteomic analysis of adult Ascaris suum fluid compartments and secretory products. PLoS Negl Trop Dis. 2014;5(8):e2939. doi: 10.1371/journal.pntd.0002939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Soblik H, et al. Life cycle stage-resolved proteomic analysis of the excretome/secretome from Strongyloides ratti—identification of stage-specific proteases. Mol Cell Proteomics. 2011;10(12):M111.010157. doi: 10.1074/mcp.M111.010157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jefferies JR, Campbell AM, van Rossum AJ, Barrett J, Brophy PM. Proteomic analysis of Fasciola hepatica excretory-secretory products. Proteomics. 2001;1:1128–1132. doi: 10.1002/1615-9861(200109)1:9<1128::AID-PROT1128>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 7.Robinson MW, Menon R, Donnelly SM, Dalton JP, Ranganathan S. An integrated transcriptomics and proteomics analysis of the secretome of the helminth pathogen Fasciola hepatica. Mol Cell Proteomics. 2009;8:1891–1907. doi: 10.1074/mcp.M900045-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Knudsen GM, Medzihradszky KF, Lim KC, Hansell E, McKerrow JH. Proteomic analysis of Schistosoma mansoni cercarial secretions. Mol Cell Proteomics. 2005;4:1862–75. doi: 10.1074/mcp.M500097-MCP200. [DOI] [PubMed] [Google Scholar]
- 9.Liu F, et al. Excretory/secretory proteome of the adult developmental stage of human blood fluke. Schistosoma japonicum. Mol Cell Proteomics. 2009;8:1236–1251. doi: 10.1074/mcp.M800538-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pérez-Sánchez R, Ramajo-Hernández A, Ramajo-Martín V, Oleaga A. Proteomic analysis of the tegument and excretory-secretory products of adult Schistosoma bovis worms. Proteomics. 2006;6(Suppl 1):S226–236. doi: 10.1002/pmic.200500420. [DOI] [PubMed] [Google Scholar]
- 11.Mathieson W, Wilson RA. A comparative proteomic study of the undeveloped and developed Schistosoma mansoni egg and its contents: the miracidium, hatch fluid and secretions. Int J Parasitol. 2010;40:617–628. doi: 10.1016/j.ijpara.2009.10.014. [DOI] [PubMed] [Google Scholar]
- 12.Cass CL, et al. Proteomic analysis of Schistosoma mansoni egg secretions. Mol Biochem Parasitol. 2007;155:84–93. doi: 10.1016/j.molbiopara.2007.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Curwen RS, Ashton PD, Johnston DA, Wilson RA. The Schistosoma mansoni soluble proteome: a comparison across four life-cycle stages. Mol Biochem Parasitol. 2004;138:57–66. doi: 10.1016/j.molbiopara.2004.06.016. [DOI] [PubMed] [Google Scholar]
- 14.Wilson RA. Proteomics at the schistosome mammalian host interface: any prospects for diagnostics or vaccines? Parasitology. 2012;139:1178–1194. doi: 10.1017/S0031182012000339. [DOI] [PubMed] [Google Scholar]
- 15.Tritten L, et al. Detection of circulating parasite-derived microRNAs in filarial infections. PLoS Negl Trop Dis. 2014;8(7):e2971. doi: 10.1371/journal.pntd.0002971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cheng G, Luo R, Hu C, Cao J, Jin Y. Deep sequencing-based identification of pathogen-specific microRNAs in the plasma of rabbits infected with Schistosoma japonicum. Parasitology. 2013;140:1751–1761. doi: 10.1017/S0031182013000917. [DOI] [PubMed] [Google Scholar]
