Abstract
Background
The genetic mechanisms for families who meet the clinical criteria for Lynch syndrome (LS) but do not carry pathogenic variants in the mismatch repair (MMR) genes are still undetermined. We aimed to study the potential contribution of genes other than MMR genes to the biological and clinical characteristics of Norwegian families fulfilling Amsterdam (AMS) criteria or revised Bethesda guidelines.
Methods
The Hereditary Cancer Biobank of the Norwegian Radium Hospital was interrogated to identify individuals with a high risk of developing colorectal cancer (CRC) for whom no pathogenic variants in MMR genes had been found in routine diagnostic DNA sequencing. Forty-four cancer susceptibility genes were selected and analyzed by using our in-house designed TruSeq amplicon-based assay for targeted sequencing. RNA splicing- and protein-dedicated in silico analyses were performed for all variants of unknown significance (VUS). Variants predicted as likely to affect splicing were experimentally analyzed by resorting to minigene assays.
Results
We identified a patient who met the revised Bethesda guidelines and carried a likely pathogenic variant in CHEK2 (c.470 T > C, p.I157T). In addition, 25 unique VUS were identified in 18 individuals, of which 2 exonic variants (MAP3K1 c.764A > G and NOTCH3 c.5854G >A) were analyzed in the minigene splicing assay and found not to have an effect on RNA splicing.
Conclusions
Among high-risk CRC patients that fulfill the AMS criteria or revised Bethesda guidelines, targeted gene sequencing identified likely pathogenic variant and VUS in other genes than the MMR genes (CHEK2, NOTCH3 and MAP3K1). Our study suggests that the analysis of genes currently excluded from routine molecular diagnostic screens may confer cancer susceptibility.
Electronic supplementary material
The online version of this article (10.1186/s12881-018-0533-9) contains supplementary material, which is available to authorized users.
Keywords: Lynch syndrome, Gene panel testing, CHEK2, RNA splicing mutations
Background
Heredity represents a major cause of colorectal cancer (CRC) with at least 20% of the cases estimated to develop due to genetic factors and about 5% being linked to inherited variants in cancer-predisposing genes [1–4]. Currently, patients with CRC are referred to germline mismatch repair (MMR) testing based on the identification of high-risk phenotypic features (i.e. early age of onset, family history, clinical criteria), but beyond microsatellite instability (MSI) and MMR immunohistochemistry (IHC) testing for Lynch syndrome (LS), no systematic approach to hereditary risk assessment exists [5].
LS is caused by a defective MMR system due to presence of germline defects in at least one of the MMR genes, MLH1, MSH2, MSH6, PMS2 or to deletions of the 3′ portion of the EPCAM gene [6]. LS is clinically classified according to the Amsterdam (AMS) criteria and/or the Bethesda guidelines, both relying in clinical information and family history. The Bethesda guidelines also take into account the MSI signature characteristic of MMR-deficient tumors [7–10]. LS patients have an increased lifetime risk of CRC (70–80%), endometrial cancer (50–60%), stomach cancer (13–19%), ovarian cancer (9–14%), cancers of the small intestine, the biliary tract and brain as well as carcinoma of the ureters and renal pelvis [11].
However, a high proportion of cases who meet the clinical criteria for LS (~ 60%) do not carry pathogenic variants in the MMR genes and have been reported as familial colorectal cancer type X (FCCTX) or Lynch-like syndrome (LLS) according to their MSI status [12–16]. The genetic mechanisms are undetermined in the majority of these families [14].
DNA sequencing (DNA-seq) studies using multigene panels have reported that as much as ~ 18% of patients diagnosed with CRC below the age of 50 years have pathogenic variants in several genes that are not traditionally associated with CRC (ATM, CHEK2, BRCA1, BRCA2, CDKN2A and PALB2) [5, 17]. Notably, there is a need to determine whether these variants contribute to hereditary CRC risk via the combination of low- and moderate-penetrance susceptibility alleles [5, 17, 18].
Given the high frequency and wide spectrum of pathogenic variants, it has been suggested that genetic counseling and testing with a multigene panel should be considered for all patients with early-onset CRC [17, 19–23]. Importantly, the identification of high-risk CRC patients is a major issue, because morbidity and mortality from CRC and extracolonic cancers in these patients and their relatives can be decreased by early screening and intensive surveillance [19, 24–26].
In an effort to discover inherited genetic variants that influence biological and clinical characteristics of familial CRC developed in unrelated high-risk patients, who previously tested negative for pathogenic variants in MMR genes, we examined 44 cancer associated genes using next generation sequencing (NGS), and applied minigene-based assay to analyze the impact of a subset of genetic variants on RNA splicing.
Methods
Study population
The Hereditary Cancer Biobank of the Norwegian Radium Hospital was used to identify unrelated high-risk CRC individuals from families that fulfilled the AMS criteria or the revised Bethesda guidelines [7–10, 27]. By the standard diagnostic clinical techniques, all study subjects were demonstrated not to carry pathogenic variants or large genomic rearrangements in MMR genes (MLH1, MSH2, MSH6 or PMS2).
Ethical approval for the study was granted by the Norwegian Data Inspectorate and Ethical Review Board (ref 2015/2382). All examined patients signed an informed consent for their participation in the study.
Targeted sequencing
Genomic DNA was isolated from peripheral blood samples and targeted sequencing was carried out using a TrueSeq amplicon based assay v.1.5 on a MiSeq apparatus, as previously described [28, 29]. The 44-gene panel used in this study includes genes associated with cancer predisposition as described in a prior study [28, 29].
Sequencing data analysis
Paired-end sequence reads were aligned to the human reference genome (build GRCh37) using the BWA-mem algorithm (v.0.7.8-r55) [30]. The initial sequence alignments were converted to BAM format and subsequently sorted and indexed with SAMtools (v.1.1) [30]. Genotyping of single nucleotide variants (SNV) and short indels was performed by GATK’s HaplotypeCaller. Filtering of raw genotype calls and assessment of callable regions/loci were done according to GATK’s best practice procedures, as described more detailed previously [28].
Variants were annotated using ANNOVAR (version November 2015) [31] and were queried against a range of variant databases and protein resources, namely dbSNP (build 147) [32], 1000 Genome Project phase3 [33], Exome Aggregation Consortium (ExAC) (http://exac.broadinstitute.org, accessed August 2015) [34], Genome Aggregation Database (gnomAD) (http://gnomad.broadinstitute.org, accessed October 2017) [34], Norwegian Germline Variations Database (http://norgene.no/vcf-miner/, accessed October 2017), ClinVar (May 2016) [35], UniProt Knowledgebase (release March 2016) [36] and the Pfam protein domain database (v29, December 2015) [28, 37].
Nomenclature and classification of genetic variants
The nomenclature guidelines of the Human Genome Variation Society (HGVS) were used to describe the detected genetic variants [38]. The recurrence of the identified variants was established by interrogating four databases (in their latest releases as of November 2016): the Leiden Open Variation Database (LOVD), the Universal Mutation Database (UMD), ClinVar and the Human Gene Mutation Database (HGMD). The variants were classified according to the 5-tier classification system into the following categories: class 5 (pathogenic), class 4 (likely pathogenic), class 3 (uncertain variants or variants of unknown significance, VUS), class 2 (likely not pathogenic) and class 1 (not pathogenic) [3].
In silico analyses of VUS
Two types of bioinformatics methods were used to predict the impact of selected variants on RNA splicing. First, we used MaxEntScan (MES) and SSF-like (SSFL) to predict variant-induced alterations in 3′ and 5′ splice site strength, as described by Houdayer et al. 2012 [39], except that here both algorithms were interrogated by using the integrated software tool Alamut Batch version 1.5, (Interactive Biosoftware, http://www.interactive-biosoftware.com). For prediction of variant-induced impact on exonic splicing regulatory elements (ESR), we resorted to ΔtESRseq- [40], ΔHZei- [41], and SPANR-based [42] as described by Soukarieh et al. [43]. Score differences (Δ) between variant and wild-type (WT) cases were taken as proxies for assessing the probability of a splicing defect. More precisely, we considered that a variant mapping at a splice site was susceptible of negatively impacting exon inclusion if ΔMES≥15% and ΔSSFL≥5% [39], whereas an exonic variant located outside the splice sites was considered as a probable inducer of exon skipping if negative Δ scores (below the thresholds described below) were provided by all the 3 ESR-dedicated in silico tools. We chose the following thresholds: <− 0.5 for ΔtESRseq-, <− 10 for ΔHZei-, and < − 0.5 for SPANR-based scores. In addition, we evaluated the possibility of variant-induced de novo splice sites by taking into consideration local changes in MES and SSFL scores. In this case, we considered that variants located outside the splice sites were susceptible of creating a competing splice site if local MES scores were equal to or greater than those of the corresponding reference splice site for the same exon.
In silico protein impact predictions of missense variants were performed with Align-GVGD (the VUS were predicted as deleterious when the values were from C35 or higher), SIFT, and MAPP using Alamut Batch version 1.4.4 (Interactive Biosoftware) and additionally with PolyPhen-2 and MutationTaster [44–48].
Cell-based minigene splicing assays
In order to determine the impact of selected exonic variants on splicing, we performed functional assays based on the comparative analysis of the splicing pattern of WT and mutant reporter minigenes, as follows. First, genomic regions containing the exon of interest (internal exons only) and at least 150 nucleotides of the flanking introns were amplified by PCR [49] using patients’ DNA as template and primers indicated in Additional file 1: Table S1. Next, representative minigenes were created by inserting the PCR-amplified fragments into a previously linearized pCAS2 vector [43]. All constructs were sequenced to ensure that no unwanted mutations had been introduced into the inserted fragments during PCR or cloning. Then, WT and mutant minigenes were transfected into HeLa cells grown in 12-well plates (at ~ 70% confluence) using the FuGENE 6 transfection reagent (Roche Applied Science). Twenty-four hours later, total RNA was extracted using the NucleoSpin RNA II kit (Macherey Nagel) and, the minigenes’ transcripts were analyzed by semi-quantitative RT-PCR using the OneStep RT-PCR kit (Qiagen), as previously described [43]. The sequences of the RT-PCR primers are shown in Additional file 1: Table S1. Later, RT-PCR products were separated by electrophoresis on 2.5% agarose gel containing EtBr and visualized by exposure to UV light under saturating conditions using the Gel Doc XR image acquisition system (Bio-Rad), followed by gel-purification and Sanger sequencing for proper identification of the minigenes’ transcripts. Finally, splicing events were quantitated by performing equivalent fluorescent RT-PCR reactions followed by capillary electrophoresis on an automated sequencer (Applied Biosystems), and computational analysis by using the GeneMapper v5.0 software (Applied Biosystems).
