Abstract
Mammalian models, most notably the mouse and ferret, have been instrumental in the assessment of avian influenza virus pathogenicity and transmissibility, and have been used widely to characterize the molecular determinants that confer H5N1 virulence in mammals. However, while H7 influenza viruses have typically been associated with conjunctivitis and/or mild respiratory disease in humans, severe disease and death is also possible, as underscored by the recent emergence of H7N9 viruses in China. Despite the public health need to understand the pandemic potential of this virus subtype, H7 virus pathogenesis and transmission has not been as extensively studied. In this review, we discuss the heterogeneity of H7 subtype viruses isolated from humans, and the characterization of mammalian models to study the virulence of H7 subtype viruses associated with human infection, including viruses of both high and low pathogenicity and following multiple inoculation routes. The use of the ferret transmission model to assess the influence of receptor binding preference among contemporary H7 influenza viruses is described. These models have enabled the study of preventative and therapeutic agents, including vaccines and antivirals, to reduce disease burden, and have permitted a greater appreciation that not all highly pathogenic influenza viruses are created equal.
1 Introduction
The emergence in Spring 2013 of a low pathogenic avian influenza (LPAI) H7N9 virus in Southeast Asia, capable of causing severe human disease and death, resulted in rapid global recognition of the pandemic potential of H7 viruses. H7N9 viruses are second only to H5N1 viruses in total confirmed human cases and fatalities from avian influenza viruses, though different epidemiologic profiles make it difficult to draw extensive parallels between these viruses (Cowling et al. 2013), necessitating increased investigation of H7 subtype virus virulence. However, this was not the first instance of human infection caused by an H7 subtype virus; prior to this outbreak, there were >100 confirmed or presumed human infections with H7 viruses in both Europe and the Americas (Belser et al. 2009a). Similar to H5N1, human-to-human transmission of H7 viruses has been rare but documented (Abdel-Ghafar et al. 2008; Koopmans et al. 2004; Qi et al. 2013).
There are numerous features of viruses within the H7 subtype associated with human infection that set this subtype apart from other avian influenza viruses with pandemic potential. While H5 viruses have caused the greatest number of confirmed human infections among all avian influenza virus subtypes, with a great deal of clade-specific diversity among viruses associated with human infection, all cases have resulted from viruses possessing only the N1 neuraminidase (NA) (Abdel-Ghafar et al. 2008). Similarly, human infections with H6 and H9 viruses have been restricted to the N1 and N2 NA subtypes, respectively (Arzey et al. 2012; Peiris et al. 1999; Yuan et al. 2013); H10 viruses with N7 and N8 subtypes have both been detected in humans (Arzey et al. 2012; WHO 2013). In contrast, human disease has resulted from H7 viruses possessing four NA subtypes, indicating a greater compatibility of the H7 hemagglutinin (HA) with several different NA configurations and suggesting that there is not a clear association of NA subtypes with virulent H7 viruses (Fig. 1) (Belser et al. 2009a). Furthermore, while human infection with H5, H6, and H9 viruses has been limited to Eurasian lineage strains, among all avian-origin viruses H7 viruses represent the subtype with the greatest number of documented human infections from both Eurasian and North American lineages. The diversity of these H7 avian influenza viruses resulting in human disease, encompassing both highly pathogenic (HPAI) and LPAI viruses and with select strains possessing atypical molecular determinants of virulence, also sets this virus subtype apart from other avian subtypes with pandemic potential. These features underscore the difficulty of studying the subtype as a whole, frequently requiring the inclusion of several virus isolates when establishing experimental designs to eliminate strain-specific differences inherent among this virus subtype.
Fig. 1.
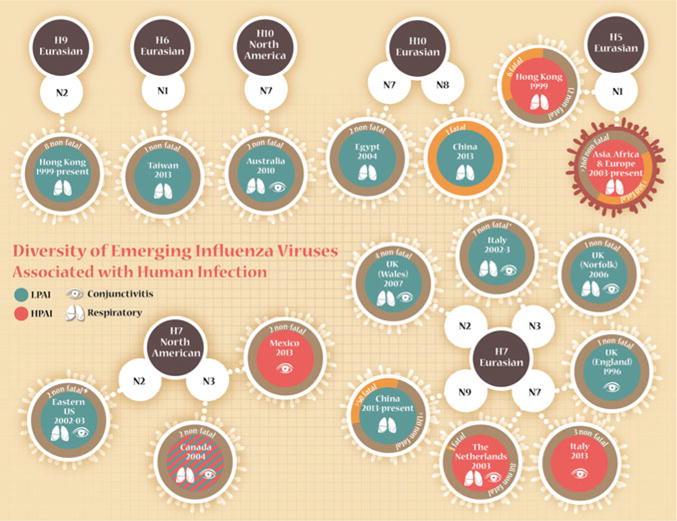
Diversity of emerging influenza viruses associated with human infection. Confirmed or presumed (serologic evidence only) cases of avian influenza virus infection in humans since 1996. *, includes cases with serologic evidence only. Red border surrounding H5N1 human cases from 2003 to present indicates that there is substantial clade-specific and geographic diversity among this virus subtype not represented in this image (Abdel-Ghafar et al. 2008)
Due to the pronounced heterogeneity among H7 subtype viruses associated with human infection, there is an urgent public health need to develop mammalian models for the study of pathogenicity and transmissibility, and to assess lineage-specific and strain-specific diversity of viruses within this subtype. Furthermore, the frequent detection of conjunctivitis with occasional and generally mild respiratory illness caused by H7 viruses in humans necessitates the development of models which reflect the multiple exposure routes exploited by this virus subtype. In this review, we discuss the pathogenicity and transmissibility of Eurasian and North American lineage H7 subtype viruses associated with human exposure and infection in the two most frequently employed mammalian models in this research, the mouse and ferret. Mice are well suited to study the pathogenesis of avian influenza viruses, as these viruses typically replicate well in the murine respiratory tract, unlike human viruses that generally require prior host adaptation. Ferrets support the replication of both human and avian strains, and represent the best small animal model for the coincident study of influenza virus pathogenesis and transmissibility. The development of these models has greatly enabled the investigation of the molecular determinants of virulence associated with HPAI and LPAI Eurasian and North American lineage H7 viruses, and provided the ability to assess existing and novel vaccine and antiviral approaches to mitigate human infection and illness from this virus subtype (Table 1).
Table 1.
Virulence of H7 viruses in mammalian models following intranasal inoculation
| Location | Year(s) | IVPIa | Subtype | Mouse virulence | Ferret virulence | References |
|---|---|---|---|---|---|---|
| Pakistan | 1995-2002 | HPAI | H7N3 | Lethality detected among selected strains | Moderate disease, no lethality reported | (Aamir et al. 2009; Abbas et al. 2010) |
| Italy | 1999-2000 | HPAI | H7N1 | Lethality detected among selected strains | Moderate disease, no lethality reported | (Cox et al. 2009; Rigoni et al. 2007; Whiteley et al. 2007) |
| Northeast US | 1994-2006 | LPAI | H7N3 | Mild to moderate disease, no lethality reported | Mild to moderate disease, no lethality reported | (Belser et al. 2007; Joseph et al. 2007) |
| Chile | 2002 | HPAIb | H7N3 | Mild to moderate disease, no lethality reported | Not examined | (Belser et al. 2013a; Joseph et al. 2007) |
| The Netherlands | 2003 | HPAI | H7N7 | Lethality detected among selected strains | Lethality detected among selected strains | (Belser et al. 2007; de Wit et al. 2005; Ekiert et al. 2011; Joseph et al. 2007) |
| Canada | 2004 | HPAIb | H7N3 | Lethality detected among selected strains | Moderate disease, no lethality reported | (Belser et al. 2007, 2008; Joseph et al. 2007, 2008; Song et al. 2009) |
| Mexico | 2012 | HPAI | H7N3 | Lethality detected among selected strains | Moderate disease, no lethality reported | (Belser et al. 2013a) |
| China | 2013-present | LPAI | H7N9 | Lethality detected among selected strains | Lethality detected among selected strains | (Belser et al. 2013b; Mok et al. 2013; Watanabe et al. 2013; Zhang et al. 2013; Zhu et al. 2013) |
IVPI, intravenous pathogenicity index
LPAI viruses were also isolated from these outbreaks and tested in these models
2 Eurasian Lineage Viruses
2.1 Historical H7 Viruses
The first descriptions of fowl plague, now recognized as highly pathogenic avian influenza, date to the late nineteenth century. The first documented isolation of an influenza virus was of the H7N7 subtype in 1902 from an outbreak of fowl plague virus (FPV) in Italy, predating the first isolation of a human influenza virus by thirty years (Horimoto and Kawaoka 2001). Documented human infection with a fowl plague virus dates to 1959, with additional human cases infrequently detected, typically following laboratory or occupational exposure (Belser et al. 2009a). While belonging to a separate phylogenetic clade, these historical H7 influenza viruses exhibit closest sequence similarity to the current Eurasian lineage (Bulach et al. 2010). Eurasian lineage H7 viruses isolated throughout the 20th century have exhibited great strain-specific diversity; most viruses examined have been found to infect mice without prior adaptation, with select H7N1 and H7N3 viruses causing lethal disease in this species (Joseph et al. 2007). Interestingly, many of these viruses elicit broadly cross-reactive antibodies to contemporary Eurasian and North American viruses (Joseph et al. 2007).
Equine influenza viruses of the H7N7 subtype were first isolated in 1956 (Timoney 1996). Despite only causing mild respiratory symptoms in horses, equine H7N7 influenza viruses were found to be lethal in mice without prior adaptation, with the capacity for enhanced neurovirulence in this species following murine adaptation (Christensen et al. 2000; Kawaoka 1991; Shinya et al. 2005, 2007). This is likely due to the presence of a multibasic amino acid HA0 cleavage site among these viruses, a molecular determinant of virulence frequently detected in viruses that exhibit a high pathogenicity phenotype in mice. While they have not been isolated from horses since 1978, serologic evidence supporting the continued circulation of equine H7N7 viruses in horses has been reported (Abd El-Rahim and Hussein 2004; Mancini et al. 2012; Timoney 1996; Webster 1993).
2.2 H7N7 Viruses
The first well-documented case of direct transmission of an H7 subtype avian influenza virus from an avian to a human host occurred in 1996 in England (Kurtz et al. 1996). The causative strain, a LPAI H7N7 virus, possessed a close phylogenetic relationship with H7 viruses isolated from poultry in the area (Banks et al. 1998). Transmission likely occurred following ocular exposure to infected ducks, and the exposed woman developed conjunctivitis; no subsequent human cases were detected (Belser et al. 2009a). The virus isolated from the human case replicated in the respiratory tract of inoculated mice without prior adaptation but did not cause severe disease in this model (Belser et al. 2009b). An outbreak of HPAI H7N7 virus in Italy resulting in three confirmed cases of human conjunctivitis represents the most recent detection of this virus (ProMED-mail 2013); research investigating the virulence of these isolates in mammalian models is ongoing.
In 2003, an outbreak of HPAI H7N7 virus in the Netherlands necessitated the culling of >30 million birds, and resulted in 89 virologically confirmed human cases, representing the largest instance of HPAI H7 subtype infection in humans to date (Fouchier et al. 2004; Koopmans et al. 2004). The majority of human infections presented as conjunctivitis, though respiratory symptoms were occasionally detected; there was one fatality, due to acute respiratory distress syndrome (Fouchier et al. 2004). Two molecular features of viruses isolated from this outbreak were similar to those identified in HPAI H5N1 viruses: the presence of a multibasic amino acid HA cleavage site and, in the virus isolated from the fatal case (A/Netherlands/219/03, NL/219), the presence of a lysine at position 627 in PB2 (Fouchier et al. 2004). With the exception of E627K, many of the known molecular markers of virulence and human adaptation present among human isolates were detected in poultry isolates before potential avian-to-human transmission events (Jonges et al. 2011), underscoring the need for early intervention during outbreaks of influenza virus in poultry.
Viruses isolated from the 2003 Netherlands outbreak were found to be highly infectious in mice following intranasal inoculation, replicating efficiently throughout the murine respiratory tract and demonstrating the capacity for systemic spread to extrapulmonary tissues, including the brain (Belser et al. 2007; de Wit et al. 2005; Joseph et al. 2007). Most viruses from this outbreak caused substantial morbidity in mice before eventual recovery, with select viruses (including the fatal case isolate NL/219) possessing a lethal dose comparable to H5N1 viruses which exhibit a high pathogenicity phenotype in this species (Belser et al. 2007; de Wit et al. 2005; Joseph et al. 2007). HPAI H7N7 viruses from 2003 also replicated to high titer following intranasal inoculation in ferrets, exhibiting systemic spread to extrapulmonary tissues and causing fever, weight loss, nasal discharge, and transient lymphopenia (Belser et al. 2007). Similar to mice, NL/219 virus exhibited a high pathogenicity phenotype in ferrets, with ferrets losing >20% of their initial body weight and exhibiting neurological symptoms during the course of infection, necessitating euthanasia.
