Abstract
Human antibody responses to pathogens, like Staphylococcus aureus, are important indicators for in vivo expression and immunogenicity of particular bacterial components. Accordingly, comparing the antibody responses to S. aureus components may serve to predict their potential applicability as antigens for vaccination. The present study was aimed at assessing immunoglobulin G (IgG) responses elicited by non-covalently cell surface-bound proteins of S. aureus, which thus far received relatively little attention. To this end, we applied plasma samples from patients with the genetic blistering disease epidermolysis bullosa (EB) and healthy S. aureus carriers. Of note, wounds of EB patients are highly colonized with S. aureus and accordingly these patients are more seriously exposed to staphylococcal antigens than healthy individuals. Ten non-covalently cell surface-bound proteins of S. aureus, namely Atl, Eap, Efb, EMP, IsaA, LukG, LukH, SA0710, Sle1 and SsaA2, were selected by bioinformatics and biochemical approaches. These antigens were recombinantly expressed, purified and tested for specific IgG responses using human plasma. We show that high exposure of EB patients to S. aureus is mirrored by elevated IgG levels against all tested non-covalently cell wall-bound staphylococcal antigens. This implies that these S. aureus cell surface proteins are prime targets for the human immune system.
Introduction
Staphylococcus aureus is a Gram-positive bacterial pathogen that colonizes about one third of the healthy human population1. The pathology caused by S. aureus may range from mild skin infections to life-threatening bacteremia. Current treatment of S. aureus infections relies on antibiotics, but the emergence of highly drug-resistant lineages2 has reignited the interest in alternative treatment options, including passive and active immunization3–7.
Surface-exposed and secreted proteins of S. aureus play pivotal roles in the colonization and subversion of the human host8. Accordingly, such proteins have been considered as possible antigens for vaccination against S. aureus infections9,10. However, the previous efforts to develop a vaccine against S. aureus have met with little success, as exemplified by trials based on capsular polysaccharides or important virulence factors, such as fibronectin binding protein (FnBP), collagen binding protein (CnBP), or clumping factor A (ClfA)5,9,11. Most likely, this relates to the broad spectrum of virulence and immune evasion factors that S. aureus employs to thrive and survive in the human host. Therefore, it has been suggested that potentially successful vaccines need to address multiple staphylococcal virulence factors and defense mechanisms5.
The S. aureus genome encodes about 2700 proteins, from which about 120 have been observed more than once in the extracellular and cell surface proteomes10,12,13. Since the pioneering experiments published by Etz et al. and Vytvytska et al. in 200214,15, diverse immunological and proteomics-based studies have addressed the antibody responses against S. aureus9,16. In these studies the antigenicity of non-covalently cell wall-associated proteins received relatively little attention. These non-covalently cell wall-associated proteins include proteins with specific cell wall-binding domains, ‘secretable expanded repertoire adhesive molecules’ (SERAMs) and typical cytoplasmic proteins that are bound to the cell wall through as yet undefined mechanisms17. Members of this group are tissue adhesins, toxins and immune evasion factors. Since the functions of these proteins could be neutralized by effective antibody responses, they might be attractive targets for vaccination, provided that they are immunogenic. Recent reports have shown high immune responses against some members of this group, including IsaA, Efb and Atl18,19. Yet, in animal models it was shown that antibodies against these antigens provide only limited protection against challenges with S. aureus6,20.
Healthy immune-competent individuals display differing antibody responses to a vast array of S. aureus antigens, possibly reflecting their history of close encounters with multiple different S. aureus lineages21,22. Anti-staphylococcal antibody levels can increase strongly during bacteremia21,23, and it has been proposed that continuous exposure to different staphylococcal antigens might improve the effectiveness of the immune response22. Patients with the genetic blistering disease epidermolysis bullosa (EB) develop wounds that are highly susceptible to S. aureus colonization. Especially the chronic wounds of EB patients are heavily colonized with S. aureus and usually contain several different types of this pathogen24–27. This severe colonization of the wounds of EB patients is reflected in the very high anti-staphylococcal immunoglobulin G (IgG) levels in their plasma and blister fluid compared to the respective IgG levels in the plasma of healthy age-matched volunteers19,28. Importantly, also IgG4 responses against various S. aureus antigens were elevated in the plasma of EB patients, which is consistent with their long-term and/or repeated exposure to these antigens28. Remarkably, S. aureus bacteremia is infrequently observed in adult EB patients, suggesting that their anti-staphylococcal immune responses may be protective against invasive S. aureus infections19. Of note, in previous studies on the antibody responses of EB patients to S. aureus antigens, the non-covalently cell wall-bound proteins were underrepresented19,28.
The aim of the present exploratory study was to assess to what extent non-covalently cell wall-bound proteins of S. aureus are immunogenic and whether the respective IgG titers are elevated in plasma samples from EB patients. Based on a bioinformatics inventory and on data from our previous proteomics analyses of the S. aureus surfacome29, 10 non-covalently cell wall-bound proteins of S. aureus were selected, produced in Lactococcus lactis, and purified. The purified proteins were used to assess specific IgG levels in plasma samples from EB patients and healthy volunteers.
Materials and Methods
Bacterial strains, plasmids and growth conditions
Strains and plasmids used in this study are listed in Table 1. L. lactis strains were grown at 30 °C in M17 broth (Oxoid Limited, Hampshire, UK) supplemented with 0.5% glucose (wt/vol) (GM17). When necessary the medium was supplemented with chloramphenicol (5 µg/ml) or erythromycin (5 µg/ml) for plasmid selection. S. aureus strains were grown at 37 °C, 250 rpm in Tryptone Soy Broth (TSB; Oxoid). E. coli strain MC1061 was grown at 37 °C, 250 rpm in Lysogeny broth (LB; Becton Dickinson, Breda, The Netherlands). When necessary, the medium was supplemented with ampicillin (100 µg/ml) for plasmid selection.
Table 1.
Strains and plasmids used in this study.
| Strain or plasmid | Relevant phenotype(s) or genotype(s) | Source or reference |
|---|---|---|
| Strains | ||
| L. lactis PA1001 | MG1363 pepN::nisRK, allows nisin-inducible expression, ΔacmA ΔhtrA | 46 |
| E. coli MC1061 | araD139 Δ(araA-leu)7697 ΔlacX74 galK16 galE15(GalS) λ- e14- mcrA0 relA1 rpsL150(strR) spoT1 mcrB1 hsdR2 | (Novagen, Madison, Wis) |
| S. aureus NCTC 8325 | Propagating strain for typing phage 47 | 64 |
| S. aureus Newman | NCTC 8178 clinical isolate | 65 |
| S. aureus Newman Δspa Δsbi | S. aureus Newman spa sbi mutant | 31 |
| Plasmids | ||
| pRE-USPnlic | ampR; camR; pRExLIC fused with E. coli pBR322 replicon | 33 |
| pERL | eryR; pERL fused with pSH71 replicon | 33 |
| pNG4210 | camR pNG400 derivative, containing BamHI/EcoRI-XbaI/NotI cloning sites, his6 followed by a Stop codon | 35 |
| pNZ:LIC:efb | Fusion of pRE-USPnlic with pERL containing efb (SAOUHSC_01114, aa 30–165) | This study |
| pNZ:LIC:eap | Fusion of pRE-USPnlic with pERL containing eap (SAOUHSC_02161, aa 31–584) | This study |
| pNZ:LIC:sle1 | Fusion of pRE-USPnlic with pERL containing sle1 (SAOUHSC_00427, aa 26–334) | This study |
| pNZ:LIC:sa0710 | Fusion of pRE-USPnlic with pERL containing sa0710 (SAOUHSC_00773, aa 25–279) | This study |
| pNZ:LIC:atl1 | Fusion of pRE-USPnlic with pERL containing atl1 (SAOUHSC_00994, aa 199–775) | This study |
| pNZ:LIC:atl2 | Fusion of pRE-USPnlic with pERL containing atl2 (SAOUHSC_00994, aa 776–1256) | This study |
| pNG4210:lukG | pNG4210 containing lukG with C-terminal his6 (SAOUHSC_02241, aa 30–338) | This study |
| pNG4210:lukH | pNG4210 containing lukH with C-terminal his6 (SAOUHSC_02243, aa 33–351) | This study |
| pNG4210:ssaA2 | pNG4210 containing ssaA2 with C-terminal his6 (SAOUHSC_02571, aa 28–267) | This study |
| pNG4210:emp | pNG4210 containing emp with C-terminal his6 (SAOUHSC_00816, aa 27–340) | This study |
| pNG4210:ftsL | pNG4210 containing ftsL with C-terminal his6 (USA300HOU_1120, aa 66–133) | 35 |
Aa, amino acid residue; ampR, ampicillin resistance gene; camR, chloramphenicol resistance gene; eryR, erythromycin resistance gene; PnisA, nisin-inducible promoter; usp45ss, signal sequence of usp45.