Results
Clinical characteristics and family history
Upon querying the Hereditary Cancer Biobank of the Norwegian Radium Hospital for cases that fulfill the AMS and/or the revised Bethesda guidelines, we identified 34 unrelated potential high-risk CRC individuals who did not carry pathogenic variants in MMR genes. The median age at first CRC diagnosis was 51.5 years (range: 34–86 years).
Pedigree information showed that 13 (38%) families fulfilled the AMS I and/or II criteria and the revised Bethesda guidelines while 21 (62%) met the revised Bethesda guidelines only (Table 1). Fifteen (44%) patients had tumors with MSI and/or MMR IHC data available, of which 2 (13%) were MSI-high and/or MMR deficient. Clinical, family and tumor data information is detailed in Table 1.
Table 1.
Summary of International Classification of Diseases, 9th Revision (ICD9), gender, age at diagnosis, clinical criteria and tumor molecular characteristics of the familial CRC families
| Patient_ID | Gender | ICD9 diagnosis (age) | AMS criteria | Revised Bethesda | Tumor molecular characteristics |
|---|---|---|---|---|---|
| 3222 | F | CC (54), Hyperplastic polyp (55/61/62/63/65), BC (70) | 0 | Y | MMR IHC proficient |
| 3308 | F | CC (43), BC (51/52) | 0 | Y | MMR IHC proficient |
| 3387 | F | BC (40), OC (70), CC (80) | 0 | Y | MMR IHC proficient |
| 3426 | M | MM (39) | I & II | Y | na |
| 4932 | F | CC (34), EC (40), Hyperplastic polyp (43), BT (46) | I & II | Y | na |
| 5324 | F | M (52), CC (59), SMC (na), BC (72) | 0 | Y | na |
| 6174 | F | Hyperplastic polyp (63/67), BC (65) | I & II | Y | MMR IHC proficient |
| 6977 | F | TC (66) | 0 | Y | MMR IHC proficient |
| 9876 | F | M (45), BC (54) | 0 | Y | na |
| 9998 | F | Hyperplastic polyp (45), CC (45) | II | Y | MMR IHC proficient |
| 10,675 | F | BC (51), Hyperplastic polyp (59), TC (60) | II | Y | na |
| 12,954 | F | Hyperplastic polyp (69), ML (70) | II | Y | na |
| 13,072 | M | Hyperplastic polyp (63/64/65), CC (65/67) | 0 | Y | na |
| 14,930 | F | Hyperplastic polyp (86), CC (86) | 0 | Y | MMR IHC proficient |
| 18,843 | F | BC (44), CC (49), SMC (na) | 0 | Y | na |
| 19,411 | M | PC (70) | 0 | Y | MSH6 IHC deficient |
| 19,673 | F | BC (40/42) | II | Y | na |
| 20,612 | F | Hyperplastic polyp (59/65), EC (70) | 0 | Y | na |
| 21,368 | F | OC (62) | 0 | Y | na |
| 22,295 | F | Hyperplastic polyp (53), M (58) | 0 | Y | na |
| 23,761 | F | Hyperplastic polyp (40/42/44), BC (50) | 0 | Y | na |
| 23,910 | F | M (43), Hyperplastic polyp (49), BC (63), BT (63) | 0 | Y | na |
| 24,140 | F | CC (45/67), BC (56) | 0 | Y | na |
| 24,447 | F | BC (57/66), CC (66) | 0 | Y | MLH1/PMS2 IHC deficient and MSI |
| 11,705 | F | THC (53), KC (53/63) | II | Y | MMR IHC proficient |
| 12,673 | F | OC (23), SMC (36), RC (62) | II | Y | na |
| 13,393 | M | RC (48), CST (58) | I & II | Y | MMR IHC proficient and MSS |
| 14,963 | F | Hyperplastic polyp (69), BC (62) | 0 | Y | na |
| 19609a | F | CC (42), M (44), BC (57) | 0 | Y | MMR IHC proficient |
| 22,953 | F | BC (53) | II | Y | na |
| 24,789 | F | CC (43), RC (65), BC (72) | I & II | Y | MMR IHC proficient and MSS |
| 25,167 | M | CC (55) | I & II | Y | MMR IHC proficient |
| 5597 | M | Hyperplastic polyp (53/54), SC (55), KC (62) | 0 | Y | MMR IHC proficient |
| 8913 | F | Hyperplastic polyp (59), BC (61), TC (69) | 0 | Y | na |
CRC colorectal, ICD9 diagnosis International Classification of Diseases, 9th Revision, CC colon cancer, BC breast cancer, AMS Amsterdam criteria, 0 not fulfill the AMS criteria, Y yes, MMR mismatch repair, IHC immunohistochemistry, MSI microsatellite instabily, MSS microsatellite stable, na not available, OC ovary cancer, MM multiple myeloma, EC endometrial cancer, BT brain tumor, M melanoma, SMC other malignant of the skin, TC trachea, bronchus, lung cancer, ML malignant neoplasms of lymphoid, PC prostate cancer, THC thyroid cancer, KC kidney cancer, RC rectum cancer, CST malignant neoplasm of connective and soft tissue, SC stomach cancer
a Patient carrying CHEK2 c.470 T > C, p.I157T
Germline findings
Given that the families that fulfilled the AMS criteria and/or the Bethesda guidelines did not carry pathogenic variants in the MMR genes, we hypothesized that other genes could be implicated in the genetic determinism of these phenotypes.
In order to pursue this hypothesis, we collected DNA samples from all probands and performed high-throughput sequencing of a panel of 44 cancer-associated genes. For the 34 samples, mean depth of coverage ranged from 127 to 507 with the fraction of target bases with coverage ≥25 ranging from 80% to 93. The NGS results revealed that each individual carried an average of 26 SNV (between 19 and 33 per individual) in the set of 44 cancer susceptibility genes, most of which were common polymorphisms (allele frequency ≥ 1% in the general population) according to the ExAC database, and some being classified as benign or likely benign (class 1 or class 2) according to either ClinVar or the American College of Medical Genetics and Genomics (ACMG) guidelines [35, 50] (Table 2).
Table 2.
Characterization of germline variants found among Norwegian familial CRC individuals
| Patient_ID | VUS (Class 3) | Benign or Likely Benign variants (Class 1 or 2) | Polymorphisms | Total variants/patient |
|---|---|---|---|---|
| 3222 | NOTCH3 NM_000435: c.5854G >A, p.V1952 M(rs115582213)b POLE NM_006231: c.3046G > A, p.V1016 M (rs147692158) |
ATM NM_000051: c.5071A > C, p.S1691R (rs1800059) BRCA1 NM_007300: c.5019G > A, p.M1673I (rs1799967) PALB2 NM_024675: c.2993G > A, p.G998E (rs45551636) PALB2 NM_024675: c.2014G > C, p.E672Q (rs45532440) |
rs459552, rs659243, rs2240308, rs1799966, rs16942, rs16941, rs169547, rs4986764, rs1805107, rs506504, rs832582, rs5868032, rs1042821, rs3219484, rs1044009, rs152451, rs2228006, rs1805321, rs4796033, rs1042522, rs861539, rs13125836 |
28 |
| 3308 | NBN NM_002485: c.1720 T > A, p.L574I (rs142334798)POLE NM_006231:c.4523G > A, p.R1508H (rs142508245) | BARD1N M_000465: c.1075_1095del, p.L359-P365delLPECSSP (rs28997575) | rs459552, rs659243, rs11528010, rs144848, rs169547, rs4986764, rs1805107, rs506504, rs702689, rs832582, rs5868032, rs1799977, rs1042821, rs3219484, rs1805794, rs1044009, rs2228006, rs5744934, rs5744751, rs4796033, rs1042522 | 24 |
| 3387 | na | CDKN2AN M_000077: c.442G > A, p.A148T (rs3731249) | rs459552, rs659243, rs1801516, rs2240308 rs2070094, rs2229571, rs11528010, rs144848, rs169547, rs4986764, rs1805107, rs506504, rs1126497, rs702689, rs832582, rs5868032, rs3219489, rs1044009, rs2228006, rs1805321, rs5744934, rs1042522 |
23 |
| 3426 | na | na | rs459552, rs659243, rs1801516, rs2240308, rs2070094, rs2229571, rs1048108, rs11528010, rs144848, rs169547, rs4986764, rs1805107, rs506504, rs1126497, rs702689, rs832582, rs5868032, rs1805794, rs1044009, rs2228006, rs1805321, rs1042522, rs13125836 | 23 |
| 4932 | NOTCH3 NM_000435: c.5854G >A, p.V1952 M (rs115582213)a STK11NM_00045:c.841C > A, p.P281T (rs377208033) | na | rs459552, rs659243, rs1801516, rs2240308, rs2229571, rs11528010, rs1799966, rs16942, rs16941, rs799917, rs144848, rs169547, rs1805107, rs506504, rs12642536, rs702689, rs832582, rs5868032, rs3219489, rs1805794, rs1044009, rs2228006, rs1805321, rs861539 | 26 |
| 5324 | na | na | rs459552, rs659243, rs2240308, rs2070094, rs2229571, rs144848, rs169547, rs4986764, rs1805107, rs506504, rs1126497, rs12642536, rs832582, rs1799977, rs3219489, rs1044009, rs2228006, rs1805321, rs5744934 | 19 |
| 6174 | na | PMS2 NM_000535: c.1454C > A, p.T485 K (rs1805323) | rs459552, rs659243, rs2240308, rs2070094, rs2229571, rs1048108, rs1799966, rs16942, rs16941, rs799917, rs4986850, rs169547, rs4986764, rs1805107, rs506504, rs1126497, rs12642536, rs702689, rs832582, rs5868032, rs1799977, rs3219484, rs1805794, rs1044009, rs2228006, rs1805321, rs1042522, rs861539 | 29 |