Due to the high incidence of conjunctivitis among infected individuals, several studies were conducted to assess the ocular tropism of HPAI H7N7 viruses in mammalian models. Underscoring the ability of H7 viruses to both use the eye as a site of virus replication and as a portal of entry to establish a productive respiratory infection, virus was detected in ocular and respiratory tissues of both mice and ferrets when inoculated by the ocular route with isolates from the Netherlands outbreak (Belser et al. 2009c, 2012a). Furthermore, NL/219 virus maintained a lethal phenotype in mice (but not ferrets) following ocular inoculation. Notably, HPAI H7N7 viruses were detected in ferret ocular tissue following traditional intranasal inoculation, indicating that influenza viruses can spread from nasal to ocular tissues more readily than previously considered (Belser et al. 2012a, 2013c).
Limited human-to-human transmission was reported during the Netherlands outbreak (Koopmans et al. 2004), raising the need to model transmissibility of HPAI H7N7 viruses. NL/219 virus was not capable of transmission to naïve contacts in the ferret model, similar to HPAI H5N1 viruses. However, a virus isolated from a conjunctivitis case (A/Netherlands/230/03, NL/230) transmitted in a ferret direct contact model following either intranasal or ocular inoculation of ferrets, though airborne transmission was not detected (Belser et al. 2008, 2012a).
2.3 H7N1, H7N2, and H7N3 Viruses
H7N1, H7N2, and H7N3 influenza viruses have been responsible for frequent poultry outbreaks in Europe and Asia in recent years (Alexander 2007; Brown 2010). While reduced in scope compared with the Netherlands outbreak in 2003, these epornitics have resulted in large culling operations leading to human occupational exposure and occasional infection (Fig. 1) (2007; Nguyen-Van-Tam et al. 2006; Puzelli et al. 2005). Suspected and confirmed human cases have been reported following several of these outbreaks, with disease ranging from mild to severe (Dudley 2008; Eames et al. 2010; Nguyen-Van-Tam et al. 2006; Puzelli et al. 2005).
Numerous poultry outbreaks of LPAI (H7N1, H7N3) and HPAI (H7N1) viruses have been reported in Italy since 1999. Viruses isolated from the 1999–2000 HPAI H7N1 outbreak in Italy generally demonstrated similar pathogenicity in mice as HPAI H7N7 viruses from the Netherlands, with frequent detection of extrapulmonary spread and lethality reported among select isolates (Rigoni et al. 2007; Whiteley et al. 2007). However, while systemic spread of virus was detected in ferrets (albeit later in infection, with virus detected day 6 p.i. and not day 3 p.i. as compared with HPAI H7N7 viruses), the lethal phenotype observed with selected HPAI H7N1 viruses does not appear to be maintained in this model (Cox et al. 2009; Whiteley et al. 2007). Virus was not isolated from any exposed individuals from these outbreaks, though a percentage (<4 %) of tested poultry workers exhibited seroconversion to H7 virus, albeit at very low titers (Di Trani et al. 2012; Puzelli et al. 2005).
The first introduction of LPAI H7N3 virus to Pakistan occurred in 1995, with viruses acquiring a HPAI phenotype within a year of circulation in chickens (Abbas et al. 2010). Over the following decade, H7N3 viruses caused sporadic outbreaks in geographically distinct regions of the country, suggesting a virus reservoir in poultry. HPAI H7N3 viruses isolated from Pakistan between 1995 and 2002 replicated efficiently in the lungs of inoculated mice, but did not spread to extrapulmonary tissues; lethality in this model was observed among the most recently isolated strains (Aamir et al. 2009). Similar to HPAI H7N1 viruses, ferrets inoculated by the intranasal or ocular route with an HPAI H7N3 virus associated with lethality in mice exhibited moderate signs of illness but did not succumb to infection. It is important to note that while the majority of mammalian pathotyping of H7 viruses occurs with strains associated with human infection or large poultry outbreaks, viruses that are detected during routine surveillance have been shown to replicate efficiently in mammalian models. As an example, a LPAI H7N2 virus from northern China isolated in 2002 was found to replicate efficiently in both pulmonary and extrapulmonary tissues in mice (Li et al. 2006), underscoring the ability of H7 avian influenza viruses to replicate in mammals without prior adaptation and emphasizing the continuous need for surveillance of galliform poultry for strains which could possess pandemic potential.
Conjunctivitis is a frequent symptom among suspected and virologically confirmed cases of H7N1, H7N2, and H7N3 virus infection (Dudley 2008; Puzelli et al. 2005). Similar to HPAI H7N7 viruses, virus was recovered from eye swabs of intranasally (HPAI H7N1) or ocularly (HPAI H7N3) inoculated ferrets, though the detection of other avian and human influenza viruses recovered from ferret ocular samples indicates that this ability is not specific to H7 subtype viruses (Aamir et al. 2009; Belser et al. 2012a; Cox et al. 2009). While mice inoculated by the ocular route with influenza viruses typically do not display macroscopic ocular signs of disease, ocular symptoms were nonetheless detected among HPAI H7N1 and H7N3 viruses following intranasal inoculation in mice (Aamir et al. 2009; Cox et al. 2009; Belser et al. 2009c). These studies further support the ability of Eurasian lineage H7 viruses to employ the ocular system during in vivo mammalian infection.
Unfortunately, mammalian pathotyping of avian H7 subtype viruses associated with all human outbreaks has not been performed, and the transmissibility of these viruses has not been well examined in mammalian models. HPAI H7N1 viruses were reported to transmit in a direct contact mouse model (Rigoni et al. 2010), though there is a paucity of data establishing mice as a reliable transmission model for influenza viruses. While human-to-human transmission has not been confirmed among the outbreaks discussed here, contact tracing of suspected cases from an outbreak of LPAI H7N2 virus in Wales indicates that limited independent chains of person-to-person transmission cannot be ruled out (Eames et al. 2010). Future studies are needed to better identify the capacity of these viruses to transmit between mammals, as studied previously using viruses from the Netherlands outbreak.
2.4 LPAI H7N9 Viruses
Eurasian lineage LPAI H7N9 viruses had been previously detected during surveillance activities, but not until 2013 were these viruses associated with human infection (Gerloff et al. 2013; Gonzalez-Reiche et al. 2012; Perez-Ramirez et al. 2010). Human infections, totaling over 440 confirmed cases with over 160 fatalities, have been characterized by severe pulmonary disease and acute respiratory distress syndrome (Gao et al. 2013b). The majority of human infections have resulted from exposure to infected poultry, though evidence for limited human-to-human transmission has been reported (Li et al. 2013; Qi et al. 2013). Interestingly, the older median age among H7N9 cases compared with H5N1 cases, and the relatively few reports of mild cases among H7N9-infected individuals, indicates potential differences in susceptibility to severe illness for these avian viruses (Cowling et al. 2013).
The unexpected severe disease following human infection with a LPAI virus warranted swift assessment of the virulence of H7N9 viruses in mammalian models. Mice inoculated by the intranasal route with the first human isolates A/Anhui/1/13 (Anhui/1) and A/Shanghai/1/13 generally showed severe morbidity and comparable lethality as HPAI H5N1 and H7N7 viruses, though strain-specific differences in virulence have been detected and not all human isolates possess a lethal phenotype in this model (Baranovich et al. 2013; Belser et al. 2013b; Mok et al. 2013; Watanabe et al. 2013; Zhang et al. 2013). While extrapulmonary spread of virus is frequently associated with a lethal outcome in infected mice, viral titers in the brain were only sporadically reported following H7N9 infection (Belser et al. 2013b; Zhang et al. 2013). In contrast, avian isolates tested from this outbreak (which lack a lysine at position 627 in PB2) were not highly pathogenic in mice (Zhang et al. 2013). This is in accord with other genetically related LPAI H7N9 viruses isolated in prior years from avian species which possess comparable infectivity and replicative ability in murine respiratory tract tissues but did not maintain the high virulence observed with human isolates in this species (Belser et al. 2013b; Mok et al. 2013; Watanabe et al. 2013).
Ferrets were used extensively to characterize the pathogenicity of LPAI H7N9 viruses isolated from humans. Intranasally inoculated ferrets exhibited modest signs of disease, including weight loss, fever, sneezing and lethargy, but generally did not maintain the lethal phenotype observed in the mouse model (Belser et al. 2013b; Watanabe et al. 2013; Zhu et al. 2013). H7N9 viruses replicated efficiently throughout the ferret respiratory tract; virus was frequently but not uniformly detected in the lungs, with moderate inflammation and inflammatory infiltrates detected in bronchioles and alveoli (Belser et al. 2013b; Watanabe et al. 2013; Zhu et al. 2013). Lymphopenia and leukopenia, often detected among severe human cases, were also observed in infected ferrets, though levels of circulating white blood cells generally returned to pre-infection levels following the acute phase of infection (Belser et al. 2013b; Gao et al. 2013a; Xu et al. 2013b). While nonhuman primates are not typically utilized for the study of H7 influenza viruses, cynomolgus macaques inoculated with Anhui/1 virus in one study exhibited generally comparable levels of pathogenicity as observed in ferrets (Watanabe et al. 2013). Taken together, these studies found that LPAI H7N9 viruses exhibit similar disease in the ferret as HPAI H7N7 viruses, but cause a less severe infection compared to highly virulent HPAI H5N1 viruses (Maines et al. 2005).
The association of LPAI H7N9 viruses with severe respiratory disease in humans is in disaccord with the mild ocular and/or respiratory disease most typically observed following infection with H7 viruses (Belser et al. 2009a). However, varying the inoculation route was found to modulate H7N9 virulence, as intratracheal inoculation of ferrets with Anhui/1 virus lead to heightened morbidity and mortality compared to traditional intranasal inoculation (Kreijtz et al. 2013). While LPAI H7N9 viruses have not exhibited an ocular tropism in humans, swollen eyes were reported at the onset of mild respiratory illness in one H7N9 virus-positive poultry worker wearing respiratory but not ocular protection during the time of occupational exposure (Lv et al. 2013), suggesting that the virus may have employed the ocular route to cause human disease. In support of this, virus was detected in the respiratory tract of mice inoculated by the ocular route, as well as in the eye washes of ferrets inoculated by the intranasal route (Belser et al. 2013b). Collectively these results indicate that, similar to other influenza viruses, H7N9 virus can utilize multiple exposure routes to mount a productive infection in mammals, with severity of disease influenced by the route of infection.
The presence among several H7N9 isolates of amino acid changes known to increase binding to α2-6 linked sialic acids, notably Q226L in the HA protein, suggested that these viruses were capable of enhanced transmissibility compared to other avian-origin viruses (Gao et al. 2013b). H7N9 isolates were found to transmit efficiently between either ferrets or guinea pigs when placed in direct contact, a capacity not generally present among avian influenza viruses (Belser et al. 2013b; Gabbard et al. 2013; Maines et al. 2006; Zhu et al. 2013). However, while the incidence of H7N9 virus transmission between ferrets in the absence of direct or indirect contact (i.e. respiratory droplet or airborne transmission) was greater than that observed among H7N9 viruses isolated from avian species, transmission was overall less efficient than seasonal or pandemic influenza viruses in this model, with delayed kinetics of virus spread to contact ferrets when transmission did occur (Belser et al. 2013b; Maines et al. 2006, 2009; Richard et al. 2013; Watanabe et al. 2013; Xu et al. 2013a; Zhang et al. 2013; Zhu et al. 2013). While selected studies have observed efficient transmission of H7N9 viruses by respiratory droplets, serial passage of H7N9 virus in ferrets did not enhance virus transmissibility, further indicating that, despite the efficient transmissibility detected in a direct contact model, these viruses are not yet capable of sustained airborne mammalian transmission and would require additional mammalian adaptation to achieve this property (Richard et al. 2013; Zhang et al. 2013).
3 North American Lineage Viruses
3.1 Historical H7 Viruses
North American lineage H7 viruses, similar to those within the Eurasian lineage, have been detected numerous times throughout the twentieth century, most frequently from avian species. The majority of these viruses examined have been found to infect mice without prior adaptation, though rarely have they been associated with lethal disease in this species following intranasal inoculation (Goff et al. 2013; Joseph et al. 2007). In 1979–1980, an H7N7 virus caused the death of ~500 seals on the New England coast, subsequently resulting in five cases of conjunctivitis among laboratory workers with occupational exposure to the virus (one virologically confirmed, four suspect cases) (Lang et al. 1981; Webster et al. 1981). This virus only caused mild respiratory disease in mice, and did not infect experimentally inoculated ferrets or rats (Scheiblauer et al. 1995). However, mammalian passage of a laboratory variant of this virus was capable of causing lethal disease in mice and ferrets, indicating that the H7N7 virus could acquire neurotropic and pathogenic properties following host adaptation.
3.2 LPAI H7 Viruses in the United States
LPAI H7N2 virus was first isolated from northeastern United States live bird markets (LBM) in 1994, and persisted for over a decade before the virus was eradicated from those markets in 2006 (Senne 2010; Senne et al. 2003). During this time, the largest outbreak of LPAI in the United States occurred in 2002 in Virginia and the surrounding area, due to an H7N2 virus with high genetic sequence identity to the LBM circulating strains (Senne 2007). These viruses were associated with one confirmed case of human infection (A/New York/107/03, NY/107) and one suspected case (among 80 tested poultry workers) with occupational exposure who seroconverted to H7N2 virus during the Virginia outbreak (CDC 2004; Ostrowsky et al. 2012). Despite the elimination of H7N2 viruses from the northeastern United States LBM system, H7 subtype viruses continue to circulate across the United States, as H7N3, H7N7, and H7N9 subtype viruses have been isolated from wild birds and galliform poultry over the past decade (Pasick et al. 2012; Senne 2007, 2010). While these viruses have not caused any confirmed human cases, seroprevalence studies have reported statistically significant elevated titers against H7 influenza virus in veterinarians compared to healthy control subjects, suggesting that occupational exposure may be occurring (Myers et al. 2007).