- 17.Hoy AM, et al. Parasite-derived microRNAs in host serum as novel biomarkers of helminth infection. PLoS Negl Trop Dis. 2014;8(2):e2701. doi: 10.1371/journal.pntd.0002701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Manzano-Román R, Siles-Lucas M. MicroRNAs in parasitic diseases: potential for diagnosis and targeting. Mol Biochem Parasitol. 2012;186(2):81–86. doi: 10.1016/j.molbiopara.2012.10.001. [DOI] [PubMed] [Google Scholar]
- 19.Buck, A. H. et al. Exosomes secreted by nematode parasites transfer small RNAs to mammalian cells and modulate innate immunity. Nat Commun. 25(5), 5488, 10.1038/ncomms6488 (2014). [DOI] [PMC free article] [PubMed]
- 20.Turchinovich A, Samatov TR, Tonevitsky AG, Burwinkel B. Circulating miRNAs: cell-cell communication function? Front Genet. 2013;28(4):119. doi: 10.3389/fgene.2013.00119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hu G, Drescher KM, Chen XM. Exosomal miRNAs: biological properties and therapeutic potential. Front Genet. 2012;3(56):1–9. doi: 10.3389/fgene.2012.00056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fernandez-Messina L, Gutierrez-Vazquez C, Rivas-Garcia E, Sanchez-Madrid F, de la Fuente H. Immunomodulatory role of microRNAs transferred by extracellular vesicles. Biol Cell. 2015;107:61–77. doi: 10.1111/boc.201400081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Théry C. Exosomes: secreted vesicles and intercellular communications. F1000 Biol Rep. 2011;3:15. doi: 10.3410/B3-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Colombo M, Raposo G, Théry C. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu Rev Cell Dev Biol. 2014;30:255–289. doi: 10.1146/annurev-cellbio-101512-122326. [DOI] [PubMed] [Google Scholar]
- 25.Silverman JM, Reiner NE. Exosomes and other microvesicles in infection biology: organelles with unanticipated phenotypes. Cell Microbiol. 2011;13(1):1–9. doi: 10.1111/j.1462-5822.2010.01537.x. [DOI] [PubMed] [Google Scholar]
- 26.Marcilla A, et al. Extracellular vesicles from parasitic helminths contain specific excretory/secretory proteins and are internalized in intestinal host cells. PLoS One. 2012;7(9):e45974. doi: 10.1371/journal.pone.0045974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang L, et al. Exosome-like vesicles derived by Schistosoma japonicum adult worms mediates M1 type immune- activity of macrophage. Parasitol Res. 2015;114:1865–1873. doi: 10.1007/s00436-015-4373-7. [DOI] [PubMed] [Google Scholar]
- 28.Sotillo J, et al. Extracellular vesicles secreted by Schistosoma mansoni contain protein vaccine candidates. Int J Parasitol. 2015;46(1):1–5. doi: 10.1016/j.ijpara.2015.09.002. [DOI] [PubMed] [Google Scholar]
- 29.Nowacki FC, et al. Protein and small non-coding RNA-enriched extracellular vesicles are released by the pathogenic blood fluke Schistosoma mansoni. JEV. 2015;4(1):28665. doi: 10.3402/jev.v4.28665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miller, I. V. & Grunewald, T. G. Tumour-derived exosomes: Tiny envelopes for big stories. Biol Cell. 10.1111/boc.201400095 (2015). [DOI] [PubMed]
- 31.Mathivanan S, Ji H, Simpson RJ. Exosomes: Extracellular organelles important in intercellular communication. J Proteomics. 2010;73:1907–1920. doi: 10.1016/j.jprot.2010.06.006. [DOI] [PubMed] [Google Scholar]
- 32.Silverman JM, et al. Leishmania exosomes modulate innate and adaptive immune responses through effects on monocytes and dendritic cells. J Immunol. 2010;185(9):5011–5022. doi: 10.4049/jimmunol.1000541. [DOI] [PubMed] [Google Scholar]
- 33.Silverman, J. M. & Reiner, N. E. Leishmania exosomes deliver preemptive strikes to create an environment permissive for early infection. Front Cell Infect Microbiol.9(1), 26 10.3389/fcimb.2011.00026 (2012). [DOI] [PMC free article] [PubMed]
