SUMMARY
Canonical Wnt signaling coordinates many critical aspects of embryonic development, while dysregulated Wnt signaling contributes to common diseases, including congenital malformations and cancer. The nuclear localization of β-catenin is the defining step in pathway activation. However, despite intensive investigation, the mechanisms regulating β-catenin nuclear transport remain undefined. In a patient with congenital heart disease and heterotaxy, a disorder of left-right patterning, we previously identified the guanine nucleotide exchange factor, RAPGEF5. Here, we demonstrate that RAPGEF5 regulates left-right patterning via Wnt signaling. In particular, RAPGEF5, regulates the nuclear translocation of β-catenin independently of both β-catenin cytoplasmic stabilization and the importin β1/Ran mediated transport system. We propose a model whereby RAPGEF5 activates the nuclear GTPases, Rap1a/b, to facilitate the nuclear transport of β-catenin, defining a parallel nuclear transport pathway to Ran. Our results suggest new targets for modulating Wnt signaling in disease states.
Keywords: RAPGEF5, Rap, GTPase, Wnt signaling, β-catenin, nuclear transport, Ran independent, heterotaxy, congenital heart disease, Xenopus
INTRODUCTION
Canonical Wnt signaling is a highly conserved regulatory pathway for many aspects of embryonic development and adult tissue homeostasis. When dysregulated, Wnt signaling contributes to numerous diseases, including congenital malformations, neurodegeneration, diabetes, and multiple cancers (Clevers and Nusse, 2012; MacDonald et al., 2009; Moon et al., 2004; Niehrs, 2012). In the defining step of the pathway, nuclear translocation of β-catenin is essential. In fact, Wnt ligand binding inhibits the cytoplasmic β-catenin destruction machinery allowing β-catenin to accumulate and enter into the nucleus (MacDonald et al., 2009; Niehrs, 2012).
However, despite intensive investigation, the regulators of β-catenin nuclear transport have been enigmatic. β-catenin lacks a nuclear localization signal (NLS) and does not require the Ran-GTPase-regulated importin/karyopherin transport carriers to access the nuclear compartment (Fagotto, 2013; Fagotto et al., 1998; Wiechens and Fagotto, 2001; Xu and Massague, 2004; Yokoya et al., 1999). Nevertheless, there are clear parallels with the Ran based system: 1) translocation of β-catenin occurs through Nuclear Pore Complexes (NPCs), 2) the armadillo (ARM) repeats in β-catenin are structurally similar to the ARM and HEAT repeats found in importin-α and importin-β family members, and β-catenin competes for NPC-binding sites with importin-β1, 3) nuclear import is energy dependent, and 4) import of β-catenin is inhibited by nonhydrolyzable GTP analogs, suggesting the involvement of an unknown GTPase (Fagotto, 2013; Fagotto et al., 1998; Jamieson et al., 2014; Lowe et al., 2015; Xu and Massague, 2004). Together, these data demonstrate that β-catenin uses a non-classic pathway for nuclear transport; however, the molecular regulators of this process are unknown.
We previously identified the guanine nucleotide exchange factor, RAPGEF5 as a candidate disease gene in a patient with congenital heart disease and heterotaxy (Fakhro et al., 2011). Heterotaxy is a disorder of left-right (LR) patterning. Normally, our internal organs, such as the heart, are asymmetrically positioned across the LR axis, and patients with heterotaxy can have an especially severe form of congenital heart disease (Brueckner, 2007; Sutherland and Ware, 2009). However, no developmental function for RAPGEF5 had been established. Here we show that RAPGEF5, regulates left-right patterning via Wnt signaling. Importantly, RAPGEF5 is critical for the nuclear localization of β-catenin independently of cytoplasmic stabilization and the importin α/β1 mediated transport system. We propose a model whereby RAPGEF5 activates nuclear Raps to facilitate the nuclear transport of β-catenin, identifying new targets for modulating Wnt signaling in disease.
RESULTS
Rapgef5 is required for left – right development
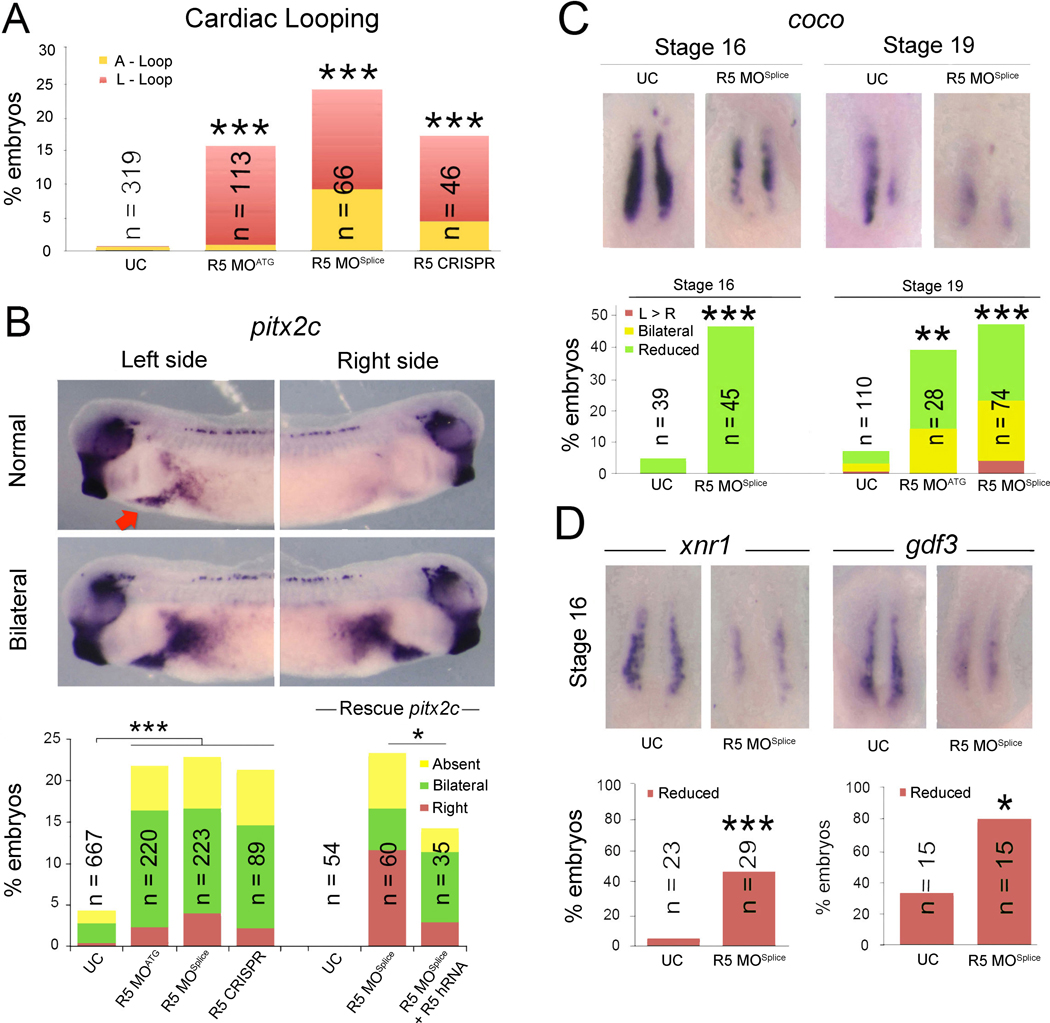
To investigate whether RAPGEF5 is required for normal left-right development, we first examined organ situs in Rapgef5 depleted Xenopus embryos. The frog model, Xenopus, is ideal for these studies as both gain and loss of function experiments are rapid and efficient; for example, three days after manipulating fertilized Xenopus eggs, we can readily assess cardiac situs by simple inspection of Xenopus tadpoles (Blum et al., 2009). In vertebrates, the developing cardiac tube initially forms in the midline and then normally loops to the right (D-loop). However, defects in left-right patterning result in morphological abnormalities, including leftward (L) or ambiguous (A) loops (Brueckner, 2007; Sutherland and Ware, 2009). Knockdown of rapgef5 using translation (R5 MOATG) or splice (R5 MOSplice) blocking morpholino oligonucleotides (MOs), as well as CRISPR mediated F0 depletion (Bhattacharya et al., 2015), resulted in abnormal cardiac looping and confirmed a role for Rapgef5 in left-right development (Fig. 1A, Fig. S1A).
Figure 1. Rapgef5 depletion disrupts left-right development.
(A) Percentage of Rapgef5 depleted embryos with abnormal cardiac looping (A or L loops). (B) Pitx2c is expressed in the left lateral mesoderm of stage 28 control Xenopus embryos (red arrow, lateral view dorsal to the top), but is abnormally, typically bilaterally, expressed following MO or CRISPR mediated depletion of Rapgef5. Co-injection of human RAPGEF5 mRNA can rescue pitx2c in Rapgef5 morphants. (C) coco expression in the LRO of control and Rapgef5 depleted embryos at stages 16 and 19. Ventral view with anterior to the top. Graphs depict the percentage of embryos displaying abnormal coco expression. Note the reduced expression in rapgef5 morphants at both stages. (D) xnr1 and gdf3 expression is reduced in the LRO of rapgef5 morphants at stage 16, prior to the onset of cilia driven flow. Ventral views with anterior to the top. A single asterisk indicates statistical significance of P<0.05, while double and triple asterisks indicate P<0.01 and P<0.005, respectively. Also see Figure S1 and Figure S2.
Left-right symmetry is first broken at the left-right organizer (LRO). Here motile cilia generate a leftward fluid flow that is detected by laterally positioned immotile sensory cilia and translated into asymmetric gene expression (Boskovski et al., 2013; Kennedy et al., 2007; McGrath et al., 2003; Tabin and Vogan, 2003; Yoshiba and Hamada, 2014). In particular, leftward flow results in the asymmetric expression of coco (dand5, cerl2) in the LRO, and subsequent activation of TGF-β family members and pitx2c expression in the left lateral plate mesoderm (Kawasumi et al., 2011; Logan et al., 1998; Schweickert et al., 2010; Vonica and Brivanlou, 2007). Knockdown of Rapgef5 resulted in abnormalities in both pitx2c and coco expression demonstrating that rapgef5 is required at the level of the LRO (Fig. 1B–C). To test the efficacy and specificity of our Rapgef5 depletion strategies, we employed multiple tests including 1) RT-PCR, 2) western blot, 3) control MO and CRISPR injection, and 4) rescue with human RAPGEF5 mRNA (Fig. 1B, Fig. S1, Fig. S2A).
Importantly in the LRO, coco expression is normally symmetric prior to cilia driven flow (Schweickert et al., 2010) (Fig. 1C, far left panel, Stage (st) 16); however, in morphants coco expression was reduced even at these pre-flow stages (st 16, Fig. 1C), suggesting a defect in the establishment of the LRO itself. This was confirmed by assaying expression of additional LRO marker genes, xnr1 and gdf3 (Hanafusa et al., 2000; Lustig et al., 1996; Sampath et al., 1997; Vonica and Brivanlou, 2007), both of which were reduced before initiation of flow in morphants (Fig. 1D). Thus, Rapgef5 is required for correct establishment of the LRO and subsequent LR development.