LPAI H7 viruses from North America have typically not caused severe disease in experimentally inoculated mammalian models. In mice, virus replicates efficiently in both the nose and lung following intranasal inoculation, with extrapulmonary spread generally not observed (Belser et al. 2007, 2013a; Joseph et al. 2007). However, LPAI H7 viruses are highly infectious in this model, and morbidity can range from mild to moderate depending on the strain used to infect. Similar results are detected in the ferret model, though viral titers are generally lower in the lung compared with the nasal turbinates and trachea (Belser et al. 2007, 2008, 2013a). Ferret morbidity is generally mild and transient, with few lymphohematopoietic perturbations and an absence of extrapulmonary virus spread.
Similar to Eurasian lineage H7 viruses, many LPAI H7 viruses from the North American lineage can employ the mammalian eye as a site of primary virus replication as well as a portal of entry to spread to the respiratory tract (Belser et al. 2009c Belser et al. 2012, 2013a). Ocular inoculation of mice with LPAI H7N3 and H7N9 viruses resulted in detectable virus titers in both eye and respiratory tract tissues in the absence of significant morbidity, as did ferrets inoculated by the ocular route with the LPAI H7N2 virus NY/107 (Belser et al. 2012a, 2013a). These findings indicate that avian isolates not associated with human disease are still capable of using the eye as a portal of entry to cause a productive mammalian infection.
Unlike the majority of Eurasian lineage viruses (with the exception of LPAI H7N9 viruses), selected contemporary North American viruses possess the ability to bind both α2-3 and α2-6 linked sialic acids, as will be discussed in more detail in the subsequent section. As a binding preference for α2-6 linked sialic acids is a feature of human and pandemic influenza viruses, continued evaluation of the transmissibility of North American H7 viruses has become essential to monitor the true pandemic potential of these viruses. Notably, several LPAI H7N2 viruses have been found capable of transmission to naïve contacts in the presence of direct contact. NY/107 virus was found to transmit efficiently in a direct contact model following either intranasal or ocular inoculation of ferrets (Belser et al. 2008, 2012a). A chicken LPAI H7N2 virus from 2003, but not a turkey isolate from 2002, was also capable of limited transmission in a direct contact model, despite both viruses exhibiting a similar receptor-binding profile as NY/107 virus, indicating that receptor binding preference alone does not confer a transmissible phenotype (Belser et al. 2008). However, no wild-type LPAI H7 North American strains have been found capable of transmission between ferrets by the airborne route to date.
3.3 HPAI H7N3 Viruses
HPAI H7N3 viruses have caused numerous poultry outbreaks in both North and South America (Lopez-Martinez et al. 2013; Senne 2007, 2010). Unlike Eurasian HPAI H5N1 and H7N7 viruses which typically possess traditional multibasic amino acid HA cleavage sites, these HPAI H7N3 viruses all appear to have arisen from LPAI precursor viruses by nonhomologous recombination between the HA and other viral genes or host cell genetic sequences (Berhane et al. 2009; Maurer-Stroh et al. 2013; Pasick et al. 2005; Suarez et al. 2004). To date, H7 viruses are the only known subtype to use this mechanism as a way to acquire a virulent phenotype. Several of these epornitics have resulted in human infection, underscoring the need to study the virulence of these viruses which pose a threat to both avian and human health and to elucidate why H7 subtype viruses are most susceptible to this form of recombination. Unfortunately, viruses from these unique outbreaks have been infrequently studied in mammalian models, limiting our understanding of how the H7 HA cleavage site influences viral pathogenicity.
An outbreak of LPAI H7N3 virus occurred in Chile in May 2002, representing the first isolation of H7 influenza virus in poultry or wild birds in the continent of South America (Suarez et al. 2004). Recombination between the HA and nucleoprotein genes of the LPAI virus resulted in the generation of a HPAI virus within a month; no human cases were reported during the 7 months of eradication efforts (Max et al. 2007). Despite exhibiting differential infectivity and virulence in poultry, both HPAI and LPAI viruses from this outbreak replicated efficiently in the lungs of mice in the absence of substantial morbidity or mortality (Belser et al. 2013a; Jones and Swayne 2004; Joseph et al. 2007).
In 2004, an H7N3 virus detected in British Columbia, Canada acquired a high pathogenicity phenotype in chickens following a nonhomologous recombination event between the HA and matrix genes (Hirst et al. 2004). This outbreak necessitated the culling of over 19 million birds, and resulted in 55 suspected and two virologically confirmed human infections (Tweed et al. 2004). While both human cases presented with conjunctivitis and influenza-like illness following direct conjunctival contact with poultry, virus isolated from one human case was found to be LPAI whereas the other human isolate (A/Canada/504/04, Can/504) was found to be HPAI (Hirst et al. 2004; Tweed et al. 2004); one additional basic amino acid was present in the cleavage region of the HPAI virus compared with the LPAI strain (Hirst et al. 2004). Similar to the H7N3 viruses from Chile, both HPAI and LPAI H7N3 viruses associated with human infection, as well as a closely related LPAI chicken isolate from this outbreak and a mallard LPAI H7N3 virus isolated in 2001, replicated efficiently in the lungs of mice but did not spread to extrapulmonary tissues following intranasal inoculation (Belser et al. 2007; Joseph et al. 2008; Song et al. 2009). However, a HPAI chicken isolate from this outbreak was capable of lethality and extrapulmonary spread to the brain following intranasal murine inoculation, indicating strain-specific differences in virulence among these HPAI H7N3 viruses (Belser et al. 2008; Joseph et al. 2007). Both HPAI and LPAI chicken isolates from the 2004 H7N3 outbreak replicated efficiently in ferret upper and lower respiratory tract tissues (Joseph et al. 2008). Unlike mice, ferrets inoculated by the intranasal route with Can/504 virus exhibited substantial weight loss before recovery, indicating species-specific differences in mammalian virulence among HPAI H7N3 viruses from this outbreak. A subsequent outbreak of HPAI H7N3 in Saskatchewan, Canada in 2007 also emerged from a nonhomologous recombination event with a genetic sequence of eukaryotic origin, but did not result in confirmed human infection (Pasick et al. 2010); the causative virus has not been extensively studied in mammalian models.
The most recent outbreak of HPAI H7N3 virus occurred in Jalisco, Mexico in 2012, resulting in the culling of over 22 million birds and the detection of two confirmed human cases among poultry workers, both presenting with conjunctivitis (Kreijtz et al. 2013; Lopez-Martinez et al. 2013). Similar to other HPAI H7N3 viruses from North America, isolates from this outbreak also possess an extended HA cleavage site, with the insertion likely acquired from host 28S ribosomal RNA (Lopez-Martinez et al. 2013; Maurer-Stroh et al. 2013). Mice inoculated with virus isolated from a human conjunctivitis case (A/Mexico/InDRE7218/12, (Mex/7218)) exhibited greater morbidity and mortality compared with previously examined North American HPAI H7N3 viruses, causing severe morbidity and lethal disease in mice intranasally inoculated with high (106 EID50) doses of virus (Belser et al. 2013a). Similar to mice, Mex/7218 virus replicated to high titer throughout the ferret respiratory tract following intranasal inoculation, including the lung, but did not spread systemically to multiple organs. However, extrapulmonary spread of virus was detected in the brain and gastrointestinal tract of infected ferrets, although all ferrets survived viral challenge (Belser et al. 2013a).
Conjunctivitis was documented among all human infections with North American H7N3 viruses, underscoring a need to examine the pathogenicity of these viruses following ocular exposure. Mice inoculated by the ocular route with HPAI H7N3 human isolates from Canada and Mexico did not exhibit significant morbidity or mortality, but did possess detectable virus in both ocular and respiratory tract tissues post-inoculation (Belser et al. 2009c, 2013a). Similarly, ferrets inoculated by the ocular route with Can/504 virus became productively infected in the absence of substantial morbidity, with virus detected in samples collected from both the respiratory tract and eye (Belser et al. 2012a).
Just as North American lineage LPAI H7N2 viruses have exhibited limited transmissibility in a direct contact ferret model, LPAI and HPAI H7N3 viruses from several of these outbreaks have also demonstrated this property. Low levels of the HPAI H7N3 virus Can/504 isolated from a human conjunctivitis case, were transmitted to ferrets in a direct contact model (Belser et al. 2008). The HPAI H7N3 virus Mex/7218, also isolated from a conjunctivitis case, efficiently transmitted to naïve ferrets in a direct contact model, though a genetically related North American LPAI H7N9 virus did not (Belser et al. 2013a). Strikingly, transmission in the presence of direct contact was also reported with a mallard LPAI H7N3 virus in the absence of prior host adaptation, underscoring the need for continued surveillance and evaluation of H7 viruses from wild birds (Song et al. 2009). However, similar to other North American H7 viruses, airborne transmission of H7N3 viruses has not been reported to date.
4 Molecular and Host Determinants of Pathogenicity and Transmissibility
4.1 Molecular Correlates of Pathogenicity
The majority of research studies investigating molecular determinants associated with mammalian virulence of avian influenza viruses have been conducted with H5 and not H7 subtype viruses. However, it appears that many features associated with H5N1 virulence are maintained in H7 subtype viruses, especially with regard to the virus HA. Similar to results obtained using H5N1 strains, reassortant viruses bearing surface glycoproteins derived from a virulent H7 virus on the backbone of a nonlethal H7 virus exhibit moderately enhanced virulence without increases in lethality in mammalian species (Munster et al. 2007; Salomon et al. 2006). The introduction of glycosylation sites proximal to the receptor binding pocket of the HA has been shown to facilitate adaptation of H7 and H5 viruses from wild birds to poultry and modulate virulence in mammalian hosts (Banks and Plowright 2003; Belser and Tumpey 2013; de Wit et al. 2010). Building upon prior studies conducted with H5N1 viruses has thus enabled greater precision in the identification of molecular markers of H7 subtype virulence.
Previous studies on H5N1 mammalian virulence have indicated that while a multibasic amino acid HA cleavage site is necessary (Hatta et al. 2001; Horimoto and Kawaoka 1994; Suguitan et al. 2012), this molecular feature alone is not sufficient for a virulent phenotype in mammals (Maines et al. 2005). In this vein, studies with H7 viruses have demonstrated that the presence of a multibasic amino acid cleavage site does not necessarily confer a virulent phenotype in either avian or mammalian species (Belser et al. 2007; Senne et al. 1996). While the composition of HA cleavage sites among Eurasian lineage LPAI and HPAI H7 viruses generally resemble that of H5N1 viruses, there is a greater diversity among North American H7 viruses. Insertion of additional basic amino acids at the cleavage site of a LPAI H7N2 virus, but not mutation of residues already present at the cleavage site, resulted in viruses which were highly pathogenic in chickens, demonstrating the capacity of an H7 virus to acquire a pathogenic phenotype, in contrast to H5 viruses which can acquire a virulent phenotype by either mutation or insertion (Lee et al. 2006). Molecular modeling of the insertional event resulting in a HPAI H7N3 virus isolated from Canada in 2004 revealed an enlarged loop formation resulting in greater accessibility of the HA cleavage site, likely leading to increased cleavage by furin-like proteases resulting in enhanced pathogenicity (Hirst et al. 2004). Future study is needed to better understand the contribution and mechanism of HA cleavage site insertions as they pertain to mammalian pathogenesis, especially among North American lineage viruses.
The presence of a lysine at position 627 in PB2 has long been identified as a critical factor of mammalian adaptation and virulence among influenza viruses, including HPAI H5N1 viruses (Hatta et al. 2001; Subbarao et al. 1993). As mentioned in preceding sections, several studies have demonstrated a similar role for this position among H7 subtype viruses. An E627K substitution in a HPAI H7N7 virus isolated from the Netherlands was identified as a major determinant of mammalian pathogenicity, with viruses bearing this mutation exhibiting increased viral load, extrapulmonary spread to the brain, and heightened mortality in mice; this enhanced virulence was absent in mice inoculated with virus bearing the reciprocal K627E mutation (de Jong et al. 2013; Munster et al. 2007). Similarly, the presence of a lysine at this position in LPAI H7N9 human isolates and a mouse-adapted H7N7 equine virus resulted in increased PB2 polymerase activity, viral loads, and disease severity in mice (Mok et al. 2014; Shinya et al. 2007; Zhang et al. 2013, 2014). Enhanced virulence of HPAI H7N7 viruses bearing E627K was also observed in ferrets compared with virus bearing glutamic acid at this position (de Jong et al. 2013). However, H7N9 viruses bearing either a glutamic acid or lysine at position 627 exhibited comparable transmissibility in the ferret model, indicating E627K does not solely confer an enhanced transmissibility phenotype (Zhang et al. 2013). These studies are supported by in vitro assays in human cell types revealing that the E627K mutation confers enhanced replicative ability and polymerase activity compared with K627E (de Wit et al. 2010). While the presence of E627K yields heightened virulence in mice, it should be noted that select viruses lacking this mutation are still capable of extrapulmonary spread and murine lethality (Aamir et al. 2009; Rigoni et al. 2007), demonstrating the polygenic nature of mammalian virulence and underscoring that PB2 position 627 is only one of several molecular determinants that contribute to mammalian pathogenicity. In support of this, additional mutations in the H7 viral polymerase independent of position 627 have been demonstrated to modulate virulence in a mouse model (Gabriel et al. 2005, 2007; Yamayoshi et al. 2013). Disruption of protein–protein binding between PA and PB1 was also shown to attenuate murine virulence of an H7N7 virus (Manz et al. 2011).