Isolation of S. aureus cell wall fragments
Cell wall fragments (CWFs) from S. aureus were isolated as described previously30. In short, S. aureus Newman cells were collected by centrifugation, glass-beads were added (0.1 µm beads, Biospec Products, Bartlesville, USA), and cells were disrupted for 2 min in a Precellys 24 homogenizer (Bertin Technologies, Saint Quentin en Yvelines Cedex, France). The resulting CWFs were collected by centrifugation and boiled at 96 °C for 10 min in 4% sodium dodecyl sulphate. This step was repeated twice. CWFs were subsequently washed six times with Phosphate Buffered Saline (PBS) and stored at −20 °C until further use.
Identification of non-covalently cell surface-bound proteins
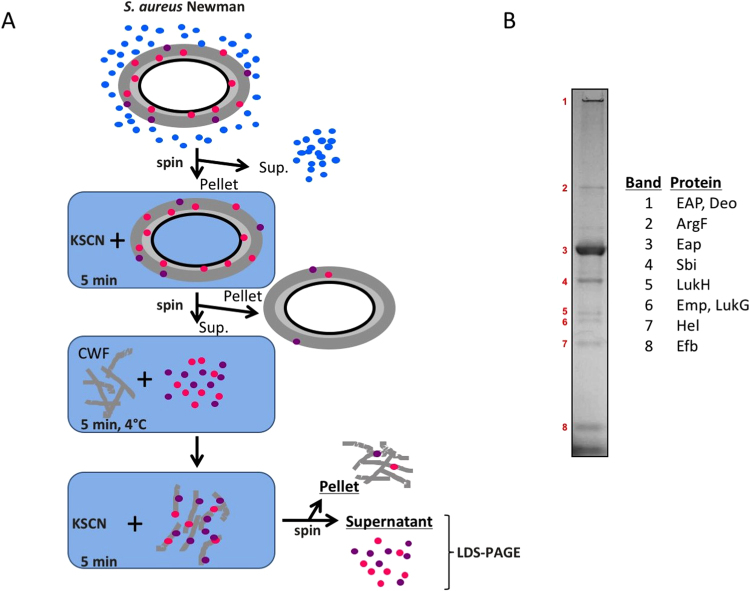
Non-covalently cell wall-bound proteins of S. aureus were identified by using the amino acid sequences of known domains for non-covalent cell wall-binding (i.e. PROSITE PS51780, PS51782, PS51781, PS51109) in BLAST searches against the sequenced S. aureus Newman strain. The actual identification of expressed non-covalently cell wall-bound proteins was accomplished as schematically represented in Fig. 1A. Upon overnight culturing in TSB, S. aureus Newman cells were incubated with 2 M potassium thiocyanate (KSCN) for 5 min leading to the release of non-covalently cell wall-bound proteins. Liberated non-covalently cell wall-bound proteins in the soluble fraction were either TCA-precipitated or dialyzed against PBS using a 3,500 Molecular weight cut-off (MWCO) membrane (Spectrum laboratories Inc. USA) as described before31. Secreted proteins present in S. aureus Newman growth medium fractions were also collected, either by TCA precipitation or dialysis of the spent growth medium against PBS. Dialyzed non-covalently cell wall-bound proteins were added to prepared S. aureus CWFs and incubated for 5 min at 4 °C unless stated otherwise. CWFs containing non-covalently bound proteins were washed with PBS and non-covalently cell wall-bound proteins were released into the supernatant fraction by incubation with 2 M KSCN as described above. Released proteins were collected either by TCA precipitation or dialyzed overnight against demineralized water. Released proteins and cell pellets were separated by lithium dodecyl sulphate (LDS) - PAGE and the respective gels were stained with SimplyBlue SafeStain (Life Technologies, Grand Island, NY. USA). Protein bands were cut from the gels (Fig. 1B) and identified by Mass Spectrometry (MS) as described previously32.
Figure 1.
Identification of non-covalently cell wall-bound S. aureus proteins. (A) Schematic representation of the experimental set-up for identification of non-covalently cell wall-bound proteins. S. aureus cells were first separated from the growth medium by centrifugation (spin). Pelleted S. aureus cells were treated with KSCN to release the non-covalently cell wall-bound proteins. KSCN-extracted proteins were re-bound to cell wall fragments (CWF) and, subsequently, released again by KSCN incubation. Upon centrifugation, the resulting pellet and supernatant fractions were analyzed by LDS-PAGE (B). Upon Simply Blue safe staining of the gel, protein bands were excised and identified by MS as indicated.
Expression of non-covalently cell wall-bound proteins in L. lactis
PCR was performed with the Pwo DNA polymerase (Roche Diagnostics, Woerden, The Netherlands), using chromosomal DNA of S. aureus NCTC 8325 as template. Primers listed in Table 2 were designed to amplify gene sequences without the region coding for the natural secretion signals and with 5′ end extensions for ligase-independent cloning (LIC)33. Briefly, SwaI-digested pRE-USP plasmid and PCR fragments were treated with T4 DNA polymerase (20 °C, 20 min; 75 °C, 20 min; Roche Diagnostics) before incubation for 5 min at room temperature (3:1 vector:insert). Z-Competent E. coli MC1061 cells (Zymo Research, Orange, CA, USA) were transformed with the plasmid:vector mixtures. Correct plasmids were confirmed by DNA sequencing (Eurofins DNA, Germany). For cloning in L. lactis, Vector Backbone Exchange was performed by mixing ~300 ng of pERL vector with ~300 ng of the pRE-USP harboring the gene of interest, both digested with SfiI (New England Biolabs, Ipswich, UK). Ligation was performed using T4 DNA Ligase (New England Biolabs) and the resulting vector was introduced into electrocompetent L. lactis PA100134. For insertion of genes into plasmid pNG4210, primers (Table 2) were designed to amplify the respective sequences without the region coding for the natural secretion signals and with 5′ end extensions encoding BamHI (forward) or NotI (reverse) restriction sites. Briefly, digested PCR products and linearized plasmid were separated by agarose gel electrophoresis, and selected DNA fragments were gel-extracted and purified. Ligation of digested plasmid and PCR fragments was performed using T4 DNA ligase and the resulting plasmid was introduced into electrocompetent L. lactis PA1001 as described before35. For the expression of cloned genes, L. lactis cultures were induced in the exponential phase of growth (0.5 O.D. at 600 nm) by the addition of nisin (final concentration 3 ng/ml, Sigma-Aldrich, St. Luis, MO). After 4 h or overnight culturing, the cells were separated from the growth medium by centrifugation. Proteins in the nisin-induced growth medium fractions were precipitated with TCA (10% W/V) and resuspended in LDS gel loading buffer (Life Technologies). Cells in LDS sample buffer were disrupted with 0.1 µm glass beads in a Precellys 24 homogenizer. Both cellular and growth medium fractions were analyzed by LDS-PAGE (Life Technologies) and proteins were either visualized using protein staining with the SimplyBlue SafeStain (Life Technologies), or by blotting onto nitrocellulose membranes (Protan nitrocellulose transfer paper, Whatman, Germany) and subsequent immunodetection. For immunodetection, mouse anti-his tag primary antibodies (Life Technologies) and fluorescently labeled secondary antibodies (goat anti-mouse IRDye 800 CW, LI-COR Biosciences, Lincoln, NE. USA) were used. Antibody binding was visualized with an Odyssey infrared imaging system (LI-COR Biosciences).
Table 2.