| 6977 | na | BARD1 NM_000465: c.1075_1095del, p.L359-P365delLPECSSP (rs28997575) BARD1NM_000465: c.1972C > T, p.R658C (rs3738888) BRCA2 NM_000059: c.9976A > T, p.K3326X (rs11571833) | rs459552, rs659243, rs1801516, rs2240308, rs1799966, rs16942, rs16941, rs799917, rs144848, rs169547, rs4986764, rs1805107, rs506504, rs1126497, rs12642536, rs702689, rs832582, rs3219489, rs1044009, rs2228006, rs5744934, rs1042522, rs861539, rs28908468 | 27 |
| 9876 | PSMC3IP NM_016556: c.136G > A, p.V46 M (rs757057684) RAD51B NM_133509: c.1063G > A, p.A355T (rs61758785) | RAD51D NM_002878: c.698A > G, p.E233G (rs28363284) | rs459552, rs659243, rs2240308, rs2070094, rs2229571, rs169547, rs4986764, rs1805107, rs506504, rs702689, rs832582, rs5868032, rs1799977, rs1042821, rs1805794, rs1044009, rs2228006, rs1726801 | 21 |
| 9998 | na | MSH6 NM_000179: c.2633 T > C, p.V878A (rs2020912) | rs459552, rs659243, rs1801516, rs2240308, rs2070094, rs2229571, rs1048108, rs144848, rs169547, rs4986764, rs1805107, rs506504, rs1126497, rs12642536, rs702689, rs832582, rs5868032, rs1799977, rs1044009, rs2228006, rs1805321, rs5744751, rs1042522 | 24 |
| 10,675 | na | PMS2 NM_000535: c.1531A > G, p.T511A (rs2228007) | rs459552, rs659243, rs1801516, rs2070094, rs2229571, rs11528010, rs1799966, rs16942, rs16941rs799917, rs144848, rs169547, rs4986764, rs1805107, rs506504, rs1126497, rs702689, rs832582,rs5868032, rs1799977, rs1805794, rs1044009, rs2228006, rs10254120, rs1042522 | 26 |
| 12,954 | MUTYH NM_012222: c.812G > A, p.R271Q (rs149866955) RAD51C NM_058216: c.790G > A, p.G264S (rs147241704) | PMS2 NM_000535: c.1789A > T, p.T597S (rs1805318) | rs459552, rs659243, rs2240308, rs2229571, rs144848, rs169547, rs4986764, rs1805107, rs506504, rs12642536, rs702689, rs832582, rs5868032, rs1044009, rs2228006, rs1805321, rs1726801, rs4796033, rs1042522 | 22 |
| 13,072 | BRCA1 NM_007300: c.4315C > T, p.L1439F (rs781260818) | PMS2 NM_000535: c.1531A > G, p.T511A (rs2228007) | rs459552, rs659243, rs1801516, rs2240308, rs2070094, rs2229571, rs11528010, rs1799966, rs16942, rs16941, rs799917, rs144848, rs169547, rs4986764, rs1805107, rs506504, rs1126497, rs12642536, rs702689, rs832582, rs5868032, rs1799977, rs1805794, rs1044009, rs2228006, rs5744751, rs1042522 | 29 |
| 14,930 | na | PALB2 NM_024675: c.925A > G, p.I309V (rs3809683) | rs459552, rs659243, rs2240308, rs2070094, rs2229571, rs1048108, rs1799966, rs16942, rs16941, rs799917, rs4986850, rs169547, rs4986764, rs1805107, rs506504, rs1126497, rs12642536, rs702689, rs832582, rs5868032, rs1799977, rs3219489, rs1805794, rs1044009, rs152451, rs2228006, rs1805321, rs5744934, rs1042522 | 30 |
| 18,843 | MSH6 NM_000179.2: c.2195G > A, p.R732Q (rs749746725) | BRCA1 NM_007300: c.3119G > A, p.S1040 N (rs4986852) | rs459552, rs659243, rs2240308, rs2070094, rs2229571, rs11528010, rs144848, rs169547, rs4986764, rs1805107, rs506504, rs1126497, rs12642536, rs832582, rs5868032, rs1799977, rs3219489, rs1044009, rs2228006, rs1805321, rs5744934, rs5744751, rs1042522, rs861539 | 26 |
| 19,411 | na | BARD1 NM_000465: c.1972C > T, p.R658C (rs3738888) PALB2 NM_024675: c.1010 T > C, p.L337S (rs45494092) | rs459552, rs659243, rs2240308, rs2070094, rs2229571, rs1799966, rs16942, rs16941, rs799917, rs4986850, rs144848, rs169547, rs1805107, rs506504, rs1126497, rs12642536, rs702689, rs832582, rs5868032, rs1805794, 1,044,009, rs2228006, rs1805321, rs5744751, rs1042522, rs861539 | 28 |
| 19,673 | AXIN2 NM_004655: c.344A > G, p.N115S (rs370257532) | APC NM_001127510: c.7504G > A, p.G2502S (rs2229995) PMS2 NM_000535: c.1454C > A, p.T485 K (rs1805323) | rs459552, rs659243, rs2240308, rs2070094, rs2229571, rs1799966, rs16942, rs16941, rs799917, rs144848, rs169547, rs4986764, rs1805107, rs506504, rs702689, rs832582, rs5868032, rs1042821, rs1805794, rs1044009, rs2228006, rs1805321, rs4796033, rs1042522, rs861539, rs3218536 | 29 |
| 20,612 | na | CDKN2A NM_000077: c.442G > A, p.A148T (rs3731249) | rs459552, rs659243, rs2240308, rs2070094, rs2229571, rs11528010, rs1799966, rs16942, rs16941, rs799917, rs169547, rs4986764, rs1805107, rs506504, rs12642536, rs702689, rs5868032, rs1799977, rs1805794, rs1044009, rs2228006, rs1805321, rs10254120, rs5744934, rs4796033, rs1042522, rs861539 | 28 |
| 21,368 | MAP3K1 NM_005921: c.764A > G, p.N255S (rs56069227) | ATM NM_000051: c.2572 T > C, p.F858 L (rs1800056) | rs459552, rs659243, rs2240308, rs2229571, rs11528010, rs1799950, rs169547, rs1805107, rs506504, rs1126497, rs702689, rs832582, rs5868032, rs1799977, rs1044009, rs2228006, rs1805321, rs5744934, rs4796033, rs1042522 | 22 |
| 22,295 | na | BRCA1 NM_007300: c.5019G > A, p.M1673I (rs1799967) BRIP1 NM_032043: c.890A > G, p.K297R (rs28997570) | rs459552, rs659243, rs2240308, rs2070094, rs2229571, rs1048108, rs1799966, rs16942, rs16941, rs799917, rs144848, rs169547, rs1805107, rs506504, rs12642536, rs702689, rs832582, rs5868032, rs1799977, rs1042821, rs3219489, rs1805794, rs1044009, rs2228006, rs1805321, rs5744934, rs1042522, rs861539 | 30 |
| 23,761 | na | na | rs459552, rs659243, rs2240308, rs2070094, rs2229571, rs11528010, rs1799966, rs16942, rs16941, rs799917, rs1799950, rs144848, rs169547, rs4986764, rs1805107, rs506504, rs1126497, rs702689, rs832582, rs5868032, rs1799977, rs1044009, rs2228006, rs1805321, rs1802683, rs1042522 | 26 |
| 23,910 | na | BRCA2 NM_000059: c.6100C > T, p.R2034C (rs1799954) MSH2 NM_000251: c.965G > A, p.G322D (rs4987188) MSH6 NM_000179: c.2633 T > C, p.V878A (rs2020912) PALB2 NM_024675: c.2794G > A, p.V932 M (rs45624036) PMS2 NM_000535: c.1454C > A, p.T485 K (rs1805323) BARD1 NM_000465: c.1670G > C, p.C557S (rs28997576) | rs459552, rs659243, rs2240308, rs2070094, rs2229571, rs11528010, rs169547, rs4986764, rs1805107, rs506504, rs1126497, rs702689, rs832582, rs5868032, rs1799977, rs1042821, rs1805794, rs2228006, rs10254120, rs1042522, rs13125836, rs3218536 | 28 |
| 24,140 | na | BRCA1 NM_007300: c.5019G > A, p.M1673I (rs1799967) PMS2 NM_000535: c.1531A > G, p.T511A (rs2228007) | rs459552, rs659243, rs2240308, rs2070094, rs2229571, rs1048108, rs1799966, rs16942, rs16941, rs799917, rs144848, rs169547, rs1805107, rs506504, rs1126497, rs702689, rs832582, rs5868032, rs1799977, rs1042821, rs3219489, rs1805794, rs1044009, rs2228006, rs10254120, rs5744751, rs1042522 | 29 |
| 24,447 | CHEK2 NM_007194: c.74 T > C, p.V25A (rs587780188) | NOTCH3 NM_000435: c.3399C > A, p.H1133Q (rs112197217) | rs459552, rs659243, rs1801516, rs2240308, rs2070094, rs2229571, rs1799966, rs16942, rs16941, rs799917, rs169547, rs1805107, rs506504, rs1126497, rs702689, rs832582, rs5868032, rs1805794, rs1044009, rs2228006, rs1805321, rs1042522 | 24 |
| 11,705 | ATM NM_000051: c.4375G > A, p.G1459R (rs145667735) MSH2 NM_000251: c.1284C > G, p.H428Q (rs776034412) | MSH2 NM_000251: c.965G > A, p.G322D (rs4987188) PMS2 NM_000535: c.1454C > A, p.T485 K (rs1805323) | rs459552, rs659243, rs1801516, rs2229571, rs11528010, rs169547, rs4986764, rs1805107, rs506504, rs1126497, rs702689, rs832582 rs5868032, rs1799977, rs3219489, rs1805794, rs1044009, rs2228006, rs1805321, rs5744751, rs1042522, rs861539, rs13125836 |
27 |
| 12,673 | na | ATM NM_000051: c.2572 T > C, p.F858 L (rs1800056) PMS2 NM_000535: c.1454C > A, p.T485 K (rs1805323) | rs459552, rs659243, rs2240308, rs2070094, rs2229571, rs1048108, rs1799950, rs169547, rs4986764, rs1805107, rs506504, rs1126497, rs12642536, rs702689, rs832582, rs5868032, rs1799977, rs1044009, rs2228006, rs1805321, rs1802683, rs4796033, rs1042522, rs861539 | 26 |
| 13,393 | NBN NM_002485.4: c.643C > T, p.R215W (rs34767364) | BARD1 NM_000465: c.1972C > T, p.R658C (rs3738888) BRIP1 NM_032043: c.577G > A, p.V193I (rs4988346) PMS2 NM_000535: c.1531A > G, p.T511A (rs2228007) ATM NM_000051: c.4258C > T, p.L1420F (rs1800058) NOTCH3 NM_000435.2: c.3058G > C, p.A1020P (rs35769976) NOTCH3 NM_000435: c.3547G > A, p.V1183 M (rs10408676) | rs459552, rs659243, rs2240308, rs2070094, rs2229571, rs1799966, rs16942, rs16941, rs799917, rs169547, rs4986764, rs1805107, rs506504, rs702689, rs832582, rs5868032, rs1805794, rs1044009, rs2228006, rs1805321, rs1042522 | 28 |