The influenza non-structural (NS1) protein possesses several varied functions which contribute to block type-1 interferon-mediated host antiviral responses (Garcia-Sastre 2001). A mouse-adapted H7N7 virus with a complete deletion of the NS1 gene was nonpathogenic in wild-type mice, suggesting that enhanced synthesis of IFN in infected mouse lungs led to virus attenuation in this species (Kochs et al. 2007). Comparison of NS1 proteins encoded by different subtypes in vitro revealed that an H7 NS1 induced lower levels of proinflammatory cytokines, chemokines, and levels of apoptosis compared to an H5 NS1 (Lam et al. 2011); this is in agreement with in vivo studies which detect proinflammatory cytokines and chemokines in respiratory tract tissues of mice and ferrets inoculated by the intranasal route with H7 subtype viruses, albeit at lower levels than virulent HPAI H5N1 viruses (Belser et al. 2007, 2013c). However, a recombinant H7N7 virus bearing the NS gene from a fatal human case did not enhance the virulence of an H7N7 virus isolated from a conjunctivitis case, indicating that this gene does not represent a major determinant of virulence among H7 strains (Munster et al. 2007).
Dysregulation of innate host immune responses and other host factors following H5N1 virus infection in mammals has been shown to contribute to the high pathogenicity phenotype of this virus subtype (Belser and Tumpey 2013), but there are only limited studies conducted in the context of mammalian H7 virus infection. H7N7 virus infection of different inbred laboratory mouse strains resulted in divergent survival outcomes, demonstrating a role for the genetic background of the host in H7 mammalian virulence (Srivastava et al. 2009). The interferon-induced resistance factor Mx1 has been shown to contribute to interferon-mediated protection against both avian and human influenza viruses, including the H7 subtype, as viruses which cause lethal disease in standard laboratory mouse strains (which lack a functional Mx1 gene) do not maintain a lethal phenotype in mice carrying a wild-type functional Mx1 gene (Koerner et al. 2007; Tumpey et al. 2007). However, Mx1-positive mice lacking functional toll-like receptor (TLR)-7 signaling or which were depleted of plasmacytoid dendritic cells exhibit a decrease in resistance to a mouse-adapted HPAI H7N7 virus isolate of seal origin (SC35M), indicating TLR7-mediated detection of H7 virus via plasmacytoid dendritic cells represents a source of interferon which confers antiviral protection in the host (Kaminski et al. 2012). Lymphopenia has been reported following H7 virus infection in mice and ferrets (Belser et al. 2007); studies using SC35M virus found that the high polymerase activity of this strain was responsible for the severe lymphocyte depletion and impaired immune response observed in SC35M virus-infected mice but not in the non-mouse-adapted counterpart (Gabriel et al. 2009). Collectively, these studies indicate that while several molecular determinants of avian influenza virulence are maintained between H5 and H7 viruses, there are numerous ways in which H7 subtype viruses differentially interact with the mammalian host.
4.2 Receptor Binding and Correlates of Transmissibility
The distribution of host cell glycoconjugates containing terminal sialic acid moities, which serve as the receptors for all influenza viruses, greatly contributes to the tissue tropism and pandemic potential of influenza viruses in humans. Avian influenza viruses, such as HPAI H5N1 viruses, typically exhibit preferential binding to sialic acids linked to galactose by an α2-3 linkage; in humans, these receptors are generally located in the lower respiratory tract. In contrast, human influenza viruses preferentially bind to α2-6 linked sialic acids, which are present in greater abundance in the epithelia of the human upper respiratory tract (Matrosovich et al. 2000). Thus, the receptor binding site of the HA governs the ability of influenza viruses to bind sialic acids in an α2-3 linked conformation, α2-6 linked conformation, or both. It is generally believed that a switch in receptor-binding specificity from an avian-like to a human-like preference is necessary for a virus to achieve sustained human-to-human transmission and cause a pandemic, though an increase in α2-6 binding preference alone is not sufficient for efficient airborne mammalian transmission of avian influenza viruses (Maines et al. 2011). Analysis of contemporary Eurasian and North American H7 influenza viruses revealed that despite maintaining a strong preference for α2-3 linked sialic acids, many possess weak but detectable binding to α2-6 linked sialic acids (Gambaryan et al. 2012). The ferret model is most commonly used to model the transmissibility of human and avian influenza viruses in a laboratory setting due to similar lung physiology, receptor distribution, and virus attachment patterns between ferrets and humans (Fig. 2) (Belser et al. 2009b).
Fig. 2.
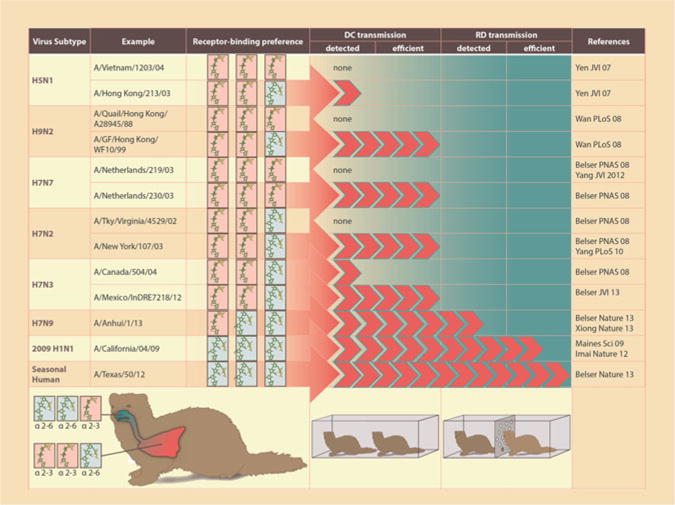
Contribution of receptor binding preference on virus transmissibility in the ferret model. Virus receptor binding is depicted as maintaining a strong avian binding preference (three α2-3 icons), maintaining an avian binding preference with detectable binding to human receptors (two α2-3 icons, one α2-6 icon), enhanced binding to human receptors while maintaining binding to avian receptors (two α2-6 icons, one α2-3 icon) or strong human binding preference (three α2-6 icons) and is not meant to be quantitative. “DC transmission” indicates one inoculated ferret and one naïve ferret co-housed in the same cage, sharing food, water, and bedding; “RD transmission” indicates one inoculated ferret and one naïve ferret housed in adjacent cages separated by a perforated side-wall (3–5 mm in diameter) allowing air exchange only in the absence of direct or indirect contact
With the notable exception of LPAI H7N9 viruses, Eurasian lineage H7 viruses associated with human infection have generally maintained an avian-like α2-3 sialic acid receptor binding preference, similar to the binding preference observed with HPAI H5N1 viruses isolated from humans (Belser et al. 2008; Gambaryan et al. 2008, 2012; Yang et al. 2012). As discussed in previous sections, these Eurasian lineage H7 viruses have generally not transmitted well between ferrets in direct contact or respiratory droplet models. However, divergent patterns of transmissibility in the ferret model and attachment patterns in respiratory tract mammalian cells between viruses isolated from the H7N7 2003 Netherlands outbreak (Fig. 2) point to subtle differences in receptor binding not identified in glycan arrays (Belser et al. 2008; de Wit et al. 2010), highlighting the complexity between closely related strains and warranting further study of additional determinants of influenza virus receptor binding specificity.
Numerous contemporary North American H7 subtype viruses possess increased binding to α2-6 linked sialosides while maintaining strong binding to α2-3 linked sialosides, with select strains further showing reduced binding to α2-3 linked sialosides similar to human influenza viruses (Belser et al. 2008; Gambaryan et al. 2008; Yang et al. 2010). Viruses which possess increased binding to α2-6 linked sialic acids have transmitted with moderate to high efficiency in a ferret direct contact model, though strain-specific differences remain (Belser et al. 2008). Furthermore, limited transmission in a direct contact model has been reported among H7 viruses which maintain an avian α2-3 sialic acid binding preference (Song et al. 2009), indicating that additional factors contribute to the transmissibility of H7 viruses.
HPAI H7N9 viruses isolated from China possess the highest affinity for α2-6 linked sialic acids among all avian viruses associated with human infection, although unlike human influenza viruses, maintain strong binding to α2-3 linked sialic acids (Tharakaraman et al. 2013; Xiong et al. 2013). As mentioned earlier, this enhanced binding to human receptors is likely aided by the presence of a glutamine to leucine switch at position 226 of the HA among H7N9 viruses, a substitution previously associated with human adaptation (Gao et al. 2013b). However, strain-specific differences in both receptor-binding preference and transmission efficiency have been reported among H7N9 human isolates (Belser et al. 2013b; Watanabe et al. 2013; Zhang et al. 2013), further underscoring the polygenic nature of both of these properties. Decreased NA activity of the H7N9 virus compared to a human pandemic virus, and reduced virus replication efficiency at 33 °C compared to pandemic viruses, were also suggested to contribute to the reduced airborne transmissibility of this virus (Belser et al. 2013b; Xiong et al. 2013).
5 Use of Mammalian Models to Develop Vaccines and Antivirals Against H7 Viruses
As discussed throughout this review, H7 influenza viruses associated with human infection exhibit a wide range of pathogenic and transmissible phenotypes, posing unique challenges in designing appropriate strategies to both reduce disease severity and prevent infection. Mammalian models have proven invaluable in assessing the immunogenicity and efficacy of novel candidate vaccines against this virus subtype. While pre-existing influenza virus-specific CD8+ T-cell memory may provide some degree of protection against virulent viruses such as H7 (Christensen et al. 2000; Zhong et al. 2010), there is a clear need to develop effective vaccines and antivirals to protect a serologically naïve human population in the event of an H7 outbreak or pandemic. The poor immunogenicity observed with vaccine candidates against avian influenza viruses in general, and H7 viruses in particular, further highlights the importance of generation and testing of vaccine approaches to this virus subtype as well as improved means to identify appropriate correlates of protection to measure H7 vaccine effectiveness in mammalian species (Krammer and Cox 2013).
Numerous inactivated vaccine candidates have been generated against both Eurasian and North American H7 strains in pandemic preparedness efforts, generally showing promise in mammalian models despite variable immunogenicity. A β-propiolactone-inactivated whole virus vaccine against a Eurasian H7N1 virus was found to elicit humoral antibody responses in mice following vaccination (Hovden et al. 2009). Formalin-inactivated whole virus vaccines against both North American and Eurasian H7 viruses were found to be effective in mice following heterologous virus challenge, restricting pulmonary virus replication and conferring protection from lethal challenge, even in the absence of an adjuvant (Jadhao et al. 2008; Pappas et al. 2007). However, the addition of adjuvant was found to greatly enhance the protection of an H7N7 classical subunit preparation in mice (de Wit et al. 2005), an important feature given the generally poor induction of neutralizing antibodies by this subtype in mammals and potential obstacles involved in eliciting broadly cross-reactive antibodies that can neutralize viruses from both Eurasian and North American lineages (Joseph et al. 2007).
Several live attenuated influenza vaccine (LAIV) candidates against H7 viruses have also been developed and evaluated in mammalian models for safety, immunogenicity, and efficacy. Both North American H7N3 and Eurasian H7N7 LAIV candidates were found to protect mice and ferrets from homologous and heterologous H7 viruses (Joseph et al. 2008; Min et al. 2010). Furthermore, sera from H7N3 and H7N7 LAIV immunized ferrets was found to cross-react against Anhui/1 virus, suggesting that these vaccine candidates may offer some degree of protection against H7N9 strains (Talaat et al. 2009; Xu et al. 2013c); based on results obtained from these studies in mammalian models, both of these candidate vaccines have been used in clinical trials in humans (Xu et al. 2013c).
While the inactivated and LAIV candidates discussed above have relied on production techniques previously established for the generation of seasonal influenza virus vaccines, alternate approaches are also under investigation. In recent years, there has been renewed interest in producing egg-free inactivated influenza vaccines, especially for viruses with pandemic potential. H7N1 vaccines have been generated in cell culture (Cox et al. 2009), and those that have undergone further evaluation have been found to elicit protective antibody responses against homologous and heterologous viruses in both mice and ferrets (Legastelois et al. 2007; Whiteley et al. 2007). Non-infectious recombinant virus-like particles (VLPs) have been generated with both Eurasian and North American lineage H7 HAs (either alone or co-expressed with other subtypes) and have been evaluated for their ability to induce serum antibody responses and confer protection against viral challenge in both mice and ferrets (Smith et al. 2013; Szecsi et al. 2006; Tretyakova et al. 2013). The use of well-characterized mammalian models for H7 virus infection has allowed for the evaluation of vaccine approaches against conserved HA epitopes, as mice vaccinated with chimeric HA constructs expressing an H3 stalk vaccine developed cross-reactive antibodies which protected against murine heterosubtypic challenge with H7 viruses (Krammer et al. 2013; Margine et al. 2013). Further demonstrating the ability of antibodies raised against H7 viruses to exhibit cross-protection, mice immunized with a recombinant Newcastle disease virus expressing a North American H7 HA were protected against a lethal heterologous H7 challenge. This was also true of mice immunized with a recombinant vaccine expressing a Eurasian H7 HA on the surface of baculovirus subsequently challenged with a murine-passaged homologous virus exhibiting enhanced virulence (Goff et al. 2013; Rajesh Kumar et al. 2013). Future study will allow for a better evaluation of these novel approaches to effectively elicit protective antibody and cell-mediated immune responses toward H7 influenza viruses in humans.