- 34.Théry C, Amigorena S, Raposo G, Clayton A. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr Protoc Cell Biol. 2006;3:22. doi: 10.1002/0471143030.cb0322s30. [DOI] [PubMed] [Google Scholar]
- 35.Simpson RJ, Kalra H, Mathivanan S. ExoCarta as a resource for exosomal research. J Extracell Vesicles. 2012;16:1. doi: 10.3402/jev.v1i0.18374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Freeman AK, Morrison DK. 14-3-3 Proteins: diverse functions in cell proliferation and cancer progression. Semin Cell Dev Biol. 2011;22(7):681–7. doi: 10.1016/j.semcdb.2011.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.D’Acquisto F, Perretti M, Flower RJ. Annexin-A1: a pivotal regulator of the innate and adaptive immune systems. Br J Pharmacol. 2008;155(2):152–69. doi: 10.1038/bjp.2008.252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Madureira PA, et al. The role of the annexin A2 heterotetramer in vascular fibrinolysis. Blood. 2011;118(18):4789–97. doi: 10.1182/blood-2011-06-334672. [DOI] [PubMed] [Google Scholar]
- 39.Kaczmarek E, et al. Identification and characterization of CD39/vascular ATP diphosphohydrolase. J Biol Chem. 1996;271(51):33116–33122. doi: 10.1074/jbc.271.51.33116. [DOI] [PubMed] [Google Scholar]
- 40.Wang D, et al. Isoform-specific regulation by NG,NG-dimethylarginine dimethylaminohydrolase of rat serum asymmetric dimethylarginine and vascular endothelium-derived relaxing factor/NO. Circ Res. 2007;101:627–635. doi: 10.1161/CIRCRESAHA.107.158915. [DOI] [PubMed] [Google Scholar]
- 41.Wang Y, et al. Kynurenine is a novel endothelium-derived relaxing factor produced during inflammation. Nat Med. 2010;16(3):279–285. doi: 10.1038/nm.2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.González-Amaro, R. & Sánchez-Madrid, F. Cell adhesion molecules: selectins and integrins. Crit Rev Immunol. 19(5–6), 389–429. Review (1999). [PubMed]
- 43.Lennon NJ, et al. Dysferlin interacts with annexins A1 and A2 and mediates sarcolemmal wound-healing. J Biol Chem. 2003;278(50):50466–73. doi: 10.1074/jbc.M307247200. [DOI] [PubMed] [Google Scholar]
- 44.Britton C, Winter AD, Gillan V, Devaney E. microRNAs of parasitic helminths - Identification, characterization and potential as drug targets. Int J Parasitol Drugs Drug Resist. 2014;4(2):85–94. doi: 10.1016/j.ijpddr.2014.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kramer, M. F. Stem-loop RT-qPCR for miRNAs. Curr Protoc Mol Biol. Chapter 15:Unit 15.10 10.1002/0471142727.mb1510s95 (2011). [DOI] [PMC free article] [PubMed]
- 46.Shi R, Sun YH, Zhang XH, Chiang VL. Poly(T) adaptor RT-PCR. Methods Mol Biol. 2012;822:53–66. doi: 10.1007/978-1-61779-427-8_4. [DOI] [PubMed] [Google Scholar]
- 47.Trocoli Torrecilhas AC, et al. Trypanosoma cruzi: parasite shed vesicles increase heart parasitism and generate an intense inflammatory response. Microb Infect. 2009;11(1):29–39. doi: 10.1016/j.micinf.2008.10.003. [DOI] [PubMed] [Google Scholar]
- 48.Gómez-Arreaza A, et al. Extracellular functions of glycolytic enzymes of parasites: Unpredicted use of ancient proteins. Mol Biochem Parasitol. 2014;193:75–81. doi: 10.1016/j.molbiopara.2014.02.005. [DOI] [PubMed] [Google Scholar]
- 49.Figuera L, et al. Plasminogen binding proteins in secreted membrane vesicles of Leishmania mexicana. Mol Biochem Parasitol. 2013;187:14–20. doi: 10.1016/j.molbiopara.2012.11.002. [DOI] [PubMed] [Google Scholar]