| 14,963 | PALB2 NM_024675: c.232G > A, p.V78I (rs515726085) | PALB2 NM_024675: c.2590C > T, p.P864S (rs45568339) STK11 NM_000455: c.1062C > G, p.F354 L (rs59912467) | rs659243, rs1801516, rs2240308, rs2070094, rs2229571, rs11528010, rs1799966, rs16942, rs16941, rs799917, rs1799950, rs144848, rs169547, rs4986764, rs1805107, rs506504, rs1126497, rs12642536, rs702689, rs5868032, rs1799977, rs3219489, rs1805794, rs1044009, rs2228006, rs1805321, rs5744751, rs1042522, rs861539, rs3218536 | 33 |
| 19,609a | na | BRCA2 NM_000059: c.4258G > T, p.D1420Y (rs28897727) POLE NM_006231: c.2083 T > A, p.F695I (rs5744799) | rs459552, rs659243, rs2240308, rs2229571, rs1799966, rs16942, rs16941, rs799917, rs144848, rs169547, rs4986764, rs1805107, rs506504, rs702689, rs832582, rs5868032, rs1799977, rs3219484, rs1805794, rs1044009, rs2228006, rs1805321, rs5744934, rs1042522, rs13125836 | 28 |
| 22,953 | NOTCH3 NM_000435: c.5208G > C, p.E1736D (rs200331646) MSH2 NM_000251: c.128A > G, p.Y43C (rs17217723) RAD51B NM_133510: c.515 T > G, p.L172 W (rs34094401) | BRCA2 NM_000059: c.2971A > G, p.N991D (rs1799944) CDH1 NM_004360: c.1774G > A, p.A592T (rs35187787) | rs459552, rs659243, rs2240308, rs2229571, rs11528010, rs1799966, rs16942, rs16941, rs799917, rs169547, rs4986764, rs1805107, rs506504, rs12642536, rs702689, rs832582, rs5868032, rs1042821, rs3219489, rs1805794, rs1044009, rs2228006, rs1805321, rs1726801 | 29 |
| 24,789 | APC NM_001127510: c.4334C > T, p.T1445I (rs760686348) PALB2 NM_024675: c.1250C > A, p.S417Y (rs45510998) | BARD1 NM_000465: c.1972C > T, p.R658C (rs3738888) PALB2 NM_024675: c.2993G > A, p.G998E (rs45551636) PALB2 NM_024675: c.2014G > C, p.E672Q (rs45532440) POLE NM_006231: c.776G > A, p.R259H (rs61732929) NOTCH3 NM_000435: c.3399C > A, p.H1133Q (rs112197217) | rs459552, rs659243, rs2240308, rs2229571, rs1799966, rs16942, rs16941, rs799917, rs169547, rs4986764, rs1805107, rs506504, rs1126497, rs12642536, rs702689, rs832582, rs5868032, rs1799977, rs3219489, rs1044009, rs152451, rs2228006, rs1042522 | 30 |
| 25,167 | NBN NM_002485.4: c.643C > T, p.R215W (rs34767364) | ATM NM_000051: c.2119 T > C, p.S707P (rs4986761) | rs459552, rs659243, rs2240308, rs2229571, rs11528010, rs169547, rs4986764, rs1805107, rs506504, rs12642536, rs702689, rs832582, rs5868032, rs1799977, rs1805794, rs2228006, rs1805321, rs1726801, rs4796033, rs1042522 | 22 |
| 5597 | MAP3K1 NM_005921: c.2816C > G, p.S939C (rs45556841) | ATM NM_000051: c.5071A >C, p.S1691R (rs1800059) | rs459552, rs659243, rs1801516, rs2070094, rs2229571, rs11528010, rs1799966, rs16942, rs16941, rs799917, rs4986850, rs144848, rs169547, rs4986764, rs1805107, rs506504, rs1126497, rs12642536, rs702689, rs832582, rs5868032, rs1042821, rs1805794, rs1044009, rs2228006, rs1805321, rs1042522 | 29 |
| 8913 | RAD51B NM_133509: c.1063G > A, p.A355T (rs61758785) EPCAM NM_002354: c.267G > C, p.Q89H (rs146480420) | PMS2 NM_000535: c.1454C > A, p.T485 K (rs1805323) | rs459552, rs659243, rs2240308, rs2070094, rs2229571, rs1048108, rs11528010, rs1799966, rs16942, rs16941, rs799917, rs144848, rs169547, rs4986764, rs1805107, rs506504, rs1126497, rs702689, rs832582, rs5868032, rs1805794, rs1044009, rs2228006, rs5744751, rs1042522, rs861539, rs13125836 | 30 |
a Recently classified as Benign by ACMG Guidelines, 2015
b Patient ID carrying CHEK2 c.470 T > C, p.I157T
Importantly, we identified a likely pathogenic variant in a moderate-penetrance gene (CHEK2 c.470 T > C, p.I157T) in a female patient diagnosed with colon cancer at 42 years, melanoma at 44 years and BC at 57 years with a proficient IHC MMR profile and fulfilling the revised Bethesda guidelines (Patient 19,609) (Table 1).
The CHEK2 c.470 T > C has been classified as pathogenic according to the ACMG guidelines [51], and has a lower allele frequency (1.89*10–3) in the Norwegian population, compared to the non-Finnish European population (5.4*10–3) (http://norgene.no/vcf-miner/ and gnomAD database, respectively) [34, 35, 50]. The variant is reported in ClinVar as “conflicting interpretations of pathogenicity, risk factor” (Variation ID: 5591). When the revised Bethesda guidelines were considered, the mutation detection rate was thus 4.8% (1/21).
Overall, 25 unique VUS were found in 18 out of the 34 patients (Table 2). The detected VUS were distributed among 17 different genes: MAP3K1 (in 2 patients), NBN (in 3 patients), NOTCH3 (in 3 patients), RAD51B (in 3 patients), MSH2 (in 2 patients), PALB2 (in 2 patients), POLE (in 2 patients) and the remaining were found in APC, ATM, AXIN2, BRCA1, CHEK2, EPCAM, MSH6, MUTYH, RAD51C and STK11 (Table 2). The minor allele frequency (MAF) values of these variants were very low or no frequency data have been reported.
Protein and splicing-dedicated in silico analyses
The 25 unique VUS were analyzed by using five in silico prediction tools with different underlying algorithms to estimate the impact of the variants on the structure and function of the corresponding proteins.
Concordances between the 5 prediction tools were found for 2 out of the 25 VUS, suggesting a potentially damaging effect on protein level for the variants: MUTYH c.812G > A (p.R271Q) and MSH2 c.128A > G (p.Y43C) (Table 3). In the other hand, 6 out of 25 VUS were consistently predicted as benign: NBN c.1720 T > A (p.L574I),
Table 3.
In silico data obtained for the variants of unknown significance (VUS) identified in our study of familial CRC individuals
| Selected variants (VAR) | Reference splice site-dedicated analyses | Cryptic splice site-dedicated analyses | ESR-dedicated analyses | Protein-dedicated analyses | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Patient_ID | Genomic position (GRCh37) | Gene | Exon | Nucleotide change (cNomen) | Predicted protein change (pNomen) | Nearest reference | MES scores | SSFL scores | Potential local splice effect | Local MES scores | ∆tESRseq | ∆Hzei | ΔΨ | |||||||||||
| splice site | WT | Var | VAR vs WT | WT | Var | VAR vs WT | WT | Var | ||||||||||||||||
| Distance | Type | ∆ (%) | ||||||||||||||||||||||
| (nt) | (3’ or 5’ss) | Δ (%) | AGVGD | SIFT | MAPP | PolyPhen-2 | MutationTaster | |||||||||||||||||
| 3222 & 4932 | chr19:15273335 C>T | NOTCH3 | 32 | c.5854G>A | p.V1952M | 39 | 3’ | 11.5 | 11.5 | 0 | 89.2 | 89.2 | 0 | - | - | - | -1.78501 | -11.15 | -0.89 | C15 | Deleterious | bad | probably damaging | disease causing |
| 3222 | POLE | 25 | c.3046G>A | p.V1016M | -15 | 5’ | 9.1 | 9.1 | 0 | 82.5 | 82.5 | 0 | - | - | - | -0.74478 | 1 | -0.43 | C0 | Deleterious | bad | benign | disease causing | |
| 3308 | chr8:90965597 A>T | NBN | 11 | c.1720T>A | p.L574I | -126 | 5’ | 8.0 | 8.0 | 0 | 82.4 | 82.4 | 0 | - | - | - | 1.97427 | 33.38 | 0.18 | C0 | Tolerated | good | benign | polymorphism |
| chr12:133219838 C>T | POLE | 35 | c.4523G>A | p.R1508H | -29 | 5’ | 7.9 | 7.9 | 0 | 73.8 | 73.8 | 0 | - | - | - | -0.600279 | -3.17 | -0.08 | C0 | Tolerated | good | benign | disease causing | |
| 4932 | chr19:1221318 C>A | STK11 | 6 | c.841C>A | p.P281T | -22 | 5’ | 6.0 | 6.0 | 0 | 79.9 | 79.9 | 0 | - | - | - | -0.17437 | 7.7 | -0.72 | C0 | Tolerated | good | benign | disease causing |
| 5597 | chr5:56177843 C>G | MAP3K1 | 14 | c.2816C>G | p.S939C | 447 | 3’ | 12.0 | 12.0 | 0 | 100.0 | 100.0 | 0 | - | - | - | -0.486881 | -16.1 | 0 | C0 | Deleterious | good | benign | polymorphism |
| 9876 | chr17: 40729320 C>T | PSMC3IP | 3 | c.136G>A | p.V46M | 1 | 3’ | 12.5 | 11.7 | -6 | 87.5 | 83.6 | -4 | - | - | - | 2.0413 | 74.24 | -0.3 | C0 | Deleterious | - | possibly damaging | disease causing |
| 9876 & 8913 | chr14:69061228 G>A | RAD51B | 11* | c.1063G>A | p.A355T | 27 | 3’ | 11.8 | 11.8 | 0 | 80.2 | 80.2 | 0 | - | - | - | -1.24035 | -50.64 | - | C0 | Deleterious | - | benign | polymorphism |
| 12954 | chr1: 45797950_C>T | MUTYH | 10 | c.812G>A | p.R271Q | 33 | 3’ | 9.5 | 9.5 | 0 | 86.8 | 86.8 | 0 | - | - | - | -2.31042 | 0.88 | 0.09 | C35 | Deleterious | bad | possibly damaging | disease causing |