Primer sequences used in this study.
| Primers | 5′ → 3′ nucleotide sequence | |
|---|---|---|
| efb.fw | ATGGTGAGAATTTATATTTTCAAGGTAGCGAAGGATACGGTCCAAG | |
| efb.rev | TGGGAGGGTGGGATTTTCATTATTTAACTAATCCTTGTTTTAATACATTATC | |
| eap.fw | ATGGTGAGAATTTATATTTTCAAGGTGCAGCTAAGCCATTAGATAAATC | |
| eap.rev | TGGGAGGGTGGGATTTTCATTATTTATTTTTTTTTGATTTAGTGTATTG | |
| sle1.fw | ATGGTGAGAATTTATATTTTCAAGGTGCTACAACTCACACAGTAAAAC | |
| sle1.rev | TGGGAGGGTGGGATTTTCATTAGTGAATATATCTATAATTATTTACTTGGT | |
| sa0710.fw | ATGGTGAGAATTTATATTTTCAAGGTCAACAACATGGCACACAAG | |
| sa0710.rev | TGGGAGGGTGGGATTTTCATTAGTGGATGTAATTATATTTTCCTG | |
| atl(1).fw | ATGGTGAGAATTTATATTTTCAAGGTGCTTCAGCACAACCAAG | |
| atl(1).rev | TGGGAGGGTGGGATTTTCATTATTTTACAGCTGTTTTTGG | |
| atl(2).fw | ATGGTGAGAATTTATATTTTCAAGGTGCTTATACTGTTACTAAACCACAAAC | |
| atl(2).rev | TGGGAGGGTGGGATTTTCATTATTTATATTGTGGGATGTCG | |
| lukG.fw | ATATGGATCCAAGATTAATTCTGAAATCAAACAAGTTTCTG | BamHI |
| lukG.rev | ATATGCGGCCGCTTTCTTTTCATTATCATTAAGTACTTTTAC | NotI |
| lukH.fw | ATATGGATCCGACTCTCAAGACCAAAATAAGAAAG | BamHI |
| lukH.rev | ATATGCGGCCGCTCCTTCTTTATAAGGTTTATTGTCATC | NotI |
| ssaA2.fw | ATATGGATCCTCTGAGCAAGATAACTACGGTTATAATCC | BamHI |
| ssaA2.rev | ATATGCGGCCGCGTGAATGAAGTTATAACCAGCAGCTTGG | NotI |
| emp.fw | ATATGGATCCTCAGTGACAGAGAGTGTTGAC | BamHI |
| emp.rev | ATATGCGGCCGCTACTCGTGGTGCTGGTAAG | NotI |
Underlined are the LIC cloning sequences/restriction sites.
Protein purification and activity measurements
When expressed proteins remained cell-associated, they were liberated from the cells either with 2 M KSCN or 6 M urea, as required. Next, the protein-containing soluble fractions were separated from the cell fraction by centrifugation. Subsequently, his-tagged proteins were purified from the respective supernatant fractions using the HisLink Protein Purification resin (Promega Corporation, Madison, WI. USA), in the absence or presence of either 2 M KSCN or 6 M urea. The HisLink binding and washing buffer was composed of 0.1 M HEPES 7.5 pH, 0.5 M NaCl and 10 mM imidazole. The elution buffers were essentially the same, but contained 200 mM or 400 mM imidazole. The IsaA and FtsL proteins were purified as described previously35,36.
Rebinding of isolated proteins to S. aureus cells
Overnight growth cultures of S. aureus Newman ΔspaΔsbi were resuspended to 1 optical density measured at 600 nm in 800 µl of PBS (pH 7) or 50 mM sodium acetate (pH 5) and incubated with 1–3 µg of histidine-tagged fusion proteins for 10 min. After incubation, cell pellets and supernatants were processed, and localization of tagged proteins was assessed by LDS-PAGE and Western blotting as described above.
Enzyme-linked immunosorbent assay (ELISA)
Plasma samples were previously donated by patients with EB from the Dutch Epidermolysis Bullosa Registry (DEBR), and by healthy volunteers from the Netherlands. The EB patients included six patients with junctional EB (EB01, EB02, EB09, EB15, EB53, EB60), one patient with EB simplex (EB11), and one patient with dystrophic EB (EB51)19. ELISA plates were coated overnight at 4 °C with histidine-tagged fusion proteins (100ng/well) diluted in carbonate coating buffer (50 mM sodium carbonate, pH 9.6)6. Subsequently, the plates were blocked with PBS containing 5% skim milk. Patient and healthy control plasma samples were processed as previously described19. Serial dilutions of plasma (1000- to 2,000,000-fold) were prepared in PBS-Tween 20/5% skim milk. Specific anti-human IgG secondary antibodies coupled to horseradish peroxidase (dilution 1:8,000; Southern Biotechnology, Birmingham, AL) were used according to the manufacturer’s recommendations. Horseradish peroxidase activity was quantified by measuring the hydrolysis of the substrate (O-Phenylenediamine, Sigma-Aldrich) at OD492 in a plate reader (Biotek Powerwave XS2, USA). The raw ELISA data are provided in Supplementary Information Table 1. Titers were calculated in arbitrary units (AU) through extrapolation using linear regression for data points from known dilutions giving OD492 values between 0.1 and 1.0. All calculated R2 linear regression values (Pearson product moment correlation coefficient) were above 0.98. IgG titers in plasma samples of EB patients were averaged and normalized by adjusting the averaged IgG titers in the control plasma samples of healthy volunteers to a single arbitrary level (AU = 10) and adjusting accordingly the averages for the EB patient samples. In brief, obtained EB and control averages were multiplied by a numeric factor that resulted in the average of all controls equal to 10 AU. After plotting normalized values, the differences in the average IgG levels measured for plasma samples from EB patients and healthy control individuals were compared for the different analyzed proteins. Statistical significance of the ELISA data was tested with a Mann-Whitney U test for two independent samples (Supplementary Information Table 1).
Ethics statements
Blood donations from EB patients were collected with approval of the medical ethics committee of the University Medical Center Groningen (approval no. NL27471,042,09). The Independent Ethics Committee of the Foundation ‘Evaluation of Ethics in Biomedical Research’ (Assen, the Netherlands), approved the protocol for blood donations from healthy volunteers, which is registered by QPS Groningen (code 04132-CS011). Blood donations were obtained after written informed consent from all eight EB patients and the six healthy volunteers included in this study, and adhering to the Helsinki Guidelines19.
Results and Discussion
Selection of non-covalently cell wall-bound proteins
To pinpoint a panel of non-covalently cell wall-bound proteins of S. aureus, we performed an extensive bioinformatics analysis using the genome sequence of S. aureus strain Newman and, in addition, we surveyed the results of our previous analysis of the cell surface proteome of this S. aureus strain29. The results are summarized in Table 3. Specifically, the bioinformatics approach identified several SERAMS, in particular the extracellular adhesive protein Eap, the extracellular matrix protein Emp, the extracellular fibrinogen-binding protein Efb, and coagulase37. Furthermore, we retrieved all known S. aureus Newman proteins containing the conserved cell wall-binding domains LysM (PROSITE: PS51782)38,39, GW (PROSITE: PS51780)40, SH3B (PROSITE: PS51781)39,41 and G5 (PROSITE: PS51109)42. Except for the amidase from phage phiNM2 and the transmembrane protein EbpS, all other identified proteins carried a predicted signal peptide for export from the cytoplasm (indicated as S in Table 3). Of note, Eap, Atl, IsaA, SsaA2, Sle1, LukG and LukH have also previously been identified as non-covalently cell wall-bound proteins16,29,43,44. Further, Sle1 was shown to be localized in the vicinity of the S. aureus cross-wall43,45, while Atl was found to be preferably bound to the septal region44.
Table 3.