Depletion of Rapgef5 impairs canonical Wnt signaling
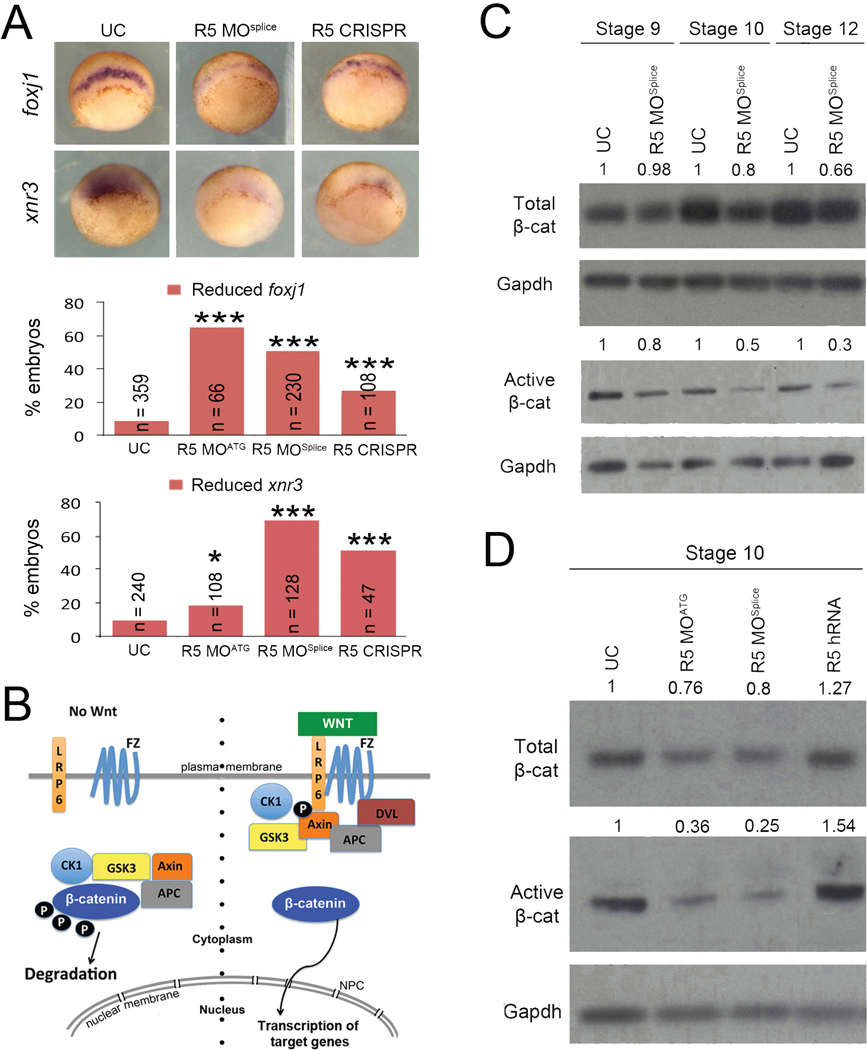
As the LRO fails to form normally in rapgef5 morphants, we next examined the preceding development of this structure. The Spemann-Mangold organizer forms at the dorsal blastopore lip and serves to pattern dorsal structures, including the LRO (De Robertis et al., 2000; Harland and Gerhart, 1997; Khokha et al., 2005; Niehrs, 2004). There, we discovered that foxj1 and xnr3 transcripts, known direct targets of Wnt signaling (Caron et al., 2012; McKendry et al., 1997; Smith et al., 1995; Stubbs et al., 2008; Walentek et al., 2012), were reduced in gastrulating rapgef5 morphants (Fig. 2A). On the other hand, numerous additional dorsal, ventral, and pan-mesodermal markers appear unaffected or only mildly affected compared to the changes seen in foxj1 and xnr3 (Fig. S3A). We note that our Rapgef5 depletion becomes marked only after stage 10, when many of these mesodermal markers are already established (Fig. S2B). Again, injection of a control morpholino or control CRISPR had no effect on foxj1 expression, while injection of human RAPGEF5 mRNA rescued the foxj1 patterning defect in rapgef5 morphants, demonstrating the specificity of our knockdown strategies (Fig. S3B,C). Based on the reduction of foxj1 and xnr3 expression in rapgef5 morphants, we hypothesized that Rapgef5 may play a role in Wnt signaling.
Figure 2. Depletion of Rapgef5 impairs canonical Wnt signaling.
(A) Depletion of Rapgef5 using MOs or CRISPR impairs foxj1 and xnr3 expression in the dorsal blastopore lip of stage 10 embryos. (B) Simplified schematic of the canonical Wnt signaling pathway. Left side: in the absence of Wnt ligand, a destruction complex containing Axin and Gsk3 phosphorylates β-catenin, which marks it for cytoplasmic destruction. Right side: Once the pathway is activated by Wnt ligand binding to Frizzled and Lrp receptors, phosphorylation of β-catenin by GSK3 is inhibited allowing β-catenin to accumulate in the cytoplasm and translocate into the nucleus to initiate transcription of Wnt target genes. (C, D) Levels of total and active β-catenin protein are essentially unchanged in Rapgef5 depleted embryos at stage 9 as assayed by western blot. However, both forms of β-catenin are reduced at stages 10 and 12. Note that levels of active β-catenin are much more severely affected. Conversely, overexpression of human RAPGEF5 mRNA results in a mild increase in total β-catenin levels and a more pronounced increase in active β-catenin levels. Triple asterisks indicates P<0.005. Also see Figure S3.
Wnt signaling plays diverse roles in embryonic development, including formation of primary axes, as well as in common human diseases such as cancer (Clevers and Nusse, 2012; Jamieson et al., 2014; MacDonald et al., 2009; Moon et al., 2004; Niehrs, 2012; Yamaguchi, 2001). β-catenin plays a central role in the canonical Wnt pathway (Fagotto et al., 1997; Funayama et al., 1995; Heasman et al., 1994; McCrea et al., 1993). In the absence of Wnt ligand, cytoplasmic β-catenin is phosphorylated and marked for degradation by a highly efficient destruction complex consisting of GSK3, CK1, APC and Axin (Clevers and Nusse, 2012; Davidson et al., 2005; Kofron et al., 2007; Liu et al., 2002; MacDonald et al., 2009; Niehrs, 2012; Taelman et al., 2010; Yost et al., 1996). This destruction complex maintains cytoplasmic β-catenin at low levels so that it cannot accumulate in the nucleus, repressing canonical Wnt signaling (Fig. 2B). When the Wnt ligand binds to the Frizzled receptor and the LRP6 co-receptor, the destruction complex is inactivated, and β-catenin accumulates and translocates into the nucleus to induce transcription of Wnt target genes (Bhanot et al., 1996; Davidson et al., 2005; Fagotto et al., 1997; Funayama et al., 1995; He et al., 1997; Heasman et al., 1994; MacDonald et al., 2009; Mao et al., 2001a; Mao et al., 2001b; Niehrs, 2012; Pinson et al., 2000; Tamai et al., 2000; Yang-Snyder et al., 1996) (Fig. 2B). Therefore, we began our analysis of the Wnt pathway by examining β-catenin protein levels from whole embryo lysates in rapgef5 morphants. By western blot, β-catenin protein levels were reduced in rapgef5 morphants at stages 10 and 12 compared to uninjected controls (Fig. 2C,D). Conversely, β-catenin levels were increased by overexpression of human RAPGEF5 (Fig. 2D). Importantly, while levels of total β-catenin were mildly affected, the active (unphosphorylated) pool of β-catenin that translocates into the nucleus was much more severely affected (Fig. 2C,D). Together these observations lead us to postulate that Rapgef5 may be required for the cytoplasmic stabilization of β-catenin.
Rapgef5 acts downstream of cytoplasmic stabilization
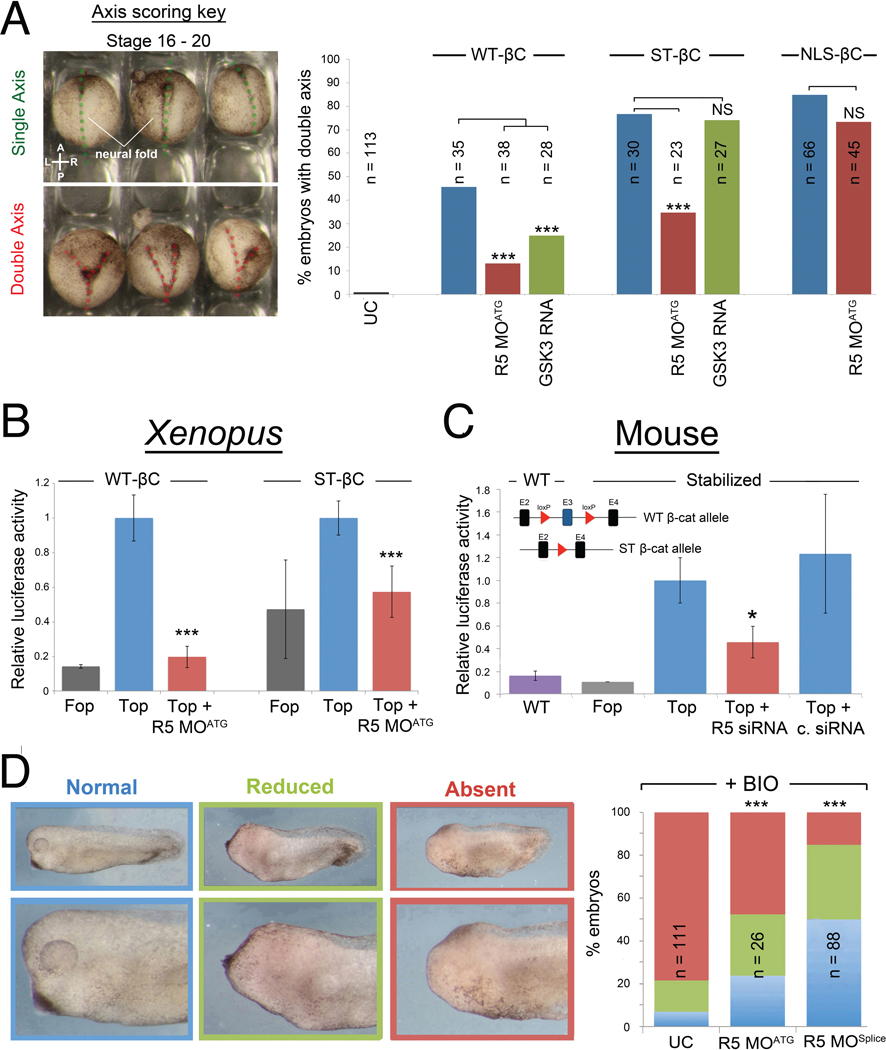
To test whether Rapgef5 functions in cytoplasmic stabilization, we first asked if Rapgef5 depletion could counteract the activity of wild type (WT) and stabilized (ST) β-catenin proteins. The degradation machinery phosphorylates β-catenin at specific serine/threonine residues via GSK3; modification of these residues to alanine (S33A, S37A, T41A) prevents phosphorylation and produces a “stabilized” form of β-catenin that evades the degradation machinery (Liu et al., 2002; Taelman et al., 2010; Yost et al., 1996). Overexpression of β-catenin can induce a second embryonic axis in Xenopus embryos (Funayama et al., 1995; Guger and Gumbiner, 1995), which can be easily scored by simple inspection (Fig. 3A, left panel). Co-injection of either rapgef5 MO or GSK3 mRNA significantly reduces the ability of WT β-catenin mRNA to induce secondary axes in Xenopus embryos (Fig. 3A), suggesting that Rapgef5 acts at the level of β-catenin signaling rather than upstream in the pathway. As expected, co-injection of GSK3 mRNA has no effect on the ability of stabilized β-catenin to induce a secondary axis. However, surprisingly, co-injection of rapgef5 MO did reduce the ability of stabilized β-catenin to induce secondary axes (Fig. 3A) suggesting that Rapgef5 acts independently of β-catenin stabilization.
Figure 3. Rapgef5 acts downstream of β-catenin cytoplasmic stabilization.