| chr17:56787304 G>A | RAD51C | 5 | c.790G>A | p.G264S | -48 | 5’ | 8.6 | 8.6 | 0 | 75.4 | 75.4 | 0 | New Acceptor Site? | 0 | 2.5 | -1.71397 | -59.31 | -0.21 | C0 | Tolerated | good | benign | disease causing | |
| 13072 | chr17:41234463 G>A | BRCA1 | 12 | c.4315C>T | p.L1439F | -43 | 5’ | 6.6 | 6.6 | 0 | 85.2 | 85.2 | 0 | - | - | - | -0.261141 | 8.83 | -2.67 | C0 | Tolerated | good | benign | polymorphism |
| 18843 | chr2:48027317 G>A | MSH6 | 4** | c.2195G>A | p.R732Q | -978 | 5’ | 8.9 | 8.9 | 0 | 81.6 | 81.6 | 0 | - | - | - | -0.805417 | -13.15 | 0 | C0 | Tolerated | good | benign | disease causing |
| 19673 | chr17:63554395 T>C | AXIN2 | 2 | c.344A>G | p.N115S | 460 | 3’ | 11.1 | 11.1 | 0 | 93.3 | 93.3 | 0 | New Acceptor Site? | 11.1 | 7.2 | -0.162849 | -2.19 | -0.05 | C0 | Tolerated | good | benign | disease causing |
| 21368 | chr5:56155672 A>G | MAP3K1 | 3 | c.764A>G | p.N255S | -71 | 5’ | 7.5 | 7.5 | 0 | 78.5 | 78.5 | 0 | New Acceptor Site? | 4.7 | 8.8 | -1.18661 | 6.7 | -0.04 | C0 | Tolerated | good | benign | polymorphism |
| 24447 | chr22:29130636 A>G | CHEK2 | 2 | c.74T>C | p.V25A | 80 | 3’ | 1.7 | 1.7 | 0 | 85.5 | 85.5 | 0 | - | - | -0.166788 | 36.67 | -0.07 | C0 | Tolerated | good | benign | polymorphism | |
| 11705 | chr11:108160467 G>A | ATM | 29 | c.4375G>A | p.G1459R | -62 | 5’ | 8.9 | 8.9 | 0 | 87.5 | 87.5 | 0 | - | - | 0.552026 | -27.26 | -0.09 | C15 | Deleterious | bad | probably damaging | disease causing | |
| chr2:47672694 C>G | MSH2 | 8 | c.1284C>G | p.H428Q | 8 | 3’ | 10.1 | 10.1 | 0 | 87.3 | 87.3 | 0 | - | - | 0.703136 | 22.05 | 0.05 | C0 | Tolerated | good | possibly damaging | disease causing | ||
| 13393 & 25167 | chr:8_90983460 G>A | NBN | 6 | c.643C>T | p.R215W | 59 | 3’ | 6.2 | 6.2 | 0 | 86.8 | 86.8 | 0 | - | - | -1.00071 | -16.87 | -0.09 | C0 | Tolerated | good | probably damaging | polymorphism | |
| 14963 | chr16:23647635 C>T | PALB2 | 4** | c.232G>A | p.V78I | 21 | 3’ | 10.0 | 10.0 | 0 | 90.3 | 90.3 | 0 | - | - | -0.783039 | -29.17 | 0.8 | C0 | Tolerated | good | benign | polymorphism | |
| 22953 | chr19:15278214 C>G | NOTCH3 | 29 | c.5208G>C | p.E1736D | 9 | 3’ | 7.8 | 7.8 | 0 | 84.2 | 84.2 | 0 | - | - | 0.0501789 | -10.44 | 0.22 | C35 | Deleterious | bad | benign | disease causing | |
| chr2:47630458 A>G | MSH2 | 1* | c.128A>G | p.Y43C | -84 | 5’ | 10.1 | 10.1 | 0 | 90.6 | 90.6 | 0 | New Donor Site? | 0 | 1.2 | 1.50425 | 4.7 | 2.11 | C55 | Deleterious | bad | probably damaging | disease causing | |
| chr14:68352648 T>G | RAD51B | 6 | c.515T>G | p.L172W | -58 | 5’ | 9.5 | 9.5 | 0 | 83.7 | 83.7 | 0 | - | - | 1.62772 | 67.77 | 1.31 | C0 | Deleterious | bad | possibly damaging | polymorphism | ||
| 24789 | chr5:112175625 C>T | APC | 15* | c.4334C>T | p.T1445I | 2376 | 3’ | 7.5 | 7.5 | 0 | 93.6 | 93.6 | 0 | - | - | -1.61023 | -90.13 | - | C0 | Tolerated | good | benign | polymorphism | |
| chr16:23646617 G>T | PALB2 | 4** | c.1250C>A | p.S417Y | -435 | 5’ | 8.9 | 8.9 | 0 | 87.5 | 87.5 | 0 | - | - | -0.551724 | -22.83 | 0.05 | C0 | Deleterious | bad | probably damaging | disease causing | ||
| 8913 | chr2:47601029 G>C | EPCAM | 3 | c.267G>C | p.Q89H | 83 | 3’ | 7.0 | 7.0 | 0 | 90.1 | 90.1 | 0 | - | - | -0.40237 | -28.93 | 0 | C0 | Tolerated | bad | possibly damaging | disease causing | |
In order to predict the biological impact of the 25 VUS, RNA splicing- and protein-dedicated bioinformatics analyses were performed as described under Materials and Methods. The stars indicate exons that could not be tested in our minigene assay, either because of their terminal position (*, 1st or last exons) or because of their large size (**). Results shown in bold were considered as predictive of a potential variant-induced negative biological effect. MES, MaxEntScan; SSFL, Splice Site Finder-Like; nt, nucleotide; 3’ or 5’ss, 3’ splice site or 5’ splice site; ESR, exonic splicing regulators; AGVGD, align-GVGD (C0, C15, C25, C35, C.45, C55, or C65 with C65 most likely to interfere with function and C0 least likely), SIFT, Sorting Intolerant From Tolerant (tolerated or deleterious), MAPP, Multivariate Analysis of Protein Polymorphism (good or bad), PolyPhen-2, Polymorphism Phenotyping v2 (benign, possibly damaging or probably damaging), MutationTaster (polymorphism or disease causing), CRC: colorectal cancer.
In order to predict the biological impact of the 25 VUS, RNA splicing- and protein-dedicated bioinformatics analyses were performed as described under Materials and Methods. The stars indicate exons that could not be tested in our minigene assay, either because of their terminal position (*, 1st or last exons) or because of their large size (**). Results shown in bold were considered as predictive of a potential variant-induced negative biological effect. MES, MaxEntScan; SSFL, Splice Site Finder-Like; nt, nucleotide; 3′ or 5’ss, 3′ splice site or 5′ splice site; ESR, exonic splicing regulators; AGVGD, align-GVGD (C0, C15, C25, C35, C.45, C55, or C65 with C65 most likely to interfere with function and C0 least likely), SIFT, Sorting Intolerant From Tolerant (tolerated or deleterious), MAPP, Multivariate Analysis of Protein Polymorphism (good or bad), PolyPhen-2, Polymorphism Phenotyping v2 (benign, possibly damaging or probably damaging), MutationTaster (polymorphism or disease causing), CRC: colorectal cancer
In order to predict their biological impact, RNA splicing- and protein-dedicated bioinformatics analyses were performed as described under Materials and Methods. Results shown in bold were considered as predictive of a potential variant-induced negative biological effect. MES, MaxEntScan; SSFL, Splice Site Finder-Like; nt, nucleotide; 3′ or 5’ss, 3′ splice site or 5′ splice site; ESR, exonic splicing regulators; AGVGD, align-GVGD (C0, C15, C25, C35, C.45, C55, or C65 with C65 most likely to interfere with function and C0 least likely), SIFT, Sorting Intolerant From Tolerant (tolerated or deleterious), MAPP, Multivariate Analysis of Protein Polymorphism (good or bad), PolyPhen-2, Polymorphism Phenotyping v2 (benign, possibly damaging or probably damaging), MutationTaster (polymorphism or disease causing)
BRCA1 c.4315C > T (p.L1439F), MAP3K1 c.764A > G (p.N255S), CHEK2 c.74 T > C (p.V25A), PALB2 c.232G > A (p.V78I) and APC c.4334C > T (p.T1445I). Discrepancies were pronounced for the variants in the POLE (n = 2), STK11, MAP3K1, PSMC3IP, RAD51C, MSH6, AXIN2, MSH2, NBN, NOTCH3, RAD51B, PALB2 and EPCAM genes (Table 3).
Two out of the 25 VUS were bioinformatically predicted to affect RNA maturation by potentially modifying splicing signals (Table 3). More specifically, according to our in silico results, NOTCH3 c.5854G >A (identified in Patients 3222 and 4932) was predicted to potentially induce exon 32 skipping by alteration of exonic splicing regulatory elements, whereas MAP3K1 c.764A > G (detected in Patient 21,368) was predicted to introduce a deletion of the first 131 nucleotides of exon 3 (r.634_764del) due to the creation of a putative new acceptor splice site. Skipping of NOTCH3 exon 32 would produce a transcript with a frameshift deletion of 98 nucleotides (NOTCH3 r.5816_5913del), potentially leading to the production of a carboxy-terminally truncated NOTCH3 protein p.(Lys1940Glyfs*14). The MAP3K1 r.634_764del transcript would be expected to be degraded by nonsense mediated decay and/or result in a very short MAP3K1 protein p.(Val212Leufs*45). The NOTCH3 c.5854G >A was identified in two patients (Patients 3222 and 4932) that fulfilled the revised Bethesda guidelines and AMS criteria, respectively while the MAP3K1 c.764A > G (Patient 21,368) in a patient which family fulfilled the revised Bethesda guidelines (Table 1).
Minigene splicing assays
Because patient RNA was not available, we decided to experimentally assess the impact of these 2 variants (NOTCH3 c.5854G >A and MAP3K1 c.764A > G) might have on RNA splicing by performing cell-based minigene splicing assays.