- 50.Mebius MM, et al. Interference with the Host Haemostatic System by Schistosomes. PLoS Pathog. 2013;9(12):e1003781. doi: 10.1371/journal.ppat.1003781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vasconcelos EG, et al. Partial purification and immunohistochemical localization of ATP diphosphohydrolase from Schistosoma mansoni. Immunological cross-reactivities with potato apyrase and Toxoplasma gondii nucleoside triphosphate hydrolase. J Biol Chem. 1996;271(36):22139–22145. doi: 10.1074/jbc.271.36.22139. [DOI] [PubMed] [Google Scholar]
- 52.Da’dara AA, Bhardwaj R, Ali YB, Skelly PJ. Schistosome tegumental ecto-apyrase (SmATPDase1) degrades exogenous pro-inflammatory and pro-thrombotic nucleotides. PeerJ. 2014;18(2):e316. doi: 10.7717/peerj.316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lin YL, He S. Sm22.6 antigen is an inhibitor to human thrombin. Mol Biochem Parasitol. 2006;147:95–100. doi: 10.1016/j.molbiopara.2006.01.012. [DOI] [PubMed] [Google Scholar]
- 54.Kuchay SM, Chishti AH. Calpain-mediated regulation of platelet signaling pathways. Curr Opin Hematol. 2007;14(3):249–54. doi: 10.1097/MOH.0b013e3280ef68f8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shimoda M, Khokha R. Proteolytic factors in exosomes. Proteomics. 2013;13(10–11):1624–1636. doi: 10.1002/pmic.201200458. [DOI] [PubMed] [Google Scholar]
- 56.McKerrow JH, Caffrey C, Kelly B, Loke P, Sajid M. Proteases in parasitic diseases. Annu. Rev. Pathol. Mech. Dis. 2006;1:497–536. doi: 10.1146/annurev.pathol.1.110304.100151. [DOI] [PubMed] [Google Scholar]
- 57.Liu S, et al. Expression profile of the Schistosoma japonicum degradome reveals differential protease expression patterns and potential anti-schistosomal intervention targets. PLoS Comput Biol. 2014;10(10):e1003856. doi: 10.1371/journal.pcbi.1003856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Horn M, et al. Trypsin- and chymotrypsin-like serine proteases in Schistosoma mansoni – ‘The undiscovered country’. PLoS Negl Trop Dis. 2014;8(3):e2766. doi: 10.1371/journal.pntd.0002766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fajtová P, et al. Prolyl Oligopeptidase from the blood fluke Schistosoma mansoni: From functional analysis to antischistosomal inhibitors. PLoS Negl Trop Dis. 2015;9(6):e0003827. doi: 10.1371/journal.pntd.0003827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rinaldi G, et al. RNA interference targeting leucine aminopeptidase blocks hatching of Schistosoma mansoni eggs. Mol Biochem Parasitol. 2009;167(2):118–126. doi: 10.1016/j.molbiopara.2009.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.McCarthya E, et al. Leucine aminopeptidase of the human blood flukes, Schistosoma mansoni and Schistosoma japonicum. Int J Parasitol. 2004;34:703–714. doi: 10.1016/j.ijpara.2004.01.008. [DOI] [PubMed] [Google Scholar]
- 62.Clegg J, Smithers S. The effects of immune rhesus monkey serum on schistosomula of Schistosoma mansoni during cultivation in vitro. Int J Parasitol. 1972;2(1):79–98. doi: 10.1016/0020-7519(72)90036-7. [DOI] [PubMed] [Google Scholar]
- 63.Tritten, L. & Geary, T. G. MicroRNAs of filarial nematodes: a new frontier in host-parasite interactions. In: Non-coding RNAs and Inter-Kingdom Communication, eds A. L. Leitão & F. J. L. Enguita, Springer International Publishing AG, Cham, Switzerland, pp. 207–223 (2016).