In the absence of a well-matched vaccine, such as the early stages of an outbreak or pandemic, antivirals represent the first line of defense against influenza viruses. Unfortunately, the sensitivity of H7 viruses to currently available antiviral drugs has not been extensively examined, with a paucity of studies performed in in vivo models. Similar to many avian and human influenza viruses, the HPAI H7N7 NL/219 virus was found to be resistant to the M2 inhibitor amantadine in a mouse model (Ilyushina et al. 2007). The neuraminidase inhibitor oseltamivir was found to be effective against H7N9 virus in mice in a dose-dependent manner, with mice protected from death and the emergence of drug-resistant variants (Baranovich et al. 2013). However, the presence of a lysine at position 292 of the NA (detected among select H7N9 clinical isolates) resulted in oseltamivir resistance without a loss of virulence or transmissibility in vivo (Hai et al. 2013). Oseltamivir has been prescribed during H7 influenza outbreaks resulting in conjunctivitis (Koopmans et al. 2004; Tweed et al. 2004); in support of this, mice inoculated by the ocular route with HPAI H7N7 Eurasian lineage or H7N3 North American lineage viruses receiving prophylactic oseltamivir treatment exhibited reductions in viral titer in both respiratory and ocular tissues (Belser et al. 2012b). Interference with host cell signaling pathways may also represent a possible avenue to attenuate virulence in vivo, as mice treated with morpholino oligomers or inhibitors of the JNK or NF-κB pathways exhibited reduced viral loads in the lungs following H7N7 virus infection (Gabriel et al. 2008; Haasbach et al. 2013; Nacken et al. 2012). Serine protease inhibitors have furthermore shown antiviral activity in mice following H7N7 challenge (Bahgat et al. 2011). The successful use of convalescent plasma for the treatment of human H5N1 infection has indicated that passive immunotherapy warrants further investigation as a therapeutic agent (Zhou et al. 2007); monoclonal antibodies with neutralizing activity against H7 viruses have increased survival and limited virus replication in mice following lethal H7 virus challenge, indicating that antibody-based therapy, administered either prophylactically or as a treatment, could provide an additional approach to contain the spread of a pandemic H7 virus in the absence of an effective vaccine (Ekiert et al. 2011; He et al. 2013).
6 Conclusions
Influenza viruses within the H7 subtype have caused a broad spectrum of human infection, from conjunctivitis and mild influenza-like illness to severe disease, acute respiratory distress syndrome, and death. The viruses responsible for these human infections have similarly demonstrated a wide range of virulence and transmissible phenotypes in mammalian models (Table 1). This diversity highlights that there is no one “representative” H7 strain and that laboratory study of this subtype necessitates the inclusion of multiple isolates to identify and control for strain-specific differences. Just as mammalian models for the study of H7 virus pathogenesis and transmission have become more refined to better address specific questions pertinent to this virus subtype, such as investigating the apparent ocular tropism frequently associated with this virus subtype in humans, the development and evaluation of both existing and nascent vaccines and antiviral treatments must be attuned to the different manifestations of disease possible following human infection with this subtype. There is a need for continued analysis of the clinical relevance of mutations present among H7 viruses which confer a resistant phenotype in vitro, especially as they pertain to viral fitness and transmissibility (Sleeman et al. 2013). The frequent interspecies transmission of Eurasian and North American H7 viruses from aquatic birds to poultry, and detection of a poultry-like receptor binding site among contemporary viruses within this subtype, indicates that this subtype readily infects gallinaceous poultry and underscores both the need for continued surveillance of this virus subtype and preparation for future cases of human infection (Gambaryan et al. 2012). Collectively, these studies show the continued requirement for the study of H7 viruses and the con-current reminder to use personal protective equipment that includes both respiratory and eye protection in all instances of potential laboratory or occupational exposure to LPAI and HPAI H7 viruses (Morgan et al. 2009).
Acknowledgments
The authors thank Alissa Eckert for graphical assistance. The findings and conclusions in this report are those of the authors and do not necessarily reflect the views of the funding agency.
Contributor Information
Jessica A. Belser, Influenza Division, MS G-16, National Center for Immunization and Respiratory Diseases, Centers for Disease Control and Prevention, 1600 Clifton Rd. NE, Atlanta, GA 30333, USA
Terrence M. Tumpey, Influenza Division, MS G-16, National Center for Immunization and Respiratory Diseases, Centers for Disease Control and Prevention, 1600 Clifton Rd. NE, Atlanta, GA 30333, USA
References
- Avian influenza A/(H7N2) outbreak in the United Kingdom. Euro Surveill. 2007;12(5):E070532.1. [PubMed] [Google Scholar]
- Aamir UB, Naeem K, Ahmed Z, Obert CA, Franks J, Krauss S, Seiler P, Webster RG. Zoonotic potential of highly pathogenic avian H7N3 influenza viruses from Pakistan. Virology. 2009;390:212–220. doi: 10.1016/j.virol.2009.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abbas MA, Spackman E, Swayne DE, Ahmed Z, Sarmento L, Siddique N, Naeem K, Hameed A, Rehmani S. Sequence and phylogenetic analysis of H7N3 avian influenza viruses isolated from poultry in Pakistan 1995-2004. Virol J. 2010;7:137. doi: 10.1186/1743-422X-7-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abd El-Rahim IH, Hussein M. An epizootic of equine influenza in Upper Egypt in 2000. Rev Sci Tech. 2004;23:921–930. doi: 10.20506/rst.23.3.1539. [DOI] [PubMed] [Google Scholar]
- Abdel-Ghafar AN, Chotpitayasunondh T, Gao Z, Hayden FG, Nguyen DH, de Jong MD, Naghdaliyev A, Peiris JS, Shindo N, Soeroso S, et al. Update on avian influenza A (H5N1) virus infection in humans. N Engl J Med. 2008;358:261–273. doi: 10.1056/NEJMra0707279. [DOI] [PubMed] [Google Scholar]
- Alexander DJ. Summary of avian influenza activity in Europe, Asia, Africa, and Australasia, 2002–2006. Avian Dis. 2007;51:161–166. doi: 10.1637/7602-041306R.1. [DOI] [PubMed] [Google Scholar]
- Arzey GG, Kirkland PD, Arzey KE, Frost M, Maywood P, Conaty S, Hurt AC, Deng YM, Iannello P, Barr I, et al. Influenza virus A (H10N7) in chickens and poultry abattoir workers, Australia. Emerg Infect Dis. 2012;18:814–816. doi: 10.3201/eid1805.111852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahgat MM, Blazejewska P, Schughart K. Inhibition of lung serine proteases in mice: a potentially new approach to control influenza infection. Virol J. 2011;8:27. doi: 10.1186/1743-422X-8-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banks J, Plowright L. Additional glycosylation at the receptor binding site of the hemagglutinin (HA) for H5 and H7 viruses may be an adaptation to poultry hosts, but does it influence pathogenicity? Avian Dis. 2003;47:942–950. doi: 10.1637/0005-2086-47.s3.942. [DOI] [PubMed] [Google Scholar]
- Banks J, Speidel E, Alexander DJ. Characterisation of an avian influenza A virus isolated from a human—is an intermediate host necessary for the emergence of pandemic influenza viruses? Arch Virol. 1998;143:781–787. doi: 10.1007/s007050050329. [DOI] [PubMed] [Google Scholar]
- Baranovich T, Burnham AJ, Marathe BM, Armstrong J, Guan Y, Shu Y, Peiris JM, Webby RJ, Webster RG, Govorkova EA. The neuraminidase inhibitor oseltamivir is effective against A/Anhui/1/2013 (H7N9) influenza virus in a mouse model of acute respiratory distress syndrome. J Infect Dis. 2013 doi: 10.1093/infdis/jit554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belser JA, Blixt O, Chen LM, Pappas C, Maines TR, Van Hoeven N, Donis R, Busch J, McBride R, Paulson JC, et al. Contemporary North American influenza H7 viruses possess human receptor specificity: Implications for virus transmissibility. Proc Natl Acad Sci USA. 2008;105:7558–7563. doi: 10.1073/pnas.0801259105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belser JA, Bridges CB, Katz JM, Tumpey TM. Past, present, and possible future human infection with influenza virus A subtype H7. Emerg Infect Dis. 2009a;15:859–865. doi: 10.3201/eid1506.090072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belser JA, Davis CT, Balish A, Edwards LE, Zeng H, Maines TR, Gustin KM, Martinez IL, Fasce R, Cox NJ, et al. Pathogenesis, transmissibility, and ocular tropism of a highly pathogenic avian influenza A (H7N3) virus associated with human conjunctivitis. J Virol. 2013a;87:5746–5754. doi: 10.1128/JVI.00154-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belser JA, Gustin KM, Maines TR, Pantin-Jackwood MJ, Katz JM, Tumpey TM. Influenza virus respiratory infection and transmission following ocular inoculation in ferrets. PLoS Pathog. 2012a;8:e1002569. doi: 10.1371/journal.ppat.1002569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belser JA, Gustin KM, Pearce MB, Maines TR, Zeng H, Pappas C, Sun X, Carney PJ, Villanueva JM, Stevens J, et al. Pathogenesis and transmission of avian influenza A (H7N9) virus in ferrets and mice. Nature. 2013b;501:556–559. doi: 10.1038/nature12391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belser JA, Lu X, Maines TR, Smith C, Li Y, Donis RO, Katz JM, Tumpey TM. Pathogenesis of avian influenza (H7) virus infection in mice and ferrets: enhanced virulence of Eurasian H7N7 viruses isolated from humans. J Virol. 2007;81:11139–11147. doi: 10.1128/JVI.01235-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belser JA, Maines TR, Gustin KM, Katz JM, Tumpey TM. Kinetics of viral replication and induction of host responses in ferrets differs between ocular and intranasal routes of inoculation. Virology. 2013c;438:56–60. doi: 10.1016/j.virol.2013.01.012. [DOI] [PubMed] [Google Scholar]
- Belser JA, Sleeman K, Pearce MB, Katz JM, Gubareva LV, Tumpey TM. Oseltamivir inhibits H7 influenza virus replication in mice inoculated by the ocular route. Antimicrob Agents Chemother. 2012b;56:1616–1618. doi: 10.1128/AAC.06101-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belser JA, Szretter KJ, Katz JM, Tumpey TM. Use of animal models to understand the pandemic potential of highly pathogenic avian influenza viruses. Adv Virus Res. 2009b;73:55–97. doi: 10.1016/S0065-3527(09)73002-7. [DOI] [PubMed] [Google Scholar]
- Belser JA, Tumpey TM. H5N1 pathogenesis studies in mammalian models. Virus Res. 2013 doi: 10.1016/j.virusres.2013.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belser JA, Wadford DA, Xu J, Katz JM, Tumpey TM. Ocular infection of mice with influenza A (H7) viruses: a site of primary replication and spread to the respiratory tract. J Virol. 2009c;83:7075–7084. doi: 10.1128/JVI.00535-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berhane Y, Hisanaga T, Kehler H, Neufeld J, Manning L, Argue C, Handel K, Hooper-McGrevy K, Jonas M, Robinson J, et al. Highly pathogenic avian influenza virus A (H7N3) in domestic poultry, Saskatchewan, Canada, 2007. Emerg Infect Dis. 2009;15:1492–1495. doi: 10.3201/eid1509.080231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown IH. Summary of avian influenza activity in Europe, Asia, and Africa, 2006–2009. Avian Dis. 2010;54:187–193. doi: 10.1637/8949-053109-Reg.1. [DOI] [PubMed] [Google Scholar]
- Bulach D, Halpin R, Spiro D, Pomeroy L, Janies D, Boyle DB. Molecular analysis of H7 avian influenza viruses from Australia and New Zealand: genetic diversity and relationships from 1976 to 2007. J Virol. 2010;84:9957–9966. doi: 10.1128/JVI.00930-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CDC. Update: influenza activity—United States, 2003–2004 season. MMWR Morb Mortal Wkly Rep. 2004;53:284–287. [PubMed] [Google Scholar]