(A) The ability to induce secondary axes in development can be used as a read out of Wnt signaling activity. Uninjected embryos have a single axis (dotted green line) while embryos injected with β-catenin mRNA can have a second embryonic axis (two dotted red lines) that are readily detected at st 16–19 embryos. Injection of WT, stabilized (ST), or NLS tagged β-catenin can induce secondary axes. Rapgef5 depletion significantly decreases the percentage of secondary axes induced by WT and ST β-catenin. Injection of GSK3 mRNA reduces secondary axes induced by WT β-catenin but has no effect on ST β-catenin. The ability of NLS-β-catenin to induce secondary axes is unaffected by reduction of Rapgef5 levels. (B) Rapgef5 knockdown reduces luciferase activity in embryos injected with WT or ST β-catenin mRNA in a TOPFlash assay. Data are represented as mean ± SD. (C) Wnt signaling activity is reduced in ST β-catenin MEFs (Δexon3) following siRNA depletion of Rapgef5. The schematic depicts the Catnblox(ex3) mouse allele. Exon 3 (E3), containing the GSK3 phosphorylation sites that target β-catenin for cytoplasmic degradation, is flanked by loxp sites allowing for its conditional removal and production of a stabilized (ST) β-catenin allele. WT: WT cells; FOP: ST β-catenin cells transfected with FOPFlash negative control; TOP: ST β-catenin cells transfected with TOPFlash reporter plasmid; TOP + R5 siRNA: ST β-catenin cells transfected with Rapgef5 siRNA and TOPFlash reporter plasmid; Top + C. siRNA: ST β-catenin cells transfected with control siRNA and TOPFlash reporter plasmid. A Renilla luciferase transfection control was included in each treatment to allow normalization. Data are represented as mean ± SEM. (D) Pharmacological inhibition of GSK3 by the addition of BIO between stages 9 – 11 results in increased β-catenin signaling and loss of anterior development in Xenopus embryos. Depletion of Rapgef5 can counteract this effect and rescue development of the head demonstrating that Rapgef5 regulates Wnt signaling downstream of GSK3. A single asterisk indicates P<0.05, while double and triple asterisks indicate P<0.01 and P<0.005, respectively.
An alternative method to assay Wnt signaling, TOPFlash luciferase assays place luciferase expression under the control of TCF/LEF binding sites (Molenaar et al., 1996; van de Wetering et al., 1997; van de Wetering et al., 1991; Veeman et al., 2003). In this case, activation of Wnt signaling leads to the accumulation of nuclear β-catenin that complexes with TCF/LEF to activate luciferase expression. Consistent with our induced secondary axis experiments, Rapgef5 depletion reduced the luciferase signal of both WT and stabilized β-catenin in TOPFlash assays in Xenopus embryos (Fig. 3B). To see if this result was conserved to mammals, we depleted Rapgef5 using siRNAs in primary mouse embryonic fibroblasts. In mice, the specific serine/threonine residues that are phosphorylated by GSK-3 and lead to degradation of β-catenin are encoded on a single exon. Excision of this exon creates cells that express a genetically stabilized β-catenin allele (Harada et al., 1999). Even in cells with the stabilized β-catenin allele, depletion of Rapgef5 led to decreased luciferase activity (Fig. 3C).
To further test whether Rapgef5 regulates Wnt signaling independently of β-catenin degradation, we pursued a parallel strategy to inhibit GSK3 activity pharmacologically using the chemical inhibitor BIO (Sato et al., 2004). Wnt signaling must be attenuated to form the head (Glinka et al., 1998; Yamaguchi, 2001), so activation of Wnt signaling with BIO between stages 9 and 12 leads to a dramatic reduction in head development. Knockdown of rapgef5 using either R5 MOATG or R5 MOSplice counteracted the effect of BIO and rescued development of the head (Fig. 3D). Taken together, these studies in both frogs and mammals indicate that Rapgef5 regulates Wnt signaling independently of the well-established β-catenin cytoplasmic destruction machinery, suggesting that Rapgef5 acts downstream in the Wnt signaling pathway (Fig. 2B).
Nuclear translocation of β-catenin is impaired in rapgef5 morphants
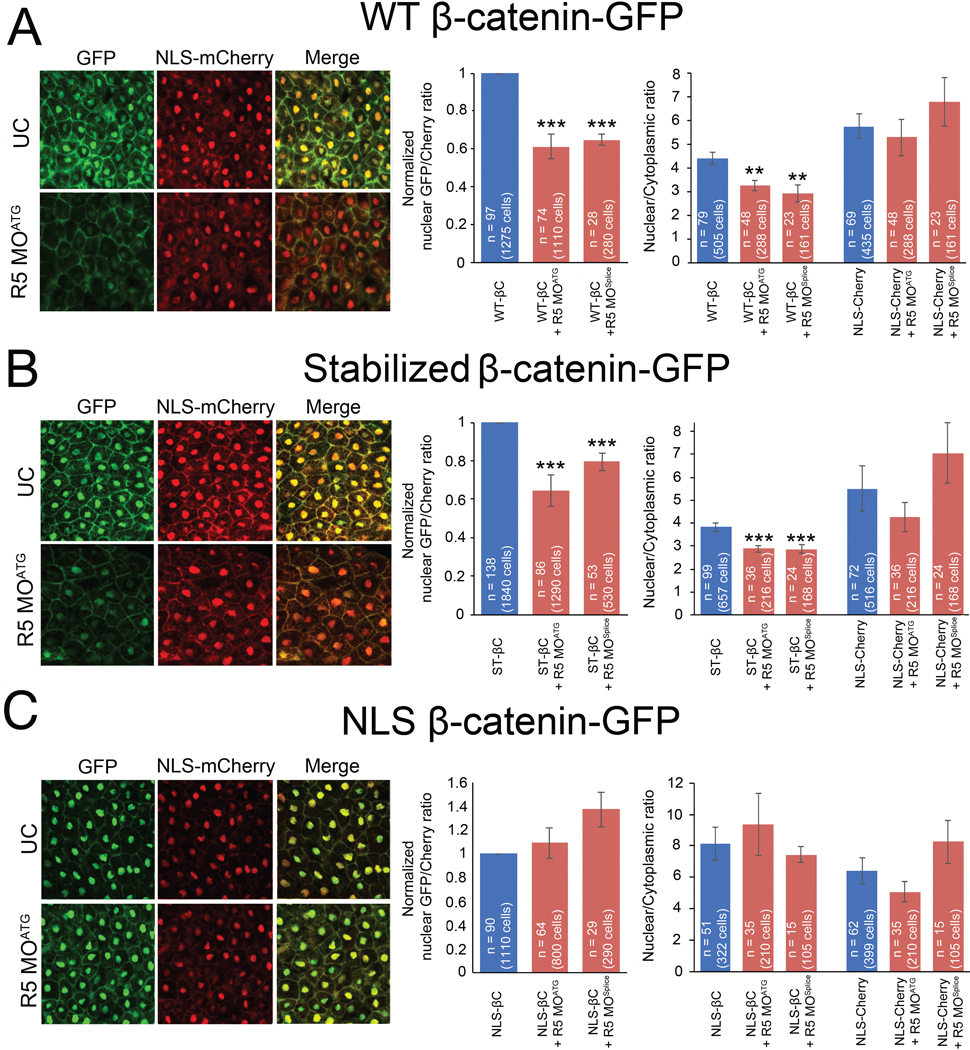
Our initial experiments with whole embryo lysates indicated that β-catenin levels were lower with Rapgef5 depletion (Fig. 2C,D); therefore, we had initially hypothesized that Rapgef5 plays a role in β-catenin degradation. Given the results described above, we sought alternative explanations and began with studies to examine β-catenin localization. To directly visualize β-catenin, we injected GFP tagged WT and stabilized β-catenin mRNA at the one cell stage and imaged the dorsal blastopore lip at stage 10. Concurrently, to enable ratiometric analysis with an importin-β1 transported cargo, we co-injected mCherry tagged with a N-terminal classic nuclear localization signal from SV40 large T antigen (“PKKKRKV”; NLS-mCherry). In control embryos, the GFP tagged β-catenin localized robustly to dorsal nuclei and the plasma membrane; however, levels of nuclear β-catenin were significantly diminished in rapgef5 morphants whether we used the WT or stabilized variant of β-catenin. We examined this result in two ways. First, we compared GFP and RFP signals in the cytoplasm compared to the nucleus. In both WT β-catenin-GFP and stabilized β-catenin-GFP injected embryos, the nuclear:cytoplasmic ratio of β-catenin was reduced with depletion of Rapgef5 while no effect was observed with the NLS-mCherry (Fig. 4A,B far right panels). Second, we compared the nuclear signal from β-catenin (green channel) to nuclear signal of NLS-mCherry (red) and observed a reduction in the β-catenin-GFP/mCherry ratio in Rapgef5 depleted embryos (Fig. 4A,B middle panels). As a control, we examined the levels of β-catenin (both nascent and GFP tagged forms) by Western blot, demonstrating that Rapgef5 depletion had no significant effect on levels of injected GFP-β-catenin (Fig. S4).
Figure 4. Rapgef5 is required for the nuclear localization of β-catenin.
(A,B) GFP tagged WT and ST β-catenin localize to the plasma membrane and nucleus in the dorsal blastopore lip of stage 10 control embryos but this nuclear localization is lost in rapgef5 morphants (note loss of nuclear GFP signal in merged images). (C) NLS β-catenin-GFP localizes normally into nuclei even in the absence of Rapgef5. Graphs in the center panel represent the ratio of nuclear localized GFP relative to nuclear localized NLS-Cherry control. Controls were normalized to ease comparison. The graphs on the right display ratiometric analysis of nuclear vs cytoplasmic GFP (β-catenin) and NLS-mCherry levels. Nuclear localized β-catenin is reduced in Rapgef5 morphants relative to controls for WT and stabilized β-catenin but not NLS-β-catenin. Localization of NLS-mCherry is unaltered. All data represented as mean ± SE. A double asterisk indicates P<0.01, a triple asterisk p<0.005.
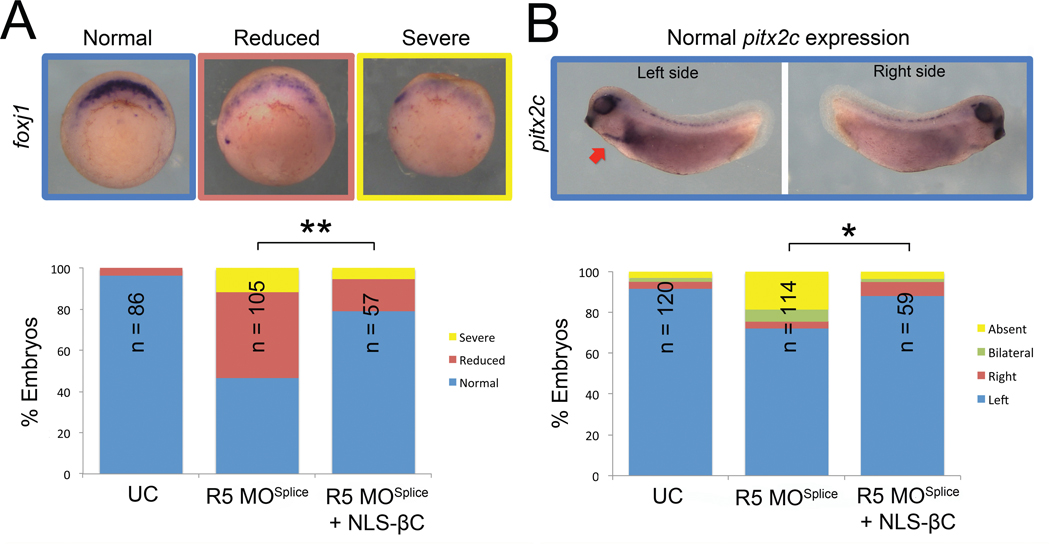
We noted in these experiments that our NLS-mCherry faithfully localizes to the nucleus even when β-catenin nuclear localization is impaired due to Rapgef5 depletion. We wondered if adding the NLS signal from SV40 to β-catenin might “rescue” the nuclear localization phenotype in embryos depleted of Rapgef5. In both the secondary axis assay (Fig. 3A) and the visualization of GFP signal (Fig. 4C), addition of a classic NLS signal to β-catenin makes it insensitive to Rapgef5 depletion. Furthermore, in rapgef5 morphants, co-injection of NLS-β-catenin rescued expression of the Wnt target gene foxj1 and LR patterning (pitx2c) (Fig. 5A,B). Taken together, depletion of Rapgef5 has little effect on the Ran/importin-β1 nuclear transport pathway, which can rescue β-catenin nuclear translocation and LR development upon the addition of a classic NLS-signal. Therefore, Rapgef5 may define a parallel nuclear transport system that is critical for β-catenin.
Figure 5. NLS β-catenin rescues foxj1 transcription and LR patterning defects in Rapgef5 morphants.