Identified non covalently cell surface-bound proteins.
| Uniprot ID | Gene names | AA | S e | Protein name |
|---|---|---|---|---|
| Proteins with known conserved non covalently cell wall binding domains | ||||
| LysM (PS51782) a | ||||
| A6QH29 | NWMN_1389 ebpS | 486 | Elastin-binding protein EbpS | |
| A0A0H3K686 | NWMN_0055 spa | 520 | S | Immunoglobulin G binding protein A |
| A0A0H3K6S5 | NWMN_0724 SA0710c | 279 | S | Uncharacterized protein |
| A0A0H3K7F5 | NWMN_0634 | 265 | S | Secretory antigen SsaA-like protein |
| A0A0H3KAZ4 | NWMN_0429 sle1d | 334 | S | N-acetylmuramoyl-L-alanine amidase AAA |
| A0A0H3KG37 | NWMN_1157 lytN | 383 | S | Cell-wall hydrolase LytN |
| GW (PS51780) a | ||||
| A0A0H3K7X7 | NWMN_0922 atl | 1256 | S | Bifunctional autolysin |
| SH3B (PS51781) a | ||||
| A0A0H3K875 | NWMN_1039 | 481 | Phage amidase [Bacteriophage phiNM2] | |
| A0A0H3K8J7 | NWMN_1534 | 291 | S | Probable cell wall amidase LytH |
| G5 (PS51109) a | ||||
| A0A0H3KAI3 | NWMN_2392 | 1501 | S | Uncharacterized protein |
| Identified by surfacome profiling | ||||
| A0A0H3K7X7 | NWMN_0922 atl | 1256 | S | Bifunctional autolysin |
| A6QIG7 | NWMN_1877 chp | 149 | S | Chemotaxis inhibitory protein precursor |
| A0A0H3KA75 | NWMN_0166 coa | 636 | S | Coagulase |
| A6QF98 | NWMN_0758 empd | 340 | S | Extracellular matrix protein-binding protein emp precursor |
| A0A0H3KF27 | NWMN_2399 fnbA | 741 | S | Fibronectin binding protein A |
| A0A0H3K6R0 | NWMN_0687 | 646 | S | Lipoteichoic acid synthase |
| A6QK59 | NWMN_2469 isaA | 233 | S | Probable transglycosylase IsaA precursor |
| A6QJQ7 | NWMN_2317 sbi | 436 | S | Immunoglobulin-binding protein sbi precursor |
| A0A0H3KCA1 | NWMN_1066 | 109 | S | Uncharacterized protein Ehp |
| A0A0H3K6X4 | NWMN_0362 | 203 | S | Uncharacterized protein |
| A0A0H3KET4 | NWMN_0585 | 168 | S | Uncharacterized protein |
| A0A0H3K7N7 | NWMN_0757 | 508 | S | Secreted von Willebrand factor-binding protein |
| A0A0H3KEG7 | NWMN_2199 ssaA2d | 267 | S | Secretory antigen SsaA |
| LDS-PAGE: Gel band MS | ||||
| 1b A6QIG2 | NWMN_1872 eapd | 584 | S | 65 kDa membrane protein precursor |
| A6QDC3 | NWMN_0083 deoB | 392 | Phosphopentomutase | |
| 2 A6QG68 | NWMN_1078 argF | 333 | Ornithine carbamoyltransferase | |
| 3 A6QIG2 | NWMN_1872 eapd | 584 | S | 65 kDa membrane protein precursor |
| 4 A6QJQ7 | NWMN_2317 sbi | 436 | S | Immunoglobulin-binding protein Sbi precursor |
| 5 A0A0H3KHT5 | NWMN_1928 lukHd | 351 | S | Leukocidin/hemolysin toxin family S subunit |
| 6A6QF98 | NWMN_0758 empd | 340 | S | Extracellular matrix protein-binding protein Emp precursor |
| A0A0H3K9N1 | NWMN_1927 lukGd | 338 | S | Leukocidin/hemolysin toxin family F subunit |
| 7A0A0H3K5Z1 | NWMN_0249 held | 296 | S | 5′-nucleotidase, lipoprotein e(P4) family protein |
| A0A0H3KIY0 | NWMN_2444 | 85 | Uncharacterized protein | |
| 8A6QG59 | NWMN_1069 efbd | 165 | S | Fibrinogen-binding protein precursor |
aPROSITE ID of Motif; bExtracted band number from LDS-PAGE; cGene locus of NWMN_0724 homolog in S. aureus N315, used instead of gene name; dGene name of homologous protein if none was found in the Uniprot record; Se Secretion signal predicted by SignalP and/or according to Uniprot.
To detect the actually produced non-covalently cell wall-bound proteins of S. aureus Newman and to ensure that the identified proteins do indeed behave as non-covalently cell wall-bound proteins, we extracted these proteins with the chaotrope KSCN from the staphylococcal cells, reattached them to isolated S. aureus CWFs, and re-extracted the proteins with KSCN (schematically represented in Fig. 1A). The proteins thus obtained were separated by LDS-PAGE. Eight dominant bands were detected, excised from the gel and identified by MS (Fig. 1B). The identifiers and characteristics of the identified proteins are summarized in Table 3. In this respect, it is noteworthy that our unpublished proteomics data indicate that only about 16% of the KSCN-extractable proteins are specifically cell wall-bound proteins.
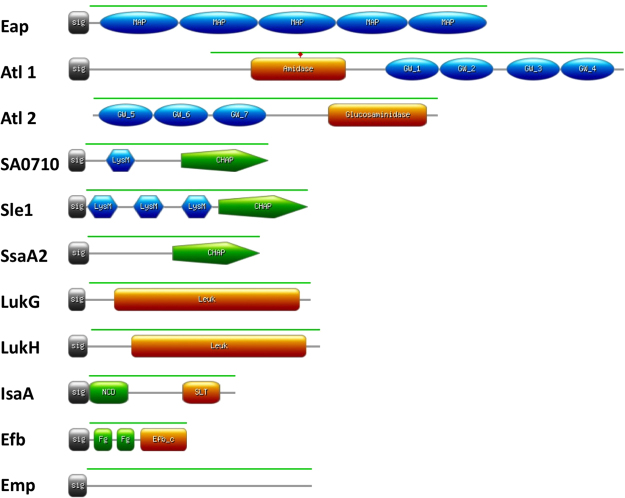
For our further studies on human antibody responses against non-covalently cell wall-bound proteins of S. aureus, we made a selection of 10 representative proteins. The inclusion criteria for these 10 proteins were identification by bioinformatics and/or biochemical analysis. Exclusion criteria were a lack of identification in previous biochemical or proteomics analyses29, the absence of a predicted signal peptide, the presence of an LPxTG motif for covalent cell wall binding, and known IgG-binding properties that would interfere with our further analyses. The domain structure of the selected proteins, highlighting domains potentially involved in cell wall binding, is represented in Fig. 2. It should be noted that Atl is synthesized in a pre-pro-form which, upon export, is cleaved into two moieties with an amidase domain (here termed Atl1) and a glucosamidase domain (here termed Atl2). Accordingly, the Atl2 moiety of Atl does not have its own signal peptide for export from the cytoplasm.
Figure 2.
Motif composition of non-covalently cell wall-bound proteins of S. aureus. The proteins shown are: the extracellular adherence protein Eap (SAOUHSC_02161); the bifunctional autolysin Atl (SAOUHSC_00994), of which the Atl1 (N-acetylmuramoyl-L-alanine amidase) and Atl2 (Endo-beta-N-acetylglucosaminidase) domains were separately expressed; the CHAP and LysM domain-containing protein SA0710 (SAOUHSC_00773); the N-acetylmuramoyl-L-alanine amidase Sle1 (SAOUHSC_00427); the staphylococcal secretory antigen SsaA2 (SAOUHSC_02571); the gamma-hemolysin subunit B LukG (SAOUHSC_02241); the leukocidin LukH (SAOUHSC_02243); the probable transglycosylase IsaA (SA2356); the fibrinogen-binding protein Efb (SAOUHSC_01114); and the extracellular matrix protein-binding protein Emp (SAOUHSC_00816). Sig, signal peptide; MAP, MAP repeat profile (PROSITE: PS51223); amidase, N-acetylmuramoyl-L-alanine amidase (Pfam: PF01510); glucosaminidase, endo-beta-N-acetylglucosaminidase (Pfam: PF01832); GW, GW domain profile (PROSITE: PS51780); LysM, LysM domain profile (PROSITE:PS51782); CHAP, CHAP domain profile (PROSITE: PS50911); Leuk, Leukocidin/Hemolysin toxin family (Pfam: PF07968); NCD, N-terminal conserved domain; SLT, Transglycosylase SLT domain (Pfam: PF01464); Efb_c, extracellular fibrinogen binding protein C terminal (Pfam: PF12199); Fg, fibrinogen-binding motifs63. The green line represents amino acid residues selected for cloning and expression.