- Christensen JP, Doherty PC, Branum KC, Riberdy JM. Profound protection against respiratory challenge with a lethal H7N7 influenza A virus by increasing the magnitude of CD8(+) T-cell memory. J Virol. 2000;74:11690–11696. doi: 10.1128/jvi.74.24.11690-11696.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowling BJ, Jin L, Lau EH, Liao Q, Wu P, Jiang H, Tsang TK, Zheng J, Fang VJ, Chang Z, et al. Comparative epidemiology of human infections with avian influenza A H7N9 and H5N1 viruses in China: a population-based study of laboratory-confirmed cases. Lancet. 2013;382:129–137. doi: 10.1016/S0140-6736(13)61171-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox RJ, Major D, Hauge S, Madhun AS, Brokstad KA, Kuhne M, Smith J, Vogel FR, Zambon M, Haaheim LR, et al. A cell-based H7N1 split influenza virion vaccine confers protection in mouse and ferret challenge models. Influenza Other Respi Viruses. 2009;3:107–117. doi: 10.1111/j.1750-2659.2009.00082.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jong RM, Stockhofe-Zurwieden N, Verheij ES, de Boer-Luijtze EA, Ruiter SJ, de Leeuw OS, Cornelissen LA. Rapid emergence of a virulent PB2 E627K variant during adaptation of highly pathogenic avian influenza H7N7 virus to mice. Virol J. 2013;10:276. doi: 10.1186/1743-422X-10-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wit E, Munster VJ, Spronken MI, Bestebroer TM, Baas C, Beyer WE, Rimmelzwaan GF, Osterhaus AD, Fouchier RA. Protection of mice against lethal infection with highly pathogenic H7N7 influenza A virus by using a recombinant low-pathogenicity vaccine strain. J Virol. 2005;79:12401–12407. doi: 10.1128/JVI.79.19.12401-12407.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wit E, Munster VJ, van Riel D, Beyer WE, Rimmelzwaan GF, Kuiken T, Osterhaus AD, Fouchier RA. Molecular determinants of adaptation of highly pathogenic avian influenza H7N7 viruses to efficient replication in the human host. J Virol. 2010;84:1597–1606. doi: 10.1128/JVI.01783-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Trani L, Porru S, Bonfanti L, Cordioli P, Cesana BM, Boni A, Di Carlo AS, Arici C, Donatelli I, Tomao P, et al. Serosurvey against H5 and H7 avian influenza viruses in Italian poultry workers. Avian Dis. 2012;56:1068–1071. doi: 10.1637/10184-041012-ResNote.1. [DOI] [PubMed] [Google Scholar]
- Dudley JP. Public health and epidemiological considerations for avian influenza risk mapping and risk assessment. Ecol Soc. 2008;13:21. [Google Scholar]
- Eames KT, Webb C, Thomas K, Smith J, Salmon R, Temple JM. Assessing the role of contact tracing in a suspected H7N2 influenza a outbreak in humans in Wales. BMC Infect Dis. 2010;10:141. doi: 10.1186/1471-2334-10-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekiert DC, Friesen RH, Bhabha G, Kwaks T, Jongeneelen M, Yu W, Ophorst C, Cox F, Korse HJ, Brandenburg B, et al. A highly conserved neutralizing epitope on group 2 influenza A viruses. Science. 2011;333:843–850. doi: 10.1126/science.1204839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fouchier RA, Schneeberger PM, Rozendaal FW, Broekman JM, Kemink SA, Munster V, Kuiken T, Rimmelzwaan GF, Schutten M, Van Doornum GJ, et al. Avian influenza A virus (H7N7) associated with human conjunctivitis and a fatal case of acute respiratory distress syndrome. Proc Natl Acad Sci USA. 2004;101:1356–1361. doi: 10.1073/pnas.0308352100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabbard JD, Dlugolenski D, Van Riel D, Marshall N, Galloway SE, Howerth EW, Campbell PJ, Jones C, Johnson S, Byrd-Leotis L, et al. Novel H7N9 influenza virus shows low infectious dose, high growth and efficient contact transmission in the guinea pig model. J Virol. 2013 doi: 10.1128/JVI.02959-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriel G, Abram M, Keiner B, Wagner R, Klenk HD, Stech J. Differential polymerase activity in avian and mammalian cells determines host range of influenza virus. J Virol. 2007;81:9601–9604. doi: 10.1128/JVI.00666-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriel G, Dauber B, Wolff T, Planz O, Klenk HD, Stech J. The viral polymerase mediates adaptation of an avian influenza virus to a mammalian host. Proc Natl Acad Sci USA. 2005;102:18590–18595. doi: 10.1073/pnas.0507415102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriel G, Klingel K, Planz O, Bier K, Herwig A, Sauter M, Klenk HD. Spread of infection and lymphocyte depletion in mice depends on polymerase of influenza virus. Am J Pathol. 2009;175:1178–1186. doi: 10.2353/ajpath.2009.090339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriel G, Nordmann A, Stein DA, Iversen PL, Klenk HD. Morpholino oligomers targeting the PB1 and NP genes enhance the survival of mice infected with highly pathogenic influenza A H7N7 virus. J Gen Virol. 2008;89:939–948. doi: 10.1099/vir.0.83449-0. [DOI] [PubMed] [Google Scholar]
- Gambaryan AS, Matrosovich TY, Philipp J, Munster VJ, Fouchier RA, Cattoli G, Capua I, Krauss SL, Webster RG, Banks J, et al. Receptor-binding profiles of H7 subtype influenza viruses in different host species. J Virol. 2012;86:4370–4379. doi: 10.1128/JVI.06959-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gambaryan AS, Tuzikov AB, Pazynina GV, Desheva JA, Bovin NV, Matrosovich MN, Klimov AI. 6-sulfo sialyl Lewis X is the common receptor determinant recognized by H5, H6, H7 and H9 influenza viruses of terrestrial poultry. Virol J. 2008;5:85. doi: 10.1186/1743-422X-5-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao HN, Lu HZ, Cao B, Du B, Shang H, Gan JH, Lu SH, Yang YD, Fang Q, Shen YZ, et al. Clinical findings in 111 cases of influenza A (H7N9) virus infection. N Engl J Med. 2013a;368:2277–2285. doi: 10.1056/NEJMoa1305584. [DOI] [PubMed] [Google Scholar]
- Gao R, Cao B, Hu Y, Feng Z, Wang D, Hu W, Chen J, Jie Z, Qiu H, Xu K, et al. Human infection with a novel avian-origin influenza A (H7N9) virus. N Engl J Med. 2013b;368:1888–1897. doi: 10.1056/NEJMoa1304459. [DOI] [PubMed] [Google Scholar]
- Garcia-Sastre A. Inhibition of interferon-mediated antiviral responses by influenza A viruses and other negative-strand RNA viruses. Virology. 2001;279:375–384. doi: 10.1006/viro.2000.0756. [DOI] [PubMed] [Google Scholar]
- Gerloff NA, Jones J, Simpson N, Balish A, Elbadry MA, Baghat V, Rusev I, de Mattos CC, de Mattos CA, Zonkle LE, et al. A high diversity of eurasian lineage low pathogenicity avian influenza A viruses circulate among wild birds sampled in Egypt. PLoS One. 2013;8:e68522. doi: 10.1371/journal.pone.0068522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goff PH, Krammer F, Hai R, Seibert CW, Margine I, Garcia-Sastre A, Palese P. Induction of cross-reactive antibodies to novel H7N9 influenza virus by recombinant Newcastle disease virus expressing a North American lineage H7 subtype hemagglutinin. J Virol. 2013;87:8235–8240. doi: 10.1128/JVI.01085-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Reiche AS, Morales-Betoulle ME, Alvarez D, Betoulle JL, Muller ML, Sosa SM, Perez DR. Influenza a viruses from wild birds in Guatemala belong to the North American lineage. PLoS One. 2012;7:e32873. doi: 10.1371/journal.pone.0032873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haasbach E, Reiling SJ, Ehrhardt C, Droebner K, Ruckle A, Hrincius ER, Leban J, Strobl S, Vitt D, Ludwig S, et al. The NF-kappaB inhibitor SC75741 protects mice against highly pathogenic avian influenza A virus. Antiviral Res. 2013;99:336–344. doi: 10.1016/j.antiviral.2013.06.008. [DOI] [PubMed] [Google Scholar]
- Hai R, Schmolke M, Leyva-Grado VH, Thangavel RR, Margine I, Jaffe EL, Krammer F, Solorzano A, Garcia-Sastre A, Palese P, et al. Influenza A(H7N9) virus gains neuraminidase inhibitor resistance without loss of in vivo virulence or transmissibility. Nat Commun. 2013;4:2854. doi: 10.1038/ncomms3854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatta M, Gao P, Halfmann P, Kawaoka Y. Molecular basis for high virulence of Hong Kong H5N1 influenza A viruses. Science. 2001;293:1840–1842. doi: 10.1126/science.1062882. [DOI] [PubMed] [Google Scholar]
- He F, Kumar SR, Syed Khader SM, Tan Y, Prabakaran M, Kwang J. Effective intranasal therapeutics and prophylactics with monoclonal antibody against lethal infection of H7N7 influenza virus. Antiviral Res. 2013;100:207–214. doi: 10.1016/j.antiviral.2013.08.003. [DOI] [PubMed] [Google Scholar]
- Hirst M, Astell CR, Griffith M, Coughlin SM, Moksa M, Zeng T, Smailus DE, Holt RA, Jones S, Marra MA, et al. Novel avian influenza H7N3 strain outbreak, British Columbia. Emerg Infect Dis. 2004;10:2192–2195. doi: 10.3201/eid1012.040743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horimoto T, Kawaoka Y. Reverse genetics provides direct evidence for a correlation of hemagglutinin cleavability and virulence of an avian influenza A virus. J Virol. 1994;68:3120–3128. doi: 10.1128/jvi.68.5.3120-3128.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horimoto T, Kawaoka Y. Pandemic threat posed by avian influenza A viruses. Clin Microbiol Rev. 2001;14:129–149. doi: 10.1128/CMR.14.1.129-149.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hovden AO, Brokstad KA, Major D, Wood J, Haaheim LR, Cox RJ. A pilot study of the immune response to whole inactivated avian influenza H7N1 virus vaccine in mice. Influenza Other Respi Viruses. 2009;3:21–28. doi: 10.1111/j.1750-2659.2009.00075.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilyushina NA, Govorkova EA, Russell CJ, Hoffmann E, Webster RG. Contribution of H7 haemagglutinin to amantadine resistance and infectivity of influenza virus. J Gen Virol. 2007;88:1266–1274. doi: 10.1099/vir.0.82256-0. [DOI] [PubMed] [Google Scholar]
- Jadhao SJ, Achenbach J, Swayne DE, Donis R, Cox N, Matsuoka Y. Development of Eurasian H7N7/PR8 high growth reassortant virus for clinical evaluation as an inactivated pandemic influenza vaccine. Vaccine. 2008;26:1742–1750. doi: 10.1016/j.vaccine.2008.01.036. [DOI] [PubMed] [Google Scholar]
- Jones YL, Swayne DE. Comparative pathobiology of low and high pathogenicity H7N3 Chilean avian influenza viruses in chickens. Avian Dis. 2004;48:119–128. doi: 10.1637/7080. [DOI] [PubMed] [Google Scholar]
- Jonges M, Bataille A, Enserink R, Meijer A, Fouchier RA, Stegeman A, Koch G, Koopmans M. Comparative analysis of avian influenza virus diversity in poultry and humans during a highly pathogenic avian influenza A (H7N7) virus outbreak. J Virol. 2011;85:10598–10604. doi: 10.1128/JVI.05369-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph T, McAuliffe J, Lu B, Jin H, Kemble G, Subbarao K. Evaluation of replication and pathogenicity of avian influenza a H7 subtype viruses in a mouse model. J Virol. 2007;81:10558–10566. doi: 10.1128/JVI.00970-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph T, McAuliffe J, Lu B, Vogel L, Swayne D, Jin H, Kemble G, Subbarao K. A live attenuated cold-adapted influenza A H7N3 virus vaccine provides protection against homologous and heterologous H7 viruses in mice and ferrets. Virology. 2008;378:123–132. doi: 10.1016/j.virol.2008.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaminski MM, Ohnemus A, Cornitescu M, Staeheli P. Plasmacytoid dendritic cells and Toll-like receptor 7-dependent signalling promote efficient protection of mice against highly virulent influenza A virus. J Gen Virol. 2012;93:555–559. doi: 10.1099/vir.0.039065-0. [DOI] [PubMed] [Google Scholar]
- Kapczynski DR, Pantin-Jackwood M, Guzman SG, Ricardez Y, Spackman E, Bertran K, Suarez DL, Swayne DE. Characterization of the 2012 highly pathogenic avian influenza H7N3 virus isolated from poultry in an outbreak in Mexico: pathobiology and vaccine protection. J Virol. 2013;87:9086–9096. doi: 10.1128/JVI.00666-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaoka Y. Equine H7N7 influenza A viruses are highly pathogenic in mice without adaptation: potential use as an animal model. J Virol. 1991;65:3891–3894. doi: 10.1128/jvi.65.7.3891-3894.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochs G, Koerner I, Thiel L, Kothlow S, Kaspers B, Ruggli N, Summerfield A, Pavlovic J, Stech J, Staeheli P. Properties of H7N7 influenza A virus strain SC35M lacking interferon antagonist NS1 in mice and chickens. J Gen Virol. 2007;88:1403–1409. doi: 10.1099/vir.0.82764-0. [DOI] [PubMed] [Google Scholar]