As shown in Fig. 1 we found that NOTCH3 c.5854G >A and MAP3K1 c.764A > G did not modify the splicing pattern of the minigenes’ transcripts. These data thus disagree with the in silico predictions and suggest that either the exon 32 of NOTCH3 and the exon 3 of MAP3K1 are refractory to splicing mutations (the predictions thus being incorrect) or that the minigenes used in our study do not fully reproduce the splicing pattern of the mutant exons in NOTCH3 and MAP3K1 bona fide transcripts (the predictions being eventually correct). Complementary studies using RNA from NOTCH3 c.5854G >A and MAP3K1 c.764A > G carriers need to be performed to verify the pertinence of these results.
Fig. 1.
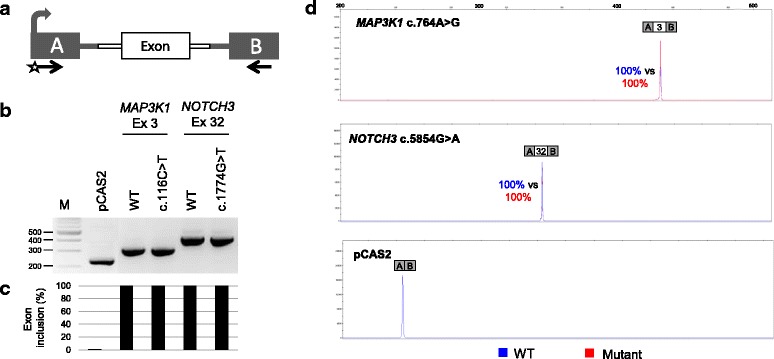
Evaluation of variant-induced splicing alterations by using a cell-based minigene assay. a Structure of pCAS2 minigenes used in the splicing reporter assay. The bent arrow indicates the CMV promoter, boxes represent exons, lines in between the boxes indicate introns, and arrows below the exons represent primers used in RT-PCR reactions. The minigenes were generated by inserting a genomic fragment containing the exon of interest together with its flanking intronic sequences into the intron of pCAS2, as described under Materials and Methods. b Analysis of the splicing pattern of pCAS2 minigenes carrying variants identified in this study. Wild-type (WT) and mutant constructs, as indicated, were introduced into HeLa cells and the transcripts of the minigenes were analyzed by RT-PCR 24 h post-transfection. The image shows the results of a representative experiment in which the RT-PCR products were separated on a 2.5% agarose gel stained with EtBr and visualized by exposure to ultraviolet light. M, 100 bp DNA ladder (New England Biolabs). c Quantification of splicing events observed in the minigene splicing assay. The relative levels of exon inclusion indicated under the gel are based on RT-PCR experiments equivalent to those shown in B but performed with a fluorescent forward primer and then separated on an automated sequencer. Quantification results were obtained by using the GeneMapper v5.0 software (Applied Biosystems) and correspond to the average of two independent fluorescent-RT-PCR experiments. d Representative fluorescent RT-PCR experiment. The panel shows superposed peaks corresponding to the WT and mutant products (in blue and red, respectively), as indicated
Discussion
The major unexpected finding in our Norwegian high-risk CRC cohort was the detection of a likely pathogenic variant in CHEK2 (c.470 T > C, p.I157T), a moderate-penetrance gene not traditionally associated with CRC, in an individual with a LS-evocative personal/family history and a high number of Class 3 variants in BC- and CRC- associated genes. Interestingly, the CHEK2 (c.470 T > C, p.I157T) has an allele frequency of 1.89*10–3 in the Norwegian population (http://norgene.no/vcf-miner/), and is reported in ClinVar as having conflicting interpretations of pathogenicity/being a risk factor (Variation ID: 5591). Importantly, there is no systematic classification for most of the genetic variants found by NGS, and, in more general terms, the impact of low- to moderate-penetrance pathogenic variants with respect to clinical management is not fully understood [52]. Co-segregation or case-control studies for further evaluation will be key in understanding whether such germline variant may have a modifying effect, since we do not yet have evidence-based guidelines for the majority of these genes.
On the other hand, CHEK2 germline variants have been described to confer an elevated risk of BC (relative risk = 3.0) [53]. However, the presence of pathogenic variants in CHEK2 is not frequently associated with cancer in high-risk BC families, prompting speculation that there may be several low-penetrance or moderate-penetrance BC risk genes segregating independently within these families [23, 54, 55]. Co-segregation analyses may add clues in our understanding whether this germline variant is implicated in CRC predisposition. Finally, we did not find pathogenic variants in POLE in our cohort, which is in contrast to what has been described in families with high burden of CRC adenomas and carcinomas in addition to extra-colonic cancers [56].
According to the Prospective LS Database (PLSDB), a total of 125 Norwegian families had a demonstrated pathogenic variant in either MLH1 (n = 21), MSH2 (n = 52), MSH6 (n = 36), or PMS2 (n = 16) [25]. On the other hand, a large portion of high-risk CRC families without pathogenic variant in MMR or EPCAM genes may be explained by a polygenic model involving a combination of multiple genomic risk factors, including the effect of either low-penetrance susceptibility alleles [57], high-penetrance genes which have not been tested, or the effect of environmental factors. In addition, emerging data suggest that CRC cases negative for pathogenic MMR variants may contain a significantly higher number of copy-neutral loss of heterozygosity (cnLOH) regions, some located within well-known oncogenes and tumor suppressor genes, compared to cases of sporadic CRC [58]. These genomic variations, which were not investigated in this study, may provide an additional explanation for high-risk CRC phenotypes without MMR or EPCAM pathogenic variants.
Recent NGS studies described the presence of heterozygous pathogenic BRCA1/2 or APC variants as well as biallelic MUTYH alterations in individuals with clinical features resembling those of LS [5, 22]. More precisely, those studies reported that 7% of patients with CRC carried pathogenic variants in non-LS genes, including 1.0% with BRCA1/2 mutations, and nearly two thirds of probands with high-penetrance non-LS mutations lacked clinical histories suggestive of their respective syndromes [5].
From 34 high-risk CRC individuals, our NGS panel testing identified one patient that carried a pathogenic variant in a gene with reportedly moderate penetrance. Our finding is in line with the mutation frequency (6%) in non-LS cancer susceptibility genes for individuals undergoing LS genetic testing [21] and 4% of patients with BC tested negative for BRCA1/2 genes [23]. Our results may have implications for an appropriate genetic counseling and follow-up of the patients and family members.
Besides the likely pathogenic CHEK2 variant, we identified a total of 25 variants in our cohort for which there were not so much data as to their clinical significance. We thus undertook bioinformatics analyses in an attempt to predict the biological impact of these Class 3 variants, both at the RNA and protein level, the ultimate goals being: (i) to discriminate pathogenic from non-pathogenic alterations in this set of variants and (ii) to further pinpoint the genetic determinants of high risk CRC in our cohort. On one hand, our RNA splicing-dedicated bioinformatics evaluation predicted that 2 out of the 25 VUS identified in this study (NOTCH3 c.5854G >A, p.V1952 M and MAP3K1 c.764A > G, p.N255S) could potentially affect RNA splicing. These two variants were then experimentally analyzed by performing minigene splicing assay. Our results revealed that neither variant altered the splicing pattern of the representative minigenes, suggesting that they do not affect the splicing of NOTCH3 or MAP3K1 transcripts. Additional experiments based on the analysis of RNA from carriers of these variants will be important to verify our minigene results. On the other hand, our protein-dedicated bioinformatics analysis yielded 8 consistent predictions (2 VUS predicted as deleterious and 6 as benign) and several conflicting results that were not explored further.
In this scenario, not only functional tests, but also co-segregation studies will be key to understanding whether the VUS detected in this work are non-pathogenic or otherwise have a causal or a modifying effect. Importantly, we do not yet have evidence-based guidelines for the majority of the genes carrying the VUS identified in this study and, in more general terms, the impact of low- to moderate-penetrance pathogenic variants with respect to clinical management is not fully understood. Most of these variants may in the future be reclassified as deleterious or benign, but in the meantime, they cannot be used to make clinical decisions [59]. Informed (re)classification of VUS in cancer-associated genes may cater to more appropriate risk-management, and may provide significant clues for the identification of additional patients carrying such uncommon variants.
NGS panel testing may benefit patients with a personal or family history compatible with more than one recognized CRC inherited syndrome. The CRC risk management strategy for these individuals is not yet available and there is a need to identify new high-, moderate-, and low- penetrance gene variants that may affect the risk of CRC or LS-associated tumors in non-MMR pathogenic carriers. The identification of such gene variants in combination with family history may contribute to more intensive surveillance and improved prevention [23].
Conclusions
Our study provides information on genetic locus that might possibly be related to cancer susceptibility, demonstrating that genes presently not routinely tested may be important for capturing cancer predisposition in these patients. In addition, we stratified 25 VUS by the use of RNA splicing- and protein-dedicated in silico analyses. Further studies are necessary for making reliable estimates of cancer risk for the VUS found in this study and allowing appropriate genetic counseling for the patients and their relatives.
Surveillance for early cancer detection is essential to ensure optimal survival for patients afflicted with familial cancers. Our findings pinpoint the need of more studies to unravel the mechanisms underlying the development of CRC in high-risk patients and the identifying for new cancer predisposition genes.
Additional file
Table S1. Primers used in the pCAS2 minigene splicing assay. (DOCX 15 kb)
Acknowledgements
We thank the families for their participation and contribution to this study.
Funding
This work was supported by the Radium Hospital Foundation (Oslo, Norway), Helse Sør-Øst (Norway), the French Association Recherche contre le Cancer (ARC), the Groupement des Entreprises Françaises dans la Lutte contre le Cancer (Gefluc), the Association Nationale de la Recherche et de la Technologie (ANRT, CIFRE PhD fellowship to H.T.) and by the OpenHealth Institute.
Availability of data and materials
All data generated or analyzed during this study are included in the manuscript.
Abbreviations
- ACMG
American college of medical genetics and genomics
- AMS
Amsterdam criteria
- BC
Breast cancer
- CRC
Colorectal cancer
- IHC
Immunohistochemistry
- LS
Lynch syndrome
- MAF
Minor allele frequency
- MMR
Mismatch repair genes
- MSI
Microsatellite instability
- NGS
Next generation sequencing
- PLSDB
Prospective lynch syndrome database
- SNV
Single nucleotide variant
- VUS
Variants of unclassified significance
Authors’ contributions
All authors have taken part in the different steps of the study: MDV, PM and EH designed the study, AM, HT performed in silico splicing predictions and the minigene assays, POE performed validation experiments, MM, AN and EHF performed in silico protein predictions, SN, DV performed the sequence analysis. MDV drafted the manuscript and all have read, revised and approved the manuscript.
Ethics approval and consent to participate
Ethical approval for the study was granted by the Norwegian Data Inspectorate and Ethical Review Board (ref 2015/2382). All examined patients signed an informed consent for their participation in the study.