Cloning, production and isolation of non-covalently cell wall-bound proteins in L. lactis
Genes for the selected non-covalently cell wall-bound proteins of S. aureus were cloned into nisin-inducible expression vectors and introduced into L. lactis strain PA1001 for expression. In the case of Atl, the Atl1 and Atl2 moieties were expressed separately, each being secreted with the lactococcal Usp45 signal peptide. Of note, the L. lactis PA1001 strain lacks the genes for the major extracellular protease HtrA and the autolysin AcmA, which minimizes proteolysis and cell lysis, respectively36,46. Next, expression of the cloned genes was induced with nisin and the subcellular localization of the respective S. aureus proteins was determined. To this end, cells were separated from the growth medium by centrifugation and the respective fractions were analyzed by LDS-PAGE and Western blotting using anti-His6 antibodies. As expected, all proteins were largely cell wall-bound (data not shown). By incubation of the cells with 2 M KSCN (Efb, Eap and Atl1) or 6 M urea (all seven other proteins), the expressed proteins were released, consistent with disruption of their non-covalent interactions with the cell wall. Upon centrifugation, the released proteins were purified from the resulting supernatant fractions using Ni-NTA agarose beads, and their potential to re-bind to cells of S. aureus Newman Δspa Δsbi was confirmed (Fig. 3; data not shown for IsaA). Notably, Efb did not re-bind to S. aureus cells under the standard assay conditions (pH 7), but its binding to the cells could be demonstrated at a lowered pH of 5 (Fig. 3). Altogether, these findings imply that the purified proteins have retained their cell wall-binding capabilities. Of note, the cell wall binding of SA0710 had not been experimentally verified so far, despite the fact that this protein has a LysM domain.
Figure 3.
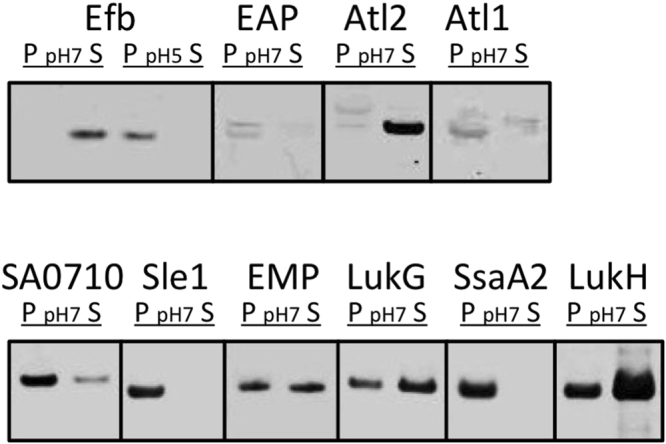
Binding of purified non-covalently cell wall-bound proteins to S. aureus cells. Non-covalently cell wall-bound proteins were expressed in L. lactis and their binding to whole cells of S. aureus was assessed upon incubation at pH 7, or pH 5 in the case of Efb as indicated. Please note that the different groupings of blots (marked by boxes) were cropped per investigated protein from different parts of the same Western blot, or from different Western blots that were similarly processed and scanned. P, cell pellet fraction; S, supernatant fraction.
Human IgG responses against non-covalently cell wall-bound proteins of S. aureus
To assess whether EB patients and healthy volunteers mount immune responses against the selected non-covalently cell wall-bound proteins of S. aureus, we applied an ELISA approach. The membrane protein FtsL was included in this analysis as a control, because it is surface-exposed, but not bound to the cell wall29. As shown in Fig. 4A and Supplementary Information Table 1, all investigated human plasma samples contained IgGs against all investigated proteins. Importantly, the levels of IgGs against non-covalently cell wall-bound proteins in plasma samples from EB patients were, on average, about 10-fold higher than those from healthy carriers. In this respect, the largest differences were observed for Eap, Atl2 and IsaA, and the smallest for Sle1 and Emp. Only, the FtsL-specific IgG levels in plasma samples from EB patients and healthy volunteers did not differ significantly (Fig. 4A). In this respect, it should be noted that for many, but not all, S. aureus antigens elevated IgG levels were previously observed in plasma from EB patients19,28. For example, no significant differences in IgG levels of EB patients and healthy volunteers were previously observed for the ClfA, ClfB and IsdH proteins bound to the staphylococcal cell wall.
Figure 4.
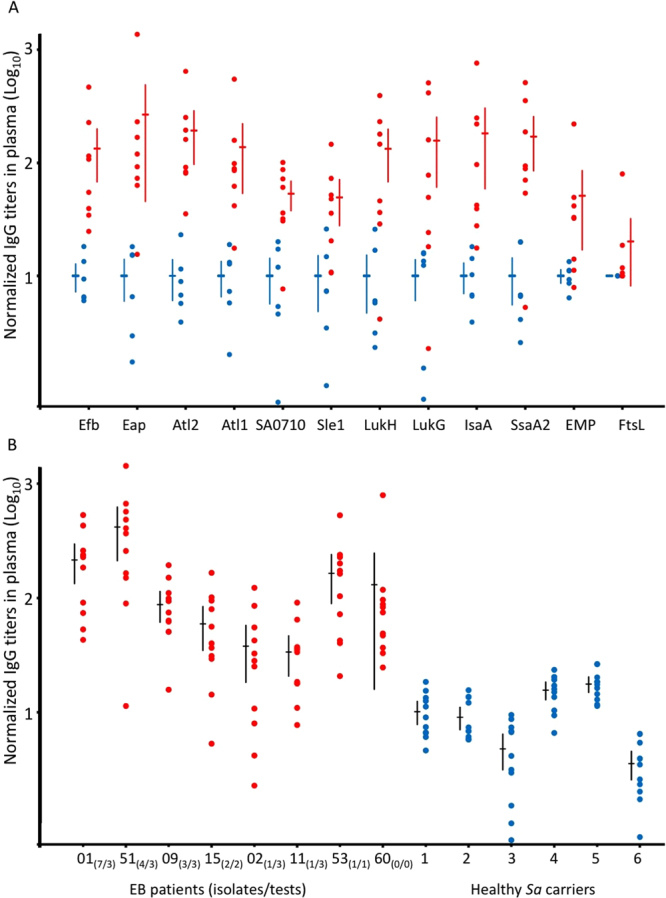
Binding of human IgGs to purified non-covalently cell wall-bound S. aureus proteins. Levels of IgGs specific for purified cell wall-bound S. aureus proteins were compared by ELISA using plasma samples of patients with EB (red bullets) or healthy S. aureus carriers (blue bullets). (A) Normalized IgG titers in different plasma samples plotted per purified S. aureus antigen. (B) Normalized IgG titers in different plasma samples plotted per EB patient and healthy carrier. In brief, obtained EB and control averages were multiplied by a numeric factor that resulted in the average of all controls equal to 10 AU. After plotting normalized values, the differences in the average IgG levels measured for plasma samples from EB patients and healthy control individuals were compared for the different analyzed proteins (isolates/tests). Per patient, the number of S. aureus types identified per number of sampling time points are indicated in parentheses19,24,26. Sa, S. aureus.
We further inspected the overall IgG responses to all non-covalently cell wall-bound proteins per plasma sample, excluding FtsL. As shown in Fig. 4B, the normalized average IgG levels against the eleven remaining S. aureus antigens were higher for the eight plasma samples from EB patients than for the six healthy volunteers. Further, the carriage of multiple S. aureus strains by EB patients correlated with higher normalized average IgG levels (Fig. 4B). Thus, the highest normalized average IgG levels were observed for patients EB01 and EB51 who, respectively, were previously shown to carry 7 and 4 different S. aureus types at 3 time points of sampling19,28.
During S. aureus colonization and invasion, immune-competent individuals may rapidly mount antibody responses to a large panel of antigens. The antibody response profiles of individuals usually reflects the history of previous encounters with S. aureus22. Accordingly, they may change after every new encounter21, which could explain the strong variations in the profiles observed for different individuals28,47,48. Importantly, this was previously shown to even be the case upon controlled nasal inoculation of healthy human volunteers with S. aureus49.