- Koerner I, Kochs G, Kalinke U, Weiss S, Staeheli P. Protective role of beta interferon in host defense against influenza A virus. J Virol. 2007;81:2025–2030. doi: 10.1128/JVI.01718-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koopmans M, Wilbrink B, Conyn M, Natrop G, van der Nat H, Vennema H, Meijer A, van Steenbergen J, Fouchier R, Osterhaus A, et al. Transmission of H7N7 avian influenza A virus to human beings during a large outbreak in commercial poultry farms in the Netherlands. Lancet. 2004;363:587–593. doi: 10.1016/S0140-6736(04)15589-X. [DOI] [PubMed] [Google Scholar]
- Krammer F, Cox RJ. The emergence of H7N9 viruses: a chance to redefine correlates of protection for influenza virus vaccines. Expert Rev Vaccines. 2013;12:1369–1372. doi: 10.1586/14760584.2013.850036. [DOI] [PubMed] [Google Scholar]
- Krammer F, Margine I, Hai R, Flood A, Hirsh A, Tsvetnitsky V, Chen D, Palese P. H3 stalk-based chimeric hemagglutinin influenza virus constructs protect mice from H7N9 challenge. J Virol. 2013 doi: 10.1128/JVI.03183-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreijtz JH, Kroeze EJ, Stittelaar KJ, de Waal L, van Amerongen G, van Trierum S, van Run P, Bestebroer T, Kuiken T, Fouchier RA, et al. Low pathogenic avian influenza A(H7N9) virus causes high mortality in ferrets upon intratracheal challenge: A model to study intervention strategies. Vaccine. 2013 doi: 10.1016/j.vaccine.2013.06.071. [DOI] [PubMed] [Google Scholar]
- Kurtz J, Manvell RJ, Banks J. Avian influenza virus isolated from a woman with conjunctivitis. Lancet. 1996;348:901–902. doi: 10.1016/S0140-6736(05)64783-6. [DOI] [PubMed] [Google Scholar]
- Lam WY, Yeung AC, Chan PK. Apoptosis, cytokine and chemokine induction by non-structural 1 (NS1) proteins encoded by different influenza subtypes. Virol J. 2011;8:554. doi: 10.1186/1743-422X-8-554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang G, Gagnon A, Geraci JR. Isolation of an influenza A virus from seals. Arch Virol. 1981;68:189–195. doi: 10.1007/BF01314571. [DOI] [PubMed] [Google Scholar]
- Lee CW, Lee YJ, Senne DA, Suarez DL. Pathogenic potential of North American H7N2 avian influenza virus: a mutagenesis study using reverse genetics. Virology. 2006;353:388–395. doi: 10.1016/j.virol.2006.06.003. [DOI] [PubMed] [Google Scholar]
- Legastelois I, Garcia-Sastre A, Palese P, Tumpey TM, Maines TR, Katz JM, Vogel FR, Moste C. Preparation of genetically engineered A/H5N1 and A/H7N1 pandemic vaccine viruses by reverse genetics in a mixture of Vero and chicken embryo cells. Influenza Other Respi Viruses. 2007;1:95–104. doi: 10.1111/j.1750-2659.2007.00015.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Zhou L, Zhou M, Chen Z, Li F, Wu H, Xiang N, Chen E, Tang F, Wang D, et al. Preliminary report: epidemiology of the avian influenza a (H7N9) outbreak in China. N Engl J Med 2013 [Google Scholar]
- Li Y, Li C, Liu L, Wang H, Wang C, Tian G, Webster RG, Yu K, Chen H. Characterization of an avian influenza virus of subtype H7N2 isolated from chickens in northern China. Virus Genes. 2006;33:117–122. doi: 10.1007/s11262-005-0042-8. [DOI] [PubMed] [Google Scholar]
- Lopez-Martinez I, Balish A, Barrera-Badillo G, Jones J, Nunez-Garcia TE, Jang Y, Aparicio-Antonio R, Azziz-Baumgartner E, Belser JA, Ramirez-Gonzalez JE, et al. Highly pathogenic avian influenza A(H7N3) virus in poultry workers, Mexico, 2012. Emerg Infect Dis. 2013;19 doi: 10.3201/eid1909.130087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv H, Han J, Zhang P, Lu Y, Wen D, Cai J, Liu S, Sun J, Yu Z, Zhang H, et al. Mild illness in avian influenza A(H7N9) virus-infected poultry worker, Huzhou, China, April 2013. Emerg Infect Dis. 2013;19 doi: 10.3201/eid1911.130717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maines TR, Chen LM, Matsuoka Y, Chen H, Rowe T, Ortin J, Falcon A, Nguyen TH, le Mai Q, Sedyaningsih ER, et al. Lack of transmission of H5N1 avian-human reassortant influenza viruses in a ferret model. Proc Natl Acad Sci U S A. 2006;103:12121–12126. doi: 10.1073/pnas.0605134103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maines TR, Chen LM, Van Hoeven N, Tumpey TM, Blixt O, Belser JA, Gustin KM, Pearce MB, Pappas C, Stevens J, et al. Effect of receptor binding domain mutations on receptor binding and transmissibility of avian influenza H5N1 viruses. Virology. 2011;413:139–147. doi: 10.1016/j.virol.2011.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maines TR, Jayaraman A, Belser JA, Wadford DA, Pappas C, Zeng H, Gustin KM, Pearce MB, Viswanathan K, Shriver ZH, et al. Transmission and pathogenesis of swine-origin 2009 A(H1N1) influenza viruses in ferrets and mice. Science. 2009;325:484–487. doi: 10.1126/science.1177238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maines TR, Lu XH, Erb SM, Edwards L, Guarner J, Greer PW, Nguyen DC, Szretter KJ, Chen LM, Thawatsupha P, et al. Avian influenza (H5N1) viruses isolated from humans in Asia in 2004 exhibit increased virulence in mammals. J Virol. 2005;79:11788–11800. doi: 10.1128/JVI.79.18.11788-11800.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancini DA, Mendonca RM, Pereira AS, Kawamoto AH, Vannucchi CI, Pinto JR, Mori E, Mancini Filho J. Influenza viruses in adult dogs raised in rural and urban areas in the state of Sao Paulo, Brazil. Rev Inst Med Trop Sao Paulo. 2012;54:311–314. doi: 10.1590/s0036-46652012000600004. [DOI] [PubMed] [Google Scholar]
- Manz B, Gotz V, Wunderlich K, Eisel J, Kirchmair J, Stech J, Stech O, Chase G, Frank R, Schwemmle M. Disruption of the viral polymerase complex assembly as a novel approach to attenuate influenza A virus. J Biol Chem. 2011;286:8414–8424. doi: 10.1074/jbc.M110.205534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margine I, Krammer F, Hai R, Heaton NS, Tan GS, Andrews SA, Runstadler JA, Wilson PC, Albrecht RA, Garcia-Sastre A, et al. Hemagglutinin Stalk-Based Universal Vaccine Constructs Protect against Group 2 Influenza A Viruses. J Virol. 2013;87:10435–10446. doi: 10.1128/JVI.01715-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matrosovich M, Tuzikov A, Bovin N, Gambaryan A, Klimov A, Castrucci MR, Donatelli I, Kawaoka Y. Early alterations of the receptor-binding properties of H1, H2, and H3 avian influenza virus hemagglutinins after their introduction into mammals. J Virol. 2000;74:8502–8512. doi: 10.1128/jvi.74.18.8502-8512.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurer-Stroh S, Lee RT, Gunalan V, Eisenhaber F. The highly pathogenic H7N3 avian influenza strain from July 2012 in Mexico acquired an extended cleavage site through recombination with host 28S rRNA. Virol J. 2013;10:139. doi: 10.1186/1743-422X-10-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Max V, Herrera J, Moreira R, Rojas H. Avian influenza in Chile: a successful experience. Avian Dis. 2007;51:363–365. doi: 10.1637/7631-042806R1.1. [DOI] [PubMed] [Google Scholar]
- Min JY, Vogel L, Matsuoka Y, Lu B, Swayne D, Jin H, Kemble G, Subbarao K. A live attenuated H7N7 candidate vaccine virus induces neutralizing antibody that confers protection from challenge in mice, ferrets, and monkeys. J Virol. 2010;84:11950–11960. doi: 10.1128/JVI.01305-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mok CK, Lee HH, Chan MC, Sia SF, Lestra M, Nicholls JM, Zhu H, Guan Y, Peiris JM. Pathogenicity of the novel A/H7N9 influenza virus in mice. MBio. 2013;4 doi: 10.1128/mBio.00362-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mok CK, Lee HH, Lestra M, Nicholls JM, Chan CW, Sia SF, Zhu H, Poon LL, Guan Y, Peiris JM. Amino-acid substitutions in polymerase basic protein 2 gene contributes to the pathogenicity of the novel A/H7N9 influenza virus in mammalian hosts. J Virol. 2014 doi: 10.1128/JVI.02740-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan O, Kuhne M, Nair P, Verlander NQ, Preece R, McDougal M, Zambon M, Reacher M. Personal protective equipment and risk for avian influenza (H7N3) Emerg Infect Dis. 2009;15:59–62. doi: 10.3201/eid1501.070660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munster VJ, de Wit E, van Riel D, Beyer WE, Rimmelzwaan GF, Osterhaus AD, Kuiken T, Fouchier RA. The molecular basis of the pathogenicity of the Dutch highly pathogenic human influenza A H7N7 viruses. J Infect Dis. 2007;196:258–265. doi: 10.1086/518792. [DOI] [PubMed] [Google Scholar]
- Myers KP, Setterquist SF, Capuano AW, Gray GC. Infection due to 3 avian influenza subtypes in United States veterinarians. Clin Infect Dis. 2007;45:4–9. doi: 10.1086/518579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nacken W, Ehrhardt C, Ludwig S. Small molecule inhibitors of the c-Jun N-terminal kinase (JNK) possess antiviral activity against highly pathogenic avian and human pandemic influenza A viruses. Biol Chem. 2012;393:525–534. doi: 10.1515/hsz-2011-0270. [DOI] [PubMed] [Google Scholar]
- Nguyen-Van-Tam JS, Nair P, Acheson P, Baker A, Barker M, Bracebridge S, Croft J, Ellis J, Gelletlie R, Gent N, et al. Outbreak of low pathogenicity H7N3 avian influenza in UK, including associated case of human conjunctivitis. Euro Surveill. 2006;11(E060504):2. doi: 10.2807/esw.11.18.02952-en. [DOI] [PubMed] [Google Scholar]
- Ostrowsky B, Huang A, Terry W, Anton D, Brunagel B, Traynor L, Abid S, Johnson G, Kacica M, Katz J, et al. Low pathogenic avian influenza A (H7N2) virus infection in immunocompromised adult, New York, USA, 2003. Emerg Infect Dis. 2012;18:1128–1131. doi: 10.3201/eid1807.111913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pappas C, Matsuoka Y, Swayne DE, Donis RO. Development and evaluation of an Influenza virus subtype H7N2 vaccine candidate for pandemic preparedness. Clin Vaccine Immunol. 2007;14:1425–1432. doi: 10.1128/CVI.00174-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasick J, Berhane Y, Hisanaga T, Kehler H, Hooper-McGrevy K, Handel K, Neufeld J, Argue C, Leighton F. Diagnostic test results and pathology associated with the 2007 Canadian H7N3 highly pathogenic avian influenza outbreak. Avian Dis. 2010;54:213–219. doi: 10.1637/8822-040209-Reg.1. [DOI] [PubMed] [Google Scholar]
- Pasick J, Handel K, Robinson J, Copps J, Ridd D, Hills K, Kehler H, Cottam-Birt C, Neufeld J, Berhane Y, et al. Intersegmental recombination between the haemagglutinin and matrix genes was responsible for the emergence of a highly pathogenic H7N3 avian influenza virus in British Columbia. J Gen Virol. 2005;86:727–731. doi: 10.1099/vir.0.80478-0. [DOI] [PubMed] [Google Scholar]
- Pasick J, Pedersen J, Hernandez MS. Avian influenza in North America, 2009-2011. Avian Dis. 2012;56:845–848. doi: 10.1637/10206-041512-Reg.1. [DOI] [PubMed] [Google Scholar]
- Peiris M, Yuen KY, Leung CW, Chan KH, Ip PL, Lai RW, Orr WK, Shortridge KF. Human infection with influenza H9N2. Lancet. 1999;354:916–917. doi: 10.1016/s0140-6736(99)03311-5. [DOI] [PubMed] [Google Scholar]
- Perez-Ramirez E, Gerrikagoitia X, Barral M, Hofle U. Detection of low pathogenic avian influenza viruses in wild birds in Castilla-La Mancha (south central Spain) Vet Microbiol. 2010;146:200–208. doi: 10.1016/j.vetmic.2010.05.008. [DOI] [PubMed] [Google Scholar]
- ProMED-mail. Avian Influenza (90): ITALY (EMILIA-ROMAGNA) Poultry, HPAI H7N7, Human, Public health. ProMED mail; 2013. [Google Scholar]
- Puzelli S, Di Trani L, Fabiani C, Campitelli L, De Marco MA, Capua I, Aguilera JF, Zambon M, Donatelli I. Serological analysis of serum samples from humans exposed to avian H7 influenza viruses in Italy between 1999 and 2003. J Infect Dis. 2005;192:1318–1322. doi: 10.1086/444390. [DOI] [PubMed] [Google Scholar]