Depletion of Rapgef5 reduces the expression of the Wnt responsive gene foxj1 and alters LR patterning (pitx2c). Co-injection of 50pg of NLS-β-catenin mRNA significantly rescues (A) expression of foxj1 at stage 10 and (B) pitx2c expression in Rapgef5 depleted embryos. A single asterisk indicates P<0.05, a double asterisk P<0.01.
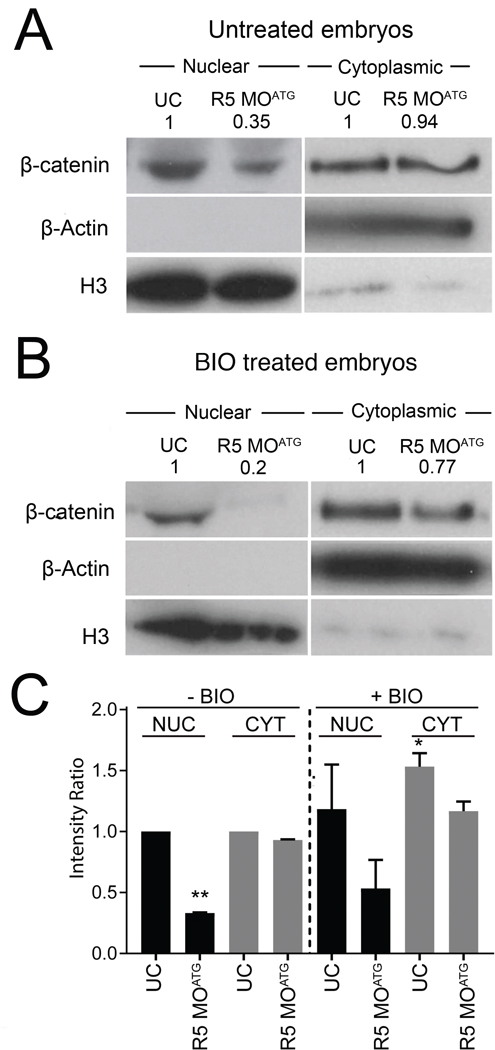
We next sought to evaluate the endogenous levels of β-catenin in different cellular compartments. We isolated the nuclear and cytoplasmic fractions and examined the levels of β-catenin by Western blot (Fig. 6). In wild type Xenopus embryos at stage 10, β-catenin is present in both nuclear and the cytoplasmic/plasma membrane fractions (where it plays a critical role in cell adhesion (Brembeck et al., 2006) (Fig. 6A). In rapgef5 morphants, levels of β-catenin were significantly reduced in the nucleus despite a relatively minor reduction in the cytoplasm (Fig. 6A,C). If we treat the embryos with the GSK-3 inhibitor, BIO, and normalize the intensities of β-catenin to control embryos without BIO treatment, inhibition of the degradation machinery increases the amount of β-catenin in both the cytoplasmic and nuclear fractions (Fig. 6B,C). However, when we deplete Rapgef5, the amount of β-catenin in the nucleus is substantially reduced despite there being more cytoplasmic β-catenin than in wildtype embryos (Fig. 6C). From these studies, we conclude that rapgef5 depletion inhibits the nuclear localization of β-catenin even if the cytoplasmic levels of endogenous β-catenin are high due to inhibition of the cytoplasmic destruction machinery.
Figure 6. Depletion of Rapgef5 impairs nuclear translocation of endogenous β-catenin.
(A) Nuclear/cytoplasmic fractionation reveals that levels of endogenous β-catenin are reduced in the nuclei of Rapgef5 morphants, while cytoplasmic levels are essentially normal. (B) BIO treatment does not rescue the nuclear localization of endogenous β-catenin. (C) Quantification of average β-catenin levels in nuclear and cytoplasmic fractions of control and morphant embryos with or without BIO treatment (error bars represent S.E.M.). All nuclear treatments (black bars) are relativized to the BIO untreated nuclear control and all cytoplasmic treatments (grey bars) are relativized to the BIO untreated cytoplasmic control. Note the reduction of nuclear localized β-catenin in Rapgef5 depleted embryos with or without BIO treatment. The BIO treated rapgef5 morphants have less nuclear localized β-catenin than BIO untreated UC controls, despite having higher levels of cytoplasmic β-catenin. The cellular compartment markers (H3 for nuclear fraction and β-actin for cytoplasmic fraction) displayed in A and B are overexposed in order to demonstrate the degree of purification. For quantitation of β-catenin (C), shorter exposures of the gel (unsaturated) were used in which these markers serve to normalize the amount of protein loaded for each compartment. A single asterisk indicates P<0.05, a double asterisk P<0.01.
A Rapgef5 centered nuclear translocation system
If rapgef5 regulates nuclear translocation of β-catenin, then its expression and activity must be consistent with a role at this location. We first examined its developmental expression pattern and subcellular localization. Using anti-sense mRNA in situ hybridization, we find that both Rapgef5 and paralogs of its target GTPases, Rap1 and Rap2 (Quilliam et al., 2002; Rebhun et al., 2000), are expressed in multiple tissues throughout development, including the blastopore lip and LRO (Fig. S5). By immunohistochemistry, Rapgef5 protein is localized specifically to the nuclei of both whole mount and paraffin sectioned wild type Xenopus embryos at stages 10 and 28 (Fig. S6A) and was reduced in rapgef5 morphants (Fig. S6A panel 2,3). Rapgef5 is also localized specifically to the nuclei of human RPE cells and mouse embryonic fibroblasts suggesting an evolutionary conserved nuclear function (Fig. S6B,C). Consistent with previously documented roles in cell adhesion and migration (Caron, 2003), Rap proteins localize near the plasma membrane as well as in the cytoplasm based on immunofluorescence or expression of fluorescently tagged proteins (Fig. S6D). Intriguingly, Rap1B, Rap2A and Rap2B also localize to the nucleus, while Rap1A accumulated in a reticular pattern that encompasses the perinuclear region suggesting a distribution throughout the nuclear envelope and ER (Fig. S6D). These results suggest an evolutionary conserved nuclear RAP system.
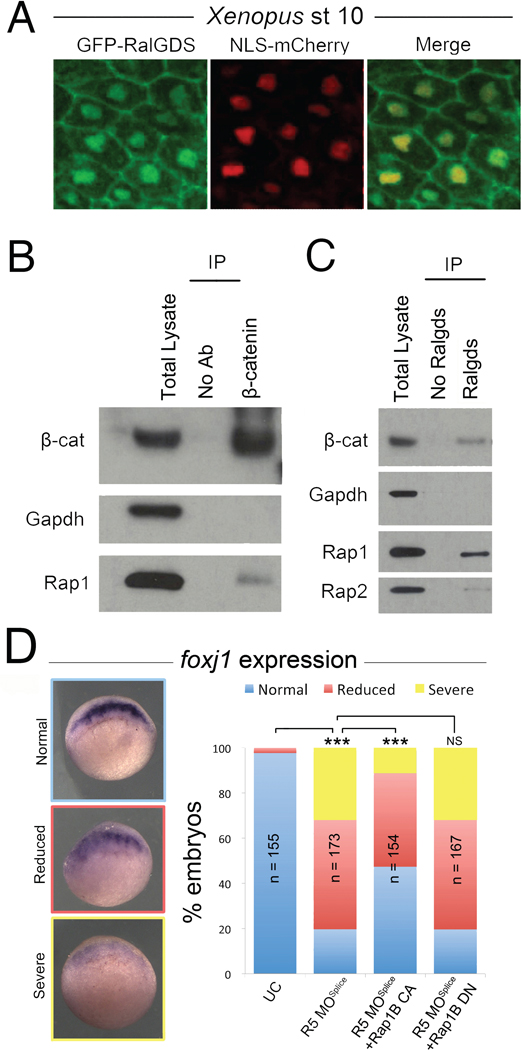
Based on their nuclear localization and previously reported biochemical activity (de Rooij et al., 2000; Quilliam et al., 2002; Rebhun et al., 2000), we propose a working model whereby Rapgef5 maintains nuclear Rap proteins in their active GTP bound state, which is crucial for the nuclear translocation of β-catenin (Fig. S7D). Indeed, previous reports have suggested that Rap1 localizes to the nucleus, influences nuclear β-catenin levels, and alters gene transcription profiles (Bivona and Philips, 2005; Goto et al., 2010; Lafuente et al., 2007). As a preliminary test of our model (Fig. S7D), we asked if nuclear Raps are in their active GTP bound conformation. GFP-RBDRalGDS is an active Rap sensor, consisting of the Ras binding domain of RalGDS fused to GFP, and binds specifically to the active form of Rap1-GTP and Rap2-GTP but not the inactive GDP form to allow visualization of subcellular localization (Bivona and Philips, 2005; Liu et al., 2010). In addition to the previously described signals at the plasma membrane, we confirmed high levels of active Raps within Xenopus nuclei (Fig. 7A). Our model would also predict that active Raps and β-catenin physically interact. In support of this, we found that Rap1 co-immunoprecipitated with β-catenin, and importantly that β-catenin is pulled down specifically with RBDRalGDS, indicating that active Rap1/2 binds β-catenin in stage 10 Xenopus embryos (Fig. 7B,C), results consistent with a previous report in human cancer cell lines (Goto et al., 2010). We confirmed the specificity of RBDRalGDS for active GTP bound Raps by its selective ability to pull down a Rap1b mutant (G12V), that mimics the GTP bound form (CA – constitutively active) but not a Rap1b mutant (S17N) which mimics the GDP bound form (DN – dominant negative) (Fig. S7A,B).
Figure 7. Active Raps and β-catenin interact.
(A) The GFP-RBDRalGDS active Rap sensor reveals the presence of active Raps in the nucleus and plasma membrane of dorsal blastopore lip cells at stage 10. (B) Rap1 protein co-immunoprecipitates with β-catenin. (C) Immunoprecipitation of active Rap proteins from stage 10 Xenopus embryos using the RalGDS sensor. β-catenin immunoprecipitates with active Raps demonstrating an interaction, BC; β-catenin. (D) Injection of CA (constitutively active) Rap1b mRNA can partially rescue loss of foxj1 expression in rapgef5 morphants, while DN (dominant negative) Rap1b mRNA does not. A triple asterisk indicates P<0.0001. Also see Figure S7.
There are five Rap paralogs (Rap1a, Rap1b, Rap2a, Rap2b, Rap2c), and many of these proteins are localized to the nucleus (Fig. S6D). In order to identify a subset of Raps required in canonical Wnt signaling, we overexpressed constitutively active (CA, G12V) and dominant negative (DN, S17N) mutant forms of many of these Raps and examined β-catenin protein levels. In this context, total β-catenin levels were significantly reduced following over-expression of DN Rap1a or Rap1b mutants and increased when we expressed the CA forms (Fig. S7C). No significant difference was observed between Rap2a CA and DN injected embryos (Fig. S7C). RBDRalGDS readily bound the CA but not the DN form of Rap1b, confirming that the Rap mutants function as predicted (Fig. S7A). To determine whether Rap1 proteins mediate Rapgef5’s function in canonical Wnt signaling, we next over-expressed Rap1b DN and CA mRNAs in embryos depleted of Rapgef5 and assayed expression of the Wnt responsive gene, foxj1. Importantly, the CA form of Rap1b partially rescued the foxj1 phenotype of Rapgef5 morphants, while the DN form had no affect (Fig. 7D). Taken together, we conclude 1) Rapgef5 and multiple Rap proteins localize to the nucleus, 2) at least a subset of the Raps in the nucleus are in the active GTP form, 3) active Rap proteins physically interact with β-catenin, 4) The GTP bound state of Rap1a and Rap1b differentially regulates both β-catenin levels and transcription of Wnt target genes downstream of Rapgef5.