Consent for publication
Not Applicable.
Competing interests
The authors declare that they have no competing interests
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Footnotes
Electronic supplementary material
The online version of this article (10.1186/s12881-018-0533-9) contains supplementary material, which is available to authorized users.
Contributor Information
Mev Dominguez-Valentin, Phone: +4740381634, Email: mev_dv@yahoo.com.
Sigve Nakken, Email: sigven@ifi.uio.no.
Hélène Tubeuf, Email: helene.tubeuf@etu.univ-rouen.fr.
Daniel Vodak, Email: danielvo@ifi.uio.no.
Per Olaf Ekstrøm, Email: per.olaf.ekstrom@rr-research.no.
Anke M. Nissen, Email: anke.nissen@mgz-muenchen.de
Monika Morak, Email: monika.morak@gmx.de.
Elke Holinski-Feder, Email: elke.holinski-feder@mgz-muenchen.de.
Alexandra Martins, Email: alexandra.martins@univ-rouen.fr.
Pål Møller, Email: moller.pal@gmail.com.
Eivind Hovig, Email: ehovig@ifi.uio.no.
References
- 1.Llor X, Pons E, Xicola RM, Castells A, Alenda C, Pinol V, Andreu M, Castellvi-Bel S, Paya A, Jover R, et al. Differential features of colorectal cancers fulfilling Amsterdam criteria without involvement of the mutator pathway. Clin Cancer Res. 2005;11(20):7304–7310. doi: 10.1158/1078-0432.CCR-05-0965. [DOI] [PubMed] [Google Scholar]
- 2.Balmana J, Castells A, Cervantes A, Group EGW Familial colorectal cancer risk: ESMO Clinical Practice Guidelines. Ann Oncol. 2010;21(Suppl 5):v78–v81. doi: 10.1093/annonc/mdq169. [DOI] [PubMed] [Google Scholar]
- 3.Buchanan DD, Tan YY, Walsh MD, Clendenning M, Metcalf AM, Ferguson K, Arnold ST, Thompson BA, Lose FA, Parsons MT, et al. Tumor mismatch repair Immunohistochemistry and DNA MLH1 Methylation testing of patients with endometrial cancer diagnosed at age younger than 60 years optimizes triage for population-level Germline mismatch repair gene mutation testing. J Clin Oncol. 2014;32(2):90. doi: 10.1200/JCO.2013.51.2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Patel SG, Ahnen DJ. Familial colon cancer syndromes: an update of a rapidly evolving field. Curr Gastroenterol Rep. 2012;14(5):428–438. doi: 10.1007/s11894-012-0280-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yurgelun MB, Kulke MH, Fuchs CS, Allen BA, Uno H, Hornick JL, Ukaegbu CI, Brais LK, McNamara PG, Mayer RJ, et al. Cancer susceptibility gene mutations in individuals with colorectal cancer. J Clin Oncol. 2017;35(10):JCO2016710012. doi: 10.1200/JCO.2016.71.0012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kuiper RP, Vissers LE, Venkatachalam R, Bodmer D, Hoenselaar E, Goossens M, Haufe A, Kamping E, Niessen RC, Hogervorst FB, et al. Recurrence and variability of germline EPCAM deletions in lynch syndrome. Hum Mutat. 2011;32(4):407–414. doi: 10.1002/humu.21446. [DOI] [PubMed] [Google Scholar]
- 7.Vasen HF, Mecklin JP, Khan PM, Lynch HT. The international collaborative group on hereditary non-polyposis colorectal cancer (ICG-HNPCC) Dis Colon Rectum. 1991;34(5):424–425. doi: 10.1007/BF02053699. [DOI] [PubMed] [Google Scholar]
- 8.Moller P, Evans G, Haites N, Vasen H, Reis MM, Anderson E, Apold J, Hodgson S, Eccles D, Olsson H, et al. Guidelines for follow-up of women at high risk for inherited breast cancer: consensus statement from the biomed 2 demonstration Programme on inherited breast cancer. Dis Markers. 1999;15(1–3):207–211. doi: 10.1155/1999/920109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.RodriguezBigas MA, Boland CR, Hamilton SR, Henson DE, Jass JR, Khan PM, Lynch H, Perucho M, Smyrk T, Sobin L, et al. A National Cancer Institute workshop on hereditary nonpolyposis colorectal cancer syndrome: meeting highlights and Bethesda guidelines. J Natl Cancer I. 1997;89(23):1758–1762. doi: 10.1093/jnci/89.23.1758. [DOI] [PubMed] [Google Scholar]
- 10.Umar A, Boland CR, Terdiman JP, Syngal S, de la Chapelle A, Ruschoff J, Fishel R, Lindor NM, Burgart LJ, Hamelin R, et al. Revised Bethesda guidelines for hereditary nonpolyposis colorectal cancer (lynch syndrome) and microsatellite instability. J Natl Cancer I. 2004;96(4):261–268. doi: 10.1093/jnci/djh034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kobayashi H, Ohno S, Sasaki Y, Matsuura M. Hereditary breast and ovarian cancer susceptibility genes (review) Oncol Rep. 2013;30(3):1019–1029. doi: 10.3892/or.2013.2541. [DOI] [PubMed] [Google Scholar]
- 12.Lindor NM. Familial colorectal cancer type X: the other half of hereditary nonpolyposis colon cancer syndrome. Surg Oncol Clin N Am. 2009;18(4):637. doi: 10.1016/j.soc.2009.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rodriguez-Soler M, Perez-Carbonell L, Guarinos C, Zapater P, Castillejo A, Barbera VM, Juarez M, Bessa X, Xicola RM, Clofent J, et al. Risk of cancer in cases of suspected lynch syndrome without Germline mutation. Gastroenterology. 2013;144(5):926. doi: 10.1053/j.gastro.2013.01.044. [DOI] [PubMed] [Google Scholar]
- 14.Dominguez-Valentin M, Therkildsen C, Da Silva S, Nilbert M. Familial colorectal cancer type X: genetic profiles and phenotypic features. Mod Pathol. 2015;28(1):30–36. doi: 10.1038/modpathol.2014.49. [DOI] [PubMed] [Google Scholar]
- 15.da Silva FC, Ferreira JRD, Torrezan GT, Figueiredo MCP, Santos EMM, Nakagawa WT, Brianese RC, de Oliveira LP, Begnani MD, Aguiar S, et al. Clinical and molecular characterization of Brazilian patients suspected to have lynch syndrome. PLoS One. 2015;10(10):e0139753. doi: 10.1371/journal.pone.0139753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Amendola LM, Jarvik GP, Leo MC, McLaughlin HM, Akkari Y, Amaral MD, Berg JS, Biswas S, Bowling KM, Conlin LK, et al. Performance of ACMG-AMP variant-interpretation guidelines among nine Laboratories in the Clinical Sequencing Exploratory Research Consortium (vol 98, pg 1067, 2016) Am J Hum Genet. 2016;99(1):247. doi: 10.1016/j.ajhg.2016.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pearlman R, Frankel WL, Swanson B, Zhao W, Yilmaz A, Miller K, Bacher J, Bigley C, Nelsen L, Goodfellow PJ, et al. Prevalence and Spectrum of Germline cancer susceptibility gene mutations among patients with early-onset colorectal cancer. JAMA Oncol. 2016;3(4):464–71. 10.1001/jamaoncol.2016.5194. [DOI] [PMC free article] [PubMed]
- 18.Picelli S, Lorenzo Bermejo J, Chang-Claude J, Hoffmeister M, Fernandez-Rozadilla C, Carracedo A, Castells A, Castellvi-Bel S, Memebers of EC-GOGotSGA, Naccarati A et al. Meta-analysis of mismatch repair polymorphisms within the cogent consortium for colorectal cancer susceptibility. PLoS One. 2013;8(9):e72091. doi: 10.1371/journal.pone.0072091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ballester V, Boardman L. Next generation multigene panel testing: the next step for identification of hereditary colorectal cancer syndromes? Gastroenterology. 2015;149(3):526–528. doi: 10.1053/j.gastro.2015.07.025. [DOI] [PubMed] [Google Scholar]
- 20.Kurian AW, Hare EE, Mills MA, Kingham KE, McPherson L, Whittemore AS, McGuire V, Ladabaum U, Kobayashi Y, Lincoln SE, et al. Clinical evaluation of a multiple-gene sequencing panel for hereditary cancer risk assessment. J Clin Oncol. 2014;32(19):2001–2009. doi: 10.1200/JCO.2013.53.6607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yurgelun MB, Masciari S, Joshi VA, Mercado RC, Lindor NM, Gallinger S, Hopper JL, Jenkins MA, Buchanan DD, Newcomb PA, et al. Germline TP53 mutations in patients with early-onset colorectal cancer in the colon cancer family registry. JAMA Oncol. 2015;1(2):214–221. doi: 10.1001/jamaoncol.2015.0197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yurgelun MB, Allen B, Kaldate RR, Bowles KR, Judkins T, Kaushik P, Roa BB, Wenstrup RJ, Hartman AR, Syngal S. Identification of a variety of mutations in cancer predisposition genes in patients with suspected lynch syndrome. Gastroenterology. 2015;149(3):604–613. doi: 10.1053/j.gastro.2015.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tung N, Lin NU, Kidd J, Allen BA, Singh N, Wenstrup RJ, Hartman AR, Winer EP, Garber JE. Frequency of Germline mutations in 25 cancer susceptibility genes in a sequential series of patients with breast cancer. J Clin Oncol. 2016;34(13):1460–1468. doi: 10.1200/JCO.2015.65.0747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moller P, Seppala T, Bernstein I, Holinski-Feder E, Sala P, Evans DG, Lindblom A, Macrae F, Blanco I, Sijmons R, et al. Cancer incidence and survival in lynch syndrome patients receiving colonoscopic and gynaecological surveillance: first report from the prospective lynch syndrome database. Gut. 2015;66(3):464–472. doi: 10.1136/gutjnl-2015-309675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moller P, Seppala T, Bernstein I, Holinski-Feder E, Sala P, Evans DG, Lindblom A, Macrae F, Blanco I, Sijmons R, et al. Incidence of and survival after subsequent cancers in carriers of pathogenic MMR variants with previous cancer: a report from the prospective lynch syndrome database. Gut. 2017;66(9):1657–64. 10.1136/gutjnl-2016-311403. [DOI] [PMC free article] [PubMed]
- 26.Moller P, Seppala TT, Bernstein I, Holinski-Feder E, Sala P, Gareth Evans D, Lindblom A, Macrae F, Blanco I, Sijmons RH, et al. Cancer risk and survival in path_MMR carriers by gene and gender up to 75 years of age: a report from the prospective lynch syndrome database. Gut. 2017; 10.1136/gutjnl-2017-314057. [Epub ahead of print] [DOI] [PMC free article] [PubMed]
- 27.Lynch HT, Riley BD, Weissman SM, Coronel SM, Kinarsky Y, Lynch JF, Shaw TG, Rubinstein WS. Hereditary nonpolyposis colorectal carcinoma (HNPCC) and HNPCC-like families: problems in diagnosis, surveillance, and management. Cancer Am Cancer Soc. 2004;100(1):53–64. doi: 10.1002/cncr.11912. [DOI] [PubMed] [Google Scholar]
- 28.Dominguez-Valentin M, Nakken S, Tubeuf H, Vodak D, Ekstrom PO, Nissen AM, Morak M, Holinski-Feder E, Martins A, Moller P, et al. Potentially pathogenic germline CHEK2 c.319+2T>a among multiple early-onset cancer families. Familial Cancer. 2018;17(1):141–53. 10.1007/s10689-017-0011-0. [DOI] [PubMed]
- 29.Dominguez-Valentin M, Evans DGR, Nakken S, Tubeuf H, Vodak D, Ekstrøm PO, Nissen AM, Morak M, Holinski-Feder E, Martins A, Møller P, Hovig E. Genetic variants of prospectively demonstrated phenocopies in BRCA1/2 kindreds. Hered Cancer Clin Pract. 2018;15(16):4. 10.1186/s13053-018-0086-0. [DOI] [PMC free article] [PubMed]
- 30.Li L, Chen HC, Liu LX. Sequence alignment algorithm in similarity measurement. 2009 International Forum on Information Technology and Applications. PRO. 2009;1:453–6.