In previous studies, we have reported that patients with EB develop wounds that are highly susceptible to S. aureus colonization19,24–26. Accordingly, it was shown that these patients mounted significantly higher immune responses against S. aureus antigens than healthy carriers19,28. In these studies, compared to healthy control individuals, elevated IgG levels were observed in plasmas of EB patients for three out of eleven tested cell wall-associated antigens (i.e. Efb, IsaA and IsdA), and for seven out of seventeen tested secreted antigens. Minor differences were observed for IgG responses against secreted superantigens19. In the present exploratory study, we show that EB patients carry significantly increased IgG levels against all eleven tested antigens that are non-covalently bound to the S. aureus cell wall, including Efb and IsaA. This novel observation highlights the strong immunogenicity of these antigens compared to other staphylococcal antigens that are covalently bound to the cell wall or secreted into the host environment. When IgG levels were compared for EB patients carrying multiple S. aureus types versus EB patients carrying only one S. aureus type, significant differences were observed for 4 out of 10 tested antigens, again including Efb and IsaA19. This general difference upon colonization with multiple S. aureus types was also observed in the present study, supporting the view that patients exposed to different S. aureus types are challenged with more staphylococcal antigens than patients carrying only one S. aureus type. Another novel finding is that EB patients showed a much larger variation in IgG levels against S. aureus antigens than healthy carrier controls. For example, the level of IgG against LukG in EB patient 02 was lower than the respective IgG levels in four of the healthy control individuals, whereas EB patient 53 showed the highest of all presently recorded IgG levels against LukG. Additionally, the comparison of plasma samples from EB patients and healthy control individuals revealed no significantly different IgG levels for other surface-exposed proteins of S. aureus, like FtsL. Thus, while it has been previously shown that EB patients have in general higher levels of antibodies against S. aureus antigens, our present study shows that (i) not all surface-exposed S. aureus proteins will elicit a similar antibody response in each EB patient, and that (ii) different EB patients similarly exposed to S. aureus may generate highly diverse patterns of antibody responses against particular S. aureus antigens. This relates most likely to their S. aureus contact history, which was previously shown to be variable over time19,24,26.
The species S. aureus is known to display high genomic plasticity. Although, the genes for some virulence factors are (almost) invariantly (Efb, Eap, Emp, IsaA) or frequently (Atl, LukG, LukH, SsaA2, Sle1) present in S. aureus isolates, their amino acid sequence identity and expression levels can show substantial inter-strain variations13,17,21,29,50–52. This diversity could be a determinant for variations in the host immune responses to S. aureus. Our results with Eap and Efb, which present large inter-lineage amino acid sequence variation51, showed a highly variable immune response in IgG titers in EB patients compared to healthy carriers. Interestingly, previous studies on different (not EB-related) patient cohorts showed either lower levels of anti-Eap and Efb antibodies in patients48,53,54 or, on the contrary, higher levels in infected patients than in healthy S. aureus carriers23,47,55,56. Nonetheless, consistently higher IgG titers against Atl14 and IsaA19 in sera of S. aureus-infected or colonized patients have been previously reported, which is in agreement with our own results.
In the context of our present study, it is noteworthy that except for very severe cases, patients with EB appear to suffer infrequently from invasive staphylococcal disease, especially if one considers their high rates of colonization with S. aureus28. This has led to the proposal that the elevated levels of anti-staphylococcal IgGs could potentially be protective. Importantly, none of the patients who participated in this and our previous studies was treated for S. aureus bacteremia in the 5 years prior to donating the investigated plasma samples. Additional support for the idea that high anti-staphylococcal IgG levels in EB patients could be protective comes from studies with monoclonal antibodies against IsaA, showing protection against S. aureus infections in murine S. aureus infection models6,7,57. At present, it is not clear whether these findings for anti-IsaA antibodies can be extrapolated to other non-covalently cell wall-bound proteins. This idea is tempting in view of the present results, but it has to be noted that vaccination studies in murine models with an octa-valent vaccine, including Atl and IsaA, did not lead to protection against S. aureus challenges18. Yet, immune responses directed against other presently investigated antigens could be beneficial, not only by promoting opsonophagocytosis, but also by interfering with the biological activity of the different antigens as was recently shown for monoclonal antibodies against the staphylococcal complement inhibitor SCIN58. Clearly, potentially beneficial effects of using non-covalently cell surface-bound proteins as antigens for vaccination need to be thoroughly validated in immunization experiments. This is important, because strong immune responses against S. aureus antigens may not be (fully) protective. This is underscored by the observation that S. aureus carriers producing antibodies that neutralize superantigens can still develop sepsis, although they do have an improved prognosis59. Also, in animal experiments high antibody titers against S. aureus antigens, such as IsaA, are not necessarily protective against infections by this pathogen18,60. In this context, it is noteworthy that Hawkins et al. showed that natural exposure of humans to S. aureus elicited a strong antibody response against ClfA, but this response did not prevent the ClfA-mediated binding of S. aureus cells to fibrinogen, the natural ligand of ClfA61. In contrast, immunization with a ClfA-containing vaccine induced functional antibodies that prevented S. aureus from binding to fibrinogen. Another recent observation that encourages further research towards the development of an anti-staphylococcal vaccine was reported by Stentzel et al., who showed that immunoglobulin replacement therapy in STAT3 hyper-IgE syndrome patients with low S. aureus-specific serum IgG levels did not only increase the S. aureus-specific IgG levels, but also resulted in an attenuated clinical course of disease62.
Conclusion
In the present study, we assessed the immunogenicity of ten non-covalently cell surface-bound proteins of S. aureus, using plasma samples from patients with EB and healthy volunteers. Surface-exposed and secreted proteins of S. aureus have previously been studied as potential vaccine targets9,10. However, while most studies focused on covalently cell wall-bound proteins, the less-studied non-covalently cell wall-bound proteins could also be promising vaccine targets16. Therefore, we applied a combined bioinformatics and biochemical approach to select ten non-covalently cell wall-bound proteins of S. aureus for further analyses. These included Eap, Efb, EMP, IsaA, LukG, LukH, SA0710, Sle1 and SsaA2, as well as two separated domains of Atl. These potential antigens were expressed in L. lactis, purified and tested for antigenicity using human plasma samples. Remarkably, our present results show that the high exposure of EB patients to S. aureus was mirrored by significantly elevated IgG levels against all tested non-covalently cell wall-bound antigens. This is a novel finding, which suggests that these antigens on the cell surface of S. aureus are prime targets for the human immune system.