DISCUSSION
Integrating the sum of our data, we develop a model where RAPGEF5 activates nuclear Raps, which are necessary for the translocation of β-catenin to the nucleus. This model parallels the classic Ran based transport system, which maintains a nuclear pool of Ran-GTP through the activity of a chromatin-bound Ran-GEF, RCC1 (Kutay et al., 1997; Lange et al., 2007; Lowe et al., 2015; Xu and Massague, 2004). Structurally, β-catenin’s 12 Arm repeats are similar to the HEAT repeats found on importin-β family members suggesting that it might directly interact with the FG-nups of the NPC central channel (Fagotto, 2013; Xu and Massague, 2004). Consistent with this idea, importin-β1 can inhibit the translocation of-β-catenin into the nucleus suggesting competition for NPC binding sites (Fagotto, 2013; Fagotto et al., 1998; Kutay et al., 1997; Xu and Massague, 2004). Finally, like the Ran based transport system, β-catenin’s nuclear translocation is energy dependent and depletion of ATP/GTP or addition of nonhydrolyzable GTP analogs (Fagotto et al., 1998) inhibits nuclear import, suggesting the involvement of a GTPase.
However, β-catenin nuclear import is independent of the Ran based transport system, and the identity of the GTPase was unknown. We propose that the requirement of RAPGEF5 for β-catenin nuclear translocation implicates a Rap GTPase in this role (Fig. S7D). Supporting this idea is our observation that addition of a classic NLS signal to β-catenin decouples β-catenin import from Rapgef5 and allows it to use the importin/Ran system for nuclear translocation, rescuing both Wnt target gene expression (foxj1) and LR development (pitx2c) in rapgef5 morphants. Furthermore, active Raps are co-localized with Rapgef5 in the nucleus and are able to bind β-catenin. Finally, overexpression of CA Rap1b can at least partially rescue the effect of Rapgef5 depletion on Wnt target gene expression while DN Rap1b cannot. Our original observation was that β-catenin levels are reduced in total cell lysates from Rapgef5 depleted embryos, suggesting a role in β-catenin stabilization. However, integrating the sum of our data, we conclude that Rapgef5 depletion partitions β-catenin out of the nucleus and into the cytoplasm where the degradation machinery is active. Of note, maternal stores of Rapgef5 protein prevent us from affecting β-catenin until the onset of gastrulation, so in contrast to the case when β-catenin is directly depleted (Heasman et al., 1994; Heasman et al., 2000; Khokha et al., 2005), expression of many Wnt responsive genes such as gsc and chordin are only minimally affected. Nevertheless, the Wnt responsive genes xnr3 and foxj1 are particularly sensitive and provided a critical clue to an affected pathway, although additional studies are necessary to evaluate other pathways as well.
Elucidating the precise mechanism of β-catenin nuclear transport requires further study, and peculiar differences with the Ran based transport system, or roles for additional rap paralogs, may be discovered. For example, while Rapgef5 is the only Rapgef currently known to localize specifically to the nucleus, there are multiple Rapgefs and Rapgaps and further studies should examine whether these establish a differential in Rap GTPase activity similar to that observed in the Ran system. Importantly, while Rap2 paralogs also localize to the nucleus, overexpression of CA and DN Rap2a did not elicit an effect on β-catenin levels similar to that caused by injection of CA and DN Rap1. Rap2a has previously been implicated in the stabilization of β-catenin at the level of the plasma membrane, and a stabilized β-catenin mutant localized to the nucleus efficiently following Rap2 depletion, suggesting that Rap2 paralogs may not play a critical role in the nuclear transport of β-catenin (Choi and Han, 2005; Park et al., 2013). Importantly, how active Rap1 affects nuclear transport of β-catenin is uncertain. One possibility is that Rap1-GTP differentially binds β-catenin compared to the Rap1-GDP form, or alternatively they bind equally, but Rap1-GTP is localized differentially in the nucleus. In addition, whether the binding between active Raps and β-catenin is direct or indirect warrants further investigation.
Currently, whether Rapgef5 activity contributes to the import of β-catenin through NPCs or functions in a pathway that mediates β-catenin retention in the nucleus remains uncertain (Fig. 7D). Additionally, Ran-GTP acts to release karyopherin/importins from their NLS-bearing cargos; whether a β-catenin cargo is present or not remains to be seen. Importantly, Rapgef5 is localized to the nuclei of all examined cells in the embryo suggesting that its role in cell signaling is equally broad. Indeed, while our investigation has focused exclusively on Rapgef5’s function in canonical Wnt signaling, other pathways may also be affected. Further studies will need to address whether the function of Rapgef5 is restricted to β-catenin or if it also effects the nuclear localization of other Ran independent nuclear localizing proteins, such as APC, p120 catenin, and Smad2.
We began our studies investigating a disease candidate gene identified in a patient with Heterotaxy and congenital heart disease. By using our high-throughput animal model, Xenopus, we identified an LR patterning defect in Rapgef5 depleted embryos, and given our desire to understand the pathophysiology of our patient, we discovered a role of Rapgef5 in the nuclear localization of β-catenin, an essential step in the Wnt signaling pathway. Wnt signaling has a broad impact across embryonic development, stem cells, and multiple diseases. In particular, aberrant Wnt signaling and Rap activity are associated with numerous cancers. Of note, increased levels of nuclear Rapgef5 have recently been linked with hereditary nonpolyposis colorectal cancer, a tumor with a well-established connection to the Wnt pathway (Chen et al., 2013). Thus, further exploration of Rapgef5 and its effectors may provide novel therapeutic targets and be of considerable relevance to human disease.
STAR METHODS
Contact for Reagent and Resource Sharing
Further information and requests for reagents may be directed to, and will be fulfilled by the corresponding author Mustafa K. Khokha (mustafa.khokha@yale.edu)
Experimental Model and Subject Details
Xenopus
X. tropicalis were maintained and cared for in our aquatics facility, in accordance with Yale University Institutional Animal Care and Use Committee protocols. Embryos were produced by in vitro fertilization and raised to appropriate stages in 1/9MR + gentamycin as per standard protocols (del Viso and Khokha, 2012).
Mice
Production of the mouse Catnblox(ex3) allele, carrying a loxP flanked β-catenin exon 3, was as previously described (Harada et al., 1999). Heterozygous Catnblox(ex3) female mice were mated to a pCAGGCre-ER™ male (Hayashi and McMahon, 2002) to produce tamoxifen inducible pCAGG+/cre;Catnb+/lox(ex3) embryos. Murine embryonic fibroblasts (MEFs) were obtained by standard protocols. Briefly, the eviscerated trunk of E12.5 embryos was isolated, finely minced with a clean scalpel, trypsin and DNAse treated, and plated in MEF media. All mice were of mixed genetic background and housed in the New Hunt’s House Biological Services Unit at King’s College London and all work was approved by the Ethical Review Board at King’s College London and performed in accordance with United Kingdom Home Office License 70/7441.
Method Details
Morpholino oligonucleotides, mRNA and CRISPRs
Injections of Xenopus embryos were carried out at the one-cell stage using a fine glass needle and Picospritzer system, as previously described (Khokha et al., 2002). We obtained ATG blocking (R5 MOATG, 5’ –CTTTAAAAGTCTGACACCCATGAGC– 3’), splice blocking (R5 MOSplice, 5’ – GCCTAAGGAAAAACTCTTACCTCCA – 3’), and scrambled control morpholino oligonucleotides from Gene Tools LLC and injected 8ng at the one cell stage to deplete Rapgef5. CRISPR sgRNAs containing the following rapgef5 target sequences (5’ – GGACTTCCTTCTCACATACA - 3’) were designed from the v7.1 model of the Xenopus tropicalis genome. sgRNAs and Cas9 protein was used to genetically knockdown rapgef5 or tyrosinase in F0 embryos. Briefly stated, one cell embryos were injected with 1.5ng Cas9 Protein (PNA-Bio) and 400pg of targeting sgRNA and raised to desired stages as previously described in detail (Bhattacharya et al., 2015). Full length human RAPGEF5 (clone HsCD00511998) was subcloned into the pCSDest2 vector using Gateway recombination techniques. Capped mRNAs were generated in vitro by first linearizing using appropriate restriction enzymes and then transcribing with the mMessage machine kit (Ambion), following the manufacturer’s instructions. β-catenin-GFP (Addgene #16839) (Miller and Moon, 1997), Stabilized β-catenin-GFP (Addgene #29684) (Taelman et al., 2010), NLS β-catenin-GFP (Addgene #16838) (Miller and Moon, 1997), NLS-mCherry (Addgene #49313) (Kirchmaier et al., 2013), TOPFlash (#12456) and FOPFlash (#12457) (Veeman et al., 2003) plasmids were obtained from Addgene. GFP-Rap1A, GFP-Rap1B, mCherry-Rap1B, GFP-XRap2, GFP-Rap2B and GFP-RBDRalGDS plasmids were generous gifts from Prof. Philip Stork at OHSU, Prof. Han at the Pohang University of Science and Technology, South Korea and Prof. Mark Philips at NYU. Constitutively active (G12V) and dominant negative (S17N) mutants for each Rap paralog were produced using the GeneArt strings DNA fragment service of Life Technologies and were subsequently cloned into pcDNA3.
Cardiac Looping
Stage 45 Xenopus embryos were paralyzed with benzocaine and scored with a light microscope. Looping was determined by position of the outflow tract. A D-loop was defined as the outflow tract going to the right, an L-loop was to the left, and an A-loop was midline.
RT-PCR
The R5 MOSplice morpholino was targeted to the splice donor site in intron 2 of rapgef5. Retention of intron 2 was confirmed by RT-PCR, using the following primers: Forward:5’ TCCACTGTACAAGTGAAGGAAGA 3’, reverse: 5’ TGTCTTGTACCTCATCCAGG 3’.
In Situ Hybridization
Digoxigenin-labeled antisense probes for chordin: Tneu011C22, coco: TEgg007d24, fgf8: Tneu007i10, foxj1: Tneu058M03, gdf3: Tgas137g21, gsc: TNeu077f20, noggin: TNeu122a14, not; TEgg044k18, otx2; TGas114a06, pitx2; TNeu083k20, Rap1A: TEgg114f09, Rap2A: TGas143d22, Rap2B; TGas123a15, t; TGas116l23, rapgef5; IMAGE:6494175, vent; TNeu119b08, xnr1 TGas124h10, and xnr3; Tgas011k18 were in vitro transcribed with T7 High Yield RNA Synthesis Kit (E2040S) from New England Biolabs. Embryos were collected at the desired stages, fixed in MEMFA for 1–2 hours at room temperature and dehydrated into 100% ETOH. Briefly summarized, whole mount in situ hybridization of digoxigenin-labeled antisense probes was performed overnight, the labelled embryos were then washed, incubated with anti-digoxigenin-AP Fab fragments (Roche 11093274910), and signal was detected using BM-purple (Roche 11442074001), as previously described in detail (Khokha et al., 2002).
Immunofluorescence
Xenopus embryos and mammalian cells were collected at desired time points and fixed in 4% paraformaldehyde/PBS (Xenopus = overnight (ON) at 4°C, cells = 15 mins at RT). Stage 10 Xenopus embryos were also embedded in paraffin and cut into 10 µm sections. Paraffin was removed using histoclear before beginning the IF. All samples were washed three times in PBS + 0.1% TritonX-100 before incubating in PBS + 0.1% Tween-20 + 3% BSA blocking solution for 1 hr at RT. Samples were then placed in blocking solution + primary antibody ON at 4°C. Samples were washed three times in PBS + 0.1% TritonX-100 before incubating in blocking solution + secondary antibody for 2hrs at RT. Samples were washed three times in PBS + 0.1% Tween-20, one of which included a 5 min incubation in dapi (1:5000), and washed again. Anti-RAPGEF5 (abcam, ab129008, 1:1000 dilution) was used as primary antibody. Alexa 488, 594, 647, and Texas red conjugated anti-mouse and rabbit secondary antibodies were obtained from Thermo Fisher Scientific. Alexa647 phalloidin (Molecular Probes, 1:40) was also used to stain cell membranes in some cases. Mammalian cells and Xenopus sections were mounted in Pro-Long Gold (Invitrogen) before imaging on a Zeiss 710 confocal microscope.