- 31.Auclair J, Vaissiere T, Desseigne F, Lasset C, Bonadona V, Giraud S, Saurin JC, Joly MO, Leroux D, Faivre L, et al. Intensity-dependent constitutional MLH1 promoter methylation leads to early onset of colorectal cancer by affecting both alleles. Genes Chromosomes Cancer. 2011;50(3):178–185. doi: 10.1002/gcc.20842. [DOI] [PubMed] [Google Scholar]
- 32.Sherry ST, Ward MH, Kholodov M, Baker J, Phan L, Smigielski EM. Sirotkin K: dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29(1):308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Genomes Project C. Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, Korbel JO, Marchini JL, McCarthy S, McVean GA, et al. A global reference for human genetic variation. Nature. 2015;526(7571):68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, O'Donnell-Luria AH, Ware JS, Hill AJ, Cummings BB, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536(7616):285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Landrum MJ, Lee JM, Riley GR, Jang W, Rubinstein WS, Church DM, Maglott DR. ClinVar: public archive of relationships among sequence variation and human phenotype. Nucleic Acids Res. 2014;42(D1):D980–D985. doi: 10.1093/nar/gkt1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Apweiler R, Bairoch A, Wu CH, Barker WC, Boeckmann B, Ferro S, Gasteiger E, Huang HZ, Lopez R, Magrane M, et al. UniProt: the universal protein knowledgebase. Nucleic Acids Res. 2004;32:D115–D119. doi: 10.1093/nar/gkh131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Finn RD, Bateman A, Clements J, Coggill P, Eberhardt RY, Eddy SR, Heger A, Hetherington K, Holm L, Mistry J, et al. Pfam: the protein families database. Nucleic Acids Res. 2014;42(D1):D222–D230. doi: 10.1093/nar/gkt1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.den Dunnen JT, Antonarakis SE. Mutation nomenclature extensions and suggestions to describe complex mutations: a discussion. Hum Mutat. 2000;15(1):7–12. doi: 10.1002/(SICI)1098-1004(200001)15:1<7::AID-HUMU4>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 39.Antoniou AC, Kuchenbaecker KB, Soucy P, Beesley J, Chen XQ, McGuffog L, Lee A, Barrowdale D, Healey S, Sinilnikova OM, et al. Common variants at 12p11, 12q24, 9p21, 9q31.2 and in ZNF365 are associated with breast cancer risk for BRCA1 and/or BRCA2 mutation carriers. Breast Cancer Res. 2012;14(1):1–18. doi: 10.1186/bcr3121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Di Giacomo D, Gaildrat P, Abuli A, Abdat J, Frebourg T, Tosi M, Martins A. Functional analysis of a large set of BRCA2 exon 7 variants highlights the predictive value of hexamer scores in detecting alterations of exonic splicing regulatory elements. Hum Mutat. 2013;34(11):1547–1557. doi: 10.1002/humu.22428. [DOI] [PubMed] [Google Scholar]
- 41.Erkelenz S, Hillebrand F, Widera M, Theiss S, Fayyaz A, Degrandi D, Pfeffer K, Schaal H. Balanced splicing at the tat-specific HIV-1 3’ss A3 is critical for HIV-1 replication. Retrovirology. 2015;12:29. doi: 10.1186/s12977-015-0154-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Susswein LR, Marshall ML, Nusbaum R, Vogel Postula KJ, Weissman SM, Yackowski L, Vaccari EM, Bissonnette J, Booker JK, Cremona ML, et al. Pathogenic and likely pathogenic variant prevalence among the first 10,000 patients referred for next-generation cancer panel testing. Genet Med. 2016;18(8):823–832. doi: 10.1038/gim.2015.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Soukarieh O, Gaildrat P, Hamieh M, Drouet A, Baert-Desurmont S, Frebourg T, Tosi M, Martins A. Exonic splicing mutations are more prevalent than currently estimated and can be predicted by using in Silico tools. PLoS Genet. 2016;12(I):1–26. doi: 10.1371/journal.pgen.1005756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tavtigian SV, Deffenbaugh AM, Yin L, Judkins T, Scholl T, Samollow PB, de Silva D, Zharkikh A, Thomas A. Comprehensive statistical study of 452 BRCA1 missense substitutions with classification of eight recurrent substitutions as neutral. J Med Genet. 2006;43(4):295–305. doi: 10.1136/jmg.2005.033878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc. 2009;4(7):1073–1082. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- 46.Stone EA, Sidow A. Physicochemical constraint violation by missense substitutions mediates impairment of protein function and disease severity. Genome Res. 2005;15(7):978–986. doi: 10.1101/gr.3804205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, Kondrashov AS, Sunyaev SR. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7(4):248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schwarz JM, Cooper DN, Schuelke M, Seelow D. MutationTaster2: mutation prediction for the deep-sequencing age. Nat Methods. 2014;11(4):361–362. doi: 10.1038/nmeth.2890. [DOI] [PubMed] [Google Scholar]
- 49.Gaildrat P, Killian A, Martins A, Tournier I, Frebourg T, Tosi M. Use of splicing reporter minigene assay to evaluate the effect on splicing of unclassified genetic variants. Methods Mol Biol. 2010;653:249–257. doi: 10.1007/978-1-60761-759-4_15. [DOI] [PubMed] [Google Scholar]
- 50.Landrum MJ, Lee JM, Benson M, Brown G, Chao C, Chitipiralla S, Gu B, Hart J, Hoffman D, Hoover J, et al. ClinVar: public archive of interpretations of clinically relevant variants. Nucleic Acids Res. 2016;44(D1):D862–D868. doi: 10.1093/nar/gkv1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, Grody WW, Hegde M, Lyon E, Spector E, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17(5):405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yadav S, Fulbright J, Dreyfuss H, Reeves A, Campian S, Thomas V, Zakalik D. Outcomes of retesting BRCA-negative patients using multigene panels. J Clin Oncol. 2015;33(28):319–28. doi: 10.1007/s10689-016-9956-7. [DOI] [PubMed] [Google Scholar]
- 53.Easton DF, Pharoah PD, Antoniou AC, Tischkowitz M, Tavtigian SV, Nathanson KL, Devilee P, Meindl A, Couch FJ, Southey M, et al. Gene-panel sequencing and the prediction of breast-cancer risk. N Engl J Med. 2015;372(23):2243–2257. doi: 10.1056/NEJMsr1501341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Meijers-Heijboer H, van den Ouweland A, Klijn J, Wasielewski M, de Snoo A, Oldenburg R, Hollestelle A, Houben M, Crepin E, van Veghel-Plandsoen M, et al. Low-penetrance susceptibility to breast cancer due to CHEK2(*)1100delC in noncarriers of BRCA1 or BRCA2 mutations. Nat Genet. 2002;31(1):55–59. doi: 10.1038/ng879. [DOI] [PubMed] [Google Scholar]
- 55.Consortium CBCC-C. CHEK2*1100delC and susceptibility to breast cancer: a collaborative analysis involving 10,860 breast cancer cases and 9,065 controls from 10 studies. Am J Hum Genet. 2004;74(6):1175–1182. doi: 10.1086/421251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hansen MF, Johansen J, Bjornevoll I, Sylvander AE, Steinsbekk KS, Saetrom P, Sandvik AK, Drablos F, Sjursen W. A novel POLE mutation associated with cancers of colon, pancreas, ovaries and small intestine. Familial Cancer. 2015;14(3):437–448. doi: 10.1007/s10689-015-9803-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Obermeier K, Sachsenweger J, Friedl TW, Pospiech H, Winqvist R, Wiesmuller L. Heterozygous PALB2 c.1592delT mutation channels DNA double-strand break repair into error-prone pathways in breast cancer patients. Oncogene. 2016;35(29):3796–806. 10.1038/onc.2015.448. [DOI] [PMC free article] [PubMed]
- 58.Villacis RA, Basso TR, Canto LM, Pinheiro M, Santiago KM, Giacomazzi J, de Paula CA, Carraro DM, Ashton-Prolla P, Achatz MI, et al. Rare germline alterations in cancer-related genes associated with the risk of multiple primary tumor development. J Mol Med (Berl). 2017; [DOI] [PubMed]
- 59.Pinto P, Paulo P, Santos C, Rocha P, Pinto C, Veiga I, Pinheiro M, Peixoto A, Teixeira MR. Implementation of next-generation sequencing for molecular diagnosis of hereditary breast and ovarian cancer highlights its genetic heterogeneity. Breast Cancer Res Treat. 2016;159(2):245–256. doi: 10.1007/s10549-016-3948-z. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Primers used in the pCAS2 minigene splicing assay. (DOCX 15 kb)
Data Availability Statement
All data generated or analyzed during this study are included in the manuscript.