Electronic supplementary material
Acknowledgements
We thank the anonymous patients with EB from the Dutch Epidermolysis Bullosa Registry for blood donations, Joris de Groot for his contribution in the early phase of the project, and Magda van der Kooi-Pol for collecting the plasma samples from EB patients in a previous study. F.R.P. received a scholarship from CONACyT (169643) and was supported in parts by the Graduate School for Medical Sciences of the University of Groningen. M.F.J. and J.M.v.D. received support from the Eleven Flowers Fund, and the Ubbo Emmius Fund of the University of ”Groningen. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author Contributions
F.R.P., J.M.v.D. and G.B. conceived and designed the experiments. F.R.P., J.N., D.G.A.M.K., D.d.G., S.E. and G.B. performed the experiments. F.R.P., J.M.v.D. and G.B. analyzed the data. J.D., M.F.J., J.M.v.D. and G.B. contributed reagents, materials and analysis tools. F.R.P., J.M.v.D. and G.B. wrote the manuscript. All authors have reviewed and approved the final manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Francisco Romero Pastrana, Jolanda Neef, Jan Maarten van Dijl and Girbe Buist contributed equally to this work.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-21724-z.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Wertheim HF, et al. The role of nasal carriage in Staphylococcus aureus infections. Lancet Infect. Dis. 2005;5:751–762. doi: 10.1016/S1473-3099(05)70295-4. [DOI] [PubMed] [Google Scholar]
- 2.Boucher HW, Corey GR. Epidemiology of Methicillin-Resistant Staphylococcus aureus. Clin. Infect. Dis. 2008;46:S344–S349. doi: 10.1086/533590. [DOI] [PubMed] [Google Scholar]
- 3.Missiakas, D. & Schneewind, O. Staphylococcus aureus vaccines: Deviating from the carol. J. Exp. Med. 213, 1645–1653. [DOI] [PMC free article] [PubMed]
- 4.Proctor RA. Recent developments for Staphylococcus aureus vaccines: clinical and basic science challenges. Eur. Cell. Mater. 2015;30:315–326. doi: 10.22203/eCM.v030a22. [DOI] [PubMed] [Google Scholar]
- 5.Sause WE, Buckley PT, Strohl WR, Lynch AS, Torres VJ. Antibody-Based Biologics and Their Promise to Combat Staphylococcus aureus Infections. Trends Pharmacol. Sci. 2016;37:231–241. doi: 10.1016/j.tips.2015.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van den Berg S, et al. A human monoclonal antibody targeting the conserved staphylococcal antigen IsaA protects mice against Staphylococcus aureus bacteremia. Int. J. Med. Microbiol. 2015;305:55–64. doi: 10.1016/j.ijmm.2014.11.002. [DOI] [PubMed] [Google Scholar]
- 7.Lorenz U, et al. Functional Antibodies Targeting IsaA of Staphylococcus aureus Augment Host Immune Response and Open New Perspectives for Antibacterial Therapy. Antimicrob. Agents Chemother. 2011;55:165–173. doi: 10.1128/AAC.01144-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nizet V. Understanding how leading bacterial pathogens subvert innate immunity to reveal novel therapeutic targets. J. Allergy Clin. Immunol. 2007;120:13–22. doi: 10.1016/j.jaci.2007.06.005. [DOI] [PubMed] [Google Scholar]
- 9.Holtfreter S, Kolata J, Bröker BM. Towards the immune proteome of Staphylococcus aureus – The anti-S. aureus antibody response. Int. J. Med. Microbiol. 2010;300:176–192. doi: 10.1016/j.ijmm.2009.10.002. [DOI] [PubMed] [Google Scholar]
- 10.Sibbald MJJB, et al. Mapping the Pathways to Staphylococcal Pathogenesis by Comparative Secretomics. Microbiol. Mol. Biol. Rev. 2006;70:755–788. doi: 10.1128/MMBR.00008-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim HK, et al. IsdA and IsdB antibodies protect mice against Staphylococcus aureus abscess formation and lethal challenge. Vaccine. 2010;28:6382–6392. doi: 10.1016/j.vaccine.2010.02.097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hempel K, Herbst F-A, Moche M, Hecker M, Becher D. Quantitative Proteomic View on Secreted, Cell Surface-Associated, and Cytoplasmic Proteins of the Methicillin-Resistant Human Pathogen Staphylococcus aureus under Iron-Limited Conditions. J. Proteome Res. 2011;10:1657–1666. doi: 10.1021/pr1009838. [DOI] [PubMed] [Google Scholar]
- 13.Ziebandt A-K, et al. Proteomics uncovers extreme heterogeneity in the Staphylococcus aureus exoproteome due to genomic plasticity and variant gene regulation. PROTEOMICS. 2010;10:1634–1644. doi: 10.1002/pmic.200900313. [DOI] [PubMed] [Google Scholar]
- 14.Etz H, et al. Identification of in vivo expressed vaccine candidate antigens from Staphylococcus aureus. Proc. Natl. Acad. Sci. 2002;99:6573–6578. doi: 10.1073/pnas.092569199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vytvytska O, et al. Identification of vaccine candidate antigens of Staphylococcus aureus by serological proteome analysis. PROTEOMICS. 2002;2:580–590. doi: 10.1002/1615-9861(200205)2:5<580::AID-PROT580>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 16.Glowalla E, Tosetti B, Krönke M, Krut O. Proteomics-Based Identification of Anchorless Cell Wall Proteins as Vaccine Candidates against Staphylococcus aureus. Infect. Immun. 2009;77:2719–2729. doi: 10.1128/IAI.00617-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dreisbach A, van Dijl JM, Buist G. The cell surface proteome of Staphylococcus aureus. PROTEOMICS. 2011;11:3154–3168. doi: 10.1002/pmic.201000823. [DOI] [PubMed] [Google Scholar]
- 18.van den Berg S, et al. Active Immunization with an Octa-Valent Staphylococcus aureus Antigen Mixture in Models of S. aureus Bacteremia and Skin Infection in Mice. PLoS ONE. 2015;10:e0116847. doi: 10.1371/journal.pone.0116847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van der Kooi-Pol MM, et al. High Anti-Staphylococcal Antibody Titers in Patients with Epidermolysis Bullosa Relate to Long-Term Colonization with Alternating Types of Staphylococcus aureus. J. Invest. Dermatol. 2013;133:847–850. doi: 10.1038/jid.2012.347. [DOI] [PubMed] [Google Scholar]
- 20.Nair N, et al. Amidase, a cell wall hydrolase, elicits protective immunity against Staphylococcus aureus and S. epidermidis. Int. J. Biol. Macromol. 2015;77:314–321. doi: 10.1016/j.ijbiomac.2015.03.047. [DOI] [PubMed] [Google Scholar]
- 21.Kolata J, et al. Distinctive patterns in the human antibody response to Staphylococcus aureus bacteremia in carriers and non-carriers. PROTEOMICS. 2011;11:3914–3927. doi: 10.1002/pmic.201000760. [DOI] [PubMed] [Google Scholar]
- 22.Bröker BM, van Belkum A. Immune proteomics of Staphylococcus aureus. PROTEOMICS. 2011;11:3221–3231. doi: 10.1002/pmic.201100010. [DOI] [PubMed] [Google Scholar]
- 23.Verkaik NJ, et al. Heterogeneity of the humoral immune response following Staphylococcus aureus bacteremia. Eur. J. Clin. Microbiol. Infect. Dis. 2010;29:509–518. doi: 10.1007/s10096-010-0888-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van der Kooi-Pol MM, et al. High genetic diversity of Staphylococcus aureus strains colonizing patients with epidermolysis bullosa. Exp. Dermatol. 2012;21:463–466. doi: 10.1111/j.1600-0625.2012.01502.x. [DOI] [PubMed] [Google Scholar]
- 25.van der Kooi-Pol MM, et al. Topography of Distinct Staphylococcus aureus Types in Chronic Wounds of Patients with Epidermolysis Bullosa. PLoS ONE. 2013;8:e67272. doi: 10.1371/journal.pone.0067272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van der Kooi-Pol MM, Duipmans JC, Jonkman MF, van Dijl JM. Host–pathogen interactions in epidermolysis bullosa patients colonized with Staphylococcus aureus. Int. J. Med. Microbiol. 2014;304:195–203. doi: 10.1016/j.ijmm.2013.11.012. [DOI] [PubMed] [Google Scholar]
- 27.García-Pérez AN, et al. From the wound to the bench: exoproteome interplay between wound-colonizing Staphylococcus aureus strains and co-existing bacteria. Virulence. 2017;0:1–35. doi: 10.1080/21505594.2017.1395129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Swierstra J, et al. IgG4 Subclass-Specific Responses to Staphylococcus aureus Antigens Shed New Light on Host-Pathogen Interaction. Infect. Immun. 2015;83:492–501. doi: 10.1128/IAI.02286-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dreisbach A, et al. Profiling the surfacome of Staphylococcus aureus. PROTEOMICS. 2010;10:3082–3096. doi: 10.1002/pmic.201000062. [DOI] [PubMed] [Google Scholar]
- 30.Steen A, et al. Cell Wall Attachment of a Widely Distributed Peptidoglycan Binding Domain Is Hindered by Cell Wall Constituents. J. Biol. Chem. 2003;278:23874–23881. doi: 10.1074/jbc.M211055200. [DOI] [PubMed] [Google Scholar]
- 31.Sibbald MJJB, et al. Synthetic Effects of secG and secY2 Mutations on Exoproteome Biogenesis in Staphylococcus aureus. J. Bacteriol. 2010;192:3788–3800. doi: 10.1128/JB.01452-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sibbald MJJB, et al. Partially overlapping substrate specificities of staphylococcal group A sortases. PROTEOMICS. 2012;12:3049–3062. doi: 10.1002/pmic.201200144. [DOI] [PubMed] [Google Scholar]
- 33.Geertsma ER, Poolman B. High-throughput cloning and expression in recalcitrant bacteria. Nat. Methods. 2007;4:705–707. doi: 10.1038/nmeth1073. [DOI] [PubMed] [Google Scholar]
- 34.Leenhouts, K. & Venema, G. Lactococcal plasmid vectors. Plasmids Pract. Approach Oxf. Univ. Press Oxf. U. K. 65–94 (1993).