Mammalian cell culture and siRNA knockdown of Rapgef5
Mammalian embryonic fibroblasts (MEFs) were grown in DMEM supplemented with 10% FBS, L-glutamine, Penicillin streptomycin, hepes and β-mercaptoethanol. To knockdown Rapgef5, cells were transfected with non-overlapping Rapgef5 siRNAs (Santa Cruz # sc-152703), or with control siRNA (Santa Cruz sc-36869) using lipofectamine RNAiMAX transfection reagent (Thermo Fisher Scientific #13778100) as per the manufacturer’s instructions. Cells were incubated with siRNA overnight in serum free medium before being returned to MEF media for 36 hours.
TOPFlash Assays
Xenopus embryos were injected with 200pg of GFP-WT-β-catenin or GFP-Stabilized-β-catenin plasmids, along with 100pg of either the TOPFlash or FOPFlash (negative control) plasmids at the one cell stage. 20pg of Renilla plasmid was co-injected as an internal control. A subset of TOPFlash and β-catenin injected embryos were also injected with either 8ng of the Rapgef5 splice morpholino or 200pg of Gsk3 mRNA. Pools of 10 embryos were collected and lysed in 100µl passive lysis buffer.
In the mammalian cell culture experiments, Rapgef5 was depleted in pCAGG+/cre;Catnb+/lox(ex3) MEFs (carrying a tamoxifen inducible β-catenin gain of function allele) by siRNA knockdown. β-catenin was stabilized by the tamoxifen induced deletion of exon 3 after 36 hours and cells were collected for analysis at 48hrs. Luciferase was quantified using the Dual Luciferase Reporter Assay System (Promega #E1910). Briefly, samples were lysed in 100ul of luciferase assay reagent and levels of TOPFlash driven firefly luciferase activity were measured using a Promega Glomax luminometer. 100ul of Stop & Glo reagent was then added to allow quantification of the Renilla luciferase control, as described in detail in the manufacturer’s instructions. Each experiment was carried out in technical and biological triplicate.
Double axis assay and β-catenin localization experiments
For the secondary axis assay, embryos were injected at the one cell stage with 150pg of WT β-catenin-GFP, Stabilized β-catenin-GFP or NLS β-catenin-GFP mRNAs. A subset of each group was then injected with either 8ng of the R5 MOATG morpholino or with full length GSK3 mRNA. Embryos were collected at stage 19 and scored for the presence of a secondary axis. To examine the subcellular localization of β-catenin in Xenopus embryos, 150pg of WT β-catenin-GFP, Stabilized β-catenin-GFP or NLS β-catenin-GFP mRNAs were co-injected with NLS-cherry mRNA at the one cell stage. Samples were collected at stage 10 and fixed in 4% PFA overnight. The dorsal blastopore lip was dissected, mounted on coverslips in Pro-Long Gold (Invitrogen) and imaged with a Zeiss 710 confocal microscope. Image analysis was carried out using ImageJ.
Pharmacological inhibition of Gsk3
To inhibit GSK3α/β function during anterior development in WT Xenopus embryos and morphants, 6-bromoindirubin-3'-oxime (BIO, Sigma-Aldrich B1686) was added to the media at a concentration of 10 µM between stages 8 and 11. Media was then changed and the embryos allowed to develop. Anterior development was scored as normal, reduced or absent as presented in Fig. 2. For the nuclear/cytoplasmic fractionation experiments BIO was added at stage 4 (1 µM concentration) and the embryos were collected at stage 10.
Co-Immunoprecipitation and active Rap pulldown
Pools of stage 10 Xenopus embryos were lysed in NP-40 buffer (150 mM NaCl, 1.0% NP-40, 50 mM Tris, pH 8.0, 10 µl/embryo) on ice. Samples were centrifuged for 10 mins at 13,000 rpm and the supernatant removed to a clean tube. Supernatant was then pre-incubated with protein G beads (Bio-Rad, # 1614023) for 1 hour, centrifuged to collect supernatant, and incubated with primary antibody (Anti-β-catenin H-102, Santa Cruz, sc-7199) overnight. Protein G beads were then added to the sample solution and incubated for 3 hrs at 4oC. Lysate was centrifuged and the supernatant discarded. The beads were washed and centrifuged with 0.1% PBS-Tween six times. 2x SDS loading dye was added to the beads and heated to 95oC for five mins. The sample was centrifuged again and the lysate moved to a clean tube. For the active Rap1 and Rap2 pull down experiments, lysate was incubated with GST-RalGDS-RBD fusion protein overnight at 4o C to allow binding, free protein was washed away, and active Rap1 and Rap2, as well as interacting proteins, were then eluted. The active rap detection kit (Cell signaling Technologies #8818) was used as per the manufacturer’s instructions. Samples were analyzed by Western blot.
Nuclear/cytoplasmic protein fractionation
Pools of 30 embryos were collected at stage 10 for each treatment including controls in eppendorf tubes, (a maximium of 35 and minimum of 20 embryos can be used with the solution volumes outlined in this protocol, but the protein concentrations will vary). All centrifugation steps were done at 4°C. 1 ml of Buffer E1 (see below for details for E1, E2, E3) was added to the embryos, and they were homogenized using a pipette. Lysates were centrifuged at 13,000 rpm at 4°C for 5 min. Approximately 600–650 µl of the supernatant was transferred to a new eppendorf tube (avoiding the lipid fraction) and placed on ice (cytoplasmic fraction). The remaining supernatant was discarded, and any lipids were removed from the tube walls using a Kimwipe, talking care not to disturb the pellet (nuclear fraction).
The nuclear and cytoplasmic fractions were then purified. For the cytoplasmic fraction, 11 µl of buffer CERII from the NE-PER Nuclear and Cytoplasmic Extraction Reagents Kit (Thermo-Scientific #78833) was added and tubes were vortexed for 15 sec, incubated on ice for 5 min and centrifuged for another 5 min. The supernatant (~ 500 µl) was transferred to a clean tube again avoiding lipid contamination. This cytoplasmic fraction was stored at −80°C until use.
The nuclear fraction pellet was resuspended in 1 ml of buffer E1 by pipetting (some material might remain insoluble), incubating on ice for 5 min and centrifuging for 5 min. The supernatant was discarded following centrifugation and any lipids were removed using Kimwipes without touching the pellet. This washing step was then repeated. After the second centrifugation the pellet was resuspended in 1 ml of buffer E2 and centrifuged for 5 min. The supernatant was discarded. This step was repeated once. Subsequently, the nuclear pellet was resuspended in 500 µl of buffer E3 and centrifuged for 5 min. The supernatant was discarded and the pellet was finally resuspended in 150 µl of Buffer E3. This is the nuclear fraction.
In these experiments, the fractions come from the same number of cells - we lyse a defined set of Xenopus embryos, and then fractionate into nuclear and cytoplasmic ratios. Once these fractions are made, we load the same amount of protein per lane and verify by loading controls (actin - cytoplasm, histone - nuclear). Quantifications are normalized to the amounts of respective loading controls. The loading controls (histone and actin) displayed in Figure 6 are overexposed in order to demonstrate the degree of purification in the fractionation. For quantitation of β-catenin we used shorter (unsaturated) exposures. Protein concentrations of the nuclear and cytoplasmic fractions were determined using DC Protein Assay (Biorad # 5000112). Protein fractions were stored at −80°C. All buffers were supplemented with proteinase inhibitor (Complete, Roche # 11697498001) and PMSF (0.2 mM final concentration) prior to use. Buffer E1 was also supplemented with DTT (1 mM final concentration).
Buffer compositions: Buffer E1: 50 mM HEPES-KOH, 140 mM mL NaCl, 1 mM EDTA (pH 8.0), 10% Glycerol (v/v), 0.5% Igepal CA-630 (v/v), 0.25% Triton X-100 (v/v). Buffer E2: 10 mM Tris-HCl (pH 8.0), 200 mM NaCl, 1 mM EDTA (pH 8.0), 0.5 mM EGTA (pH 8.0). Buffer E3: 10 mM Tris-HCl (pH 8.0), 200 mM NaCl, 1 mM EDTA (pH 8.0), 0.5 mM EGTA (pH 8.0), 1% (v/v) Sodium deoxycholate (from a 10% [w/v] solution), 5% (v/v) Sodium N-lauroylsarcosine (from a 10% [w/v] solution).
Western blotting
For total embryo western blots, pools of 10 control and treated embryos were collected at stage 9, 10 or 12 and placed in 100 ul of 1 × RIPA buffer. Embryos were then crushed using a pestle and spun down twice to separate lysate from lipids and debris. Embryos for nuclear and cytoplasmic analysis fractionations were isolated as described above. Western blotting was carried out following standard protocols and 45 µg of protein was loaded in each lane in 4–12% Tris-Bis gels. Anti-β-catenin (Santa Cruz sc-7199, 1:1000 in 5% milk), anti-active-β-catenin (Millipore 05–665), anti-GAPDH (Ambion, AM4300 1:5000 dilution), anti-β-actin (cytoplasmic marker, santa cruz Sc-4778), anti-Histone H3 (nuclear marker, Abcam, ab1791; 1:2000 dilution), anti-RAPGEF5 (abcam, ab129008, 1:200 dilution), anti-Rap1 (Enzo, ADI-KAP-GP125, 1:250) and anti-Rap2 (Abcam #ab166785, 1:500) primary antibodies were used. Anti-mouse or anti-rabbit HRP conjugated secondary antibodies were used (Jackson Immuno Research Laboratories, 715–035–150 or 211–032–171 1:15000 dilution). Bands were quantified using ImageJ and relativized to controls. Injections and protein fractionations were repeated 3 times. The cellular compartment markers (H3 for nuclear fraction and β-actin for cytoplasmic fraction) displayed in Fig. 6A,B are overexposed in order to demonstrate the degree of purification. Note: the nuclear fractions are challenging to resuspend and sometimes migrate or transfer unevenly. The H3 blot presented in Figure 6C was run as a single lane. For quantitation of β-catenin (Fig. 6C), shorter exposures of the gel (unsaturated) were used in which these markers serve to normalize the amount of protein loaded for each compartment. Western blot images show representative results and graphs show the averages of the different replicates.
Quantification and Statistical Analysis
For Xenopus experiments we estimated 20–25 samples per experimental condition were necessary for statistical significance given the magnitude of the changes expected. Typically, many more samples were obtained for each experiment except as noted in the figures. Statistical significance is reported in the figures and legends. In all figures, statistical significance was defined as P<0.05. In Figures 1, 2, 5, S1 and S3 the in situ results were analyzed by chi-squared test or Fishers exact test as appropriate for sample size. In Figure 3 the double axis assay data was examined using fishers exact test, the BIO treatment by Chi-squared and the TOPFlash by t-test. For Figure 4 a t-test (two-tailed, type two) was used to determine significance of the ratiometric analysis, and in Figure 6 the western blot data was analyzed by Anova Sidak.
Supplementary Material
Highlights.
The heterotaxy gene RAPGEF5, affects left-right patterning via Wnt signaling
RAPGEF5 regulates nuclear translocation of β-catenin independently of stabilization
RAPGEF5 may mediate an importin-β1/Ran independent nuclear transport mechanism
Acknowledgments
We are grateful to the patients and families who inspired this work. Thanks to S. Kubek and M. Slocum for animal husbandry and to G. Griffin, W. Barrell, S. Gonzalez Malagon, S. Rao and members of the Khokha, Liu and Gallagher labs for technical assistance and advice. We thank P. Lusk for critical comments on the manuscript. Thanks to the Center for Cellular and Molecular Imaging at Yale for confocal imaging. We also thank Prof. Philip Stork at OHSU, Prof. Han at the Pohang University of Science and Technology, South Korea and Prof. Mark Philips at NYU for providing plasmids and thank Xenbase (http://www.xenbase.org/, RRID:SCR_003280) and Addgene for providing access to many of the bioinformatic resources and reagents used in this work. This work was supported by the NIH (R01HD081379 and R01HL124402 to MKK) and BBSRC (grant BB/1021922/1, BB/E013872/1 to KJL). MKK is a Mallinckrodt Scholar.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
JNG, FdV, ARD, and MKK conceived and designed the embryological experiments in Xenopus. JNG and KJL designed experiments in mouse embryonic fibroblasts. AR screened RAPGEF5 for biological activity in Xenopus. JNG, FdV, ARD, HW, AR and SK performed the Xenopus experiments. JNG performed mouse experiments. JNG and MKK wrote the manuscript, which was read and approved by all authors.