- 64.Tritten L, Clarke D, Timmins S, McTier T, Geary TG. Dirofilaria immitis exhibits sex- and stage-specific differences in excretory/secretory miRNA and protein profiles. Vet Parasitol. 2016;232:1–7. doi: 10.1016/j.vetpar.2016.11.005. [DOI] [PubMed] [Google Scholar]
- 65.Bartel DP. MicroRNA target recognition and regulatory functions. Cell. 2009;136(2):215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.de Souza Gomes M, Muniyappa MK, Carvalho SG, Guerra-Sá R, Spillane C. Genome-wide identification of novel microRNAs and their target genes in the human parasite Schistosoma mansoni. Genomics. 2011;98(2):96–111. doi: 10.1016/j.ygeno.2011.05.007. [DOI] [PubMed] [Google Scholar]
- 67.Simões MC, et al. Identification of Schistosoma mansoni microRNAs. BMC Genomics. 2011;12:47. doi: 10.1186/1471-2164-12-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Marco A, et al. Sex-Biased Expression of MicroRNAs in Schistosoma mansoni. PLoS Negl Trop Dis. 2013;7(9):e2402. doi: 10.1371/journal.pntd.0002402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tritten L, et al. Loa loa and Onchocerca ochengi miRNAs detected in host circulation. Mol Biochem Parasitol. 2014;198:14–17. doi: 10.1016/j.molbiopara.2014.11.001. [DOI] [PubMed] [Google Scholar]
- 70.Wang J, et al. C. elegans ciliated sensory neurons release extracellular vesicles that function in animal communication. Curr Biol. 2014;24:1–7. doi: 10.1016/j.cub.2014.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hassani K, Olivier M. Immunomodulatory impact of Leishmania-induced macrophage exosomes: A comparative proteomic and functional analysis. PLoS Negl Trop Dis. 2013;7(5):e2185. doi: 10.1371/journal.pntd.0002185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tucker, M. S., Karunaratne, L. B., Lewis, F. A., Freitas, T. C. & Liang, Y. S. Schistosomiasis. Curr Protoc Immunol.103, Unit 19.1 10.1002/0471142735.im1901s103 (2013). [DOI] [PubMed]
- 73.Lässer C, Eldh M, Lötvall J. Isolation and characterization of RNA-containing exosomes. J Vis Exp. 2012;9(59):e3037. doi: 10.3791/3037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Xu H, Yang L, Freitas MA. A robust linear regression based algorithm for automated evaluation of peptide identifications from shotgun proteomics by use of reversed-phase liquid chromatography retention time. BMC Bioinformatics. 2008;9:347. doi: 10.1186/1471-2105-9-347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Xu H, Freitas MA. MassMatrix: a database search program for rapid characterization of proteins and peptides from tandem mass spectrometry data. Proteomics. 2009;9(6):1548–1555. doi: 10.1002/pmic.200700322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Conesa A, et al. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005;21(18):3674–3676. doi: 10.1093/bioinformatics/bti610. [DOI] [PubMed] [Google Scholar]
- 77.Myhre S, Tveit H, Mollestad T, Laegreid A. Additional gene ontology structure for improved biological reasoning. Bioinformatics. 2006;22:2020–2027. doi: 10.1093/bioinformatics/btl334. [DOI] [PubMed] [Google Scholar]
- 78.Berriman M, et al. The genome of the blood fluke Schistosoma mansoni. Nature. 2009;460(7253):352–358. doi: 10.1038/nature08160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Protasio AV, et al. A systematically improved high quality genome and transcriptome of the human blood fluke Schistosoma mansoni. PLoS Negl Trop Dis. 2012;6(1):e1455. doi: 10.1371/journal.pntd.0001455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Dillies MA, et al. French StatOmique Consortium. A comprehensive evaluation of normalization methods for Illumina high-throughput RNA sequencing data analysis. Brief Bioinform. 2013;14(6):671–83. doi: 10.1093/bib/bbs046. [DOI] [PubMed] [Google Scholar]
- 81.Chen C, et al. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 2005;33(20):e179. doi: 10.1093/nar/gni178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Mitchell PS, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci USA. 2008;105(30):10513–8. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.