- Qi X, Qian YH, Bao CJ, Guo XL, Cui LB, Tang FY, Ji H, Huang Y, Cai PQ, Lu B, et al. Probable person to person transmission of novel avian influenza A (H7N9) virus in Eastern China, 2013: epidemiological investigation. BMJ. 2013;347:f4752. doi: 10.1136/bmj.f4752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajesh Kumar S, Syed Khader SM, Kiener TK, Szyporta M, Kwang J. Intranasal immunization of baculovirus displayed hemagglutinin confers complete protection against mouse adapted highly pathogenic H7N7 reassortant influenza virus. PLoS One. 2013;8:e63856. doi: 10.1371/journal.pone.0063856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard M, Schrauwen EJ, de Graaf M, Bestebroer TM, Spronken MI, van Boheemen S, de Meulder D, Lexmond P, Linster M, Herfst S, et al. Limited airborne transmission of H7N9 influenza A virus between ferrets. Nature. 2013;501:560–563. doi: 10.1038/nature12476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigoni M, Shinya K, Toffan A, Milani A, Bettini F, Kawaoka Y, Cattoli G, Capua I. Pneumo- and neurotropism of avian origin Italian highly pathogenic avian influenza H7N1 isolates in experimentally infected mice. Virology. 2007;364:28–35. doi: 10.1016/j.virol.2007.02.031. [DOI] [PubMed] [Google Scholar]
- Rigoni M, Toffan A, Viale E, Mancin M, Cilloni F, Bertoli E, Salomoni A, Marciano S, Milani A, Zecchin B, et al. The mouse model is suitable for the study of viral factors governing transmission and pathogenesis of highly pathogenic avian influenza (HPAI) viruses in mammals. Vet Res. 2010;41:66. doi: 10.1051/vetres/2010038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salomon R, Franks J, Govorkova EA, Ilyushina NA, Yen HL, Hulse-Post DJ, Humberd J, Trichet M, Rehg JE, Webby RJ, et al. The polymerase complex genes contribute to the high virulence of the human H5N1 influenza virus isolate A/Vietnam/1203/04. J Exp Med. 2006;203:689–697. doi: 10.1084/jem.20051938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheiblauer H, Kendal AP, Rott R. Pathogenicity of influenza A/Seal/Mass/1/80 virus mutants for mammalian species. Arch Virol. 1995;140:341–348. doi: 10.1007/BF01309867. [DOI] [PubMed] [Google Scholar]
- Senne DA. Avian influenza in North and South America, 2002–2005. Avian Dis. 2007;51:167–173. doi: 10.1637/7621-042606R1.1. [DOI] [PubMed] [Google Scholar]
- Senne DA. Avian influenza in North and South America, the Caribbean, and Australia, 2006–2008. Avian Dis. 2010;54:179–186. doi: 10.1637/8921-050809-Review.1. [DOI] [PubMed] [Google Scholar]
- Senne DA, Panigrahy B, Kawaoka Y, Pearson JE, Suss J, Lipkind M, Kida H, Webster RG. Survey of the hemagglutinin (HA) cleavage site sequence of H5 and H7 avian influenza viruses: amino acid sequence at the HA cleavage site as a marker of pathogenicity potential. Avian Dis. 1996;40:425–437. [PubMed] [Google Scholar]
- Senne DA, Suarez DL, Pedersen JC, Panigrahy B. Molecular and biological characteristics of H5 and H7 avian influenza viruses in live-bird markets of the northeastern United States, 1994–2001. Avian Dis. 2003;47:898–904. doi: 10.1637/0005-2086-47.s3.898. [DOI] [PubMed] [Google Scholar]
- Shinya K, Suto A, Kawakami M, Sakamoto H, Umemura T, Kawaoka Y, Kasai N, Ito T. Neurovirulence of H7N7 influenza A virus: brain stem encephalitis accompanied with aspiration pneumonia in mice. Arch Virol. 2005;150:1653–1660. doi: 10.1007/s00705-005-0539-4. [DOI] [PubMed] [Google Scholar]
- Shinya K, Watanabe S, Ito T, Kasai N, Kawaoka Y. Adaptation of an H7N7 equine influenza A virus in mice. J Gen Virol. 2007;88:547–553. doi: 10.1099/vir.0.82411-0. [DOI] [PubMed] [Google Scholar]
- Sleeman K, Guo Z, Barnes J, Shaw M, Stevens J, Gubareva LV. R292K substitution and drug susceptibility of influenza A(H7N9) viruses. Emerg Infect Dis. 2013;19:1521–1524. doi: 10.3201/eid1909.130724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith GE, Flyer DC, Raghunandan R, Liu Y, Wei Z, Wu Y, Kpamegan E, Courbron D, Fries LF, 3rd, Glenn GM. Development of influenza H7N9 virus like particle (VLP) vaccine: Homologous A/Anhui/1/2013 (H7N9) protection and heterologous A/chicken/Jalisco/CPA1/2012 (H7N3) cross-protection in vaccinated mice challenged with H7N9 virus. Vaccine. 2013;31:4305–4313. doi: 10.1016/j.vaccine.2013.07.043. [DOI] [PubMed] [Google Scholar]
- Song H, Wan H, Araya Y, Perez DR. Partial direct contact transmission in ferrets of a mallard H7N3 influenza virus with typical avian-like receptor specificity. Virol J. 2009;6:126. doi: 10.1186/1743-422X-6-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava B, Blazejewska P, Hessmann M, Bruder D, Geffers R, Mauel S, Gruber AD, Schughart K. Host genetic background strongly influences the response to influenza a virus infections. PLoS One. 2009;4:e4857. doi: 10.1371/journal.pone.0004857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suarez DL, Senne DA, Banks J, Brown IH, Essen SC, Lee CW, Manvell RJ, Mathieu-Benson C, Moreno V, Pedersen JC, et al. Recombination resulting in virulence shift in avian influenza outbreak, Chile. Emerg Infect Dis. 2004;10:693–699. doi: 10.3201/eid1004.030396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subbarao EK, London W, Murphy BR. A single amino acid in the PB2 gene of influenza A virus is a determinant of host range. J Virol. 1993;67:1761–1764. doi: 10.1128/jvi.67.4.1761-1764.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suguitan AL, Jr, Matsuoka Y, Lau YF, Santos CP, Vogel L, Cheng LI, Orandle M, Subbarao K. The multibasic cleavage site of the hemagglutinin of highly pathogenic A/Vietnam/1203/2004 (H5N1) avian influenza virus acts as a virulence factor in a host-specific manner in mammals. J Virol. 2012;86:2706–2714. doi: 10.1128/JVI.05546-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szecsi J, Boson B, Johnsson P, Dupeyrot-Lacas P, Matrosovich M, Klenk HD, Klatzmann D, Volchkov V, Cosset FL. Induction of neutralising antibodies by virus-like particles harbouring surface proteins from highly pathogenic H5N1 and H7N1 influenza viruses. Virol J. 2006;3:70. doi: 10.1186/1743-422X-3-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talaat KR, Karron RA, Callahan KA, Luke CJ, DiLorenzo SC, Chen GL, Lamirande EW, Jin H, Coelingh KL, Murphy BR, et al. A live attenuated H7N3 influenza virus vaccine is well tolerated and immunogenic in a Phase I trial in healthy adults. Vaccine. 2009;27:3744–3753. doi: 10.1016/j.vaccine.2009.03.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tharakaraman K, Jayaraman A, Raman R, Viswanathan K, Stebbins NW, Johnson D, Shriver Z, Sasisekharan V, Sasisekharan R. Glycan receptor binding of the influenza A virus H7N9 hemagglutinin. Cell. 2013;153:1486–1493. doi: 10.1016/j.cell.2013.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timoney PJ. Equine influenza. Comp Immunol Microbiol Infect Dis. 1996;19:205–211. doi: 10.1016/0147-9571(96)00006-9. [DOI] [PubMed] [Google Scholar]
- Tretyakova I, Pearce MB, Florese R, Tumpey TM, Pushko P. Intranasal vaccination with H5, H7 and H9 hemagglutinins co-localized in a virus-like particle protects ferrets from multiple avian influenza viruses. Virology. 2013;442:67–73. doi: 10.1016/j.virol.2013.03.027. [DOI] [PubMed] [Google Scholar]
- Tumpey TM, Szretter KJ, Van Hoeven N, Katz JM, Kochs G, Haller O, Garcia-Sastre A, Staeheli P. The Mx1 gene protects mice against the pandemic 1918 and highly lethal human H5N1 influenza viruses. J Virol. 2007;81:10818–10821. doi: 10.1128/JVI.01116-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tweed SA, Skowronski DM, David ST, Larder A, Petric M, Lees W, Li Y, Katz J, Krajden M, Tellier R, et al. Human illness from avian influenza H7N3, British Columbia. Emerg Infect Dis. 2004;10:2196–2199. doi: 10.3201/eid1012.040961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T, Kiso M, Fukuyama S, Nakajima N, Imai M, Yamada S, Murakami S, Yamayoshi S, Iwatsuki-Horimoto K, Sakoda Y, et al. Characterization of H7N9 influenza A viruses isolated from humans. Nature. 2013;501:551–555. doi: 10.1038/nature12392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster RG. Are equine 1 influenza viruses still present in horses? Equine Vet J. 1993;25:537–538. doi: 10.1111/j.2042-3306.1993.tb03009.x. [DOI] [PubMed] [Google Scholar]
- Webster RG, Geraci J, Petursson G, Skirnisson K. Conjunctivitis in human beings caused by influenza A virus of seals. N Engl J Med. 1981;304:911. doi: 10.1056/NEJM198104093041515. [DOI] [PubMed] [Google Scholar]
- Whiteley A, Major D, Legastelois I, Campitelli L, Donatelli I, Thompson CI, Zambon MC, Wood JM, Barclay WS. Generation of candidate human influenza vaccine strains in cell culture—rehearsing the European response to an H7N1 pandemic threat. Influenza Other Respi Viruses. 2007;1:157–166. doi: 10.1111/j.1750-2659.2007.00022.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO. Avian influenza A (H10N8) 2013. [Google Scholar]
- Xiong X, Martin SR, Haire LF, Wharton SA, Daniels RS, Bennett MS, McCauley JW, Collins PJ, Walker PA, Skehel JJ, et al. Receptor binding by an H7N9 influenza virus from humans. Nature. 2013;499:496–499. doi: 10.1038/nature12372. [DOI] [PubMed] [Google Scholar]
- Xu L, Bao L, Deng W, Dong L, Zhu H, Chen T, Lv Q, Li F, Yuan J, Xiang Z, et al. Novel avian-origin human influenza A(H7N9) can be transmitted between ferrets via respiratory droplets. J Infect Dis. 2013a doi: 10.1093/infdis/jit474. [DOI] [PubMed] [Google Scholar]
- Xu L, Bao L, Deng W, Zhu H, Chen T, Lv Q, Li F, Yuan J, Xiang Z, Gao K, et al. The mouse and ferret models for studying the novel avian-origin human influenza A (H7N9) virus. Virol J. 2013b;10:253. doi: 10.1186/1743-422X-10-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Q, Chen Z, Cheng X, Xu L, Jin H. Evaluation of Live Attenuated H7N3 and H7N7 Vaccine Viruses for Their Receptor Binding Preferences, Immunogenicity in Ferrets and Cross Reactivity to the Novel H7N9 Virus. PLoS One. 2013c;8:e76884. doi: 10.1371/journal.pone.0076884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamayoshi S, Yamada S, Fukuyama S, Murakami S, Zhao D, Uraki R, Watanabe T, Tomita Y, Macken C, Neumann G, et al. Virulence-Affecting Amino Acid Changes in the PA Protein of H7N9 Influenza A Viruses. J Virol. 2013 doi: 10.1128/JVI.03155-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Carney PJ, Donis RO, Stevens J. Structure and receptor complexes of the hemagglutinin from a highly pathogenic H7N7 influenza virus. J Virol. 2012;86:8645–8652. doi: 10.1128/JVI.00281-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Chen LM, Carney PJ, Donis RO, Stevens J. Structures of receptor complexes of a North American H7N2 influenza hemagglutinin with a loop deletion in the receptor binding site. PLoS Pathog. 2010;6:e1001081. doi: 10.1371/journal.ppat.1001081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J, Zhang L, Kan X, Jiang L, Yang J, Guo Z, Ren Q. Origin and molecular characteristics of a novel 2013 avian influenza A(H6N1) virus causing human infection in Taiwan. Clin Infect Dis. 2013;57:1367–1368. doi: 10.1093/cid/cit479. [DOI] [PubMed] [Google Scholar]
- Zhang H, Li X, Guo J, Li L, Chang C, Li Y, Bian C, Xu K, Chen H, Sun B. The PB2E627K mutation contributes to the high polymerase activity and enhanced replication of H7N9 influenza virus. J Gen Virol. 2014 doi: 10.1099/vir.0.061721-0. [DOI] [PubMed] [Google Scholar]
- Zhang Q, Shi J, Deng G, Guo J, Zeng X, He X, Kong H, Gu C, Li X, Liu J, et al. H7N9 influenza viruses are transmissible in ferrets by respiratory droplet. Science. 2013;341:410–414. doi: 10.1126/science.1240532. [DOI] [PubMed] [Google Scholar]
- Zhong W, Liu F, Dong L, Lu X, Hancock K, Reinherz EL, Katz JM, Sambhara S. Significant impact of sequence variations in the nucleoprotein on CD8 T cell-mediated cross-protection against influenza A virus infections. PLoS One. 2010;5:e10583. doi: 10.1371/journal.pone.0010583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou B, Zhong N, Guan Y. Treatment with convalescent plasma for influenza A (H5N1) infection. N Engl J Med. 2007;357:1450–1451. doi: 10.1056/NEJMc070359. [DOI] [PubMed] [Google Scholar]
- Zhu H, Wang D, Kelvin DJ, Li L, Zheng Z, Yoon SW, Wong SS, Farooqui A, Wang J, Banner D, et al. Infectivity, transmission, and pathology of human-isolated H7N9 influenza virus in ferrets and pigs. Science. 2013;341:183–186. doi: 10.1126/science.1239844. [DOI] [PubMed] [Google Scholar]