References
- Bhanot P, Brink M, Samos CH, Hsieh JC, Wang Y, Macke JP, Andrew D, Nathans J, Nusse R. A new member of the frizzled family from Drosophila functions as a Wingless receptor. Nature. 1996;382:225–230. doi: 10.1038/382225a0. [DOI] [PubMed] [Google Scholar]
- Bhattacharya D, Marfo CA, Li D, Lane M, Khokha MK. CRISPR/Cas9: An inexpensive, efficient loss of function tool to screen human disease genes in Xenopus. Developmental biology. 2015;408:196–204. doi: 10.1016/j.ydbio.2015.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bivona TG, Philips MR. Analysis of Ras and Rap activation in living cells using fluorescent Ras binding domains. Methods. 2005;37:138–145. doi: 10.1016/j.ymeth.2005.05.022. [DOI] [PubMed] [Google Scholar]
- Blum M, Beyer T, Weber T, Vick P, Andre P, Bitzer E, Schweickert A. Xenopus, an ideal model system to study vertebrate left-right asymmetry. Dev Dyn. 2009;238:1215–1225. doi: 10.1002/dvdy.21855. [DOI] [PubMed] [Google Scholar]
- Boskovski MT, Yuan S, Pedersen NB, Goth CK, Makova S, Clausen H, Brueckner M, Khokha MK. The heterotaxy gene GALNT11 glycosylates Notch to orchestrate cilia type and laterality. Nature. 2013;504:456–459. doi: 10.1038/nature12723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brembeck FH, Rosario M, Birchmeier W. Balancing cell adhesion and Wnt signaling, the key role of beta-catenin. Curr Opin Genet Dev. 2006;16:51–59. doi: 10.1016/j.gde.2005.12.007. [DOI] [PubMed] [Google Scholar]
- Brueckner M. Heterotaxia, congenital heart disease, and primary ciliary dyskinesia. Circulation. 2007;115:2793–2795. doi: 10.1161/CIRCULATIONAHA.107.699256. [DOI] [PubMed] [Google Scholar]
- Caron A, Xu X, Lin X. Wnt/beta-catenin signaling directly regulates Foxj1 expression and ciliogenesis in zebrafish Kupffer’s vesicle. Development. 2012;139:514–524. doi: 10.1242/dev.071746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caron E. Cellular functions of the Rap1 GTP-binding protein: a pattern emerges. Journal of cell science. 2003;116:435–440. doi: 10.1242/jcs.00238. [DOI] [PubMed] [Google Scholar]
- Chen W, Yuan L, Cai Y, Chen X, Chi Y, Wei P, Zhou X, Shi D. Identification of chromosomal copy number variations and novel candidate loci in hereditary nonpolyposis colorectal cancer with mismatch repair proficiency. Genomics. 2013;102:27–34. doi: 10.1016/j.ygeno.2013.02.003. [DOI] [PubMed] [Google Scholar]
- Choi SC, Han JK. Rap2 is required for Wnt/beta-catenin signaling pathway in Xenopus early development. The EMBO journal. 2005;24:985–996. doi: 10.1038/sj.emboj.7600571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clevers H, Nusse R. Wnt/beta-catenin signaling and disease. Cell. 2012;149:1192–1205. doi: 10.1016/j.cell.2012.05.012. [DOI] [PubMed] [Google Scholar]
- Davidson G, Wu W, Shen J, Bilic J, Fenger U, Stannek P, Glinka A, Niehrs C. Casein kinase 1 gamma couples Wnt receptor activation to cytoplasmic signal transduction. Nature. 2005;438:867–872. doi: 10.1038/nature04170. [DOI] [PubMed] [Google Scholar]
- De Robertis EM, Larrain J, Oelgeschlager M, Wessely O. The establishment of Spemann’s organizer and patterning of the vertebrate embryo. Nature reviews Genetics. 2000;1:171–181. doi: 10.1038/35042039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Rooij J, Rehmann H, van Triest M, Cool RH, Wittinghofer A, Bos JL. Mechanism of regulation of the Epac family of cAMP-dependent RapGEFs. The Journal of biological chemistry. 2000;275:20829–20836. doi: 10.1074/jbc.M001113200. [DOI] [PubMed] [Google Scholar]
- del Viso F, Khokha M. Generating diploid embryos from Xenopus tropicalis. Methods Mol Biol. 2012;917:33–41. doi: 10.1007/978-1-61779-992-1_3. [DOI] [PubMed] [Google Scholar]
- Fagotto F. Looking beyond the Wnt pathway for the deep nature of beta-catenin. EMBO Rep. 2013;14:422–433. doi: 10.1038/embor.2013.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagotto F, Gluck U, Gumbiner BM. Nuclear localization signal-independent and importin/karyopherin-independent nuclear import of beta-catenin. Current biology : CB. 1998;8:181–190. doi: 10.1016/s0960-9822(98)70082-x. [DOI] [PubMed] [Google Scholar]
- Fagotto F, Guger K, Gumbiner BM. Induction of the primary dorsalizing center in Xenopus by the Wnt/GSK/beta-catenin signaling pathway, but not by Vg1, Activin or Noggin. Development. 1997;124:453–460. doi: 10.1242/dev.124.2.453. [DOI] [PubMed] [Google Scholar]
- Fakhro KA, Choi M, Ware SM, Belmont JW, Towbin JA, Lifton RP, Khokha MK, Brueckner M. Rare copy number variations in congenital heart disease patients identify unique genes in left-right patterning. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:2915–2920. doi: 10.1073/pnas.1019645108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funayama N, Fagotto F, McCrea P, Gumbiner BM. Embryonic axis induction by the armadillo repeat domain of beta-catenin: evidence for intracellular signaling. J Cell Biol. 1995;128:959–968. doi: 10.1083/jcb.128.5.959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glinka A, Wu W, Delius H, Monaghan AP, Blumenstock C, Niehrs C. Dickkopf-1 is a member of a new family of secreted proteins and functions in head induction. Nature. 1998;391:357–362. doi: 10.1038/34848. [DOI] [PubMed] [Google Scholar]
- Goto M, Mitra RS, Liu M, Lee J, Henson BS, Carey T, Bradford C, Prince M, Wang CY, Fearon ER, et al. Rap1 stabilizes beta-catenin and enhances beta-catenin-dependent transcription and invasion in squamous cell carcinoma of the head and neck. Clinical cancer research : an official journal of the American Association for Cancer Research. 2010;16:65–76. doi: 10.1158/1078-0432.CCR-09-1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guger KA, Gumbiner BM. beta-Catenin has Wnt-like activity and mimics the Nieuwkoop signaling center in Xenopus dorsal-ventral patterning. Developmental biology. 1995;172:115–125. doi: 10.1006/dbio.1995.0009. [DOI] [PubMed] [Google Scholar]
- Hanafusa H, Masuyama N, Kusakabe M, Shibuya H, Nishida E. The TGF-beta family member derriere is involved in regulation of the establishment of left-right asymmetry. EMBO Rep. 2000;1:32–39. doi: 10.1093/embo-reports/kvd008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada N, Tamai Y, Ishikawa T, Sauer B, Takaku K, Oshima M, Taketo MM. Intestinal polyposis in mice with a dominant stable mutation of the beta-catenin gene. The EMBO journal. 1999;18:5931–5942. doi: 10.1093/emboj/18.21.5931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harland R, Gerhart J. Formation and function of Spemann’s organizer. Annu Rev Cell Dev Biol. 1997;13:611–667. doi: 10.1146/annurev.cellbio.13.1.611. [DOI] [PubMed] [Google Scholar]
- Hayashi S, McMahon AP. Efficient recombination in diverse tissues by a tamoxifen-inducible form of Cre: a tool for temporally regulated gene activation/inactivation in the mouse. Developmental biology. 2002;244:305–318. doi: 10.1006/dbio.2002.0597. [DOI] [PubMed] [Google Scholar]
- He X, Saint-Jeannet JP, Wang Y, Nathans J, Dawid I, Varmus H. A member of the Frizzled protein family mediating axis induction by Wnt-5A. Science. 1997;275:1652–1654. doi: 10.1126/science.275.5306.1652. [DOI] [PubMed] [Google Scholar]
- Heasman J, Crawford A, Goldstone K, Garner-Hamrick P, Gumbiner B, McCrea P, Kintner C, Noro CY, Wylie C. Overexpression of cadherins and underexpression of beta-catenin inhibit dorsal mesoderm induction in early Xenopus embryos. Cell. 1994;79:791–803. doi: 10.1016/0092-8674(94)90069-8. [DOI] [PubMed] [Google Scholar]
- Heasman J, Kofron M, Wylie C. Beta-catenin signaling activity dissected in the early Xenopus embryo: a novel antisense approach. Developmental biology. 2000;222:124–134. doi: 10.1006/dbio.2000.9720. [DOI] [PubMed] [Google Scholar]
- Jamieson C, Sharma M, Henderson BR. Targeting the beta-catenin nuclear transport pathway in cancer. Semin Cancer Biol. 2014;27:20–29. doi: 10.1016/j.semcancer.2014.04.012. [DOI] [PubMed] [Google Scholar]
- Kawasumi A, Nakamura T, Iwai N, Yashiro K, Saijoh Y, Belo JA, Shiratori H, Hamada H. Left-right asymmetry in the level of active Nodal protein produced in the node is translated into left-right asymmetry in the lateral plate of mouse embryos. Developmental biology. 2011;353:321–330. doi: 10.1016/j.ydbio.2011.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy MP, Omran H, Leigh MW, Dell S, Morgan L, Molina PL, Robinson BV, Minnix SL, Olbrich H, Severin T, et al. Congenital heart disease and other heterotaxic defects in a large cohort of patients with primary ciliary dyskinesia. Circulation. 2007;115:2814–2821. doi: 10.1161/CIRCULATIONAHA.106.649038. [DOI] [PubMed] [Google Scholar]
- Khokha MK, Chung C, Bustamante EL, Gaw LW, Trott KA, Yeh J, Lim N, Lin JC, Taverner N, Amaya E, et al. Techniques and probes for the study of Xenopus tropicalis development. Dev Dyn. 2002;225:499–510. doi: 10.1002/dvdy.10184. [DOI] [PubMed] [Google Scholar]
- Khokha MK, Yeh J, Grammer TC, Harland RM. Depletion of three BMP antagonists from Spemann’s organizer leads to a catastrophic loss of dorsal structures. Dev Cell. 2005;8:401–411. doi: 10.1016/j.devcel.2005.01.013. [DOI] [PubMed] [Google Scholar]
- Kirchmaier S, Lust K, Wittbrodt J. Golden GATEway cloning--a combinatorial approach to generate fusion and recombination constructs. PLoS One. 2013;8:e76117. doi: 10.1371/journal.pone.0076117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kofron M, Birsoy B, Houston D, Tao Q, Wylie C, Heasman J. Wnt11/beta-catenin signaling in both oocytes and early embryos acts through LRP6-mediated regulation of axin. Development. 2007;134:503–513. doi: 10.1242/dev.02739. [DOI] [PubMed] [Google Scholar]
- Kutay U, Izaurralde E, Bischoff FR, Mattaj IW, Gorlich D. Dominant-negative mutants of importin-beta block multiple pathways of import and export through the nuclear pore complex. The EMBO journal. 1997;16:1153–1163. doi: 10.1093/emboj/16.6.1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafuente EM, Iwamoto Y, Carman CV, van Puijenbroek AA, Constantine E, Li L, Boussiotis VA. Active Rap1, a small GTPase that induces malignant transformation of hematopoietic progenitors, localizes in the nucleus and regulates protein expression. Leukemia & lymphoma. 2007;48:987–1002. doi: 10.1080/10428190701242341. [DOI] [PubMed] [Google Scholar]