- 35.Neef, J. et al. Versatile vector suite for the extracytoplasmic production and purification of heterologous His-tagged proteins in Lactococcus lactis. Appl. Microbiol. Biotechnol. 1–12, 10.1007/s00253-015-6778-8 (2015). [DOI] [PMC free article] [PubMed]
- 36.Neef J, Koedijk DGAM, Bosma T, Dijl JMvan, Buist G. Efficient production of secreted staphylococcal antigens in a non-lysing and proteolytically reduced Lactococcus lactis strain. Appl. Microbiol. Biotechnol. 2014;98:10131–10141. doi: 10.1007/s00253-014-6030-y. [DOI] [PubMed] [Google Scholar]
- 37.Chavakis T, Wiechmann K, Preissner KT, Herrmann M. Staphylococcus aureus interactions with the endothelium. The role of bacterial “Secretable Expanded Repertoire Adhesive Molecules” (SERAM) in disturbing host defense systems. Thromb. Haemost. 2005;94:278–285. doi: 10.1160/TH05-05-0306. [DOI] [PubMed] [Google Scholar]
- 38.Buist G, Steen A, Kok J, Kuipers OP. LysM, a widely distributed protein motif for binding to (peptido)glycans. Mol. Microbiol. 2008;68:838–847. doi: 10.1111/j.1365-2958.2008.06211.x. [DOI] [PubMed] [Google Scholar]
- 39.Ponting CP, Aravind L, Schultz J, Bork P, Koonin EV. Eukaryotic Signalling Domain Homologues in Archaea and Bacteria. Ancient Ancestry and Horizontal Gene Transfer. J. Mol. Biol. 1999;289:729–745. doi: 10.1006/jmbi.1999.2827. [DOI] [PubMed] [Google Scholar]
- 40.Baba T, Schneewind O. Targeting of muralytic enzymes to the cell division site of Gram-positive bacteria: repeat domains direct autolysin to the equatorial surface ring of Staphylococcus aureus. EMBO J. 1998;17:4639–4646. doi: 10.1093/emboj/17.16.4639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Whisstock JC, Lesk AM. SH3 domains in prokaryotes. Trends Biochem. Sci. 1999;24:132–133. doi: 10.1016/S0968-0004(99)01366-3. [DOI] [PubMed] [Google Scholar]
- 42.Bateman A, Holden MTG, Yeats C. The G5 domain: a potential N-acetylglucosamine recognition domain involved in biofilm formation. Bioinformatics. 2005;21:1301–1303. doi: 10.1093/bioinformatics/bti206. [DOI] [PubMed] [Google Scholar]
- 43.Frankel MB, Schneewind O. Determinants of Murein Hydrolase Targeting to Cross-wall of Staphylococcus aureus Peptidoglycan. J. Biol. Chem. 2012;287:10460–10471. doi: 10.1074/jbc.M111.336404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zoll S, et al. Ligand-Binding Properties and Conformational Dynamics of Autolysin Repeat Domains in Staphylococcal Cell Wall Recognition. J. Bacteriol. 2012;194:3789–3802. doi: 10.1128/JB.00331-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Romero Pastrana F, Neef J, van Dijl JM, Buist G. A Lactococcus lactis expression vector set with multiple affinity tags to facilitate isolation and direct labeling of heterologous secreted proteins. Appl. Microbiol. Biotechnol. 2017;101:8139–8149. doi: 10.1007/s00253-017-8524-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bosma T, et al. Novel Surface Display System for Proteins on Non-Genetically Modified Gram-Positive Bacteria. Appl. Environ. Microbiol. 2006;72:880–889. doi: 10.1128/AEM.72.1.880-889.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Colque-Navarro P, Jacobsson G, Andersson R, Flock J-I, Möllby R. Levels of Antibody against 11 Staphylococcus aureus Antigens in a Healthy Population. Clin. Vaccine Immunol. 2010;17:1117–1123. doi: 10.1128/CVI.00506-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dryla A, et al. Comparison of Antibody Repertoires against Staphylococcus aureus in Healthy Individuals and in Acutely Infected Patients. Clin. Diagn. Lab. Immunol. 2005;12:387–398. doi: 10.1128/CDLI.12.3.387-398.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Holtfreter S, et al. Human Immune Proteome in Experimental Colonization with Staphylococcus aureus. Clin. Vaccine Immunol. 2009;16:1607–1614. doi: 10.1128/CVI.00263-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hussain M, et al. eap Gene as Novel Target for Specific Identification of Staphylococcus aureus. J. Clin. Microbiol. 2008;46:470–476. doi: 10.1128/JCM.01425-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.McCarthy AJ, Lindsay JA. Genetic variation in Staphylococcus aureus surface and immune evasion genes is lineage associated: implications for vaccine design and host-pathogen interactions. BMC Microbiol. 2010;10:173. doi: 10.1186/1471-2180-10-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Romero Pastrana F, et al. Noninvasive optical and nuclear imaging of Staphylococcus-specific infection with a human monoclonal antibody-based probe. Virulence. 2017;0:1–11. doi: 10.1080/21505594.2017.1403004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Colque-Navarro P, Palma M, Söderquist B, Flock J-I, Möllby R. Antibody Responses in Patients with Staphylococcal Septicemia against Two Staphylococcus aureus Fibrinogen Binding Proteins: Clumping Factor and an Extracellular Fibrinogen Binding Protein. Clin. Diagn. Lab. Immunol. 2000;7:14–20. doi: 10.1128/cdli.7.1.14-20.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Royan S, et al. Identification of the secreted macromolecular immunogens of Staphylococcus aureus by analysis of serum. FEMS Immunol. Med. Microbiol. 2000;29:315–321. doi: 10.1111/j.1574-695X.2000.tb01539.x. [DOI] [PubMed] [Google Scholar]
- 55.Joost I, et al. Antibody response to the extracellular adherence protein (Eap) of Staphylococcus aureus in healthy and infected individuals. FEMS Immunol. Med. Microbiol. 2011;62:23–31. doi: 10.1111/j.1574-695X.2011.00783.x. [DOI] [PubMed] [Google Scholar]
- 56.Verkaik NJ, et al. Anti-Staphylococcal Humoral Immune Response in Persistent Nasal Carriers and Noncarriers of Staphylococcus aureus. J. Infect. Dis. 2009;199:625–632. doi: 10.1086/596743. [DOI] [PubMed] [Google Scholar]
- 57.Lorenz U, et al. Human antibody response during sepsis against targets expressed by methicillin resistant Staphylococcus aureus. FEMS Immunol. Med. Microbiol. 2000;29:145–153. doi: 10.1111/j.1574-695X.2000.tb01517.x. [DOI] [PubMed] [Google Scholar]
- 58.Hoekstra, H. et al. A human monoclonal antibody that specifically binds and inhibits the staphylococcal complement inhibitor protein SCIN. Virulence 1–13, 10.1080/21505594.2017.1294297 (2017). [DOI] [PMC free article] [PubMed]
- 59.Holtfreter S, et al. Staphylococcus aureus Carriers Neutralize Superantigens by Antibodies Specific for Their Colonizing Strain: A Potential Explanation for Their Improved Prognosis in Severe Sepsis. J. Infect. Dis. 2006;193:1275–1278. doi: 10.1086/503048. [DOI] [PubMed] [Google Scholar]
- 60.Koedijk DGAM, et al. Differential epitope recognition in the immunodominant staphylococcal antigen A of Staphylococcus aureus by mouse versus human IgG antibodies. Sci. Rep. 2017;7:8141. doi: 10.1038/s41598-017-08182-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hawkins J, et al. A recombinant Clumping factor A containing vaccine induces functional antibodies to Staphylococcus aureus that are not observed after natural exposure. Clin. Vaccine Immunol. 2012;19:1641–50. doi: 10.1128/CVI.00354-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Stentzel S, et al. Reduced Immunoglobulin (Ig) G Response to Staphylococcus aureus in STAT3 Hyper-IgE Syndrome. Clin. Infect. Dis. 2017;64:1279–1282. doi: 10.1093/cid/cix140. [DOI] [PubMed] [Google Scholar]
- 63.Ko Y-P, et al. Coagulase and Efb of Staphylococcus aureus Have a Common Fibrinogen Binding Motif. mBio. 2016;7:e01885–15. doi: 10.1128/mBio.01885-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Novick R. Properties of a cryptic high-frequency transducing phage in Staphylococcus aureus. Virology. 1967;33:155–166. doi: 10.1016/0042-6822(67)90105-5. [DOI] [PubMed] [Google Scholar]
- 65.Duthie ES, Lorenz LL. Staphylococcal Coagulase: Mode of Action and Antigenicity. J. Gen. Microbiol. 1952;6:95–107. doi: 10.1099/00221287-6-1-2-95. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.