- Lange A, Mills RE, Lange CJ, Stewart M, Devine SE, Corbett AH. Classical nuclear localization signals: definition, function, and interaction with importin alpha. The Journal of biological chemistry. 2007;282:5101–5105. doi: 10.1074/jbc.R600026200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Li Y, Semenov M, Han C, Baeg GH, Tan Y, Zhang Z, Lin X, He X. Control of beta-catenin phosphorylation/degradation by a dual-kinase mechanism. Cell. 2002;108:837–847. doi: 10.1016/s0092-8674(02)00685-2. [DOI] [PubMed] [Google Scholar]
- Liu C, Takahashi M, Li Y, Dillon TJ, Kaech S, Stork PJ. The interaction of Epac1 and Ran promotes Rap1 activation at the nuclear envelope. Molecular and cellular biology. 2010;30:3956–3969. doi: 10.1128/MCB.00242-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logan M, Pagan-Westphal SM, Smith DM, Paganessi L, Tabin CJ. The transcription factor Pitx2 mediates situs-specific morphogenesis in response to left-right asymmetric signals. Cell. 1998;94:307–317. doi: 10.1016/s0092-8674(00)81474-9. [DOI] [PubMed] [Google Scholar]
- Lowe AR, Tang JH, Yassif J, Graf M, Huang WY, Groves JT, Weis K, Liphardt JT. Importin-beta modulates the permeability of the nuclear pore complex in a Ran-dependent manner. Elife. 2015:4. doi: 10.7554/eLife.04052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lustig KD, Kroll K, Sun E, Ramos R, Elmendorf H, Kirschner MW. A Xenopus nodal-related gene that acts in synergy with noggin to induce complete secondary axis and notochord formation. Development. 1996;122:3275–3282. doi: 10.1242/dev.122.10.3275. [DOI] [PubMed] [Google Scholar]
- MacDonald BT, Tamai K, He X. Wnt/beta-catenin signaling: components, mechanisms, and diseases. Developmental cell. 2009;17:9–26. doi: 10.1016/j.devcel.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao B, Wu W, Li Y, Hoppe D, Stannek P, Glinka A, Niehrs C. LDL-receptor-related protein 6 is a receptor for Dickkopf proteins. Nature. 2001a;411:321–325. doi: 10.1038/35077108. [DOI] [PubMed] [Google Scholar]
- Mao J, Wang J, Liu B, Pan W, Farr GH, 3rd, Flynn C, Yuan H, Takada S, Kimelman D, Li L, et al. Low-density lipoprotein receptor-related protein-5 binds to Axin and regulates the canonical Wnt signaling pathway. Mol Cell. 2001b;7:801–809. doi: 10.1016/s1097-2765(01)00224-6. [DOI] [PubMed] [Google Scholar]
- McCrea PD, Brieher WM, Gumbiner BM. Induction of a secondary body axis in Xenopus by antibodies to beta-catenin. J Cell Biol. 1993;123:477–484. doi: 10.1083/jcb.123.2.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrath J, Somlo S, Makova S, Tian X, Brueckner M. Two populations of node monocilia initiate left-right asymmetry in the mouse. Cell. 2003;114:61–73. doi: 10.1016/s0092-8674(03)00511-7. [DOI] [PubMed] [Google Scholar]
- McKendry R, Hsu SC, Harland RM, Grosschedl R. LEF-1/TCF proteins mediate wnt-inducible transcription from the Xenopus nodal-related 3 promoter. Developmental biology. 1997;192:420–431. doi: 10.1006/dbio.1997.8797. [DOI] [PubMed] [Google Scholar]
- Miller JR, Moon RT. Analysis of the signaling activities of localization mutants of beta-catenin during axis specification in Xenopus. J Cell Biol. 1997;139:229–243. doi: 10.1083/jcb.139.1.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molenaar M, van de Wetering M, Oosterwegel M, Peterson-Maduro J, Godsave S, Korinek V, Roose J, Destree O, Clevers H. XTcf-3 transcription factor mediates beta-catenin-induced axis formation in Xenopus embryos. Cell. 1996;86:391–399. doi: 10.1016/s0092-8674(00)80112-9. [DOI] [PubMed] [Google Scholar]
- Moon RT, Kohn AD, De Ferrari GV, Kaykas A. WNT and beta-catenin signalling: diseases and therapies. Nature reviews Genetics. 2004;5:691–701. doi: 10.1038/nrg1427. [DOI] [PubMed] [Google Scholar]
- Niehrs C. Regionally specific induction by the Spemann-Mangold organizer. Nature reviews Genetics. 2004;5:425–434. doi: 10.1038/nrg1347. [DOI] [PubMed] [Google Scholar]
- Niehrs C. The complex world of WNT receptor signalling. Nat Rev Mol Cell Biol. 2012;13:767–779. doi: 10.1038/nrm3470. [DOI] [PubMed] [Google Scholar]
- Park DS, Seo JH, Hong M, Choi SC. Role of the Rap2/TNIK kinase pathway in regulation of LRP6 stability for Wnt signaling. Biochemical and biophysical research communications. 2013;436:338–343. doi: 10.1016/j.bbrc.2013.05.104. [DOI] [PubMed] [Google Scholar]
- Pinson KI, Brennan J, Monkley S, Avery BJ, Skarnes WC. An LDL-receptor-related protein mediates Wnt signalling in mice. Nature. 2000;407:535–538. doi: 10.1038/35035124. [DOI] [PubMed] [Google Scholar]
- Quilliam LA, Rebhun JF, Castro AF. A growing family of guanine nucleotide exchange factors is responsible for activation of Ras-family GTPases. Progress in nucleic acid research and molecular biology. 2002;71:391–444. doi: 10.1016/s0079-6603(02)71047-7. [DOI] [PubMed] [Google Scholar]
- Rebhun JF, Castro AF, Quilliam LA. Identification of guanine nucleotide exchange factors (GEFs) for the Rap1 GTPase. Regulation of MR-GEF by M-Ras-GTP interaction. The Journal of biological chemistry. 2000;275:34901–34908. doi: 10.1074/jbc.M005327200. [DOI] [PubMed] [Google Scholar]
- Sampath K, Cheng AM, Frisch A, Wright CV. Functional differences among Xenopus nodal-related genes in left-right axis determination. Development. 1997;124:3293–3302. doi: 10.1242/dev.124.17.3293. [DOI] [PubMed] [Google Scholar]
- Sato N, Meijer L, Skaltsounis L, Greengard P, Brivanlou AH. Maintenance of pluripotency in human and mouse embryonic stem cells through activation of Wnt signaling by a pharmacological GSK-3-specific inhibitor. Nat Med. 2004;10:55–63. doi: 10.1038/nm979. [DOI] [PubMed] [Google Scholar]
- Schweickert A, Vick P, Getwan M, Weber T, Schneider I, Eberhardt M, Beyer T, Pachur A, Blum M. The nodal inhibitor Coco is a critical target of leftward flow in Xenopus. Current biology : CB. 2010;20:738–743. doi: 10.1016/j.cub.2010.02.061. [DOI] [PubMed] [Google Scholar]
- Smith WC, McKendry R, Ribisi S, Jr, Harland RM. A nodal-related gene defines a physical and functional domain within the Spemann organizer. Cell. 1995;82:37–46. doi: 10.1016/0092-8674(95)90050-0. [DOI] [PubMed] [Google Scholar]
- Stubbs JL, Oishi I, Izpisua Belmonte JC, Kintner C. The forkhead protein Foxj1 specifies node-like cilia in Xenopus and zebrafish embryos. Nature genetics. 2008;40:1454–1460. doi: 10.1038/ng.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutherland MJ, Ware SM. Disorders of left-right asymmetry: heterotaxy and situs inversus. American journal of medical genetics Part C, Seminars in medical genetics. 2009;151C:307–317. doi: 10.1002/ajmg.c.30228. [DOI] [PubMed] [Google Scholar]
- Tabin CJ, Vogan KJ. A two-cilia model for vertebrate left-right axis specification. Genes Dev. 2003;17:1–6. doi: 10.1101/gad.1053803. [DOI] [PubMed] [Google Scholar]
- Taelman VF, Dobrowolski R, Plouhinec JL, Fuentealba LC, Vorwald PP, Gumper I, Sabatini DD, De Robertis EM. Wnt signaling requires sequestration of glycogen synthase kinase 3 inside multivesicular endosomes. Cell. 2010;143:1136–1148. doi: 10.1016/j.cell.2010.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamai K, Semenov M, Kato Y, Spokony R, Liu C, Katsuyama Y, Hess F, Saint-Jeannet JP, He X. LDL-receptor-related proteins in Wnt signal transduction. Nature. 2000;407:530–535. doi: 10.1038/35035117. [DOI] [PubMed] [Google Scholar]
- van de Wetering M, Cavallo R, Dooijes D, van Beest M, van Es J, Loureiro J, Ypma A, Hursh D, Jones T, Bejsovec A, et al. Armadillo coactivates transcription driven by the product of the Drosophila segment polarity gene dTCF. Cell. 1997;88:789–799. doi: 10.1016/s0092-8674(00)81925-x. [DOI] [PubMed] [Google Scholar]
- van de Wetering M, Oosterwegel M, Dooijes D, Clevers H. Identification and cloning of TCF-1, a T lymphocyte-specific transcription factor containing a sequence-specific HMG box. The EMBO journal. 1991;10:123–132. doi: 10.1002/j.1460-2075.1991.tb07928.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veeman MT, Slusarski DC, Kaykas A, Louie SH, Moon RT. Zebrafish prickle, a modulator of noncanonical Wnt/Fz signaling, regulates gastrulation movements. Current biology : CB. 2003;13:680–685. doi: 10.1016/s0960-9822(03)00240-9. [DOI] [PubMed] [Google Scholar]
- Vonica A, Brivanlou AH. The left-right axis is regulated by the interplay of Coco, Xnr1 and derriere in Xenopus embryos. Developmental biology. 2007;303:281–294. doi: 10.1016/j.ydbio.2006.09.039. [DOI] [PubMed] [Google Scholar]
- Walentek P, Beyer T, Thumberger T, Schweickert A, Blum M. ATP4a is required for Wnt-dependent Foxj1 expression and leftward flow in Xenopus left-right development. Cell Rep. 2012;1:516–527. doi: 10.1016/j.celrep.2012.03.005. [DOI] [PubMed] [Google Scholar]
- Wiechens N, Fagotto F. CRM1- and Ran-independent nuclear export of beta-catenin. Current biology : CB. 2001;11:18–27. doi: 10.1016/s0960-9822(00)00045-2. [DOI] [PubMed] [Google Scholar]
- Xu L, Massague J. Nucleocytoplasmic shuttling of signal transducers. Nature reviews Molecular cell biology. 2004;5:209–219. doi: 10.1038/nrm1331. [DOI] [PubMed] [Google Scholar]
- Yamaguchi TP. Heads or tails: Wnts and anterior-posterior patterning. Current biology : CB. 2001;11:R713–724. doi: 10.1016/s0960-9822(01)00417-1. [DOI] [PubMed] [Google Scholar]
- Yang-Snyder J, Miller JR, Brown JD, Lai CJ, Moon RT. A frizzled homolog functions in a vertebrate Wnt signaling pathway. Current biology : CB. 1996;6:1302–1306. doi: 10.1016/s0960-9822(02)70716-1. [DOI] [PubMed] [Google Scholar]
- Yokoya F, Imamoto N, Tachibana T, Yoneda Y. beta-catenin can be transported into the nucleus in a Ran-unassisted manner. Mol Biol Cell. 1999;10:1119–1131. doi: 10.1091/mbc.10.4.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshiba S, Hamada H. Roles of cilia, fluid flow, and Ca2+ signaling in breaking of left-right symmetry. Trends Genet. 2014;30:10–17. doi: 10.1016/j.tig.2013.09.001. [DOI] [PubMed] [Google Scholar]
- Yost C, Torres M, Miller JR, Huang E, Kimelman D, Moon RT. The axis-inducing activity, stability, and subcellular distribution of beta-catenin is regulated in Xenopus embryos by glycogen synthase kinase 3. Genes Dev. 1996;10:1443–1454. doi: 10.1101/gad.10.12.1443. